Abstract
Non-canonical inflammasome activation by cytosolic LPS is a critical component of the host response to Gram-negative bacterial infection, however we lack a comprehensive understanding of the cellular processes that control this pathway. Using a genome-scale arrayed siRNA screen to discover inflammasome regulators in macrophages, we identified the mitochondrial Nme4 gene, which codes for the multi-functional nucleoside diphosphate kinase D (NDPK-D), as a regulator of both non-canonical and canonical inflammasomes. We show that Nme4 is required for both mitochondrial DNA synthesis and cardiolipin exposure on the mitochondrial surface in response to priming signals, leading to broad inflammasome activation defects in Nme4-deficient cells. In addition, Nme4 is required for TRAF6 mitochondrial recruitment and ROS production, which supports the large-scale TLR-induced gene program via Nme4-dependent TLR signaling responses. Nme4 knock-out mice are protected from endotoxin-induced shock, consistent with attenuated ROS production and glycolytic commitment. Our findings suggest that in response to microbial challenge, Nme4-dependent TRAF6 mitochondrial recruitment triggers an energetic fitness checkpoint required to engage and support the transcriptional gene program necessary to support inflammasome activation.
Keywords: inflammasome, macrophage, LPS, IL-1, caspase-11, ROS
Introduction
Innate immune cells such as macrophages promote host defense by sensing pathogens via pattern recognition receptors (PRRs)1. While macrophages classically respond to lipopolysaccharide (LPS) from extracellular Gram-negative bacteria via Toll-like receptor (TLR)-42,3, more recent reports indicate intracellular detection of bacterial LPS relies on murine caspase-11 (caspases-4 and -5 in human cells) to induce pyroptosis and IL-1 release4-7. The host inflammatory response to such intracellular threats is mediated through multi-protein inflammasome complexes8. While several inflammasomes have been described to date9-12, the canonical NLRP3 inflammasome has been the most heavily studied13-21. Minimally comprised of the NLRP3 sensor protein, the adaptor protein ASC, and caspase-1, it can be activated by a diverse array of stimuli including both pathogen and danger signals as well as a variety of cell stress inducers including ATP, Nigericin, crystal particles, reactive oxygen species (ROS) and oxidized mitochondrial DNA (mtDNA)13,14,17,22. Although a direct activating ligand has not been identified for NLRP3, canonical inflammasome activation invokes ASC oligomerization and caspase-1 activation and cleavage, which in turn cleaves the immature forms of the IL-1 family proteins and the recently identified pore-forming Gasdermin proteins8,13,23-25.
The aforementioned cytosolic LPS sensing through caspase-11 is considered the non-canonical inflammasome pathway, as caspase-11 activation by LPS occurs upstream of NLRP3 activation4. Here, IL-1α maturation and secretion correlates with Gasdermin cleavage and pyroptosis that occurs independently from the poorly defined parallel activation of canonical NLRP3 and IL-1β release4. Two major checkpoints precede inflammasome activation by cytosolic LPS: an inflammatory priming signal, such as a TLR ligand, elevates the expression of key inflammasome genes while concomitant ‘licensing’ events post-translationally modify inflammasome components which may also be enriched at the mitochondrial outer membrane (MOM)16,18,26. It has been suggested that such recruitment is dependent on increased levels of the mitochondrial lipid cardiolipin at the MOM18,20,26. While numerous requirements for inflammasome activation have been described, we still lack a comprehensive understanding of the cellular regulators of these critical inflammatory processes.
In addition to the observed recruitment of inflammasome effectors to the mitochondrial membrane, cellular metabolic pathways have been implicated in several aspects of inflammasome activation. During priming, PRR signaling induces succinate accumulation via TCA cycle disruption to support HIF1α-dependent Il1b transcription27. Mitochondrial ROS also contributes to activation of the canonical NLRP3 inflammasome28,29. It was recently reported that this ROS production is required for enhancing mtDNA synthesis through the mitochondrial nucleotide salvage pathway and that oxidized forms of this mtDNA released from the mitochondria can activate NLRP321. Additionally, macrophage priming facilitates metabolic reprograming30-33, known as glycolytic commitment, to elevate both aerobic glycolysis and glucose uptake and to reduce oxidative phosphorylation (OXPHOS) by rechanneling the electron transport chain (ETC) for ROS production34. Despite increasing insight into the critical role of mitochondrial responses and metabolic reprogramming during immune activation, how these events relate to and regulate the mitochondrial stress accompanying inflammasome activation is not clearly understood. Moreover, the relationship between the signaling and transcriptional events induced by PRR activation, and their relative impact on inflammasome priming and licensing, remain poorly defined.
In an effort to better describe the regulation of the non-canonical inflammasome, we conducted a genome-scale arrayed siRNA screen for regulators of IL-1α release in macrophages exposed to cytosolic LPS. We identified a requirement for nucleoside diphosphate kinases (NDPKs), the sole enzyme in the last step of the nucleoside salvage pathway in the mitochondria, and further elucidated the role of the Nme4 gene (coding for NDPK-D, also known as NM23-H4) in inflammasome activation. We show that Nme4 can regulate both canonical and non-canonical inflammasomes by supporting both cardiolipin enrichment on the MOM and mtDNA synthesis. We also find that Nme4 is required for priming-induced ROS production to support PRR-activated TLR signaling and transcriptional responses. This novel role for Nme4 and ROS in stimulus-dependent transcription is proportional to the scale of the gene program induced, and establishes a link between the macrophage metabolic capacity and the LPS-driven inflammatory response. Moreover, while Nme4 is also required to support the glycolytic shift during macrophage activation, increased glycolysis is not required for the Nme4 and ROS supported acute transcriptional responses. Our data help delineate the mitochondrial and metabolic processes critical for inflammasome activation and suggest that Nme4 may coordinate a critical mitochondrial fitness checkpoint required to support a robust inflammasome response.
Results
A genome-wide siRNA screen identifies nucleoside diphosphate kinases as positive regulators of the non-canonical inflammasome
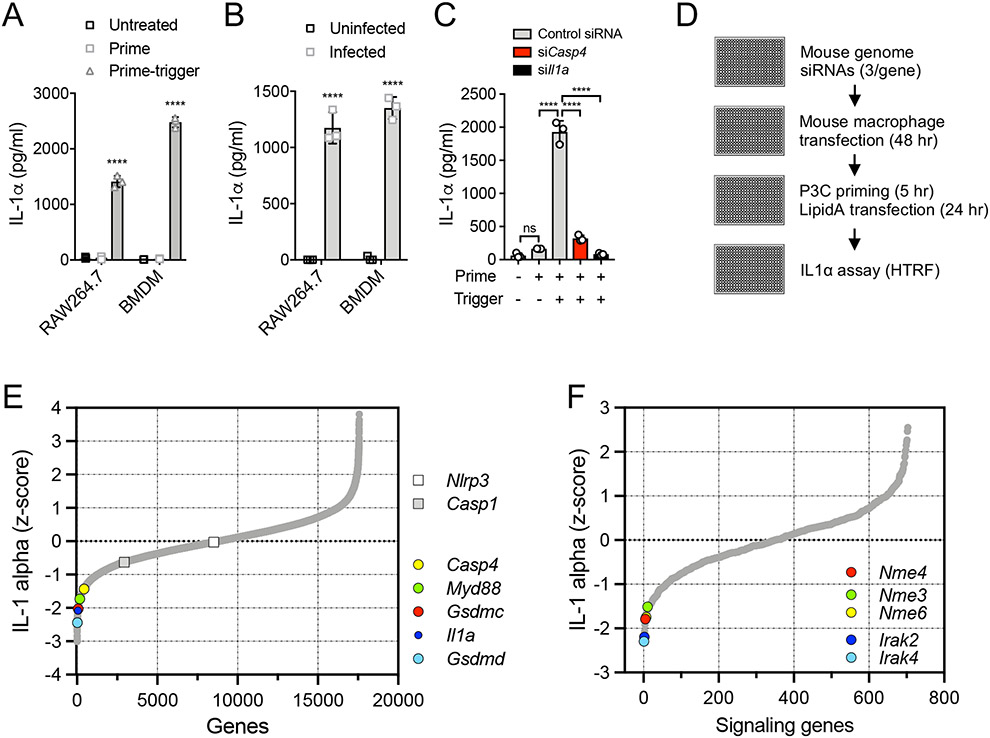
To discover regulators of the macrophage non-canonical inflammasome response to cytosolic bacteria, we first established a prime-trigger assay in which macrophages were treated with the TLR2 agonist, Pam3CSK4 (P3C), and then transfected with the immunostimulatory component of LPS, Lipid-A. This assay induced comparable levels of IL-1α to those induced by the intracellular Gram-negative bacterium Burkholderia cenocepacia, both in the RAW264.7 mouse macrophage cell line and in primary mouse bone marrow-derived macrophages (BMDM) (Fig. 1 A, B). A GFP-expressing RAW264.7 cell line was employed35, which permitted the development of a robust siRNA delivery protocol using GFP fluorescence reduction as a measure of transfection efficiency in 384-well format (Fig. S1A). When applied to the prime-trigger assay, delivery of Casp4 siRNA (targeting mouse caspase-11) or Il1a siRNA strongly reduced the cytosolic LPS-driven release of IL-1α (Fig. 1C), but had no significant effect on TNFα secretion (Fig. S1B).
Figure 1: A genome-wide RNAi screen identifies mitochondrial Nme genes as positive regulators of the non-canonical inflammasome.
(A) RAW264.7 cells and BMDM were primed with 100 nM or 1 μg/ml P3C (respectively) for 5 hr and triggered by 100 nM LipidA transfection for 18 hr. Secreted IL-1α was measured by ELISA. (B) RAW264.7 cells and BMDM were infected with B. cenocepecia (MOI 10) for 18 hr. Secreted IL-1α was measured by ELISA. (C) RAW264.7 cells, transfected with non-targeting control, Il1a or Casp4 siRNA, were primed and triggered as in (A). (D) Arrayed siRNA screen workflow. (E) Phenotypic distribution of the genome-wide screen highlighting known inflammasome components. (F) Phenotypic distribution of a pilot set of signaling genes showing Irak4 and Irak2 as a expected priming-dependent hits, and identification of the Nme genes as a positive regulators of prime-trigger-induced IL-1α release. (A-C) Data are representative of three independent experiments and expressed as mean ± SD, (n=3); (A, B) Two-Way ANOVA and (C) One-Way ANOVA followed by Tukey’s multiple comparison test; (A, B) untreated vs. treated cells; ****p < 0.0001.
To identify regulators of the non-canonical inflammasome, a genome-wide arrayed siRNA screen was conducted. To mitigate siRNA seed-based off-target effects, we used three independent siRNAs/gene36 and a screen-optimized Homogeneous Time-Resolved Fluorescence (HTRF) assay for IL-1α secretion (Fig. 1D). We identified numerous known pathway regulators among the strongest hits, including Myd88 required for the TLR priming step, and Casp4, Gsdmd and Il1a required for cytosolic LPS detection and IL-1α release (Fig. 1E, Table S1). As expected, the canonical inflammasome regulators Casp1 and Nlrp3, which should be dispensable for IL-1α release through the non-canonical pathway, did not show a screen phenotype (Fig. 1E). Analysis of a subset of signaling genes, targeting kinases, receptors and other cell signaling mediators, revealed a dependence on specific kinases in inflammasome activation (Fig 1F, Table S2). As expected, the kinases Irak4 and Irak2 were strong hits due to their requirement for the TLR2-dependent priming step in the screen assay (Fig. 1F, Table S2). Among the other genes positively regulating the non-canonical inflammasome response, we identified 3 members of the nucleoside diphosphate kinase (NDPK) Nme gene family, Nme3, Nme4 and Nme6 (Fig. 1F, Table S2). We noted that the products of each of these genes are the only NDPKs localized to mitochondria37-39,40, which is considered an important cellular organelle in inflammasome activation18-20,26. Indeed, we could detect 92 genes that code for mitochondrial proteins in the top 5th percentile of screen hits (Fig. S2A, B, Table S3). In particular, Nme4, coding for the mitochondrial protein NDPK-D, has been previously implicated in a number of mitochondrial functions which could impact the inflammasome response21,41-44.
