Abstract
Analysis of oxylipins by liquid chromatography mass spectrometry (LC-MS) is challenging because of the small mass range occupied by this diverse lipid class, the presence of numerous structural isomers, and their low abundance in biological samples. Although highly sensitive LC-MS/MS methods are commonly used, further separation is achievable by using drift tube ion mobility coupled with high-resolution mass spectrometry (DTIM-MS). Herein, we present a combined analytical and computational method for the identification of oxylipins and fatty acids. We use a reversed-phase LC-DTIM-MS workflow able to profile and quantify (based on chromatographic peak area) the oxylipin and fatty acid content of biological samples while simultaneously acquiring full scan and product ion spectra. The information regarding accurate mass, collision-cross section values in nitrogen (DTCCSN2) and retention times of the species found are compared to an internal library of lipid standards as well as the LIPID MAPS Structure Database by using specifically developed processing tools. Features detected within the DTCCSN2 and m/z ranges of the analyzed standards are flagged as oxylipin-like species, which can be further characterized using drift time alignment of product and precursor ions distinctive of DTIM-MS. This not only helps identification by reducing the number of annotations from LIPID MAPS, but also guides discovery studies of potentially novel species. Testing the methodology on Salmonella enterica serovar Typhimurium infected murine bone-marrow derived macrophages and thrombin activated human platelets yields results in agreement with literature. This workflow has also annotated features as potentially novel oxylipins, confirming its ability in providing further insights into lipid analysis of biological samples.
Oxylipins represent a large network of bioactive lipid mediators (Figure S1), possessing both physiological and pathological roles.1 These lipids derive from oxidation of polyunsaturated fatty acids (PUFA), either enzymatically by cyclooxygenases (COXs), lipoxygenases (LOXs) and cytochrome P450s, or non-enzymatically via free-radical peroxidation processes.1,2 Within the class of oxylipins, several isomers can be found which exert distinct biological functions and have a different biochemical origin. For example, the six isoforms of HETEs can derive from oxidation of arachidonic acid either by LOXs (5-, 12- and 15-HETE) or COXs (11- and 15-HETE), while 8- and 9-HETE derive mostly from non-enzymatic autoxidation. Hence, identification of the correct isomers is fundamental to fully understand both their path of generation and function. Oxidation products of C20 PUFAs such as arachidonic acid are commonly known as eicosanoids, which are generated in cells and tissues in response to physiological and pathological changes2 and play crucial roles in mediating inflammation and homeostasis as well as diseases such as cancer, atherosclerosis and Alzheimer’s.3–5 Oxylipins are either released by cells/tissues to act as free mediators, or re-esterified into phospholipids and integrated into the cell membrane of platelets and leukocytes where they can interact with coagulation factors or act pro- or anti-inflammatory.6 Oxylipins are found in low nanomolar concentrations in biological samples, therefore their analysis requires highly selective and sensitive methods. To achieve this, ultra-high performance liquid chromatography (UHPLC) using reversed-phase columns in combination with triple quadrupole mass spectrometers coupled with negative electrospray ionization are commonly used.7,8 Although this methodology allows both sensitive and selective analysis, targeted approaches are time-consuming in their development and will miss lipids that are not specifically being analyzed. Ion mobility coupled with high-resolution mass spectrometry (IM-MS) can be used to achieve more specificity in untargeted approaches, allowing measurements of both accurate mass and collision cross-section (CCS) values of the ions to provide greater certainty in terms of identification. In recent years, IM-MS has developed as an emerging technique in lip-idomics,9,10 however, oxylipins analysis by IM-MS has been limited to some eicosanoids using travelling-wave IM,11,12 leukotrienes and protectins using differential IM,13 and some oxylipins using drift tube IM (DTIM-MS).14 In this study we describe a comprehensive analytical and computational workflow for the analysis of oxylipins and fatty acids (FAs) using UHPLC-DTIM-QTOF, in which size and shape of the ions affect their mobility when travelling through a drift tube filled with low pressure nitrogen gas, allowing their separation and the calculation of their CCS values for nitrogen (DTCCSN2). Annotation and identification of oxylipins and FAs was enhanced through an extension of the KniMet pipeline for the processing of metabolomics data15 with a combined annotation method comprising the LIPID MAPS Structure Database (LMSD)16,17 and an internally generated library containing 47 unique lipids. The UHPLC-DTIM-MS method presented in this study also allows quantification of oxylipins in biological samples, and its combination with data-independent acquisition enables lipid identification based on drift time alignment between diagnostic product ions and their precursors. The computational pipeline as well as the data obtained for the 47 lipid standards is publicly available at https://github.com/sonial/KniMet and could contribute to the creation and expansion of CCS databases.18 Together, this method can be used for both discovery and quantification of oxylipins and FAs in biological samples. The approach is here demonstrated on human thrombin activated platelets and Salmonella enterica serovar Typhimurium infected murine bone-marrow derived macrophages (BMDMs).
Materials and Methods
Materials
Lipid standards were purchased from Santa Cruz Biotechnology (Heidelberg, Germany) or Cayman Chemical (supplied by Cambridge Bioscience, Cambridge, UK). All solvents were Chromasolv LC-MS grade from Honeywell. Ul-trapure LPS from Escherichia coli O111:B4 was from Invi-voGen. All other reagents were purchased from Sigma Aldrich (Merck KGaA, Darmstadt, Germany) unless otherwise stated.
Mice
Wild type C57BL/6 mice were obtained from Charles River, UK. All animals were housed under specific pathogen-free conditions and all work involving live animals complied with the University of Cambridge Ethics Committee regulations under Home Office Project License number 80/2572.
Cell isolation and culture
BMDMs were retrieved from the bone marrow of legs of wild type C57BL/6 mice and cultured in BMDM growth media (Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 20% L929 conditioned media and 5 mM L-glutamine). After centrifugation, both isolated cells and medium were divided into 2 groups to subject only one of them to infection with a solution of S. Typhimurium strain SL1344 in BMDM growth media with multiplicity of infection equal to 1. This resulted in 4 classes of samples: infected and uninfected cells, and infected and uninfected medium (see Supporting Materials for further details).
Thrombin stimulation of isolated human platelets
All experiments were performed in accordance with the Cambridge Human Biology Research Ethics Committee; informed consent was obtained from donors as applicable, according to the Declaration of Helsinki. Platelets from three donors were isolated and activated using 0.2 U/mL thrombin as described previously19 (see Supporting Materials for further details). Platelets were divided in 4 groups and differently treated with thrombin, aspirin, ethanol, or untreated.
Lipid extraction
Chilled 2.5 mL isopropanol/hexane/acetic acid extraction solution (20:30:2, v/v/v) and 10 ng internal standard (added as 1 ng/μL methanol solution, PGE2-d4, 12-HETE-d8, and arachidonic acid-d8 for platelet experiments, or deuterated primary COX & LOX LC-MS mixture CAY19228-1 for BMDM experiments) were added to 1 mL isolated platelets, cells, or cell culture media and kept on ice for 10 min. Samples were vortexed, 2.5 mL of chilled hexane added, and vortexed again. The organic layer was recovered after centrifugation for 5 min at 4°C at 900 g, while the remaining portion of the samples were re-extracted by addition of an equal volume of hexane. The aqueous layer was subjected to a modified Bligh & Dyer extraction. Briefly, 3.75 mL chloroform:methanol (1:2, v/v) were added to the aqueous layer, samples were vortexed and 1.25 mL chloroform added. Following vortexing, 1.25 mL water was added, samples were vortexed again and centrifuged at 4°C at 900 g for 5 min. The organic layer was combined with lipid extracts from the first extraction step, dried under nitrogen gas, resuspended in 100 μL of methanol and analyzed by LC-DTIM-MS.
