Abstract
Prostaglandins (PGs) are important proinflammatory lipid mediators, the significance of which is highlighted by the widespread and efficacious use of non-steroidal anti-inflammatory drugs (NSAIDs) in the treatment of inflammation. 4-Octyl itaconate (4-OI), a derivative of the Krebs cycle-derived metabolite itaconate, has recently garnered much interest as an anti-inflammatory agent. Here we show that 4-OI limits PG production in macrophages stimulated with the Toll-like receptor 1/2 (TLR1/2) ligand Pam3CSK4. This decrease in PG secretion is due to a robust suppression of COX2 expression by 4-OI, with both mRNA and protein levels decreased. Dimethyl fumarate (DMF), a fumarate derivative used in the treatment of multiple sclerosis (MS), with properties similar to itaconate, replicated the phenotype observed with 4-OI. We also demonstrate that the decrease in COX2 expression and inhibition of downstream prostaglandin production occurs in an NRF2-independent manner. Our findings provide a new insight into the potential of 4-OI as an anti-inflammatory agent and also identifies a novel anti-inflammatory function of DMF.
Introduction
Prostaglandins are key lipid mediators, which exert a wide variety of physiological roles. Their synthesis begins with the release of arachidonic acid by cytoplasmic phospholipase A2 (cPLA2) from membrane phospholipids, after which it is converted to prostaglandin H2 (PGH2) by the cyclooxygenase enzymes. COX1 is constitutively and ubiquitously expressed and is known to play several homeostatic roles, while COX2 is inducible by inflammatory stimuli including Toll-like receptor (TLR) ligands and cytokines. PGH2 can then be converted into various prostaglandins (PGs) and thromboxanes by a range of synthase enzymes (1).
The widespread and effective use of NSAIDs, which inhibit COX enzymes, highlights the clinical importance of blocking prostaglandin production in inflammation (2). The proinflammatory effects of PGE2 in particular are well characterised. The capacity of PGE2 to induce a wide range of physiological and often pathological phenotypes is largely due to its binding to four different receptors (EP1-4), which vary in their tissue expression and downstream signal transduction pathways (3). PGE2 has been shown to activate mast cells (4), T helper 1 (Th1) cells (5, 6) and Th17 cells (7–9). PGE2 has been implicated in inflammatory diseases such as psoriasis (9) and rheumatoid arthritis (10), in pain responses (11, 12) and recently in aging (13). However PGE2 has also been shown to exert anti-inflammatory functions, particularly in the environment of the lung (14–16). Another prostaglandin, PGD2, has been reported to contribute to the allergic response. Engagement of PGD2 with its receptors facilitates chemotaxis and activation of eosinophils and Th2 cells during allergic disease (17–19). Thromboxanes, which are also downstream of COX activity, promote vasoconstriction and platelet aggregation (20).
Itaconate is a metabolite that has emerged in recent years as an important immunomodulator (21). It is synthesised via the decarboxylation of the Krebs cycle intermediate cis-aconitate by the enzyme aconitate decarboxylase 1 (ACOD1, also known as IRG1), encoded by immune responsive gene 1 (Irg1) (22). The expression of IRG1 is predominantly restricted to macrophages and several other immune cell types and is markedly upregulated upon stimulation with TLR ligands (23), thereby leading to an increase in intracellular itaconate. 4-Octyl itaconate (4-OI) is a cell-permeable derivate that is commonly used in the study of itaconate and has been shown to be converted into itaconate intracellularly in macrophages (24).
An important aspect of itaconate biology is that both the endogenous metabolite and 4-OI have been shown to function as cysteine modifiers (25). This cysteine alkylation was originally termed 2,3-dicarboxypropylation and is also referred to as itaconation. This post-translational modification (PTM) is the basis of many of the anti-inflammatory functions associated with itaconate and 4-OI (24, 26–30). 4-OI has been shown to modify cysteine residues on KEAP1 (25), which functions as a negative regulator of the master antioxidant transcription factor NRF2. Modifications of these cysteine residues cause KEAP1 to be degraded, which liberates NRF2 and permits translocation to the nucleus (31), where NRF2 induces transcription of antioxidant genes (32) and inhibits transcription of certain pro-inflammatory cytokines (33). The ability of 4-OI to activate NRF2 has been implicated in several of its anti-inflammatory and protective functions (25, 34–38).
Dimethyl fumarate (DMF) is a derivative of another Krebs cycle metabolite, fumarate, that is clinically approved for the treatment of multiple sclerosis (MS) (39). Like 4-OI, DMF is also a potent cysteine modifier (cysteine alkylation by fumarate is termed succination) (40), and DMF shares some of the same targets as 4-OI, such as GAPDH (27, 41), gasdermin D (28, 30, 42) and importantly KEAP1 (25, 43), meaning that DMF is also a potent NRF2 activator. Therefore these two metabolite derivates often have similar effects on biological pathways.
The effect of itaconate and its derivatives on PG production has not been studied to date. Here we show that 4-OI greatly reduces PG production in pro-inflammatory macrophages through transcriptional suppression of COX2. We demonstrate that PG production and COX2 transcription is unchanged by the deletion of IRG1, indicating a difference with endogenous itaconate. However, DMF replicates the decrease in COX2 expression and PG secretion observed with 4-OI. Finally we show that 4-OI and DMF reduce COX2 expression in an NRF2-independent manner. We have therefore uncovered a novel anti-inflammatory role of 4-OI and DMF in macrophages.
Materials and Methods
Reagents
4-Octyl itaconate was initially supplied by Professor Richard Hartley and results were later confirmed with commercially available 4-OI (Sigma Aldrich). Pam3CSK4, dimethyl fumarate, diethyl maleate, indomethacin and NS-398 (Sigma Aldrich) were also used. Antibodies used were anti-β-actin (Sigma Aldrich), anti-COX2 (Abcam), anti-phospho-cPLA2 (Ser505), anti-cPLA2, anti-NRF2, anti-KEAP1, anti-ATF4, anti-phospho-NF-kB p65 (Ser536), anti-NF-kB p65, anti-phospho-p38 MAPK (Thr180/Tyr182), anti-p38 MAPK, anti-phospho p44/42 MAPK (Erk1/2) (Thr202/Tyr 204) and anti-p44/42 MAPK (Erk1/2) (Cell Signaling). Anti-mouse IgG and anti-rabbit IgG secondary horseradish peroxidase-conjugated antibodies (Jackson Immunoresearch) were also used. A PGE2 ELISA kit was used (Enzo Life Sciences). The Silencer Select siRNAs against NRF2 (s70522), ATF4 (s62689) and Anxa1 (s69299), as well as the Silencer Select negative control, were used (ThermoFisher Scientific).
Mice and BMDM generation
Bone marrow-derived macrophages (BMDMs) were isolated from C57BL/6J mice (Harlan UK). Legs from NRF2 knockout, KEAP1 knockdown and matched wild type mice were kindly provided by Professor Albeena Dinakova-Kostova (University of Dundee). The animals were housed under specific pathogen-free conditions in keeping with Irish and European Union regulations. All experiments carried out required prior ethical approval by Trinity College Dublin Animal Research Ethics Committee and Health Products Regulatory Authority. Mice were euthanised in a carbon dioxide chamber, after which cervical dislocation was used to confirm death. The ends of the tibia, femur and hip bones were cut and the bone marrow was flushed. The cells were then differentiated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (vol/vol) foetal calf serum (FCS), 1% penicillin/streptomycin and 20% L929 supernatant for six days. After this time the macrophages were counted and replated for use in experiments.
Human PBMC isolation
Thirty mL of whole blood was layered onto 20 mL Lymphoprep (Stemcell Technologies) in a 50 mL conical tube and centrifuged for 20 min at 2,000 rpm with no brake. The peripheral blood mononuclear cells (PBMCs) were then isolated from the middle layer and washed twice in PBS. PBMCs were cultured in RPMI supplemented with 10% (vol/vol) FCS and 1% penicillin/streptomycin for use in experiments.
siRNA transfection
The media on the cells was replaced with 500 μL DMEM containing no FCS or penicillin/streptomycin. The required amount of Lipofectamine RNAiMAX transfection reagent (Thermo Fischer Scientific) and the siRNAs were diluted in DMEM containing no FCS or penicillin/streptomycin and pre-incubated together for 15 minutes. 500 μL of this mix was then added to each well so that the final dilution of Lipofectamine RNAiMAX was 5 μL/mL and the final concentration of siRNA was 50 nM.
