Abstract
The nematode worm Caenorhabditis elegans has emerged as an important model organism in the study of the molecular mechanisms of protein misfolding diseases associated with amyloid formation because of its small size, ease of genetic manipulation, and optical transparency. Obtaining a reliable and quantitative read-out of protein aggregation in this system, however, remains a challenge. To address this problem, we here present a fast time-gated fluorescence lifetime imaging (TG-FLIM) method and show that it provides functional insights into the process of protein aggregation in living animals by enabling the rapid characterization of different types of aggregates. Specifically, in longitudinal studies of C. elegans models of Parkinson’s and Huntington’s diseases, we observed marked differences in the aggregation kinetics and the nature of the protein inclusions formed by α-synuclein and polyglutamine. In particular, we found that α-synuclein inclusions do not display amyloid-like features until late in the life of the worms, whereas polyglutamine forms amyloid characteristics rapidly in early adulthood. Furthermore, we show that the TG-FLIM method is capable of imaging live and non-anaesthetized worms moving in specially designed agarose microchambers. Taken together, our results show that the TG-FLIM method enables high-throughput functional imaging of living C. elegans that can be used to study in vivo mechanisms of protein aggregation and that has the potential to aid the search for therapeutic modifiers of protein aggregation and toxicity.
A variety of human diseases, including neurodegenerative disorders such as Parkinson’s and Alzheimer’s diseases, are characterized by the misfolding of protein species and their subsequent aggregation into amyloid fibrils.1,2 The nematode Caenorhabditis elegans is a particularly useful model organism through which to study these diseases3–8 and to screen for small molecule inhibitors of the protein aggregation process.9,10 C. elegans has a simple body plan of 959 somatic cells, its genetics are well-characterized, and at least 40% of its genes have known human homologues.11 Furthermore, it has a relatively short lifespan of only 2–3 weeks and is optically transparent, making it a highly suitable system for longitudinal imaging studies of protein aggregation. Despite these advantages, obtaining quantitative read-outs for amyloidogenic protein aggregation in vivo remains challenging.12 Fluorescence intensity measurements are prone to artifacts, and classifying aggregates by counting protein inclusions relies on arbitrary choices for intensity and size cut-offs. Furthermore, reliable discrimination between amyloid-like and amorphous aggregates is generally not possible. The use of thioflavin T, a dye that becomes fluorescent upon intercalation into the cross β-structure of amyloid aggregates, and that is commonly used for the study of protein aggregation in vitro, is not compatible with live worm imaging because it affects protein homeostasis in the nematodes.13
To address some of these issues, we have previously established a read-out for the state of protein aggregation based on fluorescence lifetime imaging microscopy (FLIM) of a fluorophore covalently linked to the amyloidogenic protein of interest.14 FLIM not only informs on the location but also on the molecular environment of the fluorescent probes, providing fully quantitative read-outs.15–17 We have shown that a reduction in lifetime from the reporter fluorophore correlates with the degree of aggregation of the protein to which it is attached and that this provides a quantitative measure of the degree of protein aggregation in vitro, in live cells, and in C. elegans.14 The decrease in lifetime is thought to be associated with fluorescence energy transfer to intrinsic energy states associated with the amyloid fibrils.14 Conjugated organic fluorophores18–20 and intrinsic protein fluorescence21 have also been used successfully as FLIM sensors for protein aggregation, as has the amyloid-binding dye heptamer-formyl thiophene acetic acid (hFTAA).22 Conventional FLIM measurements are slow, however, as they are based on time-correlated single photon counting (TCSPC)23,24 which involves acquisition times on the order of 2 min for a single field of view. This method therefore requires the use of anaesthetized or fixed animals, greatly limiting the throughput of the technique and preventing studies of freely moving, live animals.
In this work, we specifically set out to address these problems and to establish a method that improves throughput and physiological relevance and permits studies of moving animals. The method makes use of time-gated FLIM (TG-FLIM),25 a fast and quantitative imaging modality which provides unprecedented throughput for FLIM measurements of nonparalyzed animals. Unlike TCSPC, which is usually performed in conjunction with laser scanning confocal microscopy (LSCM), TG-FLIM is a wide-field technique and thus heavily parallelizes the FLIM measurements. The fluorescence decay is measured by collecting the fluorescence signal with nanosecond-wide temporal gates, which are temporally shifted to enable the fluorescence decay to be sampled. Gating and time shifting are achieved with a high-rate imager (HRI), and lifetime data are thus obtained in each pixel from a sequential set of images gated at different time delays (Figure 1a, see Supporting Information). TG-FLIM has been successfully used for high-throughput imaging of protein−protein interactions and biosensors in cells,26,27 but not so far in the context of protein aggregation, or in worm models of disease. Here, we describe a novel approach for aggregation studies in C. elegans models of protein misfolding diseases based on TG-FLIM. We show that we can monitor intracellular aggregation in the worms over their entire lifespan using TG-FLIM with excellent repeatability and precision in the measured lifetimes. Our approach reveals differences in the kinetics of protein aggregation and the types of species appearing in the worms for two disease models. Furthermore, we provide details of the quantification of TG-FLIM data from live, moving animals and introduce motion correction algorithms for the TG-FLIM data analysis.
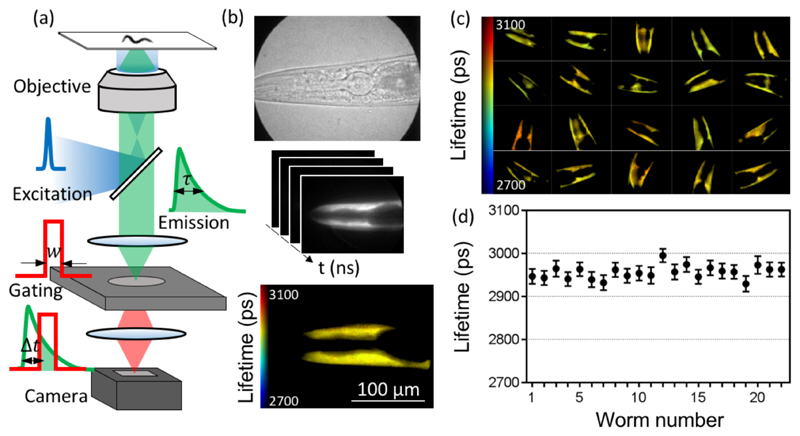
Figure 1. TG-FLIM imaging of live C. elegans expressing YFP in the body wall muscle cells.
