Summary
Nuclear RNA interference pathways work together with histone modifications to regulate gene expression and enact an adaptive response to transposable RNA elements. In the germline, nuclear RNAi can lead to trans-generational inheritance (TEI) of gene silencing. We identified and characterized a family of nuclear Argonaute-interacting proteins (ENRIs) that control the strength and target specificity of nuclear RNAi in C. elegans, ensuring faithful inheritance of epigenetic memories. ENRI-1/2 prevent misloading of the nuclear Argonaute NRDE-3 with small RNAs that normally effect maternal piRNAs, which prevents precocious nuclear translocation of NRDE-3 in the early embryo. Additionally, they are negative regulators of nuclear RNAi triggered from exogenous sources. Loss of ENRI-3, an unstable protein mostly expressed in the male germline, misdirects the RNAi response to transposable elements and impairs TEI. The ENRIs determine the potency and specificity of nuclear RNAi responses by gating small RNAs into specific nuclear Argonautes.
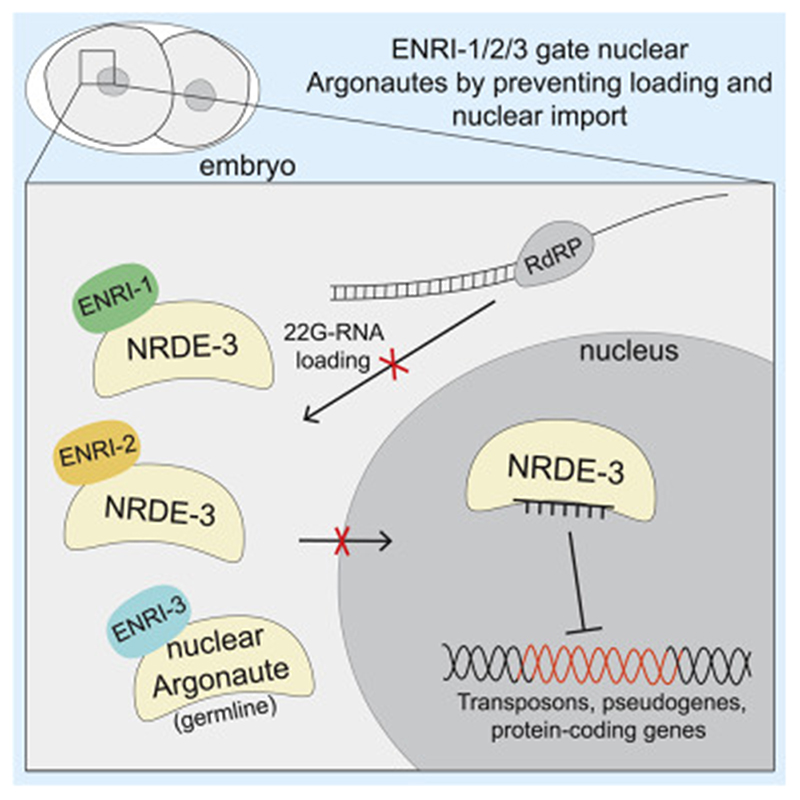
Graphic abstract.
Introduction
RNA interference (RNAi) pathways comprise a diverse network of gene regulatory mechanisms, the specificity of which is determined by small RNAs. When loaded onto Argonaute proteins, small RNAs regulate gene expression, antagonize viral infection and transposable elements, and preserve genome integrity. Small RNAs can be endogenous or be derived from exogenous sources such as experimental or viral dsRNA. Once loaded and guided to their targets via complementary base-pairing between the mRNA target and small RNA, Argonautes effect gene silencing either by slicing mRNA targets through their endonuclease activity, or through recruitment of gene silencing effectors (Burkhart et al., 2011; Gu et al., 2012; Guang et al., 2010; Guang et al., 2008; Hall et al., 2002; Meister et al., 2004; Verdel et al., 2004). Distinct classes of small RNAs that differ in their biogenesis and structure associate with different groups of Argonautes. This functional specialization is reflected through the phylogeny of Argonautes, which segregate as distinct clades (Yigit et al., 2006). Small RNA ‘sorting’ into specific Argonautes occurs in a wide range of species but has been best characterized in Drosophila melanogaster, where Ago1 and Ago2 are loaded with miRNAs and siRNAs, respectively (Hammond et al., 2001; Okamura et al., 2004; Tomari et al., 2007). Functional specialization and sorting of small RNAs is less clear in mammals as in several other species.
In C. elegans, RNAi is initiated when long double stranded triggers are processed into siRNAs and loaded into the ‘primary’ subset of Argonautes. Endogenous siRNAs are often formed through the action of an RNA-dependent RNA polymerase (RdRP) which uses target mRNAs, including coding loci, pseudogenes, transposable elements, and unannotated loci, as templates to generate siRNAs (Duchaine et al., 2006; Thivierge et al., 2011; Vasale et al., 2010). In the embryo, ERGO-1 is the primary Argonaute that binds to 26 nucleotide-long primary endogenous siRNAs (26G-RNAs), while the spermatogenic 26G-RNAs are loaded onto ALG-3/4 (Conine et al., 2010; Han et al., 2009; Vasale et al., 2010). Successful target recognition by these Argonautes results in the generation of abundant ‘secondary’ siRNAs named 22G-RNAs by the RdRPs RRF-1 and EGO-1. 22G-RNAs program the WAGO Argonautes, which make up 12 out of the 27 Argonautes expressed in C. elegans. Argonautes are limiting for the silencing activities of RNAi, and endogenous and exogenous RNAi cascades compete for the shared WAGOs (Duchaine et al., 2006; Gu et al., 2009; Lee et al., 2006; Yigit et al., 2006). When 26G endo-siRNA biogenesis is lost, a pool of WAGOs is freed, resulting in an enhanced RNAi (eri) response (Duchaine et al., 2006; Thivierge et al., 2011; Yigit et al., 2006).
Evidence across a wide range of species supports the functional importance of RNAi mechanisms in the nucleus (Guang et al., 2008; Hall et al., 2002; Verdel et al., 2004; Volpe et al., 2002). In C. elegans, a specialized subset of WAGOs can localize to the nucleus to trigger transcriptional gene regulation (nuclear RNAi). NRDE-3 is loaded in the cytoplasm and translocates to the nucleus where it associates with nascent chains of pre-mRNA and recruits other NRDE proteins to effect silencing through the accumulation of silencing H3K9me3 and the formation of heterochromatin (Burkhart et al., 2011; Guang et al., 2010; Guang et al., 2008). HRDE-1 plays an analogous role in the germline, where it is required for the inheritance of silencing phenotypes triggered by the exo-RNAi or the piRNA pathways (Ashe et al., 2012; Buckley et al., 2012; Burton et al., 2011; Lee et al., 2012). piRNAs, also referred to as 21U-RNAs, target a number of protein coding genes and transposable elements. Similar to 26G-RNAs, target recognition by 21U-RNAs triggers the biogenesis of 22G-RNAs that silence targets through transcriptional mechanisms (Ashe et al., 2012; Shirayama et al., 2012). In order to provide a faithful memory of non-self and deleterious DNA sequences to progeny, information about the epigenetic status and small RNAs must be passed through the gametes into embryos. While it has been shown that piRNAs/21U-RNAs are maternally inherited in C. elegans (de Albuquerque et al., 2015), how this response is established and regulated in the embryo is unknown.
Here, we used a combination of proteomics, biochemistry, cell biology, and genetics to identify a family of nuclear Argonaute interacting proteins that maintains genome integrity and determines the potency and specificity of nuclear RNAi. They do so by gating Argonautes from small RNAs in specific cell types and at specific moments. Their loss in the germline impacts on long-term epigenetic inheritance and transposable element silencing, and results in the misloading of embryonic NRDE-3 with small RNAs that normally effect maternal piRNAs.
Results
NRDE-3 proteomics reveal interactions with an uncharacterized protein family
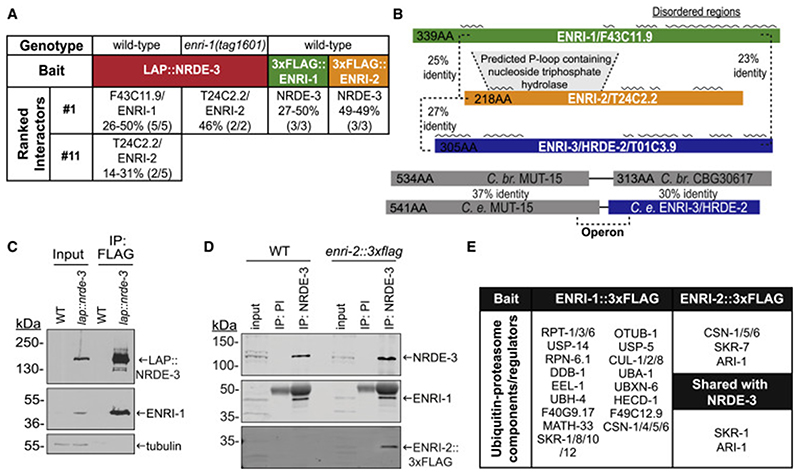
To examine the protein-protein interactions of the nuclear Argonaute NRDE-3, a LAP (GFP::3xFLAG)-tagged NRDE-3 was immunoprecipitated (IP) from embryonic extracts (Figure S1A). Recovered proteins were analyzed using Multi-Dimensional Protein Identification Technology (MuDPIT) (Washburn et al., 2001). From 44 candidate interacting proteins, the uncharacterized protein F43C11.9 was consistently detected in all replicates, and with a greater peptide coverage than any other protein (Figure 1A, Table S1). We renamed this protein for its mutant phenotype (see Figure 2), Enhanced Nuclear RNAi 1 (ENRI-1) (Figure 1A). Interestingly, T24C2.2, a protein detected in 2/5 NRDE-3 purifications is similar (25% identity) to ENRI-1, suggesting that they may represent members of a protein family that have redundant or compensatory functions (Figure 1A, 1B). In agreement with this, in the absence of ENRI-1, T24C2.2 becomes the top ranked interacting partner of NRDE-3 (Figure 1A and Table S1). For this reason and in light of partial functional compensation (below), we renamed T24C2.2 as ENRI-2. ENRI-1 and ENRI-2 also share homology with HRDE-2 (T01C3.9), a protein recently reported to be involved in trans-generational epigenetic inheritance (TEI) (Spracklin et al., 2017). This protein will be identified herein as ENRI-3. In C. elegans, ENRI-3 is encoded by the downstream ORF of the bicistronic locus that encodes MUT-15 (Figure 1B), a protein involved in the biogenesis of ERGO-1 class 26G-RNAs and in the broader accumulation of WAGO 22G-RNAs (Zhang et al., 2011). The syntenic locus is predicted to encode a single ORF in C. briggsae. The ENRI proteins bear several predicted disordered regions, and while they do not contain a distinctive recognizable domain, they bear some similarity with a P-loop containing nucleoside triphosphate hydrolase motif, which is most recognizable in ENRI-2 (Figure 1B).
