Abstract
The major hallmark of Alzheimer’s disease is the deposition of plaques of amyloid fibrils formed from amyloid-β (Aβ) peptides. Kinetic studies have contributed significantly towards a mechanistic understanding of amyloid fibril self-assembly, however dynamic features of the aggregation process cannot be captured using ensemble methods. Here we present an assay for imaging Aβ42 aggregation dynamics at the single fibril level, allowing for the quantitative extraction of concentration and temperature dependent kinetic parameters. From direct observation of elongation using TIRF and super-resolution optical microscopy, we find that Aβ42 fibril growth is strongly polarized, with fast and slow growing ends arising from different elongation rates, but also from a growth incompetent state, which dominates the process at the slow growing end. Our findings reveal the surprising complexity of the Aβ42 fibril elongation reaction at the microscopic level.
Introduction
The self-assembly of misfolded proteins into amyloid fibrils is an important event in the progression and pathology of increasingly prevalent neurodegenerative disorders such as Alzheimer’s disease (AD).1 While plaques resulting from the accumulation of fibrillar aggregates of amyloid beta (Aβ) peptides are a hallmark of this disease, the precise identity of the toxic species and the origin of their cytotoxicity have yet to be fully resolved.2 The two main Aβ isoforms involved are Aβ40 and Aβ42, with the latter being more aggregation prone3 and exhibiting greater pathogenicity.4 Intensive efforts have been made to solve the high-resolution structure of Aβ42 fibrils,5–9 providing insight into the origin of their mechanical rigidity10 and degradation resistance. Such models have also helped elucidate their prion-like propagation properties and cross-seeding propensities, which have been of particular research interest recently.7,11 Research into the chemical kinetics of aggregation, on the other hand, has shed new light on the pathways involved in the conversion of soluble Aβ42 monomers to oligomers and fibrils and is of major relevance for understanding and delineating the pathobiology of AD.12 Recent progress in this latter field has hinged on the development of highly reproducible kinetic assays13 and sophisticated theoretical models.14,15
Kinetic studies are typically performed using ensemble methods such as surface plasmon resonance (SPR),16 quartz crystal microbalances (QCM),17 or, most widely, fluorometric assays employing the amyloid fibril specific dye thioflavin T (ThT). Such macroscopic ensemble methods generally yield combined rate parameters which represent a convolution of multiple phenomena including nucleation, dissociation, elongation, and secondary processes.14 Obtaining information on the elementary microscopic reaction steps requires a deconvolution of these processes, which is challenging and subject to considerable uncertainty. The development of new methods to study the aggregation process at the level of individual molecules and fibrils is therefore highly desirable. Current in vitro methods generally fall into two classes, ex situ and in situ. Ex situ, amyloid fibril elongation rates have been inferred from length distributions obtained via atomic force microscopy (AFM) or electron microscopy18–20 at discrete time points. In situ measurements of elongation reactions in real-time have also been demonstrated using AFM.21–23 In situ AFM provides nanometre resolution of aggregate morphology, but its major disadvantages stem from the catalytic effects of high surface energy mica substrates and tip-sample interactions.22,24 Total internal reflection fluorescence microscopy (TIRFM) enables real-time observation under less perturbing conditions, and has provided unique insights into the dynamic nature of fibril growth such as fibril polarity and discontinuous, or stop- and-go, growth.25–27 TIRFM assays have however so far yielded largely qualitative data. Although Aβ42 fibrils are the predominant form of aggregates in AD plaques, the kinetics of single fibrils of this isoform have not so far been studied in detail experimentally.
In our previous work, we have established fluorescent labelling and super-resolution imaging strategies that enabled us to investigate the templated growth of fibrils of α-synuclein, a protein central to Parkinson’s disease, from preformed seeds.28,29 Leading on from this work, here we adapt single molecule imaging methods which have been used in other contexts to study cytoskeletal biopolymer growth kinetics.30,31 We establish an assay to observe the rapid growth of Aβ42 fibrils directly and in real time, under minimally perturbing and reproducible conditions. We observe asymmetrical growth from seeds thus revealing an intrinsic structural difference between the two ends of an Aβ42 fibril. From concentration and temperature dependent measurements of individual fibrils we observe that the Aβ42 elongation reaction exhibits Michaelis–Menten type behaviour, and conversion of the monomer from the soluble to fibrillar state is separated by an enthalpic energy barrier. Intermittent, or stop-and-go, growth is also observed which we attribute to a long-lived kinetically trapped state, blocking the further addition of monomer at the fibril end.
Results and discussion
TIRF microscopy reveals the dynamics of Aβ42 fibril elongation
To enable the quantitative observation of Aβ42 fibril growth dynamics, we developed an elongation assay in which preformed seed fibrils are tethered above the glass surface of a passivated flow cell followed by addition of monomeric peptide (Fig. 1a, see Methods for details). While ThT fluorescence has previously been used to image single fibril growth,32 labelling the peptide covalently with an extrinsic fluorophore allowed us to unambiguously distinguish new growth from the seed fibril, and also to perform super-resolution microscopy.33 To minimize the effect of the fluorophore on the fibril structure and aggregation kinetics, we used a low fraction of Aβ42 peptide labelled at the N-terminus relative to unlabelled Aβ42. Short seed fibrils, fluorescently labelled with HiLyte Fluor 647, were immobilised above a glass coverslip by anti-Aβ42 antibodies. Seeds were formed from a mixture of 1 : 19 labelled : unlabelled Aβ42 monomers (i.e. a 5% label fraction). We verified via electron microscopy that addition of this small fraction of fluorophore labelled monomers led to the formation of fibrils with an indistinguishable morphology compared to unlabelled fibrils (Fig. S1, ESI†). Furthermore, we ensured that at these labelling ratios, the rates of nucleation and fibril elongation were equivalent to those observed for the unlabelled peptide using ThT assays at concentrations similar to those used in the present experiments (see Fig. S9, ESI†). The rapid aggregation kinetics of Aβ42 in the presence of interfaces13,34 means that passivation of the glass coverslip was essential to prevent non-specific monomer adsorption onto the surface. The passivation layer prevented de novo fibril nucleation and substantially reduced fluorescence background. After tethering and passivation, monomeric Aβ42, of which 5% was labelled with HiLyte Fluor 488, was added and the surface was imaged every 1–2 minutes via two colour TIRF microscopy.
