Summary
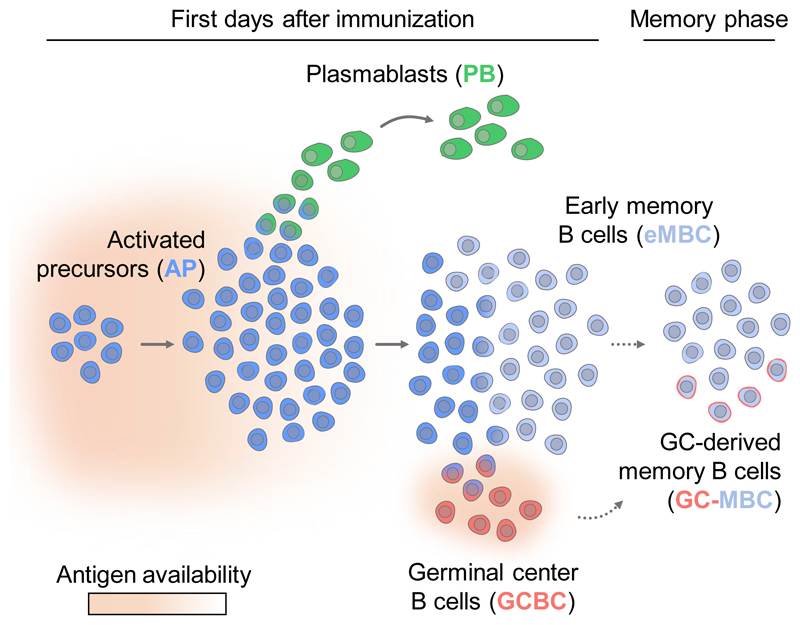
Cell fate decisions during early B cell activation determine the outcome of responses to pathogens and vaccines. We examined the early B cell response to T-dependent antigen in mice by single-cell RNA sequencing. Early after immunization, a homogeneous population of activated precursors (APs) gave rise to a transient wave of plasmablasts (PBs), followed a day later by the emergence of germinal center B cells (GCBCs). Most APs rapidly exited the cell cycle, giving rise to non-GC-derived early memory B cells (eMBCs) that retained an AP-like transcriptional profile. Rapid decline of antigen availability controlled these events; provision of excess antigen precluded cell cycle exit and induced a new wave of PBs. Fate mapping revealed a prominent contribution of eMBCs to the MBC pool. Quiescent cells with an MBC phenotype dominated the early response to immunization in primates. A reservoir of APs/eMBCs may enable rapid readjustment of the immune response when failure to contain a threat is manifested by increased antigen availability.
Graphical abstract.
Introduction
An efficient immune response to infection requires the generation of antibodies against the invading pathogen. Humoral immune responses to T cell-dependent antigens start with activation of naive B cells by cognate antigen, which, in synergy with signals provided by T cell help, leads to a burst of proliferation and initiation of one of three distinct molecular programs (Taylor et al., 2012a). Some activated B cells differentiate rapidly into plasmablasts (PBs), a terminally differentiated cell type specializing in secretion of high amounts of antibodies. Other activated B cells induce a unique molecular program required for participation in the germinal center (GC) reaction, an iterative process that involves hypermutation of immunoglobin genes, competition of the mutated B cell clones for antigen and T cell help, and selective survival and expansion of B cell clones with the highest affinity. These GC B cells (GCBCs) can then differentiate into antibody-secreting plasma cells (PCs) and GC-derived memory B cells (GC-MBCs). Finally, some activated B lymphocytes give rise to early MBCs (eMBCs) without participation in the GC reaction (Kaji et al., 2012; Taylor et al., 2012b; Toyama et al., 2002).
The events occurring during the course of the GC reaction and leading to the generation of later waves of PCs and GC-MBCs have been investigated intensively (reviewed by De Silva and Klein, 2015; Mesin et al., 2016). However, the cell fate decisions that take place early in the response are much less understood. Activated B cells migrate to the interfollicular zone in the first day after immunization, where they interact with T cells and proliferate (Kerfoot et al., 2011). Over the next days, some putative progeny of these cells migrate back to the follicles to populate GCs, whereas others give rise to extrafollicular PBs (Chan et al., 2009; Coffey et al., 2009; Kerfoot et al., 2011). A single naive B cell can give rise to all three “effector lineages,” although cell death limits the contribution of many clones to only one or two subsets (Taylor et al., 2015). A candidate common activated precursor (AP) population that shares phenotypic features with naive B cells, MBCs, and GCBCs emerges around day 2 after immunization (Schwickert et al., 2011; Taylor et al., 2012b; Zhang et al., 2017), before generation of the first GCBCs (Kitano et al., 2011; Taylor et al., 2012b), and persists for the first week of the immune response. The choice between the three lineages is, at least in part, regulated by B cell receptor (BCR) affinity and the amount of available T cell help. The expression of high-affinity BCRs and the abundance of signals associated with T cell help favor PB development over GCBC and eMBC fates (O’Connor et al., 2006; Paus et al., 2006; Taylor et al., 2015; Zhang et al., 2017). Low-affinity B cells fail to get access to T cell help and do not contribute to the GCBC compartment in the presence of competitor B cells with higher BCR affinity (Abbott et al., 2018; Dal Porto et al., 2002; Dosenovic et al., 2018; Schwickert et al., 2011; Shih et al., 2002a). Disengagement from T cell help has been suggested to favor GCBC over PB differentiation (Zhang et al., 2017). In addition, the cytokine BAFF regulates the generation and maintenance of eMBCs (Lau et al., 2021; Müller-Winkler et al., 2021). These results suggest that the initial B cell activation leads to a burst of proliferation and emergence of tripotent APs whose differentiation into GCBCs, PBs, and eMBCs is, at least in part, regulated by the strength of BCR signaling and the amount of available T cell help. However, the exact timing, hierarchy, and mechanisms of cell fate decisions in early B cell activation remain poorly understood.
We used single-cell RNA sequencing (scRNA-seq) to dissect the events that take place during the first days of the B cell response to T-dependent antigens. This analysis mapped the trajectories of AP differentiation to the three “effector lineages.” Unexpectedly, most APs gave rise to eMBCs, whereas PB differentiation was restricted to a transient wave very early in the response. These events were driven by limited access of activated B cells to antigen, and provision of antigen excess resulted in a delayed transition of eMBCs to quiescence while prolonging PB differentiation. In contrast to GCBCs and PBs, eMBCs retained a high transcriptional similarity to APs. Thus, APs that lose contact with antigen are kept in reserve as eMBCs that can be re-recruited rapidly into the response upon increased antigen availability.
Results
Dissection of early B cell activation by scRNA-seq
We sought to characterize the events that occur during the first steps of differentiation toward GCBC, early PB, and eMBC lineages by scRNA-seq. To have sufficient numbers of antigen-specific B cells early in the response for this analysis, we utilized IghB1-8hi (B1-8hi) mice expressing a rearranged immunoglobulin heavy chain that, when combined with immunoglobulin λ light chains (Igλ), recognizes the hapten 4-hydroxy-3-nitrophenylacetyl (NP) (Shih et al., 2002b). We transferred splenocytes from B1-8hi mice into congenically distinguishable wild-type (WT) ovalbumin (OVA)-primed recipients and, on the next day, immunized mice intraperitoneally (i.p.) with NP-OVA in alum. A population of GL7+CCR6+ cells, matching the cell surface phenotype of the common AP (Schwickert et al., 2011; Taylor et al., 2012b), represented the majority of cells by day 2.5 (Figure 1A). First IRF4hiCD138+ PBs were also evident on day 2.5 (Figure 1A), whereas BCL6+GL7+CCR6− GCBCs emerged around day 4 after immunization and increased in frequency at later time points (Figures 1A and 1B). As reported previously (Schwickert et al., 2011; Taylor et al., 2012b), the CCR6+ population exhibited moderate IgD downregulation (Figure S1A) and did not contain any BCL6hi cells (Figure 1B).
Figure 1. Analysis of early B cell activation by flow cytometry and scRNA-seq.
(A) OVA-primed WT mice (CD45.2) were injected with 1 × 107CD45.1/2 heterozygous B1-8hi splenocytes (~5 × 105 NP-specific B cells) and the next day were immunized i.p. with NP-OVA in alum. Surface expression of CCR6, GL7, and CD138 and intracellular expression of IRF4 is shown for Igλ+ (NP-specific) or Igλ−(control) B1-8hi B cells (gated as live CD45.1+CD19+) at the indicated time points. Shown are representative results of two independent experiments.
(B) BCL6 expression is shown in Igλ+CCR6+ APs, Igλ+CCR6-GL7+ GCBCs, and Igλ− B1-8hi B cells at day 4 after immunization. Experimental setup as in (A).
(C–G) scRNA-seq was performed with Igλ+CD19+CD45.1+ B1-8hi B cells sorted and pooled from recipient mice at day 3.5 (5 mice) and 4 (5 mice) after immunization. Experimental setup as in (A), but 4 × 107 CD45.1 B1-8hi splenocytes were transferred into each recipient.
(C) UMAP and PCA plots showing the distribution of different cell populations.
(D) mRNA expression of the indicated genes is highlighted on a UMAP plot as in (C).
(E) Dot plots showing expression of GCBC and PB signature genes in the scRNA-seq data. The gene signatures characteristic of GCBCs and PBs were defined based on the Immgen database (Yoshida et al., 2019; STAR Methods).
(F) RNA velocity analysis was performed as described in STAR Methods. The velocity field was plotted on top of the UMAP embedding as in (C).
(G) Cells were assigned to the different cell cycle phases as described in STAR Methods, and the resulting assignments are shown on a UMAP plot.
See also Figure S1.
The timing of emergence of the first PBs and GCBCs suggested that scRNA-seq analysis of antigen-specific B cells pooled from multiple mice at day 3.5 and 4 after immunization (that exhibited a wide distribution in the frequency of GCBCs; Figure S1B) should allow us to capture the developmental transitions to all “effector lineages.” We therefore sorted Igλ+ B1-8hi B cells at these timepoints and performed scRNA-seq analysis using the 10X Genomics protocol, which allowed us to analyze the transcriptomes of 2,133 activated B cells. Visualization of the resulting dataset with the uniform manifold approximation and projection (UMAP) algorithm identified three groups of cells (Figures 1C–1E). Analysis of gene expression signatures revealed that the smallest population corresponded to PBs, as judged by expression of Irf4, Prdm1 (BLIMP1), and Xbp1, encoding lineage-defining transcription factors for PBs and PCs, and downregulation of B cell program-associated genes (Figures 1D, 1E, and S1C). The second small population corresponded to GCBCs, as evidenced by their expression of Bcl6 as well as Gcsam and Rgs13 (encoding putative signal transducers highly expressed by GCBCs (Hwang et al., 2013; Schenten et al., 2006)), high expression of Aicda (encodes for activation-induced cytidine deaminase [AID]), and proliferation-related genes (Figure 1D and 1E). Finally, the largest group of cells expressed Ccr6 (Figure 1D) and, thus, corresponded to the CCR6+ cell population identified by flow cytometry (Figure 1A). PBs were fully separated from the GCBC and CCR6+ cell populations, whereas GCBCs, also exhibiting a high degree of separation from the other populations, were connected with the CCR6+ cells in UMAP and principal-component analysis (PCA) (Figure 1C). Re-analysis of the scRNA-seq data after removal of cell cycle effects did not change the overall population structure of the dataset (Figure S1D).
A large fraction of activated B cells starts to exit the cell cycle in the first days of the response to contribute to the MBC pool
To get insight into the developmental trajectories of these populations, we performed an RNA velocity analysis that takes into account the amount of spliced and unspliced transcripts to predict likely future states of cells in a scRNA-seq dataset (La Manno et al., 2018). The direction of velocity vectors suggested that the transcriptomes of all three populations, including the CCR6+ group, were diverging from each other (Figure 1F), an observation that held true after subtraction of cell-cycle-related variability (Figure S1E). This result appeared to be inconsistent with the previous notion that the whole CCR6+ population corresponded to APs. Moreover, analysis of the cell cycle status of the cells demonstrated that only a small fraction of CCR6+ cells, positioned close to GCBCs on UMAP plots, exhibited a proliferation signature and that the majority of cells in the CCR6+ population were in G1 or G0 phase of the cell cycle (Figures 1D and 1G). This observation could be consistent with two scenarios. First, going against the predictions of directionality suggested by RNA velocity (Figures 1F and S1E), it is conceivable that the CCR6+ population could represent cells that were very early in the response and that just started to enter the cell cycle. Alternatively, and in line with the developmental trajectories suggested by RNA velocity, it is also possible that this population largely contained cells that began to exit the cell cycle very early in the response. To distinguish between these possibilities, we analyzed the proliferation history of B1-8hi B cells at different time points using CellTrace Violet (CTV) dilution. Nearly all Igλ+ B cells completely diluted CTV already at day 2.5 after immunization (Figures 2A and 2B), indicating that the absence of a cell cycle signature in the majority of CCR6+ cells likely reflected withdrawal from the cell cycle of cells that had just undergone a burst of proliferation. Indeed, staining for Ki67 and 5-ethynyl-2’-deoxyuridine (EdU) incorporation experiments revealed that although, at day 2.5 after immunization, CCR6+ APs were as proliferative as GCBCs at day 7, some of the APs started their transit to a quiescent Ki67− G0 state already at day 4, and the majority of them downregulated Ki67 and stopped incorporating EdU by day 7 (Figures 2C and 2D). Similar Ki67 downregulation was observed when immunization was performed with soluble antigen intravenously (i.v.) (Figure S2A). We conclude that many activated B cells start to exit the cell cycle and therefore withdraw from immediate participation in the immune response early after immunization.
Figure 2. Early cell cycle exit defines the separation of the eMBC lineage.
(A–D) OVA-primed WT mice were transferred with congenically distinguishable CTV-labeled B1-8hi splenocytes and immunized i.p. with NP-OVA in alum. Mice were injected with EdU 14 and 4 h before analysis at the indicated time points.
(A) Experimental design.
(B) CTV dilution by Igλ+ and Igλ− B1-8hi B cells.
