Abstract
Evolutionary development of the human brain is characterized by the expansion of various brain regions. Here, we show that developmental processes unique to humans are responsible for malformations of cortical development (MCDs), which result in developmental delay and epilepsy in children. We generate a human cerebral organoid model for Tuberous Sclerosis (TSC) and identify a specific neural stem cell type, CLIP cells. In TSC, CLIP cells over-proliferate, generating excessive interneurons, brain tumors, and cortical malformations. EGF receptor inhibition reduces tumor burden, identifying potential treatment options for TSC and related disorders. The identification of CLIP cells reveals the extended interneuron generation in the human brain as a vulnerability for disease. In addition, this work demonstrates that analyzing MCDs can reveal fundamental insights into human-specific aspects of brain development.
Malformations of cortical development (MCDs) comprise varied neurodevelopmental disorders that cause over 40% of medically refractory childhood seizures (1). Several MCDs including hemimegalencephaly, focal cortical dysplasia IIb, and tuberous sclerosis (TSC) are caused by mutations in mTOR pathway members, but their disease mechanisms remain elusive. TSC is a rare autosomal dominant disorder caused by mutation of either TSC1 (hamartin) or TSC2 (tuberin), which form a complex and inhibit the mTOR kinase. Patients suffer from debilitating, often drug resistant neuropsychiatric symptoms, including intractable epileptic seizures, autism spectrum disorder (ASD), and intellectual disability (ID) (2). Most patients have focal dysplastic regions (cortical tubers) in the cortex, which consist of dysmorphic neurons, giant cells, and dysmorphic astrocytes (3, 4). In addition, 80% of patients display subependymal nodules (SEN), benign tumors that form along the proliferative niches at the lateral ventricle and can develop into subependymal giant cell astrocytomas (SEGAs) (5). Analysis of human primary tissues suggested a common cell-of-origin for cortical tubers and SEN/SEGAs based on shared transcriptomic alterations (6); however, the nature of this cell remains unclear. In mice, TSC pathogenesis is initiated by inactivation of the second allele of either TSC1 or TSC2 (7–12), and similar results have been obtained in spheroids (13). Genetic analysis in patients, however, revealed loss of the second allele in most SEN/SEGA, but only few cortical tubers (6, 14–16), challenging the previously suggested two-hit model (7, 17). We hypothesized that these inconsistencies arise because cell types and processes specific to the human brain are critical for disease initiation. To identify those human-specific features, we generated human cerebral organoids (18) from patient-derived induced pluripotent stem cells (iPSCs) and compared our results to human primary material.
Cerebral organoids recapitulate TSC
To model the brain pathology of TSC, we derived iPSCs from patients with known TSC2 mutations who suffer from drug-resistant epilepsy and show cortical tubers and subependymal tumors (Fig. 1A and Fig. S1A-C). Isogenic TSC2+/+ lines were acquired directly from the germline mosaic first patient and generated by scarless Crispr-based genome editing for the second patient (Fig. S1D, E and H). Both patient mutations result in an early stop codon in regions commonly mutated in TSC (Fig. S1F and G). To study subependymal tumors, we cultured organoids in a high-nutrient (H-) medium that promotes proliferation (Fig. 1B, indicated by pictogram with H in staining panels). To examine the formation of cortical tubers, which emerge in less proliferative cortical regions, we transferred organoids to a low-nutrient (L-) medium adapted from a published formulation (19) to 3D culture (Fig. 1B, indicated by pictogram with L in staining panels; see Materials and Methods for details).
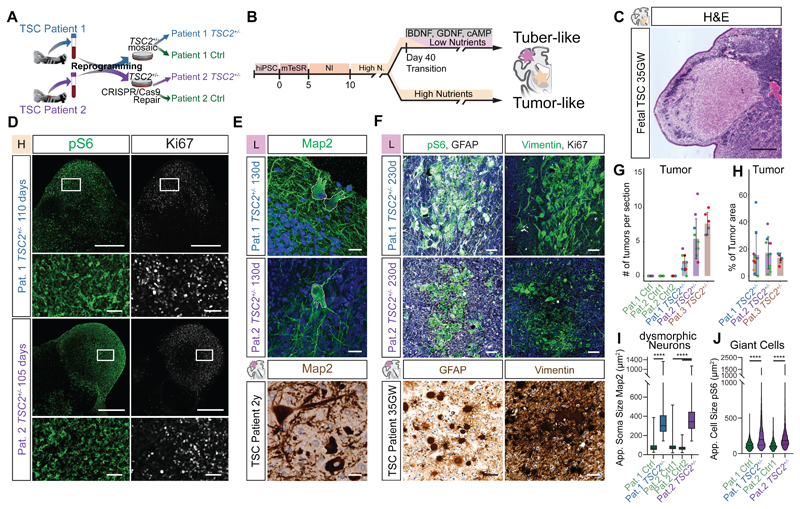
Fig 1. TSC2+/--derived organoids recapitulate histopathology of TSC.
A Ctrl and TSC2+/- cell lines derived from two patients (see Materials and Methods for details).
B High- and Low-nutrient organoid protocols used to model distinct TSC phenotypes.
C H&E staining of 35GW fetal brain depicts histopathology of a fetal SEN.
