Abstract
Recent studies have brought forward the critical role of emergent properties in shaping microbial communities and the ecosystems they are part of. Emergent properties - patterns or functions that cannot be deduced linearly from the properties of the constituent parts - underlie important ecological characteristics such as resilience, niche expansion, and spatial self-organisation. While it is clear that emergent properties are a consequence of interactions within the community, their non-linear nature makes mathematical modelling imperative for establishing the quantitative link between community structure and function. As the need for conservation and rational modulation of microbial ecosystems is increasingly apparent, so is the consideration of the benefits and limitations of the approaches to model emergent properties. Here we review ecosystem modelling approaches from the viewpoint of emergent properties. We consider the scope, advantages, and limitations of Lotka-Volterra, consumer-resource, trait-based, individual-based, and genome-scale metabolic models. Future efforts in this research area would benefit from capitalising on the complementarity between these approaches towards enabling rational modulation of complex microbial ecosystems.
Introduction
Microbial communities profoundly contribute to human and planetary health [1–5]. Yet, the quantitative principles underlying community composition and assembly, and the link between community diversity and function, remain largely unknown. Studying the emergent properties of microbial communities requires relating interactions across spatio-temporal scales, as well as their evolutionary dynamics. Unravelling, and ultimately predicting, community dynamics and emergent properties is a topical challenge in microbial ecology.
Higher-order units - from microbial consortia to entire ecosystems - feature macroscopic properties that emerge from microscopic interactions [6, 7]. Such emergent properties refer to any pattern or function that cannot be deduced as the sum of the properties of the constituent parts (Fig. 1). Examples in microbial systems include resilience to abiotic and biotic perturbations [8], stable co-existence [9, 10], and biochemical abilities [11–15]. Emergent properties typically arise when community members reach some threshold level of community size and connectivity [16–18]. Conditions such as high flow rates or high media viscosity can obstruct connectivity and hence limit the onset of emergent properties [19, 20]. Finding the threshold connectivity is often difficult, even in a defined community, due to the non-linear nature of emergence. The threshold is property-specific and may also (non-linearly) depend on environmental factors [21]. Hence, descriptions of emergent properties have been largely qualitative, although synthetic biology approaches are starting to characterise emergent properties quantitatively [14, 22].
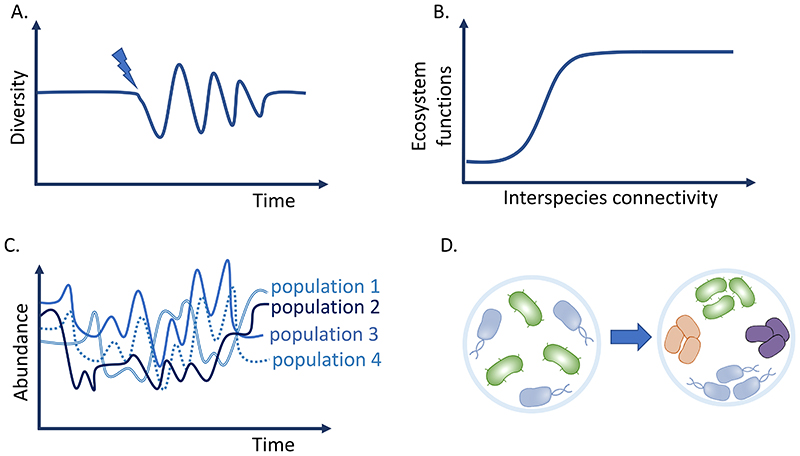
Figure 1. Examples of emergent properties arising from community complexity.
A. Stability and resilience. Interactions among community members can buffer against biotic and/or abiotic perturbations leading back to stable compositional state [82, 113, 136]. Blue arrow indicates a perturbation event. B. Phenotypes. Emergent functions such as substrate utilization, biomass production, cross-protection, result from the cooperation between community members. [104, 110, 139]. C. Persistence. Strains that would otherwise face competitive exclusion can coexist via, for instance, the intermediary of a cross-fed metabolite secreted by another community member [84, 88, 175]. D. Self-organisation. Balance between competitive and cooperative interactions can lead to spatial patterning, such as the formation of clonal patches [30, 47, 141].
Natural communities are difficult to study in situ due to their inherent dynamic nature and heterogeneity of habitats. Further, abiotic variables influencing the communities are difficult to measure or control in situ. Laboratory experiments, often considering interactions between 2 or 3 community members, are amenable to quantitative analysis but limited in capturing emergent properties of natural, more diverse microbial communities. Defined communities with larger membership can be constituted in vitro [23] yet remain limited to culturable microbes [24]. Mathematical models are therefore indispensable for linking community composition and connectivity to emergent functions. Models can bridge principles learned from simple laboratory systems to complex natural ecosystems; a continuity that is very difficult to achieve in an experimental setting.
Complex natural systems like microbial communities are never fully closed, and more than one model will be plausible for any data available from the system of interest [25, 26]. The choice of the model depends on the research question and the data available to estimate the parameters. When experimental data is limited, models can be set-up using first principles [27]. Such models can be used to probe system responses to perturbations that are inaccessible to experimentation or observations. In comparison to statistical analyses, models can help in generating mechanistic hypotheses and establishing causal relations. Examples include identification of bacteria conferring colonisation resistance against Clostridium difficile [28], and emergence of stability through competition [29, 30]. Nevertheless, models are often most useful in complementation with statistical patterns, e.g., a recent study using metabolic modelling uncovered polarisation between cooperation and competition in microbial communities [31].
Two general approaches have been commonly used to model community dynamics and emergent properties: ecological models with species or cells as basic units and interactions among them as the focus, and genome-scale metabolic models that have intra-cellular reactions as main units and nutrient generation/consumption as the focus. As the use of genome scale metabolic models has been reviewed previously [32–37], we focus on four commonly used ecological models. We provide an overview of their advantages in capturing emergent properties, discuss adaptations that challenge their limitations, and comment on their complementarity towards predictive modelling of complex ecosystems.
Ecological models
Metagenomics has enabled culture-free mapping the microbial diversity across a broad range of habitats. Statistical approaches have helped to reconstruct co-occurrence networks from which inter-species interactions are often inferred [38–41]. However, co-occurrence does not capture asymmetric relations, nor implies direct interactions [24, 41, 42]. Classical ecological models from macrofaunal ecosystems have therefore been adapted to microbial communities to unravel ecological mechanisms underlying emergent properties. These adaptations consider the following differences between macrofaunal/floral systems and microbial communities:
-
A)
Large community membership. With the number of individual cells in microbial communities often in the range of 1012, the use of concepts from statistical physics, such as central limit theorem, is justified [43].
-
B)
Short generation times. Microbial generation times are often in the range of minutes-hours, creating possibilities for easier generation of empirical data for model parameterisation and validation. Short generation times combined with rapid horizontal gene transfer, mean fast dynamics and evolution of interactions, which can alter the balance between processes such as migration and competitive exclusion. Conversely, short generation times also confound the separation of ecological from evolutionary dynamics and thus simulation intervals need to be accordingly considered.
-
C)
Trophic layer separation. Microbial communities not only exhibit biomass transfers between members via classic, predator-prey-like, trophic interactions, but also, and primarily, via secreted molecules through cross-feeding [44]. As many of the secreted common goods create one-to-many and reciprocal interactions, microbial communities cannot be structured according to the classic ecological notion of layered food webs.
A wide variety of mathematical models have been developed to address various ecological properties (Fig. 2): compositional stability [9, 10], response to abiotic perturbations or invasions [8, 28], environment-dependency of inter-species interactions [45, 46], spatio-temporal patterns of colonisation and succession [24, 47, 48], and the conflict between collective versus individual interest (i.e., the public goods problem, Black Queen Hypothesis) [49, 50]. Choosing the right model depends on the question(s), the assumptions made, and the amount of information known or feasibly measurable. We review the following models: Lotka-Volterra, MacArthur Consumer-Resource, Trait-based, Individual based, and genome-scale metabolic models (which can be regarded as metabolism-centred trait-based models). We note that boundaries between these model types are not always clear-cut, and adaptations to one model type can take it into the realm of another. For the sake of clarity, we structure this review in accordance with the common formulations of these modelling types.
