Abstract
Sequence features of genes and their flanking regulatory regions are determinants of RNA transcript isoform expression and have been used as context-independent, plug-and-play modules in synthetic biology. However, genetic context, including the adjacent transcriptional environment, also influences transcript isoform expression levels and boundaries. We used synthetic yeast strains with stochastically repositioned genes to systematically disentangle sequence from contextual effects. Profiling 120 million full-length transcript molecules across 612 genomic perturbations, we observed sequence-independent alterations to gene expression levels and transcript isoform boundaries that were influenced by neighboring transcription. We identified features of transcriptional context that could predict these alterations and used these features to engineer a synthetic circuit where neighboring transcription controlled transcript length. This demonstrates how positional context can be leveraged in synthetic genome engineering.
Gene regulatory sequence features, like promoters and terminators, are considered to be primary drivers of transcript isoform boundaries and expression levels (1–4). In synthetic biology, promoters and terminators are assembled along with coding DNA sequences (CDSs) into transcriptional units (TUs). Promoters and terminators are characterized and distributed as standardized parts. These are used analogously to plug-and-play modules in electronics as if they would function identically in any context (5–7). However, this is not always the case. TUs in eukaryotic genomes exhibit a high degree of interdependence (8) due to the physical effects of transcribing neighboring genes (9–11). For example, transcriptional interference between neighboring genes can influence the selection of isoform boundaries and impact expression levels (12–14). Quantifying the extent to which isoform expression and boundaries are driven by factors beyond DNA sequence could improve rational genome design. However, this has been difficult to test systematically, as existing technologies cannot achieve the scale required to observe genes in many alternative genetic and transcriptional contexts.
Synthetic yeast genomes create genetic diversity
To overcome this challenge, the synthetic yeast genome (Sc2.0) was designed to encode a Cre-dependent system known as SCRaMbLE (synthetic chromosome rearrangement and modification by LoxP-mediated evolution) that can generate stochastic genomic rearrangements on-demand directly in its genome (15). These rearrangements occur at 34 bp loxPsym sites inserted 3 bp downstream of the stop codon of all non-essential CDSs. Upon induction of Cre recombinase, loxPsym sites can recombine in either orientation, producing duplications, deletions, inversions, and translocation events (Fig. 1A). Due to the placement of the loxPsym sites, rearranged CDSs retain their native promoters but can be decoupled from downstream sequences like 3’-untranslated regions (UTRs).
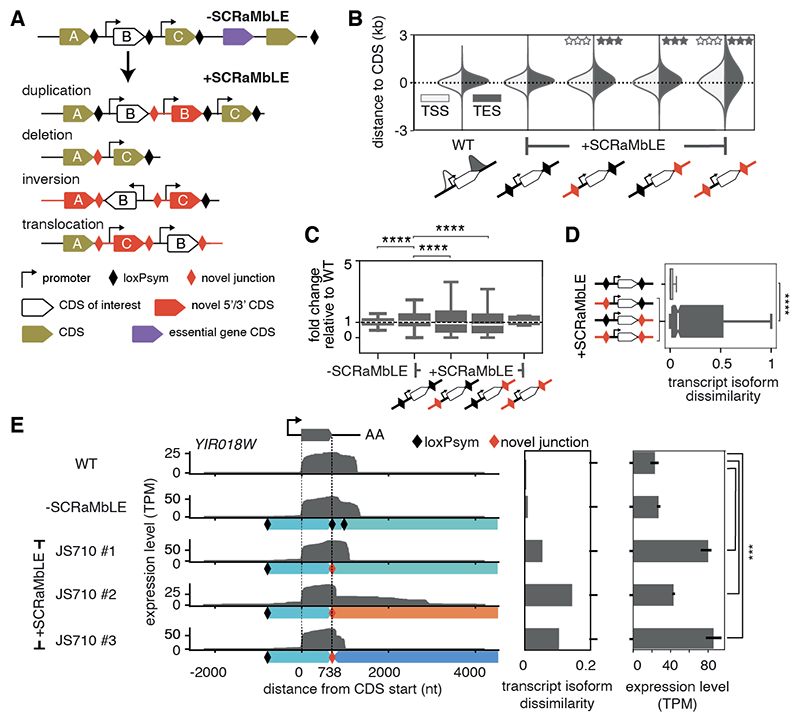
Fig. 1. Genome rearrangement alters transcript isoform expression and boundaries.
