SARS-CoV-2 across Africa
The impact of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has been hard to track in African countries, largely because of patchy data. Wilkinson et al. curated viral genomes collected in 2021 from several countries across the continent. Outbreaks during 2020 in each African country were initiated by imported cases, mostly from Europe. As the pandemic developed, case numbers in African countries were likely many times higher than reported, and subsequent waves of the pandemic appear to have been more severe. Consequently, high-transmission variants have emerged that have spread within the continent, and African countries must be included in global control efforts. —CA
The early phases of the COVID-19 pandemic in Africa were dominated by European imports, followed by the local generation of variants.
Abstract
The progression of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic in Africa has so far been heterogeneous, and the full impact is not yet well understood. In this study, we describe the genomic epidemiology using a dataset of 8746 genomes from 33 African countries and two overseas territories. We show that the epidemics in most countries were initiated by importations predominantly from Europe, which diminished after the early introduction of international travel restrictions. As the pandemic progressed, ongoing transmission in many countries and increasing mobility led to the emergence and spread within the continent of many variants of concern and interest, such as B.1.351, B.1.525, A.23.1, and C.1.1. Although distorted by low sampling numbers and blind spots, the findings highlight that Africa must not be left behind in the global pandemic response, otherwise it could become a source for new variants.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 in Wuhan, China (1, 2). Since then, the virus has spread to all corners of the world, causing almost 150 million cases of COVID-19 and more than 3 million deaths by the end of April 2021. Throughout the pandemic, it has been noted that Africa accounts for a relatively low proportion of reported cases and deaths—by the end of April 2021, there had been ~4.5 million cases and ~120,000 deaths on the continent, corresponding to less than 4% of the global burden. However, emerging data from seroprevalence surveys and autopsy studies in some African countries suggest that the true number of infections and deaths may be severalfold higher than reported (3, 4). In addition, a recent analysis has shown that in many African countries, the second wave of the pandemic was more severe than the first wave (5).
The first cases of COVID-19 on the African continent were reported in Nigeria, Egypt, and South Africa between mid-February and early March 2020, and most countries had reported cases by the end of March 2020 (6–8). These early cases were concentrated among airline travelers returning from regions of the world with high levels of community transmission. Many African countries introduced early public health and social measures, including international travel controls, quarantine for returning travelers, and internal lockdown measures, to limit the spread of the virus and give health services time to prepare (5, 9). The initial phase of the epidemic was then heterogeneous, with relatively high case numbers reported in North Africa and southern Africa, and fewer cases reported in other regions.
From the onset of the pandemic, genomic surveillance has been at the forefront of the COVID-19 response in Africa (10). Rapid implementation of SARS-CoV-2 sequencing by various laboratories in Africa enabled genomic data to be generated and shared from the early imported cases. In Nigeria, the first genome sequence was released just 3 days after the announcement of the first case (6). Similarly, in Uganda, a sequencing program was set up rapidly to facilitate virus tracing, and the collection of samples for sequencing began immediately upon confirmation of the first case (11). In South Africa, the Network for Genomic Surveillance in South Africa (NGS-SA) was established in March 2020, and within weeks, genomic analysis was helping to characterize outbreaks and community transmission (12).
Genomic surveillance has also been critical for monitoring ongoing SARS-CoV-2 evolution and detection of new SARS-CoV-2 variants in Africa. Intensified sampling by NGS-SA in the Eastern Cape Province of South Africa in November 2020, in response to a rapid resurgence of cases, led to the detection of B.1.351 (501Y.V2) (13). This variant was subsequently designated a variant of concern (VOC) by the World Health Organization (WHO), owing to evidence of increased transmissibility (14) and resistance to neutralizing antibodies elicited by natural infection and vaccines (15–17).
In this study, we performed phylogenetic and phylogeographic analyses of SARS-CoV-2 genomic data from 33 African countries and two overseas territories to help characterize the dynamics of the pandemic in Africa. We show that the early introductions were predominantly from Europe, but that as the pandemic progressed, there was increasing spread between African countries. We also describe the emergence and spread of a number of key SARS-CoV-2 variants in Africa and highlight how the spread of B.1.351 (501Y.V2) and other variants contributed to the more severe second wave of the pandemic in many countries.
SARS-CoV-2 genomic data
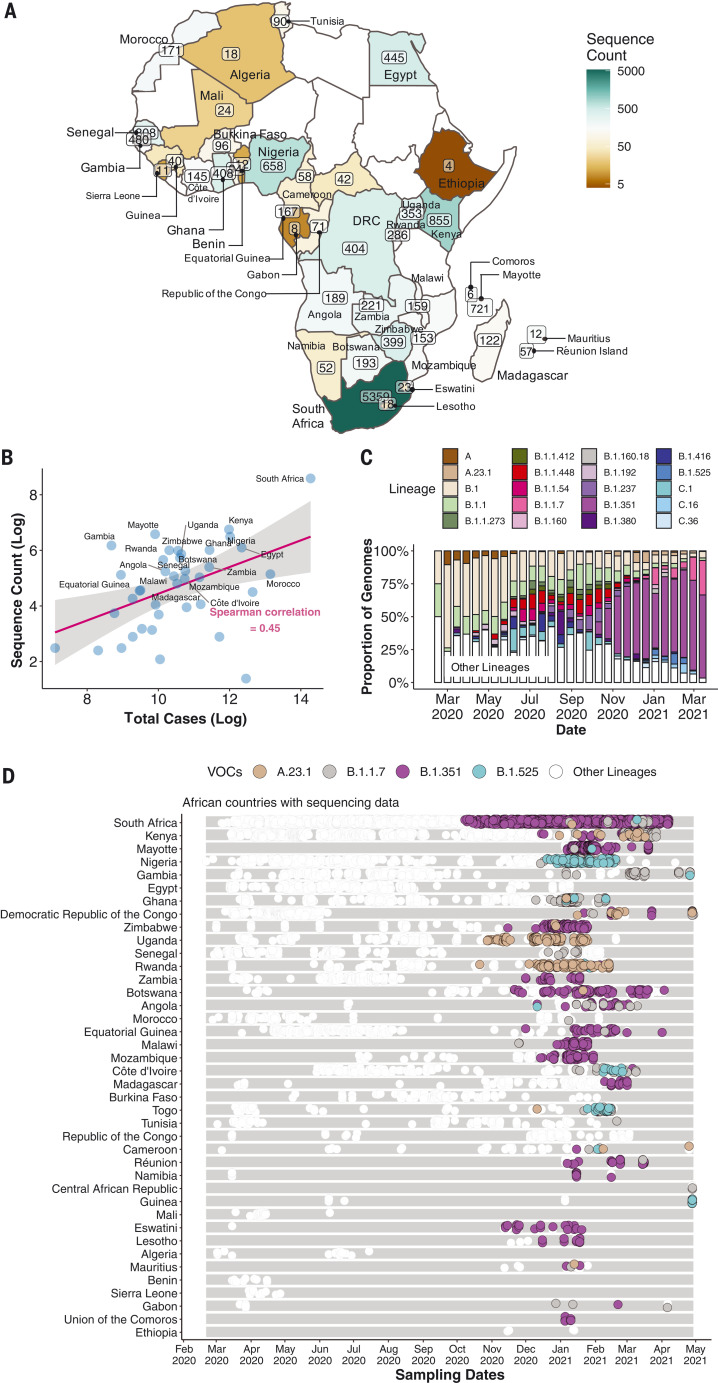
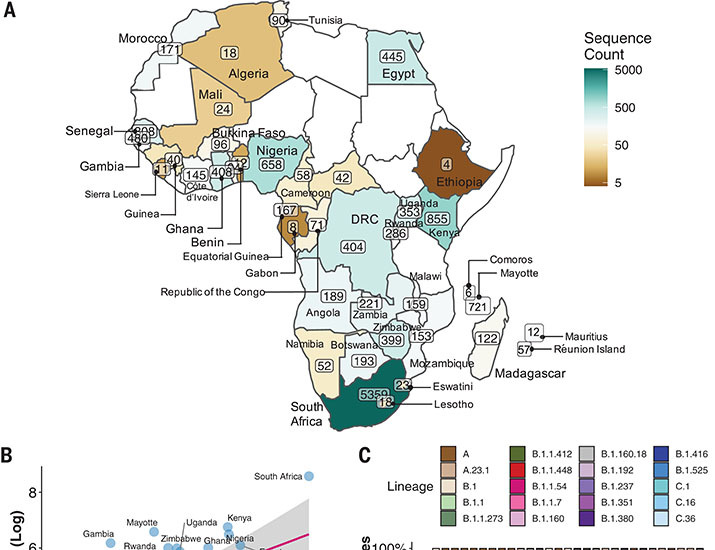
By 5 May 2021, 14,504 SARS-CoV-2 genomes had been submitted to the GISAID database (18) from 38 African countries and two overseas territories (Mayotte and Réunion) (Fig. 1A). Overall, this corresponds to approximately one sequence per ~300 reported cases. Almost half of the sequences were from South Africa (n = 5362), consistent with it being responsible for almost half of the reported cases in Africa. Overall, the number of sequences correlates closely with the number of reported cases per country (Fig. 1B). The countries and territories with the highest coverage of sequencing (defined as genomes per reported case) are Kenya (n = 856, one sequence per ~203 cases), Mayotte (n = 721, one sequence per ~21 cases), and Nigeria (n = 660, one sequence per ~250 cases). Although genomic surveillance started early in many countries, few have evidence of consistent sampling across the whole year. Half of all African genomes were deposited in the first 10 weeks of 2021, suggesting intensified surveillance in the second wave after the detection of B.1.351 (501Y.V2) and other variants (Fig. 1, C and D).
