Abstract
Differences in cardiac and aortic structure and function are associated with cardiovascular diseases and a wide range of other types of disease. Here we analyzed cardiovascular magnetic resonance images from a population-based study, the UK Biobank, using an automated machine learning-based analysis pipeline. We report a comprehensive range of structural and functional phenotypes for the heart and aorta across 26,893 participants and explore how these phenotypes vary according to sex, age and major cardiovascular risk factors. We extended this analysis with a phenome-wide association study, in which we tested for correlations of a wide range of non-imaging phenotypes of the participants with imaging phenotypes. We further explored the associations of imaging phenotypes with early-life factors, mental health and cognitive function using both observational analysis and Mendelian randomization. Our study illustrates how population-based cardiac and aortic imaging phenotypes can be used to better define cardiovascular disease risks as well as heart-brain health interactions, highlighting new opportunities for studying disease mechanisms and developing image-based biomarkers.
Cardiac and aortic structure and function are associated with cardiovascular diseases (CVDs)1,2 and a wide range of other types of disease3–6. Quantitative phenotypes derived from cardiovascular magnetic resonance (CMR) images enable us to assess cardiac and aortic structure and function in a non-invasive way and provide important biomarkers for the determination of pathological states in CVDs. For example, the left ventricular ejection fraction (LVEF) is an important clinical biomarker for the diagnosis and treatment of heart failure1. The left ventricular myocardial mass (LVM) is widely used for classifying hypertrophy and predicting risks of cardiovascular events7. Although CMR imaging phenotypes clearly play an important role in disease research and diagnosis, extracting these phenotypes demand significant involvement of experienced image analysts. This has become a limiting factor for applying CMR in large-scale studies and exploiting imaging phenotypes at a population level.
Large-scale imaging studies potentially provide a wealth of information for investigating disease risk factors and discovering early-stage image-based biomarkers. Recent large-scale imaging studies collecting CMR images include the Framingham Heart Study (offspring cohort: 1,114 subjects)8, MESA (5,004 subjects)9, DETERMINE (655 subjects)10, Dallas Heart Project (2,921 subjects)11 and UK Digital Heart Project (~2,000 subjects)12, to name a few. They illustrated the potential informativeness of CMR in large-scale studies, but lacked the power to explore a wide range of individual phenotypes simultaneously.
UK Biobank is a population-based prospective study for investigating risk factors of common adult diseases in middle and old age13–16. It recruited 500,000 women and men initially aged 40-69 years old between 2006-10 for long-term follow-up, from whom extensive sociodemographic, life-style and health-related information were collected serially along with a range of physical measurements and a growing range of genetic and biomarker data. Currently, UK Biobank is in the process of acquiring comprehensive multi-modal images of different organ systems from 100,000 participants using a highly standardised protocol, including magnetic resonance (MR) scans of the heart, brain and abdomen, ultrasound scans of carotid arteries, whole body dual-energy X-ray absorptiometry (DXA) scan of bones and joints, retinal photographs and optical coherence tomography (OCT) images14. These will add additional imaging phenotypes for understanding the determinants of diseases.
Deriving quantitative imaging phenotypes at this scale forms a major challenge. Recently, an image analysis pipeline has been developed for UK Biobank brain MR images, which generates ~4,350 imaging phenotypes of brain structure and function for 10,000 subjects17. The derived brain imaging phenotypes, along with the breadth of life-style and health information collected by UK Biobank, provide a valuable resource for studying the influence of ageing, progression of neuropathology and identifying early-stage image-based biomarkers for diseases18. An initial genome-wide association study (GWAS) has been performed19, which identified 148 clusters of associations between single nucleotide polymorphisms (SNPs) and brain imaging phenotypes that replicate at p < 0.05, providing insights into the genetic architecture relevant to the brain.
Here we present cardiac and aortic structural and functional imaging phenotypes for 26,893 subjects and demonstrate association studies enabled by these imaging phenotypes at a population level. The phenotypes were derived from UK Biobank CMR images using an automated machine learning-based analysis pipeline, built upon previously proposed cardiac and aortic image segmentation methods using convolutional neural networks20, 21. The pipeline evaluates comprehensive imaging phenotypes for the heart and aorta, including global phenotypes of the four cardiac chambers and two aortic sections: the left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), ascending aorta (AAo) and descending aorta (DAo), as well as regional phenotypes of the LV myocardial wall thickness and strain. We report associations of the cardiac and aortic imaging phenotypes with sex, age and traditional cardiovascular risk factors. We then conducted a large, population-based phenome-wide association study to relate the cardiac and aortic phenotypes to non-imaging phenotypes. We discovered a wide range of highly significant associations with life style, early-life factors, mental health and cognitive function of the participants.
Results
Image phenotyping
Figure 1 summarises the image analysis steps performed on short-axis, long-axis and aortic cine images (see detail in Methods). In total, 82 quantitative imaging phenotypes characterising the structure and function of the heart and aorta (Table 1) were generated for each subject. After running the analysis pipeline and performing quality control (Supplementary Table 1), imaging phenotypes were available for 26,893 subjects. Supplementary Table 2 describes the basic participant characteristics of the population. Supplementary Tables 3-6 report the summary statistics of the derived imaging phenotypes.
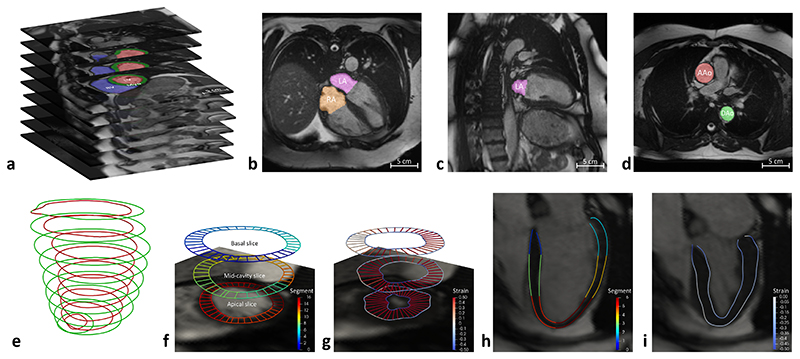
Figure 1. Automated CMR image analysis pipeline.
a) LV and RV volumes are derived from short-axis image segmentation (red: LV cavity; green: myocardium; blue: RV cavity). b, c) LA and RA volumes are derived from long-axis image segmentation (purple: LA cavity; orange: RA cavity), as illustrated using a 4 chamber view (b) and a 2 chamber view (c). d) AAo and DAo cross-sectional areas are derived from aortic image segmentation (red: AAo; green: DAo). e) Myocardial wall thickness is measured using the distance between LV endocardial contour (red) and epicardial contour (green). f) For measurement of myocardial wall thickness, three short-axis image slices are selected, including a basal slice at 25% of the LV length (the distance from the mitral annular plane to the apex of the LV), a mid-cavity slice at 50% and an apical slice at 75% of the LV length. The endocardial and epicardial contours are divided into 16 AHA segments, which are coded by different colors, as indicated by the color bar. g) Motion tracking is performed on short-axis image slices, warping the contours to each time frame across a cardiac cycle. Circumferential and radial strains (color-coded on the contours) are calculated using the change of length of the line segments. h) On the long-axis 4 chamber view image, the endocardial and epicardial contours are divided into 6 segments (coded by different colors). i) Motion tracking is performed on the long-axis 4 chamber view image. Longitudinal strain is calculated using the change of length of the line segments.
