Abstract
Protein abundance affects the evolution of protein genotypes, but we do not know how it affects the evolution of protein phenotypes. Here we investigate the role of protein abundance on the evolvability of green fluorescent protein (GFP) towards the novel phenotype of cyan fluorescence. We evolve GFP in E.coli through multiple cycles of mutation and selection, and show that low GFP expression facilitates the evolution of cyan fluorescence. A computational model whose predictions we test experimentally helps explain why: Lowly expressed proteins are under stronger selection for proper folding, which facilitates their evolvability on short evolutionary time scales. The reason is that high fluorescence can be achieved by either few proteins that fold well, or by many proteins that fold less well. In other words, we observe a synergy between a protein’s scarcity and its stability. Because many proteins meet the essential requirements for this scarcity-stability synergy, it may be a widespread mechanism by which low expression helps proteins evolve new phenotypes and functions.
Introduction
Expression level, or abundance, is one of a protein’s most fundamental attributes. It varies by several orders of magnitude across the proteome, and systematically differs between functional classes of proteins1–3. The more abundant a protein is, the more likely it is to be essential, present in multiple tissues, and interacting with many other proteins4. Protein expression also plays an important role in protein evolution5–8. For example, natural selection stabilizes protein expression during evolution, such that protein abundance correlates phylogenetically across divergent taxa9. Abundance is also the strongest predictor of the rate at which protein genotypes (sequences) evolve. Specifically, sequences of highly expressed proteins evolve slowly4,10,11,12.
We know much about how protein abundance affects the evolution of protein genotypes but next to nothing about how it affects the evolution of protein phenotypes. Here we study the effect of protein abundance on protein evolvability – a protein’s ability to evolve a new phenotype. Existing pertinent evidence is indirect and mixed. On the one hand, sequence-based evidence suggests that highly expressed proteins may be less evolvable than lowly expressed proteins. For example, in both Drosophila melanogaster and Arabidopsis thaliana, highly expressed proteins experience fewer adaptive (beneficial) amino acid changes13,14. This may be a consequence of the generally slower sequence evolution of highly expressed proteins that is caused by misfolding toxicity14. However, such sequence-based evidence need not be relevant for the evolution of new phenotypes, because proteins can evolve novel phenotypes with very little sequence change15,16.
On the other hand, highly expressed proteins can be more stable compared to lowly expressed proteins17–19, and protein stability promotes protein evolvability20,21. It increases mutational robustness and enables proteins to accrue mutations that bring forth new phenotypes – neo-functionalizing mutations – which are often destabilizing22. For example, a cytochrome P450 enzyme that is engineered for greater stability is more likely to evolve the ability to catalyze novel chemical reactions20. Similarly, stabilizing amino acid changes in populations of the antibiotic resistance protein TEM-1 beta lactamase facilitate the evolution of resistance against the antibiotic cefotaxime5. This kind of evidence, however, supports a positive role for protein abundance only indirectly, via the effect abundance has on stabilizing mutations5,20.
Here, we provide direct experimental evidence for the role of protein abundance in protein evolvability. To this end, we performed laboratory evolution experiments to study how the abundance of green fluorescent protein (GFP) affects its evolution towards the novel phenotype of cyan fluorescence in E. coli. For these experiments, we deliberately chose a protein that is non-native to E.coli for two reasons. First, because the protein has not evolved to interact with the E.coli proteome, it allows us to study the evolvability of a single protein independently of interactions with other proteins. Second, because the protein is not essential for survival, it allowed us to help minimize the role of misfolding toxicity in our experiments (Table S1). Our results show that high expression diminishes evolvability of a protein, and it does so for reasons that are unrelated to the misfolding toxicity of highly expressed proteins. We demonstrate that under strong directional selection, the deleterious effects of a destabilizing mutation that reduces fluorescence intensity can be compensated for by increased protein abundance. As a result, proteins that are highly expressed allow more genetic variants that cause folding defects, which reduces their evolvability. To our knowledge, this is the first study that investigates the role of abundance in the evolution of a single protein towards a novel phenotype.
Results
Low protein expression promotes evolvability
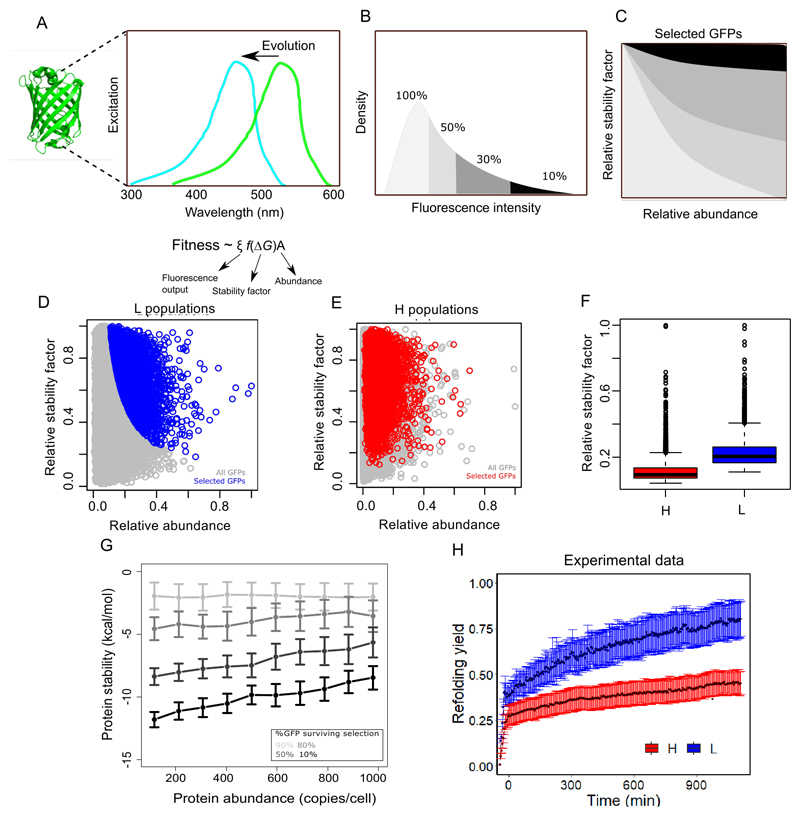
To study the influence of protein abundance on phenotypic evolution, we evolved populations of green fluorescent protein (GFP) in E.coli towards the new color phenotype of cyan fluorescence, as quantified by fluorescence emission in the AmCyan channel of an Aria III cell sorter (BD Biosciences, λex=405nm, λem=510±25nm, Figure 1A, Methods). During experimental evolution, we transcribed plasmid-encoded GFP either from the high expression acp promoter or the low expression arg promoter2,23. Protein expression driven by these promoters lies in a biologically sensible range2,24, and differs by approximately three-fold (Methods, Figure 1B). We performed four replicate evolution experiments for high and low GFP expression (8=4×2 replicates), and refer to the respective four replicate populations as H and L populations. For each population, we performed six rounds (‘generations’) of directed evolution. In each generation, we generated ~104-105 GFP variants through PCR-based mutagenesis, and used fluorescence-activated cell sorting (FACS) to select cells whose cyan fluorescence lay in the top 0.05% of the population (Figure1A, Methods).
Figure 1. Low GFP expression promotes evolution of a novel cyan fluorescence phenotype.