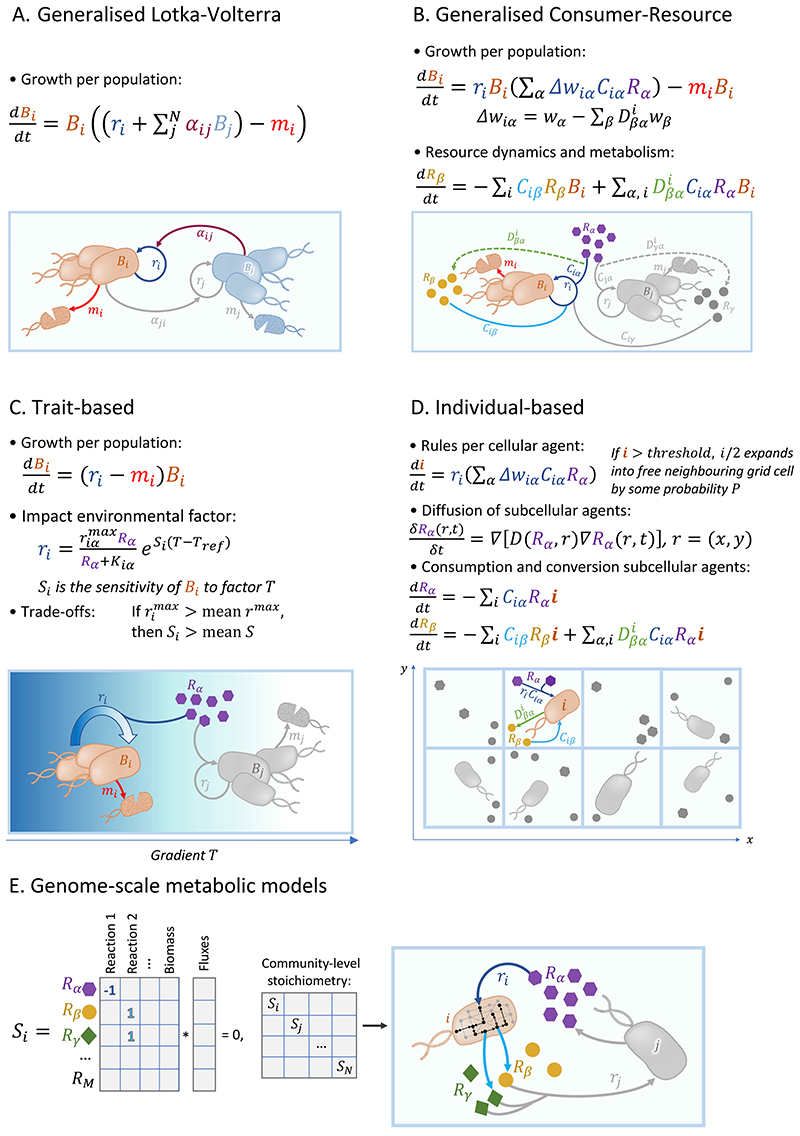
Figure 2. Model types commonly used for studying emergent properties of microbial communities.
Shown equations in A-D are examples, and could be varied to be more realistic by, for instance, making growth rates non-linearly dependent on resource availability (e.g., via Monod-like kinetics, as in panel C), or by introducing species-specific carrying capacities to make growth logistic. Parts of the equation that correspond to elements in the conceptual diagram are matched by colour, whereas grey elements are those not explicitly present in the shown equations. See Supplementary Table 1 for further details including notations used. A uniform background colour represents a homogenous (and closed) environment. In panels A & B, populations and/or resources are shown as groups for the purpose of illustration, but the model assumes spatiotemporally homogeneous distribution (effectively rendering the system like a well-stirred reactor). We further note that the boundaries between different model types are not rigid, and each model type can be extended beyond their typical application.
Lotka-Volterra Models
The Lotka-Volterra model is the most basic ecological model, originally used to describe macrofaunal predator-prey dynamics, such as those of between the Canadian lynx and snowshoe hare. This was among the first mathematical approaches to describe complex ecological dynamics [51–53]. The interspecies interaction coefficients in the model (Fig. 2 A) can be adjusted to reflect different types of interactions, such as competition between the moose and snowshoe hare [54]. The classic Lotka-Volterra model has been instrumental in studying two-species interactions and can be solved both numerically and analytically [53]. However, the classic model is limited in that it generates oscillations that are neutrally stable (i.e., do not have a characteristic amplitude or period) instead of true limit cycles. Small modifications in the model can thus lead to qualitatively distinct outcomes [55]. Further, the classic model does not capture interactions involving more than two species. Hence, of relevance to microbial communities is the generalised Lotka–Volterra model, which can accommodate any number of species.
The Lotka-Volterra model consists of a set of nonlinear, coupled, first-order differential equations. The growth rate of a species population is described as a function of its intrinsic growth rate and the linear effects exerted by the other populations. The Lotka-Volterra model requires relatively few parameters, intrinsic growth rates for each species (as a vector) and interaction coefficients for all species pairs (as a matrix). The advantage of having few parameters is that they can be inferred from (co-)culture data [56], steady state survivor data [57], or from correlations observed in metagenomic datasets [24, 42, 58–66]. The interaction parameters are easy to interpret (sign indicates either positive or negative effects of species on one another, and magnitude indicates effect strength). This simplicity and phenomenological nature makes Lotka-Volterra models easy to generalise [66]. In addition, equilibrium conditions and their stability can be identified analytically. Lotka-Volterra has therefore been widely used for connecting species interactions to community dynamics in various complex communities including the gut microbiome [24, 28, 30, 42, 58, 59, 63, 64, 67] and cheese fermentation communities [68].
The model’s simplicity also has its drawbacks. First, interactions are static in time and space, which contrasts with natural systems wherein changes in the abiotic environment or species densities can shift interactions from, e.g., mutualism to competition [69–72]. Second, positive feedback loops in the system can lead to unrestrained population growth [70]. Consequently, fitting data to a Lotka-Volterra model may underestimate the prevalence of mutualism. A third limitation is the assumption that community dynamics can be described only using pair-wise interactions, whereas the interaction between a pair can be altered by the presence of a third species (i.e., higher order interactions) [69, 73, 74]. However, there is a debate on the added value of including higher-order interactions in modelling [56, 75, 76]. Further, in its usual form, Lotka-Volterra does not account for stochastic processes such as genetic drift or environmental fluctuations. Lastly, small-molecule-mediated interactions such as metabolite exchange or quorum sensing are not explicitly modelled [77, 78], limiting the study of emergence of community-level biochemical properties [11]. This omission can also lead to qualitatively wrong conclusions [69].
Some of the limitations of Lotka-Volterra models can be addressed through more parameters. Unrestrained growth in a mutualistic scenario can be circumvented by replacing linear species interaction terms with non-linear saturating functions. The net sign of species interactions can be allowed to vary through time by, for example, adding a joint carrying capacity [24, 29, 70, 79]. Higher-order interactions can also be included [74, 80]. Each of these additions comes with the difficulty in meaningful interpretations of the parameters. Also, the number of interaction parameters scales quadratically with community membership. To account for higher-order interactions without significantly increasing the parameter space, pairwise interaction coefficients can be made to represent the net apparent effects of both higher-order and pairwise interactions [81].
The requirement for a priori assignment of interaction types has been addressed through combining linear regression with bootstrap aggregation to estimate the topology of the interaction network in the presence of measurement noise [59]. Similarly, the confidence intervals for inferred interaction coefficients can be estimated using stochastic fitting [66]. Environmental heterogeneity has been modelled as fluctuations in mortality rates [82], and by using time-varying interactions [64]. Another means to incorporate stochasticity is through adding a noise term to the equations [79, 83], or by randomly drawing interaction coefficients and/or intrinsic growth rates from predefined distributions [30, 56, 66, 84]. Indirect, metabolite-mediated, interactions can also be explicitly modelled as a quadratic species-metabolite interaction model [84]. This allows for the emergence of stable co-existence and diversity via cross-feeding, a phenomenon that is generally missing from Lotka-Volterra type models.
In sum, generalised Lotka-Volterra and its extensions are useful when a coarse-grained understanding of an ecosystem is needed or when measurements of species’ traits are not available. As for emergent properties, generalised Lotka-Volterra models and its adaptations are often used to model community diversity, stable coexistence, and multistability.
MacArthur Consumer-Resource Models
An alternative approach to Lotka-Volterra is resource-explicit model, where a population’s growth rate depends on chemical mediators such as nutrients, toxins, or signals, whose concentrations are coupled to population dynamics. Explicit modelling of metabolic by-products allows for the microbes as active modulators of their environment. In contrast to Lotka-Volterra, where the net species interactions are a priori assumptions, and researchers must grapple with what they mean mechanistically, resource-explicit models are defined “mechanistically” by how species consume and deplete resources, excrete by-products, and are hindered or helped by signals. Thus, species are not denoted as “competitors” or “mutualists” a priori, but these labels are outcomes of the model, and may be time- and environment-dependent.
