Abstract
Background
Congenital Dyserythropoietic Anaemia type I (CDA-I) is a hereditary anaemia caused by biallelic mutations in the widely expressed genes CDAN1 and C15orf41. Little is understood about either protein and it is unclear in which cellular pathways they participate.
Methods
Genetic analysis of a cohort of CDA-I patients identifies novel pathogenic variants in both known causative genes. We analyse the mutation distribution and the predicted structural positioning of amino acids affected in Codanin-1, the protein encoded by CDAN1. Using western blotting, immunoprecipitation and immunofluorescence we determine the effect of particular mutations on both proteins and interrogate protein interaction, stability and sub-cellular localisation.
Results
We identify five novel CDAN1 mutations and one novel mutation in C15orf41 and uncover evidence of further genetic heterogeneity in CDA-I. Additionally, population genetics suggests CDA-I is more common than currently predicted. Mutations are enriched in six clusters in Codanin-1 and tend to affect buried residues. Many missense and in-frame mutations do not destabilise the entire protein. Rather C15orf41 relies on Codanin-1 for stability and both proteins, which are enriched in the nucleolus, interact to form an obligate complex in cells.
Conclusion
Stability and interaction data suggest C15orf41 may be the key determinant of CDA-I and offer insight into the mechanism underlying this disease. Both proteins share a common pathway likely to be present in a wide variety of cell types, however, nucleolar enrichment may provide a clue as to the erythroid specific nature of CDA-I. The surprisingly high predicted incidence of CDA-I suggests better ascertainment would lead to improved patient care.
Introduction
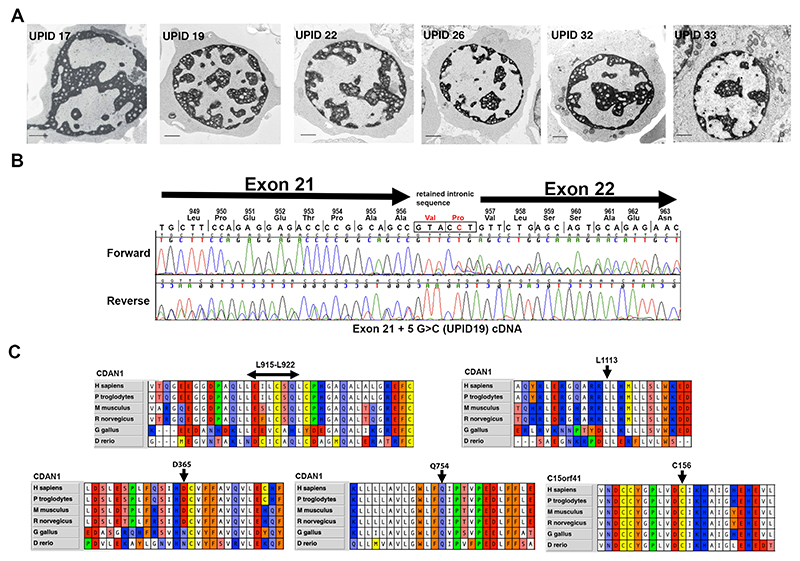
Congenital dyserythropoietic anaemia type I (CDA-I) (MIM 607465 and #224120) is an autosomal recessive macrocytic anaemia, characterised by ineffective erythropoiesis and morphological abnormalities of erythroblasts.[1] The anaemia is usually mild to moderate, however CDA-I is variable and some patients are dependent on blood transfusions. There are also limb abnormalities in a number of cases.[2] Advances in genetic diagnosis over the last ~20 years have led to the four major types of CDA (CDA-I to CDA-IV) being increasingly genetically defined and CDA-I has been shown to be caused by biallelic mutations in CDAN1 and C15orf41.[3,4] In the absence of proven pathogenic mutations in either of these genes, diagnosis of CDA-I relies on the presence of characteristic ultrastructural abnormalities of erythroblast nuclei detected by electron microscopy (EM). In affected cells heterochromatin is abnormally electron dense with electron lucent patches, this has been termed spongy or “Swiss Cheese” heterochromatin [5] and Figure 1.
Figure 1. Novel pathogenic mutations.
(A) Transmission Electron Microscopy showing the pathognomonic chromatin abnormalities in intermediate erythroblasts for patients as labelled. (B). Chromatograms showing aberrant splicing caused by the ivs21+5G>C variant in patient UPID19. (C) Alignments of Codanin-1 and C15orf41 protein sequences showing evolutionary conservation of amino acids in the regions flanking the missense changes as indicated.
