Abstract
The recent improvements in mass spectrometry instruments and new analytical methods are increasing the intersection between proteomics and big data science. In addition, bioinformatics analysis is becoming increasingly complex and convoluted, involving multiple algorithms and tools. A wide variety of methods and software tools have been developed for computational proteomics and metabolomics during recent years, and this trend is likely to continue. However, most of the computational proteomics and metabolomics tools are designed as single-tiered software application where the analytics tasks can’t be distributed, limiting the scalability and reproducibility of the data analysis. In this paper we summarise the key steps of metabolomics and proteomics data processing, including the main tools and software used to perform the data analysis. We discuss the combination of software containers with workflows environments for large scale metabolomics and proteomics analysis. Finally, we introduce to the proteomics and metabolomics communities a new approach for reproducible and large-scale data analysis based on BioContainers and two of the most popular workflow environments: Galaxy and Nextflow.
1. Introduction
Large-scale identification and quantification of proteins and metabolites provides a unique snapshot of a biological system of interest at a given time point [1]. MS-based high-throughput technologies have resulted in an exponential growth in the dimensionality and sample size [2]. This increase has two major directions: I) the number of samples processed, powered by new mass spectrometers; and II) the number of molecules (metabolites, peptides, and proteins) identified alongside each sample [3]. In addition, the data analysis in MS-based metabolomics and proteomics is becoming more complex, including several convoluted steps to go from the spectra identification to the final list of relevant molecules. This scenario creates major challenges for software developers and the bioinformatics community: I) software and data analysis scalability; II) software availability and findability; and III) reproducibility of the data analysis [3, 4].
Computational proteomics and metabolomics have been dominated by desktop and monolithic software for the past decades, which hampered high throughput analysis in High-Performance Computing systems (HPCS) and cloud environments [5, 6]. Furthermore, many of these tools are proprietary closed-source solutions, often run only on MS Windows or from vendor’s hardware, and use proprietary binary formats for data intake. These are barriers for reproducible science. During the last decade, open source software and distributed solutions have slowly made their way in these computational fields, with an ecosystem of computational tools flourishing in proteomics and metabolomics (see reviews in both fields [6, 7]). While the irruption of open source and distributed frameworks into the aforementioned omics fields is positive for the scalability, portability, and reproducibility of data analysis in this fields, it often comes at the cost of an increased technical complexity: installing, maintaining and executing these analysis software is usually complex and requires advanced software expertise, which is often a rare skill among scientific practitioners. This is further complicated by the fact that reproducibility and collaboration demand the installation of these tools on different computational environments (local computers, HPC, cloud, collaborators cluster, etc), often requiring different installation processes and software dependencies to be fulfilled [8].
Software containers, such as Docker containers, simplify the distribution and execution of software, by providing a way to isolate the software desired and its dependencies. Once a container for a specific tool has been built, it can be distributed as easily as if it was a single file, by depositing it in an online container registry. Then, the container can directly execute the enclosed software without any additional installation processes. The same container can be executed on different operating systems. In the past few years, the use of software containers and software packaging systems has markedly increased in the field of Bioinformatics [8, 9]. In particular, the BioContainers [8] (http://biocontainers.pro) and BioConda [10] (http://bioconda.github.io) communities have expanded the availability of containers and adequately packaged bioinformatics tools respectively, providing today thousands of tools in a format that can be used in local workstations, HPC and cloud environment seamlessly [9]. These software containers reduce the technical entry barrier for setting up scientific open source software and for making setups portable across multiple environments.
While containers and software packages simplify installation and increase portability of bioinformatics tools, they still leave to the scientist the task of combining tools together to create bioinformatics analysis workflows and pipelines [11]. This is a complex task and demands the use of the Linux command line environment; the underlying file system and data streams. In addition, if the analysis needs to run in distributed architectures (e.g. HPC clusters or Cloud), the bioinformatician will need to combine the workflow design (what tools to run with which data inputs and parameters) with the execution logic (e.g. job scheduler, data filesystem). To facilitate workflow design and execution on different distributed architectures, the bioinformatics community has developed various workflow systems [11].
A workflow system is a software that facilitates the setup of sequential and parallel steps of tool executions, by providing abstraction layers that deal with the connectivity between tools, tool execution, error handling, re-execution policies and adaptive layers to deal with different execution environments (single machine, containers, HPC, clouds, etc), among others. Without a workflow system, the developer or bioinformatician needs to write logic (usually on a bash script) to deal with all these functionalities, besides having to take care of designing the analysis flow itself. For a more detailed observation of what are workflow systems, see [11]. During the past 10 years, open source workflow environments have started to consolidate in the field of bioinformatics. The first popular workflow environment systems in bioinformatics where Taverna (now Apache Taverna) (https://taverna.incubator.apache.org) and Galaxy (https://galaxyproject.org/) [12], released in 2003 and 2005 respectively. The list is increasing every year with prominent examples such as Nextflow (https://www.nextflow.io/) [13], Cromwell (https://software.broadinstitute.org/wdl), toil (http://toil.ucsc-cgl.org) and Snakemake (https://snakemake.readthedocs.io/en/stable/) [14], among others. Besides workflow systems, different workflow languages have appeared, such as CWL or WDL, among others.
In this manuscript, we discuss the combination of software containers with workflows environments for large scale metabolomics and proteomics analysis. The combination of software containers and workflows environments promises to make scientific analysis pipelines scalable, reproducible, portable and accessible to scientists that do not have any expertise in the use of complex computational infrastructure and command line environments. We will introduce to the metabolomics and proteomics communities a new approach for reproducible and large scale data analysis based on BioContainers and two of the most popular workflows environments: Galaxy [12] and Nextflow [13].
2. Current approaches for computational mass spectrometry
In proteomics, the most common strategy for the interpretation of data-dependent acquisition (DDA) MS/MS spectra consists of comparing the experimental spectra to a set of ideal spectra (also called theoretical spectra), extrapolated from the predicted fragmentation of peptides derived from a protein database [15]. During this process, every spectrum obtained by the mass spectrometer needs to be compared with all the theoretical spectra within the same precursor mass. As data from larger cohorts and more complex samples is generated, data analysis running time increases [16]. During recent years, algorithms and tools have been developed to perform the identification step, such as Andromeda [17], MSGF+ [18] or MSFragger [19]. Even though most of these algorithms have become robust and reliable, analysis of large scale experiments will still be computationally intensive and take considerable execution time [20]. After the identification process, the resulting peptide-spectrum matches can be reliably controlled by false discovery rates filters (such as FDR) (Figure 1). Finally, the list of quantified protein is ensemble based on the identified peptides by using protein inference algorithms [21, 22]. The list of quantified proteins is provided to the downstream statistical analysis step, which reports the final relevant proteins. In data independent acquisition (DIA) methods precursor ions are selected according to their abundances, DIA aims to implement a parallel fragmentation of all precursor ions, regardless of their intensity or other characteristics, enabling the establishment of a complete record of the sample [23]. Different software have been implemented to analyse DIA datasets, such as OpenSWATH [24] and Skyline [25].
