Abstract
Huntington’s disease is the most frequent autosomal dominant neurodegenerative disorder, for which we have no approved disease-modifying treatments. The molecular pathogenesis of Huntington’s disease is complex, with toxicity arising from full length expanded huntingtin and N-terminal fragments of huntingtin, which are both prone to misfolding due to proteolysis, from aberrant intron-1 splicing, and from somatic expansion of the CAG repeat in the HTT gene. Potential therapies for Huntington’s disease include those targeting huntingtin DNA and RNA, clearance of huntingtin protein, DNA repair pathways, and other treatment strategies targeting inflammation and cell replacement. The early termination of the tominersen antisense oligonucleotide trials sent a strong signal that it is timely to reflect on lessons learned, where the field stands now, and our challenges and opportunities for the future.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative condition caused by a CAG repeat expansion in the first exon of the HTT gene, encoding for a mutant huntingtin (mHTT) protein1. Symptoms involve motor, cognitive and psychiatric areas, generally appearing during mid-life with slowly progressive decline over the course of two decades2. There is currently no approved disease-modifying treatment for HD3.
In this Review, we initially explore the effects of the HTT mutation and toxic species in HD pathogenesis relevant for the development of disease-modifying treatments. Next, we describe the potential implications derived from impairing the function of the wild-type protein through non-allele selective compared to allele-selective approaches targeting the disease-causing mutation or its by-products.
We review therapies targeting HTT DNA or RNA including recent results of trials using antisense oligonucleotides (ASO) followed by strategies targeting other disease mechanisms such as DNA repair, or downstream effects of mHTT protein. Subsequently, we focus on restoration of neuronal death through cell replacement. Finally, we will comment on methods for efficient delivery of these therapies to the brain, enrichment and target engagement/response biomarkers and on aspects of clinical trial design that should be considered in future HD trials.
HD pathogenesis
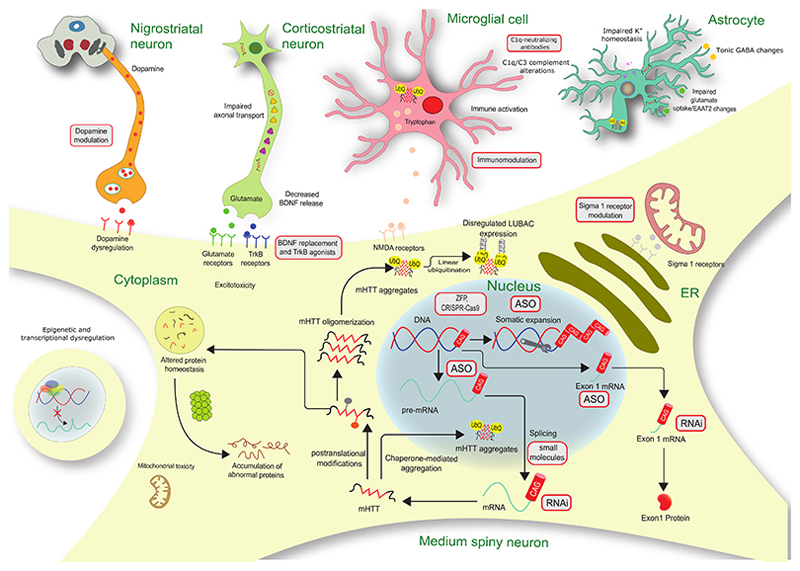
HD is primarily thought to arise predominantly by a toxic gain-of-function of the mHTT protein4,1 (Figure 1). The CAG repeat expansion encodes for an enlarged polyglutamine (polyQ) tract in the N-terminal fragment of the protein3 causing mHTT to fold abnormally and accumulate in brain cells2. The expression of mHTT leads to progressive disruption of neuronal physiology. Subsequently, there is detectable atrophy in the basal ganglia nuclei, including the caudate, putamen, globus pallidus and subthalamic nuclei as well as in subcortical white matter before the emergence of motor symptoms. As the disease progresses, significant neuronal loss in cortical, thalamic and hypothalamic areas and eventually throughout the entire brain is also observed5 (Figure 1).
Figure 1. Diagram of Huntington’s disease pathogenesis and possible therapeutic targets.
Gray boxes with red border indicate potential therapies. ASO=antisense oligonucleotide. BDNF=brain-derived neurotrophic factor. ER=endoplasmic reticulum. LUBAC=linear ubiquitin chain assembly complex. mHTT=mutant Huntingtin. NMDA=N-methyl-D-aspartate. RNAi=RNA interference. TrkB=tyrosine receptor kinase B. ZFP=Zinc-finger protein. Adapted from Bates et al (2015)2 with permission
The underlying HD disease mechanism is complex and widespread and includes deregulation of the ubiquitin/proteasome and autophagy systems contributing to toxic protein accumulation2,6. There is also oxidative stress with peripheral and central inflammation from the earliest stages of the disease 7.
In addition, mHTT affects transcriptional regulation8 and axonal transport resulting in decreased signalling mediated by brain-derived neurotrophic factor (BDNF)2. Moreover, there are alterations in cortical glutamatergic and striatal dopaminergic neurotransmission as well as evidence for synaptic dysfunction in HD2.
Genome-wide association (GWA) studies have shown that age at onset and disease progression in HD is associated with loci in DNA repair genes9 whose proteins are known to influence the extent of somatic expansion of the CAG repeat. Consistently, extreme increases in CAG repeat length (up to 1,000 CAG repeats) are present in the striatum of post-mortem HD brains10.
HTT lowering as a therapeutic approach
In contrast to most neurodegenerative conditions, in HD a single mutation is responsible for all cases of the disease1. In addition, there is substantial evidence supporting the toxic effects of mHTT, and CSF mHTT levels are correlated with disease stage and function in HD gene-carriers11.
Animal models carrying the HD mutation show protein inclusions and brain atrophy alongside progressive neurological phenotypes12. Consistently, numerous studies using different approaches in multiple mouse models to decrease mHTT have shown that this strategy is well-tolerated and effective. Mutant HTT-lowering improves motor performance13,14,15,16,17 and cognitive function17. There is increased survival and decreased weight loss in response to HTT-lowering therapies15. Moreover, biochemical deficits13, synaptic dysfunction16,13,18 and transcriptional dysregulation can be also ameliorated through mHTT-lowering, together with reduced atrophy rates16,17 and decreased intraneuronal protein aggregates13,14.
Exon 1 toxic species
A highly toxic HTT exon 1 protein containing the polyQ tract is present exclusively in mutation carriers19. Incomplete splicing of HTT mRNA generates a short polyadenylated HTT mRNA comprising exon 1 and the 5’ part of intron 1, leading to production of the exon 1 fragment19. This exon 1 protein is constantly generated and accumulates in striatal neurons20, is associated with faster neurological phenotypes in HD mouse models, and has also been detected in juvenile onset HD post-mortem brains19. These findings suggest that a highly toxic exon 1 protein may be a primary pathogenic species. However, the presence of this toxic fragment could pose a challenge to some HTT-lowering therapies. It may not be lowered by splicing modulators (branaplam, PTC518), existing ASOs (tominersen, WVE-120101, WVE-120102) or RNA interference (RNAi) (VY-HTT01) agents in clinical development, as these target HTT downstream from exon 1 HTT mRNA. Alternatively, therapies targeting DNA or HTT exon 1 mRNA may be effective in decreasing the exon 1 protein (Table S1)
Panel 1 - Challenges of delivery and distribution to the HD brain
The need to deliver molecules or cells directly to the brain is driven by protective properties of the blood-brain barrier (BBB), and by the desirability of targeting some therapies to specific brain regions (see Figure S1 and Table S1). The GENERATION HD1 study (NCT03761849) developed by Roche delivered tominersen, a non-allele selective ASO targeting HTT mRNA via lumbar intrathecal injection; advantages of this route included widespread clinical familiarity and availability of the approach, and disadvantages included unequal distribution of the ASO predominantly to cortical over deeper brain structures and the potential discomfort of repeated injections21. Intrathecal infusion catheters could circumvent the latter, but not the former and carry appreciable rates of complications22. Intraventricular catheters, inserted using contemporary navigation methods, have decreased long-term complications and are better positioned for efficient delivery to the striatum, although distribution, tolerability and safety should be determined empirically for individual therapeutic agents23. Direct intraparenchymal delivery of molecules to the striatum has benefitted from iterative improvements of convection-enhanced delivery techniques, especially within oncology, with a number of delivery systems in use for a variety of neurological applications24, whilst development of optimised devices for delivering cells has lagged behind25 and needs further attention. Stereotactic neurosurgery carries potential challenges, including risks of surgery (infection, bleeding, death), establishing appropriate technical expertise, intervention fidelity, and regulatory considerations,24,26 some of which may be addressable in the future through non-surgical approaches 27.