The molecular basis of the heterochromatin defects remains unknown. However, the specificity of the abnormality strongly suggests that both proteins known to cause CDA-I function in a common pathway. Some progress has been made in understanding the function of both proteins, yet despite this their specific functions and the pathway(s) in which they operate remain poorly understood.[6] Codanin-1 is a relatively large protein (~132 kDa) and the absence of evolutionarily conserved domains, apart from a putative B-domain (see Figure 3), or any structural information has been a hurdle to understanding its function. It is known that Codanin-1 plays a role in nucleosome assembly through regulation of the histone chaperone ASF1, [7,8] is cell cycle regulated and may localise to heterochromatin. [9] C15orf41 is predicted to function as a nuclease [4] but the specific activity remains to be shown. Given that both known causative proteins are widely expressed, the erythroid specific nature of CDA-I is surprising and it has been hypothesised that this may arise from the disruption of the connection between cell-cycle dynamics and terminal erythroid maturation.[10]
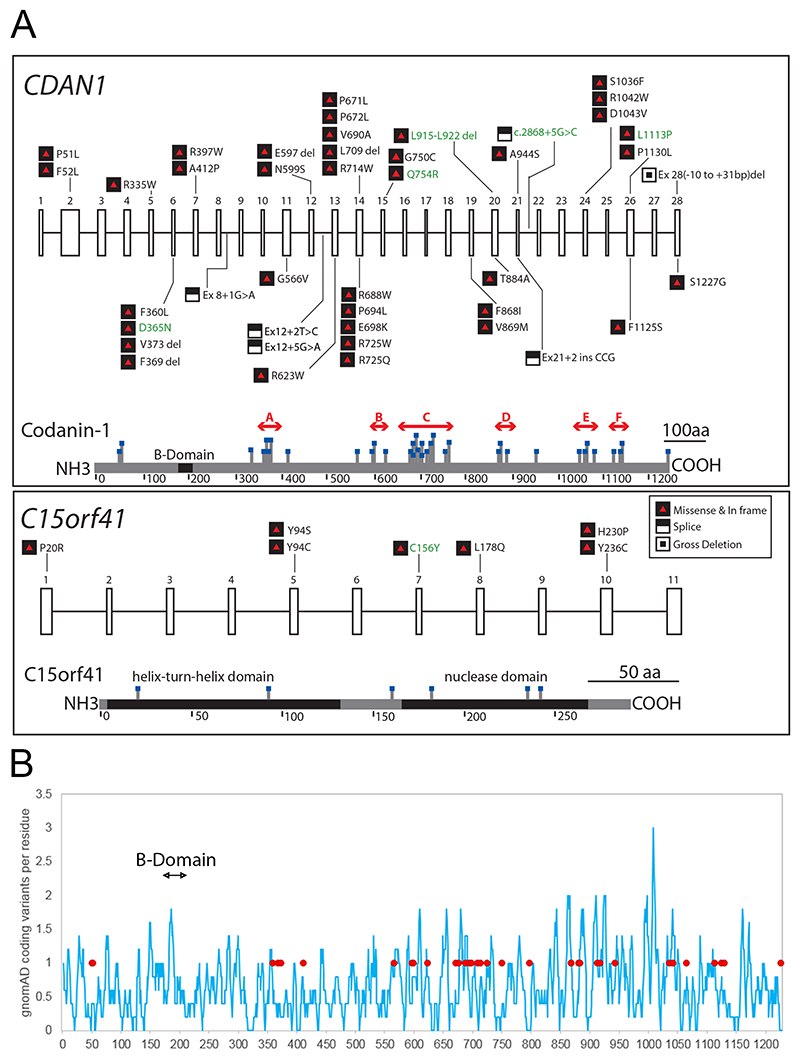
Figure 3.
A. Pathogenic variants in CDAN1 and C15orf41 causing CDA-I. Published variants are shown in black text and novel changes reported here are in green. Exons in each gene are numbered. Missense and in frame changes are depicted by black and red symbols and loss of function changes (premature stop and frameshift) by white and black symbols, changes are denoted by protein position. Splicing changes and deletions are shown according to the closest exon. Lower sections of each box show a representation of each protein with missense and in-frame deletions marked. For Codanin-1 we have identified mutation clusters A to F (red arrows), highlighting likely functional domains in the protein. B. The distribution of missense variants in the general population showing missense and in-frame indels per residue, smoothed over a 10 residue window across the length of the protein (blue line). Pathogenic mutations are shown by closed red circles and the location of the putative B-domain is indicated.
In this work we report six novel mutations causative of CDA-I, four of which are missense or in frame changes in Codanin-1, present in patients with EM-positive CDA-I. By combining these novel mutations with previously reported missense mutations and in frame deletions we find six regions of Codanin-1 are mutational hotspots for CDA-I. Analysis of patients with CDA-I caused by CDAN1 mutations suggests a number of the reported missense and in frame mutations are unlikely to completely abrogate protein function. Supporting this, we find protein levels to be unaffected in primary patient erythroblasts. Finally, we report that both Codanin-1 and C15orf41 are enriched in the nucleolus, that they form a stable complex where C15orf41 appears to be the critical partner.
Methods
Ethical Approval
This study was approved by the Wales Research Ethics Committee (REC5) (13/WA/0371) with written consent from patients and/or parents.
Patient Recruitment
Patients were recruited to the Unexplained Anaemia clinic, which is part of the Oxford Molecular Diagnostics Centre (https://www.oxford-translational-molecular-diagnostics.org.uk/content/unexplained-anaemia). Patients were assessed and analysed using the Oxford Red Cell Panel Targeted Resequencing strategy as previously described.[11]
Differentiation of CD34+ cells
CD34+ cells were isolated (as described in Supplementary Methods) and differentiated using a modified version of a published three-phase protocol [24] or using a two-phase liquid culture. [12,13]
ChlP-seq
ChIP-seq was carried out using ChIP Kit (17-295, Upstate Cell Signaling, now Millipore) following the manufacturer’s instructions. See Supplementary Methods for full method.
Next Generation Capture-C
3C Libraries were generated according to the published protocol.[14] See Supplementary Methods for a full description.
Library Preparation and Sequencing
Library preparation was performed with ILLUMINA’S TSCA v1.5 kit (FC-130-1001) using 250 ng genomic DNA, following manufacturer’s instructions. Samples were pooled (average 26 samples) and loaded at 20 pM on a MiSeq using a v3 600-cycle reagent kit sequencing 2x301 paired-end reads (Illumina).
Sanger Sequencing
Sequencing was performed on an ABI3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA) using 200− 500 ng DNA template, BigDye (Applied Biosystems) reaction mix, and 3.2 pmol of sequencing primer.
Optical Mapping Using Bionano Saphyr Technology
High molecular weight DNA from patient derived lymophoblastoid cell lines was isolated using the Bionano plug lysis protocol, using the Bionano Prep Blood and Cell Culture DNA isolation kit. Genomic DNA was barcoded using the Bionano Prep DLS labelling kit, in accordance with the manufacturer’s instructions. Labelled samples were analysed using the Saphyr device, optical maps were assembled and structural variants analysis was conducted using Bionano Access software.
RNA isolation and RT-PCR
2x106cells were pelleted and washed with PBS. The pellet was fixed in 500 μL TRIzol (Invitrogen 15596018) and snap frozen on dry ice before storage at -80°C. RNA was extracted using Direct-zol RNA MiniPrep kit (Zymo Research R2050) following manufacturers’ instructions (excepting for the DNase I treatment where the in-column DNA digestion was performed for 30min instead of 15). Total RNA was quantified using Qubit RNA BR Assay kit (Invitrogen Q10211). For RT-qPCR 1 μg of RNA was used to generate cDNA using High Capacity cDNA RT Kit (Applied Biosystems 4368814) according to manufacturers’ instructions.
