Summary
Inactivation of ARID1A and other components of the nuclear SWI/SNF protein complex occurs at exceedingly high frequency in a variety of human malignancies, suggesting a widespread role for SWI/SNF in tumor suppression. However, the underlying mechanisms remain poorly understood. Here we show that ARID1A-containing SWI/SNF complex (ARID1A/SWI/SNF) operates as inhibitor of the pro-oncogenic transcriptional coactivators YAP and TAZ. By a combination of gain and loss of function approaches in several cellular contexts, we uncovered that YAP/TAZ are instrumental for unfolding the effects of SWI/SNF inactivation, such as cell proliferation, acquisition of stem cell-like traits, and liver tumorigenesis. We found YAP/TAZ in complex with SWI/SNF; this interaction is mediated by ARID1A and alternative to YAP/TAZ association to their DNA-binding platform TEAD. Cellular mechanotransduction regulates the association between ARID1A/SWI/SNF and YAP/TAZ. The ARID1A/SWI/SNF-YAP/TAZ inhibitory interaction is predominant in cells experiencing low mechanical signaling, where loss-of-ARID1A rescues YAP/TAZ association to TEAD. At high mechanical stress, nuclear F-actin binds to ARID1A/SWI/SNF preventing the formation of the ARID1A/SWI/SNF-YAP/TAZ complex, in favor of TEAD/YAP/TAZ association. We propose that a dual requirement must be met to fully empower YAP/TAZ responses: promotion of YAP/TAZ nuclear accumulation, e.g., by loss of Hippo signaling, and inhibition of ARID1A/SWI/SNF, as obtained by either genetic inactivation or raised cell mechanics. This work offers a molecular framework whereby mechanical signals emerging at the tissue level and genetic lesions conspire to activate YAP/TAZ to induce cell plasticity and tumorigenesis.
Organs must be endowed with tissue-level checkpoints to preserve cell fates, repair wounds and avoid cancer. YAP/TAZ recently emerged as fundamental sensors by which cells read structural and architectural features of their tissue microenvironment through mechanotransduction1. Although YAP is sufficient to trigger several hallmarks of cancer, the normal microenvironment of adult tissues inhibits YAP/TAZ, such that emergence of a solid tumor must entail a successful combination of YAP/TAZ activation and removal of YAP/TAZ inhibitors.
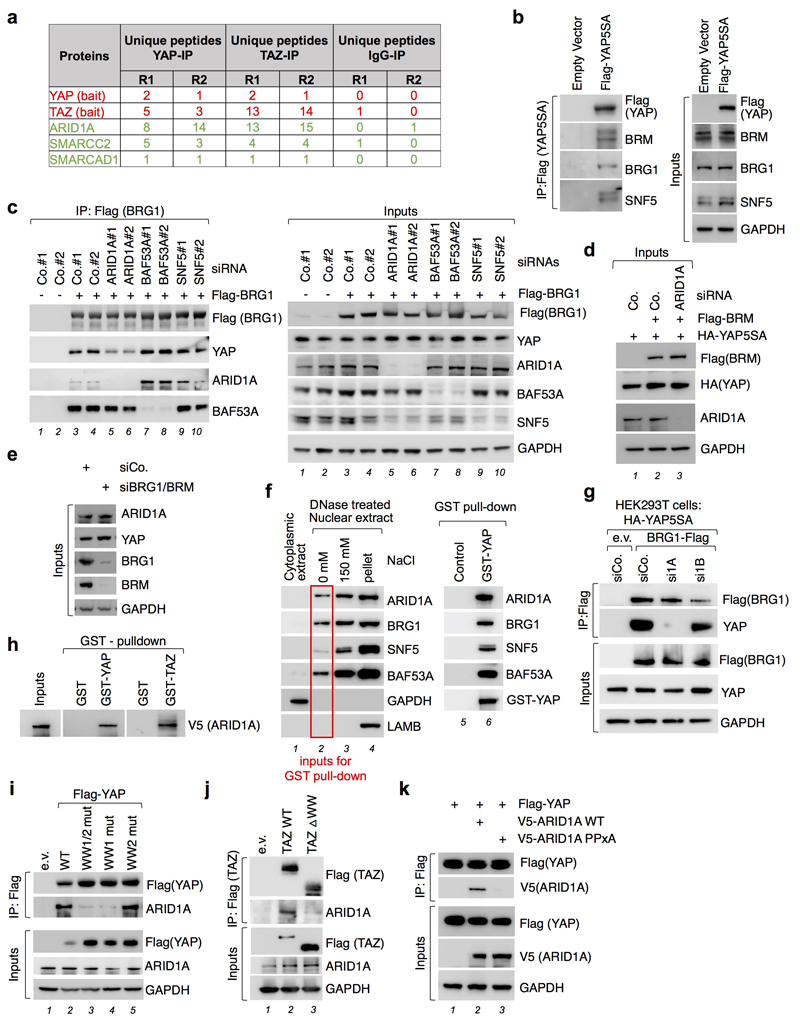
We started this investigation searching for nuclear factors interacting with YAP/TAZ by ChIP-MS2, as the nuclear layer of YAP/TAZ regulation has been so far largely overlooked in comparison with the body of knowledge on YAP/TAZ regulation in the cytoplasm3,4. The association of YAP/TAZ with several components of the SWI/SNF chromatin-remodeling complex attracted our attention (Extended Data Fig. 1a and Extended Data Table 1). SWI/SNF contains a core ATP-ase involved in nucleosome remodeling, either BRG1 or BRM, and other co-factors, such as ARID1A, whose function is less understood5. YAP/TAZ associated with ARID1A, but not with ARID1B (Extended Data Table 1), known to define alternative SWI/SNF complexes5. In several co-immunoprecipitation (co-IP) experimental set ups, we found YAP in complex with ARID1A, BRG1, BRM, and other SWI/SNF components (Fig. 1a-c and Extended Data Fig. 1b, c), also in the absence of chromatin (Extended Data Fig. 1f).
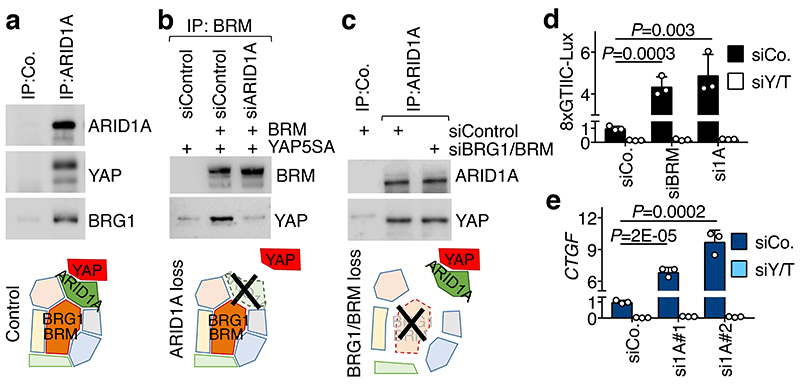
Fig. 1. YAP interacts with SWI/SNF trough ARID1A.
a, Endogenous ARID1A (top band) binds endogenous YAP and BRG1 in CoIP experiments in MCF10AT cells.
b, Binding of YAP to BRM requires ARID1A in HEK293T cells.
c, Binding of YAP to ARID1A does not require BRG1/BRM. For a-c: bottom panels represent schematics of the corresponding experimental results; See also Extended Data Fig. 1d-e.
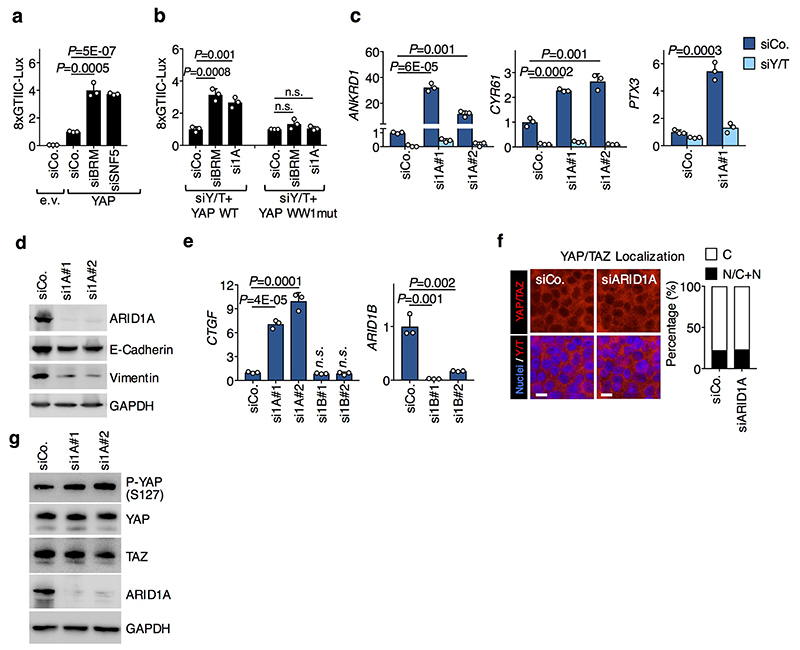
d, Luciferase assay with the 8xGTIIC-Lux reporter in HEK293 cells transfected as indicated.
e, CTGF expression in MCF10A cells transfected with the indicated siRNAs. These inductions occurred without triggering EMT (Extended Data Fig. 2d). For d-e: data are presented as mean + s.d. of n=3 biologically independent samples; P values were determined by unpaired two-sided t-test.
Shown are representative experiments, repeated independently two times for a-c, and three times for d-e, all with similar results.
YAP associates to SWI/SNF through ARID1A. Indeed, depletion of ARID1A, but not of ARID1B, impaired the ability of YAP to be incorporated into BRG1 or BRM-containing SWI/SNF complexes (Fig. 1b and Extended Data Fig. 1c, g). Conversely, depletion of BRM/BRG1 was inconsequential for the association of endogenous ARID1A and YAP proteins (Fig. 1c). Purified recombinant YAP (or TAZ) and ARID1A proteins directly interact in vitro (Extended Data Fig. 1h) through physical association of their WW domain and PPxY motifs respectively6 (Extended Data Fig. 1i-k).
We next assessed the functional relevance of SWI/SNF for YAP-dependent transcription. SWI/SNF inactivation caused induction of the YAP/TEAD luciferase reporter (8xGTIIC7) through activation of endogenous YAP/TAZ (Fig. 1d) and also strongly enhanced the activity of co-transfected exogenous wild-type YAP (Extended Data Fig. 2a); instead, WW-mutant YAP was insensitive to SWI/SNF depletion (Extended Data Fig. 2b). SWI/SNF inactivation by depletion of ARID1A (but not ARID1B) also induced the expression of several YAP/TAZ direct target genes, in a manner that is rescued by concomitant YAP/TAZ depletion (Fig. 1e and Extended Data Fig. 2c,e). Of note, SWI/SNF depletion neither affects the subcellular localization of YAP/TAZ nor YAP phosphorylation level and stability (Extended Data Fig. 2f, g), consistently with SWI/SNF acting downstream of the classic modality of YAP/TAZ regulation dictated by Hippo kinases3. Taken together, these findings indicate that SWI/SNF directly binds and inhibits nuclear YAP/TAZ, thus representing a new layer of YAP/TAZ regulation.