A. Experimental design. We evolved eight replicate populations (four at high and four at low GFP expression) for six rounds (‘generations’) of directed evolution, where mutagenesis alternated with selection in each generation. Specifically, we evolved green fluorescent protein towards cyan fluorescence, which requires a shift in excitation wavelength from 488 nm to 405 nm. B. Green fluorescence intensity for the three replicate ancestral populations that express GFP at a high (H) or a low (L) level (two-tailed unpaired t-test, n= 3, p= 0.0004, Cohen’s d=31.4±25.4, t=38.54, 95%CI= 609.8, 754.1, df= 2.11). C. Fold-change in cyan fluorescence compared to the ancestor after six generations of directed evolution in four H and four L populations (two-tailed unpaired t-test, n= 4, p= 0.0015, Cohen’s d=5.74±3.92, t=8.12, 95%CI= 9.2,19.1, df=3.79 ). D. Absolute cyan fluorescence after six generations in four H and four L populations (two-tailed unpaired t-test, n= 4, p= 0.002, Cohen’s d=3.74±2.88, t=38.54, 95%CI= 400.5, 1101.4, df=5.3). In panels B-D, red and blue represent the H and L populations, respectively. The boxes represent interquartile range while the solid line represents the median. The whiskers extend to the lowest and the highest value in the dataset. Each circle represents an ancestral (panel B) or evolved (panel C and D) population. ‘*’denotes a significant difference between H and L populations at p < 0.01 based on the unpaired t-test, Methods).
After six generations of evolution, cyan fluorescence had increased in both H and L populations (Figure 1C). However, L populations evolved significantly higher cyan fluorescence than H populations (p<0.001, unpaired t-test, Statistical analysis, Methods, Table S2, Figure 1C). By the end of the sixth generation, cyan fluorescence had increased up to ~17 fold in L populations, compared to only ~4-fold in H population (Figure 1C, Table S2). Remarkably, even though L populations had started out at ~3 fold lower absolute green fluorescence due to their lower GFP expression (Figure 1B), they evolved ~2.5 fold higher absolute cyan fluorescence than H populations (Figure 1D, p<0.001, linear mixed effects model, see Methods; Figure S1). In sum, low expression facilitated the evolution of a new color phenotype in GFP.
L populations harbor fewer non-synonymous and neo-functionalizing variants
We next wanted to identify the genetic basis of the higher evolvability in L populations. We first focused on beneficial non-synonymous mutations, i.e., mutations that change an amino acid of GFP and that improve cyan fluorescence. Some of these mutations may have been ‘neo-functionalizing’, i.e., they may have been responsible for the color-shift from green to cyan fluorescence. We hypothesized that GFP in L populations might have evolved faster if these populations accumulated more such variants. To find out, we used single-molecule real-time sequencing (SMRT) to genotype ~1000 to 4000 GFP molecules for each replicate population at every generation of directed evolution (Methods), and counted the average number of non-synonymous variants. To our surprise, not L but H populations accumulated significantly more non-synonymous variants (Figure 2A, p = 0.003, unpaired t-test, Methods).
Figure 2. L populations harbor fewer nonsynonymous and neo-functionalizing variants.
A. The average number of non-synonymous variants per GFP molecule is significantly lower in L (blue) than in H (red) populations (two-tailed unpaired t-test, n=6, p=0.003, Cohen’s d=2.95±1.87, t=5.12, 95%CI= 0.28, 0.86, df= 5Methods). B. The average number of neo-functionalizing variants per GFP molecule is significantly lower in L populations (blue) than in H (red) populations for six generations of directed evolution (two-tailed unpaired t-test, n=6, p=0.007, Cohen’s d=2.52±1.73, t=4.37, 95%CI= 0.096, 0.37, df= 5, Methods). Not all sequenced GFP molecules harbor the neo-functionalizing variant, which is why the average is lower than one in all experiments. In both panels each dashed line corresponds to data from one replicate population, and the solid line represents the mean of the four replicate populations.
To identify neo-functionalizing variants among the non-synonymous variants, we focused on variants that rose to a high frequency (f>0.4) in at least one H or L population, reasoning that variants responsible for a large increase in cyan fluorescence are likely to sweep to high frequency during evolution. There were nine such variants (A65S, D133N, S147P, K156R, K166N, S175G, T203S, A206T, and G232A). We engineered them individually into ancestral GFP populations, and analyzed their effects on green and cyan fluorescence when they are expressed in both the H and L genetic background (Methods). The increase in cyan fluorescence was not the same when a given variant was expressed in the H or L genetic background (Table S3). We suspect that the effects of expression level are not restricted to the reproductive fitness of an individual carrying the variant, as recently demonstrated25, but also extend to the fluorescence output for some variants. Despite these differences in the extent of the fluorescent increase, all nine variants exceptD133N in the H background increased the intensity of cyan fluorescence (Table S3). However, only two of these nine variants (A65S, and T203S) increased cyan fluorescence while significantly decreasing green fluorescence, due to an excitation shift from 405 nm to 488 nm (unpaired t-test p=0.02,0.006, 0.008,0.003 for T203S and A65S in H and L background respectively, Table S3, Figure S2). These were the only two neo-functionalizing variants. We expected them to rise to higher frequency in L populations, but to our surprise, the opposite was the case. They rose to higher frequencies in H populations (Figure 2B, p = 0.007, unpaired t-test, Methods). This observation shows that the greater cyan fluorescence of L populations is not caused by a higher frequency of neo-functionalizing mutations.
H populations evolve lower folding stability
Because our experiments showed that the greater evolvability of L populations is not caused by a greater accumulation of beneficial neo-functionalizing variants (Figure 2B), we next focused on deleterious variants. The most prominent class of such mutations impair protein folding20,26, and we hypothesized that such variants are retained when GFP is highly expressed. Because past work has linked high protein expression to protein misfolding toxicity, we first suspected that the cause may involve a general toxicity of misfolded GFP to E.coli host cells. However, we found that such toxicity does not play a role in our experiments, because the growth rates of H and L populations were indistinguishable during adaptive evolution towards cyan fluorescence (Table S1).
To identify a mechanism that may cause H populations to retain more destabilizing deleterious mutations than L populations, we developed a computational model for the directed evolution of GFP. The model’s structure and parameters are motivated by our experimental design (Supplementary Information). The model (Figure 3A) uses a ‘fitness’ function Fcell that represents the fluorescence intensity of a cell and relates it to the abundance of GFP and its biophysical properties. Specifically, Fcell is proportional to the fluorescence output ξ of GFP (i.e., the product of its quantum yield and its extinction coefficient), its folding stability (ΔG ), and its expression level (A), i.e.,
| (Equation 1) |
Figure 3. Modeled H populations retain more destabilizing mutations.
A. We simulated the evolution of GFP from green (excitation λ=488 nm) to cyan (excitation λ=4o5 nm) fluorescence, using a fitness function that relates the fluorescence intensity of GFP molecules to their fluorescence output (the product of extinction coefficient and quantum yield), their stability, and their abundance. B. We modeled selection by allowing a fixed percentile of fluorescent cells to survive, just as in our experiments. In this hypothetical example, the stringency of selection varies from 100% survival (light gray, no selection) to 10% survival (black). C. For GFP molecules to survive selection in the model, the product of the relative stability factor and abundance must exceed a minimum threshold that is determined by the strength of selection. In this schematic figure, the shade of gray corresponds to the percentile of cells that survive selection in panel B. We calculated relative protein stability and abundance by dividing their values by their respective maxima in the population. D. Relative stability factor versus relative abundance for GFPs prior to selection (in gray), and for GFPs surviving selection when only the top 10% of cells in terms of fluorescence intensity survived selection, for L populations (blue). E. As in D, but for H populations (red). F. The relative stability factor is significantly higher in L populations (blue) compared to H populations (red) (p ~10-16, two-sided Wilcoxon’s rank sum test, effect size calculated as Z/√N=0.93, n=1000 independently selected cells). The boxes in the box plots ranges from the first quantile to the third quantile. The median is represented by a line across the box. The whiskers represent 1.5 times of the interquartile range. The circles located above the top whisker are outliers whose values are higher than 1.5 times the interquartile range (third quartile – first quartile) above the first quartile. G. Theoretically predicted relationship between folding stability of GFP variants in evolving populations (vertical axis) and their abundance in simulations (horizontal axis). Gray color saturation is proportional to the strength of selection, ranging from a selection strength of 0.1 (90% of GFP population survives, light gray) to 0.9 (10% of GFP population survives, black). We selected cells from the replicates of our original population of 10,000 cells until we had collected n=10,000 survivors. H. Experimentally measured refolding yield (y-axis) during 1000 minutes (x-axis) for GFP variants in L and H populations after six generations of directed evolution. Black lines (composed of black circles) denote the average over four replicate populations, while colored error bars show one standard deviation. L populations show a significantly higher refolding yield than H populations (p ~ 0.0001; two-sided unpaired t-test, effect size calculated as Z/√N=1.23).