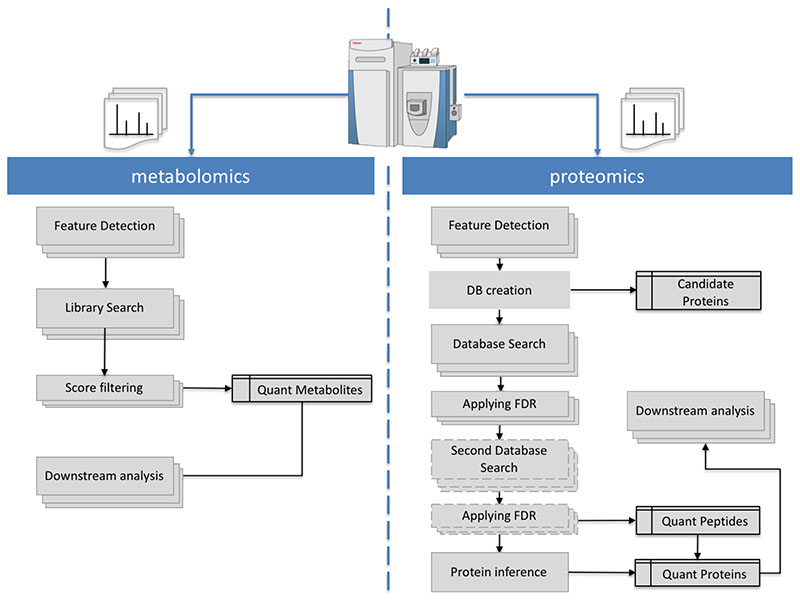
Figure 1. Mass spectrometry metabolomics and proteomics bioinformatics workflows.
The Proteomics lane (right) represent a Database search Label-free analysis workflow including Feature detection on MS1 spectra, protein database creation, database search, statistical analysis and final protein inference step. The metabolomics workflow represents a common spectral search workflow.
Similarly, computational metabolomics is mainly based on the comparison of the metabolites spectra against a well-curated database of previously identified metabolites (Spectral library strategy) (Figure 1). Spectral libraries such as METLIN (https://metlin.scripps.edu/) and MassBank (https://massbank.eu/MassBank/) contain information about mass and structure of small molecules, although MS/MS spectra are available for only a share of the small molecules in the database. The basic analytical workflow yields thousands of molecular features within minutes of data acquisition. But, similarly to proteomics, only a fraction of detected masses can be matched a molecule in the database, or more commonly to several possible molecular formulas [26, 27]. A statistical validation and manual curation can only be achieved by a matched MS/MS spectrum and/or by another compound-specific property such as retention time, which is then compared to a synthesised standard compound. In principle, quantitative analysis in metabolomic experiments is very similar to the label-free quantitation approaches based on extracted ion chromatograms in proteomic workflows. Feature alignment and detection is followed by quantitation and then perhaps identification of a compound [26].
3. Examples of established tools for computational mass spectrometry
Many established tools for proteomics and metabolomics data analysis are monolithic desktop applications and online tools. In this type of bioinformatic tools, all the analysis steps (Figure 1) are encapsulated into one application, which is used as a black box, with little understanding from users on the intermediate analysis steps. Most of these tools perform the whole data analysis on a single server machine, from raw data through protein identification and quantitation and on to statistical analysis of the results. From a bioinformatics perspective there are clear disadvantages with this approach: lack of flexibility, lack of transparency due to closed source code in some cases and scalability issues (both for desktop-based Windows software and online tools) for large datasets. We list here a few emblematic and popular software tools that illustrate these points, hampering the scalability and reproducibility of data analysis in metabolomics and proteomics.
3.1. MaxQuant (Proteomics)
MaxQuant [28] is one of the most frequently used platforms for mass-spectrometry (MS)-based proteomics data analysis. The platform includes a database search engine (Andromeda) to perform the peptide identification and a set of algorithms and tools designed for quantitative label-free proteomics, MS1-level labelling, and isobaric labelling techniques. In 2013, the MaxQuant team published a detailed documentation of the running time and input/output operations for each step of the analysis [29]. The results showed bottlenecks in overall performance and time-consuming algorithms related to peptide features detection in the MS1 data as well as the fragment spectrum search. The MaxQuant algorithms are efficiently parallelized on multiple processors and scale well from desktop computers to a server with many cores. However, all the framework and algorithms have been designed as a monolithic tool where all the steps of the data processing cannot be easily distributed in HPC or Cloud architectures [29].
3.2. Skyline (Proteomics)
Skyline [25] is an open source platform for targeted and data-independent proteomics and metabolomics data analysis. It runs on Microsoft Windows and supports the raw data formats from multiple mass spectrometric vendors. It contains a graphical user interface to display chromatographic data for individual peptide or small molecule analytes. Skyline supports multiple workflows, including selected reaction monitoring (SRM) / multiple reaction monitoring (MRM), parallel reaction monitoring (PRM), data-independent acquisition (DIA/SWATH) and targeted data-dependent acquisition. Because both SRM and DIA data are based on the analysis of MS/MS chromatograms (selected and extracted respectively), the processing (chromatogram peak integration) and visualization of data acquired using these two methods are very similar within Skyline. In a recent publication, the Skyline team has recognized that one of the areas to work in the future is the parallelization and distribution of computation and processing in HPC and cloud architectures [30]. These developments will be vital in obtaining the robust, sensitive quantitative measurements required to understand better the systems biology of cells, organisms, and disease states.
3.3. XCMS Online (Metabolomics)
XCMS-2 [31] has become arguably the most widely used free software tool for pre-processing untargeted metabolomics data. XCMS-2 is publicly available software that can be used within the R statistics language. It can provide structural information for unknown metabolites. This “similarity search” algorithm has been developed to detect possible structural motifs in the unknown metabolite which may produce characteristic fragment ions and neutral losses to related compounds contained in reference databases, even if the precursor masses are not the same.