Western blotting
Cells were lysed in sample buffer [0.125 M Tris pH 6.8, 10% (vol/vol) glycerol, 0.02% SDS] and subsequently incubated at 95°C for a duration of five minutes. The samples were resolved on SDS-polyacrylamide gels, alongside the Spectra BR protein ladder (Thermofisher Scientific) so that proteins could be identified by molecular weight. The protein was then transferred to polyvinylidene fluoride membrane and membranes were blocked for one hour in 5% (w/v) dried milk in Tris-buffered saline/Tween (TBST). The blots were then incubated overnight at 4°C with the primary antibody. Following incubation for one hour with the secondary antibody, as well as three washes in TBST before and after secondary antibody, the blots were developed using chemiluminescent substrate (Thermofisher Scientific).
Real time PCR
Cells were lysed and the RNA was extracted using the PureLink RNA minikit (Ambion). cDNA was subsequently prepared using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), according to manufacturer’s instructions. Real-time quantitative PCR (qPCR) was then performed with the resulting cDNA using a 7500 Fast Real-Time PCR System with PowerUp SYBR Green Master Mix (Applied Biosystems). All genes were normalised to Rps18 expression for mouse or RPS13 for human. The sequences of the primer pairs for murine genes that were used are as follows; Rsp18, 5’-GGA TGT GAA GGA TGG GAA GT-3’ (forward) and 5’-CCC TCT ATG GGC TCG AAT TT-3’ (reverse); Ptgs2, 5’CGG ACT GGA TTC TAT GGT GAA A-3’ (forward) and 5’CTT GAA GTG GGT CAG GAT GTA G-3’ (reverse); Ptges, 5’GGA AGA AGG CTT TTG CCA ACC-3’ (forward) and 5’-CGA AGC CGA GGA AGA GGA AA-3’ (reverse); Nqo1, 5’-GCT GCA GAC CTG GTG ATA TT-3’ (forward) and 5’-ACT CTC TCA AAC CAG CCT TT-3’ (reverse); Il1b, 5’-GGA AGC AGC CCT TCA TCT TT-3’ (forward) and 5’-TGG CAA CTG TTC CTG AAC TC-3’ (reverse); Il6, 5’-CCA CAG TCC TTC AGA GAG ATA CA-3’ (forward) and 5’-CCT TCT GTG ACT CCA GCT TAT C-3’ (reverse); Tnf, 5’-GCC TCT TCT CAT TCC TGC TT-3’ (forward) and 5’-TGG GAA CTT CTC ATC CCT TTG-3’ (reverse); Il10, 5’-AGG CGC TGT CAT CGA TTT-3’ (forward) and 5’-CAC CTT GGT CTT GGA GCT T-3’ (reverse); Tgfb1, 5’-CGA AGC GGA CTA CTA TGC TAA A-3’ (forward) and 5’-TCC CGA ATG TCT GAC GTA TTG-3’ (reverse); Hmox1, 5’-CCT CAC AGA TGG CGT CAC TT-3’ (forward) and 5’-GCT GAT CTG GGG TTT CCC TC-3’ (reverse); Nos2, 5’-TTC ACC CAG TTG TGC ATC GAC CTA-3’ (forward) and 5’-TCC ATG GTC ACC TCC AAC ACA AGA-3’ (reverse); Ccl2, 5’-GTT GGC TCA GCC AGA TGC A-3’ (forward) and 5’-AGC CTA CTC ATT GGG ATC ATC TTG-3’ (reverse); Cd86, 5’-TCT CCA CGG AAA CAG CAT CT-3’ (forward) and 5’-CTT ACG GAA GCA CCC ATG AT-3’ (reverse). The sequences of the primer pairs for human genes that were used are as follows; RPS13, 5’- TCA CCG TTT GGC TCG ATA TT-3’ (forward) and 5’-GGC AGA GGC TGT AGA TGA TT-3’ (reverse); PTGS2 5’-TGC GCC TTT TCA AGG ATG GA-3’ (forward) and 5’-CCC CAC AGC AAA CCG TAG AT-3’ (reverse); IL1B, 5’-AGC TGA TGG CCC TAA ACA GA-3’ (forward) and 5’-TGT CCA TGG CCA CAA CAA CTG A-3’ (reverse); IL6, 5’-TCT GGA TTC AAT GAG GAG ACT TG-3’ (forward) and 5’-CTC AAA TCT GTT CTG GAG GTA CT-3’ (reverse); TNF, 5’-CCA GGG ACC TCT CTC TAA TCA-3’ (forward) and 5’-TCA GCT TGA GGG TTT GCT AC-3’ (reverse); IL10, 5’-CTG TCA TCG ATT TCT TCC CTG T-3’ (forward) and 5’-TGC CTT TCT CTT GGA GCT TAT T-3’ (reverse); TGFB1, 5’-CTG CAC TAT TCC TTT GCC C-3’ (forward) and 5’-TCT TCT TCA CTA TCC CCC AC-3’ (reverse); HMOX1, 5’-CCC AGC CCT ACA CCC GCT AC-3’ (forward) and 5’-GGT GGC ACT GGC AAT GTT GG-3’ (reverse).
ELISA
Cell supernatants were harvested and PG concentrations were quantified using an ELISA kit for PGE2 (Enzo Life Sciences), according to the manufacturer’s instructions. This item is sold as a PGE2-specific ELISA kit, however, our data indicated that other COX-derived oxylipins were also likely to be detected with this kit. Therefore we refer to results obtained using this ELISA kit as measuring PGs instead of PGE2.
Oxylipin analysis
Immediately upon harvesting, cell supernatants were snap frozen in liquid nitrogen. Samples were spiked with 2.1-2.9ng of PGE2-d4, PGD2-d4, 20-HETE-d6 and TXB2-d4 standards (Cayman Chemical). Lipids were extracted by adding a 2.5 ml solvent mixture (1 M acetic acid/isopropanol/hexane; 2:20:30, v/v/v) to 1 ml supernatants in a glass extraction vial and vortexed for 30 sec. 2.5ml hexane was added to samples and after vortexing for 30 seconds, tubes were centrifuged (500 g for 5 min at 4 °C) to recover lipids in the upper hexane layer (aqueous phase), which was transferred to a clean tube. Aqueous samples were re-extracted as above by addition of 2.5 ml hexane, and upper layers were combined. Lipid extraction from the lower aqueous layer was then completed according to the Bligh and Dyer technique. Specifically, 3.75ml of a 2:1 ratio of methanol:chloroform was added followed by vortexing for 30 secs. Subsequent additions of 1.25ml chloroform and 1.25ml water were followed with a vortexing step for 30 seconds, and the lower layer was recovered following centrifugation as above and combined with the upper layers from the first stage of extraction. Solvent was dried under vacuum and lipid extract was reconstituted in 100μl HPLC grade methanol. Lipids were separated by liquid chromatography (LC) using a gradient of 30-100% B over 20 minutes (A: Water:Mob B 95:5 + 0.1% Acetic Acid, B: Acetonitrile: Methanol – 80:15 + 0.1% Acetic Acid) on an Eclipse Plus C18 Column (Agilent), and analysed on a Sciex QTRAP® 6500 LC-MS/MS system. Source conditions: TEM 475°C, IS -4500, GS1 60, GS2 60, CUR 35. Lipids were detected using MRM monitoring with the following parent to daughter ion transitions: PGE2 and PGD2 [M-H]- 351.2/271.1, 15-deoxy-PGJ2 [M-H]- 315.2/271.1, TXB2 [M-H]- 369.2/169.1. Deuterated internal standards were monitored using precursor to product ions transitions of: TXB2-d4 [M-H]- 373.2/173.1, PGE2-d4 and PGD2-d4 [M-H]-355.2/275.1, and 20-HETE-d6 [M-H]- 325.2/281.1. Chromatographic peaks were integrated using Multiquant 3.0.2 software (Sciex). The criteria for assigning a peak was signal:noise of at least 5:1 and with at least 7 points across a peak. The ratio of analyte peak areas to internal standard was taken and lipids quantified using a standard curve made up and run at the same time as the samples.
