Abstract
Helios, a member of the Ikaros family of transcription factors, is predominantly expressed in developing thymocytes, activated T cells, and regulatory T cells (Tregs). Studies in mice have emphasized its role in maintenance of Treg immunosuppressive functions by stabilizing Foxp3 expression and silencing the Il2 locus. However, its contribution to human immune homeostasis and the precise mechanisms by which Helios regulates other T cell subsets remain unresolved. Here, we investigated a patient with recurrent respiratory infections and hypogammaglobulinemia and identified a germline homozygous missense mutation in IKZF2 encoding Helios (p.Ile325Val). We found that HeliosI325V retains DNA binding and dimerization properties but loses interaction with several partners, including epigenetic remodelers. Whereas patient Tregs showed increased IL-2 production, patient conventional T cells had decreased accessibility of the IL2 locus and consequently reduced IL-2 production. Reduced chromatin accessibility was not exclusive to the IL2 locus but involved a variety of genes associated with T cell activation. Single-cell RNA sequencing of peripheral blood mononuclear cells revealed gene expression signatures indicative of a shift toward a proinflammatory, effector-like status in patient CD8+ T cells. Moreover, patient CD4+ T cells exhibited a pronounced defect in proliferation with delayed expression of surface checkpoint inhibitors, suggesting an impaired onset of the T cell activation program. Collectively, we identified a previously uncharacterized, germline-encoded inborn error of immunity and uncovered a cell-specific defect in Helios-dependent epigenetic regulation. Binding of Helios with specific partners mediates this regulation, which is ultimately necessary for the transcriptional programs that enable T cell homeostasis in health and disease.
Introduction
The Ikaros family of transcription factors comprises five members: Ikaros (encoded by IKZF1), Helios (IKZF2), Aiolos (IKZF3), Eos (IKZF4), and Pegasus (IKZF5). Three of these zinc finger proteins—Ikaros, Helios, and Aiolos—have been shown to be involved in the intricate gene-regulatory network that governs hematopoiesis, cell fate decisions, and cellular function, particularly in cells of the adaptive arm of immunity (1, 2). As transcription factors, they bind to their consensus DNA sequences through N-terminal zinc finger domains, whereas homodimerization and heterodimerization to other family members are facilitated by the two C-terminal zinc finger domains (3). In addition, they bind and recruit other transcriptional modulators as well as chromatin modifiers, such as the nucleosome remodeling (NuRD) complex (4, 5). These interactions mediate their activating and repressive effects on target genes, including lineage-defining factors and cell cycle regulators (4).
Although Ikaros and Aiolos are expressed broadly in hematopoietic cells, several studies using different mouse models have revealed a critical role for Ikaros and Aiolos in the regulation of B cell differentiation, activation, and proliferation (6–9). Ikaros has additionally been shown to play an important role during T and natural killer (NK) cell development (10–12). Helios, on the other hand, is primarily expressed in T cells, particularly in developing thymocytes, activated T cells, regulatory T cells (Tregs), and mucosa-associated invariant T (MAIT) cells (13–19). In both mice and humans, about 70 to 80% of the Treg pool expresses Helios (20). Thornton et al. (21) have shown that, in mice, Helios+ Tregs have more stable Foxp3 expression, thereby maintaining Treg-specific demethylated regions and supporting full suppressive capacity. In line with this, mice with a conditional deletion of Helios in Tregs eventually developed systemic autoimmunity as a result of a reduction in the viability and stability of Helios-deficient Tregs due to increased apoptosis and defective Treg effector functions (22, 23).
Helios is up-regulated upon CD4+ and CD8+ T cell activation and proliferation in both murine and human cells, suggesting roles beyond Tregs (15, 19, 24). Yet, despite the almost complete early postnatal lethality of Helios null mice on a C57BL/6 background, viable adult mice on a mixed B6/129 background were smaller in size but had no defects in T cell or Treg development or function (25). It has been suggested that other Ikaros family members can compensate for the Helios deficiency in developing thymocytes. On the other hand, the lack of Helios in activated T cells might be balanced by its loss in Tregs, explaining the absence of overt immune activation in these mice (26). Although all these studies have been informative, the precise roles of Helios in different immune cell subsets and their relevance to human disease have remained elusive.
Human germline autosomal dominant mutations in IKZF1 have been identified, leading to a spectrum of clinical manifestations. Mutations in the DNA binding or dimerization domains of Ikaros, leading to haploinsufficiency, have been associated with loss of B cells and concomitant hypogammaglobulinemia, predisposition to B cell acute lymphoblastic leukemia, and/or autoimmunity (27–31). Dominant-negative mutations have been found to be more severe, with profound defects in T cells and myeloid cells (32). Very recently, patients with a germline monoallelic mutation in IKZF3 have been reported, demonstrating impaired Ikaros function, hence phenocopying B cell defects characteristic of patients with dominant-negative mutations in IKZF1 (33, 34). Here, we characterize a previously unknown, inborn error of immunity (IEI) caused by a biallelic missense mutation in IKZF2, identified in an immunodeficient patient with T cell aberrations. We study the pathobiological consequences of the germline IKZF2 mutation and identify impaired interaction of Helios with several chromatin remodelers and transcription factors, resulting in global epigenetic changes translating to an aberrant T cell phenotype and function.
Results
Identification of a homozygous missense variant in IKZF2
We studied a 17-year-old Iranian male index patient who was born to a consanguineous family (Fig. 1A). He has a failure to thrive, with a very low body weight (20 kg) and a short stature (112 cm), both measured at the age of 13 (body mass index z score of –2.11, below the third percentile). He consistently suffered from joint pains, with radiography of the left wrist at age 13 showing evidence of osteopenia with fraying of the distal ulna and radius, and a bone age of 9.5 years. He was additionally diagnosed with hypothyroidism and is on substitution therapy. The patient has a history of recurrent infections, including sinusitis, otitis media, and lower respiratory tract infections with productive coughs and bronchiectasis, leading to multiple pneumonias that were accompanied by fevers, and required several hospitalizations since childhood. For most of these infectious episodes, no causative agent was isolated; however, the patient benefited from antibiotics. Laboratory results indicated consistent leukopenia (white blood cell count of 2.89 × 109/liter, reference: 4 × 109/liter to 11 × 109/liter) and hypogammaglobulinemia with low antibody titers against tetanus and diphtheria vaccinations (table S1), for which he is given a monthly dose of intravenous immunoglobulin. No autoantibodies were present (table S1), and both direct and indirect Coombs tests were negative.
Fig. 1. Identification of a homozygous missense variant in IKZF2.
(A) Family pedigree of the patient under study. The 17-year-old male patient (II-4) is the only affected member (filled square) but has a sibling who died in utero (small circle). (B) Chromatograms from Sanger sequencing showing the segregation of the C>T variant in IKZF2 in the four family members, leading to an isoleucine to valine amino acid change. (C) Illustration of the Helios transcription factor, with four N-terminal zinc fingers responsible for DNA binding to the consensus sequence and two C-terminal zinc fingers that form the homo/heterodimerization domain. Location of the missense variant (ENST00000457361.1:c.973A>G, ENSP00000410447.1:p.Ile325Val) is shown (red arrow). IKbs, Ikaros binding site. (D) Conservation of the isoleucine amino acid at position 325 of the human transcript, across several species (red rectangular box).
To investigate the underlying molecular etiology in our patient, we performed whole-exome sequencing as well as segregation analysis and identified a homozygous missense variant in the IKZF2 gene (ENST00000457361.1:c.973A>G, ENSP00000410447.1:p.Ile325Val), encoding the Helios transcription factor (Fig. 1B and table S2). The variant lies in the region between the two main zinc finger domains of the protein: the DNA binding domain and homo/heterodimerization domain (Fig. 1C). Genome3D (35) predicts this region to contain another nonclassical zinc finger domain (FYVE/PHD or Zinc/ RING finger) with high confidence (90%) where the variant lies, which could play a role in the mediation of specific protein interactions (36). There have been no reported homozygous cases of this variant in the gnomAD (37) database (last accessed 14 July 2021). The missense variant is predicted to be damaging according to functional prediction tools (table S2), and the isoleucine at this position is highly conserved across species (Fig. 1D). As a measurement, the probability of being loss-of-function (LoF) intolerant (pLI) score (38) of IKZF2 is extremely high (0.99), indicating essentiality of this gene. Collectively, this suggested that the p.I325V homozygous variant in Helios has a high possibility for being disease causing.