In 2017, Weber and co-workers conducted a survey [32] on software data usage in metabolomics and found that LC-MS data analysis in metabolomics is performed in 84% of the cases using open-source tools. The predominant open-source software is XCMS (70%), followed by Mzmine and MZmine2 (26%). Interestingly, most of the usage of XCMS is through the Online XCMS Portal (https://xcmsonline.scripps.edu/), a popular Web application that helps the user to go through each step of the data analysis. XCMS Online directly searches the experimental mass spectra into METLIN online data using the traditional precursor ion selection window and additionally a distance matrix score to obtain good spectral matches. While the Online version of XCMS cannot be used in a component-based manner, the underlying XCMS R package can be executed by parts. For the reader interested in the wider landscape of Metabolomics tools and their usage in different scenarios, Spicer et. al [6] provides guidance.
4. Moving from desktop applications to distributed HPC and cloud architectures: A proposal for developers and bioinformaticians
The field of MS-based computational proteomics and metabolomics heavily relies on monolithic desktop applications or online tools, which prevents the analysis of large amounts of data in HPC clusters and cloud architectures. In order to overcome these problems, four areas of software engineering and algorithm development need to be adopted soon in computational MS-based proteomics and metabolomics: I) component-based and modularized applications; II) standard practices for tool packaging and deployment, III) standardised input/output file formats for exchange between tools and iv) workflow systems.
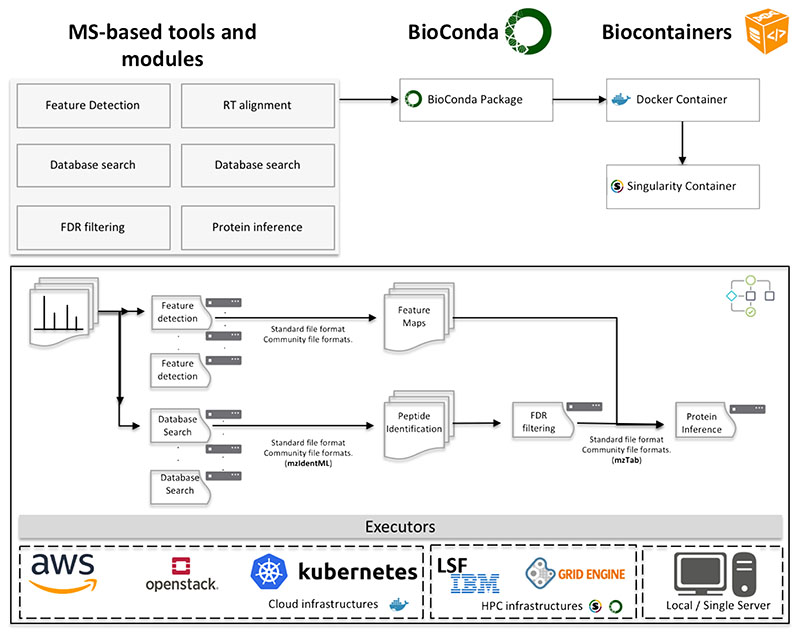
Figure 2 shows a proposal for the next generation of computational proteomics tools and algorithms. Each of the data processing step should be designed as an independent module component, that can be executed in an independent computing node. Multiple tools and modules can be packaged into the same framework but all of them should be executable independently and interchangeably (through adequate exchange file formats). A component-based software framework is a branch of software development that emphasizes the separation of concerns with respect to the wide-ranging functionality available throughout a given software system. The component-based development allows the data analyst to replace and substitute components of the workflow by new tools or a new version of the same tool without impacting the development process or the larger structure of the workflow.
Figure 2.
The proposed roadmap to scale metabolomics and proteomics data analysis includes the packaging and containerization of the specific tool and software using BioConda and BioContainers. The design of bioinformatics workflows that use the specific containers and abstract the execution from the compute environment (e.g. Cloud or HPC). A very important step of this design is the use of standard file formats that enable to communicate different tools and steps of the workflow.
4.1. Examples of component-based applications
In MS-based proteomics and metabolomics at least three popular frameworks have been designed as component-based frameworks: OpenMS (Proteomics and Metabolomics) [33], Trans-proteomics pipeline (Proteomics) [34], and MzMine (Metabolomics) [35]. OpenMS and OpenSWATH provide a set of computational tools which can be easily combined into analysis pipelines even by non-experts and can be used in proteomics workflows. These applications range from useful utilities (file format conversions, peak picking) to wrappers for known applications like peptide identification search engines. These two frameworks have been used recently to analyse big datasets [36].
MZmine is an open-source software toolbox for LC-MS metabolomics data processing, including different tools for data processing (e.g. peak noise detection) and visualization. In 2010, a critical assessment of the tool detected that MZmine was a build in a monolithic design, thus limiting the possibility of expanding the software with new methods developed by the scientific community. MZmine2 was built in multiple data processing modules, with an emphasis on usability and support for high-resolution spectra processing. MZmine2 includes the identification of peaks using online databases, MSn data support, improved isotope pattern support, scatter plot visualization.
These two frameworks (OpenMS, Mzmine), allow the bioinformaticians and developers to execute each step of the data analysis independently. For example, OpenMS allows to execute the identification step using MSGF+ tool with the command: MSGFPlusAdapter -iniid.config -database database.fasta -in run1.mzML -out run1.idXML, the following command can be parallelised and run simultaneously N MSRun in a distributed system with M compute nodes. Although these tools have been fully implemented as component-based frameworks, they have been less successful at using standard file formats between each component.
4.2. Standard file formats for better compatibility between components
Standard file formats enable the usage of a common persistence (e.g. file) representation of the data that is analysed (e.g. spectra, peptides). The proposed approach in Figure 2 reduces the need for components to translate from one file format to another, diminishing input-output (IO) and data transformation operations. Standard file formats enable the development of new tools based on a common representation, making the input and output parameters of tools common to any software component.
The Human Proteome Organization (HUPO) and the Proteomics Standards Initiative (PSI) have developed for 15 years file formats and common representations for MS-based proteomics and metabolomics data, from the spectra to protein expression [37]. For mass spectra, the mzML specification is the most stable, robust and mature file format, representing not only the MS/MS signal, but also chromatograms and instrument metadata [38]. For peptide/protein identification results, the mzIdentML file format not only captures the peptide and protein identifications, but also the software metadata (e.g. FDR thresholds, search parameters) used to perform the analysis [39]. Finally, mzTab file format stores the information of quantitative data for proteomics and metabolomics experiments [40].