(C) EdU incorporation by CCR6+ (AP-eMBC) or CCR6−GL7+ (GCBC) Igλ+ B1-8hi B cells at the indicated time points. Shown are representative contour plots (left) and quantification (right).
(D) Ki67 expression by cells gated as in (C). Shown are representative histograms indicating gating of Ki67lo cells (left), frequencies of Ki67lo cells (center), and Ki67 median fluorescence intensity (MFI) (right).
(B–D) Representative results of two independent experiments (B and D) and one experiment (C). **p < 0.01, ***p < 0.001, ****p < 0.0001 (one-way ANOVA with Holm-Sidak’s multiple comparisons test). Horizontal lines indicate the mean and error bars represent SD.
(E) UMAP plots as in Figures 1C and 1D, highlighting expression of MBC-related genes.
(F) Dot plots showing expression of MBC signature genes (top) (Laidlaw et al., 2017) and MBC transcription factor genes (bottom) (He et al., 2017) in the AP-eMBC, GCBC, and PB populations of the scRNA-seq data (day 3.5–4; Figure 1C).
(G) OVA-primed WT mice were transferred with B1-8hi cells, and EdU was injected on days 1, 2, and 3 after immunization. EdU incorporation was analyzed 4 days, 4 weeks, and 8 weeks after immunization. Expression of IgM and IgG1 by EdU+ cells is shown. Horizontal lines indicate the mean and error bars represent SD.
(H) OVA-primed WT mice were transferred with S1pr2-ERT2Cre R26tdTomato/+ B1-8hi cells, immunized i.p. with NP-OVA in alum, and treated with tamoxifen from day 2 every two days for the duration of the experiment. The frequency of tdTomato+ cells among AP-eMBCs and GCBCs is shown for the indicated time points. Horizontal lines indicate the mean and error bars represent SD. Shown are representative results of two independent experiments.
See also Figure S2.
The early cell cycle exit of activated B cells appeared to be consistent with their differentiation to eMBCs. Indeed, the CCR6+ cells expressed many MBC signature genes, including those shared by memory and naive B cells (e.g., Sell, Cd38, Klf2, and Hhex) and those that are upregulated in MBCs (e.g., Bhlhe41, Zbtb32, and Zeb2) (Bhattacharya et al., 2007; He et al., 2017; Jash et al., 2016; Laidlaw et al., 2020; Figures 2E and 2F). In fact, CCR6 itself is an MBC marker (Bhattacharya et al., 2007; Elgueta et al., 2015) that is expressed starting from the earliest stages of GC-MBC differentiation (Suan et al., 2017). The CCR6+ cell population did not exhibit high expression of the MBC subset markers Nt5e (CD73), Pdcd1lg2 (PD-L2), and Cd80 (Figure S2B; Zuccarino-Catania et al., 2014), in line with the reported absence of their expression by eMBCs (Taylor et al., 2012b; Weisel et al., 2016).
To test whether cells withdrawing from the cell cycle early in the response contributed to the long-term MBC pool, we employed pulse-chase EdU incorporation experiments (Weisel et al., 2016). We injected B1-8hi B cell-transferred mice with EdU daily for the first 3 days after NP-OVA immunization and subsequently analyzed whether any cells retained the EdU label long after the peak of the response (Figure 2G). The presence of the EdU label would indicate that a cell was proliferating at the time of EdU administration but thereafter underwent few divisions and, thus, did not dilute the label below the detection limit. The majority (~70%) of Igλ+ B1-8hi B cells incorporated EdU after three injections (Figure 2G). Mice analyzed 1 or 2 months later still contained 20% of Igλ+ B1-8hi B cells that retained the EdU label (Figure 2G), consistent with observations made by Weisel et al. (2016). The median fluorescence intensity of the EdU signal in EdU+ B cells decreased about 3.5-fold between the 4-day and 2-month time points (Figure S2C), indicating that these cells went through a minimal number of cell divisions during this period. Because EdU labeling was performed prior to emergence of a sizable number of GCBCs, and because the GC reaction involves vigorous proliferation resulting in rapid EdU loss, the EdU-retaining MBCs were likely generated by the GC-independent pathway. Similar results, but with a more homogeneous initial labeling of AP-eMBCs, were obtained when bromodeoxyuridine (BrdU) supplied in the drinking water was used for labeling (Figure S2D). Label-retaining MBCs were predominantly negative for CD73, CD80, and PD-L2 (Figure S2E) and were mostly unswitched but included some IgG1+ B cells (Figure 2G). Label-retaining MBCs were found in lymph nodes (LNs), bone marrow (BM), and blood, but the spleen was the main repository of these cells (Figure S2D). Label retention experiments with a shorter chase window demonstrated that a substantial loss of EdU still took place between days 4 and 7 after immunization, indicating that the AP-eMBC population underwent some residual proliferation in this time window (Figure S2F). The loss of EdU became much less prominent between days 7 and 10 (Figure S2F). Together with the scRNA-seq results, this suggested that CCR6+ B cells, by far the largest population at day 3.5–4 of the response in our model system, consisted of a majority of cells that began to withdraw from the response to turn into eMBCs and of a minority of APs that remained highly proliferative.
To test whether long-lived MBCs exit the cell cycle entirely or transit to a slow self-renewal-associated mode of proliferation, we measured BrdU incorporation by B1-8hi MBCs 4 months after immunization. On average, 5% of MBCs incorporated BrdU after 11 days of labeling at this late time point, indicating that at least some MBCs were indeed self-renewing (Figure S2G).
To test the actual contribution of non-GC derived cells to the overall MBC pool, we took advantage of the S1pr2-ERT2Cre R26tdTomato system that allows highly specific tamoxifen-inducible labeling of cells progressing through the GCBC state (Shinnakasu et al., 2016). S1pr2 was highly and selectively expressed in early GCBCs in our scRNA-seq datasets (Figures 1D and 1E). Fate mapping of S1pr2-ERT2Cre R26tdTomato/+ IghB1-8hi/+ B cells in NP-OVA-immunized recipient mice treated continuously with tamoxifen confirmed ~80% tdTomato labeling of GCBCs from day 4 after immunization (Figure 2H). In contrast, even at day 35 after immunization, only ~6% of MBCs expressed tdTomato (Figure 2H). The frequency of tdTomato+ GC-MBCs was not increased when a lower number of NP-specific B1-8hi B cells (5 × 103 rather than > 5 × 105) were transferred (data not shown), indicating that the high frequency of non-GC-derived MBCs is not caused by an unphysiologically high frequency of naive precursor cells. We conclude that a large fraction of APs early after immunization withdraws from participation in the response and that these non-GC derived eMBCs can make a large contribution to the long-lived MBC pool.
Early lineage split of PBs takes place in a transient wave
A single naive B cell can give rise to GCBCs, PBs, and eMBCs (Taylor et al., 2015) and thus has to progress through a proliferative tripotent AP stage. Although our scRNA-seq experiment at day 3.5–4 after immunization suggested a possible trajectory of the AP-GCBC and AP-eMBC transitions, the PB population remained separated from the APs (Figure 1F). To test whether the divergence of the PB lineage took place even earlier in the response, we performed scRNA-seq with transferred Igλ+ B1-8hi B cells at day 2.5 after immunization (Figure 3). Analysis of 7,324 cells identified two populations corresponding to CCR6+ APs and to differentiating PBs (Figures 3A and S3A). In line with the results obtained at this early time point by flow cytometry (Figure 1A), this analysis did not reveal cells with the GCBC signature (Figure S3A). In contrast to the day 3.5–4 dataset, the majority of cells in the CCR6+ cell population exhibited a pro-liferation signature (Figures 3A and S3A). Moreover, a possible transition from APs to PBs could be inferred (Figure 3A), which was further supported by the direction of RNA velocity vectors (Figure S3B). Consistent with recent reports suggesting that antibody class switching occurs primarily prior to the GC reaction (Roco et al., 2019; Sundling et al., 2021), APs at day 2.5 already expressed genes encoding enzymes, which are required for class-switch recombination (Aicda, Ung, and Apexl) (Figure 3B), and underwent substantial switching to IgG1 at this early time point, whereas the PB compartment exhibited slightly lower frequencies of switched cells (Figure 3C).
Figure 3. Early lineage split of PBs takes place in a transient wave.
(A) scRNA-seq was performed as in Figure 1 but with B1-8hi cells sorted at day 2.5 after immunization. The assignment of cells to different cell types (left) and phases of the cell cycle (right) is shown on a UMAP plot.
(B) Expression of class-switch recombination-related genes is shown on a UMAP plot as in (A).
(C) Expression of IgM and IgG1 on Igλ+ B1-8hi APs (CCR6+) and PBs (IRF4+CD138+) at day 2.5 after immunization. Experimental setup as in Figure 1A. Pooled data from three independent experiments. *p < 0.05 (paired two-tailed Student’s t test).
(D–F) Analysis of the combined scRNA-seq data of day 2.5 and 3.5–4.
(D) UMAP plot showing the contribution of cells from the different time points and expression of the indicated marker genes.
(E) Trajectory analysis with the Slingshot package (STAR Methods). Trajectories are plotted on a UMAP plot (left), smoothed expression of the marker genes in cells sorted by pseudotime along the indicated trajectories is shown (center and right).
(F) RNA velocity analysis of the combined day 2.5/3.5–4 dataset was performed and visualized as in Figure 1F.
(G) Unprimed WT mice were transferred with congenically distinguishable B1-8hi splenocytes, immunized i.p. with NP-OVA in alum, and analyzed at day 2.5 and 5 after immunization. Top: expression of CD138 and IRF4 by Igλ+ B1-8hi B cells was used to identify PBs. Bottom: expression of CD22 and MHC class II on day 2.5 PBs, day 5 PBs and non-PB Igλ+ B1-8hi cells (day 5). Shown are representative results of two independent experiments.
See also Figure S3.
To gain insight into molecular changes taking place in activated Bcell populations between days 2.5 and 3.5–4 after immunization, we performed pairwise comparisons of populations between the two time points. Comparison of PBs detected at day 2.5 and day 3.5–4 revealed upregulation of gene sets associated with the unfolded protein response as well as upregulation of the PC signature at the later time point (Figure S3C), suggesting ongoing maturation of these cells. Comparison of the AP-eMBC population between these two time points confirmed downregulation of gene sets associated with proliferation as well as metabolic activity (Figure S3C).
To map the trajectories of AP differentiation to the three "effector lineages,” we merged the datasets of the two time points for further bioinformatics analysis. The cells of day 2.5 "closed the gap” between the different cell populations identified at day 3.5–4, allowing us to envision a possible course of differentiation for all three lineages (Figure 3D). Indeed, the trajectory analysis based on the Slingshot package (Street et al., 2018) mapped the probable paths of PB, GCBC, and eMBC differentiation and identified expected gradients of gene expression along these trajectories (Figure 3E). The RNA velocity analysis likewise suggested centrifugal divergence of GCBCs and PBs from the AP-eMBC population (Figure 3F). Thus, scRNA-seq-mediated dissection of early B cell activation predicted trajectories of GCBC, PB, and eMBC differentiation from APs.
To find transcriptional regulators that may influence the differentiation of these three lineages, we next performed SCENIC analysis, an approach that uses information about co-expression of genes encoding transcription factors and their predicted target genes in a scRNA-seq dataset (Aibar et al., 2017). This analysis identified several known regulators of antigen-dependent B cell differentiation, including Xbp1 for PBs as well as Bach2 (Muto et al., 2004) and Mef2b (Brescia et al., 2018) for GCBCs (Figure S3D). In addition, it predicted the transcription factors Foxp1, Bhlhe41, Chd2, and Ets1 as possible regulators of eMBCs, the transcription factor Irf7 as a candidate regulator of PBs, and the unfolded protein response-related transcription factor Atf6 as a possible regulator of APs and PBs (Figure S3D).
Analysis of the merged dataset described above (Figure 3D) suggested the unexpected scenario that PBs may predominantly differentiate from APs in a transient wave very early after immunization. To further test this hypothesis, we took advantage of the fact that PBs downregulate expression of Cd22 (encoding a cell surface lectin) and Ciita (encoding a central regulator of major histocompatibility complex [MHC] class II genes) (Minnich et al., 2016; Figures 3E, S3E, and S3F). Flow cytometry analysis confirmed that expression of CD22 and MHC class II was high in activated B cells but lost in PBs at day 5 after immunization (Figure 3G). PBs at day 2.5 expressed intermediate levels of both cell surface proteins, providing a “missing link” between day 5 PBs and activated B cells (Figure 3G), consistent with the scRNA-seq-based observations described above (Figure 3D). These results indicate that, in our experimental setting, the majority of the non-GC-derived PBs are generated in a narrow time window very early after immunization and that the contribution of APs to the PB compartment at later time points is limited.
eMBCs remain transcriptionally similar to APs
We next compared the magnitude of transcriptional changes underlying differentiation of the precursors to PB, GCBC, and eMBC lineages. As a first approach to this problem, we calculated the diffusion pseudotime (DPT), a metric that attempts to quantify the transcriptional changes that take place during differentiation. Progression of cells along the putative GCBC and PB trajectories was associated with prominent changes in DPT, suggesting rapid transcriptional divergence of these cells from APs (Figure 4A). Gene set enrichment analysis (GSEA) of genes, ranked by their correlation with DPT, confirmed that the pseudotime reflected the expected biological changes, such as the induction of signatures associated with the unfolded protein response and protein secretion for PBs and DNA repair and proliferation for GCBCs (Figures 4B and S4A). In contrast to PB and GCBC trajectories, DPT exhibited very limited change within the AP-eMBC population, which was only revealed when PB and GCBC populations were excluded from the analysis (Figures 4A and S4B). This low heterogeneity within the AP-eMBC population suggested that eMBCs may retain high transcriptional similarity to APs. To further test this hypothesis, we compared Euclidean distances between cells that belong to different populations, focusing the analysis on the day 3.5–4 dataset to avoid any possible distortion caused by batch effects. In all of our previous analyses (Figures 1C and 3E), APs and eMBCs could not be clearly separated from each other, suggesting that they represent a continuum of transcriptionally similar cell states and that no distinct populations corresponding to APs and eMBCs can be defined. Nevertheless, to avoid sampling Euclidean distances between neighboring cells in the AP-eMBC population, we took advantage of an analysis performed with the Seurat package (Stuart et al., 2019) that splits the AP-eMBC population into three clusters (Figures 4C and 4D). In line with our hypothesis, these three AP-eMBC clusters were more related to each other than to GCBCs or PBs (Figure 4D). Analysis of Euclidean distances in the PCA space between pairs of cells in these clusters confirmed homogeneity of the AP-eMBC population, whereas GCBCs and PBs diverged away from the AP-eMBC group and from each other (Figures 4E and S4C). In fact, the median distances between cells in the different AP-eMBC clusters were lowerthan the intracluster distances within GCBCs and PBs (Figures 4E and S4C).
