Abstract
Drug repurposing provides a rapid approach to meet the urgent need for therapeutics to address COVID-19. To identify therapeutic targets relevant to COVID-19, we conducted Mendelian randomization (MR) analyses, deriving genetic instruments based on transcriptomic and proteomic data for 1,263 actionable proteins that are targeted by approved drugs or in clinical phase of drug development. Using summary statistics from the Host Genetics Initiative and the Million Veteran Program, we studied 7,554 patients hospitalized with COVID-19 and >1 million controls. We found significant Mendelian randomization results for three proteins (ACE2: P=1.6×10-6, IFNAR2: P=9.8×10-11, and IL-10RB: P=2.3×10-14) using cis-eQTL genetic instruments that also had strong evidence for colocalization with COVID-19 hospitalization. To disentangle the shared eQTL signal for IL10RB and IFNAR2, we conducted phenome-wide association scans and pathway enrichment analysis, which suggested that IFNAR2 is more likely to play a role in COVID-19 hospitalization. Our findings prioritize trials of drugs targeting IFNAR2 and ACE2 for early management of COVID-19.
Keywords: Mendelian randomization, drug repurposing, COVID-19, actionable druggable genome
Introduction
The global COVID-19 pandemic is responsible for substantial mortality, morbidity and economic hardship. Even with efficacious vaccines against the SARS-CoV-2 virus, it unknown how long it will take to achieve herd immunity, to what extent protection will diminish over time, or if future mutations will enable SARS-CoV-2 to evade immune responses stimulated by current vaccines. Hence, there is a need to rapidly identify drugs that can minimize the burden of COVID-19. Although large randomized trials have begun to successfully identity drugs that can be repurposed to address COVID-19,1–3 most drugs evaluated so far have failed to show efficacy and have been largely confined to hospitalized or critically-ill patients. It is a priority, therefore, to identify additional drugs that can be repurposed for early management in COVID-19.
Large-scale human genetic studies are now widely used to inform drug development programs. Drug target-disease pairs supported by human genetics have a greater odds of success in drug discovery pipelines.4 For example, identification of variants in PCSK9 associated with lower risk of coronary disease led to the successful development of PCSK9 inhibitors, which are now licensed for prevention of cardiovascular events.5 The value of human genetics for drug discovery and development has also been realized for infectious diseases. Human genetic studies showed that genetic variation in the CCR5 gene provides protection against infection by human immunodeficiency virus (HIV) type-1. These findings were key for the development of Maraviroc, an antagonist of CCR5, approved by the FDA for the treatment of patients with HIV-1.6
Genetic variants acting in “cis” on druggable protein levels or gene expression that encode druggable proteins can provide powerful tools for informing therapeutic targeting, as they mimic the on-target (beneficial or harmful) effects observed by pharmacological modification.7 Such Mendelian randomization (MR) analyses have been used to suggest repurposing opportunities for licensed drugs.8 MR analysis that focuses on actionable druggable genes, defined as genes that encode the protein targets of drugs that are licensed or in the clinical phase of drug development, could therefore serve as a swift and robust strategy to identify drug-repurposing opportunities to prevent the complications and mortality due to COVID-19.
To identify further potential repurposing opportunities to inform trials of COVID-19 patients, we conducted large-scale MR and colocalization analyses using gene expression and soluble protein data for 1,263 actionable druggable genes that encode protein targets for approved drugs or drugs in clinical development. By combining trans-ancestry genetic data from 7,554 hospitalized COVID-19 patients and more than 1 million population-based controls from the COVID-19 Host Genetics Initiative9 (HGI) and the Million Veteran Program10 (MVP), we provide support for two therapeutic strategies.
Results
Overall analysis plan
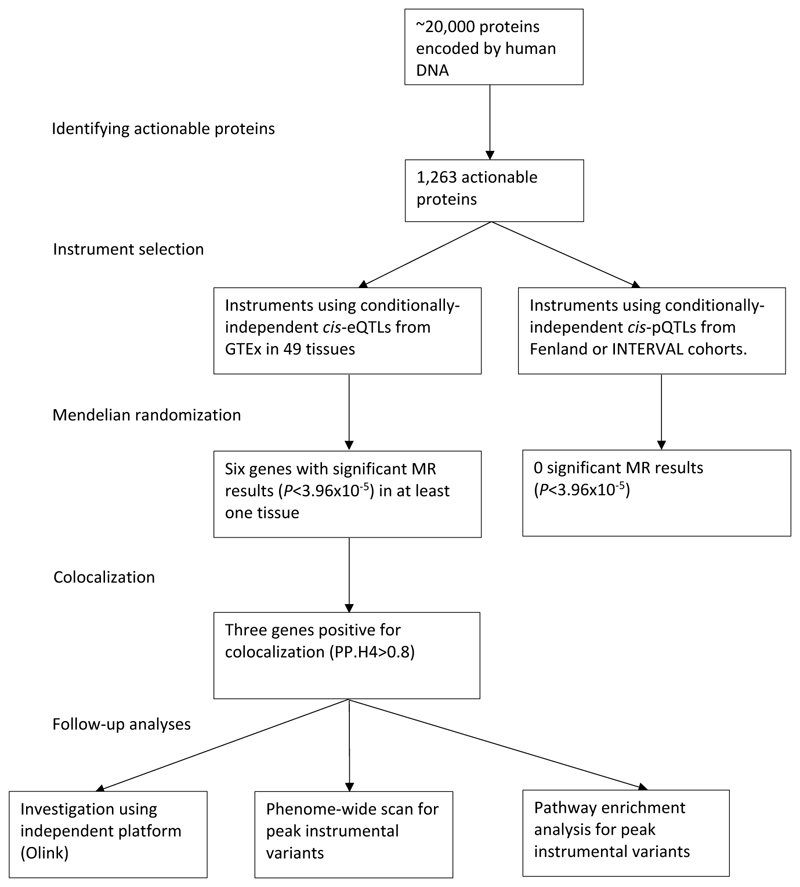
Figure 1 describes the overall scheme of the analyses. First, we identified all proteins that are therapeutic targets of approved or clinical-stage drugs. Next, we selected conditionally-independent genetic variants that act locally on plasma levels of these proteins or tissue-specific gene expression that encode these proteins. We proposed that these variants were instrumental variables and conduct two-sample MR analyses using a trans-ancestry meta-analysis of 7,554 cases from MVP and publicly available data (HGI outcome B2 from release 4 version 1, downloaded October 4th 2020, Supplementary Table 1). Given that all MR analyses relies on several assumptions, some11 unverifiable, we conducted a multi-stage strategy to minimize confounding and biases. For MR results that passed our significance threshold after accounting for multiple testing, we performed colocalization to ensure MR results were not due to confounding by linkage disequilibrium (LD). Those with evidence of colocalization were investigated further using an independent proteomics platform (Olink). Finally, we conducted phenome-wide scans and pathway enrichment of relevant variants to reduce risks of horizontal pleiotropy and other biases due to MR violations as well as to understand potential biological mechanisms.
Figure 1. Outline of the analyses performed.
Using multiple data sources, this study tested cis-pQTL and cis-eQTL proposed instruments for actionable druggable proteins against COVID-19 hospitalization summary statistics meta-analyzed from the Host Genetics Initiative and the Million Veteran Program. Significant MR associations that also showed evidence for colocalization were investigated further with an independent platform (Olink), phenome-wide scans of relevant variants, and pathway enrichment.
Actionable druggable proteins
Using data available in ChEMBL version 26, we identified 1,263 human proteins as “actionable” (i.e. therapeutic targets of approved or clinical-stage drugs) (Supplementary Table 2). Of these, we noted 700 proteins that are targets for drugs with potential relevance to COVID-19 from cell-based screening, registers of clinical trials against COVID-19 or approved immunomodulatory/anticoagulant drugs (given the clear role of these pathways in COVID-19 outcomes), or have biological evidence for the role of the protein in SARS-CoV-2 infection (Supplementary Table 3).
Genetic proposed instruments for actionable druggable proteins
Using GTEx version 8 (V8)12, we identified all conditionally-independent expression quantitative trait loci (eQTLs) in 49 tissues that act in cis (within 1 Mb on either side of the encoded gene), that covered 1,016 of the 1,263 druggable genes in at least one tissue (Supplementary Table 2 and 4). We also selected cis-pQTLs for plasma proteins measured using the SomaScan platform in 3,301 participants of the INTERVAL study13 (Supplementary Table 5) and 10,708 Fenland cohort participants14 (Supplementary Table 6) that covered a total of 67 proteins. In total 1,021 proteins had genetic proposed instruments using either eQTLs or pQTLs, and 62 had proposed instruments using both.