D pS6 and Ki67 staining on 110- and 105-day-old TSC2+/--derived organoids in high-nutrient medium identifies SEN-like structures. Lower panel shows higher magnification of inset.
E Map2 staining on 130-day-old organoids in L-medium shows dysmorphic neurons (top), with comparable morphology to those in a resected tuber of a 2-year-old patient (bottom). Nuclear counterstain with DAPI or hematoxylin.
F pS6 and GFAP identifies GCs in 230-day-old organoids comparable to GCs in patient tubers. Giant cells in organoids express Vimentin, as shown in patients. GCs can be distinguished from tumors by their lower expression of Ki67. Nuclear counterstain with DAPI or hematoxylin.
G Tumors identified as pS6- and Ki67-positive areas are found in TSC2+/--derived organoids of all three patients. Control organoids of patient 1 and two clones of repaired patient 2 showed no tumors. Color of dots mark independent batches of experiments (See Fig. S3E for summary of replicates).
H Percentage of tumor area of the total organoid area reveals similar tumor burden for organoids derived from three TSC2+/- patients. Color of dots mark independent batches of experiments (See Fig. S3F for summary of replicates).
I. Area of the soma in Map2+ neurons shows that dysmorphic neurons in TSC2+/--derived organoids are roughly 4-fold larger then Map2+ neurons in control organoids (Pat.1 Ctrl vs. TSC2+/- p<0.0001; Pat.2 Ctrl1 vs. TSC2+/- and Pat.2 Ctrl2 vs. TSC2+/- both p<0.0001; test: Ordinary One-Way ANOVA; See Fig. S4D for summary of replicates).
J. Cell area of pS6-positive cells shows enlarged pS6 cells in TSC2+/--derived organoids. (Pat.1 Ctrl vs.TSC2+/- p<0.0001, Pat.2 Ctrl vs. TSC2+/- p<0.0001, Pat.1 Ctrl vs. Pat.2 Ctrl p>0.9999, Pat.1 TSC2+/- vs. Pat.2 TSC2+/- p>0.9999; Kruskal-Wallis test with Dunn's multiple comparisons test; see Fig. S4E for summary of replicates)
Scale bars: C and D: 500μm; E: 20μm; inset D and F: 50μm
We found no obvious differences between genotypes within the first 90 days of culture (Fig. S2) corresponding to early phases of neurodevelopment, consistent with previous results (13). 110 days after embryoid body (EB) formation, however, nodular aggregates of cells expressing the proliferative marker Ki67 and the mTOR activation marker phospho-S6 (pS6) formed in TSC2+/- organoids cultured in H-medium (TSC2+/- H-organoids) (Fig. 1D, G, H and Fig. S3A to F). These structures morphologically resembled SENs (20–22) (Fig. 1C). We validated the emergence of SEN-like tumors in organoids derived from a third TSC patient (Fig. 1G, H, S3C and D). SEN/SEGAs have been proposed to originate from an uncharacterized population of neural stem cells (NSCs) (23, 24). To test for an NSC origin of SEN-like tumors in organoids we stained for NSC markers. Expression of Nestin, ASCL1, and SOX2 (Fig. S3B, G and H) demonstrated the NSC identity of SEN-like tumors in organoids.
To determine whether we could recapitulate the pathological cell types found in cortical tubers, we analyzed organoids cultured for 120-150 days in L-medium. In organoids derived from TSC2+/- cells we found neurons with an enlarged soma and thickened processes similar to dysmorphic neurons in cortical tubers (Fig. 1E, I and Fig. S4A-D). After prolonged maturation in L-medium (~230 days), clusters of enlarged pS6 positive cells appeared (Fig. 1F, J and Fig. S4E-I). The morphology and expression of markers like GFAP and Vimentin were reminiscent of giant cells (GCs) (3, 5), which had characteristically low proliferation rates (Fig. 1F).
Dysmorphic astrocytes, a cell type previously identified in patient tubers, share marker expression with GCs but are morphologically distinct (4). In organoids, we identified individual enlarged GFAP expressing cells morphologically similar to dysmorphic astrocytes with characteristic thickened and prolonged processes (Fig. S4J). GCs expressed the neural progenitor marker Nestin (24) (Fig. S4F) and almost all enlarged GFAP cells (GCs and dysmorphic astrocytes) in organoids expressed SOX2 suggesting a neural progenitor identity (Pat. 1: 99.7%, Pat. 2: 96%, Fig. S4J and K).
Both cortical and tumor lesions were detected in H- and L-medium organoids (Fig. S3I and S4L); however, the use of two different culturing conditions favored the emergence of the two specific phenotypes. Thus, organoids derived from TSC2+/- hiPSCs recapitulate the major histopathological features found in the brain of TSC patients.