Nme4/NDPK-D regulates the activation of both canonical and non-canonical inflammasomes
NDPK-D has been localized to both the mitochondrial intermembrane space and also to the matrix, where it has been shown to catalyze the last γ-phosphate transfer step in the mitochondrial nucleotide salvage pathway38,44. It has been recently proposed that the synthesis of new mitochondrial DNA (mtDNA), which requires this pathway as a nucleotide source, is a pre-requisite for the generation of TLR-induced oxidized-mtDNA, which in turn supports activation of the canonical NLRP3 inflammasome21.
NDPK-D has also been shown to associate with the mitochondrial phospholipid cardiolipin, and facilitates its transfer between the mitochondrial inner- and outer membranes (MIM and MOM, respectively)41. Under resting conditions, cardiolipin localizes to the inner leaflet of the MIM, however mitochondrial stress can induce cardiolipin transfer to the mitochondrial surface which enables release of stress signals such as cytochrome C45. Recently, cardiolipin exposure on the mitochondrial surface has been suggested to support canonical NLRP3 inflammasome activation through nucleation of a complex of key inflammasome regulators18,26. Taken together, these studies suggest that Nme4/NDPK-D could support inflammasome activation through both mitochondrial nucleotide synthesis and regulation of cardiolipin exposure.
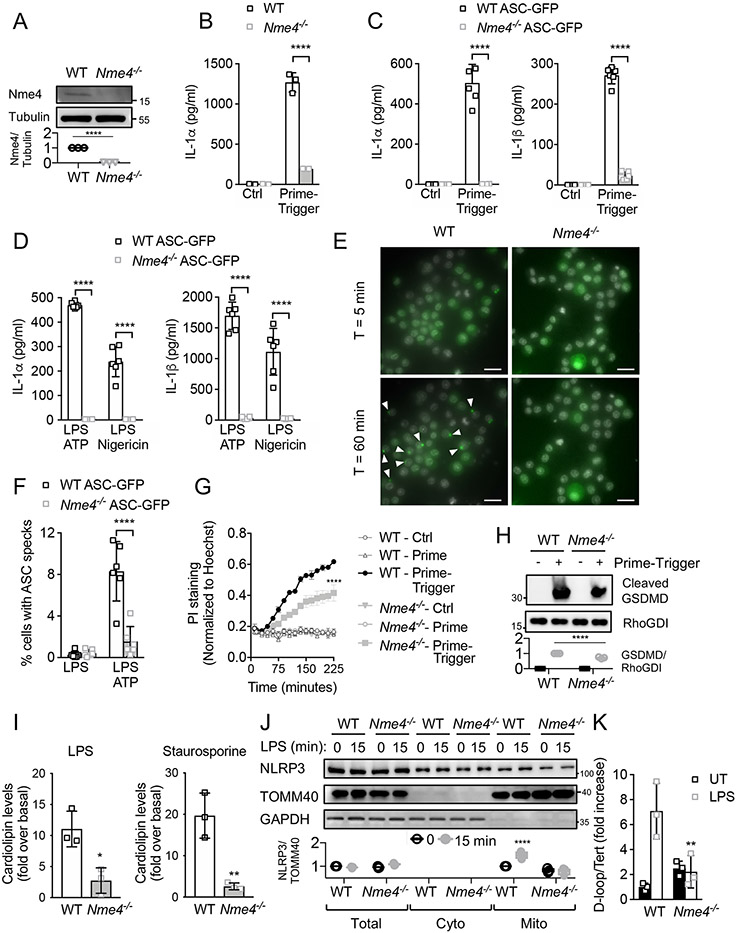
To further investigate Nme4 function, we first used CRISPR/Cas9 to target the Nme4 gene in RAW264.7 cells (Fig. 2A), which recapitulated the initial non-canonical inflammasome screen IL1α phenotype in response to a P3C/transfected KLA prime-trigger stimulus (Fig. 2B). Since RAW264.7 cells do not express sufficient ASC to support canonical NLRP3 inflammasome activation, deficiencies in IL1α secretion are likely independent of the canonical inflammasome. To assess the role of Nme4 in canonical inflammasome activation, we stably expressed ASC-GFP in wild-type and Nme4−/− RAW264.7 cells. Following cytosolic LPS (Fig. 2C), ATP or Nigericin (Fig. 2D) exposure, ASC-GFP expressing WT RAW264.7 cells support IL-1β and IL-1α release, whereas release of these cytokines is severely blunted in Nme4−/− cells, reinforcing a requirement for Nme4 in both the canonical and non-canonical inflammasome pathways. We further used these cells to assess ASC speck formation, which indicates canonical inflammasome assembly, by dynamic live cell imaging, and observed markedly reduced ASC speck formation in the absence of Nme4 (Fig. 2E, F, Movie S1), suggesting that Nme4 is essential for effective formation of the NLRP3 inflammasome complex. The requirement for Nme4 extended to the cellular pyroptotic response to cytosolic LPS, with Propidium Iodide (PI) uptake (Fig. 2G), LDH release (Fig S1C) and GSDMD cleavage (Fig 2H) all attenuated in Nme4−/− cells.
Figure 2: Nme4 is required for canonical and non-canonical inflammasome responses.
(A) Nme4 expression analysis from isolated mitochondria of WT or Nme4−/− RAW264.7 cells. Nme4 was pulled down and immunoblotted. Tubulin serves as a pulldown input reference and quantification is plotted. Prime-trigger assays in WT and Nme4−/− RAW264.7 cells (B) or WT and Nme4−/− RAW264.7 cells stably expressing ASC-GFP (C, D, E, F). IL-1α and IL-1β measurement followed 100 nM P3C priming (6 hr) then (B, C) 100 nM LipidA transfection for 18 hr or (D) 5 mM ATP or 10 μM Nigericin for 30 min. (E, F) 5 mM ATP-induced ASC-GFP speck formation in 100 nM P3C primed ASC-GFP WT or Nme4−/− cells. Representative images (ASC specks are highlighted by white arrows); scale bar: 20 μm; (E) and quantification of % speck-positive cells (F). (G, H) WT and Nme4−/− RAW264.7 cells were primed for 5 hr with 100 nM P3C then triggered by electroporation of 1μg LPS for 3 hr. PI uptake was measured (4 μg/ml) and normalized to Hoechst staining (G) while cells were immunoblotted for GSDMD cleavage (H). RhoGDI blot serves as a gel loading reference and blot quantification is plotted. (I) WT or Nme4−/− RAW264.7 cells were primed for 4 hr with LPS (100 ng/ml) or Staurosporine (2 μM) and cardiolipin levels in the mitochondrial fraction were measured by FACS analysis of Alexa Fluor 647-Annexin-V staining and normalized to untreated cells. (J) WT or Nme4−/− RAW264.7 cells were treated for 15 min with 100 ng/ml LPS and the mitochondrial fraction was isolated and analyzed for NLRP3 recruitment by SDS-PAGE. Relative NLRP3 recrutment to the mitochondrial fraction was plotted. (K) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 2 hr, followed by cytosolic and genomic DNA isolation and quantification by RT-PCR. Results are presented as cytosolic mtDNA vs. nuclear DNA. Data shown are representative of (A-E, G-H, J-K) or pooled from (F, I) at least three independent experiments. (B-D, F-G, I, K) Data expressed as mean ± SD and (I, K) as a fold induction relative to unstimulated cells. (B-D, F-H, J-K) Two-Way ANOVA followed by Sidak’s multiple comparison test; **p < 0.01, ****p < 0.0001. (A, I) Welch’s two-tailed t-Test; *p < 0.05, **p < 0.01. (C-D, F) n=6 (B, G, I, K) n=3.
Nme4 is required for both cardiolipin exposure and LPS-induced mtDNA release
Since cardiolipin has been suggested to support non-transcriptional inflammasome licensing through recruitment of components to the mitochondrial membrane26, and Nme4 has been implicated in cardiolipin externalization41,43, we hypothesized that this may contribute to defective inflammasome activation in Nme4-perturbed cells. We measured cardiolipin localization to the MOM and observed a substantially diminished LPS-induced increase in externalized cardiolipin in Nme4-deficient cells (Fig. 2I). A similarly defective response was observed when cardiolipin externalization was induced by the apoptotic stimulus staurosporine (Fig. 2I), implicating Nme4 in multiple forms of mitochondrial stress-induced cell death, in agreement with previous studies showing Nme4-mediated cardiolipin exposure sensitizes cells to apoptosis42 and mitophagy43. We also observed a marked reduction in the LPS-induced recruitment of NLRP3 to the MOM in Nme4-deficient cells (Fig. 2J), supporting a role for both Nme4 and cardiolipin exposure in the mitochondrial events that embody inflammasome licensing. We further assessed the LPS-induced cytosolic release of mtDNA in macrophages, and observed a loss of this response in the absence of Nme4 (Fig. 2K), in agreement with prior work21,46. These data support multiple roles for mitochondrial Nme4 in canonical inflammasome activation.
Mitochondrial Nme4 supports LPS-induced gene transcription
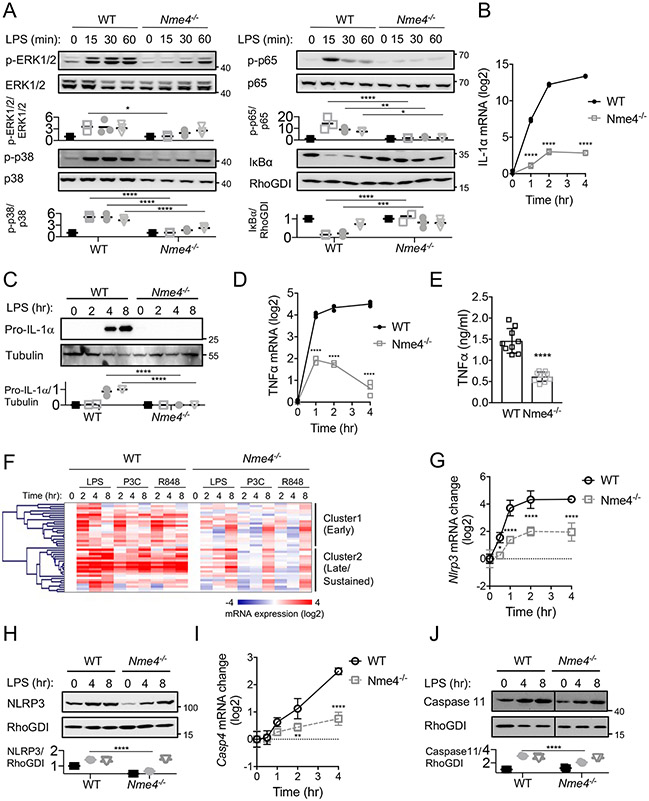
To further define the role of Nme4 in the cellular processes supporting inflammasome activation, we measured the NF-κB and MAPK signaling responses that drive large-scale gene transcription programs in response to LPS priming. We measured the nuclear translocation of NF-κB (p65/RelA), degradation of the NF-κB inhibitor IκB, and phosphorylation of the MAPKs p38 and ERK1/2, and found Nme4 deficiency resulted in diminished and delayed responses in all cases (Fig. 3A). We then tested IL-1α mRNA and protein expression and observed a diminished induction of Il1a in Nme4-deficient cells (Figs. 3B, C). Similarly weakened responses were observed for TNFα and IL-1β mRNA and protein (Fig. 3D, E, Fig. S3A), suggesting a broader role for Nme4 beyond mtDNA synthesis and cardiolipin exposure. Considering the effects of Nme4 deficiency on both LPS-induced signaling and inflammatory cytokine transcription, we tested the response of a wider gene panel to multiple TLR ligands, and found many inflammatory genes were broadly attenuated in the absence of Nme4 (Fig. 3F, Table S4). Cluster analysis revealed two classes of genes: early transient genes, including Dusp1 and Rcan1, for which expression was substantially delayed in the absence of Nme4 (Fig. 3F, Fig S3B); and later sustained genes, including Il6 and Ccl3, which were both delayed and diminished in the absence of Nme4 (Fig. 3F, Fig. S3C). A similar delay was observed for inflammasome components which are induced during priming, including NLRP3 (Fig 3G, H) and caspase11 (Fig 3I, J). In contrast, the expression of constitutively-expressed inflammasome components not induced by priming, such as caspase1 and GSDMD, showed comparable expression levels in control and Nme4−/− cells (Fig. S3D, E). These observations suggest that Nme4 is an important multi-functional regulator of mitochondrial events during macrophage activation, with an unexpected but critical role in LPS-induced transcription.
