Abstract
Tyrosyl DNA phosphodiesterase 2 (TDP2) facilitates the repair of topoisomerase II (TOP2)-linked DNA double-strand breaks and as a consequence is required for cellular resistance to TOP2 “poisons”. Recently, a deazaflavin series of compounds were identified as potent inhibitors of TDP2, in vitro. Here, however, we show that whilst deazaflavins can induce cellular sensitivity to the TOP2 poison etoposide they do so independently of TDP2 status. Consistent with this, both the cellular level of etoposide-induced TOP2 “cleavage complexes” and the intracellular concentration of etoposide was increased by incubation with deazaflavin, suggesting an impact of these compounds on etoposide uptake/efflux. In addition, deazaflavin failed to increase the level of TOP2 cleavage complexes or sensitivity to m-AMSA, a different class of TOP2 poison to which TDP2-defective cells are also sensitive. In conclusion, whilst deazaflavins are potent inhibitors of TDP2 in vitro their limited cell permeability and likely interference with drug influx/efflux limits their utility, in cells.
Introduction
TDP2 is a human 5’-tyrosyl DNA phosphodiesterase that mediates the hydrolytic removal of TOP2 peptide from the 5’-terminus of topoisomerase II (TOP2)-induced DNA double-strand breaks (DSBs) 1. As such, TDP2 influences levels of cellular sensitivity to TOP2 poisons2,3; a chemically diverse class of anti-cancer compounds that include podophylotoxins (e.g. etoposide and teniposide), anthracyclines (e.g. doxorubicin, daunorubicin), and amsacrine (m-AMSA) and all of which promote the formation of TOP2-linked DNA double-strand breaks (DSBs)4,5. TDP2 has also been identified as the host cell enzyme that can remove either Vpg from the 5’-termini of RNA molecules in certain types of enterovirus6,7 and viral reverse transcriptase from the 5’-terminus of hepatitis B circular DNA molecules8,9. Consequently, small molecule inhibitors of TDP2 have potential therapeutic value both in cancer therapy and as anti-viral drugs. We and others recently described the identification of deazaflavins as potent and specific inhibitors of TDP2 in vitro10–12. These compounds interact with a hydrophobic shelf in TDP2 to which the substrate DNA binds and gains access to the active site11. To address the cellular activity of the deazaflavins, we examined here three deazaflavines for their impact on cellular sensitivity to the TOP2 poisons etoposide and m-AMSA. In contrast to a previous report12, we report that the deazaflavines do not significantly inhibit TDP2 in cells, most likely because of poor cell permeability. In fact, surprisingly, we show here that some members of this chemical family act independently of TDP2 and increase the cellular uptake and/or retention of etoposide, resulting in phenotypes reminiscent of TDP2 inhibition but which are nevertheless “off-target”.
Results and Discussion
Deazaflavin compounds are potent small-molecule inhibitors of human TDP2, in vitro11
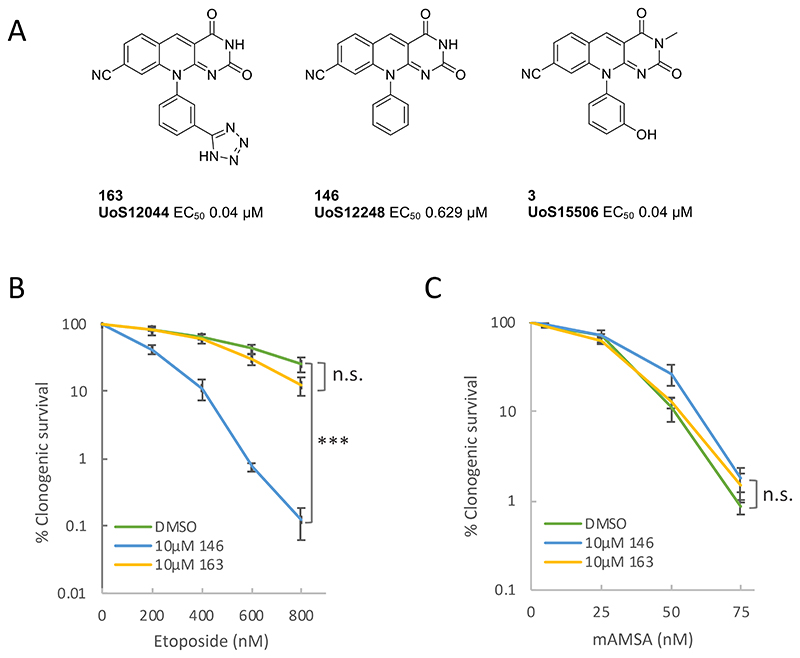
