Abstract
BACKGROUND
The U.K. 100,000 Genomes Project is in the process of investigating the role of genome sequencing in patients with undiagnosed rare diseases after usual care and the alignment of this research with health care implementation in the U.K. National Health Service. Other parts of this project focus on patients with cancer and infection.
METHODS
We conducted a pilot study involving 4660 participants from 2183 families, among whom 161 disorders covering a broad spectrum of rare diseases were present. We collected data on clinical features with the use of Human Phenotype Ontology terms, undertook genome sequencing, applied automated variant prioritization on the basis of applied virtual gene panels and phenotypes, and identified novel pathogenic variants through research analysis.
RESULTS
Diagnostic yields varied among family structures and were highest in family trios (both parents and a proband) and families with larger pedigrees. Diagnostic yields were much higher for disorders likely to have a monogenic cause (35%) than for disorders likely to have a complex cause (11%). Diagnostic yields for intellectual disability, hearing disorders, and vision disorders ranged from 40 to 55%. We made genetic diagnoses in 25% of the probands. A total of 14% of the diagnoses were made by means of the combination of research and automated approaches, which was critical for cases in which we found etiologic noncoding, structural, and mitochondrial genome variants and coding variants poorly covered by exome sequencing. Cohortwide burden testing across 57,000 genomes enabled the discovery of three new disease genes and 19 new associations. Of the genetic diagnoses that we made, 25% had immediate ramifications for clinical decision making for the patients or their relatives.
CONCLUSIONS
Our pilot study of genome sequencing in a national health care system showed an increase in diagnostic yield across a range of rare diseases. (Funded by the National Institute for Health Research and others.)
Rare diseases are a worldwide health care challenge, with approximately 10,000 disorders affecting 6% of the population in Western societies.1,2 More than 80% of rare diseases have a genetic component, and these conditions are disabling and expensive to manage. One third of children with a rare disease die before their fifth birthday.1 The adoption of next-generation sequencing has improved rates of diagnosis of rare diseases over the past decade.3-5 However, the majority of patients with rare diseases remain without a molecular diagnosis after standard diagnostic testing.3-5 To address this lack of diagnosis, the U.K. government launched the 100,000 Genomes Project in 2013 to apply whole-genome sequencing to the study of rare diseases, cancers, and infections in a national health care setting.6
To assess the effect of the whole-genome–sequencing approach on the genetic diagnosis of rare diseases in the National Health Service (NHS) in the United Kingdom, we carried out a pilot study in which we enrolled families and undertook detailed clinical phenotyping of the proband.4 We collected electronic health records from all participants and stored these together with the genomic and clinical data in a computer environment with multi-petabytes of storage (the Genomics England research environment).5 When necessary, we validated diagnostic variants in the laboratory and performed computational analyses.
METHODS
PARTICIPANTS
After approval from the national research ethics committee was obtained, we recruited participants who had been identified by health care professionals and researchers as having rare diseases (across a broad range of categories) that had not been diagnosed after receipt of usual care in the NHS, which included either no diagnostic tests (because none were available) or approved diagnostic tests that did not include genome sequencing. The participants were recruited at nine English hospitals, and written informed consent was obtained from the participants by the National Institute for Health Research (NIHR) BioResource for Rare Diseases.
To test the broad applicability of genome sequencing, we determined that participants were eligible if they had a rare disease (as defined in the United Kingdom as a disorder affecting ≤1 in 2000 persons), were likely to have a single-gene or oligogenic cause, and had not received a genomic diagnosis. Data on previous testing in probands were collected when possible; testing included single-gene tests, karyotyping, single-nucleotide polymorphism arrays, next-generation sequencing panels, and exome sequencing. Probands and, when feasible, parents or other family members were enrolled across multiple clinical specialties in the NHS. Standardized baseline clinical data were recorded with the use of Human Phenotype Ontology (HPO) terms7 guided by disease-specific data models,8 and whole blood samples were obtained for DNA extraction. In the 100,000 Genomes Project, participants are followed over their life course with the use of electronic health records (all hospital episodes, registry entries, and cause of death).
This pilot study was undertaken in partnership with the NIHR BioResource and is part of the portfolio of translational research at the NIHR Biomedical Research Centres at Barts, Cambridge University Hospitals NHS Foundation Trust, Great Ormond Street Hospital for Children NHS Foundation Trust, Manchester University NHS Foundation Trust, Moorfields Eye Hospital NHS Foundation Trust, Newcastle upon Tyne Hospitals NHS Foundation Trust, Oxford University Hospitals NHS Foundation Trust, and University College London Hospitals NHS Foundation Trust. Clinical data from the NHS and NHS Digital were used in this work.
GENOME SEQUENCING
Genome sequencing9 was performed with the use of the TruSeq DNA polymerase-chain-reaction (PCR)–free sample preparation kit (Illumina) on a HiSeq 2500 sequencer, which generates a mean depth of 32× (range, 27 to 54) and a depth greater than 15× for at least 95% of the reference human genome. Whole-genome sequencing reads were aligned to the Genome Reference Consortium human genome build 37 (GRCh37) with the use of Isaac Genome Alignment Software. Family-based variant calling of single-nucleotide variants (SNVs) and insertion or deletions (indels) for chromosomes 1 to 22, the X chromosome, and the mitochondrial genome (mean coverage, 2814×; range, 142 to 16,581) was performed with the use of the Platypus variant caller.10
DIAGNOSTIC PIPELINE
We constructed an automated analytic pipeline to filter the genome down to rare, segregating, and predicted damaging candidate variants in coding regions. To limit the possibility of overlooking or inefficiently prioritizing diagnoses, we focused initially on applied virtual gene panels (applied panels) that were based on both the recruited clinical indication or disease and the submitted HPO terms. To address the issue of which genes have sufficient evidence to show causation and be included in these applied panels, we used our PanelApp software to enable expert, crowd-sourced review and curation of genes with diagnostic-grade evidence for each of our disease categories (e.g., evidence in at least three unrelated families).11 Loss-of-function or de novo protein-altering variants affecting genes in the applied panels were classified as tier 1, other variant types such as missense variants affecting these genes were classified as tier 2, and all other filtered variants were classified as tier 3 (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). To further reduce the possibility of missing or inefficiently prioritized diagnoses, we used a phenotype-based approach with the Exomiser application12 to search across all genes in the genome for a diagnosis. Exomiser prioritizes rare, segregating, and predicted pathogenic variants in genes in which the patient phenotypes match previously referenced knowledge from human disease or model organism databases. The ontology-driven phenotype matching can identify patients who have an atypical profile for a disease. Additional details regarding the Exomiser are provided in the Diagnostic Pipeline section in the Supplementary Appendix.
Prioritization of variants and return of candidate variants for presentation to the 13 NHS Genomic Medicine Centres (GMCs) were performed with the use of decision-support systems and with assistance from clinical genetics teams from Congenica and Fabric Genomics.13,14 These variants were reviewed by NHS clinical scientists and clinicians using the guidelines of the American College of Medical Genetics and Genomics, and a diagnostic report was issued for each proband.15 Final clinical outcomes included whether a genetic diagnosis was obtained, identification of the variant or variants involved, whether the variant or variants explained all or some of the phenotypes, and whether an intervention was used.
Recruitment of the participants in the pilot study and sequencing were performed during the period from January 2014 through December 2016, while the infrastructure to collect, quality check, process, and return data was being established. Results were returned to the GMCs from May 2016 through April 2019. Now that the information pipeline has been established (post-pilot phase), results are returned to the GMCs within 6 weeks after the sample is obtained.