We refer to the function f(ΔG) as the ‘stability factor’. It relates the fluorescence intensity of individual GFP molecules to their folding stability, ΔG. It is motivated by experimental data27, has a sigmoidal form, and approaches one and zero for highly and lowly stable variants, respectively (Supplementary information, Equation S4). All three quantities (i.e., ξ, ΔG, and A) can change a cell’s fluorescence intensity. Variation in expression level A distinguishes our H and L populations. This expression level is subject to gene expression noise, which causes expression variation within a population, which we modeled through an empirically motivated log-normal distribution. Note that in our populations, ~105 GFP variants created by mutations are distributed among ~109 cells. This implies that at every instance, the fluorescence distribution of the 109 cells can be thought of as being composed of ~105 log-normal distributions. The ‘meta-distribution’ of these log-normal distributions is also log-normally distributed28 (Supplementary Information, Figure S13). Because our experimental design precluded the mutation of the GFP promoter, we did not allow mutations to change the distribution of expression level in the model. In contrast, because mutations can change both folding stability and fluorescence output, we allowed both properties to change in the model. We modeled selection by allowing a given top percentile of fluorescent cells to survive (Figure 3B), just as in our experiments. In other words, for a cell to survive, its relative fluorescence intensity – expressed as a percentage of the intensity of the highest-fluorescing cell in the population – must exceed this threshold. This also means that the product of the relative GFP stability factor and abundance must exceed this threshold (see Supplementary information for details). The stronger selection becomes, the larger the product of the relative stability factor and abundance that is necessary for a cell to survive selection (Figure 3C).
With this model, we asked whether destabilizing mutations can become enriched in H populations. To this end, we considered two populations of 10000 GFP expressing cells that differed in their average GFP expression like our experimental H and L populations (Supplementary information, section 3.5). We first made the simplifying assumption that each GFP variant in a population has the same fluorescence output. In other words, we assumed that most surviving GFPs under strong selection have at least one neo-functionalizing mutation that allowed them to emit cyan fluorescence. Under this assumption, any change in the value of cyan fluorescence is caused by changes in gene expression or by mutations that affect protein stability. In other words, the fluorescence intensity of different cells in the same population depends only on the product of GFP stability and abundance.
After one round of simulated mutagenesis (denoted by the gray circles in Figures 3D-E) we selected the top 10 percent of fluorescing cells (denoted by blue circles in Figure 3D and red circles Figure 3E) for L and H populations. Importantly, although the same percentile of fluorescent cells survived selection in both populations, the H population harbored more destabilizing mutations in the model (p < 10-16, Wilcoxon’s rank sum test; Figure 3F).
The reason is that although we selected the same percentile of fluorescent cells in both populations, the range of expression levels in surviving cells (from minimum to maximum) is broader in H populations than in L populations. This is mainly caused by the log-normality of the abundance distribution, whose skewness increases with its standard deviation. GFP abundance in H population has a higher standard deviation and is more left-skewed. Therefore, and because maximum fluorescence is more likely achieved by GFPs with high stability, the higher range of fluorescence in the H populations corresponds to a wider range of stabilities (Figure S14). Indeed, when we used a normal distribution to model protein abundance, GFP stability did not decrease with higher mean abundance (Supplementary information, Figure S9).
This observation, which rests on a single cycle of mutation and selection, extends to 10 cycles and to varying strengths of selection (Figure 3H, for simulation details see Supplementary information). In the absence of selection, evolution does not change the stability of GFP regardless of its abundance (Figure 3H). However, the stronger selection becomes, the more efficiently GFP variants with folding defects are purged. Most importantly, purging becomes less efficient as GFP abundance increases. In sum, when GFP is highly expressed during evolution, it retains more destabilizing mutations than when it is lowly expressed (Figure 3H).
To validate our theoretical results experimentally, we first asked whether H populations show a lower median abundance of GFP (relative to the maximal abundance in the same population). To find out, we compared the distribution of the relative fluorescence intensity of ancestral GFP in H and L populations. Because these populations are genetically homogeneous, any variation in fluorescence intensity within them must be caused by gene expression noise. Indeed, the fluorescence intensity of each population was log-normally distributed, but ancestral H populations displayed lower relative fluorescence intensities than ancestral L populations (p<10-16, Wilcoxon’s rank sum test, Supplementary information Figure S5).
Second and more importantly, we experimentally measured the folding stability of GFP variants in H and L populations at the end of the six generations of directed evolution. To this end we used a kinetic assay that quantifies the refolding yield of GFP populations upon combined thermal and urea-induced denaturation21. Previous experiments have shown that populations of fluorescent proteins with a higher fraction of folded and soluble proteins recover a higher percentage of their fluorescence upon thermal denaturation21. Based on our simulations, we hypothesized that the refolding yield of GFPs should be substantially higher in L populations than in H populations. This was indeed the case (Figure 3I, p<10-16; two-sample t-test, Methods). Specifically, in H populations the proportion of refolded proteins was ~0.4 ± 0.05, whereas in L populations this proportion was ~0.7 ± 0.08.
Robust H and L populations have similar evolvability
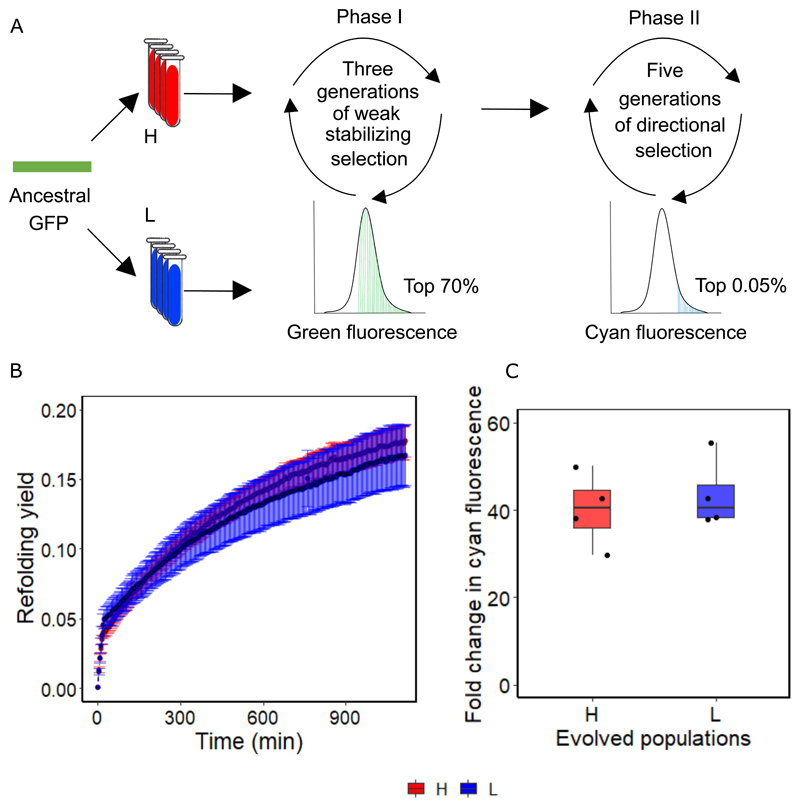
If destabilizing GFP variants impair the evolution of cyan fluorescence in H populations, removing such variants or increasing the robustness of evolving populations to such mutations might make the evolution of cyan fluorescence similar in both populations. One way to increase robustness is to subject evolving proteins to weak stabilizing selection to preserve their phenotype29,30. Under such selection, protein populations can accumulate mutations that enhance folding stability and neutralize the effect of destabilizing mutations. We therefore asked whether cyan fluorescence evolved similarly in H and L populations after subjecting both kinds of populations to weak stabilizing selection. Specifically, we performed a two-phase experiment that evolved four replicate H and L populations, first under weak stabilizing selection for green fluorescence (Figure 4A, Phase I), and then under directed evolution for cyan fluorescence (Figure 4A, Phase II).