Mendelian randomization and colocalization
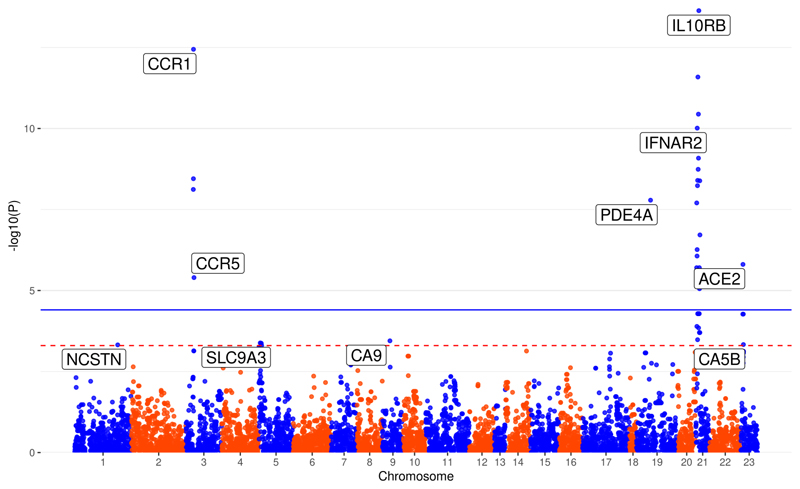
Using our (eQTL and pQTL) proposed instruments, we performed two-sample MR on transancestry summary statistics for hospitalized COVID-19 cases from MVP and HGI (Supplementary Table 1). Using GTEx cis-eQTLs as proposed instruments, we found significant (P<3.96×10-5, 0.05 Bonferroni-corrected for 1,263 actionable proteins) MR results for six genes (IL10RB, CCR1, IFNAR2, PDE4A, ACE2 and CCR5) in at least one tissue (MR results with P<3.96×10-5 shown in Table 1 and full MR results in Supplementary Table 7), and four additional genes (CA5B, CA9, NSTN and SLC9A3) with suggestive MR results (P<5.00×10-4 and P>3.96×10-5, Figure 2, Supplementary Table 7). No proposed instruments involving cis-pQTLs reached our suggestive threshold in any of the analyses (Supplementary Tables 8-9).
Table 1. Significant (P<3.96×10-5) MR results.
| Gene | Tissue | beta | S.E. | P value | P het | Variants in instrument | Colocalization |
|---|---|---|---|---|---|---|---|
| IL10RB | Muscle Skeletal | 0.5078 | 0.0665 | 2.31E-14 | 0.9732 | rs2300370, rs2834167 | 0.98, <0.01 |
| IL10RB | Nerve Tibial | 0.2859 | 0.0384 | 9.76E-14 | 0.0052 | rs13050728, rs2834167, rs2266590 |
0.98, <0.01, <0.01 |
| CCR1 | Cells Cultured fibroblasts | 0.4449 | 0.0612 | 3.60E-13 | NA | rs13095940 | <0.01 |
| IL10RB | Brain Nucleus accumbens basal ganglia |
0.2541 | 0.0363 | 2.58E-12 | 0.0019 | rs2834167, rs17860115 | 0.75, 0.98 |
| IL10RB | Brain Caudate basal ganglia | 0.2635 | 0.0398 | 3.61E-11 | 0.0003 | rs2834167, rs1051393 | 0.01, 0.97 |
| IFNAR2 | Muscle Skeletal | 0.5881 | 0.0909 | 9.75E-11 | NA | rs2300370 | 0.98 |
| IL10RB | Brain Cerebellar Hemisphere | 0.1405 | 0.0229 | 8.22E-10 | 0.0389 | rs2834167, rs2236758 | 0.01, 0.95 |
| IL10RB | Breast Mammary Tissue | 0.6490 | 0.1079 | 1.82E-09 | NA | rs12053666 | 0.95 |
| IL10RB | Brain Frontal Cortex BA9 | 0.4667 | 0.0790 | 3.55E-09 | 0.0366 | rs2834167, rs1131668 | 0.14, 0.97 |
| IL10RB | Brain Cortex | 0.1929 | 0.0328 | 3.99E-09 | 0.0354 | rs2834167, rs1131668 | 0.02, 0.96 |
| CCR1 | Esophagus Gastroesophageal Junction |
0.1776 | 0.0302 | 4.11E-09 | NA | rs13059906 | 0.05 |
| IL10RB | Brain Cerebellum | 0.1147 | 0.0197 | 5.82E-09 | 0.0239 | rs2834167, rs1131668 | <0.01, 0.96 |
| CCR1 | Esophagus Mucosa | 0.4338 | 0.0751 | 7.60E-09 | NA | rs34059564 | <0.01 |
| IFNAR2 | Esophagus Mucosa | -0.4883 | 0.0865 | 1.63E-08 | NA | rs11911133 | 0.92 |
| PDE4A | Artery Aorta | -0.5420 | 0.0965 | 1.98E-08 | 0.0202 | rs370630099, rs45524632 | 0.41, 0.61 |
| IL10RB | Testis | 0.7104 | 0.1364 | 1.92E-07 | NA | rs2284550 | 0.11 |
| IFNAR2 | Skin not Sun Exposed Suprapubic | -0.3360 | 0.0671 | 5.46E-07 | NA | rs8127500 | <0.01 |
| IFNAR2 | Pancreas | -0.4708 | 0.0957 | 8.63E-07 | NA | rs1476415 | 0.06 |
| ACE2 | Brain Frontal Cortex BA9 | 0.1121 | 0.0233 | 1.56E-06 | NA | rs4830976 | 0.95 |
| IFNAR2 | Cells Cultured fibroblasts | -0.3893 | 0.0819 | 1.98E-06 | NA | rs1131668 | 0.92 |
| IL10RB | Cells Cultured fibroblasts | -0.5197 | 0.1093 | 1.98E-06 | NA | rs1131668 | 0.96 |
| CCR5 | Lung | -0.5868 | 0.1272 | 3.99E-06 | NA | rs12639314 | 0.02 |
| IL10RB | Esophagus Gastroesophageal Junction |
0.4678 | 0.1052 | 8.80E-06 | NA | rs56079299 | 0.96 |
Significant Mendelian randomization results, P<3.96×10-5 (0.05 Bonferroni-corrected for 1,263 actionable druggable genes). Mendelian randomization estimates were calculated using inverse-variance weighting and fixed effects for instruments that contained more than one variant, and Wald-ratio for instruments with one variant. All results used cis-eQTL instruments and no results using cis-pQTL instruments yielded results P<3.96×10-5. Phet refers to the heterogeneity P value across individual-variant MR estimates within a genetic instrument calculated using the Cochrane Q method, therefore instruments containing one variant were not tested for not tested for heterogeneity. A positive beta estimate indicates that more gene expression is associated with higher risk of COVID-19 hospitalization. “Colocalization” indicates PP.H4 between eQTLs and COVID-19 hospitalization. For example, for IL10RB in skeletal muscle, the primary GWAS with rs2300370 as the peak cis-eQTL colocalizes with COVID-19 hospitalization at PP.H4=0.98, and the secondary GWAS (i.e. after adjusting for rs2300370) with rs2834167 as the peak cis-eQTL does not colocalize with COVID-19 hospitalization (PP.H4<0.01).
Figure 2. Manhattan plot of results from actionable druggable genome-wide Mendelian randomization analysis.
Mendelian randomization estimates were calculated using inverse-variance weighting and fixed effects for instruments that contained more than one variant, and Wald-ratio for instruments with one variant. Blue solid line indicates the P value threshold for significance (P<3.96×10-5, 0.05 Bonferroni-corrected for 1,263 actionable druggable genes) and red dashed line indicates a suggestive (P<5.00×10-4) threshold. Genes are labeled by their most significant MR association. For example, the results for IL10RB is most significant with cis-eQTL proposed instruments derived in skeletal muscle tissue (P=2.31x10-14), which is the point labeled. Results are plotted by the gene start position. All MR results with P value less than 5.00×10-4 used the GTEx cis-eQTLs as proposed instruments.
For three significant genes (IL10RB, IFNAR2, ACE2) there was strong evidence of colocalization (posterior probability of shared causal variant across two traits - hypothesis 4 [PP.H4] >0.80) between at least one proposed instrumental variant and our trans-ancestry meta-analysis of COVID-19 hospitalization (Table 1). Beta-coefficients of MR estimates for ACE2 were positive in all tissues (Table 1), meaning higher ACE2 expression is associated with higher risk of COVID-19 hospitalization. MR beta-coefficients for IFNAR2 and IL10RB were negative and positive, respectively, in all tissues except one for each gene (skeletal muscle for IFNAR2; cultured fibroblasts for IL10RB; Table 1).
