To The Editor
Microbial invasion of the skin and underlying soft tissues, known as skin and soft tissue infections (SSTIs), contribute to considerable burden of disease worldwide (Kaye et al. 2019; Lozano et al. 2012). Knowledge about host factors contributing to SSTI risk is important to prevent the SSTIs. The genetics of SSTI susceptibility remain largely unknown, and the only previously published genome-wide study on SSTIs is a small family-based linkage study that did not identify significant linkage to any genes for erysipelas or cellulitis susceptibility (Hannula-Jouppi et al. 2013).
A range of cardiometabolic risk factors have been associated with SSTIs (Butler-Laporte et al. 2020; Kaye et al. 2019; Winter-Jensen et al. 2020). Few studies have used genetic variants as instrumental variables (Mendelian randomization [MR]), to assess causality, which may reduce bias due to reverse causation and confounding (Davies et al. 2018). Increasing body mass index (BMI) has been found to increase the risk of SSTIs in such a framework (Butler-Laporte et al. 2020; Winter-Jensen et al. 2020), but other cardiometabolic risk factors have not been explored.
The aims of this study were to conduct the first genome-wide association study (GWAS) on susceptibility to SSTIs, explore possible biological pathways through transcriptome-wide association analyses, and perform MR analyses to investigate potential causal relationships of cardiometabolic risk factors on SSTIs.
We used two independent cohorts, where the UK Biobank served as the discovery cohort in the genome-wide association analyses, and the Trøndelag Health Study (the HUNT Study) served as the replication cohort. Subjects who had been hospitalized with a primary diagnosis of SSTI served as cases, while those who had not been hospitalized with a primary or secondary diagnosis of SSTI were considered controls (Supplementary Material and Methods).
Genome-wide association analyses were conducted using SAIGE, with age, sex, genotype chip, and ancestry-informative principal components as covariates (Zhou et al. 2018), and meta-analyses were carried out using METAL (Supplementary Materials and Methods). Associations with p-value <1e-6 and p-value <5e-8 were considered genome-wide suggestive and significant, respectively.
We used FUSION to performed transcriptome-wide association analyses by combining summary statistics from the genome-wide meta-analysis with linkage disequilibrium (European ancestry in 1000 Genomes Project) and reference gene expression panels (GTEx v7) to estimate gene expression patterns associated with SSTIs (Gusev et al. 2016). Sun-exposed skin (lower legs) was the tissue of interest for the transcriptome-wide analyses (8,609 genes tested), while all 48 general tissues from GTEx v7 were analyzed for the chromosome with genome-wide significant hits (10,518 tests). Bonferroni-corrected threshold for genome-wide significance was p-value <2.6e-6.
Two-sample MR analyses were conducted separately for results from the meta-analysis, UK Biobank and HUNT. Genetic instruments for BMI, type 2 diabetes mellitus, low-density lipoprotein cholesterol, systolic blood pressure, lifetime smoking, and sedentary lifestyle were extracted from relevant published GWASs (Supplementary Table 1). The TwoSampleMR R package (version 0.5.0) (Hemani et al. 2018) was used to carry out inverse-variance weighted MR analyses (main analyses), along with statistical test for heterogeneity, simple median, weighted median and MR Egger (sensitivity analyses).
In both UK Biobank and HUNT, cases, compared with controls, were at baseline older, had higher BMI and systolic blood pressure, and were more likely to be male, ever-smoker and self-reported diabetic (Supplementary Table 2).
