Abstract
At the heart of protein ubiquitination cascades, ubiquitin-conjugating enzymes (E2s) form reactive ubiquitin-thioester intermediates to enable efficient transfer of ubiquitin to cellular substrates. The precise regulation of E2s is thus crucial for cellular homeostasis and their deregulation is frequently associated with tumorigenesis. In addition to driving substrate ubiquitination together with ubiquitin ligases (E3s), many E2s can also auto-ubiquitinate, thereby promoting their own proteasomal turnover. To investigate the mechanisms that balance these disparate activities, we dissected the regulatory dynamics of UBE2S, a human APC/C-associated E2 that ensures the faithful ubiquitination of cell cycle regulators during mitosis. We uncovered a dimeric state of UBE2S that confers auto-inhibition by blocking a catalytically critical ubiquitin binding site. Dimerization is stimulated by the lysine-rich C-terminal extension of UBE2S that is also required for the recruitment of this E2 to the APC/C and is auto-ubiquitinated as substrate abundance becomes limiting. Consistent with this mechanism, we found that dimerization-deficient UBE2S turned over more rapidly in cells and did not promote mitotic slippage during prolonged drug-induced mitotic arrest. We propose that dimerization attenuates the auto-ubiquitination-induced turnover of UBE2S when the APC/C is not fully active. More broadly, our data illustrate how the use of mutually exclusive macromolecular interfaces enables modulation of both the activities and the abundance of E2s in cells to facilitate precise ubiquitin signaling.
Introduction
Ubiquitination triggers a vast repertoire of cellular signals that can determine the lifetimes, localization, conformations, activities, and interaction profiles of the modified proteins. It is thus pivotal that ubiquitin modifications are formed in a specific and tightly regulated manner. This is accomplished through a cascade of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) that is counteracted and refined by deubiquitinating enzymes (DUBs). Over the last two decades, structural studies revealed an astounding variety of mechanisms regulating the ubiquitination machinery, particularly at the E3 level (1–3). E3s typically catalyze the formation of an isopeptide/peptide bond between the C-terminal carboxyl group of ubiquitin and a primary amino group of a substrate (generally referred to as an ‘acceptor’). Substrate-linked ubiquitin may, in turn, function as an acceptor, promoting the assembly of ubiquitin chains. With eight primary amino groups in ubiquitin, these chains can have diverse linkages and topologies, which elicit specific signaling outcomes. The precise mechanism of linkage formation depends on the E2-E3 pair involved. RING-type ligases, which account for the majority of > 600 E3s in the human proteome (4), bring a thioester-linked ubiquitin-E2 conjugate and an acceptor into proximity, thereby facilitating the direct transfer of ubiquitin from the E2 to the acceptor. In this context, the E2 interacts with the acceptor and can dictate the selection of ubiquitination sites on a substrate and linkage specificity in ubiquitin chain formation, respetively. Identifying the factors regulating E2s is thus an important prerequiste for understanding ubiquitin signaling specificity, but has remained a considerable challenge, because it is often unclear which E2-E3 pairs cooperate in the cell. Moreover, functional redundancies exist within both enzyme families, and both act through dynamic macromolecular complexes involving weak, multi-site interactions and co-localization mechanisms that are difficult to recapitulate structurally.
The anaphase-promoting complex (APC/C), a 1.2 MDa multi-subunit E3, provides a paradigm for how combinatorial macromolecular interactions orchestrate the sequential turnover of substrates during mitosis. Landmark cryo-EM work provided detailed views of the intricate conformational dynamics of the APC/C and the specific roles of its cognate E2s, UBE2C and UBE2S, during substrate modification (5, 6): UBE2C specializes in priming substrates with short ubiquitin chains, which then undergo processive elongation, catalyzed by UBE2S. Priming follows a canonical mechanism, in which UBE2C engages the RING domain (APC11) through a conserved interface and stabilizes the active site-linked ubiquitin (known as the ‘donor’) in a catalytically critical, ‘closed’ conformation toward the E2 (7–9). In contrast, UBE2S-driven ubiquitin chain elongation occurs through an atypical mechanism, in which the RING domain stabilizes the acceptor rather than the donor ubiquitin (9, 10). Binding of UBE2S to the APC/C stimulates chain initation by UBE2C, indicating conformational coupling between the two phases of modification (11). UBE2S per se forms Lys11-linkages (12–16); however, its cooperative action with the more promiscuous UBE2C generates branches, which render the degradation of APC/C substrates particularly efficient (17). As the concentrations of substrates become limiting, UBE2S is thought to promote its own turnover through auto-ubiquitination of the non-conserved C-terminal extension (15, 16, 18). This region flanks the catalytic ‘UBC’ domain and is concomitantly required for interactions with the APC/C (8, 10, 15, 19). Moreover, we have shown that the cell cycle-dependent auto-ubiquitination of a particular lysine residue within the UBC domain confers auto-inhibition in UBE2S by preventing donor binding (18) - a mechanism that is conserved in other E2s (20).
Due to the compensatory actions of UBE2C and UBE2D with the APC/C, depletion of UBE2S causes only a mild mitotic phenotype in otherwise unperturbed mammalian cells (15, 21, 22). However, UBE2S is crucial for the release of cells from a drug-induced spindle-assembly checkpoint (SAC) arrest (22). The SAC monitors unattached or tensionless kinetochores and mediates the assembly of the mitotic checkpoint complex (MCC), a potent inhibitor of APC/CCDC20 (23). In response to microtubule-directed drugs, UBE2S contributes to SAC silencing by promoting the dissociation of the MCC components from the APC/C (10, 15) and CDC20 (22, 24). Moreover, UBE2S promotes mitotic slippage, a process in which slow degradation of cyclin B1 in the context of persistent SAC signaling ultimately results in mitotic catastrophe and aneuploidy (22).
Here we revealed that the conserved donor ubiquitin binding site of UBE2S is utilized for an additional layer of regulation, modulating the cellular concentration of this E2. Our data converge to a model in which the formation of an auto-inhibited dimer counteracts the auto-ubiquitination-induced turnover of UBE2S. We speculate that the UBE2S dimer provides an ‘inert’ storage form of the enzyme that is protected from futile cycles of degradation and poised to be activated in response to cellular demands.
Results
UBE2S can self-associate in vitro and co-localizes in cells
Oligomerization is an emerging theme in E2 regulation with diverse, enzyme-specific manifestations (25–32). For the UBC domain of UBE2S (UBE2SUBC; residues 1-156), adimeric arrangement is observed in several crystal structures (18, 33) and, in principle, compatible with the described position of a UBE2S monomer on the APC/C (8, 9). We thus investigated the possibility that UBE2S dimerizes by proximity ligation assays (PLAs), monitoring the co-localization of FLAG-tagged and HA-tagged protein variants in HCT116 cells with UBE2S knock-out (UBE2S-/- (21)) (34) (Fig. 1A, B; fig. S1A, B). Indeed, these assays revealed a significant PLA signal compared to a negative control, in which cells coexpressed HA-UBE2S and FLAG-Venus. The extent of FLAG-/HA-UBE2S co-localization was similar to that of FLAG-UBE2S with HA-ubiquitin, suggesting that UBE2S oligomers are abundant in cells. Note that the addition of HA and FLAG tags, respectively, did not interfere with UBE2S activity in vitro (fig. S1C, D).
Fig. 1. UBE2S self-associates in vitro and co-localizes in cells.
(A) Representative PLA stains for HA and FLAG epitopes in HCT116 UBE2S-/- cells co-transfected with plasmids encoding HA-UBE2S and FLAG-UBE2S. As a negative control, HA-UBE2S was co-expressed with FLAG-Venus. As a positive control, HA-ubiquitin (Ub) was coexpressed with FLAG-UBE2S. As a technical control, HA-UBE2S was expressed by itself. Bright dots indicate the co-localization of FLAG- and HA-tagged molecules. Due to varying signal intensities, the images were individually adjusted; scale bar: 10 μm. (B) Quantification of PLA signals as shown in (A). All images were treated identically. Black bars represent the median of two independent experiments. N indicates the number of analyzed cells. ‘ratio’ denotes the median signal normalized to that of the HA-UBE2S + FLAG-UBE2S experiment. Statistical significance was calculated according to Kruskal-Wallis and Dunn’s multiple comparison test. (C) Amide proton relaxation rates determined for UBE2SUBC and UBE2S at the indicated concentrations by NMR (N=1). (D) Ni2+-NTA resin-based in vitro pull-down experiment with the indicated tagged UBE2S variants, monitored by SDS-PAGE and immunoblotting (N=3 independent experiments). As negative controls, the resin was incubated with HA-UBE2S and His6-UBE2S only, respectively.
We next interrogated whether UBE2S oligomerizes in vitro. Because purified UBE2S is predominantly monomeric (12), we conducted NMR relaxation measurements to allow for the detection of weak association events, analogous to other E2s (26, 30, 31). Based on two-point spin-echo experiments, a concentration-dependent increase in the transverse relaxation rates (R2) for the amide protons in the structured regions of UBE2SUBC was observed (Fig. 1C). A similar trend was seen for R2 values for full-length UBE2S (Fig. 1C). In addition, we determined the average longitudinal (R1) and transverse (R2) relaxation rates of the nitrogen nuclei in the structured regions of UBE2SUBC at 200 μM protein concentration, yielding 0.93 +/- 0.06 and 15.0 +/- 1.6 s-1, respectively. These correspond to a rotational correlation time of 11 ns, in good agreement with the size of monomeric UBE2SUBC (35). At 600 μM protein concentration, the nitrogen R2-values were globally increased by 20 to 30%. This effect was not caused by concentration-dependent structural changes in UBE2S, since the corresponding 1H15N-HSQC spectra overlaid perfectly (fig. S1E). Instead, the increase in R2 may originate from chemical exchange processes or changes in the molecular tumbling rate; that we observed increased R2-values uniformly throughout UBE2S strongly argues for concentration-dependent changes in the molecular tumbling, indicative of oligomerization. Notably, our NMR analyses were performed under reducing conditions in which disulfide-induced aggregation was suppressed. In line with these analyses, weak self-association was detected in pull-down experiments using His6- and HA-tagged UBE2S (Fig. 1D). Our studies thus uncover a previously unrecognized ability of UBE2S to transiently oligomerize, which particularly manifests itself in the native environment of the cell.