(a) Schematic of the FLIM setup, showing the excitation pulse (blue curve), the fluorescence emission decay (green curve), and the gated detection (red curve) performed by the high-rate imager (HRI). The fluorescence lifetime (τ) is measured by gating the fluorescence decay with a gate width (w) and a set of gate positions (Δt). (b) Bright-field image acquired with the HRI camera (top), schematic FLIM stack consisting of images recorded at different time gates (middle), and the reconstructed fluorescence lifetime map obtained through such measurements from a single worm head region (bottom). (c) Fluorescence lifetime maps of 20 individual worms. Each field of view represents a 225 μm × 167 μm region in the sample plane. (d) Average lifetimes obtained from individual worms demonstrating the precision (error bars are standard deviation across pixels within the field of view) and the variability of lifetimes between individual worms in one experiment.
Results and Discussion
Time-Gated FLIM Provides a Robust Lifetime Read-out from C. elegans
A schematic of the TG-FLIM microscope setup that we have developed is shown in Figure 1a, details of which are found in the Methods section and in Supporting Information Figure 1. To assess the precision and repeatability of the method, we measured the fluorescence lifetimes of yellow fluorescent protein (YFP) when expressed in C. elegans. Figure 1c shows the head region of worms expressing YFP in body wall muscle cells. For the purpose of these experiments, the worms were anaesthetized. The average standard deviation across pixels within a single field of view was 17 ps (as shown by the error bars in Figure 1d). The variability between individual worms was estimated by comparing the mean lifetimes obtained from the worms and found to be 16 ps (standard deviation). The system exhibited excellent intraframe and interframe repeatability of the value of the fluorescence lifetimes measured in this way. The results presented in Figure 1c,d were obtained by acquiring 61 equally spaced gates with exposure times of 65 ms each. The gate width was set to 1 ns, and the time gated images were acquired every 250 ps, leading to a total acquisition time of 4.2 s per field of view. This corresponds to a ~30-fold improvement in speed compared to typical TCSPC measurements of similar quality,14 thus allowing for a much higher throughput. The results, therefore, show that TG-FLIM is a robust and quantitative read-out for biosensors in C. elegans, featuring good spatial and temporal resolution compatible with high-throughput or dynamic imaging.
Longitudinal FLIM Studies of Parkinson’s and Huntington’s Disease Models
We then explored if the method is capable of detecting age-associated protein aggregation in C. elegans models of protein misfolding diseases. To this end, we carried out a longitudinal study of the fluorescence lifetimes and the distribution of the aggregates that form in C. elegans models for Parkinson’s disease and for polyglutamine expansion disorders such as Huntington’s disease, expressing YFP-tagged α-syn5 and polyglutamine (40 glutamine residues, Q40),4 respectively, in the body wall muscle cells. We focused on imaging the head region of the worms and obtained sufficient resolution to be able to identify individual inclusions. Data were recorded across the whole lifespan of a population of nematodes, using the strain expressing only YFP as a control. We imaged a pool of ca. 20 worms for each strain (ca. 60 worms in total) on days 0, 3, 6, 10, 12, and 14 of adulthood for two independent biological replicates. The fluorescence lifetimes for each population and representative TG-FLIM images are shown in Figure 2.
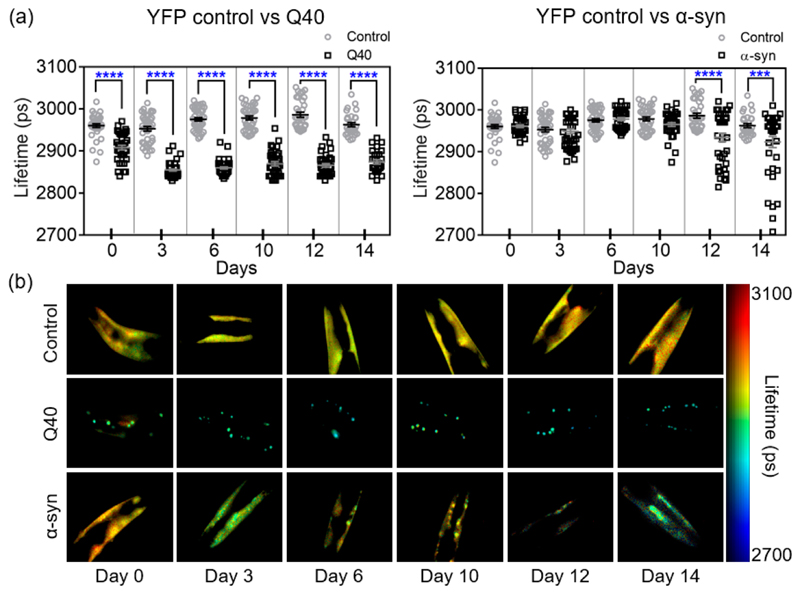
Figure 2. TG-FLIM imaging of protein aggregation in Q40 and α-syn strains across the lifespan of the animals.
(a) Fluorescence lifetimes at days 0, 3, 6, 10, 12, and 14 of adulthood for Q40 and α-syn compared to YFP controls. Squares and circles represent the average lifetime of the fluorescence from the head region of each imaged worm. Statistical analysis was performed using a one-way ANOVA, ***p < 0.005, ****p < 0.0001. The data shown are pooled from two biological replicates totalling ca. 40 worms per strain and time point. An F-test of YFP control lifetimes comparing data over the 15 days showed no significant deviation from a zero-slope, indicating that our data are compatible with a constant lifetime for the YFP control over the lifespan of the animal (p = 0.90). (b) Representative worm FLIM images for each day and each strain.
