Abstract
The variable outcome of viral exposure is only partially explained by known factors. We administered respiratory syncytial virus (RSV) to 58 volunteers, of whom 57% became infected. Mucosal neutrophil activation before exposure was highly predictive of symptomatic RSV disease. This was associated with a rapid, presymptomatic decline in mucosal interleukin-17A (IL-17A) and other mediators. Conversely, those who resisted infection showed presymptomatic activation of IL-17– and tumor necrosis factor–related pathways. Vulnerability to infection was not associated with baseline microbiome but was reproduced in mice by preinfection chemokine-driven airway recruitment of neutrophils, which caused enhanced disease mediated by pulmonary CD8+ T cell infiltration. Thus, mucosal neutrophilic inflammation at the time of RSV exposure enhances susceptibility, revealing dynamic, time-dependent local immune responses before symptom onset and explaining the as-yet unpredictable outcomes of pathogen exposure.
Introduction
The respiratory tract is by necessity an open portal, and thus is highly vulnerable to invasion by many pathogens. A complex system of defenses counters this vulnerability, but current knowledge of what governs susceptibility fails to fully explain the erratic transmission of infectious agents (1, 2). For some infections, antibodies in the respiratory mucosa are clearly protective (3). Infectious agents can also be blocked by antimicrobial peptides (4), entrapment in mucus (5), and the downstream effects of pattern recognition receptor activation (1, 2, 6), as well as by the direct inactivation by immune cells (6). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has brought into sharp focus the need for a greater understanding of the role of mucosal innate immunity in protection against respiratory infection. Better understanding of such defenses may offer ways to prevent or modulate viral disease, even that caused by novel and emerging pathogens. Among innate immune cells, neutrophils are classically ascribed roles in defense against bacteria and fungi, but their role in antiviral responses is less clear. Although neutrophils are sometimes found in the lungs of patients with severe viral infections and have a demonstrable antiviral role in animal models, information concerning how they contribute to defense and disease in human respiratory viral infections is largely lacking (7).
Respiratory syncytial virus (RSV) is the leading cause of infant hospitalization worldwide, infecting ~34 million children each year (8) and contributing prominently to morbidity and mortality in elderly and immunosuppressed adults (9, 10). Although RSV infection is essentially confined to the respiratory epithelium, the resultant neutrophilic lung inflammation can be life-threatening, particularly in at-risk children (11, 12). Although prophylaxis with palivizumab (a monoclonal anti-RSV anti-body) can prevent hospitalization of premature infants (13), no specific therapeutic intervention is currently available and no vaccine has yet been licensed that protects against RSV (14). This is despite the existence of several promising candidates (15), including enhancement of placental anti-RSV immunoglobulin G (IgG) transfer by maternal immunization. Accordingly, there is a pressing need to identify additional interventions that prevent RSV and other respiratory viral diseases.
Animal models of respiratory viral infection provide useful mechanistic insights but do not fully recapitulate human disease (6). Observational studies of hospitalized children and high-risk adults are limited by the heterogeneity of populations and disease severity and by an inability to dynamically assess the status of host immunity—specifically, the mucosa before and immediately after viral exposure. Experimental infection of human volunteers therefore provides an unparalleled opportunity in a controlled setting to identify preexisting immune factors and presymptomatic responses that correlate with protection and disease (16, 17).
All adults have RSV-specific antibodies, re-flecting multiple rounds of infection through-out the course of their lives. Nevertheless, symptomatic reinfection occurs repeatedly even in healthy people and is associated with unusually short-lived humoral and cellular memory responses (3). Such defective defense against reinfection is also observed after some other respiratory viral infections, including coronaviruses (18). Furthermore, variations in antibody levels do not accurately predict susceptibility to RSV infection in experimental studies of volunteers: Those with the lowest antibody levels are only modestly more susceptible to viral challenge (3, 19). Although mucosal IgA, circulating IgG (3), and resident memory CD8+ T cells (20) show some association with resistance to infection and reduced disease severity, these factors do not completely explain who resists and who succumbs. Additionally, although RSV viral burden approximately correlates with symptoms in experimentally infected adults (16) and moderately ill infants (21, 22), viral load may be paradoxically reduced in the most severely affected children (23).
How the very early events after viral exposure influence clinical outcomes has not been studied in either adults or children. By studying the respiratory mucosa before, during, and after human RSV challenge, we found that prior neutrophil activation in the upper respiratory tract predisposed individuals to symptomatic viral infection. The neutrophilic mucosal environment was strongly associated with a reduction in antiviral mucosal inflammatory responses immediately after viral exposure that was followed by the onset of disease. By contrast, a protective response characterized by rapid activation of type-17 inflammation was observed in those resisting infection. Moreover, studies in mice showed that recruitment of airway neutrophils before viral administration enhanced the cytotoxic CD8+ T cell response and disease severity.
Results
Characterization of the time course of responses to human RSV challenge
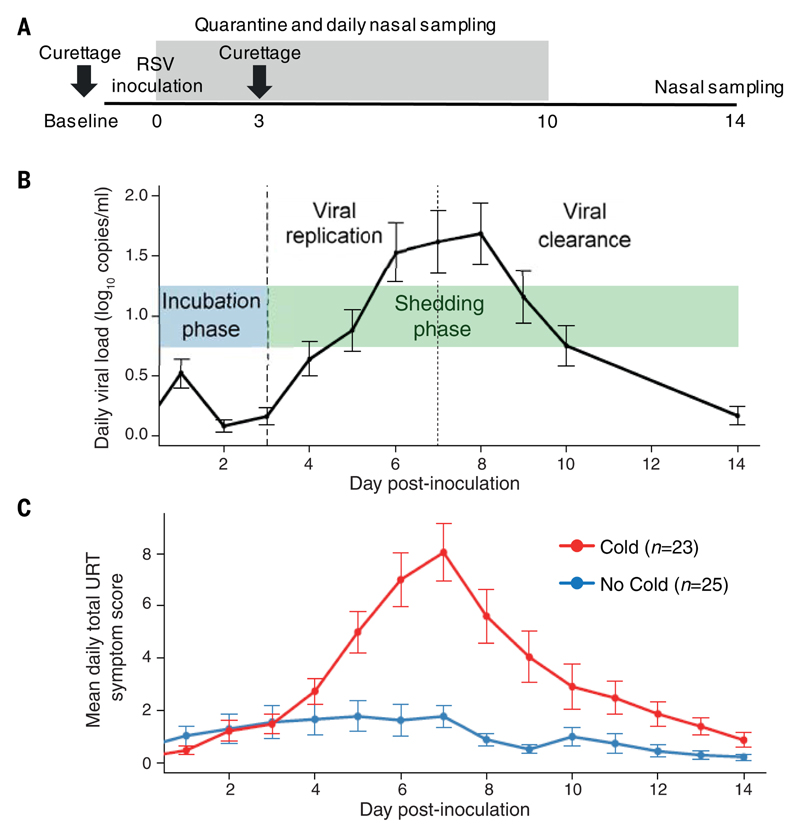
Fifty-eight healthy adult volunteers were in-oculated with RSV Memphis 37. Nasal sampling was performed at baseline (7 to 14 days before inoculation) and then repeatedly up to 14 days postinoculation (dpi) (Fig. 1A). Symptoms were quantified using a previously validated self-reported symptom scale (24). Twenty-three participants developed polymerase chain reaction (PCR)–positive RSV infection with symptoms of upper respiratory tract disease (the “Cold” group), whereas 25 showed no evidence of RSV infection and did not develop symptoms (the “No Cold” group). Ten participants had PCR evidence of RSV infection but had self-reported symptom scores that did not reach the threshold required for the Cold group. This indeterminate group was excluded from subsequent analyses, which focused on identifying factors associated with the most distinct outcomes. No significant demographic differences were observed between the Cold and No Cold groups (table S1).
Fig. 1. Kinetics of viral replication and symptoms correlate closely, with no viral replication or symptoms evident until 3 dpi.
(A) Participants were inoculated with RSV at day 0. Nasal wash and nasosorption samples were taken daily during quarantine (up to 10 dpi) and at 14 dpi. Nasal curettage was performed at baseline (7 to 14 days before inoculation) and at 3 dpi. (B) Daily viral load, measured by qPCR of nasal wash, in volunteers who developed symptomatic RSV infections (Cold, n = 23). (C) Daily upper respiratory tract (URT) symptom scores in the Cold (red, n = 23) and No Cold (blue, n = 25) groups. Data in (B) and (C) are shown as the means ± SEM.
In infected volunteers, viral load [determined by quantitative PCR (qPCR) from daily nasal washes] broadly indicated three phases: (1) a presymptomatic incubation phase during the first 3 dpi when virus was undetectable; (2) a viral replication phase from 3 to 7 dpi; and (3) a viral clearance phase from 8 dpi [previously shown to be associated with the rise in virus-specific CD8+ T cells in both the blood and lower airway (20)] (Fig. 1B). Low-level detection of virus at 1 dpi probably represented residual viral inoculum. Severity of the resultant upper respiratory tract disease (measured by a self-reported symptom score) peaked at 7 dpi, around the same time as viral load (Fig. 1C). In agreement with previous reports (16), total symptom scores positively correlated with total viral shedding throughout the study duration (P < 0.001, fig. S1) despite the known shortcomings of viral load measurement and the variability of symptom quantification.
