Abstract
Asthma is a severe and chronic disabling disease affecting more than 300 million people worldwide. Although in the past few drugs for the treatment of asthma were available, new treatment options are currently emerging, which appear to be highly effective in certain subgroups of patients. Accordingly, there is a need for biomarkers that allow selection of patients for refined and personalized treatment strategies. Recently, serological chip tests based on microarrayed allergen molecules and peptides derived from the most common rhinovirus strains have been developed, which may discriminate 2 of the most common forms of asthma, that is, allergen- and virus-triggered asthma. In this perspective, we argue that classification of patients with asthma according to these common trigger factors may open new possibilities for personalized management of asthma.
Keywords: Asthma, allergy, wheeze, rhinovirus, allergen, microarray
Asthma is a health problem of increasing importance, affecting more than 300 million people worldwide.1 It is estimated that asthma contributes to approximately 0.4 million deaths every year and the mortality might be even increased in high-risk patients.2,3 The number of disability-adjusted life-years due to asthma is approximately 23 million per year, and asthma thus accounts for approximately 1% of all disability-adjusted life-years lost.1 A considerable proportion of the asthma burden is attributed to acute exacerbations, which are associated with high morbidity and can lead to death. Acute exacerbations of asthma are an enormous problem to both adults and children, and account for approximately 50% of asthma health care costs.4 The rising prevalence of asthma and its accompanying health care costs are therefore major health and socioeconomic concerns.5
Patients suffering from asthma are often treated according to management strategies suggested by the Global Initiative for Asthma (GINA), which are regularly revised.1 GINA defines asthma as “a heterogeneous disease usually characterized by chronic airway inflammation with a history of respiratory symptoms such as wheeze, shortness of breath, chest tightness and cough that vary over time and in intensity, together with variable expiratory airflow limitations.” GINA is useful because it advocates management of asthma and respiratory disease according to published “good quality” evidence; however, suggested treatment options are limited to few drugs with little recommendations regarding the use of new biologics and other treatment options such as allergen-specific immunotherapy.6,7
Furthermore, GINA does not discriminate asthma according to the underlying trigger factors, which most certainly contribute to the phenotypic heterogeneity of asthma. Allergen exposure is a major asthma trigger factor for patients suffering from IgE-associated allergy.8,9 Likewise, viral respiratory infections, particularly those caused by rhinoviruses (RVs), represent frequent triggers for acute asthma exacerbations in allergic as well as in nonallergic subjects.10–12
Recently, new biomarkers have been developed for the diagnosis of specific IgE sensitization toward a large variety of individual allergen molecules and for the detection of RV strain–specific IgG antibody responses.
Chips Containing Microarrayed Allergen Molecules or Peptides Derived From a Comprehensive Panel of RV Strains for Serology
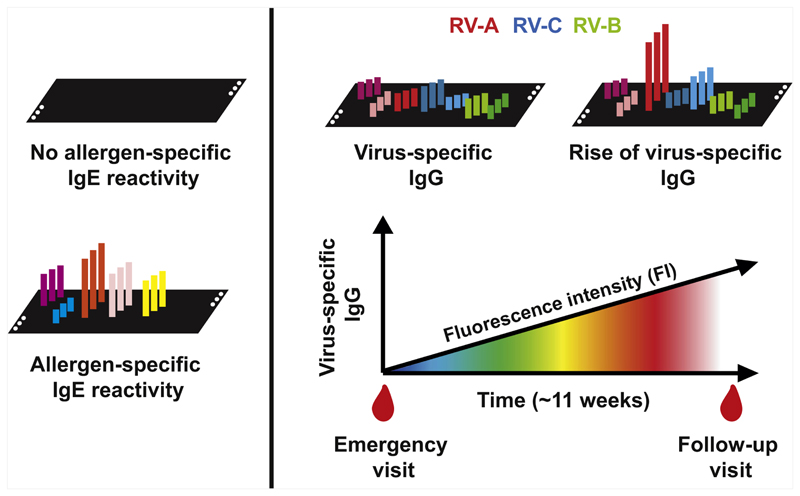
Within 2 European Union–funded research projects, that is, MeDALL (https://cordis.europa.eu/project/rcn/96850/factsheet/en) and PreDicta (https://cordis.europa.eu/project/rcn/96868/factsheet/en), 2 multiplex assays have been developed on the basis of chips containing more than 170 different allergen molecules from various allergen sources13 and microarrayed peptides from the VP1 coat proteins of the most common RV strains.14 The MeDALL allergen chip measures IgE antibodies specific for a comprehensive panel of allergen molecules comprising a spectrum of the most common allergen sources, including all important respiratory allergen molecules such as the major house-dust mite allergens, animal-derived