IL10RB and IFNAR2
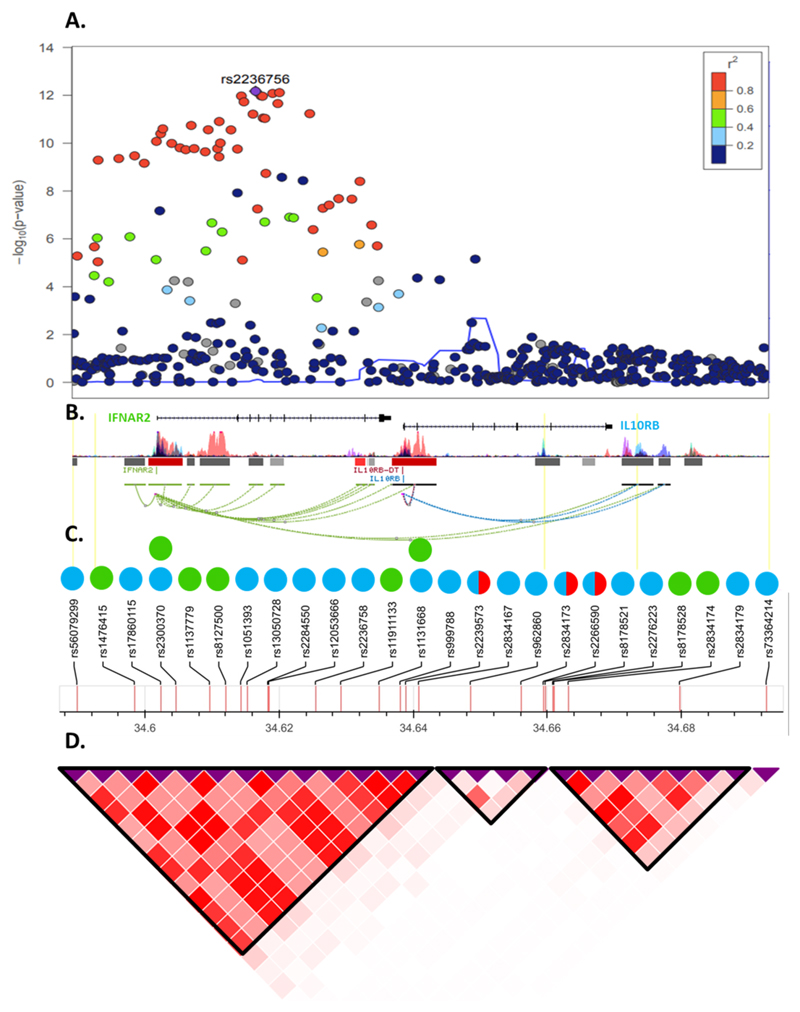
Interferon alpha receptor 2 (IFNAR2) and interleukin 10 receptor beta (IL-10RB) both act as receptors for interferons (IFN). IFNAR2 forms a complex with IFNAR1, which together act as a receptor for type I IFN (IFN-α, β, ω, κ, ε), while IL-10RB acts as a receptor for type III IFN (IFN-λ) when complexed with interferon lambda receptor-1 (IFNLR1)15, or IL-10 when complexed with IL-10RA. IL-10RB and IFNAR2 are encoded by adjacent genes and some cis-eQTLs for IL10RB are also cis-eQTLs for IFNAR2 (Supplementary Table 10, Figure 3), making it difficult to determine which gene may be responsible for the association with COVID-19 and requiring further investigation.
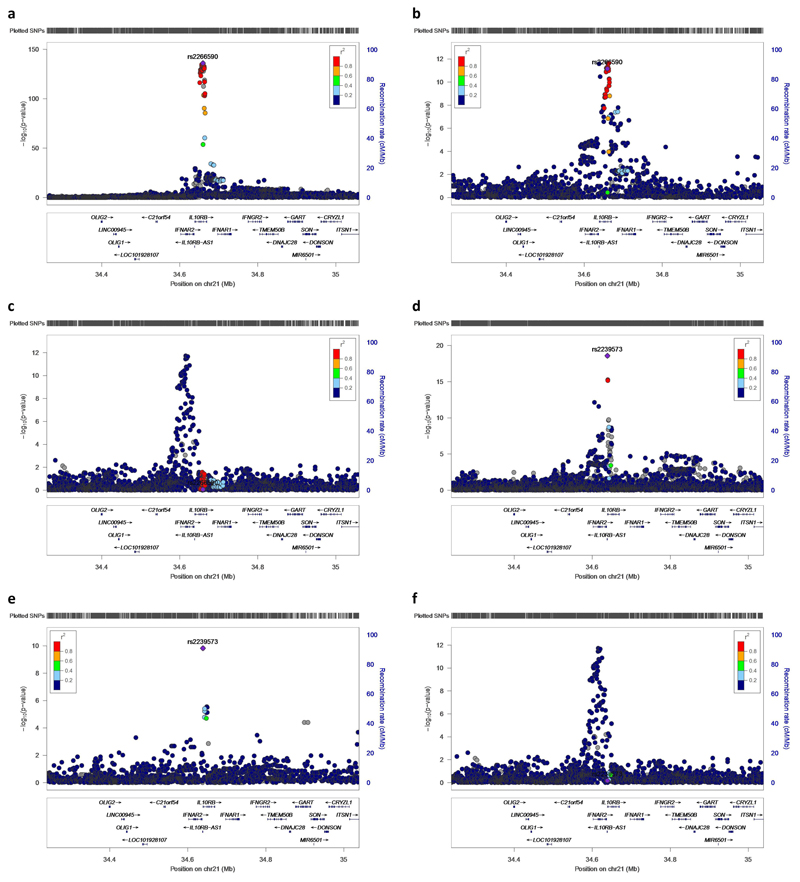
Figure 3. Genomic context, local association plot and LD structure of the IFNAR2/IL10RB region.
a, Local association plot (N=1,377,758) of the interval defined by all unique eQTLs for IL10RB or IFNAR2. Color code represents the degree of linkage disequilibrium with the most associated marker in 1000G Europeans. b, Genomic context of the region. Coding genes are represented by the refseq transcript. Bars represent epigenome Roadmap layered H3K27 acetylation markers. Connecting lines represent significant Hi-C interactions. c, Set of rsIDs used as proposed instruments for Mendelian Randomization analysis. Color code represents instruments for IL10RB (blue circles), IFNAR2 (green circles). Orange half-circles represent pQTLs for IL-10RB. d, Linkage disequilibrium structure and blocks defined using European populations from 1000G project.
All significant MR results for IFNAR2/IL10RB that colocalized with COVID-19 hospitalization contained one of nine strongly correlated (r2>0.75 in 1000G European ancestry participants) variants (rs11911133, rs1051393, rs2300370, rs56079299, rs17860115, rs13050728, rs2236758, rs12053666, and rs1131668), which are cis-eQTLs for IL10RB in eleven tissues and for IFNAR2 in four tissues (Supplementary Table 11). Within this LD block (hereafter rs13050728-LD block), rs13050728 is the eQTL most strongly associated with COVID-19 hospitalization (per T-allele odds ratio = 1.17; 95% CI = 1.12-1.23; P=1.88×10-12; Supplementary Table 10). Variants outside the rs13050728-LD block were not strongly associated with COVID-19 hospitalization (Figure 3).
pQTLs for IL10RB
Using stepwise conditional analysis on Olink measurements of plasma IL-10RB we identified two cis-pQTLs, rs2266590 (P=1.04×10-136) and rs2239573 (P= 2.66×10-19), which explained 5.4% and 1.2%, respectively, of the variance in plasma IL-10RB. rs2266590 was also an eQTL for IL10RB in three tissues and IFNAR2 in one tissue, while rs2239573 was also an eQTL for IL10RB in two tissues (Supplementary Table 11). rs2266590 and rs2239573 lie in intron 5 and 1, respectively, of the IL10RB gene and are located in separate regions of high epigenetic modification (h3k27ac marking), indicating enhancer regions (Figure 3). rs2266590 and rs2239573 were not associated with COVID-19 hospitalization (P= 0.85 for rs2266590, P=0.66 for rs2239573, Extended Data Fig. 1) and MR using these two cis-pQTLs yields a null result (P=0.74).
A third cis-pQTL (rs2834167, P=1.1×10-8) for plasma IL-10RB measured on the SomaScan platform was previously identified in 3,200 Icelanders over the age of 65.16 rs2834167 is a missense variant (Lys>Glu) and is not correlated with either of the cis-pQTLs for plasma IL-10RB measured by Olink (r2=0.01 for rs2266590, r2=0.03 for rs2239573 in 1000G EUR). Although rs2834167 was associated with IL10RB expression in 18 tissues, it was not associated with IFNAR2 expression in any tissue (Supplementary Table 11). The A allele at rs2834167, which is associated with lower IL10RB gene expression but higher plasma IL-10RB, was inversely associated with COVID-19 (per-A-allele OR= 0.91; 95%CI= 0.87-0.95; P=5.3×10-5). Because Emilsson et al.16 did not report full summary statistics we could not perform colocalization between this pQTL and COVID-19 hospitalization. However, rs2834167 as an eQTL does not colocalize (PP.H4<0.8) with COVID-19 in any tissue (Table 1). These three cis-pQTLs, while possibly functional variants altering plasma IL-10RB levels, suggest that the plasma IL-10RB levels are not likely the mediator of the association between this locus and COVID-19 hospitalization. IFNAR2 was not measured on the SomaScan or Olink platforms.