To examine the efficacy of these inhibitors in cells we chose the most potent of these; compound 163 (IC50 ~40nM; Fig.1A)10,11. However, we failed to observe any increase in sensitivity to the TOP2 poison etoposide in human A549 cells in the presence of 10μM of 163 (Fig.1B). We surmised that the high polar surface area (tPSA 140 Å2) and negatively charged tetrazole moiety (pKa 4.28) may limit passive cell permeability. In an attempt to profile compounds with improved permeability we selected an additional two compounds from the same series; compound 146 (IC50 ~600nM)10 and compound 3 (IC50 ~40nM)(Fig.1A). We anticipated that both compounds would have improved passive cell permeability compared to 163 as they are predicted to be predominantly uncharged at physiological pH and have a lower polar surface area of 85 and 97 Å2 respectively. In contrast to 163, 146 resulted in a significant increase in etoposide sensitivity in human A549 cells (Fig.1B). However, this was not the case for m-AMSA, a second TOP2 poison (Fig.1C). This discrepancy was puzzling, because although etoposide and m-AMSA are chemically distinct TOP2 poisons they both promote TOP2-induced DSBs that require TDP2 for efficient repair3.
Fig.1. Structure and activity of deazaflavins in A549 cells.
A, Structure of the deazaflavin compounds employed in this study. B, Impact of the deazaflavins 163 and 146 on the sensitivity of human A549 cells to etoposide. A549 cells in 10cm dishes were incubated with DMSO vehicle or 10μM 163 or 146 and with the indicated concentration of etoposide for 10-14 days. Visible colonies were then fixed, stained, and counted. Data are the mean +/- SEM of 3 independent experiments, and statistically significant differences (*p<0.05, **p<0.005, ***p<0.0001) between survival curves were measured by ANOVA. C, Impact of 163 and 146 on A549 sensitivity to m-AMSA. Experiments were conducted as above.
Deazaflavins induce etoposide sensitivity independently of TDP2 status
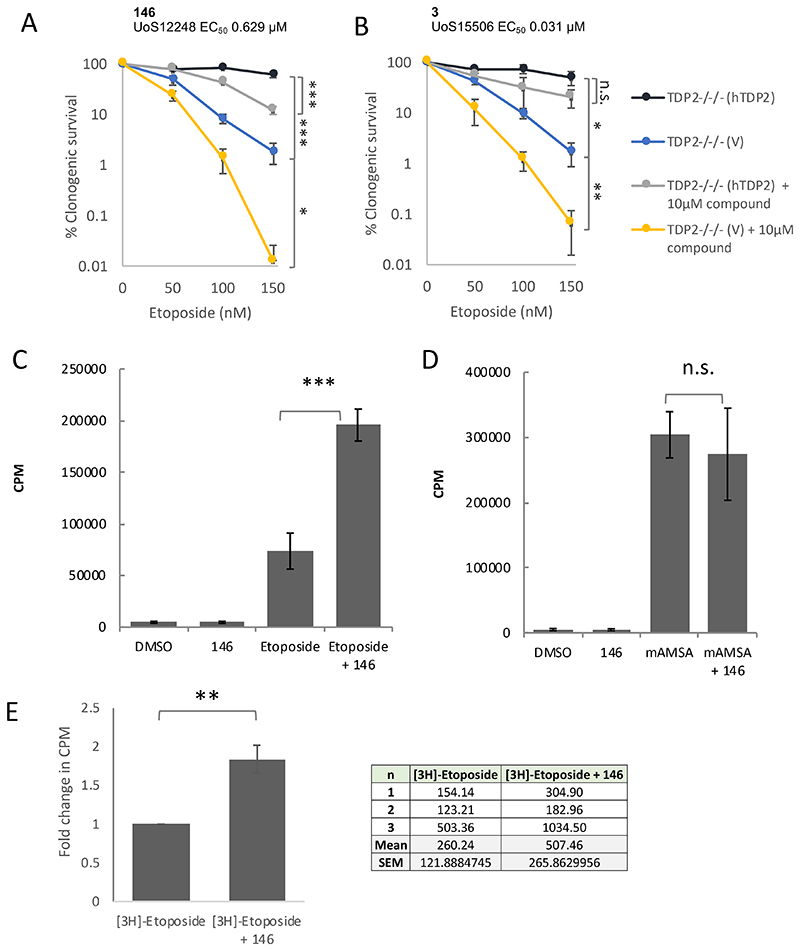
To examine the cellular specificity of the deazaflavins further, we employed TDP2-/-/- chicken DT40 cells stably transfected with empty expression vector (TDP2-/-/-(V)) or vector encoding human TDP2 (TDP2-/-/-(hTDP2))2. Consistent with the experiments described above, 146 resulted in elevated sensitivity to etoposide in (TDP2-/-/- (hTDP2) cells expressing functional human TDP2 (Fig.2A). Surprisingly, however, a similar level of increased sensitivity was observed with 3, despite this compound being ~20-fold more potent in a biochemical assay (Fig.2B). This suggested that the increased sensitivity to etoposide conferred by the two deazaflavines may be unrelated to their anti-TDP2 activity. Indeed, the two compounds also increased etoposide sensitivity in TDP2-/-/- (v) cells, which lack cellular TDP2, confirming that the cellular activity of these compounds with respect to etoposide was independent of TDP2 and thus was “off-target” (Fig.2A & 2B). Notably, we observed similar results in human cells lacking TDP2 (unpublished observations).
Fig.2. Deazaflavins can affect the cellular retention and toxicity of etoposide independently of TDP2 status.
