Abstract
Radiomics is an emerging field which extracts quantitative radiology data from medical images and explores their correlation with clinical outcomes in a non-invasive manner. This review aims to assess whether radiomics is a useful and reproducible method for clinical management of hepatocellular carcinoma (HCC) by reviewing the strengths and weaknesses of current radiomics literature pertaining specifically to HCC. From an initial set of 48 articles recovered through database searches, 23 articles were retained to be included in this review after full screening. Among these 23 studies, seven used a radiomics approach in magnetic resonance imaging (MRI). Only two studies applied radiomics to positron emission tomography-computed tomography (PET-CT). In the remaining 14 articles, a radiomic analysis was performed on computed tomography (CT). Eight studies dealt with the relationship between biological signatures and imaging findings, and can be classified as radiogenomic studies. For each study included in our review, we computed a Radiomics Quality Score (RQS) as proposed by Lambin et al. We found the RQS (mean ± standard deviation) was 8.35 ± 5.38 (out of a possible maximum value of 36). Although these scores are fairly low, and radiomics has not yet reached clinical utility in HCC, it is important to underscore the fact that these early studies pave the way for the radiomics field with a focus on HCC. Radiomics is still a very young field, and is far from being mature, but it remains a very promising technology for the future for developing adequate personalized treatment as a non-invasive approach, for complementing or replacing tumor biopsies, as well as for developing novel prognostic biomarkers in HCC patients.
Keywords: Radiomics, radiogenomics, hepatocellular carcinoma, tumor heterogeneity
Introduction
Hepatocellular carcinoma (HCC) is the most common liver cancer. HCC mostly occurs in patients with chronic liver disease such as cirrhosis or severe fibrosis. Its major causes are chronic liver disease due to chronic hepatitis B and C virus infection or metabolic liver disease, such as non-alcoholic steatohepatitis or alcoholic liver disease. HCC is poorly symptomatic at the early stages of its development, and often becomes symptomatic only at an advanced stage when curative treatments are no longer possible. Therefore, the prognosis of HCC remains unsatisfactory [1].
Recently, tumor heterogeneity in terms of biological and genomic characteristics has become a topic of interest in cancer research [2]. Tumor heterogeneity can be demonstrated not only within primary cancers and various metastases (inter-tumor heterogeneity), but also within the same tumor (intra-tumor heterogeneity). Numerous publications have shown that HCCs are extremely heterogeneous both in terms of their genotype and phenotype [3, 4]. Thus, not only can different patients develop very different types of cancer, but tumors in the same patient can also be heterogeneous. Patient prognosis depends strongly on this phenotypic expression, which could be evaluated, for example, by analyzing pathological characteristics, such as the histological grade of the tumor [5] and microscopic vascular invasion [6]. Many staging systems including clinical, biological and imaging data have been developed such as the Barcelona Clinic Liver Cancer staging system [7], the Cancer of the Liver Italian Program [8, 9], and the Okuda criteria [10]. However, beyond the size and number of lesions, none of these scoring systems include information on the tumor phenotype that affects patient survival in a significant way [11]. Diagnostic and therapeutic trends in liver cancer are changing; they now tend to be determined by significant biological and genomic tumor characteristics.
Tumor characterization via medical imaging
Among all techniques for interrogating tumor phenotype and heterogeneity, medical imaging provides several advantages [11, 12]. By allowing an evaluation of tumors as a whole, in a minimally invasive and reproducible manner, imaging is complementary to biopsies, which only provide samples that are not always representative of tumor heterogeneity [13]. Since biopsies merely aim to sample a small portion of the tumor and since it is difficult to repeat pathological assessments, they provide limited information regarding tumor heterogeneity. Conversely, medical imaging methods such as Computed Tomography (CT), positron emission tomography (PET) or magnetic resonance imaging (MRI) can capture a tumor in its entire 3D extent with features that reflect tumor heterogeneity.
Such cross-sectional imaging techniques have become essential tools for modern oncology management [14–16]. Protocols for image acquisition based on these modalities have reached such a degree of sophistication that, in order to make a therapeutic decision, tissue biopsy is often unnecessary when the diagnostic criteria for HCC are all met [17–19]. However, the methods currently used to assess the prognosis of patients with HCC based on the acquired images remain very rudimentary and are simply based on size, number of tumors and vascular invasion as subjectively analyzed by the radiologist [7–10, 20–22]. As we recognize the need to go beyond tumor size and number, given the sophistication of the acquired imaging signal, advanced image analysis tools are now required to establish biomarkers from the complex signal that can be extracted from the images [22].
Going beyond size: semantic descriptors of tumor appearance
To improve image-based tumor characterization, one possible approach is based on a qualitative or semi-quantitative analysis as performed by an expert radiologist, using standardized reading scores. Examples of characteristics generally described for HCC by radiologists include the presence of arteries in the tumor, a peri-tumoral halo or the tumor’s apparent heterogeneity. Specifically, an HCC could be encapsulated, well-limited, or homogeneously hypervascularized after contrast injection or, on the contrary, poorly limited with vascular invasion, heterogeneously enhanced, and with a larger area of necrosis. This type of image analysis is referred to as a “semantic” analysis of lesions, where images are evaluated by one or more trained radiologists on the basis of semantic descriptors of the lesion(s) that are part of the established radiologist's lexicon. Another example of a semantic approach to tumor classification is the LI-RADS classification, which provides a standardized radiological lexicon built by consensus among expert radiologists [24].
The semantic analysis approach is interesting because it often provides a pathophysiological explanation for the image descriptions. The process of quantifying visual semantic characteristics unfortunately remains quite subjective and difficult to reproduce. Its implementation also poses practical problems because this process is very time consuming and cannot easily be used with large populations or integrated into clinical practice. Furthermore, its low inter- and intra-observer reproducibility makes this analysis difficult to standardize.
