Abstract
Purpose:
Ovarian high-grade serous carcinoma (HGSC) is usually diagnosed at late stage. We investigated whether late-stage HGSC has unique genomic characteristics consistent with acquisition of evolutionary advantage compared with early-stage tumors.
Experimental Design:
We performed targeted next-generation sequencing and shallow whole-genome sequencing (sWGS) on pretreatment samples from 43 patients with FIGO stage I–IIA HGSC to investigate somatic mutations and copy-number (CN) alterations (SCNA). We compared results to pretreatment samples from 52 patients with stage IIIC/IV HGSC from the BriTROC-1 study.
Results:
Age of diagnosis did not differ between early-stage and late-stage patients (median 61.3 years vs. 62.3 years, respectively). TP53 mutations were near-universal in both cohorts (89% early-stage, 100% late-stage), and there were no significant differences in the rates of other somatic mutations, including BRCA1 and BRCA2. We also did not observe cohort-specific focal SCNA that could explain biological behavior. However, ploidy was higher in late-stage (median, 3.0) than early-stage (median, 1.9) samples. CN signature exposures were significantly different between cohorts, with greater relative signature 3 exposure in early-stage and greater signature 4 in late-stage. Unsupervised clustering based on CN signatures identified three clusters that were prognostic.
Conclusions:
Early-stage and late-stage HGSCs have highly similar patterns of mutation and focal SCNA. However, CN signature analysis showed that late-stage disease has distinct signature exposures consistent with whole-genome duplication. Further analyses will be required to ascertain whether these differences reflect genuine biological differences between early-stage and late-stage or simply time-related markers of evolutionary fitness.
Translational Relevance.
To determine whether early-stage ovarian high-grade serous carcinoma (HGSC) represents a distinct genomic entity, we collected samples from 43 patients with stage IA–IIA HGSC to identify potential differences in short genomic variants and copy-number aberrations, and compared them with a cohort of 52 late-stage (stage IIIC–IV) cases. We found no significant differences in somatic mutations or focal copy-number alterations between early-stage and late-stage cohorts. There was, however, a significant difference in both ploidy and copy-number signature exposure between early-stage and late-stage samples, with higher ploidy and signature 4 exposure in late-stage cases. Unsupervised hierarchical clustering revealed three clusters, which were prognostic. Together, our data suggest that early-stage and late-stage HGSC share fundamental genomic features, but that late-stage disease appears distinct from early-stage, with evidence of whole-genome duplication that may provide evolutionary benefit.
Introduction
High-grade serous carcinoma (HGSC) accounts for approximately 70% of all ovarian cancer cases and approximately 80% of ovarian cancer deaths. Most patients with HGSC present with advanced (FIGO stage III and IV) disease, where treatment is rarely curative. Despite the addition of anti-angiogenic agents (1, 2) and PARP inhibitor therapy (3, 4), the majority of patients with advanced disease relapse within 24 months of completion of first-line chemotherapy. In contrast, the small proportion (10%–15%) patients who present with early disease (stage I and II) have much better prognosis and are frequently cured with surgery and platinum-based chemotherapy alone (5).
HGSC is marked by near-universal TP53 mutation (6, 7) and arises from the fimbriae of the distal fallopian tube, evolving from p53 signatures (cytologically normal cells with mutant TP53), via serous intra-epithelial carcinomas (STIC) to invasive carcinomas (8) that metastasize to the ovary and throughout the peritoneal cavity. HGSC is marked by widespread copy-number change and is the archetypal C class, copy-number–driven malignancy (9). Although the genome of HGSC is highly complex, we recently described copy-number (CN) signatures, recurrent patterns of genome-wide copy-number change that were prognostic and were significantly associated with specific driver mutational processes (10).
The large studies that defined the genomic landscape of HGSC, including those from The Cancer Genome Atlas consortium (TCGA; ref. 7), the Australian Ovarian Cancer Study (11), and the International Cancer Genome Consortium (ICGC; ref. 12), all analyzed samples from patients almost exclusively with stage III or IV disease and there is little information about the genomics of early-stage HGSC. Previous analyses of genomic alterations in matched p53 signatures, STIC, ovarian, and metastatic lesions in patients with late-stage HGSC have shown clearly that there are common SNV/indel in all samples from each patient (13), and that SCNA are evident very early in disease development and are shared across samples, with an estimated 7 years required to progress from first mutation to clinical presentation with advanced disease (14).
However, it is unclear whether patients with early-stage HGSC have a distinct subtype of disease that fails to metastasize or whether these cases are genomically similar to late-stage disease but are identified essentially by chance before metastasizing. To address this, we have undertaken genomic analysis, including shallow whole-genome sequencing (sWGS) and deep sequencing of a target gene panel, of a cohort of patients with early-stage HGSC identified at three large UK centers, with comparison to late-stage samples from the BriTROC-1 study.
Materials and Methods
Study conduct, survival analyses, and patient samples
Details of the BriTROC-1 study have been reported previously (10, 15)—the study was conducted in accordance with the principles of the Declaration of Helsinki. Ethics/IRB approval was given by Cambridge Central Research Ethics Committee (Reference 12/EE/0349) and all patients gave written informed consent to participate. For all other cases, formalin-fixed paraffin-embedded (FFPE) samples were obtained from the pathology archives of participating hospitals by specialist gynecological pathologists (BK, NS, JMcD) and utilized under the auspices and ethical approval of the Imperial College Healthcare Tissue Bank (HTA license 12275, Research Ethics Committee number 17/WA/0161, Project ID R18060). Inclusion criteria were stage IA, IB, IC, or IIA ovarian or fallopian tube carcinomas of high-grade serous histology diagnosed in the previous 15 years, and patients identified through routine clinical practice. We utilized the FIGO staging criteria in use at the time of diagnosis. Exclusion criteria included non-high-grade serous pathology and samples identified at risk-reducing surgery. All samples were obtained prior to any chemotherapy treatment—the BriTROC-1 samples were those obtained at the time of diagnosis rather than relapse. At Imperial, a database of 680 patients initially revealed 45 patients with stage I to IIA HGSC. However, only 20 patients fulfilled all inclusion/exclusion criteria. Overall survival was calculated from the date of diagnosis to the date of death or the last known clinical assessment. All cases, including the early-stage cases from BriTROC-1, underwent pathologic review (CS, JMcD).