Interneuron progenitors in TSC tumors
In order to characterize the cellular composition of tumors in TSC organoids, we performed single cell transcriptomic analysis on 220-day-old organoids grown in H-medium. At this age, organoids consist almost exclusively of tumor tissues (Fig. S5A). To compare inter-tumoral heterogeneity, three organoids were dissected into three tumor regions each and barcoded separately (25). Unsupervised clustering in UMAP projection identified four main clusters: interneurons (Cl. 1), interneuron progenitors (Cl. 2), dividing interneuron progenitors (Cl. 3), and excitatory neurons (Cl. 4) (Fig. 2A). Interneurons were characterized by the expression of canonical regulators of interneuron development like DLX2, DLX5, and DLX6-AS1 (Fig. 2C, S5B). Interneuron progenitors expressed DLX2, EGFR, and progenitor markers like HES1, SLC1A3, or VIM (Fig. 2C, S5B). Dividing progenitors of the interneuron lineage were characterized by additionally expressing markers like MKI67 and TOP2A (Fig. 2C, S5B). Only a very small number of excitatory lineage cells (Cl. 4) were detected (3%, Fig. 2B), marked by the expression of NEUROD2 and NEUROD6 (Fig. S5B). Thus, tumors in TSC2+/- H-organoids consist mainly of progenitors and interneurons of ventral origin (Fig. 2C).
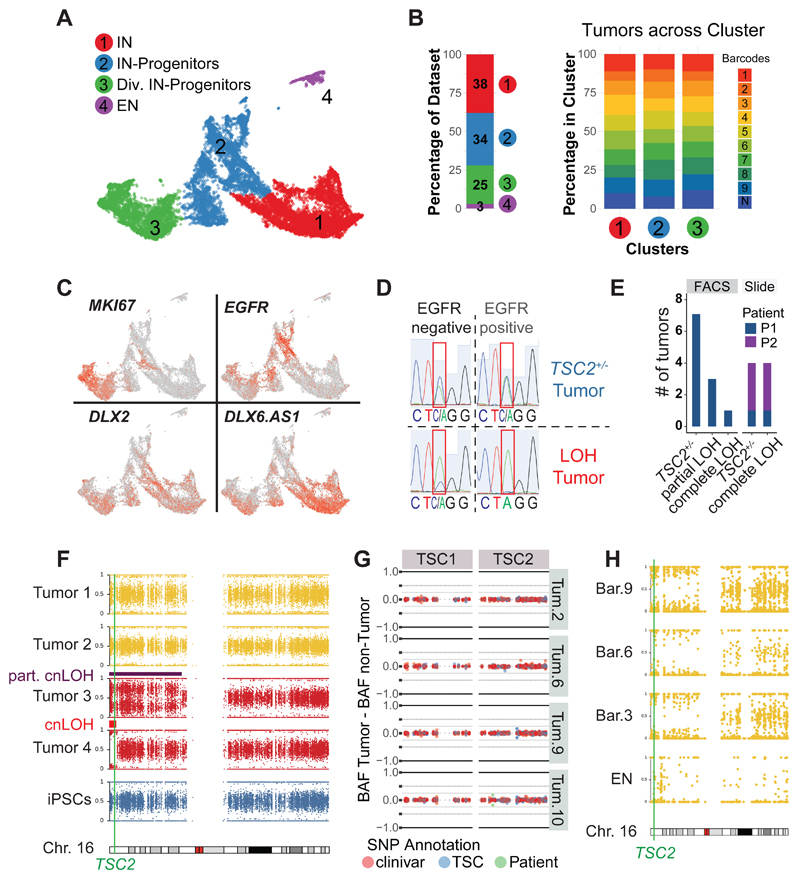
Fig 2. TSC tumors consist of interneuron progenitors and acquire cnLOH during progression.
A UMAP projection of cells isolated from 220-day-old TSC tumor organoids identified four main clusters: interneurons (Cl. 1), interneuron progenitors (Cl. 2), dividing interneuron progenitors (Cl. 3) and excitatory neurons (Cl. 4).
B Dataset composition and contribution of different tumor regions. 97% of cells in tumors were ventral cells and all nine tumor regions of three organoids were similarly distributed across ventral clusters (Cl. 1, 2 and 3).
C Expression of genes specific for dividing cells (MKI67) and interneurons (DLX2 and DLX6.AS1) in 220-day-old TSC tumors. EGFR was specifically expressed in TSC tumor progenitors.
D Genotyping example of FACS sorted tumor (EGFR+) and non-tumor (EGFR-) population, showing a heterozygous tumor and a LOH (loss-of-heterozygosity) tumor that lost the WT-allele (C) within the tumor.
E Genotyping of tumors of patient 1 in FACS sorted samples showing 7 out of 11 TSC tumors were heterozygous, three showed a partial LOH, and one tumor showed a full LOH. Genotyping tumors from stained slides confirmed the presence of heterozygous and LOH tumors (patient 1: one TSC2+/- tumor, one LOH tumor; patient 2: three TSC2+/- tumors, three LOH tumors)
F B-Allele frequency (BAF) of chromosome 16 for four tumors of Pat. 1 TSC2+/--derived organoids. While tumor 1 and 2 remain heterozygous, a shift in BAF is seen in tumors 3 and 4 compared to iPSCs. See Fig. S6L for BAF and LRR. Note that tumor 2 showed a partial cnLOH of a large section, while tumor 4 had a small complete cnLOH at the beginning of chromosome 16. Both cnLOH regions included the TSC2 gene.
