Summary Paragraph
Cytoplasmic dynein is a microtubule motor that is activated by its cofactor dynactin and a coiled-coil cargo adaptor1–3. Up to two dynein dimers can be recruited per dynactin, and interactions between them affect their combined motile behaviour4–6. Different coiled-coil adaptors are linked to different cargos7,8, and some share motifs known to contact sites on dynein and dynactin4,9–13. There is currently limited structural information on how the resulting complex interacts with microtubules and how adaptors are recruited. Here, we develop a cryo-EM processing pipeline to solve the high-resolution structure of dynein-dynactin and the adaptor BICDR1 bound to microtubules. This reveals the asymmetric interactions between neighbouring dynein motor domains and how they relate to motile behaviour. We find unexpectedly that two adaptors occupy the complex. Both adaptors make similar interactions with the dyneins but diverge in their contacts with each other and dynactin. Our structure has implications for the stability and stoichiometry of motor recruitment by cargos.
Introduction
Eukaryotic cells use cytoskeletal filaments and motor proteins to organize their cytoplasm. Dynein (cytoplasmic dynein-1) is the primary transporter of cargo towards the minus ends of microtubules (MTs) and therefore contributes to multiple cellular processes7. It is a multi-subunit dimer that must bind a 1.1 MDa cofactor, dynactin, and a coiled-coil cargo adaptor to be fully active1,2.
Once activated, dynein uses its motor domains to walk on MTs. Each motor comprises a ring of six AAA+ domains, which, unlike many other AAA+ family proteins, are non-equivalent to each other14. The ATPase cycle of the first domain (AAA1) drives motility but can be gated by the nucleotide state of AAA315–18. It’s also thought that interactions between motor domains can modulate dynein motility6, although the identity of the interactions are not known.
There is a growing family of dynein cargo adaptors7,8. Cryo-electron microscopy (cryo-EM) showed how their coiled-coils run along dynactin’s filament, positioning dynein in a conformation that relieves its intrinsic inhibition4,5,13,19. Some adaptors recruit two dyneins per dynactin, increasing the force and speed of the complex, allowing it to outcompete kinesin in a tug-of-war4–6,20,21. Our understanding of how adaptors recruit dynein and dynactin4,12,19,22 has been limited by the lack of high-resolution structural information of the motile complex.
To understand both motor interactions and adaptor recruitment, we determined a high-resolution structure of a dynein-dynactin-adaptor complex on MTs. We developed a pipeline using single-particle cryo-EM to overcome the challenge of picking and aligning complexes near strong MT signals. The resulting structures identified an asymmetry in the way the motor domains contact each other, showed the unexpected presence of two adaptors, and defined the motifs required for their recruitment.
Results
A pipeline to solve MT-bound complexes
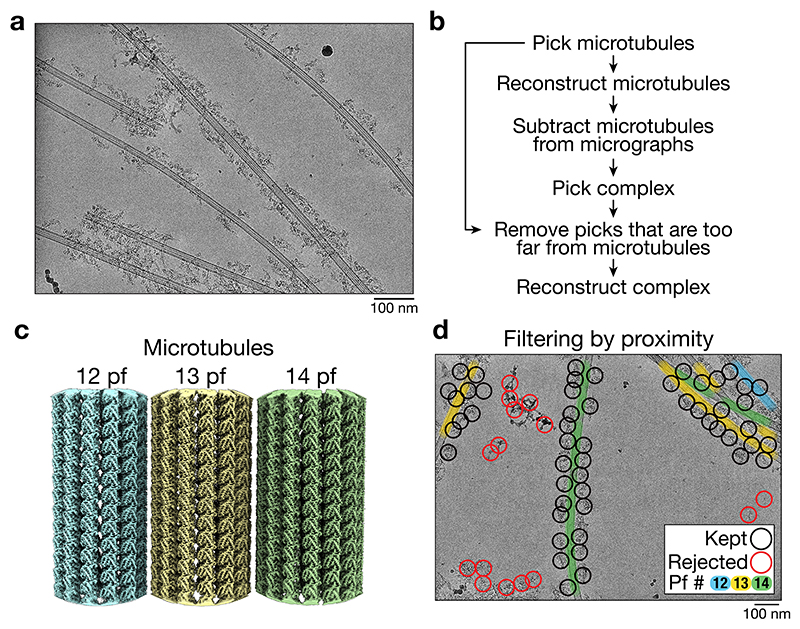
A previous cryo-electron tomography (cryo-ET) structure of dynein-dynactin on MTs was limited in resolution by the relatively low throughput of tomography data collection5. We therefore used a single-particle approach to obtain a large number of particles. We prepared dynein-dynactin complexes with the adaptor BICDR1 (also known as BICDL1, hereafter referred to as BICDR)4, and sparsely decorated them on MTs using the ATP-analogue AMPPNP (Extended Data Fig. 1a). Initially, single-particle analysis was unsuccessful due to the dominating signal of the MT5,23–26. We therefore developed a pipeline (Extended Data Fig. 1b) in which we first solved the structure of the different protofilament-number MTs (Extended Data Fig. 1c)27 and then computationally subtracted them from the micrographs28. This allowed the dynein-dynactin complexes to be aligned (Fig. 1a), and recovered those that were obscured by an overlapping MT. We also used the MT coordinates to discard particles that were too far away to be bound (Extended Data Fig. 1d). Finally, we solved the overall structure and used local refinements to obtain high resolution coverage of the majority of the complex (Fig. 1b, Extended Data Fig. 2, Extended Data Table 1, 2).
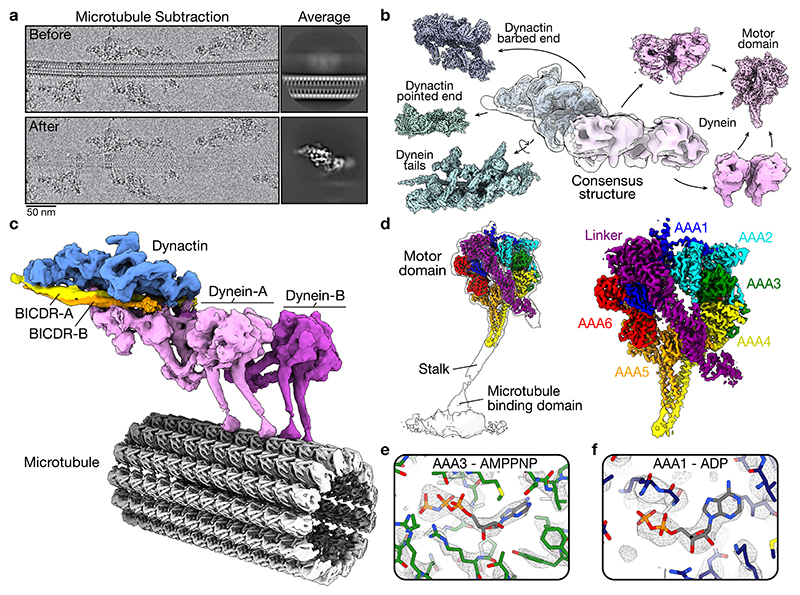
Figure 1. Cryo-EM structure of dynein-dynactin-BICDR on MTs.
a, Example of a MT before and after subtraction from cryo-EM micrographs (n = 645,193 MTs on micrographs from 15 datasets). Representative 2D averages of particles from each case are shown on the right (26,158 and 21,696 particles, respectively). b, Gallery of locally processed density maps, starting with the consensus structure in the middle. c, Composite density map of dynein-dynactin-BICDR overlaid on the 13 protofilament MT. d, Density map of the dynein motor domain overlaid on the full motor (left) with the subdomains highlighted (right). e, Density in the nucleotide pocket of AAA3 with AMPPNP docked. f, Density in the nucleotide pocket of AAA1 with ADP docked.
MT-bound dynein-dynactin-BICDR
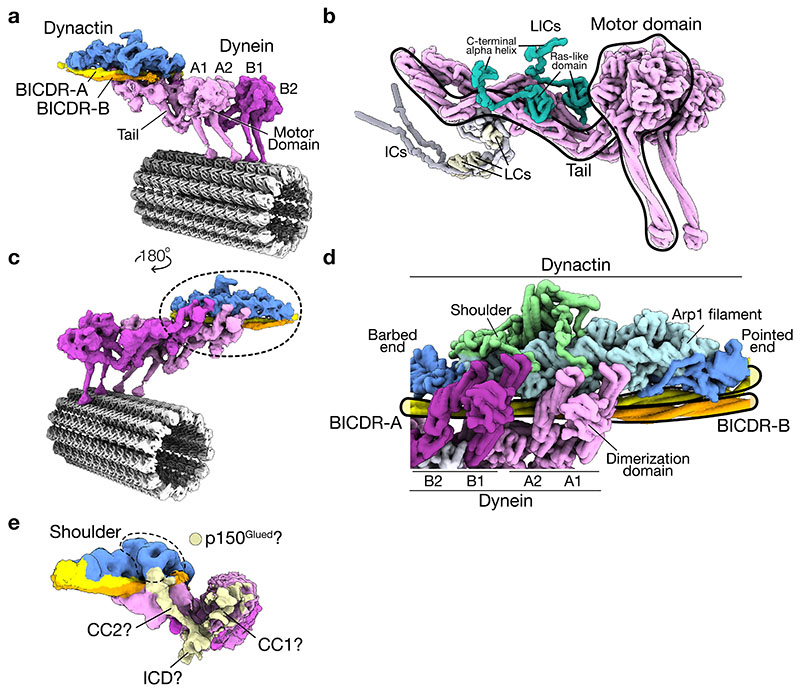
Our composite structure (Fig. 1c) shows two dynein dimers (dynein-A and B) stacked side-by-side. Each dynein heavy chain (dynein-A1, A2, B1, and B2) contains a motor domain contacting the MT and a tail extending towards dynactin (Extended Data Fig. 3a). Each tail binds a light intermediate chain (LIC) and an intermediate chain (Extended Data Fig. 3b), and sits in the grooves in dynactin’s Arp1 filament (Extended Data Fig. 3c). The main BICDR coiled-coil (hereafter BICDR-A) spans dynactin from the barbed end to the pointed end (Fig. 1c, Extended Data Fig. 3d), similar to previous structures4,19. Unexpectedly, a second BICDR (BICDR-B) is also present in the structure, which we describe below.
We also observe a density that connects to the shoulder domain of dynactin and contacts the tail and motor domain of dynein-A1 (Extended Data Fig. 3e). Given its position, this likely belongs to the p150Glued extension, including the coiled-coils (CC1 and CC2) and a globular density corresponding to the Inter-Coiled Domain (ICD)4. This suggests that p150Glued may be able to transiently dock onto the tail and motor domain of dynein-A1.
AMPPNP in AAA3 locks ADP in AAA1
All the dynein motor domains in our complex are locked in the same AMPPNP conformation, where the linker (dynein’s mechanical element) is straight (Extended Data Fig. 4a). This allowed us to average them to obtain sufficient resolution to identify which nucleotides are present (Fig. 1d). Previous studies showed that ATP binding to AAA3 blocks dynein’s mechanochemical cycle15–18. A crystal structure of S. cerevisiae dynein with AMPPNP in AAA3 and AAA1 suggests that this inhibition results from the ATP state of AAA3 preventing AAA1 from undergoing hydrolysis15.
Overall, our structure is similar to the previous one15, where the linker is docked onto AAA5 and AAA2 (Extended Data Fig. 4b, c), the density for the nucleotide in AAA3 is consistent with AMPPNP (Fig. 1e), and the AAA3 nucleotide pocket is closed (Extended Data Fig. 4d). Surprisingly, however, despite incubating the complex in saturating amounts of AMPPNP, the density at AAA1 is consistent with ADP (Fig. 1f). Additionally, the conformation of the AAA1 nucleotide pocket matches the crystal structure of ADP-dynein from D. discoideum (Extended Data Fig. 4e)29. Therefore, our MT-bound structure suggests that rather than blocking hydrolysis of AAA1, ATP in AAA3 traps AAA1 in an ADP state. In cells, this high-affinity conformation may stabilize dynein at MT plus ends and at the cortex during mitosis15, and likely affects the binding of dynein’s regulator, Lis1, which depends on AAA330,31.