Figure 1. ENRI-1 and ENRI-2 are uncharacterized NRDE-3 interacting proteins.
(A) Table indicating the proteins that were IPed in the indicated genetic backgrounds. Peptide coverage of the listed interactors is indicated in percentage, as well as the number of replicates in which the interactor was found. (B) Percent identity between each protein is listed, and predicted disordered regions marked with a black wavy line above each protein. Protein schematics are to scale. (C) Input and FLAG IPs from wild-type or lap::nrde-3 embryos were probed with an antibody to ENRI-1 to confirm the interaction observed in MudPIT. LAP::NRDE-3 was detected with a GFP antibody. (D) Wild-type and enri-2::3xflag embryo lysates were subjected to NRDE-3 IP using pre-immune (PI) serum as a negative control. Input and IPs were probed with antibodies to NRDE-3, ENRI-1, and FLAG. (E) Table depicting ubiquitin-proteasome components found to interact with ENRI-1 and ENRI-2 and those shared with LAP::NRDE-3 (bolded). See also Figure S1, Table S1 and Table S2.
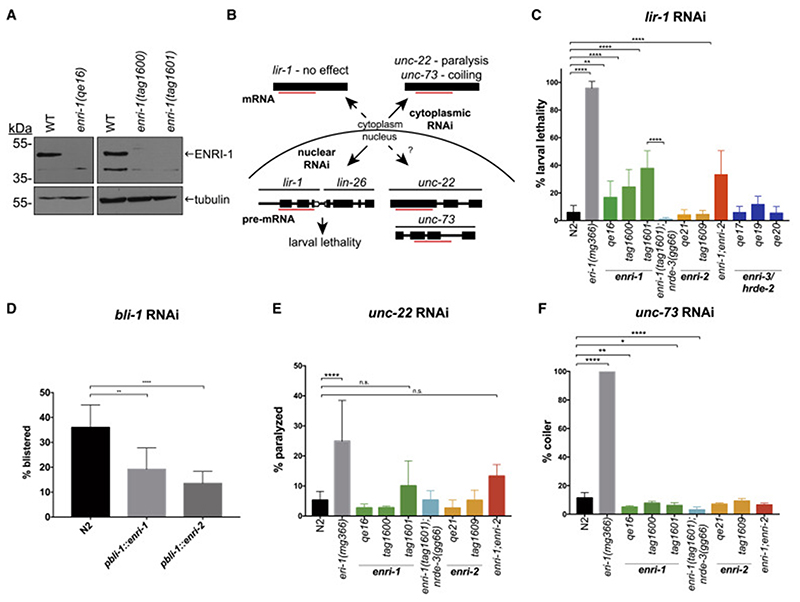
Figure 2. ENRI-1 and ENRI-2 suppress nuclear RNAi.
(A) Western blots of lysates from wild-type and enri-1 knock-out embryos probed with antibodies to ENRI-1 and tubulin. (B) Schematic detailing the contributions of cytoplasmic and nuclear RNAi and expected outcomes. (C) Quantification of animals that survive lir-1 RNAi (n >15 biological replicates). The enri-1;enri-2 double mutant comprised alleles enri-1(tag1601) and enri-2(tag1609). (D) Blistering of animals was scored following bli-1 RNAi (n > 3 biological replicates). (E) Complete paralysis of animals exposed to unc-22 RNAi was quantified (n > 6 biological replicates). (F) Animals with a coiler phenotype were quantified (n > 3 biological replicates). For all RNAi assays: Tukey’s multiple comparisons test was conducted with p-value: 0.05 – 0.0332 (*), 0.0333 – 0.0021 (**), 0.0022 – 0.0002 (***), < 0.0001 (****), and data is depicted as mean ± SD. See also Figure S2 and S3.
We generated polyclonal antisera against ENRI-1 and NRDE-3 and confirmed their specificity in western blots and IP (Figure S1B). The interaction between NRDE-3 and ENRI-1 was confirmed by co-IP on endogenous NRDE-3 and using a LAP-NRDE-3 transgenic strain (Figure 1C, D). Interaction between NRDE-3 and ENRI-2 was confirmed using a FLAG-tagged endogenous enri-2 allele generated through CRISPR (Figure 1D). We note that expression of ENRI-1 was higher in the input fraction of the transgenic LAP::NRDE-3 strain than in wild-type animals (Figure 1C). However, ENRI-1 levels remained unchanged in a nrde-3 null mutant (Figure S1C), and neither loss of enri-1 nor enri-2 affected the expression of endogenous NRDE-3 (Figure S1D).
To further detail the interactions of ENRI-1 in the nuclear RNAi pathway, we generated a strain expressing transgenic ENRI-1::3xFLAG. ENRI-1::3xFLAG was IPed from embryonic extracts and purified proteins were analyzed by MuDPIT analyses (Figure S1E, Table S2). NRDE-3 was the top interactor and the only Argonaute found in the datasets suggesting that, at least in embryos, ENRI-1 does not act through multiple Argonautes (Figure 1A, Table S2). Neither ENRI-1, nor ENRI-2::3xFLAG co-IPed with endogenously tagged ALG-2::3xFLAG or ERGO-1, two abundant embryonic Argonautes (Figure S1G, H). This data supports the selectivity of the ENRI-1/2 interaction with the nuclear Argonaute NRDE-3 in the embryo.
Interestingly, a number of proteins associated with the ubiquitin-proteasome pathway were also found to interact with ENRI-1. These include subunits of the 26S proteasome complex (RPT-1/3/6, F40G9.17, RPN-6.1), deubiquitinating enzymes (UBH-4, USP-14/5, MATH-33, OTUB-1), and components of E3 ligase complexes (DDB-1, of the CUL-4/DDB-1 E3 complex; CUL-1, SKR-1/10/8/12, of the SCF E3 ligase complex) among others (Figure 1E). We performed the same experiment with ENRI-2::3xFLAG (Figure S1F, Table S2). NRDE-3 was detected as the top interactor of ENRI-2 in embryos and young adults (Figure 1A, Table S2). ENRI-1 and ENRI-2 shared 15 putative common interactors, while ENRI-2 only shared 2 putative interactions with NRDE-3 (Figure S1I). A mass spectrometry survey for post-translational modifications (PTMs) of immunopurified ENRI-1 did not detect ubiquitylated residues but identified serines 224 and 283 as active phosphorylation sites (Figure S1J).
We conclude that ENRI-1 and ENRI-2 are predominant interactors of NRDE-3 in the C. elegans embryo. We further observed that ENRI-1 is phosphorylated and broadly interacts with the ubiquitin-proteasome machinery. The functional significance of ENRI-1 PTM and their possible interplay will be reported elsewhere.
ENRI-1 and ENRI-2 repress somatic nuclear RNAi
NRDE-3 is required for nuclear RNAi in somatic tissues (Guang et al., 2010; Guang et al., 2008). To examine possible functions for the ENRI proteins in nuclear RNAi pathways, we generated CRISPR knockout alleles for enri-1, enri-2, and enri-3 (Figure S2A). Deletion of ENRI-1 was validated by western blot (Figure 2A). enri-1 and enri-2 null mutants presented no gross phenotypes and their brood sizes at 25°C were comparable to those of wild-type animals (Figure S2B, C). enri-3 null mutants had a mortal germline phenotype when grown at 25°C, as previously reported, likely because of a breakdown in germline genome integrity (Figure S2B, C) (Spracklin et al., 2017).
To test if the enris play a role in nuclear RNAi, we tested the ability of enri mutants to silence the somatic lir-1-lin-26 nuclear-localized polycistronic RNA. Loss of lir-1 is viable, whereas loss of lin-26 results in larval lethality. Accordingly, lir-1 RNAi leads to larval lethality through the silencing of the entire nuclear lir-1-lin-26 polycistron (Bosher et al., 1999) (Figure 2B). For all RNAi assays, eri-1(mg366) was used as a positive control. Loss of ERI-1 relieves the competition between the exogenous and endogenous RNAi pathways leading to an enhanced RNAi phenotype (Duchaine et al., 2006). Notably, lir-1 RNAi in all three enri-1 alleles led to significantly enhanced larval lethality (20-40%) compared to wild-type animals (~5%) (Figure 2C). Enhanced lir-1-lin-26 silencing was dependent on the presence of NRDE-3, as a double enri-1;nrde-3 mutant remained fully viable following lir-1 dsRNA exposure (Figure 2C). Both enri-2 knockout alleles behaved similarly to wild-type in lir-1 RNAi (Figure 2C) and larval lethality in a double enri-1;enri-2 mutant was not significantly different from the enri-1(tag1601) mutant under standard RNAi conditions. However, when potency of RNAi was reduced by dilution, the enri-1;enri-2 double mutant significantly enhanced lir-1-lin-26 silencing compared to enri-1(tag1601) alone (~25% vs. ~10%) (Figure S3A). This suggests a partial compensation by enri-2 for enri-1 loss, in agreement with the biochemical replacement observed in our proteomics survey. None of the enri-3 alleles tested displayed enhanced larval lethality following lir-1 RNAi and had similar levels of lethality to wild-type animals (Figure 2C, S3A). Importantly, since mut-15 is required for exogenous RNAi (Sijen et al., 2007), we infer that mut-15 is not being disrupted in our enri-3 mutants. We next investigated the impact of enforced ectopic expression of ENRI-1 and ENRI-2 on nuclear RNAi. For this, we performed RNAi against bli-1, whose silencing is dependent on nrde-3 (Raman et al., 2017), on animals expressing ENRI-1 and ENRI-2 under the bli-1 promoter. While 35% of wild-type animals displayed a blistered phenotype, pbli-1::enri-1 and pbli-1::enri-2 animals exhibited 20% and 15% blistering, respectively (Figure 2D). Finally, transgenic expression of ENRI-1 in the eri-1(mg366) background led to a mild but significant impairment of lir-1 RNAi (Figure S3B). These results further confirm the inhibition of NRDE-3-dependent nuclear RNAi by the ENRI-1 and ENRI-2 proteins.
We next tested whether loss of enri-1 or enri-2 had an effect on the silencing of cytoplasmic RNAi targets using unc-22, unc-73 (Figure 2E, F) and dpy-11 (Figure S3C) RNAi assays. Silencing of these targets occurs predominantly in the cytoplasm as they are not completely dependent on nrde-3 (Guang et al., 2008; Zhuang et al., 2013). unc-22 RNAi leads to paralysis in animals, unc-73 RNAi triggers a coiling phenotype, and dpy-11 RNAi leads to a ‘dumpy’ phenotype, wherein the animals are shorter and fatter than wild-type animals. In unc-22 RNAi, enri-1, enri-2, and enri-1;enri-2 mutants did not behave significantly different than wild-type animals (Figure 2E). However, enri-1(qe16), enri-1(tag1600), and enri-1(tag1601);nrde-3(gg66) were marginally impaired in unc-73 silencing, while enri-2 mutants did not behave differently from wild-type animals (Figure 2F). enri-1 animals treated with dpy-11 RNAi had wild-type responses, except for a mild reduction in the number of ‘super dumpy’ worms in enri-1(qe16) (Figure S3C). In these assays, enri-2 mutants also exhibited a reduction in ‘super dumpy’ worms. The most drastic decrease in dumpy worms was in the double enri-1;nrde-3 mutant, which correlates well with previous work showing that nrde-3(gg66) is deficient in dpy-11 RNAi (Zhuang et al., 2013) (Figure S3C). Though enri-1 and enri-2 mutants are mildly impaired in silencing assays on some cytoplasmic RNAi targets, the enhanced RNAi phenotype of enri-1 and enri-1;enri-2 mutants is specific to nuclear RNAi. The different impact of enri-1 and enri-2 mutants also suggests that ENRI-1 and ENRI-2 may have partially overlapping, but different expression domains, and that nuclear RNAi output varies across cell types and physiological domains.
