Abstract
Transplantation of B cells engineered ex vivo to secrete broadly neutralizing antibodies (bNAbs) has shown efficacy in disease models. However, clinical translation of this approach would require specialized medical centers, technically demanding protocols and MHC compatibility of donor cells and recipients. Here, we report in vivo B cell engineering using two adeno-associated viral vectors, with one coding for saCas9 and the other for 3BNC117, an anti-HIV bNAb. After intravenously injecting the vectors into mice, we observe successful editing of B cells leading to memory retention and bNAb secretion at neutralizing titers of up to 6.8 μg/mL. We observed minimal CRISPR-Cas9 off-target cleavage as detected by unbiased CHANGE-Seq analysis, whereas on-target cleavage in undesired tissues is reduced by expressing saCas9 from a B cell-specific promoter. In vivo B cell engineering to express therapeutic antibodies is a safe, potent and scalable method, which may be applicable not only to infectious diseases but also in the treatment of non-communicable conditions, such as cancer and autoimmune disease.
INTRODUCTION:
Broadly neutralizing antibodies (bNAbs) against HIV can suppress viremia. In particular, combination therapy with the bNAbs 3BNC117 and 10-1074 allowed long-term suppression upon interruption of antiretroviral therapy (ART) in individuals with antibody-sensitive viral reservoirs1. Similarly, viremic individuals with dual antibody-sensitive viruses experienced diminished viremia for three months following the first of up to three dual-bNAb infusions2. However, the mean elimination half-life of the bNAbs is 16 and 23 days, respectively3, allowing the virus to rebound. Moreover, individuals with prior resistance to one of the bNAbs have mounted resistance to the second antibody, and individuals with prior resistance to both antibodies were excluded from the trials. Limited bNAb persistence may be addressed by constitutive expression from muscle following viral vector transduction4,5. However, anti-drug antibodies (ADA) may develop6, possibly because of improper glycosylation. Moreover, antibodies expressed from muscle do not undergo class switch recombination (CSR) or affinity maturation, which may be required for long-term suppression of a diverse and continuously evolving HIV infection. In order to overcome these challenges, we7,8 and others9-13 have developed B cell engineering for antibody expression. In particular, we previously combined Toll-like receptor (TLR)-mediated ex vivo activation of B cells with in vivo prime-boost immunizations, and demonstrated that engineered B cells allow immunological memory, CSR, somatic hypermutation (SHM) and clonal selection. However, cost and complexity of autologous B cell engineering ex vivo may be prohibitive. At the same time, use of engineered allogeneic B cells is challenging due to the requirement for HLA matching for receiving T cell help and avoiding graft rejection.
These challenges may be addressed using in vivo engineering. In vivo T cell engineering was previously demonstrated, using promiscuously integrating vectors14-21, episomal adeno associated viral (AAV) vectors21-24 or mRNA25,26. However, in B cells, only the specific targeting of the IgH locus, utilizing the endogenous constant exons with appropriate splicing signals, is expected to allow a well regulated expression of the antibody, first as a membrane bound B cell receptor (BCR) and then, upon antigen induced activation, also as a soluble protein, released by progeny plasmablasts and plasma cells11-13. IgH targeting is similarly required for memory retention, CSR, SHM and clonal selection7,8. Therefore, we describe here an in vivo B cell engineering protocol based on a single systemic injection of AAV vectors coding for CRISPR-Cas9 and for the desired bNAb cassette, which is targeted for integration into the IgH locus.
RESULTS:
Engineering strategy.
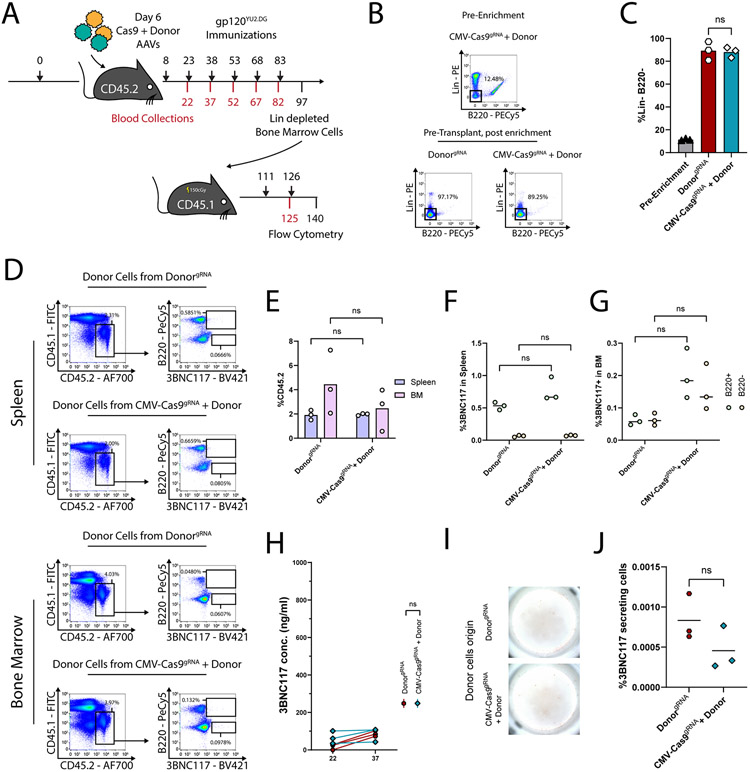
In order to promote in vivo B cell engineering, we used a pair of AAV-DJ vectors27, one coding for saCas928 and the other coding for the 3BNC117 anti-HIV bNAb29 (Fig. 1). In the first set of experiments, the saCas9 is expressed from the ubiquitously active CMV promoter, and the sgRNA, targeting saCas9 to the IgH locus, is coded on the same AAV. The bNAb, in turn, is coded as a bi-cistronic cassette under the control of an IgH-enhancer-dependent promoter and flanked by homology arms to the desired saCas9 cut-site within the J-C intron of the IgH locus7. The bNAb cassette includes the full light chain and the variable segment of the heavy chain (VH), separated by a sequence coding for a Furin cleavage site and for a 2A-peptide. A splice donor sequence follows the VH gene segment in order to allow its fusion to constant IgH exons, upon integration into the locus and subsequent transcription and splicing. Our design facilitates disruption of the endogenous IgH locus and initial bNAb expression as a membranal BCR. This allows for subsequent activation of the engineered B cells upon antigen binding, which leads to differentiation into memory and plasma cells.
Fig. 1:
Targeting an antibody to the IgH locus of B cells in order to facilitate antigen-induced activation, SHM, CSR and affinity maturation. A. Design of the two AAV vectors. One vector codes for saCas9 and an sgRNA under CMV and U6 promoters, respectively. The second vector codes for the 3BNC117 bNAb cassette flanked by homology arms for integration into the CRISPR-Cas9 cut site at the J-C intron of the IgH locus. The bNAb cassette is expressed upon integration under the control of an enhancer dependent (ED) promoter. The cassette includes the light chain in full and the variable segment of the heavy chain, separated by a sequence coding for a furin cleavage site and a 2A peptide. The variable heavy chain is followed by a splice donor sequence to allow fusion with the endogenous constant exons upon integration, transcription and splicing. An upstream polyadenylation site is provided to terminate the transcription of the endogenous variable heavy chain upon integration. B. Depiction of the IgH locus upon integration. The bNAb cassette is integrated downstream of the last J segment (J4) and upstream of the intronic enhancer (iEμ), class switch recombination locus (CSR) and the IgH Cμ exons. C. The bNAb mRNA is terminated by alternative polyadenylation sites allowing for membranal (BCR) or soluble expression, before and after differentiation into a plasma cell, respectively. D. Different isotypes of the integrated antibody may be expressed upon CSR of engineered B cells. E. SHM in the antibody coding genes may allow for affinity maturation and clonal expansion.
In vivo B cell engineering allows for high anti-HIV bNAb titers.
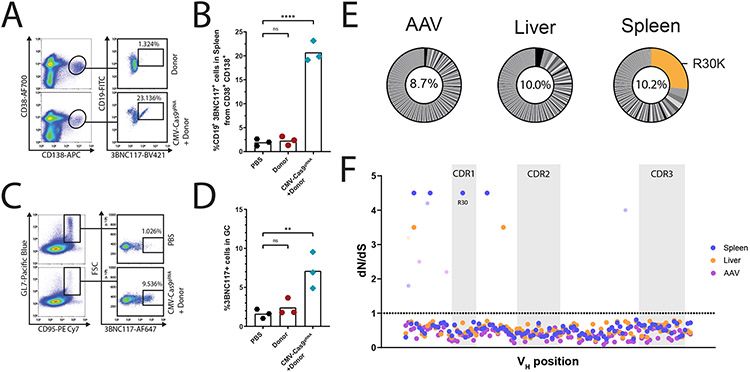
AAV injections to mice were preceded by pre-immunizations, modeling a pre-existing infection. Indeed, B cell activation is required for efficient AAV transduction30, and subsequent activation signals for the engineered B cells may benefit from prior priming of T helper cells and from presentation of appropriate immune complexes by follicular dendritic cells31. In particular, C57BL/6 mice were immunized with 20 μg of the gp120 HIV antigen, which is the target of 3BNC117. On day 6 post-immunization, each mouse was injected with 5E11 viral genomes (vg) of a bNAb coding (Donor) vector, 5E11 vg of the saCas9 coding vectors, or both (Fig. 2A). The mice then received additional immunizations on days 8, 23, 68, 98 and 128. Following the boosting regimen, mice receiving both a donor vector and an saCas9 vector had up to 5 μg/ml of the 3BNC117 bNAb in their blood, being >50x the median virus neutralization IC50 for this bNAb32,33(Fig. 2B). This is in concordance with previous reports, entailing transfer of low numbers of antigen-specific B cells from transgenic mice, demonstrating a potent immune response following immunizations34-36. Here, 3BNC117 of multiple isotypes was found in the sera, and IgG 3BNC117 accounted for as much as 1% of the total response toward gp120 (Extended Data Fig. 1). Importantly, IgG purified from treated mice can neutralize autologous YU2.DG and the heterologous tier-2 JRFL HIV pseudoviruses (Fig. 2C, Extended Data Fig. 2A). Mice injected with both a donor vector and an saCas9 vector had much higher 3BNC117 titers than mice receiving donor vector only. Nevertheless, 3BNC117 titers in mice receiving only the donor vector slightly exceeded the background levels measured in mice injected with PBS (Extended Data Fig. 2B). Indeed, integration of the antibody gene into the IgH locus was evident by RT-PCR of splenic B cell RNA from mice receiving dual vector injection but an additional, nested PCR was required to detect such integration in two of the three mice injected with the donor vector alone (Extended Data Fig. 2C-E) Very low editing frequencies without CRISPR gRNA could similarly be detected by RT-PCR only in ex vivo edited lymphocytes (Extended Data Fig. 2F-I). Notably, when using dual vector injections, high titers could be obtained not only upon immunizing the mice with the monomeric gp120 antigen of the clade B HIV strain YU2.DG, but also in independent experiments using either the clade A, BG505-based native trimer nanoparticle immunogen (MD39-ferritin)37 (Fig. 2D-E), or the stabilized soluble 2CC immunogen38, originating from the clade B HXBc2 strain (Fig. 2F-G), attesting for the breadth of the 3BNC117-expressing cells in vivo. The presence of 3BNC117-secreting cells in the bone marrow was established using ELISPOT on the bone marrow of treated mice (Fig. 2H-I) and correlated well with splenic 3BNC117 expression in these mice (Extended Data Fig. 3A-B).