Figure 3: Nme4 mediates TLR signaling and transcriptional response.
(A) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 0, 15, 30, 45, 60 min and immunoblotted for NF-κB phosphorylation (p-p65), IκB protein levels, ERK phosphorylation and p38 phosphorylation. Total protein levels for p65, ERK1/2, p38 and RhoGDI were assessed as loading controls and relative change quantification is plotted. (B-E) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for the indicated times and Il1a and Tnf mRNA were quantified by qPCR, (C) pro-IL-1α protein expression was analyzed by western blot 0, 2, 4 and 8 hr after stimulation (tubulin blot serves as a gel loading reference) and (E) TNFα secretion at 4 hr was measured by ELISA. (F) Transcriptional response to TLR ligands LPS (100 ng/ml), P3C (1 μg/ml), R848 (5 μg/ml) in WT and Nme4−/− cells 0, 2, 4 and 8 hr after stimulation. mRNA levels were assayed by Fluidigm microfluidic RT-PCR and gene expression patterns were analyzed by hierarchical clustering (Pearson uncentered). (G-J) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for the indicated times and Nlrp3 and casp4 mRNA were quantified by qPCR (G, I). NLRP3 and Caspase11 protein expression was analyzed by western blot 0, 4 and 8 hr after stimulation (RhoGDI serves as a gel loading reference) and quantification is plotted (H, J). Data shown are representative of three (A-E ,G-J) or two (F) independent experiments, and expressed as mean ± SD (A -E, G-J). (A-D, G-J) Two-Way ANOVA followed by Sidak’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n=3. (E) Welch’s two-tailed t-Test; ****p < 0.0001, n=9.
LPS-induced metabolic changes in macrophages are facilitated by Nme4
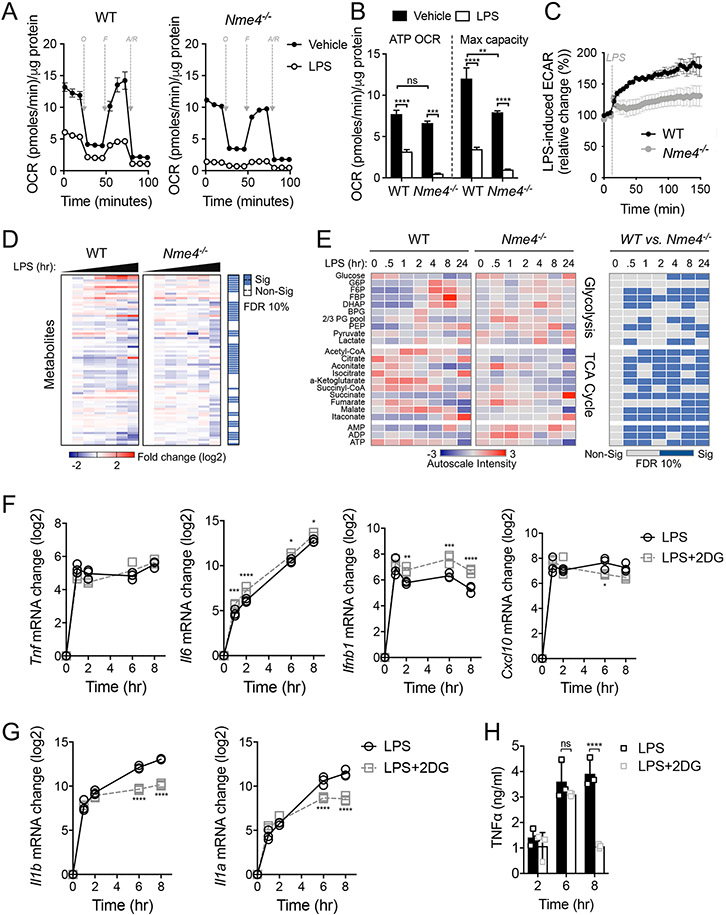
Since Nme4 is a mitochondrial protein, we tested the metabolic characteristics of Nme4-deficient cells. Using a mitochondrial stress test, we observed a comparable basal oxygen consumption rate (OCR) in the absence of Nme4 in both cell types, and detected the expected OCR reduction in response to LPS (Fig. 4A, B). The maximal respiratory capacity at the basal state was reduced in the Nme4-deficent cells, mainly due to the absence of respiratory reserve (Fig. 4B). Mitochondrial mass and membrane potential were not affected by Nme4 deficiency (Fig. S4A-B). We found that the well-established increase in extracellular acidification rate (ECAR) from acute TLR-induced glycolysis, observable within minutes of an inflammatory signal30,47,48, was markedly reduced in Nme4−/− macrophages (Fig. 4C). We then conducted a comprehensive metabolic profiling of WT and Nme4−/− macrophages (Fig. 4D) and noted a substantial difference in glycolytic and TCA metabolites (Fig. 4E). Furthermore, ADP accumulates in the Nme4−/− macrophages reflecting lower ATP levels in these cells. In the WT macrophages, TCA metabolites built up early on in response to LPS, suggesting a high energetic demand in response to LPS that is less evident in the Nme4 deficient cells. The lack of glycolytic shift in the absence of Nme4 is also reflected both by glucose accumulation and reduced flux in glycolysis intermediates compared to WT cells. Recent data have shown that the kinase TBK1 participates in the TLR-activated mitochondrial response that leads to the glycolytic shift30,48, and consistent with a potential role for Nme4 in this process, we observed reduced TBK1 phosphorylation in Nme4-deficient cells (Fig. S4C). Considering the rapid glycolytic induction by LPS in control cells, we questioned whether glycolytic commitment is an energetic pre-requisite for the robust transcriptional response. While we observed normal induction of numerous inflammatory cytokine mRNAs (Fig. 4F) in macrophages treated with the glycolysis inhibitor 2-DG (Fig. S4D), the later sustained transcription of Il1a and Il1b was reduced (Fig. 4G), consistent with prior observations27,49. Similarly, sustained secretion of TNFα, which requires glycolysis-dependent metabolic reprogramming27,30, is strongly diminished in the presence of 2-DG (Fig. 4H). Thus, while the defective glycolysis induction we observe in Nme4-deficient macrophages may limit their energetic capacity to support a sustained inflammatory state, it does not explain their early diminished transcriptional response to LPS.
Figure 4: Nme4 is required for glycolytic commitment induction in LPS-stimulated macrophages.
Representative Seahorse mitochondrial stress test measuring OCR (A) and mitochondrial OCR (B) after 24 hr of 100 ng/ml LPS administration in WT or Nme4−/− RAW264.7 cells. O = oligomycin (1 μM), F = FCCP (2 μM), R/AA = Rotenone (0.1 μM) and antimycin A (1 μM). (B) A bar graph showing quantified protein normalized mitochondrial OCR from stress test in (A). (C) Real time ECAR measurement in WT or Nme4−/− RAW264.7 cells injected with LPS to a final concentration of 100 ng/ml. (D-E) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 0, 0.5, 1, 2, 4, 8 and 24 hr and the metabolic profile was determined by LC-MS. (D) Log2 fold change on the mean and significance level between groups from an ANOVA2 comparison for time and group variance. (E) Auto-scaled intensity and significance by t-test of the normalized signal from central metabolite signals associated with glycolysis and the TCA cycle over the course of activation. (F-H) WT RAW264.7 cells were treated with LPS (100 ng/ml) in the presence or absence of 2-DG (5 mM) for the indicated times. (F) Tnf, Il6, Ifnb1, Cxcl10 and (G) Il1b and Il1a mRNA were measured by qPCR, and (H) secreted TNFα was measured by ELISA. Data are representative of two (D-E) or three independent experiments (A-C, F-H), and expressed as mean ± SEM (A, C) or mean ± SD (B, F-H). (D-E) Statistical testing was corrected for multiple comparisons using the Benjamini-Hochberg method with a false discovery rate of 10 % equating to p-values of 0.062 for (D), 0.019, 0.047, 0.053, 0.030, 0.067, 0.061 and 0.061 for 0, 0.5, 1, 2, 4, 8 and 24 hr respectively (E). (B, F-H) Two-Way ANOVA followed by Sidak’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (A-C, D-E) n=6 (D-F) n=3.
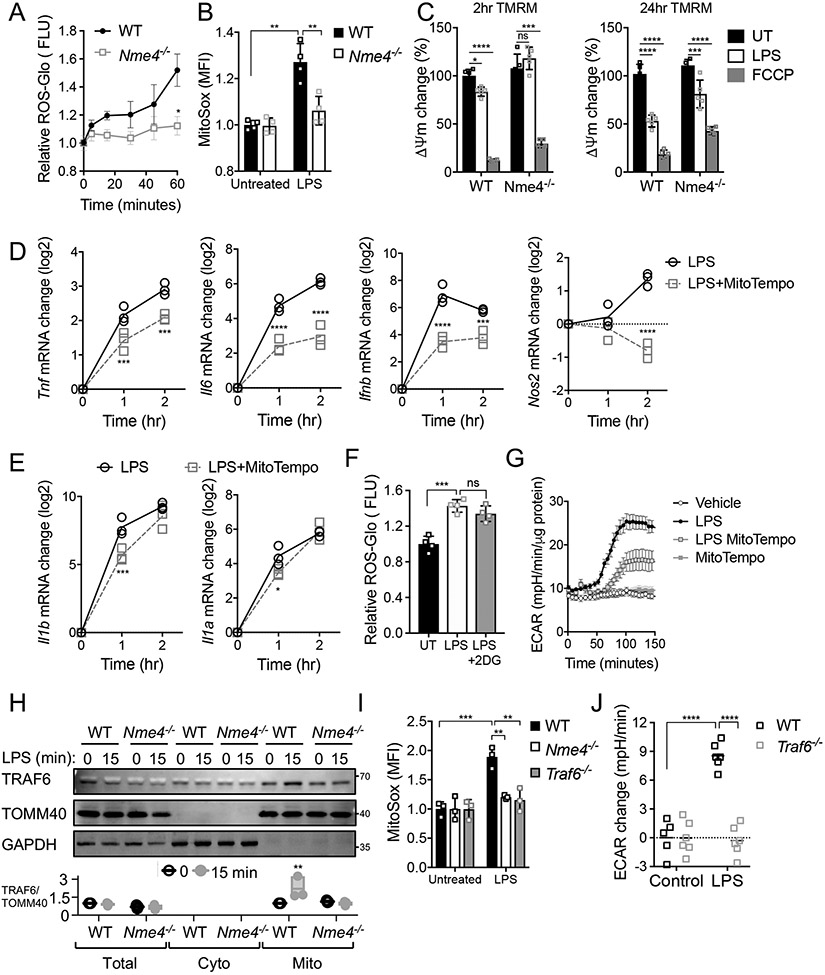
Nme4 is required for mitochondrial recruitment of TRAF6 and ROS production in response to LPS
It has been shown that activated macrophages utilize the electron transport chain for the production of reaction oxygen species (ROS) which are a critical component of the host antibacterial response30,50. We measured the early ROS response to LPS and observed a ROS induction deficiency in the absence of Nme4, both at the population (Fig. 5A) and single cell level (Fig. 5B). Mitochondrial ROS can be generated either via OXPHOS complexes I and III or through reverse electron flow in the ETC which reduces mitochondrial membrane potential in response to LPS33,34, while the macrophage inflammatory response has been shown to be driven by mitochondrial ROS produced specifically in complex III33. The dampened mitochondrial ROS in the Nme4-deficient cells is consistent with our observed reduction in TCA metabolites in these cells (Fig. 4E). Accordingly, we saw progressively reduced mitochondrial membrane potential in LPS treated macrophages which was both delayed and diminished in Nme4−/− cells (Fig. 5C), consistent with the lack of early ROS production in the absence of Nme4. Since it has been suggested that antioxidants can diminish LPS-induced inflammatory gene activation50-52, we tested whether mitochondrial ROS induction contributes to the early transcriptional response. We find that inhibition of mitochondrial ROS with the scavenger mitoTEMPO perturbs LPS-induced gene responses (Fig. 5D), and we observe a similar effect with the antioxidant N-acetylcysteine (NAC, Fig. S5A). These data support a model whereby defective ROS production underlies the acute transcriptional defect in LPS-treated Nme4−/− cells.
Figure 5: Nme4 regulates transcription via the TRAF6-Nme4-ROS pathway.