ENRI-1 and ENRI-2 bind to unloaded NRDE-3 in the cytoplasm
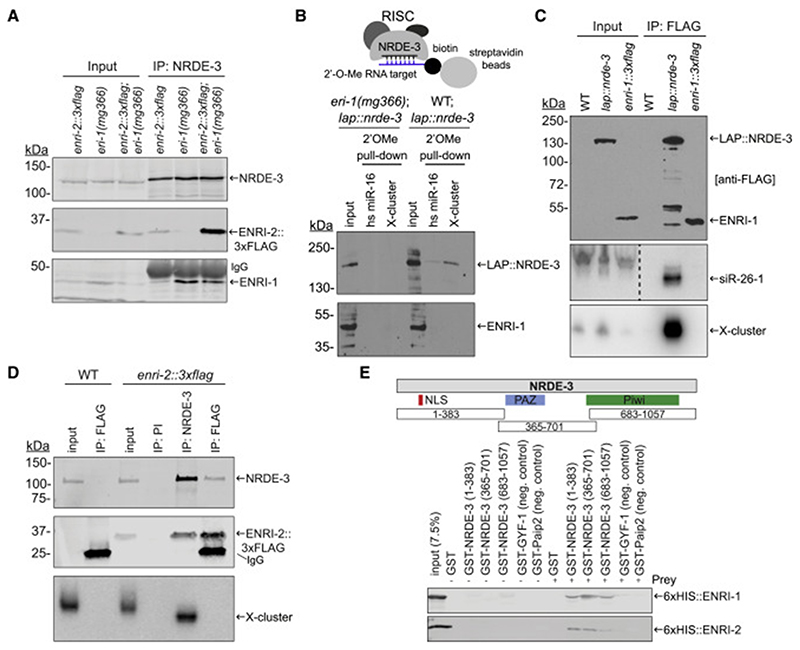
Since our proteomic data indicated that ENRI-1 is a prominent interacting partner of NRDE-3, we wondered whether ENRI-1 could also associate with 22G-RNAs. We first tested whether the interaction with ENRI-1 is dependent on the loading of NRDE-3 with small RNAs. For this, we took advantage of an eri-1 mutant (eri-1(mg366)) which fails to accumulate 26G/22G endo-siRNA as a result of defective biogenesis (Duchaine et al., 2006; Lee et al., 2006). In this background, and in the absence of exogenous siRNAs, NRDE-3 remains unloaded (Guang et al., 2008; Vasale et al., 2010). Endogenous NRDE-3 IP from the eri-1 embryos led to a striking enrichment of both ENRI-1 and ENRI-2 in comparison to wild-type (Figure 3A), suggesting that ENRI-1 and ENRI-2 specifically interact with unloaded NRDE-3. To further validate this, we captured the nuclear RISC through affinity capture using a 2’-O-methylated target analog of an abundant NRDE-3 target, the X-cluster (Duchaine et al., 2006; Guang et al., 2008). NRDE-3 was captured through pull-down with this bait, but ENRI-1 was not (Figure 3B), indicating that ENRI-1 is not a component of the nuclear RISC. Next, we tested the interaction of ENRI-1 and ENRI-2 with known NRDE-3-associated small RNAs. For this, we immunoprecipitated ENRI-1::3xFLAG, isolated the associated small RNAs and performed northern blots using radioactive probes complementary to 22G-RNAs derived from the X-cluster and sir-26-1 (which detects both 26G and the overlapping 22G sequences (Vasale et al., 2010)). These small RNAs did not co-IP with ENRI-1 (Figure 3C), and IP of endogenous ENRI-1 confirmed this result (Figure S3D). In contrast, both small RNAs could be abundantly detected in LAP::NRDE-3 IP fractions recovered under the same conditions. Similar results were observed with endogenously tagged ENRI-2::3xFLAG and endogenous NRDE-3. Both ENRI-2::3xFLAG and X-cluster 22G-RNAs co-IPed with endogenous NRDE-3, whereas no small RNAs could be detected in ENRI-2::3xFLAG IP (Figure 3D).
Figure 3. ENRI-1 and ENRI-2 interact with unloaded NRDE-3.
(A) Total lysates and NRDE-3 IPs from enri-2::3xflag, eri-1(mg366), and enri-2::3xflag;eri-1(mg366) embryos were probed with NRDE-3, FLAG and ENRI-1 antibodies. (B) Top: Schematic of the 2’O-methyl pull-down. Bottom: Input and pull-down fractions were probed with FLAG and ENRI-1 antibodies. (C) RNA extracted from FLAG IPs performed in transgenic lap::nrde-3 and enri-1::3xflag embryos was probed for 22G-RNAs mapping to siR-26-1 and X-cluster. FLAG antibody was used to detect both LAP::NRDE-3 and ENRI-1::3xFLAG. Dashed line indicates an unrelated lane was removed from the siR-26-1 northern blot. (D) RNA extracted from NRDE-3 and ENRI-2::3xFLAG IPs was probed for the 22G-RNAs mapping to the X-cluster. PI = pre-immune serum. NRDE-3 antibody and FLAG antibody were used to detect NRDE-3 and ENRI-2::3xFLAG respectively. (E) Top: schematic outlining the fragments of NRDE-3 fused to GST. Bottom: 50% of the pull-down was loaded onto a 10% gel for western blot analysis using an anti-6xHIS antibody. The other 50% was loaded onto a gel for Coomassie staining (Figure S3E, F). Each pull-down was performed at least 4 times and a representative blot for each is shown. See also Figure S3.
Altogether, these results indicate that the fraction of NRDE-3 bound to ENRI-1 and ENRI-2 is not loaded with small RNAs, and that these proteins interact with NRDE-3 prior to- and/or in the absence of loading with small RNAs.
ENRI-1 and ENRI-2 directly and specifically interact with NRDE-3 over multiple sites
The enhanced nuclear RNAi phenotype in enri-1 and -2 mutants and the fact that they associate with unloaded NRDE-3 led us to hypothesize that ENRI-1 and ENRI-2 may be preventing NRDE-3 loading through direct interaction. We designed three GST- fusions of NRDE-3 overlapping the nuclear-localization signal, the PAZ and the Piwi domains (Figure 3E). We then used them as baits in GST pull-downs against recombinant full-length ENRI-1 and ENRI-2. Both ENRI-1 and -2 bound to all three GST-NRDE-3 fragments, albeit to different degrees (Figures 3E, S3E, S3F), but neither ENRI-1 nor ENRI-2 bound to any of the GST- fusions used as controls. ENRI-1 interacted better with GST-NRDE-3 (365-701), which overlaps the PAZ domain, than with the other two fragments. We reason that the direct interaction of ENRI proteins with NRDE-3 over several of its key determinants are likely to impact on interactions with small RNAs and/or their loading machinery.
ENRI-1 and ENRI-2 preempt precocious NRDE-3 translocation in the early embryo
We next examined the expression and localization of ENRI-1 and ENRI-2 using endogenously tagged GFP fusion alleles. The expression of ENRI-1::GFP was strongest in the cytoplasm of embryos and in oocytes (Figure 4A). No nuclear localization of ENRI-1::GFP could be detected. Imaging of ENRI-2::GFP animals also revealed expression in both embryos and the female germline with a more diverse sub-cellular localization (Figure 4B, C). In early blastomeres (1-8 cell embryo) ENRI-2::GFP assumed a diffuse cytoplasmic localization while it localized to the nucleus beyond the 8-cell stage with an uneven expression across the different blastomere lineages (Figure 4B). In the germline, ENRI-2::GFP was predominantly nuclear in the maturing oocytes, thus diverging from the ENRI-1 localization (Figure 4C). These results indicate a cytoplasmic localization of ENRI-1 and ENRI-2 in the early embryo and distinct sub-cellular localization in the oocyte. It also hints towards cell-fate specific degradation and/or re-localization of ENRI-2 during later embryonic development, which may in turn reflect roles in other pathways or processes.
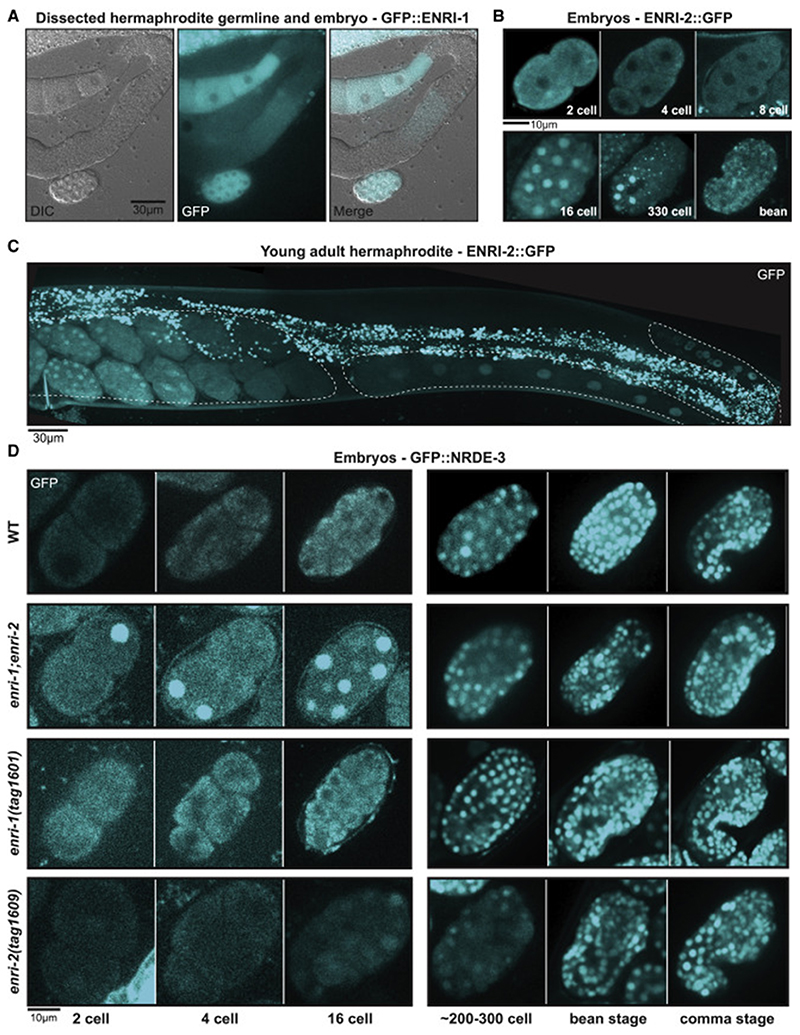
Figure 4. ENRI-1 and ENRI-2 localize to embryos and prevent premature nuclear translocation of NRDE-3.