Fig. 1. SARS-CoV-2 sequences in Africa.
(A) Map of the African continent with the number of SARS-CoV-2 sequences reflected in GISAID as of 5 May 2021. (B) Regression plot of the number of viral sequences versus the number of reported COVID-19 cases in various African countries as of 5 May 2021. Countries with >500 sequences are labeled. The shaded region indicates the 95% confidence interval. (C) Progressive distribution of the top 20 PANGO lineages on the African continent. (D) Temporal sampling of SARS-CoV-2 sequences in African countries (ordered by total number of sequences) through time, with VOCs of note highlighted and annotated according to their PANGO lineage assignment.
Genetic diversity and lineage dynamics in Africa
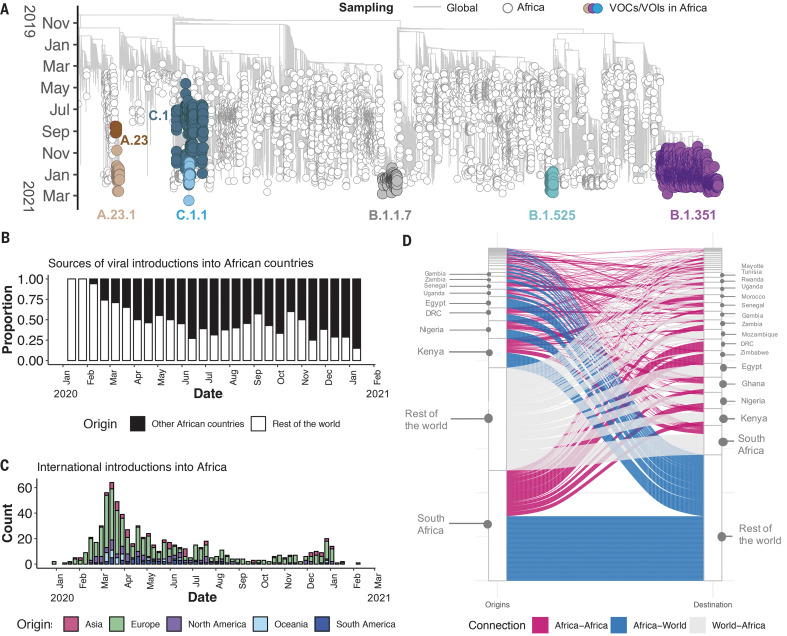
Of the 10,326 genomes retrieved from GISAID by the end of March 2021, 8746 genomes passed quality control and met the minimum metadata requirements. These genomes from Africa were compared in a phylogenetic framework with 11,891 representative genomes from around the world. Ancestral location state reconstruction of the dated phylogeny (hereafter referred to as discrete phylogeographic reconstruction) allowed us to infer the number of viral imports and exports between Africa and the rest of the world, and between individual African countries. African genomes in this study spanned the whole global genetic diversity of SARS-CoV-2, a pattern that largely reflects multiple introductions over time from the rest of the world (Fig. 2A).
Fig. 2. Phylogenetic reconstruction of the SARS-CoV-2 pandemic on the continent of Africa.
(A) Time-resolved maximum likelihood tree containing 8746 high-quality African SARS-CoV-2 near-full-genome sequences analyzed against a backdrop of global reference sequences. VOIs and VOCs are highlighted on the phylogeny. (B) Sources of viral introductions into African countries characterized as external introductions from the rest of the world versus internal introductions from other African countries. (C) Total external viral introductions over time into Africa. (D) The number of viral imports and exports into and out of various African countries depicted as internal (between African countries, in pink) or external (between African and non-African countries, in blue and gray).
In total, we detected at least 757 [95% confidence interval (CI): 728 to 786] viral introductions into African countries between the start of 2020 and February 2021, more than half of which occurred before the end of May 2020. Although the early phase of the pandemic was dominated by importations from outside Africa, predominantly from Europe, there was then a shift in the dynamics, with an increasing number of importations from other African countries as the pandemic progressed (Fig. 2, B and C). A rarefaction analysis in which we systematically subsampled genomes shows that vastly more introductions would have likely been identified with increased sampling in Africa or globally, suggesting that the introductions we identified are really just the “ears of the hippo,” or a small part of a larger problem (fig. S1).
South Africa, Kenya, and Nigeria appear as major sources of importations into other African countries (Fig. 2D), although this is likely to be influenced by these three countries having the greatest number of deposited sequences. Particularly notable is the southern African region, where South Africa is the source for a large proportion (~80%) of the importations to other countries in the region. The North African region demonstrates a different pattern to the rest of the continent, with more viral introductions from Europe and Asia (particularly the Middle East) than from other African countries (fig. S2).
Africa has also contributed to the international spread of the virus, with at least 324 (95% CI: 295 to 353) exportation events from Africa to the rest of the world detected in this dataset. Consistent with the source of importations, most exports were to Europe (41%), Asia (26%), and North America (14%). As with the number of importations, exports were relatively evenly distributed over the 1-year period (fig. S3). However, an increase in the number of exportation events occurred between December 2020 and March 2021, which coincided with the second wave of infections in Africa and with some relaxations of travel restrictions around the world.
The early phase of the pandemic was characterized by the predominance of lineage B.1. This was introduced multiple times to African countries and has been detected in all but one of the countries included in this analysis. After its emergence in South Africa, B.1.351 became the most frequently detected SARS-CoV-2 lineage found in Africa (n = 1769, ~20%) (Fig. 1C). It was first sampled on 8 October 2020 in South Africa (13) and has since spread to 20 other African countries.
As air travel came to an almost complete halt in March and April 2020, the number(s) of detectable viral imports into Africa decreased and the pandemic entered a phase that was characterized in sub-Saharan Africa by sustained low levels of within-country movements and occasional international viral movements between neighboring countries, presumably via road and rail links between these. Though some border posts between countries were closed during the initial lockdown period (table S1), others remained open to allow trade to continue. Regional trade in southern Africa was only slightly affected by lockdown restrictions and quickly rebounded to prepandemic levels (fig. S4) after the relaxation of restrictions between June 2020 and December 2020.