Susceptibility to RSV infection is associated with airway neutrophil activation
Human challenge studies are exceptional in allowing the investigation of events before symptom onset, enabling us to examine the influence of immune activation in the airway at baseline and during the early presymptomatic period on the course of infection. In agreement with our previous report (3), anti-RSV IgA titers in the nose at baseline were higher in the No Cold group (fig. S2, A and B). However, titers showed a substantial overlap with the Cold group, suggesting that nasal IgA alone did not adequately explain susceptibility unless it was at extremely low or high levels. Additionally, the frequency of RSV-specific CD8+ T cells in the lung at baseline did not distinguish between infection (Cold) or protection (No Cold) (fig. S2C), as we have previously reported (20). Thus, correlates of protection against RSV are likely multifactorial, with no single dominant protective mechanism yet defined.
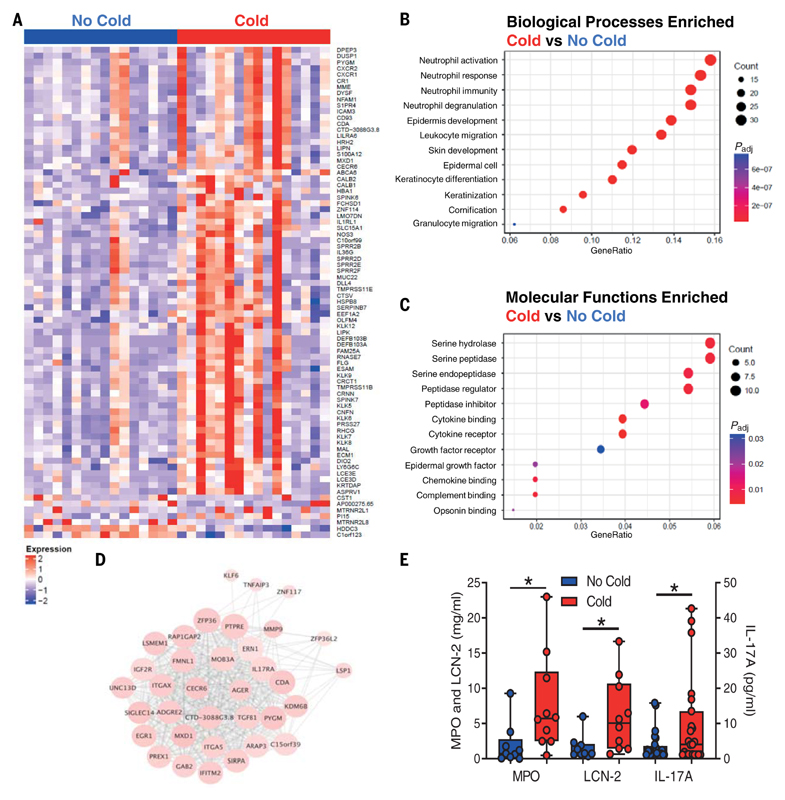
To enable the discovery of previously unanticipated factors associated with susceptibility to infection, RNA sequencing (RNA-seq) was performed on nasal tissue collected from Cold and No Cold group volunteers before inoculation. This approach identified 80 differentially expressed genes (DEGs) that distinguished the two divergent outcomes [adjusted P value (Padj) < 0.05; Fig. 2A and table S2]. Seventy-three (91%) DEGs were significantly higher in the Cold group, whereas a smaller cluster (7/80, 9%) was elevated in the No Cold group. For pathway enrichment analyses, a looser cutoff (Padj< 0.1) was applied, resulting in inclusion of a further 155 DEGs (table S2). Biological processes associated with neutrophil activation made up the most significantly enriched pathways at baseline in those susceptible to infection (Fig. 2B and table S3), with many DEGs also associated with hydrolase and peptidase activity (Fig. 2C and table S4). Furthermore, weighted gene coexpression network analysis (WGCNA) identified a cluster of highly correlated genes enriched for genes associated with neutrophil activity, including CDA, IGF2R, ITGAX, UNC13D, SIGLEC14, SIRPA, PREX1, and MMP9 (Fig. 2D). In agreement with these transcriptomic data, nasal protein levels of the neutrophil-associated mediators myeloperoxidase (MPO), lipocalin-2/neutrophil gelatinase-associated lipocalin (LCN-2), and interleukin-17A (IL-17A) were significantly higher in the Cold group relative to the No Cold group at day 0 (P = 0.010, P = 0.017, and P = 0.039, respectively; Fig. 2E and table S5). There was no difference in other cytokines or chemokines (table S5), but with IL-17 known to induce neutrophil activation and chemotaxis, these data support the hypothesis that greater airway neutrophil activity at the time of virus exposure increases susceptibility to symptomatic RSV infection (25, 26). No correlation was observed between baseline nasal antibody titers and neutrophil mediator levels, including MPO (fig. S3), suggesting that these were independent correlates of susceptibility.
Fig. 2. Preexposure airway neutrophil activation is associated with susceptibility to RSV infection.
(A) DEGs (Padj < 0.05; n = 80) were determined in baseline (7 to 14 days before inoculation) nasal curettage samples between Cold and No Cold groups by RNA-seq. (B) GO molecular functions enrichment analysis of DEGs (Padj < 0.1, n = 235). (C) GO biological processes enrichment analysis of DEGs (Padj < 0.1, n = 235). (D) WGCNA cluster of DEGs (Padj < 0.1, n = 235) enriched for those with annotations of neutrophil association. (E) Day 0 levels of MPO, LCN-2, and IL-17A in nasosorption samples between the No Cold (n = 10-25) and Cold (n = 9-23) groups. DEGs in (A) to (D) were determined by Wald tests using DESeq2. Data in (E) are shown as median and interquartiles with minimum and maximum values and were analyzed by Mann-Whitney U tests. *P < 0.05.
Early immune activation prevents symptomatic RSV infection
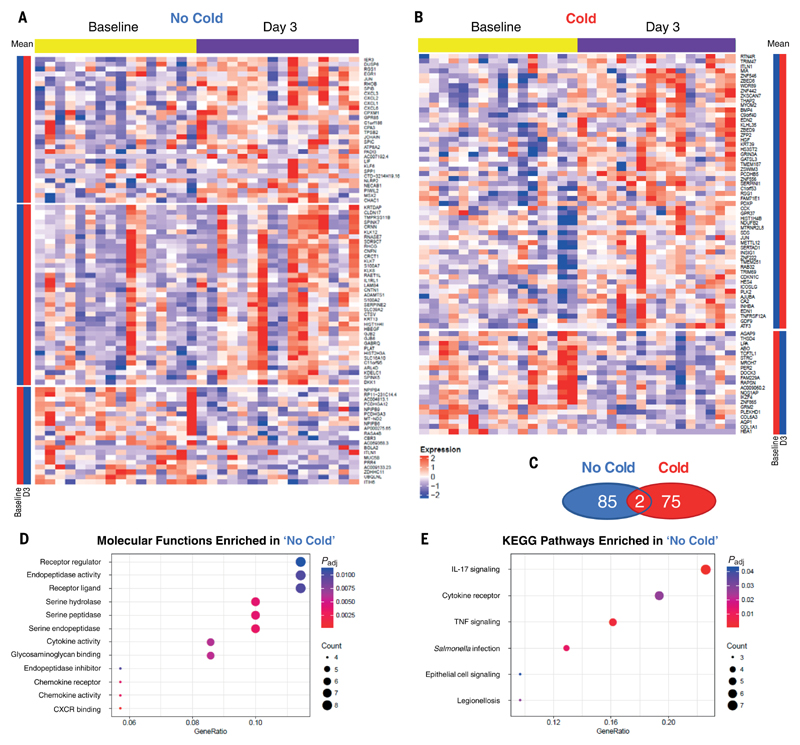
To investigate the relationship between airway neutrophil activity and subsequent mucosal inflammatory responses, nasal tissue samples collected at 3 dpi (before the phase of concerted net viral replication) were analyzed by RNA-seq and compared with matched pre-inoculation samples. Sixteen volunteers from each of the Cold and No Cold groups yielded RNA suitable for sequencing and were included in the analysis. This analysis identified 87 DEGs (P < 0.01, log2 fold change > 0.5) in the No Cold group (Fig. 3A and table S6) and 77 DEGs in the Cold group (Fig. 3B and table S7). In both the Cold and No Cold groups, most DEGs were up-regulated after inoculation (as shown by the mean expression values for each gene and K-means clustering). However, only two genes (ITLN1 and JUN) over-lapped between both groups, emphasizing the divergence of local responses associated with these two opposing outcomes of viral exposure (Fig. 3C).
Fig. 3. Early presymptomatic viral clearance is associated with activation of IL-17 signaling.
DEGs were determined by RNA-seq of nasal curettage samples at 3 dpi relative to baseline (7 to 14 days before inoculation) in the No Cold (n = 16) and Cold (n = 16) groups. Heatmaps show (A) DEGs in the No Cold group (n = 87) and (B) DEGs in the Cold group (n = 77). (C) Venn diagram of the number of overlapping DEGs in the Cold and No Cold groups. DEGs from the No Cold group were analyzed for enrichment of (D) GO molecular functions and (E) KEGG pathways. DEGs were defined as transcripts with Padj < 0.01 and log2 fold change >0.5.