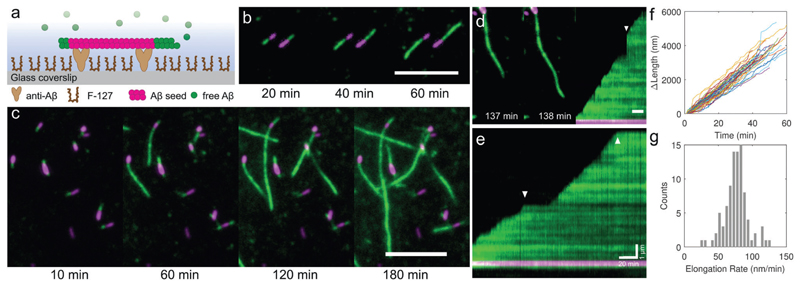
Fig. 1. A two colour TIRF elongation assay enables in situ imaging of the dynamics ofAβ42 fibril elongation from preformed seeds.
(a) Schematic of the TIRF imaging assay. Labelled seed fibrils (magenta, 5% HiLyte Fluor 647) were tethered to a coverslip via N-terminal specific anti-Aβ and the glass was passivated with Pluronic F-127 to reduce monomer adsorption and primary nucleation. Monomeric Aβ42 (green, 5% HiLyte Fluor 488, 10 μM total concentration) was added and the surface imaged over time at 37 °C using TIRF microscopy. (b and c) In situ observation of amyloid elongation dynamics from seeds. TIRF images show that the growth rates are different for each fibril end. Scale bars 5 μm. (d) Some fibrils became detached from the surface but continued to elongate, followed by their reattachment to the surface (arrow in right hand kymograph). (e) Kymographs show that growth from the fast end of the fibril is continuous and interrupted by pauses (arrows). (f) Change in length of an ensemble of fibrils over time, with paused sections removed (N = 59 fibril traces). (g) Histogram of the pause-free fast end elongation rates, 76.2 ± 17.9 nm min−1 (mean ± S.D.), for 10 μM monomeric Aβ42 at 37 °C.
TIRF microscopy provided excellent image contrast, allowing us to visualize the elongation of seeds through monomer addition over time (Fig. 1b and c, see the ESI† for videos). Background signal from the monomer solution was effectively suppressed since in TIRF the illumination region is confined to a volume extending only ~100 nm above the coverslip. An analysis of ensembles of seed fibrils revealed that the initiation of growth at each end is stochastic and that individual fibrils exhibit different growth rates. Occasionally fibrils became partially detached from the coverslip surface because they were only weakly tethered by the antibodies. They continued growing out of the plane of the coverslip and were therefore no longer excited by the evanescent illumination field (Fig. 1d). Rapid jumps in fibril length were consequently seen in the time-lapse videos that could be misinterpreted as sudden rapid increases in growth rates, but instead signified the recapture of the free fibril ends by the surface antibodies. Kymograph projections permit a clearer visualization of fibril growth dynamics and revealed that growth clearly continued at a constant slope, and hence rate, before and after reattachment (Fig. 1d, arrow on the right hand panel). Other intermittent events cannot be explained by fibril end detachment. In Fig. 1e, a representative example is shown wherein a fibril is seen to elongate at a constant rate until interrupted by a pause, before growth resumes again. During growth, the kymographs reveal elongation rates that appear constant and hence that the pool of Aβ42 monomers was not depleted significantly over the duration of the experiment. By fitting an error function to the fluorescence intensity of the growing fibril end (see ESI† and Fig. S2), we could determine the length of new growth from each seed fibril (Fig. 1f), and thus the distribution of elongation rates during the growth periods for an ensemble of fibrils (Fig. 1g). Performing such analyses over multiple fibrils yielded distributions of pause-free elongation rates, final fibril lengths, and the durations of the growth and pause phases (Fig. S3, ESI†). The excellent reproducibility from run to run is demonstrated in Fig. S4 (ESI†); the assay thus accurately reports on the effect of changing conditions on fibril elongation. We checked that the intermittency of growth and asymmetric elongation were not artefacts introduced by faults in the solid phase synthesis of the peptide, and saw these phenomena also in experiments performed with recombinantly produced Aβ42 (Fig. S8 and Supporting Methods, ESI†).
Aβ42 fibril growth is strongly polarized
TIRF imaging revealed that seed fibrils elongate asymmetrically, with some exhibiting unipolar (Fig. 1b) and others bipolar growth (Fig. 1c). All fibrils clearly featured both a fast and a slow, or non-growing, end. We could exclude flow in the cell as a contributor to polarised growth, since we found no correlation between the alignment of slow or fast end growth with the direction of the flow. For 10 μM monomeric Aβ42, the pause-free elongation rate at the fast-growing end was 76.2 ± 17.9 nm min−1 (mean ± S.D.) and the approximate slow end rate was between 10 and 20 nm min−1. A significant proportion of seeds (30%) did not elongate appreciably at all over the duration of the measurement, an observation made previously for seeded α-synuclein growth,28 and this is likely caused by a growth incompetent conformation at the fibril ends.
It has been extensively documented that Aβ40 and Aβ42 fibrils elongate asymmetrically,22,32 due to a difference in the structure of each end. Solid state NMR data show that regardless of the number of subunits that make up the fibril, interdigitation of the adjacent amino acid side chains supports a staggering of the β-strand alignment, leading to different exposed residues at either end.7,35,36 Simulations of Aβ fibril elongation have shown that the N-terminal solvent exposed end, termed the even end, supports a higher rate of monomer addition compared to the C-terminal exposed, or odd, end.37–39 Elongation of the odd end is kinetically unfavourable compared to the even end, resulting in the fibril growth asymmetry that we observe.
Elongation assays allow measurement of fast end kinetics
The reproducibility of the elongation assay allowed us to further investigate the kinetics of the fast-growing fibril end. The absolute number of molecules that associate with the growing end of each fibril per unit time can be extracted from the fibril length traces. The advantage of this approach is that it becomes possible to measure the elongation rate without the requirement to estimate the number density of seed fibrils, which greatly affects the quantification of kinetics from bulk assays.40,41 As we have observed (Fig. 1c), many seed fibrils do not elongate at all over the course of the measurement; here we present data only from those fibrils which featured growth-competent ends. From electron microscopy, we found that seed fibrils were composed of two protofilaments with a crossover distance of around 110 to 130 nm (Fig. S1, ESI†). Mass-per-length (MPL) measurements indicated that Aβ42 fibrils with a comparable crossover distance were composed of 2.44 peptides per cross-β repeat (0.47 nm).6 By assuming that the fibrils elongate only by monomer addition, we could therefore convert the pause-free elongation rate E to an absolute elongation rate by r = 2.44E/0.47 subunits per second. The fast end elongation rates we observed ranged between 2 and 15 subunits s−1.
