Abstract
Our understanding on human neurodegenerative disease was previously limited to clinical data and inferences about the underlying pathology based on histopathological examination. Animal models and in vitro experiments have provided evidence for a cell-autonomous and a non-cell autonomous mechanism for the accumulation of neuropathology. Combining modern neuroimaging tools to identify distinct neural networks (Connectomics) with target-specific Positron Emission Tomography (PET) tracers is an emerging and vibrant field of research with the potential to examine the contributions of cell-autonomous and non-cell autonomous mechanisms to the spread of pathology. The evidence provided here suggests that both cell-autonomous and non-cell autonomous processes relate to the observed in vivo characteristics of protein pathology and neurodegeneration across the disease spectrum. We propose a synergistic model of cell- autonomous and non-cell autonomous accounts that integrates the most critical factors (i.e., protein strain, susceptible cell feature and connectome) contributing to the development of neuronal dysfunction and in turn produces the observed clinical phenotypes. We believe that a timely and longitudinal pursuit of such research programs will greatly advance our understanding of the complex mechanisms driving human neurodegenerative diseases.
Introduction
Human neurodegenerative diseases are characterized by inclusions of protein aggregates comprising amyloid-β (Aβ), Tau, TDP-43 or alpha-synuclein [s 1,s 2,s 3]. A small percentage are caused by genetic mutations affecting these proteins or their cellular processing pathways, that results in the overproduction, misfolding, aggregation or reduced clearance of the pathologic proteins [s4,s5].
Although distinct clinical phenotypes relate to different types of protein aggregates [6,s7], the spectrum of neurodegenerative disease shares striking commonalities. The most common risk factor for the development is advanced age, suggesting that the biological process of aging by itself contributes considerably to pathological neurodegeneration [s8]. Further, the molecular principles of abnormal protein aggregation and clearance appear to be similar across different types of brain cells, irrespective of the disease type. Third, pathological protein aggregation is not evenly distributed across all brain regions, but each type of protein aggregate appears to be initiated in a specific location and to proceed in an ordered fashion encompassing additional brain regions with advancing disease stage [s9−s11]. Several mechanistic models have been put forward to explain the observation of gradual expansion of protein pathology in neurodegenerative disease. These include prion-like propagation [s12,s13], selective neuronal vulnerability via gene expression profiles [s14,s15] or via activity-dependent metabolic demand [s16,s17].
Advances have been made in the development of target-specific Positron-Emission-Tomography (PET) tracers to visualize and quantify local molecular alterations (e.g., amyloid imaging, tau imaging, neurotransmitter imaging). Moreover, modern imaging techniques (e.g., resting state functional magnetic resonance imaging (rs-fMRI); diffusion tensor imaging (DTI), magnetoencephalography (MEG) have matured to obtain a holistic approach to the way, brain areas are functionally and structurally connected (“Connectomics”). These developments dovetail to provide a unique opportunity to examine how protein pathology in one location may gradually spread to other brain regions, or affect connectivity between them.
In the following, we outline current concepts regarding the spatial and temporal determinants of accumulation of protein aggregates in human neurodegenerative disease and how the Connectomics approach has contributed to our understanding of the mechanistic processes underlying neuropathological spreading. We close with our perspective on the challenges that need to be addressed to fully acknowledge the underlying complexity of brain networks and molecular variations associated with human neurodegenerative disease.
1.1. Theories of Spreading
Prion-like Propagation
The term “prion-like” propagation alludes to the observation that in prion disease, such as Creutzfeld-Jakob disease, proteinaceous infectious particles (prions) increased with infectious activity and generated the formation of a conformer that can bind and convert other prion protein molecules into aberrant oligomers and insoluble aggregates [s18,s19]. This concept has been adopted to other disease-related proteins where the formation of fibrillar or oligomeric species initiates or ‘seeds’ the assimilation of non-pathological proteins into similar structural configurations. The resulting seeding process facilitates the aggregation of conformationally changed proteins that spread to interconnected neurons and adjacent glia cells [s20]. The pathological spread hypothesis is supplemented by the hypothesis that structural connectivity and synaptic activity among neurons direct the spread of pathology between cells and between regions. Where pathogenic oligomers or aggregates physically move from one neuron to another, and therefore spread into connected brain regions, it is referred to as non-cell-autonomy [s21].
Selective vulnerability via gene expression profiles
The distinct concept of selective vulnerability posits that some neurons are intrinsically more vulnerable to a pathological process associated with the disease than others. This vulnerability may be explained by gene expression profiles of neurons that are more prone to protein accumulation and in turn more likely to become dysfunctional earlier in the disease process than other neuron assemblies [14,15]. For example in vitro experiments of human motor neurons identified different gene expression profiles in neurons resistant to amyotrophic lateral sclerosis (ALS) such as oculomotor neurons, compared to spinal motor neurons vulnerable to ALS [s22]. Further, within the dopaminergic system, neuronal phenotypes could be identified that are characterized by coordinated gene expression with distinct functional and anatomical properties and projection fields more or less vulnerable for models of Parkinson’s Disease (PD) [s23]. Together, this hypothesis centers on a cell-autonomous mechanism for the progression of pathology in neurodegenerative disease. Cell-autonomy often refers to the greater susceptibility of some neurons to adverse conditions, but it also includes the ability to amplify pathogenic protein species faster: amplification of toxic oligomeric or aggregated species is distinguished from their spread. Disease progression may be enhanced by both external stress and emergence of protein aggregates in more vulnerable neurons, and the transfer of protein aggregates by non-cell-autonomous mechanisms.
Selective vulnerability via Activity-dependence
It has been proposed that highly active regions are more vulnerable to neurodegeneration. There are at least two mechanisms that may contribute to this observation. The first is that neuronal activity can enhance the propagation of pathogenic proteins from cell to cell [s24] implying that highly active, and well connected, neurons are more likely to receive or transmit the pathology. Second, highly active cells may be more vulnerable to metabolic or other cellular stress, fostering the accumulation and misfolding of proteins [s16]. Cellular stress can arise by high physiological levels of excitation or disturbances of cellular homeostasis through inflammatory reactions, oxidative stress or shifting calcium demands [s14]. Computational modeling approaches in Alzheimer’s Disease (AD) have shown that highly connected brain regions (i.e., ‘hubs’) suffer from activity dependent degeneration, suggesting that the maintenance of excessive neuronal activity renders cortical hub regions to be more vulnerable for pathological disease development [s17]. Although this proposal focuses on environmentally-induced cellular stress, it acknowledges the contribution of “intrinsic stressors” (e.g., genetic background or advancing age) [s16]. Activity-dependent stress, and activity-dependent spread, call for new tools to examine the relationships between molecular substrates of disease, brain activity and brain connectivity.