Western Blots of Patient Cells and Immunofluorescence
2.5x106 cells were pelleted, washed with ice cold PBS supplemented with 10% v:v protease inhibitor cocktail (Sigma, P8340) and subjected to subcellular fractionation using a modified published method.[15] See Supplementary Methods for full western blot protocol and immunofluorescence methods.
Mutation Cluster Analysis
The locations of the 41 known missense changes and single amino acid in-frame deletions were mapped onto the Codanin-1 protein. Over the 1227 amino length each residue has an expected mutation probability of 0.0334. We compared this figure to the observed values using a Chi squared test over a 30 amino acid sliding window using a Bonferroni correction to account for the 1227 tests performed over the entirety of the protein.
Protein Structure Predictions
Relative solvent accessibility was predicted from the Codanin-1 amino acid sequence using four different methods: NetsurfP-2.0 [ref: https://www.ncbi.nlm.nih.gov/pubmed/30785653], I-TASSER [ref: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2849174/], SPIDER3 [ref: https://academic.oup.com/bioinformatics/article/33/18/2842/3738544], and SPIDER3-Single [ref: https://www.ncbi.nlm.nih.gov/pubmed/30368831]. We compared the relative solvent accessibility of all Codanin-1 residues associated with at least one pathogenic mutation to those other residues with at least one putatively benign mutation in gnomAD v2.1. For in-frame deletions bigger than one residue, the middle residue was used. Statistical significance was assessed using the Wilcoxon rank-sum test.
U-2-OS Cells
Conditions for culture, transfection and collection of material for western blotting and immunoprecipitation from U-2-OS cells are given in Supplementary Methods. The following primary antibodies were used: Codanin-1 (Abcam ab31236, ab28392), C15orf41 in Figure. 5 A and B (Abcam, ab107352), ASF1,[7] HA-tag (Roche 1867423).
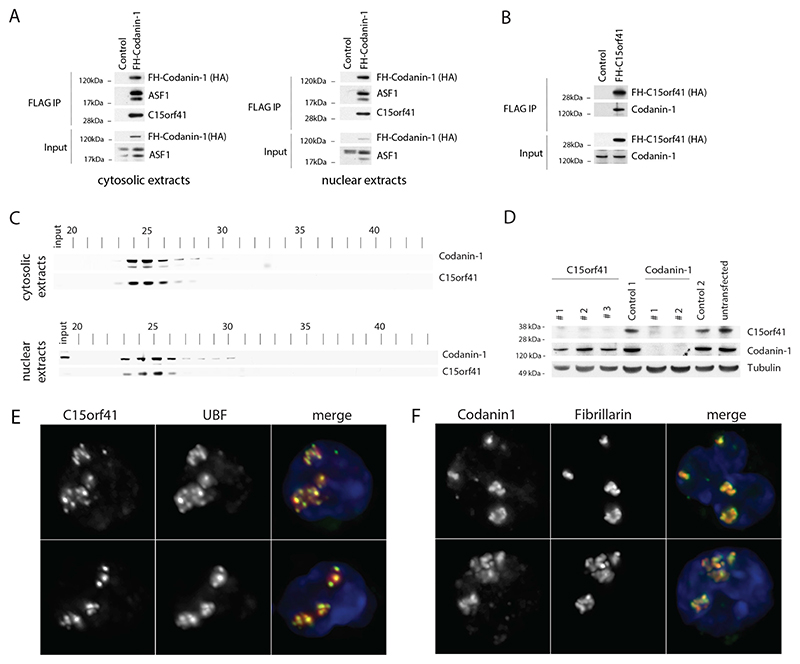
Figure 5. Codanin-1 and C15orf41 form a complex and are enriched in the nucleolus.
A. FLAG-HA-Codanin-1 (FH-Codanin-1) was immunoprecipitated from cytosolic and nuclear extracts and analyzed by western blot using HA, ASF1 and C15orf41 antibodies. B) FLAG-HA-C15orf41 (FH-C15orf41) was immunoprecipitated from whole cell lysates and analyzed by western blot using HA and Codanin-1 antibodies. C) Cytosolic and nuclear extracts from U-2-OS cells were subjected to size-exclusion chromatography and analyzed by western blot. D) U-2-OS cells transfected with the indicated siRNAs were analyzed by western blot (see also Supplementary Figure 2C). E&F) C15orf41 and Codanin-1 are enriched in the nucleolus in erythroblasts as shown by overlap with the nucleolar proteins UBF and Fibrillarin (2 representative examples of each are shown), nuclei are stained with DAPI (blue).
Gel Filtration
Size exclusion chromatography of cytosolic and nuclear extracts from U-2-OS cells was performed as previously described.[16,7] See Supplementary Methods for detailed description. The following primary antibodies were used: Codanin-1(Abcam ab31236 and Abcam ab28392) and C15orf41 in Fig. 5 C and D, Suppl. Fig. 2 C, mouse monoclonal antibody raised in house using 1-159aa of C15orf41.
Results
Novel Pathogenic Mutations
We screened a cohort of patients with unexplained anaemia, referred to the Oxford Molecular Diagnostics Centre, and identified four novel in-frame and missense changes and a loss of function (LOF) frameshift mutation in CDAN1 and one novel missense change in C15orf41 in nine patients (Figure 1 and Table 1). All novel changes are extremely rare or absent from the gnomAD database (allele frequencies listed in Table 1). Pathogenicity of novel missense and in frame changes was confirmed by finding chromatin abnormalities by EM in at least one family member (Figure 1A). Patient UPID23, who harbours an 8 amino acid deletion, is the exception to this as no EM is available, however UPID23 showed a clear response to treatment with interferon alpha, supporting the diagnosis of CDA-I (Table 1). All missense and in frame changes affect conserved residues, strongly suggesting they affect protein function (Figure 1C).
