Abstract
Phosphorus (P) acquisition is key for plant growth. Arbuscular mycorrhizal fungi (AMF) help plants acquire P from soil. Understanding which factors drive AMF-supported nutrient uptake is essential to develop more sustainable agroecosystems. Here, we collected soils from 150 cereal fields and 60 non-cropped grassland sites across a 3,000 km trans-European gradient. In a greenhouse experiment, we tested the ability of AMF in these soils to forage for the radioisotope 33P from a hyphal compartment. AMF communities in grassland soils were much more efficient in acquiring 33P and transferred 64% more 33P to plants compared to AMF in cropland soils. Fungicide application best explained hyphal 33P transfer in cropland soils. The use of fungicides and subsequent decline in AMF richness in croplands reduced 33P uptake by 43%. Our results suggest that land-use intensity and fungicide use are major deterrents to the functioning and natural nutrient uptake capacity of AMF in agroecosystems.
Introduction
With the global population growing, we need to find ways to promote crop production while minimizing environmental degradation 1,2. Understanding and harnessing the natural functions provided by species above- and below-ground in the agricultural landscape is a promising approach to address both goals, thus paving the way to an ecological intensification of agroecosystems 3,4.
Arbuscular mycorrhizal fungi (AMF) inhabit the soils of virtually all terrestrial ecosystems and form symbiotic associations with most plants, including agricultural crops 5,6. Plants deliver reduced carbon in the form of sugars and lipids to AMF in return for nutrients, especially phosphorus (P) and nitrogen, which AMF acquire through their extensive soil hyphal networks. AMF may supply up to 90% of the host plant’s P requirements, especially in nutrient-poor and undisturbed vegetation 5,7. Promoting the natural potential of AMF for crop P nutrition could therefore circumvent the adverse environmental effects of high P fertilization 8,9, reduce associated economic and environmental production costs, increase P availability in cropping systems with low fertilizer access, while contributing to other services provided by AMF such as soil aggregation 10. The AMF-symbiosis is thus one key asset to improving P-use efficiency and to the design of sustainable agroecosystems 11–13. However, whether current cropping systems support AMF functioning remains unclear, fueling the debate about the relevance of AMF for agricultural production 14–17.
The contribution of AMF to plant nutrition are context-dependent, given that AMF communities are shaped by local environmental conditions 18 and that the benefits of AMF to plant growth are cultivar dependent 19–21. Some modern crop cultivars are less efficiently colonized by AMF and have been shown to only benefit from AMF under severe P limitation 22,23 likely since crop breeding and selection have not prioritized these symbiotic associations 24. However, other studies indicate that AMF support crop yield and are a key factor to make agroecosystem more sustainable 3,10,13,17. Apart from this, the abundance and diversity of AMF communities are negatively affected by intensive management practices 25–27, especially tillage, inorganic fertilization, and pesticide use 28. Several studies indicate that AMF richness promotes plant productivity and plant P uptake 29,30. Thus, a reduction in AMF richness due to intensive management may reduce the natural nutrient uptake capacity of agricultural soils. Whether management-induced changes in AMF communities subsequently impact their ability to acquire P for growing plants is still poorly understood. Until now, broad scale assessments of AMF functioning across different environmental conditions and land use types have not been performed, contributing to our lack of understanding of the drivers of hyphal P acquisition in intensively and extensively managed plant-soil systems.
To address these knowledge gaps, we collected 210 soils originating from cropland fields and neighboring non-cropped grassland sites across a 3,000 km European north-to-south gradient (ED Fig. 1). Plantago lanceolata, well known for its associations with a wide range of AMF and hence ideal for a broad screening of the potential AMF activity across various soil conditions 31, was then grown on these soils in a greenhouse experiment in which we measured the capacity of the associated AMF to mediate the uptake of the radioisotope 33P from a labelled hyphal compartment (Fig. 1). This allowed us to 1) estimate the capacity of native AMF communities in different soils to provide plants with P, and 2) assess the drivers of hyphal P transfer by including the climatic background, biotic and abiotic soil properties, and legacy effects of land use and crop management practices in the analysis. We hypothesized that i) hyphal P transfer is strongly driven by the large gradient in climate and soil characteristics, ii) hyphal P transfer is lower in cropland compared to non-cropped grassland soils, and iii) intensive agricultural practices (pesticide use, tillage, and fertilization) and a reduction in AMF richness negatively impact hyphal P transfer. Ultimately, our research contributes to a better understanding of the main drivers of and constraints to AMF functioning in agroecosystems.
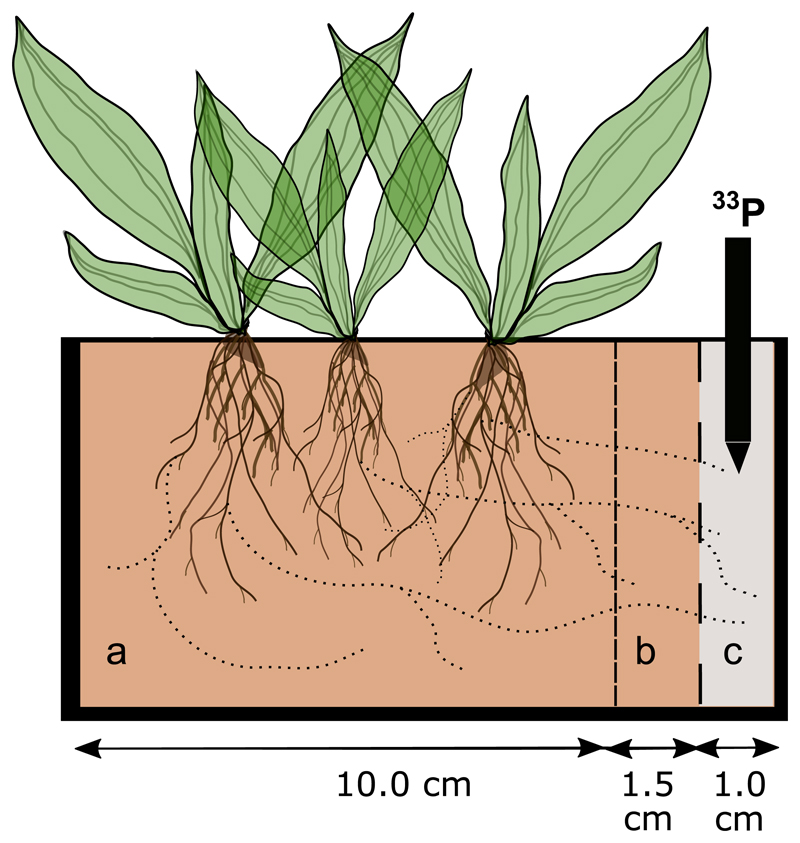
Figure 1.
Experimental set-up using Plantago lanceolata as model plant in pots (l = 12.5 cm, w = 8 cm, h = 8.5 cm) containing three compartments. The plant zone (compartment a) and the buffer zone (compartment b) were filled with the collected field soils. The compartment c contained a standardized, sterilized soil which was injected with the tracer 33P. Compartment a was separated from b using a 40 µm mesh (narrow dashed line), restricting root penetration. The mash barrier between b and c compartments had a pore size of 500 µm (wide dashed line).
Results
Effect of land use on hyphal 33P transfer and soil properties
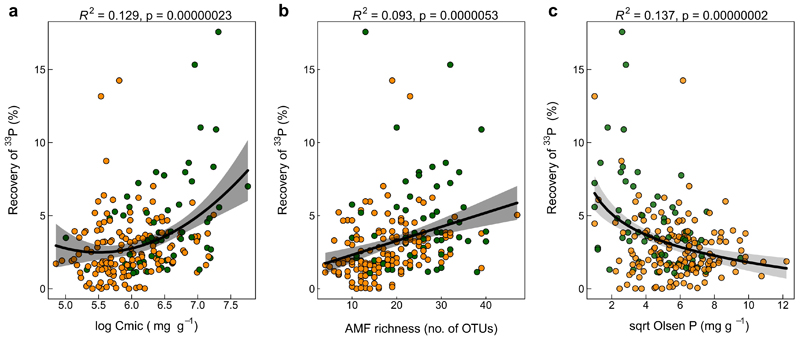
The recovery of 33P was 64% higher in pots filled with soils from grassland sites compared to the soils from croplands (p<0.001; Fig. 2). There was a large variation in hyphal 33P transfer in both systems (1.07 – 17.6% in grassland soils; 0.003-14.2% in cropland soils), which was paralleled by significant biotic and abiotic differences between the soils originating from the grassland and cropland systems (Table 1). Especially the differences in microbial biomass C (-41% in cropland soils), AMF richness (-29% in cropland soils) and available soil P (Olsen P, +60% in cropland soils) were striking, and these factors correlated significantly with hyphal 33P transfer, when considering the whole dataset (Table 1, ED Fig. 2). Among the management factors considered in croplands, the number of fungicide application events showed the strongest correlation with hyphal 33P transfer (Supplementary Table 1).