NOVEL PATHOGENIC VARIANTS
Researchers investigated coding and noncoding regions to detect novel diagnostic variants in genes matching the patients’ phenotypes, including the presence of de novo variants in highly constrained coding regions16 in the 95th percentile. We use the term novel to describe diagnostic variants we have detected that have not previously been described in the literature as causative. This is distinct from de novo variants, which are present for the first time in a family member due to either a new variant in an egg or sperm or a new mutation at conception. The variant may have been previously described. We used a new method described by Wei et al.17 to analyze mitochondrial DNA that accounts for heteroplasmy, the Genomiser to detect noncoding pathogenic variants,18 and the ExpansionHunter software tool to detect simple tandem repeat expansions.19 Finally we used a new random forest method to analyze Canvas20 and Manta21 calls and to identify potentially pathogenic copy-number and structural variants.
Gene-based burden testing to detect enrichment of rare, predicted pathogenic, and segregating variants in novel genes in specific disease cohorts relative to controls was performed on the genomes in the pilot study as well as on additional genomes from the rest of the 100,000 Genomes Project to increase power (57,002 genomes; see the Supplementary Methods in the Supplementary Appendix). The genomic and clinical data from the pilot study are freely accessible to members of a Genomics England Clinical Interpretation Partnership domain (https://www.genomicsengland.co.uk/about-gecip/).
STATISTICAL ANALYSIS
Testing was performed with the use of the R software, version 3.6.0 (R Foundation for Statistical Computing), and Stata software, version 16 (StataCorp). Further details on the individual methods used in the study are provided in the Supplementary Appendix.
RESULTS
PARTICIPANTS
We enrolled 4660 participants (2183 probands and 2477 family members), among whom 161 disorders across a broad spectrum of rare diseases were present (Table 1).22 Neurologic, ophthalmologic, and tumor syndromes were commonly represented (Table 2). Participants were recruited with varying numbers of affected and unaffected family members. We aimed to recruit family trios (both parents and a proband) or larger family structures to facilitate more effective variant prioritization, and our efforts were met with varying degrees of success. Among the recruited probands with multiple bowel polyps, 93% were singletons (i.e., probands for whom no other family member was recruited). In contrast, 12% of the probands with intellectual disability were singletons. Adult probands were more commonly enrolled than pediatric probands (age ≤18 years at recruitment) (74% vs. 26%), which is in line with the percentage of children and adults in the general population in England and Wales (79% vs. 21% [2011 census of England and Wales23]). The preponderance of adults was unusual as compared with previous sequencing projects and reflects the eligibility criterion that probands had to have undergone usual care; in many cases, usual care involved standard genetic testing (mostly single-gene or panel-based). A lower percentage of recruited probands were female than male owing to the difference among pediatric probands (232 girls and female adolescents [11%] vs. 339 boys and male adolescents [16%], P<0.001); the expected percentage of female probands was 51% (on the basis of the 2011 census of England and Wales) across most disease categories. The greater susceptibility of males than of females to recessive X-linked conditions may account for this sex bias: more than 6% of all diagnoses involved variants on the X chromosome (which represents approximately 5% of the genome). The inferred ancestry of the probands (see the Supplementary Appendix) was in line with what was expected on the basis of the general population, in which 86% of children and adults were White, 8% Asian, 3% Black, 2% mixed, and 1% other (2011 census of England and Wales). However, South Asian ancestry was significantly more common among pediatric probands than among adult probands (16% vs. 4%, P<0.001); our results indicated potential consanguinity in 43% of the 93 pediatric South Asian probands and in 1% of the other 478 pediatric probands (Table 1).
Table 1.
Demographic Characteristics of the Probands (Including Inferred Ancestry) in the 100,000 Genomes Project Pilot Study.*
| Characteristic | All Probands (N = 2183) |
Pediatric Probands (N = 571) |
Adult Probands (N = 1612) |
|---|---|---|---|
| Sex — no. (%) | |||
| Male | 1138 (52) | 339 (59) | 799 (50) |
| Female | 1045 (48) | 232 (41) | 813 (50) |
| Total | 2183 (100) | 571 (100) | 1612 (100) |
| Median age at recruitment (IQR) — yr | 35 (18–54) | 9 (5–14) | 45 (31–60) |
| Inferred ancestry — no. (%) [% consanguinity suggested in record]† | |||
| African | 50 (2) [0] | 25 (4) [0] | 25 (2) [0] |
| American with mixed ancestry | 26 (1) [23] | 12 (2) [25] | 14 (1) [21] |
| East Asian | 8 (<1) [0] | 2 (<1) [0] | 6 (<1) [0] |
| European | 1931 (88) [<1] | 438 (77) [<1] | 1493 (93) [<1] |
| South Asian | 163 (7) [36] | 93 (16) [43] | 70 (4) [25] |
| Not determined | 5 (<1) [0] | 1 (<1) [0] | 4 (<1) [0] |
| Total | 2183 (100) [3] | 571 (26) [8] | 1612 (74) [2] |
Pediatric probands were younger than 18 years of age at recruitment, and adult probands were 18 years of age or older at recruitment. IQR denotes interquartile range.
Ancestry was inferred on the basis of a random forest model trained on principal component analysis of the five super populations described for the 1000 Genomes Project.
Table 2.
Disease Categories among the Probands in the 100,000 Genomes Project Pilot Study.*
| Disease Category | All Family Structures (2183 Probands) |
Singleton (881 Probands) |
Family Duo (343 Probands) |
Family Trio (797 Probands) |
Larger Family Structure (162 Probands) |
|---|---|---|---|---|---|
| number of probands (percent) | |||||
| Cardiovascular disorder | 147 (7) | 56 (6) | 24 (7) | 49 (6) | 18 (11) |
| Ciliopathy | 69 (3) | 34 (4) | 14 (4) | 16 (2) | 5 (3) |
| Dermatologic disorder | 38 (2) | 9 (1) | 5 (1) | 22 (3) | 2 (1) |
| Dysmorphic or congenital abnormality | 20 (1) | 10 (1) | 2 (1) | 7 (1) | 1 (1) |
| Endocrine disorder | 87 (4) | 57 (6) | 14 (4) | 12 (2) | 4 (2) |
| Gastroenterologic disorder | 32 (1) | 0 | 0 | 18 (2) | 14 (9) |
| Growth disorder | 3 (<1) | 3 (<1) | 0 | 0 | 0 |
| Hematologic or immunologic disorder | 5 (<1) | 2 (<1) | 3 (1) | 0 | 0 |
| Hematologic disorder | 7 (<1) | 0 | 3 (1) | 2 (<1) | 2 (1) |
| Hearing or ear disorder | 35 (2) | 6 (1) | 5 (1) | 17 (2) | 7 (4) |
| Metabolic disorder | 93 (4) | 24 (3) | 12 (3) | 48 (6) | 9 (6) |
| Intellectual disability | 130 (6) | 10 (1) | 24 (7) | 78 (10) | 18 (11) |
| Neurologic or neurodevelopmental disorder† | 521 (24) | 193 (22) | 93 (27) | 194 (24) | 41 (25) |
| Ophthalmologic disorder | 348 (16) | 74 (8) | 62 (18) | 199 (25) | 13 (8) |
| Renal and urinary tract disorder | 176 (8) | 125 (14) | 21 (6) | 26 (3) | 4 (2) |
| Respiratory disorder | 2 (<1) | 1 (<1) | 0 | 1 (<1) | 0 |
| Rheumatologic disorder | 48 (2) | 14 (2) | 6 (2) | 25 (3) | 3 (2) |
| Skeletal disorder | 62 (3) | 15 (2) | 11 (3) | 23 (3) | 13 (8) |
| Tumor syndrome | 293 (13) | 231 (26) | 31 (9) | 27 (3) | 4 (2) |
| Other | 67 (3) | 17 (2) | 12 (3) | 34 (4) | 4 (2) |
| Total | 2183 (100) | 881 (100) | 343 (100) | 797 (100) | 162 (100) |
Singleton refers to a proband for whom no other family member was recruited, family duo to a parent–proband pair, family trio to both parents and a proband, and family quad to a proband, sibling, and parents.