(A-B) WT and Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for up to 1 hr (A) or at 1 hr (B) and early ROS production was measured using a ROS-glo assay (A) or mitoSOX (1 μM) staining and fluorescence imaging by flow (B). (C) Mitochondrial membrane potential change in WT and Nme4−/− RAW264.7 cells treated with 100 ng/ml LPS for 2 or 24 hr and stained with 200 nM TMRM (D, E) WT RAW264.7 cells were treated with 100 ng/ml LPS for the indicated times +/− mitoTempo (500 μM) and mRNA levels of the indicated genes were measured by qPCR. (F) Early ROS production in WT RAW264.7 cells treated with 100 ng/ml LPS for 1 hr +/− 5mM 2-DG. (G) Effect of mitochondrial ROS inhibition on the LPS-induced glycolytic switch. Real time ECAR measurement in WT RAW264.7 cells pretreated +/− 500 μM mitoTempo followed by 100 ng/ml LPS. (H) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 15 min and the mitochondrial fraction was isolated and analyzed by SDS-PAGE. Relative TRAF6 recruitment to the mitochondrial fraction is shown. (I) WT, Nme4−/− or Traf6−/− RAW264.7 cells were treated with 100 ng/ml LPS for 1 hr and ROS production was measured by mitoSOX (1 μM) imaging. (J) ECAR measurement in WT and Traf6−/− RAW264.7 cells +/− 15 min treatment with 100 ng/ml LPS. (A-J) Data shown are representative of at least three independent experiments and (A-F, I, J) expressed as mean ± SD, (G) mean ± SEM. (A) Welch’s two-tailed t-Test; *p < 0.05. (B-E, G-J) Two-Way ANOVA followed by Tukey’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (F) One-Way ANOVA followed by Tukey’s multiple comparison test; ***p < 0.001. (A, D, E, I) n=3, (B,C, F, G) n=4, (J) n=6.
By assessing early ROS induction by LPS in the presence or absence of 2-DG, we found that glycolysis inhibition had no significant effect on the early production of ROS (Fig. 5F) whereas ROS inhibition markedly reduced acute glycolytic commitment induced by LPS (Fig. 5G) suggesting that ROS production precedes the glycolytic shift. Accordingly, Nme4−/− cells fail to induce both ROS (Fig. 5A, B) and increases in glycolytic flux (Fig. 4C). These data indicate that Nme4 likely operates upstream of the mitochondrial ROS production that dictates the glycolytic shift and sustained transcription of Il1a and Il1b.
It has been suggested that TRAF6 recruitment to the MOM is required both for ROS production50 and glycolytic commitment48 in LPS-treated macrophages. Therefore we sought to determine whether TRAF6 is responsible for signaling to the mitochondria to engage Nme4 after LPS treatment. We detected substantial increases in TRAF6 in the mitochondrial fraction of LPS-treated wild type macrophages, while Nme4−/− cells showed lower basal levels of TRAF6 in the mitochondrial fraction and no LPS-induced increase (Fig. 5H). We measured TRAF6 translocation in the presence of a ROS inhibitor and observed no decrease in LPS-induced recruitment (Fig. S5B), suggesting that TRAF6 movement to the MOM is upstream of ROS production, as previously reported50. We then used CRISPR/Cas9 to generate Traf6−/− RAW264.7 cells and, similar to Nme4−/− macrophages, we find that these cells cannot induce an early ROS response (Fig. 5I) or a glycolytic shift after LPS treatment (Fig. 5J). Taken together, these data suggest that a mitochondrial ROS response is critical to support the acute LPS-activated gene program and is Nme4-dependent. We find that Nme4 is required for this ROS response, possibly through facilitation of TRAF6 MOM recruitment which may depend on the Nme4-supported cardiolipin enrichment on the mitochondrial surface.
Nme4 coordinates a metabolic checkpoint to support a robust cellular response to infection
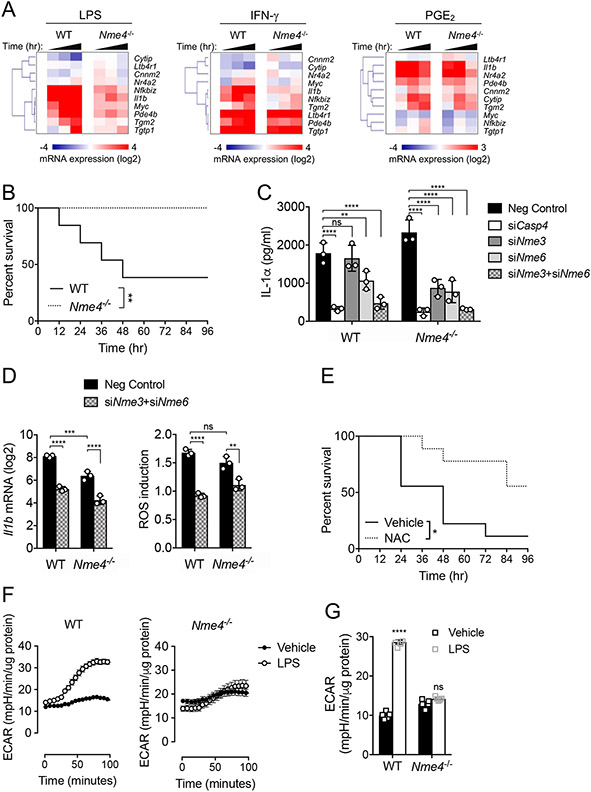
Considering that the LPS-induced gene program is severely hampered in Nme4 depleted cells, we hypothesized that Nme4-dependent mitochondrial responses may be required to initiate and support large-scale transcription in TLR-activated macrophages. To test whether the effects of Nme4 deficiency on transcription are scale dependent, we compared LPS-activated macrophages with those stimulated with either IFN-γ or PGE2, which induce substantially fewer genes (Fig. S6A, B)53. We chose a panel of 10 genes for which at least half were induced by each ligand, with several genes induced by all three. We found that all genes induced by LPS were substantially attenuated in the Nme4−/− cells, approximately half of the IFN-γ induced genes were perturbed, while all PGE2-induced genes were expressed to similar levels in control and Nme4−/− cells (Fig. 6A). These data support a model whereby the larger energetic demands placed on the cell to induce numerous genes in response to LPS likely require mitochondrial responses dependent on nucleoside diphosphate kinases.
Figure 6: Nme4 deficient mice are resistant to endotoxic shock.
(A) Nme4 acts as a mitochondrial checkpoint required to support large-scale gene program. WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS, 5 nM IFNγ or 10 μM PGE2 for 0, 1, 2, or 4 hr and mRNA levels of the indicated genes were measured by qPCR. (B) Survival of WT or Nme4−/− mice after intraperitoneal injection of 10 mg/kg LPS. (C) Primary BMDM from WT or Nme4−/− mice were transfected with negative control or Casp4, Nme3 and/or Nme6 siRNA for 48 hr and then with 1 μg/ml P3C for 6 hr and triggered by 100 nM LipidA transfection for 18 hr. IL-1α secretion was measured by ELISA. (D) Primary BMDM from WT or Nme4−/− mice were transfected with negative control or Nme3 and/or Nme6 siRNA for 48 hr and then treated with 100 ng/ml LPS for 1 hr. Il1β mRNA was quantified by qPCR and early ROS production was measured. (E) Survival of WT mice +/− intraperitoneal injection of 150 mg/kg NAC (30 min) followed by intraperitoneal injection of 10 mg/kg LPS. (F) Real time ECAR measurement in WT and Nme4−/− BMDM injected with 100 ng/ml LPS. (G) WT and Nme4−/− BMDM were treated with 100 ng/ml LPS for 24 hr followed by a glucose stress test. Quantified ECAR was normalized to protein concentration. Data shown are representative of four (A), five (B), three (C-D, F-G) and two (E) independent experiments, with n=13 WT mice and n=8 Nme4−/− mice (B) and n=9 in each group (E). (B, E) Log-Rank Mantel-Cox Test; * p < 0.02, ** p < 0.01. (F) Data (n=3 separate wells per group) represent three independent experiments, and expressed as mean ± SEM. (C-D, G) Data Data expressed as mean ± SD and analyzed by Two-Way ANOVA followed by Sidak’s multiple comparison test; **p < 0.01, ***p < 0.001, ****p < 0.0001, (C-D) n=3, (G) n=5.
Mice lacking Nme4 are resistant to endotoxin shock.
We used CRISPR/Cas9 to target the Nme4 gene locus (Fig. S7A) and generated Nme4−/− mice which appeared healthy and showed Mendelian inheritance. We tested the response of Nme4−/− mice to endotoxin-induced sepsis and observed significant resistance to septic shock in the Nme4−/− animals (Fig. 6B). Unexpectedly, we observed a normal IL-1α response to cytosolic LPS in BMDM from these mice (Fig. S7B). However, we could replicate the Nme4 dependency of the IL-1α/β responses to cytosolic LPS in BMDM when the gene was acutely perturbed by three independent siRNAs (Fig. S7C, D). We therefore hypothesized that other Nme gene family members might compensate for the loss of Nme4 in vivo. When we individually knocked down the other two Nme genes, Nme3 and Nme6, that were identified in our initial screen, we observed diminished IL-1α and IL-1β responses to cytosolic LPS in primary BMDMs (Fig. S7E, F), especially in the Nme6-perturbed cells. Thus, Nme3 and Nme6 also contribute to the cytosolic LPS response in primary macrophages, and likely compensate for Nme4 deficiency in vivo. Accordingly, we observed elevated levels of both Nme3 and Nme6 in the Nme4−/− BMDM (Fig S7G, H). Consistent with this, we found that Nme4−/− BMDM were more sensitive than WT BMDM to siRNA-mediated knockdown of either Nme3 or Nme6 (Fig. 6C). Moreover, combined knockdown of both Nme3 and Nme6 in BMDM from Nme4−/− mice reduced IL-1α to levels comparable with Casp4-depleted cells (Fig. 6C). As was observed for the Nme4−/− RAW264.7 cells, Il1b expression and ROS production were reduced when both Nme3 and Nme6 were knocked down in Nme4−/− BMDM (Fig. 6D) suggesting that inflammasome priming is defective in the absence of the mitochondrial Nme genes.
Since the Nme4−/− mice exhibit a normal IL-1α response to cytosolic LPS, the protective effect of Nme4 perturbation for in vivo endotoxin challenge might relate to the other mitochondrial functions for Nme4 observed in our earlier experiments and in prior studies54-56, including a role for Nme4 in mitophagy43. Accordingly, we found that resistance to endotoxin was also observed in mice pre-injected with the ROS scavenger N-acetyl cysteine (NAC) (Fig. 6E), supporting a role for ROS in promoting broader aspects of the inflammatory lethality of LPS in vivo. We also observed a defective glycolytic commitment in the Nme4−/− BMDM, both in the acute response to LPS (Fig. 6F) and at 24 hour after LPS stimulation (Fig. 6G). This defective glycolytic commitment in macrophages from Nme4−/− mice may also contribute to their endotoxin resistance, as glycolytic blockage by 2-DG has a similarly protective effect in this LPS challenge model57-59.
Discussion
We have utilized a genome-scale siRNA screening approach to discover new regulators of inflammasome activation, identifying the three mitochondrial proteins of the Nme gene family as novel regulators of the non-canonical inflammasome. We focused on the Nme4 gene, as it codes for a multifunctional protein (NDPK-D) that could be linked to the canonical inflammasome response through both MOM cardiolipin enrichment18,26,41 and mitochondrial DNA synthesis21,44. In this work we have further illuminated Nme4’s roles in these processes, while also uncovering unappreciated functions of Nme4 in both canonical and non-canonical inflammasome responses. We demonstrate that Nme4 is required for cardiolipin exposure on the MOM in response to a TLR priming signal, which in turn may support the recruitment of inflammasome components to the MOM during inflammasome licensing. We confirm that Nme4 is required for TLR-induced mtDNA synthesis, but also delineate additional roles for Nme4 in mitochondrial recruitment of TRAF6 and ROS production, which are required for the large-scale inflammatory gene program induced during inflammasome priming.