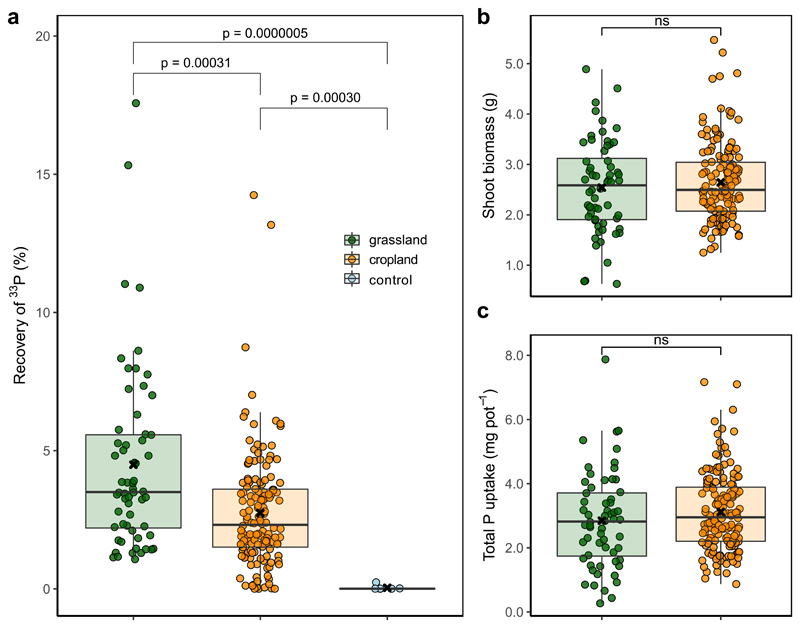
Figure 2.
Recovery of 33P in the shoot material of Plantago lanceolata plants grown in grassland (green), cropland (orange) and sterilized control soils (blue) (a). The low 33P recovery in sterilized control soils confirm that 33P recovery corresponds to hyphal activity in the field soils. Shoot biomass (b) and total P uptake per pot (c) in the grassland vs. cropland soils. Boxes mark the interquartile range, vertical lines the whiskers, bold horizontal lines the median and “x” the mean values. Bonferroni corrected p-values <0.05 (based on two-sided Wilcoxon rank test) indicate significant differences between land use systems, “ns” means no significant difference.
Table 1.
Mean values of climatic and edaphic factors (± standard error) in soils from croplands and grassland sites used in this experiment (n = 210). Differences between land use types were assessed using Wilcoxon’s rank test. The last column shows the Spearman rank correlation coefficient of all variables with the 33P recovery with p-values in brackets.
| Mean (± SEM) | p-value (Wilcoxon’s rank test) | Correlation with 33P recovery | |||
|---|---|---|---|---|---|
| Unit | Grassland | Cropland | |||
| Aridity | unitless | 0.09 (±0.05) | 0.11 (±0.03) | 0.670 | -0.14 (0.053) |
| Annual temperature | °C | 9.74 (±0.36) | 9.19 (±0.25) | 0.180 | 0.03 (0.685) |
| Clay | % | 27.3 (±1.48) | 28.2 (±0.98) | 0.740 | 0.12 (0.090) |
| Silt | % | 33.6 (±1.67) | 39.7 (±0.95) | 0.003** | 0.00 (0.983) |
| pH | unitless | 6.84 (±0.11) | 7.11 (±0.06) | 0.044* | 0.15 (0.027)* |
| Nmin | mg g-1 | 14.6 (±1.12) | 15.1 (±0.81) | 0.810 | -0.08 (0.267) |
| Olsen-P | mg g-1 | 23.9 (±2.44) | 38.2 (±2.07) | <0.001** | -0.24 (<0.001)** |
| Soil C:N | g g-1 | 8.61 (±0.33) | 8.20 (±0.17) | 0.230 | -0.07 (0.348) |
| Soil N:P | g g-1 | 0.62 (±0.59) | 0.03 (±0.00) | <0.001** | 0.20 (0.005)** |
| Soil Nmin:Pmin | g g-1 | 1.53 (±0.33) | 0.72 (±0.10) | <0.001** | 0.18 (0.010)* |
| SOC | mg g-1 | 20.2 (±1.56) | 15.1 (±0.57) | 0.003*** | 0.17 (0.013)* |
| Microbial biomass C | mg g-1 | 0.75 (±0.05) | 0.44 (±0.02) | <0.001** | 0.33 (<0.001)** |
| Root AMF Richness | No. OTUs | 24.9 (±1.07) | 17.7 (±0.64) | <0.001** | 0.41 (<0.001)** |
| Root AMF Shannon | unitless | 2.27 (±0.05) | 1.82 (±0.04) | <0.001** | 0.37 (<0.001)** |
| Soil bacteria | index | 9.74 (±0.03) | 9.88 (±0.02) | <0.001** | -0.15 (0.029)* |
| Soil fungi Shannon | index | 3.79 (±0.06) | 3.44 (±0.04) | <0.001** | 0.19 (0.006)** |
| Soil fungi Richness | No. OTUs | 198 (±5.06) | 158 (±2.84) | <0.001** | 0.20 (0.004)** |
| Soil cercozoa Shannon | index | 4.93 (±0.03) | 4.96 (±0.02) | 0.600 | -0.07 (0.329) |
| Soil archaea Shannon | index | 1.98 (±0.04) | 2.06 (±0.03) | 0.210 | -0.02 (0.745) |
| Soil archaea Richness | No. OTUs | 77.1 (±2.31) | 77.9 (±1.68) | 0.690 | -0.07 (0.308) |
P<0.05;
P<0.01
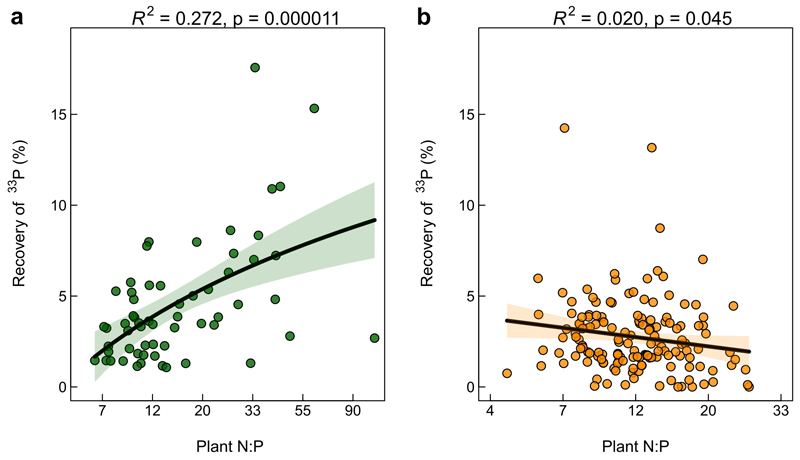
We assessed whether the observed difference in hyphal 33P transfer in the two land-use systems had implications for plant nutrition. We found that plant growth and total P uptake did not differ between land use systems (Fig. 2 b and c), despite the greater P availability in cropland soils and a strong link between available soil P and total P uptake (R2=0.404, p<0.001; Supplementary Fig. 1 b). Neither plant growth, total P uptake, nor the plant N:P ratio correlated significantly with the hyphal 33P transfer when considering both cropland and grassland together (Supplementary Table 2). However, while hyphal 33P transfer increased with increasing plant N:P ratios in grassland soils (ED Fig. 3 a), there was a weak negative correlation in cropland soils (ED Fig. 3 b).
Drivers of hyphal 33P transfer in cropland vs. grassland soils
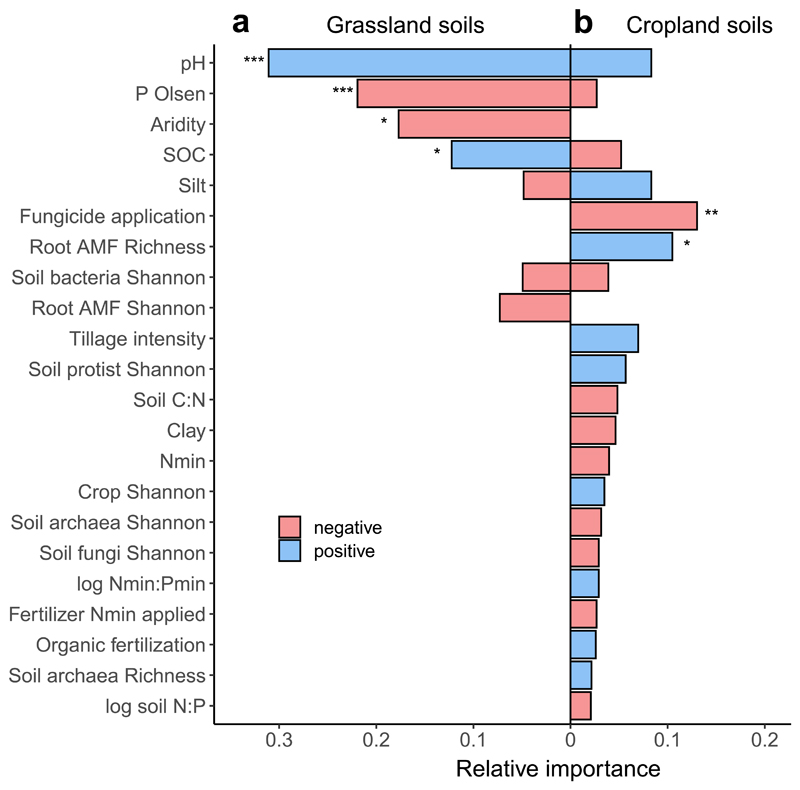
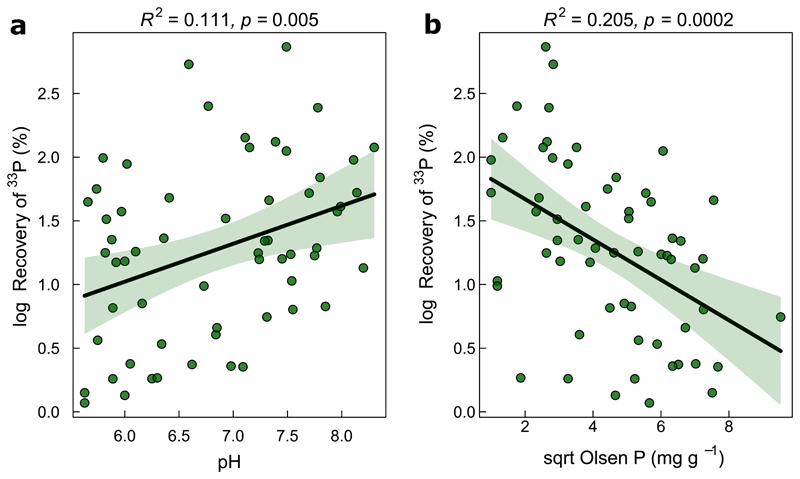
A multi-model inference approach used to assess the relative importance of climate, soil characteristics and management history on AMF functioning revealed that different predictors explained variation in 33P recovery in cropland and non-cropped grassland soils (Fig. 3). In grassland soils, the most prominent and significant predictors were soil pH, followed by available P, aridity, and SOC (Fig. 3 a, Supplementary Table 3 a). These four factors explained most variation in the model selection approach (R2adj=0.45; Supplementary Table 3 b). The strong correlation with soil pH and available P could also be observed using simple linear regression (Fig. 4).