Statistical Analysis
Statistical significance was established by the one-way or two-way ANOVA methods as indicated in the figure legends. Data are expressed as mean ± standard error of the mean. Significance was designated as follows: *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001. GraphPad Prism version 9 software was used for statistical analysis.
Results
4-OI inhibits Pam3CSK4-induced PG and thromboxane secretion by macrophages
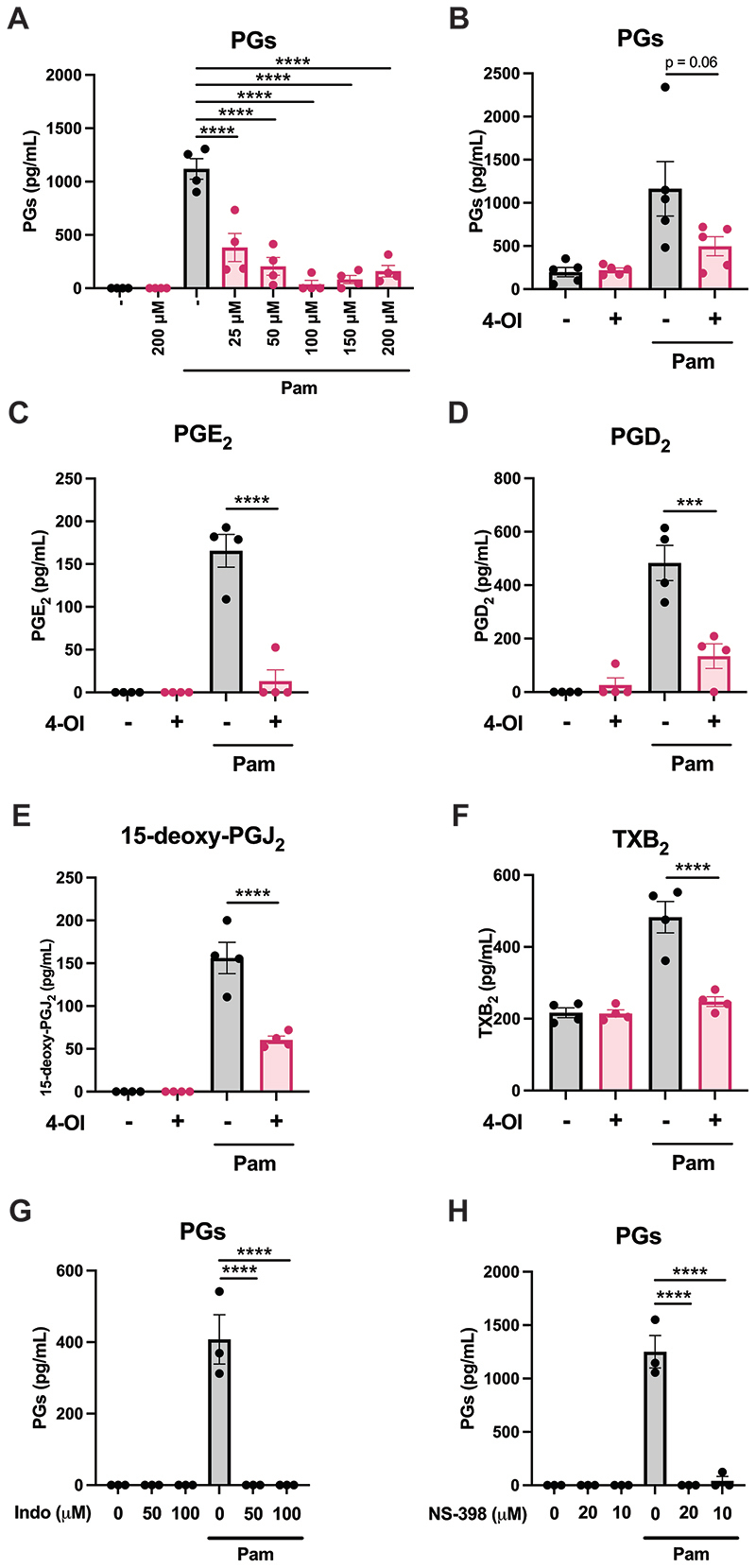
In order to investigate if 4-OI might play a role in modulation of prostaglandins, we treated murine BMDMs with 4-OI prior to stimulation with the TLR1/2 agonist Pam3CSK4 for 24 hours, which strongly induces PGs. We first used a PGE2 ELISA and observed a strong upregulation of cell supernatant PGs by Pam3CSK4, which was inhibited by 4-OI at concentrations as low as 25 μM (Figure 1A). 4-OI also reduced Pam3CSK4-induced PG secretion by human PBMCs (Figure 1B). To validate this result, we performed a lipidomic screen of oxylipins in BMDMs using quantitative tandem mass spectrometry (LC/MS/MS). Again, stimulation with Pam3CSK4 lead to a robust increase in PGE2 that was blocked by 4-OI pretreatment (Figure 1C), however the concentrations were notably lower than those measured by ELISA (Figure 1A). This is likely because of antibody-based measurements for lipid measurements displaying lower specificity and reporting on COX-derived oxylipins more broadly. From the LC/MS/MS screen we also observed that 4-OI potently inhibited Pam3CSK4-induced increases in PGD2 (Figure 1D), 15-deoxy-PGJ2 (Figure 1E) and thromboxane B2 (TXB2) (Figure 1F), in addition to PGE2. Therefore 4-OI was inhibiting all oxylipins downstream of COX activity. As confirmation that the ELISA was detecting COX-derived PGs in general, we found that the COX inhibitor indomethacin completely inhibited the Pam3CSK4-induced lipids from BMDMs detected by the ELISA (Figure 1G). In addition, we used the COX2-specific inhibitor NS-398, which also inhibited all Pam3CSK4-induced lipids detected by the ELISA (Figure 1H), indicating that PG secretion from BMDMs is almost entirely dependent on COX2.
Figure 1. 4-OI inhibits Pam3CSK4-induced prostaglandin production.
(A) BMDMs were pretreated with various concentrations of 4-OI (25-200 μM) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. The PG concentrations in the resulting supernatants were subsequently quantified by ELISA (n=4). (B) Human PBMCs were pretreated with 200 μM 4-OI for two hours prior to stimulation with Pam3CSK4 (1 μg/mL) for 24 hours. Supernatants were analysed for PG concentration by ELISA (n=5). (C-F) BMDMs were pretreated with 200 μM 4-OI for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. Supernatants were subsequently analysed by tandem mass spectrometry in order to determine (C) PGE2, (D) PGD2, (E) 15-deoxy-PGJ2 and (F) TXB2 concentrations (n=4). (G) BMDMs were pretreated with 50 μM or 100 μM indomethacin for one hour prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. The PG concentrations in the resulting supernatants were subsequently quantified by ELISA (n=3). (H) BMDMs were pretreated with 10 μM or 20 μM NS-398 for one hour prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. The PG concentrations in the resulting supernatants were subsequently quantified by ELISA (n=3). Data are mean ± S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001 by one-way ANOVA.