The most basic resource-explicit model is the MacArthur Consumer-Resource model [85]. Like Lotka-Volterra, these models were initially used in macrofaunal ecology. MacArthurian consumer-resource dynamics mathematically captures the classical theory of competitive exclusion, or, the R* resource-ratio hypothesis, viz., a species needs to be the best at consuming a certain resource in order to persist [86]. The model allows to quantify how resource use differs between species: the bigger the resource overlap, the smaller the realm of co-existence. Each species’ growth rate is proportional to the resources consumed, weighted by an energy factor, minus a maintenance requirement that includes death. Species interactions are not parameterised directly but are mediated by the depletion of (shared) resources. The resources themselves are assumed not to interact with each other but can be introduced and consumed by arbitrarily complex dynamics. Additionally, MacArthurian consumer-resource equations allow for the use of different timescales for resources and consumers [87]. In the limit where the dynamics of resources are fast, the system reduces to Lotka-Volterra dynamics.
To make the consumer-resource model applicable to microbial communities, it is necessary to extend its scope much beyond two species and two resources. For this, parameters such as uptake rates can be randomly drawn from statistical distributions [88–92]. Analytical results can be derived using generalised versions of the model as well, such as the absence of multistability for linear functional responses [93], or species having identical stoichiometries of essential nutrients [94]. To include cross-feeding, the model can be extended with stoichiometric coefficients describing how the consumption of one compound relates to secretion of another. This method captures the emergent property of stable co-existence and diversity – even for a single externally supplied resource [88, 95]. In line with empirical observations, cross-feeding critically alters the model’s qualitative behaviour, increasing the potential for co-existence over competitive exclusion [70, 88, 96].
One limitation of the model is the assumption of fixed metabolism of community members. This can be partly overcome by including dynamic metabolic adaptation, allowing each species to temporally change its preferred nutrient. Emergent properties of community self-organisation and persistent co-existence and maintenance of diversity could be thus captured [51, 108]. Another limitation is the absence of taxonomic and hierarchical metabolic structures. This has been addressed through imposing correlation structures of the consumer preference and metabolic matrix [90] [95]. To tackle missing thermodynamic and biochemical detail, which is a common limitation to many ecological models, proteomic [97] and energetic [89-91, 98] constraints have been incorporated. This allowed elucidation of emergent energy flow topologies in the community, and distinct regimes of community structure.
The MacArthur Consumer-Resource model is deterministic in nature, which contrasts with the stochastic biotic and abiotic influences on community dynamics [99]. Such probabilistic events can be simulated within the framework of consumer-resource models [89, 90, 98]. Similarly, demographic noise can be incorporated [100], capturing the emergence of apparent neutrality, i.e., giving all community members the appearance of being ecologically equivalent. Adaptation of the model to include environmental fluctuations revealed a U-shaped relationship between community diversity and disturbance intensity, which a Lotka-Volterra model failed to capture [101]. To go beyond resources that positively contribute to growth, [71] adapted the model to include toxins, describing how resources and toxins jointly influence inter-species interactions. In line with results based on game theory [45], this adaptation demonstrated how coexistence and diversity are shaped by the hostility of the environment.
Explicitly coupling the dynamics of chemicals to that of populations enables capturing interactions mediated by more than one chemical, including those with opposing effects. For example, a species could be obtaining an essential resource from several community members. The effects of environmental change can be thus studied without the need to re-parameterise the system. The downside is that the system needs to be characterised in more detail: parameters for production and uptake rates for each chemical, or chemical reaction rates need to be estimated, which can be difficult in practice [69]. Additionally, chemical interactions must be posited a priori, a considerable challenge in complex or poorly characterised communities. When choosing statistical physics to overcome this problem, it is important to explicitly include thermodynamics in parameterisation to ensure that compliance with the second law of thermodynamics [89–91, 98].
Yet, classic ecological models lack biochemical detail and do not have the resolution to explain experimental results beyond those explicitly modelled, for example pH change [88]. The MacArthur consumer-resource model and its adaptations are generally suitable to model emergent properties arising via cross-feeding such as diversity and coexistence.
Trait-based Models
The family of phenomenological models, besides the classic Lotka-Volterra and consumer-resource models, includes trait-based models wherein the focal variables are phenotypic traits rather than phylogenetic groups. The community members are defined by their traits, and the models describe how trait combinations respond to and in turn influence environmental variables [46, 102, 103]. The interactions between members and environmental variables can be directly defined and mediated by ecological trade-offs [104–108], such as stress-tolerance versus combative dominance [109]. Whereas the classic ecological models typically assume a constant environment, trait-based models focus on community dynamics along major environmental gradients. The system dynamics created by interaction between various ‘guilds’ and their environment may lead to a functional trait distribution that optimises a certain community-level functional property, such as nitrification in soil systems [110, 111], organic matter decomposition [109, 112], or resilience to invasion [113]. Trait-based models can elucidate the emergence of selection pressures and diversity patterns along modelled environmental gradients - important aspects to understanding evolutionary processes [114, 115]. Trait-based models are not confined by a priori definitions of community structure and functional groups, but instead allow for the emergence of species and groups with novel trait combinations as simulated environmental conditions change [78, 114].
The trait combinations, often referred to as ‘genes’, can be informed by (meta)genomic data [115], by theoretical considerations [116], or randomly drawn from empirical distributions [117]. Random sampling of trait combinations can even be used to simulate metagenomes and metatranscriptomes through the replacement of unfit phenotypes This strategy demonstrated the emergent property of community structure/assembly and ocean biogeochemistry [118]. Trait-based models allow community members to not only respond to, but also influence, modelled environmental gradients [46, 119]. Further, trait-based models can be applied to larger microbial communities like those in ocean systems [115, 117, 118], which are too big to be modelled via individual-based methods, and too spatio-temporally heterogenous to be captured via classic ecological models. Since trait-based models simulate how system-scale – reaching to global-scale [110] – environmental changes influence community dynamics, applications have been largely restricted to where local variations in environmental variables are considered to be of negligible importance to overall emergent functionalities [120]. Thus, trait-based models have been applied to microbial communities in oceanic [104, 108, 115–118, 121, 122] or soil systems [109–112, 119]. For systems with high local variability such as the human gut, other approaches with higher resolution (e.g., individual-based models) may be more appropriate.
Trait-based models provide a bridge between community structure and function through the environment [123]. Empirical evidence has shown that microbial communities subjected to environmental change are more likely to exhibit shifts in functional trait distributions rather than in taxonomic distributions [124, 125]. A trait-based approach may hence be more useful than a strict taxon-based approach in light of environmental heterogeneity. This is in agreement with an in silico analysis based on a Consumer-Resource-based model [126]. Unlike the classic ecological models, however, a trait-based model’s main purpose is not to elucidate interaction dynamics among community members (defined in Lotka-Volterra by direct influence on fitness, or, in MacArthur consumer-resource, through the relationship with a shared resource) nor how these relate to coexistence. Rather, trait-based models primarily consider, besides predation in few cases [117], interactions between the community and environmental heterogeneity. Therefore, the influence of inter-member interactions, both direct, e.g., antibiotics-mediated antagonism, and indirect, e.g., cross-feeding, on fitness and community dynamics may be underestimated [120, 127].
Trait-based models do not require prior imposition of trait combinations. However, they do require defining which traits are included. This can pose a problem in systems such as the gut microbiota wherein both environmental gradients and associated fitness determinants are poorly known [110]. This limitation has been addressed by, for example, changing the modelling unit from species to enzymes of interest [128]. In addition, not all relevant environmental variables or interactions between environmental variables may be known, as is the case in poorly studied communities. This problem was tackled by modelling trait distributions along interaction effects between different factors [110]. A trait-based model might be further limited in understanding community ecology under novel environmental conditions without introduction of new traits. Trait diffusion through subsequent generations could be used to overcome this limitation [108]. However, neither emergent trait distribution nor randomised initialisation fully capture evolution via natural selection, which requires de novo mutations and associated traits arising during model simulation.
In sum, if local environmental variations can be neglected, trait-based models, through defining community members as trait combinations, are well suited to simulate adaptation, emergent phenotypic structure [102], and collective function [104] along environmental gradients.