While the known role for Nme4 in the mtDNA salvage pathway depends on its nucleoside diphosphate kinase function in the final step of NTP synthesis, its cardiolipin transferase activity highlights reciprocally regulated dual functions, as cardiolipin binding inhibits NDPK-D kinase activity due to the close proximity between the lipid binding pocket and the catalytic domain42. Notably, other Nme genes exhibit a similar multi-functional nature, with Nme1 and Nme2 recently implicated as protein histidine kinases60, Nme1 regulating non-homologous end joining of DNA double-strand breaks61, Nme3 being critical for mitochondrial fusion37, and Nme6 and Nme7 regulating stem cell gene expression62. Further studies will be required to determine whether the additional role we have identified for Nme4 in TRAF6 MOM recruitment, ROS production and gene transcription is dependent on its kinase or lipid transferase function, or yet another unappreciated activity for its NDPK-D gene product.
Although our screen was initially designed to identify regulators of the non-canonical inflammasome, restoration of the canonical inflammasome by ASC expression in RAW264.7 macrophages demonstrated a requirement for Nme4 in both pathways, and our unexpected finding that Nme4 is also broadly required for transcriptional priming has obvious implications for multiple inflammasome classes. We show that Nme4 deficiency leads to a delay in both TLR-induced signaling and acute gene transcription, and a defective metabolic shift, implying that mitochondrial engagement is an early event in TLR pathway activation, consistent with recent observations47.
Mitochondria subsume a central role in the macrophage response to infection where they dictate major metabolic changes that direct ATP production from OXPHOS to aerobic glycolysis. This switch is accompanied by alteration of TCA cycle flux, including the accumulation of TCA metabolites and increased ROS production51,63. While the glycolytic commitment in TLR-activated macrophages is well established30,32,47,48,64, studies with 2DG-based glycolysis blocking have shown that the majority of TLR-induced genes are not dependent on this metabolic switch27,30,65. The primary exception to this pattern is IL-1β, whose TLR-activated transcription has been linked directly to the aforementioned TCA cycle flux alteration that increases succinate levels and activates HIF-1α through protein stabilization, which in turn directly upregulates the Il1b gene27,31,57. While we demonstrate that the acute glycolytic response is defective in Nme4-deficient macrophages, this cannot explain the broad and acute TLR-induced gene expression defect in these cells that is evident 1-2 hr after ligand activation.
Rather, our study finds that acute Nme4-dependent ROS elevation in macrophages is critical to support this early gene transcription. This ROS response requires mitochondrial recruitment of TRAF6, which is also defective in Nme4-deficient cells. Prior work has suggested that MAVS is required for TRAF6 MOM enrichment, however we observe no change in MAVS levels on the mitochondria in the absence of Nme4 (data not shown). Therefore, the Nme4-dependent loss of cardiolipin MOM enrichment may suggest that this lipid also contributes to the TRAF6 recruitment. Since we find that NF-κB and MAPK signaling are reduced and delayed in the Nme4-deficient cells, this suggests that TRAF6 recruitment to the mitochondria is required for intact TLR signaling responses and robust early gene transcription.
The requirement for ROS activation to support TLR-induced transcription is supported by the attenuation of cytokine genes by the mitochondrial ROS scavenger mitoTempo, and the general ROS scavenger NAC. ROS can be produced in macrophages via diverse sources in several cellular locations and time frames. In addition to respiration-induced ROS in the mitochondria66, ROS can also be generated as part of the pentose phosphate pathway by NADPH oxidase (NOX)66, and as a byproduct of fatty acid and protein oxidation at the ER66. It has been shown that NF-κB activity is dependent on NOX expression67 and is elevated during hydrogen peroxidase-induced oxidative stress68. Our data suggest a role for Nme-dependent mitochondrial ROS, which is critical for acute NF-κB signaling to support TLR-activated gene transcription.
Nme4−/− mice are protected from LPS shock, although Nme4−/− BMDM did not exhibit an in vitro IL-1α secretion defect in response to cytosolic LPS. It would appear that Nme4’s support of non-canonical inflammasome activation can be compensated by the other Nme genes identified in our initial screen, Nme3 and Nme6, as their perturbation in Nme4−/− BMDM uncovered both the IL-1α secretion and ROS response defect resulting from Nme deficiency. BMDM from Nme4−/− mice did however exhibit a strong defect in glycolytic commitment in response to LPS, which may contribute to their protection from endotoxin shock, and would be consistent with recent studies showing a similar protective outcome in mice treated with the glycolysis inhibitor 2DG57-59. Also, while 2DG does not block inflammatory cytokine transcription beyond the succinate/HIF-1α/IL-1β axis, it does have substantial effects on sustained secretion of inflammatory cytokine proteins27,57,69, which would be consistent with reduced susceptibility to LPS-induced shock in mice with a perturbed glycolytic commitment pathway. Nme4 has also been shown to regulate mitochondrial turnover through mitophagy43, a process also implicated in the macrophage response to infection70,71, however, we did not observe elevated mitochondrial mass in Nme4−/− cells.
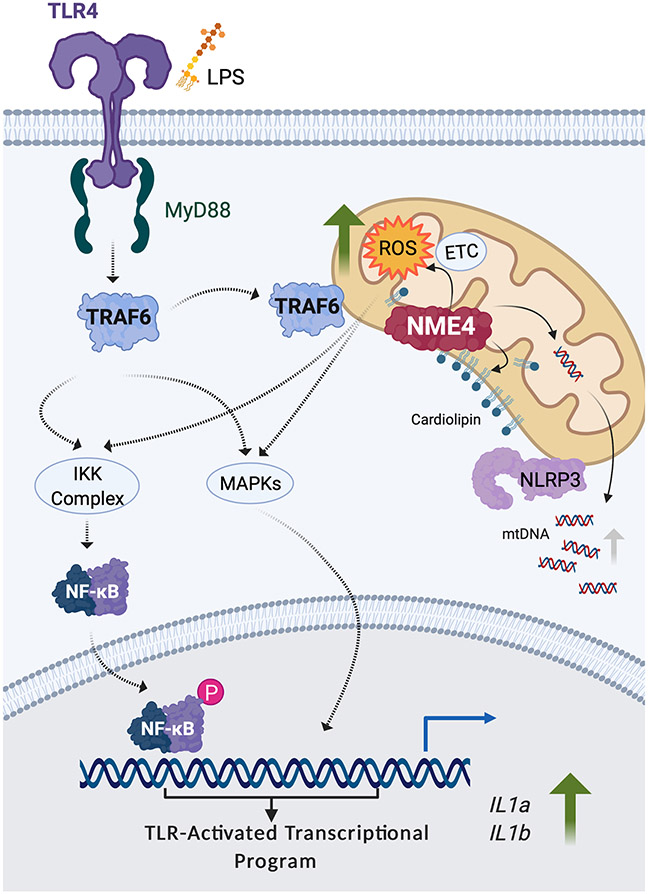
In summary, we have identified the Nme4 gene as a multifunctional regulator of inflammasome activation based on multiple mitochondrial roles for its NDPK-D protein product (Fig. 7). We show that Nme4 coordinates TLR-induced mtDNA synthesis and cardiolipin MOM exposure, which supports mitochondrial nucleation of inflammasome components. Nme4 also facilitates TRAF6 mitochondrial recruitment, supporting both ROS induction and glycolytic commitment, with the former effect on ROS being required for initiating and supporting both TLR-induced NF-κB and MAPK signaling, and the broad transcriptional program which underpins inflammasome priming. Our work suggests that Nme4-sufficiency can function as a mitochondrial fitness checkpoint which can determine the capacity of macrophage cells to support a robust metabolic and transcriptional response to infectious challenge.
Fig. 7: Nme4 supports inflammasome activation through multiple roles.
A model illustrating the multiple functions of Nme4 contributing to inflammasome activation. Nme4 facilitates recruitment of TRAF6 to the mitochondria and ROS production to support TLR-activated transcription and inflammasome gene priming. Additionally, the lipid transferase function of Nme4 mediates the cardiolipin exposure required for NLRP3 mitochondrial recruitment and inflammasome licensing, while the nucleoside diphosphate kinase activity of Nme4 supports TLR-induced mtDNA synthesis. The figure was generated with BioRender.
Methods
1. Reagents
Lipid A (Avanti Polar Lipids), Pam3CSK4, R848, nigericin and ATP (InvivoGen), LPS, NAC, mitoTempo, 2DG, oligomycin, FCCP, Rotenone, antimycin A, TMRM, FCCP, PGE2, Tributylamine were from Sigma-Aldrich. MitoTracker Green, Hoechst 33342 (ThermoFisher), LCMS-grade water, methanol, isopropanol, chloroform and acetic acid (Fisher Scientific), IFNγ (PeproTech), ELISA kits (IL-1α, IL-1β, TNFα from R&D).
2. Cell Lines
RAW264.7 (ATCC), RAW264.7 G9 cells were derived from an authenticated batch of RAW264.7 cells used by the Alliance for Cell signaling consortium35. RAW264.7 cells were maintained in complete DMEM media, comprising DMEM with 4.5 g/L glucose, 10% FBS (Gemini Bio-Products, West Sacramento, CA), 2 mM Glutamine (Lonza) and 20 mM HEPES (Lonza), hereafter complete DMEM.
3. Generation of CRISPR/Cas9-based gene edited cell lines
gRNAs targeting mouse Nme4 or Traf6 were designed using the Zhang lab online tool (https://zlab.bio/guide-design-resources) and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene #42230)72. Plasmids were electroporated into RAW264.7 cells in the presence of a GFP expressing vector (Amaxa). 16 hr later, GFP+ cells were sorted into single cells clones. Clone knockout was confirmed by genomic DNA extraction (Qiagen), gene amplification and sequencing. gRNA sequences used: gNme4#1 targeting exon 1: CAGCCTTTTCGGGCGCGTCG; gNme4#2 targeting exon 2: ATACAACGCTTTGAGAGGCG; gTraf6: GAAGCAGTGCAAACACCATG.
4. Generation of ASC-GFP RAW264.7 cell lines
The lentiviral plasmid pLEX-MCS-ASC-GFP (Addgene #73957) and packaging plasmids pCMV-VSV-G (Addgene #8454) and pCMV-delta-R8.2 (Addgene #12263) were transfected into adherent HEK293T17 cells using the TransIT-Lenti transfection system (Mirus). 72 hr later, supernatant was collected, and virus was concentrated using Lenti-X Concentrator (Takara). WT and Nme4−/− RAW264.7 cells were transduced with concentrated lentivirus for 72 hrs, and subjected to puromycin (2 μg/ml) selection for >10 days. WT RAW264.7 ASC-GFP and Nme4−/− ASC-GFP monoclonal cell lines were isolated by a limiting dilution and the clones used in this study were selected based on moderate ASC-GFP fluorescent signal and lack of spontaneous ASC speck formation in the absence of inflammasome priming and triggering stimuli.
5. Mice and generation of CRISPR/Cas9 Nme4−/− strain
All mice were maintained in specific pathogen-free facilities under 12 hr light dark cycles with access to food and water ad libitum. All procedures were approved by the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee (National Institutes of Health, Bethesda, MD). C57BL/6 were obtained from Jackson Laboratories. Nme4−/− mice were generated on the background of C57BL/6 using the two gRNAs described above to achieve a 248 bp deletion in the Nme4 loci, which codes for a frame shifted protein. Three to 4-week-old C57BL/6 female mice from Taconic Labs were superovulated by IP injection of 5 IU of pregnant mare serum gonadotropin (Prospec Protein Specialists) followed 48 hr later by 5IU of human chorionic gonadotropin (Sigma Aldrich). The females were then mated with the C57BL6 males and one cell embryos were isolated from the pregnant females. The embryos were then microinjected into pronuclei with a mixture of Cas9 (10 ng/ul) obtained from IDT and sgRNA (10 ng/ul) from Thermofisher. The microinjected embryos were transferred into oviducts of CD1 pseudo-pregnant mothers. The pups were weaned at three weeks and the ear punch biopsies were genotyped for the mutation by PCR and sequencing. Muscle tissue from the mouse femur was used to extract mitochondria which were blotted for Nme4 (the antibody was a kind gift from Dr. Marie-Lise Lacombe). Mouse BMDM were prepared by differentiation from bone marrow for 7 days in complete DMEM supplemented with 100 U/ml Penicillin-Streptomycin and 50 ng/ml M-CSF (R&D).