Interactions of the motor domains
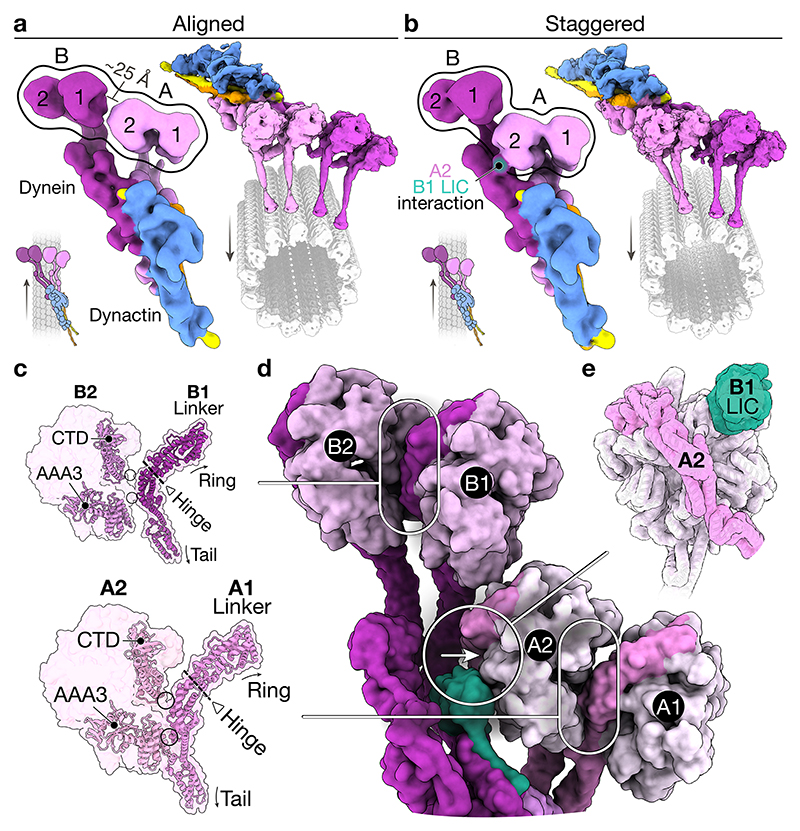
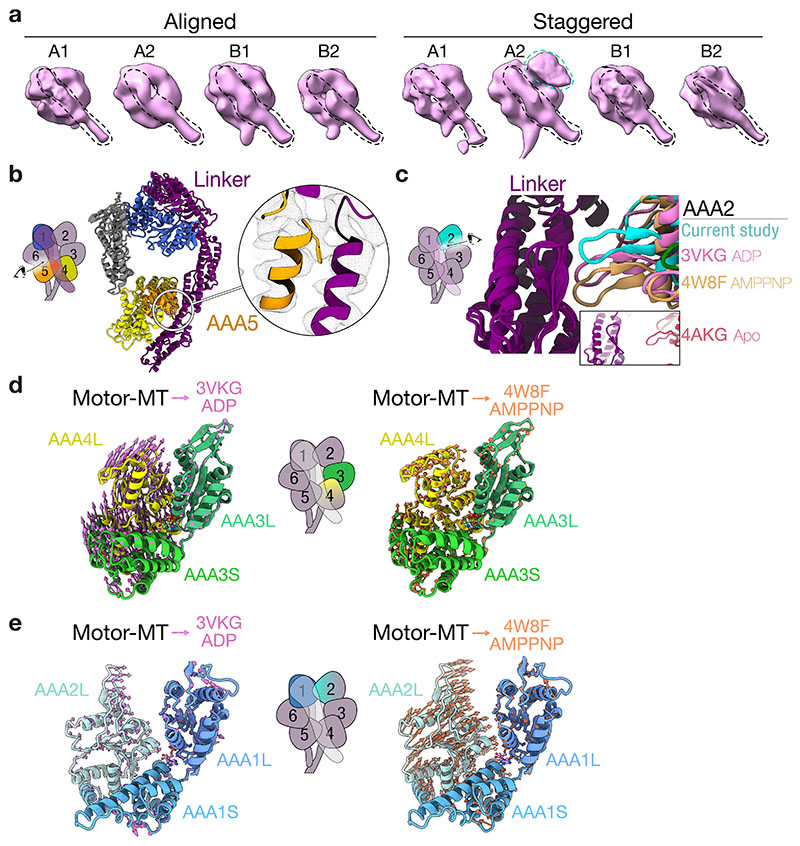
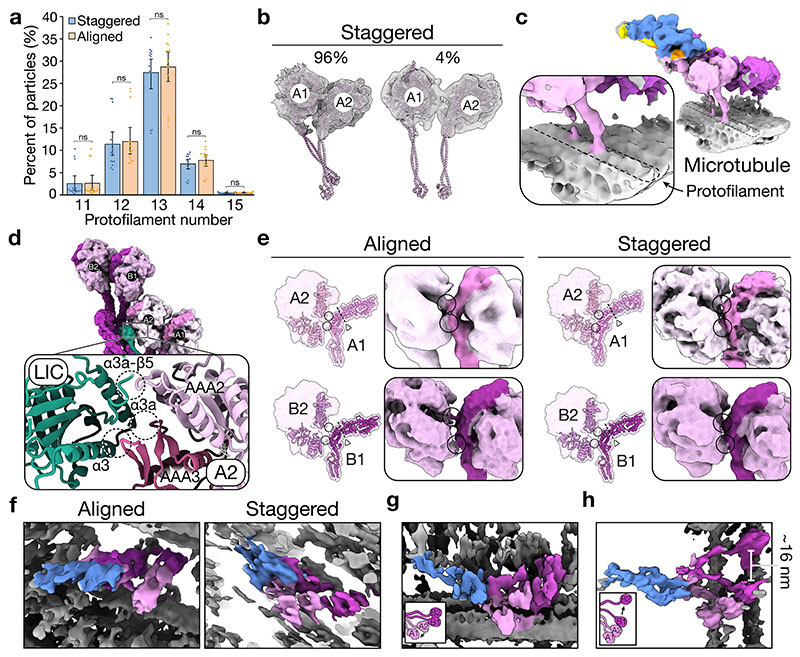
Classification of our dynein-dynactin complexes revealed that there are two main configurations of the motors: “aligned” (Fig. 2a) and “staggered” (Fig. 2b). Both bind equivalently to different protofilament-number MTs (Extended Data Fig. 5a). In the aligned state, the four stalks run parallel to each other, as observed previously by cryo-ET5. Although there are contacts between the motors within each dimer, the motor domains of the neighbouring dimers don’t interact (Fig. 2a). In the staggered state, dynein-A1’s stalk lies behind dynein-A2 in 96% of particles (Fig. 2b, Extended Data Fig. 5b). This allows the two motor domains to contact the same protofilament (Extended Data Fig. 5c). Furthermore, the motor domain of dynein-A2 is docked onto the LIC of dynein-B1 (Fig. 2b). This interaction involves AAA2 and AAA3 on the linker face of dynein-A2 with conserved sites on the LIC Ras-like domain (Extended Data Fig. 5d)32.
Figure 2. Interactions of the motor domains.
a, Top view of the density map of the aligned state (left) and composite map overlaid onto the MT (right). Arrows represent the direction of travel. b, Top view of the density map of the staggered state (left) and composite map overlaid on the MT (right). c, A molecular surface representation of adjacent motor domains in the staggered state, only showing the linker of the righthand motor. Overlaid are ribbon representations of the C-terminal domain (CTD), AAA3, and linker. The hinge in the linker is also highlighted (triangle and dotted line), and arrows point at the relative position of the ring and tail. d, Top view of a molecular surface representation of dynein in the staggered state. e, Molecular surface representation of the motor domain of dynein-A2 (pink) and the LIC of dynein-B1 (green).
To understand how motor interactions could influence dynein, we assessed how they might affect stepping. Looking down on the dynein-dynactin complex in its direction of travel (arrows in Fig. 2a, b), the motor domains A1 and B1 are on the right of A2 and B2, respectively. The stepping of each motor involves detachment from the MT followed by a priming stroke, where the linker bends at its hinge and moves the motor forward33. Within each dimer, the interface between the motor domains (i.e. dynein-A1 to A2 and B1 to B2) is similar in both the aligned and staggered states (Extended Data Fig. 5e). The linker from the right-hand motor domain (A1/B1) binds at two points on the left-hand ring (A2/B2) (Fig. 2c, d). On the linker, these interactions occur on the tail-side of the hinge. Therefore, the right-hand motors (A1/B1) are not hindered from undergoing a priming stroke. In contrast, the same interactions likely impede the left-hand motors (A2/B2) from moving. In the staggered state, dynein-A2 is further prevented from stepping by its interaction with the LIC of dynein-B1 (Fig. 2d, e). The asymmetry found in both the aligned and staggered states suggest that when stepping, dyneins likely lead off with their right-hand motors.
To assess the variability in motor domain interactions, we visualized individual dynein-dynactin-BICDR complexes by cryo-ET. Whereas some complexes lie in the aligned and staggered states described above (Extended Data Fig. 5f), we observed deviations in the positions of individual motor domains. For example, we see shifts in the position of dynein-A1 (Extended Data Fig. 5g) and a large separation (~16 nm) between dynein-B1 and B2 (Extended Data Fig. 5h). Despite this variability, there are clearly stable interactions between the motor domains which likely affect dynein’s stepping behaviour.
Two adaptors scaffold the complex
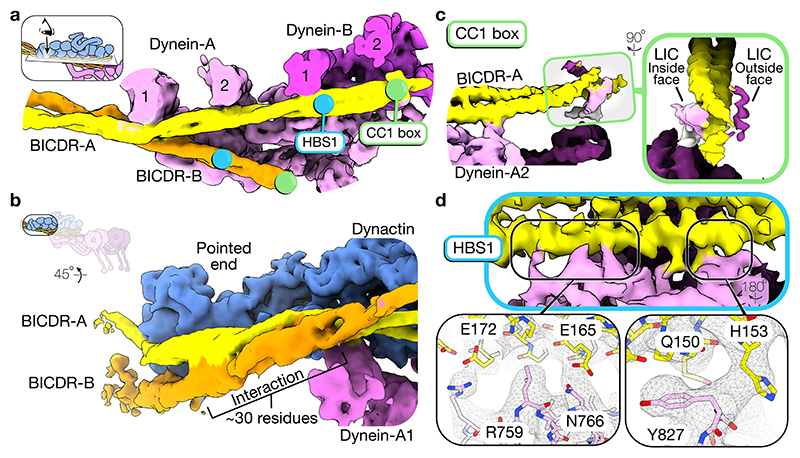
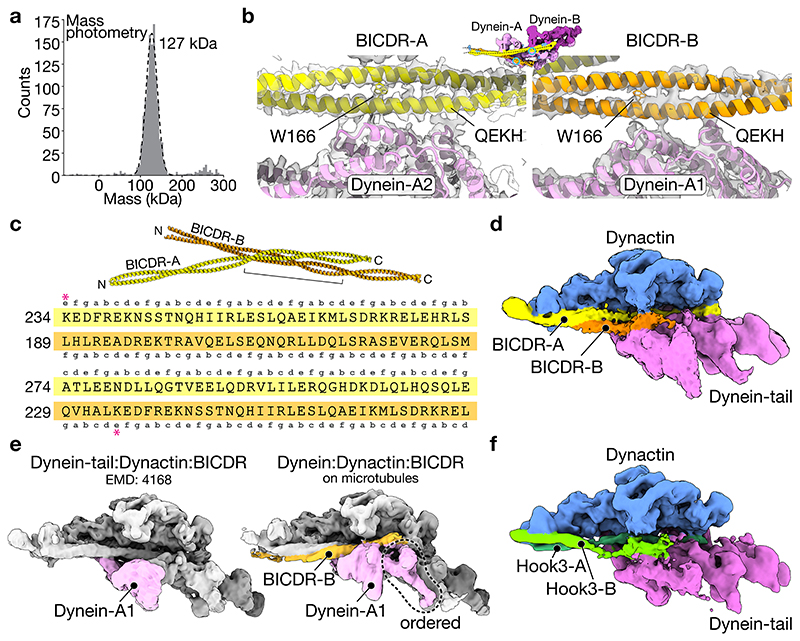
In our structure, the dyneins are scaffolded by two BICDRs that sit differently in the complex. BICDR-A runs the length of dynactin, contacting dynein-A and B (Fig. 3a). In contrast, BICDR-B only covers half the length of the complex, interacting mainly with dynein-A1. The two adaptors meet close to where BICDR-A binds dynein-A1, with their coiled-coils interacting over ~30 residues before diverging again to contact dynactin’s pointed end (Fig. 3b). Mass photometry of BICDR shows a major peak relating to a single coiled-coil (Extended Data Fig. 6a), suggesting that the interaction between BICDRs is stabilized in the complex. The registry of the BICDR coiled-coils can be determined based on a unique tryptophan (W166), which presents as a bulky density between the two alpha helices of the coiled-coil (Extended Data Fig. 6b). This shows that the interaction sites of BICDR-A and B are offset by ~47 residues (Extended Data Fig. 6c).