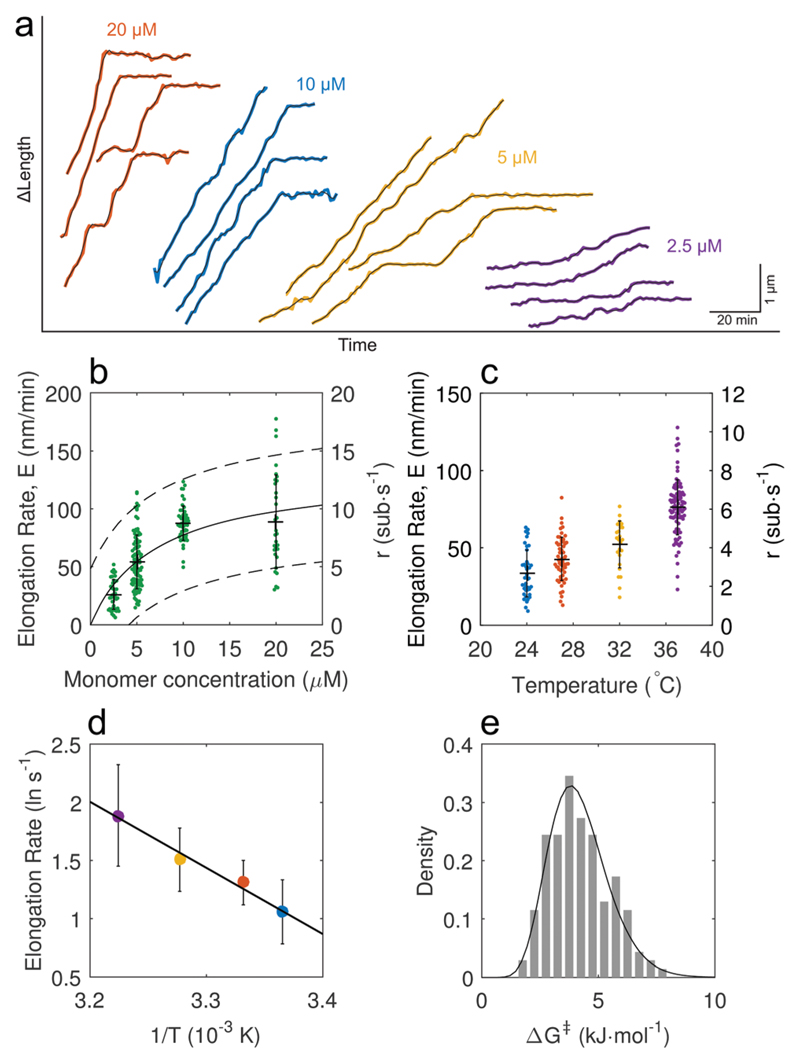
We first investigated the dependence of the elongation rate on the soluble monomer concentration (Fig. 2a), and observed a linear dependence at low concentration, followed by saturation at higher concentrations (Fig. 2b). The sublinear dependence on concentration seen here for Aβ42 is similar to that observed previously for other amyloids, including S6,42 sup35,43 and recently alpha-synuclein.44 The observed saturation is reminiscent of enzyme kinetics, where the fibril ends are analogous to the enzyme active site, while monomers play the role of the substrate.45 We thus described the concentration dependent elongation rate by a two-step Michaelis–Menten kinetic model, where m is the monomer concentration. From a fit of our data to this model we obtained a value of m1/2 = 7.2 ± 2.4 μM, the monomer concentration at half the maximal rate rmax. From a linear regression of the elongation rate in the non-saturating regime, we obtained the concentration dependent elongation rate constant k+ = (9.3 ± 1.5) × 105 M−1 s−1. This value is in the same range as earlier bulk measurements, from which k+ was estimated to be of order 3 × 106 M−1 s−1, although the latter was obtained via estimation rather than direct measurement.12 Elongation rate saturation at high monomer concentration supports the proposed two-step dock-lock process for Aβ42 fibril growth.39,46,47 In this process, elongation is described by two elementary steps: a monomer first binds reversibly to the fibril end (dock) followed by a slower conformational transition to the ordered β-sheet fibril structure (lock). The observed rate saturation is therefore a consequence of this rate limiting locking step. Extending the analogy of enzyme kinetics further, we hypothesize that if the fibril is composed of multiple subunits,6 this would result in multiple binding sites for monomers at the fibril end. The rapid elongation kinetics we observe could therefore be a consequence of cooperative binding, where the addition of one monomer at the fibril end could increase the affinity for binding of a second monomer.
Fig. 2. A quantitative analysis of single Aβ42 fibril elongation assays allows the extraction of kinetic parameters.
(a) Examples of length traces obtained from fast growing fibril ends. The elongation rate dependence on monomer concentration is clearly visible, (b) the rate of pause-free elongation has a linear concentration dependence initially and saturates at high monomer concentration. Fitting the data points to a Michaelis–Menten model yields the solid line; broken lines indicate the 95% confidence bounds of the non-linear fit (error bars denote mean ± S.D.). m1/2 = 7.2 ± 2.4 μM, maximum elongation rate Emax = 133 ± 20 nm min−1. Data points were combined from two independent repeats. N = 25, 40, 40, 18 for 2.5, 5, 10, 20 μM respectively. (c) Temperature dependence of the elongation rate at the fast end (error bars denote mean ± S.D.). N = 18, 33, 13, 32 for 24 to 37 °C at 10 μM monomer concentration. (d) The temperature dependence of the fast end elongation rate shows Arrhenius behaviour and a linear fit yields the enthalpy of activation ΔH‡ = 47 ± 17 kJ mol−1 (R2 = 0.987, ±95% CI). Error bars denote the mean ± S.D. from fits of normal distributions to the CDFs. (e) Free energy of activation ΔG‡ distribution for an ensemble of fibrils calculated from elongation rates obtained for monomer concentrations below 10 μM.