Going beyond size: quantitative descriptors of tumor appearance
Another approach to image-based tumor characterization is based on quantitative image descriptors. This type of approach is known as radiomics, and it aims to quantify the morphological appearance of the tumor, i.e. its imaging phenotype, using mathematically defined quantitative features [25, 26]. This type of quantitative information cannot be easily assessed by a radiologist, but can be computed with specialized computer algorithms. Radiomics was popularized by Lambin et al. [27] in 2012, and since then has been extensively used as a methodology to assess tumor heterogeneity, in order to establish a correlation with clinical or biological information [28].
Radiomics
The radiomics analysis pipeline consists of three main steps: 1) tumor segmentation, 2) computation of radiomic features within the segmented tumoral region and 3) feature selection, model building and classification. The details of every stage of the radiomics pipeline, their implementation details and limitations have been discussed at length elsewhere [29–32]. Here, we briefly mention a few details that are relevant to the findings of our review.
Segmentation
Radiomics requires a tumor region to be segmented in order to define the image region where quantitative descriptors are to be computed. Automatic segmentation has been a long-standing objective in the computer vision and machine learning fields [33], but remains difficult to achieve. This is why most radiomic studies still rely on manual tumor segmentation. Unfortunately, the use of manual segmentation not only makes measurements long and tedious, but also hinders measurement reproducibility. It should be noted that in studies using semantic analysis, segmentation is not necessary [34–41].
Radiomics features
Many different quantitative descriptors (features) have been proposed for radiomics [29–32]. The studies included in the present review typically use first-order, second-order and higher-order mathematical descriptors such as grayscale matrix analysis (co-occurrence) which take into account the relationships between neighboring pixels. A filtering step – using for example Gauss Laplacian filters – is usually performed prior to any analysis to reduce noise and improve performance [42–47].
Feature selection, model building, classification
One difficulty of radiomics is that it can calculate thousands of parameters for a single image. If the number of parameters is very high and the population is small – a few dozen patients – there is a significant risk of overfitting. This means that in practice there will almost always be parameters which are statistically correlated with the patient's condition. In order to limit this risk, the number of parameters must be significantly reduced before building the statistical model and, if possible, the model needs to be tested on an independent imaging dataset, obtained for instance at a different institution [33].
Limitations
The limitations of this method have been thoroughly analyzed in previous review articles about radiomics [23, 29–32, 33, 48–50]. One of the main limitations is the lack of standardization of image acquisition (such as slice thickness, choice of MRI sequences, or timing after contrast injection), which could add a strong bias to the post-imaging workflow.
Radiogenomics
Radiogenomics refers to the study of correlations between genome and molecular measurements on one hand, and radiological measurements (either quantitative or qualitative (semantic) features) on the other [51–53]. Radiomics and radiogenomics have the same objective, which is to transform radiological images into objective measurements representative of tumor heterogeneity.
Purpose
In recent years, we have seen the publication of numerous studies on radiomics with the objective of improving the diagnosis and stratification of patients with primary liver cancer [54]. The results of these studies are sometimes contradictory and complex to reproduce. Unfortunately, many of the published works show significant methodological weaknesses which have limited their impact in clinical practice. Therefore, there is a need to clarify the performance of radiomics as a prognostic and stratification tool for HCCs. We have thus conducted this systematic review to assess whether radiomics is a useful and reproducible method for HCC clinical management in terms of diagnosis, prognosis, and estimation of treatment response by reviewing the advantages and qualities of the studies included.
Review Strategy
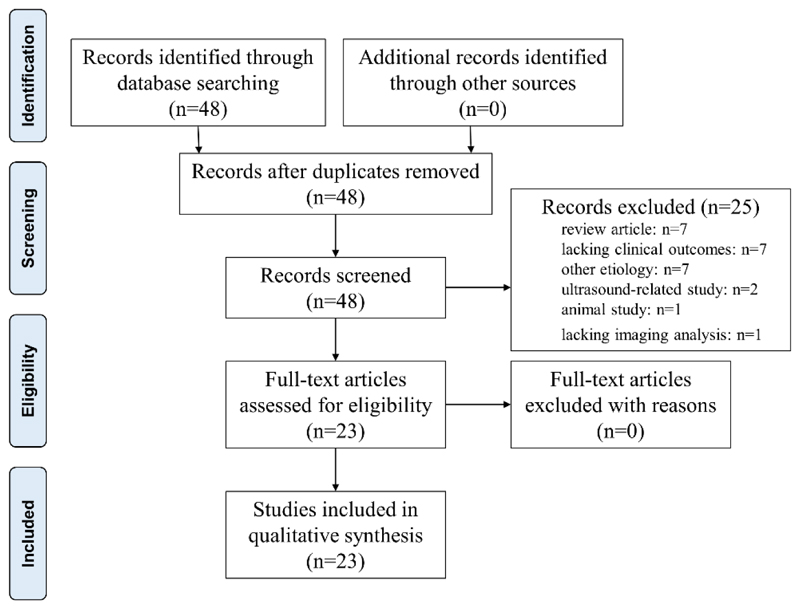
This review was conducted for all studies, published between January 1, 2007 and December 23, 2018 following PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) guidelines [55]. We used the following search strategy on PubMed and Embase: ((hepatocellular carcinoma [Title]) AND (radiomics [Title] OR radiogenomics [Title] OR omics [Title])), and a combination of associated terms from the controlled MeSH vocabulary. The final search was carried out on 23 December 2018. Inclusion criteria were (1) human studies, (2) English language studies, (3) full-text articles, and (4) studies reporting on semantic features or radiomics analyses for HCC. Exclusion criteria included (1) animal/experimental studies, (2) abstracts, reviews, and case reports, (3) only ultrasound-related studies, and (4) no investigation on clinical outcome. The existing review articles were analyzed in order to look for possible additional references. Every abstract was reviewed for initial selection, then all chosen articles were fully downloaded. Two authors (T.W, F.O), and an independent third one (B.G) when consensus was needed, individually assessed each manuscript to eliminate those which failed to meet the inclusion criteria. In accordance with the forementioned search strategy and criteria, we found 48 articles and excluded the 7 review articles, 7 articles without clinical outcomes, 7 articles focusing on other etiology, 2 ultrasound-related studies, 1 animal study, 1 article without imaging analysis. Finally, we included 23 of them in this review after a full screening (Figure 1). The articles are summarized in Table 1.