Structure-guided crosslinking defines a specific dimerization mode in solution
We reported that UBE2SUBC crystallizes as a dimer (18). The underlying ~1050 Å2 subunit interface is tightly packed and hydrophobic, as reflected in a predicted solvation free energy gain upon complex formation of -11 kcal/mol (based on the PISA service at the European Bioinformatics Institute (36)). In crystals of the wild-type protein (PDB: 1ZDN (33)), an intermolecular disulfide bond connects Cys118 of the subunits, whereas a similar dimeric arrangement is found in a crystal structure of the C118M variant (PDB: 6QH3 (18)); in the latter case, however, the relative subunit orientation is slightly tilted compared to the wild-type structure, presumably due to the introduced methionine side chain. Dimerization of UBE2SUBC in the crystal, therefore, does not depend on the formation of an intermolecular disulfide bond (18). Two additional crystal structures we determined for UBE2SUBC wild-type (at an improved resolution of 1.55 Å; PDB: 6S98) and the C118A variant (at 2.18 Å resolution; PDB: 6S96), respectively, corroborate this conclusion (Table 1): With a backbone RMSD of 1.13 Å, the disulfide-linked (wild-type) and non-covalent (C118A) crystallographic dimers are of virtually identical architecture, reflecting a recurring, energetically favored subunit interface (Fig. 2A, B).
Table 1. X-ray crystallographic data collection and refinement statistics.
Values in parentheses correspond to the highest-resolution shell.
| UBE2SUBC WT (PDB: 6S98) | UBE2SUBC C118A (PDB: 6S96) | |
|---|---|---|
| data collection | ||
| wavelength (Å) | 0.9680 | 1.033 |
| space group | P 1 21 1 | P 61 |
| unit cell parameters | ||
| α β γ (Å) | 44.8 49.05 71.93 | 120.9 120.9 45.3 |
| α β γ (°) | 90 106.03 90 | 90 90 120 |
| total reflections | 78466 (7412) | 39533 (3800) |
| unique reflections | 43484 (4292) | 19810 (1898) |
| Rpim | 2.5 (43.0) | 4.4 (40.8) |
| completeness (%) | 99.4 (98.9) | 98.8 (95.1) |
| I/σ(I) | 14.5 (2.0) | 8.6 (1.5) |
| multiplicity | 1.8 (1.7) | 2.0 (2.0) |
| Wilson B factor | 18.8 | 44.02 |
| CC ½ | 0.999 (0.638) | 0.997 (0.992) |
| refinement | ||
| resolution (Å) | 42.15 – 1.55 (1.605 – 1.55) | 34.91 – 2.18 (2.258 – 2.18) |
| Rwork / Rfree | 16.14 / 18.39 | 20.38 / 24.53 |
| no. of atoms | ||
| protein | 2358 | 2241 |
| water | 171 | 32 |
| average B-factors | ||
| protein | 29.5 | 57.2 |
| water | 32.1 | 49.8 |
| RMSD from ideality | ||
| bond lengths (Å) | 0.013 | 0.014 |
| bond angles (°) | 1.35 | 1.21 |
| Ramachandran statistics | ||
| favored (%) | 98.96 | 98.36 |
| disallowed (%) | 0.00 | 0.00 |
| MolProbity clash score | 3.35 | 9.42 |
| MolProbity overall score | 1.22 | 1.5 |
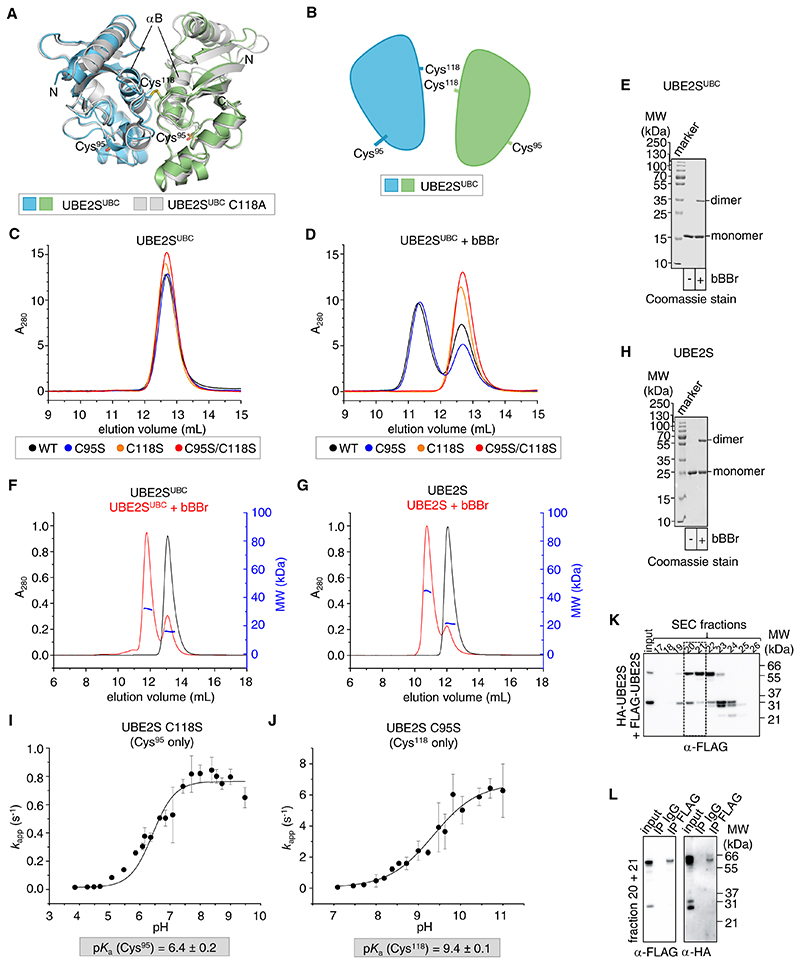
Fig. 2. Structure-guided crosslinking defines a specific dimerization mode of UBE2S in solution.
(A) Superposition of the crystal structures of UBE2SUBC wild-type (PDB: 6S98) and C118A (PDB: 6S96), determined here. Cys95 in 6S96 is oxidized to a sulfoxide. (B) Cartoon representation of the dimer (see A). (C) SEC of UBE2SUBC wild-type and variants(N=3 independent experiments). (D) SEC analogous to (B) upon bBBr-based crosslinking (N=2 independent experiments). (E) bBBr-based crosslinking of UBE2SUBC, monitored by SDS-PAGE (N=3 independent experiments). (F) SEC MALS analysis of UBE2SUBC after incubation with or without bBBr. Determined MWs: 16.5 ± 0.3 kDa (monomer) and 31.7 ± 0.5 kDa (dimer) (N=2 independent experiments). (G) SEC MALS analysis of UBE2S after incubation with or without bBBr (N=2 independent experiments). Determined MWs: 26 ± 2 kDa (monomer) and 47 ± 2 kDa (dimer). Note that SEC MALS (F, G) required higher protein concentrations than SEC (C, D), producing a larger fraction of dimer. (H) bBBr-based crosslinking of UBE2S, monitored by SDS-PAGE (N=3 independent experiments). (I) pKa-determination for the thiol group of Cys95 in UBE2S C118S, based on the reaction kinetics with DTNB. (J) pKa-determination for the thiol group of Cys118 in UBE2S C95S, determined as in (I). In (I) and (J) the mean and SD are plotted (N=5 independent experiments). (K) Immunoblot of mitotically-enriched, bBBr-treated extract from RPE-1 cells stably expressing Tet-induced HA-UBE2S and transiently expressing FLAG-UBE2S, separated by SEC (N=3 independent experiments). For the HA-immunoblot, see fig. S2A. (L) Immunoblot of control IgG and anti-FLAG immunoprecipitations from the indicated, pooled SEC fractions from (K) (N=2 independent experiments).
To test whether this arrangement mediates UBE2S dimerization in solution, we harnessed the central position of Cys118 at the interface for crosslinking experiments with the homo-bifunctional, thiol-reactive reagent dibromobimane (bBBr). This compound crosslinks thiol groups within close proximity (3.2 to 6.6 Å) (37, 38). We hypothesized that bBBr may specifically trap the Cys118-centered UBE2S dimer if it were transiently populated in solution. We thus incubated UBE2SUBC with bBBr, quenched the reaction with N-ethylmaleimide (NEM), and analyzed the sample by size exclusion chromatography (SEC). As anticipated, the addition of bBBr gave rise to a larger protein species, whereas no substantial oligomerization occured without crosslinker (Fig. 2C, D). The crosslinked species corresponds to dimeric UBE2SUBC, as shown by SDS-PAGE (Fig. 2E) and SEC-coupled multi-angle light scattering (MALS) (Fig. 2F): The experimentally determined molecular weight of 31.7 ± 0.5 kDa closely recapitulates the expected value of 34.7 kDa for a UBE2SUBC dimer (with 16.5 ± 0.3 kDa determined for the 17.4 kDa monomer). Likewise, crosslinking reactions with full-length UBE2S yielded a species of a SEC MALS-derived molecular weight of 47 ± 2 kDa in agreement with the corresponding dimer of 47.7 kDa (with 26 ± 2 kDa determined for the 23.9 kDa monomer) (Fig. 2G, H). That bBBr specifically captured a dimeric form of UBE2S is remarkable, because the E2 contains two cysteine residues, Cys118 and the catalytic Cys95, which may, in principle, promote higher-order crosslinking. In order to define the thiol groups that react with bBBr, we performed additional crosslinking experiments with single-cysteine variants of UBE2SUBC (C95S and C118S). These studies showed that the C95S variant was crosslinked with similar efficiency as the wild-type. In contrast, the C118S variant remained monomeric in the presence of crosslinker, just like the cysteine-free control (C95S/C118S) (Fig. 2C, D). bBBr-based crosslinking of UBE2S thus occurred specifically through Cys118.