Fig. 2:
In vivo engineering of B cells to express an anti-HIV bNAb. A. Experimental scheme. Immunizations are indicated in black, above the timeline. Blood collections are indicated in red, below the timeline. B. 3BNC117 IgG titers as quantified by ELISA using an anti-idiotypic antibody to 3BNC117. The black arrows indicate immunizations and the blue arrow indicates the AAV injection. Each line represents a mouse. From left to right: *; pv = 0.047, pv = 0.0201 for Two-Way ANOVA of CMV-Cas9gRNA + Donor compared to the Donor group. n=3. AUC bar graphs are available in Extended Data Fig. 2. C. Transduction neutralization of TZM.bl cells by the YU2.DG (left) and JRFL (right) HIV pseudoviruses in the presence of IgGs purified from day 136 sera. Neutralization is calculated as percent reduction from maximal luminescence per sample. The PBS control received immunizations as in (C), while the naïve control represents serum IgG from an untreated mouse. Each line represents a mouse. From left to right: *; pv = 0.0306, pv = 0.0116, **; pv = 0.0037, Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. AUC bar graphs are available in Extended Data Fig. 2. D. Experimental scheme and E. 3BNC117 IgG titers as quantified by ELISA for MD39 immunized mice. From left to right: ns; pv = 0.3724 and pv = 0.0539, ###; pv = 0.0008 for Two-Way ANOVA comparison between groups and *; pv = 0.0493, ***, pv = 0.0007 for Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to antigen respective control. F. Experimental scheme and G. 3BNC117 IgG titers as quantified by ELISA for 2CC immunized mice. #### = pv < 0.0001 for Two-Way ANOVA comparison between groups and **** = pv < 0.0001 for Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. H. A representative ELISPOT experiment of total bone marrows from 2CC immunized mice at day 82. I. Quantification of H. *; pv = 0.0317 for two-sided unpaired t-test.
In vivo engineered B cells undergo clonal expansion in germinal centers.
The frequency of 3BNC117-expressing cells reached 0.5% of total blood B cells following the later immunizations in all mice injected with both a bNAb vector and an saCas9 vector, but not in mice injected with PBS or with the bNAb vector alone (Fig. 2A, Extended Data Fig. 3C-D). Upon sacrificing the mice at day 136, 8 days after the last immunization, up to 23% of the plasmablasts in the spleen (Fig. 3A-B, Extended Data Fig. 3E) and 5-10% of germinal center (GC) lymphocytes expressed 3BNC117 (Fig. 3C-D, Extended Data Fig. 3F).
Fig. 3:
In-vivo engineered B-cells are found in lymphatic tissues 130 days following AAV injection. A. Flow cytometry plots demonstrating 3BNC117 expression among plasmablasts (CD38+, CD138+, CD19+) in the spleen at day 136. Pregated on live, singlets. B. Quantification of A. for engineered plasmablasts (CD38+ CD138+ 3BNC117+). Mean is indicated by the bars. ns; pv = 0.9892, ****; pv < 0.0001, One-way ANOVA with Tukey’s multiple comparison. C. Flow cytometry plots demonstrating 3BNC117 expression of cells with a germinal center phenotype (GL7+, CD95/Fas+) in the spleen. Pre-gated on live, singlets. D. Quantification of E. Mean is indicated by the bars, ns; pv = 0.8916, **; pv = 0.0054 One-way ANOVA with Tukey’s multiple comparison. E. Pie charts of 3BNC117 VH variants amplified from spleen and liver DNA at day 136 and from purified AAV. Orange shading indicates the R30K variant. Numbers in the middle of the pies indicate the total frequency of mutant reads in these samples. F. dN/dS values for the positions along the VH segment, based on Illumina sequencing of DNA amplified from the spleen (blue) or liver (orange) of a single mouse or AAV (purple). The dotted line represents values >1, indicative of positive selection. For dots colored with lighter shades, the assignment of a dN/dS value > 1 is not statistically significant. No position in the AAV sample reached statistical significance. Grey shading indicates CDR loops. The R30 position is indicated.
0.6% of bone marrow cells expressed CD19 and 3BNC117 (Extended Data Fig. 3G-H). However, interestingly, an additional 1.5% of bone marrow cells were CD19-, 3BNC117+ (Extended Data Fig. 3G,I).
In order to study somatic hypermutation and clonal selection, we extracted DNA from the liver and the spleen of one of the treated mice at day 136 and performed Illumina sequencing of amplified 3BNC117 VH segments. Much of the mutation repertoire was shared between the liver and the spleen and may thus reflect heterogeneity in AAV production that is subjected to little or no selection7,39. In particular, all the 3BNC117 VH variants found to be over-represented in the liver are also over-represented in the spleen. Importantly however, the inverse is not true. The CDR1 substitution R30K is the most prevalent substitution in the spleen. It accounts for more than 20% of all mutants in the spleen but is found at very low abundance in the liver or in a representative AAV batch (Fig. 3E). Indeed, bNAbs of the VRC01 family were shown to have side-chain interactions with the HIV gp120 antigen at position 3029. One may speculate that the conservative R30K substitution in 3BNC117 relieves some steric clash upon binding to monomeric gp120. Including R30K, a total of four different positions along the VH segment showed signs of positive selection in the spleen by dn/ds analysis and, as expected, none of which was enriched in the sequencing of the the representative AAV batch (Fig. 3F). We conclude that our in vivo engineering and immunization scheme has led to clonal expansion of variants stemming from either heterogeneity in AAV production or in vivo SHM. The clonal expansion is limited in span but pronounced in magnitude.
CRISPR-Cas9 cleavage is highly sequence specific but takes place also in undesired tissues.
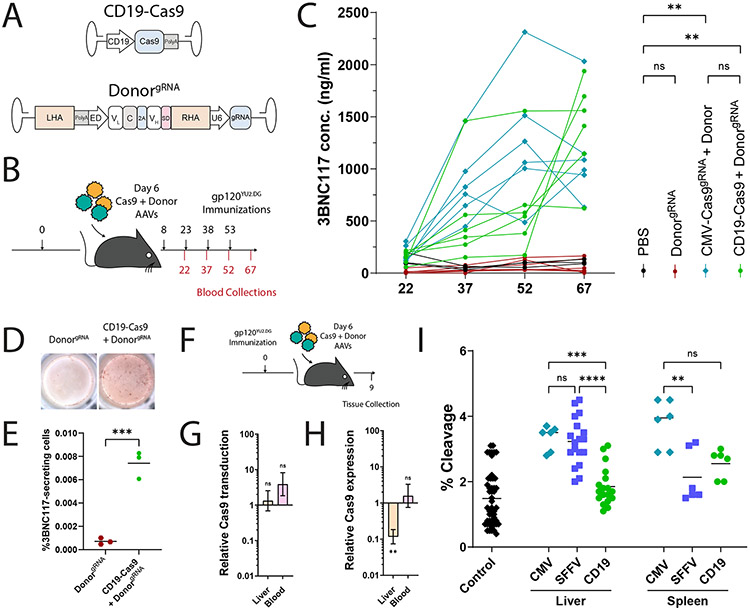
In order to assess the possible off-target effects of our in vivo engineering approach, we first quantified the copy number of the bNAb cassette in various tissues. The bNAb cassette was found at a high copy number in the liver at day 37 (Fig. 4A) and the levels were reduced by only 10 fold at day 136 (Fig. 4B), reflecting high retention of AAV episomes in the liver. High copy number was also found in the blood at day 37, but levels dropped sharply by day 136, perhaps due to multiple cell divisions. Interestingly, the AAV copy number in the bone marrow was significantly increased from day 37 to day 136, and a non-significant similar trend was also detected in the lymph nodes, indicating the possible accumulation of 3BNC117-expressing cells in these tissues (Fig. 4B). The copy number in the liver was similar whether or not the saCas9 coding AAV was co-injected to the mice. In contrast, the copy number of the bNAb cassette in the lymph nodes and in the bone marrow was found to be logs higher with saCas9 AAV co-injection, signifying the selection of 3BNC117-expressing B cells (Fig. 4C).
Fig. 4:
AAV biodistribution and saCas9 off-target cleavage analysis reveal a high safety profile. A. Donor AAV copy number quantification by qPCR in indicated tissues at day 136 from mice injected with two AAVs as in Fig. 2A. B. Relative copy number of donor AAV between day 37 and day 136 in selected tissues. C. Relative copy number of donor AAV between mice injected with two AAVs, as in Fig. 2A, and mice injected with donor AAV only, at day 136. For B. and C. Indicated are the mean of relative expression and error bars corresponding to lower and upper boundaries derived from two-sided unpaired t-test. For B., from left to right: *; pv = 0.0496, pv = 0.0139, pv = 0.0389, pv = 0.0243, **; pv = 0.0046 for comparison between the two time points and for C., from left to right: *; pv = 0.0128, pv = 0.0147 for comparison between the two mice groups. n=3 biologically independent animals. Y axis in A-C uses a log scale. LN = lymph nodes, BM = bone marrow. D. Unbiased CHANGE-seq analysis of potential saCas9 off-target cleavage with the sgRNA used in this study. Localization, annotation in the genome, number of mismatches and % read counts are indicated for each on- or off-target site. Sequence of the sgRNA with the PAM is indicated on the top. Black arrows indicate target sites used for analysis of mouse samples. Mismatches between off-target sites and intended sgRNA target are color-coded. E. On- and off-target saCas9 cleavage, of target sites indicated in D. by black arrows, in the spleen (mauve) and liver (beige) of mice injected with two AAVs, as in Fig. 2A, at day 136, as compared to uncut, naïve splenic lymphocytes DNA. For spleen and liver tissues, n=3 biologically independent animals. For the control uncut, naïve splenic lymphocyte DNA, n=1. Mean values +/− SD are indicated.
To define the genome-wide off-target activity of saCas9, we performed circularization for high-throughput analysis of nuclease genome-wide effects by sequencing (CHANGE-seq)40 on genomic DNA from C57BL/6 mice. 95% of the reads corresponded to the on-target site (Fig. 4D). We then performed targeted sequencing on four potential off-target sites, as well as on the on-target site, using genomic DNA from liver and spleen of treated mice and of a negative control mouse. Relative to control DNA from the spleen of an untreated mouse, a trend for a higher mutation rate, indicating error-prone repair of CRISPR-Cas9-induced double-stranded DNA breaks, was evident in the liver and not in the spleen, and only at the IgH on-target site rather than in any of the tested off-target sites (Fig. 4E).