Figure 3. Two BICDRs scaffold the dynein-dynactin complex.
a, Top view of the density map of the dynactin and dynein-tail region (the density for dynactin is hidden). The N-terminal motifs (CC1 box and HBS1) are highlighted on each BICDR. b, Side view of the pointed end density map showing both BICDRs. c, A close up of the N-terminus of BICDR-A, showing the density around the CC1 box. A gaussian filter was applied to better show the LICs. d, A close up of the interaction between BICDR-A and dynein-A2, highlighting the HBS1 conserved residues (Q150/H153) and C-terminal E165/E172.
To address whether two adaptors are present in the previous structure of dynein-tail/dynactin/BICDR4, we revisited the data using new 3D classification tools in cryoDRGN34. We found density for the second adaptor in a subset of particles (~5%) (Extended Data Fig. 6d). This suggests that in our new structure, which has 100% occupancy of both adaptors, the presence of MTs stabilizes the binding of BICDR-B. We propose that this results from the reduced flexibility of dynein-A1 upon MT binding. In support of this, dynein-A1 is more ordered in our structure (Extended Data Fig. 6e).
Two coiled-coils were seen previously at the pointed end of dynactin in a structure with the adaptor Hook34,13. However, at the time it was not possible to determine if they belonged to two separate adaptors or resulted from a folding back of Hook3. We can now clearly assign these coiled-coils to two Hook3s (Hook3-A and Hook3-B) using our new classifications (Extended Data Fig. 6f). Taken together, our analysis suggests that the presence of two adaptors may be found in other dynein-dynactin complexes.
Adaptor motifs define asymmetric binding
BICDR interacts with dynein and dynactin via at least three conserved motifs. At the N-terminus of BICDR, the CC1 box (AAXXG) binds an alpha helix in the extended C-terminus of the LIC (Extended Data Fig. 3b)9,11. BICDR has LIC binding sites on both sides of its coiled-coil9. Therefore, we expected that the two LIC helices from each dynein would interact with different BICDRs. In our structure, we see weak density for the LIC helix on both sides of BICDR-A (Fig. 3c, Extended Data Fig. 7a). 3D classification shows the density on the inside face connects to the LIC of dynein-A2 (Extended Data Fig. 7b), as previously observed4. This suggests the density on the outside face belongs to dynein-B. BICDR-B is too flexible to resolve the LIC helix, but 3D classification also shows a connection to the LIC of dynein-A2. (Extended Data Fig. 7c). This suggests the dynein-A2 LIC can bind both BICDR-A and BICDR-B, and disfavours our original hypothesis that each adaptor binds the LICs of a separate dynein.
In BICDR and related adaptors, there is a second conserved motif (QXX[H/Y]) downstream of the CC1 box10, which was predicted to interact with the dynein heavy chain13. Our structure shows that the highly conserved residues Q150 and H153 of BICDR-A interact with Y827 on the heavy chain of dynein-A2 (Fig. 3d). We can resolve additional contacts by E165 and E172 (Fig. 3d), suggesting a more extensive motif defines BICDR’s interaction with the heavy chain. Previously, the QXX[H/Y] motif was referred to as the CC2 box, but because it resides in the first coiled-coil segment (CC1), we will hereafter refer to the extended region as the heavy chain binding site 1 (HBS1) motif35. Based on our predicted registry of BICDR-B, its HBS1 motif contacts dynein-A1 in a similar manner to BICDR-A’s interaction with dynein-A2 (Extended Data Fig. 6b). Therefore, although the coiled-coils of the two adaptors have distinct positions in the complex, they are both anchored by HBS1 motif interactions with separate dynein-A heavy chains.
Given the anchoring interaction of HBS1 in both BICDRs, we wondered if the motif was also present in Hook3, since it was previously only described for close relatives of BICDR10. We could identify the extended HBS1 motif in a Hook3 alignment, including the C-terminal glutamates (Extended Data Fig. 7d). We ran Alphafold on fragments of the dynein heavy chain and Hook3, which predicted an interaction via HBS1 (Extended Data Fig. 7e).
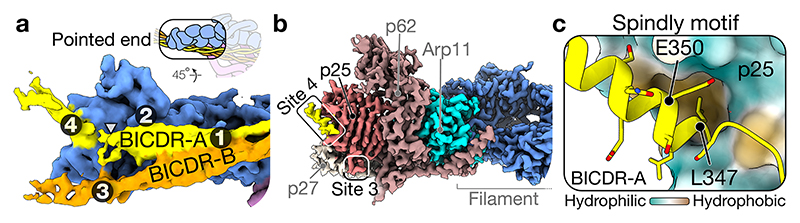
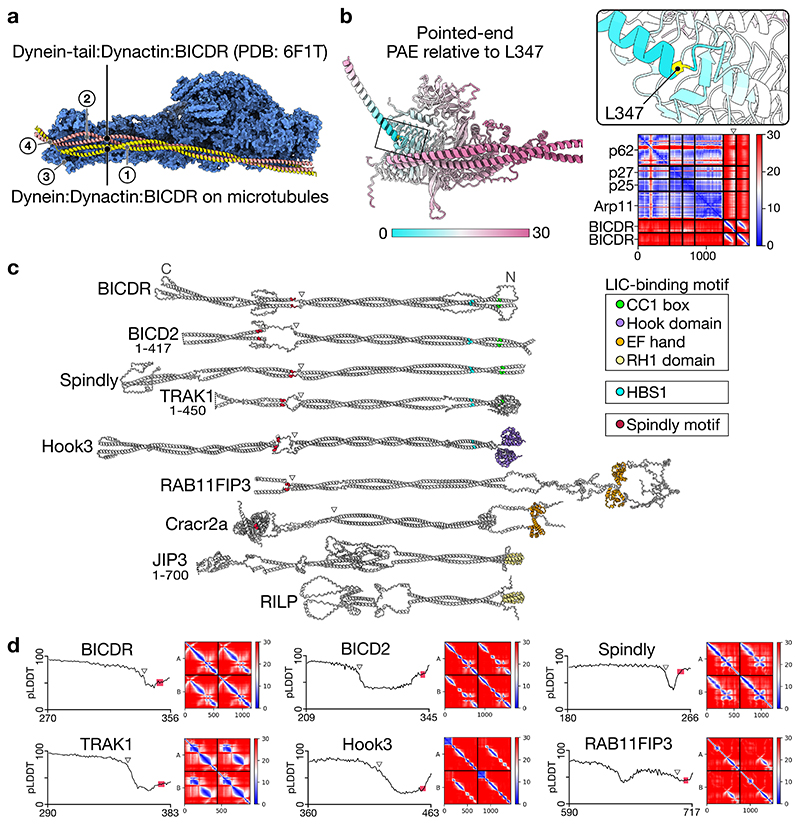
The C-termini of BICDR-A and BICDR-B diverge in the way they interact with the pointed end of dynactin (Fig. 4a). Previous work analysing the binding of different adaptors to the pointed end identified four distinct sites13. BICDR was reported to bind site 1 (p62 disordered loop), site 2 (p62 saddle region), and site 4 (p25 β-helical fold). In our structure, BICDR-A interacts similarly at site 4 (Fig. 4a), but the interactions at site 1 and site 2 are shifted to lower positions on p62, resulting from a downward rotation of the coiled-coil (Extended Data Fig. 8a). BICDR-B interacts with the pointed end at site 3 (on a loop extending from p25) (Fig. 4a).
Figure 4. Pointed end interactions of BICDR-A/B.
a, The density map of the pointed end showing BICDR-A and BICDR-B contacting four distinct sites13. The break in the coiled-coil density is highlighted (triangle). b, The density map of the pointed end showing the ordered alpha helical density at site 4. Also highlighted is the loop from p25 representing site 3. c, A molecular surface representation of the pointed end p25 subunit coloured by hydrophobicity (orange is hydrophobic, teal is hydrophilic). Overlaid is a ribbon representation of BICDR-A’s Spindly motif, highlighting the conserved residues L347 and E350.
The pointed end is the site of interaction of the Spindly motif (LΦXEΦ, where Φ is hydrophobic), which is found in many adaptors12. Our model places it at site 4, as previously predicted13. The alpha-helical density for the Spindly motif is the most ordered part of the BICDR-A C-terminus (Fig. 4b), but there is a disordered break in the coiled-coil preceding it (Fig. 4a). To provide support for our model, we ran Alphafold36 on the four-subunit pointed-end complex with a fragment of BICDR (Extended Data Fig. 8b). The resulting prediction shows high confidence for the Spindly motif interacting with site 4 as well as the preceding break in the coiled-coil. Interestingly, other cargo adaptors may also have coiled-coil breaks preceding their Spindly motifs (Extended Data Fig. 8c)35. Our structure now shows that the first Spindly motif residue (L347) sits in a hydrophobic pocket at the edge of p25 (Fig. 4c). The interaction is further stabilized by L348 and E350 interacting with S31 and N20 of p25, respectively. Due to the offset between BICDR-A and BICDR-B, the Spindly motif of BICDR-B does not contribute to its pointed end interaction.
In summary, whereas the N-terminal motifs (CC1 box and HBS1) of BICDR-A and BICDR-B bind similarly to dyneins, the C-terminal interactions with the dynactin pointed end are distinct, resulting in an asymmetry between the two adaptors.
Discussion
Cryo-EM of sparse complexes on filaments
Solving the structure of proteins sparsely decorating MTs poses unique challenges. The MT signal dominates the alignments and obscures some particles. Previously, the alignment problem was overcome by subtracting the MT from picked particles23–25, but this only works well when the proteins are in a regular array. Here, we reconstructed the MTs and subtracted them from the micrographs. An alternative approach was recently reported, where 2D averages were calculated along individual MTs and used for subtraction37, although so far this has only been used on regularly decorated samples. These approaches open the door solving other sparsely decorated complexes on cytoskeletal filaments.
Asymmetry between dynein motor domains
Despite the flexibility of the dynein-dynactin complex relative to the MT, our single-particle approach reveals two main configurations of the motor domains. In the staggered state, the stalks of the two motors of dynein-A are angled such that they can both bind the same protofilament. This ability to tightrope-walk agrees with the observation that dyneins can walk along single-protofilament polymers38. In both aligned and staggered states, there is a remarkable asymmetry between the four motor domains. Dynein-A1 and B1 appear free to undergo a priming stroke whereas dynein-A2 and B2 are constrained. Although our complex is in an AMPPNP-inhibited state, it is striking that to step forward, the right-hand motors (dynein-A1 and B1) likely move first. This asymmetry may explain the primarily right-handed pitch of dynein-dynactin trajectories on MTs6. More rarely, left-handed trajectories were observed. This is perhaps explained by the heterogeneity the motor-domain positions, as seen by our cryo-ET analysis of individual complexes. In the presence of ATP, this flexibility is likely to be higher. In summary, the asymmetry we observe likely contributes to dynein’s stepping behaviour.