allergens, pollen allergens, and mould allergens. The allergen chip was shown to be more sensitive than current forms of skin prick testing and allergen extract–based IgE serology to identify subjects with specific IgE sensitizations13,15 and most importantly, allowed precise identification of IgE sensitization to culprit allergen molecules for each individual patient. IgE testing with the allergen chip thus discriminates subjects with and without IgE sensitization. The analysis of IgE sensitizations against more than 170 allergen molecules can be performed with extremely low volumes of serum (ie, ~35 μL), which is particularly useful for an early detection of IgE sensitization in small children when limited amounts of blood is available (Fig 1, left part).13
Fig 1.
Chips containing a panel of microarrayed allergen molecules covering the most common allergen sources allow to measure IgE sensitization against each of the individual allergen molecules and thus of the culprit sensitizing allergens (left). Chips containing microarrayed peptides derived from the VP1 coat proteins of representative panels of RV-A, RV-B, and RV-C strains are useful for measuring RV-strain–specific IgG levels, cumulative strain–specific IgG levels, and increases in strain-specific IgG levels that occur some weeks after acute exacerbations of virus-triggered wheeze (right).
A further recent development is a chip containing peptides from the VP1 capsid proteins of panels of RV-A, RV-B, and RV-C strains, which can be used to measure strain-specific antibody responses in serum samples.14 Cumulative levels of RV strain–specific antibody levels were associated with the severity of asthma-related symptoms in preschool children as recently demonstrated.16 Furthermore, preschool children attending an emergency unit with acute exacerbations of wheeze showed strain-specific increases in virus-specific IgG antibodies in their follow-up serum samples obtained approximately 11 weeks later (Fig 1, right), suggesting that this increase is indicative of a wheeze attack triggered by a respiratory infection with certain RV strains. This assumption is corroborated by the observation that experimental infection with RV induced strain-specific increases in IgG antibody levels that were associated with the severity of RV-induced asthma in the tested subjects.17 Thus, the chip containing microarrayed peptides from the most common RV strains seems to discriminate between infections caused by RV strains from the 3 groups A, B, and C, and species-specific levels of antibodies and their increases in the course of infection were shown to be associated with the severity of respiratory symptoms.14,16 Using this RV chip we were previously able to identify species-specific antibody responses to RV-C–derived peptides, a species that has been difficult to propagate.18 For the latter reason, we also know less about pathogenic differences between RV-A and RV-C strains in the etiology of asthma exacerbations because no infection models for RV-C are available.
The Possible Clinical Utility of Multiplex Serum-Based Assays for Discrimination of Asthma Triggered by Allergens or Viral Infections
We would argue that the new, multiplex serum assays allow determination whether IgE reactivity against important respiratory allergen molecules (Fig 1, left) or respiratory viral infections as evidenced by increases in virus-specific IgG (Fig 1, right) are trigger factors for an individual’s asthma exacerbation. Data obtained in longitudinal birth cohorts and in studies of preschool children suffering from exacerbations of wheeze indicate that respiratory viral infections may be important trigger factors before relevant IgE sensitization to aeroallergens has occurred.19,20 However, it has been found that early multiple allergic sensitization in the first years of life is an important predictor of asthma.21–23 Thus, the importance of respiratory virus infections and allergen exposure for triggering asthma seems to vary by age. Furthermore, each of the 2 forms of asthma (ie, viral- and allergen-triggered asthma) may occur independently or in combination in the same individual.