While the YFP control remained constant over the lifespan of the animal (as examined by an F-test), we observed that the average fluorescence lifetime of Q40 dropped early in the life of the worms (between day 0 and day 3) and remained constant throughout the rest of their lifespan (Figure 2a). At day 0, the Q40 worms presented a combination of bright inclusions with lower fluorescence lifetimes than the YFP control, indicative of protein molecules being in an amyloid state,14 as well as a diffuse signal with lifetimes similar to that of the YFP control, therefore indicative of a nonamyloid state (Figures 2b and 3a,c). The fluorescence lifetime distribution of Q40 at day 0 exhibits two peaks, in agreement with the presence of the soluble and aggregated forms (Figure 3c). The diffuse signal was not observed from day 3 onward, and we infer that at this stage of the worm life all soluble protein had been incorporated in amyloid-like inclusions in agreement with the significantly lower average lifetimes of ca. 2850 ps compared to the YFP control of ca. 2950 ps (Figure 2a). These results are consistent with previous fluorescence recovery after photobleaching (FRAP) studies, where the diffuse Q40 observed in young animals was found to be relatively mobile, compared to the Q40 inclusions, which were immobile.4,28 In addition, these data are in agreement with the propensity of polyQ stretches in isolation, as well as in the context of huntingtin exon 1, to form fibrils in vitro29 and in vivo.30–32
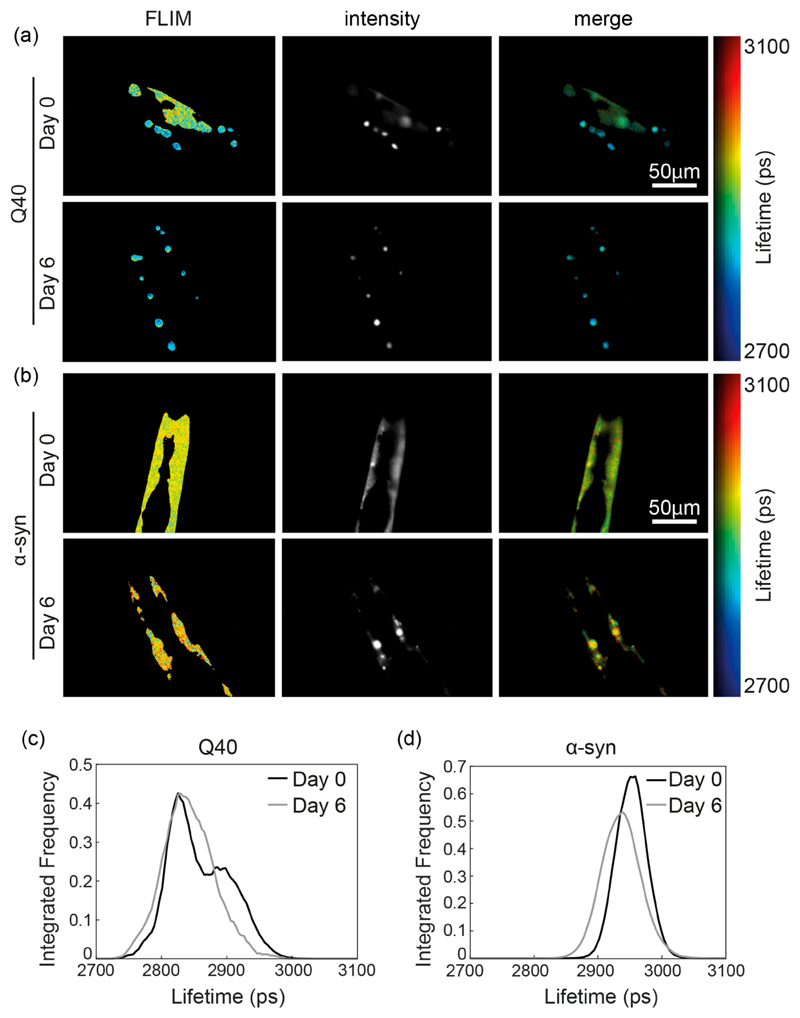
Figure 3. Representative protein aggregation of Q40 (a,c) and α-syn (b,d) at day 0 and day 6 revealed by TG-FLIM images.
(a) Representative images of Q40 worms at day 0 (top) and day 6 (bottom). Shown are the false-color FLIM maps (left), the signal intensity (middle), and the intensity-merged FLIM maps (right). The FLIM and intensity maps show that at day 0 the protein is distributed between a diffuse signal and foci, the latter having a shorter lifetime. (b) Representative images of α-syn worms at day 0 (top) and day 6 (bottom), presenting bright inclusions with comparable fluorescence lifetime as the diffuse signal. (c) Fluorescence lifetime histogram of Q40 protein, obtained from the animals shown in (a) at day 0 and day 6. (d) Fluorescence lifetime histogram of α-syn protein, obtained from the animals shown in (b).
In contrast to the Q40 animals, the fluorescence lifetime of α-syn remained similar to that of the YFP control worms until day 10, after which a reduction in lifetime was observed (Figure 2a). Detailed analysis revealed that inclusions were present in α-syn worms from day 0 onward, yet these did not correspond to highly ordered aggregates as judged from the uniform fluorescence lifetimes observed across the animals throughout most of their lifespan (Figures 2b and 3b,d). Only late in the life of the animal, after day 10, did we observe α-syn inclusions with fluorescence lifetimes indicative of well-defined amyloid fibrils (Figure 2b). Analysis of the expression levels by Western blot revealed that the α-syn concentration decreased during aging (Supporting Information Figure 2), which does not affect the lifetime values but may only lead to a small reduction in the precision of the measurement. Again, the FLIM results are consistent with FRAP data, in which immobile inclusions were observed only from day 11 onward.5
As an independent confirmation of the differences between the inclusions formed by Q40 and α-syn, we lysed the worms in detergent-containing buffer at day 6 of adulthood and observed that α-syn was fully dispersed and localized to the soluble fractions, whereas Q40 inclusions persisted and were present in the pellet fractions (Supporting Information Figure 3).