Recruiting two adaptors
The binding of two BICDRs is determined by a combination of (1) motifs that bind dynein, (2) contacts with dynactin’s pointed end, and (3) an offset interaction between the two adaptors. At the adaptor N-termini, the CC1 box and HBS1 motifs bind predominantly to the LICs and heavy chains of dynein-A. This suggests dynein-B is not required to recruit the second adaptor. At the C-termini, the two adaptors must share interactions with one pointed-end complex. BICDR-A makes multiple contacts, including via its Spindly motif. In contrast, BICDR-B only makes a single contact, suggesting its stability depends on the adaptor-adaptor interactions with BICDR-A.
Hook3 also has a LIC-binding site (the hook domain)39 and an HBS1 motif, which we identify here. This suggests that two Hook3s interact with dynein equivalently to two BICDRs. Surprisingly, the pointed end interactions differ. Whereas BICDR-A contacts sites 1/2/4, Hook3-A binds sites 1/3/4. Conversely, whereas BICDR-B contacts site 3, Hook3-B binds site 2. This results in a different offset interaction between the Hook3s, with Hook3-B going over Hook3-A, whereas BICDR-B goes under BICDR-A (Extended Data Fig. 6f). This suggests there is diversity in how pairs of adaptors are recruited. A key future question is whether adaptors from different LIC-binding site families (e.g. EF-hand and RH1-domain) (Extended Data Fig. 8c)7 also bind in pairs. Our structures suggest the key determinants of this will be the adaptor-adaptor and pointed-end interactions.
Consequences of having a second adaptor
BICDR and Hook3 transport predominantly membrane vesicles40,41. Because of their size, it is unlikely that two vesicles are recruited simultaneously to a two-adaptor complex. Instead, one vesicle is probably linked to dynein-dynactin by two connections. This bivalent tethering may increase the efficiency of motor capture and decrease the probability of release. For small molecules, such as RNAs transported by the related adaptor BicD42, there is the possibility of carrying two cargos at once, giving a greater capacity for cargo transport.
Another consequence of the second adaptor is that it reinforces the connection between dynein-A and dynactin. This will reduce their dissociation and promote long distance movement on MTs. Since both adaptors primarily contact dynein-A, this stabilization is likely to also occur in adaptors which can recruit a single dynein (e.g. BICD2)4. Many adaptors also bind kinesins43,44, leading to complexes containing opposing motors45,46. The relative number of each motor may determine the behaviour of cargo movement 47,48. We therefore speculate that different adaptors can tune the stoichiometry of the motors that they recruit, providing another layer of regulation to cargo transport.
Methods
Protein preparation
Dynactin was purified natively from pig brains19. Fresh brains were acquired from a butcher and transported in ice-cold PBS, cleaned in homogenization buffer (35 mM PIPES pH 7.2, 5 mM MgSO4, 100 μM EGTA, 50 μM EDTA), squashed in a plastic bag, and flash frozen in liquid nitrogen. Frozen brain pieces were blended and resuspended in homogenisation buffer supplemented with 1.6 mM PMSF, 1 mM DTT, and 4 complete-EDTA protease-inhibitor tablets per 500 mL (Roche). After thawing, the lysate was centrifuged in a JLA 16.250 (Beckman Coulter) at 16,000 rpm for 15 min at 4°C. The supernatant was further clarified in a Type 45 Ti (Beckman Coulter) at 45,000 rpm for 50 min at 4°C. After filtering the supernatant in a Glass Fibre filter (Sartorius) and a 0.45 μm filter (Elkay Labs), it was loaded on a column packed with 250 mL of SP-Sepharose (Cytiva) pre-equilibrated with SP buffer (35 mM PIPES pH 7.2, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA, 1 mM DTT, 0.1 mM ATP) using an Akta Pure (Cytiva). The column was washed with SP buffer with 3 mM KCl before being eluted in a linear gradient up to 250 mM KCl over 3 column volumes. The peak around ~15 mS/cm was collected and filtered in a 0.22 μm filter (Elkay Labs) before being loaded on a MonoQ 16/10 column (Cytiva) pre-equilibrated with MonoQ buffer (35 mM PIPES, 5 mM MgSO4, 100 μM EGTA, 50 μM EDTA, 1 mM DTT, pH 7.2). The column was washed with MonoQ buffer before being eluted in a linear gradient up to 150 mM KCl over 1 column volume, followed by another linear gradient up to 350 mM KCl over 10 column volumes. The peak around ~39 mS/cm was pooled and concentrated to ~3 mg/mL before being loaded on a TSKgel G4000SWXL (Tosoh Bioscience) pre-equilibrated with GF150 buffer (25 mM HEPES pH 7.2, 150 mM KCl, 1 mM MgCl2) supplemented with 5 mM DTT and 0.1 mM ATP. The peak at ~114 mL was pooled and concentrated to ~3 mg/mL. 3 μL aliquots were flash frozen in liquid nitrogen and stored at -80°C.
Human cytoplasmic dynein-1 with an N-terminal ZZ-TEV tag on the heavy chain was expressed using the baculovirus/Sf9 system1. Specifically, we used a construct with mutations in the linker (R1567E and K1610E) to help overcome the autoinhibited conformation. These mutations were previously shown not to affect motor velocity49. Fresh bacmid DNA was transfected into Sf9 cells at 0.5x106 cells/mL in 6-well cell culture plates using FuGene HD (Promega) according to the manufacturer’s protocol (final concentration 10 μg/mL). After three days, 1 mL was added to 50 mL of 1x106 cells/mL and infected for five days in a shaking incubator at 27°C. The P2 virus was isolated by collecting the supernatant after centrifugation at 4,000 rcf for 15 min and stored at 4°C. For expression, 10 mL of P2 virus was used to infect 1 L of Sf9 cells at 1.5-2x106 cells/mL for 96 hours in a shaking incubator at 27°C. Cells were harvested by centrifugation at 2,000 rcf for 10 min at 4°C, resuspended in cold PBS, and centrifuged again in 50 mL Falcon tubes (Sarstedt). The pellet was flash frozen and stored at -80°C. A pellet from 1 L expression was thawed on ice in 50 mL lysis buffer (50 mM HEPES pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 0.1 mM ATP) supplemented with 2 mM PMSF, 1 mM DTT, and 1 complete-EDTA protease-inhibitor tablets per 50 mL. Cells were lysed using a 40 mL dounce tissue grinder (Wheaton) with ~16 strokes. The lysate was clarified at 503,000 rcf for 45 min at 4°C using a Type 70 Ti Rotor (Beckman Coulter). The supernatant was incubated with 3 mL IgG Sepharose 6 Fast Flow beads (Cytiva) pre-equilibrated with lysis buffer for 4 hours in a 50 mL Falcon at 4°C on a roller at a low speed setting. The beads were then applied to a gravity flow column and washed with 150 mL of lysis buffer and 150 mL of TEV buffer (50 mM Tris-HCl pH 7.4, 150 mM KAc, 2 mM MgAc, 1 mM EGTA, 10% (v/v) glycerol, 0.1 mM ATP, 1 mM DTT). The beads were then transferred to a 5 mL centrifuge tube (Eppendorf) and filled up completely with TEV buffer. TEV protease, which was purified in-house using pRK79350, was added to a final concentration of ~60 ug/mL and incubated overnight at 4°C on a roller at a low speed setting. The beads were transferred to a gravity flow column and the flow through containing the cleaved protein was collected in a 15 mL Falcon tube. 8 mL of TEV buffer was added to the beads and collected to maximize recovery. The protein was concentrated to ~2 mg/mL and loaded onto a TSKgel G4000SWXL column preequilibrated with GF150 buffer supplemented with 5 mM DTT and 0.1 mM ATP. Peak fractions were pooled and concentrated to ~2.5 mg/mL. Glycerol was added to a final concentration of 10% from an 80% stock made in GF150 buffer. 3 μL aliquots were flash frozen and stored at -80°C.
Full-length mouse BICDR with an N-terminal ZZ-TEV tag was purified using baculovirus expression in the same way as dynein described above with the omission of ATP in the GF150 buffer.
Lyophilised porcine tubulin (Cytoskeleton) was resuspended in microtubule (MT) buffer (25 mM MES, 70 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, pH 6.5) to a final concentration of 10 mg/mL (~90 μM), aliquoted, flash frozen, and stored at -80°C. The use of a MES-based buffer increased the proportion of 13 protofilament MTs (see below)51.
Sample preparation
To polymerize MTs, tubulin was diluted in MT buffer with GTP (MilliporeSigma) such that the final concentration of tubulin was 5 mg/mL (45 μM) tubulin and GTP was 3 mM. The mixture was mixed, incubated on ice for 5 min, and MTs were polymerized at 37°C for ~1.5 hours. To stabilize the MTs, polymerization buffer supplemented with taxol (MilliporeSigma) was added such that the final taxol concentration was ~20 μM. The MTs were pelleted on a benchtop centrifuge (Eppendorf) at 20,000 rcf for 8 min at room temperature. The supernatant was discarded and the pellet was resuspended in polymerization buffer with taxol by pipetting up and down with a cut tip. The MTs were pelleted and resuspended again using an uncut tip. The concentration was approximated using Bradford reagent (Biorad) and diluted to ~0.65 mg/mL (~6 μM).
To assemble the dynein-dynactin-BICDR complex, the purified proteins were mixed in a 1:2:32 molar ratio (0.33 mg/mL dynein, 0.56 mg/mL dynactin, 0.97 mg/mL BICDR) in GF150 buffer supplemented with 1 mM DTT in 10 μL and incubated on ice for 15 min. To bind the complex to MTs, 9 μL complex was mixed at room temperature with 5 μL MTs and 9 μL binding buffer A (77 mM HEPES pH 7.2, 51 mM KCl, 13 mM MgSO4, 2.6 mM EGTA, 2.6 mM DTT, 7.68 mM AMPPNP, 13 μM taxol) such that the final concentrations of KCl and AMPPNP were 100 mM and 3 mM, respectively. After 5 min, the complex-bound MTs were pelleted at 20,000 rcf for 8 min at room temperature. The pellet was resuspended in binding buffer B lacking dynein, dynactin, or BICDR (30 mM HEPES pH 7.2, 60 mM KCl, 5 mM MgSO4, 1 mM EGTA, 1 mM DTT, 3 mM AMPPNP, 5 μM taxol, and 0.01% IGEPAL (MilliporeSigma)), using an uncut tip and left at room temperature. 3.5 μL was applied to freshly glow-discharged Quantifoil R2/2 300-square-mesh gold grids (Quantifoil) in a Vitrobot IV (ThermoFisher) at 100% humidity and 20°C, incubated for 20 s, and blotted for 0.5-1.5 s before being plunged into liquid ethane.
Cryo-EM
The samples were imaged using a FEI Titan Krios (300 kV) equipped with a K3 detector and energy filter (20 eV slit size) (Gatan) using automated data collection (ThermoFisher EPU 2.6.1) over 15 sessions. In total, 88,715 movies were acquired at 81,000 X magnification (1.11 Å/pixel, 100 μM objective aperture, ~3 sec exposure, 53 frames, -1.2 to -3.6 μm defocus range), and 9,875 movies were acquired at 42,000 X magnification (2.13 Å/pixel, 70 μM objective aperture, ~9 sec exposure, 53 frames, -1.5 to -4 μm defocus range). 1,846 of the low magnification movies were acquired at a 26° tilt of the stage. The tilted movies were acquired at early stages of the project when we expected a preferred orientation of the complex, which did not end up being an issue. In all cases, the total fluence was ~53 e-/Å2. “Datasets” here refers to separate cryo-EM data collection sessions. Each of these sessions used a separate cryo-EM grid, which originated from multiple sample preparations. Each preparation used fresh microtubules and proteins from one of two or more independent purifications.