6. Endotoxin shock
Female WT or Nme4−/− mice 8-16 weeks old were injected intraperitoneally (IP) with 10 mg/kg of body weight LPS from Salmonella enterica serotype minnesota (Sigma, L2137) dissolved at 1 mg/ml in sterile PBS. When noted, mice were pre-injected with either NAC (150 μg/kg) or saline, 30 min before an IP injection of 10 mg/kg LPS. Mice were health checked twice per day and weighed once a day for up to 5 days, after which any surviving mice were euthanized by CO2 inhalation. Survival curves were analyzed using the log-rank (Mantel-Cox) test.
7. Genome-wide siRNA screen
The RNAi screen was conducted in 384-well format using the Ambion Silencer Mouse Genome siRNA Library (#4391425), which consists of three unique, nonoverlapping, nonpooled siRNAs for each of 17,000 gene targets. siRNA reagents (2 μl, 2 μM) were stamped into 384-well white flat-bottom microplates (Corning, 3570) using a Velocity11 VPrep liquid handling system (Agilent) integrated into a BioCel robotic platform (Agilent) in columns 1–22, leaving columns 23–24 empty for negative (Ambion, Negative control #1) and positive (mouse Casp4) controls, respectively. The median value of each plate’s negative control column was used to normalize sample wells, and the positive control was used to assess transfection efficiency and assay performance.
Lipofectamine RNAiMAX Transfection Reagent (0.4 μl; Invitrogen) was added in 20 μl serum-free, antibiotic-free media to plate wells using a Thermo Scientific Matrix WellMate and Microplate Stacker. Plates were incubated for 45 min at room temperature to allow for the sufficient formation of siRNA-lipid complexes. Cells were seeded at a density of 1x104 cells per well in 20 μL media containing 20% (vol/vol) FBS without antibiotics. The final concentration of siRNA in each well was 100 nM. Cells were cultured for 48h at 37°C in 5% CO2 before addition of 10 μl of TLR ligand Pam3CSK4 for 5 hours. The medium was then removed, replaced with 15 μl growth media and cell were transfected with 10 μl of LipidA and TransIT-TKO (Mirus) for 24 hour. At the end of the LipidA treatment, 10 μl of supernatant was transferred to a 384 Greiner Bio-one non-binding low volume plate and 10 μl of IL-1α HTRF reagent (Cisbio, 62MIL1APEH) was added and incubated at room temperature overnight. The plates were read on an EnVision plate reader (PerkinElmer). Cell viability was measured by adding 20ul Cell Titer Glo to the original plate and running a luminescence read on the EnVision. The quantified IL-1α signal for each siRNA gene-targeted well was divided by the median of the negative control wells and multiplied by 100 to achieve a negative control normalized value for each well/siRNA. This normalized value was then used to generate a robust z-score by first taking the log of each value and then by subtracting the median and dividing by the mean absolute deviation. The median z-score was then used to rank genes for follow-up.
8. Calculation of mitochondria-related enrichment among screen hits
Enrichment for mitochondria related genes in hit candidates from the genome-wide screen were evaluated using the Mitochondrial Proteomics Database MitoMiner 4.0 (date accessed June 11, 2020)73. The 95th percentile of positive regulators from the genome-wide screen were analyzed through the database and assigned Integrated Mitochondrial Protein Index (IMPI) scores. IMPI scores were converted to negative values for non-mitochondrial candidates and kept positive for candidates predicted or assigned as mitochondrial.
9. BMDM RNAi
BMDMs were reverse transfected with siRNAs against screen hit genes using Viromer Green (Lipocalyx, cat# VG-01LB-00). 0.1 μl of Viromer Green transfection reagent pre-mixed with 4.9 μl of Viromer Green Buffer was mixed with 5 μl of siRNAs (0.5 μM). After incubation for 30 min at room temperature, 4 μl were added to each well of a 384-well plate (Falcon, 353962). Next, 4x104 BMDMs in 36 μl of complete DMEM were seeded per well for a final siRNA concentration of 25 nM. Plates were incubated at room temperature for 10 min to allow the cells to settle, then at 37°C in a humidified atmosphere with 5% CO2 for 48 hr. Cells were stimulated for time periods as indicated. For measurement of secreted protein level, supernatants were collected and subject to ELISA as described above. All siRNAs were from ThermoFisher with the following siRNA IDs: 102885, 74210 and 174215 for Nme3; 185507, 72411 and 74300 for Nme4, 184862 and 73403 for Nme6, 159999 and 160000 for Casp4 (caspase-11). Non-targeting negative control siRNA was from Dharmacon (NTC5).
10. Inflammasome activation (prime-trigger) assays
Non-canonical inflammasome: 5x104 RAW264.7 macrophages seeded in a 96-well plate were primed with 100 nM P3C for 5 hr, media (80 μl) was replaced and cells were triggered by transfecting 1 μM LipidA for 18 hr (20 μl). Transfection mix included 1 μl of 500 μM LipidA and 4 μl TransIT-TKO (Mirus) added to 95 μl optiMEM (Gibco). The same procedure was used for BMDM except 1 μg/ml P3C was used for priming and transfection was done using the RNAiMAX (ThermoFisher) transfection reagent (3 μl for a total of 100 μl). Canonical inflammasome: Procedures were as above except 100 ng/ml LPS for 5 hr was used for priming, and 5 mM ATP or 10 μM Nigericin for up to 2 hr was used for triggering, both for RAW264.7 and BMDM. IL-1α and IL-1β expression were measured by ELISA according to manufacturer’s instructions (R&D, #DY400, #DY401).
11. Bacterial infection
Infection of RAW264.7 cells and BMDM with B. cenocepecia (provided by Dr. David Greenberg, University of Texas Southwestern) at MOI 10 was conducted using the infection and imaging methods described previously74,75.
12. Cardiolipin exposure measurement
WT and Nme4−/− cells were incubated with 250 nM Mitotracker Green FM (Molecular Probes) for 45 min at 37°C, following the manufacturer’s instructions. After stimulation, mitochondrial isolations were performed by differential centrifugation as previously described18. Briefly, the macrophages were resuspended in mannitol-sucrose buffer and subjected to nitrogen cavitation (200 psi, 20 min at 4°C). The disruption of cells was followed by differential centrifugation steps to isolate the mitochondria. Nuclei and unlysed cells were separated from the homogenate by centrifugation at 1000g for 10 min at 4°C. The mitochondria were then pelleted from post-nuclear supernatant by centrifugation at 12,000g for 20 min at 4°C. Following isolation, cardiolipin externalization was assessed by mitochondrial Annexin V staining as previously described76. Mitochondria were incubated with Annexin V–Alexa Fluor 647 (Invitrogen) for 30 min on ice. Mitochondria were then washed twice with mannitol-sucrose buffer, fixed with 4% paraformaldehyde, and analyzed by flow cytometry on a BD LSR Fortessa.
13. Quantitative PCR
Total RNA was isolated using Direct-zol096 RNA extraction kit (Zymo Research) according to manufacturer’s instructions. RNA was reverse transcribed using iScript Reverse Transcription Supermix cDNA synthesis kit (BioRad). qPCR reactions were carried out using either SYBR Green or TaqMan assays (Applied Biosystems) with gene specific primers and FAM-conjugated probes (Life Technologies). PCR reactions were performed and analyzed in a QuantStudio 6 Flex Real Time PCR system (Applied Biosystems).
14. Fluidigm Quantitative PCR
Quantitative PCR was carried out according to the manufacturer’s instructions using the BioMark HD system (Fluidigm), with Fluidigm-designed primer sets. Ct values were automatically calculated and exported from the BioMark HD system, then normalized to either Hprt or Actb.
15. Mitochondrial fractionation
Mitochondrial fractions were isolated from RAW264.7 cells using a mitochondria isolation kit from ThermoFisher (Cat# 89874). WT and Nme4−/− RAW264.7 cells (5x106 cells per sample) were stimulated with 100 ng/ml LPS for 15 minutes. Fractionation was conducted following the manufacturer’s protocol. NLRP3 and TRAF6 abundance in the mitochondrial fraction was analyzed by western blot using the following primary antibodies: NLRP3 (AdipoGen, AG-20B-0014-C100), TRAF6 (Abcam, ab33915). TOMM40 (ProteinTech, 66658-1-Ig) and GAPDH (Abcam, ab9485) were used for normalization. Nme4 protein expression in WT and Nme4−/− RAW264.7 cells was measured by immunoprecipitation of Nme4 using a rabbit anti-Nme4 antibody (kindly provided by Dr. Marie-Lise Lacombe) with protein A/G beads (ThermoFiscer), then Nme4 was immunoblotted using a mouse anti-Nme4 antibody (Abcam ab228005).
16. Live cell imaging of RAW264.7 macrophages
WT and Nme4−/− RAW264.7 cells expressing ASC-GFP were seeded in 96-wells plates at 2.5x104 cell/well and rested overnight. Cells were primed with 100 nM LipidA for 5 hr, stained with Hoechst 33342 for 30 min, and triggered with 10 μM nigericin for 2 hr. During the triggering step, cells were imaged every 5 min at 20X magnification on a Cell Insight CX7 high-content imager (ThermoFisher), with an onstage incubator set to maintain cell at 37°C and 5% CO2. At each time point, cells and ASC specks were counted using HCS studio image analysis software (ThermoFisher), and the fraction of cells containing ASC specks was calculated.
17. Pyroptosis and GSDMD cleavage assays
WT and Nme4−/− RAW264.7 cells were primed with 100 nM P3C for 5 hr the triggered by LPS electroporation using the Neon Electroporation System (ThermoFisher). 2x106 cell were electroporated for 20 ms, 1400V in 2 pulses with 1 μg LPS in resuspension buffer R. After electroporation, cells were resuspended in optiMEM. For pyroptosis assays, 4x104 cells were seeded in a black, clear bottom 96-wells plate and stained with 4 μg/ml PI and 1 μg/ml Hoechst. The plate was centrifuged and PI uptake was measured every 15 minutes for 4 hr using a CLARIOstar plate reader (BMG-LabTech). PI uptake was normalized to the number of cells as assessed by Hoechst staining. After the PI assay, the cell supernatant was collected and LDH release was measured according to the manufacturer’s instructions (Sigma). For the GSDMD cleavage assay, primed and LPS electroporated cells (or control electroporated cells) were seeded in a 12-well plate at 5x105 cells/well for 3 hr. Whole lysates were collected and immunoblotted for GSDMD (Abcam, ab209845).
18. Western blotting
WT and Nme4−/− cells were treated with LPS for the indicated times and lysed in the presence of protease and phosphatase inhibitor cocktails (Roche). Cell extracts (20 μg protein) were boiled for 5 min in SDS-PAGE buffer, subjected to 4-12% gradient SDS-PAGE, proteins were transferred and the nitrocellulose membrane was blocked using 5% milk for 1 hr. The primary antibodies used were: phosho-p38 MAPK (Cell-Signaling, #4511), phospho-ERK1/2 (Cell-Signaling, #4370), phospho-NF-κB (Cell-Signaling, #3033), IκB (Cell-Signaling, #4814), phospho-TBK1 (Cell-Signaling, #5483), NLRP3 (Adipogen, AG-20B-0014-C100), Caspase-11 (Cell-Signaling, #14340), TRAF6 (Abcam, ab33915), RhoGDI (Sigma, R3025), pro-IL-1α, pro-IL-1β (R&D). Western blots were incubated with respective HRP-conjugated secondary antibodies (Sigma) and using ECL reagents (Bio-Rad) on the ChemiDoc imager (Bio-Rad). Immunoblot data was analyzed using ImageJ software and data from three replicate experiments were quantified for statistical analysis.
19. ROS assays
ROS measurement at the cell population level was assessed in 1x104 RAW264.7 cells seeded per-well of an opaque 384-well plate. The next day the cells were treated with 100 ng/ml LPS and ROS production was measured using the hydrogen peroxide-based ROS-glo assay kit (Promega, G8820), according to the manufacturer’s protocol.
Single-cell ROS measurements were conducted in RAW264.7 cells treated with 100 ng/ml LPS for the indicated times and stained with 1 μM MitoSOX (ThermoFisher) for 15 minutes, following two washes with PBS. Cells were then either collected and measured for fluorescence by Fortessa flow cytometer (BD Biosciences) and analyzed on FlowJo, or imaged on the Cell Insight CX7 imager (ThermoFisher) after further staining with Hoechst 33342 (ThermoFisher, R37605). Image analysis on the CX7 was done using the HCS Studio (ThermoFisher) using the spot counting protocol. Spots intensity was analyzed specifically in the cytoplasm, by segmenting the nuclei based on the Hoechst channel and defining a ring around it.