(A) Schematic showing SCRaMbLE-induced rearrangements between loxPsym sites (black diamonds) at the 3’ end of all non-essential gene CDSs in the synthetic chromosome, synIXR, inducing multiple possible rearrangements of a CDS (‘B’, here) with novel junctions (red diamonds). (B) Distributions of TSS (white) and TES (gray) distances from gene CDSs in BY4741 (WT) and +SCRaMbLE strains, divided into rearrangements with novel (red) or native (black) 5’- and/or 3’- junctions. Stars indicate significant difference in variance from WT based on Levene’s test for equality of variances (q ≤ 0.001). (C) Distribution of gene expression fold-changes compared to WT for -SCRaMbLE and +SCRaMbLE strains, divided into those with novel (red) or native (black) 5’- and/or 3’-junctions. (D) Degree of transcript isoform dissimilarity from WT for genes with novel 5’- and/or 3’-junctions (red) compared to genes in native arrangements (black) in SCRaMbLE strains. (E) Transcript expression of the YIR018W gene in different contexts: WT (top), the non-rearranged synIXR strain (-SCRaMbLE, JS94), and three contexts in a single +SCRaMbLE strain (JS710 #1, #2, and #3). Left plot: full-length transcript reads aligned by the CDS (flanked by dotted lines). Middle plot: transcript isoform dissimilarity, calculated as in D. Right plot: Salmon quantified expression levels from Illumina stranded mRNA sequencing. Genomic segments below the read-tracks are colored according to their original position on synIXR as in (16). LoxPsym sites and novel junctions are denoted by black and red diamonds, respectively. TPM: transcripts per million. Bars indicate 95% confidence interval based on 3 biological replicates. Boxplots indicate median and interquartile range (IQR), and whiskers extend to the minimum and maximum values within 1.5x IQR. Notches indicate 95% confidence intervals. Asterisks denote significance levels in Mann-Whitney U test, *** p ≤1e-3, **** p ≤1e-4.
The synthetic yeast chromosome IXR (synIXR) is 91,010 bp and contains 43 loxPsym-flanked segments. Following SCRaMbLE of synIXR, 64 strains containing 156 deletions, 89 inversions, 94 duplications and 55 additional complex rearrangements were isolated (16). Notably, these strains do not suffer from gross growth defects (Table S1). Altogether, these SCRaMbLE genomes harbor 612 novel junctions, formed by juxtaposing genomic segments that are usually separated (Fig. 1A). As some rearrangements occur more than once, there are 363 distinct novel junctions. These novel junctions represent several different types of rearrangements, specifically new convergent and tandem gene pairs, genes with alternative 3’-UTRs, and complex juxtapositions of coding sequences, ncRNAs, regulatory elements, and intergenic sequences (fig. S1A) (17). We used this genomic diversity to determine the relative contributions of DNA sequence features versus transcriptional context to establishing RNA isoform boundaries and expression levels.
We profiled the transcriptomes of the 64 previously genotyped synIXR SCRaMbLE strains, the parental strain (-SCRaMbLE, JS94), which bears synIXR without rearrangements, and two wildtype Saccharomyces cerevisiae controls using Oxford Nanopore direct RNA sequencing. This method sequences full-length, native RNA molecules from their poly(A) tail without conversion to cDNA (18). Full-length reads span novel junctions and thus allow transcripts to be mapped to their genomic origin in the rearranged genomes (fig. S2A). Additionally, both a transcript start site (TSS) and transcript end site (TES) (i.e., the isoform boundaries) can be identified for each RNA molecule. In total, we collected nearly 120 million full-length reads that passed a minimum quality threshold (Q mean ≥ 6). Genome-wide, we identified 264,899 transcript isoforms that were supported by 2 or more reads mapping within 25 nt at both ends (accounting for >77 million reads; Table S2 and S3, fig. S2B to F) (17).
To verify that direct RNA sequencing accurately reports on transcript TSSs and TESs, we compared the isoforms that we identified on the native chromosomes to 371,087 major transcript isoforms (mTIFs) identified under similar growth conditions in wildtype S. cerevisiae using transcript isoform sequencing (TIF-Seq) (19). Isoforms from direct RNA sequencing corresponded well (69% of those covering a single ORF) with mTIFs (fig. S3A) (17). Notably, we observed a 60% increase in the number of polycistronic isoforms detected with long read sequencing (4,909 isoforms) compared to TIF-Seq, particularly in the rearranged genomes, suggesting that direct RNA sequencing better captures long RNA species (fig. S3B). To determine whether the direct RNA isoforms that we measured in the SCRaMbLE strains are stable or degraded, we analyzed transcript isoforms in exonuclease mutants (rrp6Δ and xrn1Δ) constructed in a representative SCRaMbLE strain background but found no change in isoform abundance (fig. S4). Thus, these isoforms are part of the stable transcriptome in the SCRaMbLE strains.
Genomic rearrangements influence transcript isoform expression
Compared to the wildtype BY4741 (WT), individual SCRaMbLE strains produced variable numbers of transcript isoforms per gene (up to 20-fold fewer or more) (fig. S5A). We identified 3,228 unique transcript isoforms generated by 50 genes on synIXR in SCRaMbLE strains compared to the -SCRaMbLE strain. These unique isoforms were associated with either an altered TSS (in 1,313 isoforms), an altered TES (in 2,378 isoforms), or simultaneous alterations at both ends (in 2,736 isoforms).
Across all reads, we found that the location of TSSs and TESs (in relation to their CDS) were significantly more variable for genes rearranged into novel contexts than genes in their native context (Fig. 1B, Levene’s test for equality of variances, p ≤ 0.001). While novel 3’ junctions affected TES positioning only, novel 5’ junctions affected both TSS and TES positioning, even though SCRaMbLE maintains the native promoter with its CDS (Fig. 1B). To exclude that the placement of loxPsym sites 3 bp downstream of the stop codon directly affected TES positioning, we measured the change in TES positions between -SCRaMbLE (loxPsym sites) and WT (no loxPsym sites) strains. Reassuringly, this only lengthened transcripts by 34 nt (the length of the loxPsym site) on average (fig. S5B), and had no effect on the variability of transcript boundaries in unrearranged contexts (Fig. 1B). Thus, TES recognition is largely unaffected by the loxPsym site. Unexpectedly, essential genes, which are not flanked by loxPsym sites, also generated a variable number of isoforms across strains, further suggesting that isoforms are responsive to distal changes in their transcriptional neighborhood (fig. S5A).
