Abstract
Apicomplexan parasites secrete contents of the rhoptries, club-shaped organelles in the apical region, into host cells to permit their invasion and establishment of infection. The rhoptry secretory apparatus (RSA), which is critical for rhoptry secretion, was recently discovered in Toxoplasma and Cryptosporidium. It is unknown if a similar molecular machinery exists in the malaria parasite Plasmodium. In this study, we use in situ cryo-electron tomography to investigate the rhoptry secretion system in P. falciparum merozoites. We identify the presence of an RSA at the cell apex and a morphologically distinct apical vesicle docking the tips of the two rhoptries to the RSA. We also discover two additional rhoptry organizations that lack the apical vesicle. Using subtomogram averaging, we reveal different conformations of the RSA structure corresponding to different rhoptry organizations. Our results highlight previously unknown steps in the process of rhoptry secretion and indicate a regulatory role for the conserved apical vesicle in host invasion by apicomplexan parasites.
Plasmodium parasites, the causative agents of malaria, belong to the phylum Apicomplexa that comprises important disease-causing parasites including Toxoplasma and Cryptosporidium1–3. Invasive stages of these parasites, such as Plasmodium merozoites, contain a defining apical complex that harbors specialized structures and secretory organelles dedicated to parasite motility and host cell invasion4,5. Upon recognition of a red blood cell, the merozoite secretes the contents of a pair of club-shaped rhoptries into the host cytosol, permitting its invasion and development within5,6. The mechanism by which rhoptries regulate and conduct content secretion across their bounding membrane, the parasite plasma membrane (PPM), and the host plasma membrane remain largely unknown7.
Recently, multiple non-discharge (Nd) and Nd partner (NdP) proteins from T. gondii and P. falciparum were implicated in the rhoptry secretion mechanism8. Moreover, cryo-electron tomography (cryo-ET) of T. gondii and C. parvum cells has revealed an elaborate molecular assembly, called the rhoptry secretory apparatus (RSA), which in association with an apical vesicle (AV) docks the rhoptries to the parasite apex8,9. Knockdown of the Nd9 protein disrupted the RSA structure in T. gondii and abolished rhoptry secretion in both T. gondii and P. falciparum, linking the Nd-NdP proteins, the RSA, and rhoptry secretion together. However, whether P. falciparum utilizes a similar RSA and/or AV for rhoptry docking has yet to be determined. Furthermore, if the AV is an integral part of this unique eukaryotic secretion system, then two passageways – one between the rhoptry and the AV and another between the AV and the PPM – need to open for rhoptry secretion. The purpose and regulation of these proposed passageways and the role of the AV are unclear7.
Here we performed cryo-ET of P. falciparum merozoites to investigate the ultrastructure of their rhoptry secretion system. We consistently observed an RSA in all 218 merozoites imaged and an AV in nearly two-thirds of them. Through subtomogram averaging we revealed a similar RSA architecture to that of T. gondii and C. parvum9, anchoring into the PPM and forming connections to the AV held below. Furthermore, in the merozoites without an AV, we captured two additional organizations of the rhoptry pair. Subtomogram averaging of RSAs from these two AV-lacking states performed independently resolved different RSA conformations that could be important for rhoptry secretion. Together, our data support a model of the sequential events leading to rhoptry secretion, beginning with the opening of a passageway between the AV and one of the two rhoptries, followed by fusion of the two rhoptries and opening of a gate-like density in the RSA, suggesting a role for the AV in regulating rhoptry fusion to prime their secretion.
Results
The P. falciparum rhoptry tips dock at an apical vesicle
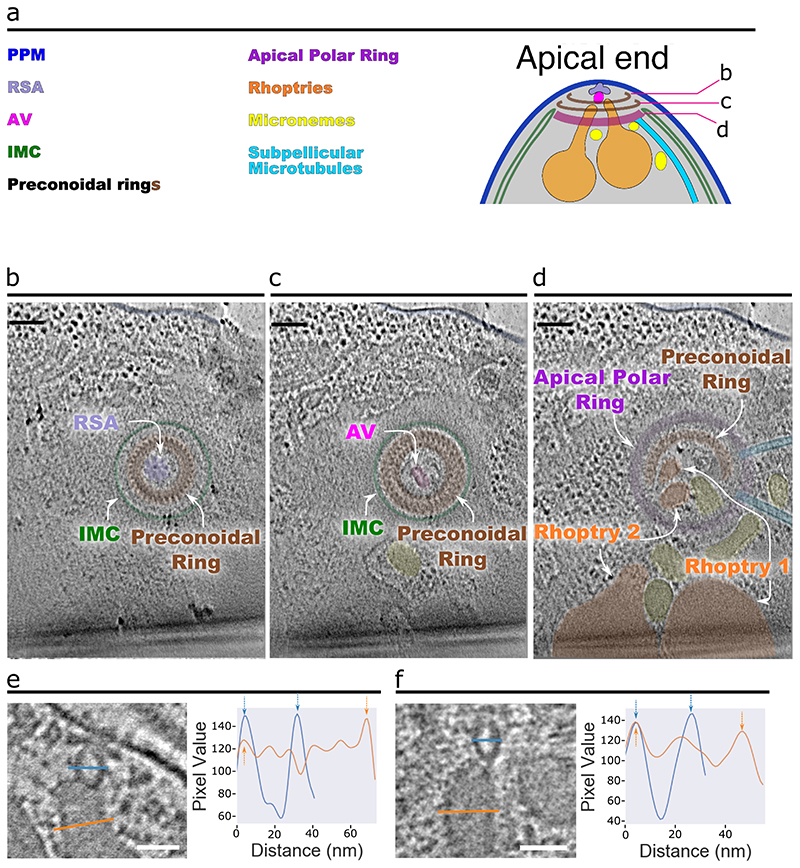
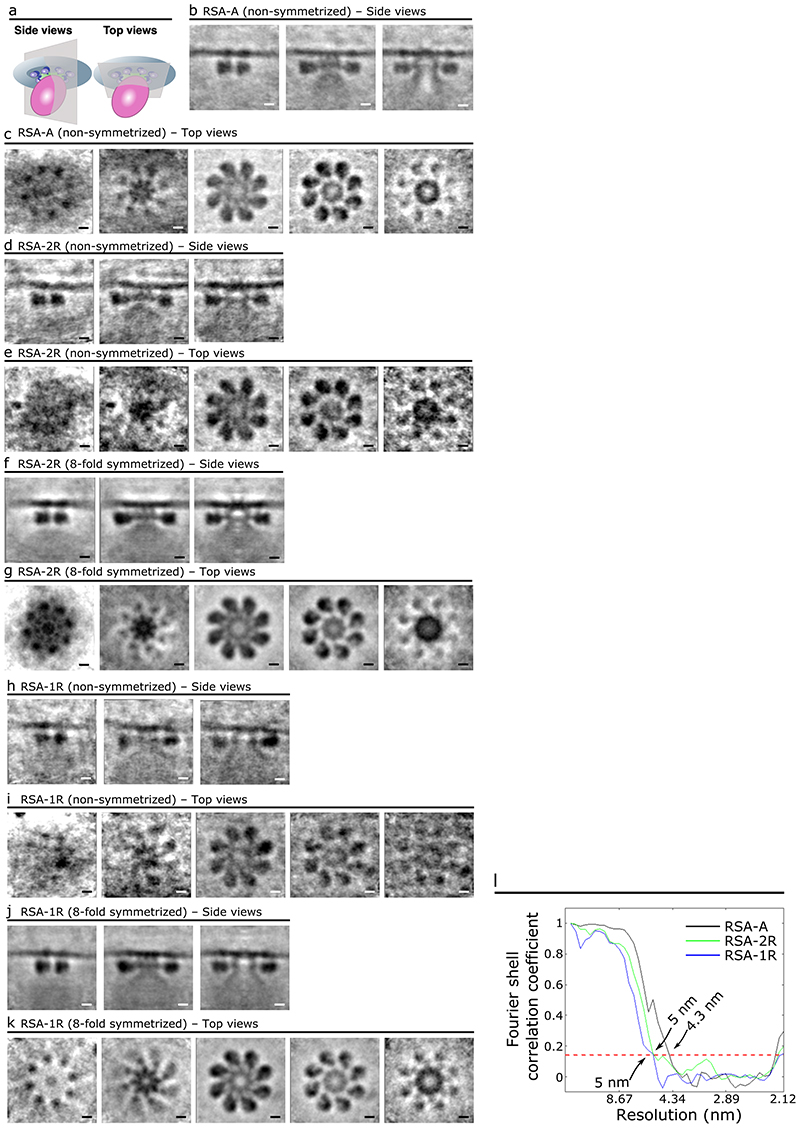
Using cryo-ET to image the apical region of P. falciparum merozoites (Fig. 1a, b), we resolved all the defining features of the apical complex, including two preconoidal rings, the apical polar ring, the inner membrane complex (IMC), rhoptries (Fig. 1b), micronemes, and subpellicular microtubules (Extended Data Fig. 1a-d). Although helical filaments conspicuously line the luminal face of T. gondii and C. parvum rhoptries9, similar filaments could not be reliably observed in P. falciparum rhoptries likely due to sample thickness and high electron density within the rhoptries. We observed a distinct AV associated with the rhoptry tips (Fig. 1b, c) that is a distinct compartment from the rhoptry as evidenced by the significantly lower electron density within the AV (Fig. 1d, e and Extended Data Fig. 1e, f). Like T. gondii and C. parvum, the AV in P. falciparum merozoites is anchored at the PPM via the RSA (see details below). Of the 39 AVs analyzed, 34 (87%) have two rhoptries docked, four (10%) have just one rhoptry docked, and one (3%) has no rhoptry docked (Fig. 1l and Extended Data Fig. 2a-c). However, the AV of P. falciparum is more variable in its size and docking parameters compared to T. gondii and C. parvum9 (Fig. 1f-k and Extended Data Fig. 3). In summary, our data reveal that the AV is a conserved feature across divergent apicomplexan parasites, docking the rhoptry tips to the RSA at the PPM.
Figure 1. P. falciparum merozoites harbor an apical vesicle that docks the rhoptries to the parasite plasma membrane.