Table 1. Quantitative imaging phenotypes of the heart and aorta, derived from short-axis, long-axis and aortic cine images.
| Anatomical structure | Quantitative imaging phenotypes |
|---|---|
| Left ventricle (LV) | End-diastolic volume, end-systolic volume, stroke volume, ejection fraction, cardiac output, myocardial mass, global and regional myocardial wall thickness at end-diastole, global and regional peak circumferential, radial and longitudinal strains |
| Right ventricle (RV) | End-diastolic volume, end-systolic volume, stroke volume, ejection fraction |
| Left atrium (LA) | Maximum volume, minimum volume, stroke volume, ejection fraction |
| Right atrium (RA) | Maximum volume, minimum volume, stroke volume, ejection fraction |
| Ascending aorta (AAo) | Maximum area, minimum area, aortic distensibility |
| Descending aorta (DAo) | Maximum area, minimum area, aortic distensibility |
Associations with sex and age
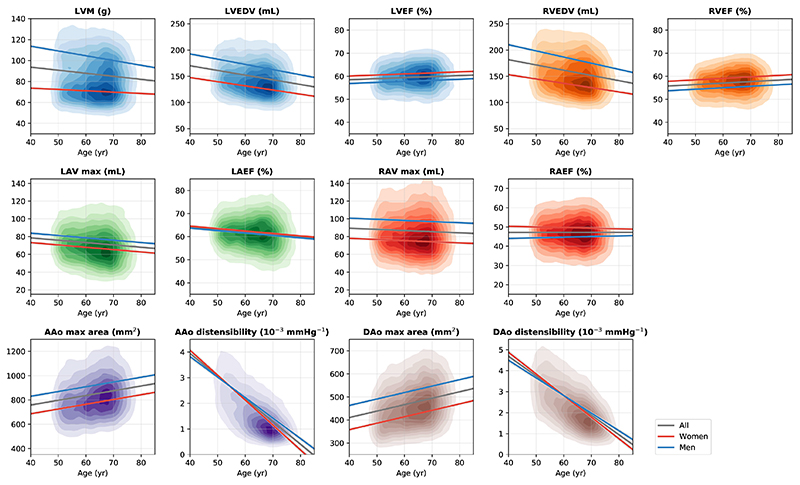
We investigated imaging phenotypes which describe cardiac and aortic size (mass, volume or area) and change (ejection fraction or distensibility) during a cardiac cycle. Two or three imaging phenotypes were selected for each of the six anatomical structures, including the LV myocardial mass (LVM), LV end-diastolic volume (LVEDV), LV ejection fraction (LVEF), RV end-diastolic volume (RVEDV), RV ejection fraction (RVEF), LA maximum volume (LAV max), LA ejection fraction (LAEF), RA maximum volume (RAV max), RA ejection fraction (RAEF), AAo maximum area, AAo distensibility, DAo maximum area and DAo distensibility.
We first characterised the associations of these phenotypes with sex and age in the population. Subjects with self-reported CVDs were excluded to mitigate the impact of established cardiac conditions on imaging phenotypes. As the majority of the subjects were Caucasian, we excluded other ethnicities to mitigate the impact of ethnicity. After these exclusions, 23,415 subjects were used with available sex and age information. Multiple linear regression models were built, using each imaging phenotype as the dependent variable and sex and age as independent variables. An interaction term sex * age was included for LVM, LVEDV, RVEDV, RAEF, AAo distensibility and DAo distensibility, where it led to a model of better fit (one-sided F-test, p < 0.05).
Figure 2 and Supplementary Table 7 report the regression results. Overall, myocardial mass (LVM: β = 53.0, p = 3×10−218), cardiac chamber volumes (LVEDV: β = 53.1, p = 2.9×10−74; RVEDV: β = 71.1, p = 7.7×10−116; LAV max: β = 10.6, p = 2.6×10−297; RAV max: β = 22.7, p = 10−324) and aortic cross-sectional areas (AAo max area: β = 143.9, p = 10−324; DAo max area: β = 104.9, p = 10−324) were greater for men than for women. Men consistently had lower cardiac chamber ejection fractions (LVEF: β = −3.2, p = 10−324; RVEF: β = −4.1, p = 10−324; LAEF: β = −0.9, p = 1.7× 10−15; RAEF: β = −8.9, p = 1.4×10−19) than women.
Figure 2. Associations of selected imaging phenotypes with sex and age.
Each graph displays a kernel density plot of an imaging phenotype plotted against age, as well as the linear regression lines for the whole population (gray), for women (red) and for men (blue). n = 23,415 subjects were included in the analysis. Detailed regression coefficients and association p-values can be found in Supplementary Table 7.
In this cross-sectional study, ageing was associated with reduced myocardial mass and cardiac chamber volumes (LVM: β = −1.0, p = 2.2× 10−12; LVEDV: β = −5.9, p = 2.5×10−136; RVEDV: β = −6.1, p = 4.7×10−130; LAV max: β = −2.0, p = 9.1× 10−44; RAV max: β = −1.0, p = 1.7×10−9), increased aortic areas (AAo max area: β = 29.3, p = 6.1× 10−146; DAo max area: β = 20.8, p = 10−324) and reduced aortic distensibilities (AAo disten.: β = −0.7, p = 10−324; DAo disten.: β = −0.8, p = 10−324). We found that women had greater aortic distensibilities than men between 40 to 50 years old (Figure 2). However, there was a strong sex and age interaction (AAo disten.: β = 0.1, p = 8.4×10−14; DAo disten.: β = 0.1, p = 10−11). Aortic distensibilities decreased more quickly with age for women than for men. LVM, LVEDV, RVEDV and RAEF also exhibited strong sex and age interactions. Myocardial mass and ventricular volumes decreased more quickly with age for men than for women (LVM: β = −2.4, p = 5.6× 10−36; LVEDV: β = −1.5, p = 8.6× 10−6; RVEDV: β = −2.6, p = 4.6×10−13).
Associations with traditional cardiovascular risk factors
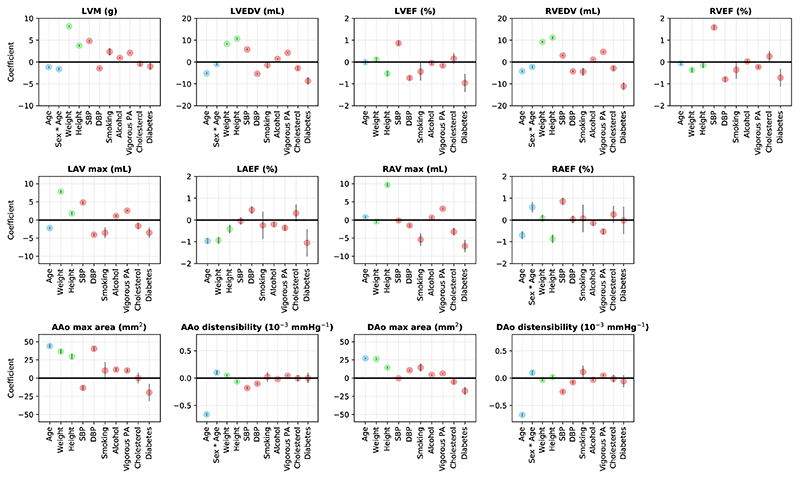
We explored the associations of cardiac and aortic imaging phenotypes with demographics (sex, age, sex * age), anthropometrics (weight, height) and cardiovascular risk factors22, including systolic blood pressure (SBP), diastolic blood pressure (DBP), current smoking status, alcohol intake, vigorous physical activity (PA) frequency, high cholesterol and diabetes. Sex, current smoking status, high cholesterol and diabetes are binary variables. The others are continuous variables.
Supplementary Table 8 reports the regression coefficients and p-values for these factors. Figure 3 plots the regression coefficients with 95% confidence intervals. As expected, higher weight or height was associated with greater cardiac chamber volumes and aortic areas (p ranges from 10−324 to 4.1×10−15, apart from weight with RAV max). Higher SBP was associated with greater myocardial mass (LVM: β = 4.8, p = 10−324), greater ventricular volumes (LVEDV: β = 5.8, p = 2.4×10−130; RVEDV: β = 3.0, p = 4.8×10−32), greater ventricular ejection fractions (LVEF: β = 0.9, p = 4.9×10−49; RVEF: β = 1.6, p = 1.4×10−161) and lower aortic distensibilities (AAo disten.: β = −0.2, p = 1.1× 10−41; DAo disten.: β = −0.3, p = 2.9×10−53). Smoking, higher alcohol intake and more frequent vigorous PA were associated with greater LVM (p ranges from 6.4×10−132 to 6.7×10−8). Smoking and vigorous PA both were associated with lower left ventricular ejection fraction (p ranges from 1.1×10−4 to 0.037). Diabetes was associated with lower myocardial mass, lower cardiac chamber volumes, lower ejection fractions and aortic areas (p ranges from 3.4×10−34 to 0.025, apart from RAEF).
Figure 3. Regression coefficients for cardiac and aortic imaging phenotypes on demographics (blue), anthropometrics (green) and cardiovascular risk factors (red).
For continuous variables, the coefficient describes the effect per standard deviation of the variable. For binary variables, the coefficient describes the effect with a change in the variable from 0 to 1. The gray bars denote the 95% confidence interval. n = 19,988 subjects were included in the analysis.