Sequencing
Details of the sequencing of BriTROC-1 samples are given elsewhere (10). For new samples, DNA was extracted from 10 × 10 μm sections using QIAmp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer's protocol. 50 to 200 ng was sheared with a Covaris LE220 focused-ultrasonicator (Covaris) to produce 100 to 200bp fragments. Libraries were generated using SureSelect XT standard protocol (Agilent Technologies) for low-input and FFPE samples. Analysis of PTEN, KRAS, RB1, BRCA2, RAD51B, FANCM, PALB2, RAD51D, TP53, RAD51C, BRIP1, CDK12, NF1, BRCA1, BARD1, and PIK3CA was performed using a custom Ampliseq panel on a HiSeq4000 system (Illumina), using paired-end 125 bp protocols. The mean coverage was >7,000×. Nine samples were used as a panel of normal controls, five of them adjacent normal tissue and four samples whole blood. sWGS was performed on a HiSeq4000 system (Illumina), using paired-end 150 bp protocols, with 250 to 300 ng input DNA according to the manufacturer's instructions. The minimum number of reads per sample was set at 5 to 10 million (mean coverage of 0.1×). Using our previous calculations (https://gmacintyre.shinyapps.io/sWGS_power/), 10 million reads with a bin size of 30kb had 80% power (a = 0.01) to detect CN change ± 2 at 30% purity assuming ploidy of 2.
Mutation calling
FASTQ files were trimmed for adapters and aligned to reference human genome hg19 using Burrows-Wheeler Alignment (BWA-MEM; ref. 16) and pre-processed using samtools and Picard to generate sorted BAM files (17). Somatic mutations were called using Mutect2 (GATK4.1.4.1; ref. 18), Varscan2 (version 2.4.2; ref. 19), Strelka2 (version 1.0.14; ref. 20), and HaplotypeCaller (21) pipelines for SNVs and small insertions and deletions (indels) on tumor-only BAM files (new early stage samples) and tumor-normal pairs for the BriTROC-1 samples, where available, using default parameters. Where multiple samples existed for individual BriTROC-1 patients, sample data in sorted BAM files were merged prior to mutation calling but individual sample identity was retained. Mutations were annotated using variant effect predictor (VEP; version 1.5.3; ref. 22). Somatic mutations were filtered by clinical significance (ClinVar, October 2020) with “benign” and “likely_benign” variants discarded. Variants were further checked for pathogenicity in the COSMIC database (GRCh37, February 2021). Germline mutations for BriTROC-1 samples were detected using similar criteria for pathogenicity using Strelka and HaplotypeCaller.
Absolute CN and CN signature calling
sWGS reads were aligned to reference human genome hg19. Relative copy numbers were obtained for predefined 30kb bins using a modified version of the QDNASeq package (23). We obtained absolute copy numbers using the sWGS-absoluteCN (swgs) pipeline—full details are given in Supplementary Information. Focal amplifications and deletions were defined according to the COSMIC definitions (see https://cancer.sanger.ac.uk/cosmic/help/cnv/overview): amplification was defined as total copy number ≥5 if average ploidy ≤2.7, or ≥9 if average ploidy >2.7. Loss was defined as total copy number 0 if average ploidy ≤2.7 or (ploidy minus 2.7) if average ploidy >2.7. Gain in Fig. 3C was defined as total copy number >2.5 but <5.0. CN signatures were calculated using the R scripts as published with previously (10).
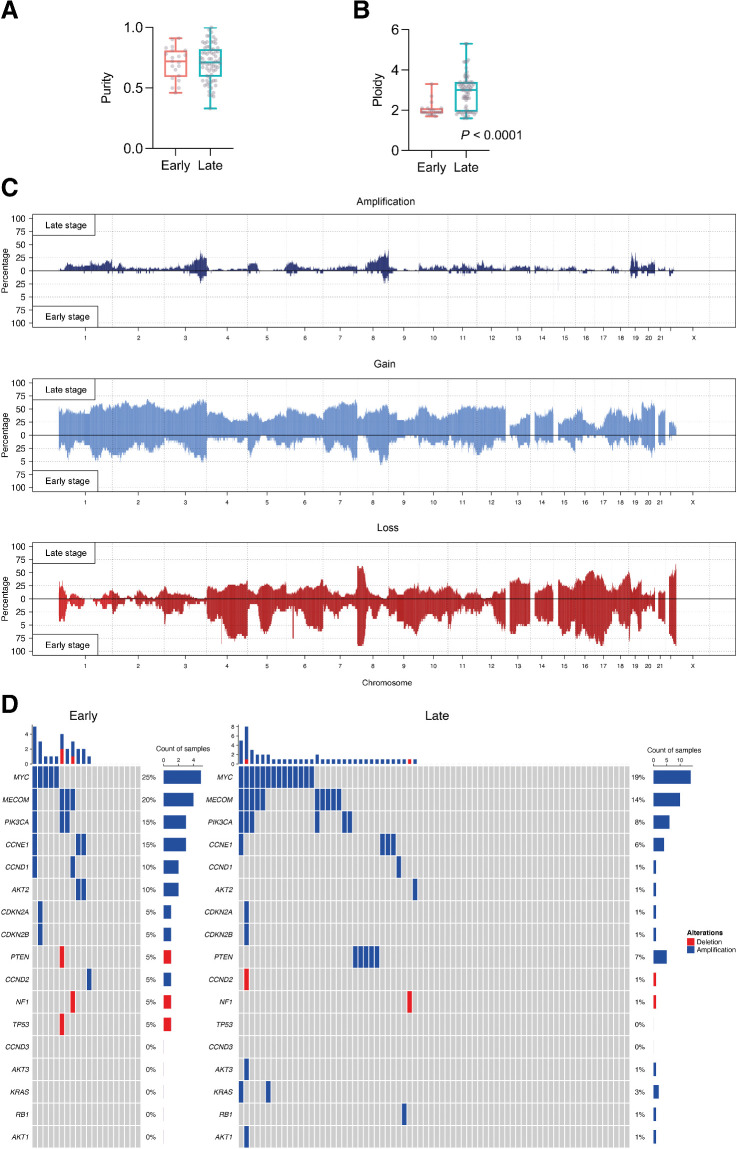
Figure 3.
Focal gene amplifications and deletions in early-stage and late-stage cohorts. A, Purity comparison of early-stage and late-stage cohorts. B, Ploidy comparison of early-stage and late-stage cohorts; Mann–Whitney test. C, Global CN amplifications, gains and losses in early-stage and late-stage cohorts. D, Estimation of focal amplifications and deletions in 17 genes of interest, determined by sWGS. The top plot shows the number of amplifications and deletions in each tumor sample.