Figure 4. Evolvability of GFP becomes indistinguishable in H and L populations when the starting population is robust to destabilizing variants.
A. We evolved eight replicate populations (four at high expression level, and four at the low expression level) in two phases. In the first phase we imposed weak stabilizing selection on green fluorescence, where we allowed the top 70% of green fluorescing cells to survive. This phase lasted three generations. In the second phase we imposed strong directional selection on cyan fluorescence, as in our main experiment (Figure 1A). B. Refolding yield measured over 1000 minutes for GFP in L and H populations after three generations of stabilizing selection. Black lines (composed of circles) denote the average refolding yield over four replicates, while error bars indicate one standard deviation of the mean. Stabilizing selection has rendered refolding yield statistically indistinguishable between H and L populations (p = 0.24, two-sample t-test, Methods). C. Fold change in cyan fluorescence, compared to ancestral GFP, after five generations of second phase of directed evolution in four H and four L populations. Stabilizing selection has rendered the fold change in cyan fluorescence statistically indistinguishable between L and H populations (two-tailed unpaired t-test, n=4, p = 0.58, Cohen’s d=0.41±1.75, t=-0.58, 95%CI= -17.89, 10.9, df= 5.98, Methods). The box represents the interquartile range, while the solid line represents the median. The whiskers extend to the lowest and the highest value in the dataset while each circle represents an evolved population. Red and blue represent H and L populations, respectively.
In phase I, which lasted for three generations, we allowed the top 70% of cells with the highest green fluorescence to survive in each generation (Figure 4A). Not unexpectedly, at the end of this phase neither green nor cyan fluorescence had changed significantly in either kind of population (unpaired t-tests p = 0.77 and p = 0.99 for green fluorescence, and p = 0.43 and p = 0.55 for cyan fluorescence of H and L populations respectively, Figure S3). To find out whether phase I had enriched our populations with foldability-enhancing mutations, we quantified the refolding yield of the H and L populations at the end of phase I15. Indeed, in contrast to our first directed evolution experiment, where H populations were less foldable (Figure 3I), phase I had rendered the refolding yield between H and L populations indistinguishable (Figure 4B). This suggests that foldability-enhancing mutations accumulated, and especially so in H populations, which had been more sensitive to foldability defects in our first directed evolution experiment.
Before continuing to phase II, we used single-molecule real-time sequencing (SMRT) to find out whether known mutations that improve folding stability had occurred in our populations. To this end, we genotyped ~1000 to 4000 evolved variants for each replicate population at the end of phase I. We found that no single variant had risen to a frequency f exceeding 0.1 in any H or L population, which is expected given the weak selection pressure of phase I. Nonetheless, those 20 variants that had reached the highest frequency – the top 20 variants – were strikingly similar between all H and L populations. First, all 160 of these variants (20 variants in each of eight populations) affected only 41 different amino acid positions. Second, the same six amino acid positions, namely, 99,101, 156, 162,166, and 238 were mutated in the top 20 variants for all H and L populations. Genetic parallelism of this kind has been observed before when foldability-enhancing mutations rose to appreciable frequency after weak selection21. Third, several amino acid positions mutated in the top 20 variants have been shown to increase fluorescence intensity by improving the foldability of GFP. For instance, mutations at positions 99 (mutated in all H and L populations), 153 and 163 (mutated in two H populations each) occur in a known GFP mutant with improved foldability31,32. Similarly, mutations at position 166 (occurring in all H and L populations) and position 64 (occurring in two L populations) can increase fluorescence of GFP up to 28 fold by improving foldability33. Furthermore, mutations at position 238 (mutated in all H and L populations) and 212 have also been implicated in improved foldability of GFP34. In sum, multiple mutations that become frequent after phase I in both H and L populations are known to increase the folding stability of GFP.
We next performed phase II directed evolution (Figure 4A) to test our central hypothesis that cyan fluorescence is less evolvable in H populations, because such populations are more susceptible to folding defects (Figure 3D&3E). After phase I evolution had enriched L and H populations with foldability-enhancing mutations and rendered their GFP molecules similar in their folding stability (Figure 4B), they should also evolve similar levels of cyan fluorescence. To find out, we used the H and L populations at the end of phase I as the starting populations for five rounds of directed evolution towards cyan fluorescence (Figure 4A, henceforth ‘phase II’). Just as in our main experiment (Figure 1A), we selected cells for survival whose cyan fluorescence lay in the top 0.05% of the population. At the end of phase II, the increase in cyan fluorescence intensities of L and H populations was identical (Figure 4C, p = 0.58, unpaired t-test). Importantly, the increase in cyan fluorescence was significantly greater in both H and L populations (~40 fold for H and ~43 fold for L) than in our main experiment (~3 fold for H and ~17 fold for L), further supporting the importance of stabilizing mutations for adaptive evolution20,22,29. Overall these results show that the evolvability of cyan fluorescence in H and L populations becomes comparable after stabilizing selection helps buffer the effect of destabilizing mutations. This reinforces our major observation that increased GFP expression in H populations allows the retention of GFP variants with folding defects, and hence inhibits the evolvability of cyan fluorescence in these populations.
Discussion
We evolved populations of green fluorescent protein (GFP) with high (H) and low (L) protein expression towards the novel phenotype of cyan fluorescence. Even though H populations accumulated more non-synonymous mutations, their high GFP expression hindered the evolution of cyan fluorescence. The reason was that highly expressed GFP populations retained more deleterious mutations that caused protein misfolding. High expression also resulted in a greater accumulation of neo-functionalizing mutations, which are often destabilizing. We theoretically predicted (Figure 3D-E) that high expression can help proteins with folding defects survive selection that would otherwise eliminate them from a population. The accumulation of such folding defects reduces a protein’s ability to evolve a new phenotype. Our experimental results support this prediction (Figure 3I). We further showed that weak stabilizing selection can help mitigate this problem by helping foldability-increasing mutations to accumulate, which eliminates the evolvability disadvantage of highly expressed proteins (Figure 4c).
Our results pertain to the role of protein abundance in the evolvability of protein phenotypes on short evolutionary time scales. We emphasize that our observations are independent from and do not contradict the misfolding avoidance hypothesis, which can help explain why the sequences of highly expressed proteins evolve slowly on long evolutionary time scales. This hypothesis posits that highly expressed proteins impose a fitness cost on a cell when misfolded, because protein misfolding can be toxic, and especially so for highly expressed proteins. Destabilizing mutations in such proteins are thus rarely tolerated, which leads to low rates of amino acid sequence evolution on longer evolutionary time scales. The selection pressure that causes this effect may be too weak to manifest itself on short evolutionary time scales. Misfolded proteins can be costly to a cell when present in large numbers and reduce the reproductive fitness of the cell. The resulting misfolding toxicity does not play a major role in our experiments, because first, the amount of GFP expressed in H and L populations is not very high (see Methods for details of expression levels). Second and more important, the growth rates of H and L populations are indistinguishable throughout our evolution experiment (Table S1). In other proteins or experiments where misfolding toxicity plays a role, it may cause an additional evolvability disadvantage for highly expressed proteins.
Protein sequence evolution is most commonly studied by comparing hundreds of protein orthologs whose sequence differences have accumulated over millions of years 4,35,36. Very few studies have demonstrated the influence of gene expression level on short-term protein evolution37. Our work shows that protein abundance can influence not only the rate of sequence evolution (Figure 2A) but also the rate of evolution of a new protein phenotype, and that it can do so even on the short time scale of laboratory evolution.