(A) ENRI-1::GFP localization in a dissected young adult hermaphrodite germline. (B) ENRI-2::GFP expression in various embryo stages. (C) ENRI-2::GFP expression in the young adult hermaphrodite. Germline outline is marked by dashed lines. (D) Localization of LAP::NRDE-3 in developing embryos in indicated genetic backgrounds. Scale bars for reference.
We then sought to determine whether loss of ENRI-1 or ENRI-2 affects the localization of NRDE-3 in the embryo. NRDE-3 localizes to the nucleus in differentiated seam cells and this localization depends on its programming by 22G-RNAs (Guang et al., 2008). However, its embryonic localization had not been studied. Towards this, we used a strain expressing a GFP::NRDE-3 fusion from a single-copy, integrated transgene (Guang et al., 2008). In a wild-type background GFP::NRDE-3 remained cytoplasmic in the early embryo and only re-localized in the nucleus in the 30-80 cells embryo (Figure 4D). Strikingly, in the absence of both enri-1 and enri-2, GFP::NRDE-3 localized to the nucleus at earlier embryonic stages (1-8 cells) than in wild-type embryo (Figure 4D). In the enri-1 single mutant alleles, GFP::NRDE-3 localization remained unchanged, whereas a faint nuclear localization could already be visible at the 2-cell stage in enri-2 mutants (Figure 4D). While differences in overall GFP signal were noticed in the single enri-2 mutant embryos, an impact on endogenous NRDE-3 expression was not supported by western blot using the NRDE-3 antibody (Figure S1D) and may thus be linked to the transgene or strain. Altogether, this data points to roles for ENRI-1 and ENRI-2 in preventing the premature nuclear translocation of NRDE-3 in the embryo.
ENRI-1 and ENRI-2 prevent maternal mis-programming of NRDE-3 in early embryo
Our model thus far points to ENRI-1 and ENRI-2 binding to unloaded NRDE-3 and preventing its nuclear translocation. To investigate the global impact of ENRI proteins on small RNA loading in NRDE-3 in embryos, we sequenced libraries generated from total small RNAs and from LAP::NRDE-3-bound small RNAs in wild-type and enri-1;enri-2 embryos. 22G-RNAs were included in libraries by using the 5’ independent library preparation method (Figure S4) and small RNA reads were normalized to the overall number of reads (RPM). Small RNAs were mapped to individual targets and changes in uniquely allocated 22G-RNAs were plotted. Sub-groups of 22G-RNAs were up- or down-regulated in total RNA libraries from enri-1;enri-2 double mutant embryos (Figures 5A, S6A), whereas no significant change was detected in 21U-RNAs or 26G-RNAs populations (Figure S5). Specifically, 22G-RNAs directed against 801 and 1074 targets were significantly upregulated and downregulated, respectively (Figure 5A). Since total 22G-RNAs populations reflect associations with all embryonic WAGOs, we looked more specifically at the identity and abundance of 22G-RNAs associated with NRDE-3. Overall 22G-RNAs directed against 275 target mRNAs were significantly over-represented, and only 89 were depleted from enri-1;enri-2 in comparison to wild-type embryonic NRDE-3 IP libraries (Figure 5B, E). This result shows that NRDE-3 binds to more 22G-RNAs overall in the enri-1;enri-2 embryo.
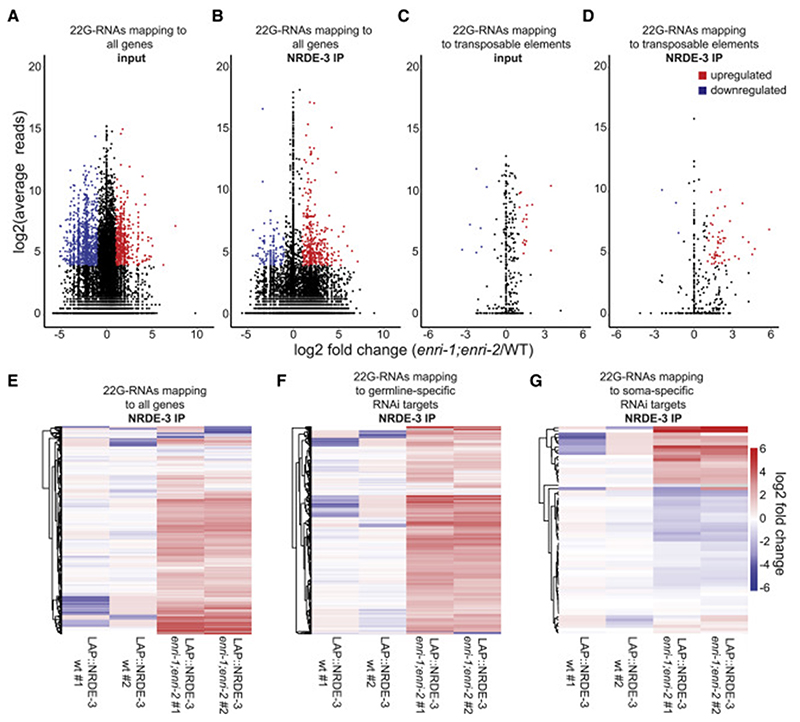
Figure 5. Loss of the enri genes leads to deregulation of NRDE-3-associated small RNAs.
(A) (B) MA plot (where M is the difference between log intensities and A is the average log intensity for a dot in the plot) of (A) input or (B) FLAG-IP 22G-RNA reads mapping antisense to individual genes from LAP::NRDE-3 (enri-1(tag1609);enri-2(tag1609)) embryos compared to 22G-RNA reads from LAP::NRDE-3 (WT). (C) (D) MA plot of (C) input or (D) FLAG-IP 22G-RNA reads mapping antisense to annotated transposable elements from LAP::NRDE-3 (enri-1;enri-2) embryos compared to 22G-RNA reads from LAP::NRDE-3 (WT). Shown in red are significantly upregulated 22G-RNAs and shown in blue are significantly downregulated 22G-RNAs. Only 22G-RNAs that displayed log2(fold-change) > 1, log2(average reads) > 4 and padj < 0.05 were considered significantly changed from wild-type. (E) Heat map of all 22G-RNAs mapping to individual genes recovered in LAP::NRDE-3 IPs. (F) Heat map of 22G-RNAs mapping to all annotated germline-specific RNAi targets recovered in LAP::NRDE-3 IPs. (G) Heat map of 22G-RNAs mapping to all annotated soma-specific RNAi targets recovered in LAP::NRDE-3 IPs. See also Figures S4, S5, and S6.
22G-RNAs target a variety of transcripts, which includes mRNAs but also repeated sequences such as transposable elements (Gu et al., 2009). 22G-RNAs directed against transposable elements were enriched in both the overall total embryonic small RNA libraries and in NRDE-3 IP associated 22G-RNAs in the enri-1;enri-2 double mutant. 22G-RNAs mapping to 17 transposable elements were significantly enriched in the enri-1;enri-2 mutant in comparison with wild-type (Figure 5C). Additionally, 22G-RNAs targeting 53 transposable elements were significantly enriched in the NRDE-3 IP (Figure 5D). This was somewhat unexpected given that transposon silencing occurs predominantly in the germline (Emmons and Yesner, 1984; Sijen and Plasterk, 2003). In the germline, thousands of genes are targeted by 22G-RNAs through the WAGO Argonautes (Claycomb et al., 2009; Gu et al., 2009). In enri-1;enri-2 mutants, we noticed an enrichment for germline-specific 22G-RNAs (Figure 5F, S6B), meaning these 22G-RNAs are normally enriched in the germline yet are abnormally upregulated in embryos in our mutants (Gu et al., 2009; Zhang et al., 2011). 22G-RNAs directed against 224 germline RNAi targets were significantly upregulated in the embryo of enri-1;enri-2 double mutant in comparison with wild-type, whereas only 6 were downregulated (Figure S6B). The same enrichment trend was visible in NRDE-3 IP libraries, where 138 of the 275 (~50%) overrepresented 22G-RNAs in NRDE-3 IPs were germline RNAi targets (Figure 5F). These results indicate that 22G-RNAs that are normally detected in the germline program NRDE-3 in the absence of ENRI-1 and ENRI-2. Finally, some changes in soma-specific 22G-RNAs could also be detected, with both up- and down-regulation of specific 22G-RNAs in enri-1;enri-2 mutant embryo (31 up- and 44 down-regulated) (Figure S6C). Similar changes were also detected in NRDE-3 IP small RNA libraries (Figure 5G).
Altogether, these findings indicate that ENRI-1 and ENRI-2 prevent precocious, inappropriate programming and nuclear translocation of NRDE-3. We propose a gating function for the ENRI-1/2 proteins to antagonize maternal 21U-RNAs and their downstream 22G-RNAs in the early embryo (see Discussion).
ENRI-3 is required for transposon silencing in the male germline
We turned to the ENRI-3 paralog to investigate the implications of the gating functions of the ENRI proteins in the germline. In hermaphrodites, ENRI-3::GFP was detected in the maturing germline of L4 larvae, with its expression peaking during spermatogenesis and in mature sperm in young adult hermaphrodites (Figure 6A). ENRI-3 localization was concentrated in distinctive perinuclear puncta around germline nuclei, and colocalizes with the P-granule marker PGL-3 (Figure 6A, Figure S7A, B, C). The strongest expression of ENRI-3 was detected in the germline cytoplasm of male animals (Figure 6B). ENRI-3 could also be detected in embryos and early larval stages, but its expression was limited to the precursor cells of the germline (Z2 and Z3) (Figure S7A).