Phenome-wide scan of rs13050728
To identify other phenotypes associated with rs13050728, we performed a phenome-wide scan of GWAS for proteins measured by Olink and SomaLogic platforms in INTERVAL participants (see methods), and publicly available data on PhenoScanner17 and GTEx. rs13050728 was associated with tryptase gamma 1 (TPSG1, P= 1.5×10-5) and vascular endothelial growth factor 2 (VEGFR2, P= 2.6×10-5, Supplementary Table 12), and both showed strong evidence of colocalization with COVID-19 hospitalization (PP.H4=0.96 for VEGFR2, PP.H4=0.96 for TPSG1, Figure 4). The C allele at rs13050728 associated with higher IFNAR2 expression in all tissues (except skeletal muscle), lower risk of COVID-19 hospitalization, and lower levels of plasma VEGFR2 and TPSG1 (Supplementary Table 12). This mimics agonistic effects of IFNAR2 through recombinant type-I IFNs, which are known to have an anti-angiogenic effect, at least in part through reduced VEGF/VEGFR2 signaling18,19, and decrease tryptase levels in a phase-2 trial using recombinant type-I IFN in patients with mastocytosis20, a condition that causes proliferation of mast cells. rs13050728 was not associated at P<3.96×10-5 (our Bonferroni corrected P value) with any phenotype beyond plasma VEGFR2 and TPSG1 and gene expression of IFNAR2 and IL10RB (Supplementary Table 12), indicating that this variant is unlikely to exhibit widespread horizontal pleiotropy. Also, the chances of substantial bias due to MR violations is low21 since the variant is not strongly associated with other risk factors that could alter the likelihood of SARS-CoV-2 testing or hospitalization of COVID-19 patients.
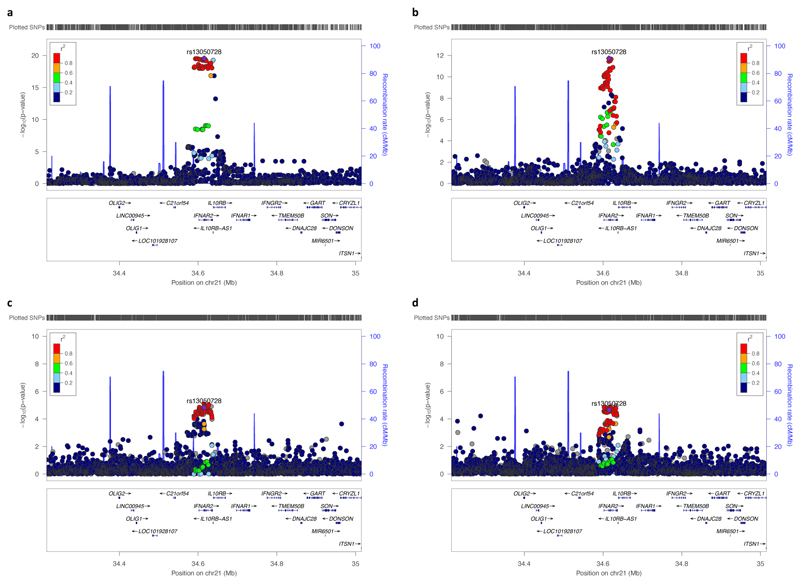
Figure 4. Regional association plots of the IFNAR2-IL10RB locus.
Regional association plots for a, IL10RB gene expression in tibial nerve tissue from GTEx (N=532), b, COVID-19 hospitalization from HGI and MVP trans-ancestry meta-analysis (N=1,377,758) c, d, Vascular endothelial growth factor receptor 2 (VEGFR2) and tryptase gamma (TPSG1), respectively, measured by SomaLogic in 3,301 INTERVAL participants. All show the correlation (1000G European ancestry) for rs13050728, the cis-eQTL most associated with COVID-19 hospitalization in the IL10RB-IFNAR2 region. All colocalize with each other (PP.H4>0.96 for all).
Pathway enrichment analysis of rs13050728
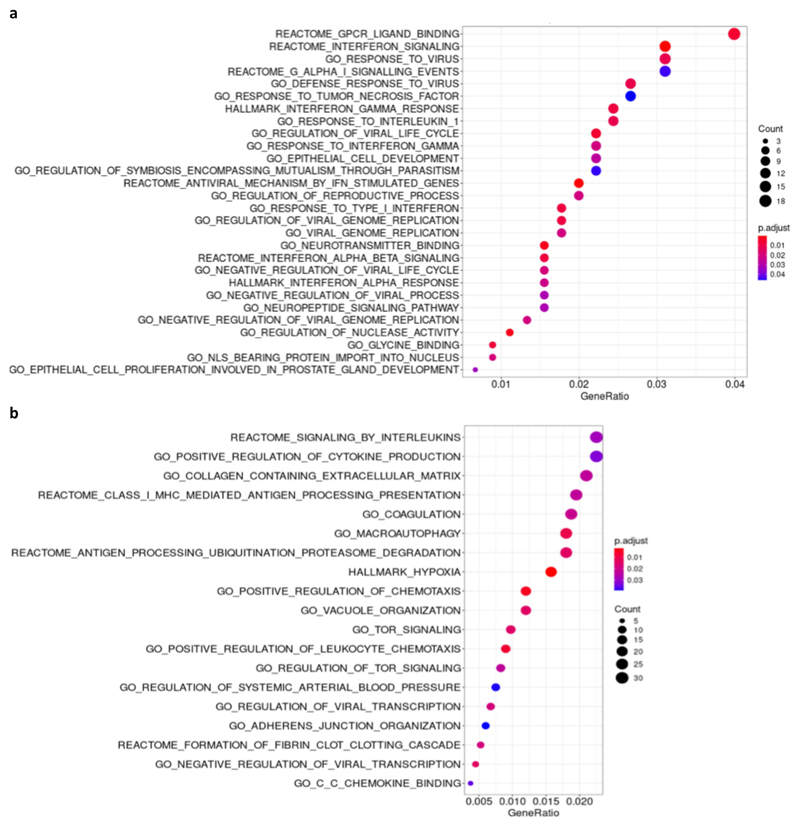
Using information from all GTEx V8 tissues we identified 476 genes whose expression levels were associated with rs13050728 at a nominal significance level (P < 0.05). Taking into consideration an adjusted P value for multiple testing within the WikiPathway corpus, only two biological pathways were significantly associated among all 624 pathways present in this database: Host-pathogen interaction of human corona viruses - IFN induction (adjusted P value = 0.0028) and Type I IFN Induction and Signaling During SARS-CoV-2 Infection (adjusted P value = 0.0098). In addition, among Gene Ontology (GO) and Reactome pathways, several gene sets were also significantly enriched. Notably, among enriched pathways were those related to IFN type I or antiviral response (Extended Data Fig. 2A).
ACE2
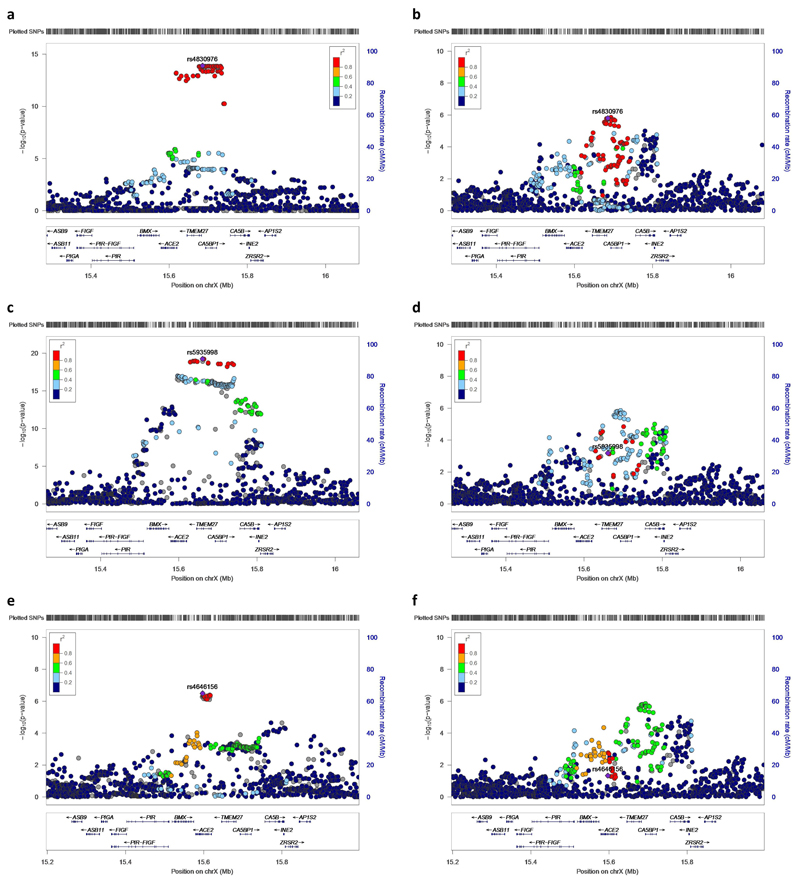
Angiotensin converting enzyme 2 (ACE2) converts angiotensin II into angiotensin (1-7) as part of the RAA system, and more importantly, is the viral receptor for SARS-CoV-2. We identified seven cis-eQTLs in seven tissues (Supplementary Table 13) for ACE2 which are strongly correlated (r2>0.75 in 1000G EUR, Supplementary Table 14) with rs4830976 being the eQTL in the region most strongly associated with COVID-19 hospitalization.