Although lineage A viruses were imported into several African countries, they only account for 1.3% of genomes sampled in Africa. Despite lineage A viruses initially causing many localized clustered outbreaks, each the result of independent introductions to several countries (e.g., Burkina Faso, Côte d’Ivoire, and Nigeria), they were later largely replaced by lineage B viruses as the pandemic evolved. This is possibly due to the increased transmissibility of lineage B viruses by virtue of the D614G (Asp614→Gly) mutation in the spike protein (19, 20). However, there is evidence of an increasing prevalence of lineage A viruses in some African countries (11). In particular, A.23.1 emerged in East Africa and appears to be rapidly increasing in prevalence in Uganda and Rwanda (11). Furthermore, a highly divergent variant from lineage A was recently identified in Angola from individuals arriving from Tanzania (21).
Emergence and spread of new SARS-CoV-2 variants
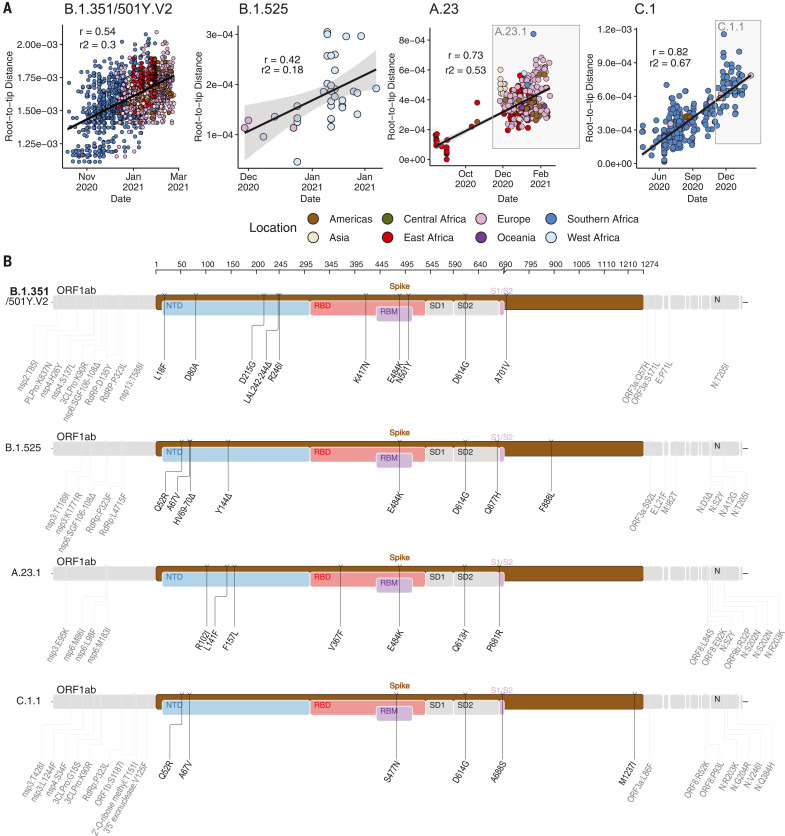
To determine how some of the key SARS-CoV-2 variants are spreading within Africa, we performed phylogeographic analyses on the VOC B.1.351, the variant of interest (VOI) B.1.525, and two additional variants that emerged and that we designated as VOIs for this analysis (A.23.1 and C.1.1). These African VOCs and VOIs have multiple mutations on the spike glycoprotein, and a molecular clock analysis of these four datasets provided strong evidence that these four lineages are evolving in a clock-like manner (Fig. 3, A and B).
Fig. 3. Genetic profile of VOCs and VOIs under investigation.
(A) Root-to-tip regression plots for four lineages of interest. C.1 and A.23 show continued evolution into VOIs C.1.1 and A.23.1, respectively. r, coefficient of correlation; r2, coefficient of determination. (B) Genome maps of four VOCs and VOIs, where the spike region is shown in detail and in color and the rest of the genome is shown in gray. ORF, open reading frame; NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; SD1, subdomain 1; SD2, subdomain 2.
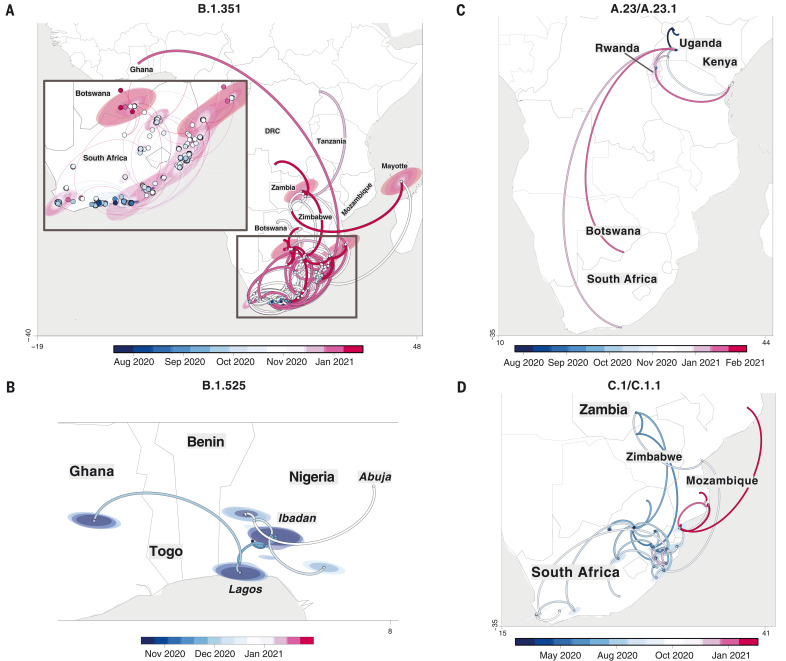
B.1.351 was first sampled in South Africa in October 2020, but phylogeographic analysis suggests that it emerged earlier, around August 2020. It is defined by 10 mutations in the spike protein, including K417N (Lys417→Asn), E484K (Glu484→Lys), and N501Y (Asn501→Tyr) in the receptor binding domain (Fig. 3B). After its emergence in the Eastern Cape, it spread extensively within South Africa (Fig. 4A). By November 2020, the variant had spread into neighboring Botswana and Mozambique, and by December 2020, it had reached Zambia and Mayotte. Within the first 3 months of 2021, further exports from South Africa into Botswana, Zimbabwe, Mozambique, and Zambia occurred. By March 2021, B.1.351 had become the dominant lineage within most southern African countries as well as the overseas territories of Mayotte and Réunion (fig. S5). Our phylogeographic reconstruction also demonstrates movement of B.1.351 into East and Central Africa directly from southern Africa. Our discrete phylogeographic analysis of a wider sample of B.1.351 isolates demonstrates the spread of the lineage into West Africa. This patient from West Africa had a known travel history to Europe, so it is possible that the patient acquired the infection while in Europe or in transit and not from other African sources (fig. S6).
Fig. 4. Phylogeographic reconstruction of the spread of four VOCs and VOIs across the African continent.
(A to D) Phylogeographic reconstruction of the spread of four VOCs and VOIs across the African continent using sequences showing strict continuous transmission across geographical regions: B.1.351 (A), B.1.525 (B), A.23/A.23.1 (C), and C.1/C.1.1 (D). Curved lines denote the direction of transmission in the counterclockwise direction. Solid lines show transmission paths as inferred by phylogeographic reconstruction and colored by date, whereas dashed lines show the known travel history of the particular case considered.
B.1.525 is a VOI defined by six substitutions in the spike protein [Q52R (Gln52→Arg), A67V (Ala67→Val), E484K, D614G, Q677H (Gln677→His), and F888L (Phe888→Leu)] and two deletions in the N-terminal domain [HV69-70Δ (deletion of His and Val at positions 69 and 70) and Y144Δ (deletion of Tyr at position 144)]. This was first sampled in the United Kingdom in mid-December 2020, but our phylogeographic reconstruction suggests that the variant originated in Nigeria in November 2020 [95% highest posterior density (HPD) 2020-11-01 to 2020-12-03] (Fig. 4B). Since then, it has spread throughout much of Nigeria and neighboring Ghana. Given sparse sampling from other neighboring countries within West and Central Africa (Fig. 1, A and C), the extent of the spread of this VOI in the region is not clear. Beyond Africa, this VOI has spread to Europe and the United States (fig. S6).