It is, therefore, of great importance to identify early on high-risk children who are likely to develop persistent asthma later on in life.24 It is estimated that approximately one-third of infants and toddlers who wheeze in the first 3 years of life continue to wheeze after the age of 3 years.25 Although most children with viral-induced wheeze lose their respiratory symptoms in their early school age, have normal spirometry, and are more commonly nonatopic, they are usually less responsive to steroid treatment.21,26 In contrast, children who become sensitized to common aeroallergens are more likely to retain their symptoms and have reduced lung function at school age.25
Although there is little doubt that exposure to both allergens and viral infections can induce asthma exacerbations, there is very little information regarding their interaction. It has proven to be very difficult to document personal allergen exposure leading to an exacerbation of asthma and perhaps this is the reason why very few reports document the interaction between naturally acquired viral infections and allergen exposure in asthma exacerbations. Green et al27 provided evidence for synergy between viral infections and allergen exposure. In their case-controlled study, the risk of being admitted to hospital was considerably increased by exposure to high levels of allergen and concurrent viral infection. Another study reported that high titers of IgE antibodies to dust mite allergen were common and significantly increased the risk for acute wheezing provoked by RV among children with asthma.28 However, regardless of the sequence of exposure to allergen and viral infection, both stimuli have been shown to affect the subsequent response to the other.29
In fact, allergic asthma is a classical type 2 (T2)-associated disease, whereas studies of the natural antibody response to RV indicate that RV infections are dominated by IgG1 and IgA antibody responses and therefore are rather reminiscent of a type 1 type of immunity.30 It is thus quite tempting to speculate that T2 asthma that is characterized by eosinophilia may be rather due to T2-dependent allergen-specific IgE sensitization, whereas asthma triggered only by certain respiratory virus infections (ie, without concomitant allergic sensitization) such as RV may belong to the T2-low or non-T2 asthma.31 In addition, other trigger factors such as pollution, exercise, stress, and obesity may be important in the different phenotypes of asthma.
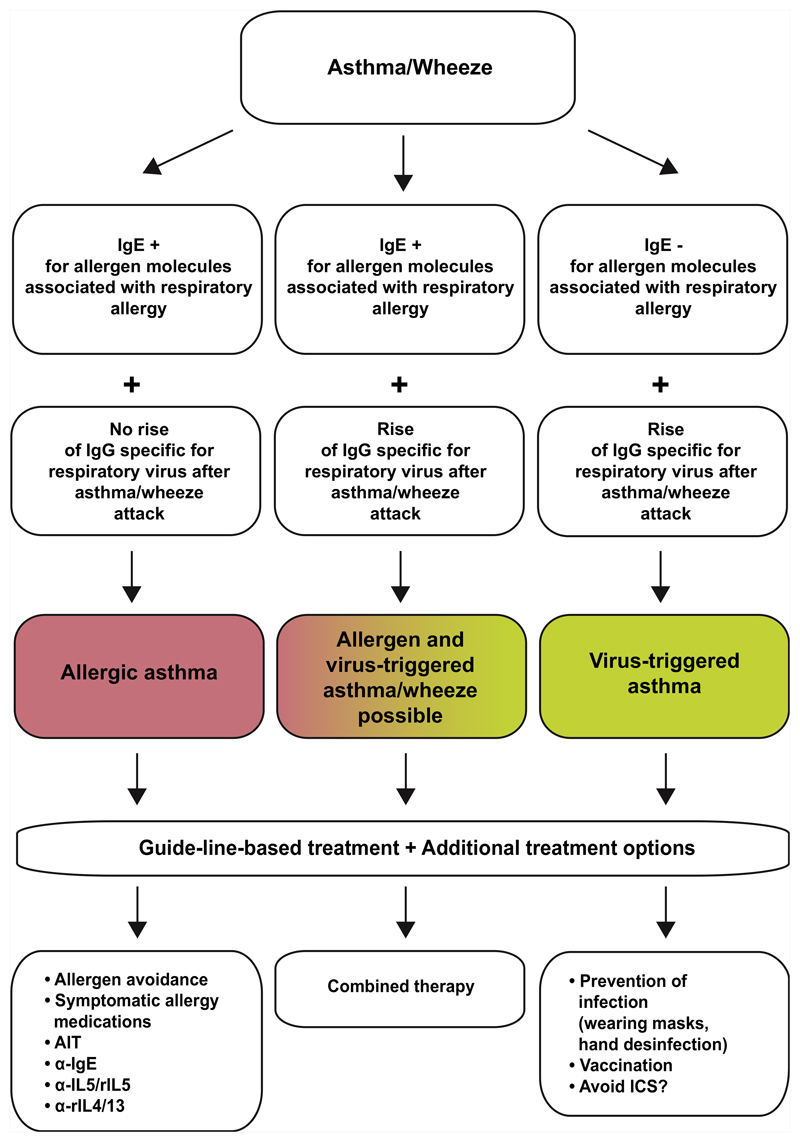