The precise molecular nature of the earlier α-syn inclusions is unclear at present, but we speculate that they could be either disordered oligomeric species as observed in the early stages of α-syn aggregation in previous studies,33,34 accumulations of monomeric α-syn with lipids or other cellular components, or perhaps droplets with liquid-like properties, which have recently attracted attention as possible precursors of amyloidogenic protein aggregation.35
Thus, this longitudinal TG-FLIM study reveals clear differences in the kinetics of the aggregation process for strains expressing Q40 and α-syn and informs on the nature of the protein inclusions in vivo as the animals age.
TG-FLIM Imaging of Live C. elegans Crawling in Agarose Microchambers
Imaging studies of C. elegans are typically performed on anaesthetized animals in order to circumvent motion artifacts at the examined length and time scales. Although the animals remain alive during the experiment, the use of anesthetics is not compatible with the observation of behavior, nor does it allow an individual animal to be monitored over long periods of time. Given the exceptional speed and precision of TG-FLIM, we set out to examine its applicability for imaging live and crawling C. elegans. To this end, we designed agarose microchambers that can hold individual worms (Figure 4a), inspired by previous studies on both C. elegans larvae and adults.36,37 The dimensions of the microchambers were chosen so that worms remained within the field of view and the depth of field of the microscope and were able to crawl freely within the microchamber as shown in the supplementary video (see Supporting Information Figure 4).
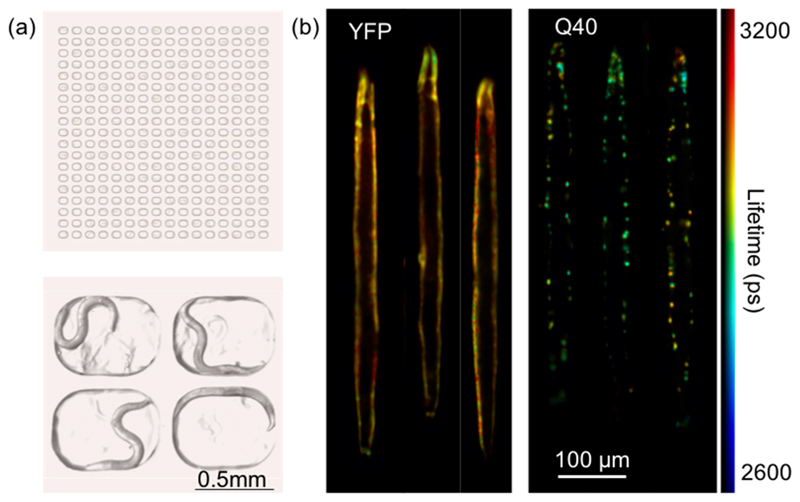
Figure 4. TG-FLIM applied to live C. elegans crawling in agarose microchambers.
(a) Schematic of the agarose-based microchamber device loaded with C. elegans (top) and an image of four chambers, each occupied by a single worm (bottom). (b) Registered and digitally stretched fluorescence lifetime maps of live crawling C. elegans. Shown here are example images of YFP and Q40 worms at day 3 of adulthood. For each worm, 10 sequential FLIM acquisitions were carried out (equaling a total acquisition time of ~5 s, therefore comparable to that used for anaesthetized worms ~4.2 s), and the fluorescence lifetime maps were averaged after digital stretching.
As a benchmark for this approach, we inserted C. elegans expressing YFP and Q40 at day 3 of adulthood in the microchambers for TG-FLIM measurements (Figure 4). To increase acquisition speed, we used only seven time gates (gate width of 1 ns with time gates acquired every 2.175 ns) compared to the 61 used for studies described in the preceding sections, thus shortening the total recording time to just 0.5 s per FLIM acquisition and minimizing movement artifacts across the TG-FLIM data set, while enabling sufficient data quality.
Although the worms did not move on the time scale of the individually recorded frames in our TG-FLIM measurements, motion over the entire acquisition sequence (0.5 s) caused significant artifacts during the FLIM reconstruction, as a consequence of the loss of spatial correspondence of image pixels between the different time gates (Supporting Information Figure 5). In order to correct for the worm movement during the data acquisition, we developed an image registration procedure that realigns each data set before FLIM analysis, similar to that used previously to remove motion artifacts from intravital imaging.38 The procedure uses a nonrigid transformation to register features in each individual frame compared to the brightest frame of the given TG-FLIM data set. We quantified the resulting standard deviations on control worms and found a lower fluorescence lifetime resolution compared to the experiments on anaesthetized animals (higher standard deviation of ca. 60 ps with seven gates, compared to 17 ps with 61 gates, with a similar level of signal in the maximum time gate). This difference can be explained by a lower sampling rate of the fluorescence decay and therefore a lower number of total photons in the decay, as well as residual errors in registration adding some noise to the fluorescence lifetime estimation.
Additionally, we implemented a digital “worm stretching” procedure inspired by the work of Christensen et al.39 This approach has two major advantages: First, it allows for the averaging of fluorescence lifetime maps of the same worm if consecutively imaged, improving the signal-to-noise ratio (SNR) of the resulting lifetime map. Second, it allows alignment of images of multiple worms along a similar template (a “stretched” form of the worm), which enables a direct comparison of multiple data sets for the assessment of the distribution of markers and functional read-outs. Being able to visualize data in a tractable way is important, especially when a high-throughput approach is taken. The digital stretching was performed by first determining the worm outline and drawing the backbone of the worm via skeletonization, using a similar approach to that used for tracking worms.40 The backbone was then used to extract the information about the curvature of the worm, which in turn allowed reconstruction of the signal from the digitally stretched worm (see Methods for details).