Next, we performed temperature dependent elongation rate measurements (Fig. 2c) for individual fibrils, which were observed to exhibit Arrhenius behaviour (Fig. 2d). We were thus able to obtain information on the free-energy barrier that must be overcome for monomer to fibril association via determination of the enthalpy of activation, ΔH‡, using the model presented by Buell et al.41 (see the ESI†). By measuring the temperature dependence of the mean pause-free elongation rate obtained from four separate elongation assays, we plotted an Arrhenius plot (Fig. 2d) and obtained an activation enthalpy of 47 ± 17 kJ mol−1 (R2 = 0.987, ±95% CI). We also calculated the free energy of activation ΔG‡ to obtain an estimate of the entropy of activation (see the ESI†). Computing the free energy of the ensemble of individual fibril elongation rates yields the activation free energy distribution, ΔG‡, between the soluble state and the fibril incorporated state (Fig. 2e). The mean value of the free energy of activation, 4.2 ± 1 kJ mol−1, combined with the enthalpy of activation from the Arrhenius plot yields a value for the entropy of activation TΔS‡, 43 kJ mol−1. The activation energy values obtained using our single fibril elongation assay indicate that the overall process of fibril growth is disfavoured: the entropy of activation is favourable, whereas the elongation enthalpy of activation is marginally less favourable. Our data agree well with previous QCM measurements41 (ΔG‡ = 5.9 kJ mol−1 and ΔH‡ = 66.1 ± 8.1 kJ mol−1) however single fibril imaging enabled the measurement of only the activation energy of the fast growing fibril end rather than the average over the fast and slow ends, leading to lower values compared to ensemble measurements. To our knowledge this is the first time TIRFM has been used to quantitatively measure such kinetic parameters of amyloid fibril elongation.
Intermittent growth is due to a temperature dependent paused state
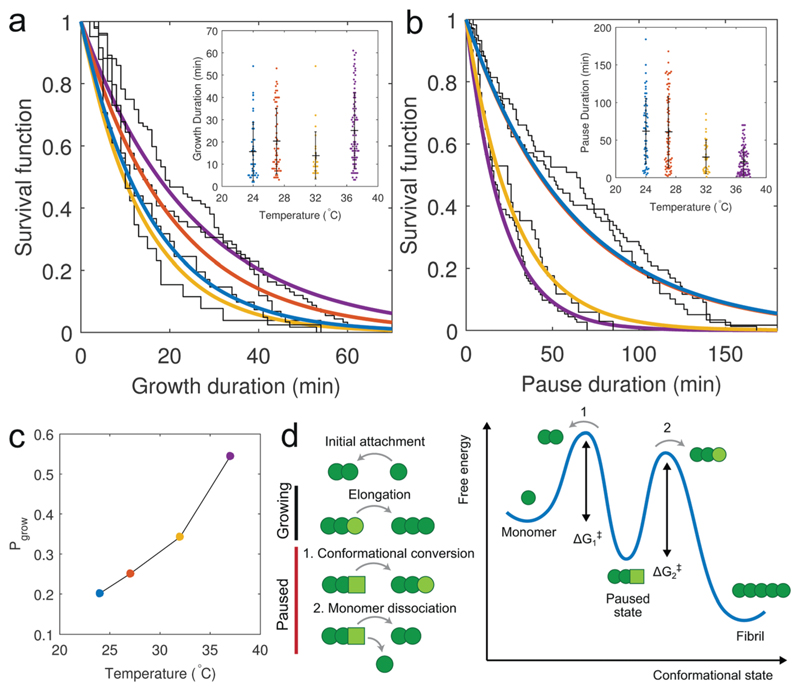
In our analysis of growth at the fast-growing fibril end we chose to disregard the pause periods and obtained kinetic parameters from the pause-free elongation rates. Recent experimental22,26,48 and computational49 studies have suggested that intermittent growth is an intrinsic feature of amyloid fibril elongation. It was suggested that the growing fibril end can be described by a two state Markov model, consisting of a growing state capable of elongation, and a paused state, where elongation is blocked. To investigate the possible origin of such intermittent growth, we measured the growth and pause durations from the temperature dependent elongation assay. Describing the growth intermittency by a two-state model allowed the distribution of growth and pause durations to be fit using single exponentials (Fig. 3a and b). While the growth duration displayed no apparent dependence on temperature (Fig. 3a, inset), we found that decreasing the temperature increased the fraction of time the fibril end spent in the paused state (Fig. 3b, inset). The temperature dependence of this duration rules out surface effects or obstacles as the cause of growth intermittency. On the other hand, the average probability that the fibril end was in the growing state, Pgrow, was estimated from the ratio of the measured growth and pause durations: and found to be dependent on temperature (Fig. 3b), with Pgrow ranging from 0.2 to 0.55 between 24 and 37 °C. We suggest that the intermittent elongation we observe is due to an additional kinetically trapped state at the fibril end that is off pathway from the normal monomer-to-fibril addition reaction. This can be reconciled with the observed temperature dependence of the pause duration, which suggests that an escape from this trapped state requires the transition across an additional energy barrier.
Fig. 3. Aβ42 fibrils exhibit intermittent growth.
(a and b) Survival functions of the growth and pause durations with data fits using single exponential functions, suggesting that the switching between the growing state, where fibril ends are competent to elongate, and the paused state, where elongation is blocked, can be described by a two-state process. The growth duration does not show a dependence on temperature, whereas the duration of the paused state exhibits an inverse dependence on temperature. (c) Decreasing the temperature increases the duration of the paused state, decreasing the growth probability, Pgrow, suggesting that the pauses in growth are due to a kinetically trapped state at the fibril end, which blocks the further attachment of monomers. (d) Schematic of the intermittent growth process and possible energy landscape reflecting entry into the paused state. Light green circle – correctly docked monomer, light green square – incorrectly docked monomer.
We propose two possible mechanisms for the observed intermittencies in growth dynamics (Fig. 3d). Initially, the fibril end may be in an elongation competent, or growing, state whereby a free monomer attaches and reversibly docks to the end. This is then followed by a conformational conversion to an irreversibly locked state; this behaviour would be consistent with our observation of Michaelis–Menten kinetics. The elongation incompetent, or paused, state, on the other hand, could arise from the incorrect (or out of register50) docking of a monomer, which is then followed either by a conversion into the elongation competent conformation, or by the dissociation of the ‘faulty’ monomer. Both scenarios would block the fibril end to prevent further monomer addition. Escape from this metastable state would require a transition across an energy barrier either from the trapped state to the correct conformational state, or to the monomer state. The existence of this additional energy barrier may explain the lower value of the enthalpy of activation ΔH‡ obtained using the pause-free elongation rates measured using our single fibril TIRFM assay (~47 kJ mol−1) compared to values obtained via ensemble QCM measurements.41 The higher energy barrier measured using ensemble methods may be due to an underestimation of the fibril elongation rate which arises from growth intermittency, because ensemble measurements represent a convolution of the pause-free elongation rate with the growth probability. If instead of using the pause-free elongation rates to generate an Arrhenius plot, we use the apparent elongation rates calculated from the final fibril lengths (Fig. S7, ESI†), we can obtain a combined enthalpy of activation of 114 ± 57 kJ mol−1 (R2 = 0.973, ±95% CI). If this apparent enthalpy barrier is the sum of the enthalpy barrier to fibril elongation and the enthalpy barrier for escape from the trapped state, the latter can be estimated from ΔHtrap = ΔHapp − ΔHel to be approximately 67 kJ mol−1.