Despite advances in the past years, standard file formats need time to evolve and consolidate. The software development process shouldn't be slower because of the development of a specific file format. Our recommendation is to replace and reuse existing file formats when possible and avoid the creation of new ones. A good example of these efforts is the peptide search engine MSGF+, that natively use mzIdentML and has been extensively used in open source workflows. A major problem in the use of standardised workflows for metabolomics and proteomics in the field of mass spectrometry is the lack of intermediate exchange formats, compared for instance to the existing formats in genomics (such as BAM, SAM, CRAM, VCF, bed, etc. to name a few). Often in younger fields like metabolomics and proteomics, tools will generate results in ad-hoc formatted files with poor specifications and often incompatible with downstream tools that would naturally pipe. This means that further tailored conversion steps are required; this slows down development, requires more maintenance, and can introduce errors or data loss.
4.3. Packaging and deployment using BioContainers
A component-based architecture like the one proposed in this manuscript (Figure 2) prompts multiple challenges in deployment and execution. Moving from single desktop applications to distributing data analysis and complex workflow systems create major challenges for the bioinformatics community: I) software availability, II) results reproducibility and III) automated software/environment deployment. Component-based pipelines require substantial effort for correct installation and configuration (e.g. conflicting dependencies, different base libraries, etc.). In addition, versioning components, key for reproducing the results of the analysis, are a burden to scientific software development groups, who are less used to proper software engineering standards.
Recently, containers and packaging technologies such as Conda (http://conda.io), Docker (http://www.docker.com) and Singularity (https://www.sylabs.io/) have emerged to overcome these challenges by automating the deployment of analysis tools inside so-called software containers. The BioContainers community [8] (http://biocontainers.pro) has created a complete architecture and solution to overcome these challenges based on community-driven BioConda packages [10] and Docker containers.
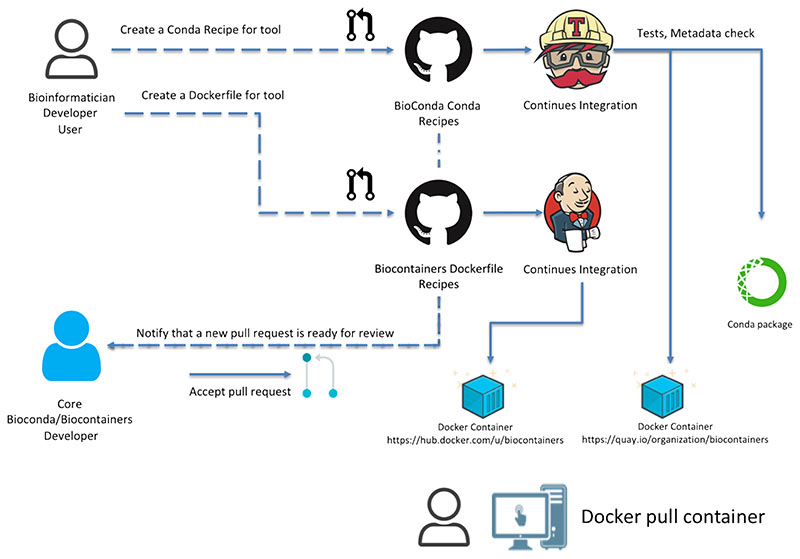
The BioContainers community has defined a set of guidelines on how to create, deploy and maintain containers (Figure 3) [9]. Each component (software tool) developer can create a Conda recipe (a set of yaml and bash scripts which describe how to consistently install a software package on Linux) or a Docker build recipe (Dockerfile), which are all stored in GitHub. Each new contribution (or recipe) is accepted through a Pull Request (PR) mechanism [41]. Pull Requests provide the opportunity for an open revision and improvement step of newly contributed or updated recipes, so that they are up to standards. This revision is done by members of the bioinformatics community that are granted permissions, reducing the burden on a small group of maintainers, which makes the model sustainable. After the PR is merged and the recipe added to the repository with a well-defined version, a continuous-integration system triggers the creation of the Conda package (in the case of a Conda recipe) and of the corresponding Docker and Singularity containers tagged at that version. Historic versions of the same package are stored both as Conda packages and containers, guaranteeing future reproducibility of older pipelines that use earlier versions of a tool. BioContainers enables users to create multi-tools containers/packages, where multiple previously packaged tools can be combined into a single container or package (https://github.com/BioContainers/multi-package-containers).
Figure 3.
BioContainers architecture from the container request by the user in GitHub to the final container deposited in DockerHub (https://hub.docker.com/u/biocontainers) and Quay.io (https://quay.io/organization/biocontainers). The BioContainers community in collaboration with the BioConda community defines a set of guidelines and protocols to create a Conda and Docker container including mandatory metadata, tests and trusted images [9]. The proposed architecture uses a continuous integration system (CI) to test and build the final containers and deposit them into public registries. All the Containers and tools can be searched from the BioContainers registry (http://biocontainers.pro/regitry).
The created package and containers include all software dependencies needed to execute the tool in question. In general, one package will contain only one tool; larger packages containing many tools are in general discouraged. This allows the end user to execute the pipeline in different compute environments, without the complexity of installation, dependency management, etc. It also makes the pipeline portable from one environment to another (e.g. HPC, Cloud or local personal computer), because everything is executed in containers. At the time of writing, BioContainers provides more than 7,000 bioinformatics containers that can be searched, tagged and accessed through a common web registry (https://biocontainers.pro/#/registry/). Importantly, the BioContainers and BioConda communities convert automatically Bioconductor packages automatically into containers.
4.4. Using workflow systems
High throughput bioinformatic genomics and transcriptomics analyses increasingly rely on pipeline frameworks to process sequence and metadata. Until recently, this was not the case for Proteomics and metabolomics, where most of the the analysis happened on single desktop machines. Modern workflows systems such as Galaxy [12] , Cromwell, CWL tool, Toil [42], Nextflow [13] or Snakemake [14] are playing an important role in porting the data analysis steps from the single desktop applications into distributed compute platforms [11]. Most of these workflow engines provide four major functionalities for data processing: I) execution in distributed architectures (HPC, Cloud); II) separation between the execution environment and workflow design; III) recovery/restart mechanisms for failed components and tasks; iv) support for automatic tool dependency resolution using Conda, Docker or Singularity technologies. The automatic tool dependency resolution (using packaging systems) allows developers and bioinformaticians to execute the workflows and pipelines without the need to install and configure each tool manually in the desired execution environment. Two different workflow systems receiving attention from the Bioinformatics community are NextFlow [13] and Galaxy [12].