We designated A.23.1 and C.1.1 as VOIs for the purposes of this analysis because they present good examples of the continued evolution of the virus within Africa (11, 13). Lineage A.23, characterized by three spike mutations [F157L (Phe157→Leu), V367F (Val367→Phe), and Q613H (Gln613→His)], was first detected in a Ugandan prison in Amuru in July 2020 (95% HPD: 2020-07-15 to 2020-08-02). From there, the lineage was transmitted to Kitgum prison, possibly facilitated by the transfer of prisoners. Subsequently, the A.23 lineage spilled into the general population and spread to Kampala, adding other spike mutations [R102I (Arg102→Ile), L141F (Leu141→Phe), E484K, and P681R (Pro681→Arg)] along with additional mutations in nsp3, nsp6, ORF8, and ORF9, prompting a new lineage classification, A.23.1 (Fig. 3, A and B). Since the emergence of A.23.1 in September 2020 (95% HPD: 2020-09-02 to 2020-09-28), it has spread regionally into neighboring Rwanda and Kenya and has now also reached South Africa and Botswana in the south and Ghana in the west (Fig. 4C). However, our phylogeographic reconstruction of A.23.1 suggests that the introduction into Ghana may have occurred via Europe (fig. S6), whereas the introductions into southern Africa likely occurred directly from East Africa. This is consistent with epidemiological data suggesting that the case detected in South Africa was a contact of an individual who had recently traveled to Kenya.
Lineage C.1 emerged in South Africa in March 2020 (95% HPD: 2020-03-13 to 2020-04-17) during a cluster outbreak before the first wave of the epidemic (13). C.1.1 is defined by the spike mutations S477N (Ser477→Asn), A688S (Ala688→Ser), and M1237I (Met1237→Ile) and also contains the Q52R and A67V mutations similar to B.1.525 (Fig. 3B). A continuous trait phylogeographic reconstruction of the movement dynamics of these lineages suggests that C.1 emerged in the city of Johannesburg and spread within South Africa during the first wave (Fig. 4D). Independent exports of C.1 from South Africa led to regional spread to Zambia (June to July 2020) and Mozambique (July to August 2020), and the evolution to C.1.1 seems to have occurred in Mozambique around mid-September 2020 (95% HPD: 2020-09-07 to 2020-10-05). An in-depth analysis of SARS-CoV-2 genotypes from Mozambique suggests that the C.1.1 lineage was the most prevalent in the country until the introduction of B.1.351, which has dominated the epidemic since (fig. S5).
The VOC B.1.1.7, which was first sampled in Kent, England, in September 2020 (22), has also increased in prevalence in several African countries (fig. S5). To date, this VOC has been detected in 11 African countries, as well as the Indian Ocean islands of Mauritius and Mayotte (fig. S7). The time-resolved phylogeny suggests that this lineage was introduced into Africa on at least 16 occasions between November 2020 and February 2021, with evidence of local transmission in Nigeria and Ghana.
Conclusions
Our phylogeographic reconstruction of past viral dissemination patterns suggests a strong epidemiological linkage between Europe and Africa, with 64% of detectable viral imports into Africa originating in Europe and 41% of detectable viral exports from Africa landing in Europe (Fig. 1C). This phylogeographic analysis also suggests a changing pattern of viral diffusion into and within Africa over the course of 2020. In almost all instances, the earliest introductions of SARS-CoV-2 into individual African countries were from countries outside Africa.
High rates of COVID-19 testing and consistent genomic surveillance in the south of the continent have led to the early identification of VOCs such as B.1.351 and VOIs such as C.1.1 (13). Since the discovery of these southern African variants, several other SARS-CoV-2 VOIs have emerged in different parts of the world, including elsewhere on the African continent, such as B.1.525 in West Africa and A.23.1 in East Africa. There is strong evidence that both of these VOIs are rising in frequency in the regions where they have been detected, which suggests that they may possess higher fitness than other variants in these regions. Although more-focused research on the biological properties of these VOIs is needed to confirm whether they should be considered VOCs, it would be prudent to assume the worst and focus on limiting their spread. It will be important to investigate how these different variants compete against one another if they occupy the same region.
Our focused phylogenetic analysis of the B.1.351 lineage revealed that in the final months of 2020, this variant spread from South Africa into neighboring countries, reaching as far north as the Democratic Republic of the Congo (DRC) by February 2021. This spread may have been facilitated through rail and road networks that form major transport arteries linking South Africa’s ocean ports to commercial and industrial centres in Botswana, Zimbabwe, Zambia, and the southern parts of the DRC. The rapid, apparently unimpeded spread of B.1.351 into these countries suggests that current land-border controls that are intended to curb the international spread of the virus are ineffective. Perhaps targeted testing of cross-border travelers, genotyping of positive cases, and the focused tracking of frequent cross-border travelers, such as long distance truckers, would more effectively contain the spread of future VOCs and VOIs that emerge within this region.
The dominance of VOIs and VOCs in Africa has important implications for vaccine rollouts on the continent. For one, slow rollout of vaccines in most African countries creates an environment in which the virus can replicate and evolve: This will almost certainly produce additional VOCs, any of which could derail the global fight against COVID-19. Conversely, with the already widespread presence of known variants, difficult decisions about balancing reduced efficacy and availability of vaccines have to be made. This also highlights how crucial it is that trials are done. From a public health perspective, genomic surveillance is only one item in the toolkit of pandemic preparedness. It is important that such work is closely followed by genotype-to-phenotype research to determine the actual relevance of continued evolution of SARS-CoV-2 and other emerging pathogens.
The rollout of vaccines across Africa has been painfully slow (figs. S8 and S9). There have, however, been notable successes that suggest that the situation is not hopeless. The small island nation of the Seychelles had vaccinated 70% of its population by May 2021. Morocco has kept pace with many developed nations and, by mid-March, had vaccinated ~16% of its population. Rwanda, one of Africa’s most resource-constrained countries, had, within 3 weeks of obtaining its first vaccine doses in early March, managed to provide first doses to ~2.5% of its population. For all other African countries, at the time of writing, vaccine coverage (first dose) was <1.0% of the general population.
The effectiveness of molecular surveillance as a tool for monitoring pandemics is largely dependent on continuous and consistent sampling through time, rapid virus genome sequencing, and rapid reporting. When this is achieved, molecular surveillance can ensure the early detection of changing pandemic characteristics. Further, when such changes are discovered, molecular surveillance data can also guide public health responses. In this regard, the molecular surveillance data that are being gathered by most African countries are less useful than they could be. For example, the time lag between when virus samples are taken and when sequences for these samples are deposited in sequence repositories is so great in some cases that the primary utility of genomic surveillance data is lost (fig. S10). This lag is driven by several factors, depending on the laboratory or country in question: (i) lack of reagents owing to disruptions in global supply chains, (ii) lack of equipment and infrastructure within the originating country, (iii) scarcity of technical skills in laboratory methods or bioinformatic support, and (iv) hesitancy by some health officials to release data. More-recent sampling and prompt reporting is crucial to reveal the genetic characteristics of currently circulating viruses in these countries.
The patchiness of African genomic surveillance data is therefore the main weakness of our study. However, there is evidence that the situation is improving, with ~50% of African SARS-CoV-2 genome sequences having been submitted to the GISAID database within the first 10 weeks of 2021. Although the precise factors underlying this surge in sequencing efforts are unclear, an important driver is almost certainly increased global interest in genomic surveillance after the discovery of multiple VOCs and VOIs since December 2020. We cannot reject that the observed increase in exports from Africa may be due to intensified sequencing activity after the detection of variants around the world. It is important to note here that phylogeographic reconstruction of viral spread is highly dependent on sampling where there is the caveat that the exact routes of viral movements between countries cannot be inferred if there is no sampling in connecting countries. Furthermore, our efforts to reconstruct the movement dynamics of SARS-CoV-2 across the continent are almost certainly biased by uneven sampling between different African countries. It is not a coincidence that we identified South Africa, Kenya, and Nigeria, which have sampled and sequenced the most SARS-CoV-2 genomes, as major sources of viral transmissions between sub-Saharan African countries. However, these countries also had the highest number of infections, which may decrease the sampling biases (Fig. 1A).