Individual-based models
Increase in computational power has enabled the usage of individual-based, also called agent-based, models. Like classic ecological models, individual-based models have been first used to study macrofaunal communities such as bird flocks, but increasingly also microbial communities [129, 130]. The individual-based model is inherently stochastic [131]. It takes a bottom-up approach by modelling every community member as an individual interacting agent with each having any number of ‘rules’ of any complexity [132]. This contrasts with the top-down models that simulate adaptations at population level, and hence cannot capture (micro)evolutionary dynamics starting from individual mutations. While top-down models can simulate the fate of mutants, both mutants and their characteristics need to be predefined. Individual-based models can simulate the complete evolutionary process via inheritable mutations [133–136]. This allows capturing the emergence of community resilience and phenotypic complexity in a changing and/or heterogeneous environment. The rules given to each individual cell can either be empirically-informed, with recent advances in single-cell microbiology and flow cytometry allowing for realistic parameterisation of single-cell behaviours [137], or based on genome-scale metabolic models [8]. The inherent discrete nature of individual-based models allows capturing discrete events that cause a critical shift in cell activity, for example cell attachment/colonisation to a certain surface [149].
Rules assigned to each agent can be responsive to local environments, e.g. within a biofilm or the surface of a decomposing leaf [138]. Individual-based models can thereby elucidate properties otherwise ‘invisible’ to models assuming no spatial dimension, such as the emergence of spatial patterns [47, 139] or large clonal patches [30]. Spatially explicit collective behaviours, such as aggregation, optimise resource utilisation and enable otherwise thermodynamically unfavourable processes [45, 140, 141], or increase resilience [8]. The inclusion of explicit spatial structure places an intrinsic upper limit on community size via grid occupancy rules. In contrast, top-down models typically resort to a priori defined carrying capacities, either explicitly as maximum population sizes or implicitly via limiting resources.
Another benefit to individual-based models is the possibility to include thermodynamic constraints on community interactions and diversity. With limited exceptions [89, 90, 98], adaptations of classic ecological models tend not to explicitly include energetics/thermodynamic constraints. Further, complete accounting of thermodynamic constraints requires incorporation of pH and chemical speciation, which can be done with an individual-based model [141]. Other important forces constraining individual cell activity/dispersal and community size are of physical/mechanical nature, such as fluid drag force and shear force. These can be also explicitly included [132, 140, 141]. Moreover, inclusion of biophysical processes, such as cell shoving, can not only increase model accuracy but also reduce computation time through confining the solution space [142, 143].
In addition to capturing individual level heterogeneity, individual-based models are well suited for simulating stochastic processes such as genetic drift, horizontal gene transfer, and cell-cell interactions like type VI secretion stabbing [136, 140, 142–144]. While changes in and dispersal of biomass are modelled via discrete individual interactions, soluble substrates/chemical species typically need to be modelled as reaction-diffusion [77]. With small molecule diffusion generally being much faster relative to biomass growth, separation of time scale enhances computational efficiency. Therefore, many individual-based models assume diffusion to be in (quasi) steady state during changes in biomass [132, 136, 137, 141, 145–147]. Another approach is to describe a reaction-diffusion system in which both bacteria and resources spread through diffusion, calculated at identical timesteps, but with the diffusion coefficient for bacteria being smaller than that for the resource [148].
The main limitation to individual-based modelling is its complexity, making it not only computationally expensive but also difficult to assess for robustness. Reduced-dimensional descriptions are essential for robustness of conclusions, for instance through setting hydrodynamic limits, moment closure, and other approaches to aggregation [149, 150]. Another strategy is to limit the number of modelled agents [8, 30], or group cell types to reduce the community-wide variation in agent-specific ‘rules’ [47, 132, 144, 151]. Similarly, dividing the total cell population into cheaters and non-cheaters is a common approach in individual-based models investigating evolutionary game theory [45, 135, 152–155]. This strategy has demonstrated the emergence of a cooperative network [45, 155]. Simplifications of individual-based models still allow studying the behaviour of individuals after perturbation departing from the binary question of whether the system is stable.
Another limitation is a priori assumptions about direct interactions, which, if incorrect, can, due to emergent (and unconstrained) feedbacks between individual and population behaviour, escalate to outcomes incongruent with population-level data. Comparison of model types in predicting emergent properties is challenging due to variations in the mechanistic and biochemical details included as well as the number of modelled dimensions. Thus, the ability of individual-based models to identify potential local processes that lead to emergence of system-wide properties also varies.
In sum, individual-based models are generally suitable to identify spatially-explicit emergent properties such as self-organisation, resilience to local perturbations, and feedbacks between individual and population behaviour influencing the spread and fixation of a mutation. These models are also suitable for identifying system-wide emergent properties, such as community cooperation and coexistence [20] – however, the robustness and stability is hard to determine and can typically not be derived analytically.
Genome-scale metabolic models
Genome-scale metabolic models are mathematical representations of the near-complete set of biochemical reactions encoded in the genome of an organism. These models can be built in a fully or semi-automated manner from a genome assembly, and ideally also by using physiological data such as nutrient requirements [156]. They can be used to simulate the metabolic phenotype of an organism – including its growth rate, intra-cellular fluxes, and secretion/uptake rates − and its response to environmental and genetic perturbations. The most common simulation approach, flux balance analysis [157], identifies an optimal metabolic state within the bounds of mass balance and reaction directionality constraints. The commonly used criterion for optimality (objective function) in case of (single) microbes is maximization of growth rate [158]. Other relevant optimality criteria, such as maximum ATP production or minimal adjustment against a reference state, can also be used.
Genome-scale metabolic models have been applied to microbial communities in several ways. Flux balance analysis can be used to identify the range of feasible metabolic exchanges and community-scale metabolic properties. When simulating a community, choosing a biologically relevant objective function is often difficult and poses a major limitation to the approach. Yet, the foundation in first principles means that these metabolic models can identify the phenotypic range within which a community must operate. This advantage can be used, for example, to identify candidate cross-fed metabolites [159], and to assess the extent of metabolic cooperation and competition [49]. Additionally, flux balance analysis can be used to predict compositional and metabolite dynamics by solving a series of steady-state problems in discrete time-steps, while the uptake / secretion of metabolites is modelled as differential equations [123]. Multiple species or genotypes can be modelled simultaneously allowing for quantitative prediction of community dynamics resulting from intracellular metabolic events [114, 124]. By further combining dynamic flux balance analysis with metabolite diffusion equations, spatial effects can also be captured as in biofilms [160, 161]. Modelling frameworks employing such a strategy, such as COMETS [160] and CROMICS [162], can thus allow investigating both spatial and temporal dynamics.
In the context of ecological models, genome-scale metabolic models are akin to trait-based models. Yet, there are several distinctions: a) a direct link to the (meta-)genome and hence the ability to simulate specific genetic changes, b) scale (typically hundreds to thousands of reactions and metabolites per species), and c) possibility to include additional molecular mechanistic details such as proteome allocation and thermodynamic constraints. These distinguishing characteristics, together with advances in automated model reconstruction [27, 163, 164], have accelerated their use in modelling microbial communities. These models have provided testable hypotheses on (nutritional) habitat range, ecosystem productivity, and cross-fed metabolites [13, 159, 165]. They have also be used to simulate community dynamics in response to altered nutrient environment or addition/removal of species [49].
One of the main limitations of genome-scale metabolic models is the absence of negative interactions beyond resource competition, such as those mediated by toxins and other antimicrobials that are prevalent in ecosystems [30, 49, 166]. Further, these models often require extensive curation to achieve high accuracy, especially for microbes that are phylogenetically distant from model organisms like Escherichia coli. This problem can get further amplified when modelling at the single cell level since the generalisation can become difficult due to model complexity [8, 162, 167, 168]. Nevertheless, due to their amenability for automated reconstruction and integrating (meta-)omics data [13], there is exciting potential for merging genome-scale metabolic models with ecological model approaches to capture emergent properties.