Next, the functional pathways to which DEGs associated with the No Cold group at day 3 significantly contributed were tested by enrichment analysis using the gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathways. GO biological processes analysis for the Cold group showed significant enrichment of several processes but none specifically related to immune responses (table S8). By contrast, in the No Cold group, GO biological processes analysis yielded several immune-related pathways that were significantly enriched. These could be categorized into three areas: (1) epidermis/skin development and bacterial defense; (2) humoral immunity, including the JCHAIN required for secretory IgA formation; and (3) leukocyte/granulocyte migration and chemotaxis (table S9). Additionally, the DEGs in the No Cold group showed the enrichment of molecular functions for protease, cytokine, and chemokine activities (Fig. 3D and table S10). KEGG analysis also revealed several pathways related to cytokine and chemokine signaling. Of these, the “IL-17 signaling pathway” was the most significantly enriched (Fig. 3E and table S11), comprising chemokines CXCL1/2/3/6, the transcription factor JUN, the major gel-forming mucin MUC5B, and the antimicrobial protein S100A7, which is induced by IL-17 and important in protection against Gram-negative bacteria (27). Other closely related KEGG terms were also enriched, including “Cytokine–cytokine receptor interaction” and “TNF signaling pathway.” No KEGG pathways were significantly enriched in the Cold group.
These findings show that the activation of an early inflammatory response is associated with protection from symptomatic RSV infection. This early protective inflammatory response is characterized by signatures of leukocyte migration and cytokine and chemokine signaling. By contrast, such inflammatory responses were not observed in volunteers who subsequently developed RSV disease.
Early RSV clearance is associated with type-17 immunity
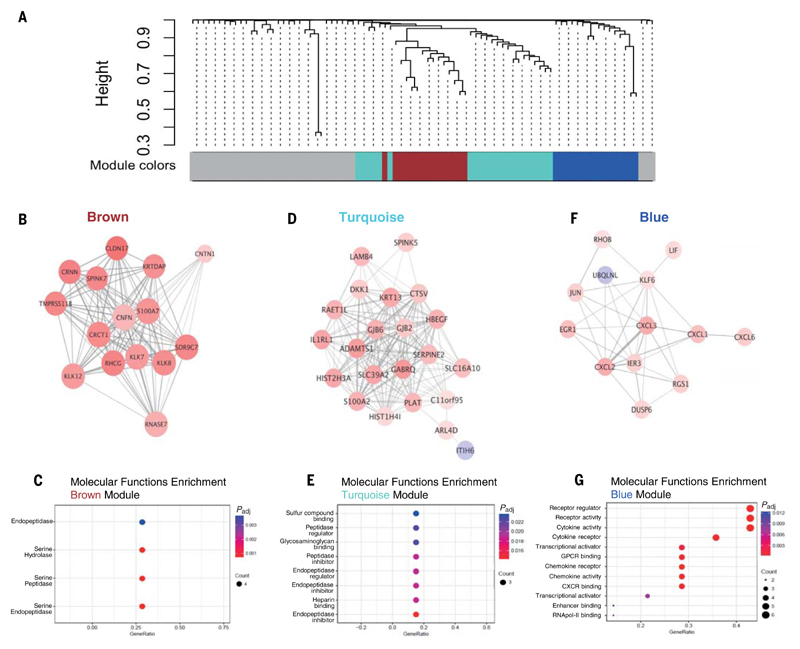
To define the core transcriptional signatures of coexpressed genes without biasing by predetermined or putative biological function knowledge, we next applied WGCNA to establish interrelated DEG networks associated with protection in No Cold individuals. Three gene clusters (depicted and named “brown,” “turquoise,” and “blue”) were identified at 3 dpi relative to baseline (Fig. 4A). The brown module mainly contained genes encoding peptidase and hydrolase activity (Fig. 4, B and C, and table S12). This was also true of the turquoise module, which clustered closely with the brown module (Fig. 4, D and E, and table S13). However, the blue module was highly enriched in IL-17 pathway components and closely corresponded to the DEGs featured in the KEGG “IL-17 signaling pathway” (Fig. 3E), namely CXCL1/2/3/6 and JUN (Fig. 4F). Indeed, GO molecular function analysis of the blue module identified enrichment of cytokine and chemokine activity and signaling (Fig. 4G and table S14). Thus, the blue module represents a signature of type-17 inflammatory gene expression induced on RSV exposure, which is associated with the prevention of infection.
Fig. 4. Network analysis reveals a module of correlated DEGs enriched for IL-17 signaling in presymptomatic individuals.
WGCNA of the differentially expressed genes (n = 87) was performed in the No Cold group at 3 dpi relative to baseline. (A) Classification of WGCNA modules. Brown module: (B) network map and (C) molecular function enrichment. Turquoise module: (D) network map and (E) molecular function enrichment. Blue module: (F) network map and (G) molecular function enrichment. DEGs were defined as transcripts with P < 0.01 and log2 fold change >0.5.
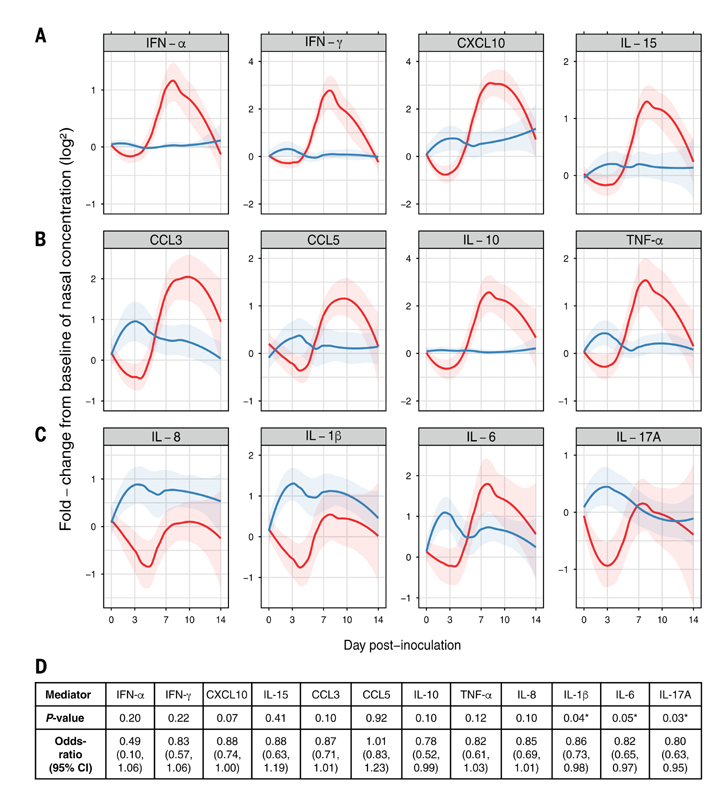
To validate these findings, cytokines and chemokines in the nasal mucosal lining fluid were quantified over time. In the Cold group, a limited response was detected until 3 dpi, after which the expression of antiviral mediators [interferon-α (IFN-α), IFN-γ,CXCL10, andIL-15] substantially increased, with maximal detection at 7 dpi (Fig. 5A), which is consistent with the peak of symptoms and viral load (Fig. 1, B and C). The chemokines CCL3 (MIP-1α) and CCL5 (RANTES), the anti-inflammatory cytokine IL-10, and tumor necrosis factor-α (TNF-α) all followed similar kinetics (Fig. 5B). The levels of all mediators in the Cold group declined during the phase of viral clearance (7 to 14 dpi). Cytokines and chemokines are well recognized to be closely associated with symptoms in a variety of inflammatory conditions, including the response to infection (28, 29). Indeed, within the Cold group, cumulative viral load and inflammatory mediator levels over the study duration were closely correlated (fig. S4), emphasizing their role in proinflammatory responses and disease triggered by infection (16). However, in the No Cold group, despite the absence of symptoms before 4 dpi, several cytokines (including IL-1ß, IL-6, IL-17A, and TNF-α) underwent a rapid burst of expression during the incubation phase shortly after virus inoculation (Fig. 5, B and C). Logistic regression analysis showed statistically significant associations between the induction of IL-1β, IL-6, and IL-17A (P = 0.04, 0.05, and 0.03, respectively; Fig. 5D) in the first 3 days after RSV exposure and protection from developing symptoms. After 3 dpi, mediators returned to baseline levels in No Cold volunteers, who did not develop the later response seen in the Cold group. Conversely, in the Cold group, almost all mediators declined relative to day 0 levels during the incubation phase despite the presence of virus (Fig. 5, A to C). Thus, a local type-17 inflammatory response early after pathogen exposure prevents the development of a symptomatic infection. Failure to mount a protective inflammatory response rapidly after inoculation is itself associated with airway neutrophil activity at the time of exposure, suggesting that neutrophilic inflammation may disrupt this early protective response.
Fig. 5. Early presymptomatic secretion of intranasal IL-1β, IL-6, and IL-17A is associated with protection from RSV infection.
(A to C) Soluble protein cytokine and chemokine mediator levels from nasosorption samples were determined on each study day and expressed as LOESS plots between Cold (red) and No Cold (blue) groups of log2 fold-change levels normalized to day 0. The 95% confidence intervals of LOESS curves are denoted by shaded areas. (D) Logistic regression analysis of mediator responses during 1 to 3 dpi measured at the protein level from nasosorption samples.
The airway microbiome is not associated with susceptibility to infection
Neutrophils are classically ascribed antibacterial and antifungal functions, and recent studies have shown perturbations of the airway bacterial microbiome in children hospitalized with RSV (30). To test for associations between the preinfection airway microbiome and susceptibility to RSV infection, 16S ribosomal RNAfrom nasopharyngeal bacteria was quantified and sequenced. Despite differential neutrophil activation in the nasal mucosa seen by multiple measures, no significant differences in bacterial load were observed between the Cold and No Cold groups at baseline (fig. S5A). Moreover, 16S sequencing demonstrated similar baseline diversity and community composition in these groups (richness, P = 0.948; Shannon–Weiner, P = 0.263; Simpson’s, P = 0.166, Bray–Curtis dissimilarity, R2 = 0.055, P = 0.087; fig. S5B). Thus, there are no significant associations between the baseline airway microbiome and nasal neutrophilic inflammation before viral exposure and thus susceptibility to RSV infection.