Figure 4. Transcriptional relatedness between populations in early B cell activation.
(A) Diffusion pseudotime (DPT), calculated as described in STAR Methods, plotted on a UMAP of the combined day 2.5/3.5–4 dataset.
(B) Gene set enrichment analysis (GSEA) of genes ranked by correlation with DPT in the indicated populations using hallmark gene sets from MSigDB (all sets that were significantly enriched at least for one population are shown) as well as BCR/CD40 signaling-related gene sets (Victora et al., 2010). Normalized enrichment score (NES) absolute value is indicated by circle size. False discovery rate (FDR) q value is indicated by color intensity (or in gray for FDR q > 0.05). Positive and negative NESs are depicted by red and blue, respectively.
(C) UMAP showing cluster assignment of cells in the day 3.5–4 dataset using the Seurat package.
(D) Cluster dendrogram for clustering shown in (C).
(E) Heatmap of median Euclidian distances in PCA space between pairs of cells in clusters defined in (C).
(F) OVA-primed WT mice were transferred with congenically distinguishable B1-8hi splenocytes and immunized i.p. with NP-OVA in alum. APs-eMBCs, GCBCs, and PBs were sorted at the indicated time points after immunization and subjected to bulk RNA-seq. Principal-component analysis (PCA) of this dataset is shown. Cell cycle-related genes were excluded from this analysis (STAR Methods).
See also Figure S4.
The analyses described above demonstrated that proliferative APs and the earliest MBCs are transcriptionally similar to each other. To test whether the long-lived eMBCs likewise retain transcriptional similarity to early APs, we performed bulk RNA-seq comparison of day 2.5 APs; day 4 AP-eMBCs, GCBCs, and PBs; and day 50 MBCs, taking advantage of the observation that the majority of B1-8hi MBCs were generated by the GC-independent pathway (Figure 2H). Although the transcriptomes of GCBCs and PBs diverged away from that of the AP-eMBC population at day 4, MBCs at day 50 retained a high transcriptional similarity to the day 2.5 and day 4 AP-eMBC populations (Figures 4F and S4D). We therefore conclude that eMBCs remain transcriptionally similar to the common APs of the three lineages.
Limited access to antigen drives cell cycle exit associated with eMBC differentiation while restraining the PB response
We next aimed to understand what makes a large fraction of the activated B cells withdraw from the response very early to contribute to the MBC pool. Because antigen and T cell help are key drivers of B cell proliferation, we tested whether limited access to these mitogenic stimuli may cause cell cycle exit of eMBCs. Provision of excessive T cell help by co-transfer of OT-II T cell receptor (TCR) transgenic T cells with B1-8hi B cells prior to NP-OVA immunization resulted in strongly increased expansion of antigen-specific B cells, a prominent increase in GCBCs, and a modest increase in PBs (Figure S5A). Nevertheless, the CCR6+ AP-eMBC population still represented a large fraction of activated B cells, and downregulation of Ki67 by these cells was not impaired in the presence of OT-II cells despite OT-II cells constituting 22% of all CD4 T cells at the time of analysis (Figure S5A). We therefore turned our attention to BCR signaling and the kinetics of antigen availability. Analysis of expression of the nuclear receptor NUR77, commonly used as a surrogate marker for recent antigen receptor signaling (Moran et al., 2011; Tan et al., 2020; Zikherman et al., 2012), revealed its rapid upregulation by the majority of Igλ+ B1-8hi B cells 12 h after immunization and its downregulation 2 days later (Figure 5A). To test whether the rapid decline in NUR77 levels could reflect decreased antigen availability, we analyzed the interaction of B1-8hi B cells with antigen in vivo. For this, we immunized mice, transferred with B1-8hi B cells, with AF488-labeled NP-BSA and quantified the antigen-BCR interaction by flow cytometry. These experiments revealed that the majority of Igλ+ B1-8hi B cells acquired high amounts of antigen 12 h after immunization (Figure 5B). Antigen uptake coincided with downregulation of Igλ, likely reflecting BCR internalization, and with upregulation of CD69 (Figure S5B). The median fluorescence intensity (MFI) of NP-BSA-AF488 exhibited an 8-fold decline already by day 2.5 and was decreased further on day 4. We conclude that cell cycle exit of a large fraction of APs coincides with a strongly reduced interaction of the activated B cells with antigen that results in decreased BCR signaling. Although we could not reliably detect NP-BSA-AF488 in the spleens by means of confocal microscopy, i.v. immunization experiments with phycoerythrin (PE) demonstrated that PE was broadly available in the spleen in the first hours after immunization but by day 4 was predominantly, if not exclusively, associated with follicular dendritic cell (FDC) clusters (Figure S5C), suggesting that, at this time point, antigen may remain accessible mostly to GCBCs.
Figure 5. Antigen availability regulates the balance between the eMBC and PB response.
(A) WT mice were transferred with congenically distinguishable B1-8hi splenocytes, immunized with NP-BSA, and analyzed at indicated time points. Intracellular expression of NUR77 by Igλ+ and Igλ− B1-8hi B cells is shown.
(B) WT (CD45.2) mice were transferred with CD45.1/CD45.2 heterozygous B1-8hi splenocytes and immunized i.p. with NP-BSA-AF488 in alum. The interaction of Igλ+ B1-8hi B cells with NP-BSA-AF488 at the indicated time points after immunization was detected by flow cytometry. Naive CD45.1 B1-8hi splenocytes were “spiked in” into the samples during preparation of single-cell suspension to control for possible antigen carryover during staining (gray histograms). Because the transferred cells at days 2.5 and 4 were much larger than the spike-in cells (Figure S5B), the NP-BSA-AF488 binding at these two time points is likely to be overestimated because of increased autofluorescence. Shown are representative results of two independent experiments.
(C–E) OVA-primed WT mice were transferred with congenically distinguishable B1-8hi splenocytes and immunized i.p. with NP-OVA in alum. On day 3.5 after immunization, mice were injected with NP-OVA i.v. or left untreated.
(C) Mice were analyzed at day 4 after i.p. immunization, with EdU being injected 4 h before analysis. EdU incorporation by CCR6+Igλ+ B1-8hi B cells is shown. Shown are representative results of two independent experiments.
(D) Mice were injected with EdU at days 1, 2, and 3 after i.p. immunization, and label retention by CCR6+Igλ+ B1-8hi B cells was analyzed at day 6 by gating on EdUhi cells. Shown are representative results of two independent experiments.
(E) The population composition of Igλ+ B1-8hi B cells was analyzed at day 6 after i.p. immunization by staining with antibodies against CD19, CD138, CCR6, and GL7. Shown are representative results of four independent experiments.
(F) Unprimed WT mice were transferred with congenically distinguishable B1-8hi splenocytes and immunized i.p. with NP-OVA in alum. On day 3.5 after immunization, mice were injected with NP-OVA i.v. or left untreated. Mice were analyzed on day 4.5 after i.p. immunization. Gating for PBs on Igλ+ B1-8hi B cells is shown. Expression of CD22 and MHC class II was used to define and quantify immature (MHC class IIintCD22int iPB) and mature (MHC class II− CD22− mPB) PBs. Shown are representative results of two independent experiments.
(C–F) *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed Student’s t test). Horizontal lines indicate the mean, and error bars represent SD.
See also Figure S5.
We reasoned that, if limited antigen availability indeed caused cell cycle exit of differentiating eMBCs, then provision of antigen excess should interfere with this process. In line with this notion, i.v. injection of additional antigen 3.5 days after immunization resulted in a strong increase in EdU incorporation by CCR6+ B1-8hi B cells half a day later (Figure 5C). Moreover, when we performed EdU labeling during the first 3 days after immunization prior to the second i.v. antigen administration, the frequencies and numbers of EdUhi label-retaining cells were decreased in the antigen-treated group. This result indicated that, when antigen availability was restored, the EdU-labeled quiescent APs-eMBCs at day 3.5 were recruited again into the cell cycle, leading to EdU loss (Figure 5D).
Consistent with enhanced proliferation, increased antigen availability resulted in an overall increase in the numbers of antigen-specific B cells (Figure 5E). Although GCBC and AP-eMBC frequencies and numbers were affected only moderately by increased antigen availability, the PB compartment was expanded strongly in response to additional antigen, accounting for up to 40% of all Igλ+ B1-8hi cells (Figure 5E). To test whether this increase in PB numbers was caused by expansion of the existing PBs or by their de novo differentiation, we utilized MHC class II and CD22 downregulation to monitor PB maturation. As expected, without provision of additional antigen, almost all PBs at day 4.5 after immunization exhibited a mature MHC class II−CD22− PB (mPB) phenotype (Figure 5F). Provision of antigen excess did not cause any changes in the number or phenotype of these mPBs but, instead, resulted in emergence of a new wave of immature MHC class IIintCD22int PBs (iPBs). Finally, comparison of the response of naive B cells to i.v. NP-OVA injection with that of activated B cells in mice that were “pre-immunized” i.p. with NP-OVA in alum 3.5 days before i.v. NP-OVA injection confirmed a ~10-fold increase in the frequency of PBs in the latter case (Figure S5D). These results demonstrate that limited antigen availability contributes to the early cell cycle exit of APs and that provision of additional antigen not only “rescues” this withdrawal from the response but also enhances differentiation to the PB lineage.
Early cell cycle exit of activated B cells takes place in polyclonal immune responses
The scRNA-seq experiments described above required transfer of high numbers of antigen-specific B cells. Thus, the observed early cell cycle exit of activated B cells could have been caused by unphysiologically high competition for antigen. To test whether this is the case, we compared the early immune response in mice transferred with approximately 5 × 105, 5 × 104, or 5 × 103 antigen-specific B1-8hi B cells, with the latter number well within the range of naive B cells specific for protein antigens (Pape et al., 2018). Despite some variability in frequencies of GCBCs and PBs, the CCR6+ AP-eMBC population remained the most prominent cell compartment at day 4 after immunization under all three conditions (Figure 6A), and a large fraction of these cells was Ki67lo/- (Figure 6A). Thus, the early cell cycle exit of the eMBCs also takes place in immune responses starting with physiological numbers of naive precursors.
Figure 6. Early cell cycle exit of eMBCs takes place with physiologically low numbers of naive B cells.
(A) OVA-primed WT mice were transferred with congenically distinguishable B1-8hi splenocytes containing an estimated number of 5 × 105, 5 × 104, or 5 × 103 NP-specific B cells. The population composition of Igλ+ B1-8hi cells and Ki67 expression in the CCR6+ population was analyzed at day 4 after i.p. immunization with NP-OVA in alum. Shown are representative results of two independent experiments.
(B) WT mice were immunized i.p. with NP-OVA in alum. NP-specific cells among Igλ+ B cells were detected by dual staining with NP-PE and NP-BSA-bio/Strep-BV650. Intracellular staining for BCL6, IRF4, and Ki67 was performed, and Ki67 expression in GCBC, PB, and AP-eMBC populations was analyzed as indicated. **p < 0.01 (paired one-way ANOVA with Geisser-Greenhouse correction). Horizontal lines indicate the mean and error bars represent SD.
(C) WT or S1pr2-ERT2cre R26tdTomato/+ mice were immunized i.p. with NP-OVA in alum, and S1pr2-ERT2cre R26tdTomato/+ mice were treated by oral gavage with tamoxifen at days 2, 4, and 6 after immunization. Expression of tdTomato at day 7 is shown for the NP-specific (NP-BSA-bio/Strep-BV650+) Igλ+ PB (TACI+-CD138+), GCBC (GL7hiIgλint), and AP-eMBC (GL7lo/intIgλhi) populations. Shown are representative results of two independent experiments.
(D) Mice were treated as in (C) but for up to 45 days (tamoxifen treatment every 2 days for the duration of the experiment). NP-specific cells among Igλ+ B cells were detected with dual staining using NP-CGG-AF488 and NP-BSA-bio/Strep-PECy7 (STAR Methods). Frequencies of tdTomato+ cells among MBCs and GCBCs at the indicated time points are shown. Shown are representative results of two independent experiments. Horizontal lines indicate the mean, and error bars represent SD.
(E) Mice were infected intranasally with IAV. Cells from mediastinal LNs pooled from nine animals at day 7 post infection were analyzed by flow cytometry. Binding of hemagglutinin (HA) probes to B-lineage cells (CD19+ and/or CD138+) and intracellular expression of IRF4, BCL6, and Ki67 in HA-specific B cells is shown. Splenic naive B cells (CD19+IgD+) of a non-infected mouse were used as a control. Shown are representative results of two independent experiments.
See also Figure S6.