Rearrangement of genes into new contexts also affected their expression levels. We used short-read Illumina sequencing for gene expression level quantification and corrected for gene copy number changes resulting from SCRaMbLE-induced duplications (fig. S6A) (17). On average, expression of genes in novel contexts tended to decrease, although it was highly variable (Fig. 1C). For example, there were 18 instances where an unexpressed gene gained detectable expression and 141 cases where gene expression was lost (Table S4) (17).
To systematically quantify the changes to a TU’s transcription profile following SCRaMbLE, we computed the cosine similarity of each TU long-read expression profile in every strain compared to WT. We refer to this metric as ‘similarity’, or ‘dissimilarity’ when we compute its inverse (1 – cosine similarity). This verified that transcript isoforms arising from novel junctions were significantly less similar to WT than those at native junctions (Fig. 1D, Mann-Whitney U test p ≤ 1e-4). For example, the CDS encoding YIR018W appears in three different genomic contexts in the SCRaMbLE strain JS710 which altered YIR018W transcript isoforms and expression levels (Fig. 1E). In particular, one context severely disrupted YIR018W TESs, leading to 3’-UTR extensions of up to 4 kb with little change in expression level (Fig. 1E, JS710 #2). Approximately 43% of all rearrangements produced transcript isoforms with less similarity to their native counterparts than this most extreme rearrangement of YIR018W, suggesting that severe transcript isoform disruptions are widespread in SCRaMbLE genomes.
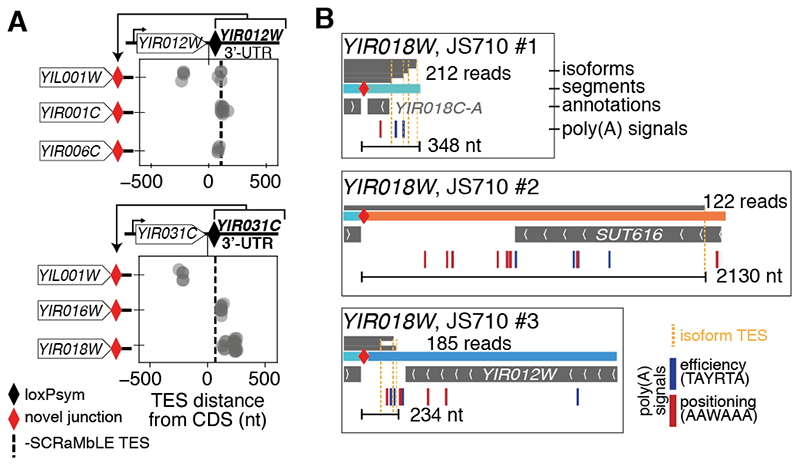
TES alterations are common in SCRaMbLE strains, as reflected by 72 novel polycistronic transcripts and 104 readthrough transcripts (≥100 bp extension), such as YIR018W. Since native 3’-UTR sequences are decoupled from the CDS by SCRaMbLE, defects in TES recognition could logically arise from the loss or gain of 3’-UTR-encoded poly(A) signal (PAS) motifs. If 3’-UTR sequences functioned as plug-and-play modules, they would produce the same TES positions when coupled to different CDSs. However, out of all the 3’-UTRs that were coupled to multiple different CDSs in the SCRaMbLE strains, only one (YIR012W) maintained the TES positioning of the control -SCRaMbLE strain (Fig. 2A). In general, neither the 3’-UTR sequence nor the CDS predicted TES positioning (fig. S7A). Densities of PAS positioning and efficiency sequence motifs downstream of CDSs were also insufficient to explain TES position (fig. S7B). For example, the lengthened YIR018W 3’-UTR isoform extended through many high-efficiency PASs (Fig. 2B, JS710 #2). Thus, a systematic and context-aware assessment of sequence-function relationships could help guide precise engineering of transcription in yeast.
Fig. 2. Isoform boundaries are influenced by factors not encoded in the CDS or 3’-UTR sequence.
(A) Examples of two 3’-UTRs (from YIR012W, top and YIR031C, bottom) rearranged to the 3’-end of three different CDSs (depicted left). The position of all isoform TESs relative to the end of the CDS are plotted for each rearrangement. The TES of the major transcript isoform without rearrangement is indicated by the dashed line. Truncated TESs potentially indicate an early termination site in the YIL001W CDS. (B) 3’-ends of YIR018W transcript isoforms (stacked gray bars with total read counts indicated) mapped to three rearrangements in the JS710 SCRaMbLE strain (as in Fig. 1E). Rearranged segments are colored based on their original location on synIXR as in (16). Annotations, and poly(A) signals (efficiency and positioning motifs, shown in blue and red, respectively) are shown below each context. The longest TES distance and total number of reads supporting the isoforms are indicated for each context.