(a) Simplified schematic of a P. falciparum merozoite. (b) 2-D slice from a tomogram of the merozoite apical complex displaying two rhoptries (orange), rhoptry tip density (cyan) two preconoidal rings (brown), an apical polar ring (purple), the parasite plasma membrane (PPM; dark blue), the inner membrane complex (IMC; green), the apical vesicle (AV; pink), and the rhoptry secretory apparatus (RSA; light blue). (c) Zoom-in view of the merozoite apex from panel b without color overlays. The orange and blue lines crossing the rhoptry and the AV, respectively, denote the locations of the pixel values shown in panel d. (d) Representative line profile plot of the inverted pixel values across the rhoptry (orange) and AV (blue). Arrows point to peaks corresponding to membranes. Shaded regions are the central 1/3 of pixels used to calculate the luminal density for the ratios shown in panel e. (e) Box plot showing the ratios of the luminal density to the corresponding membrane density (averaged) for each AV (blue) and docked rhoptry (orange). Data are presented as 25th percentile, median, and 75th percentile with whiskers representing ± 1.5x the interquartile range. Mean value represented by the dotted black line. N = 12 AVs and 12 rhoptries. (f) Summary schematic of AV dimensions and docking distance to the PPM. (g) Summary schematic of the docking parameters among the rhoptries, AV, and RSA/PPM. (h) Plot of the angle (Ψ) at which the major axis of the AV is oriented with respect to the line connecting the PPM apex to the AV centroid, measured from several cells. Data are presented as 25th percentile, median, and 75th percentile with whiskers representing ± 1.5x the interquartile range. (i) Plot of the angles at which rhoptry one (θRh1) and rhoptry two (θRh2) are docked at the AV (mean ± SD), with respect to the major axis of the AV. (j) Measurement of the shortest distance between the PPM apex and the AV membrane (mean ± SD), and (k) Plot of the major versus minor axes of the AV (mean ± SD). (l) Histogram of the number of rhoptries docked at the AV in each cell. N = 39 AVs for panels h, j-l and N = 34 AVs and 68 rhoptries for panel i. Scale bars = 100 nm.
Merozoites display multiple rhoptry fusion morphologies
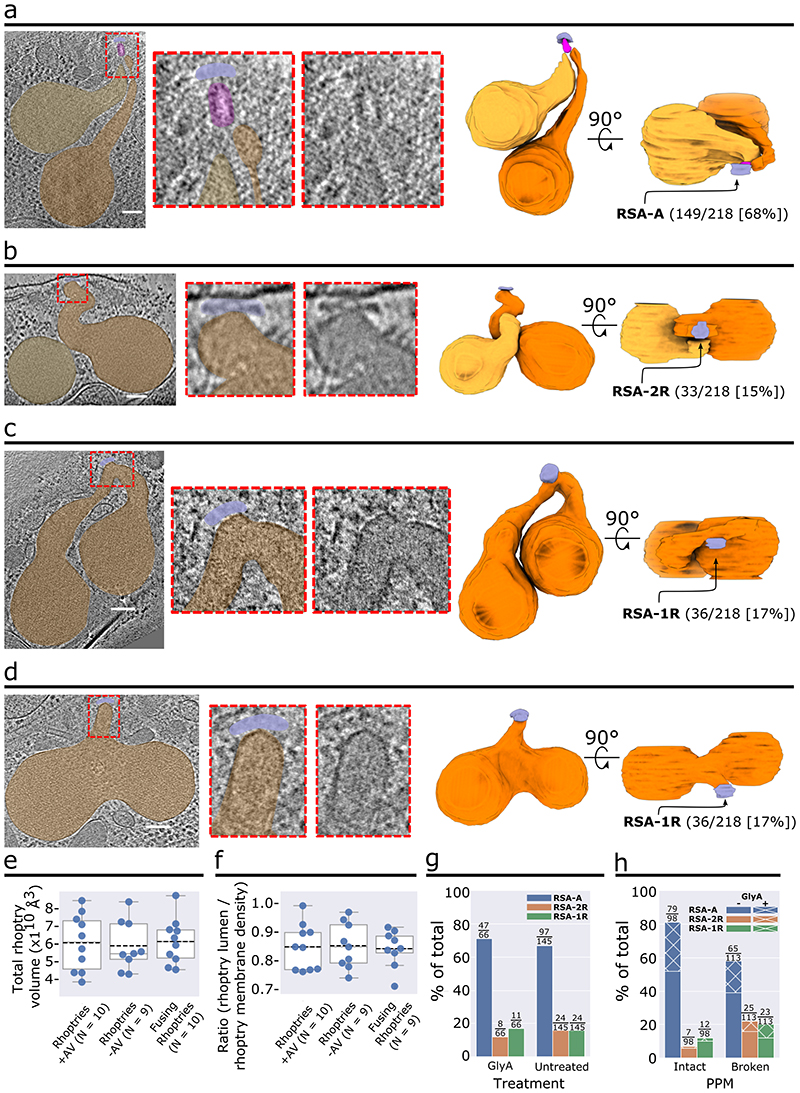
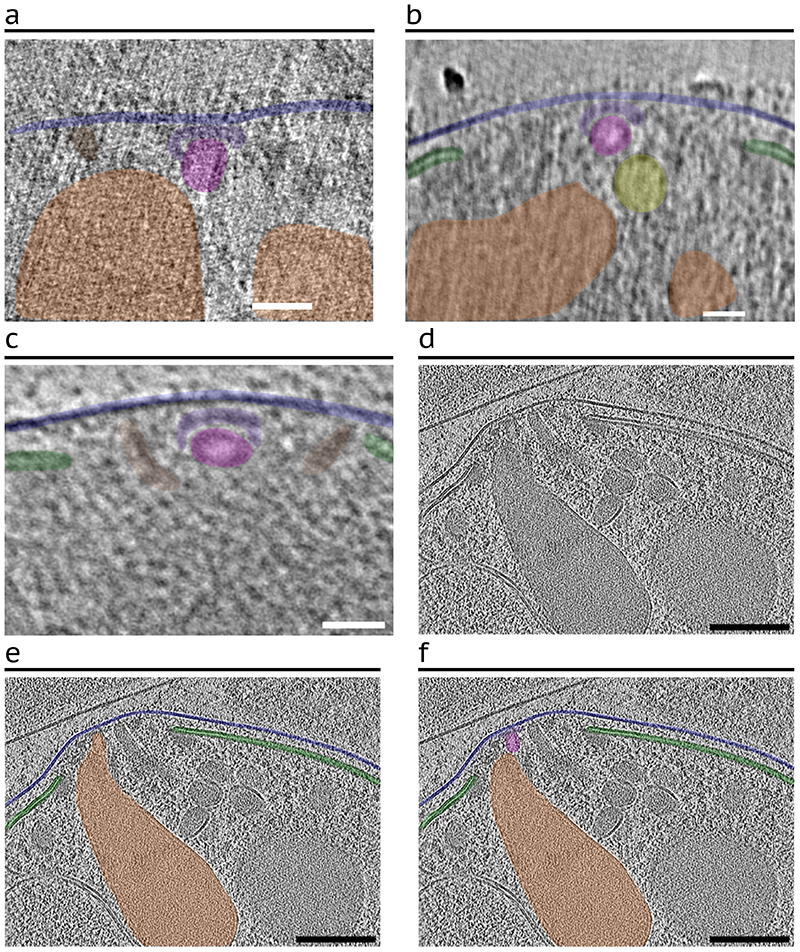
In addition to the discovery of the AV in P. falciparum (Fig. 1, 2a), we observed other rhoptry secretion system morphologies in a subset of merozoite tomograms analyzed (69/218; Fig. 2b-d). Among them, 33 (15%) lack the AV and instead have one of the two rhoptries docked directly to the RSA (Fig. 2b and Extended Data Fig. 4a-c). This likely reflects the fusion of the AV to one of the two rhoptries (Fig. 2b). Additionally, 36 (17%) merozoites display fusing rhoptries; again, these parasites lack an AV and instead the fused rhoptry tip is docked directly at the RSA. The fusion between the rhoptries ranged from fusion at the tips (Fig. 2c) to complete fusion to the base of the bulbs (Fig. 2d and Extended Data Fig. 4d-g). Importantly, no AV was observed in all cases with fused rhoptries, suggesting that disappearance of the AV precedes the fusion of the rhoptry pair.
Figure 2. Various observed morphological states of the rhoptry secretion system.
(a–d) Representative examples of a merozoite apical end displaying two separate rhoptries docked at the AV (a), two separate rhoptries with one of them docked directly at the RSA (b), and rhoptries fused at the tips (c) and along the necks (d). Left: Tomogram slice of the apical end with color overlay on the RSA (light blue), AV (pink), and rhoptries (orange). Middle: Zoom-in of the apical tip of the rhoptries, with and without color overlay. Right: 3-D segmentations of the rhoptries, AV, and RSA with the proportion of analyzed merozoites exhibiting each morphology denoted. (e) Rhoptry volumetric analysis of 3-D rhoptry segmentations among the three rhoptry morphological states and (f) Rhoptry luminal density analysis of 2-D slices of one rhoptry among the three rhoptry states from each tomogram analyzed in panel e. Data are presented as 25th percentile, median, and 75th percentile with whiskers representing ± 1.5x the interquartile range. Mean value represented by the dotted black line. (g) Plot of the proportion of P. falciparum merozoites, stratified by Glycophorin A (GlyA) treatment, and (h) Plot of the proportion of merozoites, stratified by PPM integrity, displaying each rhoptry morphology. Numbers above each bar represent the number of merozoites displaying each morphology over the total number of merozoites in each set (i.e., with or without GlyA treatment [g] or an intact or ruptured PPM [h]). N = 211 for total analyzed merozoites. Scale bars = 100 nm.
Rhoptry fusion has been previously observed during merozoite invasion of red blood cells when the rhoptries secrete their contents10,11. We therefore sought to determine if the different rhoptry morphologies observed correspond to premature secretion in the absence of host cells. We identified 9-10 tomograms of each morphological class (Fig. 2a-d) with entire rhoptries captured in the field of view. No significant difference was observed in the average rhoptry volume among the different morphological classes (Fig. 2e). Similarly, density profiles of the rhoptry lumen also show no significant difference across the different classes (Fig. 2f). These observations suggest that there is no major secretion occurring upon rhoptry fusion in the absence of a host red blood cell.
In attempts to understand the triggers for these morphological changes, we treated merozoites with the red blood cell surface protein glycophorin A (GlyA) as it was once shown to induce the secretion of two rhoptry proteins to the surface of isolated merozoites12. We found that GlyA treatment did not affect the ratio of rhoptry morphologies among 67 merozoites (Fig. 2g), suggesting an unrelated trigger for rhoptry morphology changes and AV disappearance. Interestingly, we discovered that accidental rupture of the PPM (that likely happened during cryo-ET sample preparation)13 correlated with a significant decrease in the proportion of the morphology containing an AV (from 81% to 58%; Fig. 2h). This suggests that regulation of AV’s disappearance may be influenced by the integrity of the PPM.
