Abstract
The bottom-up assembly approach for construction of synthetic cells is an effective tool for isolating and investigating cellular processes in a cell mimicking environment. Furthermore, the development of cell-free expression systems has demonstrated the ability to reconstitute the protein production, transcription and translation processes (DNA→RNA→protein) in a controlled manner, harnessing synthetic biology. Here we describe a protocol for preparing a cell-free expression system, including the production of a potent bacterial lysate and encapsulating this lysate inside cholesterol-rich lipid-based giant unilamellar vesicles (GUVs) (i.e., stable liposomes), to form synthetic cells. The protocol describes the methods for preparing the components of the synthetic cells including the production of active bacterial lysates, followed by a detailed step-by-step preparation of the synthetic cells based on a water-in-oil emulsion transfer method. These facilitate the production of millions of synthetic cells in a simple and affordable manner with a high versatility for producing different types of proteins. The obtained synthetic cells can be used to investigate protein/RNA production and activity in an isolated environment, in directed evolution, and also as a controlled drug delivery platform for on-demand production of therapeutic proteins inside the body.
Introduction
Synthetic cells are artificial cell-like particles, mimicking one or multiple functions of a living cell, such as the ability to divide, form membrane interactions, and synthesize proteins based on a genetic code1,2,3. Synthetic cells that enclose cell-free protein synthesis (CFPS) systems possess high modularity due to their ability to produce various proteins and RNA sequences following alterations in the DNA template. Presenting an attractive alternative to the current approaches of protein production, CFPS systems are based on cell lysate, purified components, or synthetic components and include all the transcription and translation machinery required for protein synthesis such as ribosomes, RNA polymerase, amino acids and energy sources (e.g., 3-phosphoglycerate and adenine triphosphate)4,5,6,7,8,9. The encapsulation of a CFPS system inside lipid vesicles enables the simple and efficient production of proteins without depending on a living cell10. Moreover, this platform allows synthesis of peptides that may degrade inside natural cells, production of proteins that are toxic to living cells, and modify proteins with non-natural amino acids11,12. Synthetic cells have been used as a model for research purposes investigating the minimal cell components required to enable cellular life from an evolutionary perspective1,13. Synthetic cells have also been used to build and implement genetic circuit and as models for directed evolution14,15,16. Other studies have focused on the ability of synthetic cells to mimic the biological activity of natural cells, aiming to replace damaged natural cells, such as beta cells in patients with diabetes17. Furthermore, the ability of these CFPS encapsulating synthetic cells to produce a variety of therapeutic proteins illustrates its potential to be incorporated into clinical use18.
Here we describe a bottom-up lab-scale protocol (Figure 1) for the production of RNA and protein-producing synthetic cells based on a CFPS system encapsulated in a lipid vesicle. This shows the potential use of synthetic cell platforms as novel drug delivery systems for the onsite production of a therapeutic protein drug in vivo19. Previous studies have investigated the optimization of the CFPS reaction and the cell lysate preparation processes4,8,20. Moreover, several techniques have been applied for cell-sized liposome preparation, such as microfluidic and polymer-based droplet stabilization methods21,22,23, which also differ in the liposomes' lipid composition24,25,26. In the presented protocol, synthetic cells are produced using a water-in-oil emulsion transfer method and the encapsulation process is carried out at low temperatures (<4 °C)5,10,24,27,28. These mild conditions have been found to be favorable for retaining the bio-functional integrity of the molecular machinery, namely ribosomes and proteins27,29,30. The lipid composition of the particles consists of both cholesterol and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). The first is found in all mammalian cell membranes and is essential for the stability, rigidity and permeability reduction of the membrane, and the latter mimics mammalian phospholipid composition11,13. The cellular transcription and translation molecular machinery are extracted from the BL21 (DE3) Escherichia coli (E. coli) strain, which is transformed with pAR1219 plasmid overexpressing T7 RNA polymerase to increase CFPS potency and protein synthesis. This system has been used to produce diagnostic and therapeutic proteins, with molecular weights of up to 66 kDa in vitro and in vivo19,31. The following protocol provides a simple and effective method for the production of the synthetic cell system, which can address a wide range of fundamental questions associated with protein synthesis in nature and can also be utilized for drug delivery applications.
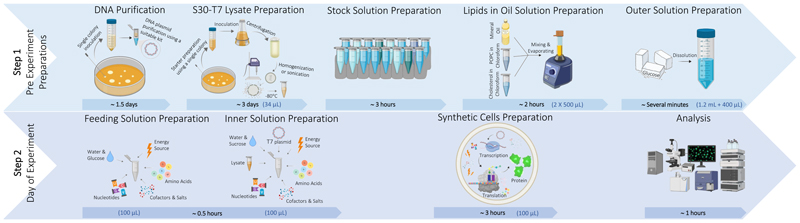
Figure 1. An illustration of the process for typical synthetic cells preparation protocol.
The process is divided into two steps. Step 1: Pre-experiment preparations including DNA plasmid purification, S30-T7 lysate preparation, stock solutions preparation required for the inner and feeding solutions, lipids-in-oil solution preparation and outer solution preparation. Step 2: Synthetic cell formation that includes feeding and inner solutions preparation, synthetic cells preparation and analysis. An example of the required volume of each ingredient for preparing 100 μL of synthetic cells solution is presented in blue in brackets.