The advancement of functional and structural MRI and electrophysiological measures of synaptic activity such as MEG has characterized the brain’s network structure that appears to employ orchestrated processes underlying human cognitive function [s25]. Understanding the complexity with which these dynamic processes among brain networks interact and produce cognitive function is a field of its own and the interested reader is referred to excellent reviews on this topic [s26,s27]. Importantly, investigating brain network dynamics (i.e., Connectomics) offers the opportunity to test how pathology in human neurodegenerative disease relates to the brain’s intrinsic network architecture and which of the aforementioned proposals of protein accumulation may best describe disease progression.
1.2. Measuring Pathology
PET imaging allows the in vivo characterization of different molecular properties in the central nervous system, such as neuroinflammation, protein aggregates, regional glucose metabolism, synaptic density and neurotransmitter release. Each of these show different aspects of pathology in neurodegenerative disease. Imaging cerebral glucose metabolism using FDG-PET is a well-established method to examine the extent and regional pattern of neuronal dysfunction. It is in widespread clinical and research use for imaging neurodegeneration, with information about location and severity of the disease [28–30], but it does not provide etiological information about the molecular mechanisms underlying the neuronal dysfunction.
Quantification of regional amyloid and tau burden in vivo has been reliant on PET imaging. Through the development of a novel class of PET tracers, targeting beta-amyloid (e.g., [C11]-PIB, [18F]-Florbetapir, [18F]-Florbetaben, [18F]-Flutemetamol), the visualization and quantification of beta-amyloid plaques is now possible and used in large-scale studies, clinical trials, and even routinely implemented in clinical practice [28,31]. PET tracers targeting tau, although not yet fully validated, have already highlighted their potential in unraveling disease mechanisms and their progression, contribution to differential diagnosis and the establishment of a link between neuropathology and resulting clinical symptoms [6,32,33]. While first-generation tau tracers such as [18F]-AV1451, [18F]-THK5317, [18F]-THK-5351, and [C11]-PBB3 expressed varying levels of off-target binding to midbrain regions, basal ganglia, choroid plexus and neuromelanin, second generation tau-PET tracers such as [18F]-PI2620, [18F]-MK-6240 or [18F]-RO9484 are hoped to overcome some of this off-target binding [s34].
Neurodegeneration has also been associated with inflammation and loss of synapses. A comprehensive discussion on the development of new inflammation biomarkers and their contribution to neurodegeneration is provided by Rodriguez-Vietez et al., (2019) in this special section.
To assess impaired nigrostriatal dopaminergic nerve terminals, a characteristic feature of early idiopathic PD, PET/SPECT markers of presynaptic dopamine transporters (e.g., [123I]FP-CIT) have been developed. While these could clearly demonstrate in-vivo loss of nigrostriatal dopamine terminals in PD, sensitivity seems to fade once loss exceeds 50 % [s35], thereby rendering this marker most sensitive to early phases of the disease. Another approach to assess dopaminergic function is using 18F-DOPA, which can estimate the dopamine turnover [s36] and has also been shown to track disease progression. Vesicular monoamine transporter 2 (VMAT2), a protein responsible for transporting monoamine neurotransmitters (dopamine, norepinephrine, serotonin) into synaptic vesicles for subsequent storage and release have been implicated in a variety of neurodegenerative disorders, including Parkinson’s and Huntington’s diseases [s37,s38].
Finally, the loss of cholinergic cells in the basal forebrain has shown to be a contributor to early memory impairments in AD [39] and was associated with global build-up of cortical amyloid deposition during the preclinical phase [40]. Measuring cortical acetylcholinesterase enzyme activity using 11C-Nmethyl-4-piperidyl acetate (MP4A) PET imaging allows the evaluation of cholinergic neurotransmitter input via examining the regional cortical activity of acetylcholinesterase enzymes [41]. Others have employed an α4β2 nicotinic acetylcholine receptors (α4β2-nAChRs) specific radioligand (-)-18FFlubatine and observed significant relationships of cholinergic deficiency and impairment in measures of episodic memory and executive function [42].
1.3. Measuring Connectomics
Connectomics refers to the organizational principle of the brain as a host of distinct neuronal networks. Brain networks can be characterized via functional or structural properties depending on the imaging modality utilized to investigate brain networks.
Electrophysiological measures of brain networks assess neuronal activity from surface sensors (EEG or MEG) with high temporal resolution and bandwidths typically of 1-100 Hz. Frequency-dependent correlations among sensors or electrodes of generalized synchronization have shown to reveal consistent dynamics and properties of brain networks [s43,s44]. These metrics can be used to identify more precise temporal dynamics of network connectivity [45], but measuring functional connectivity beyond the brain surface is restricted. This limitation is resolved by the application of functional connectivity information from resting-state functional MRI (rs-fMRI), which capitalizes on the statistical relationship of “coactivation” of brain regions. Brain data are parcellated into units (i.e., voxels, or regions that may be derived from an anatomical atlas [s46] or randomly seeded) and the time-series of the blood oxygen level-dependent (BOLD) signal across the resting-state fMRI scan is extracted from each unit. Methods of analysis typically focus on co-variance patterns that elicit brain networks. The resulting brain networks are highly reproducible, and reveal organizational principles of the human brain [e.g., s26,47].
Mapping the structural relationship between brain regions using diffusion imaging and tractography exploits water diffusion properties of fiber bundles anatomically connecting brain regions and allow the projection of cortico-cortical pathways [s48]. As this technique derives indices associated with the integrity of white matter microstructure it is referred to as ‘structural connectivity’ [s49,s50].
A novel approach to assess specific features of brain architecture is the identification of common regional gene expression profiles [s51]. Regional gene expression profiles reflect the molecular properties underlying specific brain tissue characteristics and allow the examination of spatially distinct patterns on a molecular level [s52,53].