Subtomogram averaging reveals P. falciparum RSA structure
In all tomograms of merozoites containing an AV, the AV is always anchored to the PPM via the RSA (Figs. 1b, c, 2a, and Extended Data Fig. 1). We performed subtomogram averaging using all the 149 RSAs associated with an AV (RSA-A) to enhance the structural details to an overall resolution of 4.3 nm, revealing a remarkably intricate molecular assembly (Fig. 3, Extended Data Fig. 5b, c, and Movie 1). As part of the RSA, we resolved an apical rosette of 8 membrane anchors (Anchor-I [A-I]) surrounding a central density (CD) at the PPM (Fig. 3b, c[i], and d[top]). Densities extend from each A-I towards the AV (Anchor-II [A-II] and Anchor-III [A-III]; Fig. 3b), showing a clear 8-fold arrangement in the intracellular space (Fig. 3c, d[side], f). A-II connects to A-I in the PPM (Fig. 3b), and A-III connects to the AV (Fig. 3c[v] and Extended Data Fig. 6). Furthermore, in the center of the 8-fold A-II/III densities lies a central channel (CC) that sits atop the tip of the AV (Fig. 3b[iii], c[iii], and e). The CC connects to another set of 8 anchors (Anchor-IV [A-IV]; Fig. 3b[ii], c[i]) in the PPM via the radiating spokes (rS; Fig. 3c[ii], e). We also resolved a density at the interface of CC and the AV, termed the gate density (gD; Fig. 3b[iii], e[bottom], f[bottom]). All the structural details revealed by subtomogram averaging of the P. falciparum RSA-A are summarized in 3-D in Movie 1.
Figure 3. In situ structure of the multicomponent rhoptry secretory apparatus of P. falciparum.
(a) Schematic of the RSA, AV, and PPM demonstrating the side view and top view orientations used to generate 2-D slices of the subtomogram average in panels b and c. (b) Side view slices of the RSA-A subtomogram average, from peripheral (left) to central (right) cross sections. The corresponding cross sections of the top view slices in panel c are denoted by the pink lines in b(iii). Colored outlines and overlays denote components of the RSA observed in these views: The Anchors I-IV (A-I, A-II, A-III, A-IV), the central channel (CC), the central density (CD), and the gate density (gD). (c) Top view slices of the RSA subtomogram average, from extracellular (left) to intracellular (right) cross sections. Colored outlines and overlays denote components of the RSA observed in these views: the radiating spokes density (rS) and other components as labeled in panel b. (d) 3-D segmentation of the RSA displaying outer most components from top and side views. (e) 3-D segmentation of the RSA displaying interior components from side views. (f) 3-D segmentation of the RSA from a bottom view with the AV shown (top) and not shown (bottom) to reveal the gD (bottom). Resolved to 4.3 nm resolution from 149 particles. Scale bars in panels b and c = 10 nm.
RSAs display species-specific structural variation
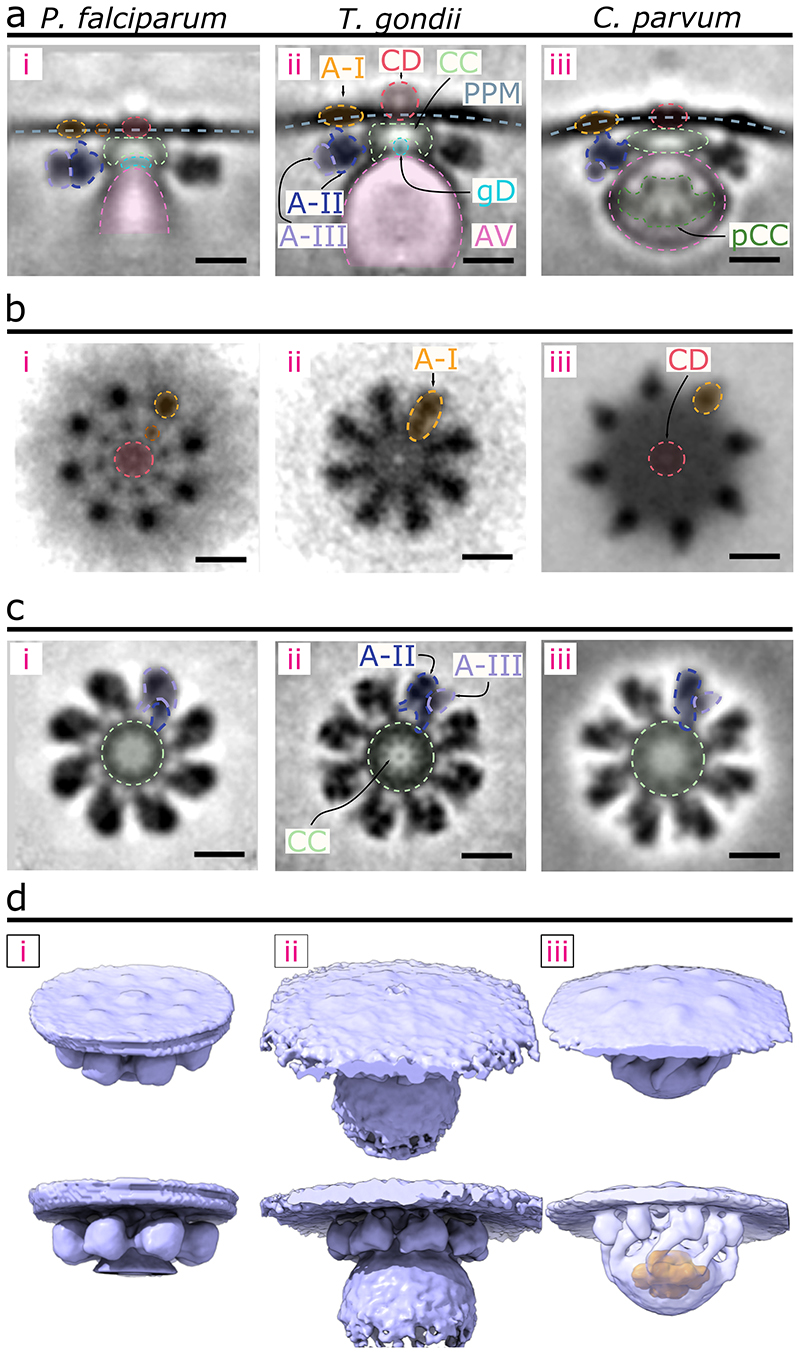
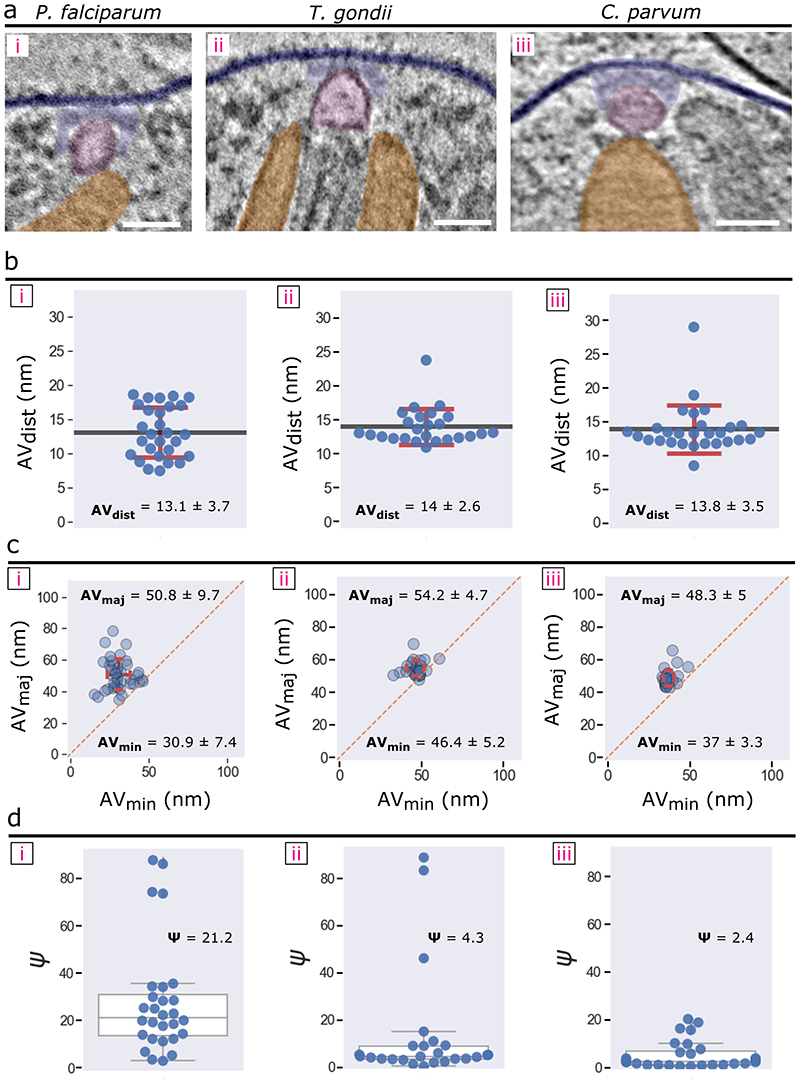
We next compared the P. falciparum RSA-A subtomogram average with that of T. gondii and C. parvum resolved previously9. Side views of the three RSA structures show a similar organization: the RSAs are anchored to the PPM intracellularly and hold the AV just below (Fig. 4a). Consistent with the greater variability in size and positioning of the P. falciparum AV compared to that of the two other organisms (Extended Data Fig. 3), only a relatively smaller portion of the AV adjacent to the RSA was resolved in P. falciparum (Fig. 4a, d). Additionally, the membrane bound by the RSA displays significant curvature in both T. gondii and C. parvum while remaining flat in P. falciparum (Fig. 4a). Top views at the PPM display a conserved rosette of particles in all three parasites, however as stated previously P. falciparum contains another inner rosette of A-IV particles (Fig. 4b). The CD in both P. falciparum and C. parvum is positioned at the PPM (Fig. 4a[i], b[i], a[iii], and b[iii]), while that of T. gondii extends into the extracellular space (Fig. 4a[ii], b[ii]). Additionally, the rosette of A-I particles appears differently shaped among the three parasites: in P. falciparum they are circular, whereas in T. gondii they appear elongated and in C. parvum teardrop shaped (Fig. 4b, d). Both P. falciparum and T. gondii have relatively globular A-II/III densities with limited or flexible (therefore largely averaged out in the subtomogram averages) contacts with the AV (Fig. 4d[i-ii, bottom]). On the other hand, the C. parvum RSA shows extensive contacts with the AV that likely contributed to the conserved AV shape and location and resulted in the preservation of the entire AV in the average (Fig. 4d[iii, bottom]). Additionally, the C. parvum RSA contains a posterior central channel within the AV (Fig. 4d[iii, bottom]) that is not seen in neither P. falciparum nor T. gondii. Together, we show that the three apicomplexan rhoptry secretion systems contain an ultrastructurally conserved secretory apparatus with notable species-specific alterations.