Neurologic and neurodevelopmental symptoms excluded intellectual disability.
CLINICAL DATA AND SEQUENCING
We collected clinical data with the use of HPO terms for each affected participant (a median of 4 [range, 1 to 61] present terms, and a median of 4 [range, 0 to 144] absent terms [phenotypes that were assessed and confirmed as definitely not observed in the proband]). We then performed genome sequencing, followed by quality assurance to check coverage, sequence quality, presence of repeat sample submissions or sample swaps, and consistency with reported family structures (see the Supplementary Appendix).
DIAGNOSTIC YIELD
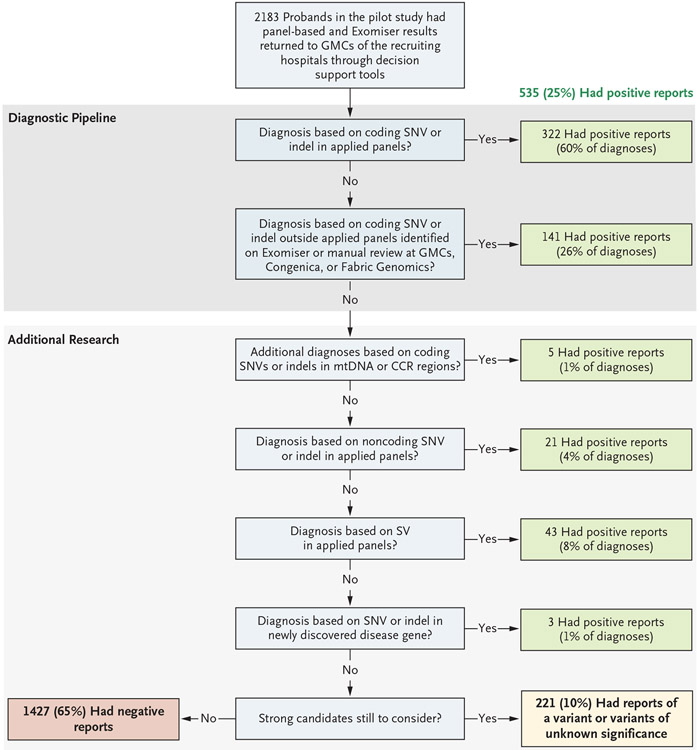
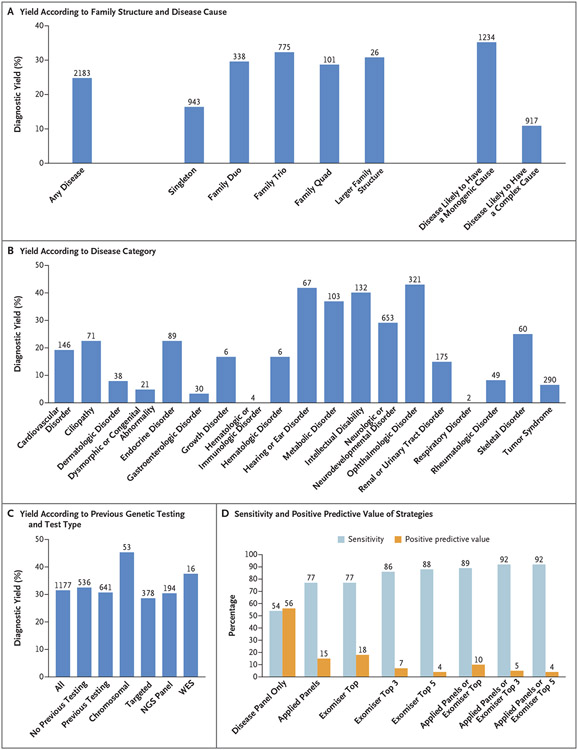
We made genetic diagnoses in 25% of the probands and deposited the genotypes into the ClinVar repository (accession numbers, SCV001759972 to SCV001760540). Of these diagnoses, 60% were made on the basis of coding SNVs or indels in the applied panels; 26% were made on the basis of coding SNVs or indels affecting well-established disease genes not included in the applied panels (diagnoses were made through phenotype-based prioritization or expert review by the study clinicians or the clinical genetics teams from Congenica or Fabric Genomics); and 14% were made on the basis of genomewide, phenotype-agnostic research analysis that investigated beyond SNVs and indels, coding regions, and disease genes in the applied panels (Fig. 1). On the basis of international guidelines,15 an additional 10% of the probands were classified as having variants of unknown significance in genes that were considered to be consistent with the phenotype on clinical review at the study site but that required further functional validation. Fewer candidate variants were returned to the GMCs after filtering (i.e., the removal of extremely unlikely candidates) in larger family structures (Table 3), which made it easier to identify causative variants and in turn led to higher diagnostic yields for family trios and quads (proband, sibling, and parents) and more complex family structures (Fig. 2A), even within a disorder (e.g., the diagnostic yield for hereditary ataxia was 21% among singletons and 32% among persons in family trios) (Table S4).
Figure 1. Overview of the Diagnostic and Research Pipeline and Source of Diagnoses.
Results from 2183 probands in the pilot study were returned for presentation to the Genomic Medicine Centres (GMCs) of the recruiting hospitals. A total of 25% of the probands received a positive diagnosis, and 10% had a variant or variants of unknown significance in genes that were determined by clinical geneticists at the recruiting site to be consistent with the phenotype but that required further functional validation. The remaining 65% of the probands received a negative report at the time but will be reassessed. The numbers and sources of these positive diagnoses are shown at each stage of the automated diagnostic pipeline, and the additional research is shown for diagnoses that were not immediately obvious. CCR denotes constrained coding region, indel insertion or deletion, mtDNA mitochondrial DNA, SNV single-nucleotide variant, and SV structural variant.
Table 3.
Candidate Variants Returned for Presentation to the NHS Genomic Medicine Centres per Proband with the Automated Virtual Panel–Based Analysis Pipeline.*
| Variable | All Family Structures (2183 Probands) |
Singleton (881 Probands) |
Family Duo (343 Probands) |
Family Trio (797 Probands) |
Larger Family Structure (162 Probands) |
|---|---|---|---|---|---|
| Median no. of variants returned after filtering (IQR)† | 221 (49–288) | 292 (258–327) | 149 (117–213) | 29 (17–136) | 22 (9–71) |
| Median no. of variants in virtual panels returned after filtering (IQR) | 1 (0–2) | 1 (0–2) | 1 (0–3) | 1 (0–2) | 0 (0–1) |
Singleton refers to a proband for whom no other family member was recruited, family duo to a parent–proband pair, family trio to both parents and a proband, and family quad to a proband, sibling, and parents.
Filtering is the removal of extremely unlikely candidate variants.