Protocol
Preparation of S30-T7 lysate
Streak plate the E. coli BL21(DE3) bacteria transformed with the T7 RNA polymerase expressing pAR1219 plasmid on a LB-agar plate supplemented with 50 μg/mL ampicillin to obtain single colonies.
Prepare a starter solution: Inoculate a single colony into 5 mL of LB-media supplemented with 50 μg/mL ampicillin in a 100 mL Erlenmeyer flask and grow overnight using a floor incubator shaker at 250 rpm and 37 °C. Prepare duplicates.
Inoculate each 5 mL starter separately into 500 mL of TB media supplemented with 50 μg/mL ampicillin in a 2 L Erlenmeyer flask with baffles and grow it using a floor incubator shaker at 250 rpm and 37 °C until it reaches OD600 of 0.8-1. Monitor periodically using a spectrophotometer.
Add 2-3 mL of 100 mM stock of IPTG (to reach 0.4 - 0.6 mM) for induction of T7 RNA polymerase expression and continue growing the culture until it reaches OD600≈4.
Transfer the solution from each Erlenmeyer flask into two 250 mL sterilized centrifuge tubes.
-
Centrifuge each at 7,000 x g for 10 min at 4 °C. Discard the supernatant.
NOTE: At this stage, the bacterial pellet can be stored at -20 °C for a few days before moving on to the next steps.
-
Re-suspend each pellet in 250 mL of cold (4 °C) S30 lysate buffer and centrifuge at 7,000 x g for 10 min at 4 °C.
NOTE: The S30 lysate buffer is used for maintaining protein stability after cell lysis is performed using the homogenizer in step 1.9. From this step forward, all steps until 1.12 should be carried out consecutively and rapidly.
NOTE: Before proceeding to the next step, pre-cool the tips and 1.5-milliliter vials that will be needed for storing the lysate
-
Discard the supernatant and re-suspend all pellets together in 15 mL of cold S30 lysate buffer. Filter the suspension using gauze pad.
NOTE: 15ml of solution is due to homogenizer min. volume. Should be noticed that bacteria concentration is also important.
Homogenize at a working pressure of 15,000 psi, with an air pressure of 4 bar (two passes) for cell breakage. Avoid solution dilution for a more concentrated and active lysate.
Add 100 μL of 0.1 M DTT (CAUTION) per 10 mL of the homogenized suspension.
Centrifuge the suspension at 24,700 x g for 30 min at 4 °C.
-
Perform the following step quickly for preserving the lysate activity: divide the supernatant one-by-one into 200 μL aliquots in precooled 1.5 mL vials and immediately snap freeze them with liquid nitrogen. Store at -80 °C for further use.
CAUTION: DTT is classified as Irritant and Harmful and should therefore be treated with care.
-
2.Preparation of lipids in oil solution
- Dissolve POPC and cholesterol in chloroform (CAUTION) separately, each to a final concentration of 100 mg/mL. Vortex each vial separately.
- Combine the components in a 2mL glass vial: add 50 μL of POPC in chloroform, 50 μL of cholesterol in chloroform and 500 μL of mineral oil. For 100 μL of synthetic cells, 2 vials of lipids in oil are required.
-
Vortex, and then heat for about 1 h at 80 °C in a chemical hood to evaporate the chloroform. Ensure that complete evaporation has occurred by following the specified time/conditions and monitoring the solution volume.NOTE: The resulting lipid-in-oil solution can be stored at room temperature for up to two weeks. For improved results, it is recommended to use a fresh preparation before each experiment. Lipid and cholesterol ratios can be altered according to the desired membrane composition. A high concentration of cholesterol can lead to the formation of aggregates in the final synthetic cell solution.CAUTION: Chloroform is classified as Irritant and Harmful and should therefore be treated with care and in areas with fume extraction.
-
3.Preparations of outer, pre-inner and feeding solutions
- Preparation of stock solutions
-
Prepare the stock solutions listed in Table 2 using ultrapure water (UPW).NOTE: Stock solutions should be prepared in advance and stored at -20 °C until further usage. Reagent 7 tends to form aggregates. Heating to 37 °C will reduce the aggregation. Slight aggregation will not affect the reaction significantly. Reagent 8 solution is milky and turbid.
-
- Outer solution
- Dissolve glucose in DNase/RNase-free H2O to a final concentration of 200 mM.
- For 100 μL of inner solution, prepare 1.6 mL of outer solution.
- Pre-inner solution
-
Add reagents 1-14 according to the amounts and concentration listed in Table 2. For example, for a final synthetic cell volume of 100 μL, prepare 100 μL of inner solution.NOTE: UPW should be added to complete the final required volume. At this stage, the mixture can be stored at 4 °C for a few hours.
-
- Feeding solution
-
Add all the reagents according to the amounts and concentration listed in Table 3.NOTE: Use 1:1 ratio of feeding solution:inner solution. For a final synthetic cell volume of 100 μL, prepare 100 μL of feeding solution. It is recommended to prepare a small excess volume of outer, inner and feeding solution.
-
-
4.
Preparation of synthetic cells
NOTE: The following volumes are adjusted for the preparation of 100 μL of synthetic cells.- Synthetic cells producing protein
- In a 15 mL tube, place 12 mL of the outer solution and slowly add, on top a layer, of 500 μL of lipids in oil solution. Incubate at room temperature for 20 min.