4-OI suppresses Pam3CSK4-induced COX2 expression
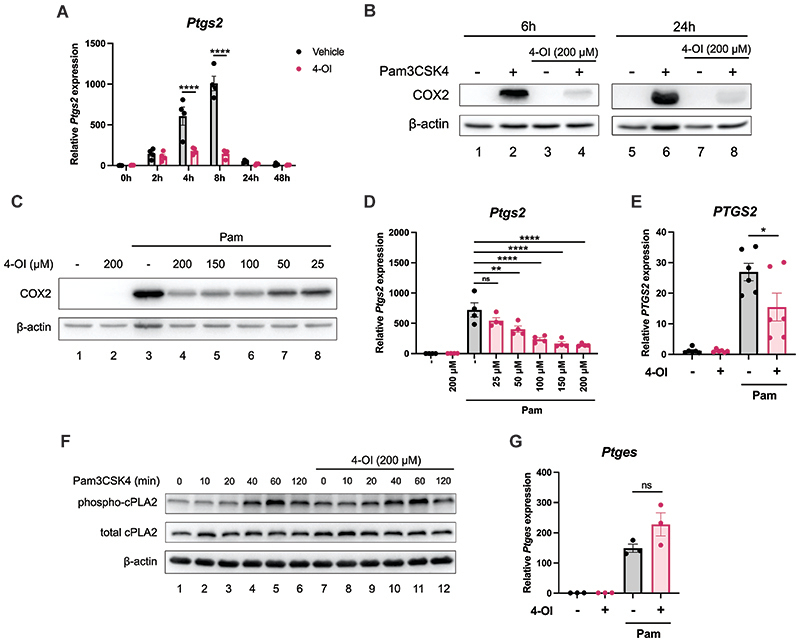
As we had observed that 4-OI suppressed all detected oxylipins downstream of COX activity, we next investigated if 4-OI had an effect on COX2 expression. COX2 is potently upregulated by TLR ligands such as Pam3CSK4, as seen by upregulation of Ptgs2 transcript, the gene that encodes COX2, as well as strong boost in COX2 protein levels. 200 μM 4-OI potently blocked Pam3CSK4-induced Ptgs2 levels at four hours and eight hours, the timepoints with the greatest induction of Ptgs2 (Figure 2A). 4-OI also greatly decreased COX2 protein levels at both six hours and 24 hours (Figure 2B). Concentrations as low as 25 μM 4-OI reduced COX2 protein at 24 hours (Figure 2C), while concentrations 50 μM or higher decreased Ptgs2 transcript at six hours (Figure 2D). 4-OI also reduced transcript levels of PTGS2 in human PBMCs (Figure 2E). The reduction of COX2 expression with 4-OI is likely the reason that 4-OI inhibits PG secretion, as COX2 is often referred to as the rate-limiting enzyme for PG synthesis. 4-OI did not alter the phosphorylation levels of cPLA2, nor did it affect total cPLA2 (Figure 2F). 4-OI also had no significant effect on the mRNA levels of Ptges (Figure 2E), the gene that encodes prostaglandin E2 synthase (PGES), the enzyme that catalyses conversion of PGH2 to PGE2. Therefore, on the PGE2 biosynthetic pathway 4-OI seems to specifically suppress COX2 expression, which leads to a decrease in downstream PG production.
Figure 2. 4-OI inhibits Pam3CSK4-induced COX2 expression.
(A) BMDMs were pretreated with 200 μM 4-OI prior to stimulation with Pam3CSK4 (100 ng/mL) for 2, 4, 8, 24 or 48 hours. The cells were lysed, mRNA was extracted and Ptgs2 expression was measured by qPCR (n=4) (B) BMDMs were pretreated with 200 μM 4-OI prior to stimulation with Pam3CSK4 (100 ng/mL) for 6 hours or 24 hours. COX2 expression was analysed by Western blotting (n=6). (C) BMDMs were pretreated with various concentrations of 4-OI (25-200 μM) prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. COX2 expression was analysed by Western blotting (n=4). (D) BMDMs were pretreated with various concentrations of 4-OI (25-200 μM) prior to stimulation with Pam3CSK4 (100 ng/mL) for six hours. After cell lysis, mRNA was extracted and Ptgs2 expression was measured by qPCR (n=4). (E) PBMCs were pretreated with 200 μM 4-OI for two hours prior to stimulation with Pam3CSK4 (1 μg/mL) for six hours. The cells were then lysed, mRNA was extracted and PTGS2 expression was measured by qPCR (n=6). (F) BMDMs were pretreated with 200 μM 4-OI prior to stimulation with Pam3CSK4 (100 ng/mL) for various timepoints (10-120 minutes). Phospho-cPLA2 and total cPLA2 expression was analysed by Western blotting (n=3). (G) BMDMs were pretreated with 200 μM 4-OI prior to stimulation with Pam3CSK4 (100 ng/mL) for four hours. After cell lysis, mRNA was extracted and Ptges expression was measured by qPCR (n=3). Data are mean ± S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001, ns = non-significant, by one-way ANOVA.
As has been previously reported (25, 27), we observed that 4-OI modulated the mRNA expression of several cytokine genes in BMDMs and PBMCs that were induced by Pam3CSK4 stimulation (Supplemental Figure 1A-E, J-N), such as yielding a reduction in Il1b, Il6 and Il10 transcripts. 4-OI also reduced mRNA levels of Nos2, which encodes nitric oxide synthase (Supplemental Figure 1G), and the chemokine Ccl2 (Supplemental Figure 1H). As expected, 4-OI increased transcript levels of the NRF2-dependent gene Hmox1 in both BMDMs and PBMCs (Supplemental Figure 1F and O respectively).
Endogenous itaconate does not affect COX2 expression or PG production
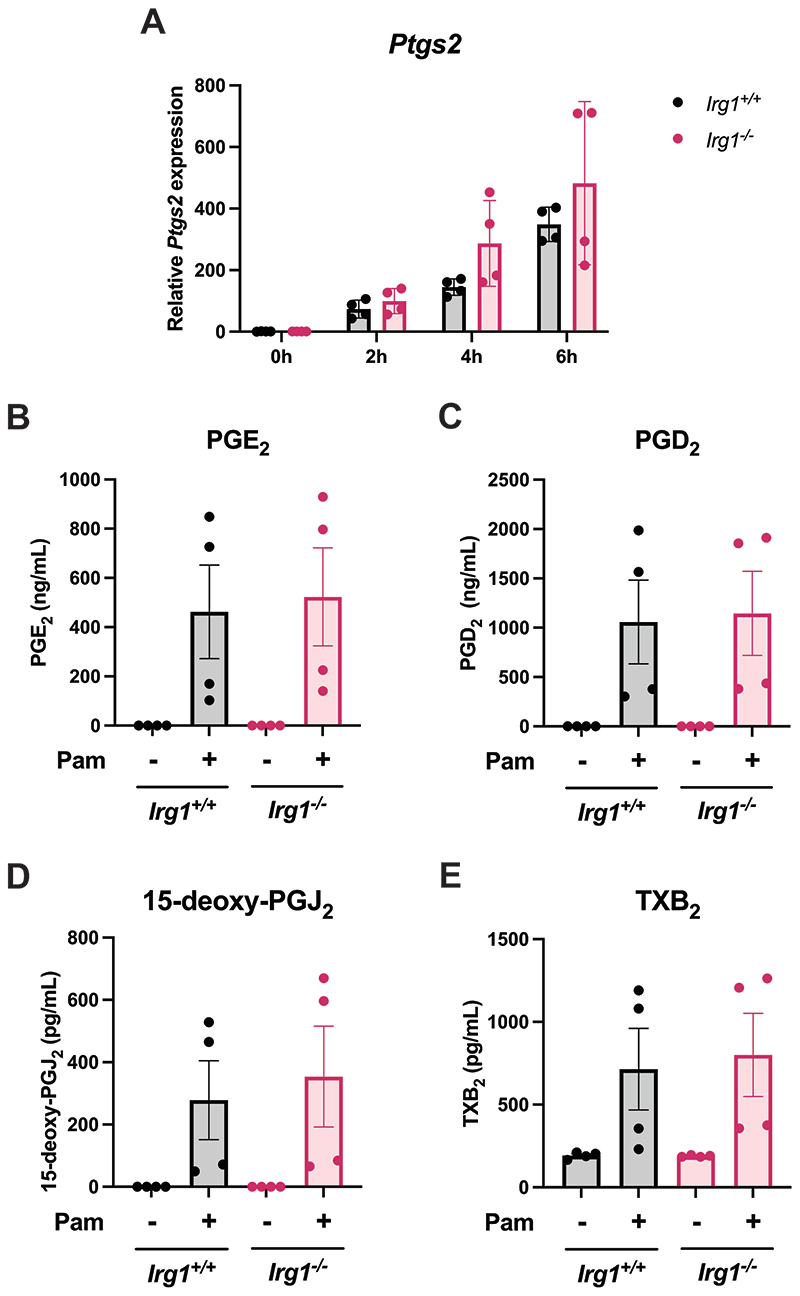
We next tested if endogenous itaconate, as well as the derivatised 4-OI, would impact COX2 expression and PG synthesis. For this we used BMDMs lacking Irg1, the gene that encodes the enzyme responsible for itaconate synthesis. However, when Ptgs2 transcript levels between Irg1+/+ BMDMs and Irg1-/- BMDMs are compared, there is no difference in COX2 induction by Pam3CSK4 (Figure 3A). In addition, there were no changes in the COX-derived oxylipins PGE2 (Figure 3B), PGD2 (Figure 3C), 15-deoxy-PGJ2 (Figure 3D) and TXB2 (Figure 3E) when measured by tandem mass spectrometry. This shows that endogenous itaconate does not affect COX2 expression or PG production.