Although these data are consistent with a transient dimerization of UBE2S in solution through the crystallographic interface, the crosslinking specificity for Cys118 could also be explained by an enhanced reactivity of Cys118 over Cys95 towards small-molecule electrophiles. To rule out this possibility we followed the reaction rates of either thiol group towards DTNB (5,5’-dithio-bis-2-nitrobenzoic acid) as a function of the pH by stopped-flow absorbance measurements. The derived pKa-values− 6.4 ± 0.2 for Cys95 (determined for the C118S variant) and 9.4 ± 0.1 for Cys118 (determined for the C95S variant) -demonstrate that Cys95 is more reactive than Cys118 (Fig. 2I, J). Therefore, the observed crosslinking specificity does not recapitulate the chemical reactivity of the cysteine residues. Instead, it reflects an inherent ability of UBE2S to self-associate in a manner that juxtaposes Cys118, but not Cys95, of two molecules; the dimer seen crystallographically represents precisely such an interaction mode.
We next investigated whether bBBr-based crosslinking can trap a UBE2S dimer in living cells. We transiently expressed FLAG-UBE2S in mitotically enriched non-transformed retina pigment epithelial (RPE-1) cells stably expressing tetracycline (Tet)-induced HA-UBE2S, exposed them to bBBr, and fractionated the cleared lysate by SEC. Indeed, immunoblotting revealed distinct species of MWs consistent with a UBE2S monomer and dimer (Fig. 2K; fig. S2A). That the later species represents a dimer (rather than UBE2S crosslinked with another cellular partner), was confirmed by co-immunoprecipitation (Fig. 2L). Together, these studies show that UBE2S transiently self-associates in vitro and in cells.
UBE2S dimerization requires the hydrophobic face of helix αB and is promoted by the C-terminal extension
Guided by the crystal structure, we characterized the interface mediating UBE2S dimerization in solution. Besides Cys118, interfacing residues include Leu107, His111, Leu114, Ile121, and His122 in helix aB (residues 109-122) of the UBC domain. Peripheral, polar contacts are provided by Glu51, which is part of a turn within the β-sheet region; Arg101 and Asp102, located in a loop adjacent to the active-site helix; as well as Asn124 and Tyr141 (Fig. 3A). We introduced mutations replacing these nine residues individually by alanine, both in UBE2SUBC and UBE2S, and subjected the purified proteins to bBBr-based crosslinking experiments. All tested variants were predominantly monomeric in the absence of crosslinker (fig. S2B), with generally reduced propensities to dimerize upon addition of bBBr compared to the wild-type; no higher-order oligomers were observed (fig. S2C). To quantify the mutational effects on the dimerization capacity we conducted kinetic measurements, exploiting the fact that bBBr fluoresces when both thiol-reactive groups have reacted (38) (Fig. 3B; fig. S3A, B). The determined crosslinking rates mirror our crystal structure-based predictions: the majority of alanine substitutions of interface residues, including of Arg101, Asp102, Leu107, His111, Leu114, His122, and Asn124, markedly reduced the crosslinking rates compared to the wild-type; only I121A and Y141A still crosslinked efficiently. The single-cysteine variants, C95S and C118S, served as controls,confirming that crosslinking specifically occurs through Cys118. In sum, these experiments establish that the crystallographic UBE2SUBC dimer and UBE2S dimerization in solution rely on a common set of contacts through the hydrophobic face of helix aB.
Fig. 3. UBE2S dimerization requires the hydrophobic face of helix αB and is promoted by the C-terminal extension.
(A) Detail of the dimerization interface in the crystal structure of UBE2SUBC wild-type (PDB: 6S98; see Fig. 2A); contacting side chains are illustrated as sticks. (B) bBBr-based crosslinking rates of 26 purified UBE2SUBC and UBE2S variants. (C) Domain organization of UBE2S. (D) bBBr-based crosslinking rates of three UBE2S variants of different length. (E) Competition experiment testing the effect of a 10-fold molar excess of a C-helix-derived peptide in trans on the bBBr-based crosslinking kinetics of UBE2S. (F) bBBr-based crosslinking rates of UBE2S1-196 and a UBE2S1-197-Ub fusion protein. In (B) and (D-F), the mean and SD are plotted (N=3 independent experiments); for raw data, see fig. S3B, S4A-C.
Moreover, we noticed that full-length UBE2S displayed a higher crosslinking propensity than UBE2SUBC – an effect that holds for all the mutated variants studied (Fig. 3B; fig. S3B). In other words, the transient dimerization of UBE2S through the UBC domain is promoted by its atypical C-terminal extension (residues 157-222). This structurally uncharacterized region is predicted to contain little secondary structure, except for a C-terminal, 23-residue a-helix, which we refer to as the ‘C-helix’ (Fig. 3C). A comparison of the bBBr-based crosslinking rates of UBE2SUBC, a construct lacking the C-helix (residues 1-196), and the full-length protein revealed that the C-helix is critical for enhancing dimerization (Fig. 3D; fig. S4A). In consequence, we predicted that the addition of a C-helix-derived peptide to UBE2S would inhibit dimerization. Indeed, the presence of the peptide reduced the crosslinking rate of UBE2S in competition experiments (Fig. 3E; fig. S4B). That a 10-fold molar excess of the peptide over UBE2S suppressed crosslinking by just ~ 40% is understandable, because the peptide competes in trans with the C-helix of UBE2S in cis. Assuming that the ~40-residue linker between the UBC domain and the C-helix is disordered, the estimated local concentration of the C-helix around the UBC domain within UBE2S is in the hundreds of micromolar range. Together, these experiments unveil a role of the C-helix in promoting UBE2S dimerization. Although the precise mechanism underlying this phenomenon remains to be elucidated and different scenarios are conceivable (fig. S5A), NMR analyses suggested that the C-helix-derived peptide can interact with the UBC domain in trans (fig. S5B, C). It is possible that such an interaction assists dimerization of UBE2S through the UBC domain either directly or allosterically (fig. S5A).
The C-helix contains numerous lysine residues that are efficiently auto-ubiquitinated in vitro and in cells, most prominently Lys197 (16, 18, 39). This prompted us to interrogate whether ubiquitination of the C-helix impacts the dimerization propensity of UBE2S. As a proxy for the Lys197-ubiquitinated state, we genetically linked ubiquitin to the C-terminus of UBE2S1-197. Compared to the corresponding unmodified form, this fusion protein displayed strongly reduced bBBr-based crosslinking rates (~60% reduction; Fig. 3F; fig. S4C). The effect may be selective to ubiquitin, because an analogous fusion of UBE2S1-197 to SUMO1 (C52S) still crosslinked efficiently (fig. S4D, E). Therefore, we propose that auto-ubiquitination of the C-helix and C-helix-enhanced dimerization are mutually exclusive and constitute disparate paths in the conformational landscape of UBE2S.
Dimerization of UBE2S confers auto-inhibition
The UBE2SUBC dimer and the functionally critical, closed UBE2SUBC-donor ubiquitin complex are structurally incompatible with each other, suggesting that dimerization confers auto-inhibition to the E2 (12, 13) (Fig. 4A, B). To test this idea we purified the bBBr-linked UBE2S dimer and monitored its ability to catalyze ubiquitin linkage formation and accept ubiquitin from the E1. In contrast to the monomeric form, the UBE2S dimer promoted neither unanchored ubiquitin chain formation nor auto-ubiquitination (Fig. 4C); it was also deficient in thioester formation with ubiquitin (Fig. 4D), implying that the dimerization-mediated auto-inhibition affects the first step of the E2 reaction cycle.
Fig. 4. Dimerization of UBE2S confers auto-inhibition.
(A)Crystal structure of UBE2SUBC with a ‘donor-like’ ubiquitin molecule (the complex is formed in trans, but recapitulates the closed state; PDB: 5BNB (13)). The side chains of Cys95, Leu107, His111, and Leu114 (which were selected for mutational studies) are shown.
(B)Crystal structure of the UBE2SUBC dimer (PDB: 6S98, see Fig. 2A) with side chains displayed as in (A). (C) Comparison of the isopeptid bond formation activities of UBE2S and its crosslinked dimeric form (N=3 independent experiments). The products of auto-ubiquitination (monomer-Ubn; highlighted by the red line) and unanchored ubiquitin chain formation (Ub2) are labeled. The asterisk denotes minor degradation of the dimer. (D) Comparison of the ubiquitin thioester formation activities of UBE2S and its crosslinked dimeric form (N=3 independent experiments). Available thioester-linked products(monomer~Ub) are indicated and sensitive to reducing agent (DTT). Both (C) and (D) include control reactions without ATP.
Notably, the monomeric and dimeric UBE2S species used in these assays were both isolated from the crosslinking reaction to ensure they were treated identically. We also verified by MS that the purified dimer was crosslinked specifically through Cys118 and did not carry additional modifications that may have caused a loss of activity (fig. S6A-C; table S1, S2). Together, these analyses demonstrate that dimeric UBE2S is auto-inhibited, in line with our structure-based predictions.