pQTLs for ACE2
Stepwise conditional analysis for plasma ACE2 measured by Olink revealed one pQTL, rs5935998 (P=1.45×10-21), which is in high LD with a previously reported cis-pQTL (rs12558179) for ACE2 (r2=0.89 in 1000G EUR)22, and a secondary suggestive signal (rs4646156, P= 3.20×10-7). rs5935998 and rs4646156 are concordant in their effect on COVID-19 hospitalization (higher ACE2 levels corresponds to higher risk of COVID-19 hospitalization for both) resulting in a strong, positive MR association (MR beta-coefficient: 0.34; 95% CI: 0.17-0.51; P=8.1×10-5). Although neither rs5935998 or rs4646156 strongly colocalized with COVID-19 hospitalization (PP.H4=0.49 for rs5935998, PP.H4=0.08 for rs4646156, Extended Data Fig. 3), the two pQTLs, while statistically independent, are mildly correlated (r2=0.20 in 1000G EUR), which can make colocalization difficult to interpret.23 One possible explanation is that these two pQTLs confer an effect on COVID-19 hospitalization that converges on the rs4830976-LD-block, as both are moderately correlated with rs4830976 (r2=0.32 for rs5935998, r2=0.42 for rs4646156 in 1000G EUR, Extended Data Fig. 3)
Phenome-wide scan of rs4830976
rs4830976 is associated (P<3.96×10-5) with and colocalized (PP.H4>0.80) with expression of nearby genes CA5B, CLTRN (also known as TMEM27), and VEGFD (Supplementary Table 15) in at least one tissue, indicating that this variant may be instrumenting on gene expression beyond ACE2. However, given the biological prior that ACE2 acts as the receptor of SARS-CoV-2, ACE2 is probably more likely than CA5B, CLTRN or VEGFD to be responsible for COVID-19 hospitalization. There were no other reported phenome-wide scan results at P<3.96×10-5 for rs4830976, which is at least in part due to the lack of reported X-chromosome results from a large proportion of GWAS.
Pathway enrichment analysis of rs4830976
Exploring the landscape of genes differentially expressed according to genotype in GTEx V8, we observed 1397 genes differentially expressed at a nominal P value less than 0.05. Over-representation analysis identified 238 significantly enriched biological pathways among differentially expressed genes (Extended Data Fig. 2B). Among these, signaling by interleukins, regulation of cytokine production, and antigen processing and presentation, might prove biologically relevant in COVID-19 infection.
Discussion
To identify drug-repurposing opportunities to inform trials against COVID-19, we conducted a large-scale MR analysis of protein and gene expression data. We first updated the “actionable” genome to an enlarged set of 1,263 human proteins and provided evidence for 700 of these as targets for drugs with some potential relevance to COVID-19. By investigating more than a thousand potential targets using several of the largest currently available human genetic datasets, we provide evidence for drug targets of type-I IFNs (IFNAR2) and ACE2 modulators (ACE2) as priority candidates for evaluation in randomized trials of early management in COVID-19.
Our finding that ACE2 may play an important role in COVID-19 is unsurprising given its well-known relevance to SARS-CoV-2. Since ACE2 acts as the primary receptor for SARS-CoV-2, increased expression of ACE2 has been hypothesized to lead to increased susceptibility to infection. ACE2 plays a vital role in the RAAS signaling pathway, providing negative regulation through the conversion of Angiotensin II to Angiotensin 1-7. This action has anti-inflammatory and cardioprotective effects24 and plays a protective role in acute respiratory distress syndrome.25 ACE2 is a single-pass membrane protein but can be cleaved from the membrane to a soluble form which retains the enzymatic function to cleave Angiotensin II. It has therefore been hypothesized that administration of human recombinant soluble ACE2 (hrsACE2) could be an effective treatment for COVID-19, through distinct mechanisms in two phases of COVID-19. First, hrsACE2 can bind the viral spike glycoprotein of SARS-CoV-2, which could prevent cellular uptake of SARS-CoV-2 by reducing binding to the membrane-bound form of ACE2 (early phase). This suggestion is supported by the finding that APN01, a hrsACE2 therapeutic, showed a strong reduction in SARS-CoV-2 viral load26 and enhanced the benefit of remdesivir27 in primate kidney epithelial (Vero) cells and human kidney organoids. In the later phase, hrsACE2 could reduce sequelae of SARS-CoV-2 infection by reducing inflammation in the lungs and other infected tissues. A case report of a hospitalized COVID-19 patient supports this hypothesis by showing that 7-day administration of APN01 was associated with a reduction in SARS-CoV-2 viral load and inflammatory markers.28 APN01 is currently being tested in a phase II trial to reduce mortality and invasive mechanical ventilation in 200 hospitalized COVID-19 patients29. Interestingly, a recent report showed that expression of a truncated ACE2 isoform, dACE2, which poorly binds with SARS-CoV-2 spike protein, is stimulated by type I, II and III IFNs in human ileum organoids30.
One of the main challenges of our analysis was to determine whether IFNAR2 or IL10RB (or both) was driving the association with COVID-19 hospitalization, given that they share cis-eQTLs used as proposed instruments for our MR analysis. Multiple lines of evidence indicate that IFNAR2 appears to be primarily responsible for the signal observed. First, our phenome-wide scan using the lead IFNAR2/IL10RB cis-eQTL reproduced known effects of type-I IFNs (the therapeutic target of IFNAR2) on VEGFR2 and TPSG1.18–20 Second, our pathway enrichment analysis using the same eQTL revealed pathways associated with type-I IFN receptor (IFNAR2) signaling. Last, three independent cis-pQTLs that are also cis-eQTLs for IL10RB did not show evidence of association with COVID-19, suggesting that plasma IL-10RB concentrations are less likely to be etiologically relevant to COVID-19.
Evidence of a role for type-I IFN in COVID-19 is rapidly emerging. Studies using in vitro (A549 pulmonary cell lines), animal (ferrets) and ex vivo (human lung tissue) models have all shown lower expression of genes encoding type-I IFNs after exposure to SARS-CoV-2 compared to other respiratory viruses.31,32 This has been confirmed in vivo by studies showing significantly impaired type-I IFN response – including almost no IFN-beta activity - in the peripheral blood of severe COVID-19 patients compared to mild to moderate COVID-19 patients.33 More importantly, lower levels of IFN alpha-2 among recently hospitalized COVID-19 patients were associated with a substantial increase in the risk of progression to critical care, supporting our observation that lower genetically-predicted IFNAR2 expression was associated with higher risk of COVID-19 hospitalization.33 Additionally, auto-antibodies for type I IFNs were found in a much higher proportion of individuals with severe COVID-19 than those with asymptomatic or mild SARS-CoV-2 infection.34
Whole exome and genome sequencing studies on severe COVID-19 patients have identified rare mutations that implicate type I IFN signaling. Zhang et al.35 found patients with severe COVID-19 were enriched for rare variants predicted to cause loss of protein function at 13 genes involved in type-I IFN response. A cases-series of four patients under the age of 35 with severe COVID-19 found a rare LOF mutation in TLR7 and decreased type I IFN signaling.36 The GenOMICC study of imputed GWAS on severe COVID-19 identified signals that lie in the IFNAR2 gene.37
Several in vitro studies have found a reduction in SARS-CoV-2 replication in multiple cell types (including animal and human) and human organoids after pre-treatment with type-I or -III IFNs when compared with controls38–41 (Supplementary Table 16). Though these in vitro studies are encouraging, evidence from randomized trials for type I IFNs in early COVID-19 stages is limited. Hung et al.42 showed that randomization to a combination of IFN beta-1b, ribavirin and lopinavir-ritonavir was superior to lopinavir-ritonavir alone in shortening the duration of viral shedding, alleviating symptoms and reducing the length of the hospital stay. Importantly, these benefits were confined to a subgroup who were hospitalized within 7 days of onset of symptoms when IFN beta-1b was administered to the intervention arm. These results, together with our genetic findings on COVID-19 hospitalization and the established role of type-I IFNs as first line of response against viral agents suggest recombinant type-I IFN as potential intervention during early stages of COVID-19. To date, there is no large randomized trial on IFN beta for early treatment of COVID-19 patients who are at high risk of hospitalization.
Trial evidence on the use of IFN-beta in late stages of COVID-19 has emerged very recently. The SOLIDARITY trial, which randomized 2,050 hospitalized COVID-19 patients to IFN beta-1a, found no effect on mortality overall (relative risk [RR]=1.16, 95% CI: 0.96-1.39), but the trial was not powered to evaluate a possible trend across subgroups of COVID-19 severity at randomization (RR=1.40, 95% CI: 0.82-2.40 for those on ventilator, RR=1.13, 95% CI: 0.86-1.50 for those not ventilated but on oxygen, and RR=0.80, 95% CI: 0.27-2.35 in those with neither).43 The Adaptive COVID-19 Treatment Trial 3 (ACTT-3) stopped enrollment of severely ill COVID-19 patients for a trial on IFN beta-1a and remdesivir due to adverse events but continued enrolling patients with less severe disease.44 The ACTT-2 found that baricitinib (an inhibitor of the JAK family of proteins, some of which are immediately downstream of IFNAR2) when administered to hospitalized COVID-19 patients was beneficial in severe cases but not in moderate disease3. These findings indicate no role for the use of IFN beta during late stages of COVID-19, when the cytokine storm is already established.