Aberrant immune cell phenotype
To investigate the consequences of the germline IKZF2 mutation on immune cell subset composition, we performed immune phenotyping on patient peripheral blood mononuclear cells (PBMCs) at 13 and 14 years of age. We observed a marked reduction of relative and absolute numbers of both B (CD19+) and NK (CD3-CD19-CD16+ CD56dim) cells (Fig. 2A), with an apparent progressive decline in B cell counts over time (table S1). When evaluating the patient B cell subsets, we noted a reduction in the frequencies of class-switched memory (CSM) and nonswitched memory (non-SM) B cells as well as plasmablasts, with a higher frequency of transitional B cells, indicative of a B cell maturation defect (Fig. 2B and fig. S1). In addition, the innate-like CD19+CD38loCD21lo B cell subset, typically expanded in chronic immune stimulation, was higher in the patient (fig. S1) (39). Within the T cell fraction, we observed a reduction in CD4+ T cells, with an inverted CD4/CD8 ratio (Fig. 2, A and C, fig. S2A, and table S1). Although the frequencies of naive and memory T cells within the patient's CD4+ T cell population were within normal range, CD8+ T cells were predominantly effector memory (CD45RA−CCR7−; mean of 73% in patient versus 25% in controls) that expressed a high level of CD57, indicative of their terminally differentiated state (Fig. 2, C and D, and fig. S2A) (40). Consistent with this, we observed a concomitant shift to highly differentiated CD8+CD27−CD28− T cells in the patient (fig. S2A). On the other hand, patient Tregs (gated on both CD127−CD25+FOXP3+ and CD25+FOXP3+) were present and showed normal expression of Helios, albeit in a lower abundance within the CD4+ T cell population compared with controls (mean of 1.56% versus 5.00%; Fig. 2E and fig. S2B). T follicular helper (TFH) cells of the patient were within the normal range (Fig. 2F and fig. S3A); however, the frequencies of other T cell populations were altered, with a higher frequency of γδ T cells in the patient (Fig. 2G and fig. S3A). We also noted a reduced frequency of the innate-like, invariant NK T (iNKT) cells and MAIT cells (Fig. 2G and fig. S3, B and C), in line with data from Hetemäki et al. (41) on two patients with a heterozygous Helios (p.R200X) mutation. Given the evident aberrations in the frequencies of several immune subsets leading to the combined immunodeficiency in the index patient in our study, we next sought to investigate the molecular mechanisms by which the biallelic variant in IKZF2 affects protein function.
Fig. 2. Aberrant immune phenotype in the index patient with biallelic Helios mutation.
(A) Flow cytometry plots of CD3 versus CD19 markers in the patient and a control. The three following graphs to the right compare the percentages of T cells (CD3+), B cells (CD19+), and NK cells (CD16+CD56dim; gated on CD3−CD19−) in lymphocytes between the patient (P; red) and controls [C; adult controls are shown in black; two age-matched controls are shown in blue (n = 8 to 16)]. (B) Graphs showing the frequency of naive B cells (IgD+CD27−), CSM B cells (IgD−CD27+), non-SM B cells (IgD+CD27+), transitional B cells (CD21++CD38+), and plasmablasts (CD38++CD27+) in patient and controls. (C) Graphs showing the frequency of CD4+ and CD8+ T cells as a percentage of CD3+ T cells (left) and the frequency of CD4+ (middle) and CD8+ (right) naïve (CCR7+CD45RA+), central memory (CM; CCR7+CD45RA−), effector memory (EM; CCR7−CD45RA−), and terminally differentiated effector memory (Temra; CCR7−CD45RA+) cells within CD4+ and CD8+ populations, respectively. (D) Frequency of CD8+CD57+ T cells as a percentage of CD3+ T cells. (E) Percentage of Tregs (CD25+FOXP3+) within the CD4+ T cell population (left), the expression of Helios shown as the mean fluorescence intensity (MFI) within the Treg population (center), and overlaid histograms showing Helios expression in the Tregs of patient versus control, including an IgG control for each in gray (left). IgG, immunoglobulin G. (F) Frequency of TFH cells (CXCR5+CD45RA−) as a percentage of CD4+ T cells. (G) Percentage of γδ T cells (left), iNKT cells (TCR-Vα24+TCR-Vβ11+; middle), and MAIT cells (CD161+TCR Vα72+; right) within the CD3+ T cell population. The patient's values were taken at two time points (13 and 14 years).
Functional and biochemical assessment of HeliosI325V
To assess the effects of the p.I325V mutation on the ability of Helios to bind DNA, we performed a chemiluminescent electromobility shift assay (EMSA). Nuclear extracts from the human embryonic kidney cell line, HEK293T, overexpressing wild-type (HeliosWT) or mutant Helios (HeliosI325V), were incubated with DNA probes containing an Ikaros consensus-binding sequence (IK-bs1), including the canonical motif GGGAA, to which Helios also binds (14, 42). Both HeliosWT and HeliosI325V showed a similar level of dimer and multimer formation on the IK-bs1 probe (Fig. 3A). We next sought to visualize the binding of HeliosWT and HeliosI325V to pericentromeric heterochromatin (PC-HC) regions and assess their cellular localization by immunofluorescence staining in NIH3T3 murine fibroblast cells. Images show the ability of HeliosI325V to localize to the nucleus and form a similar number of foci at PC-HC regions compared with HeliosWT (Fig. 3B). Therefore, we concluded that the p.I325V mutation in Helios does not alter its DNA binding capacity.
Fig. 3. Effects of p.I325V mutation on Helios function.
(A) EMSA showing the ability of Helios WT and I325V mutant to form dimers and multimers (arrows) and bind to the IK-bs1 probe, an Ikaros consensus-binding sequence. Representative image of seven independent experiments. EV, empty vector. (B) Immunofluorescence staining of NIH3T3 cells transfected with Helios WT or mutant using an anti-Helios antibody, showing the formation of foci at PC-HC regions. Graph on the right shows the quantification of number of foci per cell (n = 239 to 422). Results represent three independent experiments; unpaired t test was done showing no significant difference. (C) Co-IP experiments after cotransfection of HEK293T cells with Strep-HA-Helios WT, I325V mutant, or an EV together with FLAG-Helios-WT, mutant, or Ikaros. IP was performed with Strep-beads, and Western blot analysis was done by running both the IP and whole-cell lysate (input) on a gel and blotting with HA, FLAG, and GAPDH antibodies. Results are representative of three to five independent experiments. (D) Co-IP as in (C) after cotransfection with HA-HDAC1 and blotting with HDAC1 and Helios antibodies. Blot representative of six independent experiments.
We next assessed the ability of HeliosI325V to form dimers with the WT or mutant counterpart, as well as to other members of the Ikaros family, by coimmunoprecipitation (co-IP) experiments. HeliosI325V was able to form homo- and heterodimers with Ikaros, Aiolos, and Eos (Fig. 3C and fig. S4A). As the mutation does not lie in either the DNA binding or dimerization domains, the functional intactness of these two domains is expected. Yet, in the process of activating or repressing genes, Helios also binds to other transcription factors and chromatin modifiers, including members of the NuRD complex such as histone deacetylase 1 (HDAC1) (5). Co-IP experiments revealed a marked reduction in the ability of HeliosI325V to bind to HDAC1 (Fig. 3D). As SUMOylation of Ikaros participates in the regulation of HDAC binding (43), the SUMOylation of Helios by SUMO2 was assessed after overexpressing a SUMO2 construct with HeliosWT or HeliosI325V in HEK293T cells. Analysis by Western blot showed no defect in SUMOylation (fig. S4B). Hence, the loss in HDAC1 binding caused by the missense mutation could be due to steric hindrance, leading to our subsequent evaluation of the effects of HeliosI325V on all protein interactions.
HeliosI325V leads to disrupted interactions with epigenetic remodelers
The mediation of appropriate interactions between transcription factors and chromatin remodelers at required genomic loci during different stages of cell development is crucial for transcriptionally priming cells into a particular fate (4, 44). As many interactions are transient, or require the assembly of several proteins or complexes together, we sought to probe all these interactions by using BioID-MS, a biotinylation-proximity labeling technique, coupled with liquid chromatography–mass spectrometry (LC-MS) (45, 46). Moreover, given the loss of binding of HeliosI325V to HDAC1 (Fig. 3D), we wanted to globally identify the disrupted interactions caused by the mutation. Complementing previous attempts that identified direct physical interaction partners of Helios (5), here, we expanded Helios's interactome with 170 dynamic interactors after stringent statistical analysis [Significance Analysis of INTeractome express (SAINTexpress), Bayesian false discovery rate (BFDR) < 0.05]. We performed enrichment analysis to reveal enriched protein complexes from the complex database CORUM (Fisher's exact test, Padj < 0.05) (Fig. 4A and table S3) (47). As expected, most interactors consisted of transcription factors and histone modifiers, including known interactors from the NuRD complex (5). We also identified previously unknown interactions with members of the ATAC (Ada two A containing) complex of histone acetyltransferases and with proteins involved in cell cycle regulation and cell division (PLK1 and GTSE1) as well as DNA repair (NIPBL, BLM, BRCA1, and BRCA2) (Fig. 4A).
Fig. 4. Mutant Helios shows altered interactome.
(A) Representation of all high-confidence BioID interactors of HeliosWT and HeliosI325V, clustered into modules based on the protein complexes they are associated with using CORUM (47). Interactors that are missing, gained, or significantly reduced in HeliosI325V are indicated in red, yellow, or orange, respectively. (B) Venn diagram representing number of shared, lost, and gained interactors by HeliosI325V compared with HeliosWT. (C) GO analysis using Enrichr (48) for biological processes of the missing and significantly reduced (≥25% reduction) proteins from (A), with bars indicating the number of proteins associated with each GO term.
Comparing the detected interactors from HeliosWT with those of the mutant, we found that HeliosI325V gained 11 interactors, lost 39 interactors, and had significantly (P < 0.05, Student's t test) reduced association to 17 proteins (depicted by different colors in Fig. 4A).