The mechanism we identified for the high evolvability of lowly expressed proteins relies on a synergy between protein scarcity and stability (SSS), in the sense that low protein abundance favors stable proteins (which in turn facilitate evolvability)21. The reason is that fluorescence intensity, our focal phenotype, depends on the product of protein abundance and stability. In consequence, mutations can reduce protein stability to some extent, as long as protein abundance can compensate for the reduced stability. This is why the stability of GFP evolving in H populations can decrease to a greater extent than in L populations. Because the ability to compensate for reduced folding stability is smaller in L populations, these populations evolve genotypes with higher folding stability. The phenomenon resembles the evolution of drift-robustness, in which genotypes from small populations can evolve reduced vulnerability to genetic drift, and become less likely to accumulate small-effect deleterious mutations38.
Scarcity-stability synergy requires two further conditions to be met. The first is that directional selection must be strong, otherwise high protein abundance will not be able to compensate effectively for reduced stability (Figure 3H). Although we use threshold-based (truncation) selection in our experiments, we emphasize that a synergy between scarcity and stability can also manifest itself during other kinds of selection, as long as selection is strong. For instance, SSS also causes low evolvability for highly expressed proteins under a probabilistic form of selection, i.e., when a cell’s probability of surviving selection decreases with decreasing fluorescence (Supplementary information, Figure S11). In fact, the mere presence of gene expression noise effectively causes selection to be probabilistic. For example, in our experiments selection acted on subpopulations of ~104 genetically identical cells for each of ~105 different variants created by mutation. Each of the genetically identical cell is subject to gene expression noise, which causes different cells to express different amounts of the same protein, such that their fluorescence varies probabilistically. In other words, the population of individuals surviving selection is sampled from a meta-distribution made up of ~105 individual distributions. Cells in a subpopulation with higher mean fluorescence also have a higher probability of surviving selection, which renders selection probabilistic.
The second requirement is a heavy-tailed distribution of protein abundance per cell, which is known to be caused by gene expression noise1,2,39. In the absence of gene expression noise, scarcity-stability synergy does not lead to higher evolvability at low expression (Supplementary information, Figure S8). Also, gene expression noise must cause a heavy-tailed abundance distribution, i.e., a distribution in which a greater proportion of proteins show high abundance than in a normal distribution (Figure S9). Our model uses the log-normal distribution, a specific heavy-tailed distribution that reflects the empirically observed GFP abundance distributions for our H and L populations. However, other heavy-tailed distributions, such as a gamma distribution, also lead to a preferential accumulation of destabilizing mutations in H populations (Supplementary information, Figure S10). We note that most proteins from organisms as different as E. coli, yeast, and humans show a noise-induced heavy-tailed abundance distribution 40–44.
More generally, our observations contribute to a growing literature on the importance of gene expression noise for adaptive evolution45–47. First, gene expression noise can promote the fixation of beneficial mutations, particularly in fluctuating environments46,47. Second, expression noise can also enhance the adaptive value of beneficial mutations through a synergism between cell-to-cell expression variation and genetic variation45. A complication in distinguishing the role of gene expression noise from that of average expression is that the two are correlated – low mean gene expression entails greater expression noise45. Our results suggest that whenever gene expression noise facilitates adaptive evolution, the lower average expression of evolving proteins may be part of the reason. To disentangle the role of mean expression from that of noise remains an important task for future work.
Scarcity-stability synergy is likely to be important far beyond GFP, because it exists wherever abundance and stability contribute multiplicatively to a protein phenotype48–50. This multiplicative relationship has been shown to accurately predict the changes in bacterial growth rate with different orthologs of essential proteins, such as dihydrofolate reductase (DHFR)8, as well as resistance conferring enzymes, such as β-lactamase6.The additional requirements of a heavy-tailed protein abundance distribution and strong selection also apply to a broad range of proteins and organisms, both during experimental evolution and in the wild 21,40–44,51–53. In consequence, scarcity-stability synergy may be widespread in facilitating the evolution of new phenotypes and functions in lowly expressed proteins.
Methods
Strains, promoters, and the green fluorescent protein gene
We performed all our experiments in E. coli strain K12 MG1655. We started from a previously characterized2,24 library (Dharmacon, GE Healthcare) of 1800 E. coli promoters expressed from a low copy plasmid (Figure S4) in this strain. Each plasmid in the library contains a unique promoter region from E. coli, which constitutively drives the expression of the GFPmut2 protein, a variant of green fluorescent protein from jellyfish Aequorea aequorea54. This variant harbors three substitutions (S65A, V68L, and S72A) that result in a 100-fold increase in fluorescence. Their increased brightness results from efficient folding and from a shift in the excitation maximum from 395nm to 481nm55. We used this protein, henceforth ‘GFP’, as the starting point for all our directed evolution experiments.
From the promoter library, we chose two promoters that differed in their mean expression for our experiments. Specifically, we chose for high expression the promoter of the acpP gene, which encodes the acyl carrier protein. And we chose for low expression the promoter of argW, which encodes arginine tRNA synthase23. The two promoters differ ~3 fold in their expression level, with a less than two-fold difference in the standard deviation of expression (Table S4).
Construction of expression plasmids
We modified the two plasmids from the promoter library that carried the acpP and argW promoters by altering the upstream XbaI site of the GFP gene in both plasmids using a primer with wobble nucleotides (Table S5, “for construction of expression plasmid”). For this purpose, we amplified the GFP-coding gene together with the unique BamHI (upstream) and XbaI (downstream) restriction sites. We used Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo#F549L) to minimize copying errors during the PCR. We ligated the amplified PCR product into the BamHI-XbaI double digested plasmid backbone by using T4 DNA ligase (NEB#M0202). Figure S4 shows the relevant regions of the modified plasmid.
After electro-transforming the ligation products into E.coli competent cells, we Sanger-sequenced several of the resulting clones, and chose a correctly constructed plasmid for subsequent experiments. We measured the mean GFP expression for the newly constructed plasmids on a Fortessa cell analyzer (BD Biosciences, see ‘Fluorescence assay using flow cytometry’ for details), and found that the acpP promoter drove 2.9-fold higher expression than the argW promoter (Table S4). We split the bacterial cultures with the two ancestral plasmids to create four replicate populations, each driving GFP at either high or low expression.
Preparation of electro-competent cells for transformation
We prepared electro-competent cells using glycerol/mannitol step centrifugation56. Specifically, we cultured E. coli MG1655 cells in 5 ml SOB medium at 37°C in a shaking incubator (INFORS HT, Switzerland) at 220 rpm overnight. After overnight growth, we transferred 3 ml of culture into 300 ml SOB medium, and incubated at 37°C and 220 rpm until the culture’s OD600 had reached a value between 0.4 and 0.6 (optical path length: 1 cm, 2-4 hours). We then kept the culture on ice for 15-20 min and collected cells at 4°C by centrifuging them at 1,500 g for 15 min (Eppendorf 5810/5810 R). We re-suspended the cells in 60 ml ice-cold ddH2O and split the culture into 10 ml aliquots in 50 ml tubes. With a 10 ml pipette, we carefully added 10 ml ice-cold glycerol/mannitol solution (20% glycerol (w/v) and 1.5% mannitol (w/v)) to the bottom of every 50 ml tube. We centrifuged the tubes at 1,500 g and 4°C for 15 min without acceleration or deceleration. We then discarded the supernatant and re-suspended cells in 3.0 ml ice-cold glycerol/mannitol solution. We distributed 100 μl aliquots of the resulting suspension into pre-cooled 1.5 ml tubes, flash-froze the aliquots in liquid nitrogen, and stored them at -80°C for transformation experiments.
Electro-transformation
We mixed 10 μl of ligation product with 100 μl of electro-competent E. coli MG1655 cells, and transferred them to a 0.2 cm electroporation cuvette (EP202, Cell Projects, UK). For transformation, we provided a 15k V/cm pulse using a Micropulser electroporator (Bio-Rad). We then immediately added 1 ml pre-warmed SOC medium and transferred the culture into a 10 ml tube. Subsequently, we incubated the culture for 1.5 h at 37°C in a shaking incubator (INFORS HT, Switzerland) at 220 rpm, and used the recovered transformants for the following experiments.