The availability of molecular multiallergen tests and chips containing peptides and antigens from important respiratory viruses provides the possibility to assess recent or past acquisition of respiratory viral infection and IgE sensitization to important respiratory allergens as trigger factors for asthma and to investigate how they are related to T2, T2-low, and non-T2 asthma.31 These additional biomarkers may eventually allow us to develop a more refined asthma classification. Certainly, these biomarkers will inform whether the allergic sensitization, respiratory viral infection, or both play an important role in triggering asthma (Fig 2). First data indeed suggest that chip-based serological testing for RV infections may be more sensitive and specific than recording of upper respiratory tract symptoms and nucleic acid–based testing for RV infections often used in hospitalized patients to assume that asthma is triggered by RV infections.14
Fig 2.
How the identification of allergens and respiratory virus infections as triggers factors of asthma and wheeze could lead to personalization of asthma treatment by adding treatment options to guideline-based therapy. AIT, Allergen immunotherapy; ICS, inhaled corticosteroid.
Based on such information, different forms of individualized treatment can be considered in addition to GINA-based therapy (Fig 2). For example, the identification of certain allergens as trigger factors may provide a rationale for allergen avoidance measures32 and molecular approaches for allergen-specific immunotherapy.33 Likewise, patients with asthma with IgE sensitization to important respiratory allergens may be particularly responsive to IgE-targeting treatment strategies such as anti-IgE or selective IgE immunoadsorption.34 Furthermore, new biological treatments such as anti-T2 cytokine-based treatments may be very effective in patients suffering from severe asthma.26,31
However, one may consider measures to prevent viral infections such as wearing masks, hand disinfection, avoiding close contacts, and better ventilation of indoor environments to preclude virus-triggered asthma.35,36 Vaccination against respiratory viruses responsible for virus-triggered asthma, although currently not an option, may be another option in the future. Furthermore, in the case that asthma triggered by respiratory virus infections indeed resembles a non-T2 phenotype, the use of corticosteroids may be less effective and/or not recommended. In this context, it is of note that some of the new corticosteroids do not seem to protect against barrier damage caused by RV infections.37
For patients suffering from mixed forms with evidence for allergens and virus infections as asthma triggers, one may try to separate out “hyperresponders” to allergens or virus infections. It is possible that atopic children with moderate to severe asthma triggered by RV infections may turn out to be better candidates for a vaccine targeting RV. Although these children are currently for treatment with biologic medications (eg, omalizumab and dupilumab), these medications are at present costly, there are some poor responders, and, once started, clinicians face the challenge of deciding when to stop these treatments, which may become easier if vaccines become available.