G Targeted amplification of TSC1 and TSC2 in four tumors and matched non-tumor samples of patient 1. Detected SNPs are colored as annotation of clinivar database (red), LOVD TSC database (blue) or disease-causing SNP of patient 1 (green). The difference between BAF of the tumor and matched non-tumor sample is shown. No disease-causing SNPs increased in the tumor samples could be detected. Note that in tumor 10 there was a small shift of the patient SNP, confirmed as partial cnLOH (Fig. S6I).
H SNP mapping of scRNA seq data shown in panel A. Cells from tumors with sufficient reads (barcodes 3, 6 and 9) and excitatory neurons were aggregated per group and allelic frequencies were determined. All tumors showed cnLOH, while excitatory neurons (cluster 4, Fig 2A) remained heterozygous.
To investigate inter-tumoral heterogeneity we compared the barcoded tumor regions. The cell type composition was highly consistent among the ventral lineage clusters with all barcodes being evenly distributed (Fig. 2B and Fig. S5C, D). Whereas in the traditional view of tumorigenesis where TSC inactivation of the second allele is thought to be a prerequisite for causing the disease (7), our data suggest that a specific cell type that is sensitive to levels of mTOR signaling gives rise to TSC tumors.
Copy-neutral loss-of-heterozygosity in TSC tumors
To determine whether bi-allelic inactivation is required for the initiation of tumor lesions, we tested the mutational status of TSC2+/--derived organoids grown in H-medium at earlier stages (135 to 160-days). Tumor cells were isolated from patient 1 mutant organoids by fluorescent activated cell sorting (FACS) for EGFR (Fig. S6A-D and F-H), which was highly expressed on the interneuron progenitor cells in dissected tumors (Fig. 2C, S6E). Genotyping of the TSC2 locus indicated that many tumors remained heterozygous (Fig. 2D, E and Fig. S6I-K). In organoids from patient 1 we identified complete LOH in one tumor and partial LOH in three tumors (Fig. 2E, Fig. S6I).
Additionally, we genotyped tumor sections and confirmed LOH in a subset of tumors of patient 1 and 2 (Fig. 2E, S6J and K).
To analyze recombination events in TSC tumors, we performed whole genome sequencing (WGS) on two heterozygous and two LOH tumor samples (Fig. 2F, Fig. S6L and M). WGS revealed no major genomic rearrangements in heterozygous tumors (Fig. 2F, S6L and M). In contrast, in LOH tumors extensive regions of chromosome 16, ranging from the telomere to and beyond the TSC2 locus, had become homozygous by copy-neutral loss-of-heterozygosity (cnLOH, Fig. 2F, S6L and M), the same genomic event resulting in LOH in TSC patients (6).
To test whether heterozygous tumors acquired second hit mutations we performed targeted amplification of TSC1 and TSC2 on four tumors and matched controls of patient 1 (Fig. 2G). No other pathogenic SNPs were increased in tumor samples (Fig. 2G). Thus, a second hit at the TSC1 or TSC2 locus is not required for tumor initiation. To probe if cnLOH occurred at later stages during tumor progression, we investigated allelic frequencies in the 220-day-old scRNA data (Fig. 2H). Tumor cells aggregated per barcode showed cnLOH in all tumors (Fig. 2H). Notably, excitatory neurons (cluster 4 Fig. 2A), did not show cnLOH, further supporting an interneuron origin of TSC tumors (Fig. 2H). Thus, tumors in TSC organoids initiated from a heterozygous interneuron progenitor and acquired cnLOH only during progression.
To investigate if cnLOH was required for the formation of cortical tuber-like structures, we analyzed giant cells in 230-day-old TSC2+/- organoids. TSC2 protein expression was detected in over 98% of giant cells using an antibody recognizing only the wild type TSC2 variant (patient 1, 98.4%; patient 2, 98.7%: Fig. S7A-C). TSC2 protein was also expressed in giant cells in fetal cortical tubers (Fig. S7D), consistent with previous data (26, 27). This suggests that second-hit events are not a prerequisite for tuber formation. Thus, although previous studies in mice have defined loss-of-heterozygosity in Tsc1 (8, 9, 11, 12, 28) or Tsc2 (10) as a requirement for TSC-like phenotypes, our data demonstrate that in human tissues, bi-allelic inactivation is dispensable for disease initiation. Our observations are consistent with reports that identify bi-allelic inactivation in subependymal tumors but rarely in cortical tubers (6, 14–16, 29, 30).
Given that tumorigenesis in TSC organoids did not require cnLOH, we hypothesized that low levels of TSC1/2 complex could sensitize interneuron progenitors to further reduction of TSC1 or TSC2. We observed reduced TSC2 levels in EGFR positive interneuron progenitors in both Ctrl and in TSC2+/--derived organoids from both patients grown in H-medium (Fig. S7E to H) using immunofluorescence. To quantitate TSC1 and TSC2 protein levels we performed targeted parallel reaction monitoring mass spectrometry (tPRM-MS) on FACS sorted samples from patient 1 in H-medium. Both in Ctrl and TSC2+/--derived organoids, TSC1 and TSC2 levels were lower in EGFR-positive samples than in EGFR-negative samples (Fig. S7J). Comparing EGFR-positive populations, we observed that while TSC1 was expressed at similar levels, TSC2 was significantly more down-regulated in EGFR-positive cells in the TSC2 mutant compared to the Ctrl population (Fig. S7K). Thus, although in Ctrl organoids both components of the TSC complex were equally reduced, in TSC tumor cells loss of one functional TSC2 allele led to disproportional reduction of TSC2. These data suggest that interneuron progenitors have low levels of TSC proteins, which could sensitize them to heterozygous mutations in TSC genes.