Elevated airway neutrophils in the murine lung at the time of exposure enhance RSV disease through pulmonary cytotoxic T cells
To investigate the role of neutrophilic inflammation on early and late outcomes in respiratory viral infection, we modeled the impact of increased pulmonary neutrophils at the time of RSV challenge in mice. C57BL/6 mice were treated intranasally with recombinant CXCL1 (rCXCL1) or phosphate-buffered saline (PBS). Administration of rCXCL1 caused short-lived recruitment of neutrophils to the bronchoalveolar lavage (BAL) and lung parenchyma within 12 hours (Fig. 6A; gating strategies are shown in fig. S6), resulting in elevated levels of the neutrophil mediators MMP-9, MPO, and neutrophil elastase in BAL (Fig. 6B); this was analogous to human volunteers in the Cold group. The effect of rCXCL1 treatment was restricted to neutrophils and had no impact on either alveolar macrophage numbers in the airway or monocyte abundance in the lung (fig. S7A). Furthermore, no differences were observed in cytokine (fig. S7B) or chemokine (fig. S7C) levels in the lung before virus challenge after rCXCL1.
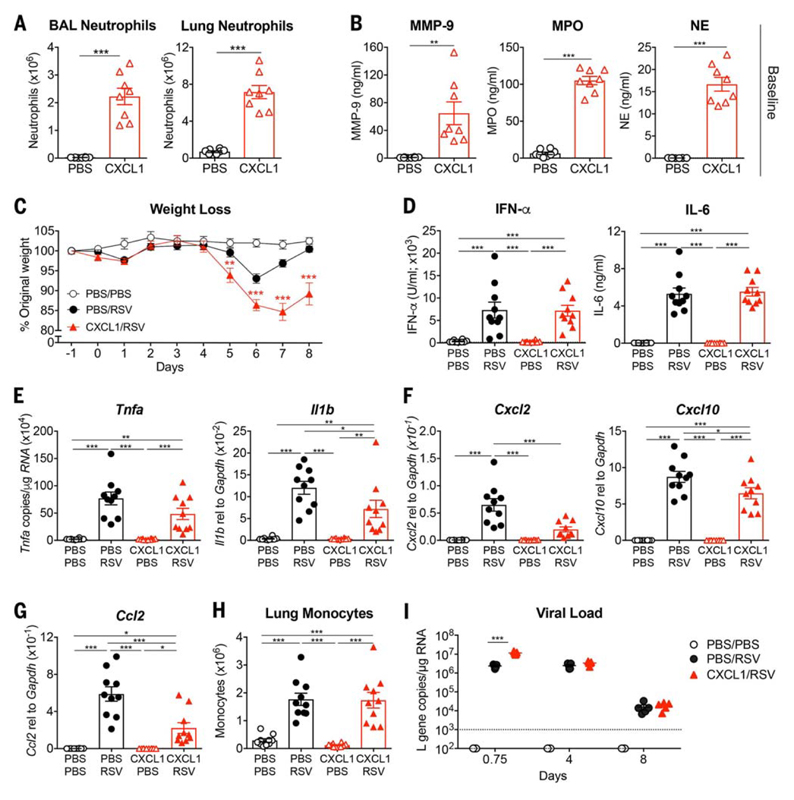
Fig. 6. Mice treated with the neutrophil chemoattractant CXCL1 before RSV infection show an early increase in viral load and develop more severe disease.
Mice were treated with mock (PBS) or 10 μg of CXCL1 intranasally. (A) At 12 hpi, BAL and lung neutrophils were quantified by flow cytometry and (B) MMP-9, MPO, and NE levels in the BAL were quantified by ELISA. Mice were treated with mock (PBS) or 5 to 10 μg of CXCL1 intranasally and after 9 to 12 hours were infected with mock (PBS) or 7.5 × 105 FFUs of RSV intranasally. (C) Weight loss as a percentage of original weight. (D) IFN-α and IL-6 quantified in the BAL using ELISA. Levels of (E) Il1b and Tnfa, (F) Cxcl2 and Cxcl10, and (G) Ccl2 were determined in lung tissue using qPCR. (H) Total number of lung monocytes quantified by flow cytometry as previously described (32). (I) RSV L gene copy numbers were quantified at 0.75, 4, and 8 dpi in lung tissue by qPCR. Data in (A) and (B) are shown as the means ± SEM of eight individual mice per group pooled from two independent experiments. Data in (C) are shown as the means ± SEM of 10 (PBS/PBS) or 14 to 16 (RSV) individual mice pooled from two (PBS/PBS) or three (RSV) independent experiments. Data in (D) to (H) are shown as the means ± SEM of 10 individual mice per group pooled from two independent experiments. Data in (I) are shown as the means ± SEM of five individual mice per group representative of at least two independent experiments, with the dotted line representing the limit of detection of the assay. The statistical significance of differences in (A) and (B) was analyzed using unpaired, two-tailed Student’s t test. The data in (C) and (I) were analyzed using two-way ANOVA with Bonferroni’s post hoc test, and only the statistically significant differences between RSV-infected groups are shown. The data in (D) to (H) were analyzed using one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
As in human challenge, increased neutrophilic inflammation before, but not shortly after (31), RSV infection led to significant enhancement of disease (measured by weight loss), with a nadir at 6 to 7 dpi (Fig. 6C). RSV infection resulted in rapid up-regulation of proinflammatory cytokines including IFN-α and IL-6 at 18 hours postinfection (hpi) (Fig. 6D). Production of these cytokines was not significantly altered by rCXCL1 pretreatment. Furthermore, rCXCL1 conditioning did not change the gene expression of Tnfa but did coincide with reduced gene expression of Il1b, Cxcl2, Cxcl10, and Cc12 in the lung tissue after RSV infection (Fig. 6, E to G). The infiltration of antiviral monocytes (32) did not change after rCXCL1 conditioning (Fig. 6H), but neutrophils and neutrophil mediators were increased in the rCXCL1-treated mice compared with mice treated with RSV alone (fig. S8). Nevertheless, rCXCL1 conditioning was associated with a modestly but significantly higher viral load at 18 hpi, which was not sustained to 4 dpi (the peak of viral replication) or 8 dpi (clearance of the virus) (Fig. 6I). Thus, neutrophil infiltration at the time of respiratory viral challenge transiently alters the inflammatory environment and is associated with a subtle early impairment in the control of virus replication during the asymptomatic period only.
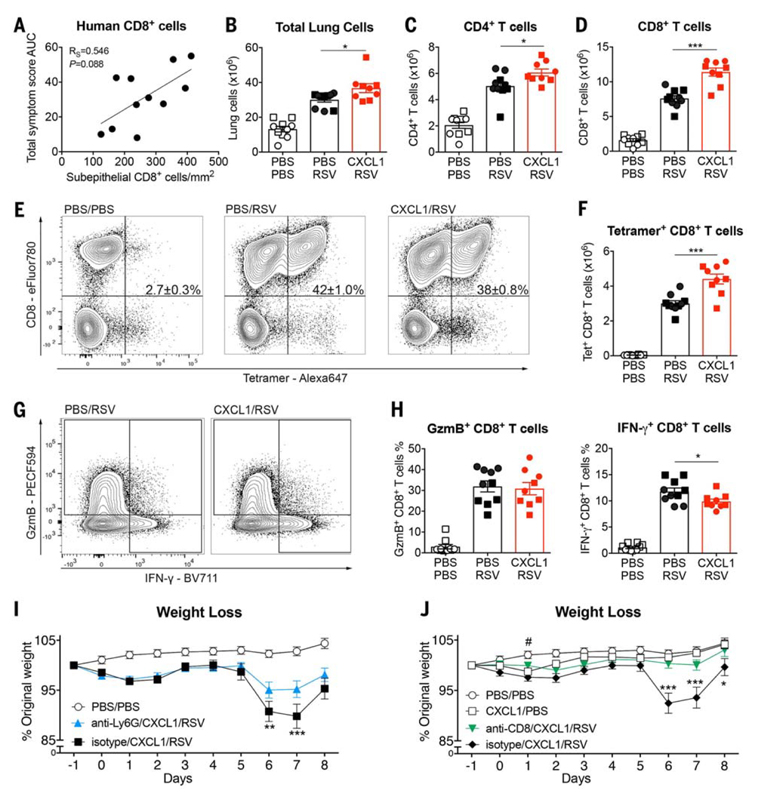
Immunopathology in the murine RSV model causes enhanced disease, which has been shown to be partially mediated by overexuberant T cell activity (6, 33, 34). In human challenge, CD8+ T cell numbers in bronchial biopsies at 7 dpi showed a nonsignificant positive association with disease severity (P = 0.088; Fig. 7A), suggesting that this may be a contributory mechanism (20). To further examine this issue, T cell responses were analyzed in mice pretreated with rCXCL1. At 8 dpi, there was significantly higher total lung cellularity in RSV-infected, rCXCL1-pretreated mice (Fig. 7B) despite the decreasing viral load (Fig. 6I) and resolution of the pulmonary neutrophil response (fig. S9A). Although CD4+ T cell numbers were somewhat increased (P < 0.05; Fig. 7C), the most marked elevation in rCXCL1-pretreated mice was in CD8+ T cell numbers (P < 0.0001; Fig. 7D), suggesting that these might be the predominant mediators of immunopathology.
Fig. 7. Mice treated with the neutrophil chemoattractant CXCL1 before RSV infection develop CD8+ T cell–driven disease.