LC-DTIM-MS analysis
5 μL of lipid extract was analyzed using an Infinity II UHPLC coupled to an Agilent 6560 IM QTOF MS (Agilent Technologies, Santa Clara, USA) using a reversed-phase ACQUITY CSH C18 column (1.7 μm, 2.1x100 mm, Waters, UK), which was held at 45°C during analysis.. LC separation was adapted from Ostermann et al.7 Briefly, the UHPLC was operated at a flow rate of 0.5 mL/min using water/solvent B (95%/5%) with 0.1% glacial acetic acid as solvent A and acetonitrile/methanol/glacial acetic acid (800/150/1, v/v/v) as solvent B. Solvent B was increased from 30 % at 0-1 min to 35 % at 4 min, 67.5 % at 12.5 min, and 100 % at 17.5 min. Following a 3.5 min of isocratic elution at 100 % solvent B, the column was re-equilibrated for 2.5 min at 30 % solvent B.20 A flow rate of 0.3 mL/min (50% solvent B) was used for flow injection experiments. Lipids were analyzed in negative IM-QTOF mode in the m/z range of 100-1000 and maximum drift time (DT) of 60 ms using nitrogen as drift gas, a trap fill time of 25 ms, trap release time of 0.5 ms and acquisition rate of 1 IM frame/s. An Agilent tuning solution was injected both before the analysis to tune the instrument in the m/z range 100-1700, and every 10 samples to perform DTCCSN2 re-calibration. An Agilent reference mix for mass re-calibration was continuously injected alongside the samples using the secondary sprayer. The Dual Agilent Jet Stream electrospray ionization source was operated at gas temperature of 325°C, drying gas 5 L/min, sheath gas temperature of 275°C and a sheath gas flow of 12 L/min. A drift gas upgrade kit maintained both drift tube and trap funnel pressure constant at 3.9 and 3.8 Torr respectively, while drift tube temperature was found to be stable at 23.5°C ± 0.3°C across all acquisition runs. Fragmentation with alternating frames was used for data-independent acquisition (IM-MS/MS) of pooled samples (QCs), with the collision energy of frame two fixed at 30V.
Creation of an Internal Database of Oxylipins
An internal database containing accurate mass, retention time (RT) and DTCCSN2 values was obtained for 47 lipid standards previously acquired in triplicates across different days using the instrumental, analytical and pre-processing method described. This data was further processed with an in-house pipeline based on the KNIME Analytics Platform21 to build a database containing [M-H]-, [M-H+CH3CO2Na]-, [2M-2H+Na]- and [2M-H]- species (Table S1 and https://github.com/sonial/KniMet). First, a list containing lipid standard names, exact molecular weight, expected RT (manually inferred from previous acquisitions), SMILES, formula and InChI was created with the aid of the ChemAxon22 and RDKit23 KNIME nodes. This list was used to annotate the individual acquisitions based on accurate mass and RT match (error ranges 10 ppm and 0.15 min, respectively) with an adapted version of the KniMet functionality “Feature Annotation”, which was modified to take into account also RT range.
A subsequent cleaning of the annotations consisted of keeping only the features annotated as different adducts of the same molecule within a RT range of ± 0.01 min from a [M-H]- conformer. This allows one to keep only co-eluting adducts and eliminate erroneous annotations derived from the large RT range required for the annotation step. The results obtained for each different acquisition were then merged in the database if they were present in at least two out of three injections with a relative standard deviation (RSD) smaller than 1% for m/z and 1.5% for both RT and DTCCSN2. This results in five peaks on average per lipid, with some of them deriving from the same adduct suggesting either different conformations or more complex ion arrangements (discussed in the section “Ion mobility allows identification of lipid clusters”).
Data processing
Data pre-processing, including mass and DTCCSN2 re-calibration and feature finding, was carried out using the packages IM-MS Reprocessor, IM-MS Browser and Mass Profiler from the MassHunter Suite (version B.08.00, Agilent Technologies, Santa Clara, USA). In particular, CCS calibration was performed in batches containing 10 samples and one calibrant, where the slope and intercept (β and TFix in IM-MS Browser) values obtained for the DT correction of the calibrant were applied to samples for calculation of DTCCSN2 (please refer to 24 and 25 for more details of CCS calibration in DTIM). Stability of the calibration was assessed by calculating the CCS for a dummy feature (DT = 28 and m/z = 313, which are the average values found in our internal database) using the β and TFix values obtained from the several calibrant infusions, with RSD equal 0.097% for the 4 calibrants infused in BMDM and 0.103% for the 5 calibrants infused in the platelets study. Feature finding was performed for signals having a minimum ion count of 100 using the maximum ion volume as a measure of abundance and an isotopic model designed for common organic molecules without halogen atoms. Moreover, only species with charge state = 1-2 were extracted, including single ion features with charge state = 1. Features were aligned if they fell within the confidence intervals of 10 ppm ± 3 mDa in m/z and in 0.3 min in RT. Furthermore, an m/z range (199-1000) was selected to include monomers, dimers and adducts. A final filter based on Q-Score (greater than 60 on a scale of 0-100; value obtained from Mass Profiler, provides an indication about data quality considering parameters such as isotopic pattern), DT (15-35 ms), and RT (0.9-20 min) or detection window (0-0.5 min) for flow injection experiments was applied. Quantification was performed using the Agilent MassHunter Quantitative Analysis software. Although the input files contained IM data, the software simply ignores the IM dimension and quantifies based on the chromatographic peak. Lipid levels were normalized to internal standard levels. On the other hand, an adaptation of the KniMet pipeline for MS-based metabolomics data processing15 was used to perform profiling of oxylipins. More specifically, features were retained if they were found in at least 70% of the samples belonging to a class with an average abundance higher than 5000. This first filtering of noise was finalized by the removal of all features whose average intensity in samples was less than twice the average intensity in uninfected medium samples in the BMDM experiment, while periodic injections of a dilution series of quality control samples (QCs, obtained pooling all samples analyzed) was used in the platelets experiment to remove features whose intensity does not vary linearly with concentration of the sample (node “QC dilution series-based Features Filtering” added to KniMet). The remaining features were then subjected to missing value imputation using k-nearest neighbor for multivariate statistical analysis, while replacement of missing values with half of the minimum value found in each sample was used for univariate analysis to avoid a possible change of the data distribution.26 Finally, signals were normalized using probabilistic quotient normalization based on all samples27 and logarithmic transformed. Identification of oxylipins and FAs was performed by accurate mass, RT and DTCCSN2 match with the internal database, with error ranges of 10 ppm for mass accuracy, 0.15 min for RT difference and 2% RSD for DTCCSN2. Moreover, lipid-like flags were added: features presenting m/z and RT within the aforementioned ranges, but different DTCCSN2 values from those present in the database, were flagged as possible new conformers, while features with m/z and DTCCSN2 within these ranges but different RT were flagged as possible new compounds. Annotation of lipids not present in our internal database was performed by comparison with a subset of the LMSD (version updated in 12/2016)16 containing only lipids belonging to the fatty acyl class (to which oxylipins belong).28 In both annotation and identification steps, species whose RT is further than 0.01 min from the relative [MH]- were removed to eliminate wrongly annotated dimers and adducts. Both annotations were performed by adapting the “Feature Annotation” functionality of KniMet. In particular, “Feature Annotation from internal IMS DB” was added to KniMet to allow annotation based also on DTCCSN2 match and, if available, RT, to flag lipid-like compounds, and to remove dimers with no monomers in a given RT range. Similarly, the “Feature Annotation - Internal2external” functionality performs annotation on a specific class of LIPID MAPS and removes dimers with no monomers in a given RT range (all created KniMet nodes available at https://github.com/sonial/KniMet).
Principal Component Analysis (PCA) was performed using SIMCA (Umetrics, Sweden), while heatmap representations were created using the function heatmap.2, part of the gplots29 library of R,30 with default parameters for both hierarchical clustering and dissimilarity calculation.