A, TDP2-/-/- chicken DT40 cells stably transfected with empty vector (TDP2-/-/- (V)) or with expression construct encoding human TDP2 (TDP2-/-/-(hTDP2) were treated for 7-10 days with the indicated concentrations of etoposide, in the absence or presence of the deazaflavine 146. Visible colonies were counted, and clonogenic survival plotted as in Fig.1. Data are the mean +/- SEM of 3 independent experiments, and statistically significant differences (*p<0.05, **p<0.005, ***p<0.0001) between survival curves were measured by ANOVA. B, The indicated TDP2-/-/- chicken DT40 cells were treated with etoposide in the absence or presence of the deazaflavine, 3, as described above. C & D, The impact of deazaflavin on induction of TOP2 cleavage complexes by etoposide (B) and m-AMSA (C). A549 cells pre-labelled with [3H]-thymidine were treated with DMSO vehicle or 146 for 1 hr and then for a further 1 hr in the presence of 30μM etoposide or 5μM m-AMSA. Covalent TOP2-DNA complexes were quantified by the amount of [3H]-thymidine labelled DNA (CPM) present in cellular protein precipitates. Data are the mean +/- SD of 3-4 independent experiments, and statistically significant differences were determined by Student’s t-test (***p=0.009). E, Impact of the deazaflavin compound 146 on etoposide uptake and/or retention in A549 cells. Cells were pre-treated with DMSO vehicle or10 μM 146 for 60 min followed by a further 60 min in the additional presence of 10 μCi [3H]-etoposide. The tritium present in each cell sample was quantified by scintillation counting and the results were normalized to the [14C]-thymidine content of each sample. The fold change in normalised [3H]-Etoposide CPM in the presence of deazaflavin, relative to that in the absence of deazaflavin, was plotted. Data are the mean of 3 independent experiments. (**p=0.002,, Student’s t-test), and the normalised 3H counts observed in each individual experiment are shown in the table.
Deazaflavins increase the level of etoposide-induced TOP2 cleavage complexes
The cytotoxicity of TOP2 poisons reflects their propensity to trap TOP2 protein-DNA intermediates known as “cleavage complexes”, in which TOP2 is covalently linked to the 5’-termini of the DNA breaks5. Given that the impact of the deazaflavin compounds tested here was etoposide-specific, we considered it likely that these compounds might impact on etoposide uptake or efflux, thereby increasing the amount of TOP2 cleavage complexes induced by etoposide. To explore this, we measured the induction of covalent TOP2 cleavage complexes in A549 cells by etoposide and m-AMSA directly, in the presence and absence of 146. In this assay, cells were pre-labelled with 3H-thymidine to label genomic DNA, lysed and sheared in hot SDS to denature proteins and dissociate non-covalent protein-DNA complexes, and covalent protein-DNA complexes then precipitated on ice by the addition of salt and centrifugation13–15 (see the Methods section for details). As expected, etoposide increased the amount of covalent protein-DNA complexes in human A549 cells >10-fold (Fig.2C). More importantly, whereas 146 did not itself measurably increase the level of covalent protein-DNA complexes, it elevated the level of etoposide-induced complexes ~2.5-fold (Fig.2C). Critically, however, similar to our clonogenic survival assays, 146 failed to increase the level of protein DNA cross links induced by m-AMSA, once again indicating that the cellular impact of the deazaflavin compounds was specific to etoposide (Fig.2D).