The challenge of allele versus non allele selectivity in HTT lowering
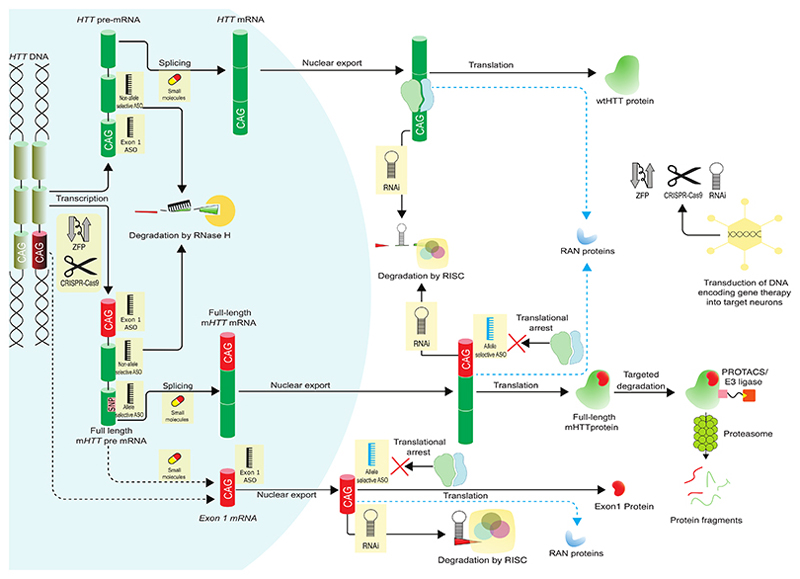
HTT-lowering therapies can be classified as non-allele selective (total HTT lowering), reducing the levels of both wild-type HTT (wtHTT) and mHTT; or allele-selective, if only mHTT is decreased4 (See Figure 2 and Table S1).
Figure 2. Production of huntingtin protein (HTT) and HTT-lowering therapeutic approaches.
Red sections of the DNA, RNA and protein represent the pathogenic CAG tract and the polyglutamine product. Yellow boxes indicate therapeutic approaches. Dotted arrows indicate proposed mechanisms for the production of alternative toxic species. Different ASO colours indicate different mechanisms of action (black, RNAse H mediated degradation; blue, translational arrest). ASO=antisense oligonucleotide. mHTT=mutant huntingtin protein. PROTACS=proteolysis-targeting chimera. RAN=repeat-associated non-ATG. RNase H=Ribonuclease H. RISC=RNA-induced silencing complex. wtHTT=wild-type huntingtin protein. ZFP=zinc-finger protein. Adapted from Wild and Tabrizi (2017)4 with permission
Depletion of wtHTT shows age-dependent pathological phenotypes in mice. Knockout of the Htt gene in rodents is embryonically lethal28 consistent with an essential role in neurodevelopment29. Inactivation on day five during the postnatal period in mice leads to progressive neurological deterioration1,30. In contrast wtHTT ablation at two months of age causes pancreatitis but does not appear to be deleterious in the adult mouse brain15,31. Importantly, CAG expansions do not alter the transcription of the wild-type allele8. Consequently, the expression of the wild-type allele is not expected to be affected by the presence of the mutation.
In human case studies (Table S2) a woman with a balanced translocation in HTT resulting in one intact and one disrupted gene that at best could produce a partial protein, displayed a normal phenotype at 46 years32. Conversely, compound heterozygous variants in HTT, expected to lead to decreased transcription of normal HTT below 50% have been associated with a Rett-like phenotype33,34. Similarly, HD individuals with decreased transcription of the wild-type allele, had earlier age at onset35 while rare cases with homozygous expansions in HTT have similar development and age at onset compared to heterozygous carriers36–39.
In summary, animal studies and case reports suggest that partial lowering of wtHTT (~50%) in the healthy adult brain is likely safe1. However, the long-term effects of decreasing total HTT levels (both wtHTT and mHTT) in the brains of HD patients are unknown3.
In contrast, allele-selective approaches lack the potential side effects from impairing the physiological function of the wild-type protein. However, there are some significant limitations with current allele-selective approaches, such as their inability to target the entire HD population with SNP-related approaches3, or decreasing the transcription of other genes sharing similar sequences with some CAG-targeting therapies3.
Nucleic acid therapeutics
Two programs of nucleic acid-based approaches for HD using gapmer ASOs to lower huntingtin have recently reported preliminary results. Gapmer ASOs are a string of nucleotides with unmodified nucleotides in the central region flanked by modified nucleotides, to increase durability of the response. After binding of the ASO to the target mRNA, the unmodified central region can recruit RNAse H enzyme leading to degradation of the transcript (Figure 2).
The Ionis/Roche ASO program uses tominersen, an ASO targeting exon 36 of human HTT mRNA. Binding of the ASO results in RNase H degradation of both the mutant and wild-type HTT transcripts15. Results from a phase 1/2a study were promising (NCT02519036) with bolus intrathecal administrations every 4 weeks at ascending dose levels from 10 to 120 mg showing no serious adverse events and around 40% reduction of mHTT in CSF after four doses21 of tominersen, being above the degree of mHTT lowering showing phenotypic improvement in animal studies21. There were dose-dependent and time-dependent increases in ventricular volume in participants receiving tominersen compared to the placebo group, without corresponding changes in caudate or whole-brain volume. Similarly, cerebrospinal fluid (CSF) neurofilament light (NFL) protein concentrations also showed time-dependent and dose-dependent increases in treated participants compared with those on placebo. Participants in the phase 1/2a study were offered to take part in the open-label extension (OLE) (NCT03342053) clinical trial with tominersen, where they received the 120 mg of the ASO every four or eight weeks during a period of 15 months. Preliminary results from the OLE study, showed increases in CSF NFL concentrations at day 141 that decreased thereafter despite continued dosing with tominersen40,41.
Based on these promising phase 1/2a data, a large multinational phase 3 study, GENERATION HD1 (NCT03761849), was initiated with intrathecal dosing of tominersen every four or eight weeks. However, based on results from the OLE study (NCT03342053) showing greater than expected CSF huntingtin-lowering, together with reduced tolerability and increased adverse events in the four-weekly dosed arms41 the phase 3 protocol was amended to two less frequent dosing frequencies: 120mg tominersen every eight or 16 weeks (NCT03761849). In March 2021, dosing in this trial was halted based on the Independent Data Monitoring Committee’s recommendation. Although there were persistent dose-dependent decreases in CSF mHTT, the group receiving tominersen every eight weeks performed worse on clinical rating scales and had higher frequencies of serious adverse events compared to placebo. There were no statistically significant differences between placebo-treated participants and the 16-weekly group regarding the clinical endpoints. Similarly, participants receiving the drug every eight weeks had significant increases in CSF NFL at week 21, decreasing thereafter despite continued dosing, while only mild CSF NFL increases were found in the 16-weekly cohort, before decreasing back to baseline. Post hoc exploratory subgroup analysis of the phase 3 study with tominersen investigated the association between disease burden and ASO exposure. The data generated by this analysis suggests that younger participants with a lower disease burden might derive benefit from less frequent or lower dose treatment with tominersen in contrast to the other subgroups. However, these analysis are preliminary and have not yet been published in a peer reviewed journal. Following these results, Roche will continue the tominersen program with a new phase 2 clinical trial exploring different doses of the ASO in younger patients with lower disease burden42–44.
The second ASO phase 1/2 program in HD used a potentially allele-specific approach to HTT-lowering developed by Wave Life Sciences. WVE-120101 (NCT03225833) and WVE-120102 (NCT03225846) are proprietary stereopure gapmer ASOs directed against SNPs unique to the mutant HTT transcript45. With this approach, mHTT mRNA is selectively targeted over wtHTT mRNA. ASOs were administered monthly via intrathecal injection with the highest dose of 32 mg. Both trials reported a lack of target engagement, with no significant mHTT lowering in the CSF at any dose43.
What can we learn from these recent ASO clinical trials? For the WVE-120101/2 trial there are no published animal studies, partly due to a lack of animal models with the relevant SNPs at the time these ASOs were developed. Failing to show any target engagement with their first two SNP-targeting ASOs, Wave is moving forward with a novel ASO based a modified chemistry, targeting a third SNP (WVE-003)(NCT05032196). In contrast to the WVE-120101/2 trials, tominersen effectively targeted the HTT mRNA based on the CSF decreases in mHTT protein. Although, the GENERATION HD1 phase 3 trial was a large study, following a phase 1/2a study and open-label phase 2 extension study, adequate data on absorption and distribution in humans was lacking. However, the transient rise in CSF NFL found with the highest ASO doses in the OLE and GENERATION HD1 studies was, in retrospect, a portent of the negative outcome of the initial tominersen studies 21,41.
The transient increase in CSF NFL before full suppression of CSF mHTT suggest an off-target inflammatory effect of the ASO damaging susceptible neurons. Furthermore, peak-dose toxicity is possible, and rodent studies have shown an initial high retention of signal in the meninges covering the spinal cord and base of the brain during one to eight hours, followed by diffuse ASO immunostaining throughout the spinal cord and brain46. However, the decrease in CSF NFL in the face of continuing dosing41 in the eight weekly cohorts suggests the potential for benefit if the initial ASO adverse effects may be reduced with a lower dose of tominersen in more resilient, younger brains with lower disease burden. However, sufficient mHTT lowering in target structures such as the caudate and putamen nuclei, will need to be achieved in the brains of HD patients.
The CSF white cell count and protein rises in the OLE study with tominersen41 also suggest an inflammatory reaction possibly due to ASO-exposure related toxicities in association with the high 120mg dose chosen to maximise ASO concentrations in the striatum after intrathecal delivery42. Patients in the phase 3 trial developed time and dose-dependent ventricular volume increases without matched brain atrophy42. In contrast to the concentrations of CSF NFL, the increases in ventricular volume did not recede while participants were receiving tominersen. However, the ventricular volume has shown to decrease gradually after ASO dosing was stopped. Interestingly, cases of hydrocephalus have been reported in infants with spinal muscular atrophy receiving nusinersen, a non-gapmer ASO47.
However, deleterious effects of too much wild-type HTT lowering with the high exposure dose of tominersen and the relatively late stages of the study participants are also important to consider, and the analyses of this invaluable dataset will inform all future HTT lowering programmes. The new dose-finding phase 2 clinical trial will determine the optimum and safest dose of tominersen to possibly change the course of the disease in younger participants with lower disease burden.