Figure 3. Endogenous itaconate does not affect COX2 expression or prostaglandin production.
(A) BMDMs from Irg1+/+ and Irg1-/- mice were stimulated with Pam3CSK4 (100 ng/mL) for two, four or six hours. The cells were lysed and mRNA extracted in order to quantify Ptgs2 by qPCR (n=4). (B-E) BMDMs from Irg1+/+ and Irg1-/- mice were stimulated with Pam3CSK4 (100 ng/mL) for 24 hours. The cell supernatants were then analysed by tandem mass spectrometry in order to determine (B) PGE2, (C) PGD2, (D) 15-deoxy-PGJ2 and (E) TXB2 concentrations (n=4). Data are mean ± S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001 by one-way ANOVA, or two-way ANOVA for (A).
DMF also reduces Pam3CSK4-induced COX2 expression and PG production
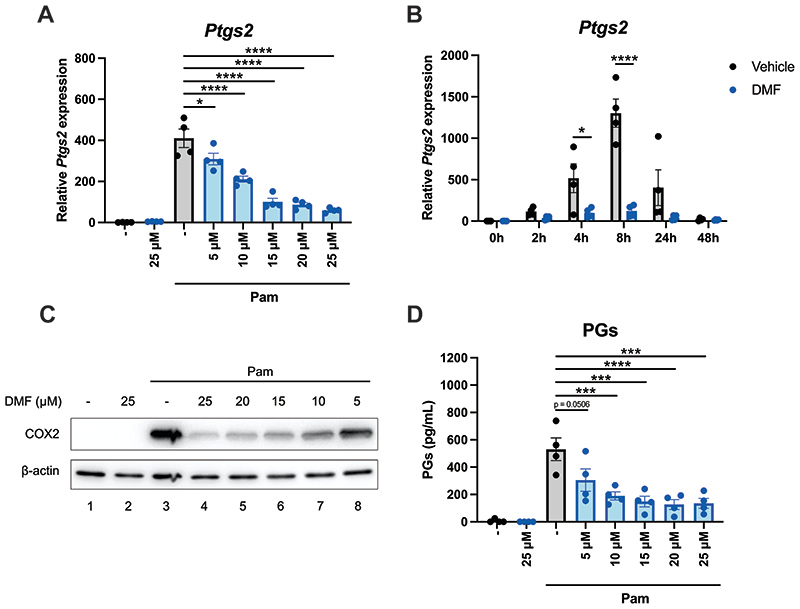
We next tested whether DMF might have similar effects on COX2 and PG production to 4-OI. Pretreatment of BMDMs with concentrations as low as 5 μM of DMF decreased levels of Ptgs2 transcript (Figure 4A). DMF significantly downregulated Ptsg2 mRNA levels at four hours and at eight hours (Figure 4B), similar to 4-OI. COX2 protein levels were also attenuated by DMF, using concentrations as low as 5 μM (Figure 4C). DMF also decreased Pam3CSK4-induced PG secretion by BMDMs (Figure 4D). Given that 4-OI and DMF are both potent cysteine modifiers and have very similar effects regarding COX2 expression and PG synthesis, it is likely that they are acting through a shared mechanism.
Figure 4. DMF decreases COX2 expression and prostaglandin production.
(A) BMDMs were pretreated with various concentrations of DMF (5-25 μM) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for four hours. After cell lysis, mRNA was extracted and Ptgs2 levels were quantified by qPCR (n=4). (B) BMDMs were pretreated with 25 μM DMF for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 2, 4, 8, 24 or 48 hours. After cell lysis, mRNA was extracted and Ptgs2 levels were quantified by qPCR (n=4). (C) BMDMs were pretreated with various concentrations of DMF (5-25 μM) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. COX2 expression was analysed by Western blotting (n=4). (D) BMDMs were pretreated with various concentrations of DMF (5-25 μM) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. The PG concentrations in the resulting supernatants were subsequently quantified by ELISA (n=4). Data are mean ± S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001 by one-way ANOVA.
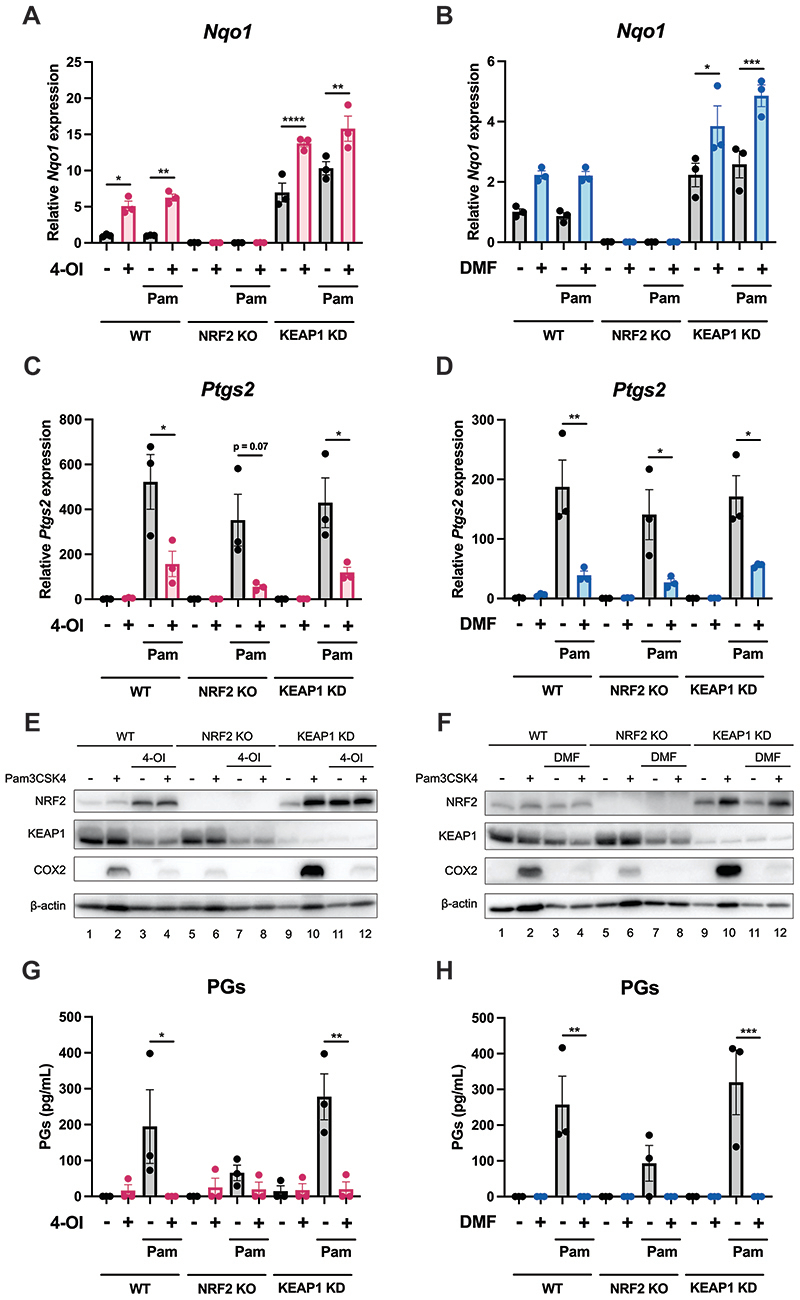
The 4-OI- and DMF-induced suppression of COX2 and PGs is NRF2-independent
4-OI and DMF are both known to be potent NRF2 activators through their ability to modify crucial cysteine residues on KEAP1, a negative regulator of NRF2. We also tested if another NRF2 activating compound, diethyl maleate, DEM, (44) would also modulate COX2. Indeed pretreatment of BMDMs with DEM resulted in decreased COX2 protein levels (Supplemental Figure 2A) and transcription (Supplemental Figure 2B), in addition to suppression of PG secretion (Supplemental Figure 2C). This indicated that perhaps NRF2 was somehow involved in the modulation of COX2 and PGs. To test this, we used BMDMs isolated from wild-type, NRF2 knockout and KEAP1 knockdown mice. As expected, expression of the NRF2-dependent gene Nqo1 was completely ablated in the NRF2 knockout BMDMs whereas the KEAP1 knockdown BMDMs displayed enhanced Nqo1 transcription compared to wild type cells (Figure 5A and B, Supplemental Figure 2D). Both 4-OI and DMF maintained the capacity to suppress Ptgs2 transcription in NRF2 knockout and KEAP1 knockdown BMDMs (Figure 5C and D respectively). 4-OI and DMF also reduced COX2 protein levels in NRF2 knockout and KEAP1 knockdown BMDMs (Figure 5E and F respectively). NRF2 knockdown using siRNA also did not abolish the capacity of 4-OI to reduce COX2 expression (Supplemental Figure 3A). Furthermore, 4-OI and DMF both blocked PG production in all genotypes (Figure 5G and H respectively). It is also worth noting that cells lacking NRF2 have lower levels of COX2 and PGs, whereas the KEAP1 knockdown cells, which exhibit augmented NRF2 activation, display elevated COX2 expression and PG production (Figure 5E-H). Interestingly, DEM, which is considered to be a well characterised NRF2 activator, could also still block Ptgs2 transcription in NRF2 knockout and KEAP1 knockdown BMDMs (Supplemental Figure 2E), indicating that its ability to reduce COX2 is separate to its NRF2-activating function. We also observed that 4-OI had no effect on NF-kB p65 phosphorylation, p38 phosphorylation or ERK phosphorylation (Supplemental Figure 3B), all of which are signalling pathways that are known to modulate ptgs2 transcription. We also investigated if ATF4 was involved in the observed effect, given that ATF4 was recently shown to bind directly to the ptgs2 promoter and induce its transcription (45). However, 4-OI actually increased ATF4 expression and ATF4 silencing had no effect on the inhibition of COX2 by 4-OI (Supplemental Figure 3C). We also tested if the effect of 4-OI on COX2 might be dependent on annexin A1, which has been shown to be modified by itaconate and itaconate derivatives (25, 28, 29) and is known to function as a negative regulator of cPLA2, which catalyses the first step of PG biosynthesis. However, silencing annexin A1 did not alter the inhibition of PG production by 4-OI (Supplemental Figure 3D). These results indicate that the capacity of 4-OI and DMF to suppress COX2 expression and downstream PG production is independent of NRF2 activation and other known signals that regulate COX2.
Figure 5. The capacity of 4-OI and DMF to reduce COX2 expression and prostaglandin production is not NRF2-dependent.