PCR mutagenesis
We used the Agilent Genemorph II mutagenesis kit to introduce random mutations into the coding region of our GFP gene. To achieve a low mutation rate (2 to 3 mutations per GFP molecule and generation), we used two-step PCRs. Specifically, we mixed 5 μl of Mutazyme buffer II, 1 μl of 40 mM dNTPs, 1 μl each of 10 μM forward and reverse primer (see Table S5, ‘for mutagenesis’), 1 μl of Mutazyme II, and 40 ng of template in a 25 μl PCR reaction volume. We used the following conditions to execute the PCR reaction: 95°C/2 min; 5 cycles of 95°C/30 s, 50°C/30 s, and 72°C/1 min; 72°C/10 min. We mixed 4 μl of the resulting PCR product with 5 μl of polymerase buffer, 2 μl of 10mM dNTP, 1 μl each of 10 μM forward and reverse primer (Table S5, ‘for mutagenesis’), and 0.25 μl of Taq polymerase (NEB#M0273S) in a 50 μl volume. We performed the PCR reaction as follows: 95°C/30 sec; 25 cycles of 95°C/20 s, 50°C/30 s, and 68°C/1 min; 68°C/2 min.
We purified the PCR products with a Qiagen PCR purification kit (Qiagen#28104), and double-digested the purified products overnight at 37°C with BamHI (NEB#R3136S) and XbaI (NEB#R0145S). We used DpnI (NEB#0176S) to digest the plasmid template. We purified the digested inserts using a PCR purification kit and verified sample purity with agarose gel electrophoresis. We used BamHI and XbaI to digest the plasmid backbone, and purified it using a QIAquick gel extraction kit. We then mixed the purified insert, the purified plasmid backbone (BamHI and XbaI), and T4 DNA ligase in a 20 μl reaction, and incubated it at 15°C overnight for ligation. To purify the ligation product, we precipitated it by mixing it with 1 µl of glycogen (Thermo#R0561), 50 µl of 7.5 M ammonium acetate (Sigma), 375 µl of ice-cold absolute ethanol, and 80 µl of ddH2O. After incubation at -20°C for 20 min, we centrifuged the mixture at 18,000 g for 20 min (Eppendorf 5810/5810 R). We then washed the precipitate twice using 800 µl of 70% cold ethanol. After drying the precipitate using a concentrator (Eppendorf 5301), we dissolved it in 10 µl of ddH2O, and used this purified ligation product for transformation.
Selection of GFP for cyan fluorescence
We implemented stringent selection for high fluorescence via two consecutive steps of cell sorting, in which only 0.05% of all cells survived selection in every generation.
After transforming E.coli cells with a mutagenized library of GFP-encoding plasmids, we incubated the transformants at 37 °C and 220 rpm for 1.5 h. We then sampled 10 µl of the recovered transformants, and diluted the culture with saline to determine the library size by plating aliquots on low salt LB agar plates (10g tryptone, 5g NaCl, and 5g yeast extract, 25g agar in 1l of water) containing 25 μg/ml kanamycin. In every generation of directed evolution, and for every replicate population, our libraries comprised 104-105 cells. We added 10 ml low salt LB medium with 25 μg/ml kanamycin to the rest of the recovered transformants (in ~900 μl culture) and incubated the culture overnight at 37°C with shaking at 220 rpm (INFORS HT, Switzerland). We added 100 μl of this overnight culture to 10 ml of M9 minimal medium supplemented with 25 μg/ml kanamycin and 0.2% arabinose, and incubated at 37°C with shaking at 220 rpm. After 22 hours of incubation, we centrifuged the culture at 8000g and 4°C for 3 minutes (Eppendorf 5810/5810 R). After discarding the supernatant we washed the cells twice with 2 ml PBS (Sigma#A9226) and re-suspended the washed cells in 2 ml cold PBS for fluorescence assisted cell sorting (FACS).
We sorted cells at 4°C on an Aria III cell sorter (BD Biosciences), using the AmCy channel (λex=405nm, λem=510/50nm), and a sorting speed of ~3×104 events/s. We used the precision of 4-Way Purity to minimize contaminating particles. We collected 50,000 cells with fluorescence in the top one percent in 500 μl cold PBS for each replicate population. To minimize cell proliferation or cell death during sorting we kept each sample on ice until all samples had been processed. After sorting, we added 1 ml of low salt LB medium and allowed the cells to recover at 37 °C for one hour in a shaking incubator at 220 rpm. Thereafter we plated 5 μl of culture on low salt LB agar plates containing kanamycin to estimate the library size after sorting. We then added 5 ml of low salt LB medium with 25 μg/ml kanamycin to the rest of the culture and incubated at 37 °C overnight at 220 rpm. We inoculated 100 μl of this overnight culture in 10 ml of M9 minimal medium supplemented with 0.2% arabinose and 25 μg/ml kanamycin, and incubated overnight at 37°C with shaking at 220 rpm.
We followed the same procedure as described above to re-sort the re-grown cells, except that we selected the top five percent of all cells in the AmCy channel. We stored part of the culture as a glycerol stock, and used the remaining culture for plasmid extraction using the QIAprep spin miniprep kit (Qiagen#27104). We used the extracted plasmids as templates for the next generation of directed evolution and SMRT sequencing.
The green (ancestral) and cyan (new) fluorescence spectra overlap to some degree but the excitation maxima are more than 80 nm apart. More importantly, however, this overlap does not affect the selection of GFP variants with neo-functionalizing mutations in the evolving GFP populations (Figure 2B). Correlations between two traits are the rule rather than the exception in evolving proteins, because ancestral and derived phenotypes are correlated in many proteins. That is, most novel protein functions are initially correlated with an ancestral function, and diverge only later through mechanisms such as gene duplication57,58.
Fluorescence assay using flow cytometry
We inoculated 100 μl of glycerol stock for every replicate population of every generation into 5 ml of M9 minimal medium supplemented with 0.2% arabinose and kanamycin. We incubated these cells at 37°C with shaking at 220 rpm for 20 hours (INFORS HT, Switzerland). We mixed 50 μl of the resulting overnight cultures thoroughly with 450 μl of PBS, and measured green (FITC channel, λex=488nm, λem=530/30nm) as well as cyan (AmCyan channel λex=405nm, λem=525/50nm) fluorescence. For this purpose, we used the Fortessa cell analyzer (BD Biosciences) at room temperature with a flow rate of ~10000 events/s. For each sample, we collected data from 105 cells.
Flow cytometry data analysis
We used FlowJo V10.4.2 (LLC) to analyze flow cytometry data. We used all 105 cells to measure the mean and variance of green and cyan fluorescence, and determined the fold change in fluorescence for every replicate population relative to its ancestral population. We used cells with plasmids encoding GFP but lacking a promoter as a negative control.
Statistical analysis
We used unpaired t-tests to compare ancestral green fluorescence (Figure 1B), fold change in cyan fluorescence (Figure 1C, 4C and S3B), absolute cyan fluorescence (Figure 1D),fold change in green fluorescence (Figure S3A), and refolding yield (Figure 3I and 4C) between H and L populations.
For the accumulation of nonsynonymous and neo-functionalizing mutations (Figure 2A and 2B) we performed an unpaired t-test where we compared the difference between L and H population in each generation, with the null expectation of no difference (mean = 0). We took this approach to avoid the routine correction for multiple testing, which considers the sample size to be 4 (for four replicate populations), and ignores that the average number of nonsynonymous and neo-functionalizing mutations are determined from ~1000-4000 different reads in each population.
We performed a Wilcoxon signed rank test to ask whether refolding yields differed significantly between H and L populations after six generations of directed evolution (Figure 3I), and after 3 generations of weak stabilizing selection (Figure 4B).