(A) Correlation between CD8+ T cell numbers in the subepithelial layer of endobronchial biopsy samples from human participants infected with RSV at 7 dpi and cumulative symptom scores over the course of infection (n = 11). (B to H) Mice were treated with mock (PBS) or 10 μg of CXCL1 intranasally 12 hours before RSV infection. Lungs were analyzed at 8 dpi. (B) Total number of lung cells, (C) lung CD4+ T cells, and (D) lung CD8+ T cells assessed by flow cytometry. (E) Representative flow cytometry plots of lung RSV tetramer–positive CD8+ T cells; the mean percentage ± SEM is indicated in the upper right quadrant. (F) Total number of lung RSV tetramer–positive CD8+ T cells as quantified by flow cytometry. (G) Representative flow cytometry plots of lung granzyme B–positive (GzmB+) and IFN-γ+ CD8+ T cells. (H) Frequency of GzmB+ and IFN-γ+ CD8+ T cells quantified by flow cytometry. (I) Anti-Ly6G or isotype control–treated mice were administered PBS or 10 μg of CXCL1 intranasally on day 0 and at 12 hours after being infected with mock (PBS) or 6 to 7.5 × 105 FFUs of RSV intranasally. Weight loss is shown as the percentage of original weight. (J) Mice treated with anti-CD8 or isotype control antibodies were administered PBS or 10 μg of CXCL1 intranasally on day 0 and at 12 hours after being infected with mock (PBS) or 6 to 7.5 × 105 FFUs of RSV intranasally. Weight loss is shown as the percentage of original weight. Data in (B) to (D), (F), and (H) are shown as the means ± SEM of 10 (PBS/PBS, PBS/RSV) or nine (CXCL1/RSV) mice pooled from two independent experiments; repeat 1, 10 μg of CXCL1 (circles) and repeat 2, 8 μg of CXCL1 (squares). Statistical significance of differences in (A) were analyzed using Spearman’s rank correlation test, where the R score is given as RS. Data in (B) to (D), (F), and (H) were analyzed using one-way ANOVA with Tukey’s post hoc test. Data in (I) and (J) were analyzed using two-way ANOVA with Bonferroni’s post hoc test. Asterisks indicate statistically significant differences between RSV-infected groups. Hash symbol indicates statistically significant differences between PBS/PBS and CXCL1/PBS groups. *#P < 0.05, **P < 0.01, ***P < 0.001.
Tetramer staining revealed no significant difference in the frequencies of RSV-specific CD8+ T cells in the lung at 8 dpi after rCXCL1 pretreatment (Fig. 7E), but the greater total influx of CD8+ T cells in rCXCL1 pretreated mice resulted in significantly increased numbers of virus-specific CD8+ T cells in the lung around the peak of disease severity (Fig. 7F). Functional markers of these RSV-specific CD8+ T cells were assessed by short-term in vitro restimulation and intracellular cytokine staining (Fig. 7G). Neither granzyme B nor IFN-γ production was enhanced by rCXCL1 pre-treatment (Fig. 7H). Depletion of neutrophils by anti-Ly6G antibody treatment in rCXCL1-pretreated mice before RSV inoculation par-tially abrogated weight loss at 6 and 7 dpi (Fig. 7I). Thus, rCXCL1-driven airway neutrophils at the time of RSV exposure are at least in part responsible for CD8+ T cell recruitment and enhanced disease. Antibody-mediated depletion of CD8+ T cells during the course of infection abrogated weight loss in RSV-infected, rCXCL1-pretreated mice at 6 to 8 dpi (Fig. 7J and fig. S8B). This strongly suggests that CD8+ T cells are the predominant effectors causing enhanced disease. These findings agreed with previous murine studies showing the role of CD8+ T cells in mediating RSV-induced weight loss (33).
Preexisting neutrophilic inflammation alters the tissue environment so that the recruitment but not cytotoxic function of CD8+ T cells to the lung is increased later in the disease course. This is associated with transient early changes in soluble mediators and viral load during the asymptomatic period, whereas peak disease is mediated by immunopathology despite the absence of elevated pulmonary neutrophils or viral load at that later time.
Discussion
Using experimental RSV infection of human volunteers and mice, we identified a mechanism of susceptibility and resistance to infection. Before exposure, airway neutrophil activation was associated with enhanced subsequent RSV disease in both mice and human volunteers. This preexisting neutrophilic inflammation was linked to a failure to induce the transient early mucosal inflammatory re-sponse to viral exposure dominated by IL-17 and related pathways, which was up-regulated in individuals who resisted infection. Our findings highlight the complex role of the mucosal microenvironment in early responses to respiratory viral infection and demonstrate that the prior state of the mucosa alters the subsequent negotiation between virus and host, which determines the outcome of viral exposure.
Associations between RSV disease and neutrophil- or IL-17–related inflammatory responses during infection have been observed in other settings. For example, IL-17 has been detected in the airway both at the height of severe symptoms and during convalescence in infants with RSV (35). However, little is known about its protective role early in the course of infection. IL-17A blockade in clinical trials of secukinumab, an anti–IL-17A monoclonal antibody, caused increased susceptibility to upper respiratory tract infections (36–38). Furthermore, in a BALB/c murine RSV model, BPZE1 (an attenuated Bordetella pertussis vaccine candidate) induced IL-17 and protected against RSV infection (39, 40). Moreover, the intranasal administration of IL-17A 2 hours after RSV challenge decreased disease severity (41). These studies support our finding that an IL-17–dominated response during the early phase of infection may be critical for protection before viral replication is established.
The pleiotropic functions of IL-17 are integral to mucosal immunity and include maintenance of barrier integrity, induction of antimicrobial proteins by epithelial cells, and recruitment of inflammatory myeloid cells (27). However, here in our controlled studies of adults during the earliest stages after virus exposure, IL-17 signaling was associated with abortive infection. The apparently “silent” incubation phase is therefore immunologically dynamic, representing a critical interval during which clinical outcome is determined. Although an early inflammatory response prevents the establishment of viral infection, subjects destined for symptomatic colds show generalized down-regulation of mucosal defense as the virus gains control. In our studies, preexisting neutrophilic inflammation appeared to prevent this early protective response and was instead associated with enhanced susceptibility to infection and disease. Although we were unable to show an association between the nasal microbiome and susceptibility to RSV infection in our volunteers, our studies do not preclude subtle influences of fungal or viral commensals, which may alter the state of the mucosa, or the effects of recent or subclinical bacterial or viral infections.
Our study has limitations. To avoid traumatic injury to nasal tissue before RSV inoculation, sampling for cytometric analysis (e.g., nasal biopsy) was not performed. The transcriptional and proteomic analyses that we report do not distinguish between elevated neutrophil abundance and greater activity of a static number of neutrophils, nor could we identify the cellular sources of cytokines such as IL-17A that we found to be elevated at baseline and during the presymptomatic viral incubation period. In future studies, we hope to resolve these issues by single-cell analysis of airway cells in the early stages of viral infection. Although our murine studies were unable to replicate all aspects of human respiratory viral infection, they permitted the examination of possible mechanisms by detailed cytometric analysis of the airway and selective recruitment of airway neutrophils through CXCL1 instillation. Enhancement of viral load at 18 hours in mice pretreated with CXCL1 showed that the induction of neutrophilic inflammation in the airway did indeed enhance RSV susceptibility, with an early impairment of antiviral immune responses translating into enhanced disease driven by CD8+ T cells.
Previous studies have demonstrated that activation of one arm of the immune response may simultaneously suppress or disable another. For example, the activation of the T helper (Th1) transcription factor T-bet limits Th2 responses by inhibiting GATA3 (42). Patients with chronic neutrophilic lung inflammation (such as bronchiectasis, chronic obstructive pulmonary disease, and some types of asthma) are known to have enhanced susceptibility to respiratory virus-induced disease (43). Similarly, in naturally acquired human influenza infection, we recently showed that inflammatory signatures characteristic of antibacterial immunity corre-lated with severe disease, whereas antiviral responses were associated with milder illness (44). In murine models of colitis, RIG-I–mediated IL-15 secretion induced by commensal viruses supported the survival of intraepithelial lymphocytes, which could in turn inhibit aberrant inflammation (45). Additionally, viral bacteriophages within Pseudomonas aeruginosa have been shown to induce TLR3- and TRIF-mediated type IIFN production with associated suppression of phagocytic responses (46). These studies thus show that antiviral immunity can suppress responses typically associated with antibacterial and inflammatory mechanisms at multiple levels. Together with our present findings, they suggest that there may be opposing mechanisms whereby neu-trophilic inflammation before viral exposure antagonizes optimal antiviral defense and results in failure to control viral infection.
We postulate that this apparent competition between alternative types of immune responses may be relevant in a number of infections with protracted incubation periods, including coronaviruses (47). The prolonged presymptomatic period that we observed during RSV infection is also evident in SARS-CoV-2 infection, in which the interval between exposure and symptom development is typically ~5.2 days (48), compared with 4 to 5 days for RSV shown here. By contrast, during infections such as influenza A, in which there is a very short incubation period [typically ~2 days (47)], intrinsic and innate responses may be rapidly overtaken by viral replication. Again, antibody response to SARS-CoV-2 (49) and other human coronaviruses (50) are apparently short-lived, a feature shared with RSV infection (3).