Lost interactors included histone remodelers, such as HDAC3, GATAD2A, GATAD2B, and KMT2D, and components of the ATAC complex (TADA2A, KAT2A, and SGF29), as well as cell cycle regulators, including GTSE1 and PLK1 (Fig. 4, A and B, and fig. S5). Enrichment analysis using Gene Ontology (GO) terms (48) indicated deregulation of transcription, histone acetylation, DNA methylation, cell cycle, and DNA repair caused by the mutant (Fig. 4C and fig. S6). Quantitative analysis of the interactome data using MaxQuant software confirmed that the mutant protein significantly loses interaction with the cell cycle regulators BRCA1 and GTSE1 and shows significantly reduced cooperation with several transcriptional and chromatin regulators (fig. S7, A and B, and table S3). Collectively, this suggests that although the p.I325V mutation lies outside of Helios zinc finger domains, with DNA binding and dimerization abilities remaining intact, specific Helios functions might be affected through an altered interactome; this is due to a disruption either in physical interactions or in the recruitment of Helios, or its partners, to the required genomic loci. Therefore, defects in the epigenetic regulation of particular loci are highly plausible, ultimately leading to transcriptional deregulation.
RNA sequencing reveals changes in transcriptional states of immune cell populations
Given the disturbance in the protein interaction network caused by HeliosI325V, we sought to systematically assess transcriptional changes that could culminate in the different cellular states observed in the patient's lymphocytes. We performed droplet-based single-cell RNA sequencing (scRNA-seq) on PBMCs from the patient, taken at 14 years of age. For comparison, we included PBMCs from four healthy controls, comprising two adults (C1, male; C2, female) and two age-matched controls (C3, male; C4, female). This analysis yielded a total of 25,081 cells passing quality control (table S4). Low-dimensional projection using uniform manifold approximation and projections (UMAP) (49) and graph-based clustering confirmed the presence of the expected PBMC populations (Fig. 5A, fig. S8, and table S5). On the basis of the expression of known marker genes, we labeled meta-cluster 1 as a monocyte cluster (CD68), meta-cluster 2 as a T and NK cell cluster (CD3D/E and KLRB1), cluster 3 as B cells (CD79A), cluster 4 as myeloid dendritic cells (DCs; CLEC10A), and cluster 6 as plasmacytoid DCs (GZMB; fig. S9A). Subpopulations of meta-clusters were further characterized (fig. S8 and table S5). As for T cells, we showed that cells of cluster 2a identified as naive T cells (CCR7 and TCF7), whereas cells of clusters 2b and 2c expressed genes associated with an NK cell, central memory, or effector T cell phenotype (GZMH, CTSW, NKG7, and KLRD1).
Fig. 5. Altered transcriptional state of Helios-mutant patient PBMCs.
(A) Low-dimensional projection (UMAP plot) of the combined scRNA-seq dataset comprising 25,081 cells from the patient (P) and four controls (two adults, C1: male, C2: female; and two age-matched, C3: male, C4: female). Numbers indicate clusters (graph-based clustering) and colors correspond to cell type (manual curation). (B) UMAP plot [same coordinates as in (A)] showing the distribution of patient cells (orange) within the clusters compared with controls (shades of gray). (C) Heatmap showing differentially expressed genes within cluster 2c between cells from the patient and those from all four controls (Padj < 0.05, |logFC| ≥ 0.25). (D) Heatmap showing differentially expressed genes between clusters 2a and 5. The complete results of the marker genes and DGE analysis for each comparison are available in tables S5 and S6. (E and F) Heatmap showing differentially expressed genes between sorted (E) naïve B cells (CD19+CD27−IgD+) and (F) Tregs (CD4+CD25hiCD127−) from PBMCs of the patient and three male healthy controls, with three replicates each (Padj < 0.05, |log2FC| ≥ 1). (G) Cytokine production assay, measuring intracellular IL-2 and IFNγ in Tregs(gated for CD4+CD25+FOXP3+) after 6 hours of stimulation with PMA and ionomycin.
With respect to lymphocytic cell frequencies, we observed an almost complete absence of patient cells in the B cell cluster 3 (Fig. 5B and fig. S9B). Moreover, a skewing of the patient T cells toward clusters 2b and 2c, and a loss of 2a, is apparent (fig. S9B). Both observations corroborate our previous findings based on flow cytometry (Fig. 2, A and C). Differential gene expression (DGE) analysis [pairwise MAST (50); Padj < 0.05, |log fold change (logFC)| ≥ 0.25; table S6] between the patient and control cells within the constituent clusters of meta-clusters 1 (i.e., 1a and 1b) and 2 (i.e., 2a, 2b, and 2c) was performed (Fig. 5C, fig. S10, A to C, and table S6). This analysis revealed an up-regulation of genes associated with inflammation, indicating that both monocytes and T cells are in an activated and effector state. For instance, within the effector T cell and NK cell cluster 2c, these genes include multiple components of the T cell receptor (TCR) activation complex (CD8A/B, CD3D/G, CD2, and CD6), the proinflammatory genes GZMH, IL32, and CCL5, an interferon (IFN)–related antiviral gene, ISG20, as well as glycolytic pathway genes PGK1 and LDHA. In addition, anti-inflammatory genes TSC22D3, TNFAIP3, and ZFP36 were up-regulated, possibly as an auto-regulatory consequence to the inflammatory response.
In contrast to the above, the "naïve T cell" cluster 2a showed a paucity of patient cells (Fig. 5B and fig. S9B). However, the isolated patient-centric cluster 5 was shown to express genes present in cluster 2a, suggesting that they are naïve CD4+ T cells of the patient (Fig. 5, A and B, and fig. S8). We confirmed this assumption by performing a principal components analysis that grouped cells from cluster 5 to the “naïve” T cell cluster 2a (fig. S9C). Analysis of the top differentially regulated genes between clusters 2a and 5 identified eight up-regulated genes (Fig. 5D and table S6). The anti-inflammatory gene TSC22D3 is up-regulated in cells of cluster 5 as well, suggesting maintenance of a suppressive state. This state is further supported by the elevated expression of protein phosphatases DUSP1 and PPP1R15A, which play a role in dampening the mitogen-activated protein kinase (MAPK) pathway and protein translation, respectively (51, 52). Upon induction by glucocorticoids, TSC22D3, encoding GILZ (glucocorticoid-induced leucine zipper), plays a key role in the inactivation of pathways including NF-κB (nuclear factor κB) (53). We noted that the patient has not been administered glucocorticoids before blood sampling. GILZ has also been shown to be induced under interleukin-2 (IL-2) deprivation and functions to prevent apoptosis (54). This is consistent with the up-regulation of the antiapoptotic gene BCL2 in cells of cluster 5. Therefore, besides the confirmed loss of B cells, transcriptomic analysis of the patient lymphocytic population revealed an increase in the inflammatory state of the predominant effector CD8+ T cell population while pointing to a suppressed naive CD4+ T cell population.
To analyze rare naive B cells and Tregs in the patient peripheral blood, we sorted naïve B cells (CD19+CD27−IgD+) and Tregs (CD4+CD25hiCD127−) and performed transcriptome analysis using SMART-seq in the patient compared with three sex-matched controls (table S7) (55). Differential analysis identified a total of 584 differentially regulated genes in naïve B cells (Fig. 5E and table S8). Pathway analysis of the 385 up-regulated genes confirmed the patient proinflammatory state, including increased granulocyte migration/chemotaxis and an up-regulation of both the IL-8 cytokine pathway (CXCL8, CXCL3, CCL4, and CCL3) and the pattern recognition receptor signaling (TLR2, FCN1, and FFAR2) (fig. S11A and table S9). Conversely, pathways with the 199 down-regulated genes indicated maturation defects of the patient B cells, with B cell receptor activation as one of the top results (fig. S11A and table S9). Down-regulated genes involve several immunoglobulin heavy and light chain genes, including IGHV1-69, IGHV3-48, IGHV1-24, FCGR2C, IGLC3, and IGLC2, as well as the pre-B cell receptor surrogate light chain, lambda-5 (encoded by IGLL1), which is critical for B cell development.
As mentioned, Helios is involved in maintaining the immunosuppressive role of Tregs by stabilizing Foxp3 (21). We found that 874 genes were differentially expressed in Tregs of the patient compared with healthy controls, of which 820 genes were down-regulated (Fig. 5F). Of these, 83 genes (9.5%), such as RPTOR, BCL6, CITED2, and WDR5, had previously been shown to be regulated by FOXP3 in Tregs (tables S8 and S9), indicating compromised Treg identity (56). Several genes involved in epigenetic and chromatin regulation were additionally found to be significantly reduced in the patient Tregs (KDM2B, KDM3A, KDM7A, KMT2B, EP300, WDR5, KANSL3, and KANSL1L), some of which we have shown significantly lose binding to HeliosI325V (KMT2D, GATAD2A, and GATAD2B) (Fig. 4A and table S8). These widespread transcriptional disturbances in the Tregs prompted us to assess their ability to repress the production of proinflammatory cytokines upon T cell stimulation. In line with the work of Kim et al. (22), we noted an increase in the production of IL-2 and IFNγ by Tregs of the patient compared with controls, confirming the proinflammatory and dysregulated state of the patient identified in our RNA-seq data (Fig. 5G and fig. S11B).