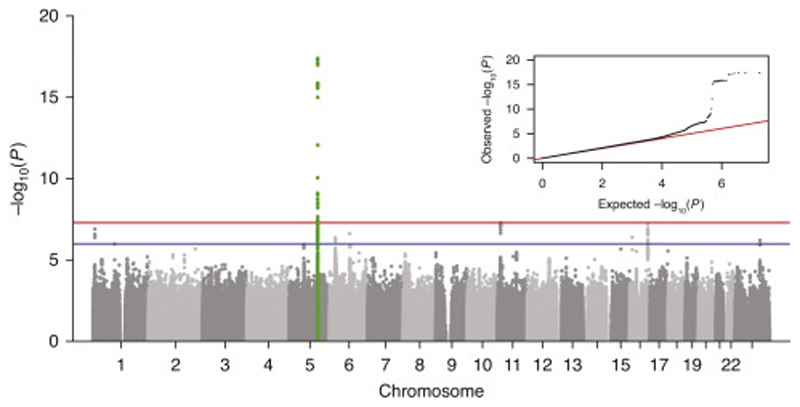
The genome-wide association analysis included 6,107 cases and 399,239 controls from UK Biobank, and 1,657 cases and 67,522 controls from HUNT. UK Biobank yielded seven suggestive loci (Supplementary Table 3 and Supplementary Figure 1), of which one was replicated in HUNT: rs3749748 in the LINC01184/SLC12A2-gene region on chromosome 5 (Supplementary Figures 2 and 3). In the meta-analysis of 7,764 cases and 466,761 controls, only the locus in LINC01184/SLC12A2 reached genome-wide significance (Figure 1), while two additional loci were close to genome-wide significance: PSMA1 on chromosome 11 and GAN on chromosome 16 (Supplementary Table 3). There was no indication of genomic inflation (Figure 1 and Supplementary Figures 1 and 2).
Figure 1. Manhattan plot of results for the meta-analysis.
Axes display the -log10 transformed p-value by chromosomal position. The blue line indicates genome-wide suggestive associations (p-value <1e-6) and the red line genome-wide significant associations (p-value <5e-8). Genome-wide significant loci (+/- 500kb of lead variant) are highlighted in green. Top right corner: Quantile-quantile plot. Axes display the observed (y-axis) and expected (x-axis) -log10 transformed p-value. The black dots represent observed p-values while the red line represents expected p-values under the null distribution. Genomic inflation factor (λ) = 1.01.
LINC01184 is part of the lincRNA class of genes that does not encode for proteins, but have still been found to modulate inflammation and infection risk (Atianand et al. 2016; Carpenter et al. 2013). SLC12A2 encodes for the protein NKCC1 which regulates transport of chloride, potassium and sodium across cell membranes, and is key in modulating ion movement across the epithelium, volume of cells, and anti-microbial activity (Matthay and Su 2007; Yang et al. 2020).
In the transcriptome-wide association analysis of skin on the lower legs, the only gene that was statistically significantly associated with SSTIs was LINC01184 (Supplementary Figure 4). A reduced expression of LINC01184 was associated with increased risk of SSTIs. The same association was observed in all tissues, but less pronounced in the brain (Supplementary Figure 4).
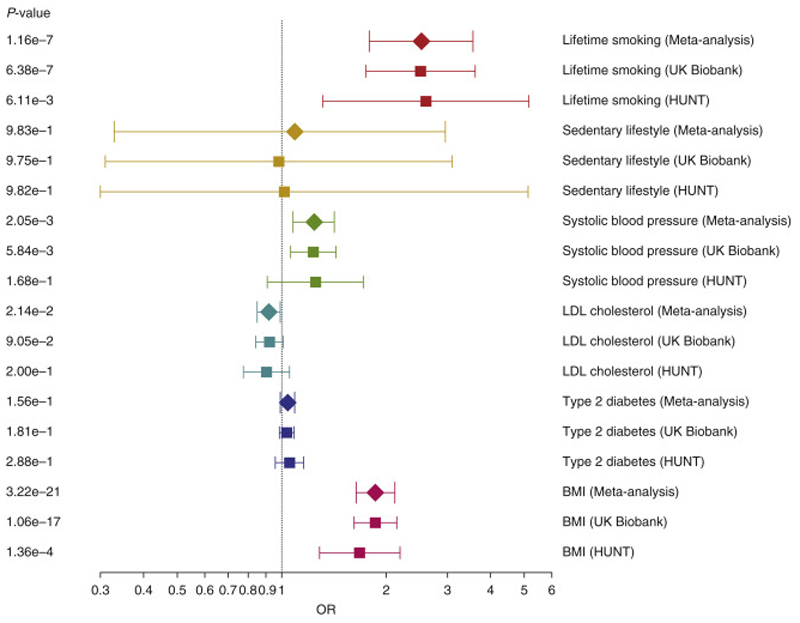
Increase in genetically predicted BMI, systolic blood pressure and smoking increased the risk of SSTIs, while increasing low-density lipoprotein cholesterol was associated with a reduced risk of SSTIs (Figure 2). Sensitivity analyses supported the findings from the inverse-variance weighted analyses (Supplementary Table 4).