MT subtraction and particle picking
Micrographs were generated from the movies after correcting for drift using MotionCorr2 52 implemented in Relion 3.153, and the contrast transfer function (CTF) parameters were estimated using CTFFIND454. MTs were picked from the micrographs using the filament option in crYOLO 1.7.555 with a model that was trained on manually picked micrographs (~100). A different model was used for low- and high-magnification micrographs. The output coordinates along each MT were spaced by 81 Å. These coordinates were used in the Microtubule Relion-based Pipeline (MiRP) to reconstruct the MTs27. Briefly, the coordinates were used to extract particles at 4x binning and a rolling average of the particles along a MT were generated to increase the contrast for subsequent classification steps. A model for 11 to 16 pf MTs was used for supervised 3D classification in Relion 3.0, 3.1, or 4.028,53. The protofilament number (pf) distribution (pooled across datasets) was 11 pf: 2.1%, 12 pf: 18.6%, 13 pf: 60.0%, 14 pf: 16.7%, 15 pf: 1.2%, 16 pf: 1.4%. After assigning the most likely pf number to each MT, the data was split and each type was processed independently. Sequential refinement of the helical parameters (Rot angle, X/Y shifts) following the MiRP protocol led to properly aligned and internally-consistent particles. The seam checking step was skipped since the misalignment of the seam was not detrimental to the quality of the subtraction described below. The data was re-extracted to generate raw particle stacks at 4x binning for local refinement in Relion 3.1 or 4.0, and then re-extracted without binning for a final local refinement.
Each pf-number MT was subtracted sequentially using the relion_particle_reposition command-line interface in Relion 4.0 and specifying the ctf, invert, subtract, and norm_radius options. The value for norm_radius was obtained from the final extraction job (found in the note.txt file as bg_radius). Dynein-dynactin-BICDR particles were then picked from the subtracted micrographs using a crYOLO model that was pre-trained on ~100 micrographs56. Complexes that were too far from the MTs were removed using the extract_if_nearby option in Starparser 1.38 (https://github.com/sami-chaaban/starparser) (doi: 10.5281/zenodo.6792794). Specifically, the closest MT coordinate was found for every complex coordinate, and those with distances that were greater than ~550 Å were removed. There was no indication of nonuniform clustering under the conditions of our assay. From this point onward, the data was processed in Relion 3.1 or Relion 4.0 unless otherwise stated.
Processing
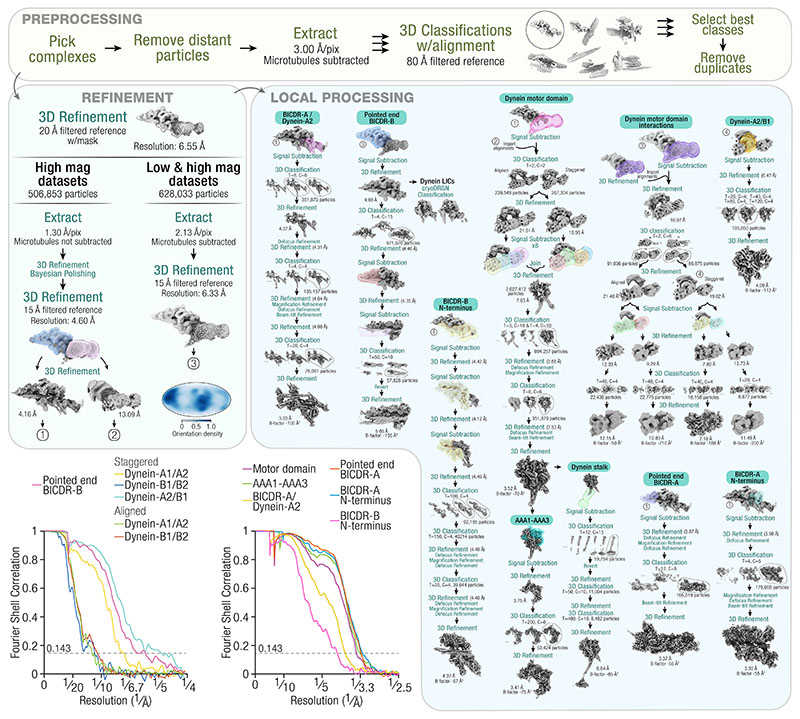
A processing pipeline can be found in Extended Data Fig. 2. Picked particles that were close to MTs were extracted with binning such that the final pixel size was 3 Å/pixel. A reference was generated using a manually built model of the full dynein-dynactin-BICDR complex based on previous structural work4,5, which was then filtered to 80 Å for 3D classification with alignment. Multiple classifications were performed in parallel, with each run varying in settings (number of classes: 6, 12, or 15, ignore CTFs until first peak: Yes or No, mask diameter: 780 Å or 820 Å, Tau fudge: 3 or 6). Particles from the best class in each run were pooled and duplicates were removed based on the rlnImageName column using the remove_duplicates option in Starparser. The number of particles remaining at this stage were 506,853 and 121,180 in the high-magnification and low-magnification set, respectively. To demonstrate the effect of microtubule subtraction on successful averaging (Fig. 1a), particles extracted from either subtracted or non-subtracted micrographs were subjected to 2D classification with the same parameters (number of classes = 8, ignore CTFs until first peak = Yes, mask diameter = 950 Å, Tau fudge = 3).
The particles were subjected to a global 3D refinement without a mask using a 60 Å filtered reference. A soft-edge mask was generated based on the resulting density and the refinement was allowed to proceed with the mask by choosing “Continue”. The high-magnification data were then re-extracted from the non-MT-subtracted micrographs at 1.17x binning (1.30 Å/pixel). Local refinement, Bayesian polishing, and a further local refinement yielded a consensus structure at 4.60 Å resolution. The dynein-tail/dynactin region was masked for local refinement and used for multiple signal subtractions to improve the local resolutions. In all cases, local refinement, defocus refinement, magnification refinement, beam-tilt refinement, and at least one round of 3D classification without alignment was performed to improve the resolution. The maps were B-factor sharpened for model building (described below). EMDA 1.1.257 was used to align the maps to the consensus dynein-tail/dynactin map56. Masks, particle numbers, resolutions, classification parameters, and B-factors can be found in Extended Data Fig. 2.
To solve the structure of the motor domain, the signal for dynein-tail/dynactin was first subtracted by using the local refinement around dynactin described above as input to a signal subtraction with a mask around all four motor domains. In parallel, a local refinement of the motor domains using the same mask was performed, and the alignments from this run were imported into the signal subtracted particles using the swap_columns option in Starparser, by passing the following columns: rlnAnglePsi, rlnAngleRot, rlnAngleTilt, rlnNormCorrection, rlnLogLikeliContribution, rlnMaxValueProbDistribution, rlnNrOfSignificantSamples, rlnOriginXAngst, and rlnOriginYAngst. This provided a more accurate set of starting alignments for a subsequent 3D classification without alignment of the subtracted particles into two classes. For each class, which represented the staggered and aligned states, four parallel signal subtractions were performed on each motor domain. The particles were then joined from all eight signal subtraction jobs. A 3D refinement yielded a structure at 7.03 Å, which was further improved to 3.52 Å after defocus refinement, magnification refinement, beam-tilt refinement, and two rounds of 3D classification without alignment. To improve the density of the nucleotide in the AAA1 domain, further signal subtraction was performed with a mask around AAA1, AAA2, and AAA3. Local refinement and 3D classification without alignment resulted in improved density for AAA1. To improve the density of the stalk, the refinement of the motor domain was signal subtracted with a mask around the stalk and MT binding domain. 3D classification without alignment identified particles with the clearest density, which were reverted to the original box size for two rounds of local refinement and 3D classification. Masks, particle numbers, resolutions, classification parameters, and B-factors can be found in Extended Data Fig. 2.
For more challenging regions (p150Glued, dynein’s stalk, the LICs, and BICDR-B), the two magnification datasets were combined: the high-magnification data were re-extracted from MT-subtracted micrographs at ~2x binning and combined with the low-magnification data (unbinned), for a shared pixel size of ~2.13 Å/pixel. Local refinement yielded a consensus structure at 6.33 Å resolution. The angular distribution plotted on a Mollweide projection is shown in Extended Data Fig. 2, which was generated in Starparser using the plot_orientations option. To better define the p150Glued connecting density, a 3D classification (Tau fudge = 20, 15 classes) was performed and the class with the clearest signal (30,570 particles) was further locally refined (13.50 Å final resolution). To show the stalks of the staggered state relative to the MT, the particles were re-centered near the MT binding domain of dynein-A2 and extracted from micrographs (with MTs) at ~3 Å/pixel. Two rounds of 3D classification were performed with a mask that includes the complex and a section of the MT wall, first with local alignment (1.8° sampling interval, local angular searches, E-step limited to 16 Å, Tau fudge = 10, 10 classes) followed by without alignment (Tau fudge = 20, 3 classes). The class with the clearest density for both the protofilaments and the stalks of dynein-A1 were chosen (3,563 particles).
The consensus structure from the combined-magnification datasets was used for signal subtraction with a mask around dynactin. This was used for cryoDRGN classification of the LICs (described below) and to resolve BICDR-B. The C-terminus of BICDR-B was resolved by first performing a signal subtraction with a mask around the pointed end. To improve the density, another signal subtraction was performed around just BICDR-B, and 3D classification without alignment identified particles with the clearest density. For the BICDR-B N-terminus, the dynactin refinement was used for signal subtraction followed by defocus refinement, magnification refinement, beam-tilt refinement, and 3D classification without alignment. Masks, particle numbers, resolutions, classification parameters, and B-factors can be found in Extended Data Fig. 2.
To better define the motor-domain interactions, the consensus refinement from the combined-magnification datasets was signal subtracted with a mask around all four motor domains. In parallel, a local refinement of the motor domains using the same mask was performed, and the alignments were imported into the signal subtracted particles using the swap_columns option in Starparser, as described above. 3D classification isolated particles from the staggered and aligned states. These were used to determine on which pf number the particles from the staggered and aligned states are bound. For this, the fetch_from_nearby option in Starparser was used, which imported the class-number from the MT particles based on their proximity in the micrographs. For each state, a mask around dynein-A1/A2 and dynein-B1/B2 was used for signal subtraction, followed by 3D classification without alignment to improve the density. Masks, particle numbers, resolutions, classification parameters, and B-factors can be found in Extended Data Fig. 2. To identify the variation in dynein-A1 in the staggered state, 3D classification was performed with a mask around dynein-A1/A2 (Tau fudge = 8, 10 classes), where one class showed the motor domains oriented in a manner that results in the stalks being parallel (3,781 particles out of 86,875).
All structures were post-processed in Relion by including the MTF of the detector. The maps were also scaled to the correct pixel size, which was calibrated by comparing the density for a tubulin dimer to a previous structure of taxol-bound tubulin58. Specifically, the maps were fit to each other using the Fit in map option in Chimera 1.1659, and the pixel size scaling with the highest correlation was used. The final pixel size of the maps from the high magnification and combined-magnification datasets are 1.24 Å/pixel (originally 1.30 Å/pixel) and 2.04 Å/pixel (originally 2.13 Å/pixel), respectively. This scaling also resulted in a match to previous dynactin structures13.
The composite map in Fig. 1c was built by summing the maps in Chimera. For the motor domains, the map with improved density for the stalk was fit into each of the four positions in the motor domain-pair maps. Although the raw data shows that there’s flexibility in the orientation of the complex relative to the MT, for illustrative purposes the composite map was overlaid onto the MT to match the configuration of the MT binding domains with the tubulin dimers 60,61. All colours were rendered in ChimeraX 1.2.562.
Model building and refinement
Building was performed in COOT 0.9.363 and refinement was done in PHENIX 1.2064. All structure predictions were performed using Alphafold through a local installation of Colabfold 1.2.065, running MMseqs266 for homology searches and Alphafold267 or Alphafold2-Multimer36 for the predictions of single or multiple chains, respectively. When used to guide model building, predictions were generated without templates and with amber relaxation68. All refinement statistics can be found in Extended Data Table 1 and Extended Data Table 2.