1.4. Multimodal Imaging Studies on Connectomics and Molecular Pathology
The advancement of target-specific PET tracers to assess the molecular pathology in combination with functional and structural neuroimaging tools, offers the opportunity to test the different hypotheses underlying the development and the progression of pathology in human neurodegenerative diseases. With the advent of tracers to target beta-amyloid pathology [s54,s55] in combination with resting-state functional imaging it was possible to demonstrate that regions maxima of beta-amyloid deposition in AD patients are found predominantly in regions of the default mode network [s56, s57, 58]. Further evidence suggested that disease specific atrophy pattern corresponded with functionally connected regions which promoted the concept of the network degeneration hypothesis [s59,s60]. This hypothesis summarizes evidence that different types of neurodegenerative pathologies apparently target specific neuronal functional networks [61–63]. In addition to studies underscoring the spatial correspondence between patterns of neurodegeneration (i.e., atrophy, metabolism) and functional brain networks, studies have shown that these functional brain networks undergo alterations in the face of disease or neuropathology [64]. For example, functional connectivity within the default mode network is disrupted as beta-amyloid deposition increases [65,s66]. However, as beta-amyloid pathology spreads extraneuronally and is ubiquitously distributed across the brain in advanced disease stage in AD, its direct relationship to network dysfunction and disease severity is less apparent [s67]. The recent development of in vivo PET tracers to examine (intraneuronal) tau-pathology, thought to be more closely linked to neuronal injury and clinical symptoms, may close this gap [s6].
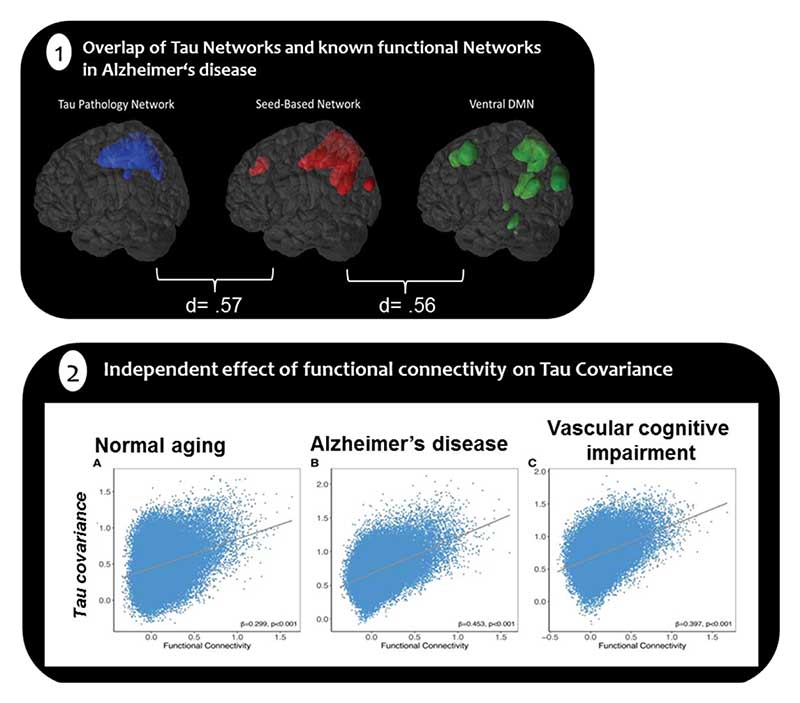
Measuring retention of [18F]-AV1451 as a proxy of underlying tau pathology in amnestic AD dementia patients (N=22), and identifying regions that independently constitute networks of tau burden, Hoenig et al. (2018) utilized seeds of the highest uptake in each tau network to map where in the brain this region is functionally connected to. These tau-seed informed functional connectivity maps match the topology of known functional connectivity networks such as the ventral default mode network and the language network (see Figure 1.1). These functional networks of tau burden have been replicated in a larger group of patients spanning the entire spectrum of AD (N=218) [68] and support the hypothesis that the co-occurrence of tau accumulation in spatially distinct regions may be linked to their functional connections [47].
Figure 1.
1) Overlap of Tau Networks and known functional Networks in Alzheimer’s Disease. Tau pathology networks correspond with functional connectivity networks quantified with a dice-coefficient of d=.57, d=.56 (adapted from Hönig et al.,). 2) Regions with high functional connectivity show covarying tau levels across normal aging, Alzheimer’s disease and vascular cognitive impairment adopted from Franzmeier et al., 2018
Further evidence on the direct relationship between measures of functional connectivity and tau pathology in AD showed that densely interconnected brain regions (i.e., graph theoretical measures of weighted degree) accrue more tau pathology suggesting that higher connectivity renders regions to be more susceptive to tau deposition [69]. In a combined [18F]-AV1451-PET and resting state fMRI- study in addition to [18F]-AV45-PET study, Franzmeier and colleagues (2018) showed that higher resting-state functional connectivity between regions was associated with higher covariance in tau-PET binding in corresponding pairs of brain regions (see Figure 1.2). These results suggested a spatial similarity between the inter-regional connectivity and the sharing of tau between regions. Importantly, hotspots of tau accumulation such as the inferior temporal lobe, were associated with higher tau-PET uptake in functionally closely connected regions, suggesting that the level of tau deposition in a brain region varies as a function of its level of functional connectivity to a tau hot-spot. Interestingly, the relationship of similarity of tau levels and strength of functional connectivity was observed in cognitively normal older adults (N=82), in patients with MCI and AD (N=23) and in patients with vascular cognitive impairment (N=36), regardless of amyloid-status [70], suggesting that association between functional connectivity and the distribution of tau is not limited to AD. Notably, these studies are of cross-sectional nature and longitudinal study approaches are needed to evaluate the progression of tau spreading via functional connectivity over time. Nevertheless, they are accumulatively suggesting a non-homogenous progression of tau pathology across independent brain networks that is closely related to functional connections between coherent brain regions.