(A-D) BMDMs from wild-type, NRF2 knockout and KEAP1 knockdown mice were pretreated with 200 μM 4-OI (A and C) or 25 μM DMF (B and D) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for six hours. The cells were lysed, mRNA was extracted and Nqo1 expression (A and B) and Ptgs2 expression (C and D) were quantified by qPCR (n=3). (E-H) BMDMs from wild-type, NRF2 knockout and KEAP1 knockdown mice were pretreated with 200 μM 4-OI (E and G) or 25 μM DMF (F and H) for two hours prior to stimulation with Pam3CSK4 (100 ng/mL) for 24 hours. COX2 expression was analysed by Western blotting (E and F) (n=3). The supernatants were analysed for PG concentration by ELISA (G and H) (n=3). Data are mean ± S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001 by one-way ANOVA.
Discussion
The itaconate derivative 4-OI has recently garnered much attention as an immunomodulator. The findings of this study suggest a role for 4-OI in the inhibition of PG production in macrophages activated with the TLR1/2 ligand Pam3CSK4. We show that 4-OI potently reduces several COX-derived oxylipins and provide evidence that 4-OI suppresses COX2 transcription. We demonstrate that while endogenous itaconate derived from IRG1 activity does not affect COX2 levels, DMF attenuated COX2 expression and PG secretion in a similar manner to 4-OI. This implies that a cysteine modification may be involved but we also provide evidence that suggests that the effect of 4-OI and DMF on PG production is not via KEAP1 degradation and NRF2 activation. Further work is required in order to fully elucidate the mechanism by which 4-OI and DMF impair COX2 transcription.
Our knowledge of how Krebs cycle activity impacts inflammation has been rapidly expanding over the last number of years. Some Krebs cycle intermediates are known to exert proinflammatory actions, such as succinate which has been shown to stabilise HIF-1α, thereby upregulating IL-1β transcription (46). Succinate has also been reported to exacerbate certain inflammatory diseases, such as arthritis (47) and type 2 diabetes (48). Another Krebs cycle intermediate, citrate, has also been shown to alter the inflammatory response. The mitochondrial export, in addition to the breakdown of citrate, have been reported to be essential for nitric oxide and prostaglandin production (49, 50). Several anti-inflammatory roles have recently been described for both itaconate (25, 30, 51) and fumarate (41, 42) and here we report a novel function for derivatives of these two Krebs cycle metabolites in macrophages.
This work also highlights the importance of bearing in mind that metabolite derivatives do not always truly represent the action of the corresponding endogenous metabolites. In the case of 4-OI, the use of this derivative replicates the biological effects of endogenous itaconate in a number of cases such as the inactivation of the NLRP3 inflammasome (24) and impairment of glycolysis (27). However there are also incidences where 4-OI and endogenous itaconate differ in their effects, such as type I interferon (IFN) production, which is boosted by endogenous itaconate (52) but inhibited by 4-OI (25). While itaconate has also been shown to modify proteins in the same manner as 4-OI, the targets are not always the same. Our previous study (25) found that, while there were some overlapping proteins modified by both 4-OI and endogenous itaconate, many targets were mutually exclusive. This could potentially be due to differences in electrophilicity between itaconate and 4-OI.
Although DMF is known to be a potent NRF2 activator (43, 53), NRF2-independent anti-inflammatory functions of DMF have begun to emerge, such as the impairment of glycolysis through GAPDH succination (41) and inhibition of pyroptosis via gasdermin D (42). It is also true for 4-OI that a number of its currently known anti-inflammatory effects are NRF2-dependent (25, 34, 35), although others are not (24, 27, 30, 51). Interestingly, DEM, which is predominantly used as an experimental tool to pharmacologically activate NRF2, also supressed COX2 expression in an NRF2-independent manner. As DEM has been shown to modify cysteines on KEAP1 in a similar way to 4-OI and DMF (44), perhaps DEM also possesses the capacity to modify other reactive cysteines in the cell with further reaching implications. An observation that adds complexity to this system is that 15-deoxy-Δ12,14-PGJ2, which is downstream of COX activity and therefore would be reduced by 4-OI, has been shown to activate NRF2 (54). Nonetheless, our data reveals another NRF2-independent anti-inflammatory role for DMF, 4-OI and even DEM.
There is also the possibility that the effect of 4-OI on COX2 and prostaglandins will contribute to the attenuation of other inflammatory markers. For example, macrophages express EP receptors (55) and therefore PGE2 can signal in an autocrine manner. We have previously shown that endogenous PGE2 acting via the EP2 receptor is required for induction of pro-IL-1β (56). Hence the inhibition of PGE2 secretion from the macrophage and subsequent binding to EP2 could potentially contribute to the 4-OI-induced decrease in pro-IL-1β that has been reported (25). PGE2 has also been shown to augment IL-6 production in macrophages (57, 58), another cytokine that is blocked by 4-OI treatment (25). Therefore inhibition of PGE2 by 4-OI could potentially be a factor in the downregulation of IL-6. Treatment of macrophages with PGE2 was shown to boost production of IL-10 in macrophages (59), an anti-inflammatory cytokine that is also downregulated by 4-OI. It is also conceivable that a reduction in PGs secreted by macrophages could contribute to the anti-inflammatory effects of 4-OI during in vivo models (24, 25, 27). Conversely, the capacity of 4-OI to attenuate the production of other proinflammatory mediators could potentially affect the induction of COX2. For example, IL-1β and nitric oxide, both of which are inhibited by 4-OI (25), have been reported to contribute to COX2 induction (60, 61).