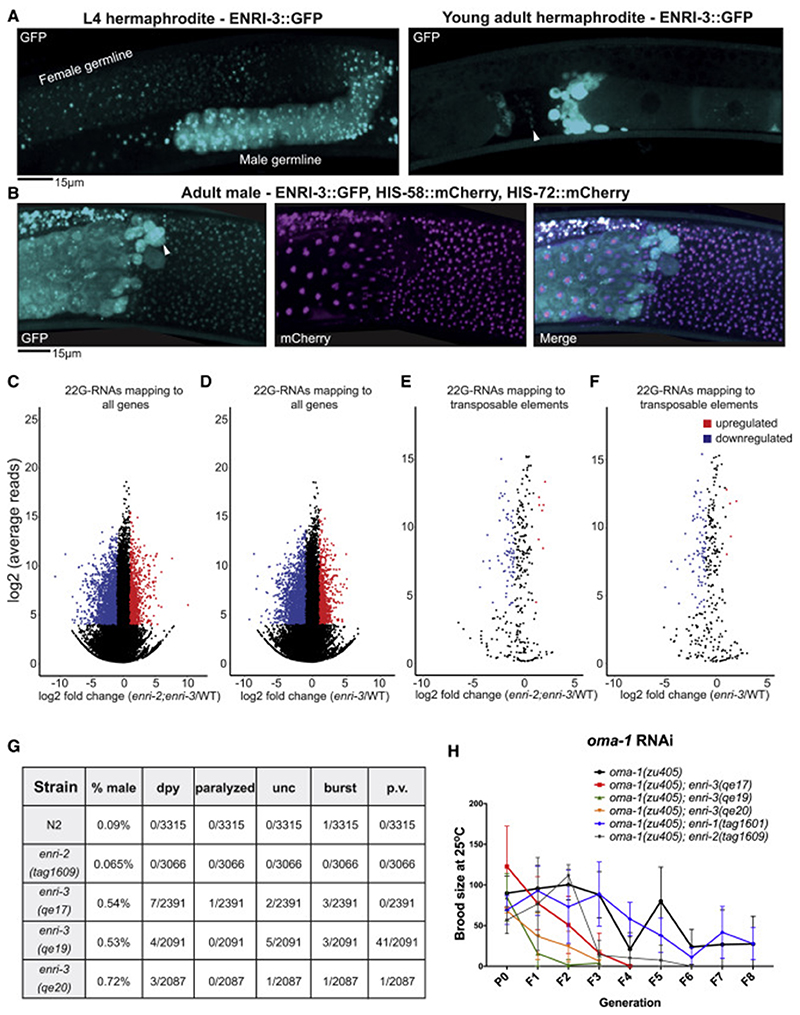
Figure 6. ENRI-3: Small RNA sorting in the male germline.
(A) Left: ENRI-3::GFP localization in the developing gonad of an L4 hermaphrodite. Region of spermatogenesis indicated as “male germline”. Right: ENRI-3::GFP localization in young adult hermaphrodites. White arrowhead indicates sperm. Bright signal to the right is likely residual bodies (anucleate cell bodies that detach from maturing spermatids). (B) ENRI-3::GFP, HIS-58::mCherry, and HIS-72::mCherry localization in adult male germline. White arrowhead indicates residual body. (C) and (D) MA plots of total 22G-RNAs mapping to individual genes in a (C) enri-2(tag1609);enri-3(qe19) double mutant or a (D) enri-3(qe17) mutant. (E) (F) MA plots of total 22G-RNAs mapping to annotated transposable elements in a (E) enri-2(tag1609);enri-3(qe19) double mutant or (F) enri-3(qe17) mutant. A filter was set such that only 22G-RNAs that displayed log2(fold-change) > 1, log2(average reads) > 4 and padj < 0.05 were considered significantly changed from wild-type. (G) A table indicating the number of worms displaying the indicates phenotypes. Worms were grown at 20°C for two generations and scored at the gravid adult stage. Dpy = dumpy, unc = uncoordinated, p.v. = protruding vulva (H) The brood size of enri mutants in the oma-1(zu405) background was scored for 8 generations following injection of P0s with oma-1 dsRNA (50ng/ul). See also Figure S6 and S7.
We generated and sequenced 22G-RNAs libraries from synchronized wild-type and enri-3(qe17) young adults and mapped the reads to protein-coding genes and transposable elements. Since a fraction of ENRI-2 is also expressed in the female germline (Figure 5B), we generated a double enri-2(tag1609);enri-3(qe19) mutant to address possible redundancy and/or compensation. enri-3 young adults displayed a profoundly altered 22G-RNAs expression profile (Figure 6D, F, S6D); 22G-RNAs targeting 2129 genes were depleted from the enri-3 libraries, while 22G-RNAs targeting 1610 genes were enriched (Figure 6D). The impact was similar in the double enri-2;enri-3 mutant; 22G-RNAs targeting 2250 genes were depleted, and 1351 enriched (Figure 6C). Strikingly, 22G-RNAs targeting transposable elements were impacted in enri-3 mutants, with a trend towards reduction (Figure 6F, S6D). 22G-RNAs targeting 75 transposable elements were significantly depleted in enri-3 young adults, whereas 22G-RNAs targeting 5 transposons were enriched (Figure 6F). Again, results were similar in the enri-2;enri-3 mutants; 22G-RNAs targeting 71 transposons were depleted while 8 were enriched (Figure 6E). The profiles of 22G-RNAs were similar between enri-3 and enri-2;enri-3 young adults, with overlap of 77% and 80% of all genes and transposable elements, respectively (Figure S7D, E). In agreement with small RNA analyzes, mRNA-seq revealed that several transposable elements were de-repressed in enri-3 mutants, even when compared to enri-1 and enri-2 mutants (Figure S6E).
This result suggests that most transposable elements are no longer silenced efficiently in the absence of ENRI-3. Along this line of thought, we detected an abnormally high incidence of spontaneous visible phenotypes in enri-3 alleles, a spectrum of phenotypes named mutator (mut). Spontaneous occurrence of protruding vulvas, dumpy worms, paralyzed worms, bursting worms, and rolling/generally uncoordinated worms was detected in enri-3 mutants, while extremely rare in wild-type animals (Figure 6G). Furthermore, the spontaneous incidence of males in the enri-3 mutant populations was also increased (Figure 6G). All together, these results suggest that a major function of ENRI-3 in the male germline is in regulating silencing of transposable elements.
Finally, we tested the impact of enri mutants on germline inheritable RNAi. For this, enri-3 alleles, enri-2(tag1609), and enri-1(tag1601) were crossed into oma-1(zu405) and we tested their ability to stably silence the oma-1 locus. oma-1 RNAi suppresses the embryonic lethality of the oma-1(zu405) temperature-sensitive gain-of-function mutant, and silencing is inheritable for many generations following initial exposure to dsRNA (Alcazar et al., 2008). enri-3 mutants fail to inherit oma-1 silencing for more than 4 generations, whereas an enri-1 null mutation had no detectable impact on oma-1 RNAi inheritance (Figure 6H). oma-1 silencing was lost earlier in the enri-2 mutant than in wild-type or in the enri-1 mutant, reverting around generation 6 (Figure 6H). These results show that ENRI-3 is required for an efficient endogenous RNAi response against transposable elements, and that ENRI-2 and ENRI-3 are required for trans-generational epigenetic inheritance of germline RNAi.
Discussion
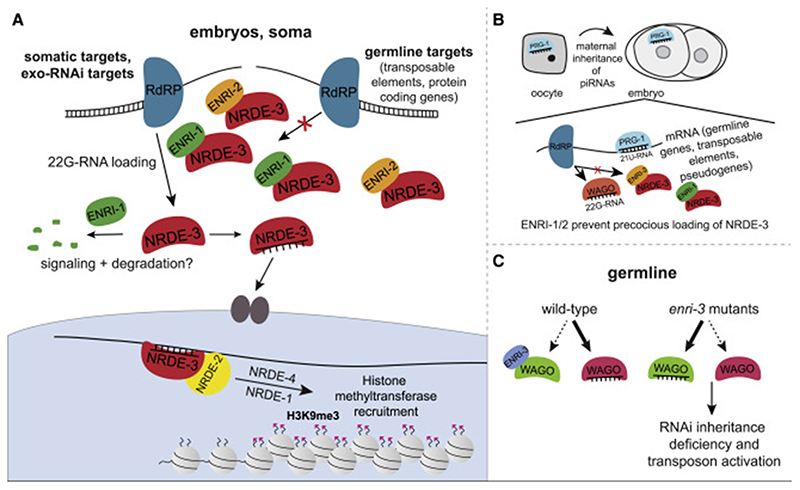
Our proteomics survey of NRDE-3 interactions unveiled an uncharacterized family of nuclear Argonaute-interacting proteins that ensures faithful and timely nuclear RNAi activity in somatic and germline tissues. Altogether, our data are consistent with a model wherein the ENRI proteins ‘gate’ nuclear RNAi activities by restricting the loading of 22G-RNAs onto nuclear Argonautes at the appropriate time and dosage, and in specific cell types (Figure 7).
Figure 7. Model of ENRI function.
(A) In the soma, ENRI-1/2 gate NRDE-3 by preventing precocious and inappropriate loading of NRDE-3 with 22G-RNAs produced downstream of the 21U-RNA and/or 26G-RNA pathways. ENRI-1/2 association with NRDE-3 could be regulated by PTMs. (B) We propose that ENRI-1 and ENRI-2 sequester NRDE-3 in the early embryo to prevent loading of 22G-RNAs downstream of germline RNAi pathways. (C) We propose that ENRI-3 gates one or many of the Argonautes acting in the germline and loss of ENRI-3 impacts multiple pathways leading to impaired inheritance of RNAi and activation of transposons.
We propose that the aberrant germline signature of 22G-RNAs associated with NRDE-3 in the absence of enri-1/2 is due to an aberrant early zygotic response to maternally inherited primary small RNAs. Precocious nuclear translocation of NRDE-3 is detected as early as the 2-cell stage in the enri mutants, before the onset of zygotic transcription which can occur as early as the 4-cell stage (Edgar et al., 1994; Seydoux and Fire, 1994; Tintori et al., 2016). piRNAs are maternally inherited in many organisms, including Drosophila, zebrafish, and C. elegans (Akkouche et al., 2013; Almeida et al., 2019; Brennecke et al., 2008; de Albuquerque et al., 2015; Houwing et al., 2007). 21U-RNAs trigger the generation of a broad range of 22G-RNAs targeting pseudogenes, transposable elements, and protein coding genes in the germline (Bagijn et al., 2012; Lee et al., 2012). Recently, the maternal contribution of the ERGO-1 pathway was examined in detail (Almeida et al., 2019). Ketting and colleagues showed that NRDE-3 is loaded with zygotically produced 22G-RNAs downstream of maternally inherited ERGO-1 class 26G-RNAs. ENRI-1/2 in the early embryo may thus serve to preempt the precocious and inappropriate loading of NRDE-3 with 22G-RNAs produced downstream of the 21U- and/or 26G-RNA pathways.