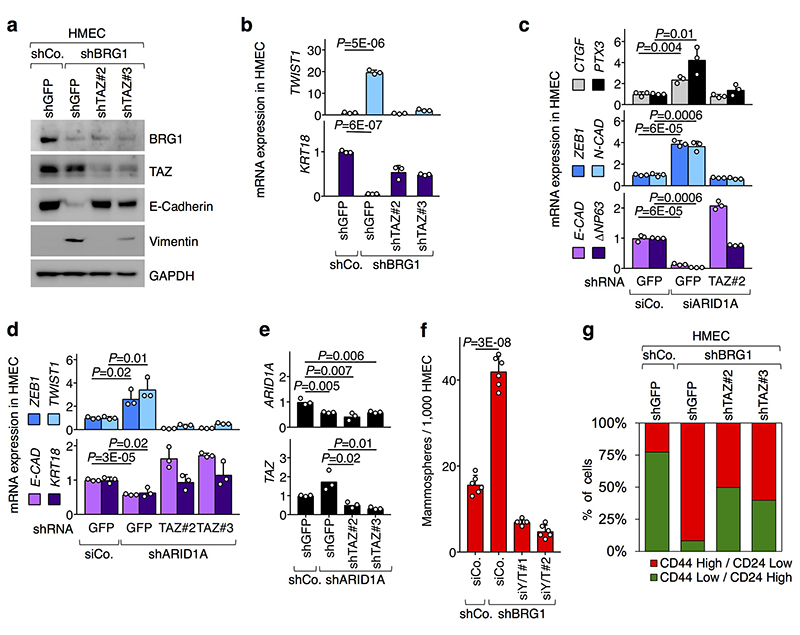
Our findings raised the intriguing possibility that YAP/TAZ regulation may contribute to SWI/SNF tumor suppressive functions. There is indeed a remarkable overlap between the biological effects of YAP/TAZ activation and of SWI/SNF inactivation, including control of cell fate plasticity, gain of stemness properties, and tumorigenesis5,8–10. It is known that loss of SWI/SNF triggers epithelial to mesenchymal transition (EMT) and entails gain of stem/progenitor-like properties in immortalized human mammary epithelial cells (HMEC)8. Crucially we found that activation of endogenous YAP/TAZ mediates the consequences of BRG1 or ARID1A inactivation in these cells (Fig. 2a, b and Extended Data Fig. 3a-g). Thus, SWI/SNF is a critical barrier that prevents activation of endogenous YAP/TAZ; loss of this control in HMECs promotes YAP/TAZ-driven induction of SC-like properties.
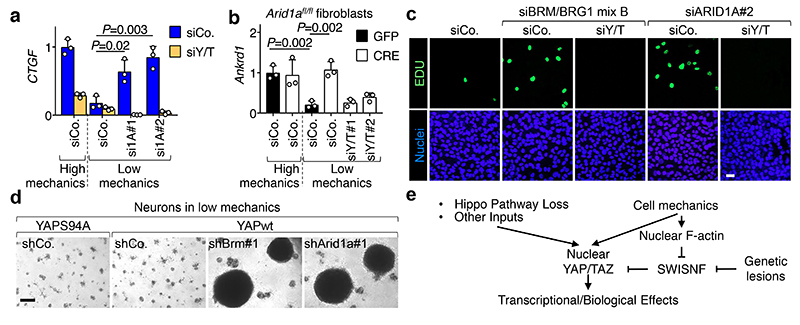
Fig. 2. Loss of SWI/SNF promotes YAP/TAZ-driven biological effects.
a, Depletion of BRG1 in HMEC causes changes in the expression of YAP/TAZ target genes (CTGF, PTX3) and the indicated markers for mesenchymal transition (ZEB1, N-CAD) and epithelial differentiation (ECAD, ΔNp63), in a TAZ-dependent manner (mean + s.d. of n=3 biologically independent samples).
b, Mammosphere formation assay (a measure of stem/progenitor-like properties) from HMEC transduced as indicated (mean + s.d. of n=6 biologically independent samples).
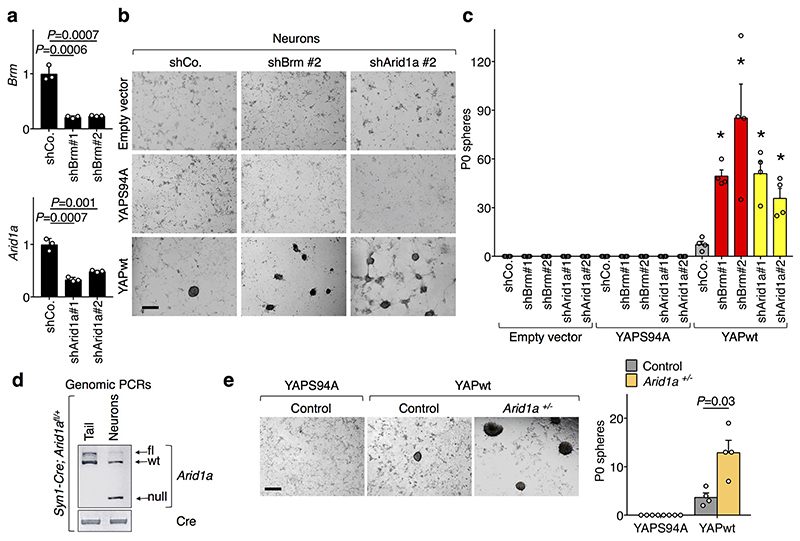
c, Neurospheres emerging from neurons infected with inducible YAP-encoding vectors and the indicated shRNAs-encoding lentiviral vectors. As negative control, we used a transcriptionally-inactive version of YAP (YAP S94A). Scale bars, 300 μM. See also Extended Data Fig. 4b, c.
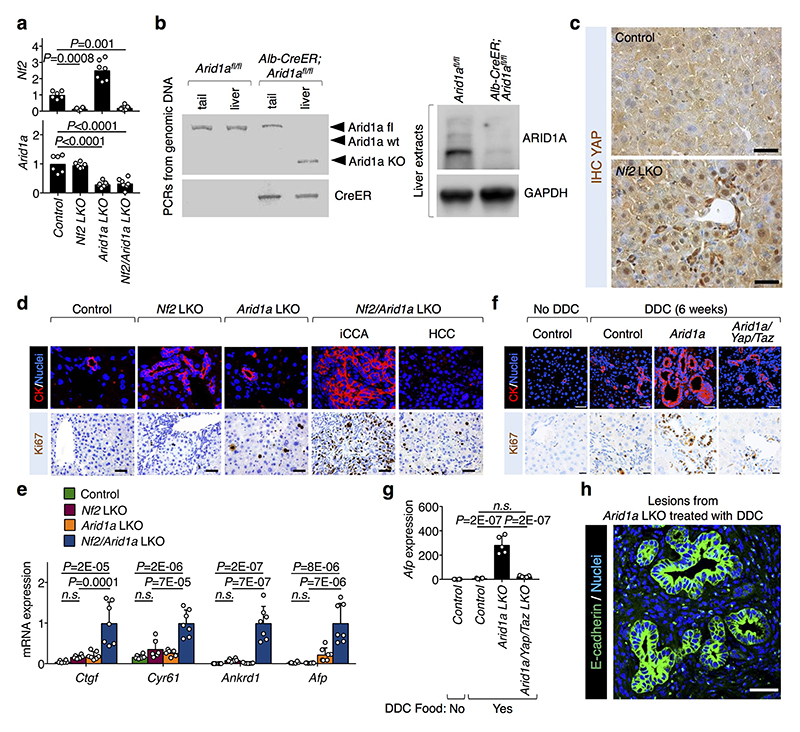
d, Gross liver images (scale bars, 1 cm) and liver-to-body weight ratio (mean + s.d.) from control (n=6 mice), and Nf2 (n=6 mice), Arid1a (n=7 mice) and Nf2/Arid1a (n=7 mice) liver mutant (LKO) mice, 4 months after tamoxifen treatment. All animals were included.
e, H&E liver sections from animals described in d (scale bars, 100 μm).
f, H&E liver sections from control (n=10), Arid1a LKO (n=12), Arid1a/Yap/Taz LKO (n=15) mice treated with tamoxifen and then DDC, compared to control mice under a normal diet (n=10) (scale bars, 100 μm).
P values were determined by unpaired two-sided t-test in a, b and one-way ANOVA with Tukey multiple comparisons test in d.
a, b, c, e, f are representative experiments, repeated independently three (a, b) or four (c) times, and for all mice (e, f), all with similar results.
In Drosophila neuroblasts, SWI/SNF are tumor suppressors because they prevent cellular de-differentiation back to a neural stem cell (NSC) state11. However, we found that the sole depletion of BRM or ARID1A is insufficient to trigger a change of fate in cultures of primary fetal hippocampal neurons (Extended Data Fig. 4a-c), possibly because mammalian neurons miss expression of specific factors required for their de-differentiation. We previously reported that fetal hippocampal neurons are void of endogenous YAP expression and that expression of exogenous YAP is sufficient to revert them into NSC-like cells12. Remarkably, inactivation of BRM or ARID1A by shRNA, or genetic deletion of ARID1A, strongly potentiated the reprogramming of YAP-expressing neurons into NSCs (Fig. 2c and Extended Data Fig. 4b-e). Thus, YAP/TAZ are central for executing key biological responses downstream of SWI/SNF inactivation.
Next, we aimed at validating the role of ARID1A/SWI/SNF as nuclear inhibitor of YAP/TAZ in vivo. Overactivation of YAP in the liver (for example downstream of inactivation of the Hippo pathway by knocking-out NF2) leads to YAP-driven tumorigenesis but only after a long latency13,14 suggesting that additional genetic or epigenetic events must be in place to uncover YAP tumorigenic potential. We hypothesized that removal of SWI/SNF might be one of these events. We used mice bearing a tamoxifen-inducible CRE recombinase under the control of the hepatocyte-specific Albumin promoter (Albumin-CRE-ERT2) to induce genetic ablations of Nf2 and Arid1a in adult hepatocytes (LKO) (Extended Data Fig. 5a, b). YAP nuclear staining was clearly induced around the portal areas of Nf2 LKO (Extended Data Fig. 5c). In spite of this, only modest induction of YAP/TAZ transcriptional activity and phenotypic effects were observed (i.e. ductular reactions with few proliferating cells) but no tumors developed up to four months after Cre activation (Fig. 2d, e and Extended Data Fig. 5d, e). Instead, at the same time point, all mice with combined knockout of Nf2 and Arid1a exhibited an impressive liver overgrowth (Fig. 2d), with widespread areas of neoplasia, ranging from early ductular atypia to full-blown cholangiocarcinomas and hepatocellular carcinoma (Fig. 2e). An extensive degree of proliferation was evident in tumors and all across the remaining hepatocytes (Extended Data Fig. 5d), also entailing hundreds-fold induction of the fetal/tumor marker Afp. Remarkably, YAP/TAZ transcriptional activity was massively induced in Nf2/Arid1a mutant livers compared to Nf2 LKO (Extended Data Fig. 5e). The liver knockout of Arid1a was inconsequential, as mice remained healthy with an ostensibly normal liver for the entire duration of our experiments (Fig. 2d-e and Extended Data Fig. 5d, e). This indicates that raising YAP/TAZ nuclear levels after Hippo pathway inactivation is insufficient for their full activation due to their nuclear inhibition by ARID1A/SWI/SNF.
Chronic tissue damage is a fundamental driver of liver carcinogenesis, causing continuous rounds of injury and compensatory proliferation15. To assess the relevance of the ARID1A-YAP/TAZ connection in this context, we fed ARID1A LKO mice a diet supplemented with the toxic compound 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) for 6 weeks. DDC induced the appearance of ductular reactions around the portal areas in wild-type livers (Fig. 2f). Strikingly, in all Arid1a LKO mice fed with DDC diet, we found areas of cholangiocarcinomatous transformation, with clear signs of atypia and massive proliferation and increased Afp expression (Fig. 2f, Extended Data Fig. 5f-h). Crucially, these lesions were absent in DDC-treated triple Arid1a/Yap/Taz mutant livers, which were similar to control DDC-treated mice (Fig. 2f and Extended Data Fig. 5f,g). Thus, inhibition of YAP/TAZ is an essential mediator of SWI/SNF tumor suppressive function in vivo.