Disruption of the dimer interface interferes with self-association of UBE2S in the cell
We next asked whether the auto-inhibited dimer represents the self-association mode of UBE2S in the cell. To address this question, we searched for mutations that specifically disrupted dimerization while leaving the recognition of the donor ubiquitin intact. Although the respective protein interfaces overlap to a considerable degree (Fig. 4A, B) and share a number of key contacts, including Cys118 (Fig. 2D, 3B (12, 13)), we identified three residues that are required for dimerization (Fig. 3B), yet are located peripherally to (Leu114) or outside (Leu107, His111) of the donor binding site. To evaluate whether these residues are indeed dispensable for donor recognition, we capitalized on our previous discovery that the UBE2S-donor interaction is detectable in trans by NMR (12). In line with the structural model of the closed state (Fig. 4A), we found that the L107A and H111A mutations left the interaction between UBE2S and ubiquitin largely intact, whereas the L114E mutation weakened, but did not abolish it (similarly to the L114A mutation (Fig. 3B)) (fig. S7A-E). Consistently, neither of the mutations interfered with UBE2S activity on the APC/C in reconstituted substrate ubiquitination assays (fig. S8A-D). Note that the reduced dimerization propensities of the mutated variants compared to the wild-type did not manifest themselves in this set-up because UBE2S is predominantly monomeric in vitro.
Based on these analyses, we used PLAs to test the effect of the selected mutations on the co-localization propensities of UBE2S in the cell. Intriguingly, the L107A, H111A, and L114E variants co-localized significantly less than the wild-type, corroborating the notion that the dimer interface characterized in vitro drives UBE2S self-association in the native cellular environment (Fig. 5A, B).
Fig. 5. Disruption of the dimer interface reduces UBE2S co-localization in the cell.
(A) Representative PLA and immunostains for HA and FLAG epitopes in HCT116 UBE2S-/- cells co-transfected with plasmids encoding the indicated HA/FLAG-tagged UBE2S variants. Due to varying signal intensities, the PLA images were individually adjusted. scale bar: 10 μm. (B) Quantification of PLA signals. Black bars represent the median from three independent experiments, as shown in (A). N indicates the number of analyzed cells; ‘ratio’ denotes the median signal normalized to that of UBE2S WT. Statistical significance according to the Kruskal-Wallis and Dunn’s multiple comparison test (left) and an unpaired, two-tailed Mann-Whitney test (right), respectively. Note that we did not observe a correlation between the cellular amount of the HA- or FLAG-tagged UBE2S variants, respectively, and the number of PLA dots.
Disruption of the dimer interface increases UBE2S turnover and suppresses mitotic slippage
We next investigated whether UBE2S dimerization regulates the cooperation of this E2 with the APC/C during mitosis and its ability to promote its own turnover (14, 15, 22). To this end we established an siRNA-and-rescue system using isogenic, Tet-inducible stable RPE-1 cell lines (hTERT RPE-1 FRT/TR) expressing siRNA-resistant UBE2S and GFP from the same mRNA to efficiently replace endogenous UBE2S without overexpressing the transgene (Fig. 6A). We then assessed the half-life of wild-type UBE2S and the dimerization-deficient variants in cells treated with cycloheximide, which revealed that the H111A mutation and to a smaller degree, the L114E and L107A mutations, enhanced UBE2S turnover in a proteasome-dependent manner (Fig. 6B-D; fig. S9A, B). Because NMR and circular dichroism analyses demonstrated that the three protein variants are well-folded (fig. S7A and S9E) have unperturbed APC/C-dependent substrate ubiquitination activities in vitro (fig. S8A-D), this finding suggests that dimerization can protect UBE2S from degradation in cells. That the H111A mutation enhanced UBE2S turnover more strongly than did the L107A or L114E mutations is consistent with the latter two mutations interfering with UBE2S auto-ubiquitination in vitro (fig. S8E, F), which may attenuate proteasomal targeting. We therefore focused on the H111A mutation to further interrogate the cellular consequences of UBE2S dimerization.
Fig. 6. Disruption of the dimer interface increases UBE2S turnover and prevents mitotic slippage.
(A) Immunoblot establishing the siRNA-and-rescue system in RPE-1 cells replacing endogenous with Tet-inducible UBE2S, expressed from the same mRNA as GFP (N=3 independent experiments). Cells were arrested in prometaphase. CSE1 serves as a loading control. (B) Immunoblot monitoring UBE2S stability in the absence of translation. Cells treated as in (A) were complemented with UBE2S wild-type and H111A, respectively, andexposed to cycloheximide (CHX) for the indicated times. To assess the contribution of proteasomal activity MG132 was added, as indicated. (C) Fluorescence imaging-based quantification of UBE2S amounts (as monitored in (B)), normalized to GFP. The data represent the mean and SD (N=3 independent experiments). (D) Half-lives (mean and SD) of the indicated UBE2S variants, derived from non-linear fitting (one-phase decay) of the data in (B) (for wild-type and H11A) and in fig. S9A, B (for L107A and L114E). Significance according to Kruskal-Wallis and Dunn’s multiple comparison test. (E and F) Automated live-cell imaging-based quantification of the release of cells treated as in (A) and expressing UBE2S wild-type and H111A, respectively, from a DMA-induced SAC arrest. The data represent the mean and SD from 9 measurements (N=3 independent experiments), normalized to the number of mitotic cells at the start (t0). (G) Immunoblot showing the amount of UBE2S wild-type and H111A, respectively, during a DMA-induced SAC arrest in cells treated as in (A). (H) Quantification of the relative amount of UBE2S H111A (normalized to the WT amount) in prometaphase, normalized to GFP; mean and SD of data as shown in (G) (N=3 independent experiments). A WT bar is illustrated as a reference only. (I) Analysis of mitotic slippage of cells as in (A), but exposed to taxol, based on live-cell imaging and single-cell analysis. The duration of the mitotic arrest was measured based on 202 cells per condition (N=2 independent experiments). Black lines indicate the median. Red dots reflect cells that did not complete mitosis during the experiment.
Although UBE2S promotes the proteasomal degradation of APC/C substrates, low concentrations of this E2 (such as upon siRNA-mediated knockdown) are sufficient to sustain faithful mitosis (15, 21, 22); however, reduced concentrations of UBE2S hinder cellular release from a drug-induced SAC arrest and mitotic slippage (22). In order to probe the role of UBE2S dimerization in this context, we blocked UBE2S-depleted cells in prometaphase by the addition of dimethylenastron (DMA), released them into fresh medium, and followed their progression to mitotic exit by automated live-cell imaging. As expected, depletion of UBE2S delayed mitotic exit and was rescued by the expression of wild-type UBE2S (fig. S9C, D). The H111A variant rescued with similar efficiency (Fig. 6E, F), indicating that the dimer interface mutation did not obstruct UBE2S activity on the APC/C, in line with our in vitro data (fig. S8A-D). We noticed, however, that the amount of the H111A variant at the start of the experiment was reduced by 75% compared to the wild-type, despite both cell lines containing equal amounts of the GFP expression control (Fig. 6G, H). This observation mirrors the shortened lifetime of the H111A variant (Fig. 6D) and may imply that the amount of UBE2S was not limiting in our experimental set-up. Alternatively, our data are consistent with the possibility that a substantial fraction of wild-type UBE2S exists in the auto-inhibited, dimeric form during release from SAC arrest.
We also interrogated the effect of the H111A mutation in UBE2S on mitotic slippage. To this end we challenged the SAC with a low concentration of taxol and quantified the duration of the ensuing mitotic arrest by live-cell imaging (Fig. 6I). As expected, the complementation of endogenous UBE2S with wild-type UBE2S strongly promoted mitotic slippage and shortened the duration of the arrest by ~50%. In contrast, cells expressing the H111A variant showed only a mild increase in mitotic slippage compared to uninduced conditions. Note that upon depletion of UBE2S and in the absence of Tet, cells with inducible expression of UBE2S H111A arrested longer than those with inducible expression of wild-type UBE2S. This effect likely reflects a small extent of leaky expression of siRNA-resistant UBE2S, which manifested more strongly for the wild-type protein due to its enhanced stability or residual expression of endogenous UBE2S.
Together, these studies show that the reduced steady-state concentration of the dimerization-deficient UBE2S variant H111A is sufficient to release cells from SAC arrest but cannot promote mitotic slippage. Hence, the ability of UBE2S to dimerize and thereby escape auto-ubiquitination-induced degradation appears to be particularly important during conditions of sustained mitotic arrest.
Discussion
Historically coined ‘ubiquitin carriers’, E2s have emerged as active determinants of ubiquitin signaling, particularly due to their ability to govern specificity in RING E3-assisted ubiquitin linkage formation. E2s thus underlie stringent control mechanisms modulating their abundance, turnover, conformational dynamics and macromolecular interactions in the cell (40). However, with few exceptions, the structural underpinnings of these regulatory layers have remained unclear. A major challenge lies in the fact that E2s typically operate through transient macromolecular assemblies that involve weak multi-site interactions with individual affinities in the high micromolar to millimolar KD-range. The overall cellular concentrations of E2s, however, fall into the nanomolar to low micromolar range, for example, 147 nM for UBE2S in HeLa cells (41). The dynamic formation of E2-mediated protein complexes thus strongly depends on co-localization effects in the cell that are hard to recapitulate in vitro.
Using a combination of sensitive structural, biochemical and cell-based approaches this study unveiled an autoinhibited, dimeric state of the APC/C-associated, chainelongating E2, UBE2S that occludes the catalytically critical binding site for the donor ubiquitin on the E2 and prevents the E1-driven transfer of ubiquitin to the active site. Although structurally distinct, this mechanism resembles an alternative auto-inhibition mode of E2s, in which donor ubiquitin binding is obstructed by the auto-ubiquitination of a conserved lysine residue, ‘Lys+5’, close to the active site in the catalytic domain (18, 20). Modulating E2 activities at the level of donor ubiquitin recognition thus provides a broad regulatory theme, of which different structural manifestations have evolved.