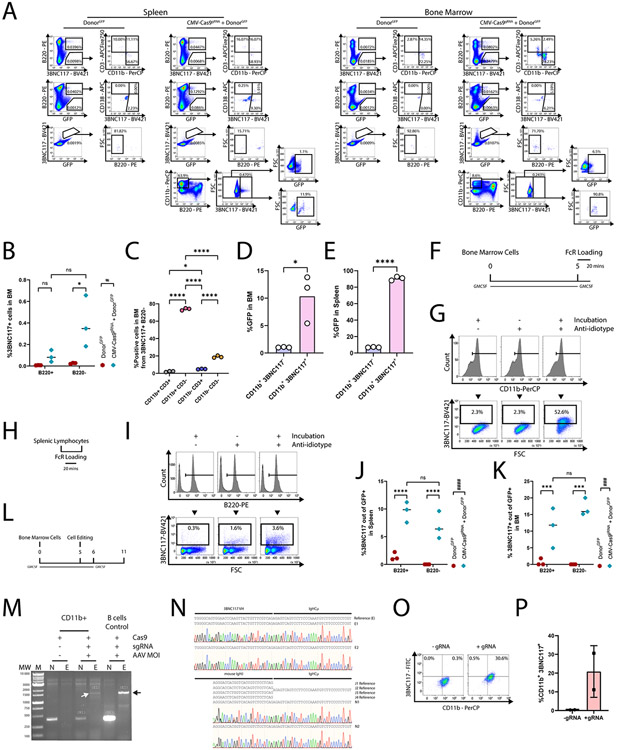
In order to better characterize the different populations of engineered cells, we next used a donor vector coding for GFP in addition to the 3BNC117 cassette (Fig. 5A). Recipient mice were immunized twice before analysis of GFP and/or 3BNC117 expression in the bone marrow and the spleen (Fig. 5B). Expectedly, GFP+, 3BNC117+ cells were enriched in the spleen (Fig. 5C, Extended Data Fig. 4A) and the bone marrow (Fig. 5D, Extended Data Fig. 4A) of mice receiving both the donor and the saCas9 vectors. Co-injection with donor and saCas9 vectors has increased the rate of B cells expressing GFP (Fig. 5E, Extended Data Fig. 4A) and, in particular, the rates of B cells expressing both GFP and 3BNC117 (Fig. 5C, Extended Data Fig. 4A) in the spleen. We estimate that, for a typical spleen of 50M cells, assuming an expansion factor of 25 fold for antigen specific B cells following a single immunization34, as low as 140 cells may have been initially engineered. The rates of GFP+, 3BNC117+ cells among B220- cells remained low (Fig. 5C, Extended Data Fig. 4A). Notably, within the GFP expressing B cells, co-injecting the saCas9 vector led to a marked increase in 3BNC117+, CD138+ plasmablasts, in both the spleen (Fig. 5F, Extended Data Fig. 4A) and the bone marrow (Fig. 5G, Extended Data Fig. 4A). In the bone marrow, we found a larger fraction of cells, stained by the anti-3BNC117 anti-idiotype antibody, to be B220− (Extended Data Fig. 4A-B). The B220− cells, stained by the anti-idiotype, were almost exclusively CD3−, and most of them were CD11b+ cells (Extended Data Fig. 4A, C), indicative of possible FcR binding of secreted 3BNC117. Indeed, the majority of the CD11b+ cells in the bone marrow, stained with the anti-idiotype, were GFP− (Extended Data Fig. 4D-E). In addition, the same anti-idiotype staining detected the ex vivo binding of soluble 3BNC117 by non-engineered CD11b+ cells at a much higher rate than by non-engineered B220+ cells (Extended Data Fig. 4F-I). Interestingly, an increase in the rate of cells, stained by the anti-idiotype, among GFP expressing cells was seen even within this B220− cell populations in the spleen (Extended Data Fig. 4A, J) and marrow (Extended Data Fig. 4A, K). This is in line with recent publications showing expression of membrane antibodies by cells of the myeloid lineage41-43. Concordantly, we were able to ex vivo engineer CD11b+ cells to express 3BNC117 from the IgH locus (Extended Data Fig. 4L-P). Cumulatively, our data imply that, pending CRISPR-Cas9 mediated on-target integration, both B and non-B cells can express the antibody on the membrane, but only B cells proliferate subsequent to antigen engagement.
Fig. 5:
Assessing expression of the transgene in different subsets of cells. A. Vector design. The donor cassette expresses a GFP, separated from the 3BNC117 cassette by a 2A peptide. B. Experimental design. C-D. Quantification of GFP+ 3BNC117+ in the spleen (C) or bone marrow (D) of recipient mice. Mean values and standard deviation are indicated. For each group, n=3 biologically independent mice. *; pv = 0.0284, ***; pv = 0.0004 for Two-Way ANOVA. E. Quantification of GFP+ cells in spleen. ####; pv < 0.0001 for Two-Way ANOVA and ****; pv < 0.0001 for Two-Way ANOVA with Tukey’s multiple comparison. F-G. Quantification of the 3BNC117+ CD138+ population from B220+, GFP+ cells in the sleen (F) *; pv = 0.0147 for unpaired two-tailed t-test, or bone marrow (G) *; pv = 0.0471 for unpaired two-tailed t-test. Mean values are indicated.
Coding the sgRNA and Cas9 on different vectors prevents cleavage in the absence of Donor DNA.
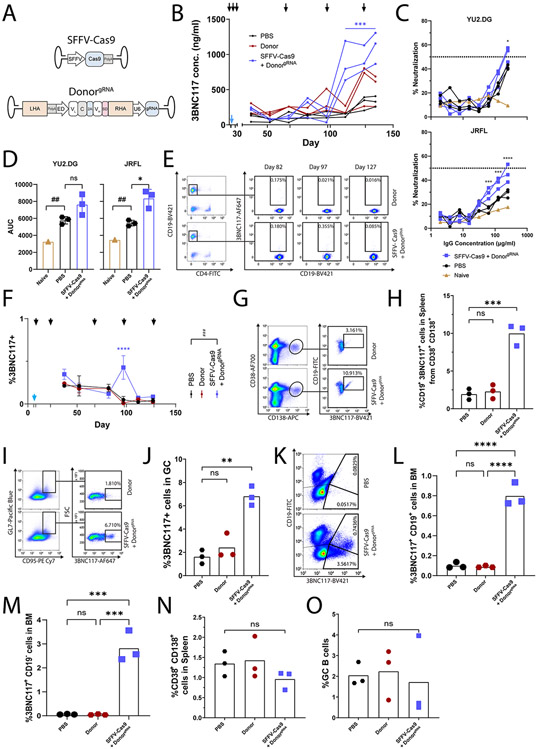
The coding of the sgRNA together with the saCas9 on the same AAV is predicted to allow DNA cleavage in many cells that are not co-transduced with the donor AAV. The resulting, non-productive, cleavage may be avoided if the sgRNA cassette is instead separated from the saCas9 gene and coded on the donor AAV (Extended Data Fig. 5A). Repeating the above mouse experiments (Fig. 2A) with this new pair of AAVs allowed high 3BNC117 titers capable of neutralizing autologous YU2.DG and heterologous JRFL HIV pseudoviruses following repeated immunizations (Extended Data Fig. 5B-D), and the frequency of 3BNC117-expressing cells reached 0.5% of total blood B cells (Extended Data Fig. 5E-F). Upon sacrificing the mice at day 136, up to 10% of splenic plasmablasts expressed 3BNC117 (Extended Data Fig. 5G-H). In addition, up to 7% of splenic B cells with a germinal center phenotype expressed 3BNC117 (Extended Data Fig. 5I-J), while 1% and 3% of the bone marrow cells expressed 3BNC117, with or without co-expressing CD19, respectively (Extended Data Fig. 5K-M). These results are of the same range as those obtained when the sgRNA was coded together with the saCas9, although direct side-by-side comparison is hindered by the use of different ubiquitously active promoters. Importantly, the overall numbers of splenic plasmablasts, germinal center B cells and bone marrow plasma cells were similar to those in the control groups (Extended Data Fig. 5N-O), mitigating concerns of B cell neoplasm (see also Extended Data Fig. 3E-F).
Driving Cas9 expression by a B cell specific promoter prevents cleavage in undesired tissues.
In order to further increase the safety of our approach, we next coded the saCas9 under the control of the CD19, B cell-specific, promoter44 (Fig. 6A). In particular, C57BL/6 mice were immunized with 20 μg of HIV gp120, and 6 days later each mouse was co-injected with one vector coding for saCas9, regulated by the CD19 promoter, and a second vector coding both the bNAb and the sgRNA (Fig. 6A-B). The mice then received up to 6 additional immunizations. Already after 4 immunizations, treated mice had up to 2 μg/ml of the 3BNC117 bNAb in their blood (Fig. 6C). 3BNC117 blood titers did not go down by 30 days later, irrespective of whether additional immunizations were administered (Extended Data Fig. 6A). Regardless of the immunization regimen, similar titers were obtained using promiscuous or B cell-specific regulation over saCas9 expression. Concordantly, 45 days after the 4th immunization, similar rates of 3BNC117-secreting cells could be detected in the bone marrow using ELISPOT (Fig. 6D-E), irrespective of whether additional immunizations were administered and irrespective of the saCas9 promoter (Extended Data Fig. 6B-D). Therefore, replacing the saCas9 promoter does not preclude the therapeutic effect, which is stable after 4 immunizations. In addition, using CD19 rather than CMV promoter, to drive saCas9 expression, reduces the engineering rate of B cell progenitors as assessed following in vitro differentiation of IL7R enriched bone marrow cells (Extended Data Fig. 7). Importantly, even when the CMV promoter is used to drive saCas9 expression, bone marrow HSPCs may not be engineered, as similarly low 3BNC117 staining and ELISA levels are obtained following syngeneic transplantation of Lin− enriched cells from mice injected with the donor vector with or without the saCas9 coding vector (Extended Data Fig. 8).
Fig. 6:
Improving safety by coding saCas9 and the sgRNA on separate AAVs and expressing saCas9 under the regulation of a B cell-specific promoter. A. Map of the AAV vectors used. saCas9 is expressed under the CD19 promoter, while the sgRNA is coded on the donor vector, outside of the homology arms. B. Experimental scheme. Mice were immunized according to the timeline in black (top), and bled as indicated in red (bottom). C. 3BNC117 IgG titers, quantified by ELISA with an anti-idiotypic antibody. Each line represents a mouse. From top to bottom and left to right: **; pv = 0.0075, and pv = 0.0055, ns; pv = 0.0876 and pv = 0.3288 for two-Way ANOVA. D. Representative ELISPOTs of bone marrows from mice, 45 days following the fourth immunization. E. Quantification of (D). ***; pv = 0.0007 for two-sided unpaired t-test. F. Experimental scheme. Mice immunized with gp120 are dosed with AAVs and tissues are collected three days following AAV injection. G. Relative transduction by the saCas9 coding AAV, calculated as the ratio of copy numbers, in the indicated tissues, between mice receiving AAVs coding for saCas9 under the CD19 or SFFV promoters. Indicated are the mean of relative expression and error bars corresponding to lower and upper boundaries derived from two-sided unpaired t–test. From left to right: ns; pv = 0.6920 and pv = 0.1441. n=3 biologically independent animals. H. Relative saCas9 mRNA expression, depicted as the ratio between saCas9 expression from the CD19 promoter and from the SFFV promoter. Indicated are the mean of relative expression and error bars corresponding to lower and upper boundaries derived from two-sided unpaired t-test. ns; pv = 0.5698, **; pv = 0.0092. n=3 biologically independent animals. I. TIDE analysis of on-target cleavage in the indicated tissues using either CMV, SFFV or CD19 driven saCas9 expression. From left to right: ns; pv > 0.9999 and pv = 0.0760, **; pv = 0.0036, ***; pv = 0.0008, ****; pv < 0.0001 for One-way ANOVA with Tukey’s multiple comparison. Each dot represents a comparison between a control sequence and an independent mouse sequence.