Growth at the slow fibril end is more sporadic than at the fast end
Having investigated the kinetics and dynamics of the fast-growing ends of the fibrils, we finally sought to investigate the kinetics of the slow end. In contrast to the fast end, where elongation was intermittent, but with an approximately equal probability of being in the growing or in the paused states at 37 °C, the growth at the slow end appeared to be much more sporadic with much longer paused periods (Fig. 4a). The relative rarity of seeds that exhibited measurable slow end growth over the timescale of the experiments precluded a kymograph analysis in the same way as was performed above for the fast end. To obtain the elongation rates of a large number of fibril slow ends, we therefore reverted to using ex situ TIRF microscopy and dSTORM superresolution microscopy of multiple fibrils at discrete time points during an elongation reaction (Fig. S5 and S6, ESI†). We incubated seed fibrils with monomeric Aβ42 for 5 hours in order to measure the length distributions of both fast and slow ends from diffraction limited TIRF images (Fig. S5, panel b, ESI†).
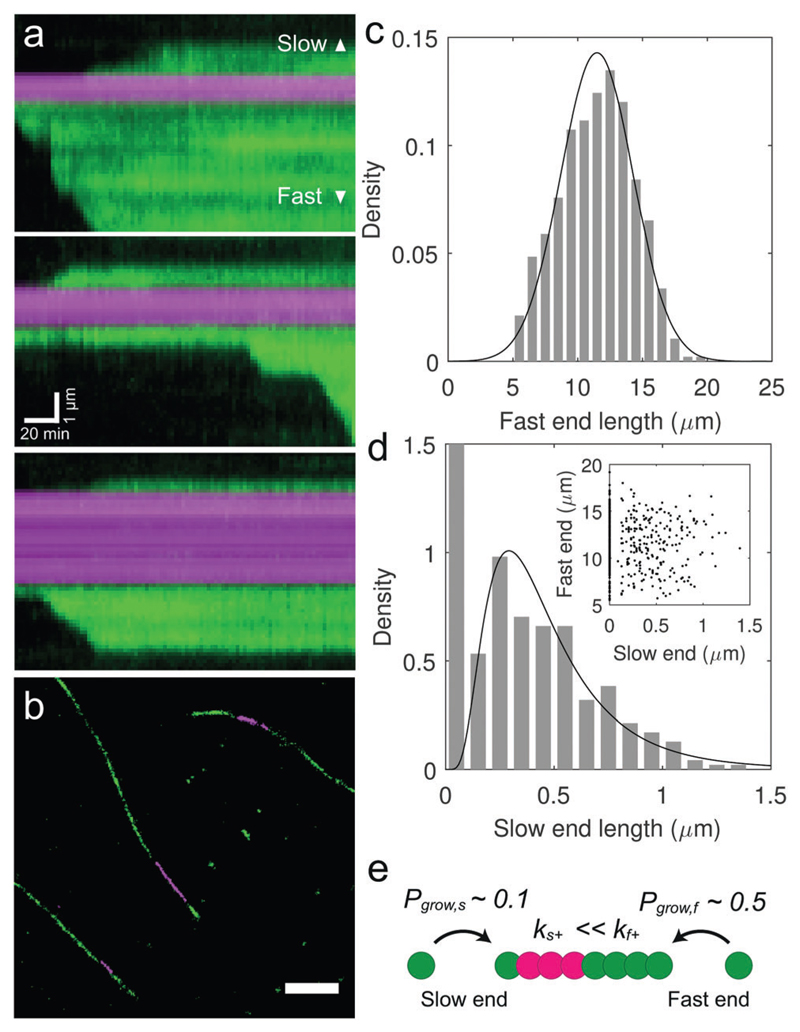
Fig. 4. Aβ42 fibrils exhibit a fast and a slow growing end.
(a) Kymographs show that slow ends elongate at a reduced rate (10–20 nm min−1) compared to the fast end (50–100 nm min−1), and grow only sporadically. (b) Ex situ imaging of Aβ42 fibrils by two colour super-resolution fluorescence microscopy permits a precise measurement of the slow end length. The panel represents a selected region from a larger field of view obtained by dSTORM imaging, as shown in Fig. S5, panel a (ESI†). Scale bar: 1 μm. (c and d) Histograms of the fast and slow end lengths after 5 h incubation of seed fibrils with 10 μM monomer. Data fits correspond to normal and log-normal distributions with mean lengths of 11.5 and 0.47 μm respectively. 51% of seeds showed no observable growth at their slow end after 5 h. Note that the y-axis was rescaled to emphasize the distribution of slow end lengths greater than zero. N = 475. (d, inset) A scatter plot of slow versus fast end length for each fibril revealed no correlation between the elongation rates at opposing ends. (e) Aβ42 fibril polarity results from different elongation rate constants, k+, and growth probabilities, Pgrow, at the fast and slow growing fibril ends.
After 5 hours, the fibrils exhibited clearly asymmetrical growth, as we had observed from the dynamic in situ assays (Fig. 4b). Multiple fields of view were imaged to cover many fibrils and the lengths of the fast and slow ends were measured to produce histograms (Fig. 4c and d). The mean lengths of fast and slow ends after 5 h were determined to be 11.5 and 0.47 μm respectively, which, combined with time lapse length measurements of the early time points acquired using dSTORM super-resolution optical microscopy (Fig. S6, ESI†), allowed us to estimate apparent elongation rates of 40–50 nm min−1 for the fast end, and 1–3 nm min−1 for the slow end. We furthermore found that 51% of the seed fibrils displayed no observable growth, suggesting the presence of growth-incompetent slow ends.