Figure 2. Diagnoses in the Rare Disease Pilot Study.
Panel A shows diagnostic yield for any disease and according to family structure and cause of disease. The diagnostic yield was 35% for diseases likely to have a monogenic cause and 11% for diseases likely to have a complex cause. The values above the bars are the numbers of probands. Singleton refers to a proband for whom no other family member was recruited, family duo to a parent–proband pair, family trio to both parents and a proband, and family quad to a proband, sibling, and parents. Panel B shows diagnostic yield according to disease category. The values above the bars are the numbers of probands. Panel C shows the diagnostic yield among probands according to previous genetic testing and most extensive testing type: chromosomal (karyotyping, array-based comparative genomic hybridization, single-nucleotide polymorphism arrays), targeted (targeted single-gene tests), next-generation sequencing (NGS) panels, or whole-exome sequencing (WES). The values above the bars are the numbers of probands. Panel D shows the performance of virtual panel-based and Exomiser-based prioritization for identifying the diagnoses. “Disease panel only” indicates the use of a single virtual panel for the recruited disease category. “Applied panels” indicates the use of all applied virtual gene panels used in the pipeline, including the recruited disease–associated panel as well as 0 or more additional panels selected on the basis of the patient’s phenotypes (Human Phenotype Ontology terms). “Exomiser top” indicates Exomiser use in the top-ranked candidate variants, “Exomiser top 3” use in the top three candidates, and “Exomiser top 5” use in the top five candidates. Sensitivity is the percentage of true positive diagnoses based on SNVs or indels that were identified, and the positive predictive value is the percentage of prioritized variants that led to a positive diagnosis. The values above the bars are percentages. In this analysis, the diagnosed variant or variants are true positives, and the other candidate variants that were returned are false positives.
We obtained a higher diagnostic yield for diseases that we considered likely to have a monogenic cause than those we considered likely to have a complex cause (35% vs. 11%) (Fig. 2A). Diseases were considered likely to have a monogenic cause if they were present in the Online Mendelian Inheritance in Man database, involved genetic testing as part of the standard diagnostic workup, and had a consensus of opinion among three clinical geneticists (who were unaware of each other’s assessments) that they had monogenic cause. Diagnostic yield was highly varied across diseases (Fig. 2B and Table S3); the diagnostic yield ranged from 40 to 55% for intellectual disability and various vision and hearing disorders and was 6% for tumor syndromes.
We obtained data on the presence or absence of previous genetic testing in 1177 participants. The number of tests per proband ranged from 0 to 16, with a median of 1 (interquartile range, 0 to 2), and approximately half the probands in this subgroup had been tested at least once. The overall diagnostic yield with the use of genome sequencing in this subgroup increased by 32%, and there was only a slight difference depending on whether previous testing had been performed (33%) or not (31%). However, many of these previous tests were not recent, dating back to the time of recruitment at the latest (2014 to 2016). The diagnostic yield provided by genome sequencing varied between 28% and 45%, depending on the type of previous testing (Fig. 2C and Table S5), which for the most part involved targeted single-gene and panel-based testing (Table S6).
DIAGNOSTIC PIPELINE
The aim of the automated diagnostic pipeline is to identify a few potentially causative candidate variants, among the millions in a whole genome, through the removal of extremely unlikely candidates (filtering) and the identification of the most likely candidates in the remainder (prioritization). This approach facilitates manual clinical interpretation and diagnostic reporting by clinicians at the GMCs.
A total of 322 (66%) of the 490 diagnoses that were based on SNVs or indels from the genomes were made with the virtual panel–based pipeline, and the positive predictive value was high given the millions of variants in the whole genomes — 291 of 1041 candidate variants (28%) returned to the GMCs proved to be diagnostic. We re-ran this analysis in December 2019 to assess the effects of updated versions of the applied panels with the latest disease gene discoveries, improved selection of the applied panel or panels on the basis of the patient’s phenotype, and advances in variant-filtering strategies (e.g., allowance for incomplete penetrance when suspected). With the use of these updated versions, the number of genetic diagnoses increased from 322 to 377 of the 490 diagnoses (77% sensitivity), and the positive predictive value was 15% (Fig. 2D). This result shows effective filtering and prioritization of the variants, with a median number of only 1 candidate variant (interquartile range, 0 to 2) included in the panels returned to the GMCs per proband (Table 3). Ongoing evolution of the applied panels with new disease genes is expected to continue to increase the diagnostic yield with this approach.
With the use of phenotype-based prioritization with the Exomiser to score and rank the most likely causative variants, diagnoses were detected in 77% of the top-ranked candidate variants, in 86% of the top three candidates, and in 88% of the top five candidates (Fig. 2D). Use of the Exomiser and applied panels was complementary — 92% of the 490 diagnoses were made with the applied panels or the Exomizer top five candidates (last blue bar in Fig. 2D). Precision phenotyping in our participants was essential for both the Exomiser and the selection of additional applied panels; without such phenotyping, only 54% of these diagnoses would have been prioritized in the virtual panel for the recruited disease and presented to the GMCs as a likely candidate (first blue bar in Fig. 2D).
RESEARCH-BASED DIAGNOSES
A total of 14% of the genetic diagnoses required further research outside the diagnostic pipeline (Fig. 1). This research involved combined analysis of the genome sequences and clinical data in our research environment and validation with the use of wet-bench orthogonal tests and computational approaches (Table S7). Additional diagnoses were made by screening for the presence of de novo variants in highly constrained coding regions.16 These diagnoses included a de novo EBF3 missense variant in a patient with hereditary ataxia. A mitochondrial genome analysis that accounted for heteroplasmy led to four new diagnoses, as well as the nine that had already been made by means of the main pipeline. Twelve probands had intronic splicing variants that were prioritized by Exomiser owing to the known pathogenic status of these variants in the ClinVar database.24 Nine diagnoses involving novel, previously undescribed noncoding variants required exploration of the whole genome and in vitro functional validation by means of reverse transcriptase–PCR, minigene, or luciferase assays.25-27 For these diagnoses, unsolved cases in probands had been queried for noncoding variants that affect genes, either alone or in compound heterozygosity with loss-of-function variants, included in the applied panels. These cases were identified with the use of Genomiser or, for probands with retinal disorders, systematic analysis of the untranslated regions, promoter, or introns. The cases in 43 additional probands were fully or partially explained by structural variants or simple tandem repeat expansions in the genes HTT or FXN in the probands with hereditary spastic paraplegia.
NEW DISEASE–GENE ASSOCIATIONS
We performed burden testing to identify new mendelian disease–gene associations and make potential genetic diagnoses in probands with unsolved cases; 828 significant disease–gene associations (Q value of <0.1) were identified, including 249 known and 579 novel genes (novel with respect to their association with disease), with a mean (±SD) number of associations of only 0.03±0.2 (range, 0 to 3) from 10,000 permutations in which the cases and controls were assigned randomly. A total of 22 candidates represent the most probable new, fully penetrant, mendelian disease genes (Table S8; ClinVar accession numbers, SCV001759972 to SCV001760540) with three recently independently confirmed diagnoses: UBAP1 in hereditary spastic paraplegia,28 FOXJ1 in non–cystic fibrosis bronchiectasis,29 and SORD in Charcot–Marie–Tooth disease.30 Diagnostic reports were issued for three probands with these genes (Fig. 1), and we are currently investigating others with the use of the online tool GeneMatcher and with functional validation studies in model organisms.