Summary
It is possible that new serological multiplex tests may improve clinical outcome by determination of allergen- and virus-triggered asthma, enabling classification of patients with asthma according to underlying trigger factors, which may help them to benefit from personalized forms of treatment while taking into account established GINA-based treatment.
Clinical trials to test whether personalization of asthma treatment based on serological identification of possible trigger factors can improve current asthma management guidelines are warranted.
Abbreviations used
- GINA
Global Initiative for Asthma
- RV
Rhinovirus
Footnotes
Disclosure of potential conflict of interest K. Niespodziana receives grant support (grant no. P29398) from the Austrian Science Foundation (FWF). K. F. Chung has received honoraria for participating in advisory board meetings of GlaxoSmithKline (GSK), AstraZeneca, Novartis, Merck, Boehringer Ingelheim, and Teva regarding treatments for asthma and chronic obstructive pulmonary disease and has also been renumerated for speaking engagements. A. Custovic reports personal fees from Novartis, Regeneron, Thermo Fisher Scientific, Philips, and Sanofi, outside the submitted work. T. Eiwegger reports other fees from DBV, grants from Innovation fund Denmark, and other fees from Regeneron, outside the submitted work; is the Co-PI or scientific lead in 3 investigator-initiated oral immunotherapy trials supported by the Allergy and Anaphylaxis Program Sickkids; is on local advisory board for ALK; and serves as associate editor for Allergy. S. L. Johnston reports board memberships of Therapeutic Frontiers and Virtus Respiratory Research and personal fees from Myelo Therapeutics GmbH, Concert Pharmaceuticals, Bayer, Synairgen, Novartis, Boehringer Ingelheim, Chiesi, Gerson Lehrman Group, resTORbio, Bioforce, Materia Medical Holdings, Pre-pBio Pharma, Pulmotect, Virion Health, and Lallemand Pharma, outside the submitted work. K. C. Nadeau reports grants and other fees from the National Institute of Allergy and Infectious Diseases; other fees from Novartis; personal fees and other fees from Regeneron; grants and other fees from FARE; grants from EAT; other fees from Sanofi, Astellas, Nestle, BeforeBrands, Alladapt, ForTra, Genentech, AImmune Therapeutics, and DBV Technologies; personal fees from AstraZeneca, ImmuneWorks, and Cour; grants from Allergenis and Ukko; and other fees from AnaptysBio, Adare Pharmaceuticals, Stallergenes-Greer, the National Heart, Lung, and Blood Institute, NIEHS, EPA, and WAO Center of Excellence, outside the submitted work. P. M. O’Byrne reports grants and personal fees from AstrZeneca and Medimmune; grants from Novartis; and personal fees from GSK and Chiesi, outside the submitted work. N. G. Papadopoulos reports personal fees from Novartis, Nutricia, HAL, Menarini/FAES Farma, Sanofi, Mylan/Meda, Biomay, AstraZeneca, GSK, MSD, ASIT Biotech, and Boehringer Ingelheim and grants from Gerolymatos International SA and Capricare, outside the submitted work. W. Pohl reports personal fees from Astra Zeneca, Chiesi, GSK, Meda, Sanofi, Novartis, Teva, and Boehringer Ingelheim, outside the submitted work. V. Siroux reports grants from Aviesan Itmo santée publique, the Scientific committee “AGIR for chronic diseases,” and the French National Research Agency (ANR), during the conduct of the study. M. van Hage reports personal fees from Thermo Fisher Scientific, ALK, and Biomay AG, Vienna, Austria, outside the submitted work. E. von Mutius reports personal fees from OM Pharma S. A., Böhringer