Representative results of the YFP control and Q40 worms are shown in Figure 4b. The fluorescence lifetime maps of the worms were digitally stretched as described above, and 10 consecutive fluorescence lifetime maps were averaged in order to achieve a comparable image quality to that obtained in our study on the anaesthetized worms. The fluorescence lifetime maps reconstructed here show no visible artifacts despite the motion of the worms during the acquisition. Additionally, we found that the fluorescence lifetimes obtained this way are in good agreement with those obtained in the experiments on immobilized animals (compare Figure 4b to Figure 2b). These measurements therefore permit functional imaging of aggregate states in entire worms, and enable their behavior to be monitored over time. Furthermore, the visualization of worms as stretched templates, as in Figure 4b, allows a direct comparison of the degree of aggregation (derived from the FLIM measurement) and of the spatial distribution of the protein deposits.
Concluding Remarks
In conclusion, we have presented here a method for the functional study of protein aggregation in live C. elegans. The method reveals differences in the aggregation kinetics and the nature of the inclusions formed during aging in models of Parkinson’s (α-syn) and polyglutamine expansion (Q40) diseases. We observed that α-syn became localized to inclusions prior to the decrease in fluorescence lifetime that is associated with amyloid formation, suggesting that the mechanism of α-syn aggregation involves the persistence of relatively disordered intermediate species prior to the formation of amyloid structure. We furthermore noticed a considerable spread in the fluorescence lifetimes of aged α-syn worms, suggesting that some animals remain largely unaffected by amyloid aggregation even at old age. By contrast, the data showed that Q40 accumulates completely into amyloid inclusions early in adulthood of the worm population. This approach could be extended to image young Q40 nematodes in larval stages at higher resolution, to find out if similar non-amyloid inclusions comprised of, e.g., oligomeric28 or liquid-like states41 are visible as precursors in this system.
For the longitudinal studies described here, we were able to perform the complete set of experiments in less than 2.5 h for each day of measurement (ca. 20 worms for each of the three strains studied here). This fast acquisition time provides a high level of reproducibility and has major advantages for high-throughput studies, as it limits the variability in worm age across the experiment while providing sufficient data to define the lifetime changes precisely. This feature constitutes a significant improvement compared to TCSPC where the measurement of ~60 worms would take over ~10 h on a single imaging day. The method also has much higher throughput than FRAP, providing information about the distribution and the aggregation states of all of the inclusions in the imaged region of the animal in one single acquisition. Crucially, TG-FLIM allows us to confirm the amyloid-like nature of the inclusions based on the ability of the FLIM sensor to distinguish between different forms of aggregation,14 whereas FRAP informs solely on diffusion, which may be similarly restricted for multiple forms of aggregates. Given its speed, we anticipate that the TG-FLIM method will provide new avenues for high-throughput studies of in vivo protein aggregation, e.g., to screen for small molecules with the ability to inhibit this process.42
We have in addition demonstrated two novel analytical approaches in combination with fast TG-FLIM to image moving nematode worms. The first one uses nonrigid transformation for gate realignment and correction of motional artifacts within the TG-FLIM dataset, which we note would not be possible with TCSPC measurements. The second uses a pseudo-templating method to allow for FLIM map averaging and for the alignment of all the imaged worms from a given population for easy evaluation of variability and phenotypical properties, e.g., the size of the worms and the spatial distributions of the fluorescent marker.
The results set the stage to apply the fast TG-FLIM approach to more advanced types of chamber devices that could support long-term culturing of C. elegans and enable protein aggregation in the same individual worm to be tracked over time. Agarose microchambers have been used to follow the development of C. elegans larvae,43 but microfluidic devices may be necessary for long-term growth, providing a continuous supply of nutrients and separation of offspring as described, e.g., by Cornaglia et al.44 The combination of our fast TG-FLIM approach with microfluidic or microchamber devices, such as the one that we have presented here, with fully automated data acquisition will greatly improve the throughput of the method compared to manual scanning, as used in the current study. This approach would constitute an invaluable tool for performing large functional screens for genetic modifiers or compounds that perturb protein aggregation. Optical sectioning capabilities45 can also be added to the imaging procedure in order to reveal the 3D organization of the system, which will provide important additional information, e.g., on the subcellular localization of protein inclusions.
Finally, the fast FLIM imaging demonstrated here can reach a speed of two FLIM frames per second (as demonstrated by the seven gates imaging experiments) and hence can lead to the observation of biologically relevant protein-protein interactions and biosensor dynamics in freely moving worms. Therefore, the use of this fast FLIM method opens up important avenues for time-dependent functional studies using other biosensors, for example, to probe Ca2+ levels for monitoring neuronal activation while having simultaneous read-outs of the associated behavior of the worms.
Methods
TG-FLIM Imaging
The TG-FLIM system was set up on an Olympus IX83 frame. The laser source was a supercontinuum laser source Fianium SC400-4, spectrally selected by a combination of two linear variable filters and set up for epifluorescence excitation by focusing the beam in the back focal plane of the microscope objective. The fluorescence image was relayed onto the photocathode of a high-rate imager (HRI, Kentech), and its phosphor screen was reimaged onto the camera sensor (PCO pixelfly, USB, PCO). The magnification of the relays was set up such that the element size of the HRI matched the resolution of the microscope and that of the camera pixel size in 2 × 2 binning mode. Details of the architecture of the microscope are shown in Supporting Information Figure 1. Imaging of worm heads was performed using a 40× objective (Olympus UApoN340 40X NA 1.35), and that of entire worms in imaging chambers was performed using a 10× (Olympus PlanFLN 10X NA 0.3) objective. The excitation wavelength was selected to be 516 nm (10 nm bandwidth), and the fluorescence was detected using a 550/49 (Semrock) filter. The instrument response function (IRF) was measured by taking an acquisition of a 1 mM solution of Erythrosin B (Sigma-Aldrich) solution in water. Additionally, the microscope was equipped with a CMOS camera (Blackfly S, BFS-U3-51S5M-C, FLIR) for bright-field measurement.