CN signature comparison
To model the presence or absence of signatures, a fixed effects Bernoulli model was used with an intercept and a coefficient for the change between early-stage and late-stage samples. The presence of a signature j in sample i is modeled by a Bernoulli with probability θij, where θ = x⊤β. x has two rows—for the intercept and the difference between the groups—and as many rows as samples. β has two rows and as many columns as the number of signatures (d = 7). The change in the differential abundance of nonzero exposures has been modeled similarly. We used a multivariate normal model based on the isometric log ratio (ILR)-transformed exposures (also called a logistic-normal model in the literature). The ILR transformation maps a d-dimensional compositional vector (the exposures) to a d − 1 dimensional vector of real values. To account for absent signatures, the transformation (and subsequently the model) only used the subset of signatures that are present in each sample. Covariates are the same as those in Bernoulli model, but this time θ = x⊤β represents the ILR-transformed probabilities. Therefore, it has six columns instead of 7, and each row can be transformed back to a seven-dimensional vector of probabilities with the inverse ILR transformation. β continues to have two rows, but only six columns, which indicate changes in log-ratios of signature exposures. The R package TMB (24) was used for inference. The model was written in C++ and a full description of the analysis is given in Supplementary Materials and Methods.
Unsupervised clustering of patients using signature exposures
Hierarchical clustering of the CN signature exposure vectors of all samples (early-stage and late-stage) used in the survival analysis was performed using the NbClust (25) package in R. The NbClust package contains 30 indices to determine the relevant number of clusters; the number of three clusters ranked the top clustering scheme from different results obtained by varying all combinations of number of clusters, distance measures, and clustering methods. A Cox proportional hazards model was fitted using the cluster labels as covariates, using the R packages survival (26) and survminer (https://rpkgs.datanovia.com/survminer/index.html). For survival analyses based on cluster, patients with >1 sample were allocated into the cluster of the sample with highest purity.
Data accessibility
All sequencing data are available via the European Genome-phenome Archive at the European Bioinformatics Institute (https://ega-archive.org) with accession number EGAS00001005567.
Statistical analyses
Unless otherwise stated above, statistical analyses were performed using Prism (v9.0.3, GraphPad) and a summary of analyses is included in the Supplementary Materials and Methods.
Results
Patients and samples
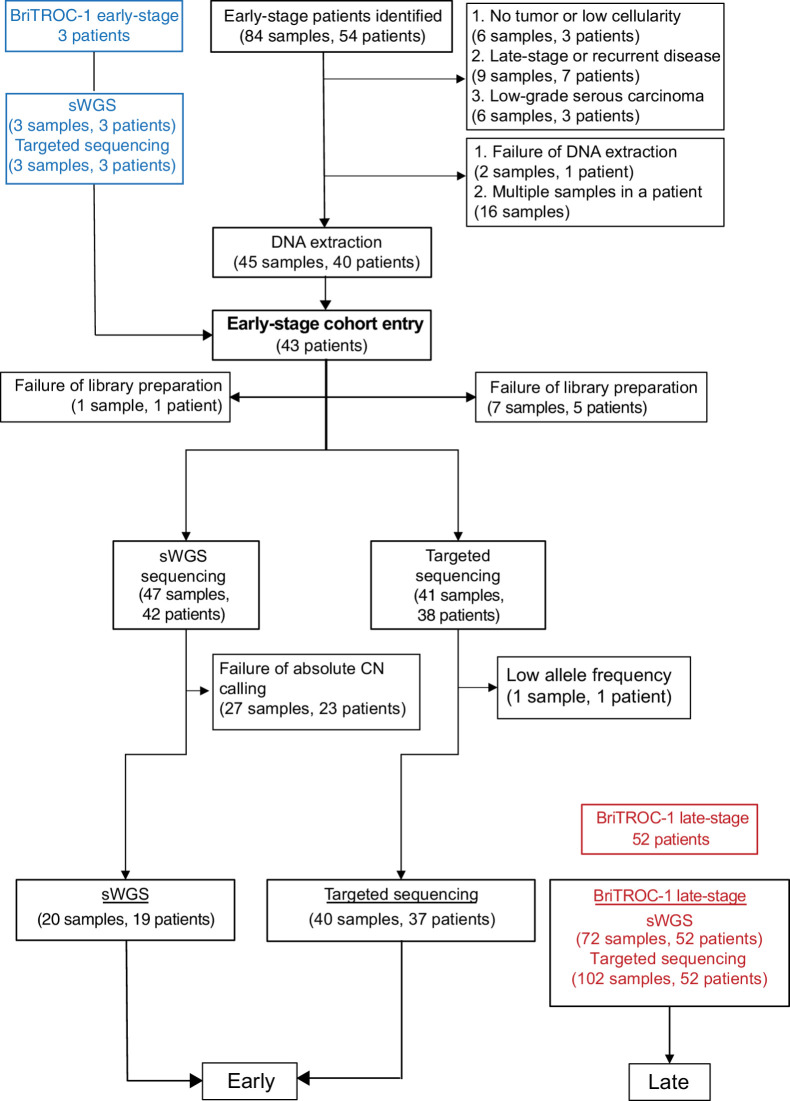
We identified 54 patients with early-stage (defined as stage IA, IB, IC, and IIA using the FIGO classification at the time of diagnosis) ovarian HGSC from the pathology archives of three large UK gynecological cancer centers (Imperial College Healthcare, University College London and Barts Health NHS Trusts). A summary of the workflow is shown in Fig. 1 and clinical details are given in Supplementary Table S1. Following pathology review, 21 samples from 13 patients were excluded, whereas two samples from one patient failed DNA extraction. In addition, we identified a further cohort of three early-stage patients recruited into the BriTROC-1 study (15), giving a total early-stage population of 43. The comparison late-stage cohort consisted of 52 patients with stage IIIC/IV disease recruited into the BriTROC-1 study (Supplementary Table S1). The early-stage patients were diagnosed more recently than the late stage (early, median 68 months, range 24–177, prior to analysis; late median 101 months, range 60–179; P = 0.0009; Supplementary Fig. S1). The median age at diagnosis for early-stage and late-stage cohorts did not differ significantly (early 61.3 years, range 40–84; late 62.3 years, range 34–76) but overall survival was, as expected, significantly longer in the early-stage cohort than for the late-stage (HR, 0.13; 95% CI, 0.07–0.26; Fig. 2A and B).