Ingelheim International GmbH, Peptinnovate Ltd, Pharmaventures Ltd, and Nestlé Deutschland AG, outside the submitted work. In addition, E. von Mutius has a patent application number LU101064, Barn dust extract for the prevention and treatment of diseases, pending; a patent publication number EP2361632: Specific environmental bacteria for the protection from and/or the treatment of allergic, chronic inflammatory and/or autoimmune disorders; with royalties paid to Protectimmun GmbH, a patent publication number EP 1411977: Composition containing bacterial antigens used for the prophylaxis and the treatment of allergic diseases; licensed to Protectimmun GmbH, a patent publication number EP1637147: Stable dust extract for allergy protection; and a patent publication number EP 1964570: Pharmaceutical compound to protect against allergies and inflammatory diseases licensed to Protectimmun GmbH. M. Zidarn reports personal fees from Takeda and Novartis, outside the submitted work. R. Valenta receives grant support from the Austrian Science Foundation (FWF) and Viravaxx, Vienna, Austria. Furthermore, R. Valenta is a recipient of a megagrant of the Government of the Russian Federation (grant no. 14.W03.31.0024) and serves as a consultant for Viravaxx, Vienna, Austria. In addition, S. L. Johnston, K. Niespodziana, and R. Valenta are authors of patent/patent applications related to the subject of the study. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.The Global Asthma Report 2018. Global Asthma Network; Auckland, New Zealand: 2019. Available at: http://www.globalasthmareport.org. [Google Scholar]
- 2.Lee H, Ryu J, Nam E, Chung SJ, Yeo Y, Park DW, et al. Increased mortality in patients with corticosteroid-dependent asthma: a nationwide population-based study. Eur Respir J. 2019;54:1900804. doi: 10.1183/13993003.00804-2019. [DOI] [PubMed] [Google Scholar]
- 3.Seth D, Saini S, Poowuttikul P. Pediatric inner-city asthma. Pediatr Clin North Am. 2019;66:967–79. doi: 10.1016/j.pcl.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Rabe KF, Vermeire PA, Soriano JB, Maier WC. Clinical management of asthma in 1999: the Asthma Insights & Reality in Europe (AIRE) study. Eur Respir J. 2000;16:802–7. doi: 10.1183/09031936.00.16580200. [DOI] [PubMed] [Google Scholar]
- 5.Van Wijk RG. Socio-economic costs of asthma. Global Atlas of Asthma. EAACI. 2013:18–20. [Google Scholar]
- 6.Heffler E, Paoletti G, Giorgis V, Puggioni F, Racca F, Del Giacco S, et al. Real-life studies of biologics used in asthma patients: key differences and similarities to trials. Expert Rev Clin Immunol. 2019;12:1–8. doi: 10.1080/1744666X.2019.1653758. [DOI] [PubMed] [Google Scholar]
- 7.Dhami S, Kakourou A, Asamoah F, Agache I, Lau S, Jutel M, et al. Allergen immunotherapy for allergic asthma: a systematic review and meta-analysis. Allergy. 2017;72:1825–48. doi: 10.1111/all.13208. [DOI] [PubMed] [Google Scholar]
- 8.Caminati M, Pham DL, Bagnasco D, Canonica GW. Type 2 immunity in asthma. World Allergy Organ J. 2018;11:13. doi: 10.1186/s40413-018-0192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froidure A, Mouthuy J, Durham SR, Chanez P, Sibille Y, Pilette C. Asthma phenotypes and IgE responses. Eur Respir J. 2016;47:304–19. doi: 10.1183/13993003.01824-2014. [DOI] [PubMed] [Google Scholar]
- 10.Wisniewski JA, McLaughlin AP, Stenger PJ, Patrie J, Brown MA, El-Dahr JM, et al. A comparison of seasonal trends in asthma exacerbations among children from geographic regions with different climates. Allergy Asthma Proc. 2016;37:475–81. doi: 10.2500/aap.2016.37.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet. 2013;381:861–7. doi: 10.1016/S0140-6736(12)62202-8. [DOI] [PubMed] [Google Scholar]