Figure 1. Study selection.
Table 1. Summary of radiomic analyses from 23 studies.
| Wu et al. [31] | Prediction of a pathological grading | - | T1- and T2-weighted MRI (7mm) | 170 | - | Q | 42 (15) | A computed radiomics signature itself or combined with clinical factors could help to classify the patients into high-grade or low-grade HCC cases. |
| Zhou et al. [32] | Prediction of a pathological grading | - | Gd-DTPA MRI (4.4mm) | 46 | Low-grade: 3.8±2.1cm, High-grade: 6.3±2.9cm | Q | 22 (8) | Two extracted features were sufficient to classify the histopathological HCC grade. |
| Miura et al. [33] | Comparison of clinicopathological properties of high- HCC | Hepatectomy or TACE | EOB-MRI | 77 | High-HCC: 3.6±1.2cm, Low-HCC: 5.3±4.9cm | Q | 17 (6) | High-HCC differs from low-HCC in PIVKA-II level, histopathological differentiation, and expression of SLCO1B3. |
| Kim et al. [34] | Prediction of MVI | Hepatectomy or LT | Contrast MRI (2-5.5 mm) | 104 | 3.6cm | S | 14 (5) | “Peritumoral enhancement” on MRI may be correlated with MVI during surgical planning for patients with HCC. |
| Hectors et al. [35] | Quantification of tumor heterogeneity, prediction of histopathology and gene expression | - | mpMRI | 32 | 4.4±3.3cm | Q | 39 (14) | First-order statistical features from mpMRI showed high intra-tumoral and inter-tumoral heterogeneity on the HCC lesions. This heterogeneity was also remarkable at genomic level. |
| Starmans et al. [36] | Diagnosis | - | T2-weighted (Fat-Sat) MRI (7mm) | 119 | >3cm | Q | 0 (0) | Patient-based (age and gender) and textural characteristics were used to differentiate between benign and malignant tumors. |
| Renzulli et al. [37] | Prediction of MVI based on TTPVI | Hepatectomy | Contrast CT (2.5mm), EOB-MRI | 125 | 3.3cm (1.8-5.2cm) | S | 8 (3) | Tumor size, “non-smooth tumor margins”, “peritumoral enhancement” and “TTPVI” were correlated with the presence of MVI in HCC. |
| Blanc et al. [38] | OS and PFS | TARE | PET/CT | 47 | 6cm (4.3-9cm) | Q | 19 (7) | Radiomics signature computed with textural features was highly correlated with survival. |
| Park et al. [39] | OS and DFS | Hepatectomy | PET/CT | 92 | 2.5cm | S | 17 (6) | High correlation was found between PET-positivity and survival. Additionally, PET positivity may also lead to determine the resection margin in order to improve survival. |
| Kuo et al. [40] | Treatment sensitivity and prediction of MVI | - | Contrast CT (2.5mm) | 30 | - | S | 19 (7) | “Tumor margin” showed a strong correlation with MVI, TNM, and the expression of a drug response gene. “Internal arteries” also showed a correlation with MVI. |
| Peng et al. [41] | Prediction of MVI | - | Contrast CT (5mm) | Training: 184 Test: 120 |
Training: MVI (+) 6.3 cm, MVI (-) 5.7cm, Validation: MVI (+) 6.4cm, MVI (-) 4.9cm | S, Q | 47 (17) | Radiological features and a radiomics signature computed with first-order statistical features showed a correlation with MVI. |
| Segal et al. [42] | OS and Prediction of MVI genes | Hepatectomy | Contrast CT | Training: 30 Test: 32 |
- | S | 42 (15) | “Internal arteries” was found as a key imaging trait to predict OS and MVI in combination with “hypodense halo”. Those features were also correlated with the expression of genes involved in the development of HCC lesions. |
| Banerjee et al. [43] | OS and RFS based on RVI | Hepatectomy or LT | Contrast CT (2.5-3mm) | 157 | 2.8cm (1.8-4.5cm) | S | 53 (19) | RVI computed with three different imaging traits was correlated with MVI. |
| Taouli et al. [44] | Prediction of MVI and aggressive phenotype | - | Contrast CT (5mm) | 38 | 5.7±3.2cm | S, Q | 19 (7) | Correlation was found between some imaging traits and the aggressive profile of the tumors. |
| Zheng et al. [45] | Recurrence prediction and OS | Hepatectomy | Contrast CT | Training: 212 Test: 107 | - | Q | 47 (17) | A radiomics score computed with textural features was sufficient to predict postoperative recurrence and survival in patients with solitary HCC. |
| Akai et al. [46] | OS and DFS | Hepatectomy | Contrast CT (5mm) | 127 | 3.7cm (2.4-7cm) | Q | 25 (9) | First-order statistical features were sufficient to predict postoperative survival. |
| Zhou et al. [47] | Recurrence prediction | Hepatectomy | Contrast CT (1.25mm) | 215 | - | Q | 25 (9) | A radiomics signature using first-order statistical features combined with clinical factors was a good predictor of early recurrence after surgery. |
| Chen et al. [48] | OS and DFS | Hepatectomy | Contrast CT (1.25mm) | 61 | - | Q | 17 (6) | Tumor prognosis could be predicted using Gabor and Wavelet filtration responses. |
| Li et al. [49] | OS and Treatment sensitivity | Hepatectomy or TACE | Contrast CT (1.25mm) | 130 | 8.0cm (5.1-18.7cm) | Q | 19 (7) | Wavelet features were correlated with postoperative survival on HCC patients, suggestive of the suitable treatment choice. |
| Raman et al. [50] | Diagnosis | - | Contrast CT (3mm) | 80 | Adenoma 7±3cm, FNH 6±3cm, HCC 8±3cm |
Q | 3 (1) | A model created using first-order statistical features could differentiate three types of common hypervascular lesions. |
| Xia et al. [51] | OS with interpretable biological meaning | Hepatectomy or LT | Contrast CT | 37 | - | Q | 22 (8) | The volume of transition between tumor and liver, and the heterogeneity of the lesion were correlated with survival. Those two features associated with six others, were correlated with prognostic genes expression. |
| Bakr et al. [52] | Prediction of MVI | - | Contrast CT (≤ 3mm) | 28 | 7.4cm | Q | 3 (1) | Textural features computed with single-phased or combined-phased images were correlated with MVI. |
| Cozzi et al. [53] | OS and local control | Radiotherapy (VMAT) | Non-contrast CT (3mm) | 138 | - | Q | 14 (5) | Survival could be predicted using a radiomics signature made by a single shape-based feature. |