We are the first to implicate a causal role for ACE2 in COVID-19 manifestations using MR techniques; we have also implicated IFNAR2 in COVID-19, concordant with recent studies37,45. However, the current study importantly complements and extends previous efforts by employing key approaches to protect against potential biases, strengthen causal inference, and enhance understanding of potential mechanisms. First, in contrast to Liu et al. and the GenOMICC study, the current study involved several measures to minimize potential biases. We used colocalization methods to minimize the chances of false positive results due to confounding by LD. We reduced the possible impact of bias due to horizontal pleiotropy by restricting our proposed instruments to variants acting in cis and performing phenome-wide scan to ensure instrumental variants were only associated/colocalized with gene expression of the tested gene or downstream phenotypes. When the possibility of horizontal pleiotropy was identified (e.g. IFNAR2 and IL10RB sharing eQTLs), we addressed it using pQTL data and pathway enrichment analysis to disentangle mechanisms, ultimately showing IFNAR2 is more likely the causal gene. Phenome-wide scans revealing effects on plasma proteins (VEGFR2 and TPSG1) that mimic known biology of type I IFN provides confidence that we are correctly instrumenting IFNAR2 and can identify on-target (harmful or beneficial) effects of administering type I IFN.
Second, our study had excellent statistical power, yielding highly significant MR associations for IFNAR2 (P=9.8×10-11), increasing confidence in the validity of the much weaker signals for IFNAR2 reported in the GenOMICC study (P=0.004), particularly as the earlier report had displayed evidence of confounding by LD (PHEIDI= 0.015).37 Indeed, compared to the analysis on COVID-19 hospitalization by Liu et al., our analysis contained more than double the number of cases.45 Third, with our rigorous instrument selection process that used comprehensive datasets on gene expression and plasma protein levels, we were able to robustly evaluate over one thousand actionable drug targets, like ACE2, which was not evaluated in the previous MR studies. Fourth, inclusion of MVP with HGI provided a more diverse population and identification of credible biological targets that were consistent across multiple ancestral groups.
Lastly, we provide an updated catalog of all actionable protein targets and drugs that are amenable to causal inference investigation through human genetics that can be applied to outcomes beyond COVID-19. For 700 proteins of the actionable genes, we also include information as to potential relevance to the treatment of COVID-19, which can help future studies to contextualize findings on COVID-19.
Our analysis also has limitations. Though we make use of instrumental variants from multiple data sources, they did not cover the entire actionable druggable genome or were derived from COVID-19 patients. Identifying the most relevant tissue or cell-type can be challenging for interpreting MR analyses of gene expression. In our case, a relevant tissue could be: one invaded by SARS-CoV-2, an organ associated with clinical complications of COVID-19, a tissue where the COVID-19-relevant protein is produced, or a tissue that would be the likely site of action for the target drug. We opted to use a data-driven strategy that incorporates all tissues available in GTEx V8. For IFNAR2, we recovered fibroblasts (the main cell type responsible for IFN-beta production), esophageal mucosa46 (a tissue invaded by SARS-CoV-2), and skeletal muscle47 (associated with the neurological manifestations of COVID-19). For ACE2, we recovered brain tissue, an organ known to be invaded by SARS-CoV-2 and associated with clinical manifestations.48,49 Lastly, this work focused on cis-variants with an effect on gene expression and protein levels. We did not consider the full complexity of gene isoforms and splice SNPs, therefore missing mediation relationships that are isoform-specific. Also, we did not consider other pathways through which variants may affect disease, such as DNA methylation, histone modification, chromatin accessibility and others.
In conclusion, our trans-ancestry MR analysis covering all actionable druggable genes identified two drug repurposing opportunities (type-I IFNs and hsrACE2) as interventions that need to be evaluated in adequately powered randomized trials to investigate their efficacy and safety for early management of COVID-19.
Methods
Identification of actionable druggable genes suitable for repurposing against COVID-19
Information about drugs and clinical candidates, and their therapeutic targets, was obtained from the ChEMBL database (release 2650, Supplementary Methods). For the purposes of our COVID-19 drug repurposing efforts, actionable proteins were defined as those that are therapeutic targets of approved drugs and clinical candidates or are potential targets of approved drugs. Therapeutic targets were identified from the drug mechanism of action information in ChEMBL and linked to their component proteins. Each protein was assigned a confidence level based on the type and size of target annotated, and the resulting list was filtered to remove non-human proteins and those with lower confidence assignments (cases where the therapeutic target consists of more than 10 proteins or the protein is known to be a non-drug-binding subunit of a protein complex). For approved drugs, additional potential human target proteins were identified from pharmacological assay data in ChEMBL with recorded affinity/efficacy measurements <= 100nM (represented by a pChEMBL value >= 7).
A total of 1,263 unique human proteins were identified as ‘actionable’ from data available in ChEMBL. These consisted of 531 proteins that are therapeutic targets of approved drugs, 381 additional proteins that are therapeutic targets of clinical candidates and 351 additional proteins that are bound by approved drugs, but not annotated as the therapeutic targets. While the biological relevance of the latter group of targets in the context of the approved drug indications may be unclear, the high affinity/efficacy measurements suggest the drug should be capable of modulating these proteins, should they be found to be relevant to COVID-19 (although likely not in a selective manner). Proteins were further annotated with biological and drug information relating to their potential role in SARS-CoV-2 infection (Supplementary methods) such as change in abundance during infection, interaction with viral proteins or the activity of drugs in antiviral cell-based assays. Of the 1,263 actionable proteins identified previously, 300 were annotated as biologically relevant in SARS-CoV-2 infection and 547 were targets of drugs with some evidence of COVID-19 relevance from cell-based assays, clinical trials or the ATC classification (Supplementary Table 2).
Selection of proposed instruments
eQTL proposed instruments
We proposed eQTL instruments using raw data from GTEx Version 8 by performing conditional analysis on normalized gene expression in European ancestry individuals in 49 tissues that had at least 70 samples. eQTLs were derived in all 49 tissues (i.e. we did not restrict it to tissues we thought most relevant to COVID-19) because the biological relevance of tissues to SARS-CoV-2 infection is still rapidly evolving. We used Matrix eQTL51 and followed the same procedure as outlined by the GTEx consortium (https://gtexportal.org/home/). Briefly, after filtering the genotypes (genotype missingness <0.05, MAF<0.01, HWE<0.000001, removing ambiguous SNPs), within each tissue, we performed GWAS between variants and gene expression adjusting for sex, the first 5 principal components of European genetic ancestry, PEER factors, sequencing platform and protocol. To identify independent eQTLs, we performed conditional analysis in regions around associations that fell below genome-wide significance, additionally adjusting for the peak variant if there exists an association reaching a P-value of 5.00×10-8. Cis-eQTLs were defined as significant (P<5.00×10-8) associations within 1Mb on either side of the encoded gene. To convert from build 38 to build 37, we used the table available from the GTEx consortium for all variants genotyped in GTEx v8 and hg19 liftover, (https://storage.googleapis.com/gtex_analysis_v8/reference/GTEx_Analysis_2017-06-05_v8_WholeGenomeSeq_838Indiv_Analysis_Freeze.lookup_table.txt.gz). In each tissue, multiple GW-significant (P<5.00×10-8) eQTLs for the same gene were combined into a single instrument using inverse-variance weighting and fixed-effects meta-analysis across variant-level MR estimates for each variant, a standard two-sample MR approach. For example, for IL10RB expression in skeletal muscle tissue, there were two conditionally-independent eQTLs (rs2300370 and rs2834167, Table 1); a variant-level MR-estimate was obtained for each by dividing the beta-coefficient for COVID-19 hospitalization by the beta-coefficient of the eQTL, and dividing the standard error of the COVID-19 hospitalization estimate by the beta-coefficient of the eQTL. The two variant-level MR estimates were then meta-analyzed using inverse-variance weighting and fixed effects to yield the final MR result. Instruments for expression of the same gene derived in different tissues were tested separately.
pQTL proposed instruments
We proposed pQTL instruments from two sources of publicly available data that reported conditionally independent pQTLs for proteins measured by the SomaLogic Inc. (Boulder, Colorado, US) SomaScan52,53 platform: (1) Sun et al.13, which reported results for 2,994 proteins in 3,301 INTERVAL participants and (2) Pietzner et al.14, which reported results for 179 proteins in 10,708 participants of the Fenland cohort. In both, we restricted proposed instrumental variants to cis-pQTLs for actionable proteins, used a P value threshold of 5×10-8 and removed variants with MAF<0.01. MR was run independently for each data source (i.e. proposed instruments for the same protein in different platforms were tested against COVID-19 hospitalization independently).
Estimates for COVID-19 hospitalization
To generate outcome summary-statistics, we meta-analyzed results from the Million Veteran Program (MVP), an ongoing, prospective cohort recruiting from 63 Veterans Health Administration (VA) medical facilities (Supplementary Methods), and the Host Genetics Initiative,9 a global collaboration to accumulate GWAS on COVID-19 infection and clinical manifestations.