Figure 3.
Relative importance of predictors for hyphal 33P transfer, measured as 33P recovery in plant shoots based on a multi-model inference analysis, in grassland (a, n=58) versus cropland soils (b, n=146). Note that the crop management predictors were used only in the cropland model. Predictors were ordered according to their total importance (i.e., sum of both land use types). Negative and positive correlations are shown in red and blue, respectively. Asterisks indicate a significant correlation at p<0.001 (***), p<0.01 (**) and p<0.05 (*) based on the averaged model coefficients. Model coefficients, standard errors, z-values and exact p-values are reported in Supplementary Tables 6 a and 7 a.
Figure 4.
Correlation of 33P recovery with soil pH (a), and available soil P (b) in the grassland sites (n=60). The green error bands mark the 95% confidence interval of the two-sided OLS regression models. F1,58 = 8.375 (a) and F1,58 = 16.21 (b). R2 corresponds to the adjusted R2 value.
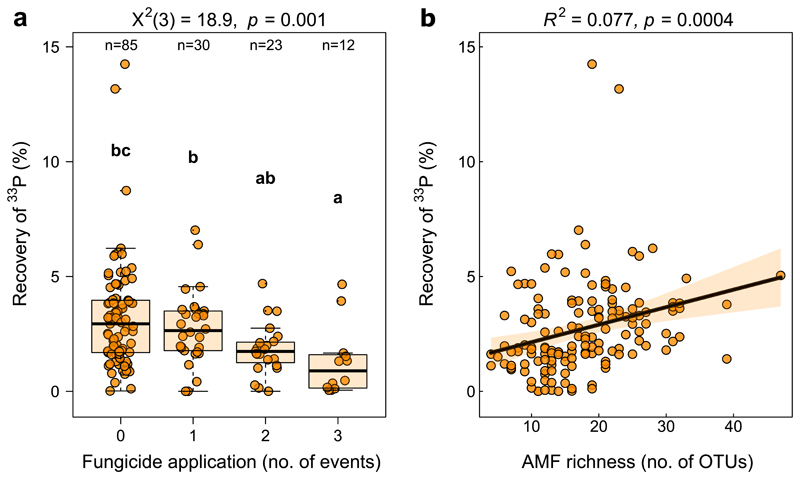
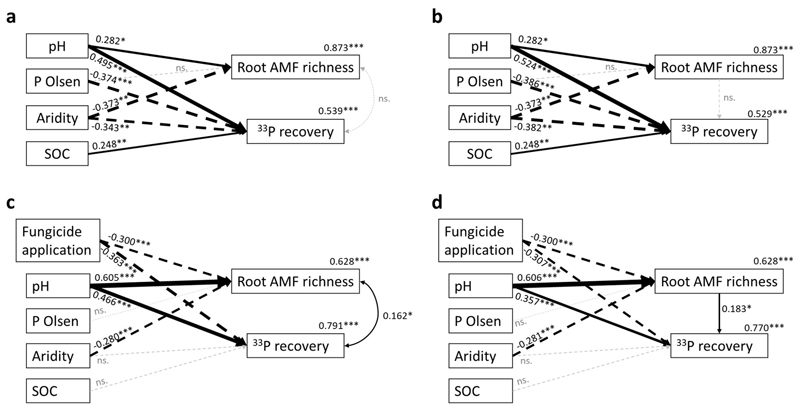
Relative to grassland soils, less variation in hyphal 33P recovery could be explained in the case of cropland soils (R2adj.=0.16, Supplementary Table 4 b) despite the larger number of observations and the inclusion of various crop management predictors used in these soils (Table 1, Supplementary Table 1). The most important predictors for hyphal 33P transfer in cropland soils were AMF richness and the number of fungicide application events during one year before sampling the soils from the fields (Figs. 3b and 5, Supplementary Table 4 a). Hyphal 33P transfer in soils without fungicide application was, on average, 2.3 times higher compared to soils that received three fungicide applications. Multiple comparison analysis supported this observation, revealing that 33P recovery rates significantly decreased with an increasing number of application events (Fig. 5 a), paralleling the results from simple linear and quantile regression analysis (Supplementary Fig. 2). Root AMF richness correlated positively with hyphal 33P transfer in cropland soils (Fig. 5 b) showing that more 33P was acquired in soils with high AMF richness. Interestingly, structural equation modelling revealed that also root AMF richness correlated negatively with fungicide application, leading to both direct and indirect effects of fungicide application on hyphal 33P recovery (ED Fig. 4 c and d). On top of that, aridity influenced 33P transfer indirectly though AMF richness, while the soil pH showed a positive direct as well as indirect relationship with 33P transfer through AMF richness.
Figure 5.
Effects of an increasing number of fungicide application events in cropland soils on 33P recovery (a). Significant differences between the number of applications are indicated with different letters examined using a Kruskal-Wallis rank test where X2 represents the model fit with the degrees of freedom in brackets (n=150). Boxes mark the interquartile range, vertical lines the whiskers, bold horizontal linesthe median and “x” the mean values. The relationship of 33P recovery and AMF richness in P. lanceolata roots (b) in the cropland soils (n=146), estimated using two-sided OLS regression (F1,144 = 13.02). The orange error band marks the 95% confidence interval. R2 corresponds to the adjusted R2 value.
Discussion
Lower hyphal 33P transfer in cropland vs. grassland soils
AMF in extensively managed or undisturbed soils are usually more abundant and diverse compared to intensively managed cropland soils that receive substantial amounts of fertilizers and pesticides 26,27,32–36. However, the functional implications of such differences have not been investigated previously. This study demonstrated that AMF communities from non-cropped grassland sites are generally more active and transfer higher amounts of 33P to host plants compared to AMF communities from cropland soils. Our observation, stemming from a vast diversity of different soil and climatic characteristics at a broad spatial scale, suggests that current cropping practices impair AMF functioning and that the capacity of AMF to support plant nutrition thus remains underexploited in European croplands.
Various mechanisms could explain the observed differences in AMF functioning between cropland and grassland soils. First, a range of studies showed that management practices associated with intensive land use, including soil tillage, pesticide and fertilizer use, reduce AMF abundance, spore abundance, AMF diversity and alter AMF community composition 37–39. Our study confirmed that both microbial biomass as well as AMF richness were reduced in intensively managed croplands, and this was linked to a reduced hyphal 33P transfer (Table 1). This suggests that hyphal P transfer is affected by intensive land-use to a similar extent as soil microbes, which have been shown to be sensitive indicators for land-use change 40–42. AMF are estimated to contribute to 20-30% of the total soil microbial biomass 24 and it is likely that the positive link between 33P recovery and microbial biomass is connected to AMF, especially because various studies showed that mycorrhizal hyphal density can correlate strongly with hyphal P transfer 43–45. Interestingly, hyphal 33P transfer was positively correlated with AMF richness in cropland soils, but not in grassland soils, perhaps indicating that functional diversity of AMF is more important under disturbed conditions like in cropland soils. Further studies are required to investigate this in more detail. It also should be noted that although earlier studies demonstrated that AMF are the main actors in the transfer of P from hyphal compartments to plants, the possibility that other microorganisms (e.g., non-mycorrhizal hyphae) might have contributed to the transfer of P to the root zone cannot be excluded. Bacteria can also facilitate or suppress AMF activity in the soil 44. Recent studies indicate that AMF fungal hyphae are colonized by specific bacterial communities 46, which in turn influence nutrient uptake, particularly from organic sources 47. Finally, P transfer via hyphal networks depends on the distinct functional traits, activity, and foraging strategies of individual strains of AMF 48–51 and such aspects need more attention in future work. While we could not make these connections in this present study, further studies should investigate whether specific microbial groups that are affected by intensive management, also influence the ability of AMF to acquire P.