Figure 4. Comparison of the RSA structures of P. falciparum, T. gondii, and C. parvum.
(a) Central slices of the side view of each RSA subtomogram average (left – P. falciparum; middle – T. gondii; and right – C. parvum) with color overlays of each corresponding component. (b) Top view of each RSA, comparing the densities observed on the PPM surface. (c) Top view of each RSA through a cross-sectional plane in the intracellular region, comparing CC, A-II, and A-III densities. (d) 3-D isosurface of each RSA, with views from outside the PPM (top row) and from the side (bottom row). The AV of C. parvum is made semi-transparent to reveal the posterior central channel density inside (orange). Scale bars = 20 nm.
RSA structure changes with rhoptry fusion
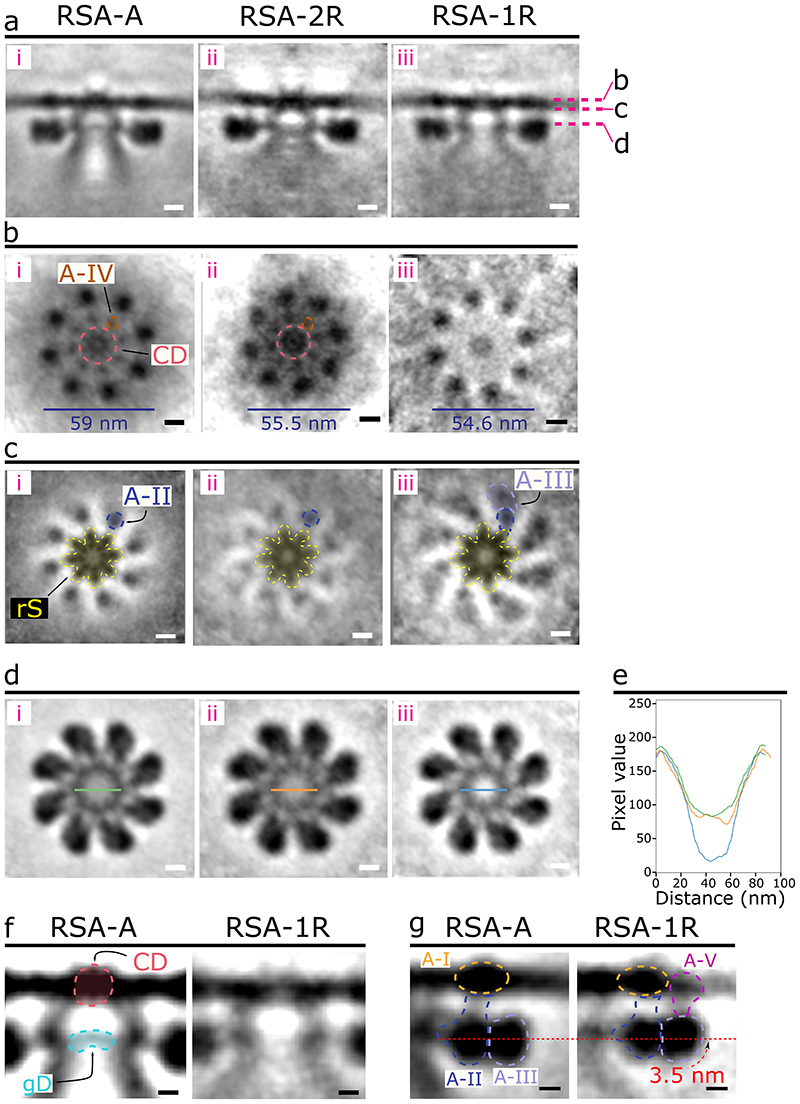
To investigate if rhoptry fusion with the AV or fusion between the two rhoptries induces conformational changes in the RSA structure, subtomogram averaging was performed independently for RSAs classified under these different AV-rhoptry morphologies: (i) those with an AV docked and two separate rhoptries (RSA-A; Fig. 2a and Movie 1); (ii) those with no AV and two separate rhoptries (RSA-2R, 33 particles resolved to an overall resolution of 5 nm; Fig. 2b, Extended Data Fig. 5d-g, and Movie 2); and (iii) those with no AV and fusing rhoptries (RSA-1R, 36 particles resolved to an overall resolution of 5 nm; Fig. 2c, d, Extended Data Fig. 5h-k, and Movie 2). The central slice from the side view of each structure (Fig. 5a) shows a similar docking of the AV to the RSA (RSA-A) or of the rhoptry tip to the RSA (RSA-2R and RSA-1R), however multiple RSA components appear different among the three structures. First, at the PPM surface, there is a significant reduction of the CD and disappearance of the A-IV densities in the RSA-1R structure (Fig. 5b[iii]). Additionally, the diameter of the A-I rosette decreases by ~3.5 nm between the RSA-A and RSA-2R structures, and by another ~1 nm between the RSA-2R and RSA-1R structures (Fig. 5b). We observed only A-II densities around the rS in the RSA-A and RSA-2R structures, whereas both A-II and A-III densities are seen in the RSA-1R structure (Fig. 5c). At a slice through the CC, we observed significantly lower electron densities within the channel of the RSA-1R structure (Fig. 5d, e [blue line]). Remarkably, from the side views of the structures, we identified the main cause for such a density decrease: loss of the gD at the interface between the CC and the fused rhoptry tip (Fig. 5f). Similarly, consistent with the top views, the CD is largely reduced in the PPM of the RSA-1R structure from the side view (Fig. 5f). Finally, the side views of the A-II/III densities display a ~3.5 nm shift towards the PPM in the RSA-1R structure (Fig. 5g), which is reflected in the top view slice through the rS density (Fig. 5c). Moreover, we observed an additional connection formed between the A-III and the PPM (termed Anchor-V [A-V]; Fig. 5g) while the A-II density connecting to the A-I appears largely reduced (Fig. 5a, g). Altogether, the results revealed intricate conformational changes of the RSA related to the sequential AV and rhoptry fusion activities. The detailed structural differences are illustrated in 3-D in Movie 3.
Figure 5. Different P. falciparum RSA conformations were resolved corresponding to different morphological states of the rhoptry secretion system.
(a) Central slices of the side view of each RSA conformation (left – RSA-A; middle – RSA-2R; right – RSA-1R). Cross sections in panels b-d are denoted by the pink dashed lines. (b) Top view of each RSA conformation from outside the PPM. The CD and A-IV densities are observed in the RSA-A (i) and RSA-2R (ii) conformations but reduced in the RSA-1R conformation (iii). The diameter of the rosette formed by the A-I densities on the PPM surface is noted by the blue lines at the bottom. (c) Top view of each RSA conformation just below the PPM. A-II and A-III densities have color overlays to highlight their change in the RSA-1R conformation. (d) Top view of each RSA through an intracellular cross section. The green, orange, and blue lines across the CC of the RSA-A, RSA-2R, and RSA-1R, respectively, denote the location of the pixel values plotted in panel e. (e) Plot of normalized, inverted pixel values across the central channel of each RSA (RSA-A, green; RSA-2R, orange; RSA-1R, blue). A large dip is seen among the pixels of the RSA-1R CC. (f) Zoom in on panel a (i and iii) highlighting changes of the gD and CD between RSA-A and RSA-1R. (g) Zoom in on panel a (i and iii) highlighting changes in A-II/III between RSA-A and RSA-1R, including a diminished A-II connection to the A-I density and 3.5 nm upward shift of the A-II/III densities (curved red arrow), as well as the presence of a new Anchor-V (A-V) in the RSA-1R conformation. RSA-2R and RSA-1R subtomogram averages resolved to 5 nm resolution from 33 and 36 particles, respectively. Scale bars = 10 nm in panels a–d and 5 nm in panels f, g.
Discussion
Previous cryo-ET and 3-D electron microscopy imaging of Plasmodium revealed important insights into parasite motility and ultrastructure of isolated parasites and during infection10,14,15. Here, we applied contrast-enhancing cryo-ET technology16 to resolve the apical complex of isolated P. falciparum merozoites (Fig. 1, Extended Data Fig. 1a-d). Similar to previous reports10,11,17–21, we observed two rhoptries with necks that extend through the preconoidal rings (Fig. 1b). We further discover that the P. falciparum rhoptry tips associate with a distinct AV in most merozoites (Fig. 1), similar to T. gondii and C. parvum9. The rhoptry tips dock at the AV like in T. gondii, rather than pre-fusing to the AV membrane like that of C. parvum (Extended Data Fig. 3a). Whereas C. parvum sporozoites contains one rhoptry22, P. falciparum merozoites have two and T. gondii tachyzoites have 8-1223,24 but only two are docked at the AV. Thus, docking at the AV, rather than being pre-fused, may represent a mode of regulation that permits secretion of multiple rhoptries at once. With this discovery of the AV in P. falciparum, we can now observe its presence in earlier cryo-ET images of merozoites10,25 where it has likely been overlooked due to its small size and flexibility (Fig. 1 and Extended Data Fig. 6) compared to that of other apicomplexan parasites8,9,23,26. Recent comparative analyses between the apicomplexan rhoptry secretion system and the extrusome system in the related, free-living Ciliata phylum27 suggests that the AV is an adaptation in Apicomplexa for parasitism7,8. By resolving the AV in P. falciparum merozoites, we provide further evidence that this enigmatic vesicle is a conserved feature among diverse apicomplexan parasites that may play a role in host cell invasion.
Our recent report discovered the RSA structures in T. gondii and C. parvum, which exhibit complex interactions with the AV and the PPM to ultimately guide the rhoptries to the parasite apex via the AV (Fig. 4)9. Here, the extensive connections observed between the P. falciparum RSA and PPM and between the RSA and the AV (Fig. 3) again support the function of the RSA as one that docks the AV to the PPM; the AV in turn guides the rhoptries to the proper site for exocytosis. Detailed differences among the RSA structures from these organisms (Fig. 4) may reflect adaptations in the molecular mechanisms for their specific parasitic activities. Further cryo-ET imaging of these parasites in the context of host cell invasion will permit the stage- and species-specific dissection of RSA-mediated rhoptry secretion, thus, to understand the roles of each common or unique structural component of the RSAs. Identification of the molecular components that comprise the RSA is also necessary to further elucidate the mechanisms of secretion. The recent discovery of Nd and NdP proteins, which are necessary for rhoptry secretion in T. gondii and P. falciparum8, provides a hopeful avenue to begin probing RSA-mediated secretion by cryo-ET. For example, knockdown of the Nd9 protein in T. gondii abolishes rhoptry secretion due to altered RSA structure was revealed by cryo-ET9. These proteins, as well as Fer228 which associates with the Nd protein complex8, are well conserved throughout Apicomplexa. Further cryo-ET analysis of related mutants and/or conditions to map molecules onto the RSA structures will permit a more comprehensive understanding of this unique molecular machine.