A starting model of the dynein motor domain was generated by using a previous structure of dynein in the phi conformation (PDB: 5NUG)49 and manually adjusting it to match the crystal structure of S. cerevisiae dynein with AMPPNP (PDB: 4W8F)15 in COOT. The model was then rebuilt into the density and challenging regions were guided by Alphafold predictions of the individual AAA+ subdomains. Nucleotides were placed in the AAA+ domains as follows (AAA1: ADP, AAA2: ATP, AAA3: AMPPNP: AAA4: AMPPNP). We modelled the diphosphate nucleotide in AAA1 as ADP assuming it was retained after purification and sample preparation. However, it is also possible that it comes from slow hydrolysis of AMPPNP69, in which case we would expect the nucleotide to be AMPPN. The resulting model was manually inspected before refinement.
The dynein-A2/B1 interaction in the staggered state was built by placing into the density the refined motor domain from above with an Alphafold prediction of the LIC (UniProt ID O43237) Ras-like domain, before refinement in COOT.
The N-terminus of the dynein tails were built using previous structures of dynein-tail/dynactin as a reference (PDB: 6F1T)4 and refined. An Alphafold prediction of the dynein intermediate chain (Q13409) WD40 domain was built into the structure and refined.
Dynactin subunits were built and refined using previous models as a reference (PDB: 6ZNL, 6F1T)4,13, as well as Alphafold predictions to guide challenging regions, and were manually inspected and refined. The pointed end complex includes actin-related protein 11 (Arp11, ACTR10), p62 (DCTN4), p25 (DCTN5), and p27 (DCTN6), as well as an N-terminal fragment of p50 (DCTN2)13. The Arp1/actin filament includes eight copies of actin-related protein 1 (ARP1, also known as ACTR1A) and one copy of β-actin4. The barbed end includes CAPZα and CAPZβ4. Based on our density, ATP-Mg was built into Arp11 and ADP-Mg was built into β-actin and the Arp1 subunits.
The structure of full-length BICDR was first predicted in Alphafold on two copies of BICDR (BICDL1: A0JNT9), and was flexibly fit into a low-resolution map of BICDR-A in COOT using the registry defined by the position of W166. The N-terminus containing the CC1 box and HBS1 was then built into high-resolution density and refined. The C-terminus containing the Spindly motif was guided by a prediction of the pointed end complex (described below). The BICDR-B model was built by flexible fitting of the BICDR-A model into a low-resolution map of BICDR-B after adjusting the registry based on the position of W166.
The LIC C-terminal alpha helices were built from a prediction of two copies of the BICDR N-terminus (BICDL1: A0JNT9) and two copies of the LIC (O43237) C-terminus, before being refined in COOT.
To generate the composite structure, a copy of the motor domain was placed in each monomer position in the composite map. The C-terminus of the dynein tails were built by first generating predictions of the dynein heavy chain (Q14204) helical bundles and placing them into the density, before stitching them together. The model was then refined in COOT into the density of each tail and stitched to the N-termini of the dynein tails and the motor domains. The dimerization domain was built by generating a prediction of two copies of the N-terminus of dynein, which was refined into the density in COOT. The complex of dynein light chain ROBL1 (DYNLRB1) and the dynein intermediate chain was flexibly fit into the density from a previous structure (PDB: 6F1T)4. The shoulder was rigid body docked from a previous structure (PDB: 6ZNL)13, which includes two copies of p150Glued (DCTN1), four copies of p50 (DCTN2), and two copies of p24 (DCTN3). The stalks of the motor domains were built by flexible fitting of the T. thermophila axonemal dynein stalk (PDB: 7K58)26 before mutating the residues to match the human sequence. The stalk was then stitched to the structure of the dynein MT binding domain (PDB: 6RZB)61. The stalk and MT binding domain were refined into the density of each motor position before being stitched to the motor domains.
Motor domain model alignments
To analyse the nucleotide pockets in AAA1 and AAA3, the large subdomains were aligned to the crystal structure of S. cerevisiae dynein-AMPPNP15 and D. discoideum dynein-ADP29 using match-maker in Chimera. The difference between our structure and the crystal structure was visualized by drawing arrows between the coordinates of corresponding residues using a custom Python script (PDBArrows 1.0) (https://github.com/sami-chaaban/PDBArrows) (doi: 10.5281/zenodo.6792805). To determine between which pairs of residues the arrows should be drawn, the sequence of the subdomains of interest were aligned using Clustal Omega 1.2.470. Pairs that were more than 12 Å apart were ignored, which likely represent residues that were poorly aligned.
Cryo-ET
Samples were prepared as for cryo-EM except that gold fiducials (BBI Solutions) pre-equilibrated in binding buffer B were added to the sample before freezing. Tomograms were acquired using a Glacios TEM (200 kV) equipped with a Falcon III detector (Thermo Fisher). SerialEM 3.9.071 was used to acquire a tilt series between -60° and +60° in 3° increments at 57,000 X magnification (2.55 Å/pixel, 70 μM objective aperture). At each tilt angle, a movie was acquired containing 13 frames with an exposure of ~3 e-/Å2/tilt. The total fluence for the tilt series was ~120 e-/Å2 and the defocus range was -3 to -6 μm. Gain and motion correction was done using alignframes from IMOD 4.10.3272, with per-frame dose weighting. Tomogram alignment and reconstruction (back projection) was done using the ETomo interface of IMOD. The tomograms were subsequently binned by 4 and processed using a Wiener-like deconvolution filter as implemented in WARP73 (https://github.com/dtegunov/tom_deconv). The resulting tomograms were visualized in ChimeraX using the Hide Dust option set to ~200. Complexes where all four motor domains were clearly defined were chosen for visualization. To colour the density, the Color Zone option was used with the atomic models of dynein and dynactin, which were rigid body fit into the density and manually adjusted in cases where the motor domains were in different conformations.
Mass photometry
To analyse the oligomerization of BICDRs in solution, we subjected the purified protein to mass photometry using the Refeyn OneMP instrument (Refeyn Ltd.). BICDR was diluted to 75 nM in GF150 buffer and 10 μL was imaged. The movies were processed using DiscoverMP 1.2.4 and the mass was estimated by fitting a Gaussian distribution to the data.
CryoDRGN classification
To help reveal densities for the flexible LIC loops, the consensus structure from the combined-magnification datasets that was signal subtracted around dynactin was used as input for multiple rounds of cryoDRGN 0.3.3 analysis34. First, the particles were binned from 400 pixels to 128 pixels (6.75 Å/pixel final pixel size) and used for training an 8-dimensional latent variable model with 3 hidden layers and 256 nodes in the encoder and decoder networks. The latent space was visualized with the analyze function on epoch 50 and 3 clusters were extracted using a gaussian mixture model (GMM). The particles from the cluster that yielded the best 3D refinement were moved forward for two more rounds of training, GMM clustering, and 3D refinement, but with binning to 256 pixels (3.33 Å/pixel final pixel size) and training with 3 hidden layers and 1024 nodes in the networks. Finally, cryoDRGN landscape analysis was performed on the latent space using a custom mask in the region encompassing the LICs, with 20 clusters extracted using ward linkage. The mean volume for each cluster was manually inspected for connecting density between the LIC Ras-like domain and the BICDR N-termini.
Previous data (dynein-tail/dynactin/BICDR and dynein-tail/dynactin/Hook3)4 was reanalysed by binning the consensus particles from 432 pixels to 256 pixels (2.26 Å/pixel final pixel size) and training an 8-dimensional latent variable cryoDRGN model with 3 hidden layers and 1024 nodes in the encoder and decoder networks. The latent space was visualized with the analyze function on epoch 50 and epoch 41 in the BICDR and Hook3 datasets, respectively, and 20 clusters were extracted using k-means analysis. The structure representing the center of each cluster was visually inspected for the presence of a second adaptor.
Cargo adaptor structure prediction
The pointed-end-BICDR complex was predicted using Colabfold 1.2.0 (10 recycles, no templates) with the following sequences: DCTN4 (Q9UJW0), DCTN6 (O00399), DCTN5 (Q9BTE1), ACTR10 (I3LHK5), and two copies of BICDR (BICDL1; A0JNT9) fragment 205-394. The predicted aligned error (PAE) with respect to L347 was mapped onto the predicted structure using a custom Python script (PointPAE 1.0) that extracts all values on the y-axis for that residue and generates an attribute file for ChimeraX visualization (https://github.com/sami-chaaban/PointPAE) (doi: 10.5281/zenodo.6792801).
Cargo adaptors were predicted by running Colabfold 1.2.0 (no templates) on two copies each of the following sequences: BICDR (BICDL1: A0JNT9), BICD2 1-423 (Q8TD16) Spindly (SPDL1: Q96EA4), TRAK1 1-450 (Q9UPV9), Hook3 (Q86VS8), RAB11FIP3 (RFIP3: O75154), Cracr2a (EFC4B: Q9BSW2), JIP3 1-700 (Q9UPT6), RILP (Q96NA2). For visualization, the models were manually linearized such that the coiled-coils run parallel to each other. Specifically, the predictions were broken up into fragments at flexible loops, rotated to be parallel, and stitched again in COOT. The PAEs and predicted local distance difference tests (pLDDTs) at the coiled-coil breaks preceding the Spindly motif can be found in Extended Data Fig. 8d.
The Hook3 HBS1 region was predicted by running Colabfold 1.2.0 (no templates) on two copies of Hook3 172-287 (Q86VS8), one copy of dynein heavy chain 576-864 (Q14204), and one copy of dynein intermediate chain (Q13409) 226-583. The PAE relative to Hook3 H200 was mapped onto the prediction as above for the pointed end complex.
Extended Data
Extended Data Fig. 1. MT subtraction from cryo-EM micrographs of dynein-dynactin-BICDR on MTs.
a, An example micrograph out of n = 66,800 that had microtubules suitable for subtraction. A pseudo-flat-field correction has been applied to normalize the intensity across the micrograph for visualization purposes (i.e. dividing the image by a gaussian-blurred copy). b, Overview of the processing pipeline to subtract MTs from cryo-EM images in order to thoroughly pick and accurately align dynein-dynactin-BICDR complexes. c, The density maps of the 12, 13, and 14 protofilament (pf) MTs. Not shown are the 11, 15, and 16 pf MTs. d, An annotated micrograph showing picked particles that were kept (black) or rejected (red) based on their proximity to the MTs.
Extended Data Fig. 2. Processing pipeline for single-particle analysis of the dynein-dynactin-BICDR complex.
(T = Tau fudge, C = number of classes). 3D classifications are without alignment unless otherwise specified. All defocus, magnification, and beam-tilt refinements were immediately followed by a 3D refinement (not shown). Plots show the gold standard Fourier shell correlation. The dotted horizontal line shows the 0.143 cut-off. An angular distribution plot is shown for the consensus refinement of the whole dataset on a Mollweide projection.
Extended Data Fig. 3. Overview of the dynein-dynactin-BICDR complex.
a, The composite density map of dynein-dynactin-BICDR overlaid on the reconstructed 13 protofilament MT, showing the position of individual dynein motor domains (dynein-A1/2, dynein-B1/2), with their tails extending towards dynactin. b, A pseudo-molecular surface representation of a dynein dimer, showing the LICs, intermediate chains (ICs) and light chains (LCs). The model was generated from our structure and the published IC/LC8/Tctex crystal structure (PDB: 2PG1)74. Additional flexible regions were added manually. c, The composite density map of the complex shown from behind, where dynein’s tails can be seen sitting in the grooves of dynactin’s Arp1 filament. d, A molecular surface representation of our model of dynactin viewed from the back, showing the position of dynactin’s Arp1 filament and barbed/pointed ends relative to the dynein tails and BICDRs. e, A 3D classification result showing density connecting the shoulder domain to a globular density near dynein-A1, which may represent the Inter-Coiled Domain (ICD) of p150Glued with adjacent coiled-coils (CC1 and CC2).