CT: computed tomography, DFS: disease-free survival, EOB-MRI: ethoxybenzyl-diethylenetriamine pentaacetic acid, FNH: focal nodular hyperplasia, HCC: hepatocellular carcinoma, LT: liver transplantation, mpMRI: multiparametric magnetic resonance imaging, MRI: magnetic resonance imaging, MVI: microvascular invasion, OS: overall survival, PET: positron emission tomography, PET/CT: computed tomography integrated with positron emission tomography, PIVKA-II: protein induced by vitamin K absence-II, Q: quantitative features, RQS: radiomics quality score, RVI: radiogenomic venous invasion, sec: seconds, S: semantic features, TACE: transarterial chemoembolization, TTPVI: two-traits predictor of vascular invasion, TARE: transarterial radioembolization, VMAT: volumetric modulated arc therapy
The 23 studies included in our review use either semantic or quantitative features. Table 2 describes the types of features used in the quantitative and semantic categories. Quantitative features are computed via specialized software and are classified as first-order, second-order and high-order descriptors, and morphological features. Semantic features are visually interpreted by radiologists and are defined as 8 features in our review: two-traits predictor of venous invasion, non-smooth tumor margin, peritumoral enhancement, tumor size, tumor-liver difference, PET-CT positivity, infiltrative pattern, and mosaic appearance.
Table 2. Summary of significant extracted features in 23 studies.
| First-order descriptors | 12 | Two-Traits Predictor of Venous Invasion | |
| Second-order descriptors | 7 | Internal arteries | 6 |
| High-order descriptors | 3 | Hypo-attenuating halos | 4 |
| Morphological feature | 1 | Non-smooth tumor margin | 3 |
| Peritumoral enhancement | 2 | ||
| Tumor size | 2 | ||
| Tumor-liver difference (estimated) | 1 | ||
| PET/CT positivity | 1 | ||
| Infiltrative pattern | 1 | ||
| Mosaic appearance | 1 | ||
PET/CT: computed tomography integrated with positron emission tomography First-order descriptors comprise of shape (compacity or sphericity), skewness, kurtosis, mean, energy, median, entropy, peak, standard deviation, intensity ratio between tumor and liver, enhancement ratio, and tumor-liver difference (computed). Second-order descriptors comprise of gray level matrices, cluster prominence, strength, and textual features variance. Morphological feature comprises of tumor margin volume.
For all articles, we analyzed separately 5 phases within the radiomics workflow: 1) data inclusion and selection criteria, 2) description and standardization of imaging acquisitions, 3) feature extraction, 4) exploratory analysis, and 5) modelling [25]. For each study, the following data were systematically recorded: first author, year of publication, type of study (retrospective or prospective), number of centers (single or multicentric), objective of the study (tumor detection, tumor characterization, prognosis, response to treatment), type of imaging modality (MRI, CT, PET), technical parameters (slice thickness, magnetic field for MRI scanner, contrast media injection), main radiomics investigated features, presence of genomic consideration, number of patients used to build the model, presence of a validation cohort.
Findings
Variations in imaging modalities and protocols
Seven out of the 23 studies used a radiomics approach on MRI images [34, 35, 56–60]. In one study, both CT and MRI were investigated [35]. MRI provides a very rich signal which can provide accurate information about tumors. However, MRI acquisitions are very difficult to standardize, with numerous acquisition parameters and many variations between manufacturers. MRI is also sensitive to many artifacts which complicate the reproducibility of measurements such as motion artifacts – cardiac or respiratory – due to long acquisition times or field homogeneity with image and signal distortion consequences.
Only two studies performed a radiomics analysis using PET-CT data [36, 61]. Blanc et al. [61] reported that a radiomics signature computed on whole-liver PET 18F-FDG imaging performed before transarterial radioembolization using Yttrium-90 predicted progression-free survival (PFS) and overall survival (OS) in patients with advanced HCC. This study is unique, as it uses an integrative whole-liver approach and underlines the importance of including not only tumor lesions, but also adjacent liver parenchyma to explore the tumor environment. In all the other articles, a radiomics analysis was carried out on CT images. An iodine contrast agent was used in all studies [35, 37–47, 62, 63] except for one study [64]. Four studies dealt with quantitative features from contrast-enhanced multiple-phase CT images [43–45, 63] and all six studies dealt with only semi-quantitative (semantic) features from contrast-enhanced multiple-phase CT images [35, 37–41]. The other four studies focused on quantitative characteristics from contrast-enhanced single-phase CT images (arterial phase in 2 [42, 47], portal phase in 2 [46, 62]).
Clinical utility of radiomic analysis in HCC
Eight studies dealt with the relationship between the biological variables and imaging findings. For these studies we used the terminology of radiogenomics – which is used often in the literature – although most refer to microscopic vascular invasion (MVI), which is not a genomic variable [35, 37, 39–41, 58, 59, 62]. Four of these articles considered MVI in their studies [34, 38–40]. In fact, MVI is the most frequent feature required to investigate the correlation with pathological characterization in our review. Among all the included articles, eight studies focused on the correlation between radiomic features and MVI [34, 35, 37–41, 63]. Bakr et al. [63] demonstrated that quantitative features which capture the lesion texture, intensities, and shape extracted from triphasic CT images had a better accuracy in MVI prediction, compared to two previously reported signatures based on semantic features, radiogenomic venous invasion [39, 40] and TTPVI [35].