HEALTH CARE OUTCOMES AFTER DIAGNOSIS
The findings from our approach ended long diagnostic odysseys for some participants and their families (the median duration of such an odyssey was 75 months, and the median number of hospital visits was 68) (Table S1), and we speculate that they will mitigate NHS resource costs (the combined cost for 183,273 episodes of hospital care among the affected participants was £87 million [$122 million]) (Table S3). In addition, 134 of the 533 genetic diagnoses (25%) were reported by clinicians to be of immediate clinical actionability — only 11 (0.2%) were described as having no benefit. As of now, the remainder of the diagnoses are of unknown usefulness. The benefits in terms of health care included 4 diagnoses that led to a suggested change in medication, 26 that led to suggested additional surveillance of the proband or relatives, 13 that allowed for clinical trial eligibility, 59 that informed future reproductive choices, and 32 that had other benefits (Table S9).
In several specific probands, diagnoses have had important clinical actionability. In a 36-year-old man with suspected choroideremia, we detected a novel CHM promoter variant causing loss of gene expression,27 a diagnosis that enabled eligibility for a gene-replacement trial. A male neonate proband presented with severe infection and transient neurologic symptoms immediately after birth and died at 4 months of age with no diagnosis but with health care costs of approximately £80,000 ($112,000) (Table S10). A diagnosis of transcobalamin II deficiency due to a homozygous frameshift in TCN2 was made from this study, which enabled predictive testing to be offered to the younger brother within 1 week after birth. The younger child, who received a positive result, received weekly hydroxocobalamin injections to prevent metabolic decompensation.
A 10-year-old girl was admitted to the intensive care unit with life-threatening chicken pox. She had undergone a diagnostic odyssey over a period of 7 years at a total cost of £356,571 ($499,199) across 307 secondary care episodes (Table S11). We were able to diagnose CTPS1 deficiency due to a homozygous, known pathogenic splice acceptor variant. A diagnosis enabled a curative bone marrow transplantation (cost of £70,000 [$98,000]), and predictive testing in her siblings showed no additional family members to be at risk.
One proband had waited until his sixth decade of life for a genomic diagnosis of an INF2 mutation causing focal segmental glomerulosclerosis. His father, brother, and uncle had all died from kidney failure. He had received two kidney transplants, had transmitted the condition to his daughter, and was concerned about whether his 15-year-old granddaughter, who was under surveillance, was at risk. After he received his genetic diagnosis, the granddaughter was tested, found to be negative, and discharged from regular medical surveillance.
DISCUSSION
Our findings show a substantial increase in yield of genomic diagnoses made in patients with the use of genome sequencing across a broad spectrum of rare disease. The enhanced diagnostic benefit was observed regardless of whether participants had undergone previous genetic testing (diagnostic yields were 31% among those who had undergone testing and 33% among those who had not). In 25% of those who received a genetic diagnosis, there was immediate clinical actionability. The standardization of procedures — from the enrollment of patients to the return of NHS-validated results to clinicians — was critical to our success. For example, the collection of clinical data with the use of disease-specific data models and HPO terms enabled diagnoses, which confirmed the value of standardization with the use of ontology terms and clinical annotation in precision medicine.31 These additional diagnoses, beyond the 264 (49% of total diagnoses) observed with the use of the single-disease virtual panel, came from the use of Exomiser and additional applied panels. The diagnostic discoveries derived by combining research, decision support, and clinical validation and assessment leveraged an additional 72 diagnoses.
Diagnostic yield was influenced by family structure, and for disorders likely to have mendelian inheritance and a single-gene etiologic factor, our yield increased to 35%: ophthalmologic, metabolic, and neurologic disorders yielded the greatest percentage of diagnoses. The scale of our data set enabled cohortwide burden testing, which identified numerous novel disease–gene associations, including three that have now been confirmed and 19 with compelling evidence that are likely to be confirmed in independent data sets.
Of the diseases we diagnosed with the use of genome sequencing, 13% were caused by mutations in noncoding sequence or mitochondrial genomes, tandem repeat expansions in persons with Huntington’s disease, and a wide range of structural variants with nucleotide resolution of breakpoints (which were identified with the use of a new random forest method). An additional 2% of the diagnoses involved coding variants in regions of low coverage on exome sequencing. Our results provide new evidence of the value of genome sequencing and mirror the findings in a previous study in which 53% of the participants who received new diagnoses from genome sequencing had previously undergone exome sequencing.5
Previous studies have shown how next-generation sequencing can lead to diagnoses, with yields of 25 to 29% with the use of exome sequencing in persons who had received no previous genetic testing.32-34 The Undiagnosed Disease Network reported a diagnostic yield of 26% with the use of a mixture of whole-exome and whole-genome sequence analysis in 382 patients,5 and another study of genome sequencing showed a yield of 42% among 50 probands with intellectual disability who had previously undergone testing.35 Among probands with a broad range of disorders (161 in total) with an unmet diagnostic need, we obtained results that were similar to those in the previous studies. Our approach is limited to diagnoses that are readily made by means of short-read genome sequencing. Fully phased, long-read sequencing better detects structural variation and delivers sequence information from parts of the genome that are poorly captured by short-read sequencing.36
The findings from our pilot study support the case for genome sequencing in the diagnosis of certain specific rare diseases in the new NHS National Genomic Test Directory.37 In patients with specific disorders, such as intellectual disability, genome sequencing is now the first-line test in the NHS (Table S12). With a new National Genomic Medicine Service, the NHS in England is in the process of sequencing 500,000 whole genomes in rare disease and cancer in health care. We hope that our findings will assist other health systems in considering the role of genome sequencing in the care of patients with rare diseases.
Supplementary Material
Acknowledgments
Supported by the NIHR, the Wellcome Trust, the Medical Research Council (MRC), Cancer Research U.K., the Department of Health and Social Care, and NHS England. The NIHR BioResource is funded by the NIHR. Drs. Caulfield and Ouwehand are NIHR senior investigators. Dr. Chinnery is a Wellcome Trust Principal Research Fellow (212219/Z/18/Z) and an NIHR Senior Investigator who receives support from the MRC Mitochondrial Biology Unit (MC_UU_00015/9), the MRC International Centre for Genomic Medicine in Neuromuscular Disease (MR/S005021/1), and the NIHR Biomedical Research Centre (BRC). Dr. Wedderburn’s work is supported by grants from Versus Arthritis (21593), the NIHR BRC at Great Ormond Street Hospital, and the MRC (MR/R013926/1). Drs. Smedley, Cacheiro, and Cipriani receive support from the National Institutes of Health (NIH, grant 5-UM1-HG006370). Dr. Smedley’s team that performed much of the analysis was supported by grants from the NIH (1R24OD011883, U54 HG006370, and 1R01HD103805-01). Dr. Arno’s work is supported by a Fight for Sight (United Kingdom) Early Career Investigator Award (5045/46), NIHR BRC at Great Ormond Street Hospital Institute for Child Health, and Moorfields Eye Charity (Stephen and Elizabeth Archer in memory of Marion Woods). The Moorfields–University College London (UCL) Institute of Ophthalmology team is additionally funded by NIHR BRC at Moorfields Eye Hospital and UCL Institute of Ophthalmology.