The developmental trajectories of tumors and tubers
To determine whether tumors and tubers have a common cell-of-origin we investigated H- and L-organoids at 110 days, when the TSC phenotypes are beginning to emerge (Fig. S8E to G). We integrated these data with the 220-day-old TSC tumor dataset (Fig. 3A). Unsupervised clustering in UMAP projection identified dorsal progenitor cells (Cl. 3, 10, 12), excitatory neurons (Cl.1, 2, 4, and14), interneuron progenitor cells, and interneurons (Cl. 5-9, 11, 13, 16, 17), and cells resembling pre-oligodendrocyte-progenitor (OPC)-like cells (31) (Cl. 15) (Fig. 3A, Fig. S8B and C). The 220-day-old TSC tumors contributed almost exclusively to the clusters containing interneuron progenitor cells and interneurons (Cl. 5-9, 11, 13, Fig. 3B). The same clusters were more abundant in 110-day-old TSC2+/- H- and L-organoids compared with control organoids: TSC2+/- H-organoids had more progenitor cells (Cl. 5, 7, 11, 13, Fig. 3B), whereas in TSC2+/- L-organoids mature interneurons were substantially increased (Cl. 9, 16, Fig. 3B). Pre-OPC-like cells (Cl. 15) were slightly increased in 110-day-old TSC organoids. However, this cell type did not show morphological changes (Fig. S9F). Thus, OPC lineages did not seem to contribute to TSC lesions in organoids.
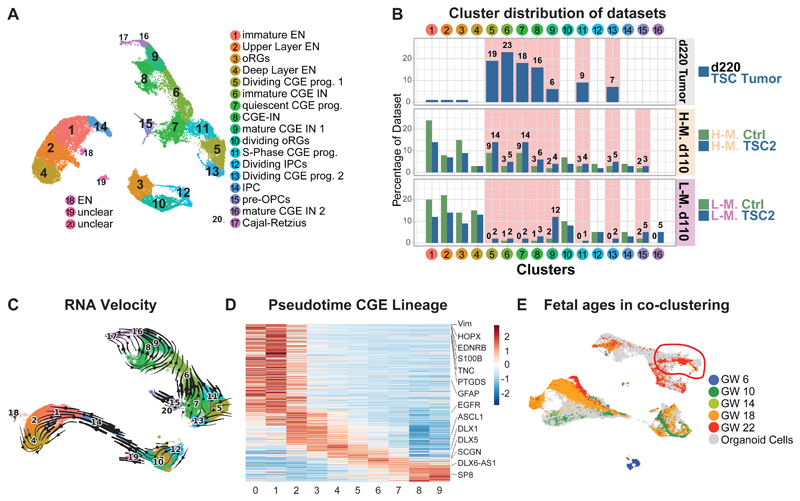
Fig 3. Late CGE progenitors give rise to TSC phenotypes.
A UMAP projection of 220-day-old TSC tumor organoids with 110-day-old Ctrl and TSC2+/--derived organoids in H- and L-medium. All cell types of the dorsal lineage were present, with radial glia (RG, Cl. 3 and 10), intermediate progenitors (IPCs, Cl. 12 and 14) and excitatory neurons (EN, Cl. 1, 2, 4, 18). Separate a lineage of CGE-derived cells was identified with quiescent CGE progenitors (Cl. 7), dividing CGE progenitor cells (Cl. 5, 11, 13) and CGE interneurons (IN, Cl. 6, 8, 9, 16, 17). Pre-OPC-like cells (Cl. 15) cluster close to quiescent CGE progenitors.
B Contribution of different datasets to clusters shown in panel A. 220-day-old TSC tumors only contributed to CGE progenitors and their progeny (Clusters 5, 6, 7, 8, 9, 11 and 13). These clusters were also increased in 110-day-old TSC2+/--derived organoids. H-medium enriched for progenitors. L-medium organoids had more mature CGE-INs (Cl. 9, and 16).
C RNA velocity projected in 2D. The cluster annotation corresponds to Fig. 3A. From the increased CGE progenitor population two trajectories emerge: a small trajectory towards pre-OPC cells (Cl. 15), and a larger trajectory towards interneurons (Cl. 6 and 9).
D Expression of genes along pseudotime in CGE lineage. Genes enriched along pseudotime were calculated and cells were binned into 10 groups (x axis, see Fig. S9B). All genes with enriched expression along the trajectory were ordered using a sliding average. Genes enriched in progenitors: top left. Genes in mature interneurons: bottom right. Selected genes were highlighted.
E UMAP of integration of organoid and fetal cells color coded for gestational ages shows GW22 cells co-clustered with quiescent CGE progenitor cluster (right top).