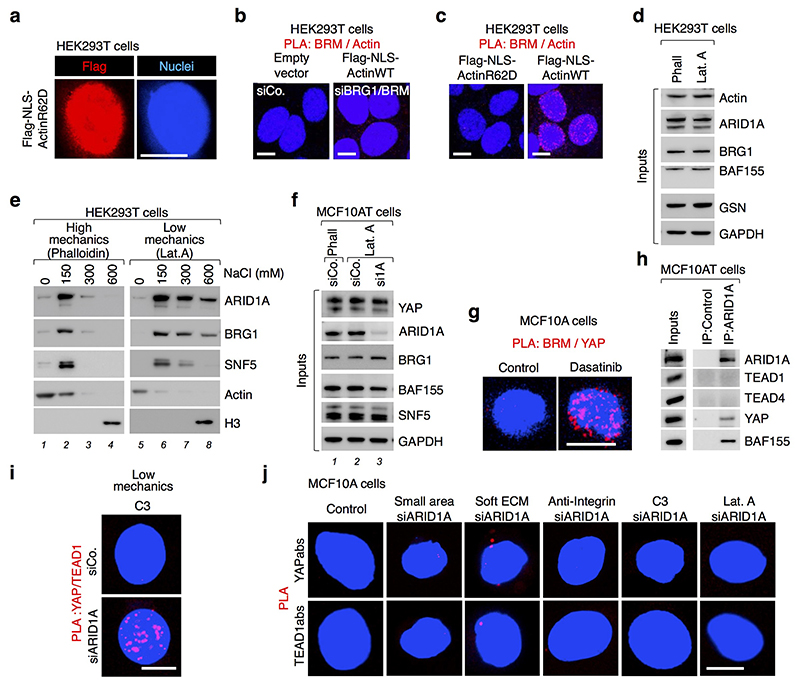
Data presented so far show that the biochemical interaction between YAP/TAZ and ARID1A/SWI/SNF represents a relevant modality to inhibit YAP/TAZ activity inside the nucleus. How is this connection regulated? Here we focused on modulation by mechanical inputs through the F-actin cytoskeleton. Indeed, YAP/TAZ respond to changes in cell shape and physical forces transmitted by the tissue that ultimately impact on the organization of the F-actin cytoskeleton1, including nuclear F-actin organization16,17. Indeed, we verified that the organization of nuclear F-actin changes dramatically in cells experiencing Low vs. High levels of mechanical signaling. By expressing actin fused to nuclear localization sequence (NLS-Actin)18 in HEK293T cells, we found that stretched cells (high mechanics) displayed a network of nuclear actin filaments crossing the whole nucleoplasm (Fig.3a, left panel), whereas a finer, almost exclusively perilaminar distribution was observed in cells confined to small adhesive areas (low mechanics) (Fig. 3a, right panel).
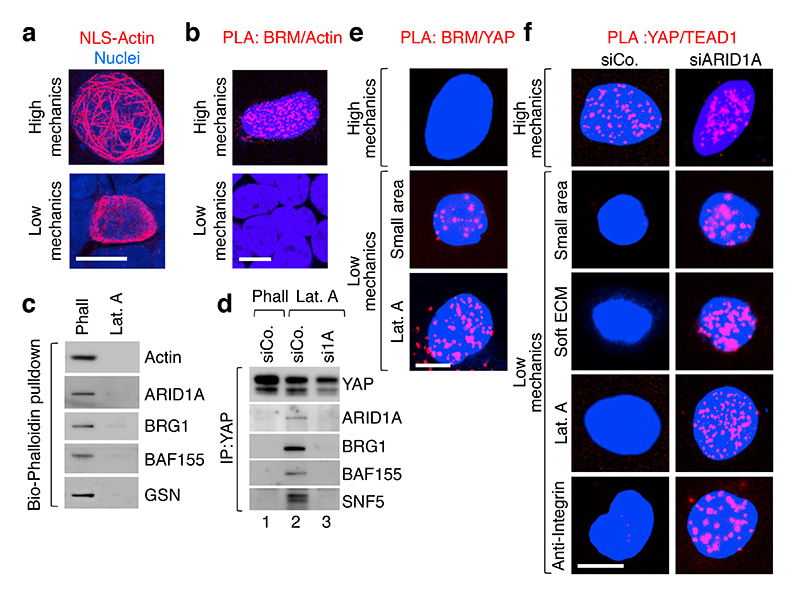
Fig. 3. Mechanical regulation of YAP/TAZ association with SWI/SNF or TEAD.
a, Visualization of Flag-tagged NLS-Actin filaments in nuclei of HEK293T cells by anti-Flag immunofluorescence (scale bars, 10 μm). No nuclear actin filaments were detected in cells transfected with a non-polymeriazable variant of actin (Extended Data Fig. 6a).
b, Representative pictures of PLA detecting the interaction between endogenous BRM and Flag-tagged NLS-Actin in the nucleus of HEK293T cells experiencing high mechanical inputs (i.e., spread cells; 94.7% PLA-positive) or confined on a small adhesive area (as in dense culture22; 0% PLA-positive). See specificity controls using BRM/BRG siRNA (Extended Data Fig. 6b).
c, Bio-Phalloidin pulldown from HEK293T cells, Phalloidin (Phall) vs. Latrunculin A (Lat.A). (See Methods and Extended Data Fig. 6d). Gelsolin serves as specificity control for purification of F-actin.
d, In Lat.A-treated MCF10AT cells, endogenous YAP binds SWI/SNF in a ARID1A-dependent manner (See also Extended Data Fig. 6f). ARID1a loaded on a separate gel.
e, Representative PLA images detecting the interaction between endogenous BRM and a version of YAP forced to enter in the nucleus (NLS-YAP) in MCF10A cells. Cells were allowed to stretch over rigid ECM (High mechanics; 0% PLA-positive) or, for Low mechanics panels, let adhering to a small area (100 μm2; 11.25% PLA-positive) or treated with Lat.A (18.75% PLA-positive). See also Extended Data Fig. 6g.
f, Representative PLA images detecting the interaction between endogenous TEAD and NLS-YAP in MCF10A cells, stretched over a rigid ECM (see Methods) or experiencing Low cell mechanics by adhesion to a small adhesive area, soft ECM or treatment with Lat.A or anti-integrin antibodies. YAP/TEAD association (High Mechanics, 46.3% PLA-positive) is lost at Low mechanics (Low Mechanics, left panels; 0% PLA-positive), but it is rescued after depletion of ARID1A (right panels; PLA-positive: small=44%; soft=49.8%; Lat.A=48.5%, anti-integrin=67%,). See also Extended Fig. 6j. All panels: representative experiments, repeated independently three times.
The SWI/SNF complex has been previously reported to associate to purified F-actin in vitro19, raising the possibility that it could be modulated by mechanical signals in cells. Remarkably, by in situ proximity-ligation assays (PLA, a method allowing investigation of protein-protein interactions while preserving the cell’s structural integrity) we found association between nuclear F-actin and endogenous SWI/SNF in nuclei of stretched cells, whereas at low mechanics (i.e., on small adhesive areas) cells were almost void of signal (Fig. 3b). A non-polymerizable form of actin20 (NLS-ActinR62D) did not interact with SWI/SNF in stretched cells (Extended Data Fig. 6c), supporting the view that only polymerized-actin can interact with SWI/SNF. In line, purification of endogenous F-actin using biotinylated(Bio)-phalloidin on streptavidin beads leads to robust co-purification of the ARID1A/SWI/SNF complex (Fig. 3c, first lane), but not in cells treated with the F-actin inhibitor Latrunculin A (Fig. 3c, second lane). Intriguingly, by sequential salt-extraction of nuclei (see Methods), ARID1A/SWI/SNF co-purified in the same fractions with F-actin when we used extracts in which F-actin was preserved with phalloidin (Extended Data Fig. 6e, lane 2); instead, in the presence of Latrunculin A, a substantial amount of ARID1A/SWI/SNF relocalized to fractions void of actin (Extended Data Fig. 6e, lane 8), paralleling PLA protein-protein interactions in cells challenged at High vs. Low mechanical stress shown in Fig. 3b.
We then asked if nuclear F-actin might interfere with the interaction between ARID1A/SWI/SNF and YAP/TAZ. As shown in Fig. 3d no ARID1A/SWI/SNF-YAP association could be detected in cell extracts prepared under conditions preserving F-actin (lane 1); SWI/SNF co-purified with endogenous YAP specifically in the absence of F-actin (lane 2), but this interaction was abolished by concomitant depletion of ARID1A (Fig. 3d, lane 3), as expected from ARID1A being required for YAP/TAZ incorporation in this pool of SWI/SNF (as per Fig. 1). These results were recapitulated in the nuclei of intact cells by PLA: YAP interacts with endogenous BRM only in mechanically inhibited cells (Fig. 3e and Extended Data Fig. 6g).
Mechanistically, we found that the inhibitory association of YAP/TAZ with ARID1A/SWI/SNF is in fact alternative to the binding of YAP/TAZ to their DNA-binding platform, TEAD, necessary for YAP/TAZ driven transcription21. We showed this first in CoIP experiments, finding that ARID1A associates to YAP, but not with TEAD proteins (Extended Data Fig. 6h). Then, we used PLA to monitor the dynamic of YAP/TEAD1 interaction in nuclei of cells cultured at High vs. Low mechanical signaling. As shown in Fig. 3f and Extended Data Fig. 6i, YAP/TEAD association in cell nuclei was severely inhibited at low mechanical regimes (left panels), when YAP/TAZ are associated to SWI/SNF; these conditions included inhibition of cellular mechanotransduction extracellularly (by culturing cells on small or soft ECM), at the level of integrin (with anti-integrin blocking antibodies) or intracellularly (by using the Rho-inhibitor C3 or inhibiting F-actin with Latrunculin A). Remarkably, in the same conditions, depletion of ARID1A was sufficient to restore YAP/TEAD association (Fig. 3f, right panels). Taken together, the results suggest the following model: in mechanically impaired cells, YAP/TAZ is sequestered within the ARID1A-containing pool of SWI/SNF complexes, away from TEAD. Conversely, in mechanically challenged cells, nuclear F-actin structures engage with ARID1A/SWI/SNF and induce YAP/TAZ detachment from that pool of SWI/SNF and their binding to TEAD.
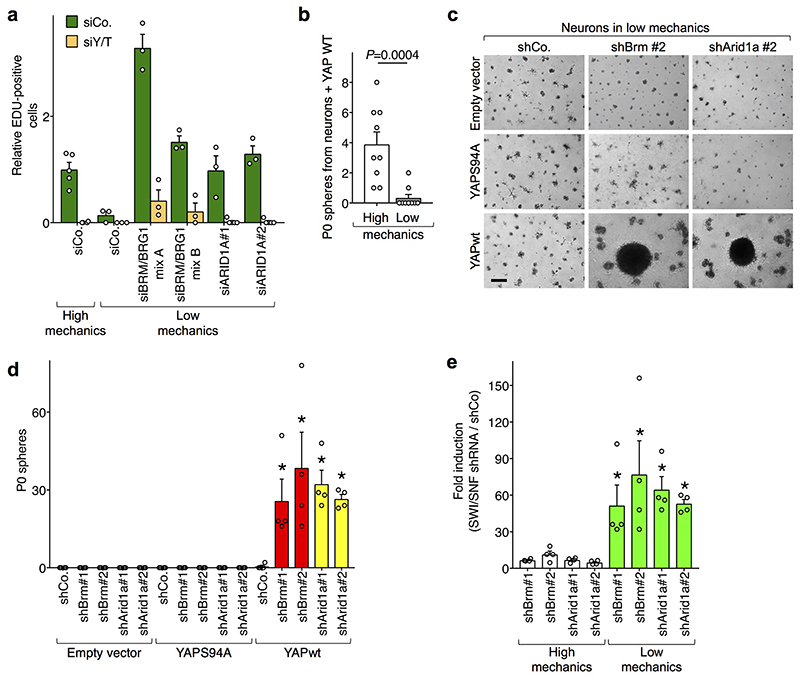
Following the above model, we next tested whether ARID1A/SWI/SNF inactivation could rescue YAP/TAZ activity in mechanical inhibited cells. In epithelial cells (MCF10A, HaCaT) experiencing low mechanical stimulation, or in which mechanotransduction was inhibited (either by attenuating ECM mechanics or inhibiting intracellular mechanotransduction), the expression of YAP/TAZ target genes was strongly downregulated, as expected7; yet, in all conditions, YAP/TAZ activity could be restored after ARID1A depletion with siRNAs. Moreover, such rescue of gene expression in mechanically impaired cells was YAP/TAZ-dependent (Fig. 4a and Extended Data Fig. 7b, c). Similar results were obtained in Arid1afl/fl mouse fibroblasts, where deletion of Arid1a was achieved by infection with Adeno-Cre (Fig. 4b and Extended Data Fig. 7d). Thus, data indicate that ARID1A functionally contributes to YAP/TAZ mechanical inhibition, and that YAP/TAZ are key mediators of the effect of ARID1A inactivation.
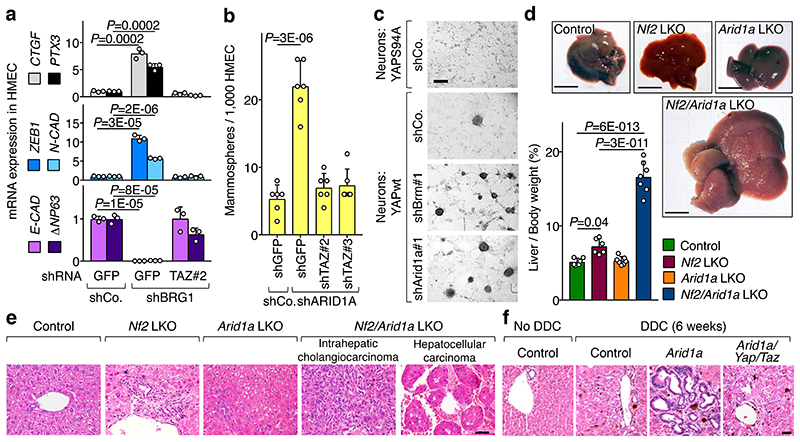
Fig. 4. Loss of SWI/SNF rescues YAP/TAZ activities and biological effects in mechanically impaired cells.
a-b, In Low mechanics conditions, ARID1A loss rescues expressions (mean + s.d. of n=3 biologically independent samples) of CTGF in MCF10A cells (a) and Ankrd1 in Arid1afl/fl fibroblasts (b) in a YAP/TAZ-dependent manner. ARID1A depletion had no effect on YAP/TAZ localization (Extended Data Fig. 7a).
c, Cell proliferation (measured by EdU incorporation) in confluent MCF10A cells transfected as indicated (scale bars, 30 μm).
d, Representative images (scale bars, 300 μM) of neurospheres emerging from neurons plated on a soft ECM and infected with inducible YAP-encoding vectors and the indicated shRNAs-encoding lentiviral vectors.
e, Model of the main conclusions.
High mechanics: stiff ECM in a, b, d or sparse monolayer in c. Low mechanics: soft ECM in a, b, d or dense monolayer in c.
P values were determined by unpaired two-sided t-test.
a-d are representative experiments, repeated independently two (c), three (a, b), four (d) times, all with similar results.
If raising cell mechanics attenuates the ARID1A/SWI/SNF-YAP/TAZ inhibitory axis through the F-actin cytoskeleton, then experimentally raising F-actin should be sufficient to overcome such inhibition. We tested this hypothesis through depletion of two F-actin severing proteins, ADF and Cofilin1, which act as cytoskeletal checkpoints of YAP/TAZ activation22; loss of ADF/Cofilin1 potently raised YAP/TAZ activity, to levels that could not be further modulated by ARID1A inactivation (Extended Data Fig. 7e). This is consistent with the results obtained in fibroblasts cultured on stiff ECM (Fig. 4b, High mechanics) and with the notion that, at maximal mechanical signaling, the ARID1A/SWI/SNF-YAP/TAZ inhibitory axis is already disabled, making ARID1A depletion in these conditions essentially inconsequential.
We next aimed to determine whether the loss of SWI/SNF paralleled with reactivation of YAP/TAZ-driven biological responses in otherwise mechanically inhibited cells. YAP/TAZ inactivation by lowered cell mechanics is a main culprit of contact-inhibition of proliferation (CIP) in a post-confluent epithelial sheet22. Indeed CIP can be revoked (leading to S-phase re-entry) through substantially Hippo-independent mechanical activation of YAP/TAZ, either by stretching the cell monolayer or remodeling the F-actin cytoskeleton22,23. Phenocopying the effects of raised cell mechanics, SWI/SNF inactivation also triggered S-phase re-entry in post-confluent epithelial sheets in a YAP/TAZ-dependent manner (Fig. 4c and Extended Data Fig. 8a).
Finally, we sought to determine the role of cell mechanics in regulating YAP-driven cell fate changes. For this, we used YAP-induced reprogramming of neurons into NSC-like cells12, and hypothesized that such reprogramming should be disabled if cells are placed on a soft ECM, where the inhibitory function of ARID1A/SWI/SNF on YAP/TAZ is preponderant. Indeed, only few neurospheres emerged from YAP-expressing wild-type neurons plated on a soft ECM when compared to stiff ECM (Extended Data Fig. 8b); more relevantly, shRNA-mediated depletion of ARID1A or BRM rescued the ability of YAP to reprogram neurons into NSC-like cells on soft ECM (Fig. 4d and Extended Data Fig. 8c-e).
The present work sheds light on the mechanisms of SWI/SNF tumor suppression, an aspect of cancer biology that remained so far enigmatic. We found that SWI/SNF binds to and inhibits YAP/TAZ; this inhibitory function is restricted to the ARID1A-containing fraction of SWI/SNF complexes. YAP/TAZ are essential and sufficient for the unfolding of complex cellular phenotypes inherent to inactivation ARID1A/SWI/SNF, occurring at exceedingly high frequency in human malignancies5. Note that, in this scenario, pools of SWI/SNF alternative to ARID1A/SWI/SNF, such as those containing ARID1B (here shown irrelevant for YAP/TAZ regulation), remain in place to carry out other SWI/SNF functions, such as chromatin remodeling. In line, in living tissues and explanted cells, ARID1A appears a largely dispensable protein, whose tumor suppressive role becomes apparent under genetic or environmental conditions leading to YAP/TAZ nuclear accumulation. Of note, others before us had noted the interaction between SWI/SNF and TAZ, but concluded that SWI/SNF positively cooperate with TAZ-induced transcription of some targets24. We have been unable to confirm the generality of those conclusions in our analyses, that included multiple redundant reagents, cellular systems, and in vivo genetic models all instead pointing to the opposite conclusion.
A second element of general interest emerging from this work concerns the mechanisms of YAP/TAZ mechanotransduction. This work suggests the existence of a pathway that could be streamlined as follows: Cell mechanics→ nuclear F-actin—| ARID1A/SWI/SNF pool —| nuclear YAP/TAZ. Biochemically, our data suggest that mechanical signals tune the ARID1A/SWI/SNF-YAP/TAZ inhibitory axis by controlling the levels and structural organization of the nuclear pool of F-actin. This nuclear pathway complements Hippo-independent and Hippo-regulated YAP/TAZ mechanotransduction occurring in the cytoplasm1,22.
Our data argue in favor of a paradigm whereby, in order to fully unleash YAP/TAZ activity, at least two requirements should be met: promotion of YAP/TAZ nuclear accumulation and SWI/SNF inhibition. Inactivation of the Hippo pathway alone is insufficient to fully empower YAP/TAZ activity in absence of concomitant inactivation of ARID1A. This implies that the response to a number of signals promoting YAP/TAZ nuclear localization - including loss of Hippo signaling - would also concomitantly require a proficient mechanical environment to surpass the ARID1A/SWI/SNF barrier (Fig. 4e). It also implies that nuclear levels of YAP/TAZ that are too low or transient to elicit any effect in normal cells may become supra-threshold after genetic or mechanical inhibition of ARID1A/SWI/SNF. More generally, the data suggest how a genetic lesion, such as loss of ARID1A in tumor cells, may serve as means to increase cellular responsiveness to an epigenetic signal, that is mechanotransduction.
Extended Experimental Procedures
Reagents and plasmids
Latrunculin A, Phalloidin, Cerivastatin and Tamoxifen were from Sigma. Doxycycline was from Calbiochem. Growth-factor reduced Matrigel was from Corning. C3 was from Cytoskeleton Inc. Dasatinib was from Selleckchem. Fasudil was from Tocris. Anti-Integrin beta-1 antibody (P5D2) was from DSHB, University of Iowa. Cre- and GFP-expressing adenoviruses were from University of Iowa, Gene Transfer Vector Core.
HA-hYAP5SA, Flag-hYAPwt, Flag-YAPS94A and Flag-hYAP5SA are cloned in pcDNA3.1 for transient expression, or in pBABE retroviral plasmids to establish stable cell lines. pBABE-Puro empty vector was used as control retroviral transduction. siRNA-insensitive Flag-YAP WW1mut, WW2mut and WW1/2mut were generated by PCR from the corresponding original cDNAs (a gift from Marius Sudol, Addgene plasmids #19046, 19047, 1904825) and subcloned in pcDNA3.1 for transient expression. pCS2-Flag mouse TAZ wt or DWW (deleted of residues 110-159) were as in Ref. 26.
pCS2-Flag-BRM was obtained by subcloning Flag-BRM from pBABEpuro Flag- human BRM (a gift from Robert Kingston, Addgene plasmid #196127) into pCS2.
pCS2-Flag-BRG1 was obtained by subcloning Flag-BRG1 from pBABEpuro-Flag-human BRG1 (a gift from Robert Kingston, Addgene plasmid #195927) into pCS2.
pcDNA6-V5-ARID1A wt was a gift from Ie-Ming Shih (Addgene plasmid # 3931128). pcDNA6-V5- ARID1A PPxA (containing the PPAY148/PPGY915 → PPAA148/PPGA915 mutations) was generated as follows: the N-terminal cDNA fragment of ARID1A containing the Y148A/Y915A mutations was synthesized by GeneScript and swapped into pcDNA6-V5-ARID1A wt by using the NheI/HpaI restriction sites.
For doxycycline-inducible expression of YAP in MCF10A cells, cDNA of NLS-YAP was subcloned in pCW-MCS, obtained by substituting the sequence between NheI and BamHI of pCW57.1 (a gift from David Root, Addgene #41393) with a new multiple cloning site (MCS).
For inducible expression of YAP in mouse neurons, FUW-tetO-wtYAP and FUW-tetO-YAPS94A (deposited as Addgene plasmids #84009 and #8401012, respectively) were used in combination with FUdeltaGW-rtTA (a gift from Konrad Hochedlinger, Addgene #1978029). Empty vector (FUW-tetO-MCS, Addgene #84008) was used as negative control.
The constructs for shControl, shArid1A, and shBrm expression in primary neurons were prepared by cloning the shCo, mouse shArid1a#1, mouse shArid1a#2, mouse shBrm#1 and mouse shBrm#2 sequences (see RNAi section) into pLKO.1-puro lentiviral vector (a gift from Bob Weinberg Addgene #845330) according to manufacturer’s protocol.
For stable shRNA infection of HMEC, we used pLKO.1-puro lentiviral vectors expressing shCo (see RNAi section), shBRG18 and shARID1A (from Sigma) in combination with pSUPER-RETRO-BLASTI vectors containing the GFP or TAZ RNAi sequences (as in Ref. 9).
Flag-NLS-Actin wt and R62D were generated by PCR from original cDNAs kindly provided by Dr. R. Treisman20 and cloned in pcDNA3.1.
For GST-pull down experiments, full-length mouse TAZ and human YAP1 were cloned in pGEX4T1. All constructs were confirmed by sequencing.
Micropatterns
The following procedure was used to make the adhesive micropatterns: a layer of photoresist (MICROPOSIT™ S1805™ G2 Positive Photoresist, Dow®) is spin coated (3000 rpm for 30 sec) on a glass substrate, functionalized with trimethoxysilylpropyl methacrylate, and cured at 120°C for 1 min. The positive resist was patterned by UV exposure for 8sec in air by irradiation with a collimated UV lamp at 365 nm (UV365, Reinraumtechnik lanz) through a quartz chromium mask with the desired pattern (arrays of 10x10 micron squares). The exposed areas, those around the squares, were removed by immersing the substrate in the developer solution MF 319 for 8 sec. To polymerize non-adhesive polyacrylamide brushes outside of the squares, a drop of acrylamide solution (8% wt/v in water with 0.225 wt/v of ammonium persulfate and 1.5% v/v tetramethylethylenediamine) is put between the patterned glass and a blank coverslip and left reacting for 30’ in air. The sandwich structure is detached immersing it for 30min in water; the functionalized pattern is then put in water overnight to completely remove unpolymerized acrylamide. The unexposed resist (the square areas) is stripped in acetone for 30sec and rinsed in water. Finally, after sterilization under UV light, the square areas are functionalized with fibronectin putting a drop of protein solution (10 μg/ml in water) on top, leaving it reacting for 1h and then rinsing in PBS.
