Abstract
Malaria, a major cause of child mortality in Africa, is engendered by Plasmodium parasites that are transmitted by anopheline mosquitoes. Fitness of Plasmodium parasites is closely linked to the ecology and evolution of its anopheline vector. However, whether the genetic structure of vector populations impacts malaria transmission remains unknown. Here, we describe a partitioning of the African malaria vectors into generalists and specialists that evolve along ecological boundaries. We next identify the contribution of mosquito species to Plasmodium abundance using Granger causality tests for time-series data collected over two rainy seasons in Mali. We find that mosquito microevolution, defined by changes in the genetic structure of a population over short ecological time-scales, drives Plasmodium dynamics in nature, whereas vector abundance, infection prevalence, temperature and rain have low predictive values. Our study demonstrates the power of time-series approaches in vector biology and highlights the importance of focusing local vector control strategies on mosquito species that drive malaria dynamics.
The Anopheles gambiae sensu lato (s.l.) species complex inhabits ecosystems ranging from the dry Saharan edge and savanna woodlands, to humid rainforests and saline environments. Population genetics and genomics studies have uncovered signatures of adaptations along ecological boundaries, suggesting an ecotype structure of the morphologically indistinguishable species complex1–5. Accumulating evidence indicates the importance of spatiotemporal structuring of vector populations for malaria transmission6, but the contribution of vector microevolution remains unknown. This question is particularly important in the light of climate-induced restructuring of vector populations worldwide. Currently used linear correlational approaches to mapping species biogeography7,8 and differences in resistance to Plasmodium are mostly based on analyses of samples collected within a limited window of time or in experimental infections9,10. However, such approaches often generate contradictory results as they are not well-suited for studying dynamics of natural complexities8,11.
Data-driven approaches such as Granger-causality (G-causality) analysis harness information contained in natural variance and offer powerful tools to decipher complex interactions that cannot be examined in the laboratory. G-causality tests are implemented by time-lagged regression based on the predictive notion of causality whereby a cause precedes and predicts the effect12. In contrast to classical epidemiological models, this quantitative approach makes minimal assumptions about the underlying biological mechanisms. Instead, it gathers the unique information contained in the dynamics (time series) of the causal variable to test how well a generated interaction network explains observed responses. Here we exploited G-causality to test whether genetic structure of a mosquito population impacts Plasmodium abundance.
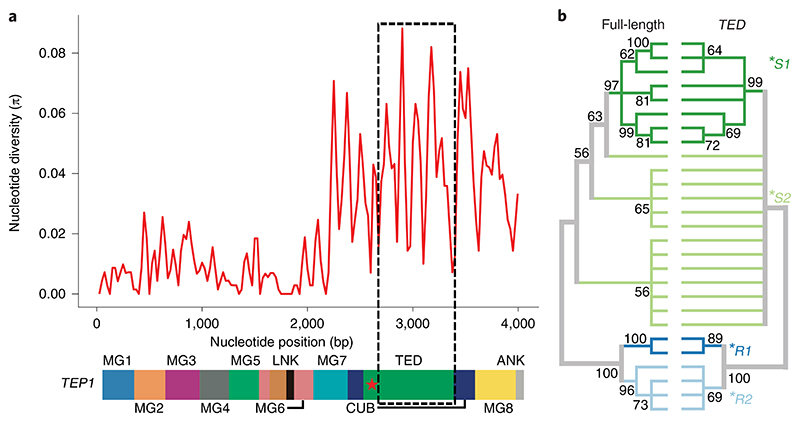
We first simplified the genetic characterization of mosquito ecotypes across sub-Saharan Africa. The Anopheles species complex is classified into species by inversions of chromosome 2 or microsatellite markers on chromosomes X and 3L13–15. The microsatellite marker on chromosome 3L encompasses a highly polymorphic key mosquito immune factor, the thioester-containing protein 1 (TEP1)16. TEP1 is a complement-like protein that mediates mosquito resistance to diverse pathogens, including bacteria17, fungi18 and Plasmodium19. As TEP1 polymorphism has been implicated in the adaptive radiation of the species complex10, we harnessed its exceptional variability to increase the genotyping resolution beyond species classification (Supplementary Fig. 1). TEP1 variation can be resolved into four allelic forms (or haplotypes) named *S1, *S2, *R1 and *R210,20. Initially, *S1 and *R1 alleles have been shown to segregate in Plasmodium-susceptible and -resistant mosquito strains, respectively21. Structural comparison and biochemical studies in vitro of the most divergent R1 and S1 forms pointed to the substitutions within the highly variable thioester domain (TED) as major contributors to the phenotypic divergence between the alleles21,22 (Fig.1a). To compare the robustness of TED as a marker for TEP1 genotyping, we sequenced full-length TEP1 and TED from laboratory and field mosquitoes and compared their amino acid sequences. Congruent topologies of the cladograms confirmed that TED contains all the information necessary for identification of TEP1 alleles (Fig.1b and Supplementary Tables 1–3).