To confirm that cnLOH was not required for the initiation of TSC phenotypes, we tested allelic frequencies in the d110 scRNA datasets. Interneuron progenitors in TSC2+/- datasets did not show cnLOH, consistent with a disease initiation from a heterozygous progenitor (Fig. S8H). This suggests that expansion of a common interneuron progenitor rather than cnLOH initiates tumor and tuber phenotypes.
To characterize the common cell-of-origin we analyzed the gene expression signatures of the cells overrepresented in TSC organoids. Expression of markers such as DLX2, DLX5, SP8, COUP-TFII (NR2F2), and SCGN (Fig. S8B and D) revealed that this lineage originated from the caudal ganglionic eminence (CGE), a region in the ventral forebrain. The quiescent CGE-progenitors (Cl. 7) also expressed markers previously not found in interneuron progenitors, such as EDNRB and PTGDS (Fig. S8D).
To investigate the developmental trajectories of these populations we performed RNA velocity and pseudotime analysis (Fig. 3C, Fig. S9A-E). RNA velocity revealed a major trajectory towards CGE interneurons and a small bifurcation of CGE progenitors towards pre-OPC-cells (Fig. 3C). Along the CGE lineage we found expression of markers for quiescent (GFAP, HOPX together with EDNRB and PTGDS), activated progenitors (EGFR and DLX2) and CGE interneurons (DLX6-AS1 and SCGN) in both Ctrl and TSC2+/- organoids (Fig. 3D, S9C). The small trajectory towards pre-OPC cells showed markers recently described for human pre-OPC lineages (31) (Fig. S9D and E). Both tumor and tuber organoids shared the trajectory from CGE-progenitors to CGE-interneurons (Fig. 3C). To test whether lesion-specific cell types emerge we investigated the trajectories within interneurons as determined by RNA velocity (Fig. 3C). We found that mature interneurons were separated into tumor- and tuber-enriched interneurons (Fig. S10A to F). Tumor interneurons were enriched in GO-terms related to ribosomal proteins and translation, whereas tuber interneurons showed specific upregulation related to synapse formation and activity (Fig. S10G to O).
Although the descriptive nature of our scRNA experiments limits their generalizability, these data indicate that interneuron progenitors increased in TSC follow defined developmental trajectories and diverge into lesion specific interneuron subtypes.
Therefore, to determine whether the common developmental trajectory is present in the human fetal brain we integrated our data with published scRNA sequencing data from different fetal ages (32) (Fig. S11A). Co-clustering revealed similar cell types in the fetal brain (Fig. S11A and B) and pseudotime analysis confirmed trajectories towards interneurons and OPC-cells (Fig. S11B and C). We found similar gene expression cascades along the neurogenic trajectory, with markers of quiescent progenitors expressed together with EDNRB and PTGDS, followed by activated progenitors and interneurons (Fig S11D to F). Thus, developmental trajectories increased in TSC patient organoids are present in the human fetal brain.
As phenotypes in TSC organoids arose at later stages of organoid development, we hypothesized that the expanded CGE progenitors might correspond to specific progenitors in the fetal brain (red circle Fig. 3E).
To test this, we sub-clustered all progenitors (Fig. S11G and H) and found that 98% of fetal cells co-clustering with the expanded CGE-progenitors originated from gestational week 22 (GW22, Fig. S11G, H and I). We calculated gene modules (33) in progenitors (Fig. S11J) and compared organoid and fetal progenitors of various stages. Quiescent CGE progenitor cells showed the highest correlation with fetal progenitors from GW22 (+0.74, Fig. S11K and L). These data suggest that the expanded CGE progenitors emerge around late mid-gestation, a time where cortical tubers and subependymal tumors are first detected in TSC patients (20). Based on their CGE-origin and late emergence, we named the expanded cell type caudal late interneuron progenitor (CLIP) cells.
CLIP cells and their progeny are abundant in tumors
To independently validate our observations in the scRNA experiments, we investigated expression of markers for CGE, MGE, and excitatory cells by immunostainings of patient tumor primary tissue and organoids.
We confirmed expression of EGFR in tumors in organoids and in surgically resected SEGAs (Fig. 4A, S12). Consistent with the scRNA sequencing results the CGE interneuron progenitor markers DLX2, COUP-TFII, and SP8 were found in tumors in organoids from three TSC patients (Fig. 4B and D, Fig. S12, S13). In contrast, NKX2.1, a marker for MGE progenitors or SATB2, specific for excitatory neurons, was not expressed (Fig. 4D, S14A and B).
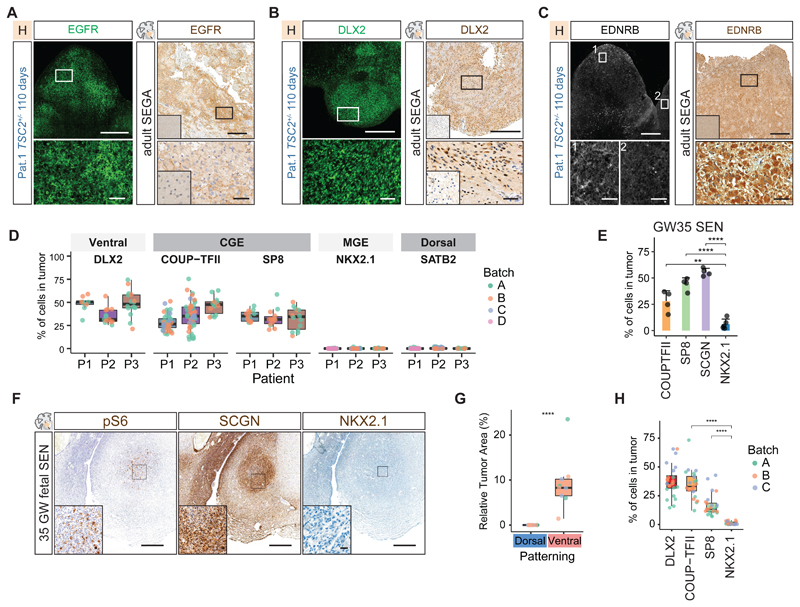
Fig 4. CGE progenitors initiate TSC tumors.