Figure 1.
REMARK diagram for early-stage and late-stage cohorts.
Figure 2.
Clinical features and mutational landscape of early-stage and late-stage cohorts. A, Diagnosis age [median 61.3 years (early stage), 62.3 years (late stage); P = NS]. B, Overall survival. Median 60.3 months for late stage, and not reached for early stage. Log-rank. HR, 0.13 (95% CI, 0.07–0.26), P < 0.0001 (Log-rank). C, Short variants (SNV and indels) for each patient in early-stage and late-stage cohorts. The top plot shows the number of mutations in each tumor sample. D, Gene mutation mapper plot of TP53 in early-stage cohort and (E) late-stage cohort. Key hotspot residues are marked. The commonest residue mutations in each cohort are marked in red.
Mutational landscape of early-stage and late-stage cohorts
Using targeted next-generation sequencing, we analyzed short variants (SNV, indels) in both cohorts (Supplementary Table S2). Mutations in TP53 were near-universal (100% late-stage patients [52/52]; 89% [34/38] early-stage patients; Fig. 2C, Supplementary Fig. S2). One early-stage case, patient ES_0007, contained two TP53 missense mutations (L139V, Y163C) at mutant allele frequencies of approximately 50% and 25%, respectively. Although these could result from the presence of two separate clones, the CN state at the TP53 locus in this case was neutral (log2 ratio shift = −0.056), suggesting that these might be bi-allelic mutations, as we have previously identified in ovarian squamous cell carcinomas arising from mature cystic teratoma (27). The four early-stage samples in which TP53 mutations were not identified underwent pathology re-review; all were still considered to be HGSC. Two had copy-number profiles consistent with HGSC, whereas two showed no CN abnormalities, suggesting very low tumor cellularity (Supplementary Fig. S3). Overall, the frequency of four key TP53 hotspot mutations (R175, R273, R248, Y220) was significantly greater in the early-stage cohort compared with late-stage cohort (Fig. 2D and E; Fisher exact test; P = 0.0029), but there was no difference in the rates of mutations in the other analyzed genes (Fig. 2C). Specifically, rates of pathologic mutations in BRCA1 and BRCA2 did not differ significantly between early-stage and late-stage patients: BRCA1 11% (4/38) early versus 17% (9/52) late; BRCA2 0% (0/38) early versus 2.0% (1/52) late.
Focal amplifications and deletions in early-stage and late-stage cohorts
We used sWGS to analyze genome-wide absolute copy number. There was no statistically significant difference in purity between the cohorts (Fig. 3A), but median ploidy was significantly greater in late-stage samples compared with early-stage samples (Fig. 3B; median early 1.9; median late 3.0; P < 0.0001, Mann–Whitney test). Global copy-number gains/losses are shown in Fig. 3C—there were generally more gains and amplifications in late-stage samples, and regions of CN loss in the early-stage samples, in keeping with the differential ploidy between the cohorts. Although there were several regions of differential gain in the late-stage cohort (e.g., chromosome 4, 6, 9, 11,12) and losses in the early-stage cohort (e.g., chromosome 4, 9, 12, 17), we found no significant differences in rates of focal amplification and deletion of 17 genes that are frequently altered in HGSC (Fig. 3D; Supplementary Fig. S4; Supplementary Table S3; refs. 7, 12). The commonest amplifications were in MYC (25% in the early-stage and 19% in the late-stage) and MECOM (20% in the early-stage and 14% in the late-stage).
CN signatures in early-stage and late-stage cohorts
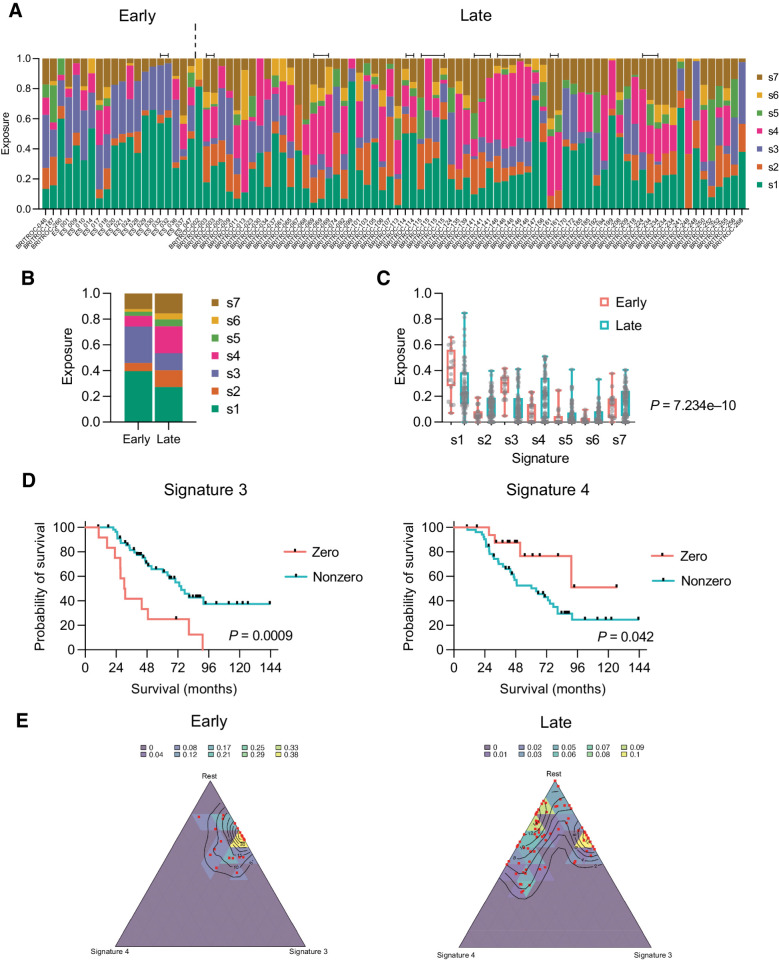
Next, we assessed the distribution of the six specific CN features—segment length, segment copy number, number of breakpoints per chromosome arm, number of breakpoints per 10Mb, copy-number change point and length of chains of oscillating copy number (Supplementary Figs. S5, 6)—and used these to generate CN signature exposures for both cohorts (Fig. 4A and B). We used a fixed-effects (Bernoulli) analysis to model the presence or absence of signatures and a fixed-effects multivariate normal distribution model, based on ILR-transformation, to compare the two cohorts (see Supplementary Materials and Methods). Overall, in the ILR analysis, there was a highly significant difference between cohorts (generalized Wald test; P = 7.234e−10), with greater signature 3 exposure in the early-stage cohort and more signature 4 in the late-stage cohort (Fig. 4C). In keeping with this, the presence of signature 3 and the absence of signature 4 were both associated with improved overall survival across all patients (Fig. 4D).