Associations with clinical outcomes
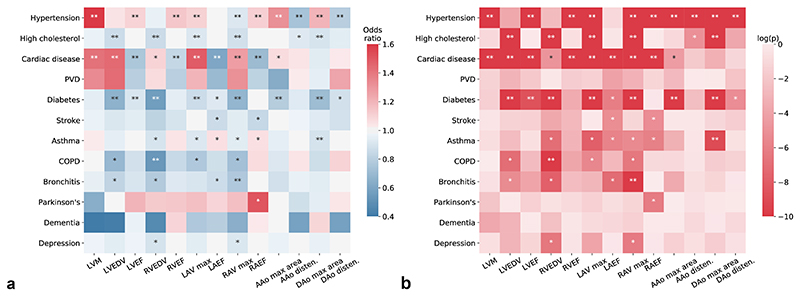
We also explored the associations of cardiac and aortic imaging phenotypes with 12 categories of common diseases, defined by self-reported disease code (Supplementary Table 9). Figure 4 and Supplementary Table 10 report the odds ratios (ORs) and p-values of each imaging phenotype as a risk factor for a common disease as the outcome in logistic regression. Greater LVM was associated with higher risk of hypertension (OR = 1.66, p = 3×10−84) and cardiac disease (OR = 1.41, p = 1.8×10−19). Greater cardiac chamber volumes were associated with higher risk of cardiac disease (p ranges from 1.5×10−63 to 0.008) and with lower risk of diabetes, asthma, COPD and bronchitis (p ranges from 1.3×10−37 to 0.004, apart from LVEDV with asthma). Greater cardiac chamber ejection fractions were associated with lower risk of cardiac disease and diabetes (p ranges from 2.3×10−107 to 0.028, apart from RAEF with diabetes). Greater aortic areas were associated with higher risk of hypertension (p ranges from 4.6×10−55 to 1.3×10−19) and lower risk of diabetes (p ranges from 3.7×10−17 to 1.8×10−10). Greater aortic distensibilities were associated with lower risk of hypertension and high cholesterol (p ranges from 6.6×10−30 to 0.022).
Figure 4. Associations of cardiac and aortic imaging phenotypes with common diseases.
(a) Odds ratio for an imaging phenotype as a risk factor for a common disease as the outcome. Sex, age, weight and height were adjusted in a logistic regression analysis. n = 25,743 subjects were included in the analysis. (b) The corresponding p-values (two-sided t-test) for odds ratios shown in (a). *: reaching the FDR threshold (pFDR = 0.017 for α = 0.05); **: reaching the Bonferroni threshold (pBonf = 3.2 ×10−4 for α = 0.05).
Phenome-wide association study
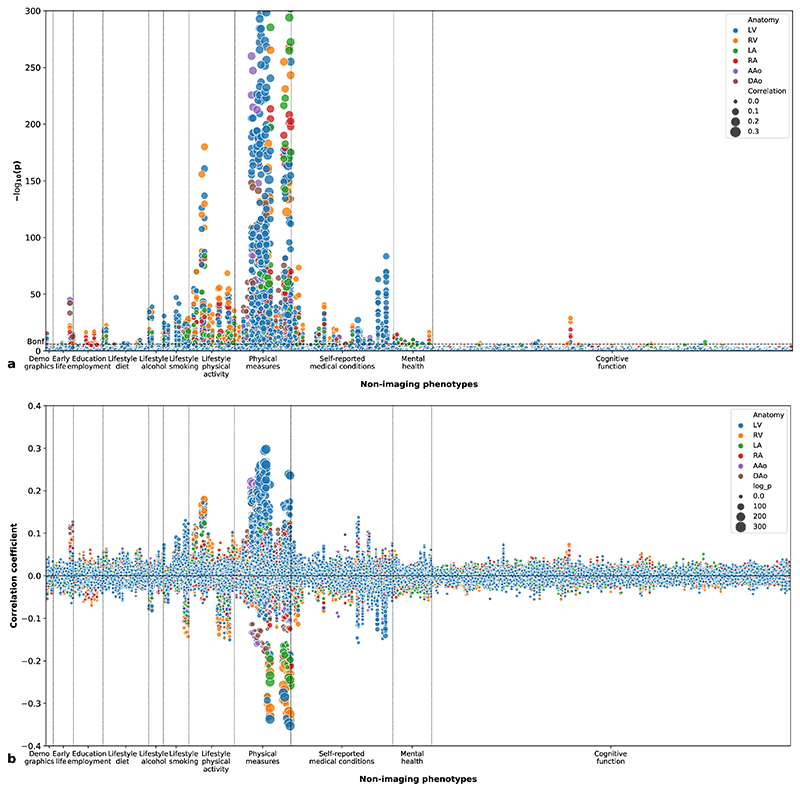
We performed a phenome-wide association study (PheWAS) to explore the correlations between imaging phenotypes and 11 categories of non-imaging phenotypes of the participants. The non-imaging phenotypes include primary demographics, early-life factors, education and employment, diet, alcohol, smoking, physical activity, physical measures, self-reported medical conditions, mental health and cognitive function. Figure 5a shows the Manhattan plot of the univariate correlation p-values (two-sided) between imaging phenotypes and non-imaging phenotypes and Figure 5b shows the correlation coefficients. Univariate correlations were performed between M = 82 imaging phenotypes and N = 555 non-imaging phenotypes for M × N = 45,510 times, with 2,617 correlations reaching the Bonferroni threshold for multiple comparisons (pBonf = 1.1×10−6 for α = 0.05) and 6,481 correlations reaching the false discovery rate (FDR) threshold (pFDR = 0.007 for α = 0.05).
Figure 5. Phenome-wide association study.
(a) Manhattan plot showing the p-values (two-sided t-test) for correlations between imaging phenotypes and non-imaging phenotypes. The height of each data point denotes the negative logarithm of the univariate correlation p-value between one imaging phenotype and one non-imaging phenotype. The area of the data point denotes the absolute value of the Pearson’s correlation coefficient. The colour of the data point denotes the anatomical structure of the imaging phenotype. The Bonferroni threshold for multiple comparisons (α = 0.05) is shown as a dashed horizontal line. n = 26,893 subjects were included in the analysis. (b) Plot showing the Pearson’s correlation coefficients between imaging phenotypes and non-imaging phenotypes. The height of each data point denotes the correlation coefficient and the area denotes the negative logarithm of the p-value (two-sided t-test).
Supplementary Table 11 lists the five most significant PheWAS associations for each anatomical structure. Higher SBP, more frequent PA and lower pulse rate were most consistently associated with larger cardiac chamber volumes. Overall health rating was associated with lower cardiac chamber volumes. Higher birth weight, whole body fat mass and hip circumference were associated with larger aortic areas.
Figure 5a also highlights associations between cardiac imaging phenotypes and mental health and cognitive performance phenotypes. Supplementary Table 12 presents five most significant PheWAS associations with mental health phenotypes. Risk taking and neuroticism score were significantly associated with cardiac and aortic phenotypes. Supplementary Table 13 reports the most significant associations with cognitive performance, assessed as fluid intelligence score, response time or number of correct trials completed in cognitive tests.
Further exploring associations with birth weight, mental health and cognitive performance
Following the mass univariate characterisation of associations using PheWAS, we selected several novel and most strongly associated non-imaging phenotypes and performed multiple linear regressions to further investigate their associations with cardiac and aortic structure and function. We adjusted for sex, age, sex * age, weight, height, SBP, DBP, current smoking status, alcohol intake, vigorous PA frequency, high cholesterol, diabetes and added the non-imaging phenotypes of interest as additional independent variables.