Transcriptional neighborhood predictably influences transcript isoform boundaries
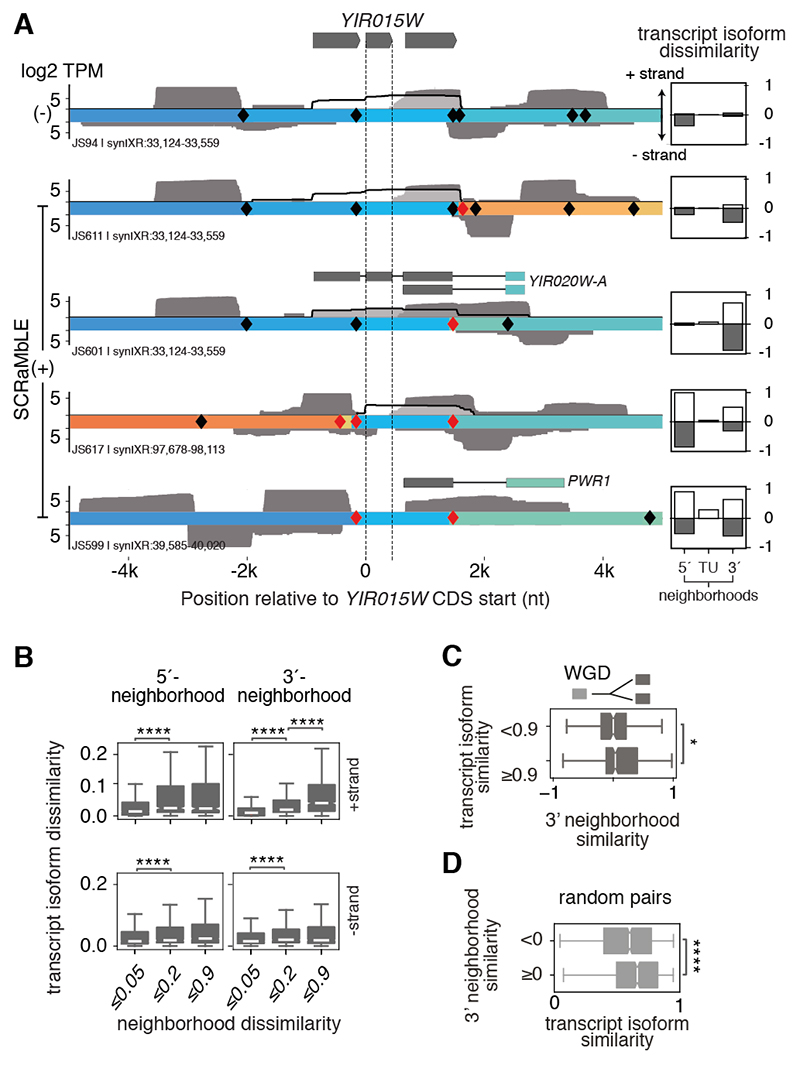
Rearrangements change not only the genetic sequence but also the transcriptional context surrounding a gene. For example, closer investigation of the YIR018W readthrough transcript (Fig. 1E, JS710 #2) shows that its context lacks the antisense transcripts present in WT and two other rearranged contexts that maintain proximal TESs (Fig. 1E). This suggests that neighboring transcription may regulate transcript isoform boundaries and expression levels (fig. S8). To systematically assess this relationship, we extended our cosine similarity metric to quantify transcriptional alterations in flanking regions for every gene, on both strands. Rearrangements that maintained the adjacent transcriptional environment also retained the local isoform properties. For example, a rearrangement that replaced the segment downstream of a polycistronic transcript encoding the essential gene YIR015W with one containing similar convergent transcription preserved the polycistron (Fig. 3A, first SCRaMbLE row). In contrast, an alternative rearrangement lacking proximal antisense transcription resulted in lengthened TESs and multiple novel downstream polycistronic transcripts (Fig. 3A, second SCRaMbLE row). Similarly, rearrangements that disrupted the upstream transcriptional environment altered the composition and expression of the polycistronic transcript (Fig. 3A, bottom rows).
Fig. 3. Transcript isoforms are altered when transcriptional neighborhoods are perturbed.
(A) Direct RNA transcript reads covering the essential nuclear RNaseP gene, YIR015W, in - SCRaMbLE and four +SCRaMbLE strains. Reads spanning YIR015W CDS are outlined in black with transparent fill; other reads within a +/-5kb region are gray. Sense and antisense reads are above and below genomic segment tracks; segments are colored according to their original position on synIXR as in (16). Gene models show novel polycistronic transcripts incorporating genes from rearranged segments. Quantification of dissimilarity relative to WT expression profiles for each strand (white and gray boxes) in the 5’ and 3’ regions flanking the TU and for the TU, itself, are displayed next to each track. (B) Transcript dissimilarity from WT assessed separately in each panel for rearrangements affecting the 5’ or 3’ transcriptional neighborhood within a 3 kb window, on either strand. (C) The transcriptional similarities of 3’ neighborhoods on both strands are compared for paralogs with more (≥0.9) or less (<0.9) transcript isoform similarity. WGD: whole genome duplication. (D) Transcript isoform similarity of randomly selected gene pairs compared based on the similarity of their downstream transcriptional environment on both strands. Data are represented as the median and interquartile range (IQR) with whiskers extending to the minimum and maximum values within 1.5x IQR. Notches indicate 95% confidence intervals. Asterisks denote significance levels in Mann-Whitney U test *p≤0.05, ****p≤1e-4.