We next tested whether early withdrawal from the response of activated B cells also takes place in a polyclonal response in NP-OVA-immunized WT mice. To this end, we analyzed expression of IRF4 and BCL6 by intracellular staining to identify GCBCs (BCL6+), PBs (IRF4+), and AP-eMBCs (BCL6lo/−IRF4lo/−) (Zhang et al., 2017). Similar to the B1-8hi B cell responses described above, many NP-specific B cells in the AP-eMBC compartment were Ki67lo/− at day 4 and day 7 after immunization, suggesting that some of these cells were transiting to the quiescent G0 state (Figure 6B). EdU labeling during the first days of the polyclonal response to NP-OVA immunization confirmed that all label-retaining cells at day 7 were found in the BCL6lo/−IRF4lo/− AP-eMBC compartment (Figure S6A). EdU incorporation experiments demonstrated that, although the AP-eMBC population at day 7 after immunization in the polyclonal setting proliferated more compared with the B1-8hi system (Figures S6B and 2C), this population was much less proliferative than GCBCs and PBs (Figure S6B). These results suggested that APs in the polyclonal response to NP-OVA immunization also slow down their proliferation early in the response, indicative of their likely eMBC differentiation. However, in these as well as other experiments with polyclonal responses described below, substantial numbers of antigen-specific B cells could be detected only at day 7 after immunization. Because GCBCs were a prominent cell population at this time point (Figure 6B), it was conceivable that some of these MBCs would already be GC derived. To quantify the contribution of GCBCs to the MBC compartment, we again took advantage of the S1pr2-ERT2Cre R26tdTomato system to label the cells that progressed through the GCBC state. We treated S1pr2-ERT2Cre R26tdTomato/+ mice with tamoxifen every 2 days for the whole duration of the experiment, starting from day 2 after NP-OVA immunization. Analysis at day 7 revealed that, although more than 70% of GCBCs were tdTomato+ at this time point, only 2%–3% of tdTomato+ cells could be detected in the PB and AP-eMBC compartments (Figure 6C). Of note, these few labeled APs-eMBCs expressed intermediate levels of tdTomato (Figure 6C), indicating a recent stop cassette excision and suggesting that, rather than representing GC-MBCs, these cells are likely APs that just have initiated GCBC differentiation. These results indicated that very few, if any, MBCs detected at day 7 after immunization originated from GCBCs. Moreover, in line with fate mapping experiments performed with B1-8hi B cells (Figure 2H), the majority (~70%) of polyclonal NP-specific Igλ+ MBCs at day 45 after immunization were likewise tdTomato− (Figure 6D). Thus, the long-lived MBC pool resulting from a polyclonal response to NP-OVA immunization included a large fraction of cells that did not participate in the GC reaction.
Finally, we investigated whether a similarly early withdrawal from the response is also observed upon pathogen encounter. To this end, we infected WT mice with influenza A virus (IAV) and analyzed hemagglutinin (HA)-specific B cells in mediastinal LNs at day 7 after infection. At this time point, the virus is largely cleared from the lungs in this model (Ginsberg and Horsfall 1952), and antigen access has likely become limited. Concordant with the observed immune responses to model antigens, BCL6lo/−IRF4lo/− cells constituted a large fraction of antigen-specific B cells, and the majority of these cells were Ki67lo (Figure 6E). Similar to the responses to NP-OVA immunization, HA-specific APs-eMBCs at day 7 after infection incorporated much less EdU compared with PBs and GCBCs (Figure S6C). We conclude that the early cell cycle exit of many activated B cells, likely manifesting their eMBC differentiation, also takes place in polyclonal responses to immunization and infection.
Evolutionary conservation of eMBC differentiation in primates
Although early B cell activation is an area of intensive investigation in mice, virtually nothing is known about these responses in primates. A recent report demonstrated that, upon nanoparticle immunization of rhesus macaques, antigen-specific GCBCs peak around day 21 after immunization, whereas the response at day 7 is dominated by BCL6− B cells, exhibiting a broad range of Ki67 expression, which the authors referred to as PBs (Havenar-Daughton et al., 2019). To test whether the pathway that leads to eMBC differentiation also exists in primates, we next analyzed the early B cell response in macaques. To this end, animals were immunized intramuscularly with nanoparticles or “soluble” vaccines containing the prefusion-stabilized variant of the respiratory syncytial virus (RSV) fusion glycoprotein (RSVF) trimer (DS-Cav1) in a squalene-in-water emulsion (SWE) adjuvant (Marcandalli et al., 2019). Analysis of antigen-specific B cells in the draining LNs of these animals at day 7 after immunization revealed the presence of relatively low numbers of BCL6hi GCBCs and IRF4hi PBs (Figures 7A and S7A). The most prominent population of antigen-specific B cells corresponded to BCL6lo/−IRF4lo/− B cells, many of which expressed the primate MBC markers CCR6, CD27, and CD95 (Kuhrt et al., 2011; Suan et al., 2017) and were mostly unswitched but included a fraction of IgG+ cells (Figures 7A and S7A). In full agreement with the results obtained in mice, these cells were largely Ki67lo/− (Figures 7A and S7A). To test how antigen availability was changed in the course of the response, we took advantage of the fact that the “soluble” vaccine was brightly labeled with AF647. Although a strong AF647 signal could be detected throughout the draining LNs as early as 2 h after immunization, the AF647 signal was detected exclusively on FDC clusters within follicles at days 3 and 7 after immunization (Figure 7B). We conclude that a large fraction of activated B cells in primates also withdraws from the response soon after the initial expansion. Like in mice, these cells upregulate memory cell markers, and emergence of these quiescent cells with an MBC phenotype coincides with limited antigen availability outside of GCs. Hence, differentiation of eMBCs appears to be evolutionarily conserved in rodents and primates.
Figure 7. Early cell cycle exit and induction of MBC marker expression on antigen-specific B cells in immunized rhesus macaques.
Rhesus macaques were immunized with AF647 fluorophore-labeled RSVF immunogens (provided as nanoparticles or soluble vaccine) by intramuscular injection to study the vaccine-draining LNs 2 h, 3 days or 7 days after immunization (STAR Methods).
(A) Flow cytometric analysis of LNs 7 days after nanoparticle immunization. B-lineage cells were gated asCD20+ and/or IRF4+, and antigen-specific B cells were identified by dual staining with RSVF-AF488 and RSVF-BV421 tetramer probes. Intracellular expression of BCL6, IRF4, Ki67, IgM, and IgG as well as surface expression of CCR6, CD27, and CD95 by antigen-specific B cell subpopulations (gated as indicated) are shown. Quantification is shown for individual draining LNs of the same immunized animal. Downregulation of Ki67 expression by antigen-specific B cells was confirmed in cryo-preserved samples from two additional animals (data not shown) at day 7 after immunization. Horizontal lines indicate the mean, and error bars represent SD.
(B) Representative images of the fluorescent soluble vaccine (red) localization in LNs stained for CD3 expression (white) to visualize LN structure. LNs after 2 h, 3 days, and 7 days after immunization are shown (left and center). Shown is a representative image of a LN follicle displaying co-localization of the vaccine (red) with CD35+ follicular dendritic cells (cyan) (right).
See also Figure S7.
Discussion
In this study, we dissected the cell fate decisions that take place early upon B cell activation. Although it is well documented that activated B cells progress through a tripotent AP state and can give rise to GCBCs, PBs, and eMBCs (Taylor et al., 2015), the hierarchy and timing of choices between these lineages was not well understood. Our scRNA-seq analysis demonstrated that, very early in the response, APs give rise to a transient wave of PBs, followed by emergence of GCBCs and eMBCs. The latter fate was taken by a very large fraction of APs, and these non-GC-derived eMBCs made a prominent contribution to the overall MBC pool at the end of the response. Although the transcriptomes of GCBCs and PBs diverged rapidly from that of APs, APs and eMBCs were transcriptionally very similar. The generation of PBs and GCBCs from a common precursor exhibiting transcriptional similarity to MBCs is an interesting parallel to the secondary immune response, where MBCs give rise to PCs and can contribute to the GC reaction.
Intriguingly, our fate mapping experiments suggested a very high contribution of eMBCs to the overall memory pool in the B1-8hi system. This contribution, although still very prominent, was lower in the case of the polyclonal response to NP-OVA immunization. These observations seem to be consistent with a recent report demonstrating that high-affinity germline-encoded recognition of PE by B cells in Ighb mice results in the generation of predominantly unswitched MBCs, whereas lack of such germline-encoded specificity in the Igha strains leads to formation of a MBC compartment in which approximately half of the cells are switched (Pape et al., 2018). In the future, it will be of interest to perform a functional comparison of eMBCs with GC-MBCs. It also remains to be seen whether eMBCs are functionally different from naive B cells or merely provide a pre-expanded pool of antigen-specific B cells.
The early cell cycle exit of APs and the short transient wave of PBs in our system was, at least in part, caused by limited antigen availability because the provision of antigen excess delayed the cell cycle exit and prolonged PB differentiation. Multiple studies have shown that common routes of immunization, many of which are also used for vaccination in humans, result in a short transient wave of antigen availability in secondary lymphoid organs (Hutchison et al., 2012; Moon et al., 2012; Moyer et al., 2020; Pape et al., 2007). When antigen is still detectable at the later time points, it has been shown to be exclusively associated with FDC networks in GCs (Ols et al., 2020; Tam et al., 2016; Tokatlian et al., 2019). We observed a similar spatiotemporal pattern of antigen distribution in this study upon immunization of rhesus macaques. These observations suggest that, in common immunization scenarios, the initial broad antigen availability induces activation of cognate B cells, but as antigen rapidly becomes limited to GCs, further B cell proliferation or PB differentiation outside of GCs is restricted. Delivery of antigen in multiple doses or with osmotic minipumps can enhance antibody responses in part by increasing the magnitude of the GC reaction (Cirelli et al., 2019; Tam et al., 2016). Our results suggest that prolonged PB generation from AP-eMBCs can also contribute to the increased antibody titers observed in these studies.
MBC function is usually considered in the context of secondary immune responses. We report here, in a variety of experimental systems, that quiescent B cells with an MBC phenotype constitute a large fraction of antigen-specific B cells early in the response and that an increase in antigen availability results in a rapid new wave of PB differentiation. These results suggest that increased antigen availability, which, in the case of responses to pathogens, can manifest failure to contain the infection, could rapidly recruit MBCs to participate in the primary response to generate PBs. Such a demand-based mechanism of PB generation should allow rapid readjustment of the balance between quiescent MBCs and the energetically and metabolically costly PB responses proportional to the pathogen level, which could provide a possible explanation for the PC-skewed potential of MBCs (Mesin et al., 2020). Of note, the metabolic costs of excessive PB responses have been shown recently to directly impair the GC branch of the humoral immune response through nutrient deprivation, interfering with the generation of high-affinity antibodies (Vijay et al., 2020).
Limitations of study
We report here that the rapid decline in antigen availability after immunization drives an early transition of APs to the quiescent eMBC state and restricts PB generation to a narrow time window early in the response. Such a decline in antigen availability is observed in many common immunization scenarios and is likely to take place in some acute infections. Nevertheless, the exact timing of all of these events and the ultimate contributions of eMBCs and GC-MBCs to the overall memory pool are likely to vary depending on the nature of the challenge, as exemplified here by delayed cell cycle exit of eMBCs and prolonged generation of PBs upon provision of additional antigen.