Deazaflavin increases etoposide uptake and/or efflux
To explain the above results, we reasoned that the cellular permeability of the deazaflavin 163 is most likely too low to inhibit intracellular TDP2, thus explaining its lack of impact on cellular sensitivity to m-AMSA. However, in the case of compounds 146 and 3 we propose that the rapid efflux of these compounds perhaps competes with the efflux of etoposide such that the intracellular level of this TOP2 poison is increased. To test this, we measured the impact of 146 on the intracellular accumulation of 3H-etoposide, directly. These experiments confirmed that 146 indeed increased the cellular concentration of etoposide by ~2-fold, most likely explaining both the ~2-fold increase in TOP2 cleavage complexes and elevated cytotoxicity induced by etoposide in the presence of 146 (Fig. 2E). We note that these data contrast to a previous report, in which it was concluded that compound 163 increased sensitivity to etoposide12. We do not know why our results differ from those of the previous report but it is possible that it is related to the use of different experimental procedures. For example, we employed clonogenic survival rather than the metabolic (ATP-based) assays employed in the previous report, which is a more direct measure of cell viability. It is important that any future deazaflavine derivatives, or other TDP2 inhibitors, that are developed are tested for cellular efficacy not only with etoposide but with other types of TOP2 poison such as mAMSA, and that they are tested in parallel in TDP2 defective cells.
In summary, we show here that the deazaflavins, whilst potent and specific TDP2 inhibitors in vitro, do not confer activity against this DNA repair protein in cells, most likely because of poor cell permeability. However, we find that some members of this class of compound inadvertently result in elevated cellular uptake and/or retention of etoposide, resulting in phenotypes that are consistent with loss of TDP2 activity but which are nevertheless independent of TDP2 and are thus “off-target”.
Methods
Cells and cell culture
Chicken DT40 B lymphoma cells were cultured in RPMI 1640 medium supplemented with 10-5 M β-mercaptoethanol, penicillin, streptomycin, 10% horse serum, and 1% chicken serum (Sigma) at 39°C. The generation of TDP2-/-/- DT40 cells and their complementation with human TDP2 (TDP2-/-/- (hTDP2) was previously described2. A549 cells were cultured in DMEM medium supplemented with penicillin, streptomycin and glutamine at 37°C.
Clonogenic survival assays
DT40 cells were seeded in 5ml medium containing 1.5% wt/vol methylcellulose (Sigma) in 6-well plates at 50, 500 and 5000 cells per well, and containing the indicated concentrations of either etoposide (Sigma-Aldrich), amsacrine hydrochloride (m-AMSA, Sigma-Aldrich), with or without the indicated deazaflavin compounds. Visible colonies were counted after 7-10 days of incubation at 39°C. A549 cells were plated in 10cm dishes at 300 or 1500 cells per dish, and 4h later treated with indicated concentrations of m-AMSA or etoposide. The cells were fixed and stained using Crystal Violet after twelve days incubation at 37°C.
Measurement of covalent protein-DNA complexes (cleavage complex assay)
The assay was conducted essentially as described13–15. Briefly, A549 cells were seeded in 10cm dishes at 5x105 cells per dish. 24h later, 3 μCi/ml [Methyl-3H]-thymidine (Perkin Elmer) was added to label DNA and after 24h the cells were rinsed in warm PBS and replaced incubated in label-free medium. The cells were treated with DMSO vehicle or 10 μM deazaflavine compound for 60 min, followed by a further 60 min in the additional presence of either 30μM etoposide or 5μM m-AMSA, as indicated. After rinsing twice in ice cold PBS, the cells were lysed in 1ml hot (65°C) lysis buffer (1.25% SDS, 5mM EDTA, 0.4mg/ml salmon sperm DNA) and their chromosomal DNA was extensively sheared by passage through a 25G needle, 15 times. Proteins were then precipitated by the addition of 0.25ml 325mM KCl and incubation of the samples on ice for 10 min. The protein precipitate was then pelleted by resuspension in 1ml wash buffer (10mM Tris-HCl, pH 7.5, 100mM KCl, 2mM EDTA, 0.1mg/ml salmon sperm DNA) at 60°C for 10 minutes. After a second cycle of protein precipitation as described above, to remove residual DNA that was not covalently linked to protein, the protein pellet was resuspended in H2O and the amount of [Methyl-3H]-thymidine labelled DNA present in the samples was quantified in a scintillation counter. The counts per minute (CPM) from [Methyl-3H]-thymidine labelled DNA in the precipitated sample was normalized to the total CPM from a non-precipitated sample.