Figure 2. Mendelian randomization analyses of cardiometabolic risk factors on risk of skin and soft tissue infection.
Forest plot of the two-sample inverse-variance weighted Mendelian randomization analyses of cardiometabolic risk factors identified as genetically correlated with skin and soft tissue infection. Each risk factor was evaluated separately using results from the meta-analysis, UK Biobank and HUNT, and the corresponding risk factors were grouped by color. The x-axis represents the increased odds ratio per standard deviation increase of the genetically predicted risk factor (per unit increase in log odds ratio for genetically proxied type 2 diabetes mellitus liability). BMI, body mass index; LDL, low-density lipoprotein.
This is the first GWAS published on SSTIs to date, with a large number of cases and controls. We were able to identify a novel locus – LINC01184/SLC12A2 – robustly associated with SSTIs in the discovery cohort and the independent replication cohort. A limitation of our study is that we did not have the power to identify more than one genome-wide significant locus, which in part may be due to non-differential misclassification of the outcome, and we thus encourage replication with meta-analysis in independent cohorts. Of note, while the minor allele frequency of rs3749748 in North-Western European populations is around 23%, it is only 4% in African-American populations (Karczewski et al. 2020). It is therefore important to evaluate populations of different ancestries than the one currently considered.
In conclusion, we have identified genetic variation in LINC01184/SLC12A2 to be strongly associated with risk of SSTIs. Interventions to reduce smoking, hypertension, overweight and obesity in the population will likely reduce the disease burden of SSTIs.
Supplementary Material
Axes display the -log10 transformed p-value by chromosomal position. The blue line indicates genome-wide suggestive associations (p-value <1e-6) and the red line genome-wide significant associations (p-value <5e-8). Genome-wide suggestive loci (+/- 500kb of lead variant) are highlighted in green. Top right corner: Quantile-quantile plot. Axes display the observed (y-axis) and expected (x-axis) -log10 transformed p-value. The black dots represent observed p-values while the red line represents expected p-values under the null distribution. Genomic inflation factor (λ) = 1.02.
Axes display the -log10 transformed p-value by chromosomal position. The blue line indicates genome-wide suggestive associations (p-value <1e-6) and the red line genome-wide significant associations (p-value <5e-8). Genome-wide suggestive loci from the discovery stage (+/- 500kb of lead variant) are highlighted in green. Top right corner: Quantile-quantile plot. Axes display the observed (y-axis) and expected (x-axis) -log10 transformed p-value. The black dots represent observed p-values while the red line represents expected p-values under the null distribution. Genomic inflation factor (λ) = 1.00.
Associations between genetic variants and skin and soft tissue infection from the meta-analysis are plotted by position (x-axis) and -log10 transformed p-values (left y-axis). rs3749748 served as sentinel variant, while the remaining variants are color coded in terms of the linkage disequilibrium (r2) to the sentinel variant. Estimated recombination rates are plotted as light blue lines (right y-axis). The European population from 1000 Genomes Project, November 2014 release, was used as reference, on genome build hg19.
Each dot represents the association between predicted gene expression in skin on lower legs with risk of SSTIs. The red line indicate statistically significant associations (p-value <2.6e-6). Top right corner: The transcriptome association statistic for LINC01184 in all 48 tissues from GTEx v7.