Table 1. Clinical Information.
| Patient ID | Sex | Ethnicity | Hb(g/L) | MCV(fL) | WBC (x109/L) | Platelet (x109/L) | Retic | Ferritin (μg/L) | EM | Gene | Alleles | Protein | Allele Freq | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPID17 | F | Caucasian | 105 | 118 | 9.3 | 596 | 4% | 328 | N | CDAN1 | c.2015C>T c.2681_2682de lAG |
p.P672L p.E894VfsX108 | 8.5x10-5 7.97x10-6 | Dgany, 2002 Novel |
| UPID19 | F | Caucasian | 120 | 96 | 2.8 | 289 | NA | 588 | Y | CDAN1 | c.2015C>T c.2868+5G>C |
p.P672L Ex21+5G>C |
8.5x10-5 4.00x10-6 | Dgany, 2002 Novel |
| UPID21 | M | Caucasian | 145 | 94 | 4.5 | 270 | NA | NA | N | CDAN1 | c.2015C>T c.3338T>C |
p.P672L p.L1113P |
8.5x10-5 1.06x10-5 | Dgany, 2002 Novel |
| UPID22 | F | Caucasian | 115 | 101.8 | 6.7 | 419 | 83 x109/L | 750 | Y | CDAN1 | c.2015C>T c.3338T>C |
p.P672L p.L1113P |
8.5x10-5 1.06x10-5 | Dgany, 2002 Novel |
| UPID23 | F | Caucasian | 138* | 104* | NA | NA | NA | NA | N | CDAN1 | c.2044C>T c.del2744_276 7 |
p.R682X p.L915_L922del |
NA 3.98x10-6 | Dgany, 2002 Novel |
| UPID26 | F | Caucasian | 121 | 103 | 8.3 | 185 | 14% | 645 | Y | C15orf41 | c.467G>A | p.C156Y | NA | Novel |
| UPID27 | F | Caucasian | 115 | 101 | 7.8 | 145 | 15% | 369 | Y | C15orf41 | c.467G>A | p.C156Y | NA | Novel |
| UPID32 ˆ | M | Romany | 90 | 87 | 4.3 | 244 | 64x109/L | NA | Y | NA | NA | NA | NA | NA |
| UPID33 ˆ | M | Caucasian /Indian | 87 | 75.2 | 5.99 | 314 | 2.5% | 2145 | Y | CDAN1 | c.1093G>A c.2261A>G |
p.D365N p.Q754R |
1.19x10-5 NA | Novel |
NA, Data not available
These values are post-interferon treatment, pre-interferon values were: Hb(g/L) 89 and MCV(fl) 103
Patient UPID 33 was 5 years of age at sampling and UPID 32 was 15 years of age at sampling.
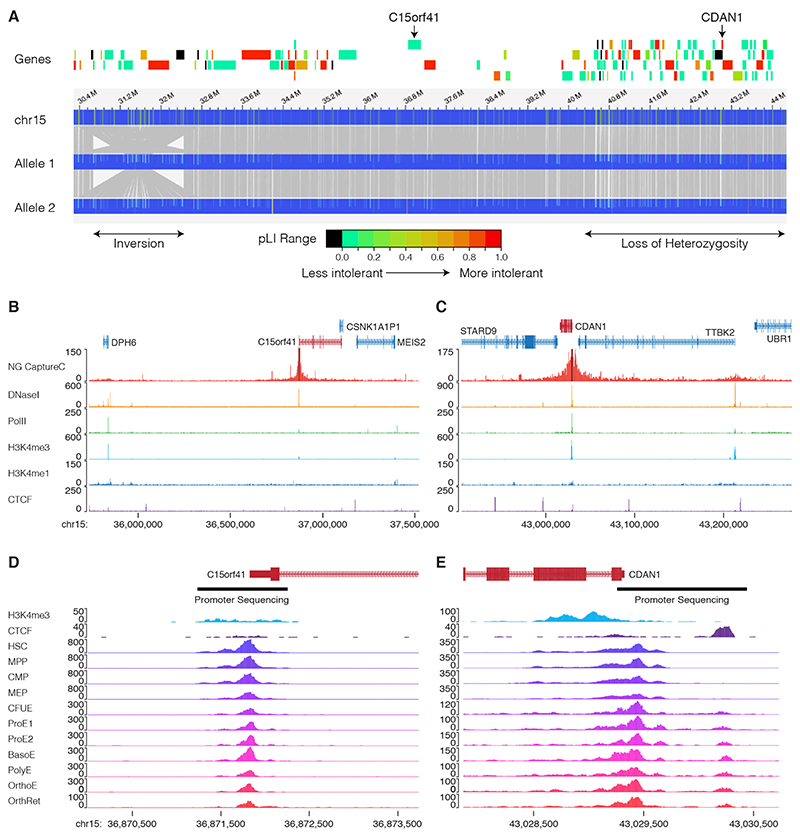
Patient UPID32 has EM positive CDA-I (Figure 1A) with splenomegaly (~16cm) but no distal limb abnormalities. There is no reported history of consanguinity and he was not responsive to two prolonged treatment periods with interferon alpha. No pathogenic mutations in the coding regions or intron/exon boundaries in CDAN1 nor C15orf41 could be identified using our targeted resequencing strategy [11] nor by whole exome sequencing (WES). Analysis of WES data revealed 705 regions of loss of heterozygosity (LOH) in this patient ranging from 1.7 kb to 10.7 Mb and encompassing a total of 740.8 Mb. One ~6 Mb LOH region (chr15:39,876,412-45,779,810 bp) includes CDAN1 (chr15:43,015,760-43,029,417). To assay for a chromosomal rearrangement in this region we analysed genomic DNA from patient UPID32 by optical mapping (Figure 2). This revealed there are no structural rearrangements within the ~6 Mb region of LOH, excluding this possibility as a cause of the patient’s CDA-I. Interestingly, there is a homozygous inversion present between ~30.4 and ~32.7 Mb (Figure 2A). This inversion has been reported as benign (nsv3955659 in [17]), suggesting it is a population specific structural variation.
Figure 2. Evidence for a third CDA-I locus.