Birth weight
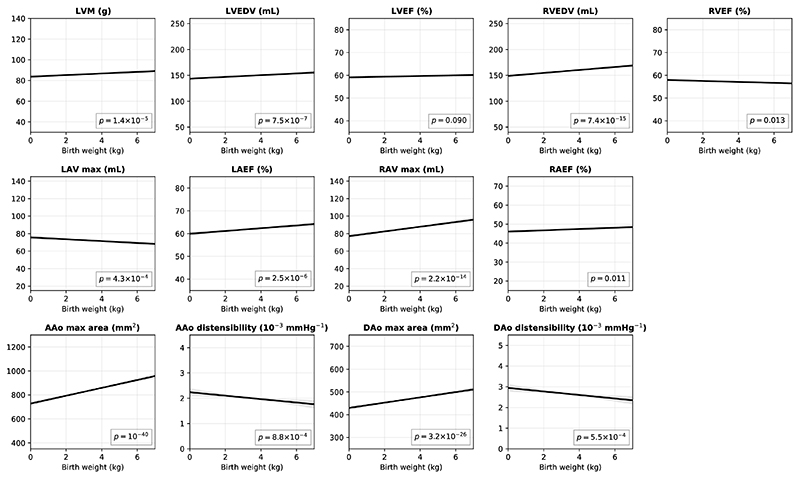
Early-life influences and birth weight are associated with later-life cardiovascular disease risk23. A previous, smaller population study concluded that these can be attributed to influences on general somatic growth24. We investigated this further. Extended Data Figure 1 shows the conditional plots of imaging phenotypes against birth weight and Supplementary Table 14 reports detailed regression coefficients and p-values. Even with control for current weight and height as covariates, birth weight was still significantly associated with myocardial mass and cardiac chamber volumes (LVM: β = 0.5, p = 1.4×10−5; LVEDV: β = 1.0, p = 7.5×10−7; RVEDV: β = 1.8, p = 7.4×10−15; LAV max: β = −0.7, p = 4.3×10−4; RAV max: β = 1.7, p = 2.2×10−14) and in particular with aortic areas. Higher birth weight was associated with greater aortic areas (AAo max area: β = 20.2, p = 10−40; DAo max area: β = 7.1, p = 3.2×10−26) and lower aortic distensibilities (AAo disten.: β = −0.04, p = 8.8×10−4; DAo disten.: β = −0.1, p = 5.5×10−4).
Risk taking and neuroticism
Symptoms potentially related to the heart and CVDs have an increased prevalence amongst people with depression and suggestive associations with other psychological states or traits, such as anxiety, stress or anger25. Here we extend these observations with evidence for more general associations of affective and psychological traits with cardiac and aortic structure and function (Supplementary Table 15). Risk taking behaviour was associated with greater LVM (β = 1.0, p = 4.4×10−6) and lower ejection fractions (LVEF: β = −0.3, p = 0.004; RVEF: β = −0.5, p = 1.6×10−7; LAEF: β = −0.5, p = 0.001; RAEF: β = −0.8, p = 5.3 ×10−7). Higher neuroticism score was associated with lower cardiac chamber volumes (LVEDV: β = −0.6, p = 0.002; RVEDV: β = −1.0, p = 9.8×10−7; LAV max: β = −0.4, p = 0.023; RAV max: β = −0.7, p = 3.7×10−4).
Fluid intelligence
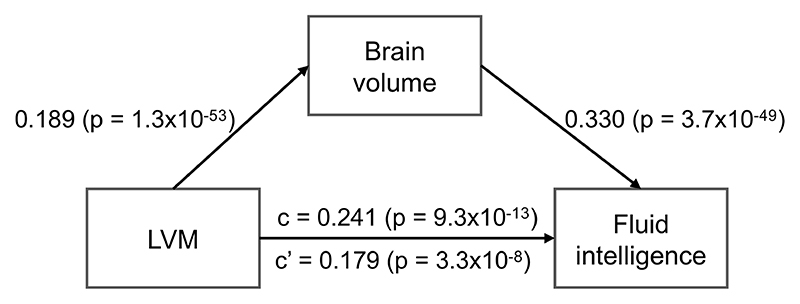
Subclinical population variation in cardiac haemodynamics has been associated with measures of processing speed and executive function in an Icelandic population26, although both the association and its potential mechanisms remain controversial27. With the greater statistical power afforded by this large population study, we have found similar and much stronger associations in a UK population (Supplementary Table 16). Higher fluid intelligence score was strongly associated with greater LVM (β = 0.8, p = 1.8×10−18) and greater cardiac chamber volumes (LVEDV: β = 1.1, p = 3.3×10−10; RVEDV: β = 2.0, p = 2×10−26; RAV max: β = 1.6, p = 1.9×10−20). We further performed a mediation analysis (Extended Data Figure 2), regressing fluid intelligence score against the LVM, using the brain volume as a mediator and adjusting for the same covariates. In this model, higher LVM was strongly associated with greater brain volume (β = 0.189, p = 1.3 ×10−53), which was associated with higher fluid intelligence score (β = 0.330, p = 3.7×10−49). Approximately one-fourth (26%) of the association between LVM and fluid intelligence score was mediated by brain volume.
Mendelian randomisation
By leveraging the SNPs associated with risk factors as instrumental variables and the genetic data from UK Biobank, we performed two-sample Mendelian randomisation analysis to investigate the relationships between risk factors and cardiac or aortic imaging phenotypes. Five risk factors, which have publicly available genetic association statistics that are needed for Mendelian randomisation, were investigated (Supplementary Tables 17). Three Mendelian methods were used, including inverse-variance weighting (IVW), weighted median (WM) and MR-Egger. Comparisons of heterogeneity statistics28 suggested that IVW and MR-Egger fitted equally well in most of the Mendelian randomisation analyses with the ratio statistic QR being close to 1. Supplementary Table 18 compares the effect sizes and p-values of Mendelian randomisation (detailed results in Supplementary Table 19) to observational analysis results. Higher SBP was associated with greater LVM (IVW: β = 4.8, p = 0.006; WM: β = 4.6, p = 0.044) and lower AAo distensibility (IVW: β = −0.3, p = 0.042; WM: β = −0.4, p = 0.048) with the same effect direction as conventional observational associations. Diabetes was associated with lower AAo max area (IVW: β = −12.0, p = 0.003) and with the same effect direction as the observational association. However, findings for SBP and diabetes were not significant after Bonferroni correction for multiple testing (pBonf = 1.4 ×10−3 for α = 0.05). Birth weight was associated with LVM, LVEDV, RVEDV and AAo max area (IVW and WM: p ranges from p = 6.2×10−8 to 0.049) and with consistent effect directions to those seen with observational analyses. The findings for birth weight using the IVW method sustained after adjustment for multiple testing and also with QR > 0.99. The effect directions of risk taking and fluid intelligence on imaging phenotypes were mostly consistent with observational associations, although the effects were not statistically significant. The MR-Egger pleiotropy test was only significant for the association between birth weight and AAo distensibility (p = 0.014).
Discussion
This study makes four major contributions. First, we presented quantitative phenotypes for six anatomical structures of the heart and aorta on a population-level CMR study, which were extracted using an automated machine learning-based analysis pipeline. We have returned the derived imaging phenotypes from the 26,893 subjects included in this study to UK Biobank to be made available for use by other researchers. A data visualisation website was built (https://heartvis.doc.ic.ac.uk), which allows researchers to explore the associations between imaging phenotypes and non-imaging phenotypes. We have shared the image analysis pipeline (https://github.com/baiwenjia/ukbb_cardiac), which we anticipate will be a generally valuable resource for CMR image analysis. Second, we demonstrated the potential informativeness of quantitative cardiac and aortic imaging phenotypes, which enabled both targeted association studies and a large-scale PheWAS. Third, based on novel observations in the PheWAS, we reported significant associations of cardiac and aortic phenotypes with a wide range of participant phenotypes, including birth weight, mental health and cognitive performance measures, illustrating the utility of generating these phenotypes even from a relatively healthy population when acquired at such a novel scale. Finally, we investigated the effects of these non-imaging phenotypes on imaging phenotypes using Mendelian randomisation and provided further evidence supporting the meaningfulness of the observational associations.
Using the imaging phenotypes, we characterised age-related changes in the heart and aorta for women and men. In this cross-sectional study, ageing was strongly associated with lower volumes of all the four cardiac chambers and with higher ejection fractions. Compared to cardiac chamber volumes and ejection fractions, aortic distensibilities were even more sensitive to age. For example, the mean distensibility of the ascending aorta in our population was substantially lower at 70 years old (1.3 ×10−3 mmHg−1) than at 50 years old (3.1 ×10−3 mmHg−1), consistent with the previous finding of an age-related tendency to increased arterial stiffness29, 30. Women showed a steeper decline of aortic distensibility with age compared to men. Some study suggested that this may be a consequence of physiological changes after menopause30.