Across the synthetic genome, TU isoforms became significantly more dissimilar to the WT as SCRaMbLE induced greater alterations to their transcriptional neighborhoods (Fig. 3B). This relationship was also apparent in the native genome. Paralogs that have maintained similar downstream transcriptional neighborhoods since the yeast whole genome duplication event ~100 million years ago retained similar transcriptional profiles. Even randomly selected gene pairs with comparable downstream transcriptional neighborhoods generated similar transcript isoforms (Fig. 3C to D). Together, these results reinforce a link between transcript isoform properties and neighboring transcription both across evolution and throughout the genome.
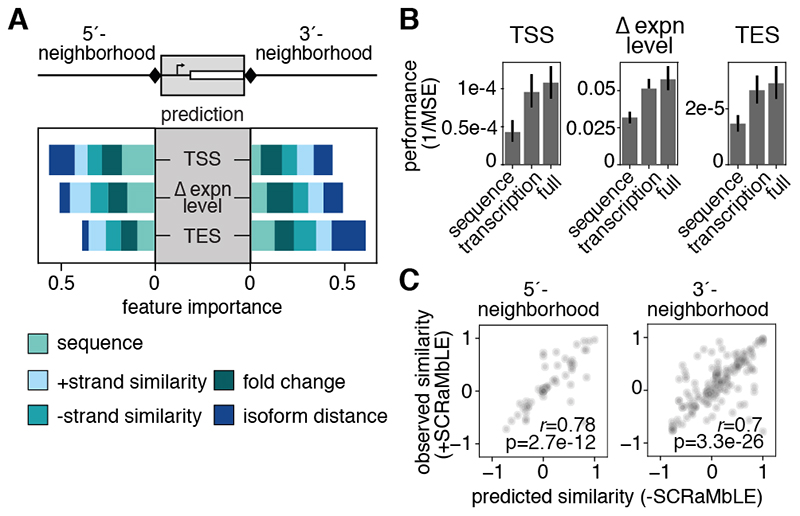
Our data suggest that both transcriptional neighborhood and genetic sequence influence isoform boundaries and expression levels. To disentangle these overlapping contributions systematically and genome-wide, we used machine learning. We trained Gradient Boosted Regression Trees (GBRT) to predict TU properties (i.e. expression level changes and TSS and TES distances from gene CDSs) in rearranged contexts, using genetic sequence and transcriptional neighborhood features for the predictions (17). Specifically, up- and downstream gene identity and orientation were used as a proxy for sequence features, and properties of the up- and downstream transcriptional environment up to 3 kb away, including gene expression levels, isoform similarity on either strand, and distance to the nearest TU, were used as transcriptional neighborhood features (17). To interpret these models, we computed the predictive value of each feature (Fig. 4A). As expected, upstream features better predicted TSSs and downstream features better predicted TESs, while both contributed equally to predicting expression level changes (Fig. 4A). Notably, models trained only on transcriptional neighborhood features performed on par with the model trained on all features (Fig. 4B) (17). Thus, changes to isoform boundaries and expression levels in novel genomic contexts are predictable solely from the transcriptional neighborhood. Observations in our dataset support individual associations learned by the GBRT model. For example, placing a TU in a highly expressed region increased its expression (fig. S9A). Likewise, TSS and TES distance from the CDS tended to increase with distance to neighboring 5’ and 3’ transcription, respectively (fig. S9B).
Fig. 4. Transcriptional neighborhood predicts transcript isoform expression levels and lengths.
(A) Averaged feature importance scores for models predicting TSS or TES positioning or expression level changes (Δ expn) for genes in the SCRaMbLE strains learned using Gradient Boosted Regression Trees (GBRT). Stacked bars show the fractional contribution of sequence features and transcriptional features (transcriptional similarity on either strand, expression level fold change, and distance to the nearest isoform) in the 5’ and 3’ neighborhoods (within 3 kb) for each prediction. The importance of all 5’ and 3’ features sum to one for each prediction task. (B) Performance of models predicting TSS or TES positioning or Δ expn trained using genomic features only (‘sequence’), features related to the transcriptional neighborhood only (‘transcription’) or all features (‘full’). Bars indicate 95% confidence interval across all models. MSE: mean squared error. (C) Observed versus predicted (from -SCRaMbLE) flanking transcriptional similarities for rearranged segments and their correlation (Pearson correlation coefficient, r). Areas of greater density are darker. Since transcript isoform coverage vectors on both strands were used, cosine similarity ranges from -1 to 1.
Engineering TU isoform properties using transcriptional neighborhoods would be impractical if the transcriptional environment (and hence the TU, itself) must be measured in each new genetic context. We therefore investigated whether changes to transcriptional neighborhoods in SCRaMbLE strains could be estimated from their transcriptional profiles in the reference, - SCRaMbLE strain. Indeed, transcriptional similarities estimated from the -SCRaMbLE strain correlated with the changes observed in SCRaMbLE strains (Fig. 4C, Pearson’s r = 0.78 and 0.7 for 5’- and 3’-neighborhoods, respectively) (17). Thus, transcript isoform properties are predictable from neighboring transcription and can be engineered by modifying the transcriptional neighborhood.