Defective proliferation and IL-2 production by patient T cells
To investigate the deleterious effects of the mutation on patient T cells, as indicated by their dysregulated transcriptome, we performed a series of in vitro experiments to functionally assess T cell activation responses. We first measured the proliferative capacity of patient T cells by stimulating the TCR with CD3/CD28 beads and monitoring the dilution of a proliferation marker. We observed a defect in the ability of the patient's CD4+ and CD8+ T cells to proliferate by the third day compared with the healthy sister and unrelated controls (mean of 30.3% versus 80.0 to 87.0% and mean of 7.8% versus 54.0 to 78.0% proliferated cells, respectively; Fig. 6, A and B, and fig. S12). Yet, in patient CD4+ T cells, this stimulation led to a normal increase in the percentage of cells up-regulating the activation marker CD25 (IL2RA) and Helios (Fig. 6C and fig. S12). Stimulation with an anti-CD3 monoclonal antibody (OKT3) alone or phytohemagglutinin (PHA) showed no proliferation in both CD4+ and CD8+ T cells of the patient at day 3, which was accompanied by a reduction in CD25 up-regulation (fig. S13, A to C).
Fig. 6. Patient T cells have defective proliferation and reduced IL-2 production.
(A) Representative histograms displaying the dilution of the violet proliferation dye (VPD450) in CD4+ and CD8+ T cells from the patient and a control after 3 and 8 days of stimulation with CD3/CD28 beads. P, patient; C, control. (B) Summary bar graphs showing the percentage of proliferated or (C) percentage of CD4+ and CD8+ T cells that have up-regulated CD25 at day 3, upon stimulation with CD3/ CD28 beads, in the patient (P; red), sister (S; orange), and controls (C; black, n = 6). Three independent experiments were done. (D) Summary graphs showing the percentage of CD4+ or CD8+ T cells that have up-regulated the checkpoint inhibitors PD-1 and TIGIT after stimulation with CD3/CD28 beads at basal state (before stimulation), days 2, 4, and 7 after stimulation. (P, patient, red; and C, controls, black, n = 3). (E) Bar graphs showing the percentage of CD4+ and memory CD8+ T cells that produced IL-2 after stimulation of PBMCs with PMA/ionomycin for 5 hours. (F) Amount of IL-2 (in micro-grams per milliliter) released into the medium after stimulating PBMCs with CD3/CD28 beads for 24 hours. The sister (orange) is included with the controls in (E) and (F) (C; n = 3 to 8). IL-2 production in CD4+ T cells of the patient was done in two independent experiments. (G) Bar graphs showing the percentage of cell death in T lymphoblasts, measured by annexin V and propidium iodide staining, after withdrawing IL-2 from the growth medium. Results are from two experiments. (H) Percentage of CD8+ T cells expressing IL-2 after stimulation of T lymphoblasts with PMA/ionomycin for 20 hours, including the T lymphoblast clone that had the IKZF2 gene CRISPR-edited to express WT alleles (P-WT, blue bar on graph). Each data point represents an independent experiment. Error bars in all summary bar graphs show the means with SEM.
By day 8 after stimulation with CD3/CD28 beads, patient CD4+ T cells have proliferated (Fig. 6A and fig. S12). As Helios is up-regulated upon T cell activation and proliferation (19), we hypothesized a delay in the onset of a downstream transcriptional program in the patient T cells. To address this, we assessed the expression of exhaustion markers and checkpoint inhibitors, such as PD-1, TIGIT, LAG3, TIM3, and AITR/GITR, which are typically up-regulated upon TCR activation. Our results indicated no increase in the expression of these markers at basal level (Fig. 6D and fig. S14, A and 0 B). However, we observed a delay in their up-regulation, particularly in the CD4+ T cells of the patient, measured at days 2, 4, and 7 after TCR activation (Fig. 6D and fig. S14, A and B), alluding to dysregulated and delayed activation of the downstream transcriptional program upon TCR stimulation. This was confirmed using IL-2-dependent expanded T lymphoblasts, where we excluded an upstream defect in proximal TCR signaling by performing intracellular calcium flux assay and assessing the phosphorylation of signaling molecules p65, p38, and ERK (extracellular signal-regulated kinase) (fig. S13, D and E). Only p-AKT, which is downstream of TCR, CD28, and IL-2 signaling, was less than fully increased upon stimulation with CD3/CD28 beads and IL-2 (fig. S13E).
We next assessed cytokine production upon T cell stimulation that is further downstream and a result of transcriptional activation. After a 5-hour stimulation of PBMCs with phorbol 12-myristate 13-acetate (PMA) and ionomycin, we noticed a decrease in IL-2–H producing CD4+ and CD8+ T cells (mean of 6.7% versus 23.2% and mean of 1.2% versus 26.9% in patient and controls cells, respectively; Fig. 6E and fig. S15). However, production of T helper cell–specific cytokines, IFNγ, IL-4, and IL-17A, by CD4+ T cells as well as IFNγ by CD8+ T cells was unaffected (fig. S16, A and B). Stimulation of patient PBMCs with CD3/CD28 beads also showed a reduction in IL-2 secretion into the supernatant when compared with controls (Fig. 6F). To address whether this reduction in IL-2 production affected cellular apoptosis, we withdrew supplementary IL-2 given to sustain the growth of T lymphoblasts. Although all control cells initiated apoptosis within 24 hours, patient T lymphoblasts showed higher levels of survival at this time point (Fig. 6G). The reduced capacity of patient T lymphoblasts to produce IL-2 was confirmed, upon PMA and ionomycin stimulation (Fig. 6H). To demonstrate causality of the HeliosI325V mutation in causing this phenotype, we generated CRISPR-Cas9–edited patient T lymphoblasts, correcting the mutation in IKZF2 and thus reconstituting it to WT (fig. S17A). We successfully generated one patient WT CD8+ clone (P-WT) and assessed its ability to produce IL-2 upon stimulation. Reconstitution of the patient mutation to WT rescued the defect in IL-2 production (Fig. 6H and fig. S17B), directly implicating the mutation in Helios (p.I325V) in the downstream control of the IL2 locus in conventional T cells. Collectively, the delay in the onset of proliferation and expression of checkpoint inhibitors, and defective IL-2 production as a result of HeliosI325V, indicated a dysregulation in the downstream transcriptional activation of genes ensuring full T cell activation. As data from our proteomic analysis demonstrated that the mutation in Helios leads to a loss or gain of interactions with chromatin remodelers and transcription factors (Fig. 4A), we next decided to assess the changes in the epigenetic state of conventional T cells of the patient upon stimulation.
Epigenetic analysis of patient T cells identifies global chromatin changes due to HeliosI325V
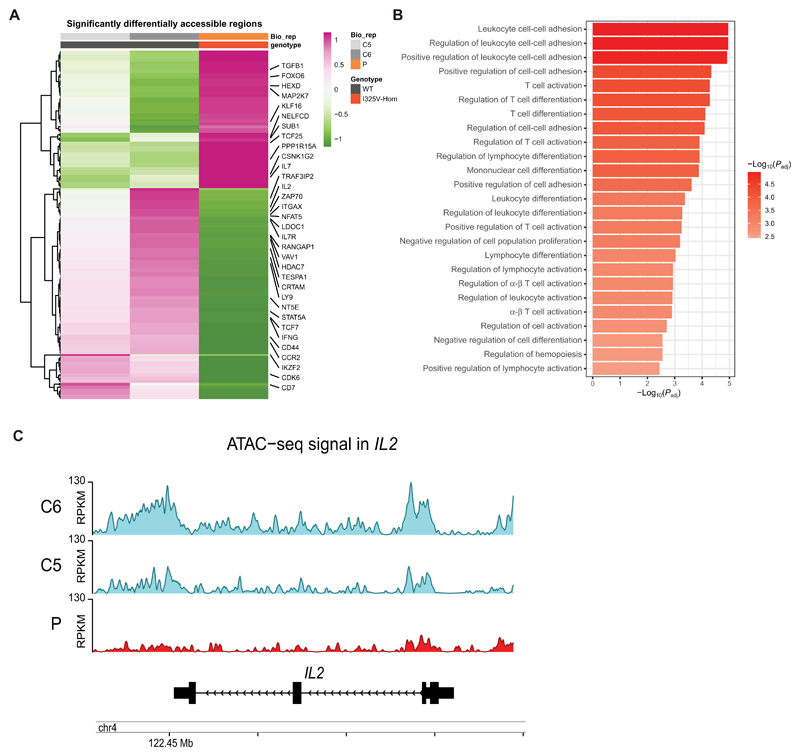
In Tregs, Helios is known to repress the Il2 locus by inducing deacetylation of core histones in the Il2 promoter (57). In conventional T cells, in which Helios is up-regulated upon T cell activation, the concerted action of multiple chromatin remodelers and transcription factors, including NFAT and the AP-1 complex, is required to initiate transcription of IL2. On the basis of our previous observations, we hypothesized an increased repression of the IL2 promoter in the patient T cells mediated by the loss of HeliosI325V interactions with activators of the IL2 promoter, including members of the ATAC complex, such as GCN5, which has been shown to be involved in IL-2 production (Fig. 4A) (58). Therefore, we performed an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) on T lymphoblasts of the patient and two male controls, upon stimulation with PMA and ionomycin for 2 hours, to initiate downstream epigenetic and transcriptional changes. Our analysis identified 21,627 peaks common to at least two samples ("consensus peaks") and 2099 peaks unique to patient-derived T lymphoblasts. As expected, most of all peaks (more than 35%) were found around promoter regions [within 1000 base pairs of an annotated transcription start site (TSS)] (fig. S18, A and B). Focusing on consensus peaks at promoters, we identified 233 differentially accessible regions between patient and control T lymphoblasts [|log2FC| > log2(1.5), |distanceToTSS| ≤ 1000], 141 of which had decreased accessibility compared with the controls (Fig. 7A and table S10). Enrichment analysis implicated many of the genes associated with these promoters in leukocyte adhesion and activation (Fig. 7B). Confirming our previous findings, we noted reduced accessibility of the IL2 promoter in the patient T lymphoblasts, indicating increased repression of this locus (Fig. 7, A and C). Other promoters with reduced accessibility included genes involved in IL-2 signaling (e.g., STAT5A) and those involved in T cell activation (e.g., IL7R, CCR2, CRTAM, IFNG, TESPA1, CD44, ZAP70, TCF7, LY9, VAV1, CD7, and IKZF2; Fig. 7A and table S10). These data corroborate our findings on reduced IL-2 production caused by HeliosI325V in patient T cells, with an evident failure to open chromatin regions required for T cell activation, which we speculate leads to the observed delay in proliferation. Collectively, these data strengthen a role for Helios in the transcriptional regulation of certain genomic loci downstream of TCR signaling in conventional T cells.