In retrospect, a full phase 2b study of tominersen would have been helpful to prepare for the phase 3 efficacy trial, a lesson that the HD field should learn from in the future.
Nucleic acid therapeutics in animal and in vitro stages of development
Allele-specific ASOs targeting SNPs are being developed by Ionis17,48 as well as ASOs targeting the CAG repeat by VICO Therapeutics49. Benefits of targeting the CAG repeat would be that a single ASO is applicable to multiple polyQ disorders50, however they risk off-target effects through binding to other CAG repeat containing RNAs.
RNAi approaches include, among others, short-interfering RNA (siRNA) or microRNA (miRNA). These molecules target mature mRNA in the cytosol triggering degradation through the RNA-induced silencing complex, eventually reducing protein expression (Figure 2) 3,51. Conjugated siRNA can be internalised into neurons, being also in development for HD. Alnylam Pharmaceuticals together with Regeneron have a programme with ALN-HTT51 targeting exon 1. In a rapidly progressing HD mouse model the siRNA was infused into the striatum with an implantable infusion system and ameliorated the molecular and motor dysfunction in this mouse model51.
Atalanta Therapeutics, together with Biogen, are developing a novel chemically-synthesized branched siRNA52, that shows widespread distribution throughout the brain after a single administration into the CSF, with promising results in lowering mHTT levels53,54.
Future perspectives for nucleic acid therapies
Challenges for the field in nucleic acid therapies lie mainly in improving safety, toxicity and delivery. New developments ensure that nucleic acid molecules still hold promise for treating HD. Tricyclo-DNA ASOs are thought to cross the BBB55. Neubase Therapeutics are developing nucleic acid ligands that target CAG-repeat expansion in the mRNA and are in early stages of development56.
What can the HD field learn from the approved RNA-targeting ASOs for neurological disorders such as nusinersen (for spinal muscular atrophy) and milasen (Batten’s disease)? Importantly, ASOs can be successful therapies. The major differences to the HD ASOs are the much lower dose requirements (12 mg for nusinersen and 63 mg for milasen57,58) and the ASOs restored wild-type protein expression instead of lowering expression of a mutant (and a wild-type) protein. Counteracting neurodegeneration by downregulating a toxic protein is much less straightforward than restoring protein expression, for which small increases of the missing protein have proven to be beneficial. In contrast, tofersen, an ASO degrading SOD1 mRNA tested in SOD1-amyotrophic lateral sclerosis at similar doses (100mg) to those used in the tominersen phase 3 clinical trial, was associated with increased CSF protein and CSF pleocytosis but not with NFL increases, while there were no imaging data59. However, concentrations of ASOs are lower in the basal ganglia and higher in cortical regions and the spinal cord after intrathecal administration, being advantageous for amyotrophic lateral sclerosis or spinal muscular atrophy, while HD requires targeting deep grey matter nuclei 60.
Gene therapy approaches
Gene therapy uses genes (transgenes) expressing a therapeutic principle to treat and prevent disease61. A major hurdle facing many current HTT-lowering therapies based on RNAi, zinc-finger proteins (ZFP) and genome editing (CRISPR) is efficient delivery of these therapeutic agents to the critical target regions in the central nervous system. Un-modified constructs do not readily cross the BBB, nor are they taken up by neurons, and currently recombinant adeno-associated viral (AAV) vectors are the most commonly used delivery method for effective cell transduction and, eventually, achieving stable expression of the transgene. Recombinant AAV vectors have a number of advantages: they are non-pathogenic, do not replicate in the host, and do not integrate into the host genome62. Delivery of viral vectors to specific brain regions can be achieved by bilateral intraparenchymal infusion. The striatum (caudate/putamen) is the most severely affected brain region in HD and is the most common target for AAV-mediated delivery, but HD affects the entire brain and effective therapies will likely require delivery to the cortex and other affected brain regions (Panel 1). Despite the potential utility, there are significant limitations to AAV delivery. Immune responses to the viral vector occur and repeated administration is not usually possible. Viral delivery is a single shot on goal, it is irreversible and thus dose correction in an individual is not possible. Tissue distribution is highly variable; decreasing as distance from the injection site increases63.
AAV RNAi Studies in HD models
In vivo animal studies of AAV-mediated RNAi therapies for HD have provided evidence for target engagement (HTT-lowering) and efficacy in transgenic mice, minipigs and non-human primate models. Early studies of AAV-mediated RNAi therapies in transgenic mouse models of HD found that 40-60% lowering of total HTT (both wtHTT and mHTT) was safe, extended lifespan, and decreased motor symptoms even when initiated in symptomatic mice64. Intracranial administration of AAV5-delivered anti-HTT miRNA into a transgenic HD minipig model reduced mHTT levels in widespread brain regions65 and resulted in sustained mHTT lowering for up to a year after injection63. These results suggested that this approach could be effective in patients with HD and support the first human trial of an AAV-mediated therapy in HD (NCT04120493). Partial lowering of wtHTT by RNAi was also safe and well-tolerated in NHPs. Reduction of HTT levels by 45% did not result in abnormal neuropathology or adverse symptoms66 and sustained total huntingtin-lowering in NHPs had no observed toxicity even 6 months post-injection67.
Clinical trials of AAV-miRNA
Clinical trials of miRNA therapy in HD are already underway (Table S1). The uniQure AMT-130 trial (NCT04120493) is evaluating the safety, tolerability, and effect on CSF biomarkers of bilateral intrastriatal injections of AAV5-miHTT in early HD. AAV5-miHTT is an engineered AAV5 vector expressing miRNA targeting total huntingtin, injected using MRI-guided convection-enhanced delivery, and a new open-label clinical trial with AMT-130 (NCT05243017) started in 2021. Voyager Therapeutics had a similar program under development for AAV-delivered anti-HTT RNAi therapy (VY-HTT01). This programme is currently on hold pending development of more efficient viral delivery (Table S1). A third AAV clinical trial in HD sponsored by Spark Therapeutics is currently being planned to evaluate an AAV1-delivered anti-HTT miRNA that has shown promise in NHPs66.
DNA targeted approaches
Therapeutic approaches directly targeting the DNA of the HTT gene have exceptional promise and correction of the underlying genetic defect should prevent all associated pathogenicity in HD (Figure 2). Zinc finger proteins (ZFPs) consist of an active effector element such as a transcriptional repressor protein or nuclease, bound to a DNA-binding element composed of multiple zinc finger peptides that enable selective targeting to specific DNA sequences68. ZFPs have been shown to selectively lower mHTT expression in cell lines without significantly altering wtHTT expression. AAV-ZFP infusions reduced mHTT levels and improved some HD-like behavioural phenotypes in HD mice13.
CRISPR (clustered regularly interspaced short palindromic repeats)-based systems have emerged as potentially versatile, specific, and efficient gene-editing technologies with potential to treat HD and other neurodegenerative disorders. CRISPR gene editing can selectively inactivate the mutant HTT gene in patient-derived fibroblasts69, in HD mice70 and in human differentiated induced pluripotent stem cells71. Animal and in vitro studies support the general feasibility of gene editing approaches in HD, but no clinical trials are yet underway. However, further testing in HD animal models and advances in both viral and non-viral delivery methods in large animals will be essential before DNA targeting CRISPR approaches are ready for testing in HD patients.
Splicing modulators
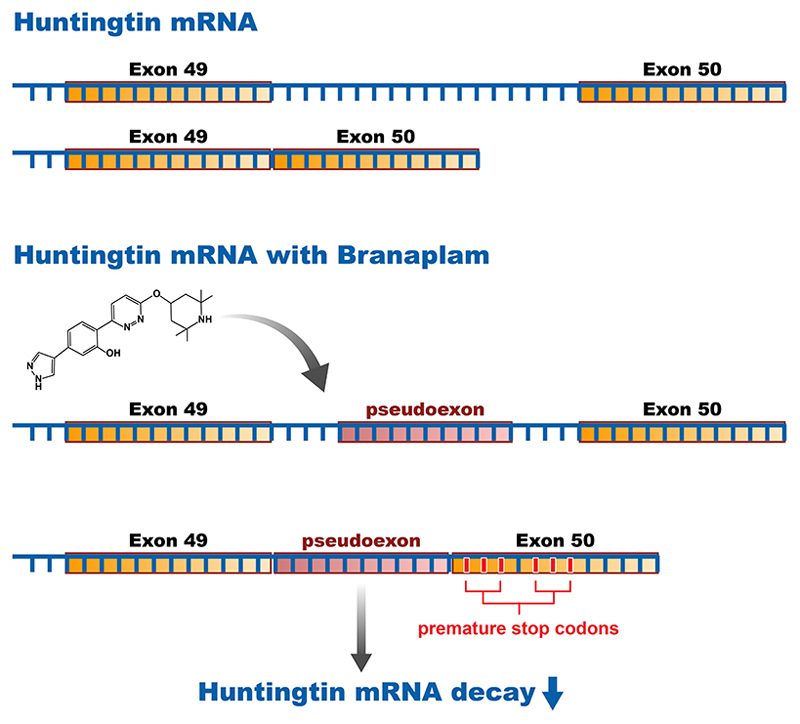
The latest therapeutic agents to emerge in HD are a new class of oral, brain-penetrant, small molecule agents targeting HTT expression, the first of which was initially identified in a phenotypic screen targeting expression levels of the spinal motor neuron-2 gene, as a treatment for spinal muscular atrophy. Branaplam (LMI070) is in phase 2 for spinal muscular atrophy (NCT02268552). Selectivity analysis showed that branaplam can decrease the expression of both copies of the HTT protein via splicing modulation. The mechanism of action seems to be mis-splicing of exon-49/50 leading to decay of the HTT mRNA and decreasing protein levels72 (Figure 3). Novartis received US FDA Orphan Drug Designation for branaplam in HD, and a phase 2b trial (NCT05111249) started enrolling in December 2021.