Acknowledgements
The Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The authors declare no conflicts of interest.
This research has been conducted using the UK Biobank Resource under Application Number ‘40135’
Funding
This study was in part funded by Samarbeidsorganet Helse Midt-Norge, NTNU, and The Research Council of Norway (grant 299765). The first author was funded in part by a Fulbright Scholarship by the U.S-Norway Fulbright Foundation. Ben Michael Brumpton, Humaira Rasheed, Laurent Thomas, Mari Løset, Kristian Hveem, and Bjørn Olav Åsvold work in a research unit funded by Stiftelsen Kristian Gerhard Jebsen; Faculty of Medicine and Health Sciences, NTNU; The Liaison Committee for education, research and innovation in Central Norway; the Joint Research Committee between St. Olavs Hospital and the Faculty of Medicine and Health Sciences, NTNU; and the Medical Research Council Integrative Epidemiology Unit at the University of Bristol which is supported by the Medical Research Council and the University of Bristol [MC_UU_12013/1]. The funding sources had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; nor in the decision to submit the article for publication. The researchers were independent from the funders, and all authors had full access to all of the data in the study and can take responsibility for the integrity of the data and the accuracy of the data analysis.
Abbreviations
- BMI
body mass index
- GWAS
genome-wide association study
- HUNT
Trøndelag Health Study
- MR
Mendelian randomization
- OR
odds ratio
- SSTIs
skin and soft tissue infections
Footnotes
Declaration of Interests
DG is employed part-time by Novo Nordisk, outside of the submitted work. The remaining authors declare no conflicts of interest.
Author Contributions
Conceptualization: TR, KVL, ES, JKD, ATD, HMF
Data Curation: TR, HMF, BMB, HR, LFT, CJW, KH, BOÅ
Formal analysis: TR, HR, LFT
Funding Acquisition: TR, ES, JKD, KH, CJW, BOÅ, JS, ATD, BMB
Investigation: TR, HR, LFT, ES, JKD, KH, CJW, BOÅ, JS, ATD, ML, BMB
Methodology: TR, HR, LFT, DG, SB, ATD, BMB
Project Administration: TR, ES, JKD, BOÅ, KH, CJW, ATD, BMB, ML
Resources: ES, JKD, BOÅ, KH, ATD, JS
Software: DG, SB, BMB, HR, LFT
Supervision: TR, ES, JKD, ATD, BMB, DG, SB, BOÅ, ML, CJW
Validation: HR, LFT, BMB, JS
Visualization: TR, HR, LFT
Writing (Original draft): TR
Writing (Review and editing): All authors
All authors made substantial contribution to the interpretation of the data, critically revised the manuscript. All authors have approved the submitted version and to be personally accountable for the author’s own contributions.
Contributor Information
Kristin V Liyanarachi, Email: kristin.v.liyanarachi@ntnu.no.
Humaira Rasheed, Email: humaira.rasheed@ntnu.no.
Laurent F Thomas, Email: laurent.thomas@ntnu.no.
Helene M Flatby, Email: helene.flatby@ntnu.no.
Jørgen Stenvik, Email: jorgen.stenvik@ntnu.no.
Mari Løset, Email: mari.loset@ntnu.no.
Dipender Gill, Email: dgill@sgul.ac.uk.
Stephen Burgess, Email: sb452@medschl.cam.ac.uk.
Cristen J Willer, Email: cristen@umich.edu.
Kristian Hveem, Email: kristian.hveem@ntnu.no.
Bjørn O Åsvold, Email: bjorn.o.asvold@ntnu.no.
Ben M Brumpton, Email: ben.brumpton@ntnu.no.
Andrew T DeWan, Email: andrew.dewan@yale.edu.
Erik Solligård, Email: erik.solligard@ntnu.no.
Jan K Damås, Email: jan.k.damas@ntnu.no.