Parameter inference and data integration
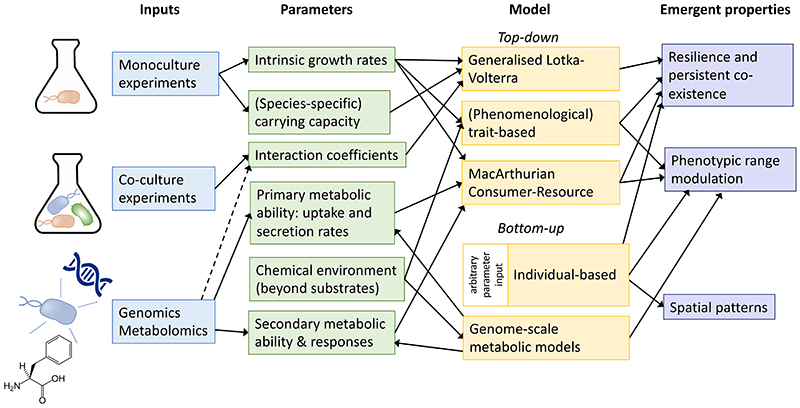
As the quality of a model depends on the accuracy and uncertainty in parameter estimates, choice of the model type and complexity is best guided by data availability (Fig. 3). The parameters required can be categorised into member-level parameters, such as carrying capacity, and interaction parameters such as cross-feeding rates. The parameters should ideally be obtained in the community-context. This remains challenging due to experimental difficulties in probing the combinatorial parameter space. Most models thereby resort to parameters obtained from monocultures or small-scale community studies. Developments in omics methods and high-throughput culturing and interaction mapping [159, 169] are helping to fill the gap. Metagenomic analyses can suggest which members of community may interact, e.g., as statistical covariation [49, 170]. Metagenomics can also help estimating in situ growth rates using read coverage over the origin of replication [171]. Meta-transcriptomics and metabolomics can be used to constrain the model structure, for example by selecting interactions consistent with the data [159, 172].
Figure 3. Choosing a suitable modelling approach.
The flow diagram indicates the model choice trajectory, starting with the available input parameters till the emergent properties that can be potentially modelled. The dotted line marks the potential for inferring interaction coefficients from omics data (e.g., co-occurrence networks predicted by genome-scale metabolic models), albeit without being indicative of direct and density-dependent interactions. Top-down models are those that consider communities in terms of population-averages, whereas bottom-up models simulate communities at individual level. The shown model types and their links to parameters and emergent properties are based on their common forms and applications. The model types can be adapted to be informed by additional parameters and be made suitable to model other emergent properties than indicated. For instance, genome-scale metabolic models can be used to study spatial patterns [160].
Discussion
Since microbial community dynamics and properties can be attributed to (interaction of) various complex networks, such as gene regulatory, metabolic, cell-environment and cell-cell, identifying which of these should be included to accurately predict the emergent property of interest is key in choosing an appropriate modelling approach (Fig. 3, Supplementary Table 2). Depending on the temporal and spatial characteristics of the property of interest, a combination of approaches is advantageous. Each approach has advantages and limitations in terms of integrating omics data, handling missing data, generating analytical solutions, predictive power, scope of applications, and amenability for generating hypotheses that can be experimentally tested. The choice of model(s) also depends on whether a general phenomenological understanding is sufficient, or if the goal is to fit data to generate predictions in a specific system. While the top-down population-based approaches (i.e., Lotka-Volterra, MacArthurian consumer-resource and phenomenological trait-based models) are generally more suited for modelling resilience and co-existence, the bottom-up approaches (i.e., individual-based and genome-scale metabolic models) are typically better suited for modelling phenotypic trait ranges and spatial patterns. Each of these approaches can be adjusted in the number of dimensions included and units modelled.
The distinction between the modelling approaches detailed in this review is not necessarily categorical. For example, adaptations of a consumer-resource model can make it fall within the category of individual-based models, whereas an individual-based model with few dimensions and low resolution can be less accurate in predicting the emergent phenotypic property of substrate utilisation than a multi-population consumer-resource model. As ecosystem stability and resilience become a pressing issue on health and environmental fronts [3, 173, 174], the need for developing predictive models cannot be overstated. With computational power becoming less limiting, and molecular and single-cell data becoming increasingly available, we envisage that fusion between different ecological and cellular-level models will, in the coming years, enable predictive modelling of emergent properties in complex ecosystems from the molecular to the ecosystem scale.
Supplementary Material
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement no. 866028), and from the UK Medical Research Council (project number MC_UU_00025/11). SM would like to thank the Swiss National Science Foundation for funding the NCCR Microbiomes and an Eccellenza project, and the ERC for Starting grant no. 715097. WH received funding from NIH through R01-GM121498, SS received funding from the Portuguese Foundation for Science and Technology (FCT) under the scope of a Ph.D grant (SFRH/BD/121695/2016) and the strategic funding of UIDB/04469/2020 unit.
Footnotes
Author contributions
NIvdB conceived the review concept, carried the literature survey, and wrote the manuscript. DM, SS, and IR contributed to the section on genome-scale metabolic models. JC, WH and SM helped with the literature survey and contributed to the manuscript structuring and writing. KRP conceived the review concept, helped with the literature survey, and wrote the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Cho I, Blaser MJ. APPLICATIONS OF NEXT-GENERATION SEQUENCING The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi ZM. Gut Microbiota: An Important Link between Western Diet and Chronic Diseases. Nutrients. 2019;11(10) doi: 10.3390/nu11102287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. New England Journal of Medicine. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 4.Glowacki RWP, Martens EC. In sickness and health: Effects of gut microbial metabolites on human physiology. PLoS Pathog. 2020;16(4):e1008370. doi: 10.1371/journal.ppat.1008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazaries L, et al. Evidence of Microbial Regulation of Biogeochemical Cycles from a Study on Methane Flux and Land Use Change. Applied and Environmental Microbiology. 2013;79(13):4031–4040. doi: 10.1128/AEM.00095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konopka A. What is microbial community ecology? Isme Journal. 2009;3(11):1223–1230. doi: 10.1038/ismej.2009.88. [DOI] [PubMed] [Google Scholar]
- 7.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–75. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 8.Bauer E, et al. BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. Plos Computational Biology. 2017;13(5) doi: 10.1371/journal.pcbi.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Hoek MJA, Merks RMH. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. Bmc Systems Biology. 2017;11 doi: 10.1186/s12918-017-0430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorter FA, Manhart M, Ackermann M. Understanding the evolution of interspecies interactions in microbial communities. Philosophical Transactions of the Royal Society B-Biological Sciences. 2020;375(1798) doi: 10.1098/rstb.2019.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Yoshinaga M, Rosen BP. The antibiotic action of methylarsenite is an emergent property of microbial communities. Molecular Microbiology. 2019;111(2):487–494. doi: 10.1111/mmi.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinidis D, et al. Adaptive laboratory evolution of microbial co-cultures for improved metabolite secretion. Mol Syst Biol. 2021;17(8):e10189. doi: 10.15252/msb.202010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H, et al. Artificial consortium demonstrates emergent properties of enhanced cellulosic-sugar degradation and biofuel synthesis. Npj Biofilms and Microbiomes. 2020;6(1) doi: 10.1038/s41522-020-00170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzman JA, et al. Bacterial growth in multicellular aggregates leads to the emergence of complex lifecycles. 2021 doi: 10.1016/j.cub.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levins R, Lewontin R. The dialectical biologist. Harvard University Press; 1985. [Google Scholar]
- 17.Diaz PI, Valm AM. Microbial Interactions in Oral Communities Mediate Emergent Biofilm Properties. J Dent Res. 2020;99(1):18–25. doi: 10.1177/0022034519880157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buerger AN, et al. Gastrointestinal dysbiosis following diethylhexyl phthalate exposure in zebrafish (Danio rerio): Altered microbial diversity, functionality, and network connectivity. Environ Pollut. 2020;265(Pt B):114496. doi: 10.1016/j.envpol.2020.114496. [DOI] [PubMed] [Google Scholar]
- 19.Kim MK, et al. Local and global consequences of flow on bacterial quorum sensing. Nat Microbiol. 2016;1:15005. doi: 10.1038/nmicrobiol.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebrahimi A, Or D. Hydration and diffusion processes shape microbial community organization and function in model soil aggregates. Water Resources Research. 2015;51(12):9804–9827. [Google Scholar]
- 21.Falconer RE, et al. Microscale heterogeneity explains experimental variability and non-linearity in soil organic matter mineralisation. PLoS One. 2015;10(5):e0123774. doi: 10.1371/journal.pone.0123774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fredrickson JK. ECOLOGY. Ecological communities by design. Science. 2015;348(6242):1425–7. doi: 10.1126/science.aab0946. [DOI] [PubMed] [Google Scholar]