Our study substantially extends findings in22 with association tests over a wider range of structural and functional phenotypes. We showed strong relationships of cardiac and aortic imaging phenotypes with SBP, smoking, alcohol intake, PA and diabetes, all of which are well described risk factors for cardiovascular diseases22. Although the percentage of current smokers is relatively low in this UK Biobank imaging subcohort (3.7%) compared to the full UK Biobank cohort (10.5%)31 or the general UK population32, we still found a relatively large effect size for smoking, highlighting its association with subclinical differences in cardiac structure and function33. In addition, due to the large sample size that we have, small signals of influence on cardiac and aortic anatomy and function are able to be detected. For example, although the effect sizes were relatively small, alcohol intake was highly significantly associated with greater myocardial mass, cardiac chamber volumes and aortic areas, which could not be established in an earlier, smaller population study22.
Additional associations between cardiac or aortic imaging phenotypes and prevalent diseases suggested their potentials for predicting the risks of diseases. Greater left ventricular mass or cavity volumes were associated with increased risk of cardiac diseases34. We also found that greater right ventricular volume was associated with increased risks for lung diseases, including asthma, COPD and bronchitis. This is probably due to the physiological interaction between the heart and the lung, consistent with previous findings35, 36. Aortic areas and distensibilities, which play important roles in the haemodynamics of the circulatory system37, were highly associated with the risk of hypertension.
The PheWAS analysis demonstrated that cardiac and aortic imaging phenotypes were significantly correlated with a wide range of non-imaging phenotypes of the participants (Supplementary Tables 20 and 21). PheWAS is a data-driven way to generate new hypotheses regarding developmental, life-style, clinical and environmental influences on disease risks through the discovery of their associations with population variation in cardiac and aortic anatomy and function. Particularly interesting were associations with early-life development, traits related to mental health and cognitive function. The large population allowed the identification of highly significant associations of aortic structure with birth weight independent of the general anthropometric measures such as adult height and weight38. We found associations between higher birth weight and greater aortic area, the diameter of which is a possible predictor for all cause mortality and incident cardiovascular events39.
The PheWAS also provided novel data for generating hypotheses relating cardiovascular function to mental health. We found that risk-taking behaviour was significantly associated with greater left ventricular myocardial mass and lower ventricular and atrial ejection fractions. This could be explained by common co-morbid behaviours such as smoking, drug and alcohol abuse40, 41, which are independently associated with cardiovascular diseases42, 43. Alternatively, neurogenic factors may contribute directly to cardiac remodelling44. Associations of risk taking with brain structure and function have been described45.
Our exploration of fluid intelligence score associations supports conclusions of an earlier, smaller study26, suggesting relationships between cardiovascular and cognitive function. Many studies have already demonstrated the importance of cardiovascular function in cognition and cognitive diseases6, 46. This may be a consequence of shared risk factors between dementia and cardiovascular diseases, such as smoking, hypertension, high cholesterol and diabetes47 or direct consequences of impaired cardiovascular function on the brain48. Our mediation analysis demonstrated a positive association between heart and brain structure, the latter of which was positively associated with general cognitive ability49.
The Mendelian randomisation analysis provided further evidence for the associations between non-imaging phenotypes and imaging phenotypes. Perhaps most interesting was the potential causal relationship discovered between birth weight and cardiac and aortic structures even in mid-later ages. This substantially extends earlier findings that heart size at birth has subtle but significant and persistent effects on cardiac structure into adulthood50. While the positive association between SBP and left ventricular mass is consistent with a previous interventional study51, it raises questions about the best interpretation of a more recent interventional study showing that intensive blood pressure (BP) control and standard BP control reduced left ventricular mass to a similar extent52. There was a negative association between diabetes and aortic area and distensibility, an observation that is useful for understanding observations such as the negative relationship of aortic distensibility to all cause mortality53.
However, there are several limitations to our analysis approach and observations. First, the central component of the image analysis pipeline, the convolutional neural network (CNN), was trained using UK Biobank images20, 21. It is possible to adapt the pipeline to images scanned on different systems or using different protocols, if training data is available for retraining or fine-tuning the network54. To apply the pipeline to images with completely unknown characteristics, however, the neural network is likely to produce less accurate segmentations. In machine learning, this problem is referred to as transfer learning or domain adaptation and machine learning methods that can reliably address this problem still remain elusive55. Second, the image analysis pipeline is automated. However, at this point, quality control for the image segmentation, was still performed manually. A future methodological extension is to develop and incorporate accurate and automated quality control to reduce the need for this manual intervention. Third, our image analysis pipeline focuses on a limited range of short-axis, long-axis and aortic cine images, which broadly characterise the cardiac and aortic structure and function. UK Biobank also collects tagged MR, aortic valve flow and T1 map images, the analysis of which will provide additional imaging phenotypes for the heart and aorta. Fourth, while Mendelian randomisation analysis may provide support for certain potential causal relationships, more reliable evidence needs to be provided by interventional research56. In the two-sample Mendelian randomisation analysis, the genetic associations of SBP, birth weight and risk taking were obtained from meta-analyses on both UK Biobank cohort and other cohorts (Supplementary Table 17), as findings excluding UK Biobank cohort were not available. This may introduce bias due to the overlap between the samples for risk factor GWAS and the samples for outcome (imaging phenotype) associations57. The summed proportion of variance in risk factors explained by all independent SNPs (r2 < 0.1) ranged from 0.25% (risk taking), 0.44% (SBP) to 10.78% (fluid intelligence) (Supplementary Tables 22-26). The relatively lower proportions of variance explained for risk taking and SBP may indicate bias due to the overlapping sample. However, since only a UK Biobank imaging subcohort (n = 22,229) was used for outcome associations, the overlap with the SBP (n = 1,006,863), birth weight (n=298,142) or risk taking GWAS cohort (n = 466,571) and the resulting bias may be presumed to be small. For the other risk factors used for Mendelian randomisation, the genetic associations were obtained from cohorts other than UK Biobank and thus without overlapping samples. Finally, the UK Biobank population is a relatively healthy cohort compared to the general UK population14. The extent to which observations are generalisable to populations with other ethnic mixes or with different frequencies of co-morbid diseases will need to be explored. The current imaging study focuses on observational associations on a cross-sectional dataset from a specific population, which may limit its generalisability for estimation of associations in other populations or making causal inferences. However, a re-imaging project for a subset of 10,000 UK Biobank subjects has just begun, which will increase its value for longitudinal studies.
In summary, we have developed and applied novel analytical methods to generate quantitative and clinically relevant phenotypes of the heart and aorta for 26,893 CMR images from the ongoing UK Biobank study. We have defined 2,617 significant associations between imaging phenotypes and non-imaging phenotypes of the participants, which provides a proof of principle for the use of quantitative imaging phenotypes for more precisely describing relationships between risk factors and cardiovascular diseases. Our findings provide novel insights into the influence of early-life factors and diabetes on cardiac and aortic structure and function, as well as the interaction between heart and cognitive phenotypes. We also demonstrate that in conjunction with data on genetic determinants of individual susceptibilities to life style or co-disease risks, causal relationships can be better defined using instruments such as Mendelian randomisation and genetic data available from UK Biobank16. A particular opportunity afforded by UK Biobank is the linkage to the long-term health outcomes of the participants through the National Health Service (NHS) record. In principle, this should enable extensions of investigations from associations to causal models for major later-life disorders. The power for such studies will increase rapidly over time. For example, of the 100,000 imaging participants, it was estimated that approximately 5,600 participants will have myocardial infarction (MI) and coronary death by 2022, rising to 9,400 by 2027 (diabetes: 8,000 rising to 13,600; stroke: 1,800 rising to 4,000; Alzheimer’s disease: 1,800 rising to 6,000)14. The derived imaging phenotypes, together with participant health information and other phenotype and genotype data, will provide a valuable resource for extension of studies on genetic architecture relevant to the heart, discovering risk factors and early-stage biomarkers for a variety of diseases and for new understanding in emerging areas of interest, such as the relationship between late-life heart and brain health.
Methods
Data
UK Biobank is an open access resource, open to bona fide scientists undertaking health-related research that is in the public good. Ethical approval is obtained from the North West Research Ethics Committee (REC reference: 11/NW/0382) and written consent is obtained from all participants. The UK Biobank CMR protocol is described in58. In brief, CMR images are acquired using clinical wide bore 1.5 Tesla scanners (MAGNETOM Aera, Syngo Platform VD13A, Siemens Healthcare, Erlangen, Germany). The image analysis pipeline consists of several parts, respectively for short-axis, long-axis and aortic cine images, which provide measures for different anatomical structures.