4.4.1. NextFlow
NextFlow (https://www.nextflow.io/), an expressive, versatile and particularly comprehensive framework for composing and executing workflows. NextFlow uses a domain-specific language (DSL) which also supports the full syntax and semantics of Groovy, a dynamic language that runs on the Java platform. One of the features that make NextFlow a powerful workflow engine is its dataflow functionality. Nextflow allows users within the workflow definition to filter data, run processes conditionally on data value or have splitting/merging pipeline steps expressed in a short, elegant syntax.
Nextflow separates the workflow definition from the execution environment, which allows users to execute the same workflow in different architectures (Cloud, HPC or a local machine). This abstraction level is guaranteed by using an execution layout that defines which type of containers will be used to execute the tools (components of the workflow) and which type of architecture will be used to execute those containers (e.g. HPC, Cloud). Currently, Nextflow supports the following technologies: Conda, Docker and Singularity; and the following execution environments: Local (the software run in single node, server), HPC clusters including (Sun Grid Engine, IBM LSF, PBS/Torque, HTCondor) and cloud providers, such as Amazon Web services (through AWS batch) or Google Cloud Platform (see full list here: https://www.nextflow.io/docs/latest/executor.html). This execution layout can be configured with workflow variables which enable to switch between architectures with no hassle.
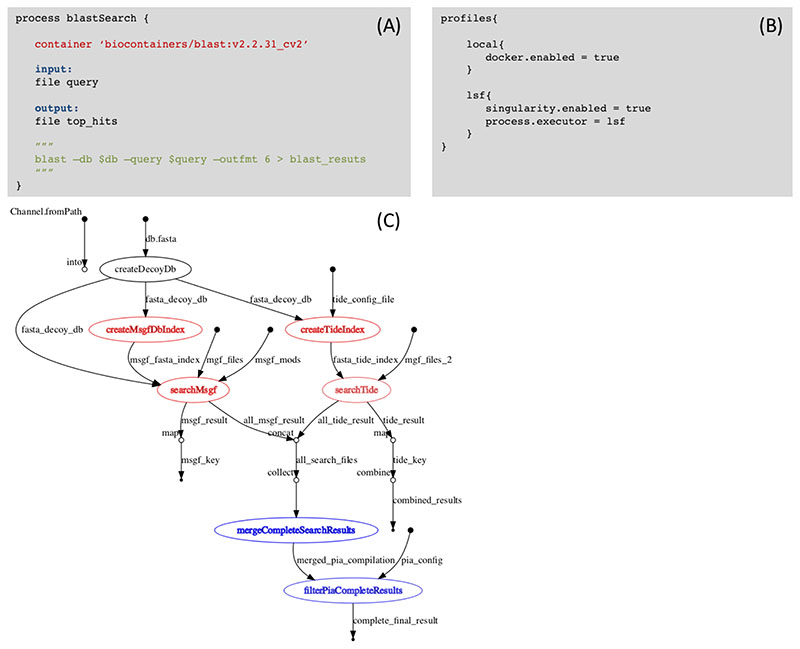
Figure 4 shows two different examples of Nextflow workflows: I) a single step workflow to perform sequence alignment using Blast (container https://biocontainers.pro/#/tools/blast) (Figure 4A-B) and II) a peptide and protein identification workflow (Figure 4C). Nextflow allows bioinformaticians to perform analysis in different architectures with the same workflow definition (https://www.nextflow.io/docs/latest/basic.html). Each step of the workflow (called process) describes which process will be performed and the input/output parameters. The container section inside the process (blastSearch) states which containers will be used; including container name (blast), version of the container (v2.2.31_cv2). Between triple quotes is the actual command which will be executed in the container (in this case blast). This is needed because one container can provide multiple tools. Finally, the Nextflow config file (https://www.nextflow.io/docs/latest/config.html) defines how the present workflow will be executed. In the example, we have defined two possible scenarios: local and lsf. If the user runs the workflow using the local configuration (command - nextflow workflow.nf -c config.nf -profile local) it will be using BioContainers Docker containers, if the user uses lsf (command - nextflow workflow.nf -c config.nf -profile lsf), then will be using Singularity BioContainers and the LSF cluster executor. Figure 4C shows the directed acyclic graph of a more complex workflow (https://github.com/bigbio/nf-workflows/tree/master/xt-msgf-nf) where two different database search engines (MSGF+ and Tide) are combined to identify more peptides and proteins. In red all the processes that can be execute in parallel for different input files (e.g. mgf files). Each of the identification process can be sent with an MS/MS spectra file to different compute node at the same time, reducing execution time of the analysis. Finally, in blue, the process that aggregate the results of multiple parallel process (e.g. protein inference step, mergeCompleteSearchResults using PIA tool [21,43] ).
Figure 4. Nextflow allows bioinformaticians to perform analysis in different architectures with the same workflow definition.
(A) The workflow step (called process) describes which process will be performed and the input/output parameters. The container section inside the blastSearch process state which containers will be use; including container name (blast), and version of the container (v2.2.31_cv2). Between triple quotes is the actual command will be executed in the container (in this case blast). This is needed because one container can provide multiple tools. (B) The Nextflow config file (https://www.nextflow.io/docs/latest/config.html) defines how the present workflow (A) will be executed. In the example, we have defined two possible scenarios: local and lsf. If the user runs the workflow using the local configuration it will be using Docker containers, if the user uses lsf, then it will be using singularity and the LSF cluster executor. (C) Directed Acyclic Graph for a peptide and protein identification workflow in Nextflow (https://github.com/bigbio/nf-workflows/tree/master/xt-msgf-nf).
In addition, it provides configuration variables to customize the computer/hardware that is required to perform each task. For example, the user can customize the type of node (Memory, number of cores) that is needed for each specific task (component tool). Another important feature of NextFlow is the simplicity of the language syntax and the support of workflow versioning, which enables better reproducibility.
4.4.2. Galaxy Project
Galaxy (https://galaxyproject.org/) is a web application workflow environment written in Python, capable of distributing jobs among a plethora of batch schedulers (PBS, LSF, GoDocker, DRMAA based schedulers, etc), local machine (through containers or conda) and cloud providers (through Kubernetes [44] and others). In the HPC case, Galaxy provides the flexibility to use either containers (Docker or Singularity) or directly Conda packages. Galaxy tool wrappers are written to point to specific package versions, for reproducibility. Galaxy provides a complete separation of concerns between the workflow logic definition and the actual execution. Both tools and workflows are versioned in Galaxy (and multiple versions of a tool can be installed on the same instance).