- To finalize the inner solution preparation: mix the inner solution ingredients on crushed ice to a final volume of 100 μL by thawing and adding S30-T7 Lysate (reagent 15) and DNA plasmid (reagent 16) to the stored mixture.
- To the second 2 mL glass vial with 500 μL of lipids in oil solution, add 100 μL of the inner solution. Pipette up and down vigorously for 1 minute and vortex for another minute on level five.
- Incubate for 10 min on crushed ice and slowly add the resulted emulsion on top of the oil phase in the 15 mL tube (from 4.1.1).
-
Centrifuge for 10 min at 100 x g and 4 °C and then centrifuge for 10 min at 400 x g and 4 °C. By the end of the centrifugation, a pellet at the bottom of the tube should be observed.NOTE: Using a swinging bucket centrifuge rotor is preferred here for acquiring a better coverage of a second layer of lipids during the water-in-oil droplets’ passage through the interphase. In case there is no observable pellet, centrifugation speed can be increased to 1000 x g. Otherwise, see the Discussion section referring to the specific gravity of the outer solution.
- Extract the pellet.
- Remove excess oil layer.
- Use a trimmed pipette tip loaded with approximately 400 μL of outer solution to extract the pellet. Release the outer solution while passing through the oil phase in order to collect only the pellet in the aqueous phase.
- Wipe the tip after the extraction of the pellet to avoid transferring oil remains and transfer the pellet to a clean 1.5 mL tube.
-
Centrifuge for 10 min at 1,000 x g and 4 °C, remove the supernatant and re-suspend the pellet in 100 μL of feeding solution (1:1 ratio of inner:feeding solutions).NOTE: A fixed angle centrifuge rotor may be used here as well.
-
For protein expression, incubate for 2 hours at 37 °C without shaking.NOTE: Optimal incubation time varies between different proteins.
- Evaluate the produced protein amount using a suitable method according to the target protein properties.
- Recommended control groups
- Inner solution and lysate activity confirmation
- Prepare a complete inner solution (with DNA & S30-T7 lysate).
-
Immediately incubate the reaction above using a floor incubator shaker at 250 rpm or a thermomixer at 1200 rpm, at a constant temperature of 37 °C for 2 h.NOTE: Adjust the incubation time to match the synthetic cells incubation time.
- Synthetic cells
- Negative control group: Prepare the protocol presented in 4.1 with inner solution without DNA.
-
Positive control: Prepare the protocol presented in 4.1 with inner solution containing a T7 plasmid encoding for a reporter gene.NOTE: Add a positive control group comprised of a reporter gene, such as sfGFP, alongside the test groups to ensure the encapsulation efficiency step.
Table 2. Inner solution corresponse.
| Final requested Inner solution volume | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Reagent | Stock conc. | Final conc. | 25 μL | 50 μL | 100 μL | 200 μL | 300 μL | 400 μL | ||||||||||||||||
| 1 | HEPES KOH pH=8 | 1 M | 55 mM | 1.375 | 2.75 | 5.5 | 11 | 16.5 | 22 | Pre-Inner solution | |||||||||||||||
| 2 | Magnesium acetate | 1 M | 14 mM | 0.35 | 0.7 | 1.4 | 2.8 | 4.2 | 5.6 | ||||||||||||||||
| 3 | Potassium acetate | 1 M | 50 mM | 1.25 | 2.5 | 5 | 10 | 15 | 20 | ||||||||||||||||
| 4 | Ammonium acetate | 5.2 M | 155 mM | 0.75 | 1.5 | 3 | 6 | 9 | 12 | ||||||||||||||||
| 5 | PEG 6000 | 50% | 3% | 1.5 | 3 | 6 | 12 | 18 | 24 | ||||||||||||||||
| 6 | 3-PGA | 0.5 M | 40 mM | 2 | 4 | 8 | 16 | 24 | 32 | ||||||||||||||||
| 7 | Amino acids - mixture I | 50 M | 2.5 mM | 1.25 | 2.5 | 5 | 10 | 15 | 20 | ||||||||||||||||
| 8 | Amino acids - mixture II | 50 M | 2.5 mM | 1.25 | 2.5 | 5 | 10 | 15 | 20 | ||||||||||||||||
| 9 | ATP | 100 mM | 1.2 mM | 0.3 | 0.6 | 1.2 | 2.4 | 3.6 | 4.8 | ||||||||||||||||
| 10 | GTP | 50 mM | 1 mM | 0.5 | 1 | 2 | 4 | 6 | 8 | ||||||||||||||||
| 11 | UTP | 100 mM | 0.8 mM | 0.2 | 0.4 | 0.8 | 1.6 | 2.4 | 3.2 | ||||||||||||||||
| 12 | IPTG | 100 mM | 1 mM | 0.25 | 0.5 | 1 | 2 | 3 | 4 | ||||||||||||||||
| 13 | Sucrose | 2 M | 200 mM | 2.5 | 5 | 10 | 20 | 30 | 40 | ||||||||||||||||
| 14 ** | H2O UPW | ** | ** | ** | ** | ** | ** | ||||||||||||||||||
| 15 | S30-T7 Lysate | 34% | 8.5 | 17 | 34 | 68 | 102 | 136 | |||||||||||||||||
| 16 * | DNA plasmid | 10 μg/mL | * | * | * | * | * | * | |||||||||||||||||
Table 3. Feeding solution composition.