Previous reports of fusion between the two rhoptries10,11,17 and formation of a pore between the rhoptry and the PPM during Plasmodium host invasion17,29,30 have led to hypotheses that the rhoptries fuse together and with the PPM during secretion31, allowing the simultaneous discharge of two rhoptries. Here, we imaged rhoptry fusion in isolated P. falciparum merozoites and were able to detect intermediate morphological states: (i) separated rhoptry pair; (ii) separated rhoptry pair with their two necks twisting around and/or bulging towards one another (Fig. 2b, Extended Data Fig. 4a-c); (iii) fused rhoptry tip (Fig. 2c); and (iv) complete fusion of the rhoptry bulb (Fig. 2d, Extended Data Fig. 4f, g). Strikingly, the AV is only present at the tips of non-fused rhoptry pairs and is no longer recognizable when the rhoptries were seen with necks twisted or fused (Fig. 2a-d and Extended Data Fig. 4). This suggests that the AV may fuse to one rhoptry and becomes unrecognizable from the neck of that rhoptry before the two rhoptries start to fuse, hinting at a role for the AV in regulating rhoptry fusion. On the other hand, the fact that the AV is present in Cryptosporidium, which has only a single rhoptry, supports additional functions of the AV in the frame of rhoptry secretion among apicomplexan parasites. A recent cryo-ET study of developing merozoites within schizonts showed images of the apical complex in which the AV was present at the rhoptry tips (Extended Data Fig. 2d)25, but was overlooked and instead marked as part of the rhoptry (Extended Data Fig. 2e, f). This indicates that AV biogenesis occurs during parasite development within the red blood cell, and that the rhoptry morphologies observed in merozoites lacking the AV are likely subsequent states to the AV-containing state. Similar observations of possible rhoptry fusion can be observed in earlier electron micrographs of T. gondii during host invasion, where the secreting rhoptries display branched morphologies that may be representative of two rhoptries fused together along the necks23,24,32. Although it has not been proven if the rhoptries are indeed fused in these data, no AV could be observed in any of the invading T. gondii parasites which again supports a role of the AV’s disappearance in rhoptry exocytosis during host invasion.
Interestingly, volumetric and density analyses revealed that no major secretion occurs across all the rhoptry morphologies captured in our cryo-ET data (Fig. 2a-f). This suggests that the fusion between the two rhoptries could be an independent event upstream of major content secretion. However, these data do not rule out the possibility of initial, small-scale secretions of specific rhoptry contents in preparation for a subsequent major secretion event. Indeed, previous studies have demonstrated that proteins important for invasion, such as Rh5, translocate from the rhoptries to the merozoite surface prior to full rhoptry secretion33,34. As GlyA-treated merozoites did not display empty rhoptries in our tomograms, it is possible that treatment only causes small-scale secretions. In the original study suggesting GlyA triggers rhoptry secretion12, this conclusion was based on the immunofluorescence detection of two rhoptry proteins on the merozoite surface after GlyA treatment, without electron microscopy inspection of the rhoptries. Concurrent with the lack of major content secretion, the different RSA conformations observed showed molecular rearrangements at the rhoptry exocytosis site correlating with the AV and rhoptry fusion activities. Importantly, the gD is absent, the CD is largely reduced, and the CC interior is less electron dense in the rhoptry-fused state (RSA-1R; Fig. 5d-f), seemingly to clear up the most direct path for rhoptry contents to leave the merozoite. Together, these data reveal previously unknown steps prior to secretion of rhoptry contents that may function as a priming mechanism to prepare the merozoite for major rhoptry secretion upon host contact. The trigger for these steps remains unknown; however, given the intimate link between calcium signaling and rhoptry secretion12,35–37, the rhoptry activities captured in our samples may be due to changes in intracellular calcium concentrations – as the proportion of merozoites with primed rhoptries (the morphologies that lack the AV) increases with PPM rupture that likely disrupts local calcium concentrations (Fig. 2h). In accordance with this, various proteins required for the secretion of rhoptries are linked to calcium, including Rasp238, Fer228, and NdP28. Further analysis is necessary to confirm whether these fusion events are linked to calcium.
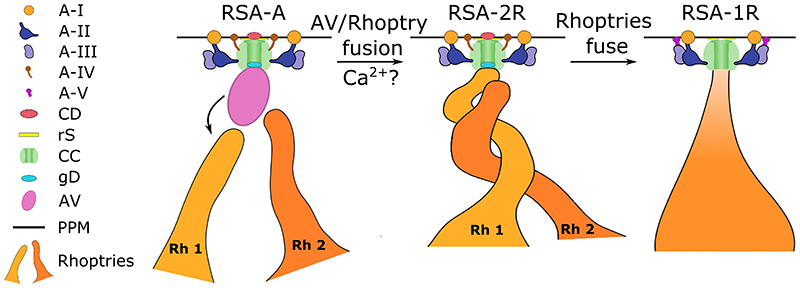
In summary, cryo-ET imaging of isolated P. falciparum merozoites has revealed a priming mechanism for the secretion of rhoptries (Fig. 6), in which AV fusion to one rhoptry may serve as a trigger to initiate rhoptry fusion. We have further resolved multiple RSA structures that correspond to the disappearance of the AV and fusion of the rhoptries, providing a structural framework to understand the mechanism at the molecular level. Through comparative analysis of the P. falciparum RSA to those of T. gondii and C. parvum, we strengthen the support for the AV’s role in host invasion in addition to the evidence for conservation of this novel secretion system across Apicomplexa. Discovery of proteins required for rhoptry secretion that localize to the AV and RSA will permit the mapping of components onto this molecular machine and the dissection of secretion mechanism to provide novel targets for the treatment of this deadly parasite.
Figure 6. Working model of the priming of rhoptry secretion in P. falciparum merozoites.
Prior to invasion signals, the AV docks to the RSA and guides the two rhoptries to their proper exocytosis site in preparation for secretion. Upon exposure to proper (unknown) signals, the AV fuses with one rhoptry (labeled Rh 1 for rhoptry 1), a process that may be mediated by calcium, creating the first passage opening for rhoptry secretion (i.e., between the rhoptry and AV). One rhoptry then docks directly at the RSA and prepares to fuse with the second rhoptry (labeled Rh 2 for rhoptry 2). During this process, the diameter of the A-I rosette in the PPM decreases and the membrane adopts a curvature around the CD. The rhoptries then fuse together partially (along the necks) or fully, resulting in a loss of the gD and a second passage opening, between the rhoptry and the RSA. This is accompanied by a further decrease in A-I rosette diameter, loss of the A-IV densities, formation of the A-V densities connecting the A-III densities to the PPM, and a large reduction of the CD density.
Methods
Culture of P. falciparum merozoites
P. falciparum 3D7 parasites were maintained at 37°C in a gas mixture of 5% O2, 5% CO2, and 90% N2 in a 2% suspension of human erythrocytes (donated from the Children’s Hospital of Philadelphia) in complete medium (RPMI-1640 completed with 27 mM NaHCO3, 11 mM glucose, 5 mM HEPES, 0.01 mM thymidine, 1 mM sodium pyruvate, 0.37 mM hypoxanthine, 10 μg/mL gentamicin (all aforementioned culture media components were purchased from Sigma Aldrich), and 5 g/L Albumax (Thermo Fisher Scientific)). Parasite cultures were synchronized over 3-4 cycles prior to merozoite isolation. Ring-stage parasite cultures were incubated with 5% (w/v) sorbitol (Sigma Aldrich) for 5 min at 37°C. Schizont-stage cultures were centrifuged in a 70% (v/v) isotonic Percoll (Sigma Aldrich) solution and the late-stage parasites were collected after treatment.
Preparation of P. falciparum merozoites and cryo-ET grid preparation
For merozoite preparation, cultures were scaled up to 60 mL at 5% hematocrit. Late-stage schizonts were purified from highly synchronous cultures by magnetic activated cell sorting (MACS). Isolated schizonts were treated with 10 μM E64 for 6-8 hours, followed by passage through 1.2 μm syringe filters (preincubated in 1% BSA + PBS for 20 minutes) to mechanically isolate merozoites39. Isolated merozoites were spun down at 2200 g for 15 minutes and resuspended in 20 μL complete media. 10 nm colloidal gold fiducials (Ted Pella, Redding, USA) were added to the suspension (for alignment during tomogram reconstruction from tilt series). 4 μL of suspended merozoites were applied onto Quantifoil 200 mesh copper R2/2 holey carbon electron microscopy grids, excess liquid blotted away, and plunge frozen into a liquid ethane/propane mixture (pre-cooled with liquid nitrogen) using an EM GP2 automatic plunger (Leica Microsystems, Wetzlar, Germany)40. The blotting chamber was set to 95% relative humidity at 24°C and blotting was done from the sample side of the grid using Whatman filter paper #1. Plunge-frozen grids were subsequently loaded into autogrid c-clip rings (Thermo Fisher). The autogrid box containing frozen grids were stored in liquid nitrogen and maintained at ≤−170°C throughout storage, transfer, and cryo-ET imaging.
Drug treatment of P. falciparum merozoites
For treatment with glycophorin A, mechanically isolated merozoites (prior to plunge freezing) were resuspended in buffer mimicking extracellular ionic conditions (140 mM NaCl, 5 mM KCl, 1 mM CaCl2) containing 1 mg/mL glycophorin A (Sigma Aldrich) and incubated at 37°C for 5 – 15 minutes12.