FLIM Lifetime Image Reconstruction
FLIM reconstruction was performed using the FLIMfit46 package (v4.12.1) from the Open Microscopy Environment (OME). Data analysis was performed by subtracting a background image acquired separately and spatially varying the IRF reference reconvolution using a lifetime of ~90 ps. A single exponential decay was fitted to the data on a single-pixel basis.
C. elegans Culturing and Sample Preparation
Nematodes were grown under standard conditions on nematode growth media (NGM) plates seeded with Escherichia coli OP50 at 20 °C. Worm strains used in these experiments were AM134 expressing YFP, AM141 expressing glutamine40-YFP (Q40),4 and OW40 expressing human wild-type α-synuclein-YFP (α-syn),5 all under control of the unc-54 promoter to drive expression in body wall muscle cells. Age-synchronized worm populations were generated by a 4 h synchronized egg lay, and animals were transferred to NGM plates containing 75 μM 5-fluoro-2′-deoxyuridine (FUDR, Sigma) at the fourth larval stage to inhibit the generation of offspring.
For imaging, worms were transferred to a drop of M9 buffer (3 g L−1 KH2PO4, 6 g L−1 Na2HPO4, 0.5 g L−1 NaCl, 1 mM MgSO4) containing NaN3 as an anesthetic on a freshly prepared pad of 2.5% agarose. A coverslip was delicately placed on top, and the sample was inverted for imaging on the inverted microscope.
Preparation of C. elegans Lysates and Western Blot
At day 6 of adulthood, 6000−9000 animals were harvested for each of the strains YFP, Q40, and α-syn by washing them off of NGM plates with M9 buffer. Worm pellets were resuspended in RIPA buffer (Sigma) supplemented with a protease inhibitor cocktail (Roche) and lysed using a cell homogenizer (Isobiotec). Lysates were fractionated by centrifugation in an Eppendorf microcentrifuge at 3000 rpm (845 g) followed by 15 000 rpm (21,130 g). The fractions were inspected for fluorescence using a Leica MZ10 F stereomicroscope.
Western blots were probed for YFP with antibody ab6556 (Abcam) and for tubulin with T6047 (Sigma) both diluted 1:5000. To examine protein expression levels during aging, animals were picked directly into SDS/urea buffer (8 M urea, 2% SDS, 50 mM DTT, 50 mM Tris pH 8.0), and amounts corresponding to the same number of worms were loaded onto the gel for each of the time points (ca. 13 worms for YFP and ca. 37 worms for α-syn). For Q40, we observed that the protein inclusions largely failed to enter the gel even under denaturing conditions, and thus we were not able to quantify the expression levels for this strain using this method.
Imaging Chambers
A silicon master with the desired micro-structures was fabricated with standard photolithography techniques. The device was designed as an array of 19 × 16 microchambers spaced by 350 μm in both directions. The chambers were shaped as rounded rectangles (700 μm × 500 μm × 80 μm depth) to fit within the field of view of the microscope when using the 10× objective (898 μm × 671 μm). Microchamber devices were made by pouring 5% high-melting agarose in S-basal buffer (5.85 g L−1 NaCl, 1 g L−1 K2HPO4, 6 g L−1 KH2PO4, 5 mg L−1 cholesterol) onto the master in a Petri dish and carefully cutting it out after solidification. To allow for sufficient oxygen supply during prolonged imaging times, slits were cut on the back of the device. A drop of E. coli OP50 resuspended in Luria Broth medium (10 g L−1 bacto-tryptone, 5 g L−1 bacto-yeast, 5 g L−1 NaCl) was applied onto the device, after which it was left to dry. Worms were washed off NGM plates with M9 buffer (3 g L−1 KH2PO4, 6 g L−1 Na2HPO4, 0.5 g L−1 NaCl, 1 mM MgSO4) and allowed to sediment, after which a ~100 μL drop of solution containing the worms was put onto the device. We observed that the worms tended to swim toward the bottom of the chambers as the drop was drying. However, spreading them with a platinum wire ensured a more homogeneous distribution with most chambers containing a single worm, or being empty. In our hands, the optimum chamber filling was achieved by loading ca. 150−200 worms onto the device, which contains 304 microchambers. As soon as the device was dry, a coverslip was put on top and a glass slide at the bottom, after which the sample was imaged in an inverted fashion.
Correction of Motion Artifacts and Digital Stretching
The correction of motion artifacts was performed by using a nonrigid transformation using the MATLAB B-spline image registration written by Dirk-Jan Kroon (MathWorks File Exchange) and initially implemented by Rueckert et al.47 The brightest image of the fluorescence decay was used as a template. Each gate was rescaled by histogram equalization prior to registration. The transformation obtained for each rescaled gate was then applied to the corresponding original data. The registered data set was then saved as an OME-tiff for subsequent FLIM analysis.
The digital stretching was performed as follows: each total intensity image was rescaled by histogram equalization followed by binarization (Otsu thresholding) and active contouring in order to obtain a faithful outline of the worm. The backbone of the worm was obtained by skeletonization and then used to calculate the position of the center of the worm along the geodesic line of the backbone. The backbone was subsequently smoothed by undersampled cubic spline interpolation. The coordinates along the geodesic line were used to estimate the angle of the worm at every point along the backbone and to rotate the image of the worm. For each position, the section of the worm image normal to the backbone was obtained and the sections were used to reconstitute the stretched image of the worm. The transformation obtained this way could then be applied to the fluorescence lifetime image and total intensity map.
Statistical Analysis
All statistical analysis (one-way ANOVA and F-test for comparing with a zero-slope curve) was performed on GraphPad Prism 7.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.9b00354.