The apparent growth rates of both the slow and fast ends measured from ex situ fibrils are clearly slower than those measured in situ (see Fig. 2b and c). This discrepancy can be explained by taking into account intermittent growth. We can estimate the value of the apparent elongation rate Eapp that we observed ex situ by multiplying the pause-free elongation rate by the growth probability Pgrow, Eapp = EΔPgrow. For a 10 μM monomer concentration, the mean pause-free elongation rate was 76 nm min−1 with a growth probability of 0.54 ± 0.2, giving rise to an apparent elongation rate of 40 ± 15 nm min−1 (mean ± S.D.), which agrees well with our measured ex situ rate. Similarly, from in situ kymographs we can see that the pause-free elongation rate of the slow end is approximately 10 to 20 nm min−1 (Fig. 4a), while the apparent ex situ growth rate is an order of magnitude slower (Fig. 4d). From a knowledge of both the apparent growth rate and the approximate pause-free rate at the slow end we could therefore estimate the growth probability at the slow end to be approximately 0.1. The presence of the slow growing fibril end therefore results not only from a reduced elongation rate compared to the fast end, but also due to a reduced probability of growth. This low probability of growth at the slow end may explain why unipolar fibril elongation was reported in previous TIRFM studies.32
It has previously been reported that polymorphisms in amyloid structure give rise to kinetic heterogeneity in fibril growth, for example for the yeast prion sup35.18 The distribution of elongation rates we observed in our elongation assays of Aβ42 raised the question of whether the observed spread of growth rates was due to the existence of a continuum of different seed fibril structures, each exhibiting different elongation rates. To examine whether this was the case, we plotted the fast end length against the slow end length, as shown in Fig. 4d (inset). If the distribution of growth rates arose from the seed fibril structure, we would expect to observe a positive correlation between the slow and fast end lengths. As the new growth at each end of the seed is templated by the fibril structure, a “rapid” fibril polymorph would have more growth at both ends compared to a “slow” polymorph. We however observed no correlation between the two ends, suggesting that the length distributions, and hence elongation rate distributions, are instead due to stochastic addition of monomers at each end of the fibril.
Conclusion
To investigate the dynamics of amyloid fibril elongation, we developed a two-colour time-lapse TIRF microscopy assay, and applied it to directly visualize Aβ42 fibril elongation from preformed seeds, thus gaining insights into the concentration and temperature dependent kinetics of the process. Single fibril imaging allowed us to directly observe dynamical features of the reaction, which cannot be captured using existing ensemble methods. Furthermore, previous in situ TIRF and AFM elongation studies used unpassivated glass and quartz coverslips, with likely catalytic effects on aggregation, or, in the case of mica or graphite, causing epitaxial fibril growth along the crystal axes of the substrate.22 Switching between different fibril structures has also been seen to occur, reflecting interactions between the growing fibril end and the mica surface.24 Weak tethering of seed fibrils to the surface via antibody binding also ensures that the fibril structure is not affected, as may be the case after immobilization using covalent coupling chemistry.51
The reproducibility of our assay allowed us to confirm directly that Aβ42 elongation is a multistep process due to the rate saturation above a limiting monomer concentration, here determined to be ca. 7 μM. It is interesting to note the differences in elongation kinetics between Aβ40 and Aβ42, the latter almost an order of magnitude faster than the former. We suggest that the presence of the extra hydrophobic residues Ile41 and Ala42 lowers the activation energy barrier to Aβ42 elongation, thus increasing the elongation rate compared to Aβ40. The measured free energy barrier to elongation (4 kJ mol−1) is also relatively small, suggesting that even minor changes to the free energy, for example by small molecule inhibitors, can modulate the aggregation rate.
From our analysis of elongation kinetics and growth intermittency we can conclude that the polarity of the Aβ42 fibril structure results in two different elongation rates at either end of the fibril, with two different growth probabilities. Our results also suggest that growth intermittency arises from a metastable trapped state with an additional energy barrier, possibly due to an incorrectly bound monomer conformation. The observation of long lived paused states, where further elongation is blocked, suggests that an alternative strategy to elongation inhibition is via stabilization of this metastable state. Instead of designing a molecule to cap the fibril end, which would necessarily require a 1 : 1 stoichiometric ratio of molecules to number of fibril ends, an inhibitor might be designed to stabilize the growth-incompetent fibril end conformation which therefore blocks further monomer addition.
One of the major difficulties in understanding the mechanism behind the inhibitory effect of a compound on amyloid aggregation is separating its effects on the primary nucleation, elongation, and secondary nucleation reactions. In addition to the results presented here, we therefore believe that our technique will provide a valuable new assay for the study of inhibitors,15,52 chaperones,53 or chaperone-mimicking agents54 designed specifically to target the elongation reaction. We also foresee the wide applicability of the assay for the investigation of the kinetics and cross-seeding propensity of other amyloid β isoforms such as Aβ40,55 Aβ42 N-terminally extended variants (NTE-Aβ42s),56 and disease related single point mutations.57
Methods
Preparation of labelled Aβ monomer and fibril seeds
Lyophilized synthetic human Aβ(1–42), hereafter referred to as Aβ42 (Bachem, UK) was disaggregated via sequential treatment with trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP) (ThermoFisher Scientific, UK), and stored as a lyophilized film in Eppendorf tubes at −80 °C. Peptide concentrations for each tube were determined by quantitative amino acid analysis (PNAC Facility, Department of Biochemistry). Synthetic Aβ42 labelled at the N-terminus with either Hilyte Fluor 488 or Hilyte Fluor 647 (Anaspec, USA), hereafter referred to as HF488 and HF647, was dissolved in 1% NH4OH and stored at −80 °C. Labelled peptides were used without further purification.
Fluorescently labelled amyloid fibril seeds were prepared by dissolving unlabelled Aβ42 in aggregation buffer (20 mM sodium phosphate buffer with 200 μM EDTA, pH 7.4) and adding HF647 labelled Aβ42 to give a molar fraction of 5% labelled to unlabelled at 50 μM total concentration (i.e. 1 :19, labelled : unlabelled Aβ42). Fibrils were formed after incubation at 37 °C for 1 week without agitation. Directly prior to starting elongation experiments, fibrils were diluted to 1 μM in PBS and sheared by pulling through a 25G needle ten times to form short seeds. Needle shearing was used rather than sonication as the former was found to generate seed fibrils with more reproducible length distributions. We verified that the presence of dyes at the concentrations stated did not affect elongation kinetics using ThT assays of labelled and unlabelled peptides (see Fig. S9, ESI†).