AMF compensate for lower P availability in grassland soils
In addition to direct effects, management can be indirectly linked with AMF-symbiosis through alterations of soil properties, such as available soil P 27. High levels of available soil P, often accompanied by low soil N:P ratios, were shown to inhibit AMF root colonization and decrease the AMF’s relative contribution to plant P nutrition 52–54. In accordance with these previous findings, we observed that AMF hyphal 33P transfer was negatively associated with available soil P levels (Table 1, ED Fig. 2 c), which were on average 60% higher in the cropland soils. In turn, regarding implications for plant nutrition and growth, our findings indicate that AMF mediated P uptake compensated for the lower available P levels in the grassland soils, allowing for a similar average total P uptake and shoot biomass. This assumption is further supported by a negative correlation between the hyphal 33P transfer and the soil available P in grassland soils, and a positive correlation with the plant N:P ratio, respectively (ED Fig. 3 a). This implies that when plant P demand in grassland is high and plant productivity is limited by P (i.e., plants with N:P ratios above 16 55), AMF supply additional P to the plant. However, this trend could not be confirmed in the cropland soils where neither a link between available soil P, nor the plant N:P ratio, and hyphal P transfer was found, possibly indicating a dysfunctional symbiosis in croplands. Verbruggen et al. (2015) argued that the selective loss of AMF communities connected to soils with high N:P ratios might leave cropland fields with AMF of reduced symbiotic quality 34. While we agree that this could be the case for a multitude of fields with high soil N:P ratios in this study, the relatively large range of soil N:P ratios in both land use systems suggests that this is not the only mechanisms at play. The observed inability of putatively P-limited plants to acquire P through AMF hyphal activity in cropland soils provides further evidence that AMF are heavily affected by crop management (e.g., fungicide application), inhibiting their potential contribution to plant P nutrition in current cropping systems.
Soil pH and P drive hyphal 33P transfer in grassland soils
Using a multi-model inference approach, we were able to identify the main drivers of AMF hyphal 33P transfer to growing plants. In grassland soils, we found that much of the observed variation in hyphal 33P transfer could be explained by soil pH, available P, SOC content and climatic factors (e.g., aridity). These results are in line with other studies demonstrating that these factors influence AMF abundance, colonization, and activity 44,52,56–58.
In addition to the above discussed influence of available soil P, our results suggest a suppression of hyphal 33P transfer with decreasing pH, emphasizing the crucial role of soil pH for AMF activity, abundance, community structure as well as the occurrence of AMF host plants 32,44,59–62. Van Aarle et al. (2002)56 found reduced growth and activity of the AMF extraradical mycelium in low pH substrates, arguing that AMF are directly stressed by acidic environments. However, it is possible that indirect effects of pH on the activity of the AMF extraradical mycelium through interactions with other microbiota might be an even more important mechanism behind the suppressiveness of low pH soils44.
Apart from the major importance of soil abiotic factors, our results indicate that increasing aridity decreased hyphal 33P transfer in grassland systems. Although we did not manipulate water availability directly and observed only the legacy effects of aridity, our observation parallels various studies that determined direct negative effects of drought on AMF abundance and extraradical hyphae 57,58. Additionally, plant communities, C inputs and SOC might be influenced by aridity, resulting in indirect effects on AMF, possibly due to reduced net photosynthesis and energy supply to AMF under water-limited conditions 63, corroborating the positive relationship between 33P transfer and SOC observed in this present study. Ultimately, these results suggest that the goals of promoting C sequestration and improving plant P nutrition through AMF go hand in hand, at least in non-cropped systems where SOC is generally higher (Table 1).
Fungicide application reduces the ability of AMF to acquire P
While a considerable amount of variation in hyphal 33P transfer in grassland soils was explained by soil pH and P availability, we found that the number of fungicide application events was the most important predictor of hyphal 33P transfer in cropland soil (Fig. 3). Interestingly, this reduced hyphal 33P transfer in cropland soils was paralleled and partially mediated by a reduced AMF richness (ED Fig. 4 d) indicating that fungicides indirectly reduce hyphal mediated P uptake by reducing AMF richness. Prior work suggests that AMF richness can promote P uptake 29, providing further evidence that a reduction of AMF richness by fungicides may have implications for plant P nutrition. A recent study found that the abundance of AMF is negatively linked to the amount of pesticide residues in agricultural soils, and certain pesticide residues could be detected even decades after their last application 64. This indicates that adverse effects of fungicides on AMF might be long-lasting, corresponding to observations by Pánková et al. (2018)65, who showed that AMF infection rates of plants were reduced up to five years after the application of fungicides in a grassland site. These earlier studies focused on AMF abundance, whereas our results add a functional component to this debate and indicate that fungicide use in real agricultural contexts suppress both AMF diversity and functioning.
Given the great variety of amounts and compounds of the applied fungicide products (Supplementary Table 5), it is remarkable that the number of fungicide application events used as rough indicator in this study could capture the adverse effects posed by fungicide use on AMF. Although fungicides are applied to combat fungal diseases such as mildew and rusts, they often have non-target effects on other fungi, including beneficial AMF. Detrimental effects of various fungicides have repeatedly been reported for AMF biomass, spore density, root colonization, and alkaline phosphatase activity in internal and external hyphae 66–70. The negative effects of some fungicides on AMF have also been previously used to assess the importance of AMF for plant community structure and diversity in grasslands (e.g., 71,72). However, the close links between fungicide application rate and AMF functioning in a wide range of cropland soils observed here have not been reported before. It further indicates that the common use of fungicides hampers the natural ability of soil organisms to provide crops with nutrients and supports the findings of Sallach et al. (2021) who showed that also fungicides applied unintentionally through wastewater or biosolids can decrease the ability of AMF to transfer P to plants 39. Future studies under controlled conditions in the field now need to specifically test to which extent fungicides suppress the ability of AMF to support crop growth and test whether such effects are persistent. Our results also call for reconsidering the design of agricultural systems to be able to make full use of the potential of AMF-symbiosis for plant nutrition. For example, applying agroecological techniques, such as crop diversification, can be a promising way to reduce disease pressure 4 and hence the need to use pesticides, while at the same time promoting AMF richness 73 which could indirectly support plant P uptake (Fig 5 B) as well as to other benefits provided by AMF. In addition to the establishment of such AMF-promoting practices, the breeding and use of AMF responsive crops, an aspect which hasn’t been directly investigated in this study, is a way to promote AMF-supported crop production that requires further consideration in future research.
In conclusion, despite the wide range of different environmental conditions along the surveyed European gradient, the results show that the capacity of AMF to support plant P nutrition is impaired in croplands compared to non-cropped grasslands, particularly by the use of fungicides. Thus, we emphasize that there is a need to reconsider the design of agricultural systems to fully exploit the natural potential of AMF-symbiosis for a sustainable crop production.
Methods
Field sites
In spring 2017 we sampled soils from 150 croplands and 60 non-cropped grassland sites across a North-South gradient in Europe covering Sweden (n=34), Germany (n=48), Switzerland (n=57), France (n=39), and Spain (n=32) (ED Fig. 1). To minimize variation in AMF caused by different crop types, we selected cereal fields planted with wheat (Triticum sp., n=119), or closely related cereals like barley (Hordeum vulgare, n=25) and oat (Avena sativa, n=6), when wheat fields were not available. Furthermore, we exclusively sampled plots where conventional tillage practices had been performed. Although 26 croplands (17%) in Switzerland and Southern Germany were organically managed, they did not statistically differ from their neighboring conventional fields in terms of hyphal 33P transfer to plants (Supplementary Fig. 3), and therefore were kept in the dataset. The non-cropped grassland sites were located in the vicinity of the croplands to cover similar soil characteristics, and served as benchmark for AMF functioning, presumably providing less disturbed AMF communities, which were also supported by a greater plant diversity as compared to the croplands. These sites comprised extensively managed grasslands and marginal land (field strips) neighboring the croplands and were characterized by a permanent, predominantly herbaceous plant cover that was not part of a crop rotation. Most of these sites were unfertilized and occasionally mowed, although exact information on fertilization, grazing and mowing was not available for all of the sites. Following the classification by the Eurostat Land Use / Cover Area Frame Survey (LUCAS) 74, we refer to these plots as grassland sites.
Environmental data and location
To characterize the variation in climate along the sampled gradient, information on mean annual temperature (MAT) and mean annual precipitation (MAP) was extracted from the WorldClim Global Climate Data 75. Aridity information was derived from the CGIAR-CSI database, where it is expressed as a function of MAP over the mean annual potential evapotranspiration 76. In line with recent studies 77,78, we subtracted the aridity index from 1, to define aridity in our analyses such that higher aridity values indicate drier conditions. Additionally, the location of the fields along the gradients was described by the geographic distance from the S-W-most sampling site using the haversine formula 79.