In somatic cells, NRDE-3 is the only known nuclear Argonaute responsible for nuclear silencing downstream of ERGO-1 and the exo-RNAi pathway (Gent et al., 2010; Guang et al., 2010; Guang et al., 2008). In the germline, however, multiple Argonautes act redundantly or in opposing pathways to effect epigenetic changes that are inherited for many generations. 21U-RNAs trigger the generation of 22G-RNAs that are loaded onto multiple germline WAGOs, including WAGO-1/2/3, PPW-1, and the nuclear Argonaute HRDE-1, but also several others. This broad silencing response is principally directed towards transposable elements, pseudogenes, and aberrant transcripts (Ashe et al., 2012; Buckley et al., 2012; de Albuquerque et al., 2015; Gu et al., 2009; Luteijn et al., 2012; Shirayama et al., 2012). Germline WAGOs are also loaded with 22G-RNAs derived from ALG-3/4 class 26G-RNAs mapping to thousands of sperm-enriched genes (Conine et al., 2010). Convergence of the RNAi pathways on the WAGOs, especially those involved in the nuclear inheritable RNAi response, is thought to embed an epigenetic memory of the ‘non-self’ in the germline. This widespread programming of an RNA-mediated silencing in the germline was proposed to be antagonized by the CSR-1 Argonaute, which targets over 4000 protein-coding genes in both the female and male germlines (Claycomb et al., 2009; Seth et al., 2013; Wedeles et al., 2013). Strikingly, the CSR-1 and WAGO targets largely overlap; an important number of CSR-1-associated 22G-RNAs are also detected in HRDE-1 and WAGO-1 IPs (de Albuquerque et al., 2015; Gu et al., 2009; Shirayama et al., 2012). HRDE-1 is loaded with CSR-1 22G-RNAs upon their overexpression (de Albuquerque et al., 2015), suggesting that the availability of the CSR-1 and WAGO Argonautes can lead to significantly distinct outcomes. It thus stands to reason that misloading (at the wrong moment, in the wrong cell type, and/or at the wrong dosage), of any of the above germline Argonautes in enri-3 animals would impact on multiple cross-talking or competing pathways. The epigenetic impact these changes bring about would be inherited and perhaps amplified over subsequent generations. This prediction is consistent with the mortal germline (mrt) phenotype observed in the enri-3 or in the hrde-1 mutants, for example.
To effectively gate nuclear Argonaute loading in specific cell lineages and at specific moments, the expression of ENRIs must be tightly regulated. Several lines of evidence suggest that the dosage of the ENRI proteins is subjected to regulation. Firstly, proteomic surveys of ENRI-1 and ENRI-2 revealed extensive interaction with the ubiquitin-proteasome pathway, including multiple subunits of the 26S proteasome complex, E3 ligases complexes, and deubiquitinating enzymes. Secondly, enforced expression of transgenic NRDE-3 leads to ENRI-1 accumulation. Thirdly, the expression of ENRI-3 peaks in the developing and mature sperm and is rapidly lost after fertilization. Consistent with this expression being at least partly established through degradative regulation, ENRI-3 is unstable in worm lysates and was barely detectable in IP samples. ENRI-3 is likely primed for destruction in the male germline so that degradation can be rapidly enacted. We speculate that one or more signaling mechanism(s) triggered by exogenous dsRNA, viral dsRNA, or the endogenous RNAi machinery to trigger ENRI degradation, thus releasing a pool of nuclear Argonautes to bolster the nuclear RNAi response.
RNAi pathways have strikingly distinct biological functions such as regulation of gene expression, chromatin organization, or antagonizing transposable elements. Regulation of the diverse RNAi responses is critical considering their extensive interplay and the multi-generational impact of nuclear Argonautes. The unveiled role of the ENRIs in gating nuclear Argonautes explains how nuclear RNAi is controlled throughout development to preserve physiological functions while ensuring a potent response in the face of genetic pathogens.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thomas F. Duchaine (thomas.duchaine@mcgill.ca)
Experimental Model and Subject Details
C. elegans were maintained using standard procedures at 16°C unless noted otherwise and fed OP50 E. coli. The strains used in this study are listed in the Key Resources Table.
Method Details
Protein purification and Multidimensional Protein Identification (MudPIT)
C. elegans pellets were homogenized in 50mM Tris-HCl pH7.5/150mM NaCl/1mM ETDA/0.1% Igepal CA-630 with Complete EDTA-free protease inhibitors (Roche), and cleared twice by 17,000xg centrifugation. Following extract preparation, FLAG-tagged proteins were purified from approximately 5mg total protein using ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich A2220) according to the manufacturer’s instructions. For MudPIT, FLAG-tagged proteins (NRDE-3 and ENRI-1) were eluted with 150ng/ul 3xFLAG peptide (Sigma-Aldrich F4799) according to the manufacturer’s instructions. ENRI-2::3xFLAG was eluted with 0.5M NH4OH pH 11. 100ul 0.5M NH4OH was added to the FLAG beads, incubated at room temperature for one minute, and spun down to collect the eluate; this was repeated twice more. In all cases, the fractions were pooled and one quarter of the total eluate was saved for western blot analysis.
ENRI-2: The NH4OH was evaporated by spinning the samples in the speed vac for 2 hours at room temperature. The fraction saved for western blot analysis was resuspended in 2X SDS Loading buffer (2X Tris/SDS (0.625 mM Tris pH 6.8, 0.125% SDS), 2% SDS, 100 mM DTT, 2.5% Glycerol, 7.5 mM Bromophenol blue).
NRDE-3 and ENRI-1: FLAG eluates saved for western blot analysis were precipitated in acetone before being resuspended in 2X SDS Loading Buffer. The NRDE-3 samples were run on a 8% SDS-PAGE gel, and the ENRI-1 and ENRI-2 samples were run on 10% SDS-PAGE gels. N2 was used as the negative control for ENRI-1 and ENRI-2 purifications. A strain expressing SUR-5::GFP was used as the negative control for the NRDE-3 purification. Only proteins found in at least two biological replicates and absent from the non-transgenic control were pursued. Five biological replicates were performed for NRDE-3 and three biological replicates were performed for ENRI-1.
Samples were prepared by sequential treatment with 5mM Tris (2-carboxyethyl) phosphine and 10mM iodoacetamide to reduce and alkylate cysteine residues followed by digestion with trypsin and lys-C proteases at 37 °C as described (Florens et al., 2006; Wohlschlegel, 2009). Digestions were stopped by the addition of 5% formic acid, desalted using Pierce C18 tips (Thermo Fisher Scientific). Desalted samples were separated on C18 reversed phase (3 uM, 100A pores, Dr. Maisch GmbH) columns, packed in house with 100uM ID and 18cm of packed resin. Digested peptides were eluted on a 140-minute water-acetonitrile gradient, with 3% DMSO in both buffers under electrospray ionization of 2.2kV. Gradient delivery was performed on an Easy nLC-1000 UHPLC at 300 nL/min, and MS/MS spectra was generated by Data Dependent acquisition strategy on a Thermo Q-Exactive mass spectrometer. Data acquisition consisted of cycles of one full MS spectrum owning resolution of 70,000 followed by MS/MS of precursor ions from the full MS scan using a resolution of 17,500.
Data analysis was performed the Integrated Proteomics pipeline 2 (Integrated Proteomics Applications, San Diego, CA) to generate peptide and protein lists that were quantified using spectra counting. In this case, MS2 spectra were searched using the ProLuCID algorithm against the WormBase C. elegans reference proteome followed by filtering of peptide-to-spectrum matches (PSMs) by DTASelect using a decoy database-estimated false discovery rate of <1%.
Immunoprecipitations
Lysates for IPs were prepared by homogenizing embryos in 50mM Tris-HCl pH7.5/150mM NaCl/1mM EDTA/0.1% Igepal CA-630 with Complete EDTA-free protease inhibitors (Roche), and cleared twice by 17,000xg centrifugation. Between 1-3mg of protein were IPed in all figures except for MudPIT experiments where 5mg was used. All FLAG IPs were performed with ANTI-FLAG M2 Affinity Gel (Sigma). 10μl of packed beads were used per 1mg of protein in the lysate and beads were incubated/rotated with the lysate for 1 hour at 4°C before being washed three times with the lysis buffer. Beads were then resuspended in 2X SDS loading buffer. ERGO-1, ENRI-1, and NRDE-3 IPs were performed by adding 10μl of rabbit serum per 1mg of protein in the lysate and the samples were incubated/rotated for 1 hour at 4°C. 10μl of packed beads (protein A sepharose CL 4B [GE healthcare]) per 1mg of protein were then incubated with the lysate for another hour at 4°C before being washed three times with lysis buffer. Beads were resuspended in 2X SDS loading buffer.
Western blotting
Antibodies used in western blots were against: ENRI-1 (1:1000) (Capralogics), tubulin (1:5000) (Abcam), FLAG (1:1000) (Sigma), GFP (1:1000) (Sigma), NRDE-3 (1:1000) (Capralogics), 6xHIS (1:1000) (Abcam), ALG-2 (1:1000) (Flamand et al., 2017), ERGO-1 (1:2000) (Thivierge et al., 2011). Secondary antibodies used were: Mouse TrueBlot ULTRA: Anti-Mouse Ig HRP (1:1000) (Rockland), Rabbit TrueBlot: Anti-Rabbit IgG HRP (1: 1000) (Rockland), IRDye 800CW Goat anti-rabbit (1:10 000) (LI-COR), IRDye 680RD Goat anti-mouse (1: 10 000) (LI-COR). Western blots from Figures 1C, 1F, 2A, 3B, 3C, S1A, S1B, S1C, and S3D were performed by transferring proteins onto nitrocellulose membranes (BioRad) in 1X Pierce Transfer Buffer (Thermo) by semi-dry transfer (1 hour, 0.2A, 20V). Membranes were blocked for 1 hour in 5% milk, and primary antibodies were detected by HRP conjugated goat anti-rabbit or goat anti-mouse secondary antibodies. All other western blots were performed by transferring proteins onto Immobilon-FL PVDF membranes (Millipore Sigma) by semi-dry transfer. Membranes were blocked for 1 hour in Odyssey blocking buffer (LICOR) and primary antibodies were detected by IRDye goat anti-rabbit or goat anti-mouse antibodies.
Plasmids and transgenics
Cloning of pBlueScript penri-1::enri-1::3xflag::enri-1 3’UTR: Full-length ENRI-1 (f43c11.9), including the upstream 1kb encompassing the promoter region, was amplified from C. elegans genomic N2 DNA using primers AAATATTTGTTGCCATTTTCTCCAG and TAGTCTTGTATGTATTCTTGTAGTC. Amplicons were cloned into pSC-A-amp/kan (Stratagene) and fully sequenced before being subcloned into pBlueScript II SK(+) (Agilent) at the SalI-XbaI sites. A NotI site was inserted upstream of the start codon using primers ATTAAGCGGCCGCATAATAATTTATGGATCTCTT and ATTAA GCGGCCGCCCCAAACATCCAATTTATACA. A 3xFLAG tag was subcloned at the NotI site and confirmed by sequencing.
Cloning of pBlueScript pbli-1::enri-1::3xflag::enri-1 3’UTR: Gibson assembly was used to generate this plasmid. Primers were designed using SnapGene software. The bli-1 promoter was amplified from genomic N2 DNA, enri-1::3xflag::enri-1 3’UTR was amplified from pBlueScript penri-1::enri-1::3xflag::enri-1 3’UTR, and pBlueScript was amplified using primers listed in Table S3. The pBlueScript vector was DpnI-digested and then all fragments were gel purified. The NEB HiFi DNA assembly kit was used to assemble the vector. 50ng of pBluescript, 38.95ng of the bli-1 promoter and 117ng of enri-1::3xflag::enri-1 3’UTR were incubated with 10μl of the NEB master mix and the volume was made up to 20μl with H2O. The fragments were incubated together for 15 minutes at 50°C.