Results and Discussion
Acquisition and data processing workflow for oxylipin and fatty acid analysis
Oxylipins are a class of lipids presenting strong structural similarities, but their polarity can vary depending on number of oxygen atoms, position of hydroxyl groups or number of double bonds. Therefore, a reversed-phase gradient method20 was used to separate oxylipins and FAs in the first dimension (Figure S2) prior to further separation by IM and accurate mass detection by QTOF MS. To profile the lipid content, features picked by Mass Profiler were processed using KniMet to perform data normalization, feature filtering, identification and annotation. For feature identification, an in-house lipid library (Table S1) was used. Features not present in the library were putatively annotated based on accurate mass using LMSD. In addition to lipid discovery, lipids were quantified using the Quantitative Analysis software if a standard curve was analyzed alongside. To confirm lipid identities, either a QC or a known sample containing oxylipins (if concentration in the QC was too low) was analyzed using data-independent acquisition, which does not require the long development time of targeted methods. Due to fragmentation taking place after the drift tube, product ions time-align to their precursors, allowing identification of diagnostic product ions9 and conferring more confidence in oxylipin identification.
Ion mobility allows identification of lipid clusters
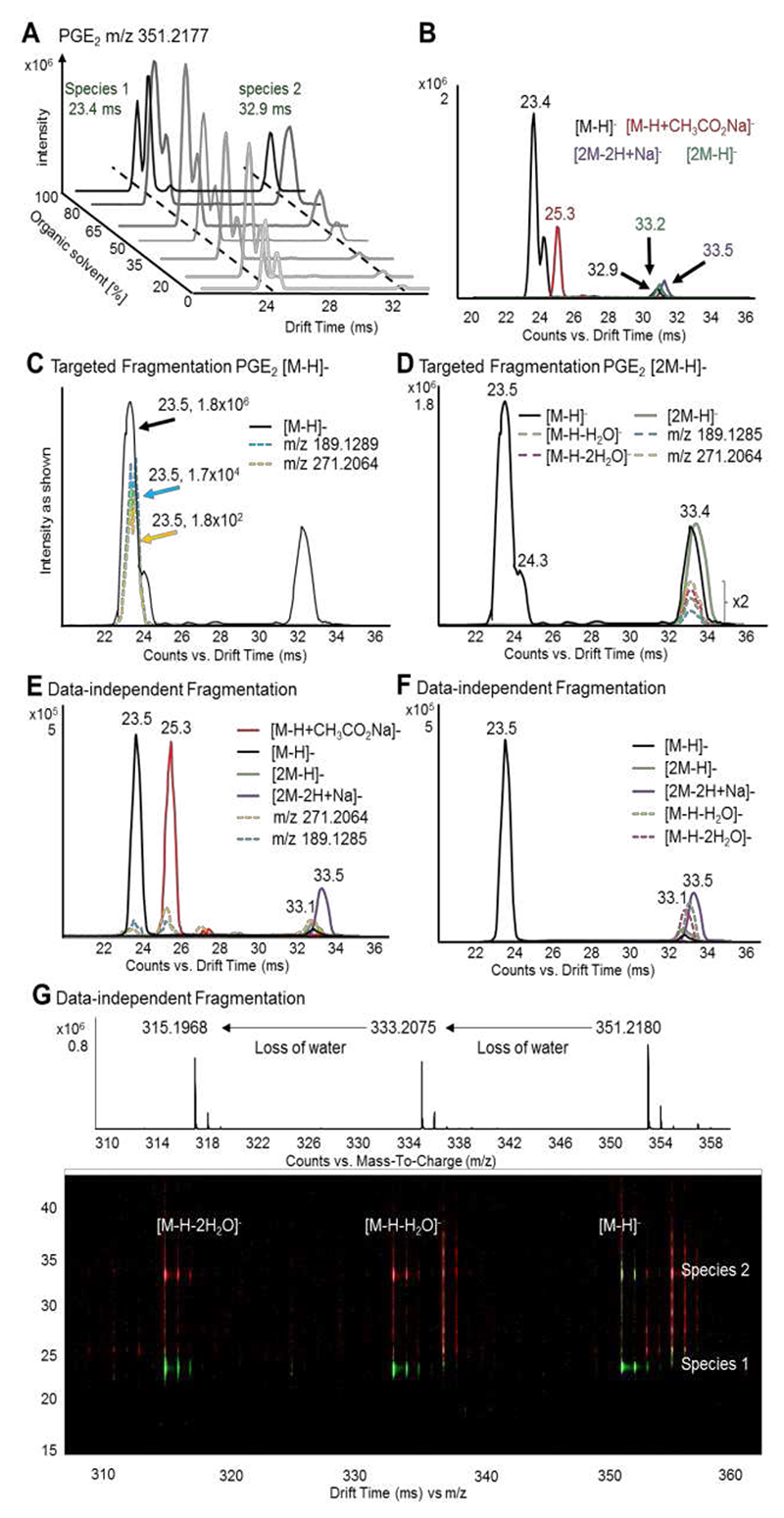
DTIM separation can be visualized in drift spectra, in which counts are plotted versus DT (ms), and driftograms, where instead DT is plotted against m/z. Multiple drift peaks were observed for all lipids analyzed in this study, with one of these peaks being dominant in intensity and hence corresponding to the most stable conformer in the gas phase. DTIM-MS analysis of the most abundant [M-H]- (referred to as species 1) conformer of oxylipins and FAs shows that they occupy both tight DTCCSN2 and mass ranges (CCS = 174-221 Å2, m/z = 296-496 for the former; CCS = 158-185 Å2, m/z = 200-328 for the latter). However, the driftograms produced by most lipids suggest the presence of multiple conformations, with several features having identical RT and m/z but different DT. Among these, low abundant species drifting in close proximity to the dominant peak were found, which could derive from either different isomers (geometric- or stereo-isomers) or different conformers of species 1.24 In contrast, the second most intense signal corresponds to a feature (species 2) drifting about 10 ms later than species 1. Although differences in DTCCSN2 values, corresponding to different conformations, have been previously described for FAs,24 the DT differences observed between species 1 and 2 (Table S1) may be too great to correspond to different conformers. The appearance of species 2 depends on the concentration of both lipid and organic solvent used, which was tested under six different conditions for PGE2 (Figure 1 A), arachidonic acid, 12-HETE and 5(6)-EET (Figure S3). Although the intensity of species 2 increased with an increase in the organic solvent content, the relationship is not linear and may also depend on the polarity and solubility of the individual lipid (Figure S4). Along with [M-H]- of species 2, co-drifting (± 0.5 ms) [2M-H]- and [2M-2H+Na]- ions were detected as well (Figure 1 B). To identify both dimers and species 2, we undertook targeted fragmentation of all species, namely [M-H]-, [2M-H]- (Figure 1 C and D), [M-H+CH3CO2Na]- [2M-2H+Na]-, [M-H-H2O]-, [M-H-2H2O]-. This led to diagnostic product ions m/z 271.2064 and m/z 189.1289 (Figure S5), which DT-align (± 0.5 ms) with their precursor ions (Figure 1 C-D). Comparable results were also obtained when applying data-independent fragmentation (Figure 1 E-F), demonstrating that diagnostic product ions derive from monomers, adducts, and dimers. Therefore, we propose that species 2 are product ions derived from lipid dimers, which are formed before IM separation and partly dissociate after the IM cell prior to mass detection. This would explain their detection as m/z corresponding to [M-H]- species but co-drifting with lipid dimers. Indeed, using the Agilent 6560 DTIM-MS, ions are focussed by a high-pressure ion funnel and accumulated by an ion trap prior to be released into the drift tube. When ions leave the drift tube, they are refocussed by a rear ion funnel, followed by m/z detection in the QTOF. These focussing and trapping procedures could result in the formation of unstable ion clusters such as dimers and solvent clusters, which partly fall apart before m/z detection and cause the aforementioned peaks with different DTs. We tested this hypothesis by altering trap fill time (20 or 30 ms) and lowering trap funnel RF voltages (-150V, -100V, -76V), but the signals remained the same (Figure S6), suggesting that more complex ion arrangements may occur during analysis. Similarly, the described ion species were present using different source conditions, including higher temperatures (data not shown). In addition, [M-H+CH3CO2Na]- species were observed, but these species drift independently from monomers and dimers (Figure 1 E, Figure S3 B, D and F, red) due to their different m/z. Moreover, using altering high/low fragmentation with DTIM it was observed that the [M-H-H2O]- and [M-H-2H2O]- species are not only products but also precursors ions, suggesting that some [M-H]- precursors lose a water molecule before fragmentation (Figure 1 G). Note, this may not be an in-source fragmentation as modifying source parameters did not remove these species (data not shown). Further discussions to identification of unspecific losses using DTIM can be found in Supporting Discussion. On the other hand, formation of sodium acetate adducts increases with increasing organic solvent content (Figure S4 C), as expected given the enhanced concentration of acetic acid in the solution. However, this does not apply to dimers (Figure S4 B and D), indicating that dimer formation occurs following ionization.