The reliability of genomic surveillance as a tool to prevent the emergence and spread of dangerous variants is dependent on the intensity with which it is embraced by national public health programs. As with most other parts of the world, the success of genomic surveillance in Africa requires that more samples are tested for COVID-19, higher proportions of positive samples are sequenced within days of sampling, and persistent analyses of these sequences are performed for concerning signals such as (i) the presence of novel nonsynonymous mutations at genomic sites associated with pathogenicity and immunogenicity, (ii) evidence of positive selection at codon sites where nonsynonymous mutations are observed, and (iii) evidence of lineage expansions. Despite limited sampling, Africa has identified many of the VOCs and VOIs that are being transmitted across the world. Detailed characterization of the variants and their impact on vaccine-induced immunity is of extreme importance. If the pandemic is not controlled in Africa, we may see the production of vaccine escape variants that may profoundly affect the population in Africa and across the world.
Supplementary Material
Acknowledgments
We acknowledge the authors from the originating laboratories and the submitting laboratories, who generated and shared, via GISAID, the genetic sequence data on which this research is based (table S4). We also acknowledge the contribution of K. Maria from the NGS-SA platform for their contribution toward the sequencing effort in Cape Town, South Africa. Similarly, we thank A. M. Elsaame, S. M. Elsayed, and R. M. Darwish from the Faculty of Medicine Ain Shams Research Institute (MASRI) for their efforts toward sequencing in Egypt. We thank S. Bane, M. Sanogo, D. Diallo, A. Combo Georges Togo, and A. Coulibaly from the University Clinical Research Centre (UCRC) at the University of Sciences, Techniques, and Technologies of Bamako for the contribution they have made toward sequencing efforts in Mali. We acknowledge the contribution of M. Moeti and A. Salam Gueye from the WHO for their contribution toward combating SARS-CoV-2 on the African continent. We further wish to extend acknowledgment to S. Lutucuta and J. Morais from the Angolan Ministry of Health for their continued hard work with regards to SARS-CoV-2 sampling, sequencing, and pandemic response in Angola. From Malawi we wish to acknowledge the work of B. Chilima, B. Mvula, and M. Chitenje from the Malawian Ministry of Health for their work on the COVID-19 response within the country.
Funding: The University of Ghana (WACCBIP) team was funded by a Wellcome/African Academy of Sciences Developing Excellence in Leadership Training and Science (DELTAS) grant (DEL-15-007 and 107755/Z/15/Z: Awandare); National Institute of Health Research (NIHR) (17.63.91) grants using UK aid from the UK government for a global health research group for genomic surveillance of malaria in West Africa (Wellcome Sanger Institute, UK) and the global research unit for Tackling Infections to Benefit Africa (TIBA partnership, University of Edinburgh); and a World Bank African Centres of Excellent grant (WACCBIP-NCDs: Awandare). Project ADAGE PRFCOV19-GP2 (2020-2022) includes 40 researchers from the Center of Biotechnology of Sfax, the University of Sfax, the University of Monastir, the University Hospital Hédi Chaker of Sfax, the Military Hospital of Tunis, and Dacima Consulting. Ministry of Higher Education and Scientific Research and Ministry of Health of the Republic of Tunisia. The Uganda contributions were funded by the UK Medical Research Council (MRC/UKRI) and the UK Department for International Development (DFID) under the MRC/DFID concordat agreement (grant agreement number NC_PC_19060) and by the Wellcome, DFID–Wellcome Epidemic Preparedness–Coronavirus (grant agreement number 220977/Z/20/Z) awarded to M.C. Work from Quadram Institute Bioscience was funded by The Biotechnology and Biological Sciences Research Council Institute Strategic Programme Microbes in the Food Chain BB/R012504/1 and its constituent projects BBS/E/F/000PR10348, BBS/E/F/000PR10349, BBS/E/F/000PR10351, and BBS/E/F/000PR10352 and by the Quadram Institute Bioscience BBSRC–funded Core Capability Grant (project number BB/CCG1860/1). The Africa Pathogen Genomics Initiative (Africa PGI) at the Africa CDC is supported by the Bill & Melinda Gates Foundation (INV018978 and INV018278), Illumina Inc, the US Centers for Disease Control and Prevention (CDC), and Oxford Nanopore Technologies. Sequences generated in Zambia through PATH were funded by the Bill & Melinda Gates Foundation. The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the Bill & Melinda Gates Foundation. Funding for sequencing in Côte d’Ivoire, Burkina Faso, and part of the sequencing in the DRC was granted by the German Federal Ministry of Education and Research (BMBF). Sequencing efforts from Morocco have been supported by Academie Hassan II of Science and Technology, Morocco. Funding for surveillance, sampling, and testing in Madagasar was provided by the WHO, the CDC (grant U5/IP000812-05), the US Agency for International Development (USAID; cooperation agreement 72068719CA00001), and the Office of the Assistant Secretary for Preparedness and Response in the US Department of Health and Human Services (DHHS; grant number IDSEP190051-01-0200). Funding for sequencing was provided by the Bill & Melinda Gates Foundation (GCE/ID OPP1211841), Chan Zuckerberg Biohub, and the Innovative Genomics Institute at UC Berkeley. The Botswana Harvard AIDS Institute was supported by the following funding: H3ABioNet through funding from the National Institutes of Health Common Fund (U41HG006941)—H3ABioNet is an initiative of the Human Health and Heredity in Africa Consortium (H3Africa) program of the African Academy of Science (AAS); DHHS–NIH–National Institute of Allergy and Infectious Diseases (NIAID) (5K24AI131928-04 and 5K24AI131924-04); Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE); a DELTAS Africa Initiative (grant DEL-15-0060—the DELTAS Africa Initiative is an independent funding scheme of the AAS’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant 107752/Z/15/Z] and the UK government; and the South African Medical Research Council (SAMRC) and the Department of Technology and Innovation as part of the Network for Genomic Surveillance in South Africa (NGS-SA) and the Stellenbosch University Faculty of Medicine & Health Sciences, Strategic Equipment Fund. D.P.M. is funded by the Wellcome Trust (Wellcome Trust grant 222574/Z/21/Z). Sequencing activities at the NICD were supported by a conditional grant from the South African National Department of Health as part of the emergency COVID-19 response; a cooperative agreement between the National Institute for Communicable Diseases of the National Health Laboratory Service and the U.S. Centers for Disease Control and Prevention (grant number 5 U01IP001048-05-00); the African Society of Laboratory Medicine (ASLM) and Africa Centers for Disease Control and Prevention through a sub-award from the Bill and Melinda Gates Foundation grant number INV-018978; the UK Foreign, Commonwealth and Development Office and Wellcome (Grant no 221003/Z/20/Z); the South African Medical Research Council (Reference number SHIPNCD 76756); the UK Department of Health and Social Care, managed by the Fleming Fund and performed under the auspices of the SEQAFRICA project. Furthermore, pandemic surveillance in South Africa and Senegal was supported in part through NIH grant U01 AI151698 for the United World Antiviral Research Network (UWARN). Support for pandemic surveillance from the Tulio de Oliveira group to other African countries is funded by the Rockefeller Foundation. Sequencing efforts in the DRC were funded by the Bill & Melinda Gates Foundation under grant INV-018030 awarded to C.B.P. and further supported by funding from the Africa CDC through the ASLM (African Society of Laboratory Medicine) for Accelerating SARS-CoV-2 Genomic Surveillance in Africa. Sequencing efforts in Rwanda were commissioned by the NIHR Global Health Research program (16/136/33) using UK aid from the UK government (funding to E.M. and N.R. through TIBA partnership) and additional funds from the government of Rwanda through RBC/National Reference Laboratory in collaboration with the Belgian Development Agency (ENABEL) for additional genomic sequencing at GIGA Research Institute–Liege/Belgium. The sequencing effort in Equatorial Guinea was supported by a public-private partnership, the Bioko Island Malaria Elimination Project, composed of the government of Equatorial Guinea Ministries of Mines and Hydrocarbons, and Health and Social Welfare, Marathon EG Production Limited, Noble Energy, Atlantic Methanol Production Company, and EG LNG. Sample collection and typing in Mali were supported by Fondation Merieux–France, and sequence efforts have been supported by the Enable and Enhance Initiative of the German Federal Government’s Security Cooperation against Biological Threats in the G5 Sahel Region. The Nigeria work was made possible by support from Flu Lab and a cohort of generous donors through TED’s Audacious Project, including the ELMA Foundation, MacKenzie Scott, the Skoll Foundation, and Open Philanthropy. Further Nigeria funding came from grants from the NIAID (www.niaid.nih.gov), NIH-H3Africa (https://h3africa.org) (U01HG007480 and U54HG007480), and the World Bank grant (worldbank.org) (ACE IMPACT project) to C.H. Analysis for the Gabon strains was supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS), Japan International Cooperation Agency (JICA), and Japan Agency for Medical Research and Development (AMED) (grant number JP20jm0110013) and a grant from AMED (grant number JP20wm0225003). Sequencing at KEMRI-Wellcome Trust Research Programme site in Kenya was supported by the National Institute for Health Research (NIHR) (project references 17/63/82 and 16/136/33), using UK aid from the UK Government to support global health research, and the UK Foreign, Commonwealth and Development Office and Wellcome Trust (grant# 102975; 220985).