Moreover, our analyses demonstrate that the functional consequences of these auto-inhibition modes are distinct. Whereas auto-ubiquitination at Lys+5 blocks catalytic activity without affecting UBE2S stability (18), mutational interference with dimerization reduces the half-life of UBE2S by promoting its proteasomal degradation. We hypothesize that dimerization confers a safe-lock to UBE2S, thereby providing a reservoir of molecules protected from auto-ubiquitination-driven turnover (Fig. 7). This may be particularly important in G2 and prometaphase, when UBE2S accumulates (15), but the APC/C is not fully engaged in substrate ubiquitination. Our results also imply that distinct, contextdependent UBE2S concentrations are required for mitotic progression: lower concentrations to release cells from a drug-induced SAC arrest and higher concentrations to promote mitotic slippage. In agreement with the latter, UBE2S is frequently overexpressed in tumor cells (42–47), which may allow them to overcome the SAC more easily and continue to proliferate without equal segregation of all chromosomes. It is, therefore, tempting to speculate that manipulating the dimerization of UBE2S holds therapeutic opportunities.
Fig. 7. Model of the conformational regulation of UBE2S, as analyzed in this study.
The catalytic UBC domain of UBE2S can form an auto-inhibited dimer, which is stabilized by the C-helix, possibly through inter-subunit contacts or allosterically. The C-helix concomitantly anchors UBE2S at the APC2/4 groove during ubiquitin chain elongation on APC/C substrates (8), stimulates chain initiation by UBE2C (11), and provides major auto-ubiquination sites that promote the proteasomal turnover of UBE2S (15, 16, 18, 39). Our model posits that the formation of the inhibited dimer stabilizes UBE2S in the cell by preventing auto-ubiquitination at the C-helix. Additional states in the conformational landscape of UBE2S not shown in this figure include the auto-inhibited, Lys+5- ubiquitinated form (18), interactions between the C-helix and the APC/C coactivators (10, 15, 19), and the architecture of the APC/C during ubiquitin chain initiation (7–10).
The particular dimeric arrangement we identified for UBE2S is not seen in other structures of E2s deposited in the PDB, indicating it may be specific to UBE2S. In line with this notion, dimerization of certain other E2s has been reported to occur through alternative interfaces and stimulate rather than inhibit catalytic activity (28, 48–51). Intriguingly, our studies demonstrate that the dimerization of the catalytic domain of UBE2S is promoted by its C-terminal extension, which is not conserved in other members of the E2 family and enables the specific APC/C-dependent functions of UBE2S (Fig. 7). Cryo-EM reconstructions of the APC/C in its chain-elongating state showed that the terminal C-helix of UBE2S is anchored within a groove at the APC2/4 interface (8, 9). This arrangement buries the C-helix, thereby excluding a simultaneous participation of this region in dimerization of UBE2S. In consequence, we posit that the dimeric state of UBE2S does not occur during APC/C-dependent chain elongation. Notably, the C-helix engages in distinct interactions with the APC/C during the activation cycle. In addition to the APC2/4 groove, the C-helix can interact with the APC/C co-activators, CDH1 and CDC20, which are positioned at the opposite face of the cullin platform, ~100 Å distant from the APC2/4 groove (10, 15, 19), as well as APC10 in Xenopus (19). To reconcile the presence of disparate binding sites for the C-helix on the APC/C, the co-activator has been proposed to recruit UBE2S to the APC/C, before the C-helix is delivered to the APC2/4 groove (10). Binding of the C-helix at the latter site activates the UBE2C-mediated priming of APC/C substrates with short ubiquitin chains, thereby providing a positive feedback mechanism for efficient chain elongation by UBE2S (11). In order to stabilize the auto-inhibited UBE2S dimer, we anticipate the C-helix being dislodged from either of its active positions. That such conformational rearrangements are possible is illustrated by the action of the APC/C inhibitor EMI1, which competes with the C-helix for binding to the APC2/4 groove (7, 19, 52).
The C-helix is enriched in lysine residues, which present major auto-ubiquitination sites of UBE2S in vitro and in cells (16, 18, 39). Our studies suggest that the attachment of ubiquitin to the C-helix interferes with dimerization. This leads us to propose that the auto-ubiquitination-induced degradation and dimerization-induced auto-inhibition of UBE2S constitute disparate pathways in the multifactorial network that regulates APC/C signaling. The cellular conditions in which individual pathways prevail and the precise order of events driving the associated conformational changes require further investigation. In particular, it will be interesting to interrogate whether additional factors control UBE2S dimerization in the cell. For instance, the key auto-ubiquitination site in UBE2S, Lys197, can also be acetylated (53), which opens regulatory opportunities through the C-helix beyond ubiquitination. Moreover, we anticipate that the formation of the UBE2S dimer is contigent upon co-localization effects triggered by the large protein complexes with which UBE2S engages(41).
The conformational transitions of UBE2S between its active monomeric form (12), the auto-inhibited dimer, and states promoting inhibitory (18) or degradative auto-ubiquitination (15) provide a paradigm for how mutually exclusive intra- and intermolecular interactions tune the concentrations and activities of E2s, thereby facilitating the precision of ubiquitin signaling. More broadly, such conformational cycling of ubiquitination enzymes explains how the cell balances two counteracting requirements of the ubiquitin system: (i) to maintain sufficient concentrations of enzymes for rapid responses when substrates ought to be modified and (ii) to prevent futile cycles of auto-ubiquitination and degradation of the enzymes when substrates are limiting. Storing enzymes in an inactive form that is poised for (re)activation provides an elegant solution to this challenge with diverse manifestations throughout the realms of cellular signaling and metabolism.
Materials and Methods
DNA constructs and antibodies
The plasmid encoding the ubiquitin-cyclin B1 NTD (residues 1-95 of cyclin B1 plus an engineered C-terminal cysteine) fusion was kindly provided by Brenda Schulman (Martinsried, Germany) (9); the plasmids encoding 3xHA-tagged UBE2S and FLAG-tagged UBE2S by Michael Rape (Berkeley, CA, USA); the plasmid encoding 2xHA-tagged ubiquitin by Steve Jackson (Cambridge, UK); the bacmid encoding the human APC/C by David Barford (Cambridge, UK) (54); and the bacmid encoding SBP-tagged CDC20 by Jonathon Pines (London, UK) (55). All other plasmids, cloning templates and primers are listed in table S3. Unless specified, ligation-free methods were used for cloning and site-directed mutagenesis (56). All antibodies used in this study are listed in table S4.
Mammalian cell culture
Cells were cultured according to standard techniques at 37°C in 5% CO2 and regularly tested for mycoplasma contamination. HCT116 UBE2S wild-type and knock-out cells with endogenously tagged cyclin B1-Venus were a gift from Chunaram Choudhary (Copenhagen, Denmark) (21). HCT116 cells were maintained in ‘DMEM, high glucose, pyruvate’ (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS) (Thermo Fisher Scientific), 1% (v/v) penicillin-streptomycin (Sigma-Aldrich), 1% (v/v) Glutamax (Thermo Fisher Scientific), and 0.5 μg/mL amphotericin B (Sigma-Aldrich); HCT116 UBE2S knock-out cells were maintained in the same media supplemented with 0.5 μg/mL puromycin (Sigma-Aldrich).
hTERT RPE-1 FRT/TR cells expressing endogenously tagged mRuby-PCNA (57) and histone3.1-iRFP were maintained in DMEM/F12 (Sigma-Aldrich) supplemented with 10% (v/v) Tet-free FBS, 1% (v/v) penicillin-streptomycin, 1% (v/v) Glutamax, 30 mM sodium bicarbonate (Thermo Fisher Scientific), and 0.5 μg/mL amphotericin B. Stable Tet-inducible cell lines expressing HA-tagged or untagged siRNA-resistant UBE2S wild-type, L107A, H111A, and L114E, respectively, were generated by electroporating the corresponding pCDNA5-FRT/TO plasmids and GFP (UBE2S-IRES-eGFP) together with a plasmid encoding flipase for integration into the single FRT site of hTERT RPE-1 FRT/TR (58); single clones were selected with 400 μg/ml neomycin (Sigma-Aldrich). The expression of wild-type UBE2S was induced with 0.02 μg/ml Tet; that of UBE2S L107A,
H111A, L114E, and HA-UBE2S, respectively, with 10 μg/ml Tet. For live-cell imaging, DMEM without phenol red and riboflavin (Thermo Fisher Scientific) supplemented with 10% (v/v) Tet-free FBS, 1% (v/v) Glutamax, 1% (v/v) penicillin-streptomycin, and 0.5 μg/ml amphotericin B was used.
Transfection
For transient transfections with plasmid DNA, HCT116 UBE2S knock-out or RPE-1 HA-UBE2S cells were seeded one day prior to transfection with Lipofectamine LTX Plus (Thermo Fisher Scientific) and cells collected after 24 (for crosslinking) and 48 hours (for PLA), respectively. Control (UGGUUUACAUGUCGACUAA) or UBE2S (ACAAGGAGGUGACGACACU) siRNAs were transfected at the indicated concentrations using RNAimax (Thermo Fisher Scientific).
Indirect immunofluorescence
Cells grown on glass coverslips were fixed with 4% (w/v) paraformaldehyde and permeabilized with an aqueous solution containing 0.1% (v/v) Triton and 0.02% (w/v) SDS in two consecutive 15-minute steps. Cells were washed with PBS, incubated in 5% horse serum (in PBS) for 1 hour, thereafter with primary antibodies in 5% (v/v) horse serum for 1 hour, and washed 3 times with 5% horse serum. Secondary antibodies were applied in 5% horse serum for 1 hour, followed by 3 washes with PBS, 0.1% (v/v) Tween 20. DNA was counter-stained with 4’,6-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich), and coverslips mounted using Vectashield (Linaris). A Delta Vision Core wide-field deconvolution fluorescence microscope with a CoolSNAPHQ2/HQ2-ICX285 camera (Applied Precision Inc.) and a UPlanSApo 40X/NA 0.95 lens (Olympus) was used for image acquisition.