Galaxy provides tool/workflow repositories, called Toolsheds (such as https://toolshed.g2.bx.psu.edu/), where users can deposit their own tools and find existing ones. Currently, more than 6,000 tools wrapped for Galaxy are available there. From a Galaxy Toolshed, users can automatically install desired versions of available Galaxy wrappers to their own Galaxy instance. Tool’s dependencies are resolved automatically by Galaxy using either Conda packages, Docker or singularity containers, depending on setup. On the same workflow, different tools can be sent to different underlying executors and rely on different dependency resolution as well.
Besides a rich and responsive user interface (UI), Galaxy enables operations through a mature REST API, Python clients (e.g. bioblend, ephemeris) and command line interface (parsec), to programmatically control the execution of tools/workflows and data upload/downloads.
Many organizations provide computing power to end users in the need of doing biological data analysis through public Galaxy instances -- in the region of 100 public instances exist today (https://galaxyproject.org/use/) -- which are normally flavoured for different research topics (examples in Table 1). Notable instances in terms of computational power are usegalaxy.org (http://usegalaxy.org) and usegalaxy.eu (http://usegalaxy.eu). Galaxy is organized in different initiatives that help to download and deploy complete solutions of galaxy tools for a specific field (e.g. Proteomics).
Table 1. Collection of public Nextflow and Galaxy workflows for proteomics and metabolomics data analysis.
| Workflow | Description | Main tools | Link |
|---|---|---|---|
| Metabolomics | processing, quantification, annotation, identification and statistics | XCMS, OpenMS, CAMERA, MetFrag, W4M Multivariate and Univariate Statistical Analysis tools | https://public.phenomenal-h2020.eu/u/phenoadmin/w/metabolomics-lcmsms-processing-quantification-annotation-identification-and-statistics-1 |
| mzQuality | Quality Control for MS data. | ms-vetfc | https://public.phenomenal-h2020.eu/u/phenoadmin/w/mzquality |
| Eco-metabolomics | MS Metabolomics for Ecology applications | Ecomet | https://public.phenomenal-h2020.eu/u/phenoadmin/w/eco-metabolomics-workflow |
| Fluxomics stationary 13C-MS iso2flux | Steady state fluxomics based on tracer data. | Iso2flux, Escher metabolic pathway viewer. | https://public.phenomenal-h2020.eu/u/phenoadmin/w/fluxomics-stationary-13c-ms-iso2flux-with-visualization |
| LC-MS XCMS 3.0 | MS Analysis with peak annotation | XCMS, CAMERA | https://galaxy.workflow4metabolomics.org/u/lecorguille/w/lcms-new-workflow-xcms-300 |
| ProteoGenomics | Proteogenomics analysis | CustomProDB, PeptideShaker, SearchGUI, msconvert, MSGF+ | http://galaxyp-proteogenomics.duckdns.org/ |
| Metaproteomics | Galaxy Metaproteomics | metaQuantome, Unipept, MetaProteomeAnalyzer | http://z.umn.edu/metaproteomicsgateway |
| IPAW Proteogenomics | ProteoGenomics workflow in Nextflow | OpenMS | https://github.com/lehtiolab/proteogenomics-analysis-workflow |
| Peptide and protein quantification workflow | OpenMS LFQ workflow | OpenMS | https://github.com/nf-core/mhcquant |
| ProteoGenomics Database (pgdb) | Nextflow for Custom proteogenomics database creation | pypgatk | https://github.com/bigbio/pgdb/ |
The Galaxy-P (Galaxy for Proteomics - http://galaxyp.org/) initiative provides workflows and tools in the fields of proteogenomics and metaproteomics (http://galaxyp.org/access-galaxy-p/). PhenoMeNal [45] (https://public.phenomenal-h2020.eu/), Galaxy-M [46], and Workflow4Metabolomics [47] (https://workflow4metabolomics.org/) are the most complete compendium of tools and workflows available in Galaxy for metabolomics researchers. Figure 5 shows an example workflow from the PhenoMeNal project for the analysis of LC-MS/MS data. This workflow combines different open source tools (XCMS, CAMERA, msnbase, MetFrag) in a component-based manner. All the PhenoMeNal workflows can be found directly in the PhenoMeNal link shown above. Table 1 shows a list of Metabolomics workflows from PhenoMeNal and Workflows4Metabolomics (to execute them, registration is needed).
Figure 5. A Galaxy workflow from PhenoMeNal H2020, used for processing LC-MS/MS data. It integrates XCMS, CAMERA, msnbase and MetFrag for matching detected fragments to potential small molecules.
Each box represents a relevant tool step and is backed by a container that can execute that process (all CAMERA steps rely of course on the same CAMERA container). Plumbing such a pipeline through a scripting language would require considerable work, including any additional logic to execute on a cluster. In the case shown here, this workflow was created by dragging and dropping tools in Galaxy, and there was no need for the analyst to be concerned about how the workflow environment is actually distributing this on a large computational infrastructure: handling the cluster and the workflow become independent and the analyst can focus on the flow of the software tools within the pipeline. The Galaxy workflow can be shared as a single file, to be imported into other Galaxy instances.
5. Towards reproducible data analysis
Reproducibility is challenging in life sciences, especially in computationally intensive domains (e.g. proteomics and metabolomics) where results rely on a series of complex analytical and bioinformatics steps that are not well captured by traditional publication approaches. While there are now several guidelines and platforms to enable reproducibility in computational biology [41, 48, 49], the approach we describe here is flexible, robust and scalable enough to guarantee the features for reproducibility research: I) managing software dependencies, II) separation between the data flow design and the execution environments, and III) virtualizing entire analyses for complete portability and preservation against time [48].
The use of BioConda and BioContainers as independent components in data analysis resolves the problem of complex software dependencies. In addition, it provides a mechanism to easily replace independent components from different technologies and programming languages (e.g. python by R package). The use of workflows in combination with container technologies allows researchers to reproduce data analysis on their own compute architecture (e.g. local PC or cloud). BioConda and BioContainers provide consistently versioned tool packages and containers, allowing users to reproduce data analysis over time.
The route to reproducibility in Galaxy is through the deposition of Galaxy tool wrappers in the Galaxy Toolshed and sharing of workflows either through GitHub or the same Toolshed. Tools and workflows available there can be installed on any Galaxy instance, and their software dependencies will be resolved automatically by the instance to either conda packages or containers, facilitating re-runs of the same workflows in different infrastructures. The deposition of modules into the Toolshed can be done either directly to the Toolshed or through a Pull Request against the IUC Tools GitHub repository (https://github.com/galaxyproject/tools-iuc/), where it will undergo a review process by one or more of the many Galaxy community members, to meet adequate standards. The latter route has the advantage, besides the review process, that the tool is left in a repository that is not owned by the original contributor, meaning that the tool can be updated in the future without necessarily needing the attention of that developer.