Fig. 1. TEP1 diversity and classification.
a, Nucleotide diversity over the TEP1 coding sequence plotted above a schematic representation of the protein domain organization. The dashed box indicates the region chosen for genotyping. b, Congruency of TEP1 allelic segregation based on neighbour-joining comparison of full-length (left) versus TED only (right) amino acid sequences, illustrated by bootstrapped (1,000 iterations) phylogenetic trees (50% cut-off). MG, macroglobulin domain; LNK, linker region; CUB, complement C1r/C1s; ANK, 2-disulfide anchor motif.
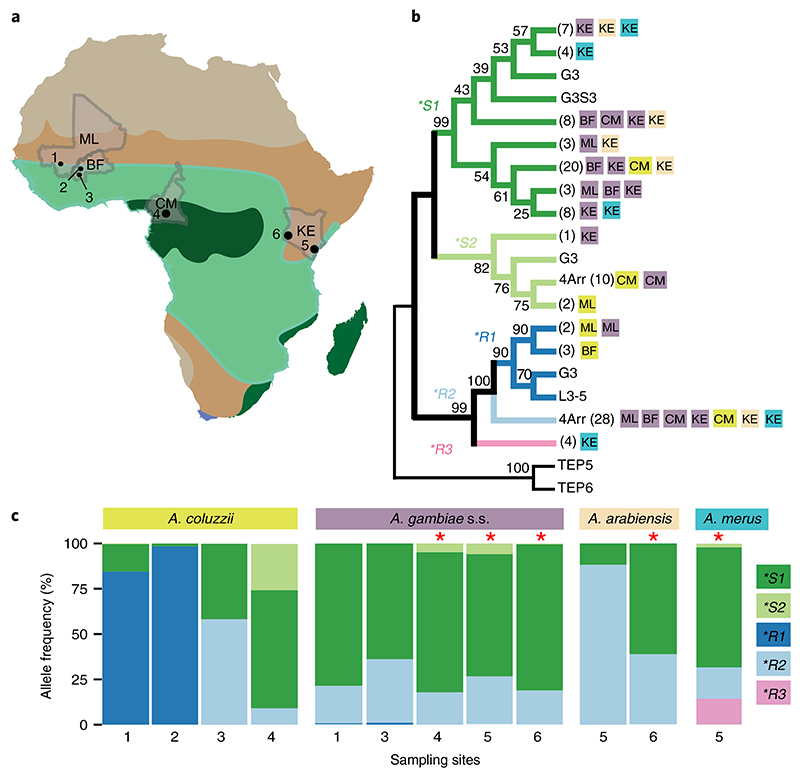
In a large-scale geographical census, we sampled and genotyped a total of 1,556 A. gambiae s.l. adults and larvae from west to east sub-Saharan Africa (Fig. 2a). The sampling sites spanned Sahelian and savanna ecosystems (Mali, Burkina Faso and Kenya), and rain-forest (Cameroon and Kenya) (Supplementary Table 4). At all sites, A. gambiae sensu stricto (s.s.) and Anopheles coluzzii were the main vector species, except for Kenya, where a temporary decline in A. gambiae s.s. tilted mosquito populations towards Anopheles arabiensis and Anopheles merus23. TEP1*S and TEP1*R alleles were identified in all species. To validate our genotyping results, we sequenced a representative set of alleles from each species and country. Marked differences were observed in the geographic and species-specific distribution of individual alleles (Fig. 2b). In addition to the known alleles, we found a previously unknown allele *R3 which was closely related to *R2 with four private small nucleotide polymorphisms in the post-β-hairpin loop (Supplementary Fig. 2). Our survey showed broad distribution of TEP1*S1 and TEP1*R2 across Africa, whereas other alleles were more geographically restricted. At the molecular level, the two most common alleles either lacked clear geographic structuring (*S1) or were highly conserved (*R2) (Fig. 2b,c and Supplementary Fig. 1). By contrast, rare alleles were species- and/or population-specific. Indeed, *R1 was only found in A. coluzzii in Mali and Burkina Faso; *S2 was enriched in A. coluzzii in Cameroon; and *R3 was restricted to the salt-water-breeding A. merus in coastal Kenya (Fig. 2c). The observed restricted allelic distribution may be caused by non-random mating resulting from local population structure or selection acting on the rare alleles. To detect signatures of non-random mating at a population level, we tested for deviations from Hardy–Weinberg equilibrium. Although significant non-random mating was detected locally for all alleles, no global pattern of inbreeding was found across species or sampling sites, providing further evidence for local selection acting at the TEP1 locus (Supplementary Fig. 3).
Fig. 2. TEP1 population genetics.
a, Location of sampling sites across African bioclimate zones (Supplementary Table 4) indicated by colour. Collected Anopheles species and sample sizes for each site are shown in brackets. Mali (ML): (1) (A. coluzzii, n = 116; A. gambiae s.s., n = 150); Burkina Faso (BF): (2) (A. coluzzii, n = 100) and (3) (A. coluzzii, n = 12; A. gambiae s.s., n = 47); Cameroon (CM): (4) (A. coluzzii, n = 210; A. gambiae s.s., n = 643) Kenya (KE): (5) (A. gambiae s.s., n = 17; A. merus, n = 49; A. arabiensis, n = 17) and (6) (A. gambiae s.s., n = 122, A. arabiensis, n = 67). b, Neighbour-joining comparison of TED amino acid sequences (n = 103) identified in the sampling sites by country (as in a) and by species (colour as in c), and compared to the laboratory strains G3, 4Arr and L3-5. Corresponding TED sequences of the closely related TEP5 and TEP6 (A. gambiae pest strain) were used as an outgroup for TEP1. Bootstrap values (1,000 iterations) are shown at the nodes of the plotted phylogenetic tree. Numbers of samples with identical sequences are shown in brackets. c, Distribution of TEP1 allelic frequencies per species across sampling sites 1–6. Significant deviations from Hardy–Weinberg equilibrium (calculated by χ2-test and indicated by the red asterisks) were observed in A. gambiae s.s. from sites 4, 5 and 6 (P=3.0×10–4, P=5.0×10–4, P=1.1 ×10–5, respectively); A. arabiensis from site 6 (P=1.5 ×10–5); and A. merus from site 5 (P=9.110–12). For details see Supplementary Table 4.
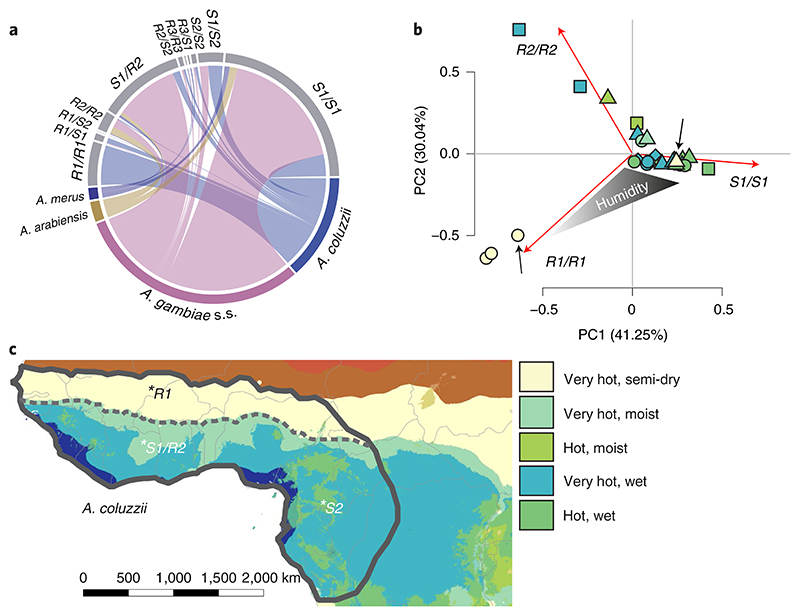
As expected from species-specific allele distribution, TEP1 genotypes were highly structured according to species. *S1/S1 and *R2/S1 were enriched in both A. gambiae s.s. and A. arabiensis; *R1/R1, *S1/S1, *S1/S2 and *R2/S2 were prominent in A. coluzzii,and *R2/S1, *R3/R3, *R3/S1 and *S1/S1 were found in A. merus (Fig.3a). Overall, the observed patterns were reminiscent of niche partitioning between generalist and specialist species. In line with our observations, previous studies have demonstrated that A. gambiae s.s. and A. arabiensis occupy a wider variety of ecosystems than the more geographically restricted A. coluzzii and A. merus24, have higher larval competitiveness25, faster larval developmental rates26 and thrive mainly during the rainy season6. Unlike specialists, generalist species are believed to be selected for dispersion rather than local competitiveness27, which prevents selection for locally adapted phenotypes. We hypothesized that ecological differences within the species complex drive local selection patterns of specialist TEP1 genotypes. To test this hypothesis, we performed principal component analysis of TEP1 genotype frequencies and observed a clear structuring of A. coluzzii into bioclimate zones according to TEP1 genotype (Fig.3b). *R1/R1 homozygotes were exclusively identified in dry biomes of the Sahel zone, whereas *R2/ S1 and *S1/S2 were mostly associated with savanna and rainforest. By contrast, no clear bioclimate structuring was observed in our samples for the generalists A. gambiae s.s. and A. arabiensis.The strongest ecotype partitioning in A. coluzzii was observed by bioclimate for *R1 and by geography for *S2 alleles, which have demonstrated roles in Plasmodium resistance20 and male fertility28, respectively. Whereas a potential trade-off between these important life-history traits was suggested for heteroallelic populations28, our results showed that these alleles occur in allopatric mosquito species. On the basis of these and previous results3,29, we propose that the observed biogeographic clines in TEP1 genotype may link TEP1 evolution to local adaptation (Fig.3c): however, other scenarios such as genetic drift or founder effects cannot be ruled out.
Fig. 3. Genetic and ecotype structures of Anopheles gambiae s.l.
a, TEP1 genotype composition of the species (sample sizes are indicated by the length of the species segment). A. gambiae s.s. (magenta) are predominantly represented by *S1/S1 and *R2/S1; A. coluzzii feature *R1/R1, *S1/S1 and *S1/S2. b, Principal component analysis of genotype frequencies in species and sampling locations (n = 15). Species are coded by shapes (A. gambiae s.s.; triangle; A. coluzzii, circle; A. arabiensis, square; A. merus, diamond), colours represent bioclimate zones39(as in colour legend, Supplementary Table 4). Arrows indicate the Mali sampling site where A. gambiae s.s. and A. coluzzii TEP*R1 breed in sympatry. c, Hypothesized map of the distribution of A. coluzzii ecotypes (delineated by grey lines) according to TEP1 genotypes over bioclimate zones39 (as in b). A. coluzzii ecotype distribution was deduced from this and previous studies3,29.
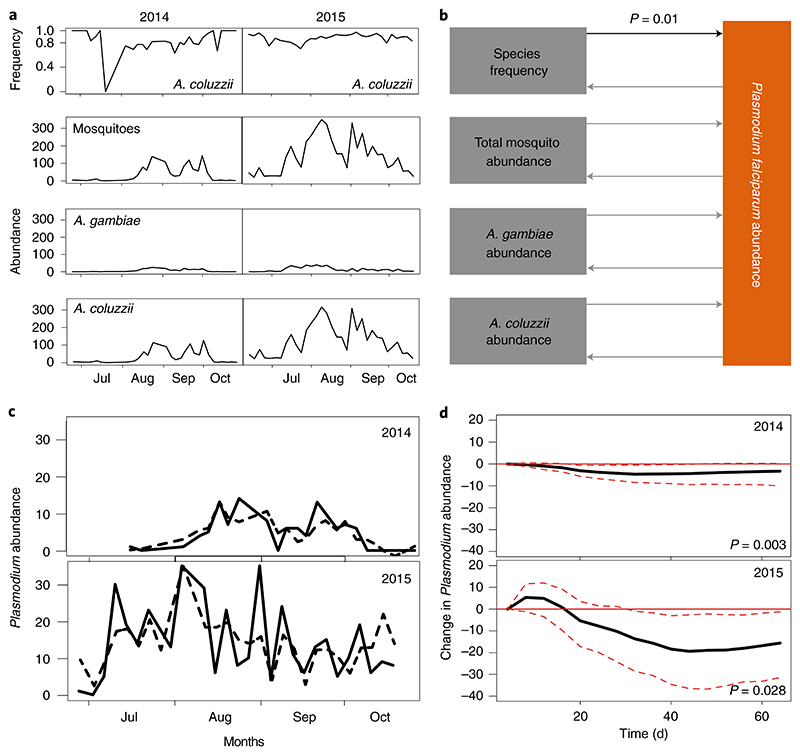
Most of the sites examined in this study featured two or more Anopheles species, with the exception of the site 2 in Burkina Faso, which was populated exclusively by Anopheles coluzzii (Fig. 2). How these sympatric vector species contribute to Plasmodium transmission, that is, whether they show similar vectorial capacity, remains unknown. Differences in vectorial capacity between A. gambiae s.s. and A. coluzzii have been examined in laboratory conditions in Mali, Burkina Faso and Cameroon with quite contradictory conclusions7,8,11. To address this question in natural settings, we revisited the exceptional Nankilabougou site in Mali, where Plasmodium-resistant A. coluzzii ecotypes (TEP1*R1) and susceptible A. gambiae (TEP1S1 and TEP1*R2) breed in sympatry (Fig. 2c, site 1). Time-series collections of adults were replicated during two rainy seasons in 2014 and 2015. Mosquito samples were genotyped for species and TEP1 as described above, whereas P. falciparum exposure was assayed in whole mosquitoes by PfCox1 -based PCR (Supplementary Table 2). Whereas the number of infected mosquitoes varied significantly during both rainy seasons, the prevalence of Plasmodium in both species was around 10% (Supplementary Fig. 4 and Supplementary Table 7). These results suggested that the two species were equally attracted to humans and had similar exposure to Plasmodium. Species genotyping confirmed the expected TEP1 allelic distribution according to mosquito species (Supplementary Fig. 5). In line with our initial characterization, A. coluzzii was predominantly TEP1*R1 and A. gambiae s.s. was represented by TEP1*S1 and TEP1*R2. Species abundances followed similar dynamics during the two rainy seasons, with consistently higher numbers of A. coluzzii than A. gambiae s.s. (Fig. 4a). To test whether vector microevolution impacted Plasmodium abundance, we used G-causality, a statistical hypothesis test for determining whether one time series can forecast another. If past (or lagged) values of a time series Y1 contain information that helps predict another signal Y2 (beyond the information contained in past values of Y2 alone), then Y1 is said to ‘G-cause’ Y2. Here, we tested whether Plasmodium abundance was G-caused by the following mosquito predictors: total mosquito abundance, abundance of A. coluzzii, abundance of A. gambiae s.s. and species frequency. We also gauged whether the environmental factors temperature and rain G-cause Plasmodium or mosquito dynamics. Every time series was modelled by a q-order vector autoregression (VAR(q)) (Methods, equations (3) and (4)) for each season separately, and a Wald test was used to determine which model had a significant contribution (Methods and Supplementary Fig. 6). The best models of both seasons (Supplementary Table 5) were subsequently summarized into one co-integrated G-causality network by Fisher’s method (Fig. 4b). Strikingly, species frequency was the only predictor that showed a significant impact on Plasmodium abundance in both seasons (adjusted P = 0.01) (Fig. 4b). Reciprocal G-causality tests revealed no impact of the number of Plasmodium-infected mosquitoes on mosquito abundance or mosquito species frequencies, indicating that Plasmodium exerts low or no selective pressure on mosquito microevolution at this sampling site. Surprisingly, the temperature and rain time series failed to predict Plasmodium abundance and species frequencies (Supplementary Table 7), suggesting that linear mathematical approaches and the time-series data collected in our study did not have sufficient power to resolve the impact of these environmental factors on mosquito populations.
Fig. 4. Granger causality analysis.
a, Time-series collections for rainy seasons in 2014 and 2015. A. coluzzii frequency, abundance of total mosquitoes, A. gambiae and A. coluzzii, and Plasmodium abundance (c, solid lines) were used to generate VAR models for each season separately. The 2014 season was sampled every 3-4 d (n = 1,091 mosquitoes; 28 time points), whereas the 2015 season was modelled as a cumulative summary of 4 d (n = 4,795 mosquitoes; 33 time points). b, Schematic representation of G-causality predictions for both seasons, arrows indicating the direction of prediction (non-significant G-causality in grey, significant in black). P values were computed using Wald-test statistics for every season, and subsequently integrated by Fisher’s method and Bonferroni correction, generating a cumulative P value for both seasons (black). c, Comparison of the observed (solid line) Plasmodium abundance in the time-series data and the predicted (dashed line) Plasmodium abundance by VAR model based on species frequency for two seasons. d, Theoretical cumulative impulse-response test. The sign of significant Granger causal interactions was determined for each season separately by estimating in silico the response to an impulse in one of the variables. A simulated 10% increase in A. coluzzii frequency in the mosquito population caused a significant drop in Plasmodium abundance in both models with a time lag of 21 d. The solid black line depicts the mean out of 1,000 bootstrap replicates, and the red dashed lines show the 95% confidence bands. Significant G-causality for every season (computed using Wald-test statistics) is given by the individual P values. For a detailed summary of the analysis, see Supplementary Table 4.
As G-causality tests do not define the sign of causal interactions (positive or negative), we explored how an increase in a causal variable affected a cumulative impulse response in the predicted variable. We simulated a theoretical increase of 10% in the frequency of A. coluzzii (the change often observed in species frequency during a rainy season) and examined in silico its effect on Plasmodium abundance. For both seasons, an increase in A. coluzzii frequency had a significant negative impact on P. falciparum abundance with a time lag of 21 d, indicating that a marked drop in the number of infected mosquitoes can be expected after one mosquito generation (Fig. 4d). Quantification of the impact of mosquito population structure on P. falciparum abundance in natural conditions has revealed significant interspecies differences in driving Plasmodium dynamics.
By exploiting the information contained in genetic fluctuations of natural populations, we demonstrated that mosquito microevolution drives Plasmodium dynamics. Although further studies of diverse ecosystems and larger datasets are needed for broader generalization of these results, our study contributes important evidence that the structure of vector populations is a crucial variable for prediction of malaria transmission. Nonlinear approaches and longer time series may identify other factors that impact Plasmodium abundance. Our genotyping results are consistent with the previously reported extensive genotypic complexity in Anopheles gambiae s.l.1–5, which is likely to be generated by differential evolutionary forces that promote alternative life-history strategies within the species complex. We found that generalist TEP1 genotypes occupy diverse environments across Africa, whereas specialized ecotypes inhabit mostly particular niches. Together with the previous reports on geo-distribution of A. coluzzii TEP1*R1 (refs. 3,29), our data suggest that this species thrives in areas with a single short-term annual rainy season and a long dry season, regulated by opposing effects of Harmattan and monsoon in West Africa. Therefore, we propose that local selection by the environment is a more likely driver of evolution of this vector competence gene than Plasmodium exposure. Further studies are needed to formally examine the role of TEP1 polymorphism on Plasmodium dynamics.
In conclusion, this report offers an advanced conceptual framework for studying natural dynamic vector–parasite interactions with potential to predict the consequences of changing environment on vector distribution and to target vector species relevant for malaria transmission.
Methods
Sample collection
Genotype mapping
Anopheles gambiae s.l. samples (n = 1,538) were collected in a cross-sectional approach in Mali, Burkina Faso, Cameroon and Kenya (Supplementary Table 4) in September and October 2009 and throughout 2012. Adult samples were collected indoors (Kenya, Mali), from mating swarms (Burkina Faso) or in the laboratory from field-collected larvae (Cameroon). Larvae were collected by spoon dipping.
Time-series collections
Collection was performed in 2014 (n = 1,091; 28 time points) and replicated in 2015 (n = 4,795; 130 time points) in Nankilabougou, Mali, where A. gambiae s.s. (TEP1*S1/S1, TEP1*R2/S1, TEP1*R2/R2) and A. coluzzii (TEP1*R1/R1) breed in sympatry. Note that bednet or ACT interventions were not performed at the site during this study. Adults were collected by CDC light traps twice a week (2014) or nightly (2015) throughout the rainy seasons. All samples were processed immediately after collection and stored in 70% ethanol before genotyping. Temperature data were collected using a data logger placed at one of the trapping sites. Rain data were obtained from the Bancumana meteorological station located 6 km from Nankilabougou.
Genotyping, sequencing and Plasmodium detection
PCR amplifications were performed on DNA extracted from whole larvae and adult legs (to prevent sperm contamination in mated females) using the DNeasy kit (QIAGEN). All samples were genotyped for species and TEP1 by previously described PCR-based methods (Supplementary Tables 1 and 2). Genotypes were identified by digestion with BamHI, HindIII and BseNI (Fermentas, according to manufacturer’s instructions) according to Supplementary Table 3. All TEP1*R1- like samples were sequenced to confirm *R1 identification. For large-scale sequence comparisons of alleles, amplicons of the TEP1–TED region (n = 103) were cloned for sequencing and compared to published sequences from the G3, L3-5 and 4Arr laboratory strains. Full-length genomic TEP1 sequences (n = 25) were obtained by PCR of overlapping fragments generated from homozygous individuals and sequenced.
For the time-series collections, DNA was extracted separately from mosquito legs and carcasses using the Nucleomag VET Kit (Marchery Nagel). The DNA isolated from legs was used for genotyping as described above. The carcass DNA served as a template to identify infected mosquitoes by PCR amplification of the P. falciparum Cox1 gene (Supplementary Table 2).
Data analysis
Sequence comparison
Sliding-window analysis of nucleotide diversity was performed by DnaSP30. Sequence comparisons were conducted with the MEGA6 Kimura (K80) model31, with invariable sites and gamma rate of evolution based on Jmodeltest32,33 for the optimal nucleotide substitution model.
Population genetics
Deviations from Hardy–Weinberg equilibrium (Supplementary Table 4) in TEP1 genotypes were examined by χ2-test in R using the HWE. chisq function from the genetics package34. This function evaluates how well the observed genotype frequencies agree with the expected genotype frequencies predicted by allele frequencies.
The fixation index FISik (ref. 35; Supplementary Fig. 3) for every subpopulation i, and every allele k, were calculated by allele frequencies p as follows:
| (1) |
where the difference of observed homozygotes P and expected homozygotes p2, is divided by the expected proportion of the allele in heterozygotes. The significance of the result can then be tested by χ2-test:
| (2) |
where N is the sample size.
Time-series analysis
We formulated vector autoregression models (VAR) of two variables, where the predicted variable y1(t) represents Plasmodium abundance and y2(t) represents one of the four considered mosquito predictors: total mosquito abundance, abundance of A. coluzzii, abundance of A. gambiae s.s. or species frequency (that is, a total of four models were built). Additionally, we also tested whether such environmental predictors as temperature and rain impact Plasmodium and mosquito dynamics. In the VAR models, the time series of one variable is trained on time-lagged values t – q of both variables, that is, for every time-point t, the value yx(t) of one variable is predicted by a historical time-lagged autoregressive term yx(t – q), and historical values yy(t – q) of the other variable. The VAR models are given by:
| (3) |
| (4) |
where q is the maximum number of lagged observations included in the model (that is, the order), and the matrix A contains the time-invariant coefficients (representing the contribution of each time-lagged observation to the predicted value y1(t) and y2(t)). The time-invariant coefficients for both variables and intercept constants c were found by linear regression. This time-series analysis was carried out in R36 using the vars package37.
Because the collection frequency of adult mosquitoes differed between 2014 and 2015 (twice a week and daily, respectively), we performed an in silico downsampling of the 2015 dataset. To this end, we summed the values of the time series over non-overlapping time windows of length k = 3–4 d. This approach effectively increased the sample sizes for every time point, thereby increasing measurement accuracy. For every time window k, we repeated this sampling procedure starting at different time points, generating k–1 different time series. Every of these generated datasets was modelled with VAR(q), and the accuracy in predicting both variables was then assessed by calculating the average goodness of fit over all q, that is, . The best VAR model was observed for k = 4 and for the datasets starting either at the earliest or at second time point (Supplementary Table 8). Given that the time series starting at the earliest time point was longer (thus containing more information), it was selected for further modelling of the 2015 season.
The model order q of VAR models have a strong impact on the conclusions that can be drawn from them. To select an appropriate time lag q, we next devised an out-of-sample validation approach, where the models that were trained on the data from one season were used to predict the measurements of another season. The model with the highest out-of-sample accuracy as determined by R2 was chosen for subsequent G-causality testing (Supplementary Table 9).
Granger-causality testing
To test whether mosquito dynamics and environmental factors Granger-cause Plasmodium abundance, the dynamics of P. falciparum abundance were modelled separately for each sampling season and for each mosquito and environmental variable (Supplementary Fig. 6). To infer Granger causality for both directions of prediction (for example, whether mosquito abundance causes Plasmodium abundance or Plasmodium causes mosquito abundance), the contribution of the slope coefficients y2(t) to equation (3) and y1(t) to equation (4), were confirmed by the Wald test:
| (5) |
| (6) |
Granger non-causality is rejected at low P values of the Wald test and the alternative hypothesis of a causal relationship is accepted. It is important to note that whereas a significant P value implies causality, the absence of significance does not prove the absence of causality.
Granger causality tests require models that are serially uncorrelated and dynamically stable. Therefore, all models where tested for correlated residuals using a portmanteau test using the serial test function of the vars package. To test for stability, the eigenvalues of the model’s characteristic polynomial were confirmed to be below 1.
The asymptotic assumption of the Wald test is violated when non-stationary time series are modelled. One approach to make a time series stationary is to difference it. However, to avoid potential bias of the Granger test caused by data transformation, we applied a method developed by Toda and Yamamoto38 for Granger causality testing in non-stationary time series. The order of integration (m) that is required to make the time series stationary was identified by the ndiffs function of the forecast package which implements the Kwiatkowski-Phillips–Schmidt–Shin, augmented Dickey–Fuller and Phillips–Perron tests. Of note, the VAR(q+m) model was only applied for hypothesis testing, and not for the described model analysis and selection.
To evaluate which interactions can be generalized beyond a single rainy season, a consensus causality network was postulated by P-value integration. To this end, P values of the Granger causality test for the same hypothesis (for example, species frequency causes Plasmodium abundance) but from different seasons were integrated by Fisher’s method. This method is commonly used in meta-analysis and yields a χ2-value, with which a cumulative P value can be derived. Because the additional replication is appreciated, the resulting P value can be lower than the input P values. As a result of the out-of-sample validation step, the hypothesis tests cannot be seen as entirely independent replicates of the hypothesis test; therefore, we introduced a false discovery rate correction (n = 2). Finally, a Bonferroni correction for multiple hypothesis testing (n = 6) with the same dataset was applied.
Impulse-response analysis
Having established that mosquito species frequency G-caused Plasmodium abundance, we tested for the sign of this interaction using a cumulative impulse response. Generally, an impulse response is the reaction of the dynamic system to an external change, which describes the reaction of the system as a function of time. To estimate the response of Plasmodium abundance to a theoretical increase in A. coluzzii frequency, cumulative impulse-response functions were generated with the irf function of the vars package with 1,000 bootstrap replicates at a 95% confidence interval. For non-zero impulse-response functions, the sign of the tested interaction is the direction of the impulse response. For example, in a negative interaction, an increase in the predictor will cause a decline in the predicted variable.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-019-0414-9.
Acknowledgements
We thank A. Telschow, F. Grziwotz and S. F. Traore for helpful suggestions; L. Spohr, S. Koppitz, D. Coulibaly, M. Coulibaly, C. Omogo, J. Shikaya, L. Abate, E. Onana, J. P. Agbor, J. Mwaura and A. Gildenhard for technical support; and M. Doumbia, B. Doumbia and C. Okech for their help with sample collections. This research was supported by funds from EC FP7 under grant agreements N°223601 (MALVECBLOK) and N°242095 (EVIMalar). E.K.R. was a DAAD fellow.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Author contributions
FC., D.M., M.D., I.M., D.S. and E.A.L. conceived and designed the study. D.S., M.D., A.Diabaté, R.K.D., P.A.-A., D.M. and I.M. designed and managed sample collections, genotyping and sequencing. E.K.R., A.B., I.M. and E.A.L. contributed molecular tools. J.T., J.P., P.A.-A., M.M., A.B., S.E.N., P.B.S., P.M., A.T., D.S., M.D., M.K.R., E.K.R., C.V.Y. and H.K. conducted sample collections, processing and genotyping. P.B., A.B., P.M., P.B.S. and E.K.R. performed sequencing. E.K.R., A.B., P.B., M.G., I.M., F.C. and E.A.L. analysed the data. M.G. and E.A.L. conceived and designed the time series. M.G., Y.R., M.D. and D.S. designed and managed time-series collections. D.C., R.M., H.K., M.G., D.S. and A.Diarra performed time-series collections and genotyping. E.A.L. and M.G. analysed time-series data. M.G. designed and carried out population genetic analyses and time-series modelling. P.C.-B. contributed to time-series analyses. M.G., E.K.R., P.C.B. and E.A.L. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data Availability
The full-length TEP1 and TEP1–TED sequences are available at the NCBI GenBank under accession numbers MF098568 to MF098592 and MF035727 to MF035924, respectively. The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 2.Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 3.Pinto J, et al. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evol Appl. 2013;6:910–924. doi: 10.1111/eva.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simard F, et al. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillies MT, Shute GT. Environmental influences and the maxillary index in Anopheles gambiae. Nature. 1954;173:409–410. doi: 10.1038/173409b0. [DOI] [PubMed] [Google Scholar]
- 6.Dao A, et al. Signatures of aestivation and migration in Sahelian malaria mosquito populations. Nature. 2014;516:387–390. doi: 10.1038/nature13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fryxell RTT, et al. Differential Plasmodium falciparum infection of Anopheles gambiae s.s molecular and chromosomal forms in Mali. Malaria J. 2012;11:133. doi: 10.1186/1475-2875-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boissière A, et al. Application of a qPCR Assay in the investigation of susceptibility to malaria infection of the M and S molecular forms of An. gambiae s.s in Cameroon. PLoS ONE. 2013;8:e54820. doi: 10.1371/journal.pone.0054820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eldering M, et al. Variation in susceptibility of African Plasmodium falciparum malaria parasites to TEP1 mediated killing in Anopheles gambiae mosquitoes. Sci Rep. 2016;10:20440. doi: 10.1038/srep20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White BJ, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci USA. 2011;108:244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnémé A, et al. Equivalent susceptibility of Anopheles gambiae M and S molecular forms and Anopheles arabiensis to Plasmodium falciparum infection in Burkina Faso. Malaria J. 2013;14:204. doi: 10.1186/1475-2875-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424. [Google Scholar]
- 13.Fanello C, Santolamazza F, Della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- 14.Wondji C, Simard F, Fontenille D. Evidence for genetic differentiation between the molecular forms M and S within the Forest chromosomal form of Anopheles gambiae in an area of sympatry. Insect Mol Biol. 2002;11:11–19. doi: 10.1046/j.0962-1075.2001.00306.x. [DOI] [PubMed] [Google Scholar]
- 15.Della Torre A, et al. Speciation within Anopheles gambiae— the glass is half full. Science. 2002;298:115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- 16.Obbard DJ, et al. The evolution of TEP1 an exceptionally polymorphic immunity gene in Anopheles gambiae. BMC Evol Biol. 2008;7:274. doi: 10.1186/1471-2148-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levashina EA, et al. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito Anopheles gambiae. Cell. 2001;104:709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 18.Yassine H, Kamareddine L, Osta MA. The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 2012;8:e1003029. doi: 10.1371/journal.ppat.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blandin S, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 20.Blandin SA, et al. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science. 2009;326:147–150. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter RHG, et al. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci USA. 2007;104:11615–11620. doi: 10.1073/pnas.0704967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le BV, Williams M, Logarajah S, Baxter RH. Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium Structural analysis of TEP1 susceptible and resistant alleles. PLoS Pathog. 2012;8:e1002958. doi: 10.1371/journal.ppat.1002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mwangangi JM, et al. Shifts in malaria vector species composition and transmission dynamics along the Kenyan coast over the past 20 years. Malaria J. 2013;8:13. doi: 10.1186/1475-2875-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinka ME, et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2011;4:89. doi: 10.1186/1756-3305-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diabaté A, et al. Larval development of the molecular forms of Anopheles gambiae (Diptera: Culicidae) in different habitats: a transplantation experiment. J Med Entomol. 2005;42:548–553. doi: 10.1093/jmedent/42.4.548. [DOI] [PubMed] [Google Scholar]
- 26.Diabaté A, et al. Evidence for divergent selection between the molecular forms of Anopheles gambiae Role of predation. BMC Evol Biol. 2008;8:5. doi: 10.1186/1471-2148-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton OJ, Phillips BL, Travis JMJ. Trade-offs and the evolution of life-histories during range expansion. Ecol Lett. 2010;13:1210–1220. doi: 10.1111/j.1461-0248.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 28.Pompon J, Levashina EA. A new role of the mosquito complement-like cascade in male fertility in Anopheles gambiae. PLoS Biol. 2015;13:1002255. doi: 10.1371/journal.pbio.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancini E, et al. Adaptive potential of hybridization among malaria vectors: Introgression at the immune locus TEP1 between Anopheles coluzzii and A. gambiae in ‘Far-West’ Africa. PLoS ONE. 2015;10:0127804. doi: 10.1371/journal.pone.0127804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Librado P, Rozas J. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 33.Posada D. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 2009;537:93–112. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- 34.Team, R. D. C. The genetics package. Bioinformatics. 2008;3:9–13. [Google Scholar]
- 35.Nei M. F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet. 1977;41:225–233. doi: 10.1111/j.1469-1809.1977.tb01918.x. [DOI] [PubMed] [Google Scholar]
- 36.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2016. [Google Scholar]
- 37.Pfaff B. VAR, SVAR and SVEC models: implementation within R package vars. J Stat Softw. 2008;27:i04. [Google Scholar]
- 38.Toda HY, Yamamoto T. Statistical inference in vector autoregressions with possibly integrated processes. J Econom. 1995;66:225–250. [Google Scholar]
- 39.Metzger MJ, et al. A high-resolution bioclimate map of the world: a unifying framework for global biodiversity research and monitoring. Glob Ecol Biogeogr. 2013;22:630–638. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full-length TEP1 and TEP1–TED sequences are available at the NCBI GenBank under accession numbers MF098568 to MF098592 and MF035727 to MF035924, respectively. The data that support the findings of this study are available from the corresponding author upon request.