Fig. 7. HeliosI325V leads to global chromatin changes and less accessibility for the IL2 locus upon T cell activation.
(A) Heatmap of significantly differentially accessible regions in controls- and patient-derived T lymphoblast cells upon T cell activation. Highlighted are key genes that have been shown to be involved in IL-2 signaling. Values are scored across rows, z scores (1 indicates that the regions of the corresponding genes are transcriptionally accessible, whereas −1 indicates that the regions are not accessible). (B) Top 25 significantly [Padj < 0.05] enriched GO Biological Process terms for genes from less chromatin accessible regions in patient-derived cells compared with normal donors. (C) Comparison of ATAC-seq signal tracks for IL2 gene locus between patient and two normal donors (ND). Reads per kilobase per million mapped (RPKM) reads normalized tracks.
Discussion
We describe a novel human IEI caused by a homozygous missense mutation in IKZF2, encoding Helios, manifesting with combined immunodeficiency. Studies in murine model systems have shown Helios expression in developing thymocytes and subsets of the T cell lineage, including Tregs, MAIT cells, and activated conventional CD4+ and CD8+ T cells (13, 14). Consistent with this, the index patient in our study had profound T cell defects, evidenced by CD4+ T cell lymphopenia, expansion of terminal effector CD8+ T cells, reduction in Tregs, and absence of invariant T cells (iNKT and MAIT cells). Despite the low expression of Helios in B cells (13, 14), our patient also exhibited a progressive loss of B cells over time, accompanied by a reduction in peripheral memory B cells and plasmablasts, and an increase in transitional B cells, indicative of a B cell maturation defect. Transcriptional analysis of the patient naïve B cells revealed a down-regulation of genes involved in B cell development and differentiation. Given the paucity of peripheral B cells in the patient, it was impossible (and beyond the scope of our study) to investigate in detail how the p.I325V mutation in Helios may affect B cell maturation and/or development and also to explore whether this is an intrinsic phenotype of HeliosI325V-mutant B cells or secondary to aberrant T-B interaction, for instance, TFH cell dysfunction.
The Ikaros family of transcription factors has key roles in hematopoiesis, immune cell development, and homeostasis. The HeliosI325V mutation is positioned between the two zinc finger domains. Consistent with this, the HeliosI325V mutant version does not affect DNA binding or homodimerization and heterodimerization to other Ikaros family members. However, we find that the p.I325V mutation in Helios does play a role in the destabilization of interactions with several binding partners. This is in line with the observation that this region, where the mutation lies, is predicted to contain a nonclassical zinc finger domain (35). Our interaction proteomics analysis expands the network of Helios interactors and identifies alterations in that network caused by the p.I325V mutation. For instance, we show that Helios interacts with cell cycle regulators, such as GTSE1 and PLK1 (59), and with transcription factors and histone modifiers, including known interactors from the NuRD complex and previously unknown interactors, such as the ATAC complex, with histone acetyltransferase activity (60). Similar to other IEIs caused by germline mutations in genes encoding for transcription factors with a key role in regulation of hematopoiesis such as GATA2 (61) or the Ikaros family transcription factor Ikaros itself (27, 62), it is possible that a phenotypic range of Heliosmutant patients with immunodeficiency may depend on the specific mutation and functional domain affected by individual Helios mutations. Future studies identifying other mutations in Helios should therefore aim to define the precise consequences of individual Helios mutations to enable genotype-phenotype correlations.
In Tregs, Helios is responsible to maintain their immunosuppressive functions (22, 23). Helios, together with Foxp3, silences the Il2 locus by promoting epigenetic modifications, in particular through histone deacetylation (57). Consistent with this, we observe an increase in IL-2 production in HeliosI325V Tregs from our patient and a significant decrease in expression of genes involved in epigenetic and chromatin regulation, confirming the role of Helios as an IL2 repressor in Tregs (57). In conventional T cells, the picture is different: HeliosI325V conventional T cells from our patient exhibit a marked decrease in IL-2 production upon stimulation with reduced accessibility to the IL2 locus. We speculate that this reduced accessibility is driven by HeliosI325V, which is in line with the observation that HeliosI325V loses interactions with crucial epigenetic remodelers, such as HDAC1 and HDAC3, as well as TADA2A, KAT2A/KAT2B, and SGF29, members of the Spt–Ada–Gcn5 acetyltransferase (SAGA)–like complexes, including the ATAC complex (60). To date, several lines of evidence support these observations. In the context of T cell activation, Gao et al. (58) have shown that a conditional knockout of KAT2A (or GCN5) in T cells leads to impaired proliferation and reduced IL-2 production upon TCR stimulation, revealing its importance in the acetylation of the Il2 promoter. Conversely, studies have shown that the Il2 promoter is silenced by Ikaros in resting CD4+ T cells to maintain an anergic state, whereas Eos is required for IL-2 production in activated T cells (63–65).
Patient conventional T cells also showed defective proliferation due to a delayed response to TCR stimulation, particularly in CD4+ T cells. Proximal TCR signaling remained largely intact, suggesting that the changes in chromatin accessibility are not exclusive of the IL2 locus but involve a variety of genes, resulting in changes in gene expression. Supporting this idea, our ATAC-seq findings revealed decreased accessibility of genes involved in T cell activation, whereas scRNA-seq analysis showed a dysregulated transcriptome, mainly in the patient expanded pool of CD8+ effector T cells, indicative of their chronically activated and terminal effector state. Together, these findings imply a role of Helios in the regulation of chromatin accessibility to certain loci in activated conventional T cells through interactions with epigenetic remodelers while pointing toward a higher level of regulation of the IL2 promoter that is cell type dependent (57). These mechanistic insights add to our understanding of how Ikaros family members exert their activating or repressive effects on gene transcription and strengthen their role in the regulation of IL-2 production.
In summary, we have identified a novel type of IEI and characterized the deleterious effects of the biallelic mutation in Helios, p.I325V, leading to a dysregulated epigenome and transcriptome in T cells. Our work highlights the importance of conserving proteomic interactions to maintain Helios function. Beyond Tregs and enabled by a point mutation leading to disrupted interactions, we have been able to further corroborate the less well-known role of Helios in conventional T cells, mediating the epigenetic changes required to ensure IL-2 production as well as timely activation and proliferation of T cells. Because of some functional redundancy between Ikaros family members and in addition to the identification of additional Helios-mutant patients mentioned above, more studies targeting specific domains and residues in Helios in both cell lines and animal models (e.g., rather than performing conditional knockouts) will be necessary to comprehensively delineate its role in epigenetic and transcriptional regulation, in the context of its interactions with various multimeric complexes (26).
Materials and Methods
Study design
The objective of this study was to investigate the role of Helios on human immune homeostasis. For this purpose, we performed an array of functional and multiomic experiments on primary material from the patient carrying a germline-encoded homozygous mutation in Helios, after genetic analysis. Furthermore, we made use of cellular models to investigate the effects of the mutation on Helios function using biochemical and proteomic approaches.
Study approval
Samples from the patient, his family members, and other healthy controls were obtained after an informed written consent and approval from the ethics committee of Tehran University of Medical Sciences and the Institutional Review Board of the Medical University of Vienna.
Immunophenotyping and Helios staining
Cryopreserved PBMCs from the patient and healthy controls were thawed and stained with fluorescent antibodies (table S9) for determining frequencies of T, B, and NK cell subpopulations. Helios and FOXP3 staining were performed using the eBioscience Foxp3/ transcription factor staining buffer set (Thermo Fisher Scientific, no. 00552300) according to the manufacturer's protocol. Stained cells were acquired with LSRFortessa or FACSCanto II (BD Biosciences). FlowJo v.10 was used to analyze the data, and Prism v.8 (GraphPad) was used to produce graphs. Gating strategies are shown in figs. S1 to S3, S12, and S16. All adult healthy controls were aged 25 to 40 with representation of both males and females. Two age-matched controls were additionally included in immunophenotyping studies and scRNA-seq.