Figure 1. DTIM allows separation of adducts and alignment of pre-cursor and product ions following fragmentation.
(A) PGE2 standard was analyzed by flow injection DTIM-MS using varying concentrations of acetonitrile:methanol. The intensity of the two most abundant [M-H]- conformer is plotted versus the DT and the concentration of organic solvent (average DTs in green). (B) Drift spectra for the conformers [M-H]- (black), [M-H+CH3CO2Na]- (red), [2M-H]- (green) and [2M-2H+Na]- (purple). (C) Targeted DTIM-MS/MS analysis of the [M-H]- species (black) enables drift-time alignment with the diagnostic product ions m/z 271.2064 (yellow) and m/z 189.1285 (light blue). (D) Similarly, fragmentation of [2M-H]- (green) results in product ions m/z 271.2064, m/z 189.1285, as well as water loss products [M-H-H2O]- (dashed green) and [M-H-2H2O]- (dashed pink) co-drifting with their precursor at 33.4 ms. Product ion peaks are magnified by factor 2. Data-independent acquisition (DIA) gives similar results, with product ions m/z 271.2064 and m/z 189.1285 as well as species [M-H-H2O]- and [M-H-2H2O]- time-aligning to their precursor ions [M-H]-, [2M-H]-, [M-H+CH3CO2Na]- (red), and [2M-2H+Na]- (purple). (E, F). The driftogram obtained from DIA (G) highlights how water-loss products deriving from both species 1 at 23.4 ms and 2 at 32.9 ms can be products (red, high fragmentation) and precursors (green, low fragmentation).
In addition to our observation using lipid standards and flow injection DTIM-MS, we also observed lipid dimers and solvent adducts using both LC-DTIM-MS analysis of lipid standards and biological samples and flow injection MS analysis of lipid standards using a triple quadrupole instrument in Q1 scan mode (Figure S7). Therefore, using LC-DTIM-MS allowed additional information regarding lipid cluster formation, which may also take place on other instruments but could potentially be overlooked.
The level of unsaturation changes the drift peak shape of fatty acids
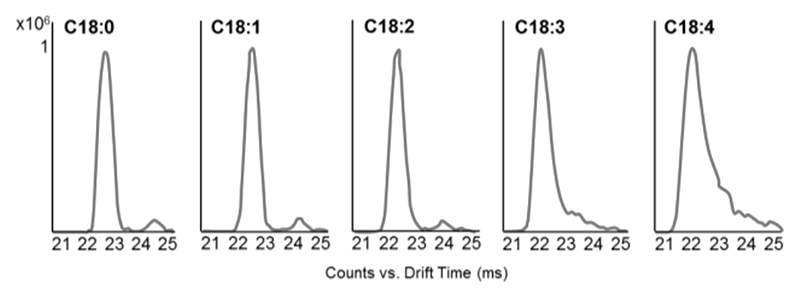
In addition to oxylipins, free FAs can be analyzed using our method. As previously shown,9,31 DT increases with both chain length and degree of saturation (Figure S8). The shape of the drift peak changes with the degree of saturation, as shown for a series of FAs with 18 carbon atoms in their chain and an increasing number of double bonds (Figure 2). The drift spectra of saturated C18:0, as well as the unsaturated species C18:1 C18:2, contain two peaks. In contrast, the drift spectra of poly-unsaturated C18:3 and C18:4 contain one tailing peak which possibly covers the equivalent to the second peak detected in C18:0, C18:1 and C18:2. Drift peak tailing suggests multiple molecular conformations of the ions, possibly due to the higher degree of unsaturation. This hypothesis finds confirmation in the recent work of Stow and co-workers, who described an increasing number of theoretical DTCCSN2 values for fatty acid conformers with different levels of unsaturation.24 Presence of multiple conformers does not affect comparability of DTCCSN2 values between different studies and laboratories, as the most abundant ones (and therefore the most stable conformers) can be used for comparison. However, as isomers are expected to drift close to the original conformer,31 peak tailing should be taken into consideration when investigating isomer formation, for example under certain biological conditions such as oxidative stress where non-enzymatic autoxidation may lead to the formation of isomers not normally observed.
Figure 2. Drift spectra of C18 fatty acids differing in the number of double bonds: the second peak gets included in the tail of the first (most abundant) peak with increasing number of double bonds in the acyl chain.
HETE-esterified phosphatidylcholines show a distinct mobility behavior
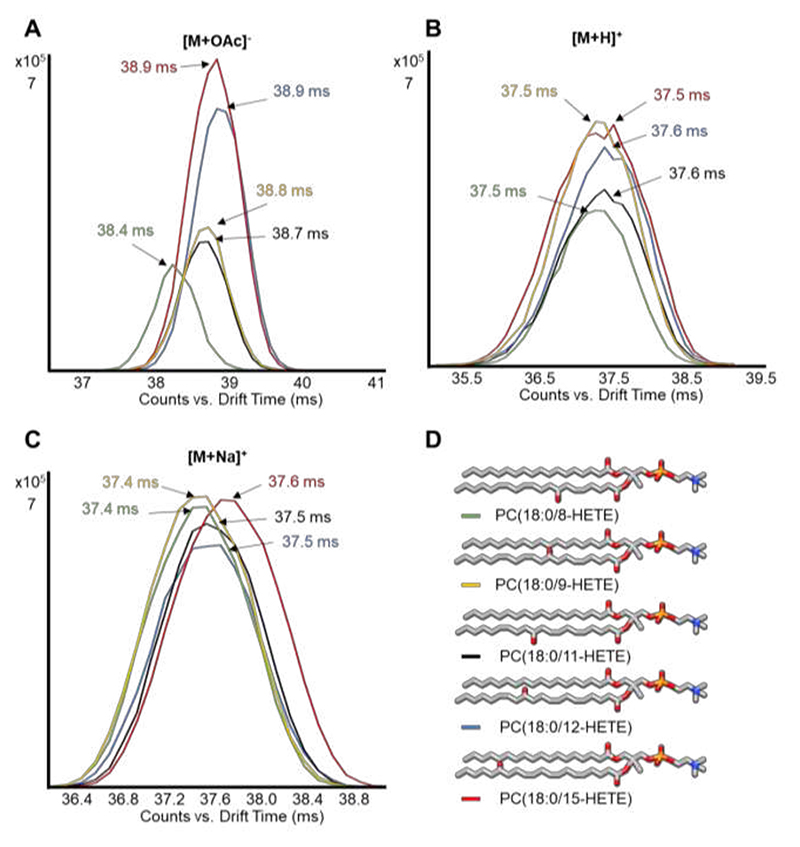
Eicosanoids not only act as free lipid mediators released by cells, but can also be re-esterified into phospholipids.6 To explore the DTIM behavior of eicosanoid-esterified phospholipids, we analyzed 8-, 9-, 11-, 12-, and 15-HETE-esterified phosphatidylcholines (PC) using flow injection DTIM-MS. Separation of the different HETE-PCs by DTIM depends on the ion type analyzed, with [M+OAc]- species presenting the greatest separation between structural isomers, followed by sodiated species, and finally protonated ions with the lowest drift separation (Figure 3). In addition, drift separation of acetate adducts demonstrated that a hydroxyl group close to the PC headgroup (such as in 8- and 9-HETE) results in faster mobility perhaps due to a smaller molecular size than that conferred by hydroxyl groups further away from the headgroup (12- and 15-HETE). The same difference in mobility, but to a smaller extent, can also be observed for the sodiated adducts. Therefore, we propose that both acetate and sodium interact with the hydroxyl group of the esterified HETE, resulting in a rather rigid structure of the molecule and distinct mobility behaviors. Although these phospholipids separate in LC,32 and baseline separation of different isomers by DTIM could not be achieved, our findings are in agreement with previous observations9 and show the potential to identify these isomeric lipids in complex samples based on DTCCSN2 values using a higher IM resolving power.