In order to assess possible effects on biodistribution and safety, different groups of mice were sacrificed for tissue analysis, 3 days after having been co-injected with the donor + sgRNA vector and with a second vector coding for saCas9 under the control of either a ubiquitous promoter or the B cell-specific CD19 promoter. Similar transduction rates were obtained for vectors coding the saCas9 under the regulation of the CD19 or SFFV promoters (Fig. 6F-G). However, the CD19 promoter significantly reduced saCas9 expression in the liver, while not reducing expression in peripheral blood mononuclear cells (PBMCs, Fig. 6H). The rates of on-target cleavage in the liver or the spleen, as measured by TIDE analysis, were significantly above background only when using the CMV or SFFV promoters, rather than the CD19 promoter, to drive saCas9 expression (Fig. 6I). Therefore, separating the coding of saCas9 and the sgRNA between the two AAVs and expressing saCas9 under a B cell-specific promoter reduce undesired cleavage to below our limit of detection while allowing high 3BNC117 titers following immunizations.
DISCUSSION:
Eliciting a specific, neutralizing antibody response to hypervariable viruses is a long-standing challenge in medicine. B cell engineering provides an opportunity to express desired therapeutic antibodies for adaptive immunity. Here, we uniquely demonstrate that B cells can be safely and robustly engineered in vivo. A single, systemic dose of dual AAV-DJ coding for CRISPR-Cas9 and donor cassettes in mice allowed for site-specific integration, with limited off-target Cas9 expression and DNA double-strand breaks. Upon immunizations, the engineered B cells underwent antigen-induced activation leading to memory retention, clonal selection and differentiation into plasma cells that secrete the bNAb at neutralizing levels.
The monoclonal bNAb titers obtained by in vivo engineering in this work are similar or higher than those obtained by ex vivo engineering followed by adoptive transfer to immunocompetent mice7,12,13, with the exception of Huang et al8. Importantly, the response of the in vivo engineered B cells to the antigen is not hindered by the endogenous polyclonal response to immunization, which can be highly potent45. In contrast to ex vivo engineering, in vivo B cell engineering is simple, fast and cost effective. It can and will be provided at the point of care, requiring no specialized facilities. Our approach further allows for CSR and clonal expansion, but the full functional consequences of these attributes will have to be tested in the prevention or treatment of infection models. Yet additional experiments may determine whether in vivo B cell engineering allows proper antibody glycosylation and expression patterns to avoid the formation of anti-drug antibodies, as seen following muscle transduction for antibody expression6. The effects of anti-AAV antibodies and T cell responses would similarly have to be assessed. In engineered B cells, autoreactivity may occur due to heterogeneity in AAV production or due to pairing of the engineered heavy chain with the endogenous light chain. Future clinical applications must aim to reduce these sources of heterogeneity, if not sufficiently eliminated by natural tolerance mechanisms46,47. Still, future modifications may include coding the bNAb as a single chain13 to reduce mispairing of the bNAb heavy chain with the endogenous light chain, potentially improving both safety and efficacy. Such single chain coding can further allow the expression of bi-specific bNAbs, which may be required to provide long-term protection from HIV resurgence1. Safety may be further improved by using more specific nucleases48,49 and by having the bNAb gene preceded by a splice acceptor rather than by a promoter, to reduce expression from off-target integration7,12. Both safety and efficacy may benefit from embedding B cell-specific targeting moieties in the AAV vector50 or in a non-viral alternative14. The therapeutic impact of our approach may best be evaluated in nonhuman primates with HIV-like infections. In the nonhuman primates as in HIV infected individuals, undergoing controlled treatment interruption, we expect a continuous and much more potent antigen induced activation, which can either replace or complement an immunization regimen in order to achieve higher antibody titers and do so in a shorter time frame. Finally, in vivo B cell engineering may have diverse future applications as it may be used to address other persistent infections as well as to treat autoimmune diseases, genetic disorders, and cancer.
MATERIALS AND METHODS:
Plasmid cloning
For the CMV-Cas9gRNA vector, pX6011 (Addgene) was cleaved with BsaI and pre-annealed, phosphorylated (PNK, NEB), sgRNA coding oligo-deoxynucleotides were ligated using T4 DNA Ligase (NEB). For the CD19-Cas9 vector, pAB2702 was cleaved using NotI and SpeI (NEB) and an saCas9 coding fragment, amplified from pX601, as well as the murine CD19 promoter, amplified from wild type C57BL/6OlaHsd genomic DNA, were assembled using Hi-Fi DNA Assembly Mix (NEB). For the SFFV-Cas9 vector, pAB270 was cleaved with NotI and SpeI (NEB). The fragment coding the SFFV promoter was amplified from GW175 (Kay Lab, Stanford) and the saCas9 was amplified from pX601. The fragments were assembled using Hi-Fi DNA Assembly Mix (NEB). For the DonorgRNA vector, the U6-gRNA fragment was amplified from ligated pX601 with the murine IgH sgRNA used in this study, and the fragment was assembled using Hi-Fi DNA Assembly Mix (NEB) into the donor vector pADN171XS3, following cleavage with SpeI (NEB). For cloning of the GFP-3BNC117-expressing donor (Fig. 3A), HiFi DNA Assembly (NEB) was performed according to manufacturer instructions using a fragment amplified from pADN171XS (Donor vector from Nahmad et al. 20203 and named “Donor” in this manuscript), a fragment amplified from pADN157CF2 (GFP expressing vector from Nahmad et al. 20203), and a homology arms bearing vector cleaved with XhoI3. A list of primers used for these reactions can be found in Supplementary Table 1. Resulting plasmid was Sanger sequenced for verification of the correct integration of the fragments into the vector.
A list of primers used for cloning can be found in Supplementary Table 1. All fragments for cloning were amplified using PrimeStar MAX (Takara).
rAAV production
rAAV-DJ were produced in HEK293T cells (ATCC) by triple transient transfection using polyethylenimine (PEI, Polysciences Inc). For each vector, fourteen 15 cm dishes were transfected at 80% confluency with pAd5 (helper plasmid), rAAV-DJ genome plasmid and vector plasmid at a 3:1:1 ratio4. In total, each plate was transfected with 41.25 μg of DNA. Purification was performed with AAVpro Extraction Kit (Takara) according to the manufacturer protocol. Titer quantification was performed by qPCR using SYBRGreen (PCR Biosystems). A list of primers used for AAV titer quantification can be found in Supplementary Table 1.
Mouse studies
Mouse experiments comply with all ethical regulations and were performed under supervision of Tel Aviv University Committee for the Use and Treatment of Laboratory Animals. In vivo engineering experiments were performed on 6-10 weeks old female CD45.2 C57BL/6OlaHsd (Envigo) mice. All mice were housed and kept at ambient temperature of 19-23°C, humidity of 45-65% and with a 12-hour light/12-hour dark cycle. Immunizations with gp120-YU2 or MD39-ferritin were performed as previously described, using 20 μg/mouse of antigen in Alum (Invitrogen)3,5. For AAV injections, mice were anesthetized with 0.1 mg/g and 0.001 mg/g Ketamine and Xylazine, respectively, and were injected i.v. with 5E11 vg/vector/100 μl/mouse in PBS. Blood samples from mice were collected in heparin. Cells and serum were separated by centrifugation. Serum was collected from the supernatant. For spleens, whole spleens were extracted from mice and mechanically crushed in PBS to be filtered in a 70 μm cell strainer (Corning). For bone marrow, cells were flushed from the posterior femur and tibia. For blood, spleen and bone marrow, cells were processed with red blood cell lysis buffer (Biolegend) and plated in 1640 RPMI (Biological Industries) supplemented with 10% HI FBS (Biological Industries) until processing. Muscle tissue was processed from femoral muscles. Right or left lungs were processed for pulmonic tissue. Right or left hemispheres were processed for brain tissue. Lobes were processed for liver tissue and whole heart was used for cardiac tissue. For lymph nodes, inguinal and cervical lymph nodes were pooled for processing.
Illumina sequencing and analysis
Total genomic DNA was extracted from fresh tissues using Gentra PureGene Tissue Kit (Qiagen). Initial PCR amplification and the subsequent barcoding PCR reaction of the 3BNC117 VH fragments or the off-target sites was performed using the proofreading PrimeStarMAX Polymerase (Takara) for 35 cycles and 8 cycles, respectively. A list of primers used for these reactions can be found in Supplementary Table 1. Following each PCR, amplicons were purified using AMPure XP beads (Beckman Coulter) at a 0.7:1 ratio. Libraries were quantified using Qubit (Invitrogen) and analyzed using an Agilent 4200 TapeStation. Combined libraries were loaded at 5pM with 25% PhiX control (Illumina) and sequencing was performed with a v2 Nano Reagent kit 2x250bp on a MiSeq machine, using the Miseq control software, at the Genomic Research Unit (GRU), Tel Aviv University. For off-target analysis, raw fastq files were submitted to Fast Length Adjustment of Short Reads (FLASH) (https://github.com/ebiggers/flash)6. The default parameters were changed to allow for lower max mismatch density ratio of 0.1. The resultant files were submitted to CRISPRpic (https://github.com/compbio/CRISPRpic)7, with a wider mutagenic window of 10 bp on either side of the DNA double-strand breaks. Presented data pools all mutation types detected.
For mutation and selection analysis, raw fastq files were submitted to FLASH using the default parameters. The resultant files submitted to Bowtie2 alignment analysis (https://github.com/BenLangmead/bowtie2)8 compared to the engineered 3BNC117 sequence, using local mode and the “xeq” parameter for match and mismatch annotations. Using a specific script, unaligned reads were filtered, as well as reads not within 80-115% of the original length and reads with more than 15% mutated bases. The primer annealing sites at both ends of the sequences were omitted from the analysis. All bases considered as mutated in this analysis had a Q score higher than 20. The Selecton software9 was used to run M8 and M8a models in order to infer positive selection and likelihood ratio test was performed between the null model (M8a) and the alternative model (M8) to determine which model better fits the data. All P-values were corrected for multiple testing using false discovery rate (FDR)10. For dn/ds and clonal expansion analyses, to reduce sequencing biases, an additional 3 nucleotides on both ends of the sequencing were removed. All alignments and phylogenies supported the M8 alternative model where positive selection is enabled.
CHANGE-seq
Genomic DNA from fresh spleens of wild type C57BL/6OlaHsd using Gentra PureGene Tissue Kit (Qiagen) and quantified using Qubit (Invitrogen) according to manufacturer instructions. CHANGE-seq was performed as previously described11. Briefly, purified genomic DNA was tagmented with a custom Tn5-transposome to an average length of 400 bp, followed by gap repair with Kapa HiFi HotStart Uracil+ DNA Polymerase (KAPA Biosystems) and Taq DNA ligase (NEB). Gap-repaired tagmented DNA was treated with USER enzyme (NEB) and T4 polynucleotide kinase (NEB). Intramolecular circularization of the DNA was performed with T4 DNA ligase (NEB) and residual linear DNA was degraded by a cocktail of exonucleases containing Plasmid-Safe ATP-dependent DNase (Lucigen), Lambda exonuclease (NEB) and Exonuclease I (NEB). In vitro cleavage reactions were performed with 125 ng of exonuclease-treated circularized DNA, 90 nM of EnGen® Sau Cas9 protein (NEB), NEB buffer 3.1 (NEB) and 270 nM of sgRNA (Synthego), in a 50 μl volume. Cleaved products were A-tailed, ligated with a hairpin adaptor (NEB), treated with USER enzyme (NEB) and amplified by PCR with barcoded universal primers NEBNext Multiplex Oligos for Illumina (NEB), using Kapa HiFi Polymerase (KAPA Biosystems). Libraries were quantified by qPCR (KAPA Biosystems) and sequenced with 151 bp paired-end reads on an Illumina MiniSeq instrument. CHANGE-seq data analyses were performed using open-source CHANGE-seq analysis software (https://github.com/tsailabSJ/changeseq).
ELISA
High binding microplates (Greiner Bio-One) were coated with 2 μg/ml of an anti-idiotipic antibody against 3BNC117 in PBS overnight at 4°C. Plates were washed with PBST, blocked for an hour with 5% BSA in PBST and washed again. For 3BNC117 IgG quantification, samples were diluted 1:50-500 fold and a standard was made using purified 3BNC117 serially diluted in PBS. For 3BNC117 isotype detection, samples were serially diluted as described in the figures. Samples and standards were incubated for an hour. Plates were then applied with HRP conjugated detection antibodies: anti-mouse IgA (Abcam), anti-mouse IgG, anti-mouse IgG1, anti-mouse IgM (Jackson ImmunoResearch) or anti mouse IgG2c (Bio-Rad Laboratories) at 2 μg/ml in PBST and were incubated for another hour. A list of antibodies used in these experiments may be found in Supplementary Table 1. Before detection with QuantaBlu (ThermoFisher) according to manufacturer protocol, plates were washed for an additional round. Detection was done in a Synergy M1 Plate reader (Biotek). When absolute quantitation is presented, the concentration of 3BNC117 was determined by reference to the dilution factor of the standard curve.
To quantify the fraction of 3BNC117 from the gp120 response, we performed Dynabead purification of sera. In short, for each sample, 5 mg of Dynabeads M-280 Tosylactivated (Invitrogen) were conjugated with 50 μg of gp120 in 0.1 M Na-Phosphate supplemented at a 1.5:1 ratio of 3 M ammonium sulphate buffer at 37°C overnight. Beads were then washed, blocked with 0.5% BSA in PBS for an hour at 37°C, washed with PBS 0.1% BSA and resuspended with 1:5 PBS diluted 50 μl of sera/5 mg conjugated Dynabeads. Binding occurred for 1 hour at 37°. Beads were washed three times with PBS and elution was performed with 0.2 M Glycine (pH 2.5) subsequently neutralized with 1 M TRIS (pH 8.5) at a 1:0.1 ratio. Resulting samples were loaded on anti-3BNC117 coated plates at a 1 μg/ml concentration, the fraction of 3BNC117 response was calculated as the standard curve derived concentration of 3BNC117 per 1 μg of purified sera per ml.
ELISPOTs
For ELISPOT assays, cells were collected from the tibia and femur bones by flushing. Red blood cell lysis was performed for 10 minutes using RBC Lysis Buffer (Biolegend) at room temperature. ELISPOT plates were prepared as previously described12. In short, Immobilon P membrane plates (Millipore) were coated with 1 μg/ml of anti-3BNC117 overnight at 4°C. Following washing and blocking with 5% BSA in PBS, cells were seeded in dilutions of 1E5-1E6 per well, in triplicate for each mouse in RPMI 1640 (Biological Industries) supplemented with P/S (Biological Industries), 55 μM β-mercaptoethanol and 10% FCS HI (Sigma). Incubation was performed for two days at 37°C and 5% CO2. Viability at the time of harvest was commonly 60-80%. After washing, plates were incubated with anti-mouse IgG (Jackson) for 1 hour, washed again and development was performed with AEC Chromogen Kit (Sigma-Aldrich). Finally, plates were dried and incubated at 4°C until acquisition on an iSpot ELISPOT reader (AID).
Neutralization Assays
Under sterile BSL2/3 conditions, the PSG3 plasmid was co-transfected into HEK293T cells along with JRFL or YU2 HIV envelope plasmids using Lipofectamine 2000 transfection reagent (ThermoFisher Scientific) to produce single-round of infection competent pseudo-viruses representing multiple clades of HIV. HEK293T cells were plated in advance overnight with DMEM medium + 10% FBS + 1% Pen/Strep + 1% L-glutamine. Transfection was done with Opti-MEM transfection medium (Gibco) using Lipofectamine 2000. Fresh medium was added 12 hours after transfection. Supernatants containing the viruses were harvested 72 hours later. In sterile 96-well plates, 25 μl of virus was immediately mixed with 25 μl of serially diluted (2×) bead protein A/G purified IgG (ThermoFisher) from mouse sera (starting at 500 μg/ml) and incubated for one hour at 37 °C to allow for antibody neutralization of the pseudoviruses. 10,000 TZM-bl cells/well (in 50 μl of media containing 20 μg/ml Dextran) were directly added to the antibody virus mixture. Plates were incubated at 37 °C for 48 hours. Following the infection, TZM-bl cells were lysed using 1× luciferase lysis buffer (25 mM Gly-Gly pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1% Triton X-100). Neutralizing ability disproportionate with luciferase intensity was then read on a Biotek Synergy 2 (Biotek) with luciferase substrate according to the manufacturer’s instructions (Promega).
qPCR
For copy number quantification of the donor AAV in tissue samples, DNA was extracted from fresh tissues using DNeasy Blood & Tissue Kit (Qiagen) with RNAse treatment on-column. Each sample was analyzed for both internal control (Albumin intron2) and the donor AAV. For quantification of donor AAV copy number per haploid genome, a standard curve was used. Standards were prepared from a PCR PrimeStar MAX (Takara) reaction using naïve C57BL/6OlaHsd mice genomic DNA for the internal control or donor AAV plasmid for the Donor sample and purified using AMPure XP beads (Beckman Coulter) at a 1:1 ratio. A list of primers used for Donor and internal control reactions can be found in Supplementary Table 1. Standard curve amplicons were quantified using Qubit (Invitrogen) and serially diluted 8 times. For AAV Cas9 quantification RNA was extracted from fresh tissues using RNeasy Mini Kit (Qiagen) with DNAse treatment on-column and post-purification using RQ1 DNAse (Promega). Reverse transcription was performed using RevertAid (ThermoFisher) and random hexamer primers. Data collection and analysis were performed on a StepOnePlus qPCR System (Applied Biosystems) using SYBRGreen (PCR Biosystems). For fold change of AAV titers and Cas9 relative expression, we used the relative quantity method13.
Flow Cytometry
Harvested cells from spleen, bone marrow or blood were resuspended in cell staining buffer (Biolegend) and incubated with 2 μg/100μl of human anti-3BNC117 and, where applicable as indicated in the legend, with 1 μg/100μl of TruStain FcX (Biolegend) for 10 minutes, washed and resuspended again in cell staining buffer containing conjugated primary antibodies. A list of antibodies and respective dilutions used in these experiments can be found in Supplementary Table 1. Secondary staining was performed in the dark, for 15 minutes, with anti-human IgG1 AF647 (Abcam) or anti-human IgK BV421 (Biolegend) or anti-human IgG1 FITC (Biolegend). For primary gp120 staining, cells were incubated with 2 μg/100μl of gp120. Then, cells were washed and data acquisition was performed on a CytoFLEX (Beckman Coulter) or Attune NxT (life Technologies) or FACS Aria III (BD Biosciences) for experiments involving cell sorting. Data collection was performed with CytExpert. Data were compiled and analyzed using Kaluza Analysis 2.1 (Beckman Coulter). Gating strategies can be found in Extended Data Fig. 9.
In-vitro B progenitor differentiation
For enrichment of IL7R+ cells, bone marrow from the tibia and femur bones of each mouse was collected at day 97 by flushing (Extended Data Fig. 7A). Red blood cell lysis was performed for 10 minutes using RBC Lysis Buffer (Biolegend) at room temperature. After washing, cells were resuspended at 2E8 cells per ml, in PBS supplemented with 10% FCS (Sigma). PE-conjugated anti-mouse IL7R (120-048-801, Miltenyi) was added 1/100 and binding was performed on ice for 30 minutes. Cells were subsequently washed and resuspended at 4E8 cells per ml, in PBS supplemented with 10% FCS. Anti-PE microbeads (Miltenyi) were supplemented 1/5 and binding was performed on ice for 30 minutes. Cells were once again washed and resuspended at 4E7 cells per ml and separated on LS or MS magnetic columns (Miltenyi).
Following separation, eluate was analyzed for IL7R and CD19 expression (Extended Data Fig. 7E-F) and in-vitro differentiation was performed as follows. Cells were seeded in RPMI 1640 (Biological Industries) supplemented with P/S (Biological Industries), 55 μM β-mercaptoethanol, 10% FCS HI (Sigma) and 10 ng/ml mouse IL7 (Peprotech) at ~2E6 cells per ml. Three days following seeding (Extended Data Fig. 7B) cells were washed and reseeded in RPMI 1640 (Biological Industries) supplemented with P/S (Biological Industries), 55μM β-mercaptoethanol, 10% FCS HI (Sigma) and 10 ng/ml mouse IL4 (Peprotech), 10 μg/ml LPS (Peprotech). Two days following seeding, cells were washed and seeded on 60 gray irradiated CD40LB feeder cells14 in RPMI 1640 (Biological Industries) supplemented with P/S (Biological Industries), 55 μM β-mercaptoethanol, 10% FCS HI (Sigma), 10 ng/ml mouse IL4 (Peprotech) and 10 ng/ml mouse IL21 (Peprotech) at 5E5 cells per ml. Two days following seeding, supernatant and cell aliquots were collected for ELISA and flow cytometry, respectively. The rest were washed and reseeded on 60 gray irradiated CD40LB feeder cells in RPMI 1640 (Biological Industries) supplemented with P/S (Biological Industries), 55 μM β-mercaptoethanol, 10% FCS HI (Sigma) and 10 ng/ml mouse IL21 (Peprotech) at 5E5 cells per ml. Finally, two days following seeding, cells and supernatant were collected for flow cytometry and ELISA, respectively.
Hematopoietic Stem and Progenitor enrichment and adoptive transfer
For bone marrow Lin− HSPCs enrichment, cells were collected from the tibia and femur bones of female CD45.2 C57BL/6JOlaHsd mice at day 97 by flushing (Extended Data Fig. 8A). Red blood cell lysis was performed for 10 minutes using RBC Lysis Buffer (Biolegend) at room temperature. Following washing, cells were magnetically enriched using the Mouse Lineage Cell Depletion Kit (Miltenyi) using LS or MS magnetic columns (Miltenyi), according to the manufacturer’s instructions.
Following separation, flow through aliquots were analyzed for Lin and B220 expression. The rest of the cells were adoptively transferred into mice as with an adaptation of a previously described protocol15. In short, recipient female, 8 weeks old, CD45.1 C57BL/6JOlaHsd mice were sub-lethally irradiated at 150 cGy and, the following day, received 6E5 cells/100 ul/mouse in PBS by retro-orbital injections.
In-vitro FcR loading
Primary bone marrow CD11b were collected five days following activation and 2E6 cells were further cultured in FBS/GMCSF supplemented DMEM with 2μg/ml of purified mouse IgK/IgG2a 3BNC117. Cells were collected after 20mins and then FcRX blocked before analysis by flow cytometry using the anti-3BNC117 anti-idiotype antibody.
For primary splenic lymphocytes, cells were collected from naïve mice and were cultured in FBS supplemented RPMI with 2 μg/ml of purified mouse IgK/IgG2a 3BNC117. Cells were collected after 20mins and then FcRX blocked before analysis by flow cytometry using the anti-3BNC117 anti-idiotype antibody.
In-vitro engineering of B cells
CRISPR-Cas9 RNP electroporations and AAV transductions of B cells was performed as described previously3. In short, total splenic lymphocytes were collected from spleens of naïve mice and activated in LPS and IL-4 for electroporation (Neon, ThermoFisher) the following day. For anti-idiotype specificity, AAV-DJ transduction was performed at 50k MOI and for AID independent integrations at 10k MOI or 100k MOI, as indicated in the figures.
Ex vivo engineering of CD11b cells
Total bone marrow cells were extracted by flushing tibia and femur. Cells were cultured in DMEM (Biological Industries) supplemented with 15% FBS (Biological Industries) and 50ng/ml of GMCSF (Peprotech) to activate and proliferate CD11b cells16. Five days following activation, 1E6 cells were electroporated with pre-generated complexes of 20pmol spCas9 (IDT) and 25 pmol sgRNA (IDT) in buffer T at 1600v, 20ms, 1pulse. Immediate AAV-DJ transduction was performed at 200k MOI. Cells were subsequently cultured for 1 day at 1E6cells/ml in GMCSF/FBS supplemented DMEM and for an additional 5 days without GMCSF before analysis flow cytometry.
Nucleic Acid Manipulations
For Reverse Transcription PCR demonstrating 3BNC117 gene integration into the IgH locus, RNA was extracted from sorted engineered B cells (3BNC117+, CD4−, CD19+) on a FACS BD AriaIII (BD Biosciences). As a positive control, we used in vitro engineered mouse splenic lymphocytes, as described previously3. In short, mouse splenic lymphocytes were activated with 10 μg/ml LPS (Santa-Cruz Biotechnology) and 10 ng/ml IL-4 (Peprotech) for 24 hours, electroporated by CRISPR-Cas9 RNP using a Neon Electroporation System (Invitrogen) and transduced at 50,000 MOI of the donor AAV-DJ vector. For RNA extraction, we used RNeasy Mini Kit (Qiagen) with DNAse treatment on-column and reverse transcription was performed using RevertAid (ThermoFisher) and Oligo dT primers. PCR on the resulting cDNA was performed for 35 cycles using PrimeStar MAX (Takara). Then, a semi-nested PCR was performed using PrimeStar MAX (Takara) for 35 cycles. A list of primers used for these reactions may be found in Supplementary Table 1. Following each PCR, resulting amplicons were analyzed by Agarose gel electrophoresis as compared to a standardizing ladder (Hylabs, GeneDireX 1Kb plus DNA ladder RTU or 100bp DNA ladder H3 RTU) and total reactions were purified using AMPure XP beads (Beckman Coulter) at a 1:1 ratio. Purified amplicons were Sanger sequenced at the DNA Sequencing Unit, Tel Aviv University and presented alignment of the chromatograms was performed using SnapGene (GSL Biotech).
For TIDE analysis of on-target cleavage, genomic DNA from tissues was extracted from fresh tissues using Gentra PureGene Tissue Kit (Qiagen). PCR was performed for 35 cycles using PrimeStar MAX (Takara). Primers used for these reactions can be found in Supplementary Table 1. For the control samples, three independent PCR were performed on independent genomic DNA samples, collected from splenic tissue of naïve C57BL/6OlaHsd mice. Resulting amplicons were purified using AMPure XP beads (Beckman Coulter) at a 1:1 ratio. Purified amplicons were Sanger sequenced at the DNA Sequencing Unit, Tel Aviv University. For each sample, multiple sequencing reactions were performed using either primers. Then, samples were compared using TIDE (https://tide.nki.nl/) 17. For control samples, we performed reciprocal sample comparisons from the independent initial PCR reactions. For all samples, along with the CMV expressed Cas9, mice received the Donor vector. Along with the SFFV or CD19 expressed Cas9, mice received the DonorgRNA vector. Control samples come from naïve splenic lymphocytes. (Fig. 6I).
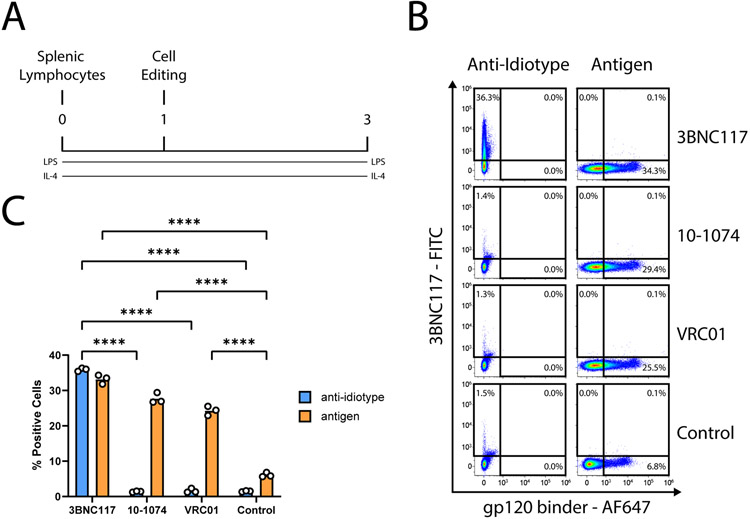
Anti-Idiotipic to 3BNC117 scFvs were generated by phage display18. Candidates were cloned into pcDNA3.1 vectors with the human kappa and IgG1 heavy chain. In short, antibodies were produced by transfection of both antibody chains into Expi293F cells (gibco) by Expifectamine (gibco) and purified using MabSelect (GE Healthcare) as previously described3. Specificity and sensitivity of the antibody were verified in-vitro using primary B cells engineered, as described previously3, to express one of the three bNAbs, 3BNC117, VRC01 or 10-1074. Both 3BNC117 and VRC01 bind to the CD4 binding site. However, importantly, only 3BNC117 binds to its anti-idiotype (Extended Data Fig. 10).
For Reverse Transcription PCR of ex vivo engineered cells, RNA was extracted, two days following treatment, from 5E5 splenic lymphocytes or 6 days following treatment from 5E5 GMCSF activated bone marrow cells. RNA extraction was performed using Quick-RNA micro prep (Zymo Research) without DNAse treatment. Reverse transcription was performed using RevertAid (ThermoFisher) and Oligo dT primers. PCR of exon-exon junctions on the resulting cDNA was performed for 30 cycles using HS Taq Mix (PCRBio), a list of primers used for these reactions can be found in Supplementary Table 1.
Statistics
Statistical analyses were performed on distinct samples using Prism (GraphPad). For Area under the curve, in each group, the mean AUC and SD were calculated and these values were compared by t-test. All t-tests were performed as two-tailed. For the TIDE analysis, each comparison between a control sample and an independently produced PCR reaction from 3 independent mice were used. Each Figure legend denotes the statistic used, central tendency and error bars.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Multiple isotypes of the 3BNC117 antibody are expressed by engineered B cells.
A-D. ELISA for each isotype. A. IgM, B. IgG1, C. IgG2c and D. IgA. All samples come from the CMV-Cas9gRNA + Donor injected mice at different time points, as indicated in each legend. Mean and SD are indicated. n=3 biologically independent animals. E-F. Area under the curve (AUC) for A-D. E. IgM, ns; pv = 0.1288, ***; pv = 0.0008, **; pv = 0.0062, F. IgG1, ns; pv = 0.131 and from top to bottom as presented in the graph, **; pv = 0.0044 and pv = 0.0013, G. IgG2c, ***; pv = 0.0007, *; pv = 0.0195, **; pv = 0.0098, H. IgA, ns; pv = 0.0587, **; = pv = 0.0013, *; pv = 0.0403, for two-sided unpaired t-test. n=3 biologically independent animals. For A-H, sample collection day is indicated. Mean values are indicated. I. Fraction of 3BNC117 IgG titers as quantified by ELISA using purified gp120 binding sera from donor injected mice immunized with gp120, at day 37. ***; pv = 0.0007 for two-sided unpaired t-test. n=3 biologically independent samples. Mean values are indicated.
Extended Data Fig. 2. bNAb genomic integration, sera titers and neutralization as a function of immunizations and co-injection of the CRISPR-Cas9 vector.
A. Area under the Curve (AUC) of Fig. 2D for YU2.DG (left) and JRFL (right) **; pv = 0.0036 (YU2.DG) and pv = 0.005 (JRFL) for unpaired t-test for CMV-Cas9gRNA + Donor to PBS comparison and ##; pv = 0.0072 (YU2.DG) and pv = 0.0063 (JRFL) for one-sample t-test for Naïve to PBS comparison. n=3 for CMV-Cas9gRNA + Donor and PBS. Naïve sample is from a single, non-immunized, non-AAV-injected mouse. Mean values +/− SD are indicated. B. Area under the curve (AUC) of Fig. 2C. From top to bottom, *; pv = 0.0185 and pv = 0.0103, **; pv = 0.0036 for two-sided unpaired t-test. n=3 biologically independent animals. Mean values +/− SEM are indicated. C. RT-PCR on RNA from sorted, 3BNC117+, CD19+, CD4− blood lymphocytes from day 37. Here, we used a reverse primer in a membranal exon of either IgHCμ or IgHCγ (all subtypes) and a forward primer on the VH of the coded 3BNC117. Numbers indicate different mice, injected with either a) PBS, b) the donor vector and the CMV-Cas9gRNA vector, or c) the donor vector only, as indicated above the gels. Control sample (C+) comes from in-vitro engineered primary mouse splenic lymphocytes, as described previously7. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. For each group, experiment was reproduced 3 times with independent samples, as indicated by the numbers. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. D. Total DNA from the previous reaction as in (C) was purified and a semi-nested PCR with the same forward primer and a reverse primer on the CH1 of the respective constant domains. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. For each group, experiments were reproduced 3 times with independent samples. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. E. Sanger sequencing alignment and chromatogram of the purified amplicon from the previous step. Reference sequences are indicated above. For the IgHCγ, each subtype reference is indicated. Sequencing of the IgHCμ amplicon of donor 3 has failed. F. Experimental design for (G-I). Splenic lymphocytes were activated with LPS and IL-4 and engineered, ex vivo, by AAV transduction and Cas9 electroporation with or without a gRNA. G. Flow cytometry of engineered splenic lymphocytes two days following treatment. Pre-gated on live, singlets. FcR block was used in the staining. Engineering parameters are indicated above each plot. H. EtBr gel electrophoresis showing products of an RT-PCR reaction with RNA from cells two days following treatment as in (F). For each sample, a control (C) reaction was performed amplifying the endogenous IgHG1 cDNA. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. The experiment was reproduced once, with similar results. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. I. Sanger sequencing of the previous amplicons, confirming the integration.
Extended Data Fig. 3. Detection of engineered B cells in the spleen, the blood and the bone marrow.
A. Flow cytometry plots demonstrating 3BNC117 expression among CD19+ CD11b− cells in the spleen at day 82 of 2CC immunized mice. Pregated on live, singlets. FcR block was used in the staining. B. Quantification of B. ***; pv = 0.0006 for two-sided unpaired t-test. C. Flow cytometry plots demonstrating 3BNC117 expression among blood B cells (CD19+, CD4−). D. Quantification of blood 3BNC117-expressing cells over time. The black arrows indicate immunizations and the blue arrow indicates AAV injection. ####; pv < 0.0001 for Two-Way ANOVA comparison between groups and *; pv = 0.0133, ****; pv < 0.0001 Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. For each group, each line represents the mean +/−SD of n=3 biologically independent mice. E. Quantification of total CD38+ CD138+ in spleens of recipient mice as in Fig. 3A. From top to bottom: ns; pv = 0. 2380 and pv = 0.9907 for One-Way ANOVA with Tukey’s multiple comparisons test. F. Quantification of total GL7+ CD95+ cells as in Fig. 3E. From top to bottom: ns; pv = 0.9857 and pv = 0.9985 for One-Way ANOVA with Tukey’s multiple comparisons test. G. Flow cytometry plots demonstrating the presence of 3BNC117-expressing B cells in the bone marrow. H-I. Quantification of G. ns; pv = 0.9965, ****; pv < 0.0001 (G) and from top to bottom: ns; pv > 0.9999, *; pv = 0.0387 and pv = 0.0372 (H) for One-Way ANOVA with Tukey’s multiple comparison.
Extended Data Fig. 4. Assessing expression of the transgene in different subsets of cells.
A. Flow cytometry examples for Fig. 5C-G and Extended Data Fig. 3B-E, J-K. B. Quantification of 3BNC117+ cells in bone marrow. #; pv = 0.0129 for Two-Way ANOVA and *; pv = 0.0255 for Two-Way ANOVA with Tukey’s multiple comparison. C. Quantification of the indicated populations from 3BNC117+ B220− cells in the bone marrow. *; pv = 0.0335, **** = pv < 0.0001 for One-Way ANOVA with Tukey’s multiple comparison. D-E. Quantification of GFP+ cells from the CD11b+ 3BNC117+ or CD11b+ 3BNC117− populations in the bone marrow (D) or spleen (E). *; pv = 0.0214, **** = pv <0.0001 for unpaired two-tailed t-test. Mean values are indicated. F. Experimental scheme for (G). Bone marrow cells were collected and activated in the presence of GMCSF. 5 days later, cells were cultured in the presence of purified mouse 3BNC117 for FcR presentation and then collected for analysis by flow cytometry. G. Analysis by flow cytometry, using the anti-3BNC117 anti-idiotypic antibody, of bone marrow cells activated by GMCSF and subsequently cultured with 3BNC117. Experimental conditions are indicated above. Pre-gated on CD11b, live, singlets. H. Experimental scheme for (I). Splenic Lymphocytes are collected from mice and cultured with purified mouse 3BNC117. Next, cells are collected for Flow Cytometry. I. Analysis by flow cytometry, using the anti-3BNC117 anti-idiotypic antibody, of splenic lymphocytes cultured with 3BNC117. Experimental conditions are indicated above. Pre-gated on B220+, live, singlets. J-K. Quantification of 3BNC117+ cells from GFP+ cells in the spleen (J) ####; pv < 0.0001 for Two-Way ANOVA and ns; pv = 0.1748, ****; pv < 0.0001 for Two-Way ANOVA with Tukey’s multiple comparison or bone marrow (K) ###; pv = 0.001 for Two-Way ANOVA and ns; pv = 0.6177, and from left to righ: ***; pv = 0.0005 and pv = 0.0005 for Two-Way ANOVA with Tukey’s multiple comparison. BM = bone marrow. L. Experimental scheme of in-vitro engineering of primary GMCSF activated bone marrow cells for (M-N). M. EtBr gel electrophoresis showing the product of an RT-PCR reaction of RNA from activated bone marrow cells, six days following treatment as in (L). For each sample, two reactions were performed (N or E). N amplifies endogenous IgHCμ mRNA. E amplifies the transgene mRNA joined by splicing to the IgHCμ exons following engineering. Indicated labeling of the amplicons (E1, E2, N1 and N2) performed for reference in (N). The experiment was performed once with expected results. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. N. Sanger sequencing of the amplicons, confirming correct integration. Amplicons are annotated E1, E2, N1 and N2 as in (M). O. Flow cytometry example of engineered GMCSF activated primary bone marrow cells. Pre-gated on B220−, live, singlets. P. quantification of O. Control cells are cells transduced with the AAV and electroporated only with the spCas9, without the sgRNA (-gRNA). For A-P and Fig. 3, FcR block was used in the staining. For each group, n=2 biologically independent samples.
Extended Data Fig. 5. Engineering B cells with the sgRNA coded on the donor AAV.
A. Vector maps of the AAVs coding for the DonorgRNA and the SFFV-Cas9. B. 3BNC117 IgG titers as quantified by ELISA over time in the SFFV-Cas9 + DonorgRNA group. The black arrows indicate immunizations and the blue arrow indicates AAV injection. Each line represents a mouse. ***; pv = 0.0005 for Two-Way ANOVA comparing the SFFV-Cas9 + DonorgRNA group to the Donor group. n=3 biologically independent mice. In this panel, the PBS and Donor control groups are the same as for Fig. 2B. C. Transduction neutralization of TZM.bl cells by the YU2.DG (top) and JRFL (bottom) HIV pseudoviruses in the presence of day 136 sera IgGs. Neutralization is calculated as percent reduction from maximal luminescence per sample. The PBS control received immunizations, while the naïve control represents serum IgG from an untreated mouse. ns = non-significant, *; pv = 0.0462 and from top to bottom: ***; pv = 0.0007 and pv = 0.0004, ****; pv < 0.0001, Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. D. Area under the Curve (AUC) for C. for YU2.DG and JRFL. ns; pv = 0.0667 (YU2.DG) and *; pv = 0.0103 (JRFL) for two-sided unpaired t-test for CMV-Cas9gRNA + Donor to PBS comparison and ##; pv = 0.0097 (YU2.DG) and pv = 0.0078 (JRFL) for two-sided one-sample t-test for naïve to PBS comparison. n=3 for CMV-Cas9gRNA + Donor and PBS. Naïve sample from a single, non-immunized, non-AAV-injected mouse. In C-D, the PBS and control group is the same as for Fig. 2C and Extended Data Fig. 2A. Mean values +/− SD are indicated. E. Representative flow cytometry analysis of 3BNC117+, CD19+, CD4− blood lymphocytes over time in the SFFV-Cas9 + DonorgRNA group. F. Quantification of E. The black arrows indicate immunizations and the blue arrow indicates AAV injection. ###; pv = 0.0006 for Two-Way ANOVA comparison between groups and ****; pv < 0.0001 for Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. For each group, each line represents the mean +/−SD of n=3 biologically independent mice. G. Representative flow cytometry analysis of 3BNC117+, CD19+, CD38+, CD138+ plasmablasts in the spleens of the SFFV-Cas9 + donorgRNA group at day 136. H. Quantification of G. Mean is indicated by the bars. ns; pv = 9892, ***; pv = 0.0005, one-way ANOVA with Tukey’s multiple comparison. I. Representative flow cytometry analysis of GL7+, Fas/CD95+ GC B cells in the spleens of the SFFV-Cas9 + DonorgRNA group at day 136. J. Quantification of I. Mean is indicated by the bars. ns; pv = 0.8916, **; pv = 0.0075, one-way ANOVA with Tukey’s multiple comparison. K. Representative flow cytometry analysis of 3BNC117+ cells in total bone marrow (BM) of the SFFV-Cas9 + DonorgRNA group at day 136. L. Quantification of 3BNC117+ CD19+ cells, ns; pv = 0.9965 and ****; pv < 0.0001 and M. Quantification of 3BNC117+ CD19− cells, ns; pv > 0.9999 and from top to bottom: ***; pv = 0.0003 and pv = 0.0003. Mean is indicated by the bars. One-way ANOVA with Tukey’s multiple comparison. N-O. Assessing overall immune homeostasis. Quantification by flow cytometry of total CD38+ CD138+ plasmablasts in spleen (N) ns; pv = 0.5622, total GL7+, Fas+ GC B cells in the spleen (O), at day 136 ns; pv = 0.9926. Mean is indicated by the bars.One-way ANOVA with Tukey’s multiple comparison. For E-O, the PBS and Donor control groups are the same as for Fig. 3 and Extended Data Fig. 3.
Extended Data Fig. 6. Long-term persistence of serum antibodies and antibody-secreting cells in the bone marrow.
A. Experimental design. Here, all mice were immunized 4 times (days 8,23,38 and 53) and selected groups received 2 additional immunizations on days 68 and 83. B. 3BNC117 IgG titers as quantified by ELISA over time. For each group, each line represents the mean +/−SD between day 22 and day 67 represents n=6 for biologically independent mice and n=3 for day 67 to 97. C-D. Representative ELISPOT experiments (C) and quantification (D) of 3BNC117-secreting cells from the bone marrow of mice of the indicated groups. Numbers in parentheses represent the number of immunizations. From top to bottom followed by left to right, ns; pv = 0.9844 and pv = 0.9963 and pv = 0.0884, *; pv = 0.0135, **; pv = 0.008, ***; pv = 0.0005 and pv = 0.0003 for One-Way ANOVA with Tukey’s multiple comparison. Mean values are indicated. Part of the data presented in panels B-D is presented also in Fig. 6C-E.
Extended Data Fig. 7. Using CD19 rather than CMV promoter, to drive saCas9 expression, reduces the engineering rate of B cell progenitors.
A. Origin of cells used in this experiment. Bone marrows were extracted 100 days following AAV injection and enriched for IL7R+. B. Experimental scheme. Enriched IL7R+ cells were grown in the presence of multiple activation factors as indicated in the representative timeline, numbers indicate days. Horizontal bars below the timeline indicate the presence of a specific factor supplemented to the growth media. C. Representative flow cytometry analysis of in-vitro differentiation of the IL7R+ enriched cells over time. Days as in (B) are indicated above the plots. D. Quantification of C. For each group, each line represents n=3 biologically independent samples. E. Representative flow cytometry analysis of IL7R enrichment from mice, as in (A). F. Quantification of E. G. Representative flow cytometry analysis of 3BNC117+ CD19+ expression by cells, following 9 days of in-vitro differentiation, as in (B). H. Quantification of G. Each dot represents cells collected from a single mouse as in (A). ns; pv = 0.9, *; pv = 0.0122 and **; pv = 0.0077 for One-Way ANOVA with Tukey’s multiple comparison. For C-G, FcR block was used in staining.
Extended Data Fig. 8. Low 3BNC117 staining and ELISA levels are obtained following syngeneic transplantation of Lin− enriched cells from mice injected with the donor vector with or without the saCas9 coding vector.
A. Experimental scheme. CD45.2 mice received either a donor AAV expressing the gRNA or both a donor AAV and an AAV expressing saCas9 and the gRNA. Immunization protocol is indicated by the bars in black. On day 97, bone marrow cells were collected, enriched for lineage negative cells (Lin−) and transplanted into, recipient CD45.1 mice. Recipient mice are sublethally irradiated before transplantation and immunized after transplantation. B-C. Representative flow cytometry analysis (B) and quantification (C) of the enriched Lin− population from the donor mice. ns; pv = 0.9627 for One-Way ANOVA with Tukey’s multiple comparison. D. Representative flow cytometry analysis (D) of spleens (top) or bone marrows (bottom) from recipient mice at day 140, as in (A) E. Quantification of D. for the CD45.2+ CD45.1− population. From top to bottom: ns; pv = 0.4595 and pv > 0.9999 for Two-Way ANOVA with Tukey’s multiple comparison. F-G. Quantification of D. for the rate of 3BNC117-expressing cells for the spleen (F) or the bone marrow (G). From top to bottom: ns; pv = 0.9994 and pv = 0.0668 (F) and pv = 0.2371 and pv = 0.0560 for Two-Way ANOVA with Tukey’s multiple comparison. H. Serum 3BNC117 IgG titers at the indicated time points. The scale of the Y-axis was chosen to correspond to the other 3BNC117 titer plots in this manuscript. ns; pv = 0.1726t for Two-Way ANOVA. I. Representative ELISPOT assay of a day 140 bone marrow from CD45.1 recipient mice. J. Quantification I. ns; pv = 0.1756 for two-sided unpaired t-test. For B-D, FcR block was used in staining.
Extended Data Fig. 9. Gating strategy for each experiment in this study.
Gating strategy for each experiment in this study.
Extended Data Fig. 10. Assessment of the specificity and sensitivity of the anti-idiotype for 3BNC117.
A. Experimental scheme. Cells are engineered a day following extraction from spleen and activation. We used three different donor AAVs, each expressing a different antibody: either 3BNC117, VRC01 or 10-1074. B. Flow cytometry of gp120 or anti-idiotype binding of engineered cells, two days following treatment. Staining procedure is indicated above the plots. FcR block was used in staining. Each row indicates a different AAV used. Untransduced cells serve as the negative control. C. Quantification of B. ****; pv < 0.0001 for Two-Way ANOVA with Tukey’s multiple comparison.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank the Veterinary Service Center, Tel Aviv University for animal husbandry. The IDRFU, GRU and SICF units, Tel Aviv University for logistic support and council. We also thank Leeor Vardi, Maoz Gelbart, Hila Kobo, David Burstein, Itai Benhar, Natalia Freund, Mark Kay, Tal Akriv and Natalia Gritsenko for reagents and feedback. This research was funded by the H2020 European Research Council grant 759296 570 (A.B.) and the Israel Science Foundation grants: 1632/16 (A.B.), 2157/16 (A.B.), The Bill and Melinda Gates Foundation: OPP1183956 (J.E.V.), National Institutes of Health: R01 AI167003-01 (A.B.) AI128836 and R01 AI073148 (D.N.), Edmond J. Safra Center for Bioinformatics fellowship (T.K. and A.S.), St. Jude Children’s Research Hospital and ALSAC, National Institutes of Health (NIH) Office Of The Director (OD) Somatic Cell Genome Editing (SCGE) initiative grant U01AI157189 (S.Q.T.), the Gertner Institute Scholarship, the Yoran Institute Scholarship, the SAIA Foundation (A.D.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
COMPETING INTERESTS:
A.D.N., D. Na., M.H.-F., I.D., and A.B. are listed as inventors on patent applications covering B cell engineering. A.D.N., and A.B. have an equity stake in and receive monetary compensation from Tabby Therapeutics LTD., a B cell engineering company. S.Q.T. is a co-inventor on patents covering the CHANGE-seq method. S.Q.T. is a member of the scientific advisory boards of Kromatid, Inc. and Twelve Bio. Other authors declare no competing interests.
DATA AVAILABILITY
Data are available in the main text, in the Extended Data Figures and Supplementary Data and Materials. Illumina sequencing data can be accessed in the SRA database under accession code PRJNA706552.
REFERENCES:
- 1.Mendoza P et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-On Y et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med (2018). doi: 10.1038/s41591-018-0186-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen YZ et al. Safety, pharmacokinetics, and immunogenicity of the combination of the broadly neutralizing anti-HIV-1 antibodies 3BNC117 and 10-1074 in healthy adults: A randomized, phase 1 study. PLoS One 14, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson PR et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med 15, 901–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs AB et al. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med 20, 296–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priddy FH et al. Adeno-associated virus vectored immunoprophylaxis to prevent HIV in healthy adults: a phase 1 randomised controlled trial. Lancet HIV 6, e230–e239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahmad AD et al. Engineered B cells expressing an anti-HIV antibody enable memory retention, isotype switching and clonal expansion. Nat. Commun 1–10 (2020). doi: 10.1038/s41467-020-19649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang D et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun 1–10 (2020). doi: 10.1038/s41467-020-19650-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusil F et al. A lentiviral vector allowing physiologically regulated membrane-anchored and secreted antibody expression depending on B-cell maturation status. Mol. Ther 23, 1734–1747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greiner V et al. CRISPR-Mediated Editing of the B Cell Receptor in Primary Human B Cells. iScience 12, 369–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voss JE et al. Reprogramming the antigen specificity of B cells using genome-editing technologies. Elife (2019). doi: 10.7554/eLife.42995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartweger H et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med 216, 1301–1310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moffett HF et al. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith TT et al. In situ programming of leukaemia-specific t cells using synthetic DNA nanocarriers. Nat. Nanotechnol 12, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal S et al. In Vivo Generation of CAR T Cells Selectively in Human CD4+ Lymphocytes. Mol. Ther 28, 1783–1794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal S, Weidner T, Thalheimer FB & Buchholz CJ In vivo generated human CAR T cells eradicate tumor cells. Oncoimmunology 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank AM et al. Combining T-cell-specific activation and in vivo gene delivery through CD3-targeted lentiviral vectors. Blood Adv. 4, 5702–5715 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank AM & Buchholz CJ Surface-Engineered Lentiviral Vectors for Selective Gene Transfer into Subtypes of Lymphocytes. Mol. Ther. - Methods Clin. Dev 12, 19–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank AM et al. CD8-Specific Designed Ankyrin Repeat Proteins Improve Selective Gene Delivery into Human and Primate T Lymphocytes. Hum. Gene Ther 31, 679–691 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Pfeiffer A et al. In vivo generation of human CD19-CAR T cells results in B-cell depletion and signs of cytokine release syndrome. EMBO Mol. Med (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michels A et al. Lentiviral and adeno-associated vectors efficiently transduce mouse T lymphocytes when targeted to murine CD8. Mol. Ther. - Methods Clin. Dev 23, 334–347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Münch RC et al. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun 6, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Breuer CB et al. In vivo engineering of lymphocytes after systemic exosome-associated AAV delivery. Sci. Rep 1–9 (2020). doi: 10.1038/s41598-020-61518-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawaz W et al. AAV-mediated in vivo CAR gene therapy for targeting human T-cell leukemia. Blood Cancer J. 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rurik JG et al. CAR T cells produced in vivo to treat cardiac injury. Science (80-. ) 375, 91–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parayath NN, Stephan SB, Koehne AL, Nelson PS & Stephan MT In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm D et al. In Vitro and In Vivo Gene Therapy Vector Evolution via Multispecies Interbreeding and Retargeting of Adeno-Associated Viruses. J. Virol 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheid JF et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science (80-. ) 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung KL et al. Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells. Mol. Ther 26, 456–467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesin L, Ersching J & Victora GD Germinal Center B Cell Dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sok D & Burton DR HIV Broadly Neutralizing Antibodies: Taking Good Care Of The 98%. Immunity 45, 958–960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garber DA et al. Durable protection against repeated penile exposures to simian-human immunodeficiency virus by broadly neutralizing antibodies. Nat. Commun 11, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor JJ, Pape KA, Steach HR & Jenkins MK Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science (80-. ) 347, 11214–11218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbott RK et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–145.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dosenovic P et al. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen binding affinity. Proc. Natl. Acad. Sci 115, 4743–4748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dey B et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steichen JM et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecomte E et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther. - Nucleic Acids 4, e260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazzarotto CR et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol (2020). doi: 10.1038/s41587-020-0555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busch S et al. Circulating monocytes and tumor-associated macrophages express recombined immunoglobulins in glioblastoma patients. Clin. Transl. Med 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong X et al. Macrophage-derived immunoglobulin m inhibits inflammatory responses via modulating endoplasmic reticulum stress. Cells 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs T et al. Expression of combinatorial immunoglobulins in macrophages in the tumor microenvironment. PLoS One 13, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau T, Bardin F, Imbert J, Chabannon C & Tonnelle C Restriction of transgene expression to the B-lymphoid progeny of human lentivirally transduced CD34+ cells. Mol. Ther 10, 45–56 (2004). [DOI] [PubMed] [Google Scholar]
- 45.McCarron MJ, Park PW & Fooksman DR CD138 mediates selection of mature plasma cells by regulating their survival. Blood 129, 2749–2759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemazee D Mechanisms of central tolerance for B cells. Nat. Rev. Immunol 17, 281–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell DM et al. Peripheral deletion of self-reactive B cells. Nature 354, 308–311 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edraki A, Mir A, Ibraheim R, Gainetdinov I & Sontheimer EJ A Compact , High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol. Cell 73, 714–726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pausch P et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science (80-. ) 337, 333–337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartmann J et al. A Library-Based Screening Strategy for the Identification of DARPins as Ligands for Receptor-Targeted AAV and Lentiviral Vectors. Mol. Ther. - Methods Clin. Dev 10, 128–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 1.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barzel A et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nahmad AD et al. Engineered B cells expressing an anti-HIV antibody enable memory retention, isotype switching and clonal expansion. Nat. Commun 1–10 (2020). doi: 10.1038/s41467-020-19649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm D et al. In Vitro and In Vivo Gene Therapy Vector Evolution via Multispecies Interbreeding and Retargeting of Adeno-Associated Viruses. J. Virol 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang D et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun 1–10 (2020). doi: 10.1038/s41467-020-19650-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magoč T & Salzberg SL FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Chang HY, Cho SW & Ji HP CRISPRpic: fast and precise analysis for CRISPR-induced mutations via prefixed index counting. NAR Genomics Bioinforma. 2, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern A et al. Selecton 2007: Advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35, 506–511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- 11.Lazzarotto CR et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol (2020). doi: 10.1038/s41587-020-0555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieland A et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature (2020). doi: 10.1038/s41586-020-2931-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Nojima T et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat. Commun 2, 411–465 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Ferrari S et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol 38, 1298–1308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santana-Magal N et al. Melanoma-Secreted Lysosomes Trigger Monocyte-Derived Dendritic Cell Apoptosis and Limit Cancer Immunotherapy. Cancer Res. 80, 1942–1956 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Brinkman EK, Chen T, Amendola M & Van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitton A, Nahary L & Benhar I Antibody Isolation From a Human Synthetic Combinatorial and Other Libraries of Single-Chain Antibodies. in Phage Display: Methods and Protocols (eds. Hust M & Lim TS) 349–363 (Springer; New York, 2018). doi: 10.1007/978-1-4939-7447-4_19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the main text, in the Extended Data Figures and Supplementary Data and Materials. Illumina sequencing data can be accessed in the SRA database under accession code PRJNA706552.