The distinct separation of pulmonary neutrophil and CD8+ T cell infiltration, which show no temporal overlap, implies an indirect relationship between these immune effectors. The early production of IL-17 may be achieved by a variety of cell types, including CD4+ Th17 and CD8+ T cytotoxic 17 T cells, γδ T cells, neutrophils, and type 3 innate lymphoid cells, which are abundant at mucosal surfaces (51–53). Several mechanisms may link neutrophilic inflammation with enhanced CD8+ T cell recruitment. The preinfection pattern of neutrophil activation is associated with the up-regulation of peptidases and hydrolases, which have been shown to modulate cytokine activity (54) in other systems, potentially altering the immediate antiviral response and signaling to natural killer and T cells (55). This is supported by the observation that Il1b, Cxc12, Cxc110, and Cc12 expression, all of which have been implicated in antiviral defense, are perturbed, either directly or through negative feedback. Enhanced CD8+ T cell migration may also result from neutrophil-derived CXCL12-containing trails, as shown in the influenza-infected mouse trachea (56). Furthermore, because TLR4 antagonists reduce influenza-induced mortality in mice (57), neutrophil-derived oxidized phospholipids (which are TLR4 agonists) may enhance susceptibility. Neutrophil proteases may also degrade anti-viral peptides such as cathelicidin (LL-37) (4), which would influence immediate susceptibility and might contribute to the early transient difference seen in viral load. The direct enhancement of antigen processing or presenta-tion of antigen by neutrophils to T cells (58) is less likely due to the low number of T cells present early after infection, when neutrophils are high, and vice versa later. However, it is possible that neutrophils influence antigen presentation by dendritic cells, synergizing with the early increase in viral load and promoting CD8+ T cell activation, differentiation, and proliferation to enhance viral clearance. The large number of potential contributors means that the precise mechanism(s) through which neutrophils enhance CD8+ T cell recruitment and therefore exacerbated RSV disease remains a major topic for future studies.
Together, our results reveal an integrated, time-dependent mucosal response that critically determines the outcome of pathogen exposure. The susceptibility to infection associated with neutrophilic inflammation before viral exposure reflects a multilayered protective network that reacts rapidly to viral encounter, determining outcome before the presentation of disease. These findings not only increase our capacity to predict previously inexplicable variations in the outcome of exposure to viral pathogens, but also suggest that manipulation of neutrophil-, peptidase-, and IL-17–related pathways may affect antiviral protection, offering additional strategies to prevent disease caused by existing and emerging viral pathogens.
Materials and Methods
Human volunteer infection study design
Healthy, nonsmoking adults 18 to 55 years of age were eligible as previously described except no prescreening for serum-neutralizing antibodies was performed (16). As an exploratory study, no power calculations were performed to determine sample size before the study began. Participants provided written informed consent and the study was approved by the UK National Research Ethics Service (study reference nos. 10/H0711/94 and 11/LO/1826). The study was registered on ClinicalTrials.gov under ID no. NCT01349543. Volunteers provided a full clinical history and underwent a physical examination and a panel of screening blood tests to exclude underlying hematological, biochemical, or immune abnormalities. All participants were screened for HIV, hepatitis B virus, and hepatitis C virus. Participants were excluded if they developed common cold symptoms or any acute illness within 6 weeks before inoculation. On the day before inoculation, a nasal lavage specimen was tested using multiplex PCR for a panel of 13 respiratory viruses and bacteria to exclude concurrent upper respiratory tract infection. Volunteers entered quarantine on study day –1, were inoculated by intranasal drops with 104 plaque-forming units of RSV strain M37 (Meridian Lifesciences, Memphis, TN, USA; GenBank sequence accession no. KM360090) the next day, and remained in quarantine as described previously (3). Volunteers provided daily assessment of their clinical symptoms in the following categories: nasal discharge, nasal obstruction, sneezing, sore throat, headache, fever, and malaise. Each category was scored by volunteers as 0 (absent), 2 (moderate), or 3 (severe). A symptomatic cold was defined as fulfilment of two of the three following conditions: (1) a cumulative symptom score ≥14, (2) nasal discharge lasting ≥3 days, and (3) a subjective assessment of having a cold.
Samples of the nasal mucosal lining fluid were collected daily using precut strips of Leukosorb (Pall Life Sciences, Portsmouth, UK), eluted in 300 μl of PBS, 1% bovine serum albumin (BSA) (w/v), 0.5% Triton X-100 (v/v), and 0.05% sodium azide (w/v) (Sigma-Aldrich, UK), and stored at –80°C for later analysis, as previously described (59). Nasal washes were performed as previously described (3). Nasal tissue samples were collected at 0 and 3 dpi using ASI-Rhinopro nasal curettage devices (Arlington Scientific, Springville, UT, USA), immediately placed in 800 μl of TRIZol buffer (Life Technologies, Paisley, UK), vortexed thoroughly, and stored at –80°C for later analysis.
Quantification of viral load
RSV load in humans was determined from daily nasal wash samples by qPCR as previously described (20).
Immunoassays
Levels of CCL3 (MIP-1a), CCL5 (RANTES), CXCL8 (IL-8), CXCL10 (IP-10), IFN-a, IFN-ß, IFN-γ, IL-1ß, IL-6, IL-10, IL-15, IL-17A, and TNF-a were assayed using MSD multiplex immunoassays (Mesoscale Diagnostics, Rockville, MD, USA).
RNA-seq of nasal curettage tissue samples
Nasal curettage samples were collected as previously described (60) and preserved in TRIzol. After collection, 0.2× chloroform was added to the samples for every 1× of TRIzol, mixed by inverting the tube 10 times, incubated for 5 min, and centrifuged for 10 min at 3500g. The supernatant was then transferred into a new tube and total RNA was extracted with an SV96 kit as per the manufacturer’s protocol (Promega, Madison, WI, USA).
cDNA libraries were prepared with the TruSeq Stranded Total RNA Library Prep Kit with the Ribo-Zero Human Sample Prep Kit (Illumina, San Diego, CA, USA). Final cDNA libraries were analyzed for size distribution using the Tapestation 2200 (Agilent Technologies), quantitated by qPCR (KAPA Library Quant Kit, KAPA Biosystems, Wilmington, MA, USA), normalized to 2 nM, and sequenced with 2 × 50-bp paired-end reads on a HiSeq 2500 (Illumina) with a minimum of 3 GB of raw RNA-seq data.
RNA-seq reads were mapped to human B38 genome with Omicsoft OSA, and read counts mapped to the ensemble genes (Release 75) were calculated with RSEM. Gene count data were normalized by DESeq2 (R package, version 1.18.1). An arbitrary filter to remove genes with low counts was applied to the normalized count data (only protein-coding genes and normalized count >35 in at least six samples were kept, which resulted in 14,670 ensembl genes). Wald tests were performed using DESeq2 to identify differential expression of these genes. DEGs were determined as those with P < 0.01 and a >0.5 log2 fold change relative to baseline for the Cold or No Cold group and Padj < 0.05 for the Cold versus the No Cold group at baseline. Log2 normalized count data were used in the heatmap. Gene set overrepresentation analysis of the GO and KEGG pathways was performed with clusterProfiler (R package, version 3.6.0). A coexpression gene network was built with WGCNA (R package, version 1.63) and visualized with Cytoscape (61).
Bronchial biopsy immunohistochemistry
Endobronchial biopsies were fixed immediately in 4% paraformaldehyde and paraffin embedded. RSV was stained using a mouse anti-RSV antibody cocktail (NCL-RSV3, Leica Biosystems, UK) at 1:50 dilution. CD8+ T cells were identified by staining with mouse anti-CD8 (M0707, Dako, Denmark) at 1:100 dilution using the EnVision peroxidase-staining method (Dako) as previously described (62). Briefly, 5-μm sections were stained according to the manufacturer’s instructions, and an irrelevant mouse IgG1 kappa antibody (MOPC21) was used as negative control for staining specificity of mouse monoclonal antibodies. RSV-infected A549 cells were used as positive staining controls. Quantification was achieved as previously described with operators blinded to sample timing and infection status (62). Briefly, slides were coded to avoid observer bias, and areas of epithelium and subepithelium were assessed using a Leitz Dialux 20 light microscope, an Apple Macintosh computer, and Image 1.5 software. Total epithelial and subepithelial areas of two to three bronchial biopsies were counted for each bronchoscopy. Cell counts were expressed as the number of cut cell profiles with visible nucleus per 1 mm2 of subepithelium and per 0.1 mm2 of epithelium. The coefficient of variation for repeat counts of positive cells by a single observer ranged from 5 to 6%.
Respiratory bacterial load and 16S sequencing
DNA extraction was performed on throat swabs using the MPBio FastDNA spin kit for soil as per the manufacturer’s instructions. Blank swabs were extracted and sequenced to control for contamination. Bacterial loads were quantified in triplicate by SYBR green qPCR as previously described (63).
Sequencing of the 16S rRNA gene was performed on the Illumina MiSeq platform using dual barcode fusion primers targeting the V4 region of the 16S rRNA gene as previously described (63). Extraction controls, PCR-negative, and mock communities were included on sequencing runs. Sequence processing was performed using QIIME, and UCLUST was used to assign taxonomic identification to operational taxonomic units using the Silva 115 NR database, as previously described (63). The operational taxonomic unit and tree files were output into the R package version 3.3.2.
Mouse infection studies
Female wild-type C57BL/6 mice (8 weeks of age) were purchased from Charles River Laboratories. All mice were housed in specific-pathogen-free conditions. All animal experiments were reviewed and approved by the Animal Welfare and Ethical Review Board within Imperial College London and approved by the UK Home Office in accordance with the Animals (Scientific Procedures) Act of 1986 and ARRIVE guidelines.