Extended Data Fig. 4. Conformation of the dynein motor domain.
a, Side view of the four motor domains from both major configurations (aligned and staggered) displaying a straight linker (dotted black line). The extra density on dynein-A2 in the staggered state (dotted green line) represents the LIC of dynein-B1. b, Front view of a ribbon representation of the motor domain showing only the linker (purple), AAA1 (blue), AAA4 (yellow), AAA5 (orange) and C-terminal domain (grey). The inset shows the conserved F3629 in AAA5 binding the linker75. c, A close-up view of the linker-AAA2 interaction overlaid with the crystal structures of D. discoideum dynein-ADP (PDB: 3VKG)29 and S. cerevisiae dynein-AMPPNP (PDB: 4W8F)15 aligned at the linker. The inset shows the crystal structure of S. cerevisiae dynein-Apo (PDB: 4AKG)75, which lacks nucleotides in AAA1 and AAA3 and the linker is undocked from AAA2. d, The domain movements in the nucleotide pocket of AAA3 represented by arrows after alignment of our structure (Motor-MT) to the crystal structure of D. discoideum dynein-ADP (left; PDB: 3VKG)29, and S. cerevisiae dynein-AMPPNP (right; PDB: 4W8F)15 at AAA3L. e, The domain movements in the nucleotide pocket of AAA1 represented by arrows after alignment of our structure (Motor-MT) to the crystal structure ofD. discoideum dynein-ADP (left; PDB: 3VKG)29 and S. cerevisiae dynein-AMPPNP (right; PDB: 4W8F)15 at AAA1L.
Extended Data Fig. 5. Motor interactions and heterogeneity.
a, Number of particles from the staggered and aligned states on different MT protofilament numbers (79,435 and 83,450 total particles, respectively). The plot shows the mean of n = 15 datasets representing independently prepared cryo-EM grids. Error bars show the 95% confidence interval (Mann Whitney U Test, two-sided; P = 0.43, 0.31, 0.24, 0.14, and 0.38, respectively) (ns = not significant). b, Front view of a 3D classification result of dynein-A in the staggered state showing a small subset of particles with parallel stalks (right), compared to the majority of particles with crossed stalks (left). A ribbon representation of the motor domain is placed in the density map to show the orientation of the stalks. c, A 3D classification result that includes the MT wall, showing the orientation of the protofilaments as well as density for the stalks of dynein-A. d, A closeup view of the interaction between dynein-A2 and the LIC of dynein-B1, highlighting the three potential interaction sites on the LIC. e, The interaction between dynein motor domains in the aligned (left) and staggered (right) states is shown as a molecular surface representation and density map, where the linker is coloured darker. The triangles and dotted lines highlight the hinge in the linker. f, Tomograms of individual dynein-dynactin-BICDR complexes on MTs in the aligned and staggered states. The dynein and dynactin densities have been coloured pink/purple and blue, respectively. g, An example tomogram where dynein-A1 is shifted away from dynein-A2. h, An example tomogram where there is a large separation between dynein-B1 and B2.
Extended Data Fig. 6. Adaptor arrangements and interactions.
a, A mass photometry result of purified BICDR on its own, showing a major peak at ~127 kDa (expected molecular weight of the dimer is 130 kDa) and a minor peak at 260 kDa (n = 1). b, The density maps of BICDR-A and BICDR-B showing the bulky density at W166 and the location of the HBS1 motif residues QEKH near dynein-A2 and dynein-A1, respectively. c, The interaction interface of the two BICDRs is shown based on their registry in the structure. Lowercase letters refer to the position in the heptad repeat of the coiled-coil, predicted with LOGICOIL76. The red asterisk at K234 shows the offset between the two BICDRs. d, A 3D classification result from the dynein-tail/dynactin/BICDR dataset which shows density for BICDR-B. e, The density map of dynein-tail/dynactin/BICDR (left) (EMD: 4168)4 and our consensus density map from the high-magnification dataset (right). Both maps were low-pass filtered to 10 Å and are shown at contour levels where the dynactin density is similar. f, A 3D classification of dynein-tail/dynactin/Hook34 showing that the second coiled-coil belongs to a second Hook3.
Extended Data Fig. 7. CC1 box interactions and the HBS1 motif of Hook3.
a, The LIC helix after fitting to the density on the inside face of the BICDR-A CC1 box relative to the registry of BICDR in our structure (left), highlighting the conserved A116, A117, and G120 of the motif. On the right is a similar view of the BICD2-LIC crystal structure (PDB: 6PSE)9. b, A 3D classification result showing the LIC of dynein-A2 connecting to the inside face of BICDR-A. c, A 3D classification result showing the LIC of dynein-A2 connecting to the inside face of BICDR-B. d, Sequence alignment of the HBS1 motif of BICDR (annotated as BICL1) and Hook3, highlighting the conserved residues and C-terminal glutamates. The UniProt codes are indicated on the left. e, An Alphafold prediction of two copies of Hook3 (fragment 172-287), the dynein heavy chain (fragment 576-864), and intermediate chain (fragment 226-583). In the middle, the PAE is displayed on the models relative to H200 (yellow), withlower values representing higher confidence. The full PAE plot is shown on the right, with the arrow pointing at H200.
Extended Data Fig. 8. Pointed end interactions and adaptor families.
a, An overlay of our structure with the previous dynein-tail/dynactin/BICDR structure (PDB: 6F1T)4, aligned at dynactin. The pointed end interaction sites are labelled 1 to 413. b, An Alphafold prediction of the pointed end complex (Arp11, p25, p27, and p62) and a C-terminal fragment of BICDR (205-394) that includes the Spindly motif. The models are coloured based on the predicted aligned error (PAE) at L347 of BICDR (yellow), with lower values representing higher confidence. The full PAE plot is also shown with the arrow pointing at L347. c, Alphafold predictions of cargo adaptors that have been manually linearized such that the coiled-coils are parallel to each other (i.e. each individual model was manually rotated at the disordered loops). White triangles depict breaks in the coiled-coil prediction preceding the Spindly motifs. The predictions are of full-length proteins unless otherwise stated. LIC-binding motifs (CC1 box, Hook domain, EF hand, RH1 domain), HBS1s, and Spindly motifs are coloured according to the legend. Only the HBS1s of BICDR, BICD2, Spindly, TRAK, and Hook3 are shown based on our analyses and previous predictions10. The orientations are C-terminus to N-terminus to match the orientation in other figures. d, The predicted local distance difference tests (pLDDTs) (left) from one chain of each of the cargo adaptors around the Spindly motif (highlighted in red) and the full PAE plots (right). White triangles point to the locations of the predicted breaks in the coiled-coils.
Extended Data Table. 1. Cryo-EM data collection, refinement, and validation statistics of the dynactin/dynein-tail regions.
| Pointed end/Spindly motif (EMDB-14559) (PDB 7Z8M) | BICDR-A Dynein-A2 Tail (EMDB-14553) (PDB 7Z8J) | Barbed end/BICDR-A (EMDB-14552) (PDB 7Z8I) | BICDR-B/Dynein-A1 Tail (EMD-14555) (PDB 7Z8K) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 81000 | 81000 | 81000 | 81000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e-/Å2) | 53 | 53 | 53 | 53 |
| Defocus range (μm) | 1.2 – 3.6 | 1.2 – 3.6 | 1.2 – 3.6 | 1.2 – 3.6 |
| Pixel size (Å) | 1.2445 | 1.2445 | 1.2445 | 1.2445 |
| Symmetry imposed | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 506853 | 506853 | 506853 | 506853 |
| Final particle images (no.) | 165019 | 78061 | 179660 | 39644 |
| Map resolution (Å) | 3.37 | 3.93 | 3.30 | 4.37 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | ||||
| Model resolution (Å) | 3.60 | 3.93 | 3.58 | 8.50 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | -55 | -100 | -55 | -67 |
| Model composition | ||||
| Non-hydrogen atoms | 17694 | 20191 | 43427 | 14475 |
| Protein residues | 2272 | 2487 | 5372 | 2918 |
| Ligands | ATP: 1 ADP: 3 MG: 4, ZN: 3 | ADP: 6, MG: 6 | ||
| B factors (Å2) | ||||
| Protein | 1.18/146.42/53.06 | 6.17/181.83/75.67 | 0.00/189.12/33.72 | 0.00/986.10/158.63 |
| Ligand | 32.92/112.93/44.97 | 7.41/30.72/16.52 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.005 (0) | 0.007 (0) | 0.005 (0) | 0.005 (0) |
| Bond angles (°) | 1.112 (0) | 1.141 (3) | 1.087 (1) | 1.111 (0) |
| Validation | ||||
| MolProbity score | 1.35 | 1.21 | 1.10 | 0.73 |
| Clashscore | 2.90 | 3.20 | 2.50 | 0.67 |
| Poor rotamers (%) | 0.27 | 0.78 | 0.13 | 0.00 |
| Ramachandran plot | ||||
| Favored (%) | 96.08 | 97.51 | 97.67 | 97.96 |
| Allowed (%) | 3.92 | 2.49 | 2.33 | 2.01 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.03 |
Table 2. Cryo-EM data collection, refinement, and validation statistics for the dynein motor domain regions.
| Dynein Motor domain (EMDB-14550) (PDB 7Z8G) | Dynein AAA1-2-3 (EMDB-14551) (PDB 7Z8H) | Dynein-A2/B1 (EMDB-14556) (PDB 7Z8L) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 81000 | 81000 | 81000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e-/Å2) | 53 | 53 | 53 |
| Defocus range (μm) | 1.2 – 3.6 | 1.2 – 3.6 | 1.2 – 3.6 |
| Pixel size (Å) | 1.2445 | 1.2445 | 1.2445 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 506853 | 506853 | 506853 |
| Final particle images (no.) | 351879 | 53424 | 105050 |
| Map resolution (Å) | 3.52 | 3.41 | 4.90 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Refinement | |||
| Model resolution (Å) | 3.82 | 3.60 | 6.99 |
| FSC threshold | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | -55 | -100 | -55 |
| Model composition | |||
| Non-hydrogen atoms | 24656 | 8811 | 16513 |
| Protein residues | 3047 | 1083 | 3311 |
| Ligands | ATP: 1 ADP: 1 AMPPNP: 2, MG: 1 | ATP: 1 ADP: 1 AMPPNP: 1, MG: 1 | ATP: 1 ADP: 1 AMPPNP: 2, MG: 1 |
| B factors (Å2) | |||
| Protein | 8.85/113.21/47.93 | 8.85/113.21/47.93 | 35.76/152.59/65.90 |
| Ligand | 8.41/57.20/36.98 | 8.41/57.20/36.98 | 47.45/57.15/51.63 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.005 (0) | 0.005 (0) | 0.012(2) |
| Bond angles (°) | 1.073 (0) | 1.128 (0) | 1.536 (3) |
| Validation | |||
| MolProbity score | 1.20 | 1.29 | 0.61 |
| Clashscore | 3.41 | 3.71 | 0.29 |
| Poor rotamers (%) | 0.27 | 0.10 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 97.70 | 97.31 | 98.79 |
| Allowed (%) | 2.30 | 2.69 | 1.21 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 |
Acknowledgements
We thank S. Scheres for help with micrograph-level signal subtraction in Relion; C.K. Lau for helpful discussions; the MRC Laboratory of Molecular Biology Electron Microscopy Facility for access and support of electron microscopy sample preparation and data collection; J. Grimmett and T. Darling for providing scientific computing resources; H.E. Foster and C. Ventura Santos for help with cryo-ET; F. Abid Ali, K. Singh, and C.K. Lau for critical reading of the manuscript. This work was supported by Wellcome [210711/Z/18/Z], the Medical Research Council, as part of United Kingdom Research and Innovation (also known as UK Research and Innovation) [MRC file reference number MC_UP_A025_1011], and the EMBO Postdoctoral Fellowship [ALTF 334-2020] to S.C. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising.