Star⋆Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AF488 Rat Anti-Mouse IgD (Clone 11-26c.2a) | BioLegend | Cat#405717; RRID: AB_10730618 |
| Biotin Rat Anti-Mouse CD21/35 (Clone CR2/CR1) | BioLegend | Cat#123405; RRID: AB_940399 |
| Biotin Recombinant Human Anti-Mouse CD169 (Siglec-1) (Clone REA197) | Miltenyi Biotec | Cat#130-105-004; RRID: AB_2655540 |
| BV711 Rat Anti-Mouse Ig, λ1,λ2 & λ3 Light chain (Clone R26-46) | BD Biosciences | Cat#744527; RRID: AB_2742301 |
| FITC Rat Anti-Mouse Ig λ Light Chain (Clone JC5-1) | Miltenyi Biotec | Cat#130-098-415; RRID: AB_2661439 |
| PerCP-Cy5.5 Mouse Anti-Mouse CD45.1 (Clone A20) | BD Biosciences | Cat#560580; RRID: AB_1727489 |
| PerCP-Cy5.5 Mouse Anti-Mouse CD45.1 (Clone A20) | BioLegend | Cat#110727; RRID: AB_893348 |
| BV605 Mouse anti-Mouse CD45.1 (Clone A20) | BioLegend | Cat#110738; RRID: AB_2562565 |
| PE-Cy7 Mouse anti-Mouse CD45.1 (Clone A20) | BioLegend | Cat#110730; RRID: AB_1134168 |
| BV510 Mouse Anti-Mouse CD45.2 (Clone 104) | BioLegend | Cat#109838; RRID: AB_2650900 |
| FITC Mouse Anti-Mouse CD45.2 (Clone 104) | BioLegend | Cat#109805; RRID: AB_313442 |
| PE Armenian Hamster Anti-Mouse CD69 (Clone H1.2F3) | BioLegend | Cat#104507; RRID: AB_313110 |
| BV786 Rat Anti-Mouse CD19 (Clone 1D3) | BD Bioscience | Cat#563333; RRID: 2738141 |
| PE-Cy7 Mouse Anti-Mouse Nur77 (Clone 12.14) | eBioscience | Cat#25-5965-80; RRID: AB_2811784 |
| APC-eFluor 780, Rat Anti-Mouse Ki67 (Clone SolA15) | eBioscience | Cat#47-5698-82; RRID: AB_2688065 |
| PE Recombinant Human Anti-Mouse CD196 (CCR6) (Clone REA277) | Miltenyi Biotec | Cat#130-103-816; RRID: AB_2655942 |
| BV605 Armenian Hamster Anti-Mouse CD196 (CCR6) (Clone 29-2L17) | BioLegend | Cat#129819; RRID: AB_2562513 |
| AF647 Rat Anti-Mouse GL7 (Clone GL7) | BioLegend | Cat#144606; RRID: AB_2562185 |
| Pacific Blue Rat Anti-Mouse GL7 (Clone GL7) | BioLegend | Cat#144614; RRID: AB_2563292 |
| PerCP-Cy5.5 Rat Anti-Mouse GL7 (Clone GL7) | BioLegend | Cat#144610; RRID: AB_2562979 |
| PE-eFluor 610 Rat Anti-Mouse/Human IRF4 (Clone 3E4) | eBioscience | Cat#61-9858-82; RRID: AB_2637108 |
| PE Recombinant Human Anti-Mouse CD22 (Clone REA1187) | Miltenyi Biotec | Cat#130-122-045; RRID: AB_2784069 |
| VioBlue Recombinant Human Anti-Mouse MHC Class II (Clone REA813) | Miltenyi Biotec | Cat#130-112-394; RRID: AB_2652908 |
| APC Recombinant Human Anti-Mouse/Human BCL6 (Clone REA373) | Miltenyi Biotec | Cat#130-122-013; RRID: AB_2801827 |
| PerCP-Cy5.5 Armenian Hamster Anti-Mouse CD3e (Clone 145-2C11) | BioLegend | Cat#100328; RRID: AB_893318 |
| PerCP-Cy5.5 Rat Anti-Mouse Ly-6G/Ly-6C (Gr-1) (Clone RB6-8C5) | BioLegend | Cat#108427; RRID: AB_893561 |
| PE-Cy7 Rat Anti-Mouse CD138 (Syndecan-1) (Clone 281-2) | BioLegend | Cat#142514; RRID: AB_2562198 |
| BV421 Rat Anti-Mouse CD138 (Syndecan-1) (Clone 281-2) | BioLegend | Cat#142523; RRID: AB_2565621 |
| PE-Vio770 Recombinant Human Anti-Mouse CD138 (Clone REA104) | Miltenyi Biotec | Cat#130-102-318; RRID: AB_2655025 |
| APC Rat Anti-Mouse CD138 (Clone 281-2) | BD Biosciences | Cat#561705; RRID: AB_10896819 |
| APC-Vio770 Recombinant Human Anti-Mouse IgG1 (Clone REA1017) | Miltenyi Biotec | Cat#130-117-104; RRID: AB_2733155 |
| Biotin Rat Anti-Mouse IgG1 (Clone RMG1-1) | BioLegend | Cat#406603; RRID: AB_315062 |
| BUV395 Rat Anti-Mouse IgD (Clone 11-26c.2a) | BD Biosciences | Cat#565988; RRID: AB_2737433 |
| FITC Recombinant Human Anti-Mouse IgM (Clone REA979) | Miltenyi Biotec | Cat#130-116-311; RRID: AB_2727466 |
| FITC Rat Anti-Mouse IgM (Clone RMM-1) | BioLegend | Cat#406505; RRID: AB_315055 |
| Biotin Rat Anti-Mouse IgM (Clone RMM-1) | BioLegend | Cat#406503; RRID: AB_315053 |
| PE Rat Anti-Mouse CD3 (Clone 17A2) | BioLegend | Cat#100205; RRID: AB_312662 |
| PE Rat Anti-Mouse CD4 (Clone GK1.5) | eBioscience | Cat#12-0041-83; RRID: AB_465507 |
| PE Armenian Hamster Anti-Mouse TCR β (Clone H57-597) | BioLegend | Cat#109208; RRID: AB_313431 |
| PE Mouse Anti-Mouse TCR gamma/delta (Clone eBioGL3 (GL-3, GL3)) | eBioscience | Cat#12-5711-82; RRID: AB_465934 |
| PE Armenian Hamster Anti-Mouse CD11c (Clone HL3) | BD Biosciences | Cat#553802; RRID: AB_395061 |
| PE Mouse Anti-Mouse NK1.1 (Clone PK136) | BioLegend | Cat#108708; RRID: AB_313395 |
| PE Rat Anti-Mouse TER-119 (Clone TER-119) | eBisociences | Cat#12-5921-82; RRID: AB_466042 |
| PE Rat Anti-Mouse TER-119/Erythroid Cells (Clone TER-119) | BioLegend | Cat#116207; RRID: AB_313708 |
| PE Rat SD. Anti-Mouse Ig κ Light Chain (Clone 187.1) | BD Biosciences | Cat#559940; RRID: AB_397384 |
| VioBright B515 Recombinant Human Anti-Mouse CD267 (TACI) (Clone REA1227) | Miltenyi Biotec | Cat#130-124-103; RRID: AB_2811602 |
| PE-Dazzle 594 Rat Anti-Mouse CD273 (B7-DC, PD-L2) (Clone TY25) | BioLegend | Cat#107215; RRID: AB_2728124 |
| PE Armenian Hamster Anti-Mouse CD80 (Clone 16-10A1) | BioLegend | Cat#104708; RRID: AB_313129 |
| PE-Cy7 Rat Anti-Mouse CD73 (Clone TY/11.8) | BioLegend | Cat#127223; RRID: AB_2716103 |
| AF647 Mouse BrdU (Clone MoBU-1) | Thermo Fisher Scientific | Cat#B35140; RRID: AB_2536440 |
| PE Armenian Hamster Anti-Mouse CD11c (Clone N418) | BioLegend | Cat#117307; RRID: AB_313776 |
| PE Rat Anti-Mouse F4/80 (Clone T45-2342) | BD Biosciences | Cat#565410; RRID: AB_2687527 |
| PE CD8a Rat Anti-Mouse (Clone 53-6.7) | BioLegend | Cat#100707; RRID: AB_312746 |
| Polyclonal Rabbit Anti-Human CD3 | Dako | Cat#A0452, RRID: AB_2335677 |
| Mouse Anti-Human CD35 (Clone E11) | BD Biosciences | Cat#555451, RRID: AB_395844 |
| Biotin Polyclonal Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | Cat#711-005-152; RRID: AB_2340585 |
| Biotin Polyclonal Donkey Anti-Mouse IgG | Jackson ImmunoResearch | Cat#715-005-150; RRID: AB_2340758 |
| PE-Cy7 Mouse Anti-Human BCL6 (Clone K112-91) | BD Biosciences | Cat#563582; RRID: AB_2738292 |
| BV510 Mouse Anti-Human CCR6 (Clone 11A9) | BD Biosciences | Cat#563241; RRID: AB_2738088 |
| APC-Cy7 Mouse Anti-Human CD14 (Clone M5E2) | BioLegend | Cat#301820; RRID: AB_493695 |
| APC-Cy7 Mouse Anti-Human CD16(Clone 3G8) | BioLegend | Cat#302018; RRID: AB_314218 |
| BV570 Mouse Anti-Human CD20 (Clone 2H7) | BioLegend | Cat#302332; RRID: AB_2563805 |
| BV650 Mouse Anti-Human CD27 (Clone M-T271) | BD Biosciences | Cat#564894; RRID: AB_2739004 |
| APC-Cy7 Mouse Anit-Human CD3 (Clone SP34-2) | BD Biosciences | Cat#557757; RRID: AB_396863 |
| PE-Cy5 Mouse Anti-Human CD95 (Clone DX2) | BD Biosciences | Cat#559773; RRID: AB_397317 |
| BV786 Mouse Anti-Human IgG (Clone G18-145) | BD Biosciences | Cat#564230; RRID: AB_2738684 |
| PerCP-Cy5.5 Mouse Anti-Human IgM (Clone G20-127) | BD Biosciences | Cat#561285; RRID: AB_10611998 |
| PE Mouse Anti-Human Ki67 (Clone B56) | BD Biosciences | Cat#556027; RRID: AB_2266296 |
| PerCP-Cy5.5 Streptavidin | BioLegend | Cat#405214; RRID: 2716577 |
| PE-Cy7 Streptavidin | eBioscience | Cat#25-4317-82; RRID: AB_10116480 |
| BV650 Streptavidin | BioLegend | Cat#405232 |
| AF488 Streptavidin | Invitrogen | Cat#S11223 |
| BV421 Streptavidin | BioLegend | Cat#405225 |
| AF405 Streptavidin | Invitrogen | Cat#S32351 |
| AF555 Streptavidin | Invitrogen | Cat#S32355 |
| AF647 Streptavidin | BioLegend | Cat#405237 |
| APC Streptavidin | Invitrogen | Cat#S868 |
| Bacterial and virus strains | ||
| Influenza A virus A/Puerto Rico/8/34 (PR8) (Mt. Sinai strain; H1N1) | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| APC-conjugated I-Ab OVA329-337 tetramers | NIH tetramer core facility | N/A |
| AF488 TFP ester | Thermo Fisher Scientific | Cat#A37570 |
| 5-Ethynyl-2′-deoxyuridine (EdU) | Sigma-Aldrich | Cat#900584 |
| 5-Bromo-2′-deoxyuridine (5-BrdU) | ChemScene | Cat#CS-3028 |
| NP7-BSA-Biotin | BioSearch Technologies | Cat#N-1026-5 |
| NP19-OVAL (Ovalbumin) | BioSearch Technologies | Cat#N-5051-100 |
| Ovalbumin (Oval) | BioSearch Technologies | Cat#O-1000-100 |
| NP29-PE (Phycoerythrin) | BioSearch Technologies | Cat#N-5070-1 |
| NP14-CGG (Chicken Gamma Globulin) | BioSearch Technologies | Cat#N-5055E-1 |
| R-Phycoerythrin | Agilent | Cat#PB32-10 |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Corn Oil | Sigma-Aldrich | Cat# C8267 |
| Prefusion-stabilized RSV F protein (DS-Cav1) | Marcandalli et al., 2019 | N/A |
| RSV-F-I53-50-NP | Marcandalli et al., 2019 | N/A |
| ProLong Diamond Antifade Mountant | Invitrogen | Cat# P36965 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat#A2153 |
| Sodium bicarbonate | Sigma-Aldrich | Cat#S5761 |
| Aluminum potassium sulfate dodecahydrate (alum) | Sigma-Aldrich | Cat#A7210 |
| Hank’s Buffered Saline Solution, HBSS with calcium and magnesium, without phenol red | Lonza | Cat#BE10-527F |
| Squalene oil-in-water emulsion (SWE) adjuvant | Vaccine Formulation Institute | N/A |
| Methanol | VWR | Cat#97065-052 |
| Critical commercial assays | ||
| Chromium single cell reagent kit version 2 | 10x Genomics | Cat#120234 |
| Chromium single cell reagent kit version 3 | 10x Genomics | Cat#1000141 |
| Single Cell Lysis Kit | Invitrogen | Cat#4458235 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Invitrogen | Cat#L34957 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit, for UV excitation | Invitrogen | Cat#L23105 |
| Foxp3 Transcription Factor Fixation/Permeabilization Concentrate and Diluent | eBioscience | Cat#00-5521-00 |
| Transcription Factor Buffer Set | BD Biosciences | Cat#562594 |
| Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat#C10632 |
| APC BrdU Kit | BD Biosciences | Cat#552598; RRID: AB_2861367 |
| CellTrace Violet Cell Proliferation Kit for Flow cytometry | Thermo Fisher Scientific | Cat#C34571 |
| Avidin/Biotin Blocking kit | Vector Laboratories | Cat#SP-2001; RRID: AB_2336231 |
| AF647 Protein Labeling Kit | Invitrogen | Cat#A20173 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| NEBNext Ultra II RNA First Strand Synthesis Module | New England Biolabs | Cat#E7771 |
| NEBNext Ultra II Directional RNA Second Strand Synthesis Module | New England Biolabs | Cat#E7550 |
| QIAseq FastSelect rRNA HMR kit | QIAGEN | Cat#334386 |
| Deposited data | ||
| Single cell RNA-seq data | This paper | GEO: GSE173673 |
| Bulk RNA-seq data | This paper | GEO: GSE175427 |
| Bulk RNA-seq data | Yoshida et al., 2019 | GEO: GSE109125 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JRj | Janvier Labs | https://www.janvier-labs.com/en/fiche_produit/c57bl-6jrj_mouse/ |
| Mouse: IghB1-8hi | Shih et al., 2002b | N/A |
| Mouse: S1pr2-ERT2Cre | Shinnakasu et al., 2016 | N/A |
| Mouse: Ai9 R26Tomato | Madisen et al., 2010 | N/A |
| Mouse: OT-II | Barnden et al., 1998 | N/A |
| Chinese-origin rhesus macaques (outbred) | Astrid Fagraeus laboratory, Karolinska Institutet | N/A |
| Software and algorithms | ||
| Prism – version 8 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| FlowJo – version 10 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo/ |
| Imaris Viewer – version 9.5.1 | Bitplane | https://imaris.oxinst.com/imaris-viewer |
| ZEN Black Edition – version 2.3 | Carl Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Caseviewer – version 2.3 | 3DHistech | https://www.3dhistech.com/caseviewer |
| Cell Ranger software – version 3.0.1 | Zheng et al., 2017 | https://github.com/10XGenomics/cellranger |
| Seurat package – version 3.0.2 | Stuart et al., 2019 | https://github.com/satijalab/seurat |
| velocyto package – version 0.1.24 | La Manno et al., 2018 | http://velocyto.org/ |
| scvelo package – version 0.1.24 | Bergen et al., 2020 | https://github.com/theislab/scvelo |
| scanpy package – version 1.4.4 | Wolf et al., 2018 | https://github.com/theislab/scanpy |
| GSEA software – version 4.1.0 | Subramanian et al., 2005 | https://www.gsea-msigdb.org/ |
| slingshot package – version 1.6.1 | Street et al., 2018 | https://www.bioconductor.org/packages/release/bioc/html/slingshot.html |
| pySCENIC package – version 0.10.0 | Aibar et al., 2017 | https://github.com/aertslab/pySCENIC |
| STAR software – version 2.7.8a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Usegalaxy.eu – Galaxy version 2.7.8a | Afgan et al., 2018) | https://usegalaxy.eu/ |
| HTseq software – version 0.5.3 | Anders et al., 2015 | https://github.com/htseq/htseq |
| DESeq2 – version 1.22.2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Other | ||
| Ensembl release 67 | Cunningham et al., 2015 | http://www.ensembl.org |
| Hallmark geneset BCR and CD40 signaling | MSigDB database | https://www.gsea-msigdb.org/gsea/msigdb/ |
| Hallmark geneset PC signature | MSigDB database | https://www.gsea-msigdb.org/gsea/msigdb/ |
| Geneset BCR and CD40 signaling | Victora et al., 2010 | N/A |
| Geneset PC signature | Shi et al., 2015 | N/A |
| NAP-10 Column | GE Healthcare Life Sciences | Cat#23227 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Taras Kreslavsky (taras.kreslavskiy@ki.se).
Materials availability
This study did not generate new unique reagents.