Our results therefore identify 4-OI and DMF as inhibitors of prostaglandin synthesis in macrophages. This work further highlights the potential of 4-OI as a therapeutic anti-inflammatory treatment and supplements the current knowledge of the anti-inflammatory effects of DMF.
Supplementary Material
Key Points.
4-OI reduces COX2 expression and inhibits prostaglandin production in macrophages
DMF also attenuates COX2 expression in macrophages and decreases prostaglandins
The 4-OI- and DMF-induced decrease in COX2 and prostaglandins is NRF2-independent
Acknowledgements
We thank Professor Richard Hartley (University of Glasgow) for the synthesis of 4-OI. We thank Professor Albeena Dinkova-Kostova (University of Dundee) for providing bones from NRF2 knockout, KEAP1 knockdown and matched wild-type mice.
Funding
The O’Neill group acknowledges the following grant support: European Research Council (834370), The Wellcome Trust (205455) and Science Foundation Ireland (12/IA/ 1531). In addition, we acknowledge Ser Cymru Project Sepsis grant funded by WG/EU-ERDF (VJT, VBO). VBO is a Royal Society Wolfson Research Merit Award Holder.
References
- 1.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vane JR. Biomedicine. Back to an aspirin a day? Science. 2002;296:474–475. doi: 10.1126/science.1071702. [DOI] [PubMed] [Google Scholar]
- 3.Woodward DF, Jones RL, Narumiya S. International Union of Basic and Clinical Pharmacology. LXXXIII: classification of prostanoid receptors, updating 15 years of progress. Pharmacol Rev. 2011;63:471–538. doi: 10.1124/pr.110.003517. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto K, Shirata N, Taketomi Y, Tsuchiya S, Segi-Nishida E, Inazumi T, Kabashima K, Tanaka S, Murakami M, Narumiya S, Sugimoto Y. Prostaglandin E2-EP3 signaling induces inflammatory swelling by mast cell activation. J Immunol. 2014;192:1130–1137. doi: 10.4049/jimmunol.1300290. [DOI] [PubMed] [Google Scholar]
- 5.Yao C, Hirata T, Soontrapa K, Ma X, Takemori H, Narumiya S. Prostaglandin E(2) promotes Th1 differentiation via synergistic amplification of IL-12 signalling by cAMP and PI3-kinase. Nat Commun. 2013;4:1685. doi: 10.1038/ncomms2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagamachi M, Sakata D, Kabashima K, Furuyashiki T, Murata T, Segi-Nishida E, Soontrapa K, Matsuoka T, Miyachi Y, Narumiya S. Facilitation of Th1-mediated immune response by prostaglandin E receptor EP1. J Exp Med. 2007;204:2865–2874. doi: 10.1084/jem.20070773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao C, Sakata D, Esaki Y, Li Y, Matsuoka T, Kuroiwa K, Sugimoto Y, Narumiya S. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat Med. 2009;15:633–640. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 8.Maseda D, Johnson EM, Nyhoff LE, Baron B, Kojima F, Wilhelm AJ, Ward MR, Woodward JG, Brand DD, Crofford LJ. mPGES1-Dependent Prostaglandin E2 (PGE2) Controls Antigen-Specific Th17 and Th1 Responses by Regulating T Autocrine and Paracrine PGE2 Production. J Immunol. 2018;200:725–736. doi: 10.4049/jimmunol.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, Aoki T, Thumkeo D, Siriwach R, Yao C, Narumiya S. T cell-intrinsic prostaglandin E2-EP2/EP4 signaling is critical in pathogenic TH17 cell-driven inflammation. J Allergy Clin Immunol. 2019;143:631–643. doi: 10.1016/j.jaci.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy JM, Wicks JR, Audoly LP. The role of prostaglandin E2 receptors in the pathogenesis of rheumatoid arthritis. J Clin Invest. 2002;110:651–658. doi: 10.1172/JCI15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamei D, Yamakawa K, Takegoshi Y, Mikami-Nakanishi M, Nakatani Y, Oh-Ishi S, Yasui H, Azuma Y, Hirasawa N, Ohuchi K, Kawaguchi H, et al. Reduced pain hypersensitivity and inflammation in mice lacking microsomal prostaglandin e synthase-1. J Biol Chem. 2004;279:33684–33695. doi: 10.1074/jbc.M400199200. [DOI] [PubMed] [Google Scholar]
- 12.Trebino CE, Stock JL, Gibbons CP, Naiman BM, Wachtmann TS, Umland JP, Pandher K, Lapointe JM, Saha S, Roach ML, Carter D, et al. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci U S A. 2003;100:9044–9049. doi: 10.1073/pnas.1332766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minhas PS, Latif-Hernandez A, McReynolds MR, Durairaj AS, Wang Q, Rubin A, Joshi AU, He JQ, Gauba E, Liu L, Wang C, et al. Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature. 2021;590:122–128. doi: 10.1038/s41586-020-03160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauvreau GM, Watson RM, O’Byrne PM. Protective effects of inhaled PGE2 on allergen-induced airway responses and airway inflammation. Am J Respir Crit Care Med. 1999;159:31–36. doi: 10.1164/ajrccm.159.1.9804030. [DOI] [PubMed] [Google Scholar]
- 15.Birrell MA, Maher SA, Dekkak B, Jones V, Wong S, Brook P, Belvisi MG. Anti-inflammatory effects of PGE2 in the lung: role of the EP4 receptor subtype. Thorax. 2015;70:740–747. doi: 10.1136/thoraxjnl-2014-206592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaslona Z, Flis E, Wilk MM, Carroll RG, Palsson-McDermott EM, Hughes MM, Diskin C, Banahan K, Ryan DG, Hooftman A, Misiak A, et al. Caspase-11 promotes allergic airway inflammation. Nat Commun. 2020;11:1055. doi: 10.1038/s41467-020-14945-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirai H, Tanaka K, Yoshie O, Ogawa K, Kenmotsu K, Takamori Y, Ichimasa M, Sugamura K, Nakamura M, Takano S, Nagata K. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J Exp Med. 2001;193:255–261. doi: 10.1084/jem.193.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujitani Y, Kanaoka Y, Aritake K, Uodome N, Okazaki-Hatake K, Urade Y. Pronounced eosinophilic lung inflammation and Th2 cytokine release in human lipocalin-type prostaglandin D synthase transgenic mice. J Immunol. 2002;168:443–449. doi: 10.4049/jimmunol.168.1.443. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura-Uchiyama C, Iikura M, Yamaguchi M, Nagase H, Ishii A, Matsushima K, Yamamoto K, Shichijo M, Bacon KB, Hirai K. Differential modulation of human basophil functions through prostaglandin D2 receptors DP and chemoattractant receptor-homologous molecule expressed on Th2 cells/DP2. Clin Exp Allergy. 2004;34:1283–1290. doi: 10.1111/j.1365-2222.2004.02027.x. [DOI] [PubMed] [Google Scholar]
- 20.Malmsten CL. Prostaglandins, thromboxanes, and leukotrienes in inflammation. Am J Med. 1986;80:11–17. doi: 10.1016/0002-9343(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 21.Hooftman A, O’Neill LAJ. The Immunomodulatory Potential of the Metabolite Itaconate. Trends Immunol. 2019;40:687–698. doi: 10.1016/j.it.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, Buttini M, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CG, Jenkins NA, Gilbert DJ, Copeland NG, O’Brien WE. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics. 1995;41:263–270. doi: 10.1007/BF00172150. [DOI] [PubMed] [Google Scholar]
- 24.Hooftman A, Angiari S, Hester S, Corcoran SE, Runtsch MC, Ling C, Ruzek MC, Slivka PF, McGettrick AF, Banahan K, Hughes MM, et al. The Immunomodulatory Metabolite Itaconate Modifies NLRP3 and Inhibits Inflammasome Activation. Cell Metab. 2020;32:468–478.:e467. doi: 10.1016/j.cmet.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diskin C, Ryan TAJ, O’Neill LAJ. Modification of Proteins by Metabolites in Immunity. Immunity. 2021;54:19–31. doi: 10.1016/j.immuni.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Liao ST, Han C, Xu DQ, Fu XW, Wang JS, Kong LY. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat Commun. 2019;10:5091. doi: 10.1038/s41467-019-13078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin W, Zhang Y, Tang H, Liu D, Chen Y, Liu Y, Wang C. Chemoproteomic Profiling of Itaconation by Bioorthogonal Probes in Inflammatory Macrophages. J Am Chem Soc. 2020 doi: 10.1021/jacs.9b11962. [DOI] [PubMed] [Google Scholar]