Management practices
To determine the legacy effects of crop management on hyphal activity in the croplands, farmers and field managers provided information on management practices including fertilizer and pesticide use, tillage intensity (all during 2016, one year before sampling), as well as the crop rotational diversity and the duration of crop cover (during ten years before sampling) (Table 1). Fertilization intensity was assessed using the total amount of mineral N (ammonium and nitrate) applied through organic and/or mineral fertilization. Organic fertilization was additionally included as a binary variable in the analyses. The use of fungicides, herbicides and insecticides was summarized with the number of their respective application events. For fungicides, we further took into consideration the total amount and number of different active compounds added per hectare. However, these parameters were strongly linked to each other. Therefore, we focused our analysis on the number of application events, since they had the strongest correlation with our output variable of interest (Supplementary Table 5). Tillage intensity was estimated by averaging the number and maximum depth of tillage events (after normalization). For the crop rotations (see Supplementary Methods for more details), we calculated the proportion of time with plant cover and the crop diversity at each site according to a Shannon diversity index 80.
Soil sampling
To reduce variation between sites related to crop growth stage, all soils were collected during wheat flowering at each site (ranging from May in Spain to August in Sweden). At each site, eight soil samples were taken in a circular pattern within a 10 m radius using a five cm diameter auger and to a depth of 20 cm. Three of the soil cores were kept intact and stored at 4 °C before preparing them for the compartment experiment. The other soil cores were homogenized and sieved to 2-mm. A portion of this soil was air-dried for further processing of soil physical and chemical analyses. Soil texture, calcium carbonate (CaCO3), pH, soil organic C (SOC), total nitrogen (N) and total phosphorus (P) were measured on the dried samples. Available phosphorus (Olsen P), and the microbial biomass C were measured on fresh soil samples within a few weeks after sampling. All above mentioned soil properties were measured following the Swiss standard protocols 81.
Soil microbial analyses
To assess the soil microbial diversity, DNA was extracted from 250 mg of each soil sample (stored at -19 °C) using the DNeasy PowerSoil-htp 96 well DNA isolation kit (Qiagen, France). Microbial diversity was further analyzed as described in Garland et al. (2021)80 (also shown in the Supplementary Methods). Briefly, amplicons of bacterial and archaeal 16S rRNA genes were generated in two steps following Berry et al. (2011)82 in two separate sequence data sets, one for each domain. The fungal ITS2 region was amplified using the PacBio SMRT Sequencing platform (Pacific Biosciences, CA) with the primers ITS1f (CTTGGTCATTTAGAGGAAGTAA) and ITS4 (TCCTCCGCTTATTGATATGC) targeting the entire ITS region (~630 bp) 83,84. Cercozoa diversity was estimated with a two-step PCR to amplify a fragment (c. 350 bp) of the V4 region of the 18S rRNA gene using the primers sets designed by Fiore-Donne et al. (2018)85 for the specific amplification of cercozoa. Finally, the diversity of soil bacterial, fungal, archaeal, and cercozoan communities was assessed using the Shannon-Weaver index of diversity, and richness was calculated as the number of observed operational taxonomic units (OTU).
AMF compartment system
To assess AMF hyphal 33P transfer rates and hyphal activity in the collected field soils, we conducted an experiment starting in March 2018 using a compartment setup (Fig. 1) following, amongst others, Schweiger et al. (1999)45 and Svenningsen et al. (2018)44. We used the radioisotope 33P to trace P transport by fungal hyphae to Plantago lanceolata plants. To this end, the plant zone (compartment a; Fig. 1) of the compartment system was separated with a mesh (pore size 40 µm), hindering the plant roots to penetrate the hyphal zone (compartments b and c; Fig. 1). The buffer zone (compartment b; Fig. 1) had the purpose to minimize possible diffusion of 33P injected into compartment c (Fig. 1) from entering the plant zone 45.
Plantago lanceolata was selected as the model plant in this experiment as it has been shown to be very AMF sensitive and unselective in terms of AMF species associations 31. In this present study, a total of 252 different AMF taxa was found to associate with P. lanceolata roots, belonging to 11 known families (See ‘Root AMF Microbiome’ methods below, ED Fig. 5). Also, very different AMF communities were found to colonize Plantago roots in soils collected from different locations. This suggests that P. lanceolata could be colonized by a great diversity of AMF, both in grasslands and croplands (with observed AMF richness values up to 40 and 47, respectively). Forming a symbiotic relationship with a wide range of AMF was an important prerequisite to capture as many of the changes induced by management as possible and to estimate the potential AMF activity across this vast diversity of soils. However, it needs to be noted that AMF-mediated P transfer might depend strongly on the plant species considered and is likely to be lower in crop cultivars that depend less strongly on AMF for their nutrition 22,23.
The different field soils were filled into the compartments a and b to a total volume of 850 cm3. To do so, three of the collected soil cores, which were kept intact and stored at 4 °C, were homogenized by gently removing stones and residues larger than 8 mm. A subsample was taken to assess the water content, available N and the exact weight of the soil at the start of the experiment. Furthermore, seven negative control pots were implemented using sterilized soils (which had been exposed to X-rays at a dose of 40.7 kGy for at least 17 hours at Steris, Däniken, Switzerland) originating from four croplands and three grassland sites representing a range of soil properties (pH 5.97 – 8.03, Olsen P 1.8 – 118 mg g-1, Sand 18.34 – 33.53%), to verify that no or little 33P would be found in plants when no AMF are present. Very small amounts of label (a median percentage of 0.24 as compared to the unsterilized pots) were detected in the plants grown in sterilized control pots (Fig. 2 a) demonstrating that fungal hyphae present in the field soil, rather than fine roots, mass flow, or microbial propagules in the greenhouse air, were responsible for the recorded 33P uptake by plants. Additionally, assessments of AMF root colonization and AMF abundance in the hyphal compartment c (Fig. 1) in a subset of the soils (n=36; methods outlined in the Supplementary Methods section) showed that roots in pots filled with unsterile field soils were strongly colonized by AMF and that AMF accessed the hyphal compartment c in the respective pots. In contrast, the sterilized control pots had little AMF root colonization and AMF abundance in the hyphal compartments was negligible (Supplementary Fig. 4 a and c).
The pots were watered to 60% of the soils’ water-holding capacity and the soil volume was adjusted after two days if necessary. The hyphal compartment c (Fig. 1), into which the 33P was injected later, was filled with a standardized, X-ray sterilized soil (58 g dry mass) from an extensively managed grassland soil at Agroscope Reckenholz, Zurich, Switzerland. Using a standardized soil in compartment c allowed for a similar sorption of 33P at the injection spot, enabling us to focus our analyses on the hyphal activity solely, rather than the soils’ P sorption capacity, which is highly variable across soils 86.
After three days of pre-incubation, five sterilized and pre-germinated seeds of Plantago lanceolata (cultivar TONIC, 2012, Agricom NZ) were planted in compartment a of each pot. To better distribute the random variation potentially caused by the genotypically non-identical seeds 21,87, five seeds were planted per pot, and seedlings were thinned to three seedlings of similar growth after one week. The pots were arranged randomly in the greenhouse. In the following 12 weeks of growth, the plants were watered three times a week to maintain a comparable water level ranging between 60 and 70% of the individual soil’s water holding capacity. Every second week the pots were newly randomized. The temperature adjustments of the greenhouse were between 16 to 18 °C at night and between 19 and 21 °C during daytime, at a humidity around 70% water vapor saturation. Daytime was set at 16 hours with 30 klx lighting.
Plant growth, 33P-injection, and analyses
After 10 weeks of plant growth, 600 µl of a carrier-free 33PO43--solution (2333 kBq ml-1), resulting in a total of 1399.8 kBq, were injected at three different locations within compartment c at a depth of 3-4 cm using a 200 µl pipette. The experiment was terminated with the harvest of Plantago shoots 12 days after 33P injection, and 12 weeks after planting. The shoots were cut above the soil surface and placed in the oven at 60 °C for 48 hours. After assessment of the dry shoot biomass, the samples were milled and used for determination of P and N contained in shoots. Total plant N was measured with an elemental analyzer (Vario Pyro Cube, Elementar GmbH, Hanau, Germany). Incinerated samples were microwave-digested with nitric acid 88. The P concentration in the extract was then determined colorimetrically using malachite green 89 and the activity of 33P with liquid scintillation counting for 10 min per sample (TRI-CARB 2500 TR, liquid scintillation analyzer, Packard Instruments, Meriden, CT), using 5 ml scintillant (Ultima Gold™ AB, PerkinElmer, USA) for about 0.05 g of shoot sample. Due to the large dataset, samples were not replicated. The measured disintegrations per minute were corrected for the background activity, the counting efficiency to account for chemical and color quenching and were back-calculated to the time point of injection to correct for radioactive decay.
Finally, the relative amount of 33P, which was recovered in the shoot material after transfer via AMF hyphae from the initially injected pool, i.e., the recovery rate, was calculated using equation (1).
| (1) |
Where r is the decay-corrected activity of 33P in each sample (Bq g-1 shoots) multiplied by the total shoot weight per pot (g), and R refers to the total injected radioactivity (Bq). Following Frossard et al. (2011)90, the recovery rate was then used as an estimate for the capacity of AMF to transfer P to growing plants, which we refer to as “hyphal 33P transfer” in this study. However, it is important to note that the 33P recovery rate is an indicator for but not an exact measurement of the relative contribution of AMF to plant P nutrition. This is because hyphae were able to take up non-radioactive P also in the compartments a and b, while 33P could only be added to the hyphal compartment c.