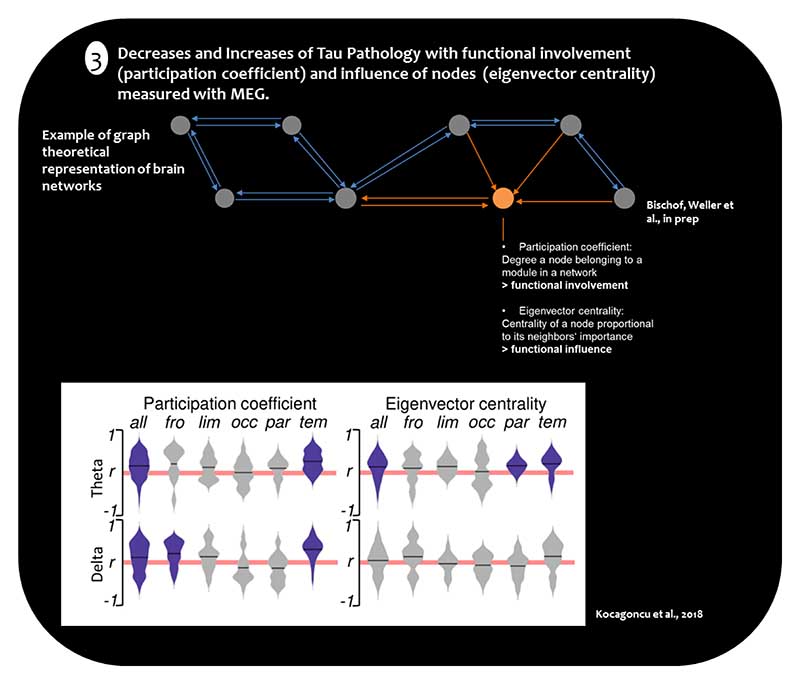
Figure 2.
3) Correlations between network measures and tau deposition. Plots display correlations of participation coefficient (functional involvement in multiple modules) and eigenvector centrality (functional influence) with the local Tau burden across functional lobes in the delta and theta bands. All - whole brain; Fro - frontal; Lim - limbic; Occ - occipital; Par - parietal; Tem - temporal. Adapted from Kocagoncu et al., 2018
Congruent evidence from the temporal dimension of brain network dynamics was recently provided by Kocagoncu et al. (2019), who used a combination of MEG and [18F]-AV1451-PET in a group of AD patients (N=12). They reported that low frequencies (i.e., delta and theta band) oscillations of cortical nodes in neural networks are associated with increased tau burden. Detailed examination of these cortical nodes showed a reduction of the functional influence of nodes in the parieto-occipital network with elevated tau pathology, whereas frontal-temporal networks increased their influence on other regions with increasing tau pathology [45]. Overall, these results point to the idea that frequency-dependent synchronous neuronal connectivity in brain regions relates to increased tau pathology. However, they also suggest a regional dissociation of up- and down-regulation of brain network connectivity in the face of tau pathology (see Figure 2). This pattern of up-regulation of functional network dynamics in frontal regions of the brain could be interpreted as a compensatory response of the brain to counteract the toxic effect of tau pathology. Corresponding with such an interpretation are data from Neitzel and coworkers (2019), who demonstrated that higher resting-state fMRI assessed hub connectivity in the left frontal cortex of the cognitive control network attenuates the adverse effect of higher entorhinal tau levels on episodic memory performance in both cognitively normal adults and patients in the prodromal phase of AD (N=125). This study provides evidence on how modulation of functional connectivity can mitigate deleterious effects of medial temporal tau pathology on the behavioral hallmarks of AD[71].
Studies focusing on structural connectivity and its relationship to beta-amyloid pathology have shown that in individuals in the preclinical phase of AD, amyloid pathology is not sufficient to impair structural network connectivity [s72], and that cognitive decline over time is related to global characteristics of reduced diffusion in white matter tracts, but independently of beta-amyloid status [73]. In contrast, measures of tau pathology, and not beta-amyloid pathology, were related to reduced regional diffusion in temporal white matter tracts in a mixed sample of cognitively normal older adults and cognitively impaired individuals (N=69) [74]. Others have shown that in a group of cognitively normal older adults (N=256) the synergistic effect of increased beta-amyloid pathology and decreased microstructural integrity of the hippocampal cingulum bundle predicted increased tau deposition in the posterior cingulate cortex, as the major cortical output area of the hippocampus, and that this was also related to progressive memory decline in this group [75]. This line of evidence suggests that beta-amyloid pathology exacerbates the spreading of tau accumulation from subcortical to cortical regions via white matter tracts connecting these regions. However, it has yet to be determined how structural connectivity across brain networks contributes to the widespread accumulation of tau pathology in advanced stages of the disease.
Together, the existing evidence on assessing functional connectivity and molecular imaging across the spectrum of AD advances our understanding on the potential mechanisms of how AD pathology evolves in various brain regions. In the context of the theories of protein propagation, many of these data support the idea of non-cell-autonomous mechanisms, where connectivity among brain regions may direct the progression of AD pathology. It is important to note however, that these studies have not specifically assessed how measures of selective regional vulnerability would influence progression of AD pathology, therefore a conclusive statement on favoring a theoretical spreading proposal over another would be premature at this point. Another interesting hypothesis suggests that mislocalized/missorted tau proteins into the somatodendritic compartments of the neuronal cell at earlier stages of pathology [76] and may promote primarily disconnection between cell, which lead to tau pathology where such disconnection emerges.
Further, the presented studies are broadly limited to the AD spectrum, and it remains an open question if functional connectivity plays an equally important role in the spreading of pathology in other human neurodegenerative disease forms, such as PD. Quantifying alpha-synuclein in PD patients in vivo has been limited by the reduced specificity of the available PET tracers (e.g, [123I]-SIL23) to selectively bind to alpha-synuclein [s77]. However, other molecular markers, such as [18F]-fluoropropyl-b-CIT or [18F]-DOPA assessing the integrity of the dopaminergic system are available. [18F]-DOPA has been recently combined with resting-state functional connectivity to examine the relationship of striatal dopamine synthesis and functional connectivity and how it relates to impulsive-compulsive behaviors in PD [78]. Hammes, Theis et al., (2019) observed that functional connectivity between the nucleus accumbens and the rostral anterior-cingulate was reduced in PD, and so was the dopamine synthesis capacity in the nucleus accumbens. Importantly, both measures were associated with the severity of impulsive-compulsive behaviors in the PD cohort (N=80), potentially representing the neurobiological constituents for the development of impulsive-compulsive behaviors in PD [79]. Moreover, multimodal imaging approaches in non-demented PD patients (N=50) revealed a reduction of the structural integrity of the cholinergic input from the basal forebrain to subcortical and cortical target areas. Importantly, this loss of structural integrity was paralleled by a reduction of functional connectivity between regions of the basal forebrain and bilateral hippocampus, middle and inferior frontal regions, thalamus and fusiform gyrus. Both reductions of connectivity and cholinergic input were associated with decreased cognitive function [80]. “Connectomics” as a measure of functional network integrity has been shown to be a sensitive parameter to capture individual differences among diverse PD populations [81]. For example, decreased global and network-specific connectivity appears to be characteristic of idiopathic PD (N=82), but only reductions in sensorimotor networks were associated with individual differences such as motor severity and dysexecutive deficits [82]. Further, connectivity alterations in the default mode network, executive networks and salience networks in drug-naïve PD patients (N=30) was a marker for the subsequent development of impulse control disorders after three years [83]. Intriguing evidence was recently provided showing that effective deep brain stimulation of the subthalamic nucleus alters the connectivity profiles of various networks, which in turn predicted clinical improvement [84]. Together these data emphasize that network connectivity can predict the phenotypical variation in PD and which highlights its potential as a primary outcome measures for disease modifying interventions.