| Final requested feeding solution volume | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Reagent | stock conc. | final conc. | 25 μL | 50 μL | 100 μL | 200 μL | 300 μL | 400 μL |
| 1 | HEPES KOH pH=8 | 1 M | 83.3 mM | 2.08 | 4.17 | 8.33 | 16.67 | 25 | 33.33 |
| 2 | Magnesium acetate | 1 M | 21.2 mM | 0.53 | 1.06 | 2.12 | 4.24 | 6.36 | 8.48 |
| 3 | Potassium acetate | 1 M | 75.5 mM | 1.89 | 3.78 | 7.55 | 15.1 | 22.66 | 30.2 |
| 4 | Ammonium acetate | 5.2 M | 236.4 mM | 1.14 | 2.27 | 4.55 | 9.09 | 13.64 | 18.18 |
| 5 | PEG - 6000 | 50% | 4.54% | 2.27 | 4.54 | 9.08 | 18.16 | 27.24 | 36.32 |
| 6 | 3-PGA | 0.5 M | 60.1 mM | 3 | 6.01 | 12.01 | 24.02 | 36.04 | 48.04 |
| 7 | Amino acids - mixture I | 50 mM | 3.8 mM | 1.89 | 3.78 | 7.56 | 15.12 | 22.68 | 30.24 |
| 8 | Amino acids - mixture II | 50 mM | 3.8 mM | 1.89 | 3.78 | 7.56 | 15.12 | 22.68 | 30.24 |
| 9 | ATP | 100 mM | 1.8 mM | 0.45 | 0.91 | 1.81 | 3.62 | 5.43 | 7.24 |
| 10 | GTP | 50 mM | 1.5 mM | 0.76 | 1.51 | 3.02 | 6.04 | 9.06 | 12.08 |
| 11 | UTP | 100 mM | 1.2 mM | 0.3 | 0.61 | 1.21 | 2.42 | 3.63 | 4.84 |
| 12 | IPTG | 100 mM | 1.5 mM | 0.38 | 0.76 | 1.51 | 3.02 | 4.53 | 6.04 |
| 13 | Glucose | 2 M | 200 mM | 2.5 | 5 | 10 | 20 | 30 | 40 |
| 14 | H2O UPW | 5.92 | 11.83 | 23.7 | 47.38 | 71.06 | 94.76 | ||
Representative Results
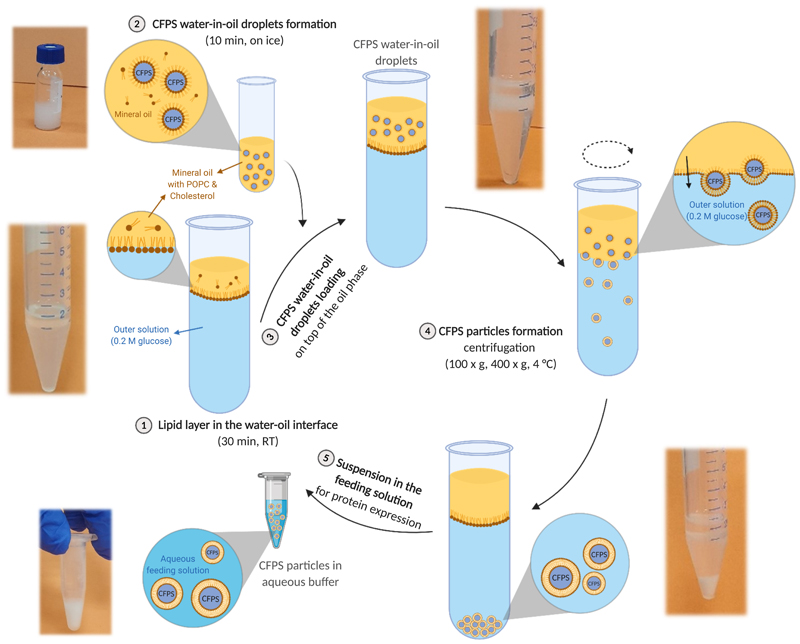
We present a protocol for the preparation of synthetic cells by encapsulating a S30-T7 CFPS system based on BL21 E. coli inside lipid vesicles. A schematic description of the preparation process that includes an image of each stage is presented in Figure 2. The success of the synthetic cell preparation process is dependent on the appropriate performance of each stage and effected by different parameters. The protocol should be adjusted to accommodate the production of a specific protein.
Figure 2. Schematic illustration of the synthetic cell preparation protocol.