We thank the personnel at NIHR BioResource for their partnership in this study; all the health care teams at Addenbrooke’s Hospital in Cambridge, Great Ormond Street Hospital NHS Foundation Trust, University College London NHS Foundation Trust, Guy’s and St. Thomas’ Hospital, Barts Health, Oxford University Hospitals NHS Foundation Trust, Manchester University NHS Foundation Trust, and the Newcastle Hospitals NHS Foundation Trust; the NHS patients and their families who made this work possible; all those across the world who have contributed to the PanelApp knowledge base and to the validation and reporting working group (Dr. Dom McMullan, Dr. Helen Firth, Dr. Steve Abbs, and Dr. Sian Ellard) for their role in supporting the development of the bioinformatics pipeline and reporting process; Dr. David Bick and Dr. Gil McVean for providing feedback on our work; Dr. Dame Sue Hill and the team at NHS England for the work to fund and establish the 13 GMCs, which enabled the NHS contribution that included the clinical return of results within the NHS in a standardized and validated format that led to the confirmation of the diagnoses, provided additional information, and led to the patient benefit reported; the Illumina Laboratory Services team at Hinxton for genome sequencing and secondary analysis; and the developers of the Human Phenotype Ontology (Monarch Initiative) and Exomiser (funded by the NIH Office of the Director [1R24OD011883]) for the support that was provided through these resources.
Appendix
The authors’ full names and academic degrees are as follows: Damian Smedley, Ph.D., Katherine R. Smith, Ph.D., Antonio Martin, M.Sc., Ellen A. Thomas, M.D., Ellen M. McDonagh, Ph.D., Valentina Cipriani, Ph.D., Jamie M. Ellingford, Ph.D., Gavin Arno, Ph.D., Arianna Tucci, M.D., Jana Vandrovcova, Ph.D., Georgia Chan, Ph.D., Hywel J. Williams, Ph.D., Thiloka Ratnaike, M.B., B.S., Ph.D., Wei Wei, Ph.D., Kathleen Stirrups, Ph.D., Kristina Ibanez, Ph.D., Loukas Moutsianas, Ph.D., Matthias Wielscher, Ph.D., Anna Need, Ph.D., Michael R. Barnes, Ph.D., Letizia Vestito, M.Sc., James Buchanan, D.Phil., Sarah Wordsworth, Ph.D., Sofie Ashford, B.Sc., Karola Rehmström, Ph.D., Emily Li, Ph.D., Gavin Fuller, M.Med.Sci., Philip Twiss, M.Sc., Olivera Spasic-Boskovic, M.Sc., Sally Halsall, Ph.D., R. Andres Floto, M.D., Ph.D., Kenneth Poole, M.D., Ph.D., Annette Wagner, M.D., Ph.D., Sarju G. Mehta, M.D., Mark Gurnell, M.D., Ph.D., Nigel Burrows, M.D., Roger James, Ph.D., Christopher Penkett, D.Phil., Eleanor Dewhurst, B.A., Stefan Gräf, Ph.D., Rutendo Mapeta, B.Sc., Mary Kasanicki, Ph.D., Andrea Haworth, M.Sc., F.R.C.Path., Helen Savage, M.Sc., Dip.R.C.Path., Melanie Babcock, Ph.D., Martin G. Reese, Ph.D., Mark Bale, Ph.D., Emma Baple, M.B., B.S., Ph.D., Christopher Boustred, Ph.D., Helen Brittain, M.D., Anna de Burca, M.B., B.S., Ph.D., Marta Bleda, Ph.D., Andrew Devereau, B.Sc., Dina Halai, M.Sc., Eik Haraldsdottir, M.Sc., Zerin Hyder, M.D., Dalia Kasperaviciute, Ph.D., Christine Patch, Ph.D., Dimitris Polychronopoulos, Ph.D., Angela Matchan, M.Sc., Razvan Sultana, Ph.D., Mina Ryten, M.D., Ph.D., Ana L.T. Tavares, M.B., B.S., Carolyn Tregidgo, Ph.D., Clare Turnbull, M.D., Ph.D., Matthew Welland, M.Sc., Suzanne Wood, M.Sc., Catherine Snow, Ph.D., Eleanor Williams, Ph.D., Sarah Leigh, Ph.D., Rebecca E. Foulger, Ph.D., Louise C. Daugherty, M.Sc., Olivia Niblock, M.Sc., Ivone U.S. Leong, Ph.D., Caroline F. Wright, Ph.D., Jim Davies, D.Phil., Charles Crichton, B.A., James Welch, B.A., Kerrie Woods, B.A., Lara Abulhoul, M.D., Paul Aurora, M.R.C.P., Ph.D., Detlef Bockenhauer, M.D., Alexander Broomfield, M.D., Maureen A. Cleary, M.D., Tanya Lam, M.B., B.S., M.P.H., Mehul Dattani, F.R.C.P., Emma Footitt, Ph.D., Vijeya Ganesan, M.D., Stephanie Grunewald, M.D., Ph.D., Sandrine Compeyrot-Lacassagne, M.D., Francesco Muntoni, M.D., Clarissa Pilkington, M.B., B.S., Rosaline Quinlivan, M.D., Nikhil Thapar, M.D., Ph.D., Colin Wallis, M.D., Lucy R. Wedderburn, F.R.C.P., Ph.D., Austen Worth, M.D., Teofila Bueser, M.Sc., Cecilia Compton, M.Sc., Charu Deshpande, M.R.C.P.C.H., Hiva Fassihi, F.R.C.P., Eshika Haque, M.Sc., Louise Izatt, Ph.D., Dragana Josifova, M.D., Shehla Mohammed, F.R.C.P., Leema Robert, M.R.C.P.C.H., Sarah Rose, M.Sc., Deborah Ruddy, Ph.D., Robert Sarkany, F.R.C.P., Genevieve Say, M.Sc., Adam C. Shaw, M.D., Agata Wolejko, M.Sc., Bishoy Habib, B.Sc., Gavin Burns, Ph.D., Sarah Hunter, M.Sc., Russell J. Grocock, Ph.D., Sean J. Humphray, B.Sc., Peter N. Robinson, M.D., Melissa Haendel, Ph.D., Michael A. Simpson, Ph.D., Siddharth Banka, M.D., Ph.D., Jill Clayton-Smith, F.R.C.P., Sofia Douzgou, F.R.C.P., Ph.D., Georgina Hall, M.Sc., Huw B. Thomas, Ph.D., Raymond T. O’Keefe, Ph.D., Michel Michaelides, F.R.C.Ophth., Anthony T. Moore, F.R.C.Ophth., Sam Malka, B.Sc., Nikolas Pontikos, Ph.D., Andrew C. Browning, M.D., Ph.D., Volker Straub, M.D., Ph.D., Gráinne S. Gorman, F.R.C.P., Ph.D., Rita Horvath, M.D., Ph.D., Richard Quinton, M.D., Andrew M. Schaefer, M.R.C.P., Patrick Yu-Wai-Man, F.R.C.Ophth., Ph.D., Doug M. Turnbull, F.Med.Sci., F.R.S., Robert McFarland, M.R.C.P.C.H., Ph.D., Robert W. Taylor, F.R.C.Path., Ph.D., Emer O’Connor, M.D., Janice Yip, M.Res., Katrina Newland, M.Sc., Huw R. Morris, F.R.C.P., Ph.D., James Polke, F.R.C.Path., Ph.D., Nicholas W. Wood, Ph.D., F.Med.Sci., Carolyn Campbell, F.R.C.Path., Carme Camps, Ph.D., Kate Gibson, B.Sc., Nils Koelling, Ph.D., Tracy Lester, Ph.D., F.R.C.Path., Andrea H. Németh, F.R.C.P., D.Phil., Claire Palles, Ph.D., Smita Patel, F.R.C.P., F.R.C.Path., Ph.D., Noemi B.A. Roy, F.R.C.Path., D.Phil., Arjune Sen, M.R.C.P., Ph.D., John Taylor, Ph.D., Pilar Cacheiro, Ph.D., Julius O. Jacobsen, Ph.D., Eleanor G. Seaby, M.D., Val Davison, F.R.C.Path., Lyn Chitty, Ph.D., M.R.C.O.G., Angela Douglas, Ph.D., F.R.C.Path., Kikkeri Naresh, F.R.C.Path., Dom McMullan, Ph.D., F.R.C.Path., Sian Ellard, Ph.D., F.R.C.Path., I. Karen Temple, Ph.D., F.R.C.Path., Andrew D. Mumford, Ph.D., F.R.C.Path., Gill Wilson, F.R.C.P., Phil Beales, F.Med.Sci., Maria Bitner-Glindzicz, M.B., B.S., Ph.D. (deceased), Graeme Black, M.D., D.Phil., John R. Bradley, D.M., Paul Brennan, F.R.C.P., John Burn, M.B., B.S., Ph.D., Patrick F. Chinnery, F.Med.Sci., Perry Elliott, M.D., Frances Flinter, M.D., Henry Houlden, M.D., Melita Irving, M.D., William Newman, M.D., Ph.D., Shamima Rahman, F.R.C.P., F.R.C.P.C.H., Ph.D., John A. Sayer, M.B., Ch.B., Ph.D., Jenny C. Taylor, Ph.D., Andrew R. Webster, F.R.C.Ophth., Andrew O.M. Wilkie, F.Med.Sci., F.R.S., Willem H. Ouwehand, F.Med.Sci., F. Lucy Raymond, M.D., Ph.D., John Chisholm, F.R.Eng., Sue Hill, Ph.D., David Bentley, D.Phil., Richard H. Scott, M.D., Ph.D., Tom Fowler, Ph.D., Augusto Rendon, Ph.D., and Mark Caulfield, F.R.C.P., F.Med.Sci.