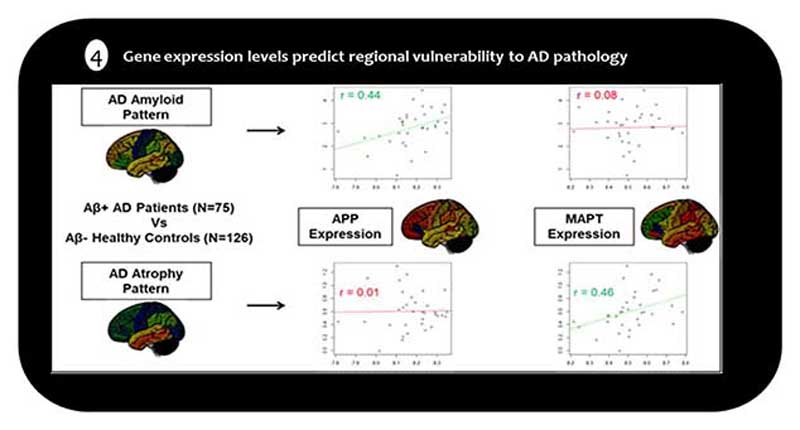
A different line of research was recently developed by characterizing regional vulnerability to AD pathology in relation to the transcriptional architecture of the human brain as revealed by brain-wide regional gene expression profiling in neurotypical controls [53]. In a hypothesis-driven analysis focusing on the genes coding for the amyloid precursor (APP) and tau proteins (MAPT), Grothe et al., (2018) demonstrated that the regional severity of amyloid deposition in AD as measured by [18F]-AV45-PET, but not neurodegeneration on structural MRI, was positively correlated with regional expression levels of APP, whereas opposite associations were observed for MAPT (see Figure 3). Further, in a follow-up study the researchers showed that regional MAPT expression levels also predicted in-vivo trajectories of spreading tau pathology in the aging brain as measured by longitudinal acquisitions of tau-sensitive [18F]-AV1451-PET in cognitively normal older individuals (N=88) [85]. Together, the studies collectively suggest that the regional vulnerability to AD pathology in the human brain may at least be partly determined by specific systemic mechanisms associated with the molecular-functional properties of the affected neural systems, and that these properties largely differ for amyloid accumulation versus neurofibrillary degeneration [53].
Similarly, regional expression profiles of the MAPT and SNCA (the gene encoding alphasynuclein) genes have also been linked to the specific pattern of functional connectivity disruptions underlying cognitive impairments in PD (N=30) and progressive supranuclear palsy (PSP) (N=56) recently [86]. Whereas reduction in global connection strength was associated with MAPT gene expression, expression of SNCA was not. Further, connectivity reduction tracked cognitive but not clinical deficits, underscoring the possible independent contributions of regional vulnerability and network integrity towards phenotypical variation in PD. Another study found that the predominantly motor-related atrophy pattern expressed by a cohort of non-demented de novo PD patients enrolled in the Parkinson’s Progression Markers Initiative (PPMI; N=149) was best predicted by regional expression levels of several genes implicated in trans-synaptic alpha-synuclein transfer [87]. Most interestingly, in another study on an overlapping sample of PD patients from the PPMI, the degree of regional atrophy correlated with the degree of functional and structural connectivity to a presumed disease epicenter in the substantia nigra, thus supporting a trans-neuronal spread model of atrophy progression in PD [88].
In combination, these findings highlight that cell-autonomous and non-cell-autonomous processes need not be mutually exclusive but may interact in determining the complex temporo-spatial progression patterns observed in neurodegenerative disease.
1.5. Summary and Outlook
Our understanding of human neurodegenerative disease was previously limited to clinical data and inferences about the underlying pathology based on histopathological confirmation only. The development of complex neuroimaging tools and the advancement of target-specific PET tracers opens the opportunity to improve our understanding of the in vivo characteristics, dynamics, and principles of the progression of pathology in human neurodegenerative disease. Overall, the evidence underscores the utility of the Connectomics approach to identify the underlying principles of the distinct temporospatial patterns characterizing pathological protein accumulation in neurodegenerative disease, and suggests that the connections between brain regions (functional or structural) may significantly contribute to the propagation of pathology. Although the causal nature of this spatial correspondence is poorly understood, data presented here suggests that susceptibility factors such as gene expression profiles may partly relate to the manifestation of pathology and neurodegeneration in distinct brain regions. Importantly, however, although specific susceptibility factors may contribute to regional pathology accumulation and neurodegeneration, they do not fully account for the observed patterns of protein aggregation or neurodegeneration observed in AD and other neurodegenerative diseases.
It is important to note that the currently existing in vivo neuroimaging tools do not have the spatial resolution to investigate cell-to-cell propagation of pathology. However integrative approaches on Connectomics, neurogenetics, and multimodal imaging (e.g., simultaneously with PET/MR) may methodologically be the most appropriate tools to capture the complex biological and functional aspects of the human brain itself, an advantage that cannot be easily translated from in vitro experiments.
Although distinct histopathological hallmarks characterize different phenotypes of human neurodegenerative disease the occurrence of co-pathologies is increasingly recognized. For example, APOE ε4-carriers showed significantly more TDP-43 pathology, which was independent of AD diagnosis or Lewy-body pathology and related to cognitive decline in a demented population [89]. Moreover, significant TDP-43 pathology was detected in half of the investigated sample of demented patients (N=1044), suggesting that multiple neuropathologies are the rule rather than the exception in late-onset dementia. The manifestation of multiple neuropathologies in human neurodegenerative disease may be particularly challenging when identifying the right target for disease modifying interventions. In the future, advances in the development of target-specific tracers of such co-pathologies (i.e., TDP-43, or alphasynuclein) may ultimately complete the picture on the emergence and interactions of in vivo copathologies.