An image of a well-performed process illustrates each stage of the protocol. This figure has been modified from Krinsky et al.19
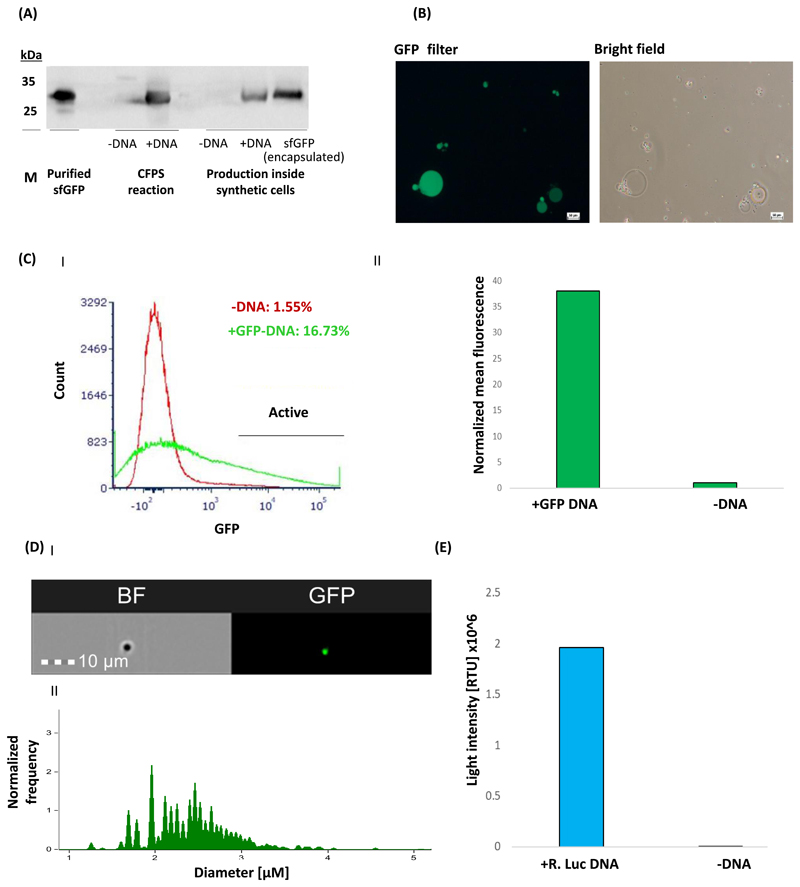
Plasmids expressing the model protein super-folder Green Fluorescent Protein (sfGFP) and Renilla Luciferase under the T7 promoter were introduced into CFPS bulk reactions and synthetic cells, and protein production was evaluated using different methods, including western blot, flow cytometry, microscopy and spectroscopy (Figure 3). A verification of the sfGFP-His6 tagged protein (~27 kDa32) production by western blot analysis is presented in Figure 3A. A sample of 30 μL of synthetic cells diluted 6 folds was mixed with 10 μL of a common SDS-PAGE sample buffer (containing the detergents sodium dodecyl sulfate (SDS) and β-mercaptoethanol) and boiled for 10 min (95 °C). We found that the combination of heat and detergents in the sample buffer is sufficient to disassemble the vesicles and to enable the running of proteins in the SDS-PAGE gel. Protein detection was performed using anti-His polyclonal primary antibody (diluted 1:12,000). As expected, while protein production is detected in samples containing sfGFP-encoding DNA templates ("+sfGFP DNA"), no protein is observed in negative control samples in which the DNA template was excluded ("-DNA"). Samples of purified sfGFP and purified sfGFP encapsulated within synthetic cells' membrane ('sfGFP (encapsulated)), are used as positive controls for the sfGFP CFPS bulk production and for its synthesis within synthetic cells, respectivley. This method can be applied to different protein types. According to Krinsky et al.19, the sfGFP production yield obtained under the detailed protocol is 380 μg/mL in CFPS solution and 5.3 μg/mL, when the CFPS solution is encapsulated inside lipid vesicles.
Figure 3. Representative results of the production of sfGFP and Renilla luciferase inside synthetic cells.
(A) A western blot analysis of sfGFP-His6 production in both CFPS bulk reactions and inside synthetic cells. Protein detection was carried out using anti-His polyclonal primary antibody (diluted 1:12,000). Purified sfGFP-His6 was used as a positive control (7.8 μg). sfGFP (encapsulated) is positive control of purified sfGFP encapsulated in synthetic cells. Samples without DNA templates were used as a negative control for the production analysis ("-DNA"). (B) Representative images of sfGFP producing synthetic cells taken using a fluorescent microscope with a bright-field and a GFP filter. Scale bars = 50 μm. (C) I&II Flow cytometry analysis of sfGFP producing synthetic cells activity (Data collected using digital, 4-laser analyzer). Samples were measured after 3 h of incubation. 10,000 events were collected for each analyzed sample. FSC (500 V) and SSC (300 V) filters were used to define the total synthetic cell population. Then, a FITC (600 V) filter was used to detect GFP fluorescence. (I) Calculation of the active synthetic cell population [%] was based on GFP fluorescence intensity threshold, defined by the “-DNA” sample (red histogram), allowing an error of ~1%. (II) Mean fluorescence intensity of sfGFP producing synthetic cells. This value was calculated from the active synthetic cell population and normalized to the “-DNA” sample by dividing the mean fluorescence of the active synthetic cells by the mean of the synthetic cells without sfGFP-DNA. (D) I&II Synthetic cell size analysis. The emission spectrum was detected by 505-560 nm and 642-745 nm for GFP and bright field signals, respectively. (I) A representative image of the analyzed synthetic cells. (II) Synthetic cells’ size distribution. Analysis was performed and the diameter distributions of the active synthetic cells was calculated based on the GFP signal. (E) Production of active Renilla luciferase inside synthetic cells. Luciferase activity was quantified with luminescence measurements after the addition of 1 μM h-coelenterazine using a plate reader and presented as light intensity [RLU] x 106
When the protein that is produced inside the synthetic cells is fluorescent, its production can be evaluated using microscopy and flow cytometry-based methods. We analyzed the synthetic cells using a fluorescent microscope with a filter for GFP fluorescence (Figure 3B). Since the CFPS components have some autofluorescent, the image acquisition parameters should be adapted according to an appropriate negative control sample (synthetic cells with no DNA template, for example).