LightShift chemiluminescent EMSA
To assess the ability of Helios variants to bind to DNA, HEK293T cells [American Type Culture Collection (ATCC), CRL-1573] were transfected with pCMV6-C-Myc-Flag-Helios WT or mutant. All HEK293T transfections were done using the Effectene Transfection Reagent (Qiagen) following the manufacturer's protocol. Gel mobility shift assays were performed with a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific), as previously described (32). IK-bs1 was used for EMSA assay (forward: 5'-BIOTIN-TCAGCTTTTGGGAATACCCTGTCA; reverse: 5'-BIOTIN-TGA-CAGGGTATTCCCAAAAGCTGA).
Immunofluorescence microscopy
To assess the ability of Helios variants to form foci around PC-HC, NIH3T3 cells adherent on glass slides in six-well plates were transfected with pCMV6-C-Myc-Flag-Helios WT or mutant using the X-tremeGENE 9 DNA Transfection Reagent (Sigma-Aldrich), following the manufacturer's protocol. After 20 hours of transfection, cells were fixed with 4% formaldehyde (Thermo Fisher Scientific, no. 28908) for 15 min, permeabilized [0.1% Triton X-100 in phosphate-buffered saline (PBS)] for 10 min, and blocked (0.1% Triton X-100 and 10% fetal bovine serum in PBS) for 30 min at room temperature. Cells were then incubated with Helios antibody (D8W4X; Cell Signaling Technology, 42427S) for 2 hours and stained with rabbit immunoglobulin G–Alexa Fluor 488 secondary antibody (Invitrogen, no. A11034) for 1 hour (both in blocking buffer in the dark) and then with 4',6-diamidino-2-phenylindole (DAPI) (Roth, no. 63351). Coverslips were mounted in Fluorescent Mounting Medium (Dako). An Axio Imager M2-1 microscope (Zeiss) was used to acquire the images with a 40x objective. The number of foci per cell for the WT and mutant was quantified using CellProfiler (https://cellprofiler.org). We used the Speckle Counting CellProfiler Pipeline provided on the website and modified it further to add a filtering step where only transfected cells were analyzed.
Coimmunoprecipitation
The ability of Helios variants to homo/heterodimerize and bind to protein interaction partners was assessed by co-IP studies. HEK293T cells were cotransfected with Strep-HA-Helios WT or mutant, together with the indicated plasmids. After 24 hours, cells were washed with PBS and lysed with IP lysis buffer [50 mM tris (pH 7.5), 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail] for 30 min on ice. Lysates were centrifuged for 10 min at 13,000 rpm; some was taken for input. The remaining lysates were incubated with Strep-Tactin Sepharose 50% suspension (IBA) by rotating at 4°C for 2 hours. Sepharose beads were then washed three times with lysis buffer and resuspended with 1.5× SDS loading buffer. Beads and input were analyzed by running on 8% SDS–polyacrylamide gel electrophoresis and Western blotting with the following primary antibodies: Helios (Cell Signaling Technology, D8W4X), HDAC1 (Abcam, ab7028), HA–horseradish peroxidase (Sigma-Aldrich, HA-7), FLAG (Sigma-Aldrich, F7425), Myc-tag (Cell Signaling Technology, 71D10), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, G-9).
Interaction proteomics
BioID and LC-MS
The method followed has been outlined in detail by Liu et al. (46). In brief, Flip-In T-Rex HEK293 cell lines were cotransfected with C-terminally MAC-tagged Helios-WT or Helios-I325V or green fluorescent protein (GFP) (with nuclear localization signal) together with the pOG44 vector for stable expression. Positive clones were selected and amplified. Tetracycline and biotin induction was done on three biological replicates for 24 hours, after which the samples were harvested. This was followed by affinity purification using a harsh lysis buffer. Lysates were cleared and loaded onto Bio-Rad spin columns with Strep-Tactin beads (IBA), followed by stringent washes and elution. The eluate was then reduced, alkylated, trypsindigested, desalted, and dried as described, followed by reconstitution in 0.1% trifluoroacetic acid and 1% acetonitrile. Analysis of the sample was performed on a Q Exactive mass spectrometer with an EASY-nLC 1000 system via an electrospray ionization sprayer (Thermo Fisher Scientific), using Xcalibur version 3.0.63. Database search was performed with Proteome Discoverer 1.4 (Thermo Fisher Scientific) using the SEQUEST search engine on the reviewed human proteome in UniProtKB/Swiss-Prot databases (www.uniprot.org, downloaded May 2020). All reported data were based on high-confidence peptides assigned in Proteome Discoverer (FDR < 0.05).
Identification of statistical confidence of interactions
SAINTexpress version 3.6.3 and Contaminant Repository for Affinity Purification [CRAPome, www.crapome.org, (66)] were used to discover statistically significant interactions from the LC-MS data. Final results represent proteins with a BFDR score lower than 0.05 and present in all three replicates. MaxQuant version 1.6.10.43 with the Andromeda search engine was used to compare differences in label-free quantitation (LFQ) intensity between WT and I325V samples, two-tailed two-sample equal variance Student's t test was performed, and P values lower than 0.05 were considered significant.
Overrepresentation analysis and visualization
Overrepresentation analysis of statistically significant interactions in GO was done with the R package Enrichr (48), and over-representation of prey proteins in CORUM database (47) was done using Fisher's exact test and multiple testing correction in an in-house R script. Interaction network was made with Cytoscape version 3.8.0.
Proliferation assay and T cell exhaustion
Cryopreserved PBMCs from the patient and controls were assessed for the ability of T cells to proliferate. PBMCs were stained with Violet proliferation dye (VPD) 450 (BD Horizon) as per the manufacturer's protocol and plated on 96-well plates. Cells were then stimulated with Dynabeads Human T-Activator CD3/CD28 (Gibco), anti-CD3 (1 μg/ml; OKT3, eBioscience), and PHA (1 μg/ml) or left unstimulated. On day 3 after stimulation, cells were stained with CD3-, CD4-, and CD8-directed antibodies and proliferation was assessed by flow cytometry using FACSCanto or LSRFortessa (BD Biosciences). For measuring the expression of exhaustion markers upon stimulation with CD3/CD28 Dynabeads, surface staining of T cells was performed on ice using antibodies against PD-1, TIGIT, LAG3, AITR/GITR, and TIM3 (table S9) at the basal state (day 0) and at different time points when cells were harvested (days 2, 4, and 7). Cells were then washed and assessed by flow cytometry.
Intracellular cytokine production and IL-2 release
IL-2 cytokine production was analyzed by stimulating 0.2 × 106 total PBMCs for 5 hours with PMA (0.2 mM) and ionomycin (1 μg/ml) and adding brefeldin A during the final 2.5 hours of the stimulation. Cells were stained for T cell surface markers (CD3, CD4, CD8, CD45RA, and CCR7) on ice for 30 min. Subsequently, cells were fixed, permeabilized, and stained with IL-2, IL-4, IL-17A, and/or IFNγ antibodies (table S7). FACSCanto II (BD Biosciences) was used for flow cytometry analysis. T lymphoblasts were also analyzed in the same way for IL-2 production after 16 hours of PMA/ionomycin stimulation and addition of brefeldin A during the final 4 hours. Surface staining was done with CD3, CD4, and CD8. LSRFortessa (BD Bio-sciences) was used for acquisition. Gating strategies are shown in the supplementary figures, and positive cytokine gates were set using the unstimulated controls. IL-2 release was assessed after harvesting cell-free supernatant after 24-hour stimulation of 0.1 × 106 PBMCs on 96-well U-bottom plates with CD3/CD28 Dynabeads using the Luminex 200 system (R&D Systems, following the manufacturer's protocol).
Generation of CRISPR-Cas9–edited patient T lymphoblasts with WT IKZF2
One million patient-derived PBMCs were stimulated on an α-CD3–coated well of a 24-well plate in T cell medium [RPMI medium (Gibco) supplemented with 5% human serum, Hepes (20 mM), penicillin/streptomycin (10 μg/ml), sodium pyruvate (1 mM), and nonessential amino acids (1x; all four from Gibco) with IL-2 (100 IU/ml)]. After 66 hours, cells were harvested and transfected using a NEPA21 electroporator with recombinant Cas9 nuclease (IDT), a single-guide RNA (sgRNA) including the patient variant "G" shown underlined (AGTCACCTACCTTGGAGCTG), and the following single-stranded oligo donor (synthesized by IDT as Ultramer DNA oligo) with the WT variant "A" shown underlined and the silent PAM change "AGG>AAG" in bold:
AGGCTGAGCTGATGCAGTCTCATATGATGGACCAAGCCATCAACAATGCAATCACCTACCTTGGAGCTGAAGCCCTTCACCCTCTGATGCAGCACCCGCC.
After transfection, cells were left to expand in T cell medium with IL-2 for a week. This was followed by single-cell cloning of CRISPR-edited cells into a 384-well plate in T cell medium with feeder cells (irradiated PBMCs from a blood donor), PHA (2 μg/ml), and IL-2 (200 IU/ml). After 10 days, clones were selected and reexpanded on a 96-well plate using feeder cells, PHA, and IL-2, after which clones were genotyped by Sanger sequencing using the same variant validation primers above.