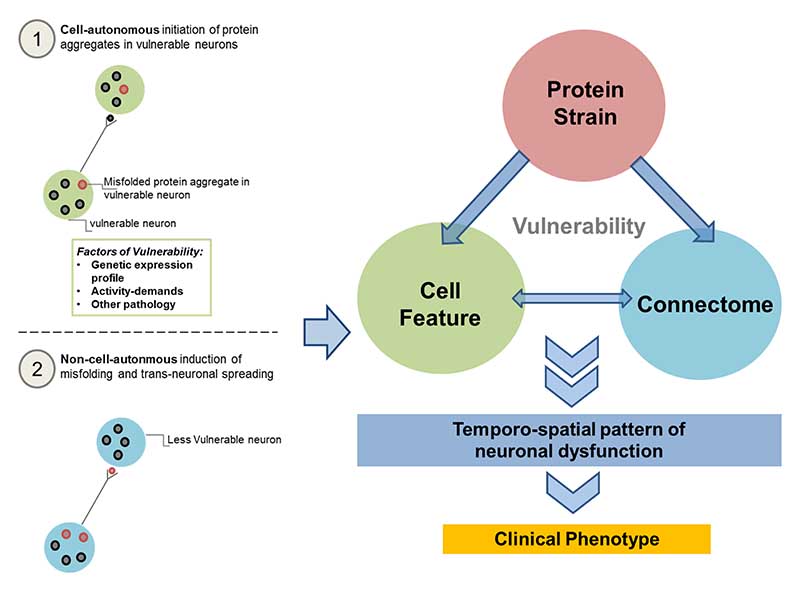
Together the existing data provides evidence for both theoretical conceptions of non-cellautonomous and cell-autonomous mechanisms underlying aggregation of pathology. To this end, we propose a synergistic model (see Figure 5) that combines cell-autonomous (Figure 5.1) and non-cellautonomous (Figure 5.2) accounts of spreading as a triad of vulnerability. This triad includes the type of misfolded protein strains, local cell features and the Connectome that in synergistic combination produce relatively prototypical temporo-spatial patterns of neuronal dysfunction and cell loss, thereby potentially explaining all observed clinical phenotypes across the neurodegenerative disease spectrum.
Figure 5.
Triad of Vulnerability: A unification of the cell-autonomous and cell-non-autonomous accounts of protein aggregation, via a triad-interaction of protein strain, cell feature and Connectome may instigate the neural vulnerability that leads to the observed pattern of neuronal dysfunction, that produces different clinical phenotypes.
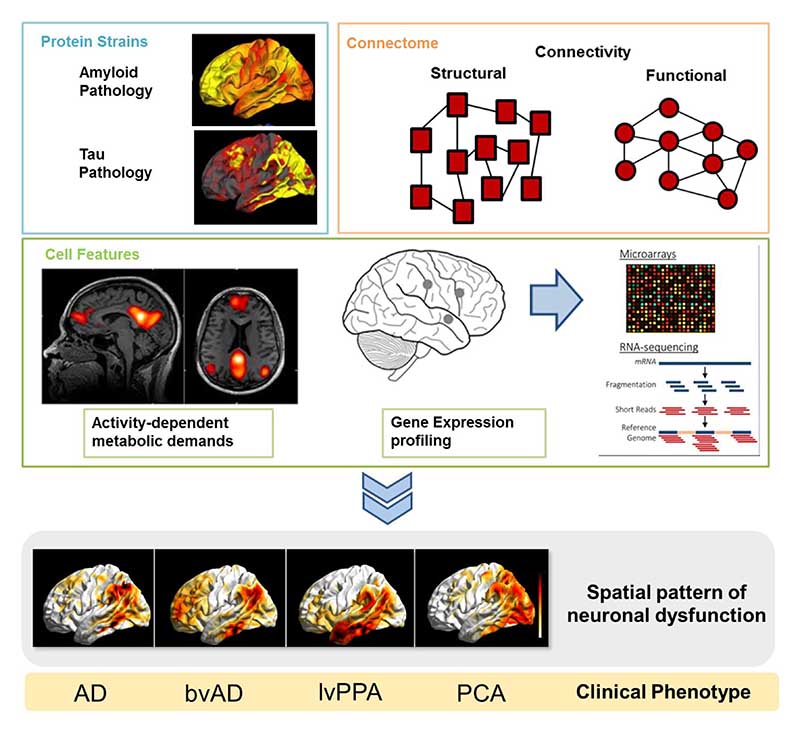
To test our proposed model, the paralleled investigation of molecular imaging of protein accumulation, structural and functional connectivity, gene expression profiling, and the characterization of activity-dependence of brain regions (see Figure 6) is needed. Such an approach allows the quantification of the factors contributing to selective vulnerability and in turn enables us to investigate its contribution to the pattern of neuronal dysfunction (see Figure 6).
Figure 6.
Example of how the synergistic triad of vulnerability may translate into the clinical phenotypes of AD. Investigations of protein accumulation and examination of the critical contribution of the connectome and cell feature such as metabolic demand or gene expression profiles may lead to the observed spatial pattern of neuronal dysfunction that in turn produces distinct clinical phenotype in AD. AD = Alzheimer’s Disease, bvAD = behavioral variant of AD, lvPPA = logopenic variant of primary progressive aphasia, PCA= Posterior Cortical Atrophy.
In summary, the combination of Connectomics and molecular imaging has revealed important insights into the development and progression of human neurodegenerative disease and the development of phenotypical variation. As we move forward, we recognize that the current hypothesis on pathological spreading and regional vulnerability may have to be investigated in concert to advance our understanding on disease progression. Although several limitations may hinder us at the moment to achieve a complete picture on the causes of development and progression of human neurodegenerative disease, the insights from clinical in-vivo neuroimaging research achieved thus far have altered our conception of target-specific intervention windows, primary and secondary outcome measures, and criteria to identify the most suitable biomarkers for disease diagnosis and disease progression [30,90].
Supplementary Material
Figure 4.
Regional expression levels of APP and MAPT genes (x-axes) correlate differentially with the regional severities of amyloid deposition (top) and neurodegeneration (bottom) in AD patients as compared to healthy controls (y-axes). Each point corresponds to one of 34 different cortical areas; r indicates Spearman correlation across these areas. Adopted from Grothe et al., 2018
Funding
The Molecular Imaging of Neurodegeneration Cologne (MINC) Symposium was partly funded by the Deutsche Forschungsgemeinschaft (DFG) awarded to Dr. Thilo van Eimeren (EI 892/5-1). The Deutsche Forschungsgemeinschaft (DFG) also awarded funding to Dr. Alexander Drzezga (DR 442/91).