Flow cell preparation
Flow cells were produced using a method adapted from single-molecule microtubule motility assays. 22 × 22 mm coverslips (#1.5 thickness, Carl Zeiss) and 25 × 75 mm microscope slides were sequentially cleaned in 70% ethanol, 1 M HCl and 1 M KOH and dried on a hotplate after extensive rinsing with ultrapure water. The cleaned glass was then exposed to trichloro-(1H,1H,2H,2H-perfluorooctyl)silane (Sigma Aldrich) under partial vacuum in a desiccator to render them hydrophobic after which they were rinsed with 70% ethanol and dried on a hot plate. Flow cells were produced by sandwiching a hydrophobic microscope slide and a coverslip together with Parafilm strips, and sealing them together by heating on a hot plate.
Elongation assay
To prepare immobilized amyloid fibril seeds, an antibody specific to the N-terminal end of amyloid-β (82E1, mouse monoclonal primary, 10323, IBL) was perfused into a flow cell and incubated to non-specifically adsorb it to the glass surface, followed by passivation with 5% (v/v) Pluronic F-127 (Invitrogen). When added to a silanized glass coverslip, the PEG–PPG copolymer surfactant forms a protein-repelling layer which effectively blocks adsorption of monomeric Aβ42. The commonly used blocking proteins casein and bovine serum albumin (BSA) proved to be ineffective at preventing Aβ42 adsorption. Following passivation and extensive washing, preformed fibril seeds were flowed into the channel and incubated for 15 min to attach to the antibody. After incubation any unbound seeds were removed by washing with PBS, then aggregation buffer (defined above). Labelled Aβ42 monomer was prepared by dissolving unlabelled Aβ42 in aggregation buffer and adding HF488 labelled Aβ42 to achieve the required labelling fraction (5%) and total concentration. A fresh aliquot of the unlabelled peptide was used for each experiment to avoid repeated freeze–thaw cycles. The dissolved peptide was not subject to additional gel filtration.
The passivated flow cells were perfused with the labelled peptide and the ends of the channels were sealed with vacuum grease to prevent evaporation. No antifade or oxygen scavenging system was used. For temperature dependent elongation measurements, a constant temperature was maintained to within 0.1 °C using a resistive foil heating element and temperature controller (TC200, Thorlabs) attached to the top side of the flow cell.
The total acquisition duration varied between 60 and 240 minutes, with images acquired every 1 to 2 minutes, depending on the Aβ42 monomer concentration and temperature.
Total internal reflection microscopy
Flow cells were imaged using a custom built multicolour TIRF microscope based on an Olympus IX-71 inverted microscope with a 100×/1.49NA objective (UAPON 100XOTIRF, Olympus).58 HF488 and HF647 dyes were excited using 488 nm and 640 nm diode lasers (iBeam SMART, Toptica and MLD, Cobolt), with detection via 525/30 nm and 676/29 nm bandpass filters, respectively (Semrock), onto an sCMOS camera (ORCA Flash 4.0, Hamamatsu). Image acquisition was controlled using Micro-Manager software and image-based autofocus was performed in the 640 nm channel before acquisition of each time point.
Supplementary Material
† Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cp03412a
Acknowledgements
We thank Q. Jeng for technical assistance and J. Skepper for TEM imaging. We also thank A. K. Buell, R. F. Laine, N. Nespovitaya, and S. Collins for helpful discussions. We thank Sara Linse for the provision of recombinant protein samples. CFK acknowledges funding from the UK Engineering and Physical Sciences Research Council, EPSRC (grants EP/L015889/1 and EP/H018301/1), the Wellcome Trust (grants 3-3249/Z/16/Z and 089703/Z/09/Z) and the UK Medical Research Council, MRC (grants MR/K015850/1 and MR/K02292X/1), MedImmune, and Infinitus (China) Ltd.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 2.Benilova I, Karran E, De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 3.Pauwels K, et al. Structural Basis for Increased Toxicity of Pathological Aβ 42:Aβ 40 Ratios in Alzheimer Disease. J Biol Chem. 2012;287:5650–5660. doi: 10.1074/jbc.M111.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuperstein I, et al. Neurotoxicity of Alzheimer's disease Aβ peptides is induced by small changes in the Aβ42 to Aβ40 ratio. EMBO J. 2010;29:3408–3420. doi: 10.1038/emboj.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lührs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt M, et al. Peptide dimer structure in an Aβ(1–42) fibril visualized with cryo-EM. Proc Natl Acad Sci U S A. 2013;112:201503455. doi: 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, et al. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silvers R, et al. Aggregation and Fibril Structure of AβM01-42 and Aβ1-42. Biochemistry. 2017;56:4850–4859. doi: 10.1021/acs.biochem.7b00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wälti MA, et al. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc Natl Acad Sci U S A. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nat Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 11.Stöhr J, et al. Purified and synthetic Alzheimer's amyloid beta (Aβ) prions. Proc Natl Acad Sci U S A. 2012;109:11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen SIa, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci U S A. 2013:1–6. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid beta-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem Neurosci. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. From Macroscopic Measurements to Microscopic Mechanisms of Protein Aggregation. J Mol Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 15.Meisl G, et al. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 16.Cannon MJ, Williams AD, Wetzel R, Myszka DG. Kinetic analysis of beta-amyloid fibril elongation. Anal Biochem. 2004;328:67–75. doi: 10.1016/j.ab.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 17.White DA, Buell AK, Knowles TPJ, Welland ME, Dobson CM. Protein aggregation in crowded environments. J Am Chem Soc. 2010;132:5170–5175. doi: 10.1021/ja909997e. [DOI] [PubMed] [Google Scholar]
- 18.DePace AH, Weissman JS. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat Struct Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- 19.Qiang W, Kelley K, Tycko R. Polymorph-Specific Kinetics and Thermodynamics of β-Amyloid Fibril Growth. J Am Chem Soc. 2013;135:6860–6871. doi: 10.1021/ja311963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arimon M, Sanz F, Giralt E, Carulla N. Template-Assisted Lateral Growth of Amyloid-β42 Fibrils Studied by Differential Labeling with Gold Nanoparticles. Bioconjugate Chem. 2012;23:27–32. doi: 10.1021/bc200077s. [DOI] [PubMed] [Google Scholar]