Cryo-electron tomography (cryo-ET)
Cryo-ET was performed on a Thermo Fisher Krios G3i 300 keV field emission cryo-transmission electron microscope. Dose-fractionated imaging was performed using the SerialEM software41 on a K3 direct electron detector42 (Gatan Inc., Pleasanton, CA, USA) operated in electron-counted mode. Motion correction of images was done using the Alignframe function in IMOD43. Imaging was done using a Volta phase plate44 to increase contrast without high defocus, and the Gatan Imaging Filter (Gatan Inc., Pleasanton, CA, USA) with a slit width of 20 eV to increase contrast by removing inelastically scattered electrons45. After initially assessing cells at lower magnifications for suitability of ice thickness and plasma membrane integrity, tilt series were collected with a span of 100° (-50° to +50°; dose-symmetric scheme46) or 120° (-60° to +60°; dose-symmetric or bi-directional scheme) with 2° increments at a magnification of 33,000x (with a corresponding pixel size of 2.65 Å) and a defocus range of -1 to -4 μm. Each tilt series was collected with a cumulative dose of around 140 e−/ Å2. Once acquired, tilt series were aligned using the 10 nm colloidal gold as fiducials and reconstructed into tomograms by our in-house automated computation pipeline utilizing the IMOD software package43.
Quantification, statistics, and reproducibility
We obtained a total of 218 P. falciparum tomograms (151 untreated and 67 GlyA-treated) from nine frozen grids prepared from six biological replicates (four untreated and two GlyA-treated) that were imaged over six multiple-day sessions. Each of the following quantifications is from a subset of 39 tomograms that resolved the feature of interest. We note that ~50% of parasites imaged had a broken plasma membrane, resulting in a greater resolution of intracellular features compared with completely intact merozoites. We also note that parasites flattened on the grid, likely due to blotting. However, this flattening did not reflect on the shape of individual rhoptries, the AV, or the RSA. Flattening could have added subtle variations to the relative positions of these features, but their organizational patterns were evident despite the presence of such a potential effect.
Rhoptry distance from the AV
Shortest distance between the AV membrane and the rhoptry tip membrane for each rhoptry.
θ (orientation parameter for rhoptries at the AV)
Orientation of rhoptry 1 (θRh1) and rhoptry 2 (θRh2) necks with respect to the major axis of the AV (see below under ‘vesicle dimensions’ for more details). Angle was measured between the major axis of the AV and the line connecting the rhoptry tip membrane to the center point of the AV major axis.
Vesicle dimensions
The AV was approximated to an ellipsoid using only two axes (major and minor) instead of three for simplicity. The longest axis of each AV in 3-D was marked as the major axis (labeled as AVmaj) while the shortest axis orthogonal to the major axis and intersecting it at the centroid was marked as the minor axis (labeled as AVmin).
Eccentricity
(1-b2/a2)1/2, where ‘a’ is the semi-major axis, and ‘b’ is the semi-minor axis.
AVdist (anchoring distance of AV)
Shortest distance between the AV membrane and the apex of the parasite plasma membrane, represented by the CD of the RSA.
Ψ (orientation parameter for AV)
Angle formed between the major axis of the AV and the line formed between the apex of the parasite plasma membrane and the centroid of the AV.
Quantifications were performed on IMOD-generated models. All model files were exported for analysis and plotting using Pandas, Numpy, Matplotlib, and Seaborn libraries in Python 3.7. The corresponding graphs report the mean ± standard deviation values. For skewed distributions (θ and Ψ) we instead report the median value alone or boxplots displaying data quartiles.
Pixel value analysis
To analyze the pixel values across line profiles through AVs, rhoptries, and RSAs, 3-D volumes were opened in IMOD and, for the AVs and rhoptries, 5 layers of voxels spanning a thickness of 5.3 nm were averaged along the Z axis. TIFF images of desired slices were subsequently imported into FIJI47 where a Gaussian filter was applied to smoothen the images. A line with a thickness of 10 pixels was drawn across the AVs and rhoptries, extending a few pixels beyond each membrane. Pixel values along these line profiles were exported for further analysis. For analysis of the RSA central channel, no filter was applied to the images in FIJI. Pixel values were inverted by subtracting each value from 255 so that electron dense regions appeared as peaks when plotted. Analysis and plotting of pixel values across each line were performed using Numpy, Matplotlib, and Seaborn libraries in Python 3.7.
Rhoptry volume analysis
29 tomograms (10 parasites with an AV, 9 lacking the AV and with two distinct rhoptries, and 10 lacking the AV and with fused rhoptries) were analyzed in EMAN248,49 and UCSF Chimera50. Tomograms were imported into the EMAN2 tomogram annotation workflow and a neural network was trained to recognize and segment density corresponding to rhoptries within the tomogram. Segmented rhoptries were subsequently imported into UCSF Chimera, where the segmentations were cleaned up using volume eraser to erase false positive noise, a dome cap was maintained at the top and bottom of rhoptry segmentations following the rhoptry curvature to account for the missing wedge effect, and the volumes of the rhoptry segmentations were measured. Plotting of the rhoptry volumes was performed using Matplotlib and Seaborn libraries in Python 3.7.
Generation of RSA subtomogram averages
Subtomogram alignment and averaging were performed using Dynamo51,52. In IMOD, we performed manual inspections of merozoite tomograms at the apical end and identified RSA ultrastructures. Subtomogram boxes were centered in the middle of the RSA central channel and manually rotated to the same orientation among RSA ultrastructures. Model points and manual orientations were imported in Dynamo using custom scripts. Subtomogram averaging was initially performed using 149 RSAs without rotational symmetry. Densities with 8-fold rotational symmetry were seen throughout the RSA in the average, from the extracellular region to the AV (Extended Data Fig. 5). We therefore utilized this symmetry during particle picking, thus yielding 8 times more subtomograms, increasing the total number to 1,192 to further enhance the signal-to-noise ratio. The result was a better-resolved average that revealed a more elaborate ensemble of components than what was directly observed in the tomograms or from the unsymmetrized average.
RSAs from cells with different rhoptry morphologies (RSA-A: two rhoptries with an AV; RSA-2R: two rhoptries without an AV; RSA-1R: fused rhoptries without an AV) were manually sorted and then aligned and averaged independently from each other using Dynamo software (1,192 subtomograms from 149 RSA-As, 264 subtomograms from 33 RSA-2Rs, and 288 subtomograms from 36 RSA-1Rs). To generate an initial reference, each group of subtomograms was averaged using the manual orientations and without any computational alignment. An ellipsoid mask that covered the RSA and part of the plasma membrane was used in conjunction with each reference that was used to align all subtomograms. The alignment procedure started with a cone range (Euler angles Z and X) of 6° and an azimuth range (Euler angle Z’) of 22.5°, followed by 6 refinement iterations, each of which used half the angular search range of the previous iteration. A new reference was generated with the best two-thirds of aligned subtomograms, and the process was repeated two more times. This process was then repeated three more times with an initial azimuth range of 6° (in total, 6 iterations of alignment). We then incorporated 8-fold rotational symmetry of the RSAs by picking 8 subtomograms from each RSA particle (by iteratively rotating each particle by 45°) and performing another subtomogram alignment with cone and azimuth ranges of 6° for 3 iterations, yielding the final average for each kind of RSA (RSA-A, RSA-2R, and RSA-1R). Fourier shell correlation plots were calculated for each subtomogram average using the “Adaptive bandpass filtering” function in Dynamo. For each average, the particles generated after applying 8-fold symmetry were split into two half-sets and refined independently of each other. Templates for independent refinement of each half-set were generated by averaging the original particles, split into half-sets, using initial manual orientations and no computational alignment. Each half-set was refined using identical parameters as mentioned previously. Finally, both half-set averages were aligned and Fourier shell correlation was computed.
Figures presentation, modeling, and segmentations
Tomograms were oriented in 3-D using IMOD’s Slicer window such that the desired tomogram section was in view for presentation. To enhance contrast, 5-10 layers of voxels spanning thicknesses of 5.3-10.6 nm were averaged around the section of interest. Figures were prepared using Inkscape 1.1. Manual rhoptry segmentations were generated in IMOD and imported into ChimeraX53 for presentation. Segmentations of the RSA-A subtomogram average was done utilizing the Segger function in ChimeraX. Subsequent visualization and animations of the RSA were also generated using ChimeraX.
1. Extended Data
Extended Data Fig. 1. The apical complex and apical vesicle of P. falciparum merozoites.
(a) Simplified schematic of a P. falciparum merozoite apical region. (b–d) 2-D slices through a tomogram of a top-down view of the merozoite apical complex with color overlays of all the observed apical end components: apical vesicle (AV; pink), parasite plasma membrane (PPM; blue), rhoptry (orange), rhoptry secretory apparatus (RSA; light blue), microneme (yellow), inner membrane complex (IMC; green), preconoidal rings (brown), apical polar ring (purple), and subpellicular microtubules (cyan). The cross-sectional planes used in b-d are arranged such that they start at the parasite apex and move inwards into the cell. Scale bar = 100 nm. (e, f) Additional examples of 2-D slices of the AV and rhoptry (left). Blue and orange lines across the AV and rhoptry, respectively, denote the locations of pixel values plotted on the right. Arrows in the pixel plots point to peaks in values corresponding to membranes. Scale bars = 50 nm.
Extended Data Fig. 2. 2-D slices through the P. falciparum merozoite apical complex.
(a–c) 2-D slices from tomograms showing either one rhoptry (a, b) or no rhoptries (c) docked at the AV. Scale bars = 50 nm. (d) 2-D slice from a P. falciparum merozoite apical end tomogram within a mature schizont, adapted from [30]. (e) The same 2-D slice from panel j with the original color overlay of the rhoptry of interest, according to [30]. (f) The same 2-D slice from panel j with an updated color overlay of the rhoptry of interest, the AV, and the RSA, based on our analysis. Scale bars = 200 nm.
Extended Data Fig. 3. Comparison of the apical vesicles from P. falciparum, T. gondii, and C. parvum.
(a) Representative 2-D slices of tomograms displaying the AV from each organism (left – P. falciparum; middle – T. gondii; and right – C. parvum), highlighting the PPM (dark blue), RSA (light blue), AV (pink), and rhoptries (orange). Scale bars = 50 nm. (b) Plots of the shortest distance (AVdist) between the PPM apex and the AV membrane from several cells each organism, showing the mean  sd. (c) Plots of the major (AVmaj) versus minor axis (AVmin) of the AV from each organism, showing mean
sd. (c) Plots of the major (AVmaj) versus minor axis (AVmin) of the AV from each organism, showing mean  sd. (d) Plots of the angle
sd. (d) Plots of the angle  at which the major axis of the AV is oriented with respect to the line connecting the PPM apex and the AV centroid each organism. The median value is shown. N = 39 AVs for P. falciparum, 25 AVs for T. gondii, and 28 AVs for C. parvum.
at which the major axis of the AV is oriented with respect to the line connecting the PPM apex and the AV centroid each organism. The median value is shown. N = 39 AVs for P. falciparum, 25 AVs for T. gondii, and 28 AVs for C. parvum.