- 12.Xepapadaki P, Bachert C, Finotto S, Jartti T, Konstantinou GN, Kiefer A, et al. Contribution of repeated infections in asthma persistence from preschool to school age: design and characteristics of the PreDicta cohort. Pediatr Allergy Immunol. 2018;29:383–93. doi: 10.1111/pai.12881. [DOI] [PubMed] [Google Scholar]
- 13.Lupinek C, Wollmann E, Baar A, Banerjee S, Breiteneder H, Broecker BM, et al. Advances in allergen-microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen-chip. Methods. 2014;66:106–19. doi: 10.1016/j.ymeth.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niespodziana K, Stenberg-Hammar K, Megremis S, Cabauatan CR, Napora-Wijata K, Vacal PC, et al. PreDicta chip-based high resolution diagnosis of rhinovirus-induced wheeze. Nat Commun. 2018;9:2382. doi: 10.1038/s41467-018-04591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skrindo I, Lupinek C, Valenta R, Hovland V, Pahr S, Baar A, et al. The use of the MeDALL-chip to assess IgE sensitization: a new diagnostic tool for allergic disease? Pediatr Allergy Immunol. 2015;26:239–46. doi: 10.1111/pai.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megremis S, Niespodziana K, Cabauatan C, Xepapadaki P, Kowalski ML, Jartti T, et al. Rhinovirus species-specific antibodies differentially reflect clinical outcomes in health and asthma. Am J Respir Crit Care Med. 2018;12:1490–9. doi: 10.1164/rccm.201803-0575OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niespodziana K, Cabauatan CR, Jackson DJ, Gallerano D, Trujillo-Torralbo B, Del Rosario A, et al. Rhinovirus-induced VP1-specific antibodies are group-specific and associated with severity of respiratory symptoms. EBioMedicine. 2014;2:64–70. doi: 10.1016/j.ebiom.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakauchi M, Nagata N, Takayama I, Saito S, Kubo H, Kaida A, et al. Propagation of rhinovirus C in differentiated immortalized human airway HBEC3-KT epithelial cells. Viruses. 2019;11 doi: 10.3390/v11030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickman M, Lupinek C, Andersson N, Belgrave D, Asarnoj A, Benet M, et al. Detection of IgE reactivity to a handful of allergen molecules in early childhood predicts respiratory allergy in adolescence. EBioMedicine. 2017;26:91–9. doi: 10.1016/j.ebiom.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston SL, Pattemore PK, Sanderson G, Smith S, Campbell MJ, Josephs LK, et al. The relationship between upper respiratory infections and hospital admissions for asthma: a time-trend analysis. Am J Respir Crit Care Med. 1996;154:654–60. doi: 10.1164/ajrccm.154.3.8810601. [DOI] [PubMed] [Google Scholar]
- 21.Illi S, von Mutius E, Lau S, Niggemann B, Grüber C, Wahn U, Multicentre Allergy Study (MAS) group Perennial allergen sensitisation early in life and chronic asthma in children: a birth cohort study. Lancet. 2006;368:763–70. doi: 10.1016/S0140-6736(06)69286-6. [DOI] [PubMed] [Google Scholar]
- 22.Belgrave DCM, Granell R, Turner SW, Curtin JA, Buchan IE, Le Souäf PN, et al. Lung function trajectories from pre-school age to adulthood and their associations with early life factors: a retrospective analysis of three population-based birth cohort studies. Lancet Respir Med. 2018;6:526–34. doi: 10.1016/S2213-2600(18)30099-7. [DOI] [PubMed] [Google Scholar]