Data Availability Statement
Data from the HUNT Study and UK Biobank are available on application. Gene expression data are available through the FUSION website (http://gusevlab.org/projects/fusion/). Summary statistics will be made available at the GWAS Catalog at the time of publication.
References
- Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. Cell. 7. Vol. 165. Elsevier Inc; 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation; pp. 1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler-Laporte G, Harroud A, Forgetta V, Richards JB. Clin Microbiol Infect. Elsevier; 2020. Elevated body mass index is associated with an increased risk of infectious disease admissions and mortality: a mendelian randomization study. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science (80.) 2013;341(6147):789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Nat Genet. 3. Vol. 48. Nature Publishing Group; 2016. Integrative approaches for large-scale transcriptome-wide association studies; pp. 245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Massinen S, Siljander T, Mäkelä S, Kivinen K, Leinonen R, et al. Genetic Susceptibility to Non-Necrotizing Erysipelas/Cellulitis. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Wang Q, Collins RL, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv. 2020 doi: 10.1038/s41586-020-2308-7. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KS, Petty LA, Shorr AF, Zilberberg MD. Current epidemiology, etiology, and burden of acute skin infections in the United States. Clin Infect Dis. 2019;68(Suppl 3):S193–9. doi: 10.1093/cid/ciz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay MA, Su X. Pulmonary barriers to pneumonia and sepsis. Nat Med. 2007;13(7):780–1. doi: 10.1038/nm0707-780. [DOI] [PubMed] [Google Scholar]
- Winter-Jensen M, Afzal S, Jess T, Nordestgaard BG, Allin KH. Eur J Epidemiol. 4. Vol. 35. Springer Netherlands; 2020. Body mass index and risk of infections: a Mendelian randomization study of 101,447 individuals; pp. 347–54. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang Q, Cao E. Nat Commun. 1. Vol. 11. Springer US; 2020. Structure of the human cation–chloride cotransporter NKCC1 determined by single-particle electron cryo-microscopy; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335–41. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Axes display the -log10 transformed p-value by chromosomal position. The blue line indicates genome-wide suggestive associations (p-value <1e-6) and the red line genome-wide significant associations (p-value <5e-8). Genome-wide suggestive loci (+/- 500kb of lead variant) are highlighted in green. Top right corner: Quantile-quantile plot. Axes display the observed (y-axis) and expected (x-axis) -log10 transformed p-value. The black dots represent observed p-values while the red line represents expected p-values under the null distribution. Genomic inflation factor (λ) = 1.02.
Axes display the -log10 transformed p-value by chromosomal position. The blue line indicates genome-wide suggestive associations (p-value <1e-6) and the red line genome-wide significant associations (p-value <5e-8). Genome-wide suggestive loci from the discovery stage (+/- 500kb of lead variant) are highlighted in green. Top right corner: Quantile-quantile plot. Axes display the observed (y-axis) and expected (x-axis) -log10 transformed p-value. The black dots represent observed p-values while the red line represents expected p-values under the null distribution. Genomic inflation factor (λ) = 1.00.
Associations between genetic variants and skin and soft tissue infection from the meta-analysis are plotted by position (x-axis) and -log10 transformed p-values (left y-axis). rs3749748 served as sentinel variant, while the remaining variants are color coded in terms of the linkage disequilibrium (r2) to the sentinel variant. Estimated recombination rates are plotted as light blue lines (right y-axis). The European population from 1000 Genomes Project, November 2014 release, was used as reference, on genome build hg19.
Each dot represents the association between predicted gene expression in skin on lower legs with risk of SSTIs. The red line indicate statistically significant associations (p-value <2.6e-6). Top right corner: The transcriptome association statistic for LINC01184 in all 48 tissues from GTEx v7.
Data Availability Statement
Data from the HUNT Study and UK Biobank are available on application. Gene expression data are available through the FUSION website (http://gusevlab.org/projects/fusion/). Summary statistics will be made available at the GWAS Catalog at the time of publication.