The percentage of studies performed for tumor characterization was 61% (14/23) [34, 35, 56–60, 36–41, 47, 63]. Rahman et al. [47] described a model that distinguished successfully different lesion types (focal nodular hyperplasia, hepatic adenomas, and HCC) and normal liver tissue with high predicted classification performance accuracy, as compared to two human readers.
Twelve out of 23 (52%) studies were conducted to aid with prognosis [36, 39, 40, 42–46, 58, 61, 62, 64]. Cozzi et al. [64] have described a radiomics method to predict tumor response and OS for patients treated with arc-based radiotherapy. The other 11 studies were related to tumor prognosis after surgical treatment. Zheng et al. [42] demonstrated that a radiomics score measured on baseline CT was a prognostic factor of the outcome in patients that underwent liver resection for HCC. They concluded that this score might be complementary to the current staging system and help to stratify individualized treatments for solitary HCC patients.
Quality assessment of radiomic studies for HCC
In order to assess the quality of the included studies, we used the Radiometric Quality Score (RQS) as published by Lambin et al [25]. The RQS – which evaluates 16 key components of the radiomics workflow – is a tool which analyzes the quality of a radiomics study. It assigns points according to 16 different criteria, for a maximum score of 36. In our work, the RQS score was evaluated by two authors (T.W. and F.O.) first separately, and then by consensus.
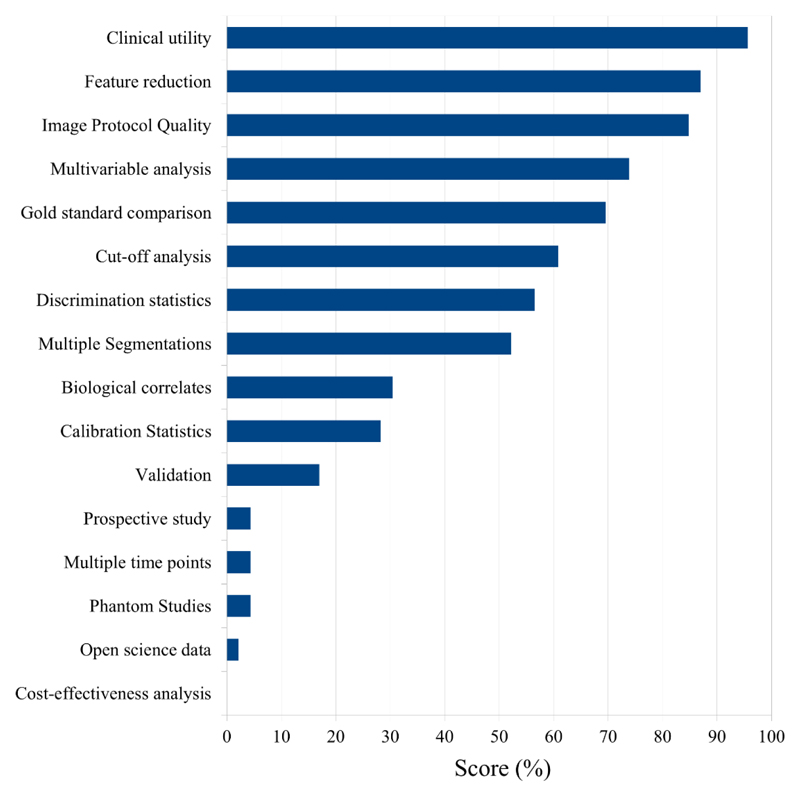
The results of the quality evaluations according to the RQS criteria are presented in Table 1 and the RQS scores are detailed by criteria in Figure 2. The RQS (mean ± standard deviation) was 8.35 ± 5.38 (representing 23% of the possible maximum value of 36). All but one study were scored below 18 (50%) due to a lack of external validation and/or to retrospective design as shown in Table 1. The main three reasons for entirely insufficient scores in the reviewed articles are the lack of prospective design except in one study [59], the lack of validation except in four studies [38, 39, 42, 56], and the lack of open-access scientific data resources except in two studies [39, 42]. Additionally, no studies have attempted to analyze the cost-effectiveness of a radiomics approach applied to a specific clinical situation.
Figure 2. Completing rate of each query item in radiomics quality score for 23 studies.
The prospective nature of the study is a major component of the RQS, representing almost 20% of the total score (7 points out of 36). A prospective study ensures that included patients could undergo a consistent imaging protocol, which would provide results that are more reliable as compared to a retrospective study. From all the studies included in the present review, all but one study [59] were retrospective evaluations.
Next, our analysis shows that existing radiomics studies in HCC have involved only relatively small patient populations, with an average number of patients of 110, with half of the studies including fewer than 100 cases. In addition, there is a general lack of validation in an independent population. Most of the studies trained the algorithm and evaluated its performance in only a small group of patients, risking overfitting. In the RQS, the type of validation performed accounts for as much as 10/36 points (nearly 28%), with the highest score being given to validation on independent datasets. In this regard, one option is the use of open-access scientific data. If such data is available for a radiomics study, researchers will be able to use the data set for a validation, reproduction, or replication with various data sets [25]. However, the lack of open-access data for HCC is a considerable limitation and results in a reduction by as many as 4 points of the total RQS.
Finally, only two articles used semi-automatic segmentation [60, 61], while all the other studies used manual tumor delineation. This could be a limitation to reproducibility and a barrier to the deployment of the method because as noted earlier, manual segmentation is very operator dependent and time consuming.
Semantic analysis of HCC
Eight articles in our review used semantic analysis, with potentially interesting results allowing the standardization of vascular invasion criteria for example [34–41]. One study examined the correlation between a quantitative and a semantic characteristic in an attempt to reduce variability between observers [38].
Discussion
The results of our analysis showed that the overall quality of the studies evaluated is low or moderate with an average RQS score of 8.35 or less than 25%. This underlines the fact that radiomics is a very recent technique that has not yet reached maturity, but also that this method is complex and that its standardization is not easy to implement.