3’-UTR lengths can be tuned by convergent transcription
To define principles of neighboring transcriptional cross-talk that could support synthetic genome design, we investigated the relationship between specific features of the model, such as intergenic distance and local expression, and transcript isoform boundaries. 3’-UTR lengths increased as a function of intergenic distance across the native yeast genome in our dataset (Fig. 5A), with 3’-UTRs of convergently transcribed genes approximately 25 nt longer for every 100 bp increment of intergenic distance. A similar trend occurred for SCRaMbLE-induced novel convergent gene pairs, although few intergenic distances increased by more than 300 nt (Fig. 5B). Additionally, 3’-UTR lengths were sensitive to downstream expression levels, with decreased levels associated with lengthened 3’-UTRs (Fig. 5C). Notably, a significant fraction (34 out of 104, hypergeometric p-value = 1.2e-7) of isoforms extended by ≥100 nt were relocated into convergent arrangements with reduced downstream gene expression.
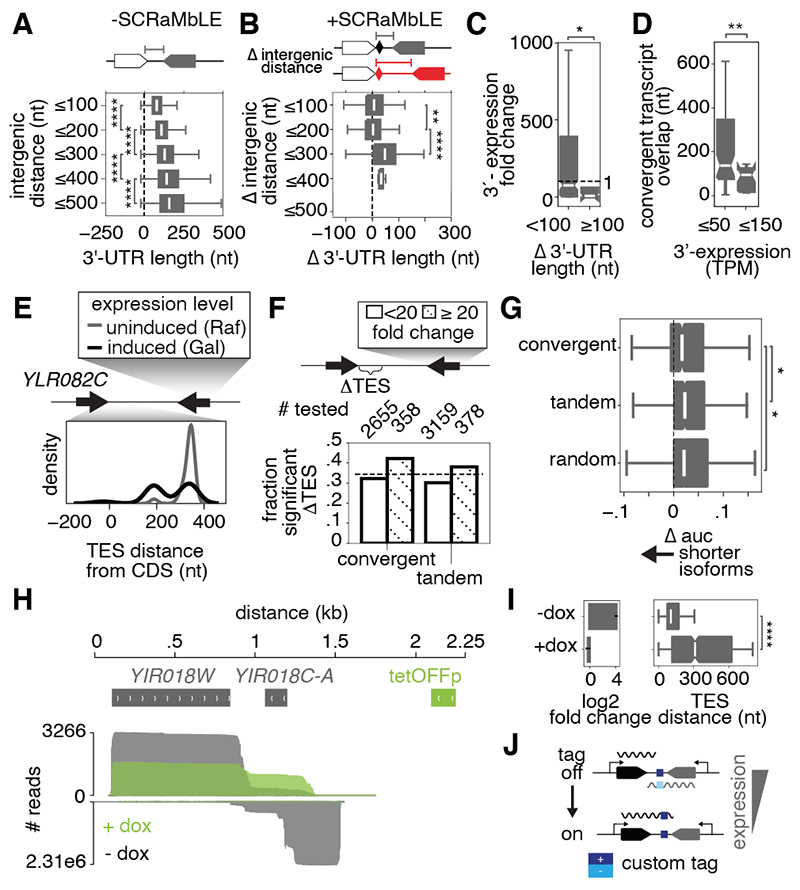
Fig. 5. Neighboring gene expression regulates and can be used to engineer 3’-UTR lengths.
(A) 3’-UTR lengths of convergent genes binned by 100 bp increments of intergenic distance in the WT genome. (B) Change in 3’-UTR lengths of convergent gene pairs plotted by increased (100 bp increments) intergenic distance after SCRaMbLE. (C) Expression fold-changes of genes convergent to those with minor (<100 nt) or major (≥100 nt) 3’-UTR extensions after rearrangement. (D) Length of overlap (nt) between novel convergent transcripts where the downstream member is lowly (≤50 TPM) or highly (≤150 TPM) expressed. TPM: transcripts per million. (E) Distribution of YLR082C TESs (relative to its CDS) when the convergent gene is over-expressed (Gal, black) or not (Raf, gray). (F) Fraction of genes in convergent and tandem pairs with significantly altered TES positions (Kolmogorov–Smirnov test, p ≤ 0.001, applied to each gene) when an adjacent gene is ≥20-fold overexpressed (hatched) or not (white) following galactose-induced transcription factor (TF) overexpression. Dashed line indicates the fraction of randomly selected genes with significantly altered TESs in galactose. Number of genes tested are indicated above the bars. (G) Change in 3’-UTR length distributions for convergent, tandem, and random gene pairs upon TF overexpression in galactose, as assessed by the change in the area under the curve (Δ auc) of TES cumulative distributions. Negative values indicate isoform shortening. (H) cDNA sequencing reads aligned to YIR018W (above) and YIR018C-A (below) in a tetracycline-repressible YIR018C-A strain in the absence (gray) and presence (green) of doxycycline. (I) Change in YIR018C-A expression (left), plotted as mean ± standard deviation, and YIR018W 3’-UTR length (right) upon doxycycline-induced inhibition of YIR018C-A expression. (J) The ability to control 3’-UTR length by altering convergent gene expression levels could be applied to embed a reversibly expressed, functional sequence tag in transcript 3’-UTRs. Only adjacent bins were tested for significance in (A) and (B). Boxplots indicate median and interquartile range (IQR), and whiskers extend to the minimum and maximum values within 1.5x IQR. Notches indicate 95% confidence intervals. Asterisks denote significance levels in Mann-Whitney U test, * p ≤0.05, ** p ≤1e-2, *** p ≤1e-3, ****p ≤1e-4.