Short-axis image analysis
The LV, myocardium and RV are segmented on short-axis cine images using a fully convolutional network20, which was trained on manual annotations of 3,975 subjects. The LV and RV volumes across a cardiac cycle are derived from segmentation, as illustrated in Figure 1a. The end-diastolic volume (EDV), end-systolic volume (ESV), stroke volume (SV) and ejection fraction (EF) are determined for both ventricles. The LV myocardial mass is calculated from the myocardial volume using a density of 1.05 g/mL. The myocardial wall thickness is measured on each image slice using the distance between endocardial contour and epicardial contour at the end-diastolic (ED) frame, illustrated in Figure 1e. The myocardium is divided into 16 AHA segments according to59. The AHA segment model is a standard model recommended by the American Heart Association (AHA), which divides the LV into 16 or 17 segments for assessing regional variations of structure and function. Based on the model, the mean wall thickness for each segment and the global mean wall thickness are calculated.
Motion tracking is performed on short-axis images using non-rigid image registration60 between successive time frames, using the MIRTK toolkit. The inter-frame displacement fields are composed to obtain the displacement at frame k with regard to the reference frame, the ED frame or frame 0. To avoid the drift effect due to the accumulation of registration errors61, motion tracking is performed twice, respectively along the forward direction (tracking starting from frame 0 to frames 1, 2, 3, …) and backward direction (tracking from frame 0 to frames T – 1, T – 2, T – 3, …, where T denotes the number of frames per cardiac cycle). The average displacement field is calculated by weighted averaging the forward field and backward field, , where u0→k(x) denotes the displacement from frame 0 to frame k at pixel x, and and denote the weights for the forward and backward displacement fields. For a frame at the early stage of a cardiac cycle (small k), the forward displacement field will have a higher weight. For a frame at the late stage of a cardiac cycle (k close to T), the backward displacement field will have a higher weight.
Three images slices are used for motion tracking, namely a basal slice at 25% of the LV length (the distance from the mitral annular plane to the apex of the LV), a mid-cavity slice at 50% and an apical slice at 75% of the LV length, according to62, 63. The myocardial contours on the three slices are divided into AHA 16 segments, illustrated in Figure 1f. Based on the displacement field from motion tracking, myocardial contours at the ED frame are warped onto each time frame of the cardiac cycle, illustrated in Figure 1g. Circumferential and radial strains are calculated for each time frame based on the change of length for each line segment according to64, using the equation , where dir denotes the direction, Ldir denotes the length of a line segment along this direction and ΔLdir denotes its change. The peak strain for each segment and the global peak strain are calculated.
Long-axis image analysis
The LA and RA are segmented on long-axis 4-chamber (4Ch) view cine images using neural networks20, illustrated in Figure 1b. The LA and RA 4Ch areas and longitudinal diameters are determined from the segmentation. The LA is also segmented on long-axis 2-chamber view (2Ch), illustrated in Figure 1c. The LA 2Ch area and longitudinal diameter are also determined. The LA volume is calculated using the biplane area-length formula , where A2Ch and A4ch denotes the atrial area on the 2Ch and 4Ch view, L denotes the longitudinal diameter averaged across two views65, 66. The RA volume is calculated using . The maximum, minimum volumes and the ejection fraction are determined for both atria.
Motion tracking is performed on long-axis 4Ch view images, using the same approach as short-axis image motion tracking. The longitudinal strain is calculated from 4Ch view motion tracking results. The myocardial contour is divided into 6 segments, basal septal (1), basal lateral (2), mid septal (3), mid lateral (4), apical septal (5) and apical lateral (6), illustrated in Figure 1h. The peak strain for each segment and the global peak strain are calculated.
Aortic image analysis
The AAo and DAo are segmented on aortic cine images using a spatio-termporal neural network21, illustrated in Figure 1d. The maximum and minimum cross-sectional areas, Amax and Amin, are derived from the segmentation. The aortic distensibility is calculated by , where Pmax denotes the central systolic blood pressure and Pmin denotes the central diastolic blood pressure. The central blood pressure is calculated from the brachial blood pressure using Vicorder (Skidmore Medical, Bristol, UK) by applying a previously described brachial-to-aortic transfer function58.
Quality control
All image segmentations are manually quality controlled by an experienced cardiologist. The segmentation screenshots for short-axis, long-axis and aortic images at ED and ES frames are visually inspected. Bad segmentations, images with insufficient coverage of the LV or missing anatomical structures are discarded. For motion tracking, subjects with failed image registration or outlier peak global strain values (positive circumferential strain, negative radial strain or positive longitudinal strain) are discarded.
Statistical analysis
Multiple linear regression is used to identify the associations between imaging phenotypes and cardiovascular risk factors, using the Python statsmodels library. The risk factors are provided by UK Biobank, including sex (31), age (derived using the date attending assessment centre (53), birth year (34) and birth month (52)), weight (21002), height (50), SBP (4080), DBP (4079), current smoking status (20116), alcohol intake, vigorous PA frequency (904), high cholesterol and diabetes. The number inside the parenthesis indicates the UK Biobank field ID. Alcohol intake in the unit of g/day is derived from self-reported alcohol intake, including average weekly intake of red wine (1568), champagne plus white wine intake (1578), beer plus cider (1588), spirits (1598) and fortified wine (1608). The quantity of each type of drink is multiplied by its standard drink size and reference alcohol content, then converted to consumption per day67. High cholesterol is derived from self-reported non-cancer illness code (20002), using illness code 1473. Diabetes is derived similarly, using illness code 1220, 1221, 1222 and 1223. Mediation analysis is performed following Baron and Kenny’s steps68 and implemented also using the Python statsmodels library.
In the PheWAS, effects such as age, sex, weight and height are regressed out of the imaging phenotypes, similar to18, as they may confound with many non-imaging phenotypes. The non-imaging phenotypes are normalised to follow the Gaussian distribution N(0,1). Univariate cross correlation is then performed between each deconfounded imaging phenotype and each normalised non-imaging phenotype. The non-imaging phenotypes are grouped into 11 categories similar to18, including primary demographics (1001), early-life factors (1002), education and employment (1007), diet summary (1004), alcohol summary (100051), smoking summary (100058), physical activity (100054), physical measure summary (1006), self-reported medical conditions (1003), mental health (1018), cognitive function (100026). The number inside the parenthesis indicates the UK Biobank category ID listed at http://biobank.ndph.ox.ac.uk/showcase/cats.cgi. We use all non-imaging phenotypes that are available to us in our UK Biobank Application 18545 and that belong to the 11 categories. Then we perform data cleaning, discarding phenotypes with more than 90% missing data and keeping only one phenotype if there are two highly correlated phenotypes with correlation coefficient > 0.9999.
Mendelian randomisation
Data sources for the genetic associations of each risk factor of interest are presented in Supplementary Table 17, including SBP, diabetes, birth weight, risk tolerance and fluid intelligence. The genetic instruments, the SNPs, were selected with p < 5 ×10−8. For fluid intelligence, due to the lack of genetic instruments, a p-value cut-off of 1 ×10−5 was used. To avoid weak-instrument bias, we calculated the F-statistics and only included genetic instruments with an F-statistic > 10. We removed correlated SNPs (r2 > 0.1) by keeping the SNP with the smallest p-value for the association with the risk factor of interest. Supplementary Tables 22-26 list the SNPs used for each risk factor. Genetic associations of imaging phenotypes were performed among genotyped Caucasian, adjusting for sex, age, height and genetic principal components. Three Mendelian randomisation methods were used56: inverse-variance weighting (IVW), weighted median (WM) and MR-Egger69, 70. We assessed the heterogeneity statistics Q for IVW, Q′ for MR-Egger and calculated the ratio statistic QR = Q′ / Q for model selection28. A QR close to 1 indicates that IVW and MR-Egger models fit the data equally well, whereas a QR much less than 1 indicates that MR-Egger is better fitting. MR-PRESSO was used to identify outlier SNPs71. Outlier SNPs identified by MR-PRESSO were excluded from the analysis. Analysis was performed using the TwoSampleMR package in R72. We adjusted for multiple testing of 5 risk factors and 7 imaging phenotypes using Bonferroni correction.
Extended Data
Extended Data Figure 1. The conditional plots of imaging phenotypes against birth weight.