- 21.Hoyer W, Cherny D, Subramaniam V, Jovin TM. Rapid self-assembly of alpha-synuclein observed by in situ atomic force microscopy. J Mol Biol. 2004;340:127–139. doi: 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 22.Kellermayer MSZ, Karsai A, Benke M, Soós K, Penke B. Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong JS, Ansaloni A, Mezzenga R, Lashuel HA, Dietler G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J Mol Biol. 2013;425:1765–1781. doi: 10.1016/j.jmb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe-Nakayama T, et al. High-speed atomic force microscopy reveals structural dynamics of amyloid β 1-42 aggregates. Proc Natl Acad Sci U S A. 2016;113:5835–5840. doi: 10.1073/pnas.1524807113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen CB, et al. Branching in Amyloid Fibril Growth. Biophys J. 2009;96:1529–1536. doi: 10.1016/j.bpj.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferkinghoff-Borg J, et al. Stop-and-go kinetics in amyloid fibrillation. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2010;82:10901. doi: 10.1103/PhysRevE.82.010901. [DOI] [PubMed] [Google Scholar]
- 27.Wordehoff MM, et al. Single Fibril Growth Kinetics of α-Synuclein. J Mol Biol. 2015;427:1428–1435. doi: 10.1016/j.jmb.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Pinotsi D, et al. Direct observation of heterogeneous amyloid fibril growth kinetics via two-color super-resolution microscopy. Nano Lett. 2014;14:339–345. doi: 10.1021/nl4041093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinotsi D, Michel CH, Buell AK, Laine RF, Mahou P, Dobson CM, Kaminski CF, Kaminski Schierle GS. Nanoscopic insights into seeding mechanisms and toxicity of α-synuclein species in neurons. Proc Natl Acad Sci U S A. 2016;113:3815–3819. doi: 10.1073/pnas.1516546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gell C, et al. Methods in Cell Biology. Vol. 95. Elsevier; 2010. pp. 221–245. [DOI] [PubMed] [Google Scholar]
- 32.Ban T, et al. Direct observation of Abeta amyloid fibril growth and inhibition. J Mol Biol. 2004;344:757–767. doi: 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 33.Pinotsi D, Kaminski Schierle GS, Kaminski CF. In: Systems Biology of Alzheimer's Disease SE – 6. Castrillo JI, Oliver SG, editors. Vol. 1303. Springer New York; 2016. pp. 125–141. [Google Scholar]
- 34.Cabaleiro-Lago C, Szczepankiewicz O, Linse S. The effect of nanoparticles on amyloid aggregation depends on the protein stability and intrinsic aggregation rate. Langmuir. 2012;28:1852–1857. doi: 10.1021/la203078w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petkova AT, et al. A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luhrs T, et al. 3D structure of Alzheimer's amyloid- (1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fawzi NL, Okabe Y, Yap EH, Head-Gordon T. Determining the Critical Nucleus and Mechanism of Fibril Elongation of the Alzheimer’s Aβ1–40 Peptide. J Mol Biol. 2007;365:535–550. doi: 10.1016/j.jmb.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han W, Schulten K. Fibril Elongation by Aβ 17–42: Kinetic Network Analysis of Hybrid-Resolution Molecular Dynamics Simulations. J Am Chem Soc. 2014;136:12450–12460. doi: 10.1021/ja507002p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwierz N, Frost CV, Geissler PL, Zacharias M. Dynamics of Seeded Aβ 40 – Fibril Growth from Atomistic Molecular Dynamics Simulations: Kinetic Trapping and Reduced Water Mobility in the Locking Step. J Am Chem Soc. 2016;138:527–539. doi: 10.1021/jacs.5b08717. [DOI] [PubMed] [Google Scholar]
- 40.Knowles TPJ, et al. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc Natl Acad Sci U S A. 2007;104:10016–10021. doi: 10.1073/pnas.0610659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buell AK, et al. Detailed Analysis of the Energy Barriers for Amyloid FibrilGrowth. Angew Chem, Int Ed. 2012;51:5247–5251. doi: 10.1002/anie.201108040. [DOI] [PubMed] [Google Scholar]
- 42.Lorenzen N, et al. Role of elongation and secondary pathways in S6 amyloid fibril growth. Biophys J. 2012;102:2167–2175. doi: 10.1016/j.bpj.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:1582–1590. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buell AK, et al. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc Natl Acad Sci U S A. 2014;111:7671–7676. doi: 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheibel T, Kowal AS, Bloom JD, Lindquist SL. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr Biol. 2001;11:366–369. doi: 10.1016/s0960-9822(01)00099-9. [DOI] [PubMed] [Google Scholar]
- 46.Esler WP, et al. Alzheimer’s Disease Amyloid Propagation by a Template-Dependent Dock-Lock Mechanism. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 47.O’Brien EP, Okamoto Y, Straub JE, Brooks BR, Thirumalai D. Thermodynamic Perspective on the Dock–Lock Growth Mechanism of Amyloid Fibrils. J Phys Chem B. 2009;113:14421–14430. doi: 10.1021/jp9050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J-X, et al. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ranganathan S, Ghosh D, Maji SK, Padinhateeri R. A minimal conformational switching-dependent model for amyloid self-assembly. Sci Rep. 2016;6:21103. doi: 10.1038/srep21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmit JD. Kinetic theory of amyloid fibril templating. J Chem Phys. 2013;138:185102. doi: 10.1063/1.4803658. [DOI] [PubMed] [Google Scholar]
- 51.Buell AK, et al. Surface attachment of protein fibrils via covalent modification strategies. J Phys Chem B. 2010;114:10925–10938. doi: 10.1021/jp101579n. [DOI] [PubMed] [Google Scholar]
- 52.Härd T, Lendel C. Inhibition of amyloid formation. J Mol Biol. 2012;421:441–465. doi: 10.1016/j.jmb.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 53.Cohen SIa, et al. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat Struct Mol Biol. 2015;22:207–213. doi: 10.1038/nsmb.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abelein A, Gräslund A, Danielsson J. Zinc as chaperone-mimicking agent for retardation of amyloid β peptide fibril formation. Proc Natl Acad Sci U S A. 2015;112:5407–5412. doi: 10.1073/pnas.1421961112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cukalevski R, et al. The Aβ4O and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem Sci. 2015;6:4215–4233. doi: 10.1039/c4sc02517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szczepankiewicz O, et al. N-Terminal Extensions Retard Aβ42 Fibril Formation but Allow Cross-Seeding and Coaggregation with Aβ42. J Am Chem Soc. 2015;137:14673–14685. doi: 10.1021/jacs.5b07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hubin E, et al. Two distinct β-sheet structures in Italian-mutant amyloid-beta fibrils: a potential link to different clinical phenotypes. Cell Mol Life Sci. 2015;72:4899–4913. doi: 10.1007/s00018-015-1983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young LJ, Ströhl F, Kaminski CF. A Guide to Structured Illumination TIRF Microscopy at High Speed with Multiple Colors. J Visualized Exp. 2016;111:e53988. doi: 10.3791/53988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.