Experimental Model and Subject Details
Mice
All mice used in this study were maintained on the C57BL/6 genetic background. The IghB1-8hi (Shih et al., 2002b), S1pr2-ERT2Cre (Shinnakasu et al., 2016), Ai9 R26Tomato (Madisen et al., 2010) and OT-II (Barnden et al., 1998) mice were described previously. WT C57BL/6J mice were obtained from Janvier Labs or bred in house. Mice analyzed in this study were at least 6 weeks old and kept under specific-pathogen-free conditions. Male and female mice were used throughout the study. Mice were bred and maintained at the Comparative Medicine Biomedicum facility of Karolinska Institutet (Stockholm, Sweden), at the Experimental Biomedicine Unit, University of Gothenburg (Gothenburg, Sweden) or at the Research Institute for Molecular Pathology (Vienna, Austria). All mouse experiments were carried out according to valid project licenses, which were approved and regularly controlled by the Swedish and/or Austrian Veterinary Authorities.
Non-human primates
This study was approved by the Local Ethical Committee on Animal Experiments. Female Chinese rhesus macaques of 5-14 years of age were housed in the Astrid Fagraeus laboratory at Karolinska Institutet according to the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care. All procedures were performed abiding to the provisions and general guidelines of the Swedish Board of Agriculture.
To maximize data collection and minimize the use of animals for these experiments, immunizations with labeled respiratory syncytial virus (RSV) prefusion-stabilized F glycoprotein trimer (DS-Cav1) immunogens were administered in multiple limbs per animal. We have previously developed and optimized this model (Liang et al., 2017; Ols et al., 2020) to emphasize the ethical considerations of conducting terminal non-human primate studies. For these experiments, intramuscular administration of nanoparticle immunogens was performed in one deltoid and one quadricep while ‘soluble’ immunogens were administered in the contralateral deltoid and quadricep in the same animal for comparison. Phosphate-buffered saline (PBS) was administered to the calves. This way six data points were collected from each animal.
Method Details
Flow cytometry
Mouse and macaque organs were harvested, and single cell suspensions were obtained by mincing through 70 μm cell strainers. Dead cells were stained using the Live/Dead Fixable Aqua Dead Cell Stain Kit for 405 nm excitation according to manufacturer’s instructions (Thermo Fisher Scientific) and excluded from further analysis. For detection of OVA-specific CD4 T cells, splenic single cell suspensions were incubated with APC-conjugated I-Ab OVA329-337 tetramers (provided by National Institutes of Health tetramer core facility) for 1 hour prior to Fc receptor blocking and further cell surface staining. For detection of NP-specific B cells in the polyclonal response in mice, splenic single cell suspensions were incubated in IMDM (Thermo Fisher Scientific) pH 3.0 on ice for 1 min, washed twice with PBS/2% FCS and subsequently incubated for 1 hour with either 7.8 ng/ml NP29-Phycoerythrin (BioSearch Technologies) and 7.8 ng/ml NP7-BSA-Biotin (BioSearch Technologies) or 7.8 ng/ml NP7-BSA-Biotin (BioSearch Technologies) and 7.8 ng/ml NP14-CGG (BioSearch Technologies) labeled in-house with Alexa Fluor 488 (AF488, See “Preparation of fluorescent antigen and tetramer RSV and HA probes”) in ice-cold PBS/2% FCS, prior to Fc receptor blocking and further cell surface staining. NP-specific B cells binding NP7-BSA-Biotin were fluorescently labeled using streptavidin-BV650 (BioLegend) or streptavidin-PECy7 (Thermo Fisher Scientific). To control for antigen carryover when determining antigen-binding B cells in vivo, spleen tissue from congenically distinguishable B1-8hi mice was ‘spiked-in’ during organ mincing and single cell suspensions were washed four times with PBS/2% FCS. For influenza experiments, mediastinal lymph nodes were stained using the antibody cocktail together with HA-APC and HA-PerCP-Cy5.5 for 1h at 4C, prior to intracellular staining.
To assess early RSVF-specific B cell responses in macaques, fresh or frozen LN cell suspensions were stained with tetramer RSVF probes in AF488 and BV421 for 20 min at 4°C prior to further cell surface and intracellular staining. For intracellular staining of single cell suspensions from mice, fixation and permeabilization was performed using the Foxp3 Fixation/Permeabilization Kit (Thermo Fisher Scientific). For intracellular staining of macaque samples, cells were permeabilized using the transcription factor buffer set (BD Biosciences).
Detection of cells positive for 5-Ethynyl-2’-deoxyuridine (EdU) was performed using the Click-iT Plus Edu A488 Flow Cytometry Assay Kit (Thermo Fisher Scientific) following manufacturer’s instructions with minor modifications. Detection of cells positive for 5-bromo-2’-deoxyuridine (BrdU) was performed using the APC BrdU Flow kit (BD Biosciences) following manufacturer’s instructions with minor modifications. Data were acquired on an LSR Fortessa Flow Cytometer (BD Biosciences) and analyzed with FlowJo software v.10 (BD).
Flow cytometry antibodies
For staining of murine samples, monoclonal antibodies specific for BCL6 (REA373 and K112-91), CCR6 (REA277 and 29-2L17), CD11c (HL3, N418), F4/80 (T45-2342), CD138 (REA104 and 281-2), CD19 (1D3), CD22 (REA1187), CD3e (145-2C11), CD4 (GK1.5), CD45.1 (A20), CD45.2 (104), CD69 (H1.2F3), GL7 (GL7), Gr-1 (RB6-8C5), IgG1 (REA1017 and RMG1-1), IgK (187.1), IgL (R26-46 and JC5-1), IgD (11-26c2a), IgM (REA979 and RMM-1), PD-L2 (TY25), CD80 (16-10A1), CD73 (TY/11.8), IRF4 (3E4), Ki67 (SolA15), MHC-II (REA813), NK1.1 (PK136), Nur77 (12.14), BrdU (MoBU-1), TCRβ (H57-597), TCRγδ (eBioGL3), TACI (REA1227), and Ter119 (Ter119) were purchased from BioLegend, BD Biosciences, Miltenyi Biotec or Thermo Fisher Scientific and were used at dilutions specified by the manufacturer or determined experimentally. For detection of OVA-specific CD4 T cells, splenic single cell suspensions were incubated with APC-conjugated I-Ab OVA329-337 tetramers (provided by National Institutes of Health tetramer core facility) for 1 hour prior to Fc receptor blocking and further cell surface staining. For staining of macaque samples, monoclonal antibodies specific for BCL6 (K112-91), CCR6 (11A9), CD14 (M5E2), CD16 (3G8), CD20 (2H7), CD27 (M-T271), CD3 (SP34-2), CD95 (DX2), IgG (G18-145), IgM (G20-127), IRF4 (3E4) and Ki67 (B56) were purchased from BioLegend, BD Biosciences or Thermo Fisher Scientific.
Adoptive transfers
For adoptive transfer experiments, unless stated otherwise, 0.6-1 × 107 of splenocytes (~3-5 × 105 NP-specific B cells) from IghB1-8hi/+ or IghB1-8hi/B1-8hi mice were injected into the tail vein of congenically distinguishable sex-matched C57BL/6 recipient mice. To address the influence of T cell help on the cell cycle exit of early MBCs, 1 × 107 B1-8hi splenocytes were intravenously transferred together with or without 1.5 × 107 OT-II splenocytes. When indicated in the figure legends, splenocytes were labeled using the CellTrace Violet Proliferation Kit (Thermo Fisher Scientific) prior to transfer. To this end, up to 50 × 106 splenocytes were incubated in 1 mL pre-warmed PBS/0.1% BSA with 2.5 μM CellTrace Violet reagent for 8 min at 37°C. The staining reaction was stopped with pre-warmed fetal calf serum.
Mouse immunizations and in vivo treatment
For B1-8hi transfer experiments, when indicated in the figure legends, C57BL/6 recipient mice were pre-immunized by intraperitoneal injection of 100 μg OVA (BioSearch Technologies) dissolved in PBS and precipitated in alum (Sigma-Aldrich) at a 1:1 ratio > 2 weeks prior to adoptive cell transfer. 12 to 24 hours after B1-8hi cell transfer, recipient mice were immunized by intraperitoneal injection of 100 μg NP19-OVA (BioSearch Technologies) or 60-80 μg fluorophore-labeled NP7-BSA-Biotin (BioSearch Technologies) precipitated in alum (Sigma-Aldrich) at a 1:1 ratio. For antigen detection by confocal microscopy, non-immunized C57BL/6 mice were intravenously injected with 100 μg phycoerythrin (PE; Agilent) in PBS. For analysis of cell cycle exit of early MBCs upon i.v. immunization, recipient mice were immunized 12 to 24 hours after B1-8hi cell transfer by intravenous injection of 100 μg NP19-OVA (BioSearch Technologies) diluted in PBS. For analysis of cell cycle exit of differentiating early MBCs and its dependence on antigen availability, excess antigen was provided 3.5 days post immunization by intravenous injection of 100 μg NP19-OVA in PBS. To study the polyclonal response to NP, mice were immunized by intraperitoneal injection of 100 μg NP19-OVA (BioSearch Technologies) precipitated in alum (Sigma-Aldrich) at a 1:1 ratio. For in vivo proliferation analysis, 1 mg 5-Ethynyl-2’-deoxyuridine (EdU, Sigma-Aldrich) diluted in PBS was intraperitoneally injected at indicated time points post immunization or mice were provided with drinking water containing 5-bromo-2’-deoxyuridine (BrdU, 1mg/ml, ChemScene) between indicated time points. In S1pr2-ERT2Crefate mapping experiments mice were treated with 4 mg Tamoxifen / 30 g body weight via oral gavage every two days for the whole duration of the experiment. Tamoxifen (Sigma-Aldrich) was dissolved in a mixture of corn oil (Sigma-Aldrich) and ethanol (10:1 ratio) at 55°C. Aliquots were stored at −20° C.
Influenza infection experiments
Mice were anesthetized with isofluorane and infected intranasally with 50 tissue culture infectious dose 50 (TCID50) of influenza A virus A/Puerto Rico/8/34 (PR8) (Mt. Sinai strain; H1N1) diluted in balanced salt solution (BSS) with 0.1% BSA. At 7 days post infection, mice were sacrificed and mediastinal lymph nodes were collected in RPMI1640.9 organs were pooled per experiment. Spleens from non-infected controls were used as a reference.
Non-human primate immunizations and terminal sample collection
For rhesus macaques studies, animals received injections of two different DS-Cav1 immunogens, one a ‘soluble’ trimer and the other a self-assembling protein nanoparticle (both previously described in Marcandalli et al., 2019), formulated in a squalene-in-water emulsion (SWE) adjuvant (Vaccine Formulation Institute). These immunogens were fluorochrome-labeled with Alexa Fluor 647 (AF647) according to the manufacturer’s instructions (Thermo Fisher Scientific). PBS injections served as internal controls. The final injection volumes were 0.5 mL and were administered on a marked injection site. Three rhesus macaques were immunized for vaccine tracking at 2 hours post injection and another four rhesus macaques were immunized for vaccine tracking at 3 and 7 days post injection. All tissues were sampled during necropsy and stored separately in RPMI1640 on ice, as previously described (Liang et al., 2017; Ols et al., 2020). All individual LNs per LN cluster (axillary, apical, inguinal, external/common iliac, mesenteric) were pooled for analysis.
Preparation of fluorescent antigen and tetramer RSV and HA probes
To detect interaction of B1-8hi B cells with antigen in vivo, recipient mice were immunized i.p. with 60-80 μg fluorescently labeled NP7-BSA-Biotin (BioSearch Technologies) in alum. For preparation of fluorescent antigen, NP7-BSA-Biotin or NP14-CGG (BioSearch Technologies) was labeled using Alexa Fluor 488 TFP ester (Thermo Fisher Scientific). In short, NP7-BSA-Biotin or NP14-CGG, respectively, were dissolved in PBS at 2 mg/ml and pH 8 was adjusted using 1 M sodium bicarbonate buffer. One mg of the protein solution was added to one vial of reactive dye and incubated rotating at room temperature for 75 min. Fluorescently labeled protein was purified using NAP-10 columns (GE Healthcare Life Sciences). The degree of labeling was determined based on measuring molar concentrations of protein and dye by absorbance at 280 and 494 nm using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). A molar ratio of 1.2 (NP7-BSA-Biotin) and 4.1 (NP14-CGG) moles of dye per mole of protein was used for experiments. The final protein concentration was confirmed using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Tetramer RSVF probes were prepared by incubation of 4-fold molar excess of avi-tag biotinylated DS-Cav1 (RSVF) protein with either streptavidin-conjugated AF488 (Invitrogen) or streptavidin-conjugated BV421 (BioLegend). Recombinant Y98F-HA was produced and labeled with streptavidin-conjugated APC (Invitrogen) or streptavidin-conjugated PerCP-Cy5.5 (BioLegend), as previously described (Angeletti et al., 2019; Whittle et al., 2014).