Figure 3.
Separation of eicosanoid-esterified phosphatidylcholines by DTIM-MS depends on both position of the hydroxyl group and adduct type. DTIM-MS analysis in both positive and negative mode of 8-, 9-, 11-, 12-, and 15-HETE esterified PCs (structures shown in D) shows enhanced separation between these isomers for [M+OAc]- conformers (A) compared to sodium adducts (C), while [M+H]+ species (B) are not distinguishable in drift space.
Quantification of oxylipins and fatty acids in S. Typhimurium activated murine BMDMs and thrombin activated human platelets
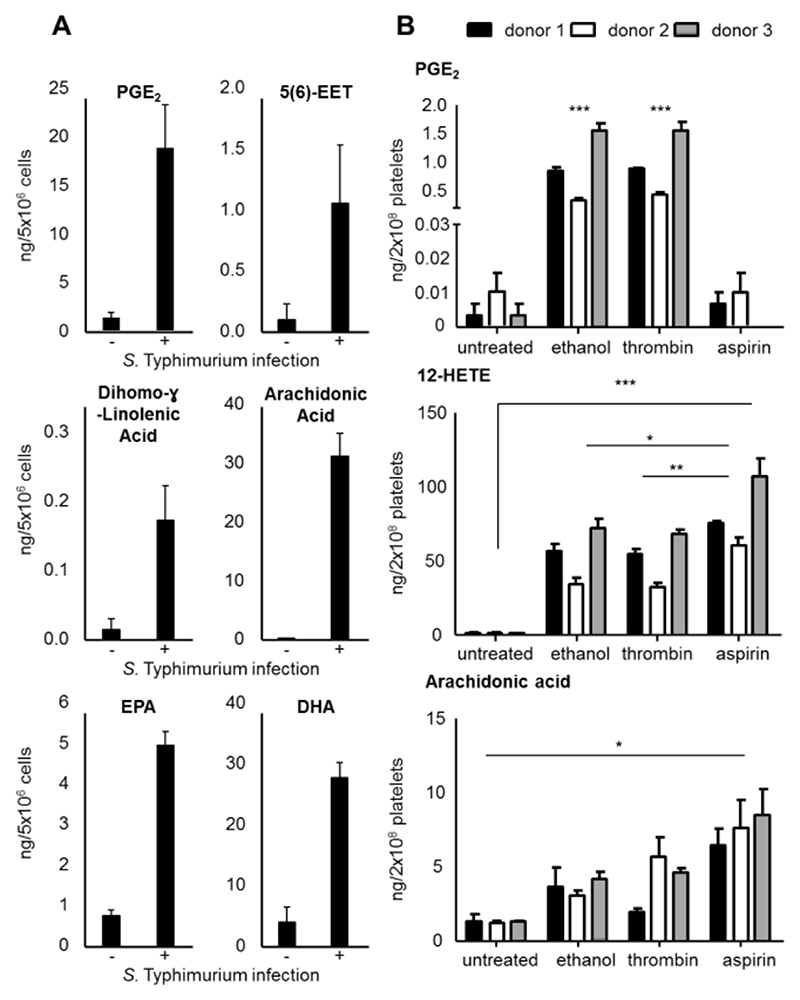
Eicosanoids are one of the least abundant lipid species found in cells and tissues, however, they are generated rapidly in response to acute cell activation during inflammatory processes or thrombus formation. As absolute quantities of oxylipins are important in many studies, we developed a workflow which can be used for both profiling and quantification. A calibration curve containing increasing amounts of 16 lipid standards as well as steady amounts of 6 deuterated analogues (Table S2) was generated. The lower limit of quantification (LLOQ) was defined as the amount of lipid required to achieve a signal-to-noise ratio greater than 9. Three deuterated standards (PGE2-d4, arachidonic acid-d8, and 12-HETE-d8) covering both oxylipins and FAs as well as different polarities, were spiked into platelet samples before lipid extraction to determine complete recovery rates, which were found to be 74% for PGE2-d4, 87% for arachidonic acid-d8 and 91% for 12-HETE-d8. This indicates low matrix and ionization effects on our approach. Lipid concentrations in samples were calculated using an external calibration curve and internal standards spiked into the samples before lipid extraction to account for both matrix effects and losses during the extraction procedure. The LLOQs ranged between 2 and 20 pg on column, equivalent to 0.6 and 11.3 nM, and regression coefficients R2> 0.98 were achieved (Figure S9, Table S2). To confirm lipid annotation from KniMet, both RTs and product ion spectra were compared to either lipid standards or spectra published on LIPID MAPS. Among the 16 compounds included in the calibration curve, seven were identified and quantified in BMDMs, with their concentration levels found to be in the low nanogram range per 5x106 cells (Figure 4 A, Table S3) as previously reported upon S. Typhimurium infection.33 Similarly, quantification of PGE2, 12-HETE and arachidonic acid in thrombin activated human platelets (Figure 4 B) agrees with the results of similar studies.19,34,35 Moreover, simultaneous treatment of platelets with thrombin and the COX inhibitor aspirin resulted in decreased prostanoid levels. In contrast, 12-HETE, which is the most abundant oxylipin amongst those quantified, was increased following both treatment with thrombin and a combination of thrombin and aspirin. This is possibly due to more arachidonic acid being available and therefore a higher turnover rate of 12-LOX. Indeed, aspirin treatment resulted in enrichment of arachidonic acid in the platelets, as a consequence of one oxylipin pathway (COX) being inhibited. Therefore, these results demonstrate that our quantification method not only reaches low oxylipin levels, but also covers the wide dynamic range required for oxylipin analysis in biological samples.
Figure 4. Quantification of oxylipins and fatty acids in S. Typhimurium infected murine BMDMs and thrombin activated human platelets analyzed by UHPLC/DTIM-MS.
(A) Lipids included in the calibration curve were quantified in BMDMs, indicating an increase of most species following infection with S. Typhimurium. (B) PGE2, 12-HETE, and arachidonic acid were quantified in platelets. PGE2 levels increased following treatment with thrombin, while its formation was inhibited by treatment with aspirin. On the other hand, both 12-HETE and arachidonic acid levels significantly increased following both thrombin alone and thrombin and aspirin treatment. Bar plots are created utilizing mean values ± standard error of the mean, * p < 0.05, ** p < 0.005, *** p < 0.0005 (Student’s t-test (BMDMs) or one-way ANOVA followed by Bonferroni correction (platelets).