The authors’ affiliations are as follows: Genomics England (D.S., K.R.S., A.M., E.A.T., E.M.M., A.T., G.C., K.I., L.M., M. Wielscher, A.N., M. Bale, E.B., C.B., H.B., M. Bleda, A. Devereau, D.H., E. Haraldsdottir, Z.H., D.K., C. Patch, D.P., A.M., R. Sultana, M.R., A.L.T.T., C. Tregidgo, C. Turnbull, M. Welland, S. Wood, C.S., E.W., S.L., R.E.F., L.C.D., O.N., I.U.S.L., C.F.W., J.C., R.H.S., T.F., A.R., M.C.), the William Harvey Research Institute, Queen Mary University of London (D.S., K.R.S., V.C., A.T., L.M., M.R.B., D.K., S. Wood, P.C., J.O.J., T.F., M.C.), University College London (UCL) Institute of Ophthalmology (V.C., G.A., M.M., A.T.M., S. Malka, N.P., P.Y.-W.-M., A.R.W.), UCL Genetics Institute (V.C., N.W.W.), GOSgene (H.J.W.), Genetics and Genomic Medicine Programme (L.V., M.R., M.D., L.C., P. Beales, M.B.-G.), National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Centre (BRC) (M.R., S. Grunewald, S.C.-L., F.M., C. Pilkington, L.R.W., L.C., P. Beales, M.B.-G.), Infection, Immunity, and Inflammation Research and Teaching Department (P.A., L.R.W.), Stem Cells and Regenerative Medicine (N.T.), and Mitochondrial Research Group (S. Rahman), UCL Great Ormond Street Institute of Child Health, UCL Ear Institute (L.V.), the Department of Renal Medicine (D. Bockenhauer), and Institute of Cardiovascular Science (P.E.), UCL, Moorfields Eye Hospital National Health Service (NHS) Foundation Trust (V.C., G.A., M.M., A.T.M., S. Malka, N.P., A.R.W.), the National Hospital for Neurology and Neurosurgery (J.V., E.O., J.Y., K. Newland, H.R.M., J.P., N.W.W., H.H.), the Metabolic Unit (L.A., S. Grunewald, S. Rahman), London Centre for Paediatric Endocrinology and Diabetes (M.D.), and the Department of Gastroenterology (N.T.), Great Ormond Street Hospital for Children NHS Foundation Trust (L.V., D. Bockenhauer, A. Broomfield, M.A.C., T. Lam, E.F., V.G., S.C.-L., F.M., C. Pilkington, R. Quinlivan, C.W., L.R.W., A. Worth, L.C., P. Beales, M.B.-G., R.H.S.), the Clinical Genetics Department (M.R., T.B., C. Compton, C.D., E. Haque, L.I., D.J., S. Mohammed, L.R., S. Rose, D.R., G.S., A.C.S., F.F., M.I.) and St. John’s Institute of Dermatology (H.F., R. Sarkany), Guy’s and St. Thomas’ NHS Foundation Trust, the Division of Genetics and Epidemiology, Institute of Cancer Research (C. Turnbull), Florence Nightingale Faculty of Nursing, Midwifery, and Palliative Care (T.B.), Division of Genetics and Molecular Medicine (M.A.S.), and Division of Medical and Molecular Genetics (M.I.), King’s College London, NIHR BRC at Moorfields Eye Hospital (P.Y.-W.-M.), NHS England and NHS Improvement, Skipton House (V.D., A. Douglas, S. Hill), and Imperial College Healthcare NHS Trust, Hammersmith Hospital (K. Naresh), London, Open Targets and European Molecular Biology Laboratory–European Bioinformatics Institute, Wellcome Genome Campus, Hinxton (E.M.M.), the Division of Evolution and Genomic Sciences, Faculty of Biology, Medicine, and Health, University of Manchester (J.M.E., S.B., J.C.-S., S.D., G.H., H.B.T., R.T.O., G. Black, W.N.), and the Manchester Centre for Genomic Medicine, St. Mary’s Hospital, Manchester University NHS Foundation Trust (J.M.E., Z.H., S.B., J.C.-S., S.D., G.H., G. Black, W.N.), Manchester, the Department of Genetic and Genomic Medicine, Institute of Medical Genetics, Cardiff University, Cardiff (H.J.W.), the Department of Clinical Neurosciences (T.R., W.W., R.H., P.F.C.), the Medical Research Council (MRC) Mitochondrial Biology Unit (T.R., W.W., P.Y.-W.-M., P.F.C.), the Department of Paediatrics (T.R.), the Department of Haematology (K.S., C. Penkett, S. Gräf, R.M., W.H.O., A.R.), the School of Clinical Medicine (K.R., E.L., R.A.F., K.P., F.L.R.), the Department of Medicine (S. Gräf), and Cambridge Centre for Brain Repair, Department of Clinical Neurosciences (P.Y.-W.-M.), University of Cambridge, NIHR BioResource, Cambridge University Hospitals (K.S., S.A., R.J., C. Penkett, E.D., S. Gräf, R.M., M.K., J.R.B., P.F.C., W.H.O., F.L.R.), and Addenbrooke’s Hospital, Cambridge University Hospitals NHS Foundation Trust (G.F., P.T., O.S.-B., S. Halsall, K.P., A. Wagner, S.G.M., N.B., M.K.), Cambridge Biomedical Campus, Wellcome–MRC Institute of Metabolic Science and NIHR Cambridge BRC (M.G.), Congenica (A.H., H.S.), Illumina Cambridge (A. Wolejko, B.H., G. Burns, S. Hunter, R.J.G., S.J.H., D. Bentley), NHS Blood and Transplant (W.H.O.), and Wellcome Sanger Institute (W.H.O.), Cambridge, the Health Economics Research Centre (J. Buchanan, S. Wordsworth) and the Wellcome Centre for Human Genetics (C. Camps, J.C.T.), University of Oxford, NIHR Oxford BRC (J. Buchanan, S. Wordsworth, J.D., C. Crichton, J.W., K.W., C. Camps, S.P., N.B.A.R., A.S., J.T., J.C.T.), the Oxford Centre for Genomic Medicine (A. de Burca, A.H.N.), and the Departments of Haematology (N.B.A.R.) and Neurology (A.S.), Oxford University Hospitals NHS Foundation Trust, Oxford Genetics Laboratories, Oxford University Hospitals NHS Foundation Trust, Churchill Hospital (C. Campbell, K.G., T. Lester, J.T.), the MRC Weatherall Institute of Molecular Medicine (N.K., N.B.A.R., A.O.M.W.) and the Oxford Epilepsy Research Group (A.S.), Nuffield Department of Clinical Neurosciences (A.H.N.), University of Oxford, and the Department of Clinical Immunology (S.P.), John Radcliffe Hospital, Oxford, Peninsula Clinical Genetics Service, Royal Devon and Exeter NHS Foundation Trust (E.B.), and the University of Exeter Medical School (E.B., C.F.W.), Royal Devon and Exeter Hospital (S.E.), Exeter, Newcastle Eye Centre, Royal Victoria Infirmary (A.C.B.), the Institute of Genetic Medicine, Newcastle University, International Centre for Life (V.S., P. Brennan), Wellcome Centre for Mitochondrial Research, Translational and Clinical Research Institute, Faculty of Medical Sciences, Newcastle University (G.S.G., R.H., A.M.S., D.M.T., R. Quin-ton, R.M., R.W.T., J.A.S.), Highly Specialised Mitochondrial Service (G.S.G., A.M.S., D.M.T., R.M., R.W.T.) and Northern Genetics Service (J. Burn), Newcastle upon Tyne Hospitals NHS Foundation Trust (J.A.S.), and NIHR Newcastle BRC (G.S.G., D.M.T., J.A.S.), Newcastle upon Tyne, the Institute of Cancer and Genomic Sciences, Institute of Biomedical Research, University of Birmingham (C. Palles), and Birmingham Women’s Hospital (D.M.), Birmingham, the Genomic Informatics Group (E.G.S.), University Hospital South-ampton (I.K.T.), and the University of Southampton (I.K.T.), Southampton, Liverpool Women’s NHS Foundation Trust, Liverpool (A. Douglas), the School of Cellular and Molecular Medicine, University of Bristol, Bristol (A.D.M.), and Yorkshire and Humber, Sheffield Children’s Hospital, Sheffield (G.W.) — all in the United Kingdom; Fabric Genomics, Oakland (M. Babcock, M.G.R.), and the Oph-thalmology Department, University of California, San Francisco School of Medicine, San Francisco (A.T.M.) — both in California; the Jackson Laboratory for Genomic Medicine, Farmington, CT (P.N.R.); and the Center for Genome Research and Biocomputing, Environ-mental and Molecular Toxicology, Oregon State University, Corvallis (M.H.).