The dark line denotes the conditional plot of an imaging phenotype against birth weight, with other variables (sex, age, sex * age, weight, height, SBP, DBP, current smoking status, alcohol intake, vigorous PA frequency, high cholesterol and diabetes) set to their mean. The grey area denotes the 95% confidence interval. n = 12,169 subjects were analysed with available birth weight information. The p-values were calculated from two-sided t-tests.
Extended Data Figure 2. Mediation model for LVM, brain volume and fluid intelligence score.
The relationship between LVM and fluid intelligence score (path c) is 26% (difference between c and c’) mediated by total brain volume. n = 18,369 subjects were analysed with available fluid intelligence information. The values are depicted as regression coefficient (two-sided t-test p-value) for standardised imaging phenotypes.
Supplementary Material
Reporting summary.
Further information on research design is available in the Life Sciences Reporting Summary linked to this paper.
Acknowledgements
We would like to thank He Gao, Deborah Schneider-Luftman, Timothy J.W. Dawes and Arinbjorn Kolbeinsson for fruitful discussion. This research was conducted using the UK Biobank Resource under Application Number 18545, using methods developed under Application Number 18545 or 2964. Images were reproduced by kind permission of UK Biobank. We wish to thank all UK Biobank participants and staff. This work is supported by the SmartHeart EPSRC Programme Grant (EP/P001009/1) and the Imperial BHF Centre of Excellence Grant (RE/18/4/34215). H.S. is supported by a research fellowship from the Uehara Memorial Foundation and the Grants-in-Aid program from the Japan Society for the Promotion of Science (20K07776). S.E.P. acknowledges support from the NIHR Barts Biomedical Research Centre and S.E.P. has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 825903 (euCanSHare project). S.E.P., S.N. and S.K.P. acknowledge the British Heart Foundation for funding the manual analysis to create a cardiovascular magnetic resonance imaging reference standard for the UK Biobank imaging resource in 5000 CMR scans (PG/14/89/31194). A.D. is funded by the Wellcome Trust seed award (206046/Z/17/Z). D.P.O. is supported by the Medical Research Council (MC-A651-53301) and British Heart Foundation (NH/17/1/32725, RG/19/6/34387, RE/18/4/34215). P.M.M. gratefully acknowledges support from the Edmond J. Safra Foundation and Lily Safra, the Imperial College Healthcare Trust Biomedical Research Centre, the EPSRC Centre for Mathematics in Precision Healthcare and the UK Dementia Research Institute.
Footnotes
Author contributions
W.B. developed the analysis pipeline and wrote the manuscript; H.S. performed manual quality control of image segmentations and provided advice on clinical research; J.H. performed Mendelian randomisation; C.F. provided support on genetic data analysis; S.W. developed the data visualisation website; G.T. and Y.G. provided advice and support on the computational methodology; F.G. provided support on data management; H.S., N.A., K.F., S.E.P., S.K.P., S.N., E.E., A.D., D.P.O. and M.R.W. provided advice and support on clinical research; W.B., P.M.M. and D.R. conceived and designed the study. All authors read, contributed to revision and approved the manuscript.
Competing interests statement
S.E.P. acknowledges consultancy fees from Circle Cardiovascular Imaging Inc., Calgary, Alberta, Canada. D.R. acknowledges consultancy fees from Circle Cardiovascular Imaging Inc., Calgary, Alberta, Canada, Heartflow, Redwood City, CA, USA and IXICO PLC, London, UK. P.M.M. acknowledges consultancy fees from Roche, Adelphi Communications, Celgene and Biogen. He has received honoraria or speakers’ honoraria from Novartis, Biogen and Roche and has received research or educational funds from Biogen, Novartis, GlaxoSmithKline and Nodthera. He is a member of the Scientific Advisory Board to the Board of Ipsen Pharmaceuticals. The remaining authors declare no competing interests.
Data Availability
The raw imaging data and non-imaging participant characteristics are available from UK Biobank via a standard application procedure at http://www.ukbiobank.ac.uk/register-apply. The image analysis code is available at https://github.com/baiwenjia/ukbb_cardiac. For PheWAS, category IDs and field IDs are defined in ukb_field_categories.py and included in perform_phenome_wide_association.py. The associations between imaging phenotypes and non-imaging phenotypes can be browsed at https://heartvis.doc.ic.ac.uk.
References
- 1.Ponikowski P, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur J Hear Fail. 2016;18:891–975. doi: 10.1002/ejhf.592. [DOI] [PubMed] [Google Scholar]
- 2.Erbel R, et al. 2014 ESC Guidelines on the diagnosis and treatment of aortic diseases. Eur Hear J. 2014;35:2873–2926. doi: 10.1093/eurheartj/ehu281. [DOI] [PubMed] [Google Scholar]
- 3.Watz H, et al. Decreasing cardiac chamber sizes and associated heart dysfunction in COPD. Chest. 2010;138:32–38. doi: 10.1378/chest.09-2810. [DOI] [PubMed] [Google Scholar]
- 4.Alonso-Gonzalez R, et al. Abnormal lung function in adults with congenital heart disease: Prevalence, relation to cardiac anatomy, and association with survival. Circulation. 2013;127:882–890. doi: 10.1161/CIRCULATIONAHA.112.126755. [DOI] [PubMed] [Google Scholar]
- 5.Gansevoort RT, et al. Chronic kidney disease and cardiovascular risk: epidemiology, mechanisms, and prevention. The Lancet. 2013;382:339–352. doi: 10.1016/S0140-6736(13)60595-4. [DOI] [PubMed] [Google Scholar]
- 6.de Bruijn RF, Ikram MA. Cardiovascular risk factors and future risk of Alzheimer’s disease. BMC Medicine. 2014;12:130. doi: 10.1186/s12916-014-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong AC, et al. LV mass assessed by echocardiography and CMR, cardiovascular outcomes, and medical practice. JACC: Cardiovasc Imaging. 2012;5:837–848. doi: 10.1016/j.jcmg.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jefferson AL, et al. Relation of left ventricular ejection fraction to cognitive aging (from the Framingham Heart Study) The Am J Cardiol. 2011;108:1346–1351. doi: 10.1016/j.amjcard.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bild DE. Multi-ethnic study of atherosclerosis: Objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 10.Kadish AH, et al. Rationale and design for the defibrillators to reduce risk by magnetic resonance imaging evaluation (DETERMINE) trial. J Cardiovasc Electrophysiol. 2009;20:982–987. doi: 10.1111/j.1540-8167.2009.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Victor RG, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. The Am J Cardiol. 2004;93:1473–1480. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 12.Bello GA, et al. Deep-learning cardiac motion analysis for human survival prediction. Nat Mach Intell. 2019;1:95–104. doi: 10.1038/s42256-019-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins R. What makes UK Biobank special? The Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 14.Sudlow C, et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Medicine. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manolio TA. UK Biobank debuts as a powerful resource for genomic research. Nat Medicine. 2018;24:1792–1794. doi: 10.1038/s41591-018-0276-3. [DOI] [PubMed] [Google Scholar]
- 16.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfaro-Almagro F, et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage. 2018;166:400–424. doi: 10.1016/j.neuroimage.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller KL, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 2016;19:1523–1536. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott LT, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai W, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson. 2018;20:65. doi: 10.1186/s12968-018-0471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai W, et al. Recurrent neural networks for aortic image sequence segmentation with sparse annotations; International Conference on Medical Image Computing and Computer-Assisted Intervention; 2018. pp. 586–594. [Google Scholar]