Profiling of oxylipins in biological samples
Although research around oxylipins, especially eicosanoids, emerged in the 1960 after the discovery of their generating enzymes, several members of this lipid class were only characterized in more recent years19,36–38 with many oxylipins possibly still remaining unknown. Hence, we developed a profiling UHPLC-DTIM-MS workflow to investigate oxylipin generation, exemplarily shown on S. Typhimurium infected murine BMDMs and thrombin activated human platelets. A three-level annotation was implemented, with features (i) identified by accurate mass, RT and DTCCSN2 match with the internal database, (ii) annotated based on accurate mass match with LMSD, or (iii) flagged as “lipid-like” features. Features were classified as lipid-like if presenting identical accurate mass but differing on either RT or DTCCSN2 values from the compounds in the database. More specifically, lipid-like features can be oxylipin category-like (e.g. prostaglandin (PG)-like) if their accurate mass and DTCCSN2 values fall within the annotation thresholds but their RT does not, hence indicating a possible new compound with different polarity within the lipid class. This is illustrated in thrombin activated platelets on the extracted ion chromatogram for m/z 351.2177, which corresponds to a number of different oxylipins, including PGE2 (Figure 5 F, G). On the other hand, if a compound is detected with accurate mass and RT comparable to those of the lipid standard but different DTCCSN2 value, it is flagged as specific lipid-like (e.g. PGF2α-like). The latter is based on the assumption that structurally similar lipids have a similar “drift-fingerprint”, i.e. a similar distribution of species with identical m/z and RT, but different DT. Hence, the discovery of a feature in a biological sample whose m/z and RT fall within the ranges of a standard, but DTCCSN2 does not, might represent a new isomer of the molecule. These oxylipin category-like or specific lipid-like flags give one more confidence in identifying new compounds or new conformers not present in the internal database than using solely accurate mass match with an external library, which commonly results in multiple putative annotations for the same feature.
Figure 5. The changes in the oxylipin profile induced by thrombin treatment of human isolated platelets can be identified using MS/MS spectra and DT alignment of product and precursor ions.
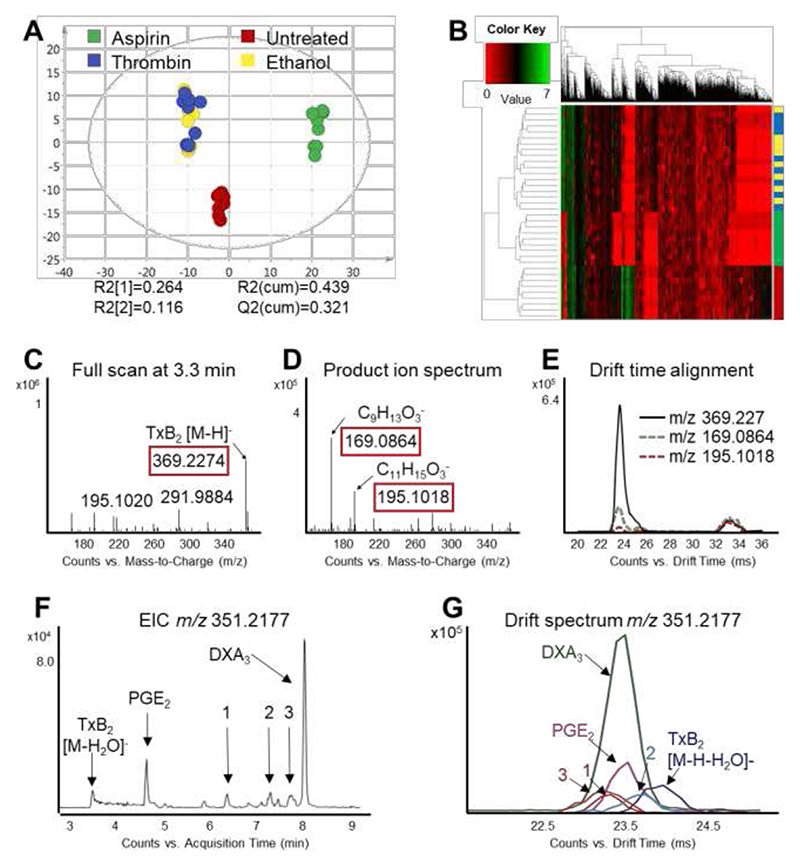
(A) Score plot of the principal component analysis (first versus second PC) of lipid extracts of isolated human platelets analyzed by LC/DTIM-MS and processed with KniMet. The four treatment groups are: thrombin plus aspirin (green), thrombin (blue), thrombin plus ethanol (yellow) and untreated (red). (B) Heatmap representation and hierarchical clustering of the distance matrix between samples in the four treatments. Separation between treatment groups can be observed, with three distinct clusters (aspirin, control and ethanol/thrombin) in both PCA and heatmap. (C) Full scan at 3.3 min shows m/z 369.2274, which is putatively annotated as TxB2 and other oxylipins. (D) Product ion spectrum at 3.3 min shows the diagnostic product ions m/z 195.1018 and m/z 169.0864 for TxB2. (E) Precursor (black) and product ions (m/z 195.1018 dashed red; m/z 169.0864, dashed green) align in DT, confirming the identification of the feature as TxB2. Unknown species 1, 2, 3 present identical m/z, different RT (F) but very similar DT (G) of TxB2, hence they are flagged as “lipid-like” features during data annotation.
Using this protocol on the BMDMs data, out of 3339 features we standard-identified 85 species, flagged 146 as lipid-like, and putatively annotated with LIPID MAPS 624 features, while 2484 features remained unknown (Table S4 and Figure S10 A). Our findings agree with previous results showing that infection with S. Typhimurium induces eicosanoid production in BMDMs.33 Among the identified lipids differentiating between infected and uninfected samples were known mediators of inflammation, such as PGE2 and 5-HETE, (Figure S11, Table S5). The high number of unknown features is due to (i) the accurate mass range of m/z 199-1000 used for feature extraction, which was chosen to include lipid dimers and adducts but might have led to the addition of features not corresponding to oxylipins; (ii) the limited size of our internal database, as well as the relatively low number of oxylipins present in the LMSD. Nonetheless, it has to be taken into consideration that each lipid characterized contains on average 5 drift peaks (discussed above), which play an important role on the overall high number of features. Considering a tighter m/z range comprising only the [M-H]- species of FAs and oxylipins present in our database (200-500), the number of features reduces to 2595 with 1775 unknowns. However, information about dimers and cluster formation would be lost using this mass range. On the other hand, data processing ignoring the mobility dimension yields 580 features, out of which only 177 are annotated with the fatty acyl portion of LMSD (Supporting Discussion, Figure S12). This noticeable reduction of annotated features further highlights the importance of IM in gaining new insights into LC-MS analysis of lipids. The high abundancy of features found using IM might raise concerns about ion formation in the source, ion movement in the drift tube or feature finding algorithms. As described above, we tested the consistency of features found across multiple injections, as well as their changing abundancy dependent on either sample concentration or solvent composition, and the results suggest that their presence is not triggered by instrument settings or data processing algorithms but that the features detected by DTIM are present in the sample.
Our oxylipins and FAs profiling method was further tested on human platelets isolated from three donors treated with the pro-coagulant thrombin to trigger oxylipin generation.39,40 Statistical analysis of the data processed with KniMet revealed differences between treatment groups (Figure 5, Figure S13 and Table S6), which are in agreement with previous findings.39,40 Out of the 2591 features remaining after filtering, 57 were identified by the internal library of standards, 109 were flagged as lipid-like, and 433 could only be annotated with LMSD, while 1992 remained unidentified (Table S7, Figure S10 B). Although putative annotation results in multiple possible identities, the combination of lipid-like flagging, annotation from the external database and finally data-independent acquisition in IM-MS/MS mode allows lipid identification, as exemplified by the identification of Thromboxane B2 (TxB2, Figure 5 C-E): the feature presenting m/z = 369.2270, RT = 3.33 min and DT = 23.89 ms was flagged as PG-like [M-H]- by the internal annotation system and putatively annotated by LMSD as 10,11-dihydro-20-dihydroxy-LTB4, 19-hydroxy-PGE1, 19-hydroxy-PGF2α, 20-hydroxy-PGF2α, and TxB2 (Table S7). Alignment in DT between precursor and product ions allowed identification as TxB2 (Figure 5 E) and confirmed our strategy of flagging compounds as structural similar to those in the internal database based on their chromatography and mobility characteristics. On the other hand, features flagged as new compounds are identical in m/z and similar in DTCCSN2 but differ in RT, as illustrated on m/z 351.2177 (Figure 5 F, G).