Finally, in order to complement the software efforts made by the BioConda and BioContainers communities, we urge software developers in the metabolomics and proteomics communities to embrace standard file formats for input and output of their software and component tools. Standard file formats not only enable better interoperability between software components, but also improve the reproducibility of the analysis [50].
6. Conclusions
Proteomics and metabolomics mass spectrometry are moving from desktop application data analysis to distributed architectures (HPC and Cloud), due to larger datasets being generated (more sample, more replicates, higher coverage, more resolution, etc). However, the most popular software used in the field, such as Skyline, MaxQuant, ProteomeDiscover and XCMS Online, are mainly developed as monolithic tools, hampering the scale up of individual steps of the analysis into distributed architectures. First, the software development and algorithmic paradigms should be changed by decoupling monolithic applications into smaller components (tasks) that can be distributed on Cloud and HPC architectures. We recommend that each of these small components support standard file formats for inputs and outputs, towards facilitating the exchange of steps in data analysis pipelines.
We presented a future paradigm for proteomics and metabolomics large scale data analysis based on BioContainers, BioConda, Docker/Singularity containers and workflow engines such as Galaxy and Nextflow. The proposed model starts through the creation of a Conda or Dockerfile recipe in BioContainers, providing the mandatory metadata and dependencies to build the container. The new recipe is built using continuous integration, where theBioContainers architecture will check the metadata, tests and push the final containers intoBioContainers public registries and packages to BioConda. The Conda-based containers are deployed to Quay.io (https://quay.io/organization/biocontainers) and the Dockerfile-based are deployed to DockerHub (https://hub.docker.com/u/biocontainers). All containers are searchable, and discoverable through the BioContainers tool registry (http://biocontainers.pro/#/registry/).
Finally, we recommend the proteomics and metabolomics community to embrace the development of bioinformatics workflows and gradually move bioinformatics pipelines and data analysis into workflow environments such as Nextflow and Galaxy. The combination of workflow environments and BioContainers will enable more reproducible and scalable metabolomics and proteomics data analysis.
Acknowledgments
Y.P.R is supported by BBSRC Grant “ProteoGenomics” [BB/L024225/1]. We would like to thanks to ELIXIR and the ELIXIR Tools and Compute platforms for the support to BioContainers Community and Architecture BioContainers
Abbreviations
- AWS
Amazon web services
- CWL
Common workflow language
- DDA
Data-dependant acquisition
- DIA
Data-independent acquisition
- DSL
Domain-specific language
- FDR
False discovery rate
- HPC
High-performance computer
- HPCS
High-performance computing systems
- HUPO
Human Proteome Organization
- IO
Input-output
- MRM
Multiple reaction monitoring
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- LC-MS
Liquid chromatography–mass spectrometry
- LSF
IBM Platform LSF
- PR
Pull request
- PRM
Parallel reaction monitoring
- PSI
Proteomics Standards Initiative
- REST API
Representational State Transfer programming interface
- SRM
Selected reaction monitoring
References
- [1].Griffiths WJ, Wang Y. Chem Soc Rev. 2009;38:1882. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- [2].Perez-Riverol Y, Bai M, da Veiga Leprevost F, Squizzato S, Park YM, Haug K, Carroll AJ, Spalding D, Paschall J, Wang M, Del-Toro N, et al. Nat Biotechnol. 2017;35:406. doi: 10.1038/nbt.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lynch C. Nature. 2008;455:28. doi: 10.1038/455028a. [DOI] [PubMed] [Google Scholar]
- [4].Kanwal S, Khan FZ, Lonie A, Sinnott RO. BMC bioinformatics. 2017;18:337. doi: 10.1186/s12859-017-1747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Perez-Riverol Y, Wang R, Hermjakob H, Muller M, Vesada V, Vizcaino JA. Biochim Biophys Acta. 2014;1844:63. doi: 10.1016/j.bbapap.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Spicer R, Salek RM, Moreno P, Canueto D, Steinbeck C. Metabolomics. 2017;13:106. doi: 10.1007/s11306-017-1242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang R, Perez-Riverol Y, Hermjakob H, Vizcaino JA. Proteomics. 2015;15:1356. doi: 10.1002/pmic.201400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].da Veiga Leprevost F, Gruning BA, Alves Aflitos S, Rost HL, Uszkoreit J, Barsnes H, Vaudel M, Moreno P, Gatto L, Weber J, Bai M, et al. Bioinformatics. 2017;33:2580. doi: 10.1093/bioinformatics/btx192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].n. null. Gruening B, Sallou O, Moreno P, da Veiga Leprevost F, MÈnager H, Søndergaard D, Röst H, Sachsenberg T, O’Connor B, Madeira F, Dominguez Del Angel V, et al. F1000Research. 2018;7 doi: 10.12688/f1000research.15140.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gruning B, Dale R, Sjodin A, Chapman BA, Rowe J, Tomkins-Tinch CH, Valieris R, Koster J, Bioconda T. Nature methods. 2018;15:475. doi: 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leipzig J. Brief Bioinform. 2017;18:530. doi: 10.1093/bib/bbw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, et al. Nucleic acids research. 2018;46:W537. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nature biotechnology. 2017;35:316. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- [14].Koster J, Rahmann S. Bioinformatics. 2012;28:2520. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- [15].Schmidt A, Forne I, Imhof A. BMC Syst Biol. 2014;8(Suppl 2):S3. doi: 10.1186/1752-0509-8-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Griss J, Perez-Riverol Y, Lewis S, Tabb DL, Dianes JA, Del-Toro N, Rurik M, Walzer MW, Kohlbacher O, Hermjakob H, Wang R, et al. Nature methods. 2016;13:651. doi: 10.1038/nmeth.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. J Proteome Res. 2011;10:1794. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- [18].Kim S, Pevzner PA. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. Nature methods. 2017;14:513. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Borges D, Perez-Riverol Y, Nogueira FC, Domont GB, Noda J, da Veiga Leprevost F, Besada V, Franca FM, Barbosa VC, Sanchez A, Carvalho PC. Bioinformatics. 2013;29:1343. doi: 10.1093/bioinformatics/btt106. [DOI] [PubMed] [Google Scholar]