Proximity ligation assay
Cells were treated as described for immunofluorescence, incubated with primary antibodies in 5% (v/v) horse serum at 4°C overnight, and washed with 5% (v/v) horse serum. The Duolink PLAs (Sigma-Aldrich) were performed according to the manufacturer’s instructions. Directly after PLA staining, the cells were incubated with the secondary antibodies at room temperature for 1 hour. Fluorescent z-stack images of randomly selected cells were acquired using a Delta Vision Core wide-field deconvolution fluorescence microscope with a UPlanSApo 100X/NA 1.4 oil immersion lens (Olympus). Only cells with low amounts of the analyzed proteins were considered. The number of PLA signals per cell was determined from maximum intensity projections using the ‘find maxima function’ (maxima within tolerance; noise tolerance: 150) in Fiji (59).
Cycloheximide pulse-chase assay
Tet-inducible UBE2S-IRES-eGFP hTERT RPE-1 FRT/TR cells were seeded in media +/- Tet, transfected with 25 nM UBE2S siRNA, and after 48 hours treated with 355 μM cycloheximide (CHX; VWR) or with CHX and 10 μM MG132 (VWR) for the indicated times. Cells were harvested in reducing LDS buffer (Thermo Fisher Scientific) and analyzed by SDS-PAGE and immunoblotting.
Analysis of UBE2S in prometaphase
Tet-inducible UBE2S-IRES-eGFP hTERT RPE-1 FRT/TR cells were transfected with 25 nM control or UBE2S siRNA and after 36 hours treated with 10 μM DMA (VWR). 12 hours later, cells were harvested by mitotic shake-off, flash frozen, re-suspended in 50 mM Tris pH 8.0, 150 mM NaCl, 2.5 mM MgCl2, 0.25 % (v/v) NP40, 5% (v/v) glycerol, 1 mM DTT and 1 mM PMSF and incubated on ice for 15 minutes (Fig. 6A). Alternatively, cells were resuspended in 8 M urea, 20 mM Tris pH 7.5, 135 mM NaCl, 1% (v/v) Triton, 10% (v/v) glycerol, 1.5 mM MgCl2, 5 mM EDTA, 50 mM ioadoacetamide, protease and phosphatase inhibitor cocktails (Roche), 10 μM MG132, and 0.25 U/μl benzonase (Sigma-Aldrich) and incubated on ice for 30 minutes (Fig. 6G). Lysates were cleared by centrifugation and proteins analyzed by SDS-PAGE and immunoblotting.
Mitotic release assay
Tet-inducible UBE2S-IRES-eGFP hTERT RPE-1 FRT/TR cells were seeded in 96-well plates in media +/- Tet, transfected with 65 nM control or UBE2S siRNA, and treated with 10 μM DMA after 36 hours. 12 hours later, cells were washed twice with PBS, and released into ‘imaging DMEM’ (see above) +/- Tet. Bright-field images were acquired every 15 or 25 minutes over a period of 24 hours on a Micro XLS wide-field screening microscope (Molecular Devices) with a 10x, 0.5 numerical aperture Plan Apo air objective (Nikon) and full environmental control (5% CO2, 37°C). Mitotic cells were identified as round cells using a customized automated analysis routine (MetaXpress, Molecular Devices) and normalized to the number of mitotic cells at the beginning of the release.
Mitotic slippage assay
Tet-inducible UBE2S-IRES-eGFP hTERT RPE-1 FRT/TR cells were seeded in 96-well plates in media +/- Tet and, in parallel, transfected with 65 nM UBE2S siRNA. After 48 hours 50 nM taxol (Sigma-Aldrich) was added to induce a prometaphase arrest, and mitotic cells monitored by live-cell imaging. The length of the arrest was manually quantified as the time between nuclear envelope breakdown and onset of chromosome segregation.
Recombinant protein preparation
The preparation of untagged, FLAG-tagged, and 3xHA-tagged UBE2S in unlabeled and isotope-enriched forms was previously described (12); mutated variants thereof as well as the UBE2S1-197-Ub and UBE2S1-197-SUMO1 C52S fusion constructs were purified analogously. Human UBA1 and ubiquitin were prepared as previously described (12); likewise APC/C, CDC20 (18) and Ub-cyclin B1 NTD (9).
His6-tagged UBE2SUBC/UBE2S was expressed in E.coli BL21 DE3 at 20 °C overnight. Cells were lysed in 50 mM Tris pH 8.0, 500 mM NaCl, 5 mM benzamidine, 8 mM β-mercaptoethanol (β-ME), 0.4% (v/v) Triton X-100, and protease-inhibitor cocktail (Roche), the protein purified by Ni-affinity chromatography (HisTrap HP; GE Healthcare) in 50 mM Tris pH 8.0, 500 mM NaCl, 71 mM imidazole, and 8 mM β-ME, and buffer-exchanged into 25 mM Tris pH 7.4, 100 mM NaCl, and 2 mM DTT, followed by SEC (HiLoad 16/600 Superdex 75; GE Healthcare) in the same buffer.
To purify the crosslinked dimer, 600 μM UBE2S was reacted with 1.8 mM bBBr (Sigma-Aldrich) at room temperature for 30 minutes, quenched with 2 mM DTT, and subjected to SEC (Superdex 75, 10/300 GL; GE Healthcare) in 25 mM Tris pH 6.8, 100 mM NaCl, and 2 mM DTT.
NMR
NMR data were recorded at 25°C on a 700 MHz Avance IIIHD spectrometer with a 1H/15N/13C cryo-probe, processed with in-house software, and analyzed with NMRViewJ (60). All samples contained 10% (v/v) D2O, 5 mM DTT, and 2 mM TCEP in addition to the specified buffer. The backbone resonance assignments for UBE2SUBC were taken from BMRB entry 17437 (12). Amide proton relaxation rates were estimated by two-point spinecho measurements with a spin-echo included in the first INEPT of an HSQC experiment (61). 15N longitudinal (R1) and transverse (R2) relaxation rates were measured as previously described (62). The relaxation delays were fitted to a mono-exponential decay (NMRViewJ). The temperature was controlled by an NMR thermometer (Bruker), based on the difference in the 2H signals of HDO and 50 mM sodium acetate-d3, with which the samples were supplemented.
To determine the dissociation constant, KD of the interaction of ubiquitin with UBE2S variants, phase-sensitive 1H-15N BEST-TROSY (63) spectra were recorded of 200 μM 15N-enriched UBE2S without or with increasing concentrations of unlabeled ubiquitin in 75 mM sodium phosphate pH 7.2, 5 mM DTT, and 1 mM EDTA. Weighted combined chemical shift perturbations, Δδ(1H15N), were calculated according to
KD-values were derived from global fitting of the data to a single-site model, given by
where [Ptot] and [Ltot] denote the concentrations of UBE2S and ubiquitin, respectively. Only resonances above the threshold of Δδ(1H15N) > 0.048 ppm in the presence of 6.5 mM ubiquitin were included in the fitting.
To test for interactions of a C-helix-derived peptide (residues 197-222 of UBE2S) with UBE2S in trans, phase-sensitive 1H-15N BEST-TROSY (63) spectra were recorded of 200 μM 15N-enriched UBE2S1-196 without or with unlabeled peptide at 0.5 to 6.5 mM concentration in 75 mM sodium phosphate pH 7.2, 5 mM DTT, and 1 mM EDTA. Weighted combined chemical shift perturbations were calculated as described above.
Circular dichroism (CD)
CD spectroscopy was performed with a JASCO J-810 spectropolarimeter in a temperature-controlled quartz cuvette with a layer thickness, d, of 0.01 cm at 20°C. The UBE2S samples were in 50 mM potassium phosphate, pH 7.9, at a concentration, [c], of 5 μM. Spectra were recorded in 0.1-nm steps within a wavelength range of 190 to 260 nm at a scanning speed of 20 nm/min, and a band width of 1 nm. 15 spectra were accumulated per sample, the reference buffer spectrum subtracted, and the molar ellipticity [Θ] calculated as
where Θ denotes the measured ellipticity in mdeg and NAS the number of amino acids.
X-ray crystallography
UBE2SUBC wild-type crystals grew at 11.7 mg/ml and 20°C in sitting drops containing 0.2 M sodium acetate trihydrate, 0.1 M Tris pH 8.5, and 30% (w/v) PEG 4000. The same conditions including 10% (v/v) glycerol were used for cryo-protection. Diffraction data were collected at beamline ID30A, ESRF, Grenoble/France.
UBE2SUBC C118A crystals grew at 22.0 mg/ml and 20°C in sitting drops containing 2.5 M potassium formate and 0.1 M sodium cacodylate pH 6.5. The same conditions including 20% glycerol were used for cryo-protection. Diffraction data were collected at beamline P13, PETRA III, DESY, Hamburg/Germany.
Data processing was performed with XDS (64); molecular replacement with Phaser (65), as implemented in ccp4 (66), using PDB entry 1ZDN (33) as a search model; refinement with Phenix (67); and model building with Coot (68).
Pull-down experiments
Equimolar amounts of His6- and HA-tagged UBE2S was incubated in 25 mM Tris pH 7.4, and 100 mM NaCl on ice for 1 hour, thereafter with Ni-NTA agarose (Macherey-Nagel) at 20°C for 15 minutes, and washed with 500 mM sodium phosphate pH 7.5, 500 mM NaCl, 30 mM imidazole, and 2 mM DTT. The supernatant was removed, the proteins were eluted with reducing loading dye and analyzed by SDS-PAGE and immunoblotting.
Crosslinking
For SEC, 40 μM of the specified protein was incubated with 60 μM bBBr at RT for 40 minutes, quenched with 5 mM NEM (Sigma-Aldrich) for 10 minutes, and SEC (Superdex 75, 10/300 GL) performed in 25 mM Tris pH 6.8, and 100 mM NaCl. Samples for SDS PAGE were prepared analogously, but quenched in reducing loading dye.