Figure 4.
CN signatures in early-stage and late-stage cohorts. A, CN signature exposures in early-stage and late-stage patients. Note that signature exposures sum to 1 in each sample. Bars above signatures indicate adjacent samples derived from the same patient. B, Mean signature exposure proportions across the early-stage and late-stage cohorts. C, Comparison of signature exposures across early-stage and late-stage cohorts; Wald test. D, Overall survival of combined early-stage and late-stage cohorts by zero versus nonzero exposures to CN signature 3 (left) and 4 (right). Log-rank (Mantel–Cox) analysis. E, Simplex plots representing exposures for CN signature 3 (right axis), signature 4 (bottom axis), and the rest of the signatures (1–S3–S4) combined (left axis) in early-stage (left) and late-stage (right) cohorts. Each red dot represents a single sample, and the contours represent the density of observed samples.
We then visualized CN signature exposures using simplex plots (Fig. 4E) comparing signature 3 (S3), signature 4 (S4), and all other signatures (1-S3-S4). In the early cohort, the sample observations (red dots) cluster towards the top of the right side of the simplex, in keeping with low or zero signature 4 exposure. For the late group, although some of the observations remain in the same place, many are located towards the left of the simplex, indicating that they have nonzero exposure to signature 4, with a relative decrease in the amount of signature 3. The relative contribution of the other signatures does not change between early and late cohorts—the distance from the observations to the top apex of the plot remains similar. Together, these suggest that, overall, signature 3 decreases in relative intensity and signature 4 increases in relative intensity in the late-stage samples, whereas the rest of the signatures remain approximately constant. However, the observations that remain towards the top right of the simplex suggest that a subset of late-stage samples have genomic features more reminiscent of early-stage. In keeping with this, the presence of signature 3 remained significantly associated with improved overall survival in the late-stage cases, whereas there was a trend for poorer survival with any exposure to signature 4 (Supplementary Fig. S7).
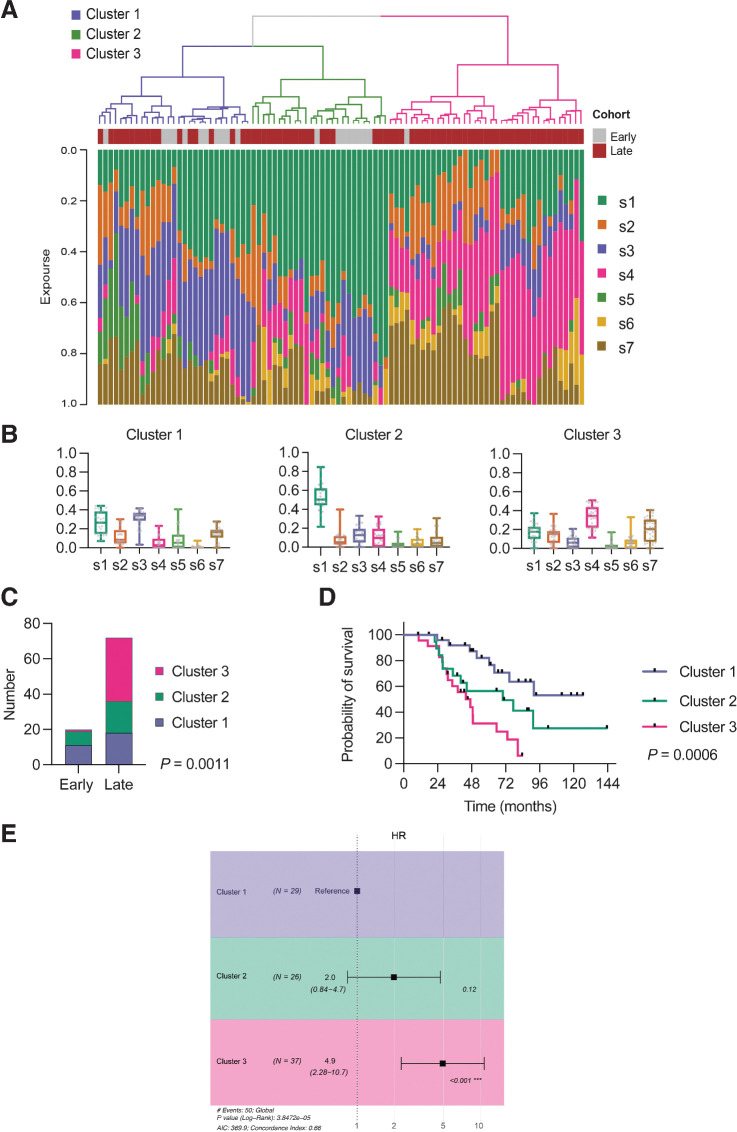
We then performed unsupervised hierarchical clustering of the CN signature exposures across both cohorts and identified three clusters (Fig. 5A; Supplementary Fig. S8). Cluster 1 had the highest exposure to CN signature 3, cluster 2 was dominated by genomes with high signature 1 exposure, whereas cluster 3 showed highest signature 4 exposure (Fig. 5B). There was a significant difference in sample distribution between clusters (P < 0.0001, Chi-squared) with nearly all early-stage samples in clusters 1 and 2, whereas late-stage samples were spread across all three clusters, with the majority in cluster 3 (Fig. 5C). In the 12 patients with >1 sample, samples clustered within the same cluster group in 11 cases (Supplementary Fig. S8). The clusters were prognostic, with a significant trend for reduced survival across the clusters (Fig. 5D), which remained significant by Cox proportional hazards (Fig. 5E).
Figure 5.
Relationship between signature exposures and clinical factors. A, Unsupervised hierarchical clustering in combined early-stage and late-stage cohorts. B, Distributions of CN signature exposures in three clusters. C, Early-stage and late-stage samples by cluster; chi-square test. D, Overall survival by cluster. Log-rank for trend. E, Forest plot of HR estimates on overall survival (OS) for clusters. Cox proportional hazards.