- [21].Uszkoreit J, Maerkens A, Perez-Riverol Y, Meyer HE, Marcus K, Stephan C, Kohlbacher O, Eisenacher M. J Proteome Res. 2015;14:2988. doi: 10.1021/acs.jproteome.5b00121. [DOI] [PubMed] [Google Scholar]
- [22].Audain E, Uszkoreit J, Sachsenberg T, Pfeuffer J, Liang X, Hermjakob H, Sanchez A, Eisenacher M, Reinert K, Tabb DL, Kohlbacher O, et al. J Proteomics. 2017;150:170. doi: 10.1016/j.jprot.2016.08.002. [DOI] [PubMed] [Google Scholar]
- [23].Koopmans F, Ho JTC, Smit AB, Li KW. Proteomics. 2018;18 doi: 10.1002/pmic.201700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT, Wolski W, Collins BC, Malmstrom J, Malmstrom L, Aebersold R. Nat Biotechnol. 2014;32:219. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- [25].MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26:966. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fischer R, Bowness P, Kessler BM. Proteomics. 2013;13:3371. doi: 10.1002/pmic.201300192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsugawa H. Current opinion in biotechnology. 2018;54:10. doi: 10.1016/j.copbio.2018.01.008. [DOI] [PubMed] [Google Scholar]
- [28].Tyanova S, Temu T, Cox J. Nat Protoc. 2016;11:2301. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- [29].Neuhauser N, Nagaraj N, McHardy P, Zanivan S, Scheltema R, Cox J, Mann M. J Proteome Res. 2013;12:2858. doi: 10.1021/pr400181q. [DOI] [PubMed] [Google Scholar]
- [30].Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, MacCoss MJ. Mass Spectrom Rev. 2017 doi: 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Benton HP, Wong DM, Trauger SA, Siuzdak G. Analytical chemistry. 2008;80:6382. doi: 10.1021/ac800795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Weber RJM, Lawson TN, Salek RM, Ebbels TMD, Glen RC, Goodacre R, Griffin JL, Haug K, Koulman A, Moreno P, Ralser M, et al. Metabolomics. 2017;13:12. doi: 10.1007/s11306-016-1147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pfeuffer J, Sachsenberg T, Alka O, Walzer M, Fillbrunn A, Nilse L, Schilling O, Reinert K, Kohlbacher O. J Biotechnol. 2017;261:142. doi: 10.1016/j.jbiotec.2017.05.016. [DOI] [PubMed] [Google Scholar]
- [34].Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, et al. Proteomics. 2010;10:1150. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pluskal T, Castillo S, Villar-Briones A, Oresic M. BMC bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].N. Cancer Genome Atlas Research. Kahles A, Lehmann KV, Toussaint NC, Huser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C, Ratsch G. Cancer Cell. 2018;34:211. [Google Scholar]; Peters S, Hains PG, Lucas N, Robinson PJ, Tully B. Journal of proteome research. 2019;18:1019. doi: 10.1021/acs.jproteome.8b00709. [DOI] [PubMed] [Google Scholar]
- [37].Deutsch EW, Orchard S, Binz PA, Bittremieux W, Eisenacher M, Hermjakob H, Kawano S, Lam H, Mayer G, Menschaert G, Perez-Riverol Y, et al. Journal of proteome research. 2017;16:4288. doi: 10.1021/acs.jproteome.7b00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Rompp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, et al. Molecular & cellular proteomics : MCP. 2011;10:R110 000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vizcaino JA, Mayer G, Perkins S, Barsnes H, Vaudel M, Perez-Riverol Y, Ternent T, Uszkoreit J, Eisenacher M, Fischer L, Rappsilber J, et al. Molecular & cellular proteomics : MCP. 2017;16:1275. doi: 10.1074/mcp.M117.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hoffmann N, Rein J, Sachsenberg T, Hartler J, Haug K, Mayer G, Alka O, Dayalan S, Pearce JTM, Rocca-Serra P, Qi D, et al. Analytical chemistry. 2019;91:3302. doi: 10.1021/acs.analchem.8b04310. [DOI] [PMC free article] [PubMed] [Google Scholar]; Griss J, Jones AR, Sachsenberg T, Walzer M, Gatto L, Hartler J, Thallinger GG, Salek RM, Steinbeck C, Neuhauser N, Cox J, et al. Molecular & cellular proteomics : MCP. 2014;13:2765. doi: 10.1074/mcp.O113.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Perez-Riverol Y, Gatto L, Wang R, Sachsenberg T, Uszkoreit J, Leprevost Fda V, Fufezan C, Ternent T, Eglen SJ, Katz DS, Pollard TJ, et al. PLoS computational biology. 2016;12:e1004947. doi: 10.1371/journal.pcbi.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, Schmidt H, et al. Nature biotechnology. 2017;35:314. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Uszkoreit J, Perez-Riverol Y, Eggers B, Marcus K, Eisenacher M. J Proteome Res. 2019;18:741. doi: 10.1021/acs.jproteome.8b00723. [DOI] [PubMed] [Google Scholar]
- [44].Moreno P, Pireddu L, Roger P, Goonasekera N, Afgan E, van den Beek M, He S, Larsson A, Ruttkies C, Schober D, Johnson D, et al. bioRxiv. 2018:488643 [Google Scholar]
- [45].Peters K, Bradbury J, Bergmann S, Capuccini M, Cascante M, de Atauri P, Ebbels TMD, Foguet C, Glen R, Gonzalez-Beltran A, Gunther UL, et al. Gigascience. 2019;8 doi: 10.1093/gigascience/giy149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Davidson RL, Weber RJ, Liu H, Sharma-Oates A, Viant MR. Gigascience. 2016;5:10. doi: 10.1186/s13742-016-0115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Giacomoni F, Le Corguille G, Monsoor M, Landi M, Pericard P, Petera M, Duperier C, Tremblay-Franco M, Martin JF, Jacob D, Goulitquer S, et al. Bioinformatics. 2015;31:1493. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gruning B, Chilton J, Koster J, Dale R, Soranzo N, van den Beek M, Goecks J, Backofen R, Nekrutenko A, Taylor J. Cell Syst. 2018;6:631. doi: 10.1016/j.cels.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cohen-Boulakia S, Belhajjame K, Collin O, Chopard J, Froidevaux C, Gaignard A, Hinsen K, Larmande P, Le Bras Y, Lemoine F. Future Generation Computer Systems. 2017;75:284. [Google Scholar]
- [50].Sandve GK, Nekrutenko A, Taylor J, Hovig E. PLoS computational biology. 2013;9:e1003285. doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]