- 23.Singer E, et al. Next generation sequencing data of a defined microbial mock community. Sci Data. 2016;3:160081. doi: 10.1038/sdata.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marino S, et al. Mathematical modeling of primary succession of murine intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):439–444. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cariboni J, et al. The role of sensitivity analysis in ecological modelling. Ecological modelling. 2007;203(1–2):167–182. [Google Scholar]
- 26.Oreskes N, Shrader-Frechette K, Belitz K. Verification, validation, and confirmation of numerical models in the earth sciences. Science. 1994;263(5147):641–646. doi: 10.1126/science.263.5147.641. [DOI] [PubMed] [Google Scholar]
- 27.Machado D, et al. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Research. 2018;46(15):7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–U207. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammarlund SP, Chacon JM, Harcombe WR. A shared limiting resource leads to competitive exclusion in a cross-feeding system. Environ Microbiol. 2019;21(2):759–771. doi: 10.1111/1462-2920.14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350(6261):663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 31.Machado D, et al. Polarization of microbial communities between competitive and cooperative metabolism. Nature Ecology & Evolution. 2021;5(2):195. doi: 10.1038/s41559-020-01353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu C, et al. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20(1):121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien EJ, Monk JM, Palsson BO. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161(5):971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang X, Lloyd CJ, Palsson BO. Reconstructing organisms in silico: genome-scale models and their emerging applications. Nature Reviews Microbiology. 2020;18(12):731–743. doi: 10.1038/s41579-020-00440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colarusso AV, et al. Computational modeling of metabolism in microbial communities on a genome-scale. Current Opinion in Systems Biology. 2021 [Google Scholar]
- 36.Garcia-Jimenez B, Torres-Bacete J, Nogales J. Metabolic modelling approaches for describing and engineering microbial communities. Comput Struct Biotechnol J. 2021;19:226–246. doi: 10.1016/j.csbj.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frioux C, et al. From bag-of-genes to bag-of-genomes: metabolic modelling of communities in the era of metagenome-assembled genomes. Computational and Structural Biotechnology Journal. 2020;18:1722–1734. doi: 10.1016/j.csbj.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaffron S, et al. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20(7):947–59. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faust K, Raes J. Microbial interactions: from networks to models. Nature Reviews Microbiology. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 40.Li JB, et al. Distinct mechanisms shape soil bacterial and fungal co-occurrence networks in a mountain ecosystem. Fems Microbiology Ecology. 2020;96(4) doi: 10.1093/femsec/fiaa030. [DOI] [PubMed] [Google Scholar]
- 41.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein RR, et al. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. Plos Computational Biology. 2013;9(12) doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbier M, et al. Generic assembly patterns in complex ecological communities. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(9):2156–2161. doi: 10.1073/pnas.1710352115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gralka M, et al. Trophic Interactions and the Drivers of Microbial Community Assembly. Current Biology. 2020;30(19):R1176–R1188. doi: 10.1016/j.cub.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Madeo D, Comolli LR, Mocenni C. Emergence of microbial networks as response to hostile environments. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratzke C, Gore J. Modifying and reacting to the environmental pH can drive bacterial interactions. Plos Biology. 2018;16(3) doi: 10.1371/journal.pbio.2004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Allison SD. Emergent properties of organic matter decomposition by soil enzymes. Soil Biology & Biochemistry. 2019;136 [Google Scholar]
- 48.Walsh AM, et al. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems. 2016;1(5) doi: 10.1128/mSystems.00052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machado D, et al. Polarization of microbial communities between competitive and cooperative metabolism. Nature ecology & evolution. 2021;5(2):195–203. doi: 10.1038/s41559-020-01353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci U S A. 2014;111(50):17941–6. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leigh ER. The ecological role of Volterra’s equations. Some mathematical problems in biology. 1968 [Google Scholar]
- 52.Nedorezov L. The dynamics of the lynx-hare system: an application of the Lotka-Volterra model. Biophysics. 2016;61(1):149–154. [Google Scholar]
- 53.Muhlbauer LK, et al. gauseR: Simple methods for fitting Lotka-Volterra models describing Gause’s “Struggle for Existence”. Ecology and Evolution. 2020;10(23):13275–13283. doi: 10.1002/ece3.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belovsky GE. Moose and Snowshoe Hare Competition and a Mechanistic Explanation from Foraging Theory. Oecologia. 1984;61(2):150–159. doi: 10.1007/BF00396753. [DOI] [PubMed] [Google Scholar]
- 55.May RM. Limit Cycles in Predator-Prey Communities. Science. 1972;177(4052):900. doi: 10.1126/science.177.4052.900. [DOI] [PubMed] [Google Scholar]
- 56.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1(5):109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 57.Voit E, Davis J, Olivenca D. Inference and validation of the structure of Lotka-Volterra models. 2021 [Google Scholar]
- 58.Bucci V, Xavier JB. Towards Predictive Models of the Human Gut Microbiome. Journal of Molecular Biology. 2014;426(23):3907–3916. doi: 10.1016/j.jmb.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisher CK, Mehta P. Identifying Keystone Species in the Human Gut Microbiome from Metagenomic Timeseries Using Sparse Linear Regression. Plos One. 2014;9(7) doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bucci V, et al. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biology. 2016;17 doi: 10.1186/s13059-016-0980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao XF, et al. Inference of Significant Microbial Interactions From Longitudinal Metagenomics Data. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li CH, et al. An expectation-maximization algorithm enables accurate ecological modeling using longitudinal microbiome sequencing data. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph TA, et al. Compositional Lotka-Volterra describes microbial dynamics in the simplex. PLoS Comput Biol. 2020;16(5):e1007917. doi: 10.1371/journal.pcbi.1007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hosoda S, Fukunaga T, Hamada M. Umibato: estimation of time-varying microbial interaction using continuous-time regression hidden Markov model. bioRxiv. 2021 doi: 10.1093/bioinformatics/btab287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Remien CH, Eckwright MJ, Ridenhour BJ. Structural identifiability of the generalized Lotka-Volterra model for microbiome studies. Royal Society Open Science. 2021;8(7) doi: 10.1098/rsos.201378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White JR. Novel methods for metagenomic analysis. University of Maryland; College Park: Ann Arbor: 2010. p. 160. [Google Scholar]
- 67.Sousa A, et al. Evolution of commensal bacteria in the intestinal tract of mice. Current Opinion in Microbiology. 2017;38:114–121. doi: 10.1016/j.mib.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Mounier J, et al. Microbial interactions within a cheese microbial community. Applied and Environmental Microbiology. 2008;74(1):172–181. doi: 10.1128/AEM.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Momeni B, Xie L, Shou WY. Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. Elife. 2017;6 doi: 10.7554/eLife.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoek TA, et al. Resource Availability Modulates the Cooperative and Competitive Nature of a Microbial Cross-Feeding Mutualism. Plos Biology. 2016;14(8) doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piccardi P, Vessman B, Mitri S. Toxicity drives facilitation between 4 bacterial species. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(32):15979–15984. doi: 10.1073/pnas.1906172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mai TSN. Ecology and Evolution. University of Groningen; 2021. Impact of metabolic plasticity on microbial community diversity and stability. [Google Scholar]
- 73.Sanchez-Gorostiaga A, et al. High-order interactions distort the functional landscape of microbial consortia. Plos Biology. 2019;17(12) doi: 10.1371/journal.pbio.3000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mickalide H, Kuehn S. Higher-Order Interaction between Species Inhibits Bacterial Invasion of a Phototroph-Predator Microbial Community. Cell Systems. 2019;9(6):521. doi: 10.1016/j.cels.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Guo XK, Boedicker JQ. The Contribution of High-Order Metabolic Interactions to the Global Activity of a Four-Species Microbial Community. Plos Computational Biology. 2016;12(9) doi: 10.1371/journal.pcbi.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meroz N, et al. Community composition of microbial microcosms follows simple assembly rules at evolutionary timescales. Nature Communications. 2021;12(1) doi: 10.1038/s41467-021-23247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zomorrodi AR, Segre D. Synthetic Ecology of Microbes: Mathematical Models and Applications. Journal of Molecular Biology. 2016;428(5):837–861. doi: 10.1016/j.jmb.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song HS, et al. Mathematical Modeling of Microbial Community Dynamics: A Methodological Review (vol 2, pg 711, 2014) Processes. 2015;3(3):699. [Google Scholar]