Cloning of pBlueScript pbli-1::enri-2::enri-2 3’UTR: The same strategy that was used to clone pBlueScript pbli-1::enri-1::3xflag::enri-1 3’UTR was used to clone enri-2. enri-2 was amplified from genomic N2 DNA. All primers are listed in Table S3.
To generate transgenic strains, injections mixes were generated that contained 90ng/μl pBluescript sur-5::gfp as an injection marker, and 20ng/μl of the transgene to be expressed. Worms were injected according to normal injection protocols (Mello and Fire, 1995). F1 animals carrying the sur-5::gfp marker were picked onto individual plates and those that passed the marker onto the next generation were considered lines.
The transgene in strain FD26 was integrated by irradiating 30 sur-5::gfp positive L4 animals in a Stratagene UV crosslinker (power 300). 200 sur-5::gfp positive F1 animals were separated onto individual plates and screened for those that had high transmission of the marker into the next progeny. 10 F2 animals per high transmitting F1 animals were transferred onto new plates and plates that had 100% inheritance were kept.
IP-northern blot
Embryo pellets were homogenized in 30mM HEPES-KOH (pH7.4)/150mM KOAc/5mM Mg(OAc)2/0.1% Igepal CA-630 with Complete EDTA-free protease inhibitors (Roche) and RiboLock RNase inhibitor (Thermo Scientific). Extracts were cleared twice by 17,000xg centrifugation. Approximately 3mg of protein was used in the IPs. 1/10th of the lysate was kept as a protein input and 1/10th of the lysate was kept for the RNA input. Proteins were purified using ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich F4799) according to the above protocol for immunoprecipitations. 1/10th of the beads were reserved for protein analysis (western) and 2x SDS loading buffer was added. 1mL of QIAzol (Qiagen) was added to the remaining beads and RNA input for RNA extraction according to the manufacturer’s protocol. 100% of the RNA isolated from the beads and 50% of the input was resolved on a 15% Urea-TBE gel. The gel was run at 200V for 1h in 0.5X TBE (45mM Tris, 45mM Boric acid, 1mM EDTA, pH8.0) and transferred to a Hybond XL nylon membrane (GE Healthcare) in 0.5X TBE on a semi-dry apparatus. RNA was UV crosslinked on the membrane and pre-hybridized in UltraHub Oligo buffer (Ambion). The StarFire probes were labeled with α32P dATP according to the manufacturer’s protocol. The probes were hybridized overnight with the membrane. In the case of the X-cluster small RNA northern blot, the two probes were used together on the same membrane (Duchaine et al., 2006). Membranes were washed twice with 0.5% SDS for 15 minutes, and once with 1X SSC/0.2% SDS for 15 minutes. Membranes were exposed to an Imaging Plate (FujiFilm) for 1 day and scanned with a Typhoon Scanner.
2’O-Methyl Pull-Down
Embryo pellets were homogenized in 25mM HEPES-KOH (pH7.4)/150mM NaCl/1mM EDTA/1mM DTT/10% glycerol/0.5% Triton X-100 with Complete EDTA-free protease inhibitors (Roche) and RiboLock RNase inhibitor (2ul/mg of protein in lysate) (Thermo Scientific). Extract were cleared twice by 17,000xg centrifugation. Extract was pre-cleared with 20μl of Dynabeads m-280 and unrelated 2’O-methyl oligonucleotide (0.1μmol anti-miR-35) for 1 hour at 4°C with rotation. The supernatant was incubated with biotinylated 2’O-methyl oligonucleotides complementary to the X-cluster (or human miR-16 as a negative control) for 1h at 25°C with rotation. Following centrifugation for 5 minutes at 13,000rpm, the supernatant was incubated with 50μl Dynabeads m-280 for 30 minutes at 4°C with rotation. Beads were washed 3 times in lysis buffer containing 0.1% Triton and 2mM DTT following by another 3 washed in lysis buffer without Triton. 2X SDS loading buffer was added to the beads.
Antibody production (ENRI-1 and NRDE-3)
f43c11.9 (enri-1) was amplified from N2 cDNA and cloned into pET-28a with EcoRI and SalI restriction sites, and transformed into BL-21 DE3 cells. Bacteria was grown at 30°C until the O.D. reached 0.5. 1mM IPTG was added and induction was performed O/N at 16°C. Cells were lysed (20mM sodium phosphate pH6.4, 500mM NaCl, 10mM imidazole, 10% glycerol) by emulsification and centrifuged at 25,000g for 20 minutes. The supernatant was incubated with 1mL Ni-NTA beads for 30 minutes at 4°C. Beads were washed sequentially with 10 volumes of lysis buffer containing 10mM, 20mM, 40mM, and 60mM imidazole and eluted 10 times with 1 volume of elution buffer (lysis buffer with 250mM imidazole). Eluted proteins were diafiltered and concentrated using AMicon Ultra-15 Centrifugal Filter Units and injected into rabbits for antibody production (Capralogics, Cambridge Massachusetts).
A fragment of nrde-3 (nucleotides 1-1149, amino acids 1-383) was amplified from N2 cDNA and cloned into pSMT3 with SacI and XhoI restriction sites and transformed into BL-21 DE3 cells. 6xHIS-SUMO-NRDE-3 was purified in the same manner as 6xHIS-ENRI-1. 6xHIS-SUMO-NRDE-3 was injected into rabbits for antibody production (Capralogics, Cambridge Massachusetts).
RNAi
unc-22, dpy-11, lir-1, unc-73 and bli-1 RNAi were all performed by feeding animals with HT115 bacteria expressing dsRNA (Timmons et al., 2001). HT115 was transformed with plasmids containing the gene to be knocked down flanked by two T7 promoters. HT115 was seeded onto NMG (nematode growth media) plates supplemented with 50μg/mL ampicillin and 1mM IPTG. IPTG induces the expression of T7 from HT115 and leads to the production of dsRNA from the gene in the plasmid. Primer sequences for unc-22, dpy-11, lir-1, and bli-1 genes were obtained from the Ahringer library (Kamath et al., 2003) (see Table S3). unc-73 bacteria was obtained from the Ahringer library. Genes were amplified and cloned into pSC-A-amp/kan (Stratagene) before being transformed into HT115. unc-22, unc-73, dpy-11 and bli-1 assays were performed by picking L4 stage animals onto at least 10 individual RNAi plates and letting their F1 progeny grow on that same plate until they reach the gravid adult stage, at which point they were scored for the expected phenotypes. Experiments were repeated at least twice with several replicates each time. lir-1 assays were performed by picking between 40-50 L1 animals onto RNAi plates and counting the number of animals that survived into the gravid adult stage. Experiments were repeated 5 times. oma-1 RNAi was performed by injecting young adult animals with 50ng/μl double-stranded oma-1 RNAi. The RNA was transcribed with the MEGAscript T7 Transcription Kit using a plasmid with T7 promoters flanking the oma-1 gene as a template.
CRISPR/Cas9
Genome editing was performed as in (Arribere et al., 2014; Ward, 2015). dpy-10 was used as a co-conversion marker. Cas9 ribonucleoprotein was injected along with a fused crRNAs and tracrRNA (sgRNA) and a dpy-10 single stranded ultramer oligonucleotide (ssODN) from IDT. The injection mixes contained 1.2μg/μl Cas9, 300mM KCl, 12.5mM HEPES pH 7.4, 50ng/μl dpy-10 sgRNA, 200ng/ul gene-specific sgRNA, and 13.75ng/μl dpy-10 repair ssODN. The mixture was injected into wild-type C. elegans according to a standard microinjection procedure (Mello and Fire, 1995). We then singled out F1 progeny with the roller or dumpy phenotypes (dpy-10 co-conversion) and genotyped them for the desired genome edits. F2 and/or F3 homozygous animals with the edits were then sequenced. In the case of the following alleles: tag1600, tag1601, tag1609, tag1654, tag1662, tag1660, Cas9 protein was isolated and purified at Max Planck Institute of Cell Biology and Genetics. The GFP tag repair templates (2xTY1-mEGFP) were amplified from a plasmid (S417) with primers modified for each enri gene (see Table S3).
GST pull-downs
Cloning and purifying GST-NRDE-3: Three fragments of nrde-3 (aa 1-383, 365-701, 683-1058) were amplified from N2 cDNA using primers listed in Table S3 and cloned into pGEX-6p-1. Plasmids were transformed into BL-21 DE3 cells and grown at 30°C until the O.D. reached 0.5. 1mM IPTG was added and induction was performed O/N at 16°C. Cells were lysed in GST extraction buffer (50mM Tris HCl pH 8.0, 100mM NaCl, 1mM EDTA, 1mM DTT, protease inhibitors, 1% Sarcosyl). Lysozyme was added to 0.5mg/ml and incubated on ice for 30 minutes. Bacteria was lysed by sonication (5 x 30 seconds). Lysate was spun for 30 min at 15 000g. To quantify the amount of GST-NRDE-3 in the lysate, 500μl of the lysate were incubated with 20μl equilibrated Glutathione Sepharose 4B-GSH beads and incubated (rotated) at 4°C for 1 hour. After washing the beads 6x in 1ml of GST extraction buffer (without Sarcosyl), beads were resuspended in 2X SDS loading dye and run alongside known amounts of BSA to quantify the amount of GST-NRDE-3 in the lysates.
Cloning and purifying 6xHIS-SUMO-ENRI-1 and 6xHIS-SUMO-ENRI-2: enri-1 and enri-2 cDNA were amplified from N2 cDNA using the primers listed in Table S3 and cloned into pSMT3. Plasmids were transformed into BL21 DE3. Bacteria was grown at 30°C until the O.D. reached 0.5. 1mM IPTG was added and induction was performed O/N at 16°C. Cells were lysed (20mM sodium phosphate pH6.4, 500mM NaCl, 10mM imidazole, 10% glycerol) by sonication and centrifuged at 15 000g for 20 minutes. The supernatant was incubated with 1mL Ni-NTA beads for 30 minutes at 4°C. Beads were washed sequentially with 10 volumes of lysis buffer containing 10mM, 20mM, 40mM, and 60mM imidazole and eluted 10 times with 1 volume of elution buffer (lysis buffer with 250mM imidazole). Fractions were pooled and concentrated with Amicon Ultra Centrifugal filters and quantified by the Bradford assay.
Pull-downs: Approximately 1μg of GST fusion proteins were incubated with 20μl Glutathione Sepharose 4B-GSH beads rotating overnight at 4°C. The next morning, beads were washed 6 times with GST extraction buffer (without Sarcosyl). 15μg of either ENRI-1 or ENRI-2 were diluted in binding buffer (PBST with 250mM KCl) to a volume of 1ml total. ENRI-1 and ENRI-2 were pre-cleared with 20μl glutathione beads for 1 hour at 4°C. After pre-clearing, ENRI-1 and ENRI-2 were added to the pre-bound glutathione beads (beads that were bound overnight) and rotated for 1 hour at 4°C. After binding, beads were washed 3 times in PBST with 250mM KCl. The beads were resuspended in 2X SDS loading buffer and loaded onto 10% SDS-PAGE gels for either Coomassie staining or western blot.