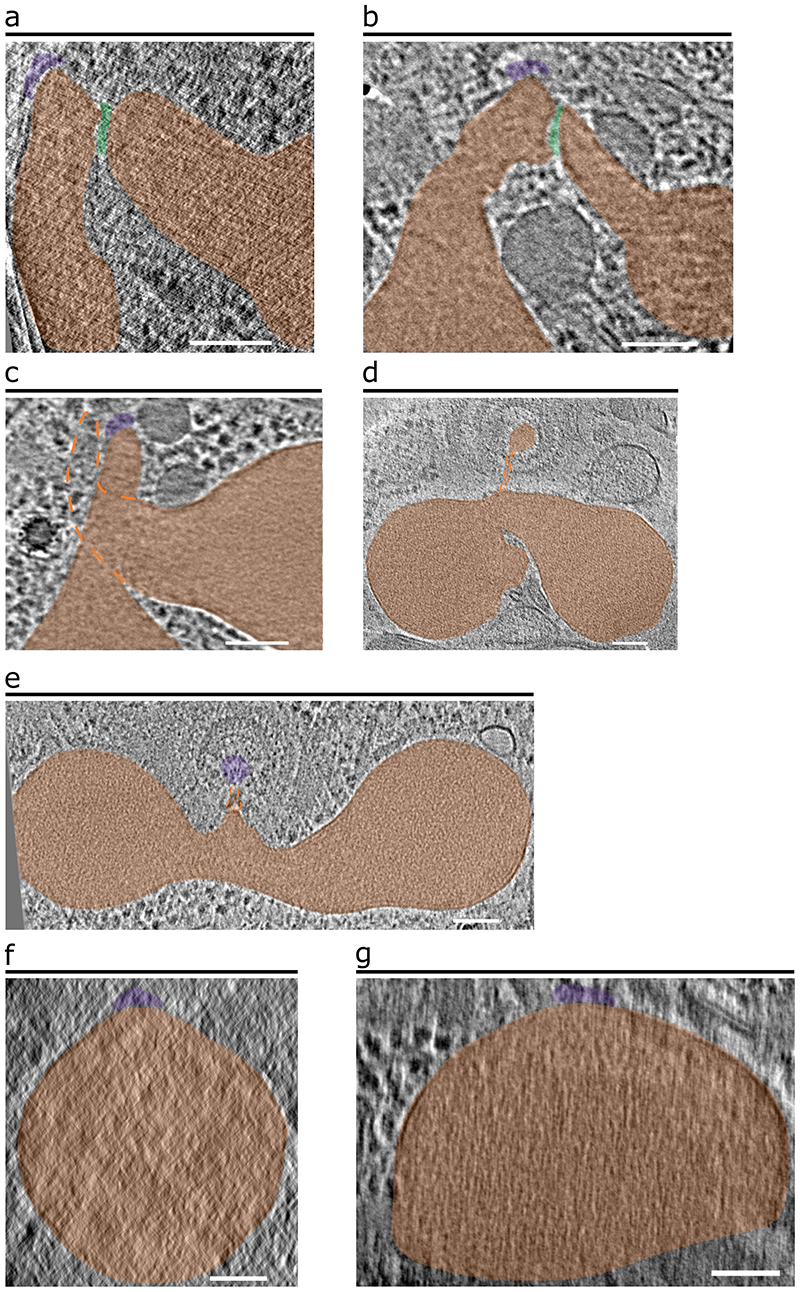
Extended Data Fig. 4. Various fusion states of the P. falciparum rhoptries.
(a–c) 2-D slices through tomograms of apical ends showing the absence of the apical vesicle at the rhoptry tips and either bulging of one rhoptry towards the other (a, b) or twisting of the rhoptries (c). Dashed orange lines in panel c denote the portion of the rhoptry neck that is out of view. A mesh of electron-dense material (green) can be observed between rhoptries (orange) that display bulging towards the other. One rhoptry is docked directly at the RSA (purple). (d, e) Examples in which the two rhoptries are fused together and docked directly at the RSA. (f, g) Examples in which only one, very large rhoptry with no discernable neck region was observed and docked directly at the RSA. Scale bars = 100 nm.
Extended Data Fig. 5. Subtomogram averages of the P. falciparum RSA.
(a) Schematic of the RSA, apical vesicle, and PPM demonstrating the top view and side view orientations used to generate 2-D slices of the subtomogram averages. (b, c) Side views (b) and top view (c) of the subtomogram average of the RSA-A structure before applying 8-fold symmetry. Left to right panels: from peripheral to central cross sections (b) or from extracellular to intracellular cross sections (c). (d–g) Side views (d, f) and top view (e, g) of the subtomogram average of the RSA-2R structure, before and after applying 8-fold symmetry. (h–k) Side views (h, j) and top views (i, k) of the subtomogram average of the RSA-1R structure, before and after applying 8-fold symmetry. (i) Gold standard Fourier shell correlation plot of the final, 8-fold symmetrized subtomogram averages of the three RSA structures. Scale bars = 10 nm.
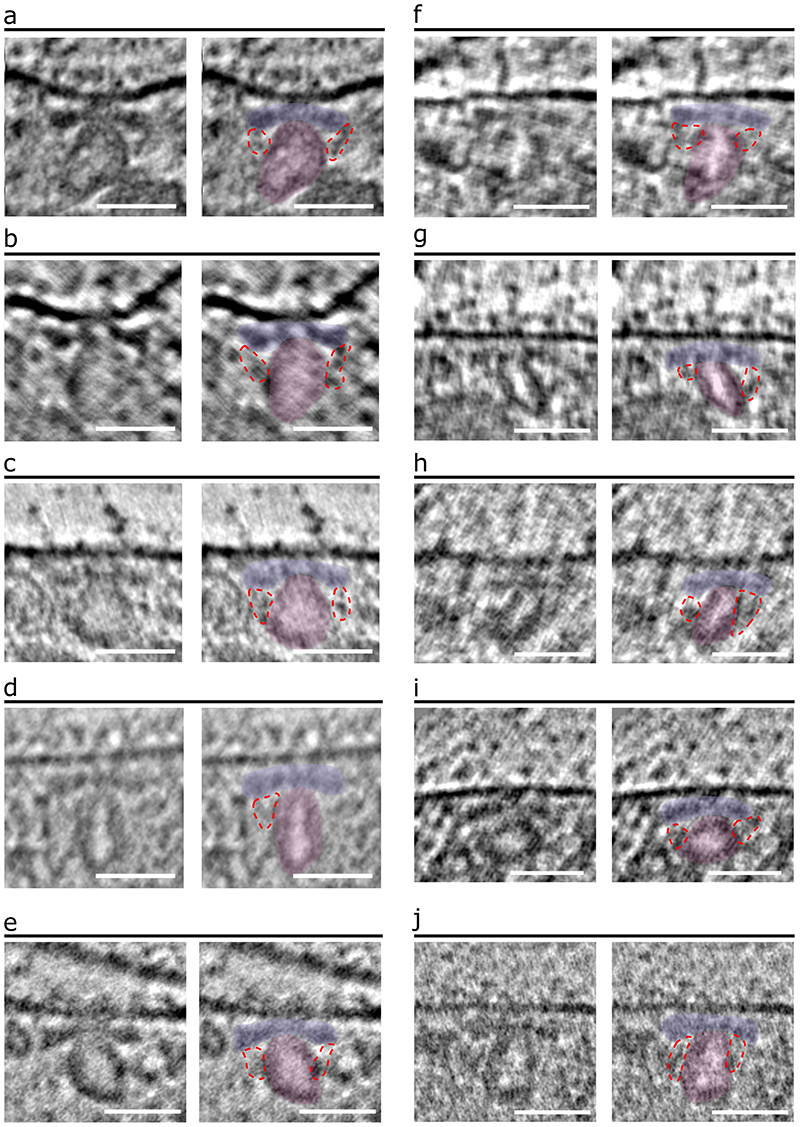
Extended Data Fig. 6. Representative cryo-ET images of the P. falciparum AV and RSA.
(a-j) 2-D slices with of merozoite apexes (left panels) with color overlays (right panels) highlighting the RSA (light blue), the AV (pink), and the connections from the RSA A- II/III densities to the AV (encircled in red dashed lines). Scale bars = 50 nm.
Supplementary Material
Acknowledgements
We thank Dr. Stefan Steimle for his technical assistance with the Titan Krios G3i cryogenic electron microscope and the Singh Center for Nanotechnology and the Beckman Center for Cryo-Electron Microscopy at the University of Pennsylvania for hosting and supporting the use of the Titan Krios; Dana Hodge in the lab of A.O.J. for training in P. falciparum culture techniques; Liam Theveny from the lab of Y.-W.C. for providing subtomogram averages of the T. gondii and C. parvum RSAs; and other members of the labs of Y.-W.C., A.O.J., and M.L. for overall support and useful discussions. This work was supported in part by a David and Lucile Packard Fellowship for Science and Engineering (2019-69645) and a Pennsylvania Department of Health FY19 Health Research Formula Fund to Y.-W.C.; by an Martin and Pamela Winter Infectious Disease Fellowship to M.M.; by the Mary L. and Matthew S. Santirocco College Alumni Society Undergraduate Research Grant to W.D.C.; by an EMBO fellowship (ALTF 58-2018) to A.G.; by an NIH/NIAID R01 AI103280, an R21 AI123808, an R21 AI130584, and an R61 DH105594 to A.O.J. who is an Investigator in the Pathogenesis of Infectious Diseases (PATH) of the Burroughs Wellcome Fund; and by a European Research Council advanced grant 833309 (KissAndSpitRhoptry) to M.L.
Footnotes
Author contributions
M.M., M.L., and Y.-W.C. conceptualized and designed the experiments. M.M. cultured and isolated parasites provided by A.O.J. M.M.C. provided a protocol and consultations for the efficient isolation of merozoites. P.M. provided further training and useful insights towards P. falciparum culture and merozoite isolation. M.M. prepared frozen grids and performed cryo-ET, with training from S.K.M., using an automated data-processing pipeline for on-the-fly tomogram reconstruction that was established by W.D.C., who also provided additional computational support during data collection, processing, and management. M.M. analyzed the tomograms, performed subtomogram averaging, and analyzed the RSA structure. W.D.C. performed data analysis of rhoptry volumes. S.K.M., A.G., M.M.C., and M.L. provided important insights for the interpretation of data. M.M. prepared the manuscript with critical inputs and revisions from all authors.
Competing interests
The authors have no competing financial interests.