Acknowledgments
We would like to thank S. Warren and I. Munro for maintaining and updating the FLIMfit package, C. Rowlands and C. Russell for fruitful conversations about the project, and the Caenorhabditis Genetics Center for strains. G.S.K.S. and C.F.K. acknowledge funding from the UK Engineering and Physical Sciences Research Council (EPSRC; grants EP/ L015889/1 and EP/H018301/1), the Wellcome Trust (grants 3-3249/Z/16/Z and 089703/Z/09/Z), the UK Medical Research Council, MRC (grants MR/K015850/1 and MR/ K02292X/1), MedImmune (Astra-Zeneca), and Infinitus, Ltd. (China). This project has furthermore received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 722380 (to C.P.), The Netherlands Organisation for Scientific Research (Rubicon fellowship 680-50-1503 to T.S.), the European Molecular Biology Organisation (long-term fellowship ALTF 72-2015 to T.S.), and the Centre for Misfolding Diseases of the University of Cambridge (T.S., M.P., C.M.D., and M.V.). R.F.L also acknowledges the support of the UK Biotechnology and Biological Sciences Research Council (BBSRC; BB/ P027431/1 and BB/R021805/1).
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- (2).Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- (3).Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–22. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Van Ham TJ, Thijssen KL, Breitling R, Hofstra RMW, Plasterk RHA, Nollen EAA. C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kraemer BC, Zhang B, Leverenz JB, Thomas JH, Trojanowski JQ, Schellenberg GD. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc Natl Acad Sci U S A. 2003;100:9980–5. doi: 10.1073/pnas.1533448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vaccaro A, Tauffenberger A, Aggad D, Rouleau G, Drapeau P, Parker JA. Mutant TDP-43 and FUS cause age-dependent paralysis and neurodegeneration in C. elegans. PLoS One. 2012;7:e31321. doi: 10.1371/journal.pone.0031321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Murakami T, Yang SP, Xie L, Kawano T, Fu D, Mukai A, Bohm C, Chen F, Robertson J, Suzuki H, Tartaglia GG, et al. ALLS mutations in FUS cause neuronal dysfunction and death in caenorhabditis elegans by a dominant gain-of-function mechanism. Hum Mol Genet. 2012;21:1–9. doi: 10.1093/hmg/ddr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Habchi J, Chia S, Limbocker R, Mannini B, Ahn M, Perni M, Hansson O, Arosio P, Kumita JR, Challa PK, Cohen SIA, et al. Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer's disease. Proc Natl Acad Sci U S A. 2017;114:E200–208. doi: 10.1073/pnas.1615613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Perni M, Challa PK, Kirkegaard JB, Limbocker R, Koopman M, Hardenberg MC, Sormanni P, Muller T, Saar KL, Roode LWY, Habchi J, et al. Massively parallel C. elegans tracking provides multi-dimensional fingerprints for phenotypic discovery. J Neurosci Methods. 2018;306:57–67. doi: 10.1016/j.jneumeth.2018.02.005. [DOI] [PubMed] [Google Scholar]
- (11).Shaye DD, Greenwald I. Ortholist: A compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Luheshi LM, Crowther DC, Dobson CM. Protein misfolding and disease: from the test tube to the organism. Curr Opin Chem Biol. 2008;12:25–31. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]
- (13).Alavez S, Vantipalli MC, Zucker DJS, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kaminski Schierle GS, Bertoncini CW, Chan FTS, van der Goot AT, Schwedler S, Skepper J, Schlachter S, van Ham T, Esposito A, Kumita JR, Nollen EAA, et al. A FRET sensor for non-invasive imaging of amyloid formation in vivo. ChemPhysChem. 2011;12:673–680. doi: 10.1002/cphc.201000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen W, Avezov E, Schlachter SC, Gielen F, Laine RF, Harding HP, Hollfelder F, Ron D, Kaminski CF. A method to quantify FRET stoichiometry with phasor plot analysis and acceptor lifetime ingrowth. Biophys J. 2015;108:999–1002. doi: 10.1016/j.bpj.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Laine R, Stuckey DW, Manning H, Warren SC, Kennedy G, Carling D, Dunsby C, Sardini A, French PMW. Fluorescence lifetime readouts of Troponin-C-based calcium FRET Sensors: a quantitative comparison of CFP and mTFP1 as donor fluorophores. PLoS One. 2012;7:e49200. doi: 10.1371/journal.pone.0049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Avezov E, Konno T, Zyryanova A, Chen W, Laine R, Crespillo-Casado A, Melo EP, Ushioda R, Nagata K, Kaminski CF, Harding HP, et al. Retarded PDI diffusion and a reductive shift in poise of the calcium depleted endoplasmic reticulum. BMC Biol. 2015;13:2. doi: 10.1186/s12915-014-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Michel CH, Kumar S, Pinotsi D, Tunnacliffe A, George-Hyslop P, Mandelkow E, Mandelkow E-M, Kaminski CF, Kaminski Schierle GS. Extracellular monomeric tau protein is sufficient to initiate the spread of tau protein pathology. J Biol Chem. 2014;289:956–967. doi: 10.1074/jbc.M113.515445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Esbjorner EK, Chan F, Rees E, Erdelyi M, Luheshi LM, Bertoncini CW, Kaminski CF, Dobson CM, Kaminski Schierle GS. Direct observations of amyloid â self-assembly in live cells provide insights into differences in the kinetics of Aâ(1 − 40) and Aâ(1−42) aggregation. Chem Biol. 2014;21:732–42. doi: 10.1016/j.chembiol.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Chen W, Young LJ, Lu M, Zaccone A, Strohl F, Yu N, Kaminski Schierle GS, Kaminski CF. Fluorescence self-quenching from reporter dyes informs on the structural properties of amyloid clusters formed in vitro and in cells. Nano Lett. 2017;17:143–149. doi: 10.1021/acs.nanolett.6b03686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pinotsi D, Buell AK, Dobson CM, Kaminski Schierle GS, Kaminski CF. A label-free, quantitative assay of amyloid fibril growth based on intrinsic fluorescence. ChemBioChem. 2013;14:846–50. doi: 10.1002/cbic.201300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nystrom S, Bäck M, Nilsson KPR, Hammarstrom P. Imaging amyloid tissues stained with luminescent conjugated oligothiophenes by hyperspectral confocal microscopy and fluorescence lifetime imaging. J Vis Exp. 2017;128:1–7. doi: 10.3791/56279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Becker W. The bh TCSPC Handbook: Scanning. Becker & Hickl GmbH; 2010. pp. 1–566. [Google Scholar]