- 79.Descheemaeker L, Grilli J, de Buyl S. Heavy-tailed abundance distributions from stochastic Lotka-Volterra models. bioRxiv. 2021 doi: 10.1103/PhysRevE.104.034404. [DOI] [PubMed] [Google Scholar]
- 80.Bairey E, Kelsic ED, Kishony R. High-order species interactions shape ecosystem diversity. Nature Communications. 2016;7 doi: 10.1038/ncomms12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji B, Herrgård MJ, Nielsen J. Microbial community dynamics revisited. Nature Computational Science. 2021;1(10):640–641. doi: 10.1038/s43588-021-00144-6. [DOI] [PubMed] [Google Scholar]
- 82.Woltz VLA, et al. Emergence of alternative stable states in microbial communities in a fluctuating environment. bioRxiv. 2019:678367 [Google Scholar]
- 83.Xu L, et al. Stochastic Generalized Lotka-Volterra Model with An Application to Learning Microbial Community Structures. arXiv preprint. 2020:arXiv:2009.10922 [Google Scholar]
- 84.Brunner JD, Chia N. Metabolite-mediated modelling of microbial community dynamics captures emergent behaviour more effectively than species-species modelling. Journal of the Royal Society Interface. 2019;16(159) doi: 10.1098/rsif.2019.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacArthur R. Species packing and competitive equilibrium for many species. Theor Popul Biol. 1970;1(1):1–11. doi: 10.1016/0040-5809(70)90039-0. [DOI] [PubMed] [Google Scholar]
- 86.Tilman D. Resource competition and community structure. Monogr Popul Biol. 1982;17:1–296. [PubMed] [Google Scholar]
- 87.Chesson P. MacArthur’s consumer-resource model. Theoretical Population Biology. 1990;37(1):26–38. [Google Scholar]
- 88.Goldford JE, et al. Emergent simplicity in microbial community assembly. Science. 2018;361(6401):469–474. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marsland R, et al. Available energy fluxes drive a transition in the diversity, stability, and functional structure of microbial communities. Plos Computational Biology. 2019;15(2) doi: 10.1371/journal.pcbi.1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marsland R, Cui WP, Mehta P. A minimal model for microbial biodiversity can reproduce experimentally observed ecological patterns. Scientific Reports. 2020;10(1) doi: 10.1038/s41598-020-60130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Estrela S, et al. Nutrient dominance governs the assembly of microbial communities in mixed nutrient environments. Elife. 2021;10 doi: 10.7554/eLife.65948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cui WP, Marsland R, Mehta P. Diverse communities behave like typical random ecosystems. Physical Review E. 2021;104(3) doi: 10.1103/PhysRevE.104.034416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haygood R. Coexistence in MacArthur-style consumer-resource models. Theoretical Population Biology. 2002;61(2):215–223. doi: 10.1006/tpbi.2001.1566. [DOI] [PubMed] [Google Scholar]
- 94.Dubinkina V, et al. Multistability and regime shifts in microbial communities explained by competition for essential nutrients. Elife. 2019;8 doi: 10.7554/eLife.49720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pacheco AR, Osborne ML, Segre D. Non-additive microbial community responses to environmental complexity. Nature Communications. 2021;12(1) doi: 10.1038/s41467-021-22426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zelezniak A, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities (vol 112, pg 6449, 2015) Proceedings of the National Academy of Sciences of the United States of America. 2015;112(51):E7156. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pacciani-Mori L, et al. Constrained proteome allocation affects coexistence in models of competitive microbial communities. Isme Journal. 2021;15(5):1458–1477. doi: 10.1038/s41396-020-00863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marsland R, et al. The Community Simulator: A Python package for microbial ecology. Plos One. 2020;15(3) doi: 10.1371/journal.pone.0230430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Obadia B, et al. Probabilistic Invasion Underlies Natural Gut Microbiome Stability. Curr Biol. 2017;27(13):1999–2006.:e8. doi: 10.1016/j.cub.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D’Andrea R, Gibbs T, O’Dwyer JP. Emergent neutrality in consumer-resource dynamics. Plos Computational Biology. 2020;16(7) doi: 10.1371/journal.pcbi.1008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mancuso CP, et al. Environmental fluctuations reshape an unexpected diversity-disturbance relationship in a microbial community. Elife. 2021;10 doi: 10.7554/eLife.67175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lajoie G, Kembel SW. Making the Most of Trait-Based Approaches for Microbial Ecology. Trends in Microbiology. 2019;27(10):814–823. doi: 10.1016/j.tim.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 103.Zakharova L, Meyer KM, Seifan M. Trait-based modelling in ecology: A review of two decades of research. Ecological Modelling. 2019;407 [Google Scholar]
- 104.Grigoratou M, et al. A trait-based modelling approach to planktonic foraminifera ecology. Biogeosciences. 2019;16(7):1469–1492. [Google Scholar]
- 105.Muscarella ME, Howey XM, Lennon JT. Trait-based approach to bacterial growth efficiency. Environmental Microbiology. 2020;22(8):3494–3504. doi: 10.1111/1462-2920.15120. [DOI] [PubMed] [Google Scholar]
- 106.Shao PS, et al. Tradeoffs among microbial life history strategies influence the fate of microbial residues in subtropical forest soils. Soil Biology & Biochemistry. 2021;153 [Google Scholar]
- 107.Malik AA, et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. Isme Journal. 2020;14(1):1–9. doi: 10.1038/s41396-019-0510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Merico A, et al. Sustaining diversity in trait-based models of phytoplankton communities. Frontiers in Ecology and Evolution. 2014;2 [Google Scholar]
- 109.Crowther TW, et al. Untangling the fungal niche: the trait-based approach. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Le Roux X, et al. Predicting the Responses of Soil Nitrite-Oxidizers to Multi-Factorial Global Change: A Trait-Based Approach. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bouskill NJ, et al. Trait-based representation of biological nitrification: model development, testing, and predicted community composition. Front Microbiol. 2012;3:364. doi: 10.3389/fmicb.2012.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kyker-Snowman E, et al. Stoichiometrically coupled carbon and nitrogen cycling in the MIcrobial-MIneral Carbon Stabilization model version 1.0 (MIMICS-CN v1.0) Geoscientific Model Development. 2020;13(9):4413–4434. [Google Scholar]
- 113.Kruk C, et al. A trait-based approach predicting community assembly and dominance of microbial invasive species. Oikos. 2021;130(4):571–586. [Google Scholar]
- 114.Litchman E, Ohman MD, Kiorboe T. Trait-based approaches to zooplankton communities. Journal of Plankton Research. 2013;35(3):473–484. [Google Scholar]
- 115.Garcia CA, et al. Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry. Philosophical Transactions of the Royal Society B-Biological Sciences. 2020;375(1798) doi: 10.1098/rstb.2019.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moreno AR, et al. Marine phytoplankton stoichiometry mediates nonlinear interactions between nutrient supply, temperature, and atmospheric CO2. Biogeosciences. 2018;15(9):2761–2779. [Google Scholar]
- 117.Follows MJ, et al. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315(5820):1843–1846. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]
- 118.Coles VJ, et al. Ocean biogeochemistry modeled with emergent trait-based genomics. Science. 2017;358(6367):1149–1154. doi: 10.1126/science.aan5712. [DOI] [PubMed] [Google Scholar]
- 119.Ratzke C, Barrere J, Gore J. Strength of species interactions determines biodiversity and stability in microbial communities. Nature Ecology & Evolution. 2020;4(3):376. doi: 10.1038/s41559-020-1099-4. [DOI] [PubMed] [Google Scholar]
- 120.Bradford MA, et al. Quantifying microbial control of soil organic matter dynamics at macrosystem scales. Biogeochemistry. 2021;156(1):19–40. [Google Scholar]
- 121.Ward BA, et al. Iron, phosphorus, and nitrogen supply ratios define the biogeography of nitrogen fixation. Limnology and Oceanography. 2013;58(6):2059–2075. [Google Scholar]
- 122.Zwart JA, Solomon CT, Jones SE. Phytoplankton traits predict ecosystem function in a global set of lakes. Ecology. 2015;96(8):2257–2264. doi: 10.1890/14-2102.1. [DOI] [PubMed] [Google Scholar]
- 123.Nemergut DR, Shade A, Violle C. When, where and how does microbial community composition matter? Frontiers in microbiology. 2014;5:497. doi: 10.3389/fmicb.2014.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Severin I, Ostman O, Lindstrom ES. Variable Effects of Dispersal on Productivity of Bacterial Communities Due to Changes in Functional Trait Composition. Plos One. 2013;8(12) doi: 10.1371/journal.pone.0080825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Staley C, et al. Core functional traits of bacterial communities in the Upper Mississippi River show limited variation in response to land cover. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Worden L. Conservation of community functional structure across changes in composition in consumer-resource models. Journal of Theoretical Biology. 2020;493 doi: 10.1016/j.jtbi.2020.110239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van der Plas F, et al. Plant traits alone are poor predictors of ecosystem properties and long-term ecosystem functioning. Nature Ecology & Evolution. 2020;4(12):1602–1611. doi: 10.1038/s41559-020-01316-9. [DOI] [PubMed] [Google Scholar]