In addition, we used flow cytometry to determine the mean fluorescence intensity of sfGFP-producing synthetic cells, and the percentage of active synthetic cells, which can produce proteins within them (Figure 3C I&II). 10,000 events were collected for each analyzed sample. The synthetic cell production, which is described in this protocol, usually yields an active population of 21-25% within a solution with an approximated concentration of 107 synthetic cells/mL. The slight fluorescence intensity presented by “-DNA” samples in Figure 3C II is due to the autofluorescence of different components in the CFPS reaction such as the S30 lysate.
Representative size analysis based on the GFP signal (in diameter) of synthetic cells prepared by the method described above shows a mean value of 2.4±0.5 μm (Figure 3D II). As the formation of water in oil emulsion in this method is an outcome of applying mechanical forces, the size distribution of the particles might be affected by different factors, such as the method and speed of pipetting the emulsion up-and-down, the model of the vortex mixer machine, etc.
To test the versatility of synthetic cells’ protein production, the expression of the reporter protein Renilla luciferase inside synthetic cells was analyzed (Figure 3E). The assay quantified Renilla luciferase activity by measuring the luminescence generated from the enzymatic reaction of luciferase and its substrate, h-coelenterazine. To induce the enzymatic reaction, a final concentration of 1 μM of h-coelenterazine was added to the synthetic cells (25 μL sample volume) just before the measurement. “-DNA” sample, not containing the luciferase-encoding DNA template was used as a negative control for testing luminescence due to non-enzymatic oxidation of the substrate. Luminescence was measured using a plate reader.
Discussion
This protocol introduces a simple and affordable method for the production of large quantities of protein-producing synthetic cells. The yield of active cells is dependent on careful and accurate execution of the protocol with emphasis on several critical steps. In the lysate preparation section of this method, it is essential to reach the appropriate bacteria density before cell lysis to achieve a sufficient amount of proteins in the bacterial lysate. Second, the lysis process should be performed at 4 °C and the lysate frozen quickly with liquid nitrogen to maintain protein activity. Moreover, in this protocol we used E. coli BL21(DE3) cells transformed with pAR1219 plasmid expressing T7 RNA polymerase. In cases where a different strain of bacteria is used, changes to the lysate concentration may be required to reach a satisfactory amount of RNA polymerase and ribosomes. Even when the same bacterial strain is used, different lysate productions may have some batch-to-batch variability.
The vesicle production section also includes a couple of significant steps. Solubilization of the lipids and removal of the chloroform from the mineral oil is important for establishing the water-in-oil emulsion. The centrifugation of the water-in-oil droplets into the outer aqueous buffer is also a key step in the protocol to generate synthetic cells with a lipid bilayer. Changes in the inner solution of the vesicles and the lipid composition might lead to a layer of droplets at the oil-water interphase without any observable pellet. To overcome this, a quick solution is to increase the centrifugation speed to 1,000 x g in the emulsion transfer step. In case this does not solve the issue, the specific gravity of the outer aqueous solution can be altered ensuring that the inner solution will have a higher specific gravity than the outer solution. Furthermore, the osmolality of the inner solution may also vary between different lysate sources and productions, ranging between 800-1100 mOsm/kg. Variability in the inner solution's osmolality is mostly due to changing concentrations of the lysate during its production process. Usually this does not lead to significant changes in protein production, yet a large dilution might lead to a reduced yield due to low concentrations of the transcription and translation enzymes in the lysate itself. Measuring the total protein concentration in each lysate batch can assist in tuning lysate concentrations in the inner solution to maintain constant values (approximately 22 mg/mL measured by Bradford assay).
Nevertheless, this method has some limitations that should be mentioned. Not all of the generated synthetic cells are active and capable of producing proteins due to incomplete encapsulation of all the required components. We measured approximately 21-25% of active cells when producing sfGFP expressing synthetic cells using flow cytometry (no significant size differences were observed between active and inactive cells). Oil and lipid excess residues occasionally remain in the bilayer lipid membrane after the transfer of the synthetic cells into the aqueous phase. Optimizing the lipid and cholesterol concentrations in the oil phase can improve this issue. It is also important to note that the size distribution of the obtained synthetic cells is quite wide in comparison to alternative microfluidic methods, with vesicles ranging from approximately 1-50 μm.
The high yield of the emulsion transfer method makes it especially suitable for synthetic cells encapsulating cell-free protein synthesis systems for therapeutic protein production. Different variations of the emulsion transfer method have been used in synthetic cell studies and included changes in the oil preparation procedure, membrane composition, sample volume, inner solution to oil ratio and centrifugation speeds5,24,28. Microfluidic and polymer-based droplet stabilization methods have also been used for synthetic cell preparation, however, encapsulation of the whole CFPS system has not been performed yet with these methods21,22,23.
The synthetic cells obtained by this method were shown in vivo to produce proteins and treat cancer in murine models19. The capabilities of these cells can be further expanded in the future beyond protein expression. Integration of other cellular processes such as cellular communication, cytoskeleton modification and cell division in synthetic cells have been recently described33,34,35,36. Substitution of the bacterial lysate with eukaryotic cell lysate will allow expression of proteins with higher complexity and post-translational modifications, and will perhaps be less immunogenic even without a cleaning procedure, therefore opening more therapeutic frontiers37.
Supplementary Material
Summary.