Etoposide uptake assay
The DNA in A549 cells was labelled for 24 hr by incubation in medium containing with 0.02μCi/ml [Methyl-14C]-thymidine (Perkin Elmer), and the cells then seeded into 10 cm dishes. 4 hr later, the cells were incubated with DMSO or 10 μM deazaflavin for 60 min followed by a further 60 min in the additional presence of 10 μCi [3H]-Etoposide (Moravek Biochemicals, Inc.). The cells were washed twice in ice-cold PBS and added to liquid scintillant. For each sample, the [3H]-Etoposide CPM was normalized using the CPM from [Methyl-14C]-thymidine.
Acknowledgements & Funding
We thank S. Ward and members of Sussex Drug Discovery Centre, L. Pearl, and T. Oliver for useful discussions. This work was funded by a CRUK Programme Grant (C6563/A16771) to K. Caldecott and a Wellcome Trust Sussex Drug Discovery Award to S. Ward (110578/Z/15/Z).
References
- (1).Cortes Ledesma F, El-Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5’-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- (2).Zeng Z, Cortes Ledesma F, El-Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5’-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gómez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilón A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, Cortes Ledesma F. TDP2-Dependent Non-Homologous End-Joining Protects against Topoisomerase II-Induced DNA Breaks and Genome Instability in Cells and In Vivo. PLoS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cuya SM, Bjornsti M-A, van Waardenburg RCAM. DNA topoisomerase-targeting chemotherapeutics: what’s new? Cancer Chemother Pharmacol. 2017;80:1–14. doi: 10.1007/s00280-017-3334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, Semler BL. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc Natl Acad Sci USA. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Maciejewski S, Nguyen JHC, Gómez-Herreros F, Cortes Ledesma F, Caldecott KW, Semler BL. Divergent Requirement for a DNA Repair Enzyme during Enterovirus Infections. MBio. 2015;7:e01931-15. doi: 10.1128/mBio.01931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cui X, McAllister R, Boregowda R, Sohn JA, Ledesma FC, Caldecott KW, Seeger C, Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS ONE. 2015;10:e0128401. doi: 10.1371/journal.pone.0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci USA. 2014:201409986. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Raoof A, Depledge P, Hamilton NM, Hamilton NS, Hitchin JR, Hopkins GV, Jordan AM, Maguire LA, McGonagle AE, Mould DP, Rushbrooke M, et al. Toxoflavins and Deazaflavins as the First Reported Selective Small Molecule Inhibitors of Tyrosyl-DNA Phosphodiesterase II. J Med Chem. 2013;56:6352–6370. doi: 10.1021/jm400568p. [DOI] [PubMed] [Google Scholar]
- (11).Hornyak P, Askwith T, Walker S, Komulainen E, Paradowski M, Pennicott LE, Bartlett EJ, Brissett NC, Raoof A, Watson M, Jordan AM, et al. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem J. 2016:BCJ20160180. doi: 10.1042/BCJ20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Marchand C, Abdelmalak M, Kankanala J, Huang S-Y, Kiselev E, Fesen K, Kurahashi K, Sasanuma H, Takeda S, Aihara H, Wang Z, et al. Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol. 2016:acschembio.5b01047. doi: 10.1021/acschembio.5b01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Caldecott K, Banks G, Jeggo P. DNA double-strand break repair pathways and cellular tolerance to inhibitors of topoisomerase II. Cancer Res. 1990;50:5778–5783. [PubMed] [Google Scholar]
- (14).Jeggo PA, Caldecott K, Pidsley S, Banks GR. Sensitivity of Chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res. 1989;49:7057–7063. [PubMed] [Google Scholar]
- (15).Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem. 1983;258:15365–15370. [PubMed] [Google Scholar]