Figure 3. Mechanism of action of a huntingtin lowering splicing modulator small molecule.
Splicing of the human HTT gene at the exon 49-50 region in the absence (upper panel) or presence (lower panel) of branaplam, a small molecule splicing modulator. Branaplam recognizes a preferred sequence motif in the intron between exon 49 and 50, defining it as an exon. Inclusion of this pseudoexon introduces in-frame stop codons in the mature HTT transcript leading to downregulation of HTT mRNA and protein.
PTC therapeutics developed a drug discovery splicing platform to identify small molecules that target the same mechanism as branaplam, finding compounds that successfully decrease the HTT mRNA and HTT protein in the brain and the periphery73. Subsequently, PTC therapeutics is conducting a phase 1 trial of PTC518, to evaluate the safety, pharmacokinetics and pharmacodynamic properties in healthy volunteers, with a phase 2 trial starting in 202274,75.
DNA repair therapeutics
Repetitive DNA is inherently unstable, and over a HD patient’s lifetime the expanded CAG repeat in the HTT gene tends to gain additional repeats, a process known as somatic instability. The speed of expansion differs between tissues and cell types, more prominent in neurons than glia, with fast expansion in striatal and cortical cells, which are most prone to degeneration in HD10. Patients with more somatic instability, as measured in blood, have earlier disease onset and faster progression, establishing somatic instability as a rate driver of HD9. The SHIELD HD study (NCT04406636) is currently underway to study the natural history of somatic instability in different biofluids of pre- and early manifest HD patients to inform future therapeutics targeting repeat expansion.
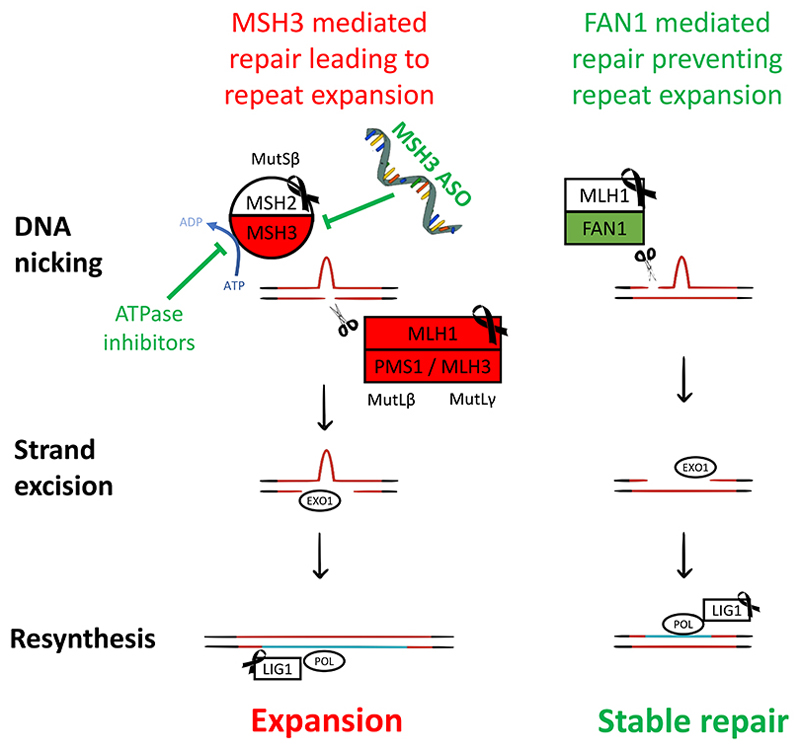
In the last few years GWA studies in thousands of HD patients have found DNA repair to be the main modifier of HD disease course9,76, and that the same genes also affect other trinucleotide repeat diseases77. One of the strongest modifier genes was FAN1, that codes for a nuclease that cuts DNA during repair of crosslinks between DNA strands9. FAN1 is protective in HD, likely through accurate repair of loopouts that form at CAG repeats78 (Figure 4) and its depletion in animal models and HD patient neurons accelerates repeat expansion78. Genetic variants decreasing FAN1 function advance onset by over 5 years, whereas those that increase its expression can delay onset over a year9.
Figure 4. The role of DNA repair in repeat expansion, and its new therapeutic targets.
Left: MutSβ initiates mismatch repair at a CAG loopout, introducing repeat expansion (turquoise). Right: FAN1 protects against repeat expansion through a mechanism that depends on its nuclease activity and binding to MLH1. MSH3 inhibition (top left), e.g., by knockdown or ATPase inhibition, is expected to slow repeat expansion and be well tolerated. Red lines = CAG repeat DNA containing a loopout. Red boxes = repair proteins promoting repeat expansion. Green boxes = protect against expansion. Black ribbon = genes whose loss of function in is associated with cancer in humans.
Mismatch repair is the most significant DNA repair pathway driving HD pathogenesis. It is initiated by the recognition of base mismatches or single strand loopouts by a MutS complex. This involves recruitment of a MutL nuclease complex to nick DNA, digestion of the error-containing strand, re-synthesis using the remaining template strand, and finally sealing the ends of the newly synthesised strand79 (Figure 4). Several components of this mismatch repair pathway influence HD onset, including MSH3, MLH1, PMS1, PMS2 and LIG19,76. Studies in HD mice suggest that MSH3, PMS1 and MLH3 drive expansion, whereas MSH6 and PMS2 may protect against it80.
Our understanding of the role that DNA repair mechanisms play in trinucleotide repeat expansion is crystalising, with opposing forces battling to either drive or prevent it, and this is already translating into new therapeutics targeting the mutation itself. Whilst MSH2, MSH6 or MLH1 depletion is associated with cancer in humans79, MSH3 is tolerant of loss of function and deletion of MSH3 in mice does not affect lifespan or cause cancer79. Strategies such as small molecule MSH3 inhibitors, acting for example through ATPase inhibition, promotors of FAN1 activity, or ASOs against MSH3 could also be considered as potential therapies for HD.
Other therapeutic approaches – pridopidine, C1q antibody
Pridopidine
Pridopidine (formerly ACR16) is a sigma-1 receptor agonist81. Pridopidine rescued mHTT-induced cell death in HD mouse neurons and human derived induced pluripotent stem cells, and restored homeostatic synaptic plasticity and calcium signalling alterations in HD neurons; responsiveness to the compound was abolished in sigma-1 receptor knockout neurons82, in HD patient lymphoblasts83, and human HD derived induced pluripotent stem cells 84. In an HD mouse model, pridopidine led to improvements in motor coordination, and ameliorated a subset of gene expression changes83.
Studies using [18F] fluspidine (an sigma-1 receptor ligand) and [18F] fallypride (a D2/D3R ligand), in healthy volunteers and in a small number of patients with HD81, suggest that at the doses tested clinically (90 mg/day), pridopidine is saturating for sigma-1 receptors and does not occupy D2/D3Rs81.
Pridopidine has been investigated as a treatment for HD in three randomized, double-blind, placebo controlled clinical trials: HART (NCT00724048, NCT01306929), MermaiHD (NCT00665223), and PRIDE-HD (NCT02006472, NCT02494778). Initial analysis suggested a small effect on motor endpoints in the HART and MermaiHD studies. PRIDE-HD85 failed its primary endpoint analysis but showed a small beneficial effect on Unified Huntington’s Disease Rating Scale (UHDRS) Total Functional Capacity (TFC) at week 52 which was more pronounced for early HD participants and supported by quantitative motor assessments. These observations led to a phase 3 study, PROOF-HD (NCT04556656), currently enrolling using the UHDRS-TFC score as the primary endpoint.
Complement (C1q-neutralizing antibody) directed therapeutics
Synaptic elimination can be mediated by components of the classical complement pathway. For example, synapses marked with activated C1q protein are subsequently eliminated by activated astrocytes. Dysregulation of this mechanism has been associated with excessive synaptic loss in affected circuits in neurodegenerative disorders including HD86 and complement proteins were elevated in a small sample of HD brains87. Annexon has developed the anti-C1q monoclonal antibody ANX-005, currently enrolling for a phase 2 open label study in HD (NCT04514367) with the aim of evaluating the pharmacokinetics and pharmacodynamic effects of this agent.
Cell replacement therapy
The underpinning concept of cell therapy is restoration of structure and function. This can be achieved through implanting cells designed to adopt the function of cells lost to the disease process, or through cellular secretion of neuroprotective or disease-modifying molecules. For many years, the focus of cell therapy in HD was to replace degenerated medium-spiny neurons (MSNs) using cells procured from normal developing foetal striatum (Figure S1). Foetal grafts can restore synaptic connectivity and function in HD animal models88. Human studies have demonstrated safety and feasibility, with some proof-of-concept evidence to suggest that foetal grafts can improve function, although solid evidence of efficacy is still required. The practical limitations of foetal-derived cells for human use has encouraged research using MSN-like neurons derived from pluripotent stem cell sources, and provided evidence of functional improvements with a view to future clinical translation88. There is significant interest in the restorative value of non-MSN cell types, including cortical-like cells and glial progenitors. The latter population has been employed in rodent models of HD and shown to confer significant phenotypic benefit, restoring deficient glial signalling, both in astrocytes and oligodendrocytes89.
Panel 2 - Biomarkers for clinical trials and the state of the field
Critical to the success of clinical trials is the ability to reliably measure the impact of treatment on the disease. Effects can be assessed at the cellular, macrostructural and behavioural levels60,90-92 (Table S3) and biomarkers (target engagement, response to intervention) are being employed in ongoing clinical trials.