Microscopy
Worms were placed directly onto imaging slides containing 1.5ul of worm immobilization mixture (25mM levamisole, 25nM sodium azide in water) and covered with a coverslip. Embryos before the 80-cell stage were dissected from adult hermaphrodites placed in the immobilization mixture. Embryos after the 80-cell stage as well as worms from the larval stage through gravid adult were picked directly from OP50 plates and placed directly into the immobilization mixture. All embryos and worms were imaged using a real-time confocal microsocpe (Olympus IX81) equipped with a spinning disk (Yokogawa CSU-X1) and Andor iXon EM camera. A cooling chamber (Warner) was added to take images at 20°C. Images were taken with either a 40X silicon oil immersion lens or a 60X silicon oil immersion lens (both Olympus UPlanSApochromat). For analysis of GFP expression, a 488nm sapphire laser was used (Coherent). Images were adjusted as necessary in Fiji (ImageJ) using BigStitcher (Horl et al., 2019), cropping, brightness and contrast tools.
Small RNA sequencing
Total RNA was extracted with Trisure reagent and DNase treated. RNA fraction was isolated by a chloroform extraction. Small RNA sequencing was performed with 5μg total RNA treated with Epicentre 5’ polyphosphatase to remove the 5’ triphosphate from 22G RNAs. After treatment, RNA is extracted by phenol/chloroform treatment and 1μg final RNA was used for the library preparation using the Illumina Truseq small RNA kit. 5’-independent libraries were sequenced with Hiseq 2500 with single end 50 bps option. Adapters were trimmed with cutadapt v1.9 with the following options: “--minimum-length 18 --discard-untrimmed -a TGGAATTCTCGGGTGCCAAGG”. Raw and collapsed libraries enrichment with 22G RNAs were analysed with the subread package on RStudio. Adaptor trimmed libraries were mapped to C. elegans ce11 genome using the STAR aligner (Dobin et al., 2013) with the following parameters: “--outFilterMultimapNmax 5000 --winAnchorMultimapNmax 10000 --outFilterMismatchNmax 2 --alignEndsType EndToEnd --outSAMtype BAM Unsorted --runThreadN --readFilesCommand 'gunzip -c'”.
Total RNA-seq
PolyA selected total RNA-seq libraries were sequenced with Hiseq 1500 with single end 50 bps option and mapped to C. elegans ce11 genome using STAR aligner with the following options: “--outFilterMultimapNmax 50 --winAnchorMultimapNmax 50 --outFilterMismatchNmax 0 --limitBAMsortRAM 31000000000 --alignIntronMax 1 --alignEndsType EndToEnd --outSAMtype BAM SortedByCoordinate --runThreadN 6 --outBAMsortingThreadN 6 --readFilesCommand 'gunzip –c”. A GTF files containing C. elegans ce11 gene annotations were downloaded from Ensemble 92. A SAF file containing transposable element annotations were generated using Repeatmasker (Smit, 2015) version open-4.0.5 in sensitive mode, run with rmblastn version 2.2.27+ using RepeatMasker database version 20140131 against ce11 genome version.
Small and total RNA library were sorted and indexed using the samtools v1.3 (Li et al., 2009). Counts against the annotations in the GTF files were generated with featureCounts v1.5.0-p1 (Liao et al., 2014) with the following parameters: “-T 8 –M –fraction –O”.
Differential gene expression analysis
For 5’-independent small RNA-seq libraries, normalized counts, adjusted p values(padj) and log2 fold change values were obtained through both a custom method using normalisation unique reads per million for individual libraries and DESEQ2 package on RStudio. Heatmaps, volcano and density plots were generated using the pheatmap.2 and ggplot2 functions with a custom script on RStudio. Germline and somatic RNAi targets were taken from (Gu et al., 2009; Zhang et al., 2011).
Disordered regions prediction
Disordered regions of the ENRI proteins were predicted using the software PONDR (www.pondr.com).
Post-translational modification analysis
8mg of total protein was used to immunoprecipitate ENRI-1::3xFLAG from embryos. Lysates for IPs were prepared by homogenizing embryos in 50mM Tris-HCl pH7.5/150mM NaCl/1mM EDTA/0.1% Igepal CA-630 with Complete EDTA-free protease inhibitors (Roche), and cleared twice by 17,000xg centrifugation. Embryo lysates were incubated with 80μl magnetic M2 FLAG beads for 1 hour (rotating) at 4°C. Beads were then washed 3 times with lysis buffer. Beads were resuspended in 2X SDS loading buffer. A western blot was performed on 10% of the IP to confirm efficacy of the IP, the other 90% was loaded onto a gel and stained with Coomassie gel. The band corresponding to ENRI-1::3xFLAG was cut and sent to the Taplin Mass Spectrometry Facility at Harvard University for post-translational modification analysis.
Quantification and statistical analysis
Statistical analyses were conducted with Tukey’s multiple comparisons test unless specifically indicated. All data represent the mean ± standard deviation of at least three independent experiments. For all RNAi assays a p-value under 0.05 was considered significant, and p-value: 0.05-0.0332 (*), 0.0333-0.0021 (**), 0.00222-0.0002 (***), <0.0001 (****). Tests for the RNAi assays were conducted with Prism 7. DESEQ2 was used for statistical analysis of small RNA and total mRNA sequencing data.
Supplementary Material
Acknowledgments
We thank Shaolin Li for generating enri-1(qe16), enri-2::3xflag, and enri-2(qe21). We thank Dana Olbert from the MPI-CBG Genome Engineering Facility for generating the strains enri-1(tag1654), enri-2(tag1662), enri-3(tag1660). We thank Scott Kennedy for sharing the strain YY179. The eri-1(mg366) and nrde-3(gg66) strains were obtained from the Caenorhabditis Genetics Center (CGC). We thank Vinay Mayya for generating and providing the GST-GYF-1 construct. AL is supported by the FRQS Doctoral Award, FRQS Master’s Award, Canada Graduate Scholarship Master’s Award, and Defi Canderel Studentship. ACB is supported by the Marie-Skladowska Curie Fellowship 2017 (grant number 747666). ANS was supported by the CIHR Frederick Banting and Charles Best Doctoral Research Award, Ruth and Alex Dworkin Scholarship, Alexander McFee Memorial Fellowship, and Defi Canderel Studentship. MF was supported by the Vanier Canadian Graduate Scholarship. We would like to thank the following Services and Facilities of the MPI-CBG for their support: Light Microscopy; Scientific Computing; Sequencing and Genotyping and Genome Engineering. This work was supported by the Human Frontier Science Program (HFSP) Organization grant to TFD, MS, and EAM, the Cancer Research UK (C13474/A18583, C6946/A14492) and the Wellcome Trust (104640/Z/14/Z, 092096/Z/10/Z) to EAM, and a Fonds de recherche du Québec – Santé (FRQS) Chercheur-boursier Senior Salary award and a Natural Sciences and Engineering Research Council of Canada (NSERC) grant to TFD.
Footnotes
Author contributions
ANS and TFD conceived and ANS performed the NRDE-3 MudPIT experiments. AL and TFD designed and conceived the following project and experiments. AL, ACB, AG, ANS, AV, SM and MNF performed the experiments. ACB performed all sequencing experiments. ACB and AL analyzed the sequencing data. AG and SM performed all imaging experiments. AL and TFD wrote the manuscript.
Declaration of Interests
EAM is a founder and director of STORM Therapeutics Ltd. This company has not contributed to this research in any way. The authors declare no competing interests.
Data and Code Availability
Data on small RNA-seq of the enri mutants and the total mRNA seq on transposons was submitted to GEO: GSE126377.
References
- Akkouche A, Grentzinger T, Fablet M, Armenise C, Burlet N, Braman V, Chambeyron S, Vieira C. Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO reports. 2013;14:458–464. doi: 10.1038/embor.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MV, de Jesus Domingues AM, Ketting RF. Maternal and zygotic gene regulatory effects of endogenous RNAi pathways. PLoS genetics. 2019;15:e1007784. doi: 10.1371/journal.pgen.1007784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science (New York, N.Y.) 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher JM, Dufourcq P, Sookhareea S, Labouesse M. RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics. 1999;153:1245–1256. doi: 10.1093/genetics/153.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science (New York, NY) 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS genetics. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BF, Placentino M, Ketting RF. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Developmental cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegans embryos. Development (Cambridge, England) 1994;120:443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- Emmons SW, Yesner L. High-frequency excision of transposable element Tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984;36:599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- Flamand MN, Gan HH, Mayya VK, Gunsalus KC, Duchaine TF. A non-canonical site reveals the cooperative mechanisms of microRNA-mediated silencing. Nucleic acids research. 2017;45:7212–7225. doi: 10.1093/nar/gkx340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods (San Diego, Calif) 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Molecular cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature genetics. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science (New York NY) 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science (New York NY) 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nature reviews. Genetics. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horl D, Rojas Rusak F, Preusser F, Tillberg P, Randel N, Chhetri RK, Cardona A, Keller PJ, Harz H, Leonhardt H, et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nature methods. 2019;16:870–874. doi: 10.1038/s41592-019-0501-0. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 32.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA (New York, N Y) 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 36.Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. The EMBO Journal. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Molecular cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Mello C, Fire A. DNA transformation. Methods in cell biology. 1995;48:451–482. [PubMed] [Google Scholar]
- 39.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & development. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raman P, Zaghab SM, Traver EC, Jose AM. The double-stranded RNA binding protein RDE-4 can act cell autonomously during feeding RNAi in C. elegans. Nucleic acids research. 2017;45:8463–8473. doi: 10.1093/nar/gkx484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr, Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Developmental cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development (Cambridge, England) 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science (New York, NY) 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2015.
- Spracklin G, Fields B, Wan G, Becker D, Wallig A, Shukla A, Kennedy S. The RNAi Inheritance Machinery of Caenorhabditis elegans. Genetics. 2017;206:1403–1416. doi: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, Conte D, Jr, Duchaine TF. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nature structural & molecular biology. 2011;19:90–97. doi: 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tintori SC, Osborne Nishimura E, Golden P, Lieb JD, Goldstein B. A Transcriptional Lineage of the Early C. elegans Embryo. Developmental cell. 2016;38:430–444. doi: 10.1016/j.devcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science (New York, NY) 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science (New York, NY) 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Ward JD. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics. 2015;199:363–377. doi: 10.1534/genetics.114.172361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Developmental cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods in molecular biology (Clifton, NJ) 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Banse SA, Hunter CP. The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics. 2013;194:117–131. doi: 10.1534/genetics.113.149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data on small RNA-seq of the enri mutants and the total mRNA seq on transposons was submitted to GEO: GSE126377.