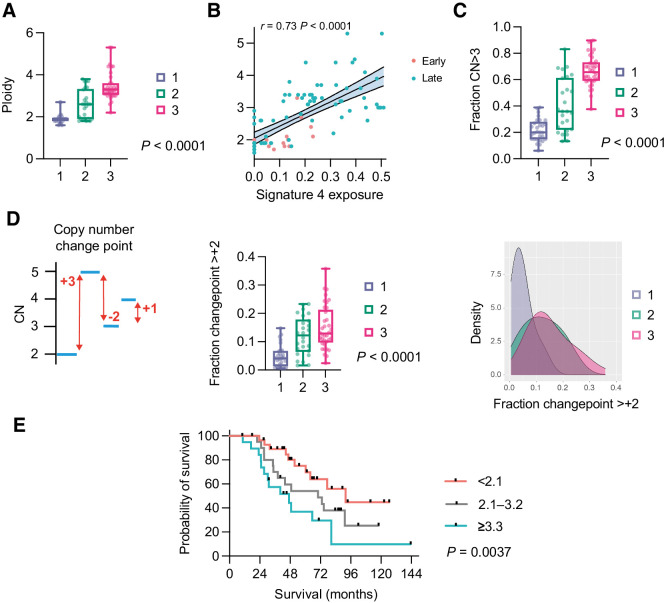
We next quantified ploidy by cluster and found highly significant differences across the whole cohort (Fig. 6A) with median ploidy of 1.9, 2.6, and 3.3 in clusters 1, 2, and 3, respectively. High ploidy can reflect whole-genome duplication (WGD), which is frequent in HGSC (28). However, identifying WGD definitively requires assessment of allele-specific copy number, which is not possible using sWGS or the limited next-generation sequencing panel that we utilized. Nonetheless, we previously identified a strong statistical association between CN signature 4 exposure and WGD (10): here, we found a very strong correlation (Spearman ρ = 0.73; P < 0.0001) between ploidy and signature 4 exposure across both cohorts (Fig. 6B) implying that the high ploidy may indeed be driven by WGD. In addition, WGD is likely to generate high ploidy states across the entire genome and we found a significantly greater proportion of segments with ploidy >3.0 in clusters 2 and 3 compared with cluster 1 (Fig. 6C) as well as in late-stage compared with early-stage samples (Supplementary Fig. S9). Analysis of copy-number changepoint (the change in CN state between adjacent segments) also indicated that clusters 2 and 3 had a significantly greater fraction of segments with changepoint of at least +2 than cluster 1, again in keeping with WGD (Fig. 6D; ref. 29).
Figure 6.
Cluster ploidy and WGD. A, Ploidy distribution of late-stage samples in three clusters. Kruskal–Wallis test. B, Correlation between CN signature 4 exposure and ploidy across both cohorts. Spearman rank correlation. C, Fraction of CN segments with absolute CN ≥3 in three clusters. Kruskal–Wallis test. D, CN changepoint. Graphic depiction of CN changepoint (left); distribution of CN changepoint ≥+2 in the three clusters. Kruskal–Wallis test (center); density distribution (right). E, Overall survival of combined early-stage and late-stage cohorts by ploidy. Log-rank for trend analysis.
Finally, it was previously shown that median ploidy for cancers with and without WGD were 3.3 and 2.1, respectively (28). Using these delineators, we analyzed overall survival of the whole cohort (Fig. 6E) and of late-stage patients (Supplementary Fig. S10) and identified significant differences in both analyses.
Discussion
Comparing genomic profiles between cancers with high levels of chromosomal instability is challenging. We have previously developed CN signatures that deconvolute CN features from whole-genome analysis to identify the underlying mutational processes that shape the genome. In this manuscript, we have used novel methods to compare CN signature exposures and reveal significant genomic differences between early-stage and late-stage HGSC. This is clinically important because the large majority of patients with HGSC have advanced disease at the time of diagnosis, reflecting the ease with which HGSC disseminates from the fallopian tube throughout the peritoneal cavity. An important question is whether early-stage HGSC is identified fortuitously through early development of symptoms or whether it has discrete characteristics that reduce the likelihood of peritoneal dissemination, and whether late-stage samples have acquired evolutionary fitness that facilitates metastatic spread.
As expected, we found that TP53 mutations were near-universal, although four early-stage samples were TP53 wild type, possibly due to a combination of low tumor cellularity and poor DNA quality from FFPE preservation. A previous study of 16 early-stage HGSC cases (30) also identified that TP53 mutations were very frequent but not universal, and two of our TP53 wild-type samples certainly had CN profiles that were highly consistent with HGSC. Together, these data suggest that a small proportion of early-stage HGSC cases may be truly TP53 wild type. The rate of missense TP53 mutations here was higher (75%) than in previous HGSC cohorts (7) and hotspot mutations (defined here as mutation at the four most commonly mutated codons, R248, R273, R175, and Y220) were more prevalent in our early-stage cohort than late-stage. However, a recent analysis of nearly 800 HGSC cases (78 stage I/II, 709 stage III/IV) has suggested no overall difference in mutation type between early and late samples (31). Crucially, we found no differences in rates of BRCA1/2 mutations between early and late cohorts. The absence of germline DNA from most of our early-stage cohort meant that we were unable to verify the germline mutation status but our BRCA1/2 mutation results are broadly in line with previous cohorts (32) and reflect the fact that these samples were obtained from routine practice rather than risk reducing surgery. When examining global CN change, there were no regions uniquely lost or gained in either cohort, suggesting that the process of dissemination is not driven by specific amplifications or deletions, and we also found no significant difference in CN of 17 key genes. Together, the SNV/indel and focal CNA data corroborate findings by Köbel and colleagues that early-stage and late-stage HGSC appear identical by IHC (33). The previous WGS analysis of early-stage HGSC (30) also identified high levels of genomic instability with few, if any, recurrent focal differences between early-stage and late-stage HGSC, and no unique mutation or focal CN alterations in the early-stage patients.