This review demonstrates the importance of being cautiously optimistic about radiomic signatures. This new field of research has led to an accumulation of experimental and analytical work, most often thorough retrospective studies. However, the consolidation and standardization of experimental methods has not been standardized or validated. This review shows that the published radiomics work on HCC adds little to scientific knowledge, and is currently not useful in clinical decision-making. However, radiomics is a very young field, far from being mature, and has many subtleties that researchers are just learning to manage. In any case, the automated calculation of oncology biomarkers based on data acquired through medical imaging remains a necessity and is a matter of urgency. Radiomics in its current conventional form is probably only one step in the development of reliable computational image biomarkers that will probably need to be specific to a particular organ and tumor type. Although the results of our review article are somewhat disappointing regarding HCC, it is important to note that these published studies pave the way of the field of radiomics with a focus on HCC. Also, they demonstrate that radiomics is a topic of current interest for the management of HCC.
For radiomics to be a promising option for personalized medicine, it becomes clear that the methods of analysis should be standardized and automated. Radiomics is particularly interesting in the case of HCC because this tumor has an extremely varied phenotype, depending not only on the tumor type but also on the underlying liver disease, and this phenotype has a direct impact on the evolution of cancer. New prospective studies integrating the potential clinical impact are therefore needed. The standardization of image acquisition methods and injection protocols is also essential to obtain more relevant results.
Future perspectives in radiomic approaches for HCC
It is possible to yield additional accuracy with a standardization of CT-scan, MRI, and PET protocols. Further developments may also potentially require higher resolution imaging modalities. As a consequence, radiomics features may become promising diagnostic and prognostic factors, in particular with a carefully conducted validation. However, it will always be preferable for radiomics studies to be conducted on large patient populations, ideally collected prospectively from multiple institutions.
One of the most important challenges radiomics has to face is the segmentation step. An ideal segmentation will define correctly the target region in the image with high reproducibility and at a low cost. However, this ideal scenario is far from being achieved. Currently used manual drawings of the tumor region lead to a high rate of disagreement among interpreters, missing crucial information because of tumor heterogeneity [65]. The increasing number of publicly available liver image datasets and the development of machine and deep learning can help in automating liver and lesion segmentation [66, 67].
While radiomics in HCC is in the early stages of development, recent work in biology has shown that variations in phenotype, such as those potentially observed through imaging, are at least as important as tumor genetics. In this context, the search for imaging biomarkers able to quantify variations in tumor phenotype remains a promising avenue for research. These new biomarkers will have to be built in concordance with the latest discoveries in HCC biology, in order to attempt capturing the changes that occur specifically at the interface between the tumor and the liver, in terms of immune and inflammatory reaction, as well as tumor heterogeneity. A better quality of radiomics analyses can be achieved using the entire tumor [68] plus the peritumoral environment with a three-dimensional analysis. An analysis of the whole liver and factors affecting its structure, its baseline signal and vascularization should also be associated with the tumor analysis [61]. This is needed because liver cancer is not an isolated cancer, but occurs, in most cases, in a pre-existing chronic liver disease. To do this, we must develop computer analysis tools specific to the tumor under investigation, while also integrating the adjacent hepatic tissue into the analysis. Furthermore, there is a need to use more complex image analysis methods – including artificial intelligence – that are more specific than the simple accumulation of a large number of very generic and non-specific features used in “classical” radiomics.
Utilization of deep learning in radiomic analysis
In the case of conventional radiomics, the features mined by the discovery algorithm are designed by experts in medical image processing. However, a new class of artificial intelligence method known as deep learning may replace this approach [69]. Deep learning radiomics automatically identify – without human intervention – the best characteristics for a specific task [70–72] without the need for tumor segmentation. However, regardless of the image analysis method used, it is essential to create public image databases of patients with chronic liver diseases, with or without cancer, and make them accessible to researchers. This will make it possible to improve patient prognosis and to anticipate response to therapy for patient stratification. Unfortunately, to our knowledge, there is only one open access database fulfilling those criteria for the liver [73].
Summary
In summary, radiomics is at its very early stages in HCC and many challenges need to be addressed. Nevertheless, recent pilot studies using radiomics in patients with HCC have shown their potential. For diagnosis, radiomics may help to characterize pathological and molecular liver lesions. For prognosis, image features could be independent prognostic factors, as they can be associated with tumor biological characteristics. By estimating treatment response, radiomics analysis may also help to pave the way for personalized medicine. Additionally, there is a need for prospective evaluations in order to allow for potential clinical applications. As shown in other cancer entities, radiomics may be an appropriate option for personalized treatment, as a non-invasive approach which can complement or replace tumor biopsy, and which can also be used to develop novel prognostic biomarkers in HCC patients.
Footnotes
Author’s contributions
Wakabayashi T and Gallix B designed the research; Wakabayashi T and Ouhmich F extracted the data; Wakabayashi T, Savadjiev P and Gallix B wrote the paper; Gonzalez C, Felli E, Saviano A, Agnus V, Baumert T.F, Pessaux P, and Gallix B edited the paper; Marescaux J supervised the paper; All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest: Thomas F. Baumert, Patrick Pessaux, Jacques Marescaux, and Benoit Gallix have received research grants from ARC, Paris and Institut hospitalo-universitaire, Strasbourg (TheraHCC IHUARC IHU201301187). Antonio Saviano and Thomas F. Baumert have received research grants from the European Union (ERC-AdG-2014-671231-HEPCIR, H2020-667273-HEPCAR). Taiga Wakabayashi, Farid Ouhmich, Cristians Gonzalez-Cabrera, Emanuele Felli, Vincent Agnus, and Peter Savadjiev declare that they have no conflict of interest.