This protocol describes the method, materials, equipment and steps for bottom-up preparation of RNA and protein producing synthetic cells. The inner aqueous compartment of the synthetic cells contained the S30 bacterial lysate encapsulated within a lipid bilayer (i.e., stable liposomes), using a water-in-oil emulsion transfer method.
Table 1. Buffer and stock solutions preparation.
| Comments | |
|---|---|
| Luria Bertani (LB) agar (1.5%) plate: | Prepare and sterilize. Add Ampicillin at a final concentration of 50 µg/mL only after cooling to room temperature. |
| 10 g/L Bacto-tryptone | |
| 10 g/L Sodium chloride (NaCl) | |
| 5 g/L Bacto-Yeast extract | |
| 15 g/L Agar agar purified | |
| 50 µg/mL Ampicillin | |
| LB media (20 mL): | Minimal media. Prepare and sterilize in advance. Just prior to inoculating the bacteria, add Ampicillin at a final concentration of 50 µg/mL. |
| 10 g/L Bacto-tryptone | |
| 10 g/L Sodium chloride (NaCl) | |
| 5 g/L Bacto-Yeast extract | |
| 50 µg/mL Ampicillin | |
| Terrific Broth (TB) media (1 L): | Rich media. Prepare and sterilize in advance. Just prior to inoculating the bacteria, add Ampicillin at a final concentration of 50 µg/mL. |
| 12 g/L Bacto-tryptone | |
| 24 g/L Bacto-Yeast extract | |
| 4% (v/v) Glycerol anhydrous | |
| 2.32 g/L K2HPO4 | |
| 12.54 g/L KH2PO4 | |
| 50 µg/mL Ampicillin | |
| S30 lysate buffer (1.5 L): | Prepare and sterilize in advance (*excluding DTT and 2-mercaptoethanol). Store at -4 °C. |
| 10 mM Tris-acetate at pH = 7.4 | Trisma-base. Prepare and adjust pH to 7.4 using Acetic acid. |
| 14 mM magnesium acetate | |
| 60 mM potassium acetate | |
| *1 mM DTT | Add just prior to use |
| *0.5 mL/L 2-mercaptoethanol | Add just prior to use |
| 1 M HEPES-KOH (pH = 8): | Dissolve HEPES to a final concentration of 1 M. Adjust to pH 8 using KOH solution. |
| HEPES | |
| Potassium hydroxide (KOH) |
Table 4. Required solution for synthetic cells preparation.
| Solution & materials required | Comments |
|---|---|
| Lipids in oil | Store at room temperature until use. Prepared according to section 2 |
| Outer solution | Prepared according to section 3.1 |
| Inner solution | Prepared according to section 3.2. |
| Prepare tests + controls samples according to section 4.1. | |
| Keep on crushed ice until use. | |
| Feeding solution | Prepared according to section 3.3. |
| Keep on crushed ice until use. |
Acknowledgments
This work was supported by ERC-STG-2015-680242.
The authors also acknowledge the support of the Technion Integrated Cancer Center (TICC); the Russell Berrie Nanotechnology Institute; the Lorry I. Lokey Interdisciplinary Center for Life Sciences & Engineering; the Israel Ministry of Economy for a Kamin Grant (52752); the Israel Ministry of Science Technology and Space – Office of the Chief Scientist (3-11878); the Israel Science Foundation (1778/13, 1421/17); the Israel Cancer Association (2015-0116); the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); the Phospholipid Research Center Grant; a Rosenblatt Foundation for cancer research, a Mallat Family Foundation Grant; and the Unger Family Foundation. A. Schroeder acknowledges Alon and Taub Fellowships. O. Adir acknowledges the Sherman and Gutwirth fellowships. G. Chen acknowledges the Sherman Fellowship. N. Krinsky acknowledges the Baroness Ariane de Rothschild Women Doctoral Program from the Rothschild Caesarea Foundation.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/60829.
Disclosures
The authors have nothing to disclose.
References
- 1.Blain JC, Szostak JW. Progress toward synthetic cells. Annual review of biochemistry. 2014;83:615–640. doi: 10.1146/annurev-biochem-080411-124036. [DOI] [PubMed] [Google Scholar]
- 2.Richmond DL, et al. Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proceedings of the National Academy of Sciences. 2011;108(23):9431–9436. doi: 10.1073/pnas.1016410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshpande S, Spoelstra WK, van Doorn M, Kerssemakers J, Dekker C. Mechanical division of cell-sized liposomes. ACS Nano. 2018;12(3):2560–2568. doi: 10.1021/acsnano.7b08411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu DV, Zawada JF, Swartz JR. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnology Progress. 2005;21(2):460–465. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 5.Stano P. Gene Expression Inside Liposomes: From Early Studies to Current Protocols. Chemistry-A European Journal. 2019 doi: 10.1002/chem.201806445. [DOI] [PubMed] [Google Scholar]
- 6.Lewandowski B, et al. Sequence-specific peptide synthesis by an artificial small-molecule machine. Science. 2013;339(6116):189–193. doi: 10.1126/science.1229753. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. Cogenerating synthetic parts toward a self-replicating system. ACS Synthetic Biology. 2017;6(7):1327–1336. doi: 10.1021/acssynbio.6b00342. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, et al. An economical and highly productive cell-free protein synthesis system utilizing fructose-1,6-bisphosphate as an energy source. Journal of Biotechnology. 2007;130(4):389–393. doi: 10.1016/j.jbiotec.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Kim TW, et al. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. Journal of Biotechnology. 2006;126(4):554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proceedings of the National Academy of Sciences. 2004;101(951):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He M, He Y, Luo Q, Wang M. From DNA to protein: No living cells required. Process Biochemistry. 2011;46(3):615–620. [Google Scholar]
- 12.Casteleijn MG, Urtti A, Sarkhel S. Expression without boundaries: cell-free protein synthesis in pharmaceutical research. International Journal of Pharmaceutics. 2013;440(91):39–47. doi: 10.1016/j.ijpharm.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Luisi PL, Ferri F, Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften. 2006;93(91):1–13. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- 14.Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. Engineering genetic circuit interactions within and between synthetic minimal cells. Nature Chemistry. 2017;9(5):431–439. doi: 10.1038/nchem.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Y, Contreras-Llano LE, Morris E, Mao M, Tan C. Minimizing Context Dependency of Gene Networks Using Artificial Cells. ACS Applied Materials & Interfaces. 2018;10(36):30137–30146. doi: 10.1021/acsami.8b10029. [DOI] [PubMed] [Google Scholar]
- 16.Arnold FH. Design by directed evolution. Accounts of Chemical Research. 1998;31(93):125–131. [Google Scholar]
- 17.Chen Z, et al. Synthetic beta cells for fusion-mediated dynamic insulin secretion. Nature Chemical Biology. 2018;14(1):86–93. doi: 10.1038/nchembio.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohr BP, Retterer ST, Doktycz MJ. While-you-wait proteins? Producing biomolecules at the point of need. Expert Review of Proteomics. 2016;13(8):707–709. doi: 10.1080/14789450.2016.1209415. [DOI] [PubMed] [Google Scholar]
- 19.Krinsky N, et al. Synthetic Cells Synthesize Therapeutic Proteins inside Tumors. Advanced Healthcare Materials. 2018;7(9):1701163. doi: 10.1002/adhm.201701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TW, Kim DM, Choi CY. Rapid production of milligram quantities of proteins in a batch cell-free protein synthesis system. Journal of Biotechnology. 2006;124(2):373–380. doi: 10.1016/j.jbiotec.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Deng NN, Yelleswarapu M, Zheng L, Huck WT. Microfluidic assembly of monodisperse vesosomes as artificial cell models. Journal of the American Chemical Society. 2016;139(2):587–590. doi: 10.1021/jacs.6b10977. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande S, Caspi Y, Meijering AE, Dekker C. Octanol-assisted liposome assembly on chip. Nature Communications. 2016;7:10447. doi: 10.1038/ncomms10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopfrich K, et al. One-Pot Assembly of Complex Giant Unilamellar Vesicle-Based Synthetic Cells. ACS synthetic biology. 2019 doi: 10.1021/acssynbio.9b00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii S, et al. Liposome display for in vitro selection and evolution of membrane proteins. Nature Protocols. 2014;9(7):1578. doi: 10.1038/nprot.2014.107. [DOI] [PubMed] [Google Scholar]
- 25.Periasamy A, et al. Cell-free protein synthesis of membrane (1,3)-beta-d-glucan) curdlan) synthase: co-translational insertion in liposomes and reconstitution in nanodiscs. Biochim Biophys Acta. 2013;1828(2):743–757. doi: 10.1016/j.bbamem.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Kalmbach R, et al. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. Journal of Molecular Biology. 2007;371(3):639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura K, et al. Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir. 2012;28(22):8426–8432. doi: 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]
- 28.Rampioni G, et al. Synthetic cells produce a quorum sensing chemical signal perceived by Pseudomonas aeruginosa. Chemical Communications. 2018;54(17):2090–2093. doi: 10.1039/c7cc09678j. [DOI] [PubMed] [Google Scholar]
- 29.Stano P, Kuruma Y, de Souza TP, Luisi PL. Biosynthesis of proteins inside liposomes. Humana Press. 2010:127–145. doi: 10.1007/978-1-60761-447-0_11. [DOI] [PubMed] [Google Scholar]
- 30.Pautot S, Frisken BJ, Weitz DA. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19(7):2870–2879. [Google Scholar]
- 31.Krinsky N, et al. A Simple and Rapid Method for Preparing a Cell-Free Bacterial Lysate for Protein Synthesis. PLoS One. 2016;11(10):0165137. doi: 10.1371/journal.pone.0165137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology. 2006;24(91):79. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 33.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proceedings of the National Academy of Sciences. 2013;110(27):11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkle D, Kahya N, Schwille P. Reconstitution and anchoring of cytoskeleton inside giant unilamellar vesicles. ChemBioChem. 2008;9(16):2673–2681. doi: 10.1002/cbic.200800340. [DOI] [PubMed] [Google Scholar]
- 35.Vleugel M, Roth S, Groenendijk CF, Dogterom M. Reconstitution of basic mitotic spindles in spherical emulsion droplets. Journal of Visualized Experiments. 2016:e54278. doi: 10.3791/54278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayoumi M, Bayley H, Maglia G, Sapra KT. Multi-compartment encapsulation of communicating droplets and droplet networks in hydrogel as a model for artificial cells. Scientific Reports. 2017;7:45167. doi: 10.1038/srep45167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnology Advances. 2012;30(5):1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.