For SEC MALS, 300 μM UBE2S was incubated with 450 μM bBBr at RT for 1 hour, quenched with 5 mM NEM, and analyzed by SEC (Superdex 75 10/300 GL) coupled to Dawn8+ and Optilab T-rEX detectors (Wyatt Technology) at RT. Molecular weights were determined at the tips of the elution peaks using ASTRA 6 (Wyatt Technology).
To analyze kinetics, 20 μM protein was incubated with 30 μM bBBr in 25 mM Tris pH 6.8, and 100 mM NaCl and the fluorescence (λex: 315 - 390 nm; λem: 420 - 480 nm) measured with a CLARIOstar plate reader (BMG Labtech). The initial ~5-minute data portion was fitted by linear regression using OriginPro (OriginLab). The peptide (residues 197-222 of UBE2S) used in competition assays was purchased from Elim Biopharm at >95% purity.
To perform cell-based crosslinking, HA-UBE2S expressing RPE-1 cells transfected with FLAG-UBE2S for 24 hours were enriched in prometaphase by 14-hour treatment with 332 nM nocodazole (Sigma-Aldrich), followed by a mitotic shake-off, washing in PBS, and incubation with 40 μM bBBr at 37°C for 20 minutes. Subsequently, cells were washed with PBS and lysed in 1% Triton X-100, 0.01% SDS in PBS supplemented with 100 mM DTT (to quench excess bBBr) on ice for 15 minutes. ~ 500 μg of the cleared supernatant after spinning was analyzed by SEC (Superdex 75 PC 3.2/30) in PBS at 4°C, collecting 50 μl fractions. For IP of FLAG-UBE2S, the SEC fractions 20 and 21 were pooled and added to control and FLAG antibodies, respectively, crosslinked to Dynabeads (Invitrogen). After 2 hours at 4°C, the resin were washed with lysis buffer, bound proteins eluted at RT by non-reducing LDS buffer (Thermo Fisher Scientific) supplemented with 100 mM DTT, and heat-denatured for 5 minutes prior to SDS-PAGE.
Mass Spectrometry
For denaturing, intact ESI-MS (electrospray ionization-mass spectrometry) analyses the purified, bBBr-linked UBE2S dimer was buffer-exchanged into 20 mM ammonium acetate, pH 8.0. 20 μl of 29 μM protein was then diluted with 60 μl of an aqueous solution containing 20% (v/v) acetonitrile and 0.2% (v/v) formic acid (FA) and injected into a LTQ XL mass spectrometer (Thermo Fisher Scientific) at 5 μl/min. The measurements were performed using Tune Plus v.2.5.0 (Thermo Fisher Scientific) at the ‘normal’ scan rate in the range of 550-2000 m/z and the spectra deconvoluted with ProMass for Xcalibur v.2.8 (Thermo Fisher Scientific).
For the MS-based mapping of the bBBr crosslinking site(s) in UBE2S, the crosslinking reaction was subjected to SDS PAGE, the band corresponding to the UBE2S dimer excised and split into two. One half was reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide, before in-gel digestion with 12.5 mg/L trypsin; the other was digested without reduction and alkylation. The extracted peptides were dissolved in an aqueous solution containing 5% (v/v) acetonitrile and 0.1% (v/v) FA, injected into a UHPLC instrument (UltiMateTM 3000 RSLC; Thermo Fisher Scientific), in-line desalted for 3 minutes on a trapping column (5-mm μ-pre-column C18PepMap100; Thermo Fisher Scientific), and separated on a 28-cm analytical column (packed in-house with C18 1.9 μm Reprosil beads; Dr. Maisch GmbH) using a 43-minute, linear, 10 to 42% gradient of mobile phase B in A at a flow rate of 0.3 μl/min (mobile phase A: 0.1% (v/v) FA in water; mobile phase B: 80% (v/v) acetonitrile and 0.08% (v/v) FA in water). The separated peptides were analyzed on-line by ESI-MS using a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). MS2 was performed for the 40 most abundant precursors observed in the MS1 scan in the range of 350 to 1600 m/z and charge states of 2 to 6. Precursor ions were isolated with a 1.4-m/z isolation window and fragmented at a normalized collision energy of 30%. The MS1 and MS2 resolutions were set to 60,000 and 15,000, respectively; a dynamic exclusion of 20 s was used. To identify protein–protein crosslinks, the raw files were searched with the pLink 2.3.0 software (http://pfind.ict.ac.cn/software/pLink (69, 70)) against a database containing UBE2S and 293 common contaminating proteins. Oxidation of methionine and carbamidomethylation of cysteine residues were considered as variable modifications. The MS1 and MS2 tolerances were set to 10 and 20 ppm, respectively; the crosslink and monolink mass additions to 188.059 and 267.985 Da, respectively. The results were filtered at FDR 1%.
pKa Determination
The pKa values of the thiol groups of Cys95 and Cys118 were derived from the reaction kinetics of 14.4 μM UBE2S C95S and C118S, respectively, with 100 μM DTNB at 10°C in 100 mM NaCl, 1 mM EDTA, and one of the following buffers (pH-range 3.8 to 11.0): 20 mM sodium acetate, 20 mM sodium phosphate, 20 mM Tris-HCl, or 20 mM glycine. At least 5 reactions under identical conditions were monitored by absorbance (λ = 412 nm) using an SFM-3000S stopped-flow instrument (Bio-Logic), the data averaged, and fitted to a single exponential function (BioKine, Bio-Logic). The rate constants from 3 technical replicates were averaged, plotted against the pH-value, and fitted to a two-state model (OriginPro):
where kapp is the experimentally derived apparent rate constant, and ksH and ks− are the rate constants of the reaction of fully protonated and deprotonated cysteines, respectively (71).
UBE2S activity assays in vitro
Thioester formation was assessed by incubating 0.25 μM UBA1, 30 μM ubiquitin, 3 mM ATP, and 9 mM MgCl2 in 25 mM Tris pH 7.0, and 100 mM NaCl on ice for 3 minutes. The reaction was then supplemented with 2 μM E2 at RT for 10 minutes and quenched with non-reducing and reducing loading dye, respectively. To assess isopeptide bond formation, 0.25 μM UBA1, 2 μM E2, 30 μM ubiquitin, 3 mM ATP, and 7.5 mM MgCl2 were incubated in 25 mM Tris pH 7.0, and 100 mM NaCl. All reactions were incubated at 30°C for the indicated times, quenched with reducing loading dye, and analyzed by SDS-PAGE and Coomassie staining.
APC/C-dependent ubiquitination reactions with recombinant enzymes were performed in 30 mM HEPES pH 7.4, 175 mM NaCl, 8 mM MgCl2, 0.05% (v/v) Tween-20, 1 mM DTT, and 5% (v/v) glycerol at 30°C and contained 0.05 μM UBA1, 0.28 μM UBE2S, 0.02 μM APC/C, 0.39 μM CDC20, 21 μM His6-ubiquitin, 0.035 μM IR dye-labelled Ub-cyclin B1 NTD, 2.6 mM ATP, 10 mM phosphocreatine, and 11 μM creatine kinase. The reactions were quenched with LDS-sample buffer (Thermo Fisher Scientific) supplemented with 100 mM DTT and subjected to SDS-PAGE and fluorescence imaging.
Statistical analyses
Prism 6.0 (Graphpad) was used, unless specified otherwise.
Supplementary Material
Acknowledgements
We thank the CMCB Light Microscopy Facility, TU Dresden; the technical staff at the ESRF and DESY; the Northern Bavaria NMR Center; Julia Haubenreisser and Doris Müller for technical assistance; Bodo Sander for advice; John Kuriyan, Alexander Buchberger, Christian Feiler, and Hermann-Josef Meyer for insightful discussions.
Funding
This work was supported by the DFG: Emmy Noether LO 2003/1-1 (S.L.), Emmy Noether MA5831/1-1 (J.M.), MA5831/3-1 (J.M.), GRK2243 (S.L.); the EMBO YIP (S.L.); the ERC under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 680042; J.M.); and a DIGSBB fellowship (A.K.).
Footnotes
Author Contributions: J.M. and S.L. designed research. A.K.L.L., A.K., K.S., D.S, O.D., J.M, and S.L. performed research. A.K.L.L., A.K., K.S., D.S, O.D., J.M., and S.L. analyzed data. A.K.L.L. and S.L. wrote the paper (original draft). A.K., K.S., O.D., H.U., and J.M. completed and edited the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
Atomic coordinates and structure factors have been deposited in the PDB under accession codes 6S98 (UBE2SUBCwild-type) and 6S96 (UBE2SUBC C118A). All other data needed to evaluate the conclusions of this paper are in the paper or the Supplementary Materials.