Acknowledgment
The authors acknowledge the support of ARC, Paris and Institut hospitalo-universitaire, Strasbourg (TheraHCC IHUARC IHU201301187), as well as the European Union (ERC-AdG-2014-671231-HEPCIR, H2020-667273-HEPCAR). In addition, the authors are grateful to Camille Goustiaux, Christopher Burel, and Guy Temporal for their assistance in proofreading the manuscript.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Hiley C, de Bruin EC, McGranahan N, Swanton C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol. 2014;15:453. doi: 10.1186/s13059-014-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin L, Liu X, Ding LW, et al. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017;77:2255–2265. doi: 10.1158/0008-5472.CAN-16-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu LC, Hsu CH, Hsu C, Cheng AL. Tumor Heterogeneity in Hepatocellular Carcinoma: Facing the Challenges. Liver Cancer. 2016;5:128–138. doi: 10.1159/000367754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martins-Filho SN, Paiva C, Azevedo RS, Alves VAF. Histological Grading of Hepatocellular Carcinoma-A Systematic Review of Literature. Front Med (Lausanne) 2017;4:193. doi: 10.3389/fmed.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazzaferro V, Llovet JM, Miceli R, Bhoori S, Schiavo M, Mariani L, Camerini T, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 8.Prospective validation of the CLIP score: a new prognostic system for patients with cirrhosis and hepatocellular carcinoma. The Cancer of the Liver Italian Program (CLIP) Investigators. Hepatology. 2000;31:840–845. doi: 10.1053/he.2000.5628. [DOI] [PubMed] [Google Scholar]
- 9.Farinati F, Rinaldi M, Gianni S, Naccarato R. How should patients with hepatocellular carcinoma be staged? Validation of a new prognostic system. Cancer. 2000;89:2266–2273. [PubMed] [Google Scholar]
- 10.Okuda K, Ohtsuki T, Obata H, Tomimatsu M, Okazaki N, Hasegawa H, Nakajima Y, et al. Natural history of hepatocellular carcinoma and prognosis in relation to treatment. Study of 850 patients. Cancer. 1985;56:918–928. doi: 10.1002/1097-0142(19850815)56:4<918::aid-cncr2820560437>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 11.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadot E, Simpson AL, Do RK, Gonen M, Shia J, Allen PJ, D’Angelica MI, et al. Cholangiocarcinoma: Correlation between Molecular Profiling and Imaging Phenotypes. PLoS One. 2015;10:e0132953. doi: 10.1371/journal.pone.0132953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman M, Bruix J. Biopsy for liver cancer: how to balance research needs with evidence-based clinical practice. Hepatology. 2015;61:433–436. doi: 10.1002/hep.27563. [DOI] [PubMed] [Google Scholar]
- 14.Hricak H. Oncologic imaging: a guiding hand of personalized cancer care. Radiology. 2011;259:633–640. doi: 10.1148/radiol.11110252. [DOI] [PubMed] [Google Scholar]
- 15.Sharma B, Martin A, Stanway S, Johnston SR, Constantinidou A. Imaging in oncology--over a century of advances. Nat Rev Clin Oncol. 2012;9:728–737. doi: 10.1038/nrclinonc.2012.195. [DOI] [PubMed] [Google Scholar]
- 16.Tirkes T, Hollar MA, Tann M, Kohli MD, Akisik F, Sandrasegaran K. Response criteria in oncologic imaging: review of traditional and new criteria. Radiographics. 2013;33:1323–1341. doi: 10.1148/rg.335125214. [DOI] [PubMed] [Google Scholar]
- 17.Elsayes KM, Hooker JC, Agrons MM, Kielar AZ, Tang A, Fowler KJ, Chernyak V, et al. 2017 Version of LI-RADS for CT and MR Imaging: An Update. Radiographics. 2017;37:1994–2017. doi: 10.1148/rg.2017170098. [DOI] [PubMed] [Google Scholar]
- 18.An C, Rakhmonova G, Choi JY, Kim MJ. Liver imaging reporting and data system (LI-RADS) version 2014: understanding and application of the diagnostic algorithm. Clin Mol Hepatol. 2016;22:296–307. doi: 10.3350/cmh.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 22.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis. 2010;30:52–60. doi: 10.1055/s-0030-1247132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savadjiev P, Chong J, Dohan A, Agnus V, Forghani R, Reinhold C, Gallix B. Image-based biomarkers for solid tumor quantification. Eur Radiol. 2019 doi: 10.1007/s00330-019-06169-w. [DOI] [PubMed] [Google Scholar]
- 24.Tang A, Bashir MR, Corwin MT, Cruite I, Dietrich CF, Do RKG, Ehman EC, et al. Evidence Supporting LI-RADS Major Features for CT- and MR Imaging-based Diagnosis of Hepatocellular Carcinoma: A Systematic Review. Radiology. 2018;286:29–48. doi: 10.1148/radiol.2017170554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 26.Cassinotto C, Dohan A, Zogopoulos G, Chiche L, Laurent C, Sa-Cunha A, Cuggia A, et al. Pancreatic adenocarcinoma: A simple CT score for predicting margin-positive resection in patients with resectable disease. Eur J Radiol. 2017;95:33–38. doi: 10.1016/j.ejrad.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee G, Lee HY, Ko ES, Jeong WK. Radiomics and imaging genomics in precision medicine. Precis Future Med. 2017;1:10–31. [Google Scholar]
- 29.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine. A review JAMA Oncol. 2016;2:1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 30.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90:20160665. doi: 10.1259/bjr.20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savadjiev P, Chong J, Dohan A, Vakalopoulou M, Reinhold C, Paragios N, Gallix B. Demystification of AI-driven medical image interpretation: past, present and future. Eur Radiol. 2019;29(3):1616–1624. doi: 10.1007/s00330-018-5674-x. [DOI] [PubMed] [Google Scholar]
- 34.Kim KA, Kim MJ, Jeon HM, Kim KS, Choi JS, Ahn SH, Cha SJ, et al. Prediction of microvascular invasion of hepatocellular carcinoma: usefulness of peritumoral hypointensity seen on gadoxetate disodium-enhanced hepatobiliary phase images. J Magn Reson Imaging. 2012;35:629–634. doi: 10.1002/jmri.22876. [DOI] [PubMed] [Google Scholar]
- 35.Renzulli M, Brocchi S, Cucchetti A, Mazzotti F, Mosconi C, Sportoletti C, Brandi G, et al. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology. 2016;279:432–442. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 36.Park JH, Kim DH, Kim SH, Kim MY, Baik SK, Hong IS. The Clinical Implications of Liver Resection Margin Size in Patients with Hepatocellular Carcinoma in Terms of Positron Emission Tomography Positivity. World J Surg. 2018;42:1514–1522. doi: 10.1007/s00268-017-4275-1. [DOI] [PubMed] [Google Scholar]