Root AMF microbiome
After the harvesting of Plantago shoots, the roots were gently separated from the surrounding soil and thoroughly rinsed with deionized water to remove any remaining rhizosphere soil. Subsamples were taken from various parts of the root structure and stored at -20°C. Details on DNA extraction, sequencing, and characterization of the root AMF microbiome are explained in detail in the Supplementary Methods section. Briefly, DNA was extracted from the root samples, and an amplicon sequencing library was generated using the AMF-specific PCR primers AMV4.5NF and AMGDR 91. The MiSeq library was prepared and sequenced in collaboration with the Genetic Diversity Center, ETH Zurich.
Sequences generated in this study will be deposited at the European Nucleotide Archive and be accessible at the time of publication. Paired-end reads were processed and denoised into amplicon sequence variants (ASVs) using the DADA2 plugin 92 implemented in QIIME2 93. The ASV reference sequences were clustered to the virtual taxa (VT) sequences of the AMF-specific taxonomy database MaarjAM 94(status June 2019) at ≥ 97% sequence similarity with the VSEARCH plug-in 95 in QIIME2. Sequences not clustering to a VT in the MaarjAM database were clustered de-novo into OTUs at ≥ 97% sequence similarity and taxonomically assigned with the SILVA database (v128)96 to detect other AMF taxa not present in the MaarjAM database. The non-VT OTU taxonomy file and count table were then filtered to remove any OTU sequences not belonging to known AMF taxa. The count table and taxonomy assignments of the VT were combined with those of the AMF-OTUs identified by SILVA in order to produce the final count table and corresponding taxonomy assignments of the profiled AMF community.
For comparisons of AMF diversity within samples (alpha diversity), the OTU table was rarefied to 1000 sequences/sample, which was sufficient to capture the AMF richness of most of the samples (Supplementary Fig. 5). Eight samples were removed from subsequent analyses due to sequence counts less than 1000. The alpha diversity measures of observed richness and Shannon Diversity were calculated on the rarefied counts using the R package phyloseq 97.
Statistical analyses
Statistical analyses were performed using the software R, version 3.6.0 98. Individual missing values in the explanatory variables (1.04% of all investigated predictor values), except for AMF richness and diversity (as these were also used as dependent variables in some models), were replaced with the median value of the respective land use type and country. Consequently, when the AMF data was included in models, the number of observations changed from 150 to 146 in the cropland soils, and from 60 to 58 in the grassland soils. For all correlations (i.e., between the different predictors and output variables) a Spearman rank correlation was applied. To assess the most important drivers of hyphal activity within the two land use systems, we performed a three-step model selection approach. First, we selected a wide range of available environmental and management data, under the condition that there is no strong co-correlation between the distinct variables. To do so, we scanned the correlations between all variables (Supplementary Tables 6 & 7) and reduced variables that were strongly correlated (i.e., Spearman rank correlation coefficient ρ > 0.8). This approach resulted in a pool of 15 predictors used to explain variation in the grassland soils and 25 for the cropland soils. To better describe variation in the cropland soils, the latter also included nine predictors describing management practices (tillage, fertilization, pesticide use, crop diversity, crop cover). The choice of final variables is indicated in the Supplementary Tables 6 and 7, and all used predictors with corresponding units are shown in Table 1 and Supplementary Table 1. The second step of the model selection involved a multi-model inference approach using the R package glmulti (version 1.0.8)99, resulting in a high number of possible linear models, ranked by the Akaine Information criterion (AICc). Using the best model, we explored whether model residuals were normally distributed and applied transformations to the data when needed. Logarithmic transformations were applied to the soil N:P ratio, Nmin:Pmin ratio, and plant N:P ratio, due to their strong non-normal distribution. Moreover, the hyphal 33P transfer rates were log-transformed for analyses focusing on the grassland systems. In the last step, the best models that fell within an AIC range of 2 were averaged to obtain a conservative estimate and relative importance of the most important predictors following Cade (2015)100 using the MuMin package (version 1.43.17)101. We used ΔAIC values to compute the Akaike weight (wi) of each of the selected models, and wi was used to compute a weighted-average model102. The relative importance of each predictor was estimated taking all the models into consideration as the sum of the weights/probabilities for the models in which the variable appears.
Structural equation modeling (SEM) was performed using the R package lavaan 103 to test the validity of the multi-model inference approach and to investigate direct relationships between the identified predictors (Olsen-P, pH, Aridity, SOC; and in the cropland soils fungicide application) and hyphal 33P transfer as well as indirect effects through changes in AMF richness. Initial SEMs comprised the complete set of possible correlation paths with hyphal 33P transfer. A stepwise removal of non-significant correlation paths was performed until a maximum of model-fit parameters (Chi-squared) was reached.
The relationships between the most important predictors and the hyphal 33P transfer were examined. Variables were transformed, if necessary, to reach a normal distribution of model error and residuals. Furthermore, we compared linear, polynomial (2nd order) and logarithmic functions and, eventually, showed the best-fitting functions (highest R2). For pair-wise comparisons between grassland and cropland soils, Wilcoxon rank test was used. For comparisons of more than two groups, a Kruskal-Wallis test followed by Dunn’s test was applied to identify which groups are different.
Extended Data
Extended Data Figure 1.
Extended Data Figure 2.
Extended Data Figure 3.
Extended Data Figure 4.
Extended Data Figure 5.
Supplementary Material
Acknowledgements
We are thankful to all the farmers and farm managers for allowing us to sample their fields and for completing our detailed questionnaires. We also thank Andrea Bonvicini, Dr. Laurie Schönholzer, Monica Macsai, Dr. Federica Tamburini, Dr. Hannes Gamper, Susanne Müller, Diane Bürge, Martin Zuber, Shuai Zhao, Vincent Somerville, Andri Brugger, Orlando Scholz, David Bugmann, Robin Heiz, Benjamin Seitz, and Miriam Roser for help with field work, the design and execution of the greenhouse experiment and lab analyses. We also thank Dr. Julian Helfenstein and the anonymous reviewers for valuable feedback on the manuscript. The Digging Deeper project was funded through the 2015-2016 BiodivERsA COFUND call for research proposals, with the national funders Swiss National Science Foundation (grant 31BD30-172466), Deutsche Forschungsgemeinschaft (317895346), Swedish Research Council Formas (contract 2016-0194), Ministerio de Economía y Competitividad (Digging_Deeper, Ref. PCIN-2016-028) and Agence Nationale de la Recherche (ANR, France; grant ANR-16-EBI3-0004-01).
Footnotes
Author contributions
M.v.d.H., S.H., F.T.M., L.P. and M.C.R. designed the study and obtained research funding. A.E., G.G., K.H., S.B., A.V-H., C.H., E.K., D.S.P., S.R., A.Sa., F.D., P.G-P. and A.Sp. contributed to data collection and analysis. J.J. and E.F. supported the design and execution of the greenhouse experiment. A.E., conducted data analysis. A.E., G.G., M.v.d.H. and J.J. were involved in the interpretation of results. A.E., G.G and M.v.d.H. drafted the manuscript, with significant contributions to the writing from all co-authors. All authors commented on and approved the final manuscript.
Competing interest
The authors declare no competing interests.
Data availability
The data that support the findings of this study are available here: 10.6084/m9.figshare.15134328.
Code availability
The code used to analyze the data is available here: 10.6084/m9.figshare.15134670.