Conclusion
In this study we present a comprehensive LC-DTIM-MS method for both profiling and quantification of oxylipins and their precursors FAs in biological samples. Using replicate acquisitions of lipid standards across different days, an average RSD of 0.19%, 0.21%, 0.20% and 0.18% for the DTCCSN2 of the species [M-H]-, [M-H+CH3CO2Na]-, [2M-H]- and [2M-2H+Na]- respectively, was obtained (Table S1). Further confirmation of the validity of our measurements comes from comparison with CCS values in the literature.12,14,24 This results in an average RSD below 2% (Table S8), which is the accepted error threshold for intra-lab CCS measurements. However, several species are found above this value, with the strongest outlier being lauric acid (RSD = 5.59%). Nonetheless, inhouse triplicate acquisitions of lauric acid resulted in highly repeatable measurements (RSD = 0.013-0.466, table S1). This high variability with the CCS reported in the literature could derive from a number of sources of uncertainty, including differences in the analytical details, which must be taken into consideration when building CCS libraries on standards and/or literature values if not enough experimental or computationally generated CCS information exists for comparison. However, the low internal RSDs allowed us to use DTCCSN2 in addition to RTs and accurate mass to identify oxylipins and FAs in two sets of biological samples. As the CCS value of a compound is a physical-chemical parameter and therefore instrument-independent, it can be used by others as additional lipid identifiers. We showed how DTCCSN2 values in combination with m/z can improve the identification process of oxylipins and FAs and allows to flag potential new members of this family. Moreover, addition of IM to the workflow allowed separation and identification of different lipid conformers, including dimers and solvent clusters, based on DT-alignment of product ions to their precursors. In addition, our study highlights the complexity of lipid analysis by LC-MS and the necessity of additional ion separation level to gain further insights into ionization, adduct formation and fragmentation mechanisms. Despite a higher number of detected features by LC-DTIM-MS compared to LC-MS alone, the results presented herein show the potential of this technique in simplifying lipid identification by using DTCCSN2 and DT-alignment of product and precursor ions as additional identifiers. To further automate the identification process, diagnostic product ions could be added to the library. Moreover, the constant improvement of in-silico tools for DTIM data analysis41–43 will allow the combination of our methodology with these computational techniques once oxylipins have been integrated in their training set.
Acknowledgment
The authors would like to thank Dr Hania Khouri and Sufyan Pandor for assisting with the Agilent Software. Work in the JLG lab is supported by the Medical Research Council, UK (MR/P011705/1), the National Institution of Health, USA (R01 ES022186 / ES / NIEHS NIH HHS / United States), and Agilent Technologies, USA (Thought Leader Award). CEB is supported by a Wellcome Trust Investigator Award (108045/Z/15/Z). VBO thanks the Wellcome Trust (203014/Z/16/Z). The RWF lab is supported by the British Heart Foundation, (RG/15/4/31268 and SP/10/011/28199).
References
- (1).Mosblech A, Feussner I, Heilmann I. Oxylipins: Structurally Diverse Metabolites from Fatty Acid Oxidation. Plant Physiol Biochem. 2009;47(6):511–517. doi: 10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- (2).Smith WL, Murphy RC. The Eicosanoids: Cyclooxygenase, Lipoxygenase, and Epoxygenase Pathways. Biochem Lipids, Lipoproteins Membr. 2002;(4):341–371. [Google Scholar]
- (3).Manev H, Chen H, Dzitoyeva S, Manev R. Cyclooxygenases and 5-Lipoxygenase in Alzheimer’s Disease. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(2):315–319. doi: 10.1016/j.pnpbp.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Greene ER, Huang S, Serhan CN, Panigrahy D. Regulation of Inflammation in Cancer by Eicosanoids. Prostaglandins Other Lipid Mediat. 2011;96(1–4):27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Theken KN, Schuck RN, Edin ML, Tran B, Ellis K, Bass A, Lih FB, Tomer KB, Poloyac SM, Wu MC, et al. Evaluation of Cytochrome P450-Derived Eicosanoids in Humans with Stable Atherosclerotic Cardiovascular Disease. Atherosclerosis. 2012;222(2):530–536. doi: 10.1016/j.atherosclerosis.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hammond VJ, O’Donnell VB. Esterified Eicosanoids: Generation, Characterization and Function. Biochimica et Biophysica Acta - Biomembranes. 2012 October;:2403–2412. doi: 10.1016/j.bbamem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ostermann AI, Willenberg I, Schebb NH. Comparison of Sample Preparation Methods for the Quantitative Analysis of Eicosanoids and Other Oxylipins in Plasma by Means of LC-MS/MS. Anal Bioanal Chem. 2015;407(5):1403–1414. doi: 10.1007/s00216-014-8377-4. [DOI] [PubMed] [Google Scholar]
- (8).Murphy RC, Barkley RM, Berry KZ, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Electrospray Ionization and Tandem Mass Spectrometry of Eicosanoids. Analytical Biochemistry. 2005 November;:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- (9).Hinz C, Liggi S, Griffin JL. The Potential of Ion Mobility Mass Spectrometry for High-Throughput and High-Resolution Lipidomics. Curr Opin Chem Biol. 2018;42:42–50. doi: 10.1016/j.cbpa.2017.10.018. [DOI] [PubMed] [Google Scholar]
- (10).Chouinard CD, Cruzeiro VWD, Beekman CR, Roitberg AE, Yost RA. Investigating Differences in Gas-Phase Conformations of 25-Hydroxyvitamin D3 Sodiated Epimers Using Ion Mobility-Mass Spectrometry and Theoretical Modeling. J Am Soc Mass Spectrom. 2017;28(8):1497–1505. doi: 10.1007/s13361-017-1673-4. [DOI] [PubMed] [Google Scholar]
- (11).Kapron J, Wu J, Mauriala T, Clark P, Purves RW, Bateman K. Simultaneous Analysis of Prostanoids Using Liquid Chromatography/High-Field Asymmetric Waveform Ion Mobility Spectrometry/Tandem Mass Spectrometry. Rapid Commun Mass Spectrom. 2006;20:1504–1510. doi: 10.1002/rcm. [DOI] [PubMed] [Google Scholar]
- (12).Di Giovanni JP, Barkley RM, Jones DNM, Hankin JA, Murphy RC. Tandem Mass Spectrometry and Ion Mobility Reveals Structural Insight into Eicosanoid Product Ion Formation. J Am Soc Mass Spectrom. 2018;29(6):1231–1241. doi: 10.1007/s13361-018-1927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jónasdóttir HS, Papan C, Fabritz S, Balas L, Durand T, Hardardottir I, Freysdottir J, Giera M. Differential Mobility Separation of Leukotrienes and Protectins. Anal Chem. 2015;87(10):5036–5040. doi: 10.1021/acs.analchem.5b00786. [DOI] [PubMed] [Google Scholar]