- 37.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 38.Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L. A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol. 2018;24:121–127. doi: 10.5152/dir.2018.17467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, Rutman AM, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taouli B, Hoshida Y, Kakite S, Chen X, Tan PS, Sun X, Kihira S, et al. Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol. 2017;27:4472–4481. doi: 10.1007/s00330-017-4844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng BH, Liu LZ, Zhang ZZ, Shi JY, Dong LQ, Tian LY, Ding ZB, et al. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18:1148. doi: 10.1186/s12885-018-5024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akai H, Yasaka K, Kunimatsu A, Nojima M, Kokudo T, Kokudo N, Hasegawa K, et al. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging. 2018;99:643–651. doi: 10.1016/j.diii.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, He L, Huang Y, Chen S, Wu P, Ye W, Liu Z, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2017;42:1695–1704. doi: 10.1007/s00261-017-1072-0. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Zhu Y, Liu Z, Liang C. Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol. 2017;90:198–204. doi: 10.1016/j.ejrad.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 46.Li M, Fu S, Zhu Y, Liu Z, Chen S, Lu L, Liang C. Computed tomography texture analysis to facilitate therapeutic decision making in hepatocellular carcinoma. Oncotarget. 2016;7:13248–13259. doi: 10.18632/oncotarget.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raman SP, Schroeder JL, Huang P, Chen Y, Coquia SF, Kawamoto S, Fishman EK. Preliminary data using computed tomography texture analysis for the classification of hypervascular liver lesions: generation of a predictive model on the basis of quantitative spatial frequency measurements--a work in progress. J Comput Assist Tomogr. 2015;39:383–395. doi: 10.1097/RCT.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 48.Scrivener M, de Jong EEC, van Timmeren JE, Pieters T, Ghaye B, Geets X. Radiomics applied to lung cancer: a review. Translational Cancer Research. 2016;5:398–409. [Google Scholar]
- 49.Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS. Rapid review: radiomics and breast cancer. Breast Cancer Res Treat. 2018;169:217–229. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]
- 50.Grossmann P, Gutman DA, Dunn WD, Jr, Holder CA, Aerts HJ. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in Glioblastoma. BMC Cancer. 2016;16:611. doi: 10.1186/s12885-016-2659-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai HX, Lee AM, Yang L, Zhang P, Davatzikos C, Maris JM, Diskin SJ. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89:20151030. doi: 10.1259/bjr.20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, Hricak H, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging. 2018;47:604–620. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong WK, Jamshidi N, Felker ER, Raman SS, Lu DS. Radiomics and radiogenomics of primary liver cancers. Clin Mol Hepatol. 2018 doi: 10.3350/cmh.2018.1007. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, Zhu S, et al. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. 2018 doi: 10.1007/s00330-018-5787-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Zhou W, Zhang L, Wang K, Chen S, Wang G, Liu Z, Liang C. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging. 2017;45:1476–1484. doi: 10.1002/jmri.25454. [DOI] [PubMed] [Google Scholar]
- 58.Miura T, Ban D, Tanaka S, Mogushi K, Kudo A, Matsumura S, Mitsunori Y, et al. Distinct clinicopathological phenotype of hepatocellular carcinoma with ethoxybenzyl-magnetic resonance imaging hyperintensity: association with gene expression signature. Am J Surg. 2015;210:561–569. doi: 10.1016/j.amjsurg.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 59.Hectors SJ, Wagner M, Bane O, Besa C, Lewis S, Remark R, Chen N, et al. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep. 2017;7:2452. doi: 10.1038/s41598-017-02706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starmans MPA, Miclea RL, van der Voort SR, Niessen WJ, Thomeer MG, Klein S. In: Medical Imaging 2018: Image Processing. Angelini ED, Landman BA, editors. Vol. 10574. Spie-Int Soc Optical Engineering; Bellingham: 2018. Classification of malignant and benign liver tumors using a radiomics approach. [Epub ahead of print] [Google Scholar]
- 61.Blanc-Durand P, Van Der Gucht A, Jreige M, Nicod-Lalonde M, Silva-Monteiro M, Prior JO, Denys A, et al. Signature of survival: a (18)F-FDG PET based whole-liver radiomic analysis predicts survival after (90)Y-TARE for hepatocellular carcinoma. Oncotarget. 2018;9:4549–4558. doi: 10.18632/oncotarget.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia W, Chen Y, Zhang R, Yan Z, Zhou X, Zhang B, Gao X. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol. 2018;63:035044. doi: 10.1088/1361-6560/aaa609. [DOI] [PubMed] [Google Scholar]
- 63.Bakr S, Echegaray S, Shah R, Kamaya A, Louie J, Napel S, Kothary N, et al. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham) 2017;4:041303. doi: 10.1117/1.JMI.4.4.041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cozzi L, Dinapoli N, Fogliata A, Hsu WC, Reggiori G, Lobefalo F, Kirienko M, et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer. 2017;17:829. doi: 10.1186/s12885-017-3847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N, Napel S. Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging (Bellingham) 2015;2:041011. doi: 10.1117/1.JMI.2.4.041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer. 2017;3:43. doi: 10.1038/s41523-017-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papp L, Potsch N, Grahovac M, Schmidbauer V, Woehrer A, Preusser M, Mitterhauser M, et al. Glioma Survival Prediction with Combined Analysis of In Vivo (11)C-MET PET Features, Ex Vivo Features, and Patient Features by Supervised Machine Learning. J Nucl Med. 2018;59:892–899. doi: 10.2967/jnumed.117.202267. [DOI] [PubMed] [Google Scholar]
- 68.Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology. 2013;266:177–184. doi: 10.1148/radiol.12120254. [DOI] [PubMed] [Google Scholar]
- 69.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 70.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, et al. Deep Learning: A Primer for Radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 71.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]