Footnotes
The authors’ full names, academic degrees, and affiliations are listed in the Appendix.
The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the National Institute for Health Research (NIHR), or the Department of Health and Social Care.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Generation Genome. Annual report of the chief medical officer. 2016. (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/631043/CMO_annual_report_generation_genome.pdf).
- 2.Ferreira CR. The burden of rare diseases. Am J Med Genet A 2019;179:885–92. [DOI] [PubMed] [Google Scholar]
- 3.Boycott KM, Rath A, Chong JX, et al. International cooperation to enable the diagnosis of all rare genetic diseases. Am J Hum Genet 2017;100:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet 2015;47:717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Splinter K, Adams DR, Bacino CA, et al. Effect of genetic diagnosis on patients with previously undiagnosed disease. N Engl J Med 2018;379:2131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomics England. The 100,000 Genomes Project protocol. 2017. (https://figshare.com/articles/journal_contribution/GenomicEnglandProtocol_pdf/4530893/4).
- 7.Köhler S, Carmody L, Vasilevsky N, et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 2019;47:D1018–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genomics England. Rare disease conditions clinical data models. 2018. (https://www.genomicsengland.co.uk/?wpdmdl=5500).
- 9.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimmer A, Phan H, Mathieson I, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014;46:912–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin AR, Williams E, Foulger RE, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet 2019;51:1560–5. [DOI] [PubMed] [Google Scholar]
- 12.Smedley D, Jacobsen JOB, Jäger M, et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc 2015;10:2004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Congenica home page (https://www.congenica.com/platform).
- 14.Fabric Genomics home page (https://fabricgenomics.com/).
- 15.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havrilla JM, Pedersen BS, Layer RM, Quinlan AR. A map of constrained coding regions in the human genome. Nat Genet 2019;51:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W, Tuna S, Keogh MJ, et al. Germline selection shapes human mitochondrial DNA diversity. Science 2019;364(6442):eaau6520. [DOI] [PubMed] [Google Scholar]
- 18.Smedley D, Schubach M, Jacobsen JOB, et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am J Hum Genet 2016;99:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res 2017;27:1895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Bai W, Yuan N, Du Z. Comprehensively benchmarking applications for detecting copy number variation. PLoS Comput Biol 2019;15(5):e1007069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol 2019;20:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genomics England. Rare disease conditions eligibility criteria. 2018. (https://www.genomicsengland.co.uk/wp-content/uploads/2018/06/Rare-Disease-Eligibility-Criteria-v1.9.0-PAR-GUI-058_approved-version-1.pdf).
- 23.Office for National Statistics. 2011. Census (https://www.ons.gov.uk/census/2011census).
- 24.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carss KJ, Arno G, Erwood M, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet 2017;100:75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowlands C, Thomas HB, Lord J, et al. Comparison of in silico strategies to prioritize rare genomic variants impacting RNA splicing for the diagnosis of genomic disorders. Sci Rep 2021;11:20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radziwon A, Arno G, Wheaton DK, et al. Single-base substitutions in the CHM promoter as a cause of choroideremia. Hum Mutat 2017;38:704–15. [DOI] [PubMed] [Google Scholar]
- 28.Farazi Fard MA, Rebelo AP, Buglo E, et al. Truncating mutations in UBAP1 cause hereditary spastic paraplegia. Am J Hum Genet 2019;104:767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallmeier J, Frank D, Shoemark A, et al. De novo mutations in FOXJ1 result in a motile ciliopathy with hydrocephalus and randomization of left/right body asymmetry. Am J Hum Genet 2019;105:1030–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortese A, Zhu Y, Rebelo AP, et al. Biallelic mutations in SORD cause a common and potentially treatable hereditary neuropathy with implications for diabetes. Nat Genet 2020;52:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med 2018;379:1452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Li N, Xu Y, et al. Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet Med 2018;20:1045–53. [DOI] [PubMed] [Google Scholar]
- 34.Vissers LELM, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med 2017;19:1055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344–7. [DOI] [PubMed] [Google Scholar]
- 36.Eichler EE. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med 2019;381:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NHS. National Genomic Test Directory. 2020. (https://www.england.nhs.uk/publication/national-genomic-test-directories/).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.