References
- 1.Foley JA, et al. Solutions for a cultivated planet. Nature. 2011;478:337–342. doi: 10.1038/nature10452. [DOI] [PubMed] [Google Scholar]
- 2.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender SF, Wagg C, van der Heijden MGA. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol Evol. 2016;31:440–452. doi: 10.1016/j.tree.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Tamburini G, et al. Agricultural diversification promotes multiple ecosystem services without compromising yield. Sci Adv. 2020;6 doi: 10.1126/sciadv.aba1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith S, Read D. Mycorrhizal Symbiosis Mycorrhizal Symbiosis. Elsevier; 2008. [DOI] [Google Scholar]
- 6.Soudzilovskaia NA, et al. Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Glob Ecol Biogeogr. 2015;24:371–382. [Google Scholar]
- 7.Van Der Heijden MGA, Bardgett RD, Van Straalen NM. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 8.Bennett EM, Carpenter SR, Caraco NF. Human impact on erodable phosphorus and eutrophication: A global perspective. Bioscience. 2001;51:227–234. [Google Scholar]
- 9.Smith VH, Schindler DW. Eutrophication science: where do we go from here? Trends Ecol Evol. 2009;24:201–207. doi: 10.1016/j.tree.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Rillig MC, Mummey DL. Mycorrhizas and soil structure. New Phytol. 2006;171:41–53. doi: 10.1111/j.1469-8137.2006.01750.x. [DOI] [PubMed] [Google Scholar]
- 11.Bender SF, van der Heijden MGA. Soil biota enhance agricultural sustainability by improving crop yield, nutrient uptake and reducing nitrogen leaching losses. J Appl Ecol. 2015;52:228–239. [Google Scholar]
- 12.Rodriguez A, Sanders IR. The role of community and population ecology in applying mycorrhizal fungi for improved food security. ISME Journal. 2015;9:1053–1061. doi: 10.1038/ismej.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oviatt P, Rillig MC. Mycorrhizal technologies for an agriculture of the middle. Plants People Planet. 2020:ppp3.10177. doi: 10.1002/ppp3.10177. [DOI] [Google Scholar]
- 14.Ryan MH, Graham JH. Little evidence that farmers should consider abundance or diversity of arbuscular mycorrhizal fungi when managing crops. New Phytologist. 2018;220:1092–1107. doi: 10.1111/nph.15308. [DOI] [PubMed] [Google Scholar]
- 15.Rillig MC, et al. Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytol. 2019;222:1171–1175. doi: 10.1111/nph.15602. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Lehmann A, Zheng W, You Z, Rillig MC. Arbuscular mycorrhizal fungi increase grain yields: a meta-analysis. New Phytol. 2019;222:543–555. doi: 10.1111/nph.15570. [DOI] [PubMed] [Google Scholar]
- 17.Thirkell TJ, Charters MD, Elliott AJ, Sait SM, Field KJ. Are mycorrhizal fungi our sustainable saviours? Considerations for achieving food security. J Ecol. 2017;105:921–929. [Google Scholar]
- 18.Davison J, et al. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science (80-.) 2015;349:970–973. doi: 10.1126/science.aab1161. [DOI] [PubMed] [Google Scholar]
- 19.Pringle A, Bever JD. Analogous effects of arbuscular mycorrhizal fungi in the laboratory and a North Carolina field. New Phytol. 2008;180:162–175. doi: 10.1111/j.1469-8137.2008.02537.x. [DOI] [PubMed] [Google Scholar]
- 20.Francis R, Read DJ. Mutualism and antagonism in the mycorrhizal symbiosis, with special reference to impacts on plant community structure. Can J Bot. 1995;73:1301–1309. [Google Scholar]
- 21.Thirkell TJ, Pastok D, Field KJ. Carbon for nutrient exchange between arbuscular mycorrhizal fungi and wheat varies according to cultivar and changes in atmospheric carbon dioxide concentration. Glob Chang Biol. 2020;26:1725–1738. doi: 10.1111/gcb.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann A, Barto EK, Powell JR, Rillig MC. Mycorrhizal responsiveness trends in annual crop plants and their wild relatives—a meta-analysis on studies from 1981 to 2010. Plant Soil. 2012;355:231–250. [Google Scholar]
- 23.Martín-Robles N, et al. Impacts of domestication on the arbuscular mycorrhizal symbiosis of 27 crop species. New Phytol. 2018;218:322–334. doi: 10.1111/nph.14962. [DOI] [PubMed] [Google Scholar]
- 24.Leake J, et al. Networks of power and influence: The role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Can J Bot. 2004;82:1016–1045. [Google Scholar]
- 25.Oehl F, et al. Impact of Land Use Intensity on the Species Diversity of Arbuscular Mycorrhizal Fungi in Agroecosystems of Central Europe. Appl Environ Microbiol. 2003;69:2816–2824. doi: 10.1128/AEM.69.5.2816-2824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang D, et al. Land use influences arbuscular mycorrhizal fungal communities in the farming-pastoral ecotone of northern China. New Phytol. 2014;204:968–978. doi: 10.1111/nph.12961. [DOI] [PubMed] [Google Scholar]
- 27.Bainard LD, et al. Plant communities and soil properties mediate agricultural land use impacts on arbuscular mycorrhizal fungi in the Mixed Prairie ecoregion of the North American Great Plains. Agric Ecosyst Environ. 2017;249:187–195. [Google Scholar]
- 28.Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. Ploughing up the wood-wide web? Nature. 1998;394:431. doi: 10.1038/28764. [DOI] [PubMed] [Google Scholar]
- 29.van der Heijden MGA, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- 30.Vogelsang KM, Reynolds HL, Bever JD. Mycorrhizal fungal identity and richness determine the diversity and productivity of a tallgrass prairie system. New Phytol. 2006;172:554–562. doi: 10.1111/j.1469-8137.2006.01854.x. [DOI] [PubMed] [Google Scholar]
- 31.Scheublin TR, Ridgway KP, Young JPW, van der Heijden MGA. Nonlegumes, Legumes, and Root Nodules Harbor Different Arbuscular Mycorrhizal Fungal Communities. Appl Environ Microbiol. 2004;70:6240–6246. doi: 10.1128/AEM.70.10.6240-6246.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oehl F, et al. Soil type and land use intensity determine the composition of arbuscular mycorrhizal fungal communities. Soil Biol Biochem. 2010;42:724–738. [Google Scholar]
- 33.De Vries FT, et al. Soil food web properties explain ecosystem services across European land use systems. Proc Natl Acad Sci U S A. 2013;110:14296–14301. doi: 10.1073/pnas.1305198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verbruggen E, Xiang D, Chen B, Xu T, Rillig MC. Mycorrhizal fungi associated with high soil N:P ratios are more likely to be lost upon conversion from grasslands to arable agriculture. Soil Biol Biochem. 2015;86:1–4. [Google Scholar]
- 35.Balami S, Vašutová M, Godbold D, Kotas P, Cudlín P. Soil fungal communities across land use types. IForest. 2020;13:548–558. [Google Scholar]
- 36.Öpik M, Mari M, Liira J, Zobel M. Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol. 2006;94:778–790. [Google Scholar]
- 37.Jansa J, et al. Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza. 2002;12:225–234. doi: 10.1007/s00572-002-0163-z. [DOI] [PubMed] [Google Scholar]
- 38.van Groenigen KJ, et al. Abundance, production and stabilization of microbial biomass under conventional and reduced tillage. Soil Biol Biochem. 2010;42:48–55. [Google Scholar]
- 39.Sallach JB, Thirkell TJ, Field KJ, Carter LJ. The emerging threat of human-use antifungals in sustainable and circular agriculture schemes. PLANTS, PEOPLE, PLANET. 2021;3:685–693. [Google Scholar]
- 40.Meyer A, et al. Different Land Use Intensities in Grassland Ecosystems Drive Ecology of Microbial Communities Involved in Nitrogen Turnover in Soil. PLoS One. 2013;8:e73536. doi: 10.1371/journal.pone.0073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsiafouli MA, et al. Intensive agriculture reduces soil biodiversity across Europe. Glob Chang Biol. 2015;21:973–985. doi: 10.1111/gcb.12752. [DOI] [PubMed] [Google Scholar]
- 42.Tardy V, et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol Biochem. 2015;90:204–213. [Google Scholar]
- 43.Sawers RJH, et al. Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 2017;214:632–643. doi: 10.1111/nph.14403. [DOI] [PubMed] [Google Scholar]
- 44.Svenningsen NB, et al. Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J. 2018;12:1296–1307. doi: 10.1038/s41396-018-0059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweiger PF, Thingstrup I, Jakobsen I. Comparison of two test systems for measuring plant phosphorus uptake via arbuscular mycorrhizal fungi. Mycorrhiza. 1999;8:207–213. [Google Scholar]
- 46.Emmett BD, Lévesque-Tremblay V, Harrison MJ. Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi. ISME J. 2021;15:2276–2288. doi: 10.1038/s41396-021-00920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang F, Zhang L, Zhou J, George TS, Feng G. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytol. 2021;230:304–315. doi: 10.1111/nph.17081. [DOI] [PubMed] [Google Scholar]
- 48.Thonar C, Schnepf A, Frossard E, Roose T, Jansa J. Traits related to differences in function among three arbuscular mycorrhizal fungi. Plant Soil. 2011;339:231–245. [Google Scholar]
- 49.Cavagnaro TR, Smith FA, Smith SE, Jakobsen I. Functional diversity in arbuscular mycorrhizas: exploitation of soil patches with different phosphate enrichment differs among fungal species. Plant, Cell and Environment. 2005;28 [Google Scholar]
- 50.Jakobsen I, Gazey C, Abbott LK. Phosphate transport by communities of arbuscular mycorrhizal fungi in intact soil cores. New Phytol. 2001;149:95–103. doi: 10.1046/j.1469-8137.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- 51.Pearson JN, Jakobsen I. The relative contribution of hyphae and roots to phosphorus uptake by arbuscular mycorrhizal plants, measured by dual labelling with 32P and 33P. New Phytol. 1993;124:489–494. [Google Scholar]