Footnotes
Compliance with Ethical Standards:
Conflict of Interest: The authors declare no conflict of interest in relation to this article.
Ethic Approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Contributor Information
Michael Ewers, Email: Michael.Ewers@med.uni-muenchen.de.
Nicolai Franzmeier, Email: Nicolai.Franzmeier@med.uni-muenchen.de.
Michel J. Grothe, Email: Michel.Grothe@dzne.de.
Merle Hoenig, Email: merle.hoenig@uk-koeln.de.
Ece Kocagoncu, Email: ek390@cam.ac.uk.
Julia Neitzel, Email: Julia.Neitzel@med.uni-muenchen.de.
James B Rowe, Email: James.Rowe@mrc-cbu.cam.ac.uk.
Antonio Strafella, Email: antonio.strafella@uhn.ca.
Alexander Drzezga, Email: alexander.drzezga@uk-koeln.de.
Thilo van Eimeren, Email: thilo.van-eimeren@uk-koeln.de.
Bibliography
- 6.Bischof GN, Endepols H, van Eimeren T, Drzezga A. Tau-imaging in neurodegeneration. Methods. 2017;130:114–23. doi: 10.1016/j.ymeth.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Hammes J, Bischof GN, Drzezga A. Molecular imaging in early diagnosis, differential diagnosis and follow-up of patients with neurodegenerative diseases. Clin Transl Imaging. 2017;5:465–71. [Google Scholar]
- 29.Teipel S, Drzezga A, Grothe MJ, Barthel H, Chételat G, Schuff N, et al. Multimodal imaging in Alzheimer’s disease: validity and usefulness for early detection. The Lancet Neurology. 2015;14:1037–53. doi: 10.1016/S1474-4422(15)00093-9. [DOI] [PubMed] [Google Scholar]
- 30.Strafella AP, Bohnen NI, Perlmutter JS, Eidelberg D, Pavese N, Van Eimeren T, et al. Molecular imaging to track Parkinson’s disease and atypical parkinsonisms: New imaging frontiers. Mov Disord. 2017;32:181–92. doi: 10.1002/mds.26907. [DOI] [PubMed] [Google Scholar]
- 31.Barthel H, Sabri O. Clinical Use and Utility of Amyloid Imaging. J Nucl Med. 2017;58:1711–7. doi: 10.2967/jnumed.116.185017. [DOI] [PubMed] [Google Scholar]
- 32.Saint-Aubert L, Lemoine L, Chiotis K, Leuzy A, Rodriguez-Vieitez E, Nordberg A. Tau PET imaging: present and future directions. [cited 2017 Mar 31];Molecular Neurodegeneration. 2017 12 doi: 10.1186/s13024-017-0162-3. [Internet], Available from: http://molecularneurodegeneration.biomedcentral.com/articles/10.1186/s13024-017-0162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passamonti L, Vázquez Rodríguez P, Hong YT, Allinson KSJ, Williamson D, Borchert RJ, et al. 18F-AV-1451 positron emission tomography in Alzheimer’s disease and progressive supranuclear palsy. Brain. 2017 doi: 10.1093/brain/aww340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grothe M, Heinsen H, Teipel SJ. Atrophy of the cholinergic Basal forebrain over the adult age range and in early stages of Alzheimer’s disease. Biol Psychiatry. 2012;71:805–13. doi: 10.1016/j.biopsych.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grothe MJ, Ewers M, Krause B, Heinsen H, Teipel SJ. Alzheimer’s Disease Neuroimaging Initiative. Basal forebrain atrophy and cortical amyloid deposition in nondemented elderly subjects. Alzheimers Dement. 2014;10:S344–353. doi: 10.1016/j.jalz.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter N, Beckers N, Onur OA, Dietlein M, Tittgemeyer M, Kracht L, et al. Effect of cholinergic treatment depends on cholinergic integrity in early Alzheimer’s disease. Brain. 2018;141:903–15. doi: 10.1093/brain/awx356. [DOI] [PubMed] [Google Scholar]
- 42.Sabri O, Meyer PM, Gräf S, Hesse S, Wilke S, Becker G-A, et al. Cognitive correlates of α4β2 nicotinic acetylcholine receptors in mild Alzheimer’s dementia. Brain. 2018;141:1840–54. doi: 10.1093/brain/awy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kocagoncu E, Quinn A, Firouzian A, Cooper E, Greve A, Gunn R, et al. Tau pathology in early Alzheimer’s disease disrupts selective neurophysiological networks dynamics. bioRxiv. 2019:524355. doi: 10.1016/j.neurobiolaging.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoenig MC, Bischof GN, Seemiller J, Hammes J, Kukolja J, Onur ÖA, et al. Networks of tau distribution in Alzheimer’s disease. Brain. 2018 doi: 10.1093/brain/awx353. [DOI] [PubMed] [Google Scholar]
- 53.Grothe MJ, Sepulcre J, Gonzalez-Escamilla G, Jelistratova I, Schöll M, Hansson O, et al. Molecular properties underlying regional vulnerability to Alzheimer’s disease pathology. Brain. 2018;141:2755–71. doi: 10.1093/brain/awy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grothe MJ, Teipel SJ. Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of atrophy, hypometabolism, and amyloid deposition in Alzheimer’s disease correspond to dissociable functional brain networks. Hum Brain Mapp. 2016;37:35–53. doi: 10.1002/hbm.23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drzezga A. The Network Degeneration Hypothesis: Spread of Neurodegenerative Patterns Along Neuronal Brain Networks. J Nucl Med. 2018;59:1645–8. doi: 10.2967/jnumed.117.206300. [DOI] [PubMed] [Google Scholar]
- 62.Chételat G, Villemagne VL, Bourgeat P, Pike KE, Jones G, Ames D, et al. Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann Neurol. 2010;67:317–24. doi: 10.1002/ana.21955. [DOI] [PubMed] [Google Scholar]