- (24).Draaijer A, Sanders R, Gerritsen HC. Handbook of Biological Confocal Microscopy. Springer US; Boston, MA: 1995. Fluorescence lifetime imaging, a new tool in confocal microscopy; pp. 491–505. [Google Scholar]
- (25).Dowling K, Dayel MJJ, Lever MJJ, French PMWMW, Hares JDD, Dymoke-Bradshaw AKL. Fluorescence lifetime imaging with picosecond resolution for biomedical applications. Opt Lett. 1998;23:810. doi: 10.1364/ol.23.000810. [DOI] [PubMed] [Google Scholar]
- (26).Alibhai D, Kelly DJ, Warren S, Kumar S, Margineau A, Serwa RA, Thinon E, Alexandrov Y, Murray EJ, Stuhmeier F, Tate EW, et al. Automated fluorescence lifetime imaging plate reader and its application to Förster resonant energy transfer readout of Gag protein aggregation. J Biophotonics. 2013;6:398–408. doi: 10.1002/jbio.201200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kumar S, Alibhai D, Margineanu A, Laine R, Kennedy G, Mcginty J, Warren S, Kelly D, Alexandrov Y, Munro I, Talbot C, et al. FLIM FRET technology for drug discovery: automated multiwell-plate high-content analysis, multiplexed readouts and application in situ. ChemPhysChem. 2011;12:609–626. doi: 10.1002/cphc.201000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Beam M, Silva MC, Morimoto RI. Dynamic imaging by fluorescence correlation spectroscopy identifies diverse populations of polyglutamine oligomers formed in vivo. J Biol Chem. 2012;287:26136–45. doi: 10.1074/jbc.M112.362764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen S, Berthelier V, Hamilton JB, O’Nuallai B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- (30).DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- (31).Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G, Davies S, Lehrach H, Wanker E. Huntingtin encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- (32).Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martínez-Sánchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, Ferrnndez-Busnadiego R. In situ architecture and cellular interactions of PolyQ inclusions. Cell. 2017;171:179–187. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- (33).Cremades N, Cohen SIA, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, Bertoncini CW, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149:1048–59. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen SW, Drakulic S, Deas E, Ouberai M, Aprile Fa, Arranz R, Ness S, Roodveldt C, Guilliams T, De-Genst EJ, Klenerman D, et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc Natl Acad Sci U S A. 2015;112:E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- (36).Bringmann H. Agarose hydrogel microcompartments for imaging sleep- and wake-like behavior and nervous system development in Caenorhabditis elegans larvae. J Neurosci Methods. 2011;201:78–88. doi: 10.1016/j.jneumeth.2011.07.013. [DOI] [PubMed] [Google Scholar]
- (37).Turek M, Besseling J, Bringmann H. Agarose microchambers for long-term calcium imaging of Caenorhabditis elegans. J Vis Exp. 2015;100:e52742. doi: 10.3791/52742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Warren SC, Nobis M, Magenau A, Mohammed YH, Herrmann D, Moran I, Vennin C, Conway JR, Mélénec P, Cox TR, Wang Y, et al. Removing physiological motion from intravital and clinical functional imaging data. Elife. 2018;7:e35800. doi: 10.7554/eLife.35800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Christensen RP, Bokinsky A, Santella A, Wu Y, Marquina-Solis J, Guo M, Kovacevic I, Kumar A, Winter PW, Tashakkori N, McCreedy E, et al. Untwisting the Caenorhabditis elegans embryo. Elife. 2015;4:e10070. doi: 10.7554/eLife.10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Peskett TR, Rau F, O’Driscoll J, Patani R, Lowe AR, Saibil HR. A liquid to solid phase transition underlying pathological huntingtin exon1 aggregation. Mol Cell. 2018;70:588–601. doi: 10.1016/j.molcel.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Perni M, Galvagnion C, Maltsev A, Meisl G, Muller MBD, Challa PK, Kirkegaard JB, Flagmeier P, Cohen SIA, Cascella R, Chen SW, et al. A natural product inhibits the initiation of α-synuclein aggregation and suppresses its toxicity. Proc Natl Acad Sci U S A. 2017;114:E1009–E1017. doi: 10.1073/pnas.1610586114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gritti N, Kienle S, Filina O, van Zon JS. Long-term time-lapse microscopy of C. elegans post-embryonic development. Nat Commun. 2016;7:12500. doi: 10.1038/ncomms12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cornaglia M, Krishnamani G, Mouchiroud L, Sorrentino V, Lehnert T, Auwerx J, Gijs MAM. Automated longitudinal monitoring of in vivo protein aggregation in neuro-degenerative disease C. elegans models. Mol Neurodegener. 2016;11:1–13. doi: 10.1186/s13024-016-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Grant DM, Elson DS, Schimpf D, Dunsby C, Requejo-Isidro J, Auksorius E, Munro I, Neil MAA, French PMW, Nye E, Stamp G, et al. Optically sectioned fluorescence lifetime imaging using a Nipkow disk microscope and a tunable ultrafast continuum excitation source. Opt Lett. 2005;30:3353–3355. doi: 10.1364/ol.30.003353. [DOI] [PubMed] [Google Scholar]
- (46).Warren SC, Margineanu A, Alibhai D, Kelly DJ, Talbot C, Alexandrov Y, Munro I, Katan M, Dunsby C, French PMW. Rapid global fitting of large fluorescence lifetime imaging microscopy datasets. PLoS One. 2013;8:e70687. doi: 10.1371/journal.pone.0070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.