Data availability
Representative tomograms showing different morphological states of P. falciparum rhoptry fusion (Fig. 2a, b, d) are available in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-26745 (Fig. 2a), EMD-26746 (Fig. 2b), and EMD-26747 (Fig. 2d). Subtomogram averages of the P. falciparum RSA in different conformations are available in the EMDB under accession codes EMD-26670 (RSA-A), EMD-26671 (RSA-2R), and EMD-26672 (RSA-1R).
References
- 1.Cowman AF, Healer J, Marapana D, Marsh K. Malaria: Biology and Disease. Cell. 2016;167:610–624. doi: 10.1016/j.cell.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 2.Blader IJ, Coleman BI, Chen C-T, Gubbels M-J. Lytic Cycle of Toxoplasma gondii: 15 Years Later. Annu Rev Microbiol. 2015;69:463–485. doi: 10.1146/annurev-micro-091014-104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guérin A, Striepen B. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host Microbe. 2020;28:509–515. doi: 10.1016/j.chom.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Dubois DJ, Soldati-Favre D. Biogenesis and secretion of micronemes in Toxoplasma gondii. Cell Microbiol. 2019;21:e13018. doi: 10.1111/cmi.13018. [DOI] [PubMed] [Google Scholar]
- 5.Ben Chaabene R, Lentini G, Soldati-Favre D. Biogenesis and discharge of the rhoptries: key organelles for entry and hijack of host cells by the Apicomplexa. Mol Microbiol. 2020 doi: 10.1111/mmi.14674. [DOI] [PubMed] [Google Scholar]
- 6.Riglar DT, et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe. 2011;9:9–20. doi: 10.1016/j.chom.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Sparvoli D, Lebrun M. Unraveling the Elusive Rhoptry Exocytic Mechanism of Apicomplexa. Trends Parasitol. 2021 doi: 10.1016/j.pt.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Aquilini E, et al. An Alveolata secretory machinery adapted to parasite host cell invasion. Nat Microbiol. 2021 doi: 10.1038/s41564-020-00854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mageswaran SK, et al. In situ ultrastructures of two evolutionarily distant apicomplexan rhoptry secretion systems. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-25309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanssen E, et al. Electron tomography of Plasmodium falciparum merozoites reveals core cellular events that underpin erythrocyte invasion. Cell Microbiol. 2013;15:1457–1472. doi: 10.1111/cmi.12132. [DOI] [PubMed] [Google Scholar]
- 11.Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. 1978;77:72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh S, Alam MM, Pal-Bhowmick I, Brzostowski JA, Chitnis CE. Distinct External Signals Trigger Sequential Release of Apical Organelles during Erythrocyte Invasion by Malaria Parasites. PLoS Pathog. 2010;6:e1000746. doi: 10.1371/journal.ppat.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepper S, Merkel M, Sartori A, Cyrklaff M, Frischknecht F. Rapid quantification of the effects of blotting for correlation of light and cryo-light microscopy images. J Microsc. 2010;238:21–26. doi: 10.1111/j.1365-2818.2009.03327.x. [DOI] [PubMed] [Google Scholar]
- 14.Kudryashev M, et al. Structural basis for chirality and directional motility of Plasmodium sporozoites. Cell Microbiol. 2012;14:1757–1768. doi: 10.1111/j.1462-5822.2012.01836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cyrklaff M, Frischknecht F, Kudryashev M. Functional insights into pathogen biology from 3D electron microscopy. FEMS Microbiol Rev. 2017;41:828–853. doi: 10.1093/femsre/fux041. [DOI] [PubMed] [Google Scholar]
- 16.Theveny LM, et al. Parasitology meets cryo-electron tomography–exciting prospects await. Trends Parasitol. 2022 doi: 10.1016/J.PT.2022.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Miller LH, Aikawa M, Johnson JG, Shiroishi T. Interaction between cytochalasin B-treated malarial parasites and erythrocytes. J Exp Med. 1979;149:172–184. doi: 10.1084/jem.149.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowman AF, Tonkin CJ, Tham W-H, Duraisingh MT. The Molecular Basis of Erythrocyte Invasion by Malaria Parasites. Cell Host Microbe. 2017:232–245. doi: 10.1016/j.chom.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liffner B, et al. PfCERLI1 is a conserved rhoptry associated protein essential for Plasmodium falciparum merozoite invasion of erythrocytes. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherling ES, et al. The Plasmodium falciparum rhoptry bulb protein RAMA plays an essential role in rhoptry neck morphogenesis and host red blood cell invasion. PLOS Pathog. 2019;15:e1008049. doi: 10.1371/journal.ppat.1008049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudlaff RM, Kraemer S, Marshman J, Dvorin JD. Three-dimensional ultrastructure of Plasmodium falciparum throughout cytokinesis. PLoS Pathog. 2020;16:e1008587. doi: 10.1371/journal.ppat.1008587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tetley L, Brown SMA, Mcdonald V, Coombsl GH. Ultrastructural analysis of the sporozoite of Cryptosporidium parvum. Microbiology. 1998;144:3249–3255. doi: 10.1099/00221287-144-12-3249. [DOI] [PubMed] [Google Scholar]
- 23.Paredes-Santos TC, De Souza W, Attias M. Dynamics and 3D organization of secretory organelles of Toxoplasma gondii. J Struct Biol. 2012;177:420–430. doi: 10.1016/j.jsb.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Dubremetz JF. Rhoptries are major players in Toxoplasma gondii invasion and host cell interaction. Cell Microbiol. 2007;9:841–848. doi: 10.1111/j.1462-5822.2007.00909.x. [DOI] [PubMed] [Google Scholar]
- 25.Bisson C, Hecksel CW, Gilchrist JB, Fleck RA. Preparing Lamellae from Vitreous Biological Samples using a Dual-Beam Scanning Electron Microscope for Cryo-Electron Tomography. J Vis Exp. 2021:e62350. doi: 10.3791/62350. [DOI] [PubMed] [Google Scholar]
- 26.Burrell A, et al. Cellular electron tomography of the apical complex in the apicomplexan parasite Eimeria tenella shows a highly organised gateway for regulated secretion. bioRxiv. 2021 doi: 10.1101/2021.06.17.448283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plattner H. Trichocysts-Paramecium’s Projectile-like Secretory Organelles: Reappraisal of their Biogenesis, Composition, Intracellular Transport, and Possible Functions. J Eukaryot Microbiol. 2017;64:106–133. doi: 10.1111/jeu.12332. [DOI] [PubMed] [Google Scholar]
- 28.Coleman BI, et al. A member of the ferlin calcium sensor family is essential for toxoplasma gondii rhoptry secretion. MBio. 2018;9 doi: 10.1128/mBio.01510-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyth O, et al. Cellular dissection of malaria parasite invasion of human erythrocytes using viable Plasmodium knowlesi merozoites. Sci Rep. 2018;8:10165. doi: 10.1038/s41598-018-28457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aikawa M, Miller LH, Rabbege JR, Epstein N. Freeze-Fracture Study on the Erythrocyte Membrane during Malarial Parasite Invasion. J Cell Biol. 1981;91:55–62. doi: 10.1083/jcb.91.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liffner B, Balbin JM, Wichers JS, Gilberger T-W, Wilson DW. The Ins and Outs of Plasmodium Rhoptries, Focusing on the Cytosolic Side. Trends Parasitol. 2021 doi: 10.1016/J.PT.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Nichols BA, Chiappino ML, Richard O’connor G. Secretion from the Rhoptries of Toxoplasma gondii during Host-Cell Invasion. J Ultrastruct Res. 1983;83:85–98. doi: 10.1016/s0022-5320(83)90067-9. [DOI] [PubMed] [Google Scholar]
- 33.Baum J, et al. Reticulocyte-binding protein homologue 5-An essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int J Parasitol. 2009;39:371–380. doi: 10.1016/j.ijpara.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Counihan NA, Kalanon M, Coppel RL, De Koning-Ward TF. Plasmodium rhoptry proteins: why order is important. Trends Parasitol. 2013;29 doi: 10.1016/j.pt.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 35.de Oliveira LS, et al. Calcium in the Backstage of Malaria Parasite Biology. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.708834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volz JC, et al. Essential Role of the PfRh5/PfRipr/CyRPA Complex during Plasmodium falciparum Invasion of Erythrocytes. Cell Host Microbe. 2016;20:60–71. doi: 10.1016/j.chom.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Weiss GE, et al. Revealing the Sequence and Resulting Cellular Morphology of Receptor-Ligand Interactions during Plasmodium falciparum Invasion of Erythrocytes. PLoS Pathog. 2015;11:e1004670. doi: 10.1371/journal.ppat.1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suarez C, et al. A lipid-binding protein mediates rhoptry discharge and invasion in Plasmodium falciparum and Toxoplasma gondii parasites. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11979-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle MJ, et al. Isolation of viable Plasmodium falciparum merozoites to define erythrocyte invasion events and advance vaccine and drug development. PNAS. 2010;107:14378–14383. doi: 10.1073/pnas.1009198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iancu CV, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2006;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- 41.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Xuong NH, et al. Future Directions for Camera Systems in Electron Microscopy. Methods Cell Biol. 2007;79:721–739. doi: 10.1016/S0091-679X(06)79028-8. [DOI] [PubMed] [Google Scholar]
- 43.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 44.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci U S A. 2014;111:15635–15640. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krivanek OL, Friedman SL, Gubbens AJ, Kraus B. An imaging filter for biological applications. Ultramicroscopy. 1995;59:267–282. doi: 10.1016/0304-3991(95)00034-x. [DOI] [PubMed] [Google Scholar]
- 46.Turoňová B, et al. Benchmarking tomographic acquisition schemes for high-resolution structural biology. Nat Commun. 2020;11 doi: 10.1038/s41467-020-14535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Chen M, et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat Methods. 2017;14:983–985. doi: 10.1038/nmeth.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Castaño-Díez D, Kudryashev M, Arheit M, Stahlberg H. Dynamo: A flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol. 2012;178:139–151. doi: 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Castaño-Díez D. The Dynamo package for tomography and subtomogram averaging: components for MATLAB, GPU computing and EC2 Amazon Web Services. Acta Cryst. 2017;D73:478–487. doi: 10.1107/S2059798317003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Representative tomograms showing different morphological states of P. falciparum rhoptry fusion (Fig. 2a, b, d) are available in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-26745 (Fig. 2a), EMD-26746 (Fig. 2b), and EMD-26747 (Fig. 2d). Subtomogram averages of the P. falciparum RSA in different conformations are available in the EMDB under accession codes EMD-26670 (RSA-A), EMD-26671 (RSA-2R), and EMD-26672 (RSA-1R).