Author contributions: Conceptualization: E.W., H.T., J.G.S., J.O., J.O.G., K.O.D., R.A.D., R.A.K., R.L., S.K.T., S.Ma., T.d.O. Methodology: A.-R.N.Z., A.Re., C.N.A., D.P.M., D.A.R., E.W., H.T., J.A.E., J.Gy., J.G.S., K.H.Y., K.O.D., L.d.O.M., M.A.B., M.C., M.G., M.M.N., M.V.T.P., P.A., T.d.O., V.F. Investigation: A.E., A.Ir., A.-R.N.Z., A.Re., A.So., A.v.G., C.A.K., C.W., D.C., D.J.N., D.P.M., D.A.R., D.S., E.M., E.N.N., E.W., F.L., G.G., H.T., J.A.E., J.E.S., J.Gy., J.G.S., J.-J.M.T., J.L., J.Nk., J.Q.M., K.H.Y., M.-M.D., M.A.B., M.C., M.G., M.Mar., M.Mas., M.S., M.V.T.P., N.A., N.H.R., N.K., P.K., R.A.A.C., R.A.D., R.G., S.A.M., S.F.A., S.Ma., S.O., T.L.V., V.F., W.P. Sampling: A.-S.K., A.N.A., A.A.A., A.A.S., A.D., A.E., A.E.O.O., A.F., A.G., A.H., A.Ib., A.Ka., A.K.Sa., A.K.Se., A.L., A.M.O., A.O.O.O., A.P., A.Ro., A.Sy., A.F.V., A.V.G., A.v.G., B.B., B.L.H., B.Ko., B.Kl., B.Ma., B.N., B.S.O., B.T., C.N.A., C.D., C.P., C.S., C.W., D.Bas., D.C., D.D., D.G., D.G.A., D.J.N., D.S., E.K.L., E.M., E.M.O., E.N.N., E.P., E.Sh., F.Ab., F.Ad., F.O.E., F.M.M., F.S.M., F.O., F.T., F.T.T., G.A.A., G.G., G.K.M., G.P.M., G.T., G.v.Z., H.C., H.A.E., H.N., I.C., I.Gaa., I.Gaz., I.K., I.M., I.O., I.Ss., J.C.A., J.-C.M., J.B.L.-D., J.E.S., J.Gi., J.Gy., J.J.L., J.K., J.L., J.M.H., J.M.M., J.Nam., J.Nak., J.T.K., K.T.A., K.M.S., L.B., M.-M.D., M.Al., M.C., M.D., M.e.H., M.G.S., M.K.K., M.Mp., M.Mar., M.Mas., M.M.D., M.N., M.Ou., M.O.A., M.R., M.S., M.W.M., M.T.Y., N.A., N.G., N.H., N.H.R., N.Ig., N.K., N.Ma., N.Nd., N.B.S., N.S., O.Ch., O.E.C., O.Fa., O.Fe., O.I., O.J., O.C.K., O.E.O., O.P., O.S., O.Te., P.E.O., P.A.B., P.C., P.C.S., P.D., P.K., P.K.Q., P.O., P.S., R.A.D., R.G., R.N., S.A., S.S.A., S.B., S.B.L., S.D., S.El., S.E.K., S.F.A., S.Gas., S.Kam., S.L., S.L.D., S.Me., S.Mo., S.M.M., S.N., S.Pi., S.S., S.To., T.L.V., T.Ma., T.S., U.C., U.G., U.J., U.R., V.G., W.K.A., W.T.C., W.P., W.H.R., Y.Bu., Y.K.T., Y.N., Z.R.D. Sequencing: A.-S.Kam., A.N.A., A.A.A., A.A.S., A.C., A.D., A.E.O.O., A.F., A.Ib., A.Is., A.Ko., A.Ka., A.K.K., A.K.Sa., A.K.Se., A.L., A.-R.N.Z., A.P., A.Sa., A.So., A.Sy., A.S.O., A.J.T., A.F.V., A.V.G., A.v.G., A.A.Y., B.B., B.D., B.L.H., B.Ko., B.Kl., B.Mc., B.N., B.T., C.N.A., C.B., C.B.P., C.D., C.M.M., C.P., C.S., D.Bas., D.D., D.G., D.G.A., D.J.B., D.L.B., D.M., D.O.O., D.P., D.S.Y.A., D.T., E.E.F., E.F.N., E.K.L., E.L., E.M.O., E.P., E.Sh., E.Si., E.S.L., F.Ab., F.Aj., F.A.D., F.D., F.M.M., F.S.M., F.O., F.T.T., F.W., G.A.A., G.G., G.P.M., G.T., G.v.Z., H.Ab., H.An., H.C., H.C.R., H.A.E., H.G., H.H.K., H.N., I.B.-B.B., I.C., I.Gaa., I.Gaz., I.K., I.M., I.Ss., J.C.A., J.B., J.-C.M., J.B.L.-D., J.F., J.Gi., J.Gy., J.J.L., J.K., J.M.H., J.M.M., J.M.N., J.Nam., J.Nak., J.Q.M., J.T.K., J.Y., K.T.A., K.M.S., K.O.D., K.S., K.A.T., L.B., L.F., L.S., L.T., M.-M.D., M.Al., M.A.B., M.C., M.D., M.e.H., M.G.S., M.I.M., M.K.K., M.Mi., M.Mp., M.Mw., M.M.D., M.M.N., M.Ow., M.Ou., M.O.A., M.V.T.P., M.W.M., M.T.Y., N.D., N.G., N.H., N.Ig., N.Is., N.Ma., N.Nd., N.Ns., N.B.S., N.S., N.T., O.Cy., O.Ch., O.E.C., O.Fa., O.I., O.J., O.Te., P.A., P.A.B., P.C.S., P.D., P.E.O., P.K., P.K.Q., P.K.M., P.O., P.S., R.A.A.C., R.G., R.N., R.O.P., S.A., S.S.A., S.B., S.B.L., S.C.S., S.D., S.El., S.En., S.E.K., S.Gar., S.Gas., S.H.Ab., S.Kas., S.L., S.L.D., S.Me., S.Mo., S.Ma., S.M.M., S.N., S.Pi., S.Pr., S.R., S.S., S.To., S.Tr., S.v.d.W., T.A., T.Ma., T.Mo., T.S., U.C., U.G., U.J., U.J.A., U.R., V.G., W.K.A., W.T.C., W.H.R., Y.Bu., Y.Be., Y.K.T., Y.N., Z.R.D. Visualization: A.C., A.Is., A.Ko., A.K.K., A.Sa., A.So., A.A.Y., B.T., C.B., C.M.M., D.Bak., D.O.O., D.P., D.A.R., D.S.Y.A., E.A.A., E.B., E.S.L., E.W., F.A.D., F.B., F.D., F.W., G.S., H.Ab., H.An., H.G., H.L., H.T., I.B.A., I.Ss., J.A.E., J.B., J.F., J.Gy., J.M.N., J.Y., K.H.Y., K.S., L.F., L.S., L.T., M.Ao., M.Al., M.G., M.T., M.V.T.P., M.R.W., N.D., N.Is., N.K., N.Ns., N.T., O.Cy., O.To., P.A., P.C.S., P.E.O., R.A.A.C., S.B., S.F.S., S.H.A., S.Kas., S.Ma., T.A., T.Mo., V.E., Y.Be. Funding acquisition: A.J.P., A.Re., A.v.G., B.Ko., C.N.A., C.A.K., C.B.P., C.W., D.C., D.J.B., D.J.N., F.L., G.A.A., G.G., G.P.M., H.C., J.E.S., J.