A Immunostaining for EGFR identified tumors in 110-day-old TSC2+/--derived organoids grown in H-medium. EGFR expression was present in adult SEGAs. Inset in primary tissue staining shows healthy control cortex negative for EGFR (See Fig. S12C for co-staining with COUP-TFII in organoids).
B DLX2 was expressed in tumors in 110-day-old TSC2+/--derived organoids and resected SEGA tissue. Inset in primary tissue staining shows healthy control cortex negative for DLX2 (Fig. S13, See Fig. S13 for EGFR and SP8 in organoid and patient tumors).
C EDNRB was expressed in tumors in 110-day-old TSC2+/--derived organoids. Zoom-in 1 and 2; tumor regions expressing EDNRB and negative region. See Fig. S16A for co-staining with PROX1 and pS6 in two patients. EDNRB expression was found in adult SEGAs. Inset in primary tissue staining shows healthy control cortex negative for EDNRB.
D Quantification of ventral (DLX2), CGE (COUP-TFII and SP8), MGE (NKX2.1) and dorsal markers (SATB2) in tumors of three patients. Tumors of at least two independent batches were quantified shown by different colors. Ventral and CGE markers are found in all tumors, MGE and dorsal markers are not found (See Fig. S14B for statistical comparison).
E Quantification of CGE markers COUP-TFII, SCGN, SP8 and MGE marker NKX2.1 in 35GW fetal SENs (mean and standard deviation, see Fig. S14D for statistical comparison).
F Immunostaining on fetal SENs. Fetal tumors characteristically expressed Vimentin (Fig. S14C) and pS6 in enlarged cells. Tumor were enriched in SCGN expressing CGE interneurons. Only few MGE cells were found.
G Dorsal and ventral patterning of tumors of patient 1. In 120-day-old TSC2+/--derived organoids tumors only appear in ventral patterned organoids containing interneuron progenitors (Student's t-test, see Fig. S15B. for overview of samples).
H Tumors in ventral patterned organoids expressed the ventral marker DLX2 and the CGE markers COUP-TFII and SP8, while NKX2.1 is almost absent (Ordinary one-way ANOVA with Tukey HSD, see Fig. S15H for statistical comparison).
Scale bars: overview images A, B, C: 500μm; Zoom-in A, B, C: 50μm; overview F: 500μm; Inset F: 20μm
To investigate the origin of TSC tumors in patients, we stained 35GW SENs. Fetal tumors like their organoid counterparts were enriched in CGE cells, whereas NKX2.1 expressing cells were rare (Fig. 4E and F, Fig. S14C and D). In contrast, NKX2.1 has been shown to be expressed in postnatal SEGAs (34). We confirmed expression of NKX2.1 (Fig. S14E) but found that postnatal tumor cells also expressed CGE markers (Fig. S14E to G). Thus, although fetal SENs consisted mostly of CGE cells, co-expression of NKX2.1 and CGE markers in post-natal SEGAs could suggest aberrant differentiation at later stages or the involvement of other lineages.
To test which lineages can generate tumors in TSC organoids, we utilized patterning protocols to generate dorsal and ventral forebrain separately (35, 36) (Fig. S15A). Dorsal patterning produced excitatory cells, whereas ventral patterned organoids contained both CGE and MGE lineages (Fig. S15C and D). Tumors were only observed in ventral patterned organoids (Fig. 4G and Fig. S15B), supporting an origin in the ventral forebrain. Notably, all tumors in ventral patterned organoids expressed abundantly the CGE markers COUP-TFII and SP8, whereas only a few cells were positive for NKX2.1 (Fig. 4H, Fig. S15D to H). This further supports the hypothesis that CGE progenitors initiate TSC tumors.
To test whether CGE progenitors in TSC tumors were CLIP cells we stained for EDNRB and PTGDS. Both patient primary tumor samples and organoid tumors contained cells that expressed these CLIP cell markers together with ventral neural stem cell (NSC) (SOX2, GAD1) and CGE markers (PROX1, COUP-TFII) (Organoids: EDNRB Fig. 4C and PTGDS Fig. S16I with PROX1, COUP-TFII, GAD1, SOX2 Fig. S16A, F and I; surgically removed SEGA: EDNRB Fig. 4C with GAD1 Fig. S16E and G). These data suggest that CLIP cells are the neural progenitors found in TSC tumors.