Cell lines and treatments
HMEC were a kindly gift of Dr. Livingston8 (DFCI), and were cultured in MEGM medium (Lonza). MCF10A and MCF10AT (also called MII) cells were gift from Dr. Miller (Karmanos). and were cultured in DMEM/F12 (Gibco) with 5% horse serum (HS), glutamine and antibiotics, freshly supplemented with insulin (Sigma), EGF (Peprotech), hydrocortisone (Sigma), and cholera toxin (Sigma). HEK293 or HEK293T cells were from ATCC and were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS), glutamine and antibiotics. HaCaT cells were a gift from Dr. Fusening (DKFZ) and were cultured in DMEM (Gibco) supplemented with 10% FBS, glutamine and antibiotics. HEK293, HEK293T, MCF10A, MCF10AT and HaCaT were authenticated by DSMZ/Eurofins Genomics. All cell lines tested negative for mycoplasma contamination.
For experiments with NLS-YAP-transduced MCF10A, cells were treated with 0.5 μg/ml doxycycline in culture media for the whole duration of the experiments.
For Stiff vs. Soft ECM experiments, cells plated on standard Fibronectin-coated tissue culture supports or on Fibronectin-coated >40KPa hydrogels (produced as described in Ref. 7) were considered as cultured on a “Stiff ECM” in the “High Mechanics” conditions, as labeled in the Figures. For experiments on Soft ECM, 5,000-10,000 cells per cm2 were seeded in drop on top of 0.7 kPa fibronectin-coated hydrogels; after attachment, the wells containing the hydrogels were filled with appropriate medium. Cells were then harvested for immunofluorescence or RNA extraction after 24 hours. For experiments with cells experiencing small cell-ECM adhesion in ultra-confluent monolayers, we plated 200000 cells per cm2 in the appropriate well (i.e., plated at approximately 150% confluency). Cells were then harvested for immunofluorescence or RNA extraction after 48 hours.. For experiments with Fibronectin-coated micropatterns, cells were seeded on Fibronectin-coated micropatterns (100μm2; “Small Area” in Figure 3); after attachment, floating cells were removed and wells were filled with medium; cells were fixed 24 hours later. These cells were compared to cells experiencing unpatterned/unconfined adhesive area (defined as stretched cells and labeled as High Mechanics).
Latrunculin A was used at a final concentration of 0.5 μM for the time indicated in the dedicated sections of these Methods. C3 was used at a final concentration of 0.5 μg/ml in culture media for 24 hours. Dasatinib was used at a final concentration of 0.1 μM for 24 hours. Fasudil was used at a final concentration of 10 μM for 24 hours. Anti-Integrin was used at a final concentration of 0.23 μg/ml for 24 hours. Cerivastatin was used at a final concentration of 5 μM for 24 hours.
Primary fibroblasts (from adult mouse ear biopsies) were cultured in DMEM (Gibco) supplemented with 20% FBS, glutamine and antibiotics. For the experiments depicted in Fig. 4b and Extended Data Fig. 7d, fibroblasts were transduced with adenoviral vectors and transfected with the indicated siRNAs (day 0), replated either on a soft or a stiff ECM (day 1), and then harvested for RNA extraction 48 hours later (day 3).
RNA interference
siRNA transfections were done with Lipofectamine RNAi-MAX (Thermo Fisher Scientific) in antibiotics-free medium according to manufacturer instructions. Sequences of siRNAs are provided in Supplementary Table 2.
Western blot
Cells were harvested in Lysis Buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM KCl, 1% Triton, 5% Glycerol, 0.5% NP40, 2 mM MgCl2, 1 μM DTT, phosphatase and protease inhibitors) and lysed at 4°C by sonication. Extracts were quantified with Bradford method. Proteins were run in 4-12% Nupage-MOPS acrylamide gels (ThermoFisher) and transferred onto PVDF membranes by wet electrophoretic transfer. Blots were blocked with 0.5% non-fat dry milk and incubated overnight at 4°C with primary antibodies. Secondary antibodies were incubated 1 hour at room temperature, and then blots were developed with chemiluminescent reagents. Images were acquired with Image Quant LAS 4000 1.2 (GE healthcare).
For Western blot: anti-YAP/TAZ (sc-101199), anti-BAF53A (sc-137062 or sc47808), anti-BRG1 (sc-10768 or sc-17796), anti-LAMIN B (sc-6216), anti-SMARCC1/BAF155 (sc-137138 or sc-9746), anti-SNF5 (sc-166165), anti-Vimentin (sc-7557-r), anti-Gelsolin (sc-57509) and anti-TEAD4 (sc-101184) were from Santa Cruz; anti-ARID1A (HPA005456), anti-SNF5 (HPA018248), anti-TAZ (HPA007415) and anti-Actin (A5316) were from Sigma; anti-YAP (ab52771), anti-Histone H3 (ab1791) and anti-BRM (ab15597) were from Abcam; anti-GAPDH (MAB347) and anti-ARID1A (04-080) monoclonal antibody were from Millipore. Anti-E-cadherin (610181) and anti-TEAD1 (610922) were from BD. Anti-phospho YAP (Ser127) (CST 4911) was from Cell signaling Technology.
Horseradish-peroxidase-conjugated anti-Flag (clone M2, A8592) was from Sigma, anti-HA (A190-107P) was from Bethyl and the anti-V5 was from Abcam (ab27671).
Unless otherwise specified, loading control for all blots were run on the same gel.
F-actin pulldown
For the experiments depicted in Fig. 3c, cells were plated in sparse conditions and treated for 4 hours with Lat. A (0.5 μM) or Biotinylated-Phalloidin (Bio-Phall, 40 ng/ml). After treatment, cells were washed with prewarmed HBSS once and harvested in “Actin-lysis buffer” (20 mM HEPES (pH 7.5), 50 mM KCl, 0.1% Triton, 5% Glycerol, 0.1% NP40, 5 mM MgCl2, 1 μM DTT, 10 μM MG115, 10 μM MG132, 1 mM ATP, 20 μM Phosphocreatine di(tris) salt (P1937, Sigma), phosphatase and protease inhibitors). All buffers were freshly prepared and pre-warmed at RT. Cells were scraped and mechanically lysed by passaging ten times through a 26G-needle syringe at room temperature. For Lat.A-treated cells, Lat. A was also present in the buffers (1 μM) used for harvesting and pull-down to avoid any F-actin re-assembly; for the Bio-Phall-treated cells, Bio-Phall was also present in the buffers used for harvesting and pull-down (40 ng/ml). Extracts were cleared by centrifugation (10000 rcf in low retention eppendorf tubes) at RT and supernatants immediately (we never froze supernatants) incubated at RT for three hours with streptavidin-conjugated resin (Sigma) and Biotinylated-Phalloidin (1 μg/ml). Phalloidin-bound complexes were then washed with Actin lysis buffer three times at room temperature, resuspended in SDS sample buffer at 95°C for 3 minutes, and then subjected to SDS-PAGE and Western blot analysis.
Sequential salt extraction
We have adapted a sequential salt extraction assay for evaluating the chromatin binding affinities of the SWI/SNF complex in HEK293T cells. All buffers were freshly-prepared and pre-warmed at room temperature before use and all procedures were carried out at room temperature. Nuclei were isolated from 10 cm-dish confluent HEK293T cells by hypotonic lysis in 5 ml Buffer 1 (20 mM HEPES (pH 7.5), 10 mM KCl, 0.1% NP40, 5% Glycerol, 5 mM MgCl2, 1 μM DTT, 10 μM MG115, 10 μM MG132, 1 mM ATP, 20 μM Phosphocreatine di(tris) salt, phosphatase and protease inhibitors) for 5 min. After centrifugation at 600 g for 3 min, supernatant was saved for western blot analysis, whereas nuclear pellet was sequentially resuspended and centrifuged at 6000 g for 3 min in buffer 1 supplemented with increasing concentrations of NaCl (from 0 to 600 mM), as indicated in Extended Data Fig. 6e. The released proteins in each fraction were directly analyzed by SDS-PAGE and western blot.
For Lat. A-treated cells (0.5 μM, 4 hours treatment), Lat. A (1 μM) was also present in all the buffers used for harvesting and the salt extraction assay, to avoid any F-actin re-assembly; for Phalloidin-treated cells, Phalloidin (50 μM) was also present in all the buffers used for harvesting and the salt extraction assay.
Coimmunoprecipitation of Endogenous proteins
For immunoprecipitations of endogenous proteins of Fig. 1a,c and Extended Data Fig. 6h, cells were plated (day 0), transfected with the indicated siRNAs (day 1) and harvested 2 days after siRNA transfection (day 4) and lysed by sonication in Lysis Buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM KCl, 1% Triton, 5% Glycerol, 0.5% NP40, 2 mM MgCl2, 1 μM DTT, phosphatase and protease inhibitors) and extracts were cleared by centrifugation. Extracts were incubated with anti-ARID1A (sc-98441; Santa Cruz) antibody or control anti-HA (sc-805; Santa Cruz), immobilized on Protein A-Sepharose beads at 4°C for 3 hours. Immunocomplexes were then washed with cold Lysis buffer three times, resuspended in SDS sample buffer, and subjected to SDS-PAGE and Western blot analysis.
For the experiments depicted in Fig. 3d, cells were plated in sparse conditions and treated and harvested as described above for F-actin pull-down. Extracts were cleared by centrifugation at RT and incubated with anti-YAP (ab52771; Abcam) antibody immobilized on Protein A-Sepharose beads for 3 hours at RT. Immunocomplexes were then washed with “Actin lysis buffer” (see above) three times at RT, resuspended in SDS sample buffer at 95°C for 3 minutes, and subjected to SDS-PAGE and Western blot analysis.
Coimmunoprecipitation of Tagged proteins
Cells were harvested and lysed by sonication in Lysis Buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM KCl, 1% Triton, 5% Glycerol, 0.5% NP40, 2 mM MgCl2, 1 μM DTT, phosphatase and protease inhibitors) and extracts were cleared by centrifugation at 4°C. Extracts were incubated three hours at 4°C with anti-Flag resin (Sigma). Immunocomplexes were then washed with cold Lysis buffer three times, resuspended in SDS sample buffer, and subjected to SDS-PAGE and Western blot analysis. Inputs were loaded according to Bradford. In particular, for Fig. 1b, Extended Data Fig. 1c and Extended Data Fig. 1g and i-k, we used lysates from HEK293T cells transfected with the indicated plasmids (for Fig. 1b: Flag-BRM was 83 ng/cm2, HA-YAP5SA was 17 ng/cm2; for Extended Data Fig. 1c: Flag-BRG1 was 83 ng/cm2, HA-YAP5SA was 17 ng/cm2; for Extended Data Fig. 1g: HA-YAP5SA was 17 ng/cm2, Flag-BRG1 was 83 ng/cm2; for Extended Data Fig. 1i: Flag-YAPwt was 83 ng/cm2, Flag-YAP WW1mut was 83 ng/cm2, Flag-YAP WW2mut was 83 ng/cm2, Flag-YAP WW1/2mut was 83 ng/cm2; for Extended Data Fig. 1j: Flag-TAZwt was 83 ng/cm2, Flag-TAZ DWW was 83 ng/cm2; for Extended Data Fig. 1k: Flag-YAP was 83 ng/cm2, V5-ARID1Awt was 166 ng/cm2, V5-ARID1A PPxA was 166 ng/cm2;) and harvested 48 hours after transfection. Where indicated, siRNAs were transfected 24 hours before DNA transfection. For Extended Data Fig. 1b, we used lysates from Empty vector-transduced MCF10A cells or MCF10A cells constitutively expressing Flag-YAP5SA.
GST Pull-Down
For the experiment of Extended Data Fig. 1h, V5-ARID1A was purified from transfected HEK293T cells: briefly, cells were transfected with pcDNA6-V5-ARID1A, harvested and lysed by sonication in Lysis Buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM KCl, 1% Triton, 5% Glycerol, 0.5% NP40, 2 mM MgCl2, 1 μM DTT, phosphatase and protease inhibitors) and extracts were cleared by centrifugation at 4°C. Extracts were incubated for 3 hours at 4°C with anti-V5 resin (Sigma). After washing three times with Lysis Buffer (2 min at RT), V5-ARID1A protein was eluted by incubation with V5 peptide (V7754, Sigma) in Lysis Buffer. V5 resin was eliminated by centrifugation. For the GST-pulldown, beads with purified proteins (GST-YAP or GST-TAZ, as indicated) were incubated with purified V5-ARID1A in Lysis Buffer for 3 hours at 4°C. After 3 washes, GST pull-downs were analyzed by western blots.