- 22.Heckbert SR, et al. Traditional cardiovascular risk factors in relation to left ventricular mass, volume, and systolic function by cardiac magnetic resonance imaging. J Am Coll Cardiol. 2006;48:2285–2292. doi: 10.1016/j.jacc.2006.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geelhoed JJM, Jaddoe VWV. Early influences on cardiovascular and renal development. Eur J Epidemiol. 2010;25:677–692. doi: 10.1007/s10654-010-9510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy R, Ghosh AK, Deanfield J, Kuh D, Hughes AD. Birthweight, childhood growth and left ventricular structure at age 60-64 years in a British birth cohort study. Int J Epidemiol. 2016:1091–1102. doi: 10.1093/ije/dyw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaddha A, Robinson EA, Kline-Rogers E, Alexandris-Souphis T, Rubenfire M. Mental health and cardiovascular disease. The Am J Medicine. 2016;129:1145–1148. doi: 10.1016/j.amjmed.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Sabayan B, et al. Cardiac hemodynamics are linked with structural and functional features of brain aging: The age, gene/environment susceptibility (AGES)-Reykjavik Study. J Am Hear Assoc. 2015;4 doi: 10.1161/JAHA.114.001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedrich MG. Interplay of cardiac and cognitive function: How much do we really understand? J Am Hear Assoc. 2015;4 doi: 10.1161/JAHA.114.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden J, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47:1264–1278. doi: 10.1093/ije/dyy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redheuil A, et al. Reduced ascending aortic strain and distensibility: Earliest manifestations of vascular aging in humans. Hypertension. 2010;55:319–326. doi: 10.1161/HYPERTENSIONAHA.109.141275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nethononda RM, et al. Gender specific patterns of age-related decline in aortic stiffness: a cardiovascular magnetic resonance study including normal ranges. J Cardiovasc Magn Reson. 2015;17:20. doi: 10.1186/s12968-015-0126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson LM, et al. Factors associated with potentially serious incidental findings and with serious final diagnoses on multi-modal imaging in the UK Biobank Imaging Study: A prospective cohort study. PLOS ONE. 2019;14:e0218267. doi: 10.1371/journal.pone.0218267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fry A, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadruz W, et al. Smoking and cardiac structure and function in the elderly. Circ Cardiovasc Imaging. 2016;9 doi: 10.1161/CIRCIMAGING.116.004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. New Engl J Medicine. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 35.Steingrub JS, Tidswell M, Higgins TL. Hemodynamic consequences of heart-lung interactions. J Intensive Care Medicine. 2003;18:92–99. doi: 10.1177/0885066602250369. [DOI] [PubMed] [Google Scholar]
- 36.Repessé X, Charron C, Vieillard-Baron A. Acute cor pulmonale in ARDS: Rationale for protecting the right ventricle. Chest. 2015;147:259–265. doi: 10.1378/chest.14-0877. [DOI] [PubMed] [Google Scholar]
- 37.Alastruey J, Xiao N, Fok H, Schaeffter T, Figueroa CA. On the impact of modelling assumptions in multi-scale, subject-specific models of aortic haemodynamics. J The Royal Soc Interface. 2016;13:20160073. doi: 10.1098/rsif.2016.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang B, Godfrey KM, Martyn CN, Gale CR. Birth weight and cardiac structure in children. Pediatrics. 2006;117:e257–e261. doi: 10.1542/peds.2005-1325. [DOI] [PubMed] [Google Scholar]
- 39.Kamimura D, et al. Increased proximal aortic diameter is associated with risk of cardiovascular events and all-cause mortality in blacks the Jackson Heart Study. J Am Hear Assoc. 2017;6 doi: 10.1161/JAHA.116.005005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Haan L, Egberts A, Heerdink E. The relation between risk-taking behavior and alcohol use in young adults is different for men and women. Drug Alcohol Dependence. 2015;155:222–227. doi: 10.1016/j.drugalcdep.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- 42.Ambrose JA, Barua RS. The pathophysiology of cigarette smoking and cardiovascular disease. J Am Coll Cardiol. 2004;43:1731–1737. doi: 10.1016/j.jacc.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 43.Ronksley PE, Brien SE, Turner BJ, Mukamal KJ, Ghali WA. Association of alcohol consumption with selected cardiovascular disease outcomes: a systematic review and meta-analysis. BMJ. 2011;342:d671. doi: 10.1136/bmj.d671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozanski A, Blumenthal JA, Kaplan J. Impact of psychological factors on the pathogenesis of cardiovascular disease and implications for therapy. Circulation. 1999;99:2192–2217. doi: 10.1161/01.cir.99.16.2192. [DOI] [PubMed] [Google Scholar]
- 45.Strawbridge RJ, et al. Genetics of self-reported risk-taking behaviour, trans-ethnic consistency and relevance to brain gene expression. Transl Psychiatry. 2018;8:178. doi: 10.1038/s41398-018-0236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu C, Fratiglioni L. A major role for cardiovascular burden in age-related cognitive decline. Nat Rev Cardiol. 2015;12:267–277. doi: 10.1038/nrcardio.2014.223. [DOI] [PubMed] [Google Scholar]
- 47.Gorelick PB, et al. Vascular contributions to cognitive impairment and dementia. Stroke. 2011;42:2672–2713. doi: 10.1161/STR.0b013e3182299496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Buchem MA, et al. The heart-brain connection: A multidisciplinary approach targeting a missing link in the pathophysiology of vascular cognitive impairment. J Alzheimer’s Dis. 2014;42:S443–S451. doi: 10.3233/JAD-141542. [DOI] [PubMed] [Google Scholar]
- 49.Royle NA, et al. Estimated maximal and current brain volume predict cognitive ability in old age. Neurobiol Aging. 2013;34:2726–2733. doi: 10.1016/j.neurobiolaging.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnott C, et al. Subtle increases in heart size persist into adulthood in growth restricted babies: the Cardiovascular Risk in Young Finns Study. Open Hear. 2015;2:e000265. doi: 10.1136/openhrt-2015-000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson HJ, et al. Left ventricular hypertrophy: reduction of blood pressure already in the normal range further regresses left ventricular mass. Heart. 2010;96:148–152. doi: 10.1136/hrt.2009.177238. [DOI] [PubMed] [Google Scholar]
- 52.Upadhya B, et al. Effect of intensive blood pressure reduction on left ventricular mass, structure, function, and fibrosis in the SPRINT-HEART. Hypertension. 2019;74:276–284. doi: 10.1161/HYPERTENSIONAHA.119.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruickshank K, et al. Aortic pulse-wave velocity and its relationship to mortality in diabetes and glucose intolerance. Circulation. 2002;106:2085–2090. doi: 10.1161/01.cir.0000033824.02722.f7. [DOI] [PubMed] [Google Scholar]
- 54.Bhuva AN, et al. A multicenter, scan-rescan, human and machine learning CMR study to test generalizability and precision in imaging biomarker analysis. Circ Cardiovasc Imaging. 2019;12 doi: 10.1161/CIRCIMAGING.119.009214. [DOI] [PubMed] [Google Scholar]
- 55.Marcus G. Deep learning: A critical appraisal. arXiv: 1801.00631. 2018 [Google Scholar]
- 56.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen SE, et al. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson. 2015;18:8. doi: 10.1186/s12968-016-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerqueira MD, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 60.Rueckert D, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Transactions on Med Imaging. 1999;18:712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 61.Tobon-Gomez C, et al. Benchmarking framework for myocardial tracking and deformation algorithms: An open access database. Med Image Analysis. 2013;17:632–648. doi: 10.1016/j.media.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Taylor RJ, et al. Myocardial strain measurement with feature-tracking cardiovascular magnetic resonance: normal values. Eur Hear J - Cardiovasc Imaging. 2015;16:871–881. doi: 10.1093/ehjci/jev006. [DOI] [PubMed] [Google Scholar]
- 63.Schuster A, et al. Cardiovascular magnetic resonance feature-tracking assessment of myocardial mechanics: Intervendor agreement and considerations regarding reproducibility. Clin Radiol. 2015;70:989–998. doi: 10.1016/j.crad.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puyol-Anton E, et al. Fully automated myocardial strain estimation from cine MRI using convolutional neural networks; IEEE International Symposium on Biomedical Imaging; 2018. pp. 1139–1143. [Google Scholar]
- 65.Tsang TS, et al. Prediction of cardiovascular outcomes with left atrial size. J Am Coll Cardiol. 2006;47:1018–1023. doi: 10.1016/j.jacc.2005.08.077. [DOI] [PubMed] [Google Scholar]
- 66.Maceira AM, Cosín-Sales J, Roughton M, Prasad SK, Pennell DJ. Reference left atrial dimensions and volumes by steady state free precession cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2010;12:65. doi: 10.1186/1532-429X-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evangelou E, et al. New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat Hum Behav. 2019;3:950–961. doi: 10.1038/s41562-019-0653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 69.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Evangelou E, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morris A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warrington NM, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Linnér RK, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davies G, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw imaging data and non-imaging participant characteristics are available from UK Biobank via a standard application procedure at http://www.ukbiobank.ac.uk/register-apply. The image analysis code is available at https://github.com/baiwenjia/ukbb_cardiac. For PheWAS, category IDs and field IDs are defined in ukb_field_categories.py and included in perform_phenome_wide_association.py. The associations between imaging phenotypes and non-imaging phenotypes can be browsed at https://heartvis.doc.ic.ac.uk.