Besides CLIP cells, TSC tumors also contained interneurons (24) (Fig. 2A and B). Expression of the CGE interneuron marker SCGN (Fig. S17A and B) in fetal SENs (Fig. 4F, and S17D) and in tumors in organoids (Fig. S17C) supported a CGE origin for these interneurons. The lineage relationship between CLIP cells and CGE interneurons was further confirmed by EdU labeling (Fig. S17E and F). After 24h, EdU labelled cells co-expressed either EGFR and SCGN or both, confirming that CLIP cells produce SCGN positive interneurons (Fig. S17E and F).
Our data indicating that CLIP cells originating from the CGE generate TSC tumors could provide an explanation for why SEN/SEGAs are typically found in the caudothalamic groove, the region where the CGE is located during fetal development.
CLIP cells initiate cortical tuber development
To determine whether CLIP cells also give rise to giant cells that make up cortical tubers, we stained tuber-like structures in L-organoids and tubers in patient-derived brain tissue. Giant cells in organoids and in fetal cortical tubers expressed markers of ventral NSCs (GAD1 Fig. 5A-C and Fig. S19A; EGFR Fig. S18A and S19D). Expression of CLIP cell markers (EDNRB Fig. 5D, F; PTGDS Fig. S18D) together with CGE markers (PROX1 and COUP-TFII, Fig. S18B-D) in giant cells in organoids underlined their CLIP cell origin. Similarly, expression of EDNRB (Fig. 5E, Fig. S19A and B) and PTGDS with CGE-markers (PROX1, COUP-TFII, Fig. S19C) further suggested that CLIP cells are also the cell-of-origin for giant cells in TSC patients.
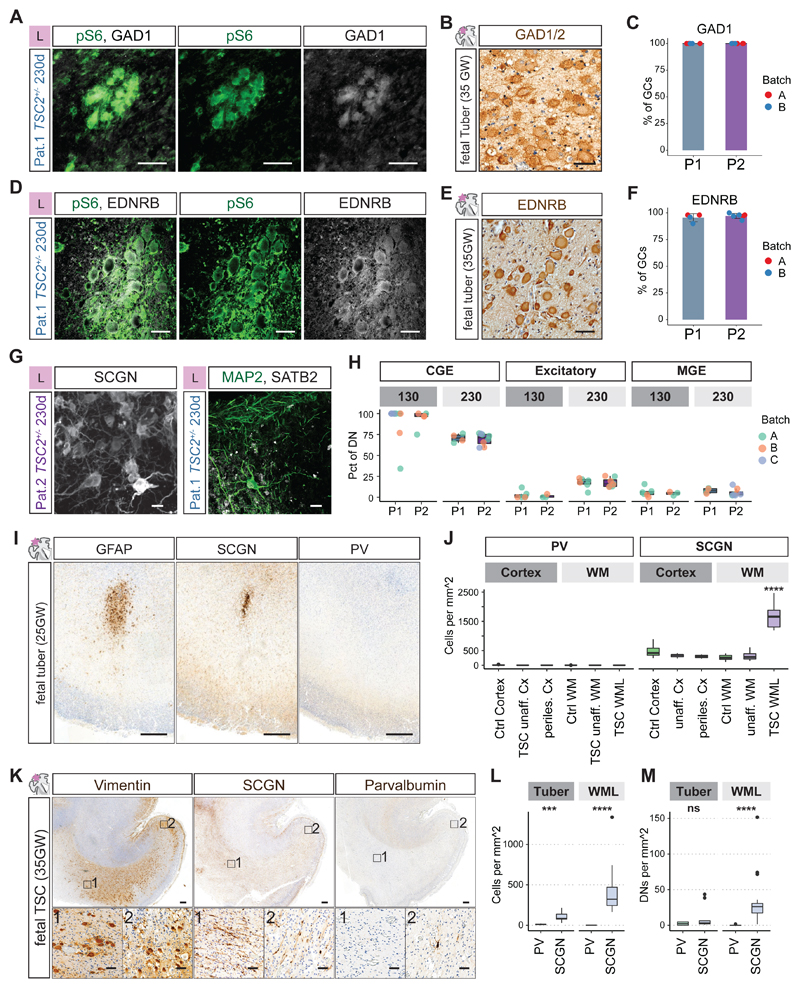
Fig 5. CLIP cells initiate cortical tuber pathogenesis.
A and B GCs in organoids (A) and fetal tubers (B) expressed the interneuron lineage marker GAD1.
C Quantification of co-expression of GAD1 and pS6 in giant cells in organoids (P1: N = 2, n = 6, mean = 100%; P2: N = 2, n = 8, mean = 100%)
D and E EDNRB was expressed in GCs in organoids and in fetal tubers.
F Quantification of co-expression of EDNRB and pS6 in GCs in organoids (P1: N = 2, n = 4, mean = 95.5%; P2: N = 2, n = 6, mean = 97.2%).
G Dysmorphic neurons at 230 days expressed the CGE-marker SCGN. Individual dysmorphic neurons expressed the excitatory marker SATB2 (See Fig. S20C-G for stainings at different timepoints).
H Quantification of expression of CGE (SCGN, COUP-TFII), excitatory (SATB2) and MGE markers (PV) in dysmorphic neurons (DNs) at early and late stages in organoids of two patients. CGE DