Abstract
All organisms are subjected each day to changes in the light intensity generated by the Earth’s rotation around its own axis. To anticipate this geo-physical variability, and to appropriately respond biochemically, most species, including mammals, have evolved an approximate 24-hour endogenous timing mechanism known as the circadian clock (CC). The ‘clock’ is self-sustained, cell autonomous and present in every cell type. At the core of the clock resides the CC-oscillator, an exquisitely crafted transcriptional-translational feedback system. Remarkably, components of the CC-oscillator not only maintain daily rhythmicity of their own synthesis, but also generate temporal variability in the expression levels of numerous target genes through transcriptional, post-transcriptional and post-translational mechanisms, thus, ensuring proper chronological coordination in the functioning of cells, tissues and organs, including the liver. Indeed, a variety of physiologically critical hepatic functions and cellular processes are CC-controlled. It is not surprising then, that the modern lifestyle (e.g. travel and jet lag, night and rotating shift work), which force ‘circadian misalignment’ have emerged as major contributors to global health problems including obesity, non-alcoholic fatty liver disease (NAFLD) and steatohepatitis (NASH). Here, we provide an overview of the CC-dependent pathways which play critical roles in mediating several hepatic functions under physiological conditions and, whose deregulations are implicated in chronic liver disease including NASH and alcoholic liver disease (ALD).
Introduction
The word circadian is Latin in origin, and translates to ‘about a day’, hence, oscillations of ~24 hours are referred as circadian rhythms. These rhythms are generated by the Earth’s 24 hours rotation, which, in turn drives the light-dark cycle. This daily change in the light intensity leads to overt rest-activity and feeding-fasting cycles, e.g. human beings are diurnal and conduct most of their activities during the day and rest at night. Teleologically, these rhythms have allowed organisms to anticipate changes in the external environment (e.g. the light-dark cycle), and to respond by adjusting CC-driven physiological functions, e.g. metabolism (1–7). Accordingly, the CC-controlled behavioral synchronization with feeding-fasting cycles generates diurnal variations in metabolic activities, which, in turn, ensures energy homeostasis. Recent investigations have established that, in mammals, the expression of numerous genes in different organs (including the liver) display circadian rhythmicity, which enables regulation of both anabolism and catabolism (1–6). Indeed, hepatocyte activities such as nutrient uptake, processing, assimilation and detoxification exhibit remarkable diurnal variations, which enable their alignment with food availability and energetic demand. Physiologically these ‘metabolic rhythms’ are generated and maintained by the dynamic interactions between the CC and timing cues e.g. light and food (time of eating and its quality). However, our modern lifestyle (jet lag, shift-work, energy-dense foods etc.), which often ‘misaligns’ CC functioning has recently emerged as prominent contributors to different metabolic diseases and carcinogenesis (8–13).
Here, we focus on how the CC regulates hepatic metabolism to maintain homeostasis and, also provide an overview of how deregulation of CC-controlled pathways could lead to the development of non-alcoholic fatty liver (NAFL) and its progression to NASH. Furthermore, we also discuss evidences linking the CC and alcohol-induced liver disease (ALD).
The anatomic and molecular organization of the mammalian CC-system
Retinal photoreceptors (rods and cones) transform photic energy to electrical impulses and convey them to the brain through retinal ganglion cells (RGC). A subset of RGCs expressing the photopigment melanopsin are intrinsically sensitive to the visible spectrum and directly relay the photic signal to a hypothalamic region called the suprachiasmatic nucleus (SCN) (7–8). Hence, anatomically, the mammalian circadian system is hierarchical, whereby the light-entrained SCN is the ‘central’ CC. In turn, the SCN-CC by utilizing humoral and neuronal mechanisms communicates the ‘time cue’ (a. k. a; ‘Zeitgeber’; ZT) to other organs, thereby enabling the synchronization of peripheral CCs (PCCs) (1–8).
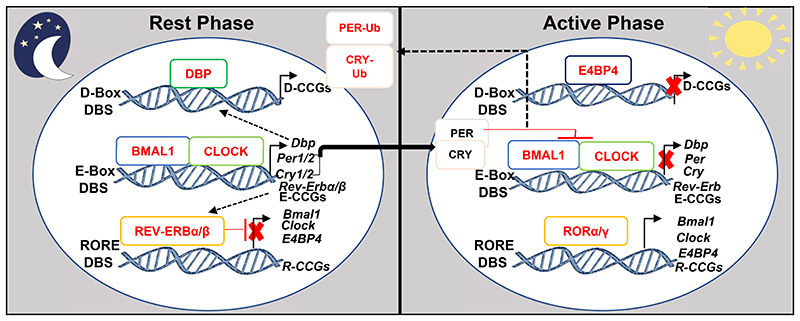
At the molecular level, the components of the central SCN and PCCs are the same and are identically organized in multiple transcriptional-translational feedback systems (Figure 1) and generate a cell autonomous self-sustained CC-oscillator with a periodicity of ~24 hours (1–6). The heart of this oscillator is constituted by a heterodimer of transcription factors (TFs), the Brain and Muscle ARNTL-Like protein1 (BMAL1) associated with the Circadian Locomotor Output Cycles Kaput (CLOCK), which activate genes containing E-Box DNA Binding Sequences (DBS) in their promoter-enhancer regions, including those of Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes. In turn, PER1/2 and CRY1/2 proteins heterodimerize to inhibit the transcriptional activity of the BMAL1/CLOCK-complex (the first-loop of the oscillator), thereby eventually suppressing their own expression (Figure 1). BMAL1/CLOCK-also binds to the E-Box DBSs present in the genes of the nuclear receptors Rev-Erbα and Rev-Erbβ to activate their transcription, while the presence of RORE DBSs in the Rev-Erbα/β genes mediate their autorepression. REV-ERBs also inhibit (through RORE DBSs) the transcription of their activators Bmal1 and Clock, thus constituting the second-loop of the CC-oscillator. At the beginning of the active phase, levels of REV-ERBs (which are repressors of transcription) decrease, while simultaneously protein levels of transcriptional activators RORα/γ increase, which then bind to RORE DBSs present in Bmal1 and Clock and activate their transcription, thereby initiating the next round of CC oscillation. Additionally, BMAL1/CLOCK-induces expression of the transactivator D-Box binding protein (Dbp). The DBP activator and the E4BP4 repressor (which is activated by RORα/γ and repressed by REV-ERBα) competes for binding to D-Box DBSs present in several CC-controlled genes (CCGs).These inter-connected feed-back loops generate circadian oscillations of the expression of ~20% of the genome, such that CCGs containing RORE DBS are transcribed during the active phase, while E-Box and D-Box DBS-bearing genes are expressed during the rest phase(1–6,14). Moreover, post-translational modifications of CC-components also aids in further fine-tuning of the CC-oscillator functioning. Thus, utilizing multiple mechanisms the CC-oscillator drives a temporally-restricted gene expression pattern, which lies at the core of generating distinct biochemical outputs in individual organs.
Figure 1. The molecular architecture of the Circadian Clock (CC)-oscillator.
The recruitment of BMAL1/ CLOCK-heterodimer to the E-Box DBS present in the promoter-enhancer elements of numerous CCGs, including Periods (Per1/2) and Cryptochromes (Cry1/2) augment their expression during the rest phase. Following accumulation, PERs and CRYs proteins dimerize and translocate to inhibit BMAL1/CLOCK-dependent transcription during the active phase. Next, post-translational modifications including ubiquitination induce proteasomal degradation of PERs and CRYs, thus, initiating the next circadian cycle. In the second loop, BMAL1/CLOCK-dependent expression of Rev-Erbα/β during the rest phase, leads to the trans-repression of several RORE-DBS-containing CCGs including, Bmal1, Clock and E4BP4. In the active phase, the reduction in REV-ERBs levels permit the RORα/γ-dependent RORE-mediated activation of CCGs including Bmal1 and Clock, which enables the turning of the circadian clock. Furthermore, DBP expression during the rest phase activates D-Box DBS containing CCGs, which are transcriptionally repressed by E4BP4 during the active phase. These coupled transcriptional-translational regulatory circuits are ubiquitously present in almost all cell types and directly control the expression of a vast number of mammalian genes.
CCG-Clock Controlled Genes. E-CCGs: E-Box DBS-containing CCGs, R-CCGs: RORE-containing CCGs, D-CCGs: D-Box-containing CCGs.
Feeding cycles and peripheral clocks
Establishment of feeding cycles as the prominent zeitgeber for peripheral tissues, including the liver (15–16), has revealed the existence of extensive cross-talk between metabolism and the CC, and the list of mechanisms through which metabolic signals influence CC functioning and, vice versa, are increasing rapidly (1–5). The dominance of feeding cycles on the liver-clock was demonstrated in ‘arrhythmic’ Cry1/2 mutant mice, in which an imposed feeding regime recovered ‘rhythmicity’ in circadian gene expression pattern (17). Furthermore, changing the feeding time from active to rest phases in mice shifts the PCCs by ~12 hours (15–16), driven by metabolic alterations acting through PPARα and CREB (18). Notably, high-fat diet (HFD)-induced ‘reprogramming’ of the hepatic CC (19) can be prevented by restricting HFD feeding during the active phase (20). One physiological example of CC-metabolism crosstalk is provided by BMAL1/CLOCK-dependent transcription of the nicotinamide phosphoribosyl transferase (NAMPT) gene, which is involved in NAD+ synthesis (21–22). CC-dictated NAMPT expression ensures not only a rhythmicity in NAD+ synthesis but also regulates the activities of NAD+ -dependent proteins (1–5), e.g. the SIRT1 deacetylase and the ADP-ribosyltransferase PARP-1. In turn, SIRT1 determines: (i) the activity of BMAL1/CLOCK-complex towards their target genes and, (ii) the stability of PER2 protein, which together maintain CC-oscillator functioning (1–5). Akin to NAD+, feedback regulation between the CC and heme biosynthesis has also been demonstrated (23–24). Thus, by controlling metabolite sensors (NAD+, heme etc.), the CC gauges the cellular energetic and redox status to reset the oscillator with metabolism. Altogether, these investigations have established metabolism as a critical modulator of PCCs.
Circadian regulation of hepatic functions
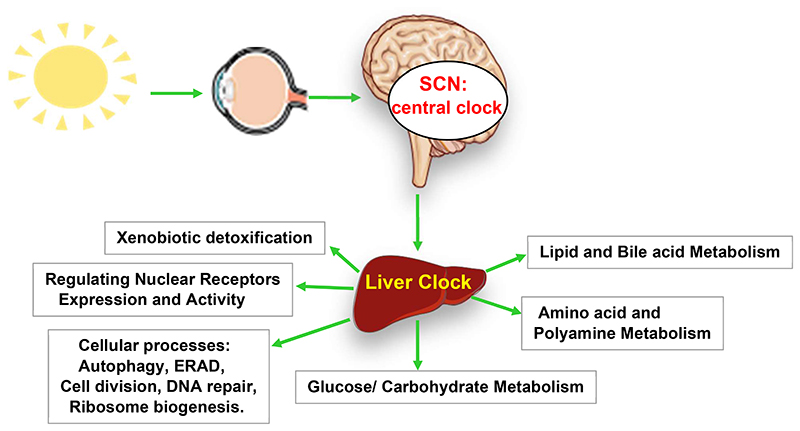
Given the centrality of the liver in maintaining whole-body physiology, several high-throughput circadian time-course studies have been performed in mouse models investigating cistrome (25–28), transcriptome (29–30), proteome (31–33), and lipidome (34–35). Circadian transcriptome analyses have revealed two broad crests of transcription in liver, corresponding to the transition between successive active and rest phases (1–5). Cistromic analyses (25–28) revealed that these two distinct mRNA repertoires are generated due to the rhythmicity of the CC-oscillator, which enables periodic recruitment/removal of TFs and coregulators to epigenetically alter the chromatin landscape of CCGs. Cellular processes like DNA repair, ribosome biogenesis, autophagy, ER-stress are also subjected to circadian regulation, but mainly at the post-translational level (31–33). Altogether, these investigations have revealed an unprecedented level of CC-control on hepatic physiology (Figure 2). Importantly. deregulation of these CC-regulated pathways/processes has been shown to contribute to the development of NAFLD and other diseases.
Figure 2. The clock controls the physiology of liver.
Light-entrained central SCN-clock synchronizes peripheral tissue clocks including that of liver. The ‘clock’ machinery in turn drives the expression of several key transcription factors, rate limiting enzymes and transport proteins to spatiotemporally regulate several biochemical processes, which, together maintain physiological homeostasis. The ‘clock’-connections to some of these processes and their connections to NAFLD and NASH have been discussed in detail.
The circadian clock and pathophysiology of NAFLD and NASH
Over the last decades, life-style changes have shifted health care priorities world-wide from infectious to metabolic diseases (36–38). In the context of liver disease, vaccination can now prevent hepatitis B virus (HBV) infection and antivirals can control chronic HBV infection (39–40,43), and recently developed direct-acting antivirals can cure chronic hepatitis C virus in a large majority of infected patients (41–43). In contrast, the prevalence of metabolic liver diseases such as NAFL and NASH are increasing dramatically in conjunction with obesity and type II diabetes (36–38,44). NAFLD, is a continuous spectrum of disease initiated by excessive triglyceride (TG) accumulation in the liver. In the absence of concomitant inflammation and hepatocytic injury, this state is largely benign and commonly referred as nonalcoholic fatty liver (NAFL) or simple steatosis (36–38, 45). However, chronic NAFL usually drives simple steatosis to steatohepatitis (NASH), which is typified by concomitant presence of both lobular inflammation and hepatocellular damage (ballooning). Moreover, NASH predisposes to fibrosis, progressing to cirrhosis and hepatocellular carcinoma (HCC) (36–38). Like every other aspect of the metabolic syndrome, development of NAFLD and NASH is highly complex which has been reviewed extensively elsewhere (36–38,46–48). Almost two decades earlier the ‘two-hit’ theory (49) was posited to explain NASH pathogenesis. This theory proclaimed that unrestrained TG deposition in the liver (first-hit; NAFL) leads to ‘secondary hits’ such as oxidative stress, which ultimately leads to NASH. However, with increasing knowledge of metabolism and associated pathologies, NAFLD is now considered as a multi-factorial systemic metabolic disorder (36–38). Indeed, investigations have revealed crucial roles for intestine, adipose tissue and muscle in NAFLD development. Importantly, insulin resistance also plays a critical, if not indispensable role in NAFLD (36,38,45).
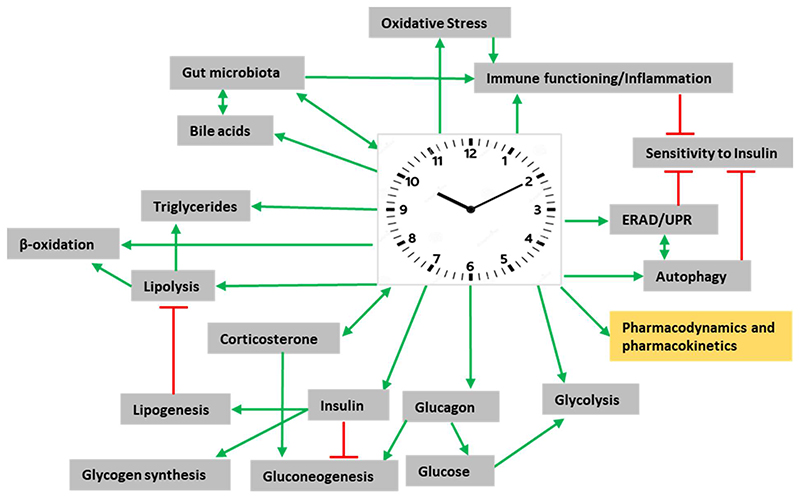
Systemic energy homeostasis is maintained by communications between numerous intra- and inter-organ signaling networks and at the core of NAFLD pathogenesis lies the inability of the liver to effectively metabolize carbohydrates and fatty acids (36–38, 50). The pathology of NAFLD is generally initiated by perturbations in free fatty acid (FFA) metabolism, which drives excessive TG accumulation in hepatocytes (36–37). Increased FFA release from adipocytes due to insulin resistance (51) and conversion of excess carbohydrates to FFA via hepatic de novo lipogenesis (DNL) (52) are two major sources of TG deposition during NAFLD development, in addition to excess caloric intake. In hepatocytes, FFA can either undergo β-oxidation or be re-esterified as TG. In turn, this pool of TG can either be exported as VLDL particles or stored in lipid droplets (36–38). The capacity to metabolize FFA through either β-oxidation or TG formation when overwhelmed (perturbation of dynamic lipid fluxes), leads to the accumulation of lipotoxic species. This buildup of lipotoxic molecules in turn damages hepatocytes through several pathways; e.g. enhanced ERand oxidative-stress, a dysfunctional unfolded protein response (UPR), and inflammosome activation to finally lead to NAFLD development (37,50,53–54). In the subsequent sections we describe some of these hepatic functions and processes which show diurnal variations and whose deregulation could predispose towards NAFLD/NASH (Figure 3).
Figure 3. Multidimensional connections of the ‘clock’ to the pathogenesis of fatty liver.
Model representing a global view of how alterations in circadian clock-controlled ‘rhythmic’ functions/pathways and processes could predispose to non-alcoholic fatty liver disease. Knowledge of the mechanisms through which the ‘clock’-system influences all these systems and essential pharmacological parameters, in turn, could be utilized to develop novel chronotherapeutics. Green arrowheads represent activation and red bar-heads represent inhibition. See text for details.
Circadian control of glucose metabolism
The liver is the principal gluconeogenic organ in mammals, and participates, along with several other organs, to maintain homeostatic blood glucose levels. The CC sustains the physiological levels of blood glucose by synchronizing tissue-specific mechanisms of glucose metabolism. Accordingly, the SCN-clock controls the feeding/fasting rhythms, while PCCs (liver, pancreatic β-cells, skeletal muscles) drive temporally coordinated gene expression programs to maintain physiological levels of glucose in blood (55).
One of the first studies indicating a role for the liver-CC-oscillator in glucose metabolism showed that Bmal1 ablation in hepatocytes reduced expression of the glucose transporter (Glut2), leading to a decreased post-absorptive glucose uptake in mutant mice (56). Post-hepatocytic entry, glucose is phosphorylated to glucose-6-phosphate (G6P), which can be either used (through glycolysis or hexose monophosphate pathway) or stored (glycogen synthesis). Remarkably, the CC influences all these processes (2,6). For example, the hepatic expression of glucokinase (Gck), which controls both glycolysis and glycogen synthesis is rhythmic reaching its zenith during the transition from the rest-phase to the active-phase (17, 57) and temporally matches the surge of postprandial insulin secretion from the pancreas. This increase in insulin secretion also leads to a pulsatile glycogen synthase kinase 3 (GSK3) activity in liver (17), which, in turn, determines: (i) the enzymatic activity of glycogen synthase, (ii) the activity of the glycosylating enzyme O-linked N-acetylglucosamine transferase (OGT), thereby, generating rhythmicity in the glycosylation levels of numerous proteins (58) and, (iii) the stability of REV-ERBα (2,4), which in turn dictates the expression of many CCGs. By controlling the expression of trans-activators Klf10 (59) and Hnf4α (60), the liver-CC further dictates transcription of several genes, which are involved in glucose metabolism.
The CC also controls glucagon-induced gluconeogenesis in liver by regulating the duration of hepatic cAMP production (61). It was demonstrated that the interaction of the CC-component CRY1 with the regulatory α-subunit of the glucagon receptor blocks hepatic cAMP accumulation during the circadian active phase, thus leading to a temporally-restricted (between rest- and active-phases) activation of the gluconeogenic transcription factor CREB (55,61). Moreover, BMAL1-regulates the expression of the Pgc1α gene (62), which is a coactivator of the gluconeogenic transcription program (2–4). Thus, by employing multiple strategies the CC controls diverse mechanisms which co-operate to maintain physiological glucose levels (1–5, 55).
Circadian regulation of liver lipid metabolism
In a seminal study, Turek et al. (63) demonstrated that Clock mutant mice are obese and have increased blood levels of cholesterol and TG. Since then, multiple genetic studies in mice models have established the CC as a critical regulator of lipid metabolism (64–66). Indeed, plasma levels of FFA, TG and cholesterol display diurnal variations, and are altered upon mutations of CC-components. Notably, the liver plays a crucial role in generating these variations in blood levels. Indeed, hepatocyte-specific ablation of Rev-Erbα/β was found to increase plasma levels of FFA, TG and cholesterol (27,66)· In this regard, a lipidomic study revealed that TG, phosphatidyl inositol and phosphatidyl choline preferentially accumulate in mouse liver during the rest phase (35). Mechanistically, the CC controls enzymes that are critically involved in regulating various steps of lipid metabolism. As example, expression of the enzyme ATP citrate lyase, which drives mitochondrial export of acetyl Coenzyme A (acetyl CoA), is maximal at the beginning of the active phase (17). Cytosolic acetyl CoA is carboxylated by acetyl CoA carboxylase (ACC1) to generate malonyl CoA an essential step in fatty acid synthesis. It is well known that AMPK inactivates ACC by phosphorylation (2–4), and CCs by controlling AMPK ‘temporally gate’ ACC activity (67). Furthermore, the liver CC by controlling the transcription of Elovl3, Elovl6, Fas etc. ‘times’ fatty acid synthesis (2–6). Moreover, the expression of enzymes regulating β-oxidation (Cpt1/2) and ketone-body production (Hmgcs2) (68–69), as well as their transcriptional regulators PPAR α and δ are also circadian in nature (60).
Hepatic TG synthesis from glycerol-3-phosphate is a multistep process and expression of several genes (Gpat2, Agpat1/2, Lipin1/2 and Dgat2) that regulate successive steps of TG synthesis is circadian in nature (35). Importantly, by controlling the transcription of Pnpla3 the CC also regulates lipid droplet dynamics (35). Altogether, in ad libitum fed mice livers, a prominent crest and trough of TG levels are observed during the rest (~ZT8) and active phases (~ZT2O). Additionally, REV-ERBα-controlled expression of Insig2 regulates the activity of SREBP1c, thereby leading to CC-command over lipogenesis (70).
Clock and metabolism of bile acids
Intestinal absorption of lipids requires bile acids (BA), which are synthesized in hepatocytes. Besides lipid absorption, recent evidences have established BA as signaling molecules (71–72). BA are physiological ligands for FXR and the G-protein coupled receptor TGR5 and can activate signaling modules such as the MAPK-pathway (71–72). By regulating these diverse signaling networks, BA not only control their own levels but also those of TG, cholesterol and glucose homeostasis (71–72). BA synthesis is controlled by a transcriptional feed-back loop consisting of the nuclear receptors FXR and SHP and hormone FGF15 (FGF19 in humans) (38,73). Hepatic expressions of both FXR and SHP (60) and the intestinal secretion of FGF15 are ‘clock’-gated (74), which, together, drive the circadian transcription of cholesterol 7α-hydroxylase (Cyp7a1), the rate-limiting enzyme in the classical BA synthesis pathway. Moreover, the CC-output regulator DBP controls Cyp7a1 transcription to restrict its temporal expression (75). Additionally, by regulating the transcription of both E4BP4 and SHP, REV-ERBα directly regulates the expression of Cyp7a1. (76) Altogether, these mechanisms cooperatively generate diurnal rhythmicity in BA levels (Figure 3), which is also observed in humans (77).
Clock-controlled cellular processes and NAFLD
Along with controlling systemic metabolism (2–5), several investigations have indicated a critical role for the CC-machinery in regulating autophagy, ER stress and oxidative stress (78–80), all of which may participate in NAFLD and in its transition to NASH (36–37). For example, in murine livers, expression of key genes controlling different steps of the autophagic process display circadian rhythms, thereby leading to an overall diurnal rhythm in autophagic activity (78–79). Consistently, hepatocytic mutation of Bmal1 impairs the entire autophagic process in murine liver. The CC also modulates the ER stress-induced activation of the UPR-driven gene expression program (78,80). Physiologically, the UPR is necessary to restore cellular secretory capacity following an accumulation of misfolded proteins in the ER and functions by degrading unfolded proteins and activating the expression of chaperones which enable protein folding (81). The CC by generating ultradian (lesser than a day) rhythms in the expression of UPR master regulators i.e. the inositol-requiring enzyme1α (IRE1α) and the X-box binding protein1 (XBP1) controls the expression of several genes within the UPR pathway (78,80). Moreover, by regulating activation of the transcription factor CREBH (82) and expression of the deadenylase CPEB4 (83), the CC extends control over the ER-stress response pathway. Deregulation in reactive oxygen species (ROS)production and scavenging have been implicated in the development of NAFLD and NASH. To avoid the dangers of excessive ROS levels, cells are dependent on anti-oxidant enzymes. Interestingly, expressions and activities of several enzymes, e.g. glutathione reductase, superoxide dismutase, glutathione peroxidase and peroxiredoxins display CC-controlled diurnal rhythms (84–85). Consistently, in peripheral tissues levels of ROS as well as peroxidized lipids/proteins vary per the light-dark cycle (86). Thus, it is evident that the ‘clock’ plays a remarkable role in regulating several cellular processes where deregulation have been strongly implicated in chronic liver diseases (Figure 3).
Circadian clock, nuclear receptors (NRs) and NAFLD
The nuclear receptor (NR) superfamily which comprises 48 members in humans, control diverse aspects of physiology including metabolism (87–89). NRs are transcription factors, which upon ligand (natural and synthetic) binding drive gene expression programs, amongst whom are pathways controlling metabolism. Investigation of the circadian expression patterns of all NRs in four mice tissues, including, the liver (60), revealed that at least 20 of the 41 transcribed NR in the murine liver are expressed in a circadian manner thereby providing a possible molecular link between the clock, NRs, and metabolism. In the liver, NRs control a broad range of crucial hepatic functions and are prominently implicated in NAFLD development (38,88–89). Here, we briefly discuss a few of these NRs that are not only known to be regulated by the ‘clock’ but also have emerged as therapeutic targets for NAFLD (see below). PPARα regulates β-oxidation and ketogenesis (90) and plays a prominent role in inflammation by trans-repressing NF-κB and AP-1 pathways (91). Importantly, genetic studies in mice indicate that through this trans-repressive activity PPARα can prevent fibrosis development which is a crucial event in NASH pathogenesis (91). Ligand activation of PPARβ/δ (which plays a prominent role in lipid catabolism) also prevents hepatic fibrogenesis (92). The Liver X receptors (LXRs) are transcriptional regulators of cholesterol metabolism and hepatic lipogenesis, and LXR activation lowers atherosclerosis by enhancing reverse cholesterol transport (89). Whereas LXR could be a possible antifibrotic target (93), LXR-activation enhances lipogenesis due to LXR-induced activation of SREBP1c activity (93). Lastly, with the establishment of pleiotropic roles of BAs in metabolic regulation, FXR has gained considerable attention as a therapeutic target for NAFLD (see below). Indeed, hepatic FXR activation reduces lipogenesis and improves fibrosis (72).
Clock, gut microbiota and NAFLD
Besides liver-restricted functions and processes, extrahepatic tissues also play a crucial role in NAFLD. Obesity-associated alterations in the gut microbiota (i.e., dysbiosis) composition and their interactions with the host (intestinal epithelial cells; IEC) have been implicated as an etiological agent in the pathogenesis of metabolic diseases, including NAFLD (94–97). A mechanism through which changes in gut microbiota composition may promote NAFLD is through increased LPS production and delivery to the liver via the portal circulation (98–99). In turn, microbiota-derived LPS can perturb hepatic lipid metabolism by modulating the production of short-chain fatty acids and altering the BA pool composition (99), which may influence intestinal and hepatic FXR activity, thus affecting both glucose and lipid homeostasis (99). Remarkably, the CC by regulating the expression of microbial pattern recognition receptors (e.g. TLRs, NOD2) provides a ‘temporal window’ during which microbiota-signals regulate gene expression to maintain homeostasis (100). Interestingly, the gut microbiota also displays ‘clock’-controlled diurnal rhythmicity (101–102). Consistently, circadian perturbations (mutation of CC-components or jet lag) lead to dysbiosis and development of metabolic pathologies (101). Furthermore, mutation of innate immune genes (Tlr5, Nlrp6, Nlrp3) which play pivotal roles in sensing gut microbiota, modulate metabolic pathologies, including NAFL(95).
Chronopharmacology: detoxification, pharmacokinetics, and dynamics
Considering its overall influence on physiology, it is hardly surprising that clinically relevant pharmacological aspects, e.g. pharmacokinetics (PK) and pharmacodynamics (PD), of many drugs are also governed by the ‘clock’, thereby introducing circadian variations in drug metabolism/detoxification and efficacy (84,103). One of the most prominent examples of circadian control over pharmacology emerges from its ability to regulate almost every step of xenobiotic detoxification in the liver, including absorption, biotransformation and elimination (84,103–104). Notably, in humans, hepatic absorption of lipophilic drugs occurs more swiftly in the morning than in the evening (103). Consistently, expression of several transport proteins which mediate xenobiotic uptake e.g. cationic and anionic transporters (Oct-1, Oatp1, Oatp1a4 etc.) display circadian rhythmicity (103). Classically, xenobiotic metabolism is grouped into three (I, II, and III) phases. Phase I involves biochemical modification of substrates by the CYP450 superfamily of enzymes. Importantly, transcript levels of several members (Cyp2a4, Cyp2a5, Cyp2b10, Cyp2e1, Cyp3a11 etc.) of this family are rhythmic, attaining in mouse liver their peak during the rest phase (84,104). In phase II, xenobiotics are rendered hydrophilic by conjugation to various small molecules.
Notably, phase II controlling genes (Sult1c1, Sult1d1, Gsta1, Gsta2 etc.) are also expressed in a circadian manner. The excretion of xenobiotics (phase III) is controlled by different transporter proteins, and several of them (Mrp2, Mdr2, Abcg2, Abcc2 etc.) oscillate rhythmically in the mouse liver. This pervasive circadian control of all these phases is molecularly achieved by hepatic CC-driven regulation of TFs, enzymes and transport proteins participating in the detoxification process (84,104). Hepatic expression of TFs (PXR, CAR and AhR), which bind and metabolize xenobiotics, is rhythmic (60). Moreover, CC-components (RORα/γ) and CC-output regulators (DBP, HLF and TEF) also transcriptionally regulate the detoxification process (105–106). Accordingly, mice with ablation of the Dbp, Hlf and Tef genes exhibit widespread deficiencies in both basal and inducible detoxification processes (106).
Circadian control of ‘pharmacology’ extends beyond the liver and has been reviewed elsewhere (84,107). ‘Timing’ is of crucial but less-appreciated factor in drug efficacy. Indeed, 56 of the top 100 best-selling drugs in the USA target the product of a circadian gene (108). However, most of them are yet to be associated and dosed as per the circadian rhythm. Importantly, clock perturbations resulting from HFD feeding can be rescued by ‘properly-timed’ pharmacological interventions (109). Taken together, recent studies (although many in rodent models) suggest that it will be highly prudent to investigate the mechanistic basis of circadian variations in PK/PD, in order to include a ‘circadian’ component for better therapeutic outcomes (84,107).
Therapeutic impact: CC and pharmacological targeting of NAFLD
While licensed pharmacological therapies are not yet available(36–38), a larger number of approaches and compounds are in preclinical and clinical development. Most therapeutic strategies aim to decrease inflammation, fibrosis, and metabolic substrate availability or to increase their disposal from the liver. Weight loss management or bariatric surgery not only improves NASH, but can also induce fibrosis regression (38, 110). Considering the key impact of CC-control in regulation of metabolism, it is likely that the molecular targets of several drug candidates are CC-regulated. For example, obeticholic acid (INT-747) which activates FXR, reverses histological features of NASH (111) and the CC is well known to control BA metabolism. Interestingly, FGF19 which is rhythmically secreted from intestine (post-feeding) has efficacy in murine models of NASH (112). Importantly, treatment with the FGF19 analog NGM282 reduces hepatic fat content in NASH patients (113). The CC-regulated NRs PPAR-α/β are activated by elafibranor (currently in phase 3 trial), which enhances lipid metabolism, insulin sensitivity and reduces inflammation (114). Furthermore, FGF21, a direct transcriptional target of PPARα reduces steatosis (37). Significantly, some other potential NASH-modulating compounds (37–38), e.g. resveratrol (SIRT1-agonist), inhibitors of acetyl-CoA carboxylase (ACC) and FAS, further strengthen the CC-connection to therapeutics.
The intimate relationship between metabolism and the CC, as well as the amenability of the CC-oscillator to a variety of ‘resetting’ signals (1–7), has spurred investigations to explore the potential of ‘clock’ modulating small molecules as a possible treatment for metabolic disorders (115–118). Using high-throughput phenotypic screening or medicinal chemistry approaches several molecules affecting the affect circadian period, phase and/or amplitude have been identified (115–118). Consistent with the molecular-genetic studies revealing a regulatory role for PERs and CRYs in CC-functioning, several compounds have been found to affect their levels and alter (mostly lengthening) circadian periods (115–117). One such compound, KL001 was found to bind CRY proteins which prevented their ubiquitination and proteasomal degradation (116,119). Consistent with the known role of CRYs in suppressing gluconeogenesis, KL001 administration was shown to improve glucose tolerance in diet-induced obese (DIO) mice (116,120). Recent investigations have also identified several modulators of CC-components RORα/γ and REV-ERBα/β with therapeutic potential in animal models of metabolic disorders (116–118). In this regard, agonists of REV-ERBs; e.g., SR9009 and SR9011 (121), and an inverse ROR agonist of (SR1555) (122) were found to improve several metabolic parameters in DIO mice. Amongst identified ROR modulators nobiletin (NOB) was demonstrated to enhance the amplitude of the CC, reduce weight gain, improve energy homeostasis and metabolic parameters in both DIO and genetically diabetic (db/db) mice (116–118, 123). Taken together, these small molecule modulators of CC-components provide an opportunity to further reveal regulatory networks in circadian functioning which could be targeted, alone or in combination, to treat metabolic liver disease.
CC and alcohol-induced liver disease (ALD)
Like NAFLD, pathogenesis of alcohol-induced liver disease (ALD) is complex and arises from interactions between metabolic, environmental and genetic risk factors in heavy alcohol consumers. In the context of alcohol-related disease, the CC has largely been investigated from a neurobehavioral perspective, noting CC disruption in alcohol use disorders and addiction (124–125). For example, rotating shift-workers have increased alcohol intake and tendencies to engage in binge drinking (126). Genetic variants in some clock genes are also associated with alcohol dependence and increased drinking in humans (127–128). Finally, transcript levels of CC genes are significantly lower in peripheral blood mononuclear cells from alcohol-dependent patients compared with healthy control subjects (129). Together, these studies suggest that CC alterations could promote alcoholic disorders and excessive alcohol consumption.
Investigations using Per2::Luciferase knock-in mice demonstrated that alcohol consumption misaligns peripheral clocks from the master SCN clock (130–131). Additionally, in liver, chronic alcohol consumption disrupts rhythmic oscillations of several CC components and CC-controlled output genes involved in regulating glucose, glycogen, cholesterol, BA and FFA metabolism (130, 132). For example, chronic alcohol intake in mice alters the diurnal rhythm in hepatic glycogen content due to dampened and/or shifted oscillations in glucose and glycogen metabolism genes (57, 132) Moreover, liver-specific BMAL1 deletion and chronic alcohol abolish day-night differences in hepatic glycogen content (132). Alcohol consumption also disrupts rhythmic oscillations in the cofactor NAD+ (130) required for numerous metabolic functions in the liver, including pathways regulated by SIRT1 and PARP-1. Furthermore, CC disruption (mutation or disrupting the lightdark cycle) enhances alcohol-induced tissue injury in mice. For example, alcohol-induced steatosis is higher in livers of Clock-mutant mice as compared to wild-type mice (133). Liver-specific deletion of BMAL1 also increases hepatic steatosis in mice treated with chronic plus binge alcohol (134). Moreover, CC-disruption through weekly 12-h shifts in the light-dark cycle, increases gut leakiness and liver injury in alcohol-fed mice (135). Importantly, whole gut and colon permeability is enhanced in night-shift but not in day-shift workers who consumed moderate amounts of alcohol (0.5 g/kg/day) for only one week (136). Collectively, these studies suggest that CC disruption may increase risk for liver disease in alcohol consumers. Importantly, as key mechanisms and linkages among alcohol-mediated CC disruption, metabolic dysregulation, and tissue injury emerge, it may become possible to pharmacologically target the CC in ALD patients.
Conclusion and future perspectives
The pathogenesis of NAFL and its progression to NASH, the most-prevalent noninfectious liver disease, is complex and multifactorial. Both genetic factors as well as the environment have been shown to play important functional roles. Perturbations in FFA metabolism, which lies at the core of NAFLD could potentially arise from deregulation of several distinct mechanisms. Remarkably, under physiological conditions most of these processes are governed by the CC-machinery. Consistently, in mice models, either mutation of CC-components (1–5) or change in the feeding time (137) are closely associated with a range of metabolic diseases including NAFL. Importantly, recent investigations have categorically established that in humans ‘circadian misalignment’ has adverse metabolic and cardiovascular consequences (138–139). Furthermore, epidemiologically, single nucleotide polymorphisms (SNPs) in the Clock gene (140) and in several CC-controlled transcriptional regulators (e.g. Pparγ, Stat3, Ppargc1α) (88) and the enzyme Pnpla3 (141) are associated with the development of obesity, metabolic syndrome, NAFLD and NASH. Most significantly, our nouveau life-style (nutrient-dense foods, timing of eating and activity) which continuously interferes with endogenous circadian rhythms is also epidemiologically correlated with increasing incidences of all the hallmarks of metabolic syndrome including, NAFLD (8–12). Given the socio-economic realities in modern societies, it is difficult to avoid circadian disruption. Thus, in addition to life-style modification, CC-targeting approaches may provide therapeutic opportunities overcoming these challenges. Furthermore, comprehensive systems level investigations of the circadian system elucidating physical- and genetic-interaction networks will reveal novel targets to prevent and treat chronic liver disease.
Acknowledgements
The authors apologize to colleagues whose work could not be cited due to space limitations.
Funding
This work was supported by the ERC-AdG-2014-671231-HEPCIR, EU H2020-667273-HEPCAR, the National Institute of Health (NIAID 1R03AI131066-01A1, NCI R21 CA209940), the Fondation ARC pour la Recherche sur le Cancer (TheraHCC IHUARC IHU201301187). The work has been published under the framework of LABEX ANR-10-LABX-0028-HEPSYS, ANR-10-LABEX-0046, Inserm Plan Cancer and, benefits from the state managed funding by the French National Research Agency as part of the investments for the future program. BS and TFB hold advanced ERC grants (694717, 671231).
Footnotes
Conflict of interest: The authors disclose no conflicts of interest.
References
- 1.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Eckel-Mahan KL, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 4.Feng D, Lazar MA. Clocks, Metabolism and Epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeGates TA, Fernandez DC, Hattar S. Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci. 2014;15:443–454. doi: 10.1038/nrn3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi JS, Hong HK, KO CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X-S, Armstrong MEG, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med. 2011;61:78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colles SL, Dixon JB, O’Brien PE. Night eating syndrome and nocturnal snacking: association with obesity, binge eating and psychological distress. Int J Obes. 2007;31:1722–1730. doi: 10.1038/sj.ijo.0803664. [DOI] [PubMed] [Google Scholar]
- 12.Sun M, Feng W, Wang F, Zhang L, Wu Z, et al. Night shift work exposure profile and obesity: Baseline results from a Chinese night shift worker cohort. Plos One. 2018;13:e0196989. doi: 10.1371/journal.pone.0196989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005;16:254–258. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damiola F, LeMinh N, Preitner N, Kornmann B, Fleury-Olela F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 17.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherji A, Kobiita A, Chambon P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci USA. 2015;112:E6683–E6690. doi: 10.1073/pnas.1519735112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckel-Mahan KL, Patel VR, deMateo S, Orozco-Solis R, Ceglia NJ. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaasik K, Lee C. Reciprocal regulation of heme biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 24.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 25.Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang B, Everett LJ, Jager J, Briggs E, Armour SM, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 30.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 31.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, et al. Circadian clock-dependent- and - independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aviram R, Manella G, Kopelman N, Neufeld-Cohen A, Zwighaft Z, et al. Lipidomic analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol Cell. 2016;62:636–648. doi: 10.1016/j.molcel.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuel VT, Shulman GI. Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metab. 2018;27:22–41. doi: 10.1016/j.cmet.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedmann SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas JT, Francque S, Staels B. Pathophysiology and mechanisms of nonalcoholic fatty liver disease. Annu Rev Physiol. 2016;78:181–205. doi: 10.1146/annurev-physiol-021115-105331. [DOI] [PubMed] [Google Scholar]
- 39.Baumert TF, Verrier ER, Nassal M, Chung RT, Zeisel MB. Host-targeting agents for treatment of hepatitis B virus infection. Curr Opin Virol. 2015;14:41–46. doi: 10.1016/j.coviro.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Zeisel MB, Lucifora J, Mason WS, Sureau C, Beck J, et al. Towards an HBV cure: state-of-the-art and unresolved questions-report of the ANRS workshop on HBV cure. Gut. 2015;64:1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 41.Chung RT, Baumert TF. Curing chronic hepatitis C- the arc of a medical triumph. N Engl J Med. 2014;370:1576–1578. doi: 10.1056/NEJMp1400986. [DOI] [PubMed] [Google Scholar]
- 42.Hoshida Y, Fuchs BC, Bardeesy N, Baumert TF, Chung RT. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J Hepatol. 2014;61:S79–S90. doi: 10.1016/j.jhep.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeisel MB, Lupberger J, Fofana I, Baumert TF. Host-targeting agents for prevention and treatment of chronic hepatitis-perspectives and challenges. J Hepatol. 2013;58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 44.Younossi ZM, Blissett D, Blissett R, Henry L, et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64:1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- 45.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heyman F, Tacke F. Immunology in the liver-from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13:88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 47.Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganz M, Szabo G. Immune and inflammatory pathways in NASH. Hepatol Int. 2013;7:771–781. doi: 10.1007/s12072-013-9468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day CP, James OF. Steatohepatitis: a tale of ‘two’ hits? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 50.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–88. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 51.Donnelly KL, Smith CI, Schwarzenberg SJ, Jessrun J, Boldt MD, et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Investig. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert JE, Ramos-Roman MA, Browning JD, Parks EJ. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014;146:726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirsova P, Ibrahim SH, Gores GJ, Malhi H. Lipotoxic lethal and sublethal stress signaling in hepatocytes: relevance to NASH pathogenesis. J Lipid Res. 2016;57:1758–1770. doi: 10.1194/jlr.R066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mota M, Banini BA, Cazanave SC, Sanyal AJ. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism. 2016;65:1049–1061. doi: 10.1016/j.metabol.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalsbeek A, LaFleur S, Fliers E. Circadian control of glucose metabolism. Mol Metab. 2014;3:372–383. doi: 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Udoh US, Swain TM, Filiano AN, Gamble KL, Young ME. Chronic ethanol consumption disrupts diurnal rhythms of hepatic glycogen metabolism in mice. Am J Physiol Gastrointest Liver Physiol. 2015;308:G964–974. doi: 10.1152/ajpgi.00081.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guillaumond F, Grechez-Cassiau A, Subramaniam M, Brangolo S, et al. Kruppel-like factor KLF10 is a link between thecircadian clock and metabolism in liver. Mol Cell Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Downes M, Yu RT, Bookout AL, He W, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 61.Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Li S, Liu T, Borigin J, Lin JD. Transcriptional coactivator PGC1-alpha integrates mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 63.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paschos GK, Ibrahim S, Song W-L, Kuneida T, Grant G, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, et al. Rev-Erbα and Rev-Erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol. 2013;366:163–169. doi: 10.1016/j.mce.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chavan R, Feillet C, Fonesca Costa S, Delorme JE, Okabe T, et al. Liver-derived ketone bodies are necessary for food anticipation. Nat Commun. 2016;7:10580. doi: 10.1038/ncomms10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemberger T, Saladin R, Vazquez M, Assimapoulos F, Staels B, et al. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- 70.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, et al. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PloS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas C, Pelliccari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signaling for metabolic disease. Nat Rev Drugdisc. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 72.Chàvez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterol. 2017;152:1679–1694. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 73.Chiang JYL. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017;1:3–9. doi: 10.1016/j.livres.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stroeve JH, Brufau G, Stellard F, Gonzalez FJ, Staels B, et al. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest. 2010;90:1457–1467. doi: 10.1038/labinvest.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 76.Duez H, Veen JN, Duhem C, Pourcet B, Touvier T, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterol. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 77.Duane WC, Levitt DG, Mueller SM, Behrens JC, et al. Regulation of bile acid synthesis in man. Presence of a diurnal rhythm. J Clin Invest. 1983;72:1930–1936. doi: 10.1172/JCI111157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaix A, Zarrinpar A, Panda S. The circadian coordination of cell biology. J Cell Biol. 2016;215:15–25. doi: 10.1083/jcb.201603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab. 2012;23:319–325. doi: 10.1016/j.tem.2012.03.004. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 81.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Z, Kim H, Qiu Y, Chen X, Mendez R, et al. CREBH couples circadian clock with hepatic lipid metabolism. Diabetes. 2016;65:3369–3383. doi: 10.2337/db16-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maillo M, Martín J, Sebastián D, Hernández-Alvarez M, García-Rocha M, et al. Circadian- and UPR-dependent control of CPEB4 mediates a translational response to counteract hepatic steatosis under ER stress. Nat Cell Biol. 2017;19:94–105. doi: 10.1038/ncb3461. [DOI] [PubMed] [Google Scholar]
- 84.Dallmann R, Brown SA, Gachon F. Chronopharmacology: new insights and therapeutic implications. Annu Rev Pharmacol Toxicol. 2014;54:339–361. doi: 10.1146/annurev-pharmtox-011613-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22:709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanabrocki EL, Murray D, Hermida RC, Scott GS, Bremner WF, et al. Circadian variation in oxidative stress markers in healthy and typeII diabetic men. Chronobiol Int. 2002;19:423–439. doi: 10.1081/cbi-120002914. [DOI] [PubMed] [Google Scholar]
- 87.Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR and the big bang. Cell. 2014;157:P255–P266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazzoccoli G, Vinciguerra M, Oben J, Tarquini R, De Cosmo S. Non-alcoholic fatty liver disease: the role of nuclear receptors and circadian rhythmicity. Liver Int. 2014;34:1133–1152. doi: 10.1111/liv.12534. [DOI] [PubMed] [Google Scholar]
- 89.Trauner M, Halilbasic E. Nuclear receptors as new perspectives for management of liver diseases. Gastroenterology. 2011;140:1120–1125.:e1-12. doi: 10.1053/j.gastro.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 90.Pawlak M, Lefevre P, Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62:720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 91.Pawlak M, Bauge E, Bourguet W, De Bosscher K, Lalloyer F, et al. The transrepressive activity of peroxisomal proliferator-activated receptor α is necessary and sufficient to prevent liver fibrosis in mice. Hepatology. 2014;60:1593–1606. doi: 10.1002/hep.27297. [DOI] [PubMed] [Google Scholar]
- 92.Iwaisako K, Haimerl M, Paik YH, Taura K, Kodama Y, et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor δ agonist. Proc Natl Acad Sci. 2012;109:E1369–E1376. doi: 10.1073/pnas.1202464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen-Naftaly M, Friedman SL. Current status of novel antifibrotic therapies in patients with chronic liver disease. Ther Adv Gastroenterol. 2011;4:391–417. doi: 10.1177/1756283X11413002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bashiardes S, Shapiro H, Rozin S, Shibolet O, Elinav E. Non-alcoholic fatty liver and the gut microbiota. Mol Metab. 2016;5:782–794. doi: 10.1016/j.molmet.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ray K. NAFLD: leaky guts: intestinal permeability and NASH. Nat Rev Gastroenterol Hepatol. 2015;12:123. doi: 10.1038/nrgastro.2015.15. [DOI] [PubMed] [Google Scholar]
- 97.Van Olden C, Groen AK, Nieuwdorp M. Role of intestinal microbiome in lipid and glucose metabolism in diabetes mellitus. Clin Ther. 2015;37:1172–1177. doi: 10.1016/j.clinthera.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 98.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 99.Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for dyslipidemia and cardiovascular disease. J Lipid Res. 2012;53:1723–1737. doi: 10.1194/jlr.R024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 101.Thaiss CA, Zeevi D, Levy M, Zilberman-Shapira G, Suez J, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 102.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baraldo M. The influence of circadian rhythms on the kinetics of drugs in humans. Expert Opin Drug Metab Toxicol. 2008;4:175–192. doi: 10.1517/17425255.4.2.175. [DOI] [PubMed] [Google Scholar]
- 104.Claudel T, Cretenet G, Saumet N, Gachon F. Crosstalk between xenobiotics metabolism and circadian clock. FEBS Lett. 2007;581:3626–3633. doi: 10.1016/j.febslet.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 105.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genom. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 106.Gachon F, Olela FF, Schaad O, Descombes P, et al. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 107.Paschos GK, Baggs JE, Hogenesch JB, FitzGerald GA. The role of clock genes in pharmacology. Annu Rev Pharmacol Toxicol. 2010;50:187–214. doi: 10.1146/annurev.pharmtox.010909.105621. [DOI] [PubMed] [Google Scholar]
- 108.Zhang R, Lahens NF, Balance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Woller A, Duez H, Staels B, Lefranc M. A mathematical model of the liver circadian clock linking feeding and fasting cycles to clock function. Cell Reports. 2016;17:1087–1097. doi: 10.1016/j.celrep.2016.09.060. [DOI] [PubMed] [Google Scholar]
- 110.Promrat K, Kleiner DE, Niemeier HM, Jackvony E, Kearns M, et al. Randomized controlled trial testing the effects of weight loss on nonalcoholic steatohepatitis. Hepatology. 2010;51:121–129. doi: 10.1002/hep.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicenter, randomized placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, et al. Modulation of the intestinal bile acid/FXR/FGF15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67:2150–2166. doi: 10.1002/hep.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harrison SA, Rinella ME, Abdelmalek MF, Trotter JF, Paredes AH, et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicenter, randomized, double-blind, placebo-controlled, phase 2 trial. Lancet. 2018;391:1174–1185. doi: 10.1016/S0140-6736(18)30474-4. [DOI] [PubMed] [Google Scholar]
- 114.Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, et al. Elafibranor, an agonist of the peroxisome proliferator activated receptor -α and -δ induces resolution of nonalcoholic steatohepatitis without fibrosis worsening. Gastroenterology. 2016;150:1147–1159.:e5. doi: 10.1053/j.gastro.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 115.Wallach T, Kramer A. Chemical chronobiology: toward drugs manipulating time. FEBS Lett. 2015;589:1530–1538. doi: 10.1016/j.febslet.2015.04.059. [DOI] [PubMed] [Google Scholar]
- 116.Cheng Z, Yoo SH, Takahashi JS. Development and therapeutic potential of small-molecule modulators of circadian systems. Annu Rev Pharmacol Toxicol. 2018;58:231–252. doi: 10.1146/annurev-pharmtox-010617-052645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci. 2013;70:2985–2998. doi: 10.1007/s00018-012-1207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Humphries PS, Bershot R, Kincaid J, Marbery E, McCluskie K, et al. Carbazole-containing sulfonamides and sulfamides: discovery of cryptochrome modulators as antidiabetic agents. Bioorg Med Chem Lett. 2016;26:757–760. doi: 10.1016/j.bmcl.2015.12.102. [DOI] [PubMed] [Google Scholar]
- 121.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, et al. Regulation of circadian behavior and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chang MR, He Y, Khan TM, Kuruvilla DS, Garcia-Ordonez R, et al. Antiobesity effect of a small molecule repressor of RORγ. Mol Pharmacol. 2015;88:48–56. doi: 10.1124/mol.114.097485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.He B, Nohara K, Park N, Park YS, Guillroy B, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 2016;23:610–621. doi: 10.1016/j.cmet.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Prosser RA, Glass JD. Assessing ethanol’s actions in the suprachiasmatic circadian clock using in vivo and in vitro approaches. Alcohol. 2015;49:321–339. doi: 10.1016/j.alcohol.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rosenwasser AM. Chronobiology of ethanol: Animal models. Alcohol. 2015;49:311–319. doi: 10.1016/j.alcohol.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 126.Virtanen M, Jokela M, Nyberg ST, Madsen IE, Lallukka T, et al. Long working hours and alcohol use: systematic review and meta-analysis of published studies and unpublished individual participant data. BMJ. 2015;350:g7772. doi: 10.1136/bmj.g7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blomeyer D, Buchmann AF, Lascorz J, Zimmermann US, Esser G, et al. Association of per2 genotype and stressful life events with alcohol drinking in young adults. PLoS One. 2013;8:e59136. doi: 10.1371/journal.pone.0059136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kovanen L, Saarikoski ST, Haukka J, Pirkola S, Aromaa A, et al. Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- 129.Huang MC, Ho CW, Chen CH, Liu SC, Chen CC, et al. Reduced expression of circadian clock genes in male alcoholic patients. Alcohol Clin Exp Res. 2010;34:1899–1904. doi: 10.1111/j.1530-0277.2010.01278.x. [DOI] [PubMed] [Google Scholar]
- 130.Filiano AN, Millender-Swain T, Johnson R, Jr, Young ME, Gamble KL, et al. Chronic ethanol consumption disrupts the core molecular clock and diurnal rhythms of metabolic genes in the liver without affecting the suprachiasmatic nucleus. PLoS One. 2013;8:e71684. doi: 10.1371/journal.pone.0071684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou P, Ross RA, Pywell CM, Liangpunsakul S, Duffield GE. Disturbances in the murine hepatic circadian clock in alcohol-induced hepatic steatosis. Scientific reports. 2014;4:3725. doi: 10.1038/srep03725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bailey SM. Emerging role of circadian clock disruption in alcohol-induced liver disease. Am J Physiol Gastrointest Liver Physiol. 2018;315:G364–G373. doi: 10.1152/ajpgi.00010.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kudo T, Tamagawa T, Shibata S. Effect of chronic ethanol exposure on the liver of clock-mutant mice. J Circadian Rhythms. 2009;7:4. doi: 10.1186/1740-3391-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang D, Tong X, Nelson BB, Jin E, Sit J, et al. The hepatic bmal1/akt/lipogenesis axis protects against alcoholic liver disease via promoting ppar alpha pathway. Hepatology. 2018;68:883–896. doi: 10.1002/hep.29878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, et al. Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol-induced hepatic pathology and inflammation. PLoS One. 2013;8:e67102. doi: 10.1371/journal.pone.0067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Swanson GR, Gorenz A, Shaikh M, Desai V, Kaminsky T, et al. Night workers with circadian misalignment are susceptible to alcohol-induced intestinal hyperpermeability with social drinking. Am J Physiol Gastrointest Liver Physiol. 2016:ajpgi 00087 02016. doi: 10.1152/ajpgi.00087.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mukherji A, Kobiita A, Damara M, Misra N, Meziane H, et al. Shifting eating to the circadian rest phase, misaligns the peripheral circadian clocks with the master SCN clock, which leads to a metabolic syndrome. Proc Natl Acad Sci. 2015;112:E6691–6698. doi: 10.1073/pnas.1519807112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Scheer FAJL, Hilton MF, Mantozoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc NatlAcad Sci. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morris CJ, Purvis TE, Hu K, Scheer FAJL. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci. 2016:E1402–E1411. doi: 10.1073/pnas.1516953113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sookian S, Castano G, Gemma C, Gianotti TF, et al. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13:4242–4248. doi: 10.3748/wjg.v13.i31.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]