Biofluids
Two particularly useful protein biomarkers have emerged recently: mHTT itself - the pathogenic agent; and NFL – released from damaged axons. Both increase with disease progression93,94, but NFL has the greater prognostic potential11, while mHTT has demonstrated specific value as an indicator of target engagement by HTT-lowering drugs3. Dose-ranging studies of future HTT-lowering therapeutics may try to use mHTT or NFL or both to define biochemically the lower and upper bounds, respectively, of a therapeutic window. There are still challenges to this approach namely because there is large inter-individual variability for mHTT.
Biomarkers reflecting specific pathways such as YKL4095 and IL-696 (neuroinflammation), tau (microtubular damage)97 or with some region-specificity like the striatal marker proenkephalin98 have shown potential in HD but have yet to be evaluated to the same extent as mHTT and NFL.
CSF huntingtin quantification remains limited by the sensitivity of current mHTT assays – many far from clinical motor onset individuals have unquantifiably low levels11,99, making therapeutic lowering difficult to assess; and the difficulty of quantifying wtHTT either independently or through inference from the total HTT level based on current assays100. More broadly, the HD biomarker field lacks well-powered replication studies, but the HDClarity bioresource has a large and expanding CSF and plasma collection for this purpose101.
MRI
Structural MRI is a well-established technique which has already been employed in clinical trials21,102. Studies have highlighted disease-related atrophy centred in the striatum and throughout the white matter in HD gene carriers at least 15 years before onset103 with progressive involvement of the cortex as symptom onset approaches104. Reduced striatal volume in gene carriers compared with controls is even evident as early as 24 years prior to expected symptom onset99 although it is not clear whether this is a developmental or neurodegenerative effect. White matter microstructure can be interrogated with diffusion MRI techniques which have shown sensitivity before clinical motor onset105.
PET
PET is generally able to provide more specific information about pathological processes in HD, although is invasive, more expensive and standardisation across multiple sites may be more difficult. Glucose metabolism is reduced, likely preceding atrophy106, and predicts clinical motor onset107. PET has also shown disease-related decreases of dopamine receptors and increased microglial activation108 as well as decreased PDE10 binding109. Novel PET ligands targeting aggregated mHTT itself have recently shown promise in animal studies110 and phase 1/2 studies in humans are ongoing.
Digital biomarkers
The UHDRS Total Motor Score (TMS) is well-established as a clinical measure to assess movement disability2, yet digital biomarkers aim to capture these motor deficits in a more objective and sensitive way. Unbiased quantitative assessment of motor symptoms using force transducers with the Q-motor suite have been used in multicentric clinical trials, being more sensitive than the UHDRS-TMS prior to clinical motor onset111. In addition, digital measures of daytime motor activity112,113, finger tapping103,114, chorea115, gait116, circadian rhythm, speech117 and sleep disturbance118 have all shown sensitivity to disease effects and have great potential for clinical trials119. Cognitive function can be assessed by the HD-CAB, a computerised assessment of multiple cognitive domains, which has been specifically developed for use in clinical trials, showing decline in HD participants after clinical motor onset, alongside acceptable reliability120. Recent developments in wearable/portable devices show great promise for capturing such data non-invasively, but more work is required to standardise devices and harmonize protocols before they are widely accepted as endpoints for clinical trials92.
Conclusions and future directions
Therapeutic approaches under development have the potential to deliver several licensable medicinal products, but uncertainties remain. Genetically validated targets have an overall higher development success rate121; therefore, both HTT and genetic modifiers targets are the best positioned. Besides target validation, drug development success depends on effective delivery to the brain and the benefit-risk relationship. There are three main approaches in the run namely ASOs, gene therapy and small molecules. Gene therapy may see revolutionary changes with the development of novel vectors (viral and non-viral) and gene editing approaches. The concept of aiming at multiple targets simultaneously deserves stronger consideration.
The pathophysiology of HD is complex (Figure 1) and the wild-type protein has important functions122. The correction of such complex mechanisms is less straightforward than correcting mutations that cause loss of protein expression for which small increases of the missing protein have proven to be beneficial57. Those characteristics of the HD mutation partially explain why the targeted interventions currently in development are not tailored to halt the disease or revert the prior pathogenic processes. Intervening when the degree of neurodegenerative changes is marked and there are already clinical manifestations may prove to be a critical limitation, although the recently halted and ongoing trials will be an invaluable dataset to inform future programmes.
Other HD features pose challenges for designing efficient clinical trials: Huntington’s Disease in adults is phenotypically indistinguishable from controls in established biomarkers or clinical readouts on average at least until 20 years of age99; the pathogenic process evolves very slowly, spanning more than 4 decades2, making the detection of an intervention effect particularly difficult. These issues may be addressed by conducting clinical trials using biomarkers (single or signature) (Panel 2) as primary endpoints in the very early phases of the disease, before the emergence of detectable clinical symptoms (Stage 0 and 1 in the Huntington Disease Integrated Staging System-HD-ISS123). Regulatory authorities consider such designs possible124, but the actual roadmap is still unclear, despite important inroads in form of guidance documents125 and fruitful collaborations between the academia and industry102. Even if traditional methods for demonstration of a clinically meaningful effect will not be applicable at those early stages, we need a mechanism to show that treatments are of value for far-from-clinical motor onset individuals126. Evidence of disease-modifying benefit will justify the putative health risks, long term compliance, and the financial effort of treating early in the course of the disease. Novel research in this area is critically needed. Achieving a meaningful reduction in the progression rate might require combination therapy (aiming at complementary targets) either in the same construct or by combining independent constructs. Such combination treatments have specific regulatory constraints that need a swift solution. The ongoing technological and regulatory progress allows optimism about the development of impactful therapies for HD in a reasonable timeframe.
Search Strategy and Selection Criteria
A literature search was done using the PubMed and Scopus electronic databases for articles published in English from 1st Jan 2016 to 10th February 2022. The search terms “Huntington’s disease”, “gene silencing”, “huntingtin lowering”, “antisense oligonucleotide”, “RNA interference”, “siRNA”, “miRNA”, “zinc finger”, “CRISPR”, “Cas9”, “AAV”, “somatic instability”, “biomarker”, “pridopidine”, “complement”, “c1q”, “cell therapy” and “cell replacement” were used. Additional articles were identified by reviewing the reference lists from relevant articles. We also included references from conference presentations, publicly available webinars and clinicaltrials.gov. We prioritised material published between 2016 and 2022, including older references only if they were essential to the field. There were no language restrictions. The final references were chosen on the basis of relevance to the topics covered in this Review.
Supplementary Material
Acknowledgements
We thank Hannah Kelly for assistance in preparation of this manuscript, Richard Newton for graphics support and Rajeev Sivasankaran, PhD for the design of Figure 3.
Footnotes
Contributors
SJT chose and designed the content of the manuscript. SJT supervised and had full oversight of project and edited all the sections of the review. All other authors contributed to the writing of the manuscript and edited the final version of the manuscript approving the final version. CEF, WvRM, AER, MDF, IMS, BRL and SJT contributed to figure preparation and all authors approved the final version of the figures.
Declaration of Interests
SJT receives research grant funding from the CHDI Foundation, Vertex Pharmaceuticals, the UK Medical Research Council, the Wellcome Trust (200181/Z/15/Z), and the UK Dementia Research Institute that receives its funding from DRI Ltd., funded by the UK MRC, Alzheimer’s Society, and Alzheimer’s Research UK. She has undertaken consultancy services for Annexon, Alphasights, Alnylam Pharmaceuticals Inc., Atalanta Pharmaceuticals (SAB), F. Hoffmann-La Roche Ltd/ Genentech, Guidepoint, Horama, Locanobio, LoQus23 Therapeutics Ltd (SAB), Novartis Pharma, PTC Therapeutics, Sanofi, Spark Therapeutics, Takeda Pharmaceuticals Ltd, Triplet Therapeutics (SAB), University College Irvine and Vertex Pharmaceuticals Incorporated. All honoraria for these consultancies were paid through the offices of UCL Consultants Ltd., a wholly owned subsidiary of University College London. SJT has a patent Application number 2105484.6 on the FAN1-MLH1 interaction and structural analogs licensed to Adrestia Therapeutics. WRM is being employed by the LUMC which has patents on exon skipping approaches for neurological disorders, some of which have been licensed to ProQR and Amylon. Remunerations are paid to LUMC. WvRM is an ad hoc consultant for Bridge Bio, received project funding from Amylon and ProQR, and funding for contract research from UniQure. She is project leader in a project to develop an ASO therapy for SCA1 together with Vico therapeutics. EJW reports grants from CHDI Foundation, and F. Hoffmann-La Roche Ltd. He has undertaken consultancy/advisory board work with Hoffman La Roche Ltd, Triplet Therapeutics, PTC Therapeutics, Takeda, Vico Therapeutics, Voyager, Huntington Study Group, Teitur Trophics, EcoR1 Capital, PTC Therapeutics, Annexon Biosciences and Remix Therapeutics. He has participated in advisory boards for Hoffmann La Roche, Triplet therapeutics and PTC therapeutics. All honoraria for these consultancies were paid through the offices of UCL Consultants Ltd., a wholly owned subsidiary of University College London. He holds a stock option for Triplet Therapeutics in part compensation for advisory board membership. AER is Chair of the executive committee of the European Huntington Disease Network (EHDN), which is funded by CHDI Foundation, with remuneration paid to CU, European co-PI for the PROOF-HD trial, with remuneration paid to CU, and serves on Scientific advisory boards for Roche, Wave pharmaceuticals, Prilenia and Triplet Therapeutics. AER has obtained research grants from the MRC, Health and Care Research Wales, EU Horizon 2020, CHDI, Jacques and Gloria Gossweiler foundation and is a chair of the Guarantors of Brain charity. BRL is on the Scientific Advisory Board of sRNAlytics (GateHouse Bio) for which he received stock options, and reports scientific consultancy fees from Teva, Roche/Genentech, Takeda, Triplet, Novartis, Spark, Scintetica, LifeEdit, Design, UniQure, Remix Therapeutics, and PTC Therapeutics. Dr Leavitt’s Laboratory has obtained previous and current research grants from CIHR, HSC, NMIN, CHDI, and uniQure. He is a founding co-Editor-in-Chief, Journal of Huntington’s Disease, Former Co-Chair of the Huntington Study Group, and is a Co-Founder and CEO of Incisive Genetics Inc., in which he has stock and stock options. Incisive Genetics Inc. is an early-stage pre-clinical biotechnology company that was founded to develop in vivo lipid nanoparticle delivery of CRISPR/Cas9 genome editing. This is not a therapeutic approach that is currently in clinical testing for HD, nor is this approach in late pre-clinical stages. The company has no products to endorse, does not have an IND for HD, nor are any commercial efforts currently underway. CS serves as Chief Medical Officer and IM-S serves as Vice President, Translational Biology, respectively, for CHDI Management, Inc., an advisor to CHDI Foundation, Inc. CHDI Foundation provided financial and scientific support to Ionis Pharmaceuticals, Inc.’s Huntington’s disease drug discovery program through a development collaboration with Ionis. Over time, CHDI Foundation was reimbursed for its financial support of Ionis’ program out of milestone payments received by Ionis from Hoffman-La Roche for progress in Roche’s tominersen program. CS received consultancy honoraria (unrelated with Huntington’s disease programs) from the following companies: Kyowa Kirin, Biocodex, Neuraly, NDA, Neuroderm, Pinteon Pharmaceuticals, vTv Therapeutics. CE-F receives support from a Wellcome Trust Collaborative Award (200181/Z/15/Z). Within the last 36 months MF has received grant funding towards his research from the CHDI Foundation, the UK Dementia Research Institute and the Rosetrees Trust. RS reports no conflicts of interest.