For the experiment of Extended Data Fig. 1f, beads with purified GST-YAP were incubated for 3 hours at 4°C with the 0 mM NaCl-fraction containing proteins released from DNAse-treated nuclei of HEK293T cells. To prepare such extracts, nuclei of HEK293T were isolated from 10 cm-dish confluent HEK293T cells by hypotonic lysis in 5 ml Buffer 1 for 5 min. After centrifugation at 600 g for 3 min, supernatant was saved for western blot analysis, whereas nuclear pellet was subjected to DNAse treatment for 30 min at 37°C in buffer 1 supplemented with 1 mM CaCl2. After centrifugation at 6000g for 3 min, supernatant was discarded and the DNAse-treated nuclear pellet was sequentially resuspended and centrifuged at 6000 g for 3 min in buffer 1 supplemented with increasing concentrations of NaCl (from 0 to 600 mM). The 0 mM NaCl-fraction was used for GST-pulldown. After 3 washes in buffer 1, GST pull-downs were then analyzed by western blot.
Identification of native YAP/TAZ complexes by Mass Spectroscopy
Live cells were cross-linked with 1% formaldehyde (Sigma) in culture medium for 10min at room temperature before harvesting. Lysis was achieved by consecutive incubations in Lysis Buffer 1 (50mM HEPES, pH7.5, 10mM NaCl, 1mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100), Lysis Buffer 2 (10mM Tris-HCl pH8, 200 mM NaCl, 1mM EDTA, 0.5 mM EGTA) and Lysis Buffer 3 (10mM Tris-HCl pH8, 200 mM NaCl, 1mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine), followed by sonication with a Branson Sonifier 450D. Immunoprecipitation was performed by incubating cleared extracts (corresponding to 2x106 cells) with 20 μg of antibody (anti-YAP: EP1674Y, Abcam; anti-TAZ: HPA007415, Sigma; pre-immune rabbit IgGs: I5006, Sigma) and 100 μl of Dynabeads-ProteinG (Invitrogen). After extensive washing, immunoprecipitates were eluted in 7.5%SDS, 200mM DTT and de-crosslinked. After alkylation with iodoacetamide (IAA), proteins were purified with SP3 beads as previously described (PMID 25358341), resuspended in 50 mM ammonium bicarbonate and digested with trypsin. Peptides were subjected to SP3 cleanup and they were eluted in 0.1% TFA. Samples were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher).
Quantitative Real-Time PCR
Cells were harvested by RNeasy Mini Kit (Qiagen) for total RNA extraction, and contaminant DNA was removed by DNase treatment. Total RNA from MEF (Fig. 4b and Extended Data Fig. 7d) and from livers (Extended Data Fig. 5e, g) was extracted using Trizol (ThermoFisher) and NucleoSpin RNA (MACHEREY-NAGEL, 740955.250), respectively. qRT-PCR analyses were carried out on retro-transcribed cDNAs with QuantStudio5 (applied Biosystems, ThermoFisher Scientific) and analyzed with QuantStudio Design & Analysis software (version 1.4.3). Expression levels are always normalized to GAPDH. PCR oligo sequences are listed in Supplementary Table 1.
Proliferation assay (EdU staining)
Cells were first transfected with indicated siRNAs in the standard culture condition. The day after, cells were re-plated in fibronectin-coated glass chamber slides. After 24 hours, EdU (10 μM) was added to the culture medium for 1 hour. Cells were then fixed in 4% PFA for 10 min at room temperature. The EdU assay were performed according manufacturer’s instructions (“Click-iT EdU Imaging Kits”, Invitrogen). Images were obtained with a Leica TCS SP5 equipped with a CCD camera and analyzed using Volocity software (PerkinElmer, version 5.5.1).
Immunofluorescence
Immunofluorescence on PFA-fixed cells and on PFA-fixed paraffin embedded tissue slices was performed as in Ref. 12.
Primary antibodies are: anti-YAP/TAZ (sc-101199, Santa Cruz), anti-cytokeratin (wide spectrum screening, ZO622; Dako), anti-E-cadherin (610181, BD), anti-Flag (F1804, Sigma). Secondary antibodies (1:200) were from Molecular Probes. Samples were counterstained with ProLong-DAPI (Molecular Probes, Life Technologies) to label cell nuclei. Confocal images were obtained with a Leica TCS SP5 equipped with a CCD camera and analyzed using Volocity software (PerkinElmer, version 5.5.1).
Immunohistochemical stainings were performed on formalin-fixed, paraffin-embedded tissue sections as described in Ref. 9. For immunohistochemistry: anti-Ki67 polyclonal antibody (clone SP6; M3062) was from Spring Bioscience; anti-YAP (13584-I-AP) was from Proteintech.
In situ proximity ligation assay (PLA)
In situ PLA was performed with Duolink in situ reagents (Sigma).
For the experiments of Fig. 3b and Extended Data Fig. 6b, c, HEK293T cells were plated in standard cell culture dish on day 1. Cells were transfected with the indicated DNA plasmids (Flag-NLS-Actin WT was 150 ng/cm2, Flag-NLS-Actin R62D was 150 ng/cm2; Empty vector was 150 ng/cm2) on day 2; on day 3 cells were transfected with indicated siRNAs. On day 4, cells were replated into fibronectin-coated glass chamberslides. Cells from each condition were plated in duplicate. After 24 hours, cells were fixed in 4% PFA for 10 min at RT. With one of the duplicates we performed the anti-Flag (F-1804; Sigma) immunofluorescence in order to check the DNA transfection efficiency: the percentage of transfected cells was used as normalizer. The other duplicate was subjected to PLA, following the manufacturer’s instructions. Primary antibodies used in the PLA are: anti-Flag (F-1804, Sigma) and anti-BRM (ab15597, Abcam).
For the experiments of Fig. 3e-f and Extended Data Fig. 6g, i, j, NLS-YAP-MCF10A cells were treated with doxycycline to induce NLS-YAP expression for the whole duration of the experiments. For Fig.3e and Extended data 6g, cells were either plated on small/micropatterned Fibronectin-coated areas or treated for 24 hours with: Lat A (0.5 μM) or Dasatinib (0.1 μM). For Fig. 3f and Extended Data Fig. 6i-j, cells were first transfected with siRNAs (siCo or siARID1a) and replated on small micropatterned/Fibronectin-coated areas, soft (0.7 KPa) hydrogels, or treated for 24 hours with Lat A (0.5 μM); C3 (0.5 μg/ml); or anti-Integrin (0.23 μg/ml). Cells at “High mechanics” are plated on unpatterned Fibronectin-coated chamberslides. Cells were then fixed in 4% PFA for 10 min at RT. Samples were subjected to PLA, following the manufacturer’s instructions. Primary antibodies used in the PLA are: anti-YAP (sc-101199, Santa Cruz) and anti-BRM (ab15597, Abcam) for Fig. 3e and Extended Data Fig. 6g; anti-YAP (ab52771, Abcam) and anti-TEAD1 (610922, BD Biosciences) for Fig. 3f and Extended Data Fig. 6i, j.
Images were acquired with a Leica TCS SP5 confocal microscope equipped with a CCD camera and analyzed using Volocity software (PerkinElmer, version 5.5.1).
The percentages of PLA-positive cells reported in the legend of Fig. 3 have been determined by manual counting of at least 90 cells for each experimental condition.
Lenti- and retro-virus preparation
Lentiviral particles were prepared by transiently transfecting HEK293T (as in Ref. 9) with lentiviral vectors (10 μg/60 cm2 dishes) together with packaging vectors pMD2-VSVG (2.5 μg) and pPAX2 (7.5 μg) by using TransIT-LT1 (Mirus Bio) according to manufacturer instructions.
Retroviral particles were prepared by transiently transfecting HEK293GP (Takara) with retroviral vectors (15 μg/60 cm2 dishes) together with pMD2-Env (5 μg/60 cm2 dishes) by using TransIT-LT1. Infections were carried out as previously described9.
Mammosphere Assays
Confluent monolayers of HMECs were trypsinized, counted and plated as single cell suspensions (in the density of 1000 cells/cm2) on ultra-low attachment plates (Costar). Cells were cultivated in DMEM/F12 supplemented with 1xB27 (Invitrogen), glutamine, antibiotics, 5 μg/ml Insulin (Sigma), 20 ng/mL EGF (Peprotech), 0.5 μg/mL Hydrocortisone (Sigma), 52 μg/mL BPE (Thermo Fisher), 20 ng/mL bFGF (Peprotech) and 4 μg/mL heparin. Mammospheres were counted after 10-14 days.
Luciferase Assays
Luciferase assays were performed in HEK293 cells with the established YAP/TAZ-responsive luciferase reporter 8xGTIIC-Lux7.
8xGTIIC-Lux reporter (50 ng/cm2) was transfected together with CMV-β-gal (75 ng/cm2) to normalize for transfection efficiency with CPRG (Roche) colorimetic assay. DNA transfections were done with TransitLT1 (Mirus Bio) according to manufacturer instructions. DNA content in all samples was kept uniform by adding pBluescript plasmid up to 250 ng/cm2. In experiments in siRNA-depleted cells (Fig. 1d and Extended Data Fig. 2a and b), cells were plated at 15% confluence (day 0), transfected with the indicated siRNAs (day 1), washed from transfection media and transfected with plasmid DNA (for Extended Data Fig. 2a: Empty vector was 2 ng/cm2, Flag-YAP wt was 2 ng/cm2; for Extended Fig. 2b: Flag-YAP wt was 2 ng/cm2, Flag-YAP WW1 mut was 21 ng/cm2) (day 2), and harvested 48 hr later (day 4).
Primary neuron isolation, infection and culturing
Preparation of neurons, transduction and culturing were performed as in Ref. 12. Briefly, neurons were isolated from hippocampi of E18-19 embryos of the indicated genotypes and plated on poly-L-lysine-coated wells (stiff conditions) or on top of a thick 0.5 cm Matrigel layer (soft conditions) in DMEM supplemented with 10% FBS, glutamine and antibiotics for hippocampal neurons (day1). After 24 hours (day 2), the medium of hippocampal preparation was changed to fresh DMEM/Neurobasal (1:1) supplemented with 5% FBS, 1X B27, glutamine and antibiotics. For reprogramming experiments neurons were infected on the following day (day 3) with FUW-tetO-wtYAP and FUdeltaGW-rtTA viral supernatants. Negative controls were provided by neurons transduced with FUdeltaGW-rtTA in combination with FUW-tetO-YAPS94A, or empty vector. After 24 hours (day 4), medium was changed and cells were incubated in Neurobasal medium supplemented with 1X B27, glutamine, antibiotics, and 5 μM Ara-C (cytosine β-D-arabinofuranoside; Sigma) for additional 7 days, at the end of which well-differentiated neurons are visible.
For the experiments of Extended Data Fig. 4e and 8b, neurons were switched to NSC medium and 2 μg/ml doxycycline for activating tetracycline-inducible gene expression. Sphere formation was evident upon YAP induction after 14 days on stiff ECM of doxycycline treatment.
For the experiments of Fig. 2c and 4d, and Extended Data Fig. 4b, c and 8c, d, after Ara-C treatment, neurons were infected with pLKO.1-shRNAs vectors. For infection of a 10 cm2 plate, we mixed 500 μl of PLKO.1-shRNA produced in NSC medium (DMEM/F12 supplemented with 1X N2, 20 ng/ml murine EGF, 20 ng/ml murine bFGF, glutamine, and antibiotics) and 1.5 ml of serum-free Neurobasal medium with 1X B27. After 24 hours of infection, treated neurons were switched to NSC medium and 2 μg/ml doxycycline for activating tetracycline-inducible gene expression. After 7 days, fresh doxycycline (final concentration of 2 μg/ml) was added. Sphere formation was evident upon YAP induction after 14 days (stiff condition) or 30-45 days (soft condition) of doxycycline treatment.
Bright field images were acquired with a Leica DM IRB microscope by using a LASV4.4 software.