- (14).Kyle JE, Aly N, Zheng X, Burnum-Johnson KE, Smith RD, Baker ES. Evaluating Lipid Mediator Structural Complexity Using Ion Mobility Spectrometry Combined with Mass Spectrometry. Bioanalysis. 2018;10(5):279–289. doi: 10.4155/bio-2017-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Liggi S, Hinz C, Hall Z, Santoru ML, Poddighe S, Fjeldsted J, Atzori L, Griffin JL. KniMet - A Pipeline for the Processing of Chromatography-Mass Spectrometry Metabolomics Data. Metabolomics. 2018;14(52) doi: 10.1007/s11306-018-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Murphy RC, Raetz CRH, Russell DW, et al. LMSD: LIPID MAPS Structure Database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS Online Tools for Lipid Research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhou Z, Tu J, Zhu ZJ. Advancing the Large-Scale CCS Database for Metabolomics and Lipidomics at the Machine-Learning Era. Curr Opin Chem Biol. 2018;42:34–41. doi: 10.1016/j.cbpa.2017.10.033. [DOI] [PubMed] [Google Scholar]
- (19).Hinz C, Aldrovandi M, Uhlson C, Marnett LJ, Longhurst HJ, Warner TD, Alam S, Slatter DA, Lauder SN, Allen-Redpath K, et al. Human Platelets Utilize Cycloxygenase-1 to Generate Dioxolane A3, a Neutrophil Activating Eicosanoid. J Biol Chem. 2016;291(26):13448–13464. doi: 10.1074/jbc.M115.700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Meckelmann SW, Hellhake S, Steuck M, Krohn M, Schebb NH. Comparison of Derivatization/Ionization Techniques for Liquid Chromatography Tandem Mass Spectrometry Analysis of Oxylipins. Prostaglandins Other Lipid Mediat. 2017;130:8–15. doi: 10.1016/j.prostaglandins.2017.02.003. [DOI] [PubMed] [Google Scholar]
- (21).Berthold MR, Cebron N, Dill F, Gabriel TR. Studies in Classification, Data Analysis, and Knowledge Organization (GfKL 2007) Vol. 11. Springer; 2007. KNIME: The Konstanz Information Miner; pp. 319–326. [Google Scholar]
- (22).Infocom Corporation. ChemAxon/Infocom Marvin Extensions Feature. [accessed Apr 19, 2018]. https://chemaxon.com/
- (23).NIBR. RDKit KNIME integration. [accessed Apr 19, 2018]. http://www.rdkit.org/
- (24).Stow SM, Causon TJ, Zheng X, Kurulugama RT, Mairinger T, May JC, Rennie EE, Baker ES, Smith RD, McLean JA, et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Anal Chem. 2017;89(17):9048–9055. doi: 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gabelica V, Alfonso C, Barran PE, Benesch JLP, Bleiholder C, Bowers MT. Recommendations for Reporting Ion Mobility Mass Spectrometry Measurements. Mass Spectrom Rev. 2019 doi: 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Di Guida R, Engel J, Allwood JW, Weber RJM, Jones MR, Sommer U, Viant MR, Dunn WB. Non-Targeted UHPLC-MS Metabolomic Data Processing Methods: A Comparative Investigation of Normalisation, Missing Value Imputation, Transformation and Scaling. Metabolomics. 2016;12(5):1–14. doi: 10.1007/s11306-016-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application In1H NMR Metabonomics. Anal Chem. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- (28).Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, Spener F, van Meer G, Wakelam MJO, Dennis EA. Update of the LIPID MAPS Comprehensive Classification System for Lipids. J Lipid Res. 2009;50(Supplement):S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, et al. gplots: Various R Programming Tools for Plotting Data. https://cran.r-project.org/package=gplots .
- (30).R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- (31).Kyle JE, Zhang X, Weitz KK, Monroe ME, Ibrahim YM, Moore RJ, Cha J, Sun X, Lovelace ES, Wagoner J, et al. Uncovering Biologically Significant Lipid Isomers with Liquid Chromatography, Ion Mobility Spectrometry and Mass Spectrometry. Analyst. 2016;141(5):1649–1659. doi: 10.1039/C5AN02062J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Morgan LT, Thomas CP, Kühn H, O’Donnell VB. Thrombin-Activated Human Platelets Acutely Generate Oxidized Docosahexaenoic-Acid-Containing Phospholipids via 12-Lipoxygenase. Biochem J. 2010;431(1):141–148. doi: 10.1042/BJ20100415. [DOI] [PubMed] [Google Scholar]
- (33).Von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, Van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, et al. Rapid Induction of Inflammatory Lipid Mediators by the Inflammasome in Vivo. Nature. 2012;490(7418):107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Aldrovandi M, Hammond VJ, Podmore H, Hornshaw M, Clark SR, Marnett LJ, Slatter Da, Murphy RC, Collins PW, O’Donnell VB. Human Platelets Generate Phospholipid-Esterified Prostaglandins via Cyclooxygenase-1 That Are Inhibited by Low Dose Aspirin Supplementation. J Lipid Res. 2013;54(11):3085–3097. doi: 10.1194/jlr.M041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Thomas CP, Morgan LT, Maskrey BH, Murphy RC, Kühn H, Hazen SL, Goodall AH, Hamali HA, Collins PW, O’Donnell VB. Phospholipid-Esterified Eicosanoids Are Generated in Agonist-Activated Human Platelets and Enhance Tissue Factor-Dependent Thrombin Generation. J Biol Chem. 2010;285(10):6891–6903. doi: 10.1074/jbc.M109.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hong S, Gronert K, Devchand PR, Moussignac R-L, Serhan CN. Novel Docosatrienes and 17S-Resolvins Generated from Docosahexaenoic Acid in Murine Brain, Human Blood, and Glial Cells. Autacoids in Anti-Inflammation. J Biol Chem. 2003;278(17):14677–14687. doi: 10.1074/jbc.M300218200. [DOI] [PubMed] [Google Scholar]
- (37).Schwab JM, Chiang N, Arita M, Serhan CN. Resolvin E1 and Protectin D1 Activate Inflammation-Resolution Programmes. Nature. 2007;447(7146):869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hong S, Lu Y, Yang R, Gotlinger KH, Petasis Na, Serhan CN. Resolvin D1, Protectin D1, and Related Docosahexaenoic Acid-Derived Products: Analysis via Electrospray/Low Energy Tandem Mass Spectrometry Based on Spectra and Fragmentation Mechanisms. J Am Soc Mass Spectrom. 2007;18(1):128–144. doi: 10.1016/j.jasms.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).O’Donnell VB, Murphy RC, Watson SP. Platelet Lipidomics. Circ Res. 2014;114(7):1185–1203. doi: 10.1161/CIRCRESAHA.114.301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Slatter DA, Aldrovandi M, O’Connor A, Allen SM, Brasher CJ, Murphy RC, Mecklemann S, Ravi S, Darley-Usmar V, O’Donnell VB. Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation. Cell Metab. 2016;23(5):930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zhou Z, Shen X, Chen X, Tu J, Xiong X, Zhu ZJ. LipidIMMS Analyzer: Integrating Multi-Dimensional Information to Support Lipid Identification in Ion Mobility - Mass Spectrometry Based Lipidomics. Bioinformatics. 2019;35(4):698–700. doi: 10.1093/bioinformatics/bty661. [DOI] [PubMed] [Google Scholar]
- (42).Zhou Z, Tu J, Xiong X, Shen X, Zhu Z-J. LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision To Support Ion Mobility–Mass Spectrometry-Based Lipidomics. Anal Chem. 2017;89(17):9559–9566. doi: 10.1021/acs.analchem.7b02625. [DOI] [PubMed] [Google Scholar]
- (43).Blaženović I, Shen T, Mehta SS, Kind T, Ji J, Piparo M, Cacciola F, Mondello L, Fiehn O. Increasing Compound Identification Rates in Untargeted Lipidomics Research with Liquid Chromatography Drift Time-Ion Mobility Mass Spectrometry. Anal Chem. 2018;90(18):10758–10764. doi: 10.1021/acs.analchem.8b01527. [DOI] [PubMed] [Google Scholar]