Single-cell RNA sequencing
Frozen PBMCs from the patient and four controls were thawed, washed in RPMI medium, and resuspended in sterile PBS with 0.04% bovine serum albumin. A single-cell suspension was obtained by passing 1 million cells into a 5-ml fluorescence-activated cell sorting (FACS) tube through a cell strainer and sorting for the live lymphocytes and monocytes based on the forward and side scatter using the FACSAria Fusion (BD Biosciences). scRNA-seq was then performed on the live samples using the 10x Genomics Chromium Controller with the Chromium Single Cell 3' Reagent Kit (v3 chemistry) following the manufacturer's instructions. After quality control, libraries were sequenced on the Illumina HiSeq 4000 platform in 2 × 75–base pair (bp) paired-end mode. Table S4 includes an overview of sequencing data and performance metrics. We used the Cell Ranger v3.0.2 software (10x Genomics) for demultiplexing and alignment to GRCh38 human reference transcriptome. The R statistics software was used to analyze the processed data. For optimal reproducibility, we used a Docker container containing R and all dependent libraries preinstalled (cancerbits/dockR:mo-ikzf2-v1 available from https://hub.docker.com/r/cancerbits/dockr). Briefly, Cell Ranger outputs were loaded into R to perform quality control {removing cells with less than 1000 genes, mitochondrial content more than 10%, or log10 doublet score > 3 [calculated using the function doubletCells from scran (66) v1.14.6]} and downstream analysis using Seurat v3.1.5 (68). Each dataset was normalized [function SCTransform (69)] to generate corrected, log-transformed counts, and Pearson residuals by regressing out mitochondrial content. We then used these normalized data to integrate all datasets using Harmony v1.0 (70). Integrated data were used for low-dimensional projection using UMAP (49) based on the top 30 principal components and for clustering cells (resolution, 0.2). DGE analysis for this rare disease dataset suffers from lack of replication. In an attempt to reduce the impact of interindividual differences (which would confound comparisons with variable number of cells from different individuals on H both sides of a comparison), we used a stratified comparison scheme in which we performed comparisons per pair of individuals and focused on consistent differences across all pairwise comparisons. Specifically, statistics from sample-wise DGE tests were combined using the minimum logFC ("worst fold change") and Fisher's method for P values. Ribosomal proteins and cluster-sample strata with less than 50 cells were excluded from DGE analysis. We used MAST as a statistical test with FDR-adjusted P value threshold of Padj < 0.05 and |logFC| ≥ 0.25. Genes identified with the same direction of change in 75% of comparisons for between-cluster comparisons and 100% of comparisons for within-cluster comparisons were recorded as differentially expressed genes (table S6).
Supplementary Material
Acknowledgments
We thank the patient and his family members for their participation in our study. We also thank the Biomedical Sequencing Facility [BSF; at the CeMM Research Center of Molecular Medicine (CeMM) of the Austrian Academy of Sciences] for their scRNA-seq service.
Funding
This study has been supported by the European Research Council Consolidator Grant iDysChart (ERC, grant agreement no. 820074) (to K.B.); a doctoral fellowship program [Cell Communication in Health and Disease (CCHD) from the Medical University of Vienna] (to K.B. and T.S.); the Austrian Science Fund FWF project T934-B30 (to B.H.); the Intramural Research Program, NIH Clinical Center, U.S. National Institutes of Health (NIH) (to S.D.R.); and the Academy of Finland (nos. 288475 and 294173), the Sigrid Juselius Foundation, the Finnish Cancer Institute, and Biocenter Finland (to M.V.).
Footnotes
Author contributions:
T.S. performed most of the experiments on patient material, analyzed, and interpreted the data. T.S., H.K., B.H., and S.G. performed the biochemical experiments. T.S. supervised S.G. M.R.S. analyzed the scRNA-seq and ATAC-seq data under the supervision of F.H. P.R. analyzed SMART-seq data. T.S. interpreted the scRNA-seq results, with input from S.Z. L.G. performed the BioID experiments and processed the mass spectrometry data. M.V. provided the laboratory resources for the BioID experiments and supervised L.G. O.Y.P. did the immunofluorescence stainings and analyzed the data on Cell Profiler. N.R. and S.Z. identified the index patient in Iran and provided clinical and immunological data. S.K.B. assisted in clinical and immunological data interpretation. J.D. identified the IKZF2 mutation; S.Z. performed the genetic segregation analysis. D.S. provided additional interpretation of scientific data. T.S., I.C., and K.B. wrote the manuscript with input from all co-authors. S.D.R. and H.K. provided the laboratory resources for some experiments and gave critical intellectual input into the study. K.B. conceptualized and coordinated the study, provided the laboratory resources, was involved in data interpretation and took overall responsibility of the study.
Competing interests:
The authors declare that they have no competing interests.
Data and materials availability
Raw and processed sequencing data for scRNA-seq, bulk RNA-seq, and ATAC-seq have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession no. EGAS00001005675. R codes used for the analysis of scRNA-seq, bulk RNA-seq, and ATAC-seq are available on GitHub: https://github.com/cancerbits/shahin2021_ikzf2. The dataset from the BioID LC-MS analysis is available in the MassIVE repository under the identifier MSV000088165.
References
- 1.John LB, Ward AC. The Ikaros gene family: Transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48:1272–1278. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Heizmann B, Kastner P, Chan S. The Ikaros family in lymphocyte development. Curr Opin Immunol. 2018;51:14–23. doi: 10.1016/j.coi.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Fan Y, Lu D. The Ikaros family of zinc-finger proteins. Acta Pharm Sin B. 2016;6:513–521. doi: 10.1016/j.apsb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgopoulos K. The making of a lymphocyte: The choice among disparate cell fates and the IKAROS enigma. Genes Dev. 2017;31:439–450. doi: 10.1101/gad.297002.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 6.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 7.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, Andrikopoulos K, Liang L, Pillai S, Morgan BA, Georgopoulos K. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 9.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30:4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinmann E, Geimer Le Lay A-S, Sellars M, Kastner P, Chan S. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol. 2008;28:7465–7475. doi: 10.1128/MCB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 13.Kelley M, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 14.Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serre K, Benezech C, Desanti G, Bobat S, Toellner K-M, Bird R, Chan S, Kastner P, Cunningham AF, Maclennan ICM, Mohr E. Helios is associated with CD4 T cells differentiating to T helper 2 and follicular helper T cells in vivo independently of Foxp3 expression. PLOS ONE. 2011;6:e20731. doi: 10.1371/journal.pone.0020731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen H-R, Bruno TC, Durham NM, Hipkiss L, Pyle KJ, Wada S, Pan F, et al. A role for the transcription factor Helios in human CD4+CD25+ regulatory T cells. Mol Immunol. 2010;47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leeansyah E, Svärd J, Dias J, Buggert M, Nyström J, Quigley MF, Moll M, Sönnerborg A, Nowak P, Sandberg JK. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLOS Pathog. 2015;11:e1005072. doi: 10.1371/journal.ppat.1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias J, Boulouis C, Gorin J-B, van den Biggelaar RHGA, Lal KG, Gibbs A, Loh L, Gulam MY, Sia WR, Bari S, Hwang WYK, et al. The CD4− CD8− MAIT cell subpopulation is a functionally distinct subset developmentally related to the main CD8 + MAIT cell pool. Proc Natl Acad Sci USA. 2018;115:E11513–E11522. doi: 10.1073/pnas.1812273115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLOS ONE. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced FOXP3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornton AM, Lu J, Korty PE, Kim YC, Martens C, Sun PD, Shevach EM. Helios+ and Helios− Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur J Immunol. 2019;49:398–412. doi: 10.1002/eji.201847935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H-J, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TAW, Chan S, Kastner P, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM. Helios controls a limited subset of regulatory T cell functions. J Immunol. 2016;196:144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross EM, Bourges D, Hogan TV, Gleeson PA, van Driel IR. Helios defines T cells being driven to tolerance in the periphery and thymus. Eur J Immunol. 2014;44:2048–2058. doi: 10.1002/eji.201343999. [DOI] [PubMed] [Google Scholar]
- 25.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183:2303–2311. doi: 10.4049/jimmunol.0901407. [DOI] [PubMed] [Google Scholar]