In MVP, 1,062 COVID-19 cases (Supplementary Table 1) were identified between March 1st and September 17, 2020 using an algorithm developed by the VA COVID National Surveillance Tool (NST). The NST classified COVID-19 cases as positive or negative based on reverse transcription polymerase chain reaction (rRT-PCR) laboratory test results conducted at VA clinics, supplemented with Natural Language Processing (NLP) on clinical documents. The algorithm to identify COVID-19 patients is continually updated to ensure new annotations of COVID-19 are captured from the clinical notes, with chart reviews performed periodically to validate the algorithm.54 COVID-19-related hospitalizations were defined as admissions from 7 days before up to 30 days after a patient’s first positive test for SARS-CoV-2 test. We tested association between all our proposed genetic instruments and COVID-19 hospitalization (versus population controls) in MVP adjusting for age, sex and the first 10 principal components in three mutually-exclusive, ancestry-specific strata separately (European, African and Hispanic ancestry) using PLINK v2 (analysis completed on October 10, 2020). We have previously provided a detailed description of the genotype data quality control process55. The MVP received ethical and study protocol approval by the Veterans Affairs Central Institutional Review Board and informed consent was obtained for all participants.
We downloaded publicly available summary statistics for the B2 outcome from Host Genetic Initiative on October 4, 2020 (release 4 version 1). In total, HGI accumulated 6,492 cases of COVID-19 hospitalization through collaboration from 16 contributing studies (Supplementary Table 1), which were asked to define cases as “hospitalized laboratory confirmed SARS-CoV-2 infection (RNA and/or serology based), hospitalization due to corona-related symptoms” versus population controls (https://docs.google.com/document/d/1okamrqYmJfa35ClLvCt_vEe4PkvrTwggHq7T3jbeyCI/view) and use a model that adjusts for age, age2, sex, age*sex, PCs, and study specific covariates (https://docs.google.com/document/d/16ethjgi4MzlQeO0KAW_yDYyUHdB9kKbtfuGW4XYVKQg/view). Summary statistics (i.e. betas and standard errors) from the four analyses, MVP-European, MVP-African, MVP-Hispanic, and COVID-19 Host Genetics Initiative (HGI summary statistics were already meta-analyzed from GWAS that contributed to the HGI consortium) were meta-analyzed using METAL software56 with inverse-variance weighting and fixed effects.
Quantile-Quantile plots of P values from genome-wide association testing in MVP did not display any inflation of results in any ancestry-specific stratum (Supplementary Figure 1). Additionally, Phet values from the meta-analysis (output from METAL’s “analyze heterogeneity” command) were not inflated (Supplementary Figure 2), indicating that there is little overall heterogeneity between estimates across ancestries within MVP and between MVP and HGI.
Mendelian randomization and colocalization
We conducted MR analyses using the R package TwoSampleMR (https://mrcieu.github.io/TwoSampleMR/). We used fixed-effects, inverse-variance weighted MR for proposed instruments that contain more than one variant, and Wald-ratio for proposed instruments with one variant. For proposed instruments with multiple variants, we also tested the heterogeneity across variant-level MR estimates, using the Cochrane Q method (mr_heterogeneity option in TwoSampleMR package). We defined significant MR results using a P value threshold of P<3.96×10-5 (0.05 Bonferroni-corrected for 1,263 actionable druggable genes) and identified a list of "suggestive" actionable druggable targets that passed a threshold of P<5.00×10-4. For statistically significant MR results, we also performed colocalization57 between each eQTL and the trans-ancestry meta-analysis on COVID-19 hospitalization using the moloc R package (https://github.com/clagiamba/moloc) with default priors (probability of shared causal variant for trait 1 and trait 2 is p1=p2=1×10-4, probability of shared causal variant across two traits is p12=1×10-5). For example, if a proposed instrument contained two variants, we performed colocalization for the primary eQTL GWAS with COVID-19 hospitalization, as well as the secondary eQTL GWAS (i.e. eQTL GWAS after adjusting for peak variant from primary GWAS) with COVID-19 hospitalization. Statistically significant MR hits with posterior probability for hypothesis-4 (PP.H4) > 0.8 (i.e. the probability of a shared causal variant) for a least one instrumental variant were then investigated further using the following analyses.
Identifying pQTLs using Olink assay
We performed stepwise conditional analysis to identify cis-pQTL proposed instruments for proteins that passed our significance and colocalization thresholds and were one of 354 unique proteins measured on four Olink58 panels (CVD1, CVD2, Inflammation, and Neuro59) in 4,998 INTERVAL participants.13 INTERVAL is a prospective cohort study of ~50,000 blood donors recruited from 25 National Health Service Blood and Transplant centers in England. Participants were genotyped using the UK Biobank Affymetrix Axiom array, followed by phasing using SHAPEIT3 and imputation on the Sanger Imputation Server using a 1000 Genomes Phase 3-UK10K imputation panel. Alleles were tested against Olink proteins using SNPTEST v2.5.2 and adjusted for age, sex, plate, time from blood draw to processing, season and the first 5 principal components. Conditional analysis was performed by adjusting for peak variants until no association fell below 5.00×10-6.
Phenome-wide scan
We conducted a phenome-wide scan for variants with the following goals. First, we want to evaluate that our proposed instruments could reproduce the known phenotype associations (e.g. disease, biomarkers) ascribed to the drug that are due to on-target effects. Secondly, we want to identify if our proposed instruments are associated with comorbidities associated with greater likelihood of SARS-CoV-2 testing or predictors of hospitalization in COVID-19 patients, as this could potentially highlight the presence of certain biases.21 Also, for genes that were the target of licensed drugs, we checked whether the disease indication was also a risk factor for COVID-19 outcomes, as this might introduce a bias analogous to confounding by indication in MR.
To accomplish these goals, we investigated proposed instruments for associations of a phenome-wide range of outcomes. We searched the GTEx12 Portal (https://gtexportal.org/home/) for gene expression, and Phenoscanner17 (http://www.phenoscanner.medschl.cam.ac.uk/) for proteins, traits and diseases. We additionally queried variants in GWAS for 354 Olink proteins (described earlier), and all the proteins measured by the SomaScan platform (described in Sun et al.13) in 3,301 INTERVAL participants.
Characterizing downstream transcriptional consequences of associated loci
In order to confirm the specificity of the identified loci and to better explore their most important downstream transcriptional consequences, we have studied the transcriptional landscape modulation associ3ated with the selected variants using GTEx V8 data with representation of 49 different tissues. For this we have used rs13050728 as the proxy of the IFNAR2/IL10RB locus and rs4830976 as the proxy of the ACE2 locus and conducted a differential gene-expression analysis for all transcripts available in GTEx V8. After fitting models for all genes, enrichment pathway analysis was conducted to retrieve the most enriched pathways using both the differentially expressed (DE) gene list (through an over-representation analysis) and a Gene Set Enrichment Analysis framework (using the R package clusterProfiler60). For enrichment analysis we have used the corpus from WikiPathways, Gene Ontology and Reactome.
Extended Data
Extended Data Fig. 1. Regional association plots for rs2266590 and rs2239573 for plasma IL-10RB, IL10RB gene expression, and COVID-19 hospitalization.
This region was investigated further using IL-10RB pQTL data because it was available on an independent proteomic platform (Olink) and results using eQTL instruments for IL10RB passed our Mendelian randomization P value threshold (0.05 Bonferroni-corrected for 1,263 actionable druggable genes) and colocalization threshold (PP.H4>0.8). a, rs2266590 as pQTL for plasma IL-10RB measured by Olink in 4,998 INTERVAL participants. b, rs2266590 as an eQTL for IL10RB expression tibial artery tissue (N=584). c, rs2266590 in COVID-19 hospitalization. d, rs2239573 as pQTL for plasma IL-10RB (after adjusting for rs2266590) measured by Olink in 4,998 INTERVAL participants. e, rs2239573 as an eQTL for IL10RB expression in whole blood (N=670). f, rs2239573 in COVID-19 hospitalization. a colocalizes with b (PP.H4=0.97), and d colocalizes with e (PP.H4=1.00). The two variants highlighted in this figure (rs2266590 and rs2239573), which are associated with gene expression and plasma protein levels of IL-10RB, are not associated with COVID-19 hospitalization (P=0.85 for rs2266590, P=0.66 for rs2239573).
Extended Data Fig. 2. Enrichment analysis of peak eQTLs for IFNAR2-IL10RB and ACE2 regions.
Results obtained from association analysis using all 49 tissues from GTEx V8 contrasted against variant genotypes in an additive model. Dotplot of over-representation analysis using all significant (p<0.05) differentially expressed (DE) genes (476 for rs13050728; 1,397 for rs4830976) for a, rs13050728, peak eQTL in the IFNAR2-IL10RB region and b, rs4830976, peak eQTL in the ACE2 region. Count = number of DE genes part of the enriched pathway. Gene ratio is the rate of DE genes represented in each pathway.
Extended Data Fig. 3. Regional association plots for cis-variants associated with ACE2 gene expression or ACE2 plasma protein levels, and their association with COVID-19 hospitalization.
a, rs4830976 as an eQTL for ACE2 expression in brain frontal cortex tissue (N=175). b, rs4830976 in COVID-19 hospitalization (N=1,377,758). c, rs5935998 as the primary pQTL for plasma ACE2 measured by Oink in 4,998 INTERVAL participants d, rs5935998 in COVID-19 hospitalization e, rs4646156 as the secondary pQTL (i.e. after adjusting for rs5935998) for plasma ACE2 measured by Oink in 4,998 INTERVAL participants. f, rs4646156 in COVID-19 hospitalization. a colocalizes with b (PP.H4=0.95), but c does not colocalize with d (PP.H4=0.49) and e does not colocalize with f (PP.H4=0.08).