In situ staining of mouse and macaque tissue
Fresh mouse spleens or macaque LN biopsies were embedded in Tissue Tek optimal cutting temperature (OCT, Sakura Finetek) media and snap frozen with dry ice before storage at −80° C. Sections of 8 μm-thickness were cut using a cryostat, mounted on super-frost plus glass slides (Thermo Fisher Scientific), air-dried for 15 min and fixed with 2% PFA (Sigma) for 20 min. Tissues were blocked and permeabilized with 2% FCS in permwash buffer (tris-buffered saline containing 1% HEPES buffer (Sigma) and 0.1% saponin (Sigma)) for 30 min. An avidin/biotin blocking kit (Vector Laboratories) was used for blocking of endogenous biotin. For staining of mouse spleen sections, monoclonal rat anti-mouse IgD (11-26c.2a, BioLegend), monoclonal rat anti-mouse CD21/35 (CR2/CR1, BioLegend) and anti-mouse CD169 (REA197, Miltenyi Biotec) were used. For staining of macaque tissue, a combination of polyclonal rabbit anti-human CD3 (Dako) and mouse anti-human CD35 (E11, BD Biosciences) diluted in permwash buffer was used. The antibodies were added as a cocktail and incubated for 1 hour at RT (mouse tissue) or overnight at 4°C (macaque tissue). For staining of macaque tissue, slides were blocked with 1% donkey serum before biotinylated secondary antibodies and streptavidin-conjugated fluorophores were added sequentially for 30 min each, with additional avidin/biotin blocking performed between each secondary antibody and fluorophore pair. Secondary antibodies were all raised in donkey and included anti-rabbit and anti-mouse (Jackson Immunoresearch). Streptavidin-conjugated fluorophores used included AF405 and AF555 (Invitrogen). After completion of staining, slides were washed with water, air-dried in the dark and mounted with Prolong Diamond anti-fade mounting media (Invitrogen) and 22x50 mm coverslips. For detection of the antigen signal in mouse spleen tissue, sections were directly imaged without prior fixation, rehydration, or staining. Images were either captured on an LSM 700 system (Carl Zeiss) with 405-, 488-, 555-, and 639 nm excitation lines at the Biomedicum Imaging Core at the Karolinska Institute or using an automated confocal slide scanner (Pannoramic MIDI II FL, 3DHistech) utilizing a FLIR Grass-hopper3 camera equipped with a Zeiss 20x Plan-Apochromat 0.8NA objective, Lumencor SOLASM light engine, and Pannoramic slide scanning software along with CaseViewer software. Images were analyzed using Imaris 9.5.1 (Bitplane), Zen 2.3 Black Edition (Carl Zeiss) software.
Cell sorting and RNA-seq library preparation
For single cell sequencing, 40 × 106 total splenocytes, isolated from female B1-8hi/+ CD45.1 mice, were transferred into C57BL6/J CD45.2 recipients, immunized intraperitoneally with 100 μg OVA precipitated at a 1:1 ratio with alum in PBS > 2 weeks prior transfer. Mice were immunized with 100 μg NP19-OVA precipitated at a 1:1 ratio with alum in PBS the day after transfer and spleens were harvested 2.5 or 3.5-4 days after immunization. For the day 3.5-4 time point, for reasons unrelated to the current study (see Rauschmeier et al., 2021), B1-8hi Bhlhe40−/− CD45.1/.2 splenocytes were co-transferred along with B1-8hi WT CD45.1 splenocytes and WT and Bhlhe40−/− B1-8hi were separated during cell sorting. Lineage depletion for CD4/TCRβ/TCRγδ/CD11c/Gr1/NK1.1/Ter119/Igκ was performed using anti-PE Micro-Beads (Miltenyi Biotec). CD19+Igλ+CD45.1 + cells were double sorted using a FACSAria III sorter. Broad gates were applied to include PBs that may start downregulating some of these surface molecules. For day 3.5-4 samples, cells were processed using 10X Genomics kit version 2 immediately after sorting. For day 2.5, cells were resuspended in cold PBS/0.04% BSA and fixed by adding 4 volumes 100% Methanol (VWR) and incubation for 30 min at −20°C. Day 2.5 cells were stored at −80°C and also processed as described (Chen et al., 2018) using 10X Genomics kit version 3. Cells were sequenced paired end with 75 bp read length on a NextSeq550 system (Illumina).
Analysis of single cell RNA-seq data
The sequenced 10X Chromium libraries from two samples were mapped to mm10 mouse genome and assigned to droplets with Cell Ranger software (version 3.0.1) with default parameters (Zheng et al., 2017). Transcriptomes of 7,324 cells with the median read coverage of 2,292 and 2,133 cells with a median read coverage of 5,341 were obtained for days 2.5 and 3.5-4, respectively. To obtain the individual embeddings at day 2.5 and day 3.5-4, the resulting read count matrices were analyzed with Seurat (version 3.0.2), (Stuart et al., 2019). We filtered the cells with the number of detected genes higher than 200 and lower than 7.000 and the percentage of reads mapped to mitochondrial genes not higher than 25. Principal component analysis was performed on 2000 genes with the most variable expression (selected by Seurat). First 6 principal components were used to perform UMAP embedding (parameter k = 100). Clustering of cells was performed based on PCA embedding (first 6 PCs taken into account). The data was scaled to regress out total number of detected features and total percentage of mitochondrial reads. Cell cycle was regressed out by a standard approach: first, for each cell, S phase score and G2M phase score were estimated with CellCycleScoring function from Seurat, next, these scores were regressed out.
To perform RNA-velocity analysis, we estimated exonic and intronic gene counts in bam files from Cell Ranger with velocyto (version 0.17.13), (La Manno et al., 2018). The repeat regions of the genome were masked. The resulting loom files were analyzed with scvelo package (version 0.1.24) (Bergen et al., 2020). The dynamical model was used to estimate velocities. Estimated velocity field was plotted on top of the UMAP embeddings.
Joined embedding for days 2.5 and 3.5-4 (WT and, for reasons unrelated to this study, Bhlhe40−/− samples) was performed with scanpy based on spliced transcripts from velocyto. First, the data were normalized and filtered with scanpy.pp.recipe_seurat workflow (Satija et al., 2015). Next, UMAP embedding was performed with default parameters. For the further analysis, we considered only WT samples. Cells were clustered with louvain algorithm (Blondel et al., 2008) and cell populations were defined as groups of clusters identified by this algorithm.
To analyze the similarity of cell transcriptomes between different clusters, we first applied BuildClusterTree and PlotClusterTree functions from Seurat package. In addition, we explored the distribution Euclidean distances between coordinate vectors in the PCA space for 1000 random pairs of cells taken from a given pair of clusters at day 3.5-4 (Figure S4C). Median distances between clusters were presented as a heatmap (Figure 4E).
To discover the genes that were significantly changing their expression along the transitions into GCBC, PB and memory cells, we performed pseudotime analysis with scanpy package (Wolf et al., 2018). Diffusion pseudotime was inferred by scanpy.tl.dpt function. Randomly chosen cell in a centrally located (among AP-eMBC cells) cluster 7 was selected as the root point. Choice of the root cell within AP-eMBC cell type did not affect the inferred pseudotime values for other cell fates (not shown). We next smoothed gene expression (read counts in individual cells) in PB and GCBC cell types by performing lowess along the pseudotime axis with stats-models.api.nonparametric.lowess function (with parameters frac = 0.01). DPT for MBC was calculated similarly but after exclusion of cells in GCBC and PB clusters from the analysis. Pearson correlation was calculated between the smoothed expression and pseudotime for each individual gene, separately among PBs, GCBCs and MBCs. Genes lists ranked by these correlations were used for GSEA analysis using software from the Broad Institute (Subramanian et al., 2005). Hallmark gene sets from MSigDB database as well as previously published gene sets for BCR and CD40 signaling (Victora et al., 2010) were used in this analysis.
As an alternative approach, differentiation trajectories were fitted with slingshot package (version 1.6.1) (Street et al., 2018). One of the centrally located clusters among AP-eMBC cells was selected as a starting point. Pseudotime was assigned to the cells along three trajectories that ended in GCBC, PB, and memory cells clusters. To explore the changes of gene expression along these differentiation trajectories, we smoothed gene expression counts with cubic splines with smooth.spline R function. Smoothed gene expression was shown on a heatmap with cells ordered according to pseudotime (Figure 3E).
The GCBC signature used in Figure 1E was generated using Immgen database (RNA-seq datasets; Yoshida et al., 2019; GEO accession number GSE109125) by ranking genes by ratio of expression in GCBCs versus maximal expression in any other mature B cell subset and filtering for genes with minimal expression in GCBC of 1000 counts. 15 top genes were included in the signature; Bcl6 was added manually to this signature. PB signature was generated in the same way, Irf4 and Prdm1 were added manually.
SCENIC analysis was performed to estimate the activity of transcription factors in individual cells and discover lineage-specific transcription factors (Aibar et al., 2017). SCENIC was applied to the joined dataset from days 2.5 and 3.5-4.
The following annotations were used to run SCENIC: 1) correspondence between genes and transcription factors that can bind near their transcription start sites https://resources.aertslab.org/cistarget/databases/mus_musculus/mm10/refseq_r80/mc9nr/gene_based/mm10__refseq-r80__10kb_up_and_down_tss.mc9nr.feather and https://resources.aertslab.org/cistarget/databases/mus_musculus/mm10/refseq_r80/mc9nr/gene_based/mm10__refseq-r80__500bp_up_and_100bp_down_tss.mc9nr.feather; 2) sequence motifs in transcription factor binding sites: https://resources.aertslab.org/cistarget/motif2tf/motifs-v9-nr.mgim0.001-o0.0.tbl; 3) list of known transcription factors https://raw.githubusercontent.com/aertslab/pySCENIC/master/resources/mm_mgi_tfs.txt. The activity of motifs in individual cells was obtained with aucell function from SCENIC and plotted on a UMAP embedding from the previous step.
To discover the genes that were differentially expressed between the time points (days 2.5 and 3.5-4), we compared gene expression between the time points within the AP-eMBC population and within the PB population. With FindMarkers function from Seurat package, we applied the Wilcoxon test to scaled gene expression values. GSEA was performed on genes ranked by fold change between the two time points using software from the Broad Institute (Subramanian et al., 2005). Hallmark gene sets from the MSigDB database as well as previously published PC gene signature (Shi et al., 2015; Figures 1 and 7) were used in this analysis. Results of GSEA were visualized with a dotplot where the size of each dot was proportional to the absolute value of normalized enrichment score (|NES|) and the color of a dot corresponded to the FDR of enrichment.
Bulk RNA-seq
For RNA-seq analysis of activated B cell populations, congenically distinguishable B1-8hi B cells were co-transferred into OVA-pre-immunized WT recipients as described above and isolated at days 2.5, 4, and 50 after NP-OVA immunization. Splenocytes were depleted using PE-conjugated Igκ, CD4,CD8, F4/80, CD3, Ter119, and CD11c antibodies and anti-PE Micro-Beads (Miltenyi Biotec), and donor (CD45.1+) antigen-specific (CD19+Igλ+) GC B cells (CCR6−GL7hi), AP-eMBC cells (CCR6+GL7int/lo), PBs (TACI+CD138+) or day 50 MBC (CD38+GL7−) were double sorted. For the second sort, 600-1000 cells were directly sorted into single cell lysis solution (Invitrogen) and processed according to manufacturer’s instructions with minor modifications, as sorted cells were incubated for 15 min in the single cell DNase/lysis solution. cDNA and second strand synthesis was performed using NEBNext Ultra II RNA First Strand Synthesis (NEB Cat E7771) and NEBNext Ultra II Directional RNA Second Strand Synthesis (NEB Cat E7550) modules. QIAseq FastSelect rRNA HMR kit (QIAGEN) was utilized to minimize rRNA contribution to the final library. Custom made Tn5 (transposase) in combination with oligo replacement and PCR amplification was utilized to generate indexed sequencing libraries (Gertz et al., 2012). Libraries were pooled and paired-end sequenced (40 cycles) using the Nextseq 2000 system (Illumina).
Bioinformatic analysis of bulk RNA-seq data
Sequence reads that passed the Illumina quality filtering were considered for alignment. Reads were aligned to the mouse transcriptome (genome assembly version of July 2007 NCBI37/mm9) using RNA STAR (Dobin et al., 2013; https://usegalaxy.eu; Galaxy Version 2.7.8a; Afgan et al., 2018). Raw counts calculation was based on the RefSeq database, which was downloaded from UCSC on January 10th, 2014. The annotation of immunoglobulin and T cell receptor genes was incorporated from the Ensembl release 67 (Cunningham et al., 2015). Genes with overlapping exons were flagged and double entries (i.e., exactly the same gene at two different genomic locations) were renamed. Genes with several transcripts were merged to consensus-genes consisting of a union of all underlying exons using the fuge software (I. Tamir, personal communication), which resulted in 24,733 gene models.
For analysis of differential gene expression, the number of reads per gene was counted using HTseq version 0.5.3 (Anders et al., 2015) with the overlap resolution mode set to ‘union’. The datasets were analyzed using the R package DESeq2 version 1.22.2 (Love et al., 2014). Sample normalizations and dispersion estimations were conducted using the default DESeq2 settings. For principal-component analysis, gene counts were transformed into variance stabilized values with the R package DESeq2. Genes from gene-ontology categories GO:0007049 (cell cycle), GO:0006259 (DNA metabolic process), GO:0005813 (centrosome), GO:0005819 (spindle) and GO:0000776 (kinetochore), as well as genes encoding histones were excluded from the principal-component analysis; the 500 most varying genes were used. Signatures from (Laidlaw et al., 2017) and (He et al., 2017) (MBCs), (Mabbott and Gray, 2014) (GCBCs) and (Shi et al., 2015) (PCs) were used in the analysis shown in Figure S4D.
Quantification and Statistical Analysis
Statistical analysis was performed with the GraphPad Prism 8 software. Two-tailed unpaired Student’s t test analysis was used to assess the statistical significance of one observed parameter between two experimental groups. Unpaired (or paired with Geisser-Greenhouse correction, as indicated in the figure legends) analysis of variance (ANOVA) was used, when more than two experimental groups were compared, and the statistical significance was determined with Holm-Sidak’s multiple comparisons test. Error bars in all figures represent SD. The statistical evaluation of the RNA-seq data is described under ‘Analysis of single cell RNA-seq data’.