- 26.Thornton AM, Shevach EM. Helios: Still behind the clouds. Immunology. 2019;158:161–170. doi: 10.1111/imm.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand W, Maffucci P, Pierce KR, Abbott JK, Voelkerding KV, South ST, et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374:1032–1043. doi: 10.1056/NEJMoa1512234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogaert DJ, Kuehn HS, Bonroy C, Calvo KR, Dehoorne J, Vanlander AV, De Bruyne M, Cytlak U, Bigley V, De Baets F, De Baere E, et al. A novel IKAROS haploinsufficiency kindred with unexpectedly late and variable B-cell maturation defects. J Allergy Clin Immunol. 2018;141:432–435.:e7. doi: 10.1016/j.jaci.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eskandarian Z, Fliegauf M, Bulashevska A, Proietti M, Hague R, Smulski CR, Schubert D, Warnatz K, Grimbacher B. Assessing the functional relevance of variants in the IKAROS Family Zinc Finger Protein I (IKZFI) in a cohort of patients with primary immunodeficiency. Front Immunol. 2019;10:568. doi: 10.3389/fimmu.2019.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunes-Santos CJ, Kuehn HS, Rosenzweig SD. IKAROS family zinc finger 1-associated diseases in primary immunodeficiency patients. Immunol Allergy Clin North Am. 2020;40:461–470. doi: 10.1016/j.iac.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehn HS, Niemela JE, Stoddard J, Mannurita SCiullini, Shahin T, Goel S, Hintermeyer M, Heredi RJimenez, Garofalo M, Lucas L, Singh S, et al. Germline IKAROS dimerization haploinsufficiency causes hematologic cytopenias and malignancies. Blood. 2021;137:349–363. doi: 10.1182/blood.2020007292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, Lenoir C, Barlogis V, Farnarier C, Vely F, Yoshida N, et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest. 2018;128:3071–3087. doi: 10.1172/JCI98164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita M, Kuehn HS, Okuyama K, Okada S, Inoue Y, Mitsuiki N, Imai K, Takagi M, Kanegane H, Takeuchi M, Shimojo N, et al. A variant in human AIOLOS impairs adaptive immunity by interfering with IKAROS. Nat Immunol. 2021;22:893–903. doi: 10.1038/s41590-021-00951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuehn HS, Chang J, Yamashita M, Niemela JE, Zou C, Okuyama K, Harada J, Stoddard JL, Nunes-Santos CJ, Boast B, Baxter RM, et al. T and B cell abnormalities, pneumocystis pneumonia, and chronic lymphocytic leukemia associated with an AIOLOS defect in patients. J Exp Med. 2021;218:e20211118. doi: 10.1084/jem.20211118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis TE, Sillitoe I, Andreeva A, Blundell TL, Buchan DWA, Chothia C, Cozzetto D, Dana JM, Filippis I, Gough J, Jones DT, et al. Genome3D: Exploiting structure to help users understand their sequences. Nucleic Acids Res. 2015;43:D382–D386. doi: 10.1093/nar/gku973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews JM, Sunde M. Zinc fingers—Folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 37.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 August 13;:531210. doi: 10.1101/531210. [Preprint] [DOI] [Google Scholar]
- 38.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorarinsdottir K, Camponeschi A, Gjertsson I, Martensson IL. CD21-/low B cells: A snapshot of a unique B cell subset in health and disease. Scand J Immunol. 2015;82:254–261. doi: 10.1111/sji.12339. [DOI] [PubMed] [Google Scholar]
- 40.Strioga M, Pasukoniene V, Characiejus D. CD8+ CD28− and CD8+ CD57+ T cells and their role in health and disease. Immunology. 2011;134:17–32. doi: 10.1111/j.1365-2567.2011.03470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hetemäki I, Kaustio M, Kinnunen M, Heikkilä N, Keskitalo S, Nowlan K, Miettinen S, Sarkkinen J, Glumoff V, Andersson N, Kettunen K, et al. Loss-of-function mutation in IKZF2 leads to immunodeficiency with dysregulated germinal center reactions and reduction of MAIT cells. Sci Immunol. 2021;6:eabe3454. doi: 10.1126/sciimmunol.abe3454. [DOI] [PubMed] [Google Scholar]
- 42.Sun L, Kerawalla H, Wu X, Lehnert MS, Uckun FM. Expression of a unique helios isoform in human leukemia cells. Leuk Lymphoma. 2002;43:841–849. doi: 10.1080/10428190290016980. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: Switching out of repression. Mol Cell Biol. 2005;25:2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 45.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Salokas K, Tamene F, Jiu Y, Weldatsadik RG, Ohman T, Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A. CORUM: The comprehensive resource of mammalian protein complexes—2019. Nucleic Acids Res. 2018;47:D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McInnes L, Healy J, Melville J. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv: 1802.03426 [stat.ML] 2018 February 9; [Google Scholar]
- 50.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seternes OM, Kidger AM, Keyse SM. Dual-specificity MAP kinase phosphatases in health and disease. Biochim Biophys Acta Mol Cell Res. 2019;1866:124–143. doi: 10.1016/j.bbamcr.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 a to the endoplasmic reticulum and promotes dephosphorylation of the a subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Marco B, Massetti M, Bruscoli S, Macchiarulo A, Di Virgilio R, Velardi E, Donato V, Migliorati G, Riccardi C. Glucocorticoid-induced leucine zipper (GILZ)/NF -kB interaction: Role of GILZ homo-dimerization and C-terminal domain. Nucleic Acids Res. 2007;35:517–528. doi: 10.1093/nar/gkl1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asselin-Labat LM, David M, Biola-Vidamment A, Lecoeuche D, Zennaro MC, Bertoglio J, Pallardy M. GILZ, a new target for the transcription factor FoxO3, protects T lymphocytes from interleukin-2 withdrawal-induced apoptosis. Blood. 2004;104:215–223. doi: 10.1182/blood-2003-12-4295. [DOI] [PubMed] [Google Scholar]
- 55.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 56.Birzele F, Fauti T, Stahl H, Lenter MC, Simon E, Knebel D, Weith A, Hildebrandt T, Mennerich D. Next-generation insights into regulatory T cells: Expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res. 2011;39:7946–7960. doi: 10.1093/nar/gkr444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baine I, Basu S, Ames R, Sellers RS, Macian F. Helios induces epigenetic silencing of Il2 gene expression in regulatory T cells. J Immunol. 2013;190:1008–1016. doi: 10.4049/jimmunol.1200792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao B, Kong Q, Zhang Y, Yun C, Dent SYR, Song J, Zhang DD, Wang Y, Li X, Fang D. The histone acetyltransferase Gcn5 positively regulates T cell activation. J Immunol. 2017;198:3927–3938. doi: 10.4049/jimmunol.1600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: Structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 60.Spedale G, Timmers HTM. Pijnappel WWMP: ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chong CE, Venugopal P, Stokes PH, Lee YK, Brautigan PJ, Yeung DTO, Babic M, Engler A, Lane SW, Klingler-Hoffmann M, Matthews JM, et al. Differential effects on gene transcription and hematopoietic differentiation correlate with GATA2 mutant disease phenotypes. Leukemia. 2018;32(1):194–202. doi: 10.1038/leu.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, Yamashita M, Okano T, Tsumura M, Nishimura S, Sakata S, et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol. 2016;140:223–231. doi: 10.1016/j.jaci.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 63.Thomas RM, Chunder N, Chen C, Umetsu SE, Winandy S, Wells AD. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4+ T lymphocytes. J Immunol. 2007;179:7305–7315. doi: 10.4049/jimmunol.179.11.7305. [DOI] [PubMed] [Google Scholar]
- 64.Bandyopadhyay S, Dure M, Paroder M, Soto-Nieves N, Puga I, Macian F. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells. Blood. 2007;109:2878–2886. doi: 10.1182/blood-2006-07-037754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rieder SA, Metidji A, Glass DD, Thornton AM, Ikeda T, Morgan BA, Shevach EM. Eos is redundant for regulatory T cell function but plays an important role in IL-2 and Th17 production by CD4+ conventional T cells. J Immunol. 2015;195:553–563. doi: 10.4049/jimmunol.1500627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, et al. The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-r, Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salzer E, Cagdas D, Hons M, Mace EM, Garncarz W, Petronczki ÖY, Platzer R, Pfajfer L, Bilic I, Ban SA, Willmann KL, et al. RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat Immunol. 2016;17:1352–1360. doi: 10.1038/ni.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Müller H, Jimenez-Heredia R, Krolo A, Hirschmugl T, Dmytrus J, Boztug K, Bock C. VCF.Filter: Interactive prioritization of disease-linked genetic variants from sequencing data. Nucleic Acids Res. 2017;45:W567–W572. doi: 10.1093/nar/gkx425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 77.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma'ayan A. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith JP, Corces MR, Xu J, Reuter VP, Chang HY, Sheffield NC. PEPATAC: An optimized ATAC-seq pipeline with serial alignments. bioRxiv. 2020 October 22;:2020.10.21.347054. doi: 10.1101/2020.10.21.347054. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2021. https://www.r-project.org/ [Google Scholar]
- 80.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, Francois R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, et al. Welcome to the tidyverse. Open J Source Softw. 2019;4:1686. [Google Scholar]
- 81.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan M, Carey VJ. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carroll TS, Liang Z, Salama R, Stark R, de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet. 2014;5:75. doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: Identification of problematic regions of the genome. Sci Rep. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.B. C. Team, B. P. Maintainer. TxDb.Hsapiens.UCSC.hg38.knownGene Annotation package for TxDb object(s) 2021.
- 85.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, Santana-Garcia W, et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2019;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan G, Lenhard B. TFBSTools: An R/Bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schep A. motifmatchr: Fast Motif Matching in R. p. 2021.
- 90.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gel B, Serra E. karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khan K, Zech M, Morgan AT, Amor DJ, Skorvanek M, Khan TN, Hildebrand MS, Jackson VE, Scerri TS, Coleman M, Rigbye KA, et al. Recessive variants in ZNF142 cause a complex neurodevelopmental disorder with intellectual disability, speech impairment, seizures, and dystonia. Genet Med. 2019;21:2532–2542. doi: 10.1038/s41436-019-0523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data for scRNA-seq, bulk RNA-seq, and ATAC-seq have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession no. EGAS00001005675. R codes used for the analysis of scRNA-seq, bulk RNA-seq, and ATAC-seq are available on GitHub: https://github.com/cancerbits/shahin2021_ikzf2. The dataset from the BioID LC-MS analysis is available in the MassIVE repository under the identifier MSV000088165.