Footnotes
Author Contributions
S.C. performed experiments/analysis and prepared figures. S.C. and A.P.C. conceived the project and wrote the manuscript.
Competing Interest
The authors declare no competing interests.
Data Availability
Atomic coordinates and cryo-EM maps have been deposited in the Protein Data Bank (PDB) / Electron Microscopy Data Bank (EMDB) under accession codes 7Z8F/14549 (composite dynein-dynactin-BICDR structure), 7Z8G/14550 (dynein motor domain), 7Z8H/14551 (dynein AAA1-3), 7Z8I/14552 (Barbed end/BICDR-A), 7Z8J/14553 (BICDR-A/dynein-A2), 7Z8K/14555 (BICDR-B/dynein-A1), 7Z8L/14556 (dynein motor domain/LIC), 7Z8M/14559 (pointed end/BICDR-A), and 15396 (consensus map). Other atomic coordinates used in this study for alignments and model building are available in the PDB (2PG1, 3VKG, 4AKG, 4W8F, 5NUG, 6F1T, 6RZB, 6PSE, 6ZNL, and 7K58). The cryo-EM map of dynein-tail/dynactin/BICDR1 used for comparisons is available in the EMDB (4168). The protein sequences used for sequence alignment and Alphafold predictions are available the Universal Protein Resource (UniProt) under accession codes Q13409, O43237, Q9UJW0, O00399, Q9BTE1, I3LHK5, A0JNT9, Q8TD16, Q96EA4, Q9UPV9, Q86VS8, O75154, Q9BSW2, Q9UPT6, Q96NA2, Q14204, Q6ZP65, A0A140LGI1, Q8BUK6, and Q6G73.
Code availability
Custom scripts, including the Starparser package, are available at https://github.com/sami-chaaban (10.5281/zenodo.6792794, 10.5281/zenodo.6792805, 10.5281/zenodo.6792801).
References
- 1.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Splinter D, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urnavicius L, et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 2018;554:202–206. doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grotjahn DA, et al. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat Struct Mol Biol. 2018;25:203–207. doi: 10.1038/s41594-018-0027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elshenawy MM, et al. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat Chem Biol. 2019;15:1093–1101. doi: 10.1038/s41589-019-0352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reck-Peterson SL, Redwine WB, Vale RD, Carter AP. The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol. 2018;19:382–398. doi: 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olenick MA, Holzbaur ELF. Dynein activators and adaptors at a glance. J Cell Sci. 2019;132:jcs227132. doi: 10.1242/jcs.227132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee I-G, et al. A conserved interaction of the dynein light intermediate chain with dynein-dynactin effectors necessary for processivity. Nat Commun. 2018;9:986. doi: 10.1038/s41467-018-03412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacristan C, et al. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol. 2018;20:800–810. doi: 10.1038/s41556-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celestino R, et al. A transient helix in the disordered region of dynein light intermediate chain links the motor to structurally diverse adaptors for cargo transport. PLOS Biology. 2019;17:e3000100. doi: 10.1371/journal.pbio.3000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gama JB, et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J Cell Biol. 2017;216:943–960. doi: 10.1083/jcb.201610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau CK, et al. Cryo-EM reveals the complex architecture of dynactin’s shoulder region and pointed end. EMBO J. 2021;40:e106164. doi: 10.15252/embj.2020106164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhabha G, et al. Allosteric communication in the dynein motor domain. Cell. 2014;159:857–868. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat Struct Mol Biol. 2015;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas MP, et al. Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proc Natl Acad Sci U S A. 2015;112:6371–6376. doi: 10.1073/pnas.1417422112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldtzvik Y, Mugnai ML, Thirumalai D. Dynamics of Allosteric Transitions in Dynein. Structure. 2018;26:1664–1677.:e5. doi: 10.1016/j.str.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elshenawy MM, et al. Lis1 activates dynein motility by modulating its pairing with dynactin. Nat Cell Biol. 2020;22:570–578. doi: 10.1038/s41556-020-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Htet ZM, et al. LIS1 promotes the formation of activated cytoplasmic dynein-1 complexes. Nat Cell Biol. 2020;22:518–525. doi: 10.1038/s41556-020-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I-G, Cason SE, Alqassim SS, Holzbaur ELF, Dominguez R. A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat Commun. 2020;11:5695. doi: 10.1038/s41467-020-19538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma M, et al. Structure of the Decorated Ciliary Doublet Microtubule. Cell. 2019;179:909–922.:e12. doi: 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walton T, Wu H, Brown A. Structure of a microtubule-bound axonemal dynein. Nat Commun. 2021;12:477. doi: 10.1038/s41467-020-20735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo S, et al. Remodeling and activation mechanisms of outer arm dyneins revealed by cryo-EM. EMBO Rep. 2021;22:e52911. doi: 10.15252/embr.202152911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao Q, et al. Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism. Nat Struct Mol Biol. 2021;28:799–810. doi: 10.1038/s41594-021-00656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook AD, Manka SW, Wang S, Moores CA, Atherton J. A microtubule RELION-based pipeline for cryo-EM image processing. Journal of Structural Biology. 2020;209:107402. doi: 10.1016/j.jsb.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimanius D, Dong L, Sharov G, Nakane T, Scheres SHW. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochemical Journal. 2021;478:4169–4185. doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kon T, et al. The 2.8 Â crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 30.DeSantis ME, et al. Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell. 2017;170:1197–1208.:e12. doi: 10.1016/j.cell.2017.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu R, Zhang J, Rotty JD, Xiang X. Dynein activation in vivo is regulated by the nucleotide states of its AAA3 domain. Current Biology. 2021;31:4486–4498.:e6. doi: 10.1016/j.cub.2021.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder CM, Ostrem JML, Hertz NT, Vale RD. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. Elife. 2014;3:e03351. doi: 10.7554/eLife.03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter AP. Crystal clear insights into how the dynein motor moves. J Cell Sci. 2013;126:705–713. doi: 10.1242/jcs.120725. [DOI] [PubMed] [Google Scholar]
- 34.Zhong ED, Bepler T, Berger B, Davis JH. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods. 2021;18:176–185. doi: 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.d’Amico E, et al. Conformational transitions of the mitotic adaptor Spindly underlie its interaction with Dynein and Dynactin. 2022:2022.02.02.478874. doi: 10.1083/jcb.202206131. at https://www.biorxiv.org/content/10.1101/2022.02.02.478874v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans R, et al. Protein complex prediction with AlphaFold-Multimer. 2021:2021.10.04.463034. at https://www.biorxiv.org/content/10.1101/2021.10.04.463034v1. [Google Scholar]
- 37.Chai P, Rao Q, Zhang K. Multi-curve fitting and tubulin-lattice signal removal for structure determination of large microtubule-based motors. 2022:2022.01.22.477366. doi: 10.1016/j.jsb.2022.107897. at https://www.biorxiv.org/content/10.1101/2022.01.22.477366v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata K, et al. A Single Protofilament Is Sufficient to Support Unidirectional Walking of Dynein and Kinesin. PLOS ONE. 2012;7:e42990. doi: 10.1371/journal.pone.0042990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder CM, Vale RD. Assembly and activation of dynein–dynactin by the cargo adaptor protein Hook3. J Cell Biol. 2016;214:309–318. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlager MA, et al. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 2010;29:1637–1651. doi: 10.1038/emboj.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielska E, et al. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock MA, et al. RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. eLife. 2018;7:e36312. doi: 10.7554/eLife.36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock WO. Bidirectional cargo transport: moving beyond tug of war. Nat Rev Mol Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu M, Holzbaur ELF. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenton AR, Jongens TA, Holzbaur ELF. Mitochondrial adaptor TRAK2 activates and functionally links opposing kinesin and dynein motors. Nat Commun. 2021;12:4578. doi: 10.1038/s41467-021-24862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canty JT, Hensley A, Yildiz A. TRAK adaptors coordinate the recruitment and activation of dynein and kinesin to control mitochondrial transport. 2021:2021.07.30.454553. doi: 10.1038/s41467-023-36945-8. at https://www.biorxiv.org/content/10.1101/2021.07.30.454553v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai A, et al. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell. 2016;164:722–734. doi: 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belyy V, et al. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat Cell Biol. 2016;18:1018–1024. doi: 10.1038/ncb3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell. 2017;169:1303–1314.:e18. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 51.Pierson GB, Burton PR, Himes RH. Alterations in number of protofilaments in microtubules assembled in vitro. J Cell Biol. 1978;76:223–228. doi: 10.1083/jcb.76.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner T, et al. Two particle-picking procedures for filamentous proteins: SPHIRE-crYOLO filament mode and SPHIRE-STRIPER. Acta Crystallogr D Struct Biol. 2020;76:613–620. doi: 10.1107/S2059798320007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:1–13. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warshamanage R, Yamashita K, Murshudov GN. EMDA: A Python package for Electron Microscopy Data Analysis. J Struct Biol. 2021;214:107826. doi: 10.1016/j.jsb.2021.107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kellogg EH, et al. Insights into the Distinct Mechanisms of Action of Taxane and Non-Taxane Microtubule Stabilizers from Cryo-EM Structures. J Mol Biol. 2017;429:633–646. doi: 10.1016/j.jmb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Redwine WB, et al. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lacey SE, He S, Scheres SH, Carter AP. Cryo-EM of dynein microtubulebinding domains shows how an axonemal dynein distorts the microtubule. eLife. 2019;8:e47145. doi: 10.7554/eLife.47145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mirdita M, et al. ColabFold: making protein folding accessible to all. Nat Methods. 2022:1–4. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirdita M, Steinegger M, Söding J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics. 2019;35:2856–2858. doi: 10.1093/bioinformatics/bty1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki Y, Shimizu T, Morii H, Tanokura M. Hydrolysis of AMPPNP by the motor domain of ncd, a kinesin-related protein. FEBS Lett. 1997;409:29–32. doi: 10.1016/s0014-5793(97)00472-9. [DOI] [PubMed] [Google Scholar]
- 70.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three-Dimensional Image Data Using IMOD. Journal of Structural Biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 73.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams JC, et al. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proceedings of the National Academy of Sciences. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Â crystal structure. Nat Struct Mol Biol. 2012;19:492–497.:S1. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vincent TL, Green PJ, Woolfson DN. LOGICOIL—multi-state prediction of coiled-coil oligomeric state. Bioinformatics. 2013;29:69–76. doi: 10.1093/bioinformatics/bts648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates and cryo-EM maps have been deposited in the Protein Data Bank (PDB) / Electron Microscopy Data Bank (EMDB) under accession codes 7Z8F/14549 (composite dynein-dynactin-BICDR structure), 7Z8G/14550 (dynein motor domain), 7Z8H/14551 (dynein AAA1-3), 7Z8I/14552 (Barbed end/BICDR-A), 7Z8J/14553 (BICDR-A/dynein-A2), 7Z8K/14555 (BICDR-B/dynein-A1), 7Z8L/14556 (dynein motor domain/LIC), 7Z8M/14559 (pointed end/BICDR-A), and 15396 (consensus map). Other atomic coordinates used in this study for alignments and model building are available in the PDB (2PG1, 3VKG, 4AKG, 4W8F, 5NUG, 6F1T, 6RZB, 6PSE, 6ZNL, and 7K58). The cryo-EM map of dynein-tail/dynactin/BICDR1 used for comparisons is available in the EMDB (4168). The protein sequences used for sequence alignment and Alphafold predictions are available the Universal Protein Resource (UniProt) under accession codes Q13409, O43237, Q9UJW0, O00399, Q9BTE1, I3LHK5, A0JNT9, Q8TD16, Q96EA4, Q9UPV9, Q86VS8, O75154, Q9BSW2, Q9UPT6, Q96NA2, Q14204, Q6ZP65, A0A140LGI1, Q8BUK6, and Q6G73.
Custom scripts, including the Starparser package, are available at https://github.com/sami-chaaban (10.5281/zenodo.6792794, 10.5281/zenodo.6792805, 10.5281/zenodo.6792801).