References and Notes
- 1.Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annual review of Biochemistry. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 2.Walden H, Rittinger K. RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nature Structural & Molecular Biology. 2018;25:440–445. doi: 10.1038/s41594-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 3.Lorenz S. Structural mechanisms of HECT-type ubiquitin ligases. Biological Chemistry. 2017;399:127–145. doi: 10.1515/hsz-2017-0184. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CAP. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle?s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barford D. Structural interconversions of the anaphase-promoting complex/cyclosome (APC/C) regulate cell cycle transitions. Current Opinion in Structural Biology. 2019;61:86–97. doi: 10.1016/j.sbi.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Watson ER, Brown NG, Peters J-M, Stark H, Schulman BA. Posing the APC/C E3 Ubiquitin Ligase to Orchestrate Cell Division. Trends in Cell Biology. 2019;29:117–134. doi: 10.1016/j.tcb.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu K-P, Zhang W, Yu S, Mercredi PY, Harrison JS, Davidson IF, et al. Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C. Cell. 2016;165:1440–1453. doi: 10.1016/j.cell.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown NG, Watson ER, Weissmann F, Jarvis MA, VanderLinden R, Grace CRR, Frye JJ, Qiao R, Dube P, Petzold G, Cho SE, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Molecular Cell. 2014;56:246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin Chain Elongation Requires E3-Dependent Tracking of the Emerging Conjugate. Molecular Cell. 2014;56:232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Chacin RC, Bodrug T, Bolhuis DL, Kedziora KM, Bonacci T, Ordureau A, Gibbs ME, Weissmann F, Qiao R, Grant GD, Cook JG, et al. Ubiquitin chain-elongating enzyme UBE2S activates the RING E3 ligase APC/C for substrate priming. Nat Struct Mol Biol. 2020:1–11. doi: 10.1038/s41594-020-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz S, Bhattacharyya M, Feiler C, Rape M, Kuriyan J. Crystal Structure of a Ube2S-Ubiquitin Conjugate. PLoS ONE. 2016;11:e0147550. doi: 10.1371/journal.pone.0147550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the Anaphase-Promoting Complex. Proceedings of the National Academy of Sciences. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proceedings of the National Academy of Sciences. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bremm A, Freund SMV, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nature Structural & Molecular Biology. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer H-J, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liess AKL, Kucerova A, Schweimer K, Yu L, Roumeliotis TI, Diebold M, Dybkov O, Sotriffer C, Urlaub H, Choudhary JS, Mansfeld J, et al. Autoinhibition Mechanism of the Ubiquitin-Conjugating Enzyme UBE2S by Autoubiquitination. Structure. 2019;27:11185–1187. doi: 10.1016/j.str.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Sako K, Suzuki K, Isoda M, Yoshikai S, Senoo C, Nakajo N, Ohe M, Sagata N. Emi2 mediates meiotic MII arrest by competitively inhibiting the binding of Ube2S to the APC/C. Nature Communications. 2014;5 doi: 10.1038/ncomms4667. [DOI] [PubMed] [Google Scholar]
- 20.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Molecular Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 21.Wild T, Larsen MSY, Narita T, Schou J, Nilsson J, Choudhary C. The Spindle Assembly Checkpoint Is Not Essential for Viability of Human Cells with Genetically Lowered APC/C Activity. Cell Reports. 2016;14:1829–1840. doi: 10.1016/j.celrep.2016.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nature Cell Biology. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biology Cb. 2015;25:R1002–18. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi M, VanderLinden R, Weissmann F, Qiao R, Dube P, Brown NG, Haselbach D, Zhang W, Sidhu SS, Peters J-M, Stark H, et al. Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation. Mol Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry. 1997;36:10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Yuan YC, Shen B, Chen DJ, Chen Y. Conformational flexibility of a ubiquitin conjugation enzyme (E2) Biochemistry. 1999;38:1415–1425. doi: 10.1021/bi981840h. [DOI] [PubMed] [Google Scholar]
- 27.Ptak C, Prendergast JA, Hodgins R, Kay CM, Chau V, Ellison MJ. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. The Journal of Biological Chemistry. 1994;269:26539–26545. [PubMed] [Google Scholar]
- 28.Varelas X, Ptak C, Ellison MJ. Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Molecular and Cellular Biology. 2003;23:5388–5400. doi: 10.1128/MCB.23.15.5388-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knipscheer P, Dijk WJV, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. The EMBO Journal. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vittal V, Wenzel DM, Brzovic PS, Klevit RE. Biochemical and structural characterization of the ubiquitin-conjugating enzyme UBE2W reveals the formation of a noncovalent homodimer. Cell Biochemistry and Biophysics. 2013;67:103–110. doi: 10.1007/s12013-013-9633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi C, Li D-F, Feng L, Hou Y, Sun H, Wang D-C, Liu W. Biochemical and structural characterization of a novel ubiquitin-conjugating enzyme E2 from Agrocybe aegeria reveals Ube2w family-specific properties. Scientific Reports. 2015;5:16056. doi: 10.1038/srep16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. The Journal of Biological Chemistry. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheng Y, Hong JH, Doherty R, Srikumar T, Shloush J, Avvakumov GV, Walker JR, Xue S, Neculai D, Wan JW, Kim SK, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Molecular & Cellular Proteomics. 2012;11:329–341. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nature Biotechnology. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 35.Rossi P, Swapna GVT, Huang YJ, Aramini JM, Anklin C, Conover K, Hamilton K, Xiao R, Acton TB, Ertekin A, Everett JK, et al. A microscale protein NMR sample screening pipeline. Journal of Biomolecular NMR. 2010;46:11–22. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein science : a publication of the Protein Society. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JS, Raines RT. Dibromobimane as a fluorescent crosslinking reagent. Analytical Biochemistry. 1995;225:174–176. doi: 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]
- 39.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Research. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Research. 2016;26:423–440. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. eLife. 2016;5:570. doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tedesco D, Zhang J, Trinht L, Lalehzadeh G, Meisner R, Yamaguchi KD, Ruderman DL, Dinter H, Zajchowski DA. The Ubiquitin-Conjugating Enzyme E2-EPF Is Overexpressed in Primary Breast Cancer and Modulates Sensitivity to Topoisomerase II Inhibition. Neoplasia. 2007;9:601–613. doi: 10.1593/neo.07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M-F, Lee K-D, Lu M-S, Chen C-C, Hsieh M-J, Liu Y-H, Lin P-Y, Chen W-C. The predictive role of E2-EPF ubiquitin carrier protein in esophageal squamous cell carcinoma. Journal of Molecular Medicine. 2009;87:307–320. doi: 10.1007/s00109-008-0430-3. [DOI] [PubMed] [Google Scholar]
- 44.Roos FC, Evans AJ, Brenner W, Wondergem B, Klomp J, Heir P, Roche O, Thomas C, Schimmel H, Furge KA, Teh BT, et al. Deregulation of E2-EPF ubiquitin carrier protein in papillary renal cell carcinoma. The American Journal of Pathology. 2011;178:853–860. doi: 10.1016/j.ajpath.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayesha AK, Hyodo T, Asano E, Sato N, Mansour MA, Ito S, Hamaguchi M, Senga T. UBE2S is associated with malignant characteristics of breast cancer cells. Tumor Biology. 2015;37:763–772. doi: 10.1007/s13277-015-3863-7. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z, Xu L. UBE2S promotes the proliferation and survival of human lung adenocarcinoma cells. BMB Reports. 2018;51:642–647. doi: 10.5483/BMBRep.2018.51.12.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Wang Y, Li Y, Yin W, Mo L, Qian X, Zhang Y, Wang G, Bu F, Zhang Z, Ren X, et al. Ube2s stabilizes beta-Catenin through K11-linked polyubiquitination to promote mesendoderm specification and colorectal cancer development. Cell Death & Disease. 2018;9 doi: 10.1038/s41419-018-0451-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 49.Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. The EMBO Journal. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proceedings of the National Academy of Sciences. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W, Shang Y, Zeng Y, Liu C, Li Y, Zhai L, Wang P, Lou J, Xu P, Ye Y, Li W. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. The EMBO Journal. 2014;33:46–61. doi: 10.1002/embj.201385315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frye JJ, Brown NG, Petzold G, Watson ER, Grace CRR, Nourse A, Jarvis MA, Kriwacki RW, Peters J-M, Stark H, Schulman BA. Electron microscopy structure of human APC/C-CDH1-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature Structural & Molecular Biology. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, Carr SA. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Molecular & Cellular Proteomics. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Yang J, Kong EH, Chao WCH, Morris EP, daFonseca PCA, Barford D. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) The Biochemical Journal. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 55.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Ent F, Löwe J. RF cloning : a restriction-free method for inserting target genes into plasmids. Journal of Biochemical and Biophysical Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Zerjatke T, Gak IA, Kirova D, Fuhrmann M, Daniel K, Gonciarz M, Müller D, Glauche I, Mansfeld J. Quantitative Cell Cycle Analysis Based on an Endogenous All-in-One Reporter for Cell Tracking and Classification. Cell Reports. 2017;19:1953–1966. doi: 10.1016/j.celrep.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nature Cell Biology. 2011;13:1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in Molecular Biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 61.Iwahara J, Tang C, Clore GM. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. Journal of Magnetic Resonance. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 63.Favier A, Brutscher B. Recovering lost magnetization: polarization enhancement in biomolecular NMR. Journal of Biomolecular NMR. 2011;49:9–15. doi: 10.1007/s10858-010-9461-5. [DOI] [PubMed] [Google Scholar]
- 64.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta crystallographica Section D, Biological Crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological Crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D, Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Yang B, Wu Y-J, Zhu M, Fan S-B, Lin J, Zhang K, Li S, Chi H, Li Y-X, Chen H-F, Luo S-K, et al. Identification of cross-linked peptides from complex samples. Nature Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 70.Sinz A, Wang K. Mapping spatial proximities of sulfhydryl groups in proteins using a fluorogenic cross-linker and mass spectrometry. Analytical Biochemistry. 2004;331:27–32. doi: 10.1016/j.ab.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 71.Koch JR, Schmid FX. Mia40 targets cysteines in a hydrophobic environment to direct oxidative protein folding in the mitochondria. Nature Communications. 2014;5:3041. doi: 10.1038/ncomms4041. [DOI] [PubMed] [Google Scholar]
- 72.Chen Z-L, Meng J-M, Cao Y, Yin J-L, Fang R-Q, Fan S-B, Liu C, Zeng W-F, Ding Y-H, Tan D, Wu L, et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nature Communications. 2019;10:3404–12. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bakos G, Yu L, Gak IA, Roumeliotis TI, Liakopoulos D, Choudhary JS, Mansfeld J. An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules. Nature Communications. 2018;9:4776. doi: 10.1038/s41467-018-07251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the PDB under accession codes 6S98 (UBE2SUBCwild-type) and 6S96 (UBE2SUBC C118A). All other data needed to evaluate the conclusions of this paper are in the paper or the Supplementary Materials.