Mice
Transgenic lines used in the experiments were gently provided by: Duojia Pan (Nf2fl/fl, Ref. 13); Zhong Wang (Arid1afl/fl: these mice have loxP sites flanking exon 8, Ref. 31); Pierre Chambon (Albumin-CreERT2, Ref. 32); Ivan De Curtis and Riccardo Brambilla (Syn1-Cre, Ref. 33). Tazfl/fl and double Yapfl/fl; Tazfl/fl conditional knock-out mice were as described in Ref. 34.
Animals were genotyped with standard procedures35 and with the recommended set of primers. Animal experiments were performed adhering to our institutional and national guidelines as approved by OPBA (University of Padova) and the Ministery of Health of Italy. For experiments in mice, the limits for the endpoint “body condition scoring” were never exceeded.
For the experiment depicted in Extended Data Fig. 4e we used control (Arid1afl/+) and Syn1-Cre; Arid1afl/+ mice. For this, we crossed Syn1-Cre females (as transgene expression in male mice results in germline recombination36) with Arid1afl/fl males. Littermate embryos derived from these crossings were harvested at E18-19 and kept separate for neurons derivation and yNSCs induction; genotypes were confirmed on embryonic tail biopsies and leftover brains. These animals were mixed strains.
Yap, Taz, Arid1a and Nf2 conditional knockouts were intercrossed with Albumin-CreERT2 to obtain the different genotypes used for the experiments in Fig. 2 and Extended Data Fig. 5 (including controls). These animals were mixed strains. For the induction of the recombination in the liver, mice of the indicated genotypes (2 month-old) received 1 intraperitoneal injection per day of 3 mg Tamoxifen (Sigma) dissolved in corn oil (Sigma) during 5 consecutive days. For the experiments depicted in Fig. 2d, e and Extended Data Fig. 5c-e, mice were sacrificed 4 months after tamoxifen treatment. For the DDC experiments (Fig. 2f and Extended Data Fig. 5f-h), 2 weeks after tamoxifen treatment, mice were fed with either normal diet (Mucedola) or the same diet containing 0.1% DDC (Sigma) for 6 weeks (DDC diet).
Statistics
The number of biological and technical replicates and the number of animals are indicated in figure legends, main text and methods. All tested animals were included. Animal ages are specified in the text and Methods. Sample size was not predetermined. Randomization was not applicable for our experiments with cell lines. Mice were randomly allocated to experimental or treatment groups to ensure equal sex/age across genotypes. Investigators were not blinded for analyses relying on unbiased measurements of quantitative parameters, with exception of pathological examination of histological section carried out by M. Fassan (a professional pathologist), who was blind to animal genotypes, sex/age or treatment. Error bars, SD or SEM are as indicated in Figure legend. Student t-test, Mann-Whitney U-test and one-way ANOVA analyses were performed with GraphPad Prism 7.0d for Mac software.
Extended Data
Extended Data Figure. 1. Interaction between YAP and the SWI/SNF complex.
a, Proteomic analyses of endogenous YAP/TAZ-binding partners reveal interaction with endogenous SWI/SNF complex components (green). In red, bait. R1 and R2 are the results from n=2 biologically independent samples. See Extended Data Table 1.
b, YAP5SA was immunoprecipitated from lysates of MCF10A cells stably expressing Flag-tagged YAP5SA by using an anti-Flag antibody, and co-precipitating endogenous components of the SWI/SNF complex were detected by Western blot. As a negative control, immunoprecipitation (IP) was repeated from cells transduced with empty vector. GAPDH serves as a loading control in inputs (right).
c, HEK293T cells were transfected with independent siRNAs for indicated genes (ARID1A in lanes 5 and 6; BAF53A in lanes 7 and 8; SNF5 in lanes 9 and 10) and control siRNAs (siCo.; lanes 1, 2, 3 and 4) and with plasmids encoding HA-YAP5SA (all lanes) and Flag-BRG1 (lanes 3 to 10), as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation and co-precipitating proteins were checked by western blot. ARID1A depletion impairs the interaction of YAP with BRG1, but it had no effect on the association of BRG1 with BAF53A (lanes 5 and 6). Depletion of BAF53A (lanes 7 and 8) or SNF5 (lanes 9 and 10) had no effect on YAP-BRG1 interaction. ARID1A blot, top band represents full-length ARID1A. Input ARID1a from a separate gel.
d, Panels are western blots of the inputs of the IP experiment shown in Fig. 1b. HEK293T cells were transfected with control siRNA or siRNA against ARID1A and with plasmids encoding HA-YAP5SA and Flag-BRM, as indicated.
e, Panels are western blots of the inputs of the IP experiment shown in Fig. 1c. HEK293T cells were transfected with control siRNAs or with a siRNA mix against BRG1 and BRM.
f, DNAse-treated nuclei preparations from HEK293T cells were subjected to sequential salt extraction and fractions were analyzed by western blot (left panels, lanes 1-4). The un-sonicated, chromatin-free 0 mM-NaCl fraction was incubated with GST-YAP or GST protein (negative control), immobilized on a glutathione-resin, and pulled down proteins were analyzed by western blot (right panels, lanes 5-6).
g, HEK293T cells were transfected with siRNAs for indicated genes and with plasmids encoding HA-YAP5SA and Flag-BRG1, as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation and co-precipitating proteins were checked by western blot.
h, Western blot of recombinant V5-ARID1A pulled down by GST-YAP or GST-TAZ, immobilized on a glutathione-resin. GST protein was used as a negative control. Input: fraction of V5-ARID1A used for the pull-down experiments.
i, HEK293T cells were transfected with plasmids encoding empty vector (e.v.) or Flag-YAP wt or WW-domain mutants, as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation and western blot analysis for endogenous ARID1A. GAPDH serves as a loading control in inputs.
j, Flag-TAZ was immunoprecipitated from lysates of HEK293T cells transfected with Flag-tagged TAZ wt or TAZ DWW by using an anti-Flag antibody, and co-precipitating endogenous ARID1A was detected by western blot only with TAZ wt. As a negative control, immunoprecipitation was repeated from HEK293T cells transfected with empty vector.
k, HEK293T cells were transfected with plasmids encoding Flag-YAP wt (all lanes) and V5-ARID1A either wt or PPxA mutant, as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation and western blot analysis for V5-ARID1A. We notice that other SWI/SNF components (such as BRG1 itself or SNF5) also carry PPxY motifs; although these components are by themselves not essential for association to YAP/TAZ, the presence of a second WW motif in YAP (although not in TAZ) raise the possibility of stronger, cooperative associations between YAP and other elements of the SWI/SNF complex.
b-c and f-k are representative experiments, repeated independently two (c, f-k) or three (b) times, all with similar results.
Extended Data Figure. 2. Effect of ARID1A depletion on YAP/TAZ levels, localization and activity.
a, The panel represents the results of luciferase assays with the 8xGTIIC-Lux reporter in HEK293 cells transfected with empty (e.v.) or YAP-expressing vectors and the indicated siRNAs. Data are normalized to siCo/empty vector-transfected cells and are presented as mean + s.d. of n=3 biologically independent samples.
b, The panel represents the results of luciferase assays with the 8xGTIIC-Lux reporter in HEK293 cells reconstituted with either YAP wt or YAP WW1mut and transfected with the indicated siRNAs. Data are normalized to siCo-transfected cells and are presented as mean + s.d. of n=3 biologically independent samples.
c, Panels are qRT-PCR analyses for the YAP/TAZ targets ANKRD1, CYR61 and PTX3 (mean + s.d. of n=3 biologically independent samples) in MCF10A cells transfected as indicated.
d, Western blot analysis for ARID1A, E-Cadherin and Vimentin from lysates of MCF10A cells transfected with the indicated siRNAs.
e, Panels are qRT-PCR analyses for CTGF (left) and ARID1B (right) expression (mean + s.d. of n=3 biologically independent samples) in MCF10A cells transfected as indicated.
f, Representative confocal images (left) and quantifications (right; >100 cells per condition) of YAP/TAZ localization in MCF10A cells transfected with the indicated siRNAs.
g, Western blot analysis for YAP, TAZ and YAP phosphorylation on the key Hippo/LATS target site (P-YAP S127) from lysates of MCF10AT cells transfected with the indicated siRNAs.
P values were determined by unpaired two-sided t-test.
All panels display representative experiments, repeated independently two (d, e, g) or three (a-c, f) times with similar results.
Extended Data Figure. 3. YAP/TAZ are required for biological effects of SWI/SNF depletion in human mammary epithelial cells.
a-b, HMECs were transduced with the indicated shRNA-encoding vectors and harvested for protein extraction (a) or RNA extraction (b). a, Western blot for BRG1, TAZ and epithelial (ECAD) and mesenchymal (Vimentin) markers (N=2). b, qRT-PCR analyses (mean + s.d. of n=3 biologically independent samples) for mesenchymal (TWIST1) and epithelial (KRT18) markers. Compendium of Fig. 2a.
c-e, HMECs were transduced with the indicated shRNA-encoding vectors and/or transfected with the indicated siRNAs and harvested for RNA extraction. Shown are qRT-PCR analyses (mean + s.d. of n=3 biologically independent samples) for the indicate genes.
f, Mammospheres (mean + s.d. of n=6 biologically independent samples) formed by HMEC transduced with the indicated shRNAs and transfected with indicated siRNAs.
g, HMEC were transduced with the indicated shRNA-encoding vectors and analyzed for their CD44/CD24 immunophenotype. Bars are quantifications of the percentage of cells displaying either CD44High/CD24Low (stem-like mesenchymal cells) or CD44Low/CD24High (differentiated epithelial cells) profile8.
P values were determined by unpaired two-sided t-test.
All panels display representative experiments, repeated independently three times with similar results.
Extended Data Figure. 4. SWI/SNF depletion potentiates YAP-induced reprogramming of neurons into NSCs.
a, Efficiency of Brm and Arid1a downregulation in neurons transduced with the indicated shRNA-encoding vectors, as measured by qRT-PCR (mean + s.d. of n=3 biologically independent samples). Shown is a representative experiment repeated twice with similar results.
b-c, Compendium of Fig. 2c. Neurons were infected with doxycycline-inducible YAP-encoding vectors or empty vector and the indicated shRNAs-encoding lentiviral vectors. Shown are representative images (b; scale bar, 300 μm) of the cultures after 14 days in NSC medium with doxycycline and quantifications of emerging (P0) neurospheres (c; mean + s.e.m. of four independent experiments; asterisk is shCo vs. shBrm (P value = 0.03) or shArid1A with YAPwt (P value = 0.03)).
d-e, Effect of ARID1A depletion on YAP-induced reprogramming of neurons. d, Syn1-Cre drives Arid1a knockout specifically in neurons as shown by genotyping. Neuron genomic DNA was compared to tail genomic DNA of the same Syn1-Cre; Arid1afl/+ mouse. Panels represent PCR bands for the indicated alleles. e, Control (Arid1a+/+) and Arid1a+/- (from Syn1-Cre; Arid1afl/+ mice) neurons were infected with inducible YAP-encoding vectors. Shown are representative images (left; scale bars, 300 μm), and the quantification (right; mean + s.e.m. of four independent experiments) of P0-neurospheres emerging from these cultures after doxycycline treatment in NSC medium. YAPS94A serves as negative control. e complements Fig. 2c and Extended Data Fig. 4b-c showing comparable results between shRNA and genetic attenuation of ARID1a.
P values were determined by unpaired two-sided t-test for a, and by two-sided Mann Whitney U test for c and e.