-J.M.T., J.L., J.M.H., J.Nk., J.O., K.O.D., M-M.D., M.C., M.I.M., M.Mas., M.V.T.P., N.A., P.C.S., P.K., P.K.M., R.A.K., S.A.M., S.El., S.Mo., S.v.d.W., T.d.O., W.P. Project administration: A.J.P., A.Re., A.F.V., A.v.G., B.Ko., C.W., D.J.B., D.J.N., E.W., F.Aj., F.T., G.A.A., G.P.M., G.S., G.T., H.C., J.C.O., J.-J.M.T., J.M.H., J.O., J.O.G., J.Y., K.O.D., M.C., M.K., M.Mar., M.P., M.V.T.P., M.R.W., N.R., O.To., P.C.S., P.K., P.K.M., R.A.K., S.A.M., S.El., S.F.S., S.Gas., S.Mo., T.d.O. Supervision: A.J.P., A.Re., B.Ko., C.W., D.J.N., E.N.N., E.W., F.T., G.A.A., G.L.K., H.C., J.B., J.M.H., J.Nk., J.O., J.O.G., K.O.D., M.Al., M.C., M.I.M., M.Mar., M.M.N., M.S., N.Mu., N.R., P.C.S., P.K., P.K.M., R.A.K., S.El., S.E.K., S.Gas., S.Me., S.Mo., S.Pa., T.d.O. Writing – original draft: A.K.Sa., A.-R.N.Z., B.Ko., D.P.M., E.W., F.T., G.L.K., H.T., J.B., J.-C.M., M.Al., M.A.B., M.C., M.G., M.Mi., N.Mu., R.L. Writing – review and editing: A.-R.N.Z., B.Ko., C.M.M., D.J.N., D.P.M., D.A.R., D.S.Y.A., D.T., E.K.L., E.L., E.S.L., E.W., H.T., J.E.S., J.G.S., L.d.O.M., M.A.B., M.C., M.e.H., P.K.Q., P.K.M., R.L., S.K.T., T.d.O., U.J.A. Author contributions are listed alphabetically. A full list of author abbreviations is included on the GitHub repository (https://github.com/krisp-kwazulu-natal/africa-covid19-genomics) (23).
Competing interests: P.C.S. is a founder and shareholder of Sherlock Biosciences and is on the board and serves as shareholder of the Danaher Corporation. The authors declare no other conflicts of interest.
Data and materials availability: All sequences that were used in the present study are listed in table S4 (accessible on the GitHub repository) along with their GISAID sequence IDs, dates of sampling, the originating and submitting laboratories, and main authors. All input files (e.g., alignments or XML files), all resulting output files, and scripts used in the study are shared publicly on GitHub (https://github.com/krisp-kwazulu-natal/africa-covid19-genomics) (23). This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party, obtain authorization from the rights holder before using such material.
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
Table S4
MDAR Reproducibility Checklist
References and Notes
- 1.Wang C., Horby P. W., Hayden F. G., Gao G. F., A novel coronavirus outbreak of global health concern. Lancet 395, 470–473 (2020). 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K. S. M., Lau E. H. Y., Wong J. Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T. T. Y., Wu J. T., Gao G. F., Cowling B. J., Yang B., Leung G. M., Feng Z., Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020). 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uyoga S., Adetifa I. M. O., Karanja H. K., Nyagwange J., Tuju J., Wanjiku P., Aman R., Mwangangi M., Amoth P., Kasera K., Ng’ang’a W., Rombo C., Yegon C., Kithi K., Odhiambo E., Rotich T., Orgut I., Kihara S., Otiende M., Bottomley C., Mupe Z. N., Kagucia E. W., Gallagher K. E., Etyang A., Voller S., Gitonga J. N., Mugo D., Agoti C. N., Otieno E., Ndwiga L., Lambe T., Wright D., Barasa E., Tsofa B., Bejon P., Ochola-Oyier L. I., Agweyu A., Scott J. A. G., Warimwe G. M., Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Kenyan blood donors. Science 371, 79–82 (2021). 10.1126/science.abe1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mwananyanda L., Gill C. J., MacLeod W., Kwenda G., Pieciak R., Mupila Z., Lapidot R., Mupeta F., Forman L., Ziko L., Etter L., Thea D., Covid-19 deaths in Africa: Prospective systematic postmortem surveillance study. BMJ 372, n334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salyer S. J., Maeda J., Sembuche S., Kebede Y., Tshangela A., Moussif M., Ihekweazu C., Mayet N., Abate E., Ouma A. O., Nkengasong J., The first and second waves of the COVID-19 pandemic in Africa: A cross-sectional study. Lancet 397, 1265–1275 (2021). 10.1016/S0140-6736(21)00632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. Oluniyi, First African SARS-CoV-2 genome sequence from Nigerian COVID-19 case. Virological (2020); https://virological.org/t/first-african-sars-cov-2-genome-sequence-from-nigerian-covid-19-case/421.