For statistical analysis of data from all rounds of directed evolution (Figure S1), we fitted a general linear model (LLM), and tested the significance of each factor with an ANOVA. We then performed Tukey-HSD (Tukey-honest significant difference) to determine the differences between relevant pairs.
We performed all statistical analyses with R software (v3.5.2).
SMRT sequencing
To prepare DNA for single molecule real-time (SMRT) sequencing59, we barcoded DNA from every replicate population (four H and four L populations) from each generation of our directed evolution experiment with a unique barcode, and pooled all barcoded samples to create a single library for sequencing. We chose the barcodes from a list of 384 barcodes recommended by Pacific BioSciences for the Sequel system. To create this library, we used a two PCR approach, where the first PCR adds a universal sequence at both ends of the target DNA. The universal sequence allows one to create many unique barcode combinations with few barcoded primers. The second PCR adds a unique barcode combination downstream of the universal sequence for DNA extracted from each of our experimental populations.
For the first PCR, we mixed 0.3 μl of Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo), 6 μl of the GC buffer, 0.6 μl of 10 mM dNTP, 1.5 μl of 10 μM forward and reverse primers each (Table S5, ‘for attaching universal sequence’) with 2 ng of template in a 30 μl reaction. In this PCR, we used the following PCR conditions: 98°C/30 s; 15 cycles of 98°C/10 s, 63.2°C/15 s and 72°C/30s; 72°C/5 min. We treated the PCR product with a mix of 0.25 μl of DpnI, 0.25 μl ExoI (NEB#M0568), and 3 μl of cutsmart buffer at 37°C for 1 hour, and then deactivated the enzymes at 85°C for 20 min. We used 2 μl of this product as a template for the second PCR, mixing it with 0.5μl of Phusion Hot Start II High-Fidelity DNA Polymerase, 10 μl of the GC buffer, 1 μl of 10 mM dNTP, 2.5 μl of 10 μM forward and reverse primers each (Table S5, ‘primers with unique barcodes’) in a 50 μl reaction. In this second PCR, we used the following PCR conditions: 98°C/30 s; 30 cycles of 98°C/10 s, 71.2°C/15 s and 72°C/30s; 72°C/5 min. We purified the resulting PCR product using a PCR purification kit, and checked the purity using agarose gel electrophoresis. We measured the concentration of purified products using a Qubit fluorometer (Thermo Fisher Scientific). We submitted the library of pooled barcoded samples to the Functional Genomics Center Zurich, where it was purified with AMPure beads, ligated with appropriate adapters, and sequenced on a single cell of the PacBio Sequel machine using P6/C4 chemistry.
Primary data analysis
We analyzed SMRT sequence data using SMRTLink, a web-based end-to-end workflow manager for PacBio Sequel Systems, and SMRT Tools, a suite of command-line tools included with SMRTLink. We chose only those circular consensus sequence (CCS) reads that resulted from three complete passes of DNA, and that had a predicted accuracy of 99 percent. We also discarded any CCS reads that were shorter than 850 base pairs to exclude partial GFP sequences. We then de-multiplexed the CCS reads with a barcode score of 80 or higher. To this end, we used the 384 Sequel barcodes provided by PacBio and searched for all possible barcode combinations in our sequence data. We could accurately identify the 136 barcode combinations that we had used, out of more than seventy thousand possible combinations. We mapped the de-multiplexed reads using the SMRT tool ‘pbalign’, which uses the ‘blasr’ algorithm60. We restricted the minimum mapping length to at least 900 base pairs, the maximum possible divergence from the ancestral sequence to 75%, and the minimum mapping accuracy to 90%. Through this procedure, we recovered 819 to 4241 reads for each population.
We then used SAMtools to convert the mapped sam files to bam format61. We used custom Python scripts and MEGA software (Molecular Evolutionary Genetics Analysis, v10.0.5)62 for all further data analysis.
Identification of SNPs
We sequenced two clones of non-mutated ancestral GFP using SMRT sequencing. The results confirmed the well-known fact that single nucleotide indels are the most common errors in SMRT sequencing30. Because most indels are sequencing artifacts, we considered only point mutations for the rest of our analysis. We restricted our analysis to changes at the amino acid level, because including silent mutations can result in overestimating the number of mutations that affect a phenotype. We further analyzed those amino acid changing mutations that reached a frequency exceeding 0.1 at the end of directed evolution30.
Identification of neo-functionalizing mutations
We used whole-plasmid PCR to engineer single mutants by designing primers that carry the corresponding mutations (Table S5). We used Phusion Hot Start II High-Fidelity DNA Polymerase to minimize copying errors during the PCR. After electro-transforming the ligation products into E.coli competent cells, we Sanger-sequenced several of the resulting clones, and chose a correctly constructed plasmid for each mutant. We measured the mean green and cyan fluorescence for these mutants on a Fortessa cell analyzer (BD Biosciences, see ‘Fluorescence assay using flow cytometry’ for details). We then used the two-fluorescence plots (cyan fluorescence vs green fluorescence) for every mutant, and contrasted them with those of the ancestral populations to identify neo-functionalizing mutations (Figure S2) that shift the fluorescence distribution towards the y-axis. In contrast, folding stability improving mutations shift the fluorescence distribution along the diagonal of the plot, indicating an increase in both green and cyan fluorescence (Figure S2).
Refolding kinetics in selected H and L populations
We grew H and L populations at the end of directed evolution in 8 ml low salt LB with 25 μg/ml of kanamycin in a 10 ml tube shaken at 37 °C and 220 rpm for 18 h. We used CelLytic™ B Cell Lysis Reagent (B7435-10 500ml, Sigma) to extract soluble proteins from the collected cells by following the manufacturer’s protocol. We studied the refolding kinetics during five hours by measuring cyan fluorescence (λex=405nm, λem=510/50nm) using a plate reader (Tecan Spark M2)21.
Stabilizing selection on green fluorescence
To allow GFP-coding genes to accumulate genetic variation before evolving them towards high cyan fluorescence, we performed three generations of directed evolution to maintain the ancestral green fluorescence phenotype. In each generation we performed stabilizing selection on green fluorescence via two consecutive steps of cell sorting. In each step we selected the top 70% of green-fluorescing cells (FITC channel with λex=488nm, λem=530/30nm). We used the same mutagenesis and library preparation protocols as for our main directed evolution experiment (see Methods, ‘PCR mutagenesis’ and ‘Selection of GFP for cyan fluorescence’).
We used the populations from the third generation of this experiment as starting points for directed evolution towards cyan fluorescence. For this second phase of directed evolution, we employed the same protocols as for the main experiment in which we evolved cyan from green fluorescence (see Methods ‘PCR mutagenesis’ and ‘Selection of GFP for cyan fluorescence’), except that we evolved populations for five instead of six generations.
Supplementary Material
Acknowledgements
This project has received funding from the European Research Council under Grant Agreement No. 739874. We would also like to acknowledge support by Swiss National Science Foundation grant 31003A_172887, by the University Priority Research Program in Evolutionary Biology, as well as by the flow cytometry facility and the functional genomics center at the University of Zurich. S.K. thanks Bharat Ravi Iyengar and Miriam Olombrada for discussions and support.
Footnotes
Author contributions
S.K., P.D. and A.W. were involved in the conceptualization of the study. S.K. and P.D. performed the experiments. P.D. performed computational modeling and formulated theoretical predictions. J.Z. provided the resources and training for working with fluorescent proteins. S.K., P.D., J.Z. and A.W. contributed to data analysis and edited the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
All data are available in the manuscript or the supplementary materials. SMRT sequencing data are available at NCBI with a BioProject ID PRJNA833567 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA833567).
Code availability
Custom code used in this study is available in a public GitHub repository (https://github.com/dasmeh/Discrete_Time_Markov_Chain_Evolution).