- 23.Jackson DJ, Evans MD, Gangnon RE, Tisler CJ, Pappas TE, Lee WM, et al. Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281–5. doi: 10.1164/rccm.201104-0660OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sly PD, Boner AL, Björksten B, Bush A, Custovic A, Eigenmann PA, et al. Early identification of atopy in the prediction of persistent asthma in children. Lancet. 2008;372:1100–6. doi: 10.1016/S0140-6736(08)61451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332:133–8. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 26.Holt PG, Sly PD. Viral infections and atopy in asthma pathogenesis: new rationales for asthma prevention and treatment. Nat Med. 2012;18:726–35. doi: 10.1038/nm.2768. [DOI] [PubMed] [Google Scholar]
- 27.Green RM, Custovic A, Sanderson G, Hunter J, Johnston SL, Woodcock A. Synergism between allergens and viruses and risk of hospital admission with asthma: case–control study. BMJ. 2002;324:763. doi: 10.1136/bmj.324.7340.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soto-Quiros M, Avila L, Platts-Mills TA, Hunt JF, Erdman DD, Carper H, et al. High titers of IgE antibody to dust mite allergen and risk for wheezing among asthmatic children infected with rhinovirus. J Allergy Clin Immunol. 2012;129:1499–505.:e5. doi: 10.1016/j.jaci.2012.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bossios A, Gourgiotis D, Skevaki CL, Saxoni-Papageorgiou P, Lötvall J, Psarras S, et al. Rhinovirus infection and house dust mite exposure synergize in inducing bronchial epithelial cell interleukin-8 release. Clin Exp Allergy. 2008;38:1615–26. doi: 10.1111/j.1365-2222.2008.03058.x. [DOI] [PubMed] [Google Scholar]
- 30.Niespodziana K, Napora K, Cabauatan C, Focke-Tejkl M, Keller W, Niederberger V, et al. Misdirected antibody responses against an N-terminal epitope on human rhinovirus VP1 as explanation for recurrent RV infections. FASEB J. 2012;26:1001–8. doi: 10.1096/fj.11-193557. [DOI] [PubMed] [Google Scholar]
- 31.Chung KF, Adcock IM. Precision medicine for the discovery of treatable mechanisms in severe asthma. Allergy. 2019;74:1649–59. doi: 10.1111/all.13771. [DOI] [PubMed] [Google Scholar]
- 32.Cipriani F, Calamelli E, Ricci G. Allergen avoidance in allergic asthma. Front Pediatr. 2017;5:103. doi: 10.3389/fped.2017.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zieglmayer P, Focke-Tejkl M, Schmutz R, Lemell P, Zieglmayer R, Weber M, et al. Mechanisms, safety and efficacy of a B cell epitope-based vaccine for immunotherapy of grass pollen allergy. EBioMedicine. 2016;11:43–57. doi: 10.1016/j.ebiom.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Incorvaia C, Riario-Sforza GG, Ridolo E. IgE depletion in severe asthma: what we have and what could be added in the near future. EBioMedicine. 2017;17:16–7. doi: 10.1016/j.ebiom.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contreras RD, Wilson AM, Garavito F, Sexton JD, Reynolds KA, Canales RA. Assessing virus infection probability in an office setting using stochastic simulation. J Occup Environ Hyg. 2020;17:30–7. doi: 10.1080/15459624.2019.1691219. [DOI] [PubMed] [Google Scholar]
- 36.Zhou SS, Lukula S, Chiossone C, Nims RW, Suchmann DB, Ijaz MK. Assessment of a respiratory face mask for capturing air pollutants and pathogens including human influenza and rhinoviruses. J Thorac Dis. 2018;10:2059–69. doi: 10.21037/jtd.2018.03.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waltl EE, Selb R, Eckl-Dorna J, Mueller CA, Cabauatan CR, Eiwegger T, et al. Betamethasone prevents human rhinovirus-and cigarette smoke-induced loss of respiratory epithelial barrier function. Sci Rep. 2018;8:9688. doi: 10.1038/s41598-018-27022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]