Our most striking observation was the difference in overall ploidy between early and late cases, which was further reflected in CN signature exposures. Comparison of CN signatures between samples and cohorts is complex because signature exposures are compositional (i.e., they sum to 1 in each sample) and are thus not independent variables: any decrease in one signature will, by definition, be mirrored by an increase in at least one other. In addition, classical statistical methods for analyzing compositional data are poor at dealing with zero proportions and many samples have zero exposure to at least one signature. However, using isometric-log ratio analysis of nonzero signature exposures, we found a significant difference between the cohorts overall, driven by signatures 3 (higher in early-stage) and 4 (higher in late-stage). The features that define signature 4 are high segment CNs and high CN changes, both of which are greater in the late cohort. The simplex analysis indicates that the late-stage genomes overall have increased signature 4, although a proportion remains “early-like” with prominent CN signature 3. The unsupervised hierarchical clustering identified three patterns in the signatures. Clusters 1 and 2 contained most of the early-stage samples, whereas the late-stage samples were divided between the clusters. However, cluster 3 contained almost exclusively late-stage samples and was associated both with higher ploidy and worse survival.
The higher CN, greater overall ploidy and greater signature 4 exposure all suggest that a proportion of the late-stage tumors had undergone WGD. WGD has been described in many solid malignancies (28) and is generally associated with poor prognosis (34). It is thought to arise from aberrant cell division (35) and potentially may mitigate the effects of mutations that would otherwise be deleterious, thus preventing cancer cell attrition (36). In pan-cancer analysis, HGSC has been shown to have one of the highest rates of WGD at approximately 40% (28).
Definitive demonstration of WGD is challenging and requires assessment of both ploidy and extent of LOH (29), which is not possible using sWGS and the targeted capture sequencing panel employed here. In practical terms, the analysis of early stage HGSC requires use of FFPE material that precludes deep WGS analysis. This is an important limitation of our findings. However, our original analysis using deep WGS analysis of HGSC specimens showed that CN signature 4 was significantly associated with WGD (10) and here we observed a strong correlation between signature 4 and ploidy, suggesting that the high ploidy samples in cluster 3 may have undergone WGD. We also confirmed other features suggestive of WGD, including that cluster 3 samples were more likely to have high CN across the whole genome and high CN changepoint between segments. Finally, the median ploidy of early and late cohorts was similar to pan-cancer analyses showing that median ploidy of WGD tumors is 3.3, compared with 2.1 in those lacking WGD (28). The commonest genomic correlate of WGD in previous analyses was mutation in TP53, which usually precedes the duplication, as well as CCNE1 amplification and loss of RB1 (28). Although the rates of CCNE1 amplification here did not differ significantly between our early-stage and late-stage cohorts, our data support the idea that high ploidy is associated with advanced HGSC and poorer prognosis, a concept first explored over 20 years ago (37). HGSC has profound levels of segregation error during cell division, an essential precursor of aneuploidy and WGD (38), and recent data suggest that WGD can emerge in hTERT-immortalized human fallopian tube epithelial cells with loss of wild-type p53 function in the presence of BRCA1 mutation and MYC overexpression, but not with TP53 mutation alone (39).
Although this cohort represents one of the largest collections of early-stage HGSC samples that have been characterized with WGS, this project has potential shortcomings. Our cohort is small, reflecting the rarity of this patient population, which limits our statistical power, in particular the ability to compare early-stage cases that relapsed to those that did not. In addition, the samples were identified retrospectively from pathology archives; thus, the extent of surgery was not defined, and chemotherapy regime given was at the discretion of the treating oncologists. The early-stage cohort was diagnosed more recently, on average, than the late stage, although this difference is unlikely to have influenced clinical management, and nearly 80% were alive 10 years following diagnosis, in keeping with data from randomized clinical trials (5). This strongly suggests that our cohort did not contain large numbers of understaged patients with occult stage III disease. The samples were all FFPE and analyzed up to 15 years following diagnosis, and it was not possible to estimate absolute copy number, and hence CN signatures, on several of the samples. Our previously developed methods allow reliable analysis of genome-wide changes in fixed material (40), but demand strict quality control criteria to ensure robust CN determination. Our estimation of the number of reads required (see Materials and Methods) was clearly insufficient in some cases, especially those with low tumor cellularity. If sWGS is to be developed for use in clinical trials, it will be imperative to establish robust criteria for sequencing depth, especially in low cellularity samples or small core biopsies. We also did not perform whole exome sequencing or deep WGS, so are unable to comment upon small variants (SNV, indel) beyond our targeted panel nor on larger scale rearrangements (12, 41).
The critical outstanding question is whether the processes that generate high ploidy are the primary drivers of rapid dissemination in HGSC, or whether high ploidy/WGD are simply time-related markers of evolutionary fitness and are thus more likely to be observed in late-stage than early-stage disease. The absence of an age difference between our two cohorts may suggest that the genomic differences are not time-related, consistent with the finding that WGD is an early event in colorectal (35) and non–small cell lung carcinomas (36). In addition, there was close clustering of samples in patients with >1 sample, again suggesting that WGD does not appear as a late event. However, definitive assessments of true rates of WGD in early HGSC compared with advanced disease will require WGS analysis of prospectively collected samples and detailed comparison between fallopian tube primary site and multisite examination of large cohorts of disseminated late-stage HGSC as well as in vitro models.
In summary, our results indicate that early and late-stage HGSC are similar but also that there may be critical differences, potentially resulting from the appearance of WGD in a subset of late-stage disease, which is associated with poor outcome. However, our data, reinforced by the striking difference in overall survival in our cohorts, highlight once again the importance of improving strategies that will allow early detection of HGSC.