- 29.Qin W, Qin K, Zhang Y, Jia W, Chen Y, Cheng B, Peng L, Chen N, Liu Y, Zhou W, Wang YL, et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat Chem Biol. 2019;15:983–991. doi: 10.1038/s41589-019-0323-5. [DOI] [PubMed] [Google Scholar]
- 30.Bambouskova M, Potuckova L, Paulenda T, Kerndl M, Mogilenko DA, Lizotte K, Swain A, Hayes S, Sheldon RD, Kim H, Kapadnis U, et al. Itaconate confers tolerance to late NLRP3 inflammasome activation. Cell Rep. 2021;34:108756. doi: 10.1016/j.celrep.2021.108756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 32.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K, Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olagnier D, Brandtoft AM, Gunderstofte C, Villadsen NL, Krapp C, Thielke AL, Laustsen A, Peri S, Hansen AL, Bonefeld L, Thyrsted J, et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat Commun. 2018;9:3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olagnier D, Farahani E, Thyrsted J, Blay-Cadanet J, Herengt A, Idorn M, Hait A, Hernaez B, Knudsen A, Iversen MB, Schilling M, et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu G, Wu Y, Jin S, Sun J, Wan BB, Zhang J, Wang Y, Gao ZQ, Chen D, Li S, Pang Q, et al. Itaconate ameliorates methicillin-resistant Staphylococcus aureus-induced acute lung injury through the Nrf2/ARE pathway. Ann Transl Med. 2021;9:712. doi: 10.21037/atm-21-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H, Xu T, Feng X, Lai Y, Yang Y, Zheng H, He X, Wei G, Liao W, Liao Y, Zhong L, et al. Itaconate prevents abdominal aortic aneurysm formation through inhibiting inflammation via activation of Nrf2. EBioMedicine. 2020;57:102832. doi: 10.1016/j.ebiom.2020.102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi Z, Deng M, Scott MJ, Fu G, Loughran PA, Lei Z, Li S, Sun P, Yang C, Li W, Xu H, et al. Immune-Responsive Gene 1/Itaconate Activates Nuclear Factor Erythroid 2-Related Factor 2 in Hepatocytes to Protect Against Liver Ischemia-Reperfusion Injury. Hepatology. 2020;72:1394–1411. doi: 10.1002/hep.31147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linker RA, Haghikia A. Dimethyl fumarate in multiple sclerosis: latest developments, evidence and place in therapy. Ther Adv Chronic Dis. 2016;7:198–207. doi: 10.1177/2040622316653307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blatnik M, Frizzell N, Thorpe SR, Baynes JW. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by fumarate in diabetes: formation of S-(2-succinyl)cysteine, a novel chemical modification of protein and possible biomarker of mitochondrial stress. Diabetes. 2008;57:41–49. doi: 10.2337/db07-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, Snyder SH. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphries F, Shmuel-Galia L, Ketelut-Carneiro N, Li S, Wang B, Nemmara VV, Wilson R, Jiang Z, Khalighinejad F, Muneeruddin K, Shaffer SA, et al. Succination inactivates gasdermin D and blocks pyroptosis. Science. 2020;369:1633–1637. doi: 10.1126/science.abb9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linker RA, Lee DH, Ryan S, van Dam AM, Conrad R, Bista P, Zeng W, Hronowsky X, Buko A, Chollate S, Ellrichmann G, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jessen C, Kress JKC, Baluapuri A, Hufnagel A, Schmitz W, Kneitz S, Roth S, Marquardt A, Appenzeller S, Ade CP, Glutsch V, et al. The transcription factor NRF2 enhances melanoma malignancy by blocking differentiation and inducing COX2 expression. Oncogene. 2020;39:6841–6855. doi: 10.1038/s41388-020-01477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S, Valeaux S, et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med. 2016;213:1655–1662. doi: 10.1084/jem.20160061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Diepen JA, Robben JH, Hooiveld GJ, Carmone C, Alsady M, Boutens L, Bekkenkamp-Grovenstein M, Hijmans A, Engelke UFH, Wevers RA, Netea MG, et al. SUCNR1-mediated chemotaxis of macrophages aggravates obesity-induced inflammation and diabetes. Diabetologia. 2017;60:1304–1313. doi: 10.1007/s00125-017-4261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFalpha- and IFNgamma-triggered inflammation. Biochim Biophys Acta. 2014;1839:1217–1225. doi: 10.1016/j.bbagrm.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Infantino V, Iacobazzi V, Palmieri F, Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem Biophys Res Commun. 2013;440:105–111. doi: 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 51.Bambouskova M, Gorvel L, Lampropoulou V, Sergushichev A, Loginicheva E, Johnson K, Korenfeld D, Mathyer ME, Kim H, Huang LH, Duncan D, et al. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature. 2018;556:501–504. doi: 10.1038/s41586-018-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swain A, Bambouskova M, Kim H, Andhey PS, Duncan D, Auclair K, Chubukov V, Simons DM, Roddy TP, Stewart KM, Artyomov MN. Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages. Nature Metabolism. 2020 doi: 10.1038/s42255-020-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gopal S, Mikulskis A, Gold R, Fox RJ, Dawson KT, Amaravadi L. Evidence of activation of the Nrf2 pathway in multiple sclerosis patients treated with delayed-release dimethyl fumarate in the Phase 3 DEFINE and CONFIRM studies. Mult Scler. 2017;23:1875–1883. doi: 10.1177/1352458517690617. [DOI] [PubMed] [Google Scholar]
- 54.Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, Kelly V, Sekizawa K, Uchida K, Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaslona Z, Serezani CH, Okunishi K, Aronoff DM, Peters-Golden M. Prostaglandin E2 restrains macrophage maturation via E prostanoid receptor 2/protein kinase A signaling. Blood. 2012;119:2358–2367. doi: 10.1182/blood-2011-08-374207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaslona Z, Palsson-McDermott EM, Menon D, Haneklaus M, Flis E, Prendeville H, Corcoran SE, Peters-Golden M, O’Neill LAJ. The Induction of Pro-IL-1beta by Lipopolysaccharide Requires Endogenous Prostaglandin E2 Production. J Immunol. 2017;198:3558–3564. doi: 10.4049/jimmunol.1602072. [DOI] [PubMed] [Google Scholar]
- 57.Hinson RM, Williams JA, Shacter E. Elevated interleukin 6 is induced by prostaglandin E2 in a murine model of inflammation: possible role of cyclooxygenase-2. Proc Natl Acad Sci U S A. 1996;93:4885–4890. doi: 10.1073/pnas.93.10.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams JA, Shacter E. Regulation of macrophage cytokine production by prostaglandin E2. Distinct roles of cyclooxygenase-1 and -2. J Biol Chem. 1997;272:25693–25699. doi: 10.1074/jbc.272.41.25693. [DOI] [PubMed] [Google Scholar]
- 59.Cilenti F, Barbiera G, Caronni N, Iodice D, Montaldo E, Barresi S, Lusito E, Cuzzola V, Vittoria FM, Mezzanzanica L, Miotto P, et al. A PGE2-MEF2A axis enables context-dependent control of inflammatory gene expression. Immunity. 2021;54:1665–1682.:e1614. doi: 10.1016/j.immuni.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molina-Holgado E, Ortiz S, Molina-Holgado F, Guaza C. Induction of COX-2 and PGE(2) biosynthesis by IL-1beta is mediated by PKC and mitogen-activated protein kinases in murine astrocytes. Br J Pharmacol. 2000;131:152–159. doi: 10.1038/sj.bjp.0703557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chun KS, Cha HH, Shin JW, Na HK, Park KK, Chung WY, Surh YJ. Nitric oxide induces expression of cyclooxygenase-2 in mouse skin through activation of NF-kappaB. Carcinogenesis. 2004;25:445–454. doi: 10.1093/carcin/bgh021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.