Plaque-purified human RSV (originally the A2 strain from ATCC, USA) was grown in HEp2 cells (from ATCC, CCL-23) (64). Mice were lightly anesthetized before intranasal administration of 100 μl containing 6 to 7.5 × 105 focus-forming units (FFUs) of RSV or PBS control. In some instances, this was preceded by an intranasal instillation of 5 to 10 μg of recombinant murine CXCL1 (BioLegend, San Diego, USA) 12 hours before infection; 500 μg of anti-Ly6G (1A8) or isotype control (2A3) intraperitoneally on –1,1, and 3 dpi; or 250 μg of anti-CD8 (YTS169.4) or isotype control (LTF-2) intraperitoneally on –1, 2, and 5 dpi (all antibodies from BioXCell, West Lebanon, NH, USA). Mice were sacrificed by a fatal dose of pentobarbital injected intraperitoneally.
To enumerate neutrophils in the BAL, 1 ml of PBS supplemented with 0.5 mM EDTA was used to lavage the lungs three times. The BAL was centrifuged at 800g for 10 min, and red blood cells were removed by lysis with ACK buffer (0.15 M NH4Cl, 1.0 mM KHCO3, and 0.1 mM Na2EDTA). The cell number was determined by Trypan blue (Thermo Scientific) exclusion of dead cells. The cellular composition was determined by spinning 1 to 2 × 105 cells onto a microscope slide (Thermo Scientific) at 1000g for 5 min using a Cytospin 4 Cytocentrifuge (Thermo Scientific). Slides were stained with hematoxylin and eosin using the Reastain Quick-Diff kit (Gentaur, Kampenhout, Belgium) according to the manufacturer’s instructions. Cells were classified as neutrophils using a microscope, and ≥300 cells were counted.
Mediators in the BAL were measured using enzyme-linked immunosorbent assay. Murine IFN-a and IL-6 were detected as previously described (32). Murine MMP-9, MPO, and NE were detected using DuoSet enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems). Data were acquired on a FLUOstar Omega plate reader (BMG Labtech) andanalyzed using MARS data analysis (BMG Labtech).
Lung tissue was homogenized using a TissueLyser LT (Qiagen), and total RNA was extracted from homogenized lung tissue supernatant using the RNeasy Mini kit including DNA removal (Qiagen). RNA concentrations were determined by NanoDrop (Thermo Scientific). RNA was converted to cDNA using the High-Capacity RNA-to-cDNA kit (Applied Biosystems). All primers and probes used are listed in supplementary materials, and all assays used QuantiTect Probe PCR Master Mix (Qiagen). To quantify RSV L gene and Tnfa levels in lung tissue (64, 65), the exact number of copies was calculated using a plasmid standard and the results were normalized to the housekeeping gene Gapdh (Applied Biosystems). For relative quantification, the expression of Ccl2, Cxcl2, Cxcl10, and Il1b (all from Applied Biosystems) were expressed relative to the expression of Gapdh. First, the ΔCt (where Ct = cycle threshold) between the target gene and the Gapdh for each sample was calculated. Then, the expression was calculated as 2–ΔCt. Analysis was performed using the 7500 Fast System SDS Software (Applied Biosystems).
Flow cytometry and restimulation experiments
For flow cytometry staining, 2.5 × 106 lung cells were incubated for 20 min at 4°C with purified rat IgG2b anti-mouse CD16/CD32 receptor antibody in PBS containing 1% BSA and 5 mM EDTA and then stained with fluorochrome-conjugated antibodies (see the supplementary materials) in PBS for 30 min at 4°C. Cells were fixed with fixation buffer (BioLegend). For RSV M tetramer staining, Alexa Fluor 647–conjugated M187–195 tetramers (H-2Db/NAITNAKII) were obtained from the National Institutes of Health (NIH) Tetramer Core Facility (Emory University, Atlanta, GA, USA). Tetramer staining was performed after Fc block and before surface staining for 30 min at room temperature in PBS containing 1% BSA and 5 mM EDTA. For intracellular staining, the cells were restimulated with 5 μg/ml RSV M187–195 peptide (A&A Labs) for 1 hour at 37°C. After the addition of Golgi Plug (BD Biosciences), the samples were incubated for another 3 hours, stained for sur-face marker expression as described above, and fixed in fixation buffer (BioLegend). Cells were stained with fluorochrome-conjugated antibodies against granzyme B and IFN-γ in the presence of purified rat IgG2b anti-mouse CD16/CD32 receptor antibody in permeabilization buffer (BioLegend) for 1 hour. Analysis was performed on an LSR Fortessa flow cytometer (BD Biosciences). Acquisition was set to 2.5 × 105 single, live, CD45+ cells. The gating strategies are shown in fig. S6. Monocyte and neutrophil enumeration, shown in Fig. 6G and figs. S7A and S8A, was performed using gating strategies described previously (32). Data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA). Total cell populations were quantified as the whole lung count × % population of CD45+ cells × % CD45+ live cells × proportion of lung tissue sampled.
Statistical analyses
Statistical analyses were performed with Prism version 7 software (GraphPad, La Jolla, CA, USA) and R version 3.3.2 software. Mediator concentrations were determined from a standard curve with a four-parameter logistic model fit as per the product literature. Limits of detection for each mediator were calculated as twice the standard deviation of the assay blank well (background signal) duplicates, and samples yielding a signal below this value were assigned a value of this limit. Daily mediator concentrations were expressed as the log2 fold change over the baseline (day 0) value for each subject and a least-squares locally estimated scatterplot smoothing (LOESS) model was applied using a polynomial degree of 1 and a span of 0.75. A grid of 100 points across the span of days 1 to 10 was used to produce the prediction curves. Pointwise 95% confidence intervals were derived using a bootstrap percentile method as previously described (3) except the derived LOESS fit was used to calculate the predicted response values. One property of this LOESS fitting is that the graphical representation of the line at each time point is influenced by the preceding and subsequent data. As such, any apparent differences between groups at baseline results from the difference between groups at day 1 (the next time point). Univariate logistic regression was performed with disease status (Cold or No Cold) as the dependent variable to identify the mediator response(s) associated with inoculation outcome. For the mouse studies, statistical analysis was performed using Prism version 7 software. Data are presented as the mean ± SEM. As indicated, Student’s t test was used to compare two groups; one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used to compare multiple groups. The weight curves were analyzed using a two-way ANOVA followed by Bonferroni’s post hoc test. P < 0.05 was considered statistically sig-nificant for all tests (*P <0.05; **P < 0.01; ***P < 0.001 in all figures).
Supplementary Material
Acknowledgments
We thank staff at the NIH Tetramer Core Facility at Emory University for providing tetramers and staff at the St. Mary’s flow cytometry and animal facilities for their technical support.
Funding
The authors gratefully acknowledge support from the Wellcome Trust (087805/Z/08/Z, 102126/B/13/Z, and 109058/Z/15/Z); The Medical Research Council HIC-Vac network (MR/R005982/1); RSV Consortium in Europe (RESCEU) Horizon 2020 Framework Grant 116019; the UK National Institute for Health Research (NIHR) Comprehensive Local Research Networks (CLRNs); European Respiratory Society and the Asociación Latinoamericana de Tórax joint long-term Research Fellowship 2019 (LTRF 20190100546); an NIHR Senior Investigator award; the Biomedical Research Centre (NIHR Imperial BRC); and the Health Protection Research Unit in Respiratory Infections at Imperial College London (NIHR HPRU RI).
Footnotes
Author contributions: P.J.M.O. and C.C. conceived the study. M.S.H., A.P., and C.C. performed infections and collected volunteer samples. M.S.H., R.S.T., A.J., L.C., P.J., and C.C. collected human data. Murine experiments were designed, performed, and analyzed by F. K., A.V., A.O., and C.J. Human data were analyzed by M.S.H., R.S.T., M.C., L.C., T.T., D.N., T.T.H., M.F.M., C.C., and P.J.M.O. The manuscript was written by M.S.H., R.S.T., C.J., C.C., and P.J.M.O. with input from all authors.
Competing interests: The authors declare no competing interests.
Data and materials availability
The RNA-seq data discussed have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE155237. Sequences from 16S rRNA sequencing have been deposited in the European Nucleotide Archive with the accession codes PRJEB28323 (ERP110514). All other data needed to evaluate the conclusions in this study are present in the main text or the supplementary materials.