A. Optical mapping of ~35 Mb on chromosome 15 for patient UPID32. The locations of the inversion and the region of loss of heterozygosity are shown (lower). Protein coding genes are shown and shaded according to their constraint metric (gnomAD) and C15orf41 and CDAN1 are labelled. Optical mapping shows there are no structural rearrangements over the CDAN1 or C15orf41 loci, however, there is a homozygous inversion between 30.4 Mb and 32.6 Mb. B,C. Capture-C showing chromatin interactions of the CDAN1 and C15orf41 loci. In each case the upper track shows Capture-C demonstrating chromatin interaction counts for each captured fragment (black bars). Lower tracks show normalised (RPKM) DNaseI-seq and ChIP seq for Polymerase II (Pol2), H3K4me4, H3K4me1 and CTCF. These data show the promoters of CDAN1 and C15orf41, despite being actively transcribed (as shown by Pol2 loading and DNaseI hypersensitivity), do not interact with any other DNaseI sensitive sites decorated with the H3K4me1 mark. This suggests that neither of these genes is controlled by distal cis-acting regulatory elements in cultured erythroblasts. D. Chromatin accessibility of the genomic landscape of CDAN1 and C15orf41 during erythroid differentiation (data from references 18 & 19) shows the extent of the gene promoters. The regions of the promoters subjected to Sanger sequencing is shown by black bars Abbreviations: HSC, Haematopoietic Stem Cell; MPP, Multipotent progenitor; CMP, common myeloid progenitor; MEP, Megakaryocyte erythroid precursor; CFUE, colony forming unit erythroid; ProE1, Proerythroblast stage 1; ProE2, Pro-erythroblast stage 2; BasoE, Basophilic erythroblast; PolyE, polychromatic erythroblast; OrthoE, orthochromatic erythroblast; OrthRet, Orthochromatic/Reticulocyte.
To determine whether the CDA-I in this patient resulted from a cis-acting mutation affecting the expression of CDAN1 or C15orf41, we searched for any cis-acting regulatory regions using a combination of erythropoietic ATAC-seq [18,19] and chromosome conformation capture (3C). We determined chromatin interactions for the CDAN1 and C15orf41 promoters using NG Capture-C [14] in cultured human erythroblasts. Neither promoter forms chromatin loops with distal open chromatin elements, suggesting both genes are entirely promoter driven in intermediate erythroblasts (Figure 2B), excluding the possibility of a long-range distal regulatory variant causing CDA-I in UPID32. We also captured from the alpha-globin gene promoters in the same experiment, as a positive control, and identified the known enhancer interactions (Supplementary Figure 3). Analysis of open chromatin showed both gene promoters are accessible from early haematopoiesis to terminal erythroid maturation (Figure 2). Sanger sequencing of both of these accessible regions found no likely pathogenic changes.
Given the clear phenotype of CDA-I in UPID32 and the absence of coding variants, cís-acting distal regulatory sites and structural variants disrupting CDAN1 and C15orf41, it is likely that there is at least one further locus underlying CDA-I, identification of which would be likely to offer insight into the pathogenic mechanism.
CDA-I Prevalence
We combined the allele frequencies of all known pathogenic missense and in frame mutations in CDAN1 and C15orf41 together with all loss of function mutations in ~140,000 individuals in the gnomAD database (https://gnomad.broadinstitute.org/). This shows the incidence of CDA-I caused by compound heterozygosity for two mutations in either gene is 4.83794 x 10-6, or ~4.84 cases per million live births, which is a five fold increase on the currently accepted incidence. Mutations used are given in Supplementary Tables 4 and 5.
This calculation is based on the likelihood of occurrence of compound heterozygosity for two missense mutations, or a missense mutation with a loss of function mutation, and assumes compound heterozygosity for two LOF mutations for either gene would be incompatible with life (see below). These calculations use a neutral model, however, the actual incidence of CDA-I is likely to be higher than this owing to population specific variation in allele frequencies.
Mutation Clustering Indicates Functional Domains
CDA-I mutations are present throughout CDAN1 and C15orf41. However, the positioning of mutations likely to lead to loss of function (LOF), such as truncations and frameshift mutations is less important for determining protein function as all LOF changes result in a lack of protein resulting from nonsense mediated decay. CDA-I mutations are present throughout Codanin-1 and C15orf41, so by focusing on the location of missense and in frame INDEL mutations rather than truncation mutations, important sub-domains within proteins can be identified. Even with the report of one novel C15orf41 missense mutation here, there are six total C15orf41 missense mutations, which is too few for a domain enrichment analysis. Codanin-1 however, has 37 missense and in frame INDEL mutations. This approach shows six regions are significantly enriched for pathogenic mutations (p<0.001) (Figure 3A) and these are therefore more likely to play a role in affecting specific protein functions and hence in the pathogenesis of CDA-I. These six areas of Codanin-1 are 340-398aa, 594-626aa, 648-754aa, 852-910aa, 1014-1071aa and 1101-1142aa (regions A-F in Figure 3A). The distribution of missense changes in the general population across the length of the Codanin-1 protein shows a uniform degree of variation (Figure 3B), suggesting there are no specific regions intolerant to missense variation. Therefore, we hypothesised the six mutational clusters represent functional domains.
Analysis of Codanin-1 Missense Mutations
No three-dimensional structure is available for Codanin-1, nor are there related structures that can be used for homology modelling. However, we can still use predicted structural properties to investigate the possible molecular mechanism underlying the pathogenic mutations. In particular, sequence-based predictors of solvent accessibility, a measure of residue burial, have been used successfully to differentiate mutations by phenotype in the absence of a structure.[20] It is well known that, in general, pathogenic mutations are enriched at buried positions within proteins, as mutations in these regions are much more likely to cause a loss of function by disrupting protein folding and stability.[21] Therefore, mutations at residues with lower predicted relative solvent accessibility values should be more likely to destabilise the protein and cause a loss of function.