Highlights.
scRNA-seq reveals early divergence of PBs, GCBCs, and MBCs from the common precursor
Unlike PBs and GCBCs, eMBCs remain transcriptionally similar to the precursors
Cell cycle exit of eMBCs is driven by the decline in antigen availability
Provision of antigen excess drives generation of a new PB wave from eMBCs
Acknowledgments
We thank T. Kurosaki and R. Shinnakasu for S1pr2-ERT2cre mice, G. Victora for gene sets for GSEA, A. Sommer’steam at the Vienna Biocenter Support Facilities GmbH (VBCF) for library preparation and Illumina sequencing, I. Mantas for help with preparation of the graphical abstract, and C. Sundling for helpful discussions and critical reading of the manuscript. We acknowledge resources provided by the Swedish National Infrastructure for Computing (SNIC) at UP-PMAX, partially funded by the Swedish Research Council through grant agreement no. 2018-05973 (projects SNIC 2019/8-289 and 2020/16-34). This study was supported by the Swedish Research Council (grants 2017-01118 to T.K. and 2017-01439 to D.A.), Cancerfonden (CAN 2018/710 to T.K.), long-term bioinformatics support from SciLifeLab and the Knut and Alice Wallenberg Foundation, Åke Wibergs Stiftelse (M18-0094 to T.K.), a stipend from the Wenner-Gren Foundations (to T.K.), a stipend from the German Research Foundation (DFG) (RE 4264/1-1 to A.R.), Boehringer Ingelheim (M.B. and R.R.), the European Research Council (ERC) grants (grants 740349-PlasmaCellControl to M.B. and 850638 to D.A.), and a grant from the Bill & Melinda Gates Foundation (OPP1156262 to N.P.K. and K.L.). A.V.A. was supported by Ministry of Science and Higher Education of the Russian Federation (agreement no. 075-15-2020784). We thank the NIH tetramer core facility for preparation of MHC tetramers, the Mammalian Protein Expression core facility at the University of Gothenburg for recombinant HA production, and the Mammalian Protein and Nanoparticle Core Labs at the Institute for Protein Design for assistance with RSV F protein and nanoparticle production.
Footnotes
Author Contributions
V.G., R.R., A.R., and Y.Y. performed all mouse experiments with the exception of IAV infections. S.O. performed experiments with non-human primates. C.G. and R.M. performed bulk RNA-seq experiments. A.V.A., Å.K.B., and C.M. performed bioinformatics analyses. A.E. performed IAV infection experiments. L.P. and N.P.K. provided reagents crucial for this study. D.A., K.L., I.A., M.B., and T.K. provided research supervision.
Declaration of Interests
N.P.K. is a co-founder, shareholder, and chair of the scientific advisory board of Icosavax, Inc., and the King laboratory has received an unrelated sponsored research agreement from Pfizer.
Supplemental Information
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.08.017.
Data and code availability
Single cell- and bulk-RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper also used some existing, publicly available data. These datasets are listed in the Key resources table.
This paper did not generate any unique code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity. 2018;48:133–146.:e6. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, Mankowski MC, Chambers M, Prabhakaran M, Hickman HD, et al. Outflanking immunodominance to target sub-dominant broadly neutralizing epitopes. Proc Natl Acad Sci USA. 2019;116:13474–13479. doi: 10.1073/pnas.1816300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;2008:P10008 [Google Scholar]
- Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, Basso K, Dalla-Favera R. MEF2B Instructs Germinal Center Development and Acts as an Oncogene in B Cell Lymphomagenesis. Cancer Cell. 2018;34:453–465.:e9. doi: 10.1016/j.ccell.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TD, Gatto D, Wood K, Camidge T, Basten A, Brink R. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J Immunol. 2009;183:3139–3149. doi: 10.4049/jimmunol.0901690. [DOI] [PubMed] [Google Scholar]
- Chen J, Cheung F, Shi R, Zhou H, Lu W, CHI Consortium PBMC fixation and processing for Chromium single-cell RNA sequencing. J Transl Med. 2018;16:198. doi: 10.1186/s12967-018-1578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell. 2019;177:1153–1171.:e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey F, Alabyev B, Manser T. Initial clonal expansion of germinal center B cells takes place at the perimeter of follicles. Immunity. 2009;30:599–609. doi: 10.1016/j.immuni.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195:1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosenovic P, Kara EE, Pettersson A-K, McGuire AT, Gray M, Hartweger H, Thientosapol ES, Stamatatos L, Nussenzweig MC. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc Natl Acad Sci USA. 2018;115:4743–4748. doi: 10.1073/pnas.1803457115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgueta R, Marks E, Nowak E, Menezes S, Benson M, Raman VS, Ortiz C, O’Connell S, Hess H, Lord GM, Noelle R. CCR6-dependent positioning of memory B cells is essential for their ability to mount a recall response to antigen. J Immunol. 2015;194:505–513. doi: 10.4049/jimmunol.1401553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gertz J, Varley KE, Davis NS, Baas BJ, Goryshin IY, Vaidyanathan R, Kuersten S, Myers RM. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22:134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginsberg HS, Horsfall FL., Jr Quantitative aspects of the multiplication of influenza A virus in the mouse lung; relation between the degree of viral multiplication and the extent of pneumonia. J Exp Med. 1952;95:135–145. doi: 10.1084/jem.95.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havenar-Daughton C, Carnathan DG, Boopathy AV, Upadhyay AA, Murrell B, Reiss SM, Enemuo CA, Gebru EH, Choe Y, Dhadvai P, et al. Rapid Germinal Center and Antibody Responses in Non-human Primates after a Single Nanoparticle Vaccine Immunization. Cell Rep. 2019;29:1756–1766.:e58. doi: 10.1016/j.celrep.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J-S, Subramaniam S, Narang V, Srinivasan K, Saunders SP, Carbajo D, Wen-Shan T, Hidayah Hamadee N, Lum J, Lee A, et al. IgG1 memory B cells keep the memory of IgE responses. Nat Commun. 2017;8:641. doi: 10.1038/s41467-017-00723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchison S, Benson RA, Gibson VB, Pollock AH, Garside P, Brewer JM. Antigen depot is not required for alum adjuvanticity. FASEB J. 2012;26:1272–1279. doi: 10.1096/fj.11-184556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang I-Y, Hwang K-S, Park C, Harrison KA, Kehrl JH. Rgs13 constrains early B cell responses and limits germinal center sizes. PLoS ONE. 2013;8:e60139. doi: 10.1371/journal.pone.0060139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jash A, Wang Y, Weisel FJ, Scharer CD, Boss JM, Shlomchik MJ, Bhattacharya D. ZBTB32 Restricts the Duration of Memory B Cell Recall Responses. J Immunol. 2016;197:1159–1168. doi: 10.4049/jimmunol.1600882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209:2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, Haberman AM. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Kuhrt D, Faith S, Hattemer A, Leone A, Sodora D, Picker L, Borghesi L, Cole KS. Naïve and memory B cells in the rhesus macaque can be differentiated by surface expression of CD27 and have differential responses to CD40 ligation. J Immunol Methods. 2011;363:166–176. doi: 10.1016/j.jim.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Duan L, Xu Y, Vazquez SE, Cyster JG. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat Immunol. 2020;21:1082–1093. doi: 10.1038/s41590-020-0713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Schmidt TH, Green JA, Allen CDC, Okada T, Cyster JG. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J Exp Med. 2017;214:639–649. doi: 10.1084/jem.20161461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AWY, Turner VM, Bourne K, Hermes JR, Chan TD, Brink R. BAFFR controls early memory B cell responses but is dispensable for germinal center function. J Exp Med. 2021;218:e20191167. doi: 10.1084/jem.20191167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Lindgren G, Sandgren KJ, Thompson EA, Francica JR, Seubert A, De Gregorio E, Barnett S, O’Hagan DT, Sullivan NJ, et al. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci Transl Med. 2017;9:eaal2094. doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbott HA, Gray D. Identification of co-expressed gene signatures in mouse B1, marginal zone and B2 B-cell populations. Immunology. 2014;141:79–95. doi: 10.1111/imm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcandalli J, Fiala B, Ols S, Perotti M, de van der Schueren W, Snijder J, Hodge E, Benhaim M, Ravichandran R, Carter L, et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell. 2019;176:1420–1431.:e17. doi: 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, Okada T, Kurosaki T, Victora GD. Restricted Clonality and Limited Germinal Center Reentry Characterize Memory B Cell Reactivation by Boosting. Cell. 2020;180:92–106.:e11. doi: 10.1016/j.cell.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich M, Tagoh H, Bönelt P, Axelsson E, Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M, Busslinger M. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17:331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Suh H, Li AV, Ockenhouse CF, Yadava A, Irvine DJ. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand Tfh cells and promote germinal center induction. Proc Natl Acad Sci USA. 2012;109:1080–1085. doi: 10.1073/pnas.1112648109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TJ, Kato Y, Abraham W, Chang JYH, Kulp DW, Watson N, Turner HL, Menis S, Abbott RK, Bhiman JN, et al. Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat Med. 2020;26:430–440. doi: 10.1038/s41591-020-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Winkler J, Mitter R, Rappe JCF, Vanes L, Schweighoffer E, Mohammadi H, Wack A, Tybulewicz VLJ. Critical requirement for BCR, BAFF, and BAFFR in memory B cell survival. J Exp Med. 2021;218:e20191393. doi: 10.1084/jem.20191393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- O’Connor BP, Vogel LA, Zhang W, Loo W, Shnider D, Lind EF, Ratliff M, Noelle RJ, Erickson LD. Imprinting the fate of antigen-reactive B cells through the affinity of the B cell receptor. J Immunol. 2006;177:7723–7732. doi: 10.4049/jimmunol.177.11.7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ols S, Yang L, Thompson EA, Pushparaj P, Tran K, Liang F, Lin A, Eriksson B, Karlsson Hedestam GB, Wyatt RT, Loré K. Route of Vaccine Administration Alters Antigen Trafficking but Not Innate or Adaptive Immunity. Cell Rep. 2020;30:3964–3971.:e7. doi: 10.1016/j.celrep.2020.02.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape KA, Catron DM, Itano AA, Jenkins MK. The humoral immune response is initiated in lymph nodes by B cells that acquire soluble antigen directly in the follicles. Immunity. 2007;26:491–502. doi: 10.1016/j.immuni.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pape KA, Maul RW, Dileepan T, Paustian AS, Gearhart PJ, Jenkins MK. Naive B Cells with High-Avidity Germline-Encoded Antigen Receptors Produce Persistent IgM+ and Transient IgG+ Memory B Cells. Immunity. 2018;48:1135–1143.:e4. doi: 10.1016/j.immuni.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus D, Phan TG, Chan TD, Gardam S, Basten A, Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J Exp Med. 2006;203:1081–1091. doi: 10.1084/jem.20060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauschmeier R, Reinhardt A, Gustafsson C, Glaros V, Artemov AV, Taneja R, Adameyko I, Månsson R, Busslinger M, Kreslavsky T. Cell-intrinsic functions of the transcription factor Bhlhe40 in activated B cells and T follicular helper cells restrain the germinal center reaction and prevent lymphomagenesis. bioRxiv. 2021 doi: 10.1101/2021.03.12.435122. [DOI] [Google Scholar]
- Roco JA, Mesin L, Binder SC, Nefzger C, Gonzalez-Figueroa P, Canete PF, Ellyard J, Shen Q, Robert PA, Cappello J, et al. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity. 2019;51:337–350.:e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Egert A, Pasparakis M, Rajewsky K. M17 a gene specific for germinal center (GC) B cells and a prognostic marker for GC B-cell lymphomas, is dispensable for the GC reaction in mice. Blood. 2006;107:4849–4856. doi: 10.1182/blood-2005-10-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, Nussenzweig MC. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, Smyth GK, Hodgkin PD, Nutt SL, Corcoran LM. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16:663–673. doi: 10.1038/ni.3154. [DOI] [PubMed] [Google Scholar]
- Shih T-AY, Shih E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002a;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- Shih T-AY, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002b;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, Kurosaki T. Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol. 2016;17:861–869. doi: 10.1038/ni.3460. [DOI] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suan D, Kräutler NJ, Maag JLV, Butt D, Bourne K, Hermes JR, Avery DT, Young C, Statham A, Elliott M, et al. CCR6 Defines Memory B Cell Precursors in Mouse and Human Germinal Centers, Revealing Light-Zone Location and Predominant Low Antigen Affinity. Immunity. 2017;47:1142–1153.:e4. doi: 10.1016/j.immuni.2017.11.022. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Lau AWY, Bourne K, Young C, Laurianto C, Hermes JR, Menzies RJ, Butt D, Kräutler NJ, Zahra D, et al. Positive selection of IgG+ over IgM+ B cells in the germinal center reaction. Immunity. 2021;54:988–1001.:e5. doi: 10.1016/j.immuni.2021.03.013. [DOI] [PubMed] [Google Scholar]
- Tam HH, Melo MB, Kang M, Pelet JM, Ruda VM, Foley MH, Hu JK, Kumari S, Crampton J, Baldeon AD, et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc Natl Acad Sci USA. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Hiwa R, Mueller JL, Vykunta V, Hibiya K, Noviski M, Huizar J, Brooks JF, Garcia J, Heyn C, et al. NR4A nuclear receptors restrain B cell responses to antigen when second signals are absent or limiting. Nat Immunol. 2020;21:1267–1279. doi: 10.1038/s41590-020-0765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Jenkins MK, Pape KA. Heterogeneity in the differentiation and function of memory B cells. Trends Immunol. 2012a;33:590–597. doi: 10.1016/j.it.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. 2012b;209:597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Pape KA, Steach HR, Jenkins MK. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science. 2015;347:784–787. doi: 10.1126/science.aaa1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, Steichen JM, Kumari S, Allen JD, Dane EL, et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama H, Okada S, Hatano M, Takahashi Y, Takeda N, Ichii H, Takemori T, Kuroda Y, Tokuhisa T. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R, Guthmiller JJ, Sturtz AJ, Surette FA, Rogers KJ, Sompallae RR, Li F, Pope RL, Chan JA, de Labastida Rivera F, et al. Infection-induced plasmablasts are a nutrient sink that impairs humoral immunity to malaria. Nat Immunol. 2020;21:790–801. doi: 10.1038/s41590-020-0678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol. 2014;88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, et al. Immunological Genome Project The cis-Regulatory Atlas of the Mouse Immune System. Cell. 2019;176:897–912.:e20. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TT, Gonzalez DG, Cote CM, Kerfoot SM, Deng S, Cheng Y, Magari M, Haberman AM. Germinal center B cell development has distinctly regulated stages completed by disengagement from T cell help. eLife. 2017;6:e19552. doi: 10.7554/eLife.19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Single cell- and bulk-RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper also used some existing, publicly available data. These datasets are listed in the Key resources table.
This paper did not generate any unique code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.