- 63.Myers N, Pasquini L, Göttler J, Grimmer T, Koch K, Ortner M, et al. Within-patient correspondence of amyloid-β and intrinsic network connectivity in Alzheimer’s disease. Brain. 2014;137:2052–64. doi: 10.1093/brain/awu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drzezga A, Becker JA, Van Dijk KRA, Sreenivasan A, Talukdar T, Sullivan C, et al. Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 2011;134:1635–46. doi: 10.1093/brain/awr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koch K, Myers NE, Göttler J, Pasquini L, Grimmer T, Förster S, et al. Disrupted Intrinsic Networks Link Amyloid-β Pathology and Impaired Cognition in Prodromal Alzheimer’s Disease. Cereb Cortex. 2015;25:4678–88. doi: 10.1093/cercor/bhu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones DT, Graff-Radford J, Lowe VJ, Wiste HJ, Gunter JL, Senjem ML, et al. Tau, amyloid, and cascading network failure across the Alzheimer’s disease spectrum. Cortex. 2017;97:143–59. doi: 10.1016/j.cortex.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cope TE, Rittman T, Borchert RJ, Jones PS, Vatansever D, Allinson K, et al. Tau burden and the functional connectome in Alzheimer’s disease and progressive supranuclear palsy. Brain. 2018;141:550–67. doi: 10.1093/brain/awx347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franzmeier N, Rubinski A, Neitzel J, Kim Y, Damm A, Na DL, et al. Functional connectivity associated with tau levels in ageing, Alzheimer’s, and small vessel disease. Brain. 2019 doi: 10.1093/brain/awz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neitzel J, Franzmeier N, Rubinski A, Ewers M. Left frontal connectivity attenuates the adverse effect of entorhinal tau pathology on memory. Neurology. 2019 doi: 10.1212/WNL.0000000000007822. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rabin JS, Perea RD, Buckley RF, Neal TE, Buckner RL, Johnson KA, et al. Global White Matter Diffusion Characteristics Predict Longitudinal Cognitive Change Independently of Amyloid Status in Clinically Normal Older Adults. Cereb Cortex. 2019;29:1251–62. doi: 10.1093/cercor/bhy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strain JF, Smith RX, Beaumont H, Roe CM, Gordon BA, Mishra S, et al. Loss of white matter integrity reflects tau accumulation in Alzheimer disease defined regions. Neurology. 2018;91:e313–8. doi: 10.1212/WNL.0000000000005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobs HIL, Hedden T, Schultz AP, Sepulcre J, Perea RD, Amariglio RE, et al. Structural tract alterations predict downstream tau accumulation in amyloid-positive older individuals. Nat Neurosci. 2018;21:424–31. doi: 10.1038/s41593-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zempel H, Mandelkow E. Lost after translation: missorting of Tau protein and consequences for Alzheimer disease. Trends in Neurosciences. 2014;37:721–32. doi: 10.1016/j.tins.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 78.Hammes J, Theis H, Giehl K, Hoenig MC, Greuel A, Tittgemeyer M, et al. Dopamine metabolism of the nucleus accumbens and fronto-striatal connectivity modulate impulse control. Brain. 2019;142:733–43. doi: 10.1093/brain/awz007. [DOI] [PubMed] [Google Scholar]
- 79.Strafella AP. Mesolimbic dopamine and anterior cingulate cortex connectivity changes lead to impulsive behaviour in Parkinson’s disease. Brain. 2019;142:496–8. doi: 10.1093/brain/awz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gargouri F, Gallea C, Mongin M, Pyatigorskaya N, Valabregue R, Ewenczyk C, et al. Multimodal magnetic resonance imaging investigation of basal forebrain damage and cognitive deficits in Parkinson’s disease. Mov Disord. 2019;34:516–25. doi: 10.1002/mds.27561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tahmasian M, Eickhoff SB, Giehl K, Schwartz F, Herz DM, Drzezga A, et al. Resting-state functional reorganization in Parkinson’s disease: An activation likelihood estimation meta-analysis. Cortex. 2017;92:119–38. doi: 10.1016/j.cortex.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lang S, Hanganu A, Gan LS, Kibreab M, Auclair-Ouellet N, Alrazi T, et al. Network basis of the dysexecutive and posterior cortical cognitive profiles in Parkinson’s disease. Mov Disord. 2019 doi: 10.1002/mds.27674. [DOI] [PubMed] [Google Scholar]
- 83.Tessitore A, De Micco R, Giordano A, di Nardo F, Caiazzo G, Siciliano M, et al. Intrinsic brain connectivity predicts impulse control disorders in patients with Parkinson’s disease. Mov Disord. 2017;32:1710–9. doi: 10.1002/mds.27139. [DOI] [PubMed] [Google Scholar]
- 84.Horn A, Reich M, Vorwerk J, Li N, Wenzel G, Fang Q, et al. Connectivity Predicts deep brain stimulation outcome in Parkinson disease. Ann Neurol. 2017;82:67–78. doi: 10.1002/ana.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sepulcre J, Grothe MJ, d’Oleire Uquillas F, Ortiz-Terán L, Diez I, Yang H-S, et al. Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat Med. 2018;24:1910–8. doi: 10.1038/s41591-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rittman T, Rubinov M, Vértes PE, Patel AX, Ginestet CE, Ghosh BCP, et al. Regional expression of the MAPT gene is associated with loss of hubs in brain networks and cognitive impairment in Parkinson disease and progressive supranuclear palsy. Neurobiol Aging. 2016;48:153–60. doi: 10.1016/j.neurobiolaging.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freeze B, Acosta D, Pandya S, Zhao Y, Raj A. Regional expression of genes mediating trans-synaptic alpha-synuclein transfer predicts regional atrophy in Parkinson disease. Neuroimage Clin. 2018;18:456–66. doi: 10.1016/j.nicl.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeighami Y, Ulla M, Iturria-Medina Y, Dadar M, Zhang Y, Larcher KM-H, et al. Network structure of brain atrophy in de novo Parkinson’s disease. Elife. 2015;4 doi: 10.7554/eLife.08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang H-S, Yu L, White CC, Chibnik LB, Chhatwal JP, Sperling RA, et al. Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: a community-based cohort study. Lancet Neurol. 2018;17:773–81. doi: 10.1016/S1474-4422(18)30251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Eimeren T, Antonini A, Berg D, Bohnen N, Ceravolo R, Drzezga A, et al. Neuroimaging biomarkers for clinical trials in atypical parkinsonian disorders: Proposal for a Neuroimaging Biomarker Utility System. Alzheimers Dement (Amst) 2019;11:301–9. doi: 10.1016/j.dadm.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.