Extended Data Figure. 5. Effect of ARID1A depletion in hepatocytes on tumor formation.
a, qRT-PCR analysis for Nf2 and Arid1a expression (mean + s.d.) in the livers from control (n=6 mice), and Nf2 (n=6 mice), Arid1a (n=7 mice) and Nf2/Arid1a (n=7 mice) liver mutant (LKO) mice, 4 months after tamoxifen treatment. All animals were included.
b, Livers from control (Arid1afl/fl) and Arid1a LKO (Alb-CreER; Arid1afl/fl) mice were harvested 2 weeks after tamoxifen treatment, and genomic DNA and proteins were extracted with standard procedures. Panels are representative results, repeated on four mice for each genotype: (left) PCR bands for the indicated alleles; (right) western blots for GAPDH (loading control) and ARID1A.
c, YAP IHC staining in control and Nf2 mutant livers. Scale bars, 40 μM. Shown are representative images independently replicated on three mice for each genotype with similar results.
d, Compendium of Fig. 2e. Representative Cytokeratin (CK; top) and Ki67 (bottom) stainings in sections of livers of the indicated genotypes (same as in Fig. 1d-e, and Extended data Figure 5a). Note intrahepatic cholangiocarcinomas (iCCA; CK-positive, Ki67-positive) and hepatocellular carcinomas (HCC; Ki67-positive, but CK-negative) only in livers from Nf2/Arid1a LKO. Scale bars, 100 μm. Shown are representative images independently replicated for all the mice of each genotype, with similar results.
e, Quantitative real-time PCR analysis of selected genes from liver of mice with the indicated genotypes. All animals were included. Data (mean + s.d. of n mice per genotype, as in Extended data Figure 5a above) are normalized to Nf2/Arid1a LKO mice.
f, Compendium of Fig. 2f. Control, Arid1a LKO, Arid1a/Yap/Taz LKO mice were treated with tamoxifen and then were fed DDC-containing diet for 6 weeks. Panels are CK (top; scale bars, 40 μM) and Ki67 (bottom; scale bars, 20 μM) stainings of liver sections from the indicated mice. Note the presence of early cholangiocarcinoma lesions (CK-positive, Ki67-positive) in the Arid1a LKO mice and their absence upon concomitant YAP/TAZ loss (i.e., in the Arid1a/Yap/Taz LKO mice). Asterisks indicate porfirin deposits, typically present in liver of animals treated with DDC. Shown are representative images, independently replicated for all the mice of each genotype (n as in Fig. 2f), with similar results.
g, Representative qRT-PCR analysis for Afp expression (mean + s.d. of n mice) in the livers from control (n=4), Arid1a LKO (n=5), Arid1a/Yap/Taz LKO (n=5) mice treated with tamoxifen and then DDC. Data are normalized to livers of mice not treated with DDC (n=4). This experiment was independently repeated three times with similar results, analyzing, in total, at least 10 mice for each genotype.
h, Representative E-cadherin staining (scale bars, 30 μm) showing that CCA lesions retain epithelial morphology in sections of livers of the indicated mice. Experiments were independently repeated on three mice, with similar results.
P values were determined by one-way ANOVA with Dunnett’s multiple comparisons test in a or with Tukey’s multiple comparisons test in e and g.
Extended Data Figure. 6. Interaction of SWI/SNF with F-actin and YAP is mutually exclusive.
a, Compendium of Fig. 3a. HEK293T cells were transfected with NLS-Flag-ACTIN R62D. Shown are representative anti-Flag IF staining to visualize Actin. Nuclei were counterstained with DAPI. Scale bars, 10 μM.
b-c, Compendium of the proximity ligation assay (PLA) of Fig. 3b. b, Negative controls of the PLA of Fig. 3b: in the absence of one of the two partners, no dots can be seen. c, In HEK293T cells, by PLA, endogenous BRM interacts with Flag-tagged NLS-Actin WT, but not with Flag-tagged NLS-Actin R62D, indicating that the association is specific for filamentous and not monomeric actin.
d, Western blots of the inputs of the experiment shown in Fig. 3c.
e, Sequential salt extraction of HEK293T cells treated with either Phalloidin or Lat.A. Shown are western blot for the indicated proteins. H3 was loaded on a different blot.
f, Western blots of the inputs of the experiment shown in Fig. 3d. MCF10AT cells were transfected with control siRNAs (siCo., lanes 1 and 2) or siRNAs against ARID1A (si1A; lane 3) and treated with Phalloidin (Phall; lane 1) or Lat.A (lanes 2 and 3), as indicated.
g, Compendium of Fig. 3e. PLA detecting the interaction between endogenous BRM and NLS-YAP in MCF10A cells. Control untreated cells, 0% PLA-positive cells; cells treated with the Src inhibitor Dasatinib (i.e., a low mechanics condition in addition to those shown in Fig. 3e), 14.5% PLA-positive cells.
h, CoIP/western blot analysis of MCF10AT lysates showing endogenous ARID1A bound to endogenous YAP but not to TEAD1 and TEAD4. As a specificity control, immunoprecipitation was repeated from the same lysates using unrelated rabbit IgG.
i-j, Compendium of Fig. 3f. i, Representative PLA images detecting the interaction between endogenous TEAD and NLS-YAP in MCF10A cells. YAP/TEAD association is lost in C3-treated cells (that is, in cells with attenuated mechanotransduction (low mechanics) upon C3-mediated inhibition of RhoGTPases), but rescued after depletion of ARID1A (PLA-positive cells: 43.4%). j, Specificity controls of single antibodies for the PLA of Fig. 3f and Extended Data Fig. 6i.
a-c, e, g-j are representative experiments, repeated independently two (e, h) or three (a-c, g, i-j) times, with similar results.
Extended Data Figure. 7. Loss of SWI/SNF restores YAP/TAZ transcriptional activity in mechanically-inhibited cells.
a, Representative confocal images (left) and quantifications (right; >100 cells per conditions) of YAP/TAZ localization in MCF10A cells transfected with the indicated siRNAs and replated on a soft ECM.
b, MCF10A cells were transfected with the indicated siRNAs, and either left untreated (Control) or treated with anti-integrin antibodies, the Rho-inhibitors C3 and Cerivastatin, the Src-inhibitor Dasatinib, or the ROCK inhibitor Fasudil. Shown are qRT-PCR analyses for CTGF expression (mean + s.d. of n=3 biologically independent samples). Anti-integrin and Fasudil were part of the same experiment and thus share the same control repeated in their corresponding panels.
c, HaCaT cells were transfected with the indicated siRNAs and replated to obtain either sparse (High mechanics) or dense monolayers (Low mechanics). Shown are qRT-PCR analyses for CTGF expression (mean + s.d. of n=3 biologically independent samples).
d, Efficiency of Arid1a downregulation in Arid1afl/fl fibroblasts after transduction with Adeno-Cre, as measured by qRT-PCR (mean + s.d. of n=3 biologically independent samples) and western blot (GAPDH serves as loading control).
e, MCF10A cells were transfected with the indicated siRNAs and replated at very high density (see Methods). Shown are qRT-PCR analyses for CTGF expression (mean + s.d. of n=3 biologically independent samples).
All panels display representative experiments, repeated independently three times with similar results.
P values were determined by unpaired two-sided t-test.
Extended Data Figure. 8. Loss of SWI/SNF enables YAP-induced biological effects in mechanically-inhibited cells.
a, MCF10A cells were transfected with the indicated siRNAs, and replated to obtain dense monolayers (Low mechanics). After 24 hours, cells were incubated for 1 hour with a pulse of EdU to label cells undergoing DNA duplication. Cells were fixed and processed for EdU staining. Bars are quantifications (mean + s.e.m. of at least n=3 biologically independent samples) of proliferation measured as EdU-positive cells. Data are normalized to sparse cells (High mechanics) transfected with control siRNA. Statistics for rescue experiments at low mechanics: siCo (n=3) vs. siBRM/BRG1 mixA, n=3, P=0.0003; vs. siBRM/BRG1 mixB, n=3, P=0.0005; vs. siARID1a#1, n=3, P=0.04; vs. siARID1a#2, n=3, P=0.002. Shown is a representative experiment, that was repeated independently twice with similar results.
b, Neurons were plated on a stiff or soft ECM and infected with inducible YAP-encoding vectors. The panel depicts quantification of neurospheres emerging from these cultures after doxycycline treatment in NSC medium (mean + s.e.m. of all biological independent samples of three experiments, n=9).
c-d, Compendium of Fig. 4d. Neurons were plated on a soft ECM and infected with inducible YAP-encoding vectors or empty vector and the indicated shRNAs-encoding lentiviral vectors. Shown are (c) representative images (scale bar, 300 μM) and (d) quantification of neurospheres (P0) emerging after doxycycline treatment (shown are mean + s.e.m. of four independent experiments; asterisk is shCo vs. shBrm, P value = 0.03, or shArid1A with YAPwt, P value = 0.03).
e, Fold changes in neurospheres emerging from cultures of YAP-induced neurons transduced with the indicated shRNAs against BRM or ARID1A, and plated either on a stiff (High mechanics) or soft (Low mechanics) ECM, with respect to the corresponding shCo-expressing cultures. Shown are mean + s.e.m. of four independent experiments. Asterisk is the comparison of shBRM or shArid1a at high mechanics with the corresponding samples at low mechanics, all P values = 0.03.
P values were determined by unpaired two-sided t-test in a, and by two-sided Mann Whitney U test in b, d and e.
Supplementary Material
Acknowledgments
We thank Atsushi Fujimura for help with neuron preparation; G. Della Giustina for micropattern fabrication; V. Guzzardo for histology; C. Frasson and G. Basso for FACS; D. Livingston for HMEC and plasmids; D.J. Pan, M. Giovannini, Z. Wang, P. Chambon, and I. De Curtis and R. Brambilla for gifts of mice; R. Treisman for Actin cDNAs; L. Naldini for plasmids. We are grateful to: Sirio Dupont for performing the initial experiments leading to biochemical identification of SWI/SNF and for the protocol to perform F-actin pull-down; Gianluca Grenci and Mona Suryana (MBI-Singapore) and the MBI microfabrication facility team for the supply of quartz masks. This work is supported by AIRC Special Program Molecular Clinical Oncology “5 per mille”, by an AIRC PI-Grant, by a MIUR-FARE grant, and by Epigenetics Flagship project CNR-MIUR grants to S.P. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (DENOVOSTEM grant agreement No 670126 to S.P.).
Footnotes
Author contributions
LC carried out experiments in vitro, LA carried out experiments on mice. Role of other coauthors: DDB, molecular biology and IFs; DDB and RLX, liver experiments; GB and FZ, molecular biology and preparation of samples for Chip-MS; LC and TP, neuronal reprogramming; SG, hydrogel preparation; GS and JK for mass spectroscopy; MF, histology and histopathological evaluations; GBr and AG, microfabrication; SP and MC conceived the initial hypothesis and experimental design, planned, discussed and organized the work. LC, LA, FZ, MC and SP wrote the MS.
Authors Information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Data Availability
Mass spectrometry data can be found in the Extended Data Table 1. Source Data for Main and Extended Data Figures can be found in the online version of the paper. For uncropped images of immunoblots see Supplementary Fig. 1. All relevant data are included in the manuscript as source data; all other data are available from the corresponding authors upon reasonable request.
References
- 1.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rafiee MR, Girardot C, Sigismondo G, Krijgsveld J. Expanding the Circuitry of Pluripotency by Selective Isolation of Chromatin-Associated Proteins. Mol Cell. 2016;64:624–635. doi: 10.1016/j.molcel.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nature cell biology. 2018;20:888–899. doi: 10.1038/s41556-018-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ege N, et al. Quantitative Analysis Reveals that Actin and Src-Family Kinases Regulate Nuclear YAP1 and Its Export. Cell Syst. 2018;6:692–708.:e613. doi: 10.1016/j.cels.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol Cell. 2016;63:277–292. doi: 10.1016/j.molcel.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 10.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eroglu E, et al. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 12.Panciera T, et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benhamouche S, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes & development. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakiri L, Wagner EF. Mouse models for liver cancer. Mol Oncol. 2013;7:206–223. doi: 10.1016/j.molonc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem. 2015;290:11209–11216. doi: 10.1074/jbc.M114.627166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosse R, Vartiainen MK. To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol. 2013;14:693–697. doi: 10.1038/nrm3681. [DOI] [PubMed] [Google Scholar]
- 18.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 19.Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 21.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nature cell biology. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 23.Benham-Pyle BW, Pruitt BL, Nelson WJ. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skibinski A, et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell reports. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 26.Azzolin L, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 27.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes & development. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan B, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao X, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes & development. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 36.Rempe D, et al. Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis. 2006;44:44–49. doi: 10.1002/gene.20183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data can be found in the Extended Data Table 1. Source Data for Main and Extended Data Figures can be found in the online version of the paper. For uncropped images of immunoblots see Supplementary Fig. 1. All relevant data are included in the manuscript as source data; all other data are available from the corresponding authors upon reasonable request.