20. Mitochondrial membrane potential and mitochondrial mass measurements
RAW264.7 cells were treated with 100 ng/ml LPS for the indicated time and stained with 200 nM TMRM or 250 nM MitoTracker Green for 30 min in phenol-free DMEM. Cells were washed twice and fluorescence was detected using a CLARIOstar plate reader (BMG-LabTech).
21. Metabolic analyses
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were examined using the XF96 Seahorse Metabolic Analyzer from Seahorse Biosciences. Briefly, 3x104 RAW264.7 cells or 1x105 BMDM were cultured overnight in complete media. The next day, media was changed to Seahorse XF media and cells were incubated for 1 hr at 37°C without CO2. Metabolic mitochondrial stress tests were performed according to the manufacturer’s protocol. Port injections and times are indicated in the figures. Mitochondrial ATP OCR and maximal respiratory capacity are defined as the percentage of OCR that is oligomycin or antimycin A/rotenone sensitive, respectively. When indicated, 100 ng/ml LPS, 10 mM 2-DG, and/or 250 μM mitoTempo was added during the assay to assess metabolic changes. Where noted, data are represented as percent of control levels calculated as the mean of the basal state. In other assays, BMDM were incubated in complete culture media supplemented with 100 ng/ml of LPS overnight.
Metabolic profile studies were done by Liquid Chromatography Mass Spectrometry (LC-MS). LCMS-grade solvents were used for all metabolomics methods. RAW264.7 cells were seeded at 5x105 in a 6-well plate and treated with 100 ng/ml of LPS for 0, 0.5, 1, 2, 4, 8, 24 hr. The experiment was stopped by removing media, washing with 1 ml of 0.9% sodium chloride and immediately immersing in 0.4 ml of ice-cold methanol for 5 minutes. Next, 0.4 ml of ice-cold water were added and cells were scraped and collected in Eppendorf tubes. 0.4 ml of ice-cold chloroform was added to each sample and samples were shaken for 30 minutes at 4°C then centrifuged at 16,000 g for 20 minutes. 550 μl of the top (aqueous) layer was collected and stored at −80°C. For LC-MS injection, samples were separated using a Sciex ExionLC™ AC system and analyzed using a Sciex 5500 QTRAP® mass spectrometer. The order of injections was randomized. To account for carry over and instrument drift, quality control (QC) and blank injections were distributed throughout the runs.
Metabolites were measured using a previously established ion pairing method with modification77. Samples were separated on a Waters Atlantis T3 column (100Å, 3 μm, 3 mm X 100 mm) and eluted with a gradient from 5 mM tributylamine, 5 mM acetic acid in 2% isopropanol, 5% methanol, 93% water (v/v) to 100% isopropanol over 15 minutes. All targets utilized negative mode with two distinct MRM pairs per metabolite. Only relative quantification was performed.
All signals were integrated using MultiQuant® Software 3.0.3. Signals with greater than 50 % missing values or a QC coefficient of variance of greater than 30 % were discarded. Remaining missing values were replaced with the lowest registered signal value. Signals were normalized using total sum normalization. Initial analysis was performed with multiple MRM signals included for each metabolite when possible. For display and pathway mapping a single MRM signal was used for each metabolite. Univariate t-tests were performed in MarkerView® Software 1.3.1. For all univariate analysis an unpaired t-test was used and a Benjamini-Hochberg correction with a false discovery of 10 % was utilized to correct for multiple comparisons.
22. Cytosolic mtDNA quantification
Cells were seeded at 2x106 in a 6-well plate and 24 hr later were treated with 100 ng/ml of LPS for 2 hr. Cells were collected in PBS and divided into two batches. Total DNA extract was prepared from one batch by suspension in 50 μM NaOH, boiling for 30 min and neutralizing with 50 μl 1M Tris-HCl ph 8. The other batch was suspended in 500 μl solubilization buffer (150 mM NaCl, 50 mM HEPES pH 7.4, and 25 μg/ml digitonin), incubated for 10 min and centrifuged at 980g for 3 min to pellet nuclei. The supernatant, containing the mtDNA, was centrifuged at 17,000g for 10 min to pellet intact mitochondria and any remain debris, and mtDNA was isolated from the supernatant using QIAQuick Nucleotide Removal Columns (Qiagen). Mitochondrial DNA was quantified using RT-PCR from 10 ng DNA and calculated as cytosolic mtDNA relative to whole cell mtDNA.
23. Statistical analysis
Data are presented as means ± standard deviation (± SD) or mean ± standard error mean (± SEM) for Seahorse assays, and are representative of at least two independent experiments. Statistical analysis was performed using GraphPad Prism 7. One-way or two-way ANOVA tests were used when multiple groups were analyzed as indicated in figure legends. Student’s t-test was used when two groups were compared.
24. Data visualization
Heatmaps were generated using Morpheus (software.broadinstitute.org/morpheus/) and MeV (mev.tm4.org) analysis and visualization and software.
Supplementary Material
Figure S1: siRNA delivery optimization in macrophages. (A) RAW264.7 G9 cells stably expressing RelA-GFP34 were transfected with a GFP siRNA or a non-targeting control. GFP intensity was measured by high-content imaging. (B) RAW264.7 cells and BMDM were infected with B. cenocepecia (MOI 10) for 18 hr. Secreted TNFα was measured by ELISA. (C) WT or Nme4−/− RAW264.7 cells were primed for 5 hr with 100 nM P3C then triggered by LPS electroporation for 3 hr and LDH release was measured. Data are representative of four (A) and three (B, C) independent experiments. (A) Welch’s two-tailed t-Test; (C) Two-Way ANOVA followed by Sidak’s multiple comparison test; ****p < 0.0001. (A) n=20, (B, C) n=3.
Figure S2: Enrichment of mitochondrial proteins among positive regulators of the non-canonical inflammasome. (A) The top 5th percentile of gene hits from the genome-wide screen were analyzed with MitoMiner data to determine mitochondrial enrichment. (D) Screen hit genes coding for mitochondrial proteins. Data plotted as gene name vs. mitochondrial protein index score generated by MinoMiner (see Methods).
Figure S3: Differential gene regulation by Nme4.
(A) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for the indicated times and Il1b mRNA was quantified by qPCR, and pro-IL-1β protein expression was analyzed by western blot 0, 4 and 8 hr after stimulation. Tubulin blot serves as a gel loading reference and quantification is plotted. (B, C) Transcriptional responses to TLR ligands LPS (100 ng/ml), P3C (1 μg/ml), R848 (5 μg/ml) in WT and Nme4−/− RAW264.7 cells at 0, 2, 4 and 8 hr after stimulation. Expression dynamics of (B) the early induced genes Dusp1 and Rcan1 (see Fig. 3F cluster 1) or (C) the sustained genes Il6 and Ccl3 (see Fig. 3F cluster 2). mRNA levels were assayed by Fluidigm microfluidic RT-PCR. (D) Caspase1 mRNA levels in WT and Nme4−/− RAW264.7 cells treated with 100 ng/ml LPS for the indicated times. (E) Protein levels of GSDMD in WT and Nme4−/− RAW264.7 cells treated with 100 ng/ml LPS for 0, 4, 6 and 18 hr. RhoGDI serves as a gel loading reference. (A-E) Data are representative of two independent experiments. (A, D) Two-Way ANOVA followed by Sidak’s multiple comparison test; ****p < 0.0001, n=3.
Figure S4: Normal mitochondrial basal function in Nme4−/− cells and 2-DG effect on glycolysis.
(A) WT and Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 1 hr and mitochondrial mass was measured using Mitotracker Green (250 nM) staining and fluorescence imaging by flow cytometry. (B) Basal mitochondrial membrane potential was measured in WT and Nme4−/− RAW264.7 cells by TMRM (200 nM) staining. (C) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 0, 5, 15, 30, 45, 60, 120 min and immunoblotted for TBK1 phosphorylation (p-TBK1). Tubulin serves as a gel loading reference and relative TBK1 phosphorylation was plotted. (D) WT RAW264.7 cells were injected with 100 ng/ml LPS and/or 10 mM 2-DG and real time ECAR levels were measured using a Seahorse machine. (A-D) Data are representative of three independent experiments; (B) Welch’s two-tailed t-Test; *p < 0.05. (B-D) n=4. Two-Way ANOVA followed by Sidak’s multiple comparison test; *p < 0.05, **p < 0.001, n=3.
Figure S5: ROS production is required for LPS-induced transcription, but not for TRAF6 recruitment to the mitochondria.
(A) Early transcription response (1, 2 hr) of WT RAW264.7 cells treated with 100 ng/ml LPS +/− NAC (10 mM) 30 min preincubation. (B) TRAF6 recruitment to the mitochondrial fraction in response to 15 min treatment of 100 ng/ml LPS +/− 30 min pre-incubation with DPI (10 μM). Relative TRAF6 recruitment to the mitochondria was plotted. (A, B) Data shown are representative of three independent experiments. (A, B) Two-Way ANOVA followed by Tukey’s multiple comparison test; * p < 0.05, ** p < 0.01, **** p < 0.0001. n=3.
Figure S6: ligand capacity of gene program stimulation in macrophages.
Gene expression induced by 100 ng/ml LPS, 5 nM IFNγ and 10 μM PGE2 at 1, 2, 4 hr in RAW264.7 cells (data replotted from Zhu et al.49).
Fig. S7: CRISPR/Cas9 Nme4−/− mouse generation and macrophage responses.
(A) Generation of CRISP/Cas9 Nme4−/− mice – a diagram and loci sequence showing the deleted region, and Nme4 expression analysis from isolated mitochondria of WT or Nme4−/− mice. (B) Primary BMDM from WT or Nme4−/− mice were primed with 1 μg/ml P3C for 6 hr and triggered by 100 nM LipidA transfection for 18 hr. Secreted IL-1α was measured by ELISA. (C-F) WT BMDM transfected with the specified siRNA were primed and triggered as in (B). IL-1α (C, E) and IL-1β (D, F) secretion were measured by ELISA. (G, H) Primary BMDM from WT or Nme4−/− mice were tested for Nme3 and Nme6 expression by qPCR. Data shown are pooled from (B) or representative of (C-H) three independent experiments and expressed as mean ± SD, (B) n=9, (C-H) n=3. (C-F) One-Way ANOVA followed by Dunnett’s multiple comparison test. *p < 0.05, **p < 0.01 ****p < 0.0001. (G-H) Welch’s two-tailed t-Test; **p < 0.01.
Acknowledgments
We thank Dr. Marie-Lise Lacombe from INSERM, Centre de Recherche Saint-Antoine, France, for the antibody against Nme4. We thank colleagues in the Laboratory of Immune System Biology for helpful discussions and critical reading of the manuscript. We thank Dr. Danielle A. Sliter (NCI) for protocol advice on mitochondrial DNA quantification, Dr. David Greenberg (UT Southwestern) for provision of B. cenocepecia strains and Dr. Lu Chen (NCATS) for assistance with data deposition to PubChem. F.S.S. was supported by the NIH grant R01AI118719. This work was generously supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Ethics declarations
The authors declare no competing interests.
Data availability
Data supporting the findings of this work are available within the paper, its supplementary information and source data files. The genome-wide screen data have been deposited to PubChem BioAssays repository (AID 1508600).