- 7.Medhat M. A., El Kassas M., COVID-19 in Egypt: Uncovered figures or a different situation? J. Glob. Health 10, 010368 (2020). 10.7189/jogh.10.010368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allam M., Ismail A., Khumalo Z. T. H., Kwenda S., van Heusden P., Cloete R., Wibmer C. K., Mtshali P. S., Mnyameni F., Mohale T., Subramoney K., Walaza S., Ngubane W., Govender N., Motaze N. V., Bhiman J. N., SA-COVID-19 Response Team , Genome sequencing of a severe acute respiratory syndrome coronavirus 2 isolate obtained from a South African Patient with coronavirus disease 2019. Microbiol. Resour. Announc. 9, e00572-e20 (2020). 10.1128/MRA.00572-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haider N., Osman A. Y., Gadzekpo A., Akipede G. O., Asogun D., Ansumana R., Lessells R. J., Khan P., Hamid M. M. A., Yeboah-Manu D., Mboera L., Shayo E. H., Mmbaga B. T., Urassa M., Musoke D., Kapata N., Ferrand R. A., Kapata P.-C., Stigler F., Czypionka T., Zumla A., Kock R., McCoy D., Lockdown measures in response to COVID-19 in nine sub-Saharan African countries. BMJ Glob. Health 5, e003319 (2020). 10.1136/bmjgh-2020-003319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inzaule S. C., Tessema S. K., Kebede Y., Ogwell Ouma A. E., Nkengasong J. N., Genomic-informed pathogen surveillance in Africa: Opportunities and challenges. Lancet Infect. Dis. 21, e281–e289 (2021). 10.1016/S1473-3099(20)30939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugembe D. L., Phan M. V. T., Ssewanyana I., Semanda P., Nansumba H., Dhaala B., Nabadda S., O’Toole Á. N., Rambaut A., Kaleebu P., Cotten M., A SARS-CoV-2 lineage A variant (A.23.1) with altered spike has emerged and is dominating the current Uganda epidemic. medRxiv 2021.02.08.21251393 (2021); 10.1101/2021.02.08.21251393. 10.1101/2021.02.08.21251393 [DOI] [PMC free article] [PubMed]
- 12.Giandhari J., Pillay S., Wilkinson E., Tegally H., Sinayskiy I., Schuld M., Lourenço J., Chimukangara B., Lessells R., Moosa Y., Gazy I., Fish M., Singh L., Sedwell Khanyile K., Fonseca V., Giovanetti M., Carlos Junior Alcantara L., Petruccione F., de Oliveira T., Early transmission of SARS-CoV-2 in South Africa: An epidemiological and phylogenetic report. Int. J. Infect. Dis. 103, 234–241 (2021). 10.1016/j.ijid.2020.11.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tegally H., Wilkinson E., Lessells R. J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J. N., von Gottberg A., Walaza S., Fonseca V., Allam M., Ismail A., Glass A. J., Engelbrecht S., Van Zyl G., Preiser W., Williamson C., Petruccione F., Sigal A., Gazy I., Hardie D., Hsiao N., Martin D., York D., Goedhals D., San E. J., Giovanetti M., Lourenço J., Alcantara L. C. J., de Oliveira T., Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 27, 440–446 (2021). 10.1038/s41591-021-01255-3 [DOI] [PubMed] [Google Scholar]
- 14.C. A. Pearson, T. W. Russell, N. Davies, A. J. Kucharski, Estimates of severity and transmissibility of novel SARS-CoV-2 variant 501Y.V2 in South Africa. CMMID Repository (2021); https://cmmid.github.io/topics/covid19/sa-novel-variant.html. 10.1101/2021.02.08.21251393 [DOI]
- 15.Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., Karim F., Ganga Y., Khan K., Bernstein M., Balazs A. B., Gosnell B. I., Hanekom W., Moosa M.-Y. S., Lessells R. J., Oliveira T., Sigal A., Escape of SARS-CoV-2 501Y.V2 variants from neutralization by convalescent plasma. medRxiv 2021.01.26.21250224 (2021); 10.1101/2021.01.26.21250224. 10.1101/2021.01.26.21250224 [DOI] [PMC free article] [PubMed]
- 16.Madhi S. A., Baillie V., Cutland C. L., Voysey M., Koen A. L., Fairlie L., Padayachee S. D., Dheda K., Barnabas S. L., Bhorat Q. E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S., Bhiman J. N., Bhorat A. E., du Plessis J., Esmail A., Groenewald M., Horne E., Hwa S.-H., Jose A., Lambe T., Laubscher M., Malahleha M., Masenya M., Masilela M., McKenzie S., Molapo K., Moultrie A., Oelofse S., Patel F., Pillay S., Rhead S., Rodel H., Rossouw L., Taoushanis C., Tegally H., Thombrayil A., van Eck S., Wibmer C. K., Durham N. M., Kelly E. J., Villafana T. L., Gilbert S., Pollard A. J., de Oliveira T., Moore P. L., Sigal A., Izu A., NGS-SA Group, Wits-VIDA COVID Group , Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 384, 1885–1898 (2021). 10.1056/NEJMoa2102214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wibmer C. K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B. E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J. N., Moore P. L., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 27, 622–625 (2021). 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- 18.Shu Y., McCauley J., GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 22, 30494 (2017). 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volz E., Hill V., McCrone J. T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F. F., Rey S. M., Nicholls S. M., Colquhoun R. M., da Silva Filipe A., Shepherd J., Pascall D. J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N. J., Pybus O. G., Robertson D. L., Thomson E. C., Rambaut A., Connor T. R., COG-UK Consortium , Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75.e11 (2021). 10.1016/j.cell.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O., Montefiori D. C., Angyal A., Brown R. L., Carrilero L., Green L. R., Groves D. C., Johnson K. J., Keeley A. J., Lindsey B. B., Parsons P. J., Raza M., Rowland-Jones S., Smith N., Tucker R. M., Wang D., Wyles M. D., Sheffield COVID-19 Genomics Group , Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19 (2020). 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Oliveira T., Lutucuta S., Nkengasong J., Morais J., Paixão J. P., Neto Z., Afonso P., Miranda J., David K., Inglês L., Raisa Rivas Carralero A. P. A. P., Freitas H. R., Mufinda F., Tessema S. K., Tegally H., San E. J., Wilkinson E., Giandhari J., Pillay S., Giovanetti M., Naidoo Y., Katzourakis A., Ghafari M., Singh L., Tshiabuila D., Martin D., Lessells R. J., A novel variant of interest of SARS-CoV-2 with multiple spike mutations is identified from travel surveillance in Africa. medRxiv 2021.03.30.21254323 (2021); 10.1101/2021.03.30.21254323. 10.1101/2021.03.30.21254323 [DOI]
- 22.Meng B., Kemp S. A., Papa G., Datir R., Ferreira I. A. T. M., Marelli S., Harvey W. T., Lytras S., Mohamed A., Gallo G., Thakur N., Collier D. A., Mlcochova P., Duncan L. M., Carabelli A. M., Kenyon J. C., Lever A. M., De Marco A., Saliba C., Culap K., Cameroni E., Matheson N. J., Piccoli L., Corti D., James L. C., Robertson D. L., Bailey D., Gupta R. K., COVID-19 Genomics UK (COG-UK) Consortium , Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 35, 109292 (2021). 10.1016/j.celrep.2021.109292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. E. James, T. Houriiyah, krisp-kwazulu-natal/africa-covid19-genomics: A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa - Code and scripts. Zenodo (2021); 10.5281/zenodo.5386379. 10.5281/zenodo.5386379 [DOI] [PMC free article] [PubMed]
- 24.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemoine F., Domelevo Entfellner J. B., Wilkinson E., Correia D., Dávila Felipe M., De Oliveira T., Gascuel O., Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456 (2018). 10.1038/s41586-018-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sagulenko P., Puller V., Neher R. A., TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 4, vex042 (2018). 10.1093/ve/vex042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambaut A., Holmes E. C., O’Toole Á., Hill V., McCrone J. T., Ruis C., du Plessis L., Pybus O. G., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020). 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemey P., Rambaut A., Welch J. J., Suchard M. A., Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010). 10.1093/molbev/msq067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). 10.1093/ve/vew007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellicour S., Rose R., Faria N. R., Lemey P., Pybus O. G., SERAPHIM: Studying environmental rasters and phylogenetically informed movements. Bioinformatics 32, 3204–3206 (2016). 10.1093/bioinformatics/btw384 [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson E., Rasmussen D., Ratmann O., Stadler T., Engelbrecht S., de Oliveira T., Origin, imports and exports of HIV-1 subtype C in South Africa: A historical perspective. Infect. Genet. Evol. 46, 200–208 (2016). 10.1016/j.meegid.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 35.Sukumaran J., Holder M. T., DendroPy: A Python library for phylogenetic computing. Bioinformatics 26, 1569–1571 (2010). 10.1093/bioinformatics/btq228 [DOI] [PubMed] [Google Scholar]
- 36.Sankoff D., Minimal mutation trees of sequences. SIAM J. Appl. Math. 28, 35–42 (1975). 10.1137/0128004 [DOI] [Google Scholar]
- 37.International Trade Statistics Database, UN Comtrade Database; https://comtrade.un.org/.
- 38.Johns Hopkins University (JHU) Center for Systems Science and Engineering (CSSE), CSSEGISandData/COVID-19, Novel coronavirus (COVID-19) cases, GitHub; https://github.com/CSSEGISandData/COVID-19.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- S. E. James, T. Houriiyah, krisp-kwazulu-natal/africa-covid19-genomics: A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa - Code and scripts. Zenodo (2021); 10.5281/zenodo.5386379. 10.5281/zenodo.5386379 [DOI] [PMC free article] [PubMed]
- Johns Hopkins University (JHU) Center for Systems Science and Engineering (CSSE), CSSEGISandData/COVID-19, Novel coronavirus (COVID-19) cases, GitHub; https://github.com/CSSEGISandData/COVID-19.
Supplementary Materials
Table S4
MDAR Reproducibility Checklist