Genome-wide, transcripts of convergent genes consistently overlapped by 85 nt on average in our dataset (fig. S10A), consistent with previous observations (20). Even genes rearranged by SCRaMbLE into novel convergent pairs produced transcripts overlapping by 85 nt on average, implying that the process of transcription itself, rather than sequence features, directs the length-restricted interdigitation of convergent 3’-UTRs. Reinforcing the observation that transcript length responds to transcriptional context, we found that the overlap length and the fraction of the intergenic space dominated by a transcript increased as the expression level of a convergent transcript decreased (fig. S10B and C). Additionally, novel convergent gene pairs with a lowly-expressed downstream gene produced significantly longer overlaps (Fig. 5D).
To confirm that 3’-UTR lengths are limited by convergent transcription in the native yeast genome, we measured the effects of perturbing gene expression on isoform boundaries of gene pairs genome-wide. We overexpressed transcription factors (MSN2, GCN4, STE12, ADR1 and HAC1) in a galactose-inducible manner and mapped the shortening of 3’ ends of genes adjacent to those induced by transcription factor overexpression (Fig. 5E) (21). Across all transcription factor overexpression strains, 449 convergent and 502 tandem gene pairs showed ≥20-fold increased expression of at least one of their members when grown in galactose (fig. S11). In line with our predictions, 42% of all genes convergent to a gene with a ≥20-fold expression-level increase in galactose had significantly altered TES positioning (Fig. 5F, Kolmogorov-Smirnov test, p ≤ 0.001, applied to each gene). Convergent genes also had significantly shorter 3’-UTR length alterations when their neighbor was overexpressed than tandem or random gene pairs (Fig. 5G, Mann-Whitney U test, p ≤ 0.05), supporting a role for convergent transcription in limiting 3’-UTR length.
Finally, to demonstrate that our model can be applied to genome engineering, we constructed a tetracycline-repressible system to reversibly control a transcript’s 3’-UTR length by tuning the expression of a downstream, convergent transcript. We chose the YIR018W/YIR018C-A convergent gene pair, as the length of the YIR018W 3’-UTR appeared sensitive to downstream convergent transcription when placed into novel contexts (Fig. 1E and fig. S8). Incorporating a P7xtetO promoter in the BY4741 YIR018C-A locus increased its expression 20-fold and shortened transcript isoforms from the convergent gene, YIR018W. Adding doxycycline to turn off the promoter returned YIR018C-A expression to WT levels and restored YIR018W transcript lengths (Fig. 5H to I). As there was no sequence alteration, the 3’-UTR alterations resulted solely from transcription changes in the downstream convergent transcript.
Discussion
We show that distal transcription influences local transcript isoform boundaries and expression levels in a predictable manner. Our observations regarding 3’-UTR extensions in convergent transcripts suggest that adjacent transcription imposes physical constraints on isoform boundaries. Along with other factors, such as PAS motifs, antisense transcription appears to play a role in TES positioning, making convergent genes sensitive to the expression level of their neighbor. We suggest that convergent transcription may slow RNA polymerase transit, thereby affecting TES selection, similar to the regulation of proximal PAS usage by nucleotide availability or mutations that slow RNA polymerase elongation (22).
Relationships between neighboring TUs could be co-opted to engineer genomes. For example, the dynamic range of gene expression changes that we observed in rearranged genetic contexts suggests that transcriptional neighborhoods could be exploited to tune expression of TUs by at least five-fold (Fig. 1C). Furthermore, local gene expression, order, orientation, and/or distance could inform the construction of synthetic circuits that interlink the regulation of neighboring TUs. Specifically, expression could be increased by placing a gene in a highly-expressed region, or TES position could be modulated by altering expression levels or distance of a neighboring convergent transcript. These design principles expand the synthetic biology toolkit, revealing the potential to embed functionalities into a reversibly expressed 3’-UTR controlled by neighboring TU expression, which we term ‘transcriptional embedding’ (Fig. 5J).
We conclude that most yeast DNA sequences do not encode simple plug-and-play properties but have evolved co-functional relationships that are perturbed outside of their native context. Evaluating the behavior of DNA sequence parts in alternative genomic contexts will provide additional tools to improve rational genome design.
Supplementary Material
One Sentence Summary.
Transcript isoform levels and boundaries can be predicted and engineered based on neighboring gene expression patterns.
Acknowledgments
We thank members of the Steinmetz lab, particularly B. Linder, D. Schraivogel, B. Rauscher, M. Bertolini, and K. Fenzl, for useful discussion and helpful comments on the manuscript. We also thank Life Science Editors for assistance with preparing the manuscript. We thank V. Benes, F. Jung and the EMBL Genomics Core Facility for performing Illumina RNA sequencing.
Funding
This work was funded by grants from the BMBF (031A460 to L.M.S.), the Volkswagen Stiftung (94769 to L.M.S.). A.N.B. was supported by a fellowship from the EMBL Interdisciplinary Postdoc (EI3POD) program under Marie Skłodowska-Curie Actions COFUND (grant number 664726). L.A.M. and J.D.B. were supported by NSF grants MCB-1616111 and MCB-1445537.