Bibliography
- 1.Akira S, Uematsu S & Takeuchi O Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Preston-Hurlburt P & Janeway CA A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Poltorak A et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Kayagaki N et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kayagaki N et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Hagar JA, Powell DA, Aachoui Y, Ernst RK & Miao EA Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franchi L, Eigenbrod T, Muñoz-Planillo R & Nuñez G The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol 10, 241–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder K & Tschopp J The inflammasomes. Cell 140, 821–832 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Latz E, Xiao TS & Stutz A Activation and regulation of the inflammasomes. Nat. Rev. Immunol 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik A & Kanneganti T-D Inflammasome activation and assembly at a glance. J. Cell Sci 130, 3955–3963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant C & Fitzgerald KA Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 19, 455–464 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Mariathasan S et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Shimada K et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen IC et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stutz A et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med 214, 1725–1736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung V et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol 9, 847–856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer SS et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou R, Yazdi AS, Menu P & Tschopp J A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Subramanian N, Natarajan K, Clatworthy MR, Wang Z & Germain RN The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Z et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinon F, Pétrilli V, Mayor A, Tardivel A & Tschopp J Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Lamkanfi M, Kanneganti T-D, Franchi L & Núñez G Caspase-1 inflammasomes in infection and inflammation. J. Leukoc. Biol 82, 220–225 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Liu X et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutterwala FS et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity 24, 317–327 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Elliott EI et al. Cutting edge: mitochondrial assembly of the NLRP3 inflammasome complex is initiated at priming. J. Immunol 200, 3047–3052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tannahill GM et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platnich JM et al. Shiga Toxin/Lipopolysaccharide Activates Caspase-4 and Gasdermin D to Trigger Mitochondrial Reactive Oxygen Species Upstream of the NLRP3 Inflammasome. Cell Rep. 25, 1525–1536.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Chung I-C et al. Mitochondrial oxidative phosphorylation complex regulates NLRP3 inflammasome activation and predicts patient survival in nasopharyngeal carcinoma. Mol. Cell Proteomics (2019). doi: 10.1074/mcp.RA119.001808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everts B et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol 15, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills EL et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baseler WA et al. Autocrine IL-10 functions as a rheostat for M1 macrophage glycolytic commitment by tuning nitric oxide production. Redox Biol 10, 12–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron AM et al. Inflammatory macrophage dependence on NAD+ salvage is a consequence of reactive oxygen species-mediated DNA damage. Nat. Immunol 20, 420–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robb EL et al. Control of mitochondrial superoxide production by reverse electron transport at complex I. J. Biol. Chem 293, 9869–9879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N et al. Development of a cell system for siRNA screening of pathogen responses in human and mouse macrophages. Sci. Rep 5, 9559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marine S, Bahl A, Ferrer M & Buehler E Common seed analysis to identify off-target effects in siRNA screens. J. Biomol. Screen 17, 370–378 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Chen C-W et al. Two separate functions of NME3 critical for cell survival underlie a neurodegenerative disorder. Proc. Natl. Acad. Sci. USA 116, 566–574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokarska-Schlattner M et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J. Biol. Chem 283, 26198–26207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boissan M, Schlattner U & Lacombe M-L The NDPK/NME superfamily: state of the art. Lab. Invest 98, 164–174 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Tsuiki H et al. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J. Cell Biochem 76, 254–269 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Epand RF, Schlattner U, Wallimann T, Lacombe M-L & Epand RM Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys. J 92, 126–137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlattner U et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J. Biol. Chem 288, 111–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kagan VE et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 23, 1140–1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milon L et al. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem 275, 14264–14272 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Garcia Fernandez M et al. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ 13, 449–455 (2002). [PubMed] [Google Scholar]
- 46.Zhan X et al. LPS-induced mitochondrial DNA synthesis and release facilitate RAD50-dependent acute lung injury. Signal Transduct. Target. Ther 6, 103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauterbach MA et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity 51, 997–1011.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Tan Y & Kagan JC Innate immune signaling organelles display natural and programmable signaling flexibility. Cell 177, 384–398.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palsson-McDermott EM et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West AP et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mills EL, Kelly B & O’Neill LAJ Mitochondria are the powerhouses of immunity. Nat. Immunol 18, 488–498 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Wardi J et al. 3-Aminobenzamide Prevents Concanavalin A-Induced Acute Hepatitis by an Anti-inflammatory and Anti-oxidative Mechanism. Dig. Dis. Sci 63, 3382–3397 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Zhu X et al. Dual ligand stimulation of RAW 264.7 cells uncovers feedback mechanisms that regulate TLR-mediated gene expression. J. Immunol 177, 4299–4310 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Victor VM, Rocha M & De la Fuente M N-acetylcysteine protects mice from lethal endotoxemia by regulating the redox state of immune cells. Free Radic. Res 37, 919–929 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Yang Z et al. PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation. J. Immunol 193, 6114–6123 (2014). [DOI] [PubMed] [Google Scholar]
- 56.de Mello RO et al. Effect of N-acetylcysteine and fructose-1,6-bisphosphate in the treatment of experimental sepsis. Inflammation 34, 539–550 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Liu L et al. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. USA 113, 1564–1569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang A et al. Specific sequences of infectious challenge lead to secondary hemophagocytic lymphohistiocytosis-like disease in mice. Proc. Natl. Acad. Sci. USA 116, 2200–2209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Z et al. Enhanced glycolytic metabolism contributes to cardiac dysfunction in polymicrobial sepsis. J. Infect. Dis 215, 1396–1406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Attwood PV & Muimo R The actions of NME1/NDPK-A and NME2/NDPK-B as protein kinases. Lab. Invest 98, 283–290 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Xue R et al. Metastasis suppressor NME1 promotes non-homologous end joining of DNA double-strand breaks. DNA Repair (Amst.) 77, 27–35 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Wang C-H et al. A shRNA functional screen reveals Nme6 and Nme7 are crucial for embryonic stem cell renewal. Stem Cells 30, 2199–2211 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Williams NC & O’Neill LAJ A role for the krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol 9, 141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haschemi A et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y et al. Bioenergetic state regulates innate inflammatory responses through the transcriptional co-repressor CtBP. Nat. Commun 8, 624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmström KM & Finkel T Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol 15, 411–421 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Han W et al. NADPH oxidase limits lipopolysaccharide-induced lung inflammation and injury in mice through reduction-oxidation regulation of NF-κB activity. J. Immunol 190, 4786–4794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schreck R, Rieber P & Baeuerle PA Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 10, 2247–2258 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C et al. HIF1α-dependent glycolysis promotes macrophage functional activities in protecting against bacterial and fungal infection. Sci. Rep 8, 3603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong Z et al. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 164, 896–910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patoli D et al. Inhibition of mitophagy drives macrophage activation and antibacterial defense during sepsis. J. Clin. Invest 130, 5858–5874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith AC & Robinson AJ MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 44, D1258–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller AH, Vayttaden SJ, Al-Khodor S & Fraser IDC Assay Development for Image-Based Quantification of Intracellular Bacterial Replication and Analysis of the Innate Immune Response to Infection. Assay Drug Dev Technol 13, 515–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al-Khodor S et al. Burkholderia cenocepacia J2315 escapes to the cytosol and actively subverts autophagy in human macrophages. Cell Microbiol. 16, 378–395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu CT et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol 15, 1197–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCloskey D, Gangoiti JA, Palsson BO & Feist AM A pH and solvent optimized reverse-phase ion-paring-LC–MS/MS method that leverages multiple scan-types for targeted absolute quantification of intracellular metabolites. Metabolomics 11, 1338–1350 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: siRNA delivery optimization in macrophages. (A) RAW264.7 G9 cells stably expressing RelA-GFP34 were transfected with a GFP siRNA or a non-targeting control. GFP intensity was measured by high-content imaging. (B) RAW264.7 cells and BMDM were infected with B. cenocepecia (MOI 10) for 18 hr. Secreted TNFα was measured by ELISA. (C) WT or Nme4−/− RAW264.7 cells were primed for 5 hr with 100 nM P3C then triggered by LPS electroporation for 3 hr and LDH release was measured. Data are representative of four (A) and three (B, C) independent experiments. (A) Welch’s two-tailed t-Test; (C) Two-Way ANOVA followed by Sidak’s multiple comparison test; ****p < 0.0001. (A) n=20, (B, C) n=3.
Figure S2: Enrichment of mitochondrial proteins among positive regulators of the non-canonical inflammasome. (A) The top 5th percentile of gene hits from the genome-wide screen were analyzed with MitoMiner data to determine mitochondrial enrichment. (D) Screen hit genes coding for mitochondrial proteins. Data plotted as gene name vs. mitochondrial protein index score generated by MinoMiner (see Methods).
Figure S3: Differential gene regulation by Nme4.
(A) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for the indicated times and Il1b mRNA was quantified by qPCR, and pro-IL-1β protein expression was analyzed by western blot 0, 4 and 8 hr after stimulation. Tubulin blot serves as a gel loading reference and quantification is plotted. (B, C) Transcriptional responses to TLR ligands LPS (100 ng/ml), P3C (1 μg/ml), R848 (5 μg/ml) in WT and Nme4−/− RAW264.7 cells at 0, 2, 4 and 8 hr after stimulation. Expression dynamics of (B) the early induced genes Dusp1 and Rcan1 (see Fig. 3F cluster 1) or (C) the sustained genes Il6 and Ccl3 (see Fig. 3F cluster 2). mRNA levels were assayed by Fluidigm microfluidic RT-PCR. (D) Caspase1 mRNA levels in WT and Nme4−/− RAW264.7 cells treated with 100 ng/ml LPS for the indicated times. (E) Protein levels of GSDMD in WT and Nme4−/− RAW264.7 cells treated with 100 ng/ml LPS for 0, 4, 6 and 18 hr. RhoGDI serves as a gel loading reference. (A-E) Data are representative of two independent experiments. (A, D) Two-Way ANOVA followed by Sidak’s multiple comparison test; ****p < 0.0001, n=3.
Figure S4: Normal mitochondrial basal function in Nme4−/− cells and 2-DG effect on glycolysis.
(A) WT and Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 1 hr and mitochondrial mass was measured using Mitotracker Green (250 nM) staining and fluorescence imaging by flow cytometry. (B) Basal mitochondrial membrane potential was measured in WT and Nme4−/− RAW264.7 cells by TMRM (200 nM) staining. (C) WT or Nme4−/− RAW264.7 cells were treated with 100 ng/ml LPS for 0, 5, 15, 30, 45, 60, 120 min and immunoblotted for TBK1 phosphorylation (p-TBK1). Tubulin serves as a gel loading reference and relative TBK1 phosphorylation was plotted. (D) WT RAW264.7 cells were injected with 100 ng/ml LPS and/or 10 mM 2-DG and real time ECAR levels were measured using a Seahorse machine. (A-D) Data are representative of three independent experiments; (B) Welch’s two-tailed t-Test; *p < 0.05. (B-D) n=4. Two-Way ANOVA followed by Sidak’s multiple comparison test; *p < 0.05, **p < 0.001, n=3.
Figure S5: ROS production is required for LPS-induced transcription, but not for TRAF6 recruitment to the mitochondria.
(A) Early transcription response (1, 2 hr) of WT RAW264.7 cells treated with 100 ng/ml LPS +/− NAC (10 mM) 30 min preincubation. (B) TRAF6 recruitment to the mitochondrial fraction in response to 15 min treatment of 100 ng/ml LPS +/− 30 min pre-incubation with DPI (10 μM). Relative TRAF6 recruitment to the mitochondria was plotted. (A, B) Data shown are representative of three independent experiments. (A, B) Two-Way ANOVA followed by Tukey’s multiple comparison test; * p < 0.05, ** p < 0.01, **** p < 0.0001. n=3.
Figure S6: ligand capacity of gene program stimulation in macrophages.
Gene expression induced by 100 ng/ml LPS, 5 nM IFNγ and 10 μM PGE2 at 1, 2, 4 hr in RAW264.7 cells (data replotted from Zhu et al.49).
Fig. S7: CRISPR/Cas9 Nme4−/− mouse generation and macrophage responses.
(A) Generation of CRISP/Cas9 Nme4−/− mice – a diagram and loci sequence showing the deleted region, and Nme4 expression analysis from isolated mitochondria of WT or Nme4−/− mice. (B) Primary BMDM from WT or Nme4−/− mice were primed with 1 μg/ml P3C for 6 hr and triggered by 100 nM LipidA transfection for 18 hr. Secreted IL-1α was measured by ELISA. (C-F) WT BMDM transfected with the specified siRNA were primed and triggered as in (B). IL-1α (C, E) and IL-1β (D, F) secretion were measured by ELISA. (G, H) Primary BMDM from WT or Nme4−/− mice were tested for Nme3 and Nme6 expression by qPCR. Data shown are pooled from (B) or representative of (C-H) three independent experiments and expressed as mean ± SD, (B) n=9, (C-H) n=3. (C-F) One-Way ANOVA followed by Dunnett’s multiple comparison test. *p < 0.05, **p < 0.01 ****p < 0.0001. (G-H) Welch’s two-tailed t-Test; **p < 0.01.
Data Availability Statement
Data supporting the findings of this work are available within the paper, its supplementary information and source data files. The genome-wide screen data have been deposited to PubChem BioAssays repository (AID 1508600).