- 128.Song HS, et al. Regulation-Structured Dynamic Metabolic Model Provides a Potential Mechanism for Delayed Enzyme Response in Denitrification Process. Front Microbiol. 2017;8:1866. doi: 10.3389/fmicb.2017.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hemelrijk CK, Hildenbrandt H. Schools of fish and flocks of birds: their shape and internal structure by self-organization. Interface Focus. 2012;2(6):726–737. doi: 10.1098/rsfs.2012.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hellweger FL, et al. Advancing microbial sciences by individual-based modelling. Nat Rev Microbiol. 2016;14(7):461–71. doi: 10.1038/nrmicro.2016.62. [DOI] [PubMed] [Google Scholar]
- 131.Griesemer M, Sindi SS. Microbial Systems Biology. Springer; 2022. Rules of Engagement: A Guide to Developing Agent-Based Models; pp. 367–380. [DOI] [PubMed] [Google Scholar]
- 132.Jayathilake PG, et al. A mechanistic Individual-based Model of microbial communities. PLoS One. 2017;12(8):e0181965. doi: 10.1371/journal.pone.0181965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clark JR, et al. Individual-based modelling of adaptation in marine microbial populations using genetically defined physiological parameters. Ecological Modelling. 2011;222(23–24):3823–3837. [Google Scholar]
- 134.Nadell CD, et al. Cutting through the complexity of cell collectives. Proceedings of the Royal Society B-Biological Sciences. 2013;280(1755) doi: 10.1098/rspb.2012.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allen B, Gore J, Nowak MA. Spatial dilemmas of diffusible public goods. Elife. 2013;2:e01169. doi: 10.7554/eLife.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Abs E, Leman H, Ferriere R. A multi-scale eco-evolutionary model of cooperation reveals how microbial adaptation influences soil decomposition. Communications Biology. 2020;3(1) doi: 10.1038/s42003-020-01198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kreft JU, et al. Mighty small: Observing and modeling individual microbes becomes big science. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(45):18027–18028. doi: 10.1073/pnas.1317472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Allison SD, Goulden ML. Consequences of drought tolerance traits for microbial decomposition in the DEMENT model. Soil Biology & Biochemistry. 2017;107:104–113. [Google Scholar]
- 139.Allison SD. A trait-based approach for modelling microbial litter decomposition. Ecology Letters. 2012;15(9):1058–1070. doi: 10.1111/j.1461-0248.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 140.Doloman A, et al. Modeling de novo granulation of anaerobic sludge. Bmc Systems Biology. 2017;11 doi: 10.1186/s12918-017-0443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gogulancea V, et al. Individual Based Model Links Thermodynamics, Chemical Speciation and Environmental Conditions to Microbial Growth. Front Microbiol. 2019;10:1871. doi: 10.3389/fmicb.2019.01871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gutierrez M, Rodriguez-Paton A. Simulating multicell populations with an accelerated gro simulator; Fourteenth European Conference on Artificial Life (Ecal 2017); 2017. pp. 186–187. [Google Scholar]
- 143.Gutierrez M, et al. A New Improved and Extended Version of the Multicell Bacterial Simulator gro. Acs Synthetic Biology. 2017;6(8):1496–1508. doi: 10.1021/acssynbio.7b00003. [DOI] [PubMed] [Google Scholar]
- 144.Momeni B, Waite AJ, Shou W. Spatial self-organization favors heterotypic cooperation over cheating. Elife. 2013;2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kreft JU, Booth G, Wimpenny JWT. BacSim, a simulator for individual-based modelling of bacterial colony growth. Microbiology-Uk. 1998;144:3275–3287. doi: 10.1099/00221287-144-12-3275. [DOI] [PubMed] [Google Scholar]
- 146.Picioreanu C, Van Loosdrecht MC, Heijnen JJ. Effect of diffusive and convective substrate transport on biofilm structure formation: a two-dimensional modeling study. Biotechnol Bioeng. 2000;69(5):504–15. doi: 10.1002/1097-0290(20000905)69:5<504::aid-bit5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 147.Lardon LA, et al. iDynoMiCS: next-generation individual-based modelling of biofilms. Environmental Microbiology. 2011;13(9):2416–2434. doi: 10.1111/j.1462-2920.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- 148.Chacon JM, Mobius W, Harcombe WR. The spatial and metabolic basis of colony size variation. ISME J. 2018;12(3):669–680. doi: 10.1038/s41396-017-0038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oyebamiji OK, et al. Gaussian process emulation of an individual-based model simulation of microbial communities. Journal of Computational Science. 2017;22:69–84. [Google Scholar]
- 150.Parise F, Lygeros J, Ruess J. Bayesian inference for stochastic individual-based models of ecological systems: a pest control simulation study. Frontiers in Environmental Science. 2015;3 [Google Scholar]
- 151.Allison S. A trait-based approach for modelling microbial litter decomposition. Ecology letters. 2012;15(9):1058–1070. doi: 10.1111/j.1461-0248.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 152.Nadell CD, Foster KR, Xavier JB. Emergence of Spatial Structure in Cell Groups and the Evolution of Cooperation. Plos Computational Biology. 2010;6(3) doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Menon R, Korolev KS. Public Good Diffusion Limits Microbial Mutualism. Physical Review Letters. 2015;114(16) doi: 10.1103/PhysRevLett.114.168102. [DOI] [PubMed] [Google Scholar]
- 154.Dobay A, et al. Interaction effects of cell diffusion, cell density and public goods properties on the evolution of cooperation in digital microbes. Journal of evolutionary biology. 2014;27(9):1869–1877. doi: 10.1111/jeb.12437. [DOI] [PubMed] [Google Scholar]
- 155.Canzian L, et al. A dynamic network formation model for understanding bacterial self-organization into micro-colonies. IEEE Transactions on Molecular, Biological and Multi-Scale Communications. 2015;1(1):76–89. [Google Scholar]
- 156.Mendoza SN, et al. A systematic assessment of current genome-scale metabolic reconstruction tools. Genome Biology. 2019;20(1) doi: 10.1186/s13059-019-1769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nature biotechnology. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Feist AM, Palsson BO. The biomass objective function. Current Opinion in Microbiology. 2010;13(3):344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blasche S, et al. Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nature Microbiology. 2021;6(2):196–208. doi: 10.1038/s41564-020-00816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dukovski I, et al. Computation Of Microbial Ecosystems in Time and Space (COMETS): An open source collaborative platform for modeling ecosystems metabolism. arXiv preprint. 2020:arXiv:2009.01734 [Google Scholar]
- 161.Varahan S, et al. Resource plasticity-driven carbon-nitrogen budgeting enables specialization and division of labor in a clonal community. Elife. 2020;9 doi: 10.7554/eLife.57609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Angeles-Martinez L, Hatzimanikatis V. Spatio-temporal modeling of the crowding conditions and metabolic variability in microbial communities. PLoS Comput Biol. 2021;17(7):e1009140. doi: 10.1371/journal.pcbi.1009140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zimmermann J, Kaleta C, Waschina S. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 2021;22(1):81. doi: 10.1186/s13059-021-02295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zorrilla F, Patil KR, Zelezniak A. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes. bioRxiv. 2021:2020.12. 31.424982. doi: 10.1093/nar/gkab815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ponomarova O, et al. Yeast Creates a Niche for Symbiotic Lactic Acid Bacteria through Nitrogen Overflow. Cell Systems. 2017;5(4):345. doi: 10.1016/j.cels.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Current biology. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 167.Borer B, et al. Modeling metabolic networks of individual bacterial agents in heterogeneous and dynamic soil habitats (IndiMeSH) Plos Computational Biology. 2019;15(6) doi: 10.1371/journal.pcbi.1007127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Labhsetwar P, et al. Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proc Natl Acad Sci U S A. 2013;110(34):14006–11. doi: 10.1073/pnas.1222569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kehe J, et al. Positive interactions are common among culturable bacteria. Science Advances. 2021;7(45) doi: 10.1126/sciadv.abi7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zmora N, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174(6):1388. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 171.Korem T, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349(6252):1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hamilton JJ, et al. Metabolic Network Analysis and Metatranscriptomics Reveal Auxotrophies and Nutrient Sources of the Cosmopolitan Freshwater Microbial Lineage acI. Msystems. 2017;2(4) doi: 10.1128/mSystems.00091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guerra CA, et al. Tracking, targeting, and conserving soil biodiversity. Science. 2021;371(6526):239–241. doi: 10.1126/science.abd7926. [DOI] [PubMed] [Google Scholar]
- 174.Shukla P, et al. IPCC, 2019: Climate Change and Land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. 2019 [Google Scholar]
- 175.Pacciani-Mori L, et al. Dynamic metabolic adaptation can promote species coexistence in competitive microbial communities. PLoS Comput Biol. 2020;16(5):e1007896. doi: 10.1371/journal.pcbi.1007896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.