- 52.Nagy R, Drissner D, Amrhein N, Jakobsen I, Bucher M. Erratum: Mycorrhizal phosphate uptake pathway in tomato is phosphorusrepressible and transcriptionally regulated (New Phytologist (2009) 181 (950-959)) New Phytol. 2009;184:1029. doi: 10.1111/j.1469-8137.2008.02721.x. [DOI] [PubMed] [Google Scholar]
- 53.Smith SE, Jakobsen I, Grønlund M, Smith FA. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011;156:1050–1057. doi: 10.1104/pp.111.174581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams A, Manoharan L, Rosenstock NP, Olsson PA, Hedlund K. Long-term agricultural fertilization alters arbuscular mycorrhizal fungal community composition and barley (Hordeum vulgare) mycorrhizal carbon and phosphorus exchange. New Phytol. 2017;213:874–885. doi: 10.1111/nph.14196. [DOI] [PubMed] [Google Scholar]
- 55.Koerselman W, Meuleman AFM. The Vegetation N:P Ratio: a New Tool to Detect the Nature of Nutrient Limitation. J Appl Ecol. 1996;33:1441. [Google Scholar]
- 56.Van Aarle IM, Olsson PA, Söderström B. Arbuscular mycorrhizal fungi respond to the substrate pH of their extraradical mycelium by altered growth and root colonization. New Phytol. 2002;155:173–182. doi: 10.1046/j.1469-8137.2002.00439.x. [DOI] [PubMed] [Google Scholar]
- 57.Staddon PL, et al. Mycorrhizal fungal abundance is affected by long-term climatic manipulations in the field. Glob Chang Biol. 2003;9:186–194. [Google Scholar]
- 58.Weber SE, et al. Responses of arbuscular mycorrhizal fungi to multiple coinciding global change drivers. Fungal Ecol. 2019;40:62–71. [Google Scholar]
- 59.Peat HJ, Fitter AH. The distribution of arbuscular mycorrhizas in the British flora. New Phytol. 1993;125:845–854. doi: 10.1111/j.1469-8137.1993.tb03933.x. [DOI] [PubMed] [Google Scholar]
- 60.Cruz-Paredes C, et al. Suppression of arbuscular mycorrhizal fungal activity in a diverse collection of non-cultivated soils. FEMS Microbiol Ecol. 2019;95 doi: 10.1093/femsec/fiz020. [DOI] [PubMed] [Google Scholar]
- 61.Jansa J, Erb A, Oberholzer H-R, Šmilauer P, Egli S. Soil and geography are more important determinants of indigenous arbuscular mycorrhizal communities than management practices in Swiss agricultural soils. Mol Ecol. 2014;23:2118–2135. doi: 10.1111/mec.12706. [DOI] [PubMed] [Google Scholar]
- 62.Davison J, et al. Temperature and pH define the realised niche space of arbuscular mycorrhizal fungi. New Phytol. 2021;231:763–776. doi: 10.1111/nph.17240. [DOI] [PubMed] [Google Scholar]
- 63.Yang H, et al. Changes in soil organic carbon, total nitrogen, and abundance of arbuscular mycorrhizal fungi along a large-scale aridity gradient. Catena. 2011;87:70–77. [Google Scholar]
- 64.Riedo J, et al. Widespread Occurrence of Pesticides in Organically Managed Agricultural Soils—the Ghost of a Conventional Agricultural Past? Environ Sci Technol. 2021 doi: 10.1021/acs.est.0c06405. [DOI] [PubMed] [Google Scholar]
- 65.Pánková H, Dostálek T, Vazačová K, Münzbergová Z. Slow recovery of arbuscular mycorrhizal fungi and plant community after fungicide application: An eight-year experiment. J Veg Sci. 2018;29:695–703. [Google Scholar]
- 66.Ipsilantis I, Samourelis C, Karpouzas DG. The impact of biological pesticides on arbuscular mycorrhizal fungi. Soil Biol Biochem. 2012 doi: 10.1016/j.soilbio.2011.08.007. [DOI] [Google Scholar]
- 67.Buysens C, Dupré de Boulois H, Declerck S. Do fungicides used to control Rhizoctonia solani impact the non-target arbuscular mycorrhizal fungus Rhizophagus irregularis? Mycorrhiza. 2015 doi: 10.1007/s00572-014-0610-7. [DOI] [PubMed] [Google Scholar]
- 68.Lekberg Y, Wagner V, Rummel A, McLeod M, Ramsey PW. Strong indirect herbicide effects on mycorrhizal associations through plant community shifts and secondary invasions. Ecol Appl. 2017;27:2359–2368. doi: 10.1002/eap.1613. [DOI] [PubMed] [Google Scholar]
- 69.Hage-Ahmed K, Rosner K, Steinkellner S. Arbuscular mycorrhizal fungi and their response to pesticides. Pest Management Science. 2019;75:583–590. doi: 10.1002/ps.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kjøller R, Rosendahl S. Effects of fungicides on arbuscular mycorrhizal fungi: differential responses in alkaline phosphatase activity of external and internal hyphae. Biol Fertil Soils. 2000;31:361–365. [Google Scholar]
- 71.Gange AC, Brown VK, Sinclair GS. Vesicular-Arbuscular Mycorrhizal Fungi: A Determinant of Plant Community Structure in Early Succession. Funct Ecol. 1993;7:616. [Google Scholar]
- 72.Hartnett DC, Wilson GWT. The role of mycorrhizas in plant community structure and dynamics: Lessons from grasslands. Plant Soil. 2002;244:319–331. [Google Scholar]
- 73.Guzman A, et al. Crop diversity enriches arbuscular mycorrhizal fungal communities in an intensive agricultural landscape. 2021 doi: 10.1111/nph.17306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eurostat. LUCAS 2018 Technical reference document C3 Classification (Land cover & Land use) 2018;2018:98. [Google Scholar]
- 75.Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–4315. [Google Scholar]
- 76.Trabucco A, Zomer R. Global Aridity Index and Potential Evapotranspiration (ET0) Climate Database v2. figshare. 2019 doi: 10.6084/m9.figshare.7504448.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.García-Palacios P, Gross N, Gaitán J, Maestre FT. Climate mediates the biodiversity-ecosystem stability relationship globally. Proc Natl Acad Sci U S A. 2018;115:8400–8405. doi: 10.1073/pnas.1800425115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berdugo M, et al. Global ecosystem thresholds driven by aridity. Science (80-.) 2020;367:787–790. doi: 10.1126/science.aay5958. [DOI] [PubMed] [Google Scholar]
- 79.Sinnott RW. Virtues of the Haversine. Sky Telescope. 1984;68(2):158. [Google Scholar]
- 80.Garland G, et al. Crop cover is more important than rotational diversity for soil multifunctionality and cereal yields in European cropping systems. Nat Food. 2021;2:28–37. doi: 10.1038/s43016-020-00210-8. [DOI] [PubMed] [Google Scholar]
- 81.Swiss federal research stations. Schweizerische Referenzmethoden der Eidgenössischen Forschungsanstalten. Boden-und Substratuntersuchungen zur Düngeberatung. 1996 [Google Scholar]
- 82.Berry D, Ben Mahfoudh K, Wagner M, Loy A. Barcoded Primers Used in Multiplex Amplicon Pyrosequencing Bias Amplification † ‘Barcode-tagged’ PCR primers used for multiplex amplicon sequencing generate a thus-far-overlooked amplification bias that produces variable terminal restriction fragment length polymorphism (T-RFLP) and pyrosequencing data from the same environmental DNA template. We propose a simple two-step PCR ap-proach that increases reproducibility and consistently recovers higher genetic diversity in pyrosequencing libraries. Appl Environ Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gardes M, White TJ, Fortin JA, Bruns TD, Taylor JW. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Can J Bot. 1991;69:180–190. [Google Scholar]
- 84.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 85.Fiore-Donno AM, et al. New barcoded primers for efficient retrieval of cercozoan sequences in high-throughput environmental diversity surveys, with emphasis on worldwide biological soil crusts. Mol Ecol Resour. 2018;18:229–239. doi: 10.1111/1755-0998.12729. [DOI] [PubMed] [Google Scholar]
- 86.Helfenstein J, Jegminat J, McLaren TI, Frossard E. Soil solution phosphorus turnover: Derivation, interpretation, and insights from a global compilation of isotope exchange kinetic studies. Biogeosciences. 2018;15:105–114. [Google Scholar]
- 87.Thirkell TJ, et al. Cultivar-dependent increases in mycorrhizal nutrient acquisition by barley in response to elevated CO 2. PLANTS, PEOPLE, PLANET. 2021;3:553–566. [Google Scholar]
- 88.Rodushkin I, Ruth T, Huhtasaari Å. Comparison of two digestion methods for elemental determinations in plant material by ICP techniques. Anal Chim Acta. 1999;378:191–200. [Google Scholar]
- 89.Ohno T, Zibilske LM. Determination of Low Concentrations of Phosphorus in Soil Extracts Using Malachite Green. Soil Sci Soc Am J. 1991;55:892–895. [Google Scholar]
- 90.Frossard E, et al. The Use of Tracers to Investigate Phosphate Cycling in Soil–Plant Systems. 2011:59–91. doi: 10.1007/978-3-642-15271-9_3. [DOI] [Google Scholar]
- 91.Sato K, Suyama Y, Saito M, Sugawara K. A new primer for discrimination of arbuscular mycorrhizal fungi with polymerase chain reaction-denature gradient gel electrophoresis. Grassl Sci. 2005;51:179–181. [Google Scholar]
- 92.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Öpik M, et al. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota) New Phytol. 2010;188:223–241. doi: 10.1111/j.1469-8137.2010.03334.x. [DOI] [PubMed] [Google Scholar]
- 95.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.R Core team. R: A Language and Environment for Statistical Computing. 2013 [Google Scholar]
- 99.Calcagno V. Model Selection and Multimodel Inference Made Easy [R package glmulti version 1.0.8] 2020 [Google Scholar]
- 100.Cade BS. Model averaging and muddled multimodel inferences. Ecology. 2015 doi: 10.1890/14-1639.1. [DOI] [PubMed] [Google Scholar]
- 101.Barton K. MuMIn: Multi-Model Inference R package version 1.43.17. Version 1. 2020:18. [Google Scholar]
- 102.Model Selection and Multimodel Inference. Springer; New York: 2002. [DOI] [Google Scholar]
- 103.Rosseel Y. Lavaan: An R package for structural equation modeling. J Stat Softw. 2012 doi: 10.18637/jss.v048.i02. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available here: 10.6084/m9.figshare.15134328.
The code used to analyze the data is available here: 10.6084/m9.figshare.15134670.