Footnotes
Author contributions: Conceptualization, A.N.B., A.L.H., and L.M.S.; Methodology, Writing – Original Draft, and Visualization, A.N.B. and A.L.H.; Software, Data Curation, and Formal Analysis, A.N.B.; Investigation, A.L.H. and S.C.M.; Writing – Review & Editing, A.N.B., A.L.H., J.D.B. and L.M.S.; Resources, L.A.M. and J.D.B.; Funding Acquisition and Supervision, A.N.B and L.M.S.
Competing interests: L.A.M. is also affiliated with Neochromosome, Inc.
Data and materials availability
Raw data can be downloaded from NCBI SRA (#PRJNA664019). Code used to perform analyses can be accessed at git.embl.de/brooks/scramble-transcriptome/. Code and trained GBRT models can be accessed on Zenodo (DOI 10.5281/zenodo.5676293) (23). A genome browser featuring both long and short read alignments is available at: https://apps.embl.de/scramble/.
References and Notes
- 1.Guo Z, Sherman F. Signals sufficient for 3’-end formation of yeast mRNA. Mol Cell Biol. 1996;16:2772–2776. doi: 10.1128/mcb.16.6.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danino YM, Even D, Ideses D, Juven-Gershon T. The core promoter: At the heart of gene expression. Biochim Biophys Acta. 2015;1849:1116–1131. doi: 10.1016/j.bbagrm.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Lubliner S, Regev I, Lotan-Pompan M, Edelheit S, Weinberger A, Segal E. Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 2015;25:1008–1017. doi: 10.1101/gr.188193.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran KA, Karim AS, Gupta A, Alper HS. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab Eng. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redden H, Alper HS. The development and characterization of synthetic minimal yeast promoters. Nat Commun. 2015;6:7810. doi: 10.1038/ncomms8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 8.Mellor J, Woloszczuk R, Howe FS. The Interleaved Genome. Trends Genet. 2016;32:57–71. doi: 10.1016/j.tig.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Meyer S, Beslon G. Torsion-mediated interaction between adjacent genes. PLoS Comput Biol. 2014;10:e1003785. doi: 10.1371/journal.pcbi.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colin J, Candelli T, Porrua O, Boulay J, Zhu C, Lacroute F, Steinmetz LM, Libri D. Roadblock termination by reb1p restricts cryptic and readthrough transcription. Mol Cell. 2014;56:667–80. doi: 10.1016/j.molcel.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proceedings of the National Academy of Sciences. 2002;99 doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greger IH, Proudfoot NJ. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai J, Lindstrom DL, Boeke AC, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Stracquadanio G, Wang Y, Yang K, Mitchell LA, Xue Y, Cai Y, Chen T, Dymond JS, Kang K, Gong J, et al. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2016;26:36–49. doi: 10.1101/gr.193433.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.supplementary material
- 18.Garalde D, Snell E, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 19.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497 doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen T, Fischl H, Howe FS, Woloszczuk R, Serra Barros A, Xu Z, Brown D, Murray SC, Haenni S, Halstead JM, O’Connor L, et al. Transcription mediated insulation and interference direct gene cluster expression switches. Elife. 2014;3:e03635. doi: 10.7554/eLife.03635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, et al. Mapping pathways and phenotypes by systematic gene overexpression. Molecular Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Yague-Sanz C, Vanrobaeys Y, Fernandez R, Duval M, Larochelle M, Beaudoin J, Berro J, Labbé S, Jacques P-É, Bachand F. Nutrient-dependent control of RNA polymerase II elongation rate regulates specific gene expression programs by alternative polyadenylation. Genes Dev. 2020;34:883–897. doi: 10.1101/gad.337212.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenodo. [DOI] [Google Scholar]
- 24.Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. PNAS. 2006;130:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen K, Gitsham P, Wishart J, Oliver SG, Zhang N. An improved tetO promoter replacement system for regulating the expression of yeast genes. Yeast. 2003;20:1255–1262. doi: 10.1002/yea.1037. [DOI] [PubMed] [Google Scholar]
- 26.Steensma HY, Ter Linde JJ. Plasmids with the Cre-recombinase and the dominant nat marker, suitable for use in prototrophic strains of Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast. 2001;18:469–472. doi: 10.1002/yea.696. [DOI] [PubMed] [Google Scholar]
- 27.Sprouffske Kathleen. growthcurver: Simple Metrics to Summarize Growth Curves. R package version 0.3.1. 2020. [Google Scholar]
- 28.Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CLV, Chandrasegaran S, Cai Y, Boeke JD, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 29.Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ. GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. Methods Mol Biol. 2016;1418:283–334. doi: 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
- 31.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Depledge DP, Srinivas KP, Sadaoka T, Bready D, Mori Y, Placantonakis DG, Mohr I, Wilson AC. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat Commun. 2019;10:754. doi: 10.1038/s41467-019-08734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System; Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016. pp. 785–794. [Google Scholar]
- 37.Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 38.Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 39.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nature Genetics. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data can be downloaded from NCBI SRA (#PRJNA664019). Code used to perform analyses can be accessed at git.embl.de/brooks/scramble-transcriptome/. Code and trained GBRT models can be accessed on Zenodo (DOI 10.5281/zenodo.5676293) (23). A genome browser featuring both long and short read alignments is available at: https://apps.embl.de/scramble/.