Authors' Disclosures
G. Macintyre reports a patent for a method of characterizing a DNA sample pending and a patent for methods for predicting treatment response in cancers pending and licensed to Tailor Bio Ltd.; in addition, G. Macintyre is a founder, director, and reports ownership of stock in Tailor Bio Ltd. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This work was funded by an Imperial/China Scholarship Council scholarship to Z. Cheng, the NIHR Imperial Biomedical Research Centre (grant no. P77646), Ovarian Cancer Action (grant no. 006), the Wellcome Trust (grant no. RG92770), and Cancer Research UK (grant nos. A15973, A15601, A18072, A17197, A19274, and A19694). Imperial samples were provided by the Imperial College Healthcare Tissue Bank, which is supported by the NIHR Imperial Biomedical Research Centre. Other infrastructure support was provided by Experimental Cancer Medicine Centres at participating sites and the Cancer Research UK Imperial Centre.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
Z. Cheng: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. H. Mirza: Formal analysis, validation, writing–original draft. D.P. Ennis: Resources, formal analysis, investigation, writing–original draft. P. Smith: Software, formal analysis, methodology, writing–original draft. L. Morrill Gavarró: Formal analysis, methodology. C. Sokota: Resources, formal analysis. G. Giannone: Formal analysis. T. Goranova: Data curation, formal analysis. T. Bradley: Data curation, formal analysis. A. Piskorz: Resources, data curation, formal analysis. M. Lockley: Resources. B. Kaur: Resources. N. Singh: Resources. L.A. Tookman: Resources. J. Krell: Resources. J. McDermott: Resources. G. Macintyre: Data curation, software, formal analysis, methodology. F. Markowetz: Conceptualization, resources, formal analysis, supervision, funding acquisition, methodology. J.D. Brenton: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, writing–original draft. I.A. McNeish: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med 2011;365:2484–96. [DOI] [PubMed] [Google Scholar]
- 2. Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med 2011;365:2473–83. [DOI] [PubMed] [Google Scholar]
- 3. González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019;381:2391–402. [DOI] [PubMed] [Google Scholar]
- 4. Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, et al. Olaparib plus Bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 5. Collinson F, Qian W, Fossati R, Lissoni A, Williams C, Parmar M, et al. Optimal treatment of early-stage ovarian cancer. Ann Oncol 2014;25:1165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high-grade serous carcinoma of the ovary. J Pathol 2010;221:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. TCGA. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol 2007;211:26–35. [DOI] [PubMed] [Google Scholar]
- 9. Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013;45:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macintyre G, Goranova T, De Silva D, Ennis D, Piskorz AM, Eldridge M, et al. Copy-number signatures and mutational processes in ovarian carcinoma. Nat Genet 2018;50:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 2008;14:5198–208. [DOI] [PubMed] [Google Scholar]
- 12. Patch A-M, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489–94. [DOI] [PubMed] [Google Scholar]
- 13. Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov 2016;6:1342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun 2017;8:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goranova T, Ennis D, Piskorz AM, Macintyre G, Lewsley LA, Stobo J, et al. Safety and utility of image-guided research biopsies in relapsed high-grade serous ovarian carcinoma-experience of the BriTROC consortium. Br J Cancer 2017;116:1294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sandmann S, de Graaf AO, Karimi M, van der Reijden BA, Hellstrom-Lindberg E, Jansen JH, et al. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci Rep 2017;7:43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling somatic SNVs and Indels with Mutect2. bioRxiv 2019:861054.
- 19. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 21. Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2018:201178.
- 22. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res 2014;24:2022–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kristensen K, Nielsen A, Berg CW, Skaug H, Bell BM. TMB: automatic differentiation and laplace approximation. J.Stat.Softw 2016;70:1–21. [Google Scholar]
- 25. Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: An R package for determining the relevant number of clusters in a data set. Journal of Statistical Software 2014;61:1–36. [Google Scholar]
- 26. Therneau T. 2020A package for survival analysis in R. < https://CRAN.R-project.org/package=survival.>.
- 27. Cooke SL, Ennis D, Evers L, Dowson S, Chan MY, Paul J, et al. The driver mutational landscape of ovarian squamous cell carcinomas arising in mature cystic teratoma. Clin Cancer Res 2017;34:7633–40. [DOI] [PubMed] [Google Scholar]
- 28. Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 2021;184:2239–54.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chien J, Sicotte H, Fan JB, Humphray S, Cunningham JM, Kalli KR, et al. TP53 mutations, tetraploidy and homologous recombination repair defects in early stage high-grade serous ovarian cancer. Nucleic Acids Res 2015;43:6945–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tuna M, Ju Z, Yoshihara K, Amos CI, Tanyi JL, Mills GB. Clinical relevance of TP53 hotspot mutations in high-grade serous ovarian cancers. Br J Cancer 2020;122:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rust K, Spiliopoulou P, Tang CY, Bell C, Stirling D, Phang THF, et al. Routine germline BRCA1 and BRCA2 testing in ovarian carcinoma patients: analysis of the Scottish real life experience. BJOG 2018;125:1451–8. [DOI] [PubMed] [Google Scholar]
- 33. Köbel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 2008;5:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 2004;5:45–54. [DOI] [PubMed] [Google Scholar]
- 35. Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Grönroos E, Endesfelder D, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014;4:175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. López S, Lim EL, Horswell S, Haase K, Huebner A, Dietzen M, et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat Genet 2020;52:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tropé C, Kaern J, Hogberg T, Abeler V, Hagen B, Kristensen G, et al. Randomized study on adjuvant chemotherapy in stage I high-risk ovarian cancer with evaluation of DNA-ploidy as prognostic instrument. Ann Oncol 2000;11:281–8. [DOI] [PubMed] [Google Scholar]
- 38. Nelson L, Tighe A, Golder A, Littler S, Bakker B, Moralli D, et al. A living biobank of ovarian cancer ex vivo models reveals profound mitotic heterogeneity. Nat Commun 2020;11:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bronder D, Wangsa D, Zong D, Meyer TJ, Wardenaar R, Minshall P, et al. TP53 loss initiates chromosomal instability in high-grade serous ovarian cancer. bioRxiv 2021:2021.03.12.435079. [DOI] [PMC free article] [PubMed]
- 40. Piskorz AM, Ennis D, Macintyre G, Goranova TE, Eldridge M, Segui-Gracia N, et al. Methanol-based fixation is superior to buffered formalin for next-generation sequencing of DNA from clinical cancer samples. Ann Oncol 2016;27:532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ewing A, Meynert A, Churchman M, Grimes G, Hollis RL, Herrington CS, et al. Structural variants at the BRCA1/2 loci are a common source of homologous repair deficiency in high grade serous ovarian carcinoma. Clin Cancer Res 2021;27:3201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available via the European Genome-phenome Archive at the European Bioinformatics Institute (https://ega-archive.org) with accession number EGAS00001005567.



![Figure 2. Clinical features and mutational landscape of early-stage and late-stage cohorts. A, Diagnosis age [median 61.3 years (early stage), 62.3 years (late stage); P = NS]. B, Overall survival. Median 60.3 months for late stage, and not reached for early stage. Log-rank. HR, 0.13 (95% CI, 0.07–0.26), P < 0.0001 (Log-rank). C, Short variants (SNV and indels) for each patient in early-stage and late-stage cohorts. The top plot shows the number of mutations in each tumor sample. D, Gene mutation mapper plot of TP53 in early-stage cohort and (E) late-stage cohort. Key hotspot residues are marked. The commonest residue mutations in each cohort are marked in red.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/121d/9365357/0d964ec6c267/2911fig2.jpg)