References
- 1.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 2.Silander OK, et al. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLoS Genet. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Molecular systems biology. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nature Reviews Genetics. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 5.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. Journal of molecular biology. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Socha RD, Chen J, Tokuriki N. The molecular mechanisms underlying hidden phenotypic variation among metallo-β-lactamases. Journal of molecular biology. 2019;431:1172–1185. doi: 10.1016/j.jmb.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 7.Dasmeh P, Serohijos AW. Estimating the contribution of folding stability to nonspecific epistasis in protein evolution. Proteins: Structure, Function, Bioinformatics. 2018;86:1242–1250. doi: 10.1002/prot.25588. [DOI] [PubMed] [Google Scholar]
- 8.Bershtein S, et al. Protein homeostasis imposes a barrier on functional integration of horizontally transferred genes in bacteria. PLoS Genet. 2015;11:e1005612. doi: 10.1371/journal.pgen.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent JM, et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010;10:4209–4212. doi: 10.1002/pmic.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yang J-R. Determinants of the rate of protein sequence evolution. Nature Reviews Genetics. 2015;16:409–420. doi: 10.1038/nrg3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. Journal of molecular medicine. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 12.Yang JR, Zhuang SM, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Molecular systems biology. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moutinho AF, Trancoso FF, Dutheil JY. The impact of protein architecture on adaptive evolution. Molecular biology and evolution. 2019;36:2013–2028. doi: 10.1093/molbev/msz134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moutinho AF, Bataillon T, Dutheil JY. Variation of the adaptive substitution rate between species and within genomes. Evolutionary Ecology. 2020;34:315–338. [Google Scholar]
- 15.Yip SH-C, Matsumura I. Substrate ambiguous enzymes within the Escherichia coli proteome offer different evolutionary solutions to the same problem. Molecular biology and evolution. 2013;30:2001–2012. doi: 10.1093/molbev/mst105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larion M, Moore LB, Thompson SM, Miller BG. Divergent evolution of function in the ROK sugar kinase superfamily: role of enzyme loops in substrate specificity. Biochemistry. 2007;46:13564–13572. doi: 10.1021/bi700924d. [DOI] [PubMed] [Google Scholar]
- 17.Serohijos AW, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cell reports. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuenberger P, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355 doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 19.Serohijos AW, Lee SR, Shakhnovich EI. Highly abundant proteins favor more stable 3D structures in yeast. Biophysical journal. 2013;104:L1–L3. doi: 10.1016/j.bpj.2012.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proceedings of the National Academy of Sciences. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Guo N, Wagner A. Selection enhances protein evolvability by increasing mutational robustness and foldability. Science. 2020;370 doi: 10.1126/science.abb5962. [DOI] [PubMed] [Google Scholar]
- 22.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS computational biology. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keseler IM, et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic acids research. 2017;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nature methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, et al. Expression level is a major modifier of the fitness landscape of a protein coding gene. Nature ecology evolution. 2022;6:103–115. doi: 10.1038/s41559-021-01578-x. [DOI] [PubMed] [Google Scholar]
- 26.Pakula AA, Sauer RT. Genetic analysis of protein stability and function. Annual review of genetics. 1989;23:289–310. doi: 10.1146/annurev.ge.23.120189.001445. [DOI] [PubMed] [Google Scholar]
- 27.Sarkisyan KS, et al. Local fitness landscape of the green fluorescent protein. Nature. 2016;533:397–401. doi: 10.1038/nature17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell RL. Permanence of the log-normal distribution. JOSA. 1968;58:1267–1272. [Google Scholar]
- 29.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Current opinion in structural biology. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Zheng J, Payne JL, Wagner A. Cryptic genetic variation accelerates evolution by opening access to diverse adaptive peaks. Science. 2019;365:347–353. doi: 10.1126/science.aax1837. [DOI] [PubMed] [Google Scholar]
- 31.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nature biotechnology. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 32.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 33.Nam SH, Oh KH, Kim GJ, Kim HS. Functional tuning of a salvaged green fluorescent protein variant with a new sequence space by directed evolution. Protein engineering. 2003;16:1099–1105. doi: 10.1093/protein/gzg146. [DOI] [PubMed] [Google Scholar]
- 34.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Current biology. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 35.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Molecular biology and evolution. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 36.Plotkin JB, Fraser HB. Assessing the determinants of evolutionary rates in the presence of noise. Molecular biology and evolution. 2007;24:1113–1121. doi: 10.1093/molbev/msm044. [DOI] [PubMed] [Google Scholar]
- 37.Maddamsetti R. Universal Constraints on Protein Evolution in the Long-Term Evolution Experiment with Escherichia coli. Genome Biology and Evolution. 2021;13 doi: 10.1093/gbe/evab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaBar T, Adami C. Evolution of drift robustness in small populations. Nature Communications. 2017;8:1–12. doi: 10.1038/s41467-017-01003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raser JM, O’shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardin J, Wilson J. A note on oligonucleotide expression values not being normally distributed. Biostatistics. 2009;10:446–450. doi: 10.1093/biostatistics/kxp003. [DOI] [PubMed] [Google Scholar]
- 41.Ham L, Brackston RD, Stumpf MP. Extrinsic noise and heavy-tailed laws in gene expression. Physical review letters. 2020;124:108101. doi: 10.1103/PhysRevLett.124.108101. [DOI] [PubMed] [Google Scholar]
- 42.Furusawa C, Suzuki T, Kashiwagi A, Yomo T, Kaneko K. Ubiquity of log-normal distributions in intra-cellular reaction dynamics. Biophysics. 2005;1:25–31. doi: 10.2142/biophysics.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casellas J, Varona L. Modeling skewness in human transcriptomes. PLoS One. 2012;7:e38919. doi: 10.1371/journal.pone.0038919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bengtsson M, Ståhlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome research. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bódi Z, et al. Phenotypic heterogeneity promotes adaptive evolution. PLoS biology. 2017;15:e2000644. doi: 10.1371/journal.pbio.2000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Qian W, Zhang J. Positive selection for elevated gene expression noise in yeast. Molecular systems biology. 2009;5:299. doi: 10.1038/msb.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acar M, Mettetal JT, Van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nature genetics. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 48.Kacser H, Burns JA, Kacser H, Fell D. The control of flux. Biochemical Society Transactions. 1995;23:341–366. doi: 10.1042/bst0230341. [DOI] [PubMed] [Google Scholar]
- 49.Chen P, Shakhnovich EI. Lethal mutagenesis in viruses and bacteria. Genetics. 2009;183:639–650. doi: 10.1534/genetics.109.106492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nature Reviews Genetics. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 51.Hoekstra HE, et al. Strength and tempo of directional selection in the wild. Proceedings of the National Academy of Sciences. 2001;98:9157–9160. doi: 10.1073/pnas.161281098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oz T, et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Molecular biology and evolution. 2014;31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jahn LJ, Munck C, Ellabaan MMH, Sommer MOA. Adaptive Laboratory Evolution of Antibiotic Resistance Using Different Selection Regimes Lead to Similar Phenotypes and Genotypes. Front Microbiol. 2017;8:816. doi: 10.3389/fmicb.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chemical reviews. 2002;102:759–782. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 55.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 56.Warren DJ. Preparation of highly efficient electrocompetent Escherichia coli using glycerol/mannitol density step centrifugation. Analytical biochemistry. 2011;413:206–207. doi: 10.1016/j.ab.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 57.Khersonsky O, Tawfik DS. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annual review of biochemistry. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 58.Aharoni A, et al. The ’evolvability’ of promiscuous protein functions. Nature Genetics. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 59.Rhoads A, Au KF. PacBio sequencing and its applications. Genomics, proteomics & bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC bioinformatics. 2012;13:1–18. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution. 2018;35:1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript or the supplementary materials. SMRT sequencing data are available at NCBI with a BioProject ID PRJNA833567 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA833567).
Custom code used in this study is available in a public GitHub repository (https://github.com/dasmeh/Discrete_Time_Markov_Chain_Evolution).