References
- 1.Leavitt BR, Kordasiewicz HB, Schobel SA. Huntingtin-Lowering Therapies for Huntington Disease. JAMA Neurol. 2020;77:764. doi: 10.1001/jamaneurol.2020.0299. [DOI] [PubMed] [Google Scholar]
- 2.Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Prim. 2015;1:1–21. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 3.Tabrizi SJ, Ghosh R, Leavitt BR. Huntingtin Lowering Strategies for Disease Modification in Huntington’s Disease. Neuron. 2019;101:801–19. doi: 10.1016/j.neuron.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Wild EJ, Tabrizi SJ. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16:837–47. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rüb U, Vonsattel JPV, Heinsen H, Korf H-W. The Neuropathology of Huntington’s disease: classical findings, recent developments and correlation to functional neuroanatomy. Adv Anat Embryol Cell Biol. 2015;217:1–146. [PubMed] [Google Scholar]
- 6.Well EM, Bader V, Patra M, et al. A protein quality control pathway regulated by linear ubiquitination. EMBO J. 2019;38:1–21. doi: 10.15252/embj.2018100730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Politis M, Lahiri N, Niccolini F, et al. Increased central microglial activation associated with peripheral cytokine levels in premanifest Huntington’s disease gene carriers. Neurobiol Dis. 2015;83:115–21. doi: 10.1016/j.nbd.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Langfelder P, Cantle JP, Chatzopoulou D, et al. Integrated genomics and proteomics define huntingtin CAG length–dependent networks in mice. Nat Neurosci. 2016;19:623–33. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J-M, Correia K, Loupe J, et al. CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell. 2019;178:887–900.:e14. doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouro Pinto R, Arning L, Giordano JV, et al. Patterns of CAG repeat instability in the central nervous system and periphery in Huntington’s disease and in spinocerebellar ataxia type 1. Hum Mol Genet. 2020;29:2551–67. doi: 10.1093/hmg/ddaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigues FB, Byrne LM, Tortelli R, et al. Mutant huntingtin and neurofilament light have distinct longitudinal dynamics in Huntington’s disease. Sci Transl Med. 2020;12:eabc2888. doi: 10.1126/scitranslmed.abc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howland D, Ellederova Z, Aronin N, et al. Large Animal Models of Huntington’s Disease: What We Have Learned and Where We Need to Go Next. J Huntingtons Dis. 2020;9:201–16. doi: 10.3233/JHD-200425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeitler B, Froelich S, Marlen K, et al. Allele-selective transcriptional repression of mutant HTT for the treatment of Huntington’s disease. Nat Med. 2019;25:1131–42. doi: 10.1038/s41591-019-0478-3. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 15.Kordasiewicz HB, Stanek LM, Wancewicz EV, et al. Sustained Therapeutic Reversal of Huntington’s Disease by Transient Repression of Huntingtin Synthesis. Neuron. 2012;74:1031–44. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N, Gray M, Lu X-HH, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat Med. 2014;20:536–41. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southwell AL, Kordasiewicz HB, Langbehn D, et al. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci Transl Med. 2018;10:eaar3959. doi: 10.1126/scitranslmed.aar3959. [DOI] [PubMed] [Google Scholar]
- 18.Carrillo-Reid L, Day M, Xie Z, et al. Mutant huntingtin enhances activation of dendritic Kv4 K+ channels in striatal spiny projection neurons. Elife. 2019;8:e40818. doi: 10.7554/eLife.40818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neueder A, Landles C, Ghosh R, et al. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-01510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Yang S, Jing L, et al. Truncation of mutant huntingtin in knock-in mice demonstrates exon1 huntingtin is a key pathogenic form. Nat Commun. 2020;11:1–15. doi: 10.1038/s41467-020-16318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, et al. Targeting huntingtin expression in patients with Huntington’s disease. N Engl J Med. 2019;380:2307–16. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 22.Necking E, Levi R, Ertzgaard P. Complications of intrathecal drug delivery therapy (ITDD): A retrospective study of 231 implantations between 1999 and 2014. Clin Neurol Neurosurg. 2021;205:106630. doi: 10.1016/j.clineuro.2021.106630. [DOI] [PubMed] [Google Scholar]
- 23.Lau JC, Kosteniuk SE, Walker T, Iansavichene A, Macdonald DR, Megyesi JF. Operative Complications with and without Image Guidance: A Systematic Review and Meta-Analysis of the Ommaya Reservoir Literature. World Neurosurg. 2019;122:404–14. doi: 10.1016/j.wneu.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Fiandaca MS, Lonser RR, Elder JB, Ząbek M, Bankiewicz KS. Advancing gene therapies, methods, and technologies for Parkinson’s Disease and other neurological disorders. Neurol Neurochir Pol. 2020;54:220–31. doi: 10.5603/PJNNS.a2020.0046. [DOI] [PubMed] [Google Scholar]
- 25.Potts MB, Silvestrini MT, Lim DA. Devices for cell transplantation into the central nervous system: Design considerations and emerging technologies. Surg Neurol Int. 2013;4:S22–30. doi: 10.4103/2152-7806.109190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam MF, Thomas MG, Lind CRP. Neurosurgical convection-enhanced delivery of treatments for Parkinson’s disease. J Clin Neurosci. 2011;18:1163–7. doi: 10.1016/j.jocn.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Terstappen GC, Meyer AH, Bell RD, Zhang W. Strategies for delivering therapeutics across the blood-brain barrier. Nat Rev Drug Discov. 2021;20:362–83. doi: 10.1038/s41573-021-00139-y. [DOI] [PubMed] [Google Scholar]
- 28.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11:155–63. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 29.Bocchi VD, Conforti P, Vezzoli E, et al. The coding and long noncoding single-cell atlas of the developing human fetal striatum. Science. 2021;372 doi: 10.1126/science.abf5759. [DOI] [PubMed] [Google Scholar]
- 30.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–6. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Liu X, Gaertig MA, Li S, Li X-J. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci U S A. 2016;113:3359–64. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambrose CM, Duyao MP, Barnes G, et al. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- 33.Lopes F, Barbosa M, Ameur A, et al. Identification of novel genetic causes of Rett syndrome-like phenotypes. J Med Genet. 2016;53:190–9. doi: 10.1136/jmedgenet-2015-103568. [DOI] [PubMed] [Google Scholar]
- 34.Rodan LH, Cohen J, Fatemi A, et al. A novel neurodevelopmental disorder associated with compound heterozygous variants in the huntingtin gene. Eur J Hum Genet. 2016;24:1826–7. doi: 10.1038/ejhg.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becanovic K, Nørremølle A, Neal SJ, et al. A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat Neurosci. 2015;18:807–16. doi: 10.1038/nn.4014. [DOI] [PubMed] [Google Scholar]
- 36.Wexler NS, Young AB, Tanzi RE, et al. Homozygotes for Huntington’s disease. Nature. 1987;326:194–7. doi: 10.1038/326194a0. [DOI] [PubMed] [Google Scholar]
- 37.Kremer B, Goldberg P, Andrew SE, et al. A Worldwide Study of the Huntington’s Disease Mutation: The Sensitivity and Specificity of Measuring CAG Repeats. N Engl J Med. 1994;330:1401–6. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 38.Dürr A, Hahn-Barma V, Brice A, Pêcheux C, Dodé C, Feingold J. Homozygosity in Huntington’s disease. J Med Genet. 1999;36:172–3. [PMC free article] [PubMed] [Google Scholar]
- 39.Squitieri F, Almqvist EW, Cannella M, Cislaghi G, Hayden MR. Predictive testing for persons at risk for homozygosity for CAG expansion in the Huntington disease gene. Clin Genet. 2003;64:524–5. doi: 10.1046/j.1399-0004.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- 40.Ducray PS, Frances N, Smart K, et al. Translational Pharmacokinetic/Pharmacodynamic (PK/PD) Modeling Strategy to Support RG6042 Dose Selection in Huntington’s Disease (HD) (S16.005) Neurology. 2019;92:S16.005 [Google Scholar]
- 41.Schobel SA. Preliminary results from a 15-month open-label extension study investigating tominersen (RG6042) huntingtin protein antisense oligonucleotide in adults with manifest Huntington’s disease; CHDI Foundation Annual Therapeutics Conference 24–27th February; 2020. [Google Scholar]
- 42.Schobel SA. Preliminary results from GENERATION HD1, a Phase III trial of tominersen in individuals with manifest HD; CHDI Foundation Annual Therapeutics Conference 27-29th April 2021; [Accessed 3rd March 2022]. https://chdifoundation.org/2021-conference/ [Google Scholar]
- 43.Kingwell K. Double setback for ASO trials in Huntington disease. Nat Rev Drug Discov. 2021;20:412–3. doi: 10.1038/d41573-021-00088-6. [DOI] [PubMed] [Google Scholar]
- 44.Boak L, McColgan P. Understanding the treatment and post-treatment effects of tominersen in the Phase III GENERATION HD1 study; CHDI Foundation Annual Therapeutics Conference 28th February-3rd March 2022; 2022. [Accessed 3rd March 2022]. https://medically.gene.com/global/en/asset-viewer.278a7153-aa96-4148-83ac-03c2d3a9d499.qr.html?cid=slpsxx2203nehdchdi2022 . [Google Scholar]
- 45.Pfister EL, Kennington L, Straubhaar J, et al. Five siRNAs Targeting Three SNPs May Provide Therapy for Three-Quarters of Huntington’s Disease Patients. Curr Biol. 2009;19:774–8. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazur C, Powers B, Zasadny K, et al. Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI insight. 2019;4 doi: 10.1172/jci.insight.129240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biogen. New warning of nusinersen-related communicating hydrocephalus. React Wkly. 2018;1714:3. [Google Scholar]
- 48.Skotte NH, Southwell AL, Østergaard ME, et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: Providing a therapeutic option for all Huntington disease patients. PLoS One. 2014;9:e107434. doi: 10.1371/journal.pone.0107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datson NA, González-Barriga A, Kourkouta E, et al. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS One. 2017;12:1–24. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kourkouta E, Weij R, González-Barriga A, et al. Suppression of Mutant Protein Expression in SCA3 and SCA1 Mice Using a CAG Repeat-Targeting Antisense Oligonucleotide. Mol Ther Nucleic Acids. 2019;17:601–14. doi: 10.1016/j.omtn.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Figlia M, Sena-Esteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci. 2007;104:17204–9. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stiles DK, Zhang Z, Ge P, et al. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp Neurol. 2012;233:463–71. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Alterman JF, Godinho BMDC, Hassler MR, et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat Biotechnol. 2019;37:884–94. doi: 10.1038/s41587-019-0205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alterman JF, Hall LM, Coles AH, et al. Hydrophobically Modified siRNAs Silence Huntingtin mRNA in Primary Neurons and Mouse Brain. Mol Ther Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imbert M, Blandel F, Leumann C, Garcia L, Goyenvalle A. Lowering Mutant Huntingtin Using Tricyclo-DNA Antisense Oligonucleotides As a Therapeutic Approach for Huntington’s Disease. Nucleic Acid Ther. 2019;29:256–65. doi: 10.1089/nat.2018.0775. [DOI] [PubMed] [Google Scholar]
- 56.Thadke SA, Perera JDR, Hridya VM, et al. Design of Bivalent Nucleic Acid Ligands for Recognition of RNA-Repeated Expansion Associated with Huntington’s Disease. Biochemistry. 2018;57:2094–108. doi: 10.1021/acs.biochem.8b00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagenacker T, Wurster CD, Günther R, et al. Nusinersen in adults with 5q spinal muscular atrophy: a non-interventional, multicentre, observational cohort study. Lancet Neurol. 2020;19:317–25. doi: 10.1016/S1474-4422(20)30037-5. [DOI] [PubMed] [Google Scholar]
- 58.Kim J, Hu C, Moufawad El, Achkar C, et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med. 2019;381:1644–52. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller T, Cudkowicz M, Shaw PJ, et al. Phase 1-2 Trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2020;383:109–19. doi: 10.1056/NEJMoa2003715. [DOI] [PubMed] [Google Scholar]
- 60.Tabrizi SJ, Flower MD, Gregory S, Ross CA, Wild EJ. Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat Rev Neurol. 2020;16:529–46. doi: 10.1038/s41582-020-0389-4. [DOI] [PubMed] [Google Scholar]
- 61.High KA, Roncarolo MG. Gene Therapy. N Engl J Med. 2019;381:455–64. doi: 10.1056/NEJMra1706910. [DOI] [PubMed] [Google Scholar]
- 62.Miniarikova J, Evers MM, Konstantinova P. Translation of MicroRNA-Based Huntingtin-Lowering Therapies from Preclinical Studies to the Clinic. Mol Ther. 2018;26:947–62. doi: 10.1016/j.ymthe.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vallès A, Evers MM, Stam A, et al. Widespread and sustained target engagement in Huntington’s disease minipigs upon intrastriatal microRNA-based gene therapy. Sci Transl Med. 2021;13:eabb8920. doi: 10.1126/scitranslmed.abb8920. [DOI] [PubMed] [Google Scholar]
- 64.Drouet V, Perrin V, Hassig R, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–85. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 65.Evers MM, Miniarikova J, Juhas S, et al. AAV5-miHTT Gene Therapy Demonstrates Broad Distribution and Strong Human Mutant Huntingtin Lowering in a Huntington’s Disease Minipig Model. Mol Ther. 2018;26:2163–77. doi: 10.1016/j.ymthe.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–73. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grondin R, Kaytor MD, Ai Y, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135:1197–209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–31. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 69.Shin JW, Kim K-H, Chao MJ, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25:ddw286. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heman-Ackah SM, Bassett AR, Wood MJA. Precision Modulation of Neurodegenerative Disease-Related Gene Expression in Human iPSC-Derived Neurons. Sci Rep. 2016;6:1–12. doi: 10.1038/srep28420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gubser Keller C, Shin Y, Monteys AM, et al. An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion. Nat Commun. 2022;13:1150. doi: 10.1038/s41467-022-28653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhattacharyya A, Trotta CR, Narasimhan J, et al. Small molecule splicing modifiers with systemic HTT-lowering activity. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-27157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.PTC Therapeutics. PTC518 Huntington’s Disease Program Update. 2021. [accessed Feb 16, 2022]. https://ir.ptcbio.com/static-files/2162ff85-c7ed-4555-8542-52fa2129f7fa .
- 75.Lokuwithana D. PTC Therapeutics slips after early stage results from Huntington’s disease trial. seekingalpha.com; 2021. [accessed Feb 16, 2022]. https://seekingalpha.com/news/3743661-ptc-therapeutics-slips-after-early-stage-results-from-huntingtons-disease-trial . [Google Scholar]
- 76.Moss DJH, Pardiñas AF, Langbehn D, et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 2017;16:701–11. doi: 10.1016/S1474-4422(17)30161-8. [DOI] [PubMed] [Google Scholar]
- 77.Bettencourt C, Hensman-Moss D, Flower M, et al. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann Neurol. 2016;79:983–90. doi: 10.1002/ana.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Porro A, Mohiuddin M, Zurfluh C, et al. FAN1-MLH1 interaction affects repair of DNA interstrand cross-links and slipped-CAG/CTG repeats. Sci Adv. 2021;7:eabf7906. doi: 10.1126/sciadv.abf7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer RR, Pluciennik A. DNA Mismatch Repair and its Role in Huntington’s Disease. J Huntingtons Dis. 2021;10:75–94. doi: 10.3233/JHD-200438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wheeler VC. Dissecting genetic modifiers of HD: towards understanding mechanism; CHDI Foundation Annual Therapeutics Conference 27-29th April; 2021. [Google Scholar]
- 81.Grachev ID, Meyer PM, Becker GA, et al. Sigma-1 and dopamine D2/D3 receptor occupancy of pridopidine in healthy volunteers and patients with Huntington disease: a [18F] fluspidine and [18F] fallypride PET study. Eur J Nucl Med Mol Imaging. 2021;48:1103–15. doi: 10.1007/s00259-020-05030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith-Dijak AI, Nassrallah WB, Zhang LYJ, Geva M, Hayden MR, Raymond LA. Impairment and Restoration of Homeostatic Plasticity in Cultured Cortical Neurons From a Mouse Model of Huntington Disease. Front Cell Neurosci. 2019;13:209. doi: 10.3389/fncel.2019.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naia L, Ly P, Mota SI, et al. The Sigma-1 Receptor Mediates Pridopidine Rescue of Mitochondrial Function in Huntington Disease Models. Neurotherapeutics. 2021;18:1017–38. doi: 10.1007/s13311-021-01022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eddings CR, Arbez N, Akimov S, Geva M, Hayden MR, Ross CA. Pridopidine protects neurons from mutant-huntingtin toxicity via the sigma-1 receptor. Neurobiol Dis. 2019;129:118–29. doi: 10.1016/j.nbd.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGarry A, Leinonen M, Kieburtz K, Geva M, Olanow CW, Hayden M. Effects of Pridopidine on Functional Capacity in Early-Stage Participants from the PRIDE-HD Study. J Huntingtons Dis. 2020;9:371–80. doi: 10.3233/JHD-200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liddelow SA, Guttenplan KA, Clarke LE, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–7. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–78. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 88.Björklund A, Parmar M. Neuronal Replacement as a Tool for Basal Ganglia Circuitry Repair: 40 Years in Perspective. Front Cell Neurosci. 2020;14:146. doi: 10.3389/fncel.2020.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benraiss A, Wang S, Herrlinger S, et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat Commun. 2016;7:11758. doi: 10.1038/ncomms11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeun P, Scahill RI, Tabrizi SJ, Wild EJ. Fluid and imaging biomarkers for Huntington’s disease. Mol Cell Neurosci. 2019;97:67–80. doi: 10.1016/j.mcn.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Wilson H, Politis M. International review of neurobiology. Elsevier; 2018. Molecular Imaging in Huntington’s Disease; pp. 289–333. [DOI] [PubMed] [Google Scholar]
- 92.Tortelli R, Rodrigues FB, Wild EJ. The use of wearable/portable digital sensors in Huntington’s disease: A systematic review. Parkinsonism Relat Disord. 2021;83:93–104. doi: 10.1016/j.parkreldis.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Byrne LM, Rodrigues FB, Johnson EB, et al. Evaluation of mutant huntingtin and neurofilament proteins as potential markers in Huntington’s disease. Sci Transl Med. 2018;10:eaat7108. doi: 10.1126/scitranslmed.aat7108. [DOI] [PubMed] [Google Scholar]
- 94.Byrne LM, Rodrigues FB, Blennow K, et al. Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington’s disease: a retrospective cohort analysis. Lancet Neurol. 2017;16:601–9. doi: 10.1016/S1474-4422(17)30124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vinther-Jensen T, Budtz-Jørgensen E, Simonsen AH, Nielsen JE, Hjermind LE. YKL-40 in cerebrospinal fluid in Huntington’s disease – A role in pathology or a nonspecific response to inflammation? Parkinsonism Relat Disord. 2014;20:1301–3. doi: 10.1016/j.parkreldis.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 96.Björkqvist M, Wild EJ, Thiele J, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–77. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vinther-Jensen T, Börnsen L, Budtz-Jørgensen E, et al. Selected CSF biomarkers indicate no evidence of early neuroinflammation in Huntington disease. Neurol Neuroimmunol Neuroinflammation. 2016;3:e287. doi: 10.1212/NXI.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niemela V, Landtblom A-M, Nyholm D, et al. Proenkephalin Decreases in Cerebrospinal Fluid with Symptom Progression of Huntington’s Disease. Mov Disord. 2021;36:481–91. doi: 10.1002/mds.28391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scahill RI, Zeun P, Osborne-Crowley K, et al. Biological and clinical characteristics of gene carriers far from predicted onset in the Huntington’s disease Young Adult Study (HD-YAS): a cross-sectional analysis. Lancet Neurol. 2020;19:502–12. doi: 10.1016/S1474-4422(20)30143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fodale V, Pintauro R, Daldin M, et al. Analysis of mutant and total huntingtin expression in Huntington’s disease murine models. Sci Rep. 2020;10:22137. doi: 10.1038/s41598-020-78790-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rodrigues FB, Owen G, Sathe S, et al. Safety and feasibility of research lumbar puncture in Huntington’s disease: the HDClarity cohort and bioresource. medRxiv. 2021:2021.07.30.21261340. doi: 10.3233/JHD-210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kinnunen KM, Schwarz AJ, Turner EC, et al. Volumetric MRI-Based Biomarkers in Huntington’s Disease: An Evidentiary Review. Front Neurol. 2021;12:712555. doi: 10.3389/fneur.2021.712555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tabrizi SJ, Reilmann R, Roos RAC, et al. Potential endpoints for clinical trials in premanifest and early Huntington’s disease in the TRACK-HD study: Analysis of 24 month observational data. Lancet Neurol. 2012;11:42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- 104.Tabrizi SJ, Scahill RI, Owen G, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12:637–49. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 105.Shaffer JJ, Ghayoor A, Long JD, et al. Longitudinal diffusion changes in prodromal and early HD: Evidence of white-matter tract deterioration. Hum Brain Mapp. 2017;38:1460–77. doi: 10.1002/hbm.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ciarmiello A, Cannella M, Lastoria S, et al. Brain white-matter volume loss and glucose hypometabolism precede the clinical symptoms of Huntington’s disease. J Nucl Med. 2006;47:215–22. [PubMed] [Google Scholar]
- 107.Ciarmiello A, Giovacchini G, Orobello S, Bruselli L, Elifani F, Squitieri F. 18F-FDG PET uptake in the pre-Huntington disease caudate affects the time-to-onset independently of CAG expansion size. Eur J Nucl Med Mol Imaging 2012 396. 2012;39:1030–6. doi: 10.1007/s00259-012-2114-z. [DOI] [PubMed] [Google Scholar]
- 108.Politis M, Pavese N, Tai YF, et al. Microglial activation in regions related to cognitive function predicts disease onset in Huntington’s disease: A multimodal imaging study. Hum Brain Mapp. 2011;32:258–70. doi: 10.1002/hbm.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fazio P, Fitzer-Attas CJ, Mrzljak L, et al. PET Molecular Imaging of Phosphodiesterase 10A: An Early Biomarker of Huntington’s Disease Progression. Mov Disord. 2020;35:606–15. doi: 10.1002/mds.27963. [DOI] [PubMed] [Google Scholar]
- 110.Liu L, Prime ME, Lee MR, et al. Imaging Mutant Huntingtin Aggregates: Development of a Potential PET Ligand. J Med Chem. 2020;63:8608–33. doi: 10.1021/acs.jmedchem.0c00955. [DOI] [PubMed] [Google Scholar]
- 111.Reilmann R, Schubert R. Motor outcome measures in Huntington disease clinical trials. Handb Clin Neuro. 2017;144:209–25. doi: 10.1016/B978-0-12-801893-4.00018-3. [DOI] [PubMed] [Google Scholar]
- 112.van Vugt JPP, Siesling S, Piet KKE, et al. Quantitative assessment of daytime motor activity provides a responsive measure of functional decline in patients with Huntington’s disease. Mov Disord. 2001;16:481–8. doi: 10.1002/mds.1097. [DOI] [PubMed] [Google Scholar]
- 113.Adams JL, Dinesh K, Xiong M, et al. Multiple Wearable Sensors in Parkinson and Huntington Disease Individuals: A Pilot Study in Clinic and at Home. Digit Biomarkers. 2017;1:52–63. doi: 10.1159/000479018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paulsen JS, Langbehn DR, Stout JC, et al. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–80. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dinesh K, Snyder CW, Xiong M, et al. A Longitudinal Wearable Sensor Study in Huntington’s Disease. J Huntingtons Dis. 2020;9:69–81. doi: 10.3233/JHD-190375. [DOI] [PubMed] [Google Scholar]
- 116.Collett J, Esser P, Khalil H, et al. Insights into gait disorders: Walking variability using phase plot analysis, Huntington’s disease. Gait Posture. 2014;40:694–700. doi: 10.1016/j.gaitpost.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 117.Skodda S, Grönheit W, Lukas C, et al. Two different phenomena in basic motor speech performance in premanifest Huntington disease. Neurology. 2016;86:1329–35. doi: 10.1212/WNL.0000000000002550. [DOI] [PubMed] [Google Scholar]
- 118.Bartlett DM, Domínguez DJF, Reyes A, et al. Investigating the relationships between hypothalamic volume and measures of circadian rhythm and habitual sleep in premanifest Huntington’s disease. Neurobiol Sleep Circadian Rhythm. 2019;6:1–8. doi: 10.1016/j.nbscr.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Killoran A. In: Neurodegenerative Diseases Biomarkers: Towards Translating Research to Clinical Practice. Peplow PV, Martinez B, Gennarelli TA, editors. New York, NY, NY: Springer US; 2022. Biomarkers in Huntington’s Disease; pp. 235–62. [Google Scholar]
- 120.Stout JC, Andrews SC, Glikmann-Johnston Y. Cognitive assessment in Huntington disease clinical drug trials. Handb Clin Neurol. 2017;144:227–44. doi: 10.1016/B978-0-12-801893-4.00019-5. [DOI] [PubMed] [Google Scholar]
- 121.Bulaklak K, Gersbach CA. The once and future gene therapy. Nat Commun. 2020;11:5820. doi: 10.1038/s41467-020-19505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saudou F, Humbert S. The Biology of Huntingtin. Neuron. 2016;89:910–26. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 123.Tabrizi SJ, Schobel S, Gantman EC, et al. Huntington’s Disease Integrated Staging System (HD-ISS): A Novel Evidence-Based Classification System For Staging. medRxiv. 2021:2021.09.01.21262503 [Google Scholar]
- 124.Dunn B, Stein P, Cavazzoni P. Approval of Aducanumab for Alzheimer Disease—The FDA’s Perspective. JAMA Intern Med. 2021;181:1276. doi: 10.1001/jamainternmed.2021.4607. [DOI] [PubMed] [Google Scholar]
- 125.FDA. Human Gene Therapy for Neurodegenerative Diseases - Draft Guidance for Industry. 2021. [accessed Nov 12, 2021]. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/human-gene-therapy-neurodegenerative-diseases .
- 126.Zhou FF, Ward K, Kennedy A, Roberts TS, Gaebler JA. Value for Whom? Incorporating Patient Perspectives into Value Assessment for Novel Cell and Gene Therapies. PIPC. 2020 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.