Interestingly, no homozygotes or compound heterozygotes for CDAN1 LOF mutations have been identified, [22] suggesting Codanin-1 may have a unique function and may be essential during development. This view is supported by evidence from Cdan1 null mice, which show embryonic lethality prior to embryonic day 6.5.[23] Because of these observations, we can deduce that missense mutations found in the homozygous state or in compound heterozygosity with a loss of function mutation are unlikely to cause a complete LOF. Following a search of the literature and review of our own data we identified 13 missense and in frame mutations present in patients in the homozygous state or in combination with a LOF allele. We can assume these 13 changes are not causing complete loss of function and so we termed them “non-LOF”. The remaining 24 in frame and missense CDAN1 mutations have an unknown effect on the protein and here are termed “mutations of unknown effect”. The status of the latter group could be determined by functional testing or identification in homozygosity or in heterozygosity with a LOF allele. Mutations in each category are listed in Supplementary Information. Interestingly, we find a highly significant tendency for the non-LOF mutations to be more solvent accessible (i.e. less buried) than the mutations of unknown effect, suggesting at least some of the latter group are likely to cause complete destabilisation of the protein. Mutations in both groups tend to be more buried than the sites of putatively benign variants observed in gnomAD. (Figure 4A and Supplementary Figure 1). Whilst the non-LOF mutations tend to be slightly more buried than the gnomAD variants, there is no statistically significant difference between the groups. This suggests the non-LOF mutations are either causing weaker destabilisation (i.e. they’re hypomorphic) or their damaging effect is due to some other molecular mechanism.
Figure 4. Effect of missense mutations on Codanin-1.
A. Pathogenic Codanin-1 mutations observed Homozygously, or compound heterozygously with a loss of function mutation (‘not LOF’), are predicted to be significantly more accessible to solvent (i.e. less buried) by the program NetSurfP-2.0 than other pathogenic mutations (‘possibly LOF’), but are not significantly different from putatively benign variants observed in the human population (gnomAD). B. Representative image of quantitative western blots using ex vivo cultured patient erythroblasts detecting Codanin-1 (upper) and C15orf41 (lower). C. Quantification of western blots shown in C showing normalized values of Codanin-1 and C15orf41 proteins compared with erythroblasts cultured from three normal individuals. Bars show the mean +/- SD. D. Box and whiskers plot showing median, quartiles and outliers of mRNA levels of CDAN1 and C15orf41 in CDA-I patients plotted relative to the levels seen in healthy controls (patient samples colour coded as in C) E. Erythroid cell counts showing that altered maturation rates do not explain the lack of protein degradation seen in this experiment. ns, not significant. * p=<0.05, **p=<0.005, significance determined by one-way ANOVA with Sidak’s multiple comparison test.
Codanin-1 and C15orf41 Expression and Stability
We tested the effects of pathogenic mutations on Codanin-1 and C15orf41 by two colour near-infrared quantitative western blot using protein extracted from erythroblasts cultured from the peripheral blood of normal individuals and CDAN1 and C15orf41 CDA-I patients using a well characterised in vitro culture system [24] (Figure 4B). To detect Codanin-1 we validated a polyclonal antibody from Bethyl Laboratories (catalogue number A304-951A) by showing it cross-reacts with Codanin-1 conjugated to mCherry when over expressed in HEK293Tcells (Supplementary Figure 2A). To detect C15orf41 we validated a polyclonal antibody supplied by Cusabio (catalogue number CSBPA897474LA01HU) by expressing a FLAG-tagged version of C15orf41 in HEK293T cells and showing colocalization of both C15orf41 antibodies with the FLAG antibody (Sigma F1804) by western blotting using the immunoprecipitated cell lysate (Supplementary Figure 2B).
Quantification of Codanin-1 and C15orf41 in three healthy control samples (NCO27, NCO28 and NCO29) showed protein levels decrease during terminal erythroid differentiation (Figure 4C). Interestingly, the Codanin-1 and C15orf41 protein levels in erythroblasts cultured from two CDA-I patients with CDAN1 mutations (UPID 15: p.F369del;D1043V and UPID 20 p.P672L; V993GfsTer13) and two with C15orf41 mutations (UPID25: p.Y94C;Y94C and UPID26: p.C156Y;C156Y) showed no reduction in either protein compared to the normal control samples. The D1043V and P672L mutations are predicted to be non-LOF and F369del is categorised as a mutation of unknown effect. We also measured CDAN1 and C15orf41 mRNA levels, which were comparable to those of healthy controls (Figure 4D), showing increased transcription does not underlie the constant protein levels in these cases. To ensure patient and control samples were stage matched, erythroid cell counts at sequential stages of differentiation in culture showed similar numbers of morphologically identified erythroid cells were present at day 10 (intermediate erythroblasts) and day 13 (late stage erythroblasts) in cells from patients and healthy donors (Figure 4E). Taken together these data show there is no overall destabilisation of Codanin-1 nor C15orf41 by the mutations we tested, confirming the prediction that D1043V and P672L changes do not destabilise the entire protein.
Protein Interaction and Nucleolar Enrichment
Because mutations in CDAN1 and C15orf41 both lead to the specific chromatin abnormalities seen in CDA-I, we hypothesised both proteins function in the same pathway. To test whether the proteins interact, we performed co-immunoprecipitation experiments. Using cell lines expressing FLAG-HA tagged Codanin-1, we found that the known interactor, ASF1, as well as C15orf41 co-purified with FLAG-HA-Codanin-1 (Figure 5A). Codanin-1 also co-purified with FLAG-HA tagged C15orf41, confirming the interaction (Figure 5B). Gel-filtration analysis revealed that the two proteins co-elute in both nuclear and cytosolicextracts, implying the majority of Codanin-1 and C15orf41 are found in a complex together. This suggested that the two proteins might form an obligate complex and we therefore addressed whether their stability might be interdependent. Whereas depletion of C15orf41 did not affect the level of Codanin-1, removal of Codanin-1 by several independent and validated siRNAs [7] resulted in a concomitant reduction in C15orf41 levels. Immunofluorescence of the two proteins in erythroblasts shows they are pan-cellular but both are enriched in the nucleolus (Figure 5 E,F).
Discussion
In this work we report five mutations in CDAN1 and one in C15orf41 causative of CDA-I, thus increasing the diagnostic range of this disease. In cases with novel missense mutations we show that patients have the pathognomonic chromatin abnormalities indicative of CDA-I, critical for demonstrating pathogenicity of novel missense and in-frame mutations. This increases the number of patients who can be diagnosed without recourse to EM examination of bone marrow biopsy, which is an invasive procedure. This is particularly helpful as EM is unavailable at the majority of referral centres.
Although causative mutations are spread throughout CDAN1, when only the locations of pathogenic missense and in-frame mutations in Codanin-1 protein are considered, it becomes clear these mutations occur in 6 distinct domains. Because complete loss of Codanin-1 is incompatible with life, [23] any missense mutation that occurs homozygously, or in tandem with a loss of function mutation, is unlikely to cause a complete LOF. When all reported patients are considered, 13 missense mutations are likely to impair protein function rather than obliterate it. Our analysis of solvent accessibility shows that these 13 mutations tend to be less buried than mutations of unknown effect. This suggests that the 13 non-LOF mutations are less likely to destabilise the entire protein and we show this is the case in erythroblasts from two CDAN1 CDA-I patients. It is interesting that knockdown of Codanin-1 destabilises C15orf41, yet in patient cells we find protein levels remain unchanged for the mutations tested. This further suggests that a specific interaction is affected in CDA-I patients. This hypothesis is supported by our previous data showing that the R714W missense mutation, present in the largest mutation cluster in Codanin-1 (cluster C), disrupts the interaction with ASF1.[7] It is unclear which other protein interactions are affected by mutations in the six domains, however, identification of further interacting proteins would allow this to be tested. Given the enrichment of mutation clusters in the C-terminal region of the protein, it may be this region that interacts with C15orf41, although this will need to be directly tested.
We find that the two mutations in C15orf41 we tested do not destabilise this protein. Since both mutations we tested were present in the homozygous state, this provides evidence that C15orf41 is an essential protein.
Codanin-1 is cell cycle regulated [9] and it is possible that some of the mutations affect post translational modifications that regulate Codanin-1 levels. Analysis of synchronised cell populations may be able to address this question. Expression of CDAN1 and C15orf41 has been shown to be tightly correlated. [25] In our study we observe a similar trend as both genes are down regulated in tandem during terminal erythroid differentiation. Russo and colleagues [25] also report no change in localisation of mutant proteins, further suggesting that mutations affect specific interactions.
We report enrichment of both proteins in the nucleolus. This observation has the potential to provide insight into the erythroid specific nature of CDA-I as mutations affecting ribosome biogenesis are known to give rise to tissue specific abnormalities, including severely impaired erythropoiesis in diseases such as Diamond Blackfan Anaemia and Shwachman-Diamond Syndrome [26]. Further studies will be required to establish whether ribosomes are affected in CDA-I. Insight into the biochemical activity of C15orf41 would greatly help with understanding the role of these proteins in the nucleolus and CDA-I disease pathology.
Identification of a third locus for CDA-I would also offer insight into disease pathology and broaden the diagnostic range. Here we show that, unless there are distal cis-acting elements at earlier stages of erythroid maturation, it is extremely unlikely that mutations in distal regulatory elements or structural rearrangements cause CDA-I in the patient. This observation argues for the presence of at least one further locus underlying this disease. We hope excluding the possibility of distal regulatory elements and defining the extent of promoter sequences for both causative genes is of use to other researchers.
As with many rare disorders, establishing an accurate estimate of incidence and prevalence of CDA-I is difficult. Over 300 cases have been reported. [10] Most are sporadic cases from diverse regions such as Western Europe, North Africa and Asia, [27] while some series are accounted for by afounder effect, particularly in the Middle East.[28] Currently, the reported incidence of CDA-I suggests a frequency of ~1:1,000,000 live births [6] although estimates as low as 0.24 cases per million live births have been made based on European epidemiological data. [22] However, it has been suggested that CDA-I may be more common than previously estimated. [29] In support of this, analysis of allele frequencies in this work suggests that CDA-I may be present at a frequency of 1 per ~207,000 live births suggesting the majority CDA-I remains undiagnosed. Because treatment with interferon alpha is effective in many cases of CDA-I, improving ascertainment will be important in best treating this disease.
Here we have expanded the diagnostic range for CDA-I and reassessed its prevelance. We prove the hypothesis that the two CDA-I causative proteins, Codanin-1 and C15orf41, are involved in a common pathway by showing that they interact and that Codanin-1 stabilises C15orf41. We postulate that mutation hotspots we have identified across Codanin-1 represent functional domains, some of which mediate Codanin-1’s interaction with C15orf41. Further, our finding of enrichment of these proteins in nucleoli is an important step towards elucidating the erythroid-specific nature of the disease. Taken together these results will improve CDA-I patient ascertainment and allow future focus to further understand the mechanisms behind this disease as well as the normal process of erythropoiesis
Supplementary Material
Acknowledgements
This work was supported by MRC (MC_uu_12009), the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Haematology Theme at Oxford University Hospitals NHS Trust and University of Oxford and the charities Blood Buddies, the Congenital Anaemia Network and Action Medical Research for Children (GN2300). D.J.D. and J.H. were funded by a Wellcome Trust Strategic Award (106130/Z/14/Z). EM work was undertaken at the Dunn School EM Facility. J.M. is supported by an MRC Career Development Award (MR/M02122X/1) and is a Lister Institute Research Prize Fellow.
Footnotes
Authorship Statement
CB, VJB, DRH, NR conceived the study and wrote the manuscript. A-AO cultured erythroblasts and performed quantitative western blots. CS cultured erythroblasts. JAM performed analysis of missense and in frame mutations. KL and KA performed immunoprecipitation and western blots. NR performed capture-C. DJD analysed capture-C, ChIP and ATAC data. JB cultured patient cells and commented on the manuscript. SB performed antibody validation. EJ performed EM studies. BX performed optical mapping. MP and RK analysed patient DNA and identified pathogenic mutations. KR, PF, JM, ES, NS, LM, QAH, NR and RR ascertained patients, provided samples and analysed clinical data. JRH, RG, AG and PJMcH provided supervision, analysed data and commented on the manuscript.
Competing Interests
J.R.H is a founder and shareholder of Nucleome Therapeutics.
Data accessibility
Erythroid sequencing data generated for this work are deposited with the GEO archive (GSE125753). Previously published open-chromatin data sets (GSE86393, GSE75384, GSE115684) were used. [(18, 19, 29]
References
- 1.Heimpel H, Wendt F. Congenital dyserythropoietic anemia with karyorrhexis and multinuclearity of erythroblasts. Helv Med Acta. 1968;34(2):103–15. [PubMed] [Google Scholar]
- 2.Wickramasinghe SN, Wood WG. Advances in the understanding of the congenital dyserythropoietic anaemias. Br J Haematol. 2005;131(4):431–46. doi: 10.1111/j.1365-2141.2005.05757.x. [DOI] [PubMed] [Google Scholar]
- 3.Dgany O, Avidan N, Delaunay J, et al. Congenital dyserythropoietic anemia type I is caused by mutations in codanin-1. Am J Hum Genet. 2002;71(6):1467–74. doi: 10.1086/344781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babbs C, Roberts NA, Sanchez-Pulido L, et al. Homozygous mutations in a predicted endonuclease are a novel cause of congenital dyserythropoietic anemia type I. Haematologica. 2013;98(9):1383–87. doi: 10.3324/haematol.2013.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimpel H, Forteza-Vila J, Queisser W, et al. Electron and light microscopic study of the erythroblasts of patients with congenital dyserythropoietic anemia. Blood. 1971;37(3):299–310. [PubMed] [Google Scholar]
- 6.Roy NBA, Babbs C. The pathogenesis, diagnosis and management of congenital dyserythropoietic anaemia type I. Br J Haematol. 2019;185(3):436–49. doi: 10.1111/bjh.15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ask K, Jasencakova Z, Menard P, et al. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBOJ. 2012;31(8):2013–23. doi: 10.1038/emboj.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamary H, Marcoux N, Noy-Lotan S, et al. Codanin-1, the Product of the Gene Mutated In Congenital Dyserythropoietic Anemia Type I (CDA I), Binds to Histone Chaperone Asf1a and Inhibits Its Nucleosome Assembly Activity. Blood. 2010;116(21):442–42. [Google Scholar]
- 9.Noy-Lotan S, Dgany O, Lahmi R, et al. Codanin-1, the protein encoded by the gene mutated in congenital dyserythropoietic anemia type I (CDAN1), is cell cycle-regulated. Haematologica. 2009;94(5):629–37. doi: 10.3324/haematol.2008.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iolascon A, Heimpel H, Wahlin A, et al. Congenital dyserythropoietic anemias: molecular insights and diagnostic approach. Blood. 2013;122(13):2162–6. doi: 10.1182/blood-2013-05-468223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy NBA, Wilson EA, Henderson S, et al. A novel 33-Gene targeted resequencing panel provides accurate, clinical-grade diagnosis and improves patient management for rare inherited anaemias. Brit J Haematol. 2016;175(2):318–30. doi: 10.1111/bjh.14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope SH, Fibach E, Sun J, et al. Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur J Haematol. 2000;64(5):292–303. doi: 10.1034/j.1600-0609.2000.90032.x. [DOI] [PubMed] [Google Scholar]
- 13.Fibach E, Manor D, Oppenheim A, et al. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73(1):100–3. [PubMed] [Google Scholar]
- 14.Davies JOJ, Telenius JM, McGowan SJ, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods. 2016;13(1):74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baghirova S, Hughes BG, Hendzel MJ, et al. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2015;2:440–5. doi: 10.1016/j.mex.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth A, Ray-Gallet D, Quivy JP, et al. Human Asf1 regulates the flow of S phase histories during replicational stress. Molecular Cell. 2005;17(2):301–11. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Levy-Sakin M, Pastor S, Mostovoy Y, et al. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10(1):1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig LS, Lareau CA, Bao EL, et al. Transcriptional States and Chromatin Accessibility Underlying Human Erythropoiesis. Cell Rep. 2019;27(11):3228–40. doi: 10.1016/j.celrep.2019.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces MR, Buenrostro JD, Wu B, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48(10):1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw ND, Brand H, Kupchinsky ZA, et al. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat Genet. 2017;49(2):238–48. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue P, Li ZL, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–73. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Iolascon A, Esposito MR, Russo R. Clinical aspects and pathogenesis of congenital dyserythropoietic anemias: from morphology to molecular approach. Haematologica. 2012;97(12):1786–94. doi: 10.3324/haematol.2012.072207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renella R, Roberts NA, Brown JM, et al. Codanin-1 mutations in congenital dyserythropoietic anemia type 1 affect HP1a localization in erythroblasts. Blood. 2011;117(25):6928–38. doi: 10.1182/blood-2010-09-308478. [DOI] [PubMed] [Google Scholar]
- 24.Scott C, Downes DJ, Brown JM, et al. Modelling erythropoiesis in congenital dyserythropoietic anaemia type I (CDA-I) bioRxiv. 2019 doi: 10.1101/744367. published. [DOI] [Google Scholar]
- 25.Russo R, Marra R, Andolfo I, et al. Characterization of Two Cases of Congenital Dyserythropoietic Anemia Type I Shed Light on the Uncharacterized C15orf41 Protein. Front Physiol. 2019;10:621. doi: 10.3389/fphys.2019.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iolascon A, Delaunay J. Close to unraveling the secrets of congenital dyserythropoietic anemia types I and II. Haematologica. 2009;94(5):599–602. doi: 10.3324/haematol.2009.005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamary H, Dgany O, Proust A, et al. Clinical and molecular variability in congenital dyserythropoietic anaemia type I. Br J Haematol. 2005;130(4):628–34. doi: 10.1111/j.1365-2141.2005.05642.x. [DOI] [PubMed] [Google Scholar]
- 29.Renella R, Wood WG. The congenital dyserythropoietic anemias. Hematol Oncol Clin North Am. 2009;23(2):283–306. doi: 10.1016/j.hoc.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Schwessinger R, Suciu MC, McGowan SJ, et al. Sasquatch: predicting the impact of regulatory SNPs on transcription factor binding from cell- and tissue-specific DNase footprints. Genome Res. 2017;27(10):1730–42. doi: 10.1101/gr.220202.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Erythroid sequencing data generated for this work are deposited with the GEO archive (GSE125753). Previously published open-chromatin data sets (GSE86393, GSE75384, GSE115684) were used. [(18, 19, 29]