Supplementary Material
Editor summary.
Large-scale Mendelian randomization and colocalization analyses using gene expression and soluble protein data for 1,263 actionable druggable genes, which encode protein targets for approved drugs or drugs in clinical development, identify IFNAR2 and ACE2 as the most promising therapeutic targets for early management of COVID-19.
Editor recognition statement.
Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reviewer recognition statement.
Nature Medicine thanks David Evans, Steven Wolinsky, and the other, anonymous, reviewers for their contribution to the peer review of this work.
Acknowledgements
We are grateful to the Host Genetic Initiative for making their data publicly available (full acknowledgements can be found here: https://www.covid19hg.org/acknowledgements/). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award #MVP035. This research was also supported by additional Department of Veterans Affairs awards grant #MVP001. This publication does not represent the views of the Department of Veteran Affairs or the United States Government. Full acknowledgements for the VA Million Veteran Program COVID-19 Science Initiative can be found in the supplementary methods. C.G. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 754490 – MINDED project. A.G., P.B. and A.R.L. are funded by the Member States of the European Molecular Biology Laboratory (EMBL). I.B.- H. received funding from Open Targets (grant agreement OTAR-044). The Fenland Study (10.22025/2017.10.101.00001) is funded by the Medical Research Council (MC_UU_12015/1); we are grateful to all the volunteers and to the General Practitioners and practice staff for assistance with recruitment; we thank the Fenland Study Investigators, Fenland Study Co-ordination team and the Epidemiology Field, Data and Laboratory teams; we further acknowledge support for genomics from the Medical Research Council (MC_PC_13046); proteomic measurements were supported and governed by a collaboration agreement between the University of Cambridge and Somalogic. J.E.P. is supported by UKRI Innovation Fellowship at Health Data Research UK (MR/S004068/2). L.R., N.H. and C.L. are supported by the Swedish Research Council. E.A. was supported by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking BigData@Heart grant n° 116074 and by the British Heart Foundation Programme Grant RG/18/13/33946. We thank Dr. Angela Wood for feedback on statistical analyses used in the paper. We thank the INTERVAL Study investigators, co-ordination team and the epidemiology field, data and laboratory teams, which were supported by core funding from the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (RG/13/13/30194; RG/18/13/33946), the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014) [The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care], and the NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014-10024). This work was also supported by Health Data Research UK, which is funded by the UK Medical Research Council, the Engineering and Physical Sciences Research Council, the Economic and Social Research Council, the Department of Health and Social Care (England), the Chief Scientist Office of the Scottish Government Health and Social Care Directorates, the Health and Social Care Research and Development Division (Welsh Government), the Public Health Agency (Northern Ireland), the British Heart Foundation, and Wellcome. J.D. holds a British Heart Foundation Professorship and a National Institute for Health Research Senior Investigator Award.
Footnotes
Author contribution
J.P.C., A.S.B. and J.M.G. conceived the study design. A.G., A.P.B. and A.R.L. defined the actionable genome, and identified and curated drug information relating to SARS-CoV-2; P.B. and I.B.-H. provided biological annotation relating to SARS-CoV-2; C.G. performed stepwise conditional analysis on GTEx raw data; D.P. tested associations for COVID-19 in MVP; L.G. and J.H.Z. performed meta-analysis of HGI and MVP; L.G. performed Mendelian randomization analysis; L.G. and C.G. performed colocalization analyses; L.G. and B.P.P. performed conditional analysis on Olink proteins; L.G. and E.A. performed phenome-wide scans; A.C.P. performed pathway enrichment analysis; J.N.D., A.S.B., and J.E.P. provided INTERVAL data; Several authors were involved in the curation of the MVP data; L.G., C.G., A.C.P., A.G., D.P., A.S.B. and J.P.C. wrote the manuscript. J.P.C. oversaw all analyses. All authors critically reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
GTEx project version 8 data are available at: https://gtexportal.org/home/. CheMBL database data are available at: https://www.ebi.ac.uk/chembl/. Fenland-Somalogic protein GWAS data are available at: https://omicscience.org/apps/covidpgwas/. Host Genetics Initiative COVID-19 hospitalization summary statistics are available at: https://www.covid19hg.org/. PhenoScanner results are available at http://www.phenoscanner.medschl.cam.ac.uk/.
References
- 1.Horby P, et al. Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beigel JH, et al. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalil AC, et al. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Lopalco L. CCR5: From Natural Resistance to a New Anti-HIV Strategy. Viruses. 2010;2:574–600. doi: 10.3390/v2020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan C, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swerdlow DI, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379:1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaziano JM, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Labrecque J, Swanson SA. Understanding the Assumptions Underlying Instrumental Variable Analyses: a Brief Review of Falsification Strategies and Related Tools. Curr Epidemiol Rep. 2018;5:214–220. doi: 10.1007/s40471-018-0152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietzner M, et al. Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nat Commun. 2020;11:6397. doi: 10.1038/s41467-020-19996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Marschall Z, et al. Effects of interferon alpha on vascular endothelial growth factor gene transcription and tumor angiogenesis. J Natl Cancer Inst. 2003;95:437–448. doi: 10.1093/jnci/95.6.437. [DOI] [PubMed] [Google Scholar]
- 19.Jia H, et al. Endothelial cell functions impaired by interferon in vitro: Insights into the molecular mechanism of thrombotic microangiopathy associated with interferon therapy. Thromb Res. 2018;163:105–116. doi: 10.1016/j.thromres.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Casassus P, et al. Treatment of adult systemic mastocytosis with interferon-alpha: results of a multicentre phase II trial on 20 patients. Br J Haematol. 2002;119:1090–1097. doi: 10.1046/j.1365-2141.2002.03944.x. [DOI] [PubMed] [Google Scholar]
- 21.Swanson SA, Tiemeier H, Ikram MA, Hernán MA. Nature as a Trialist?: Deconstructing the Analogy Between Mendelian Randomization and Randomized Trials. Epidemiology. 2017;28:653–659. doi: 10.1097/EDE.0000000000000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson CP, et al. Genetic Associations With Plasma Angiotensin Converting Enzyme 2 Concentration: Potential Relevance to COVID-19 Risk. Circulation. 2020;142:1117–1119. doi: 10.1161/CIRCULATIONAHA.120.049007. [DOI] [PubMed] [Google Scholar]
- 23.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16:e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemnes AR, et al. A potential therapeutic role for angiotensin-converting enzyme 2 in human pulmonary arterial hypertension. Eur Respir J. 2018;51 doi: 10.1183/13993003.02638-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuba K, Imai Y, Rao S, Jiang C, Penninger JM. Lessons from SARS: control of acute lung failure by the SARS receptor ACE2. J Mol Med (Berl) 2006;84:814–820. doi: 10.1007/s00109-006-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monteil V, et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.:e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteil V, et al. Human soluble ACE2 improves the effect of remdesivir in SARS-CoV-2 infection. EMBO Mol Med. 2020;13:e13426. doi: 10.15252/emmm.202013426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoufaly A, et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Recombinant Human Angiotensin-converting Enzyme 2 (rhACE2) as a Treatment forPatients With COVID-19. ( https://ClinicalTrials.gov/show/NCT04335136).
- 30.Onabajo OO, et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco-Melo D, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.:e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu H, et al. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin Infect Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastard P, et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Made CI, et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. Jama. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pairo-Castineira E, et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 38.Lokugamage KG, et al. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. Journal of Virology. 2020;94:e01410-01420. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clementi N, et al. Interferon-β-1a Inhibition of Severe Acute Respiratory Syndrome-Coronavirus 2 In Vitro When Administered After Virus Infection. J Infect Dis. 2020;222:722–725. doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinnon KH, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung IF, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Repurposed Antiviral Drugs for Covid-19 — Interim WHO Solidarity Trial Results. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.NIAID Stops COVID-19 Trial Enrollment Over Adverse Events. 2020 [Google Scholar]
- 45.Liu D, et al. Mendelian randomization analysis identified genes pleiotropically associated with the risk and prognosis of COVID-19. J Infect. 2020 doi: 10.1016/j.jinf.2020.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao F, et al. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.:e1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao L, et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song E, et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218 doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puelles VG, et al. Multiorgan and Renal Tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendez D, et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–d940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohloff JC, et al. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman AB, et al. A Natural Language Processing System for National COVID-19 Surveillance in the US Department of Veterans Affairs. 2020 [Google Scholar]
- 55.Hunter-Zinck H, et al. Genotyping Array Design and Data Quality Control in the Million Veteran Program. Am J Hum Genet. 2020;106:535–548. doi: 10.1016/j.ajhg.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giambartolomei C, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34:2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olink Target 96 & Target 48 panels for protein biomarker discovery [Google Scholar]
- 60.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GTEx project version 8 data are available at: https://gtexportal.org/home/. CheMBL database data are available at: https://www.ebi.ac.uk/chembl/. Fenland-Somalogic protein GWAS data are available at: https://omicscience.org/apps/covidpgwas/. Host Genetics Initiative COVID-19 hospitalization summary statistics are available at: https://www.covid19hg.org/. PhenoScanner results are available at http://www.phenoscanner.medschl.cam.ac.uk/.