REFERENCES AND NOTES
- 1.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. 2015;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet. 2013;381:861–873. doi: 10.1016/S0140-6736(12)62202-8. [DOI] [PubMed] [Google Scholar]
- 3.Habibi MS, et al. Impaired antibody-mediated protection and defective IgA B-cell memory in experimental infection of adults with respiratory syncytial virus. Am J Respir Crit Care Med. 2015;191:1040–1049. doi: 10.1164/rccm.201412-2256OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Currie SM, et al. Cathelicidins have direct antiviral activity against respiratory syncytial virus in vitro and protective function in vivo in mice and humans. J Immunol. 2016;196:2699–2710. doi: 10.4049/jimmunol.1502478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanin M, Baviskar P, Webster R, Webby R. The interaction between respiratory pathogens and mucus. Cell Host Microbe. 2016;19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Openshaw PJM, Chiu C, Culley FJ, Johansson C. Protective and harmful immunity to respiratory syncytial virus infection. Annu Rev Immunol. 2017;35:501–532. doi: 10.1146/annurev-immunol-051116-052206. [DOI] [PubMed] [Google Scholar]
- 7.Geerdink RJ, Pillay J, Meyaard L, Bont L. Neutrophils in respiratory syncytial virus infection: A target for asthma prevention. J Allergy Clin Immunol. 2015;136:838–847. doi: 10.1016/j.jaci.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi T, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 10.Shi T, et al. Global disease burden estimates of respiratory syncytial virus-associated acute respiratory infection in older adults in 2015: A systematic review and meta-analysis. J Infect Dis. 2019:jiz059. doi: 10.1093/infdis/jiz059. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aherne W, Bird T, Court SD, Gardner PS, McQuillin J. Pathological changes in virus infections of the lower respiratory tract in children. J Clin Pathol. 1970;23:7–18. doi: 10.1136/jcp.23.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Committee on Infectious Diseases, From the American Academy of Pediatrics: Policy statements-Modified recommendations for use of palivizumab for prevention of respiratory syncytial virus infections. Pediatrics. 2009;124:1694–1701. doi: 10.1542/peds.2009-2345. [DOI] [PubMed] [Google Scholar]
- 14.Vekemans J, et al. Respiratory syncytial virus vaccine research and development: World Health Organization technological roadmap and preferred product characteristics. Vaccine. 2019;37:7394–7395. doi: 10.1016/j.vaccine.2017.09.092. [DOI] [PubMed] [Google Scholar]
- 15.Crank MC, et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science. 2019;365:505–509. doi: 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- 16.DeVincenzo JP, et al. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med. 2010;182:1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard AJ, et al. Human microbial challenge: The ultimate animal model. Lancet Infect Dis. 2012;12:903–905. doi: 10.1016/S1473-3099(12)70292-X. [DOI] [PubMed] [Google Scholar]
- 18.Kiyuka PK, et al. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J Infect Dis. 2018;217:1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagga B, et al. Effect of preexisting serum and mucosal antibody on experimental respiratory syncytial virus (RSV) challenge and infection of adults. J Infect Dis. 2015;212:1719–1725. doi: 10.1093/infdis/jiv281. [DOI] [PubMed] [Google Scholar]
- 20.Jozwik A, et al. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun. 2015;6:10224. doi: 10.1038/ncomms10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devincenzo JP. Natural infection of infants with respiratory syncytial virus subgroups A and B: A study of frequency, disease severity, and viral load. Pediatr Res. 2004;56:914–917. doi: 10.1203/01.PDR.0000145255.86117.6A. [DOI] [PubMed] [Google Scholar]
- 22.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis. 2011;204:996–1002. doi: 10.1093/infdis/jir494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thwaites RS, et al. Reduced nasal viral load and IFN responses in infants with respiratory syncytial virus bronchiolitis and respiratory failure. Am J Respir Crit Care Med. 2018;198:1074–1084. doi: 10.1164/rccm.201712-2567OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson GG, Dowling HF, Spiesman IG, Boand AV. Transmission of the common cold to volunteers under controlled condition. I. The common cold as a clinical entity. AMA Arch Intern Med. 1958;101:267–278. doi: 10.1001/archinte.1958.00260140099015. [DOI] [PubMed] [Google Scholar]
- 25.Stoppelenburg AJ, et al. Local IL-17A potentiates early neutrophil recruitment to the respiratory tract during severe RSV infection. PLOS ONE. 2013;8:e78461. doi: 10.1371/journal.pone.0078461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amatya N, Garg AV, Gaffen SL. IL-17 signaling: The yin and the yang. Trends Immunol. 2017;38:310–322. doi: 10.1016/j.it.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aujla SJ, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden FG, et al. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest. 1998;101:643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson EG, et al. Robust cytokine and chemokine response in nasopharyngeal secretions: Association with decreased severity in children with physician diagnosed bronchiolitis. J Infect Dis. 2016;214:649–655. doi: 10.1093/infdis/jiw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Steenhuijsen Piters WA, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirsebom F, Michalaki C, Agueda-Oyarzabal M, Johansson C. Neutrophils do not impact viral load or the peak of disease severity during RSV infection. Sci Rep. 2020;10:1110. doi: 10.1038/s41598-020-57969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goritzka M, et al. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med. 2015;212:699–714. doi: 10.1084/jem.20140825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannon MJ, Openshaw PJ, Askonas BA. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J Exp Med. 1988;168:1163–1168. doi: 10.1084/jem.168.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirsebom FCM, Kausar F, Nuriev R, Makris S, Johansson C. Neutrophil recruitment and activation are differentially dependent on MyD88/TRIF and MAVS signaling during RSV infection. Mucosal Immunol. 2019;12:1244–1255. doi: 10.1038/s41385-019-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faber TE, Groen H, Welfing M, Jansen KJ, Bont LJ. Specific increase in local IL-17 production during recovery from primary RSV bronchiolitis. J Med Virol. 2012;84:1084–1088. doi: 10.1002/jmv.23291. [DOI] [PubMed] [Google Scholar]
- 36.Langley RG, et al. Secukinumab in plaque psoriasis—Results of two phase 3 trials. N Engl J Med. 2014;371:326–338. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- 37.Mease PJ, et al. Secukinumab inhibition of interleukin-17A in patients with psoriatic arthritis. N Engl J Med. 2015;373:1329–1339. doi: 10.1056/NEJMoa1412679. [DOI] [PubMed] [Google Scholar]
- 38.McInnes IB, et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (FUTURE 2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2015;386:1137–1146. doi: 10.1016/S0140-6736(15)61134-5. [DOI] [PubMed] [Google Scholar]
- 39.Schnoeller C, et al. Attenuated Bordetella pertussis vaccine protects against respiratory syncytial virus disease via an IL-17-dependent mechanism. Am J Respir Crit Care Med. 2014;189:194–202. doi: 10.1164/rccm.201307-1227OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedele G, Bianco M, Debrie AS, Locht C, Ausiello CM. Attenuated Bordetella pertussis vaccine candidate BPZE1 promotes human dendritic cell CCL21-induced migration and drives a Th1/Th17 response. J Immunol. 2011;186:5388–5396. doi: 10.4049/jimmunol.1003765. [DOI] [PubMed] [Google Scholar]
- 41.Huang H, Saravia J, You D, Shaw AJ, Cormier SA. Impaired gamma delta T cell-derived IL-17A and inflammasome activation during early respiratory syncytial virus infection in infants. Immunol Cell Biol. 2015;93:126–135. doi: 10.1038/icb.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jha A, et al. Patterns of systemic and local inflammation in patients with asthma hospitalised with influenza. Eur Respir J. 2019;54:1900949. doi: 10.1183/13993003.00949-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunning J, et al. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat Immunol. 2018;19:625–635. doi: 10.1038/s41590-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, et al. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat Immunol. 2019;20:1681–1691. doi: 10.1038/s41590-019-0513-z. [DOI] [PubMed] [Google Scholar]
- 46.Sweere JM, et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science. 2019;363:eaat9691. doi: 10.1126/science.aat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 48.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. bioRxiv. 2020:2020.05.13.092619. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–446. doi: 10.1017/S0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutton CE, Mielke LA, Mills KH. IL-17-producing γδ T cells and innate lymphoid cells. Eur J Immunol. 2012;42:2221–2231. doi: 10.1002/eji.201242569. [DOI] [PubMed] [Google Scholar]
- 52.Hamada H, et al. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J Immunol. 2009;182:3469–3481. doi: 10.4049/jimmunol.0801814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Grove KC, et al. Characterization and quantification of innate lymphoid cell subsets in human lung. PLOS ONE. 2016;11:e0145961. doi: 10.1371/journal.pone.0145961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clancy DM, et al. Extracellular neutrophil proteases are efficient regulators of IL-1, IL-33, and IL-36 cytokine activity but poor effectors of microbial killing. Cell Rep. 2018;22:2937–2950. doi: 10.1016/j.celrep.2018.02.062. [DOI] [PubMed] [Google Scholar]
- 55.Nicolás-Ávila JA, Adrover JM, Hidalgo A. Neutrophils in homeostasis, immunity, and cancer. Immunity. 2017;46:15–28. doi: 10.1016/j.immuni.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Lim K, et al. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science. 2015;349:aaa4352. doi: 10.1126/science.aaa4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shirey KA, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hufford MM, et al. Influenza-infected neutrophils within the infected lungs act as antigen presenting cells for anti-viral CD8(+) T cells. PLOS ONE. 2012;7:e46581. doi: 10.1371/journal.pone.0046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thwaites RS, et al. Absorption of nasal and bronchial fluids: Precision sampling of the human respiratory mucosa and laboratory processing of samples. J Vis Exp. 2018;131:e56413. doi: 10.3791/56413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhariwal J, et al. Mucosal type 2 innate lymphoid cells are a key component of the allergic response to aeroallergens. Am J Respir Crit Care Med. 2017;195:1586–1596. doi: 10.1164/rccm.201609-1846OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu J, et al. Airway inflammation and illness severity in response to experimental rhinovirus infection in asthma. Chest. 2014;145:1219–1229. doi: 10.1378/chest.13-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuthbertson L, et al. The impact of persistent bacterial bronchitis on the pulmonary microbiome of children. PLOS ONE. 2017;12:e0190075. doi: 10.1371/journal.pone.0190075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee DC, et al. CD25+ natural regulatory T cells are critical in limiting innate and adaptive immunity and resolving disease following respiratory syncytial virus infection. J Virol. 2010;84:8790–8798. doi: 10.1128/JVI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goritzka M, et al. Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. J Virol. 2014;88:6128–6136. doi: 10.1128/JVI.00333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data discussed have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE155237. Sequences from 16S rRNA sequencing have been deposited in the European Nucleotide Archive with the accession codes PRJEB28323 (ERP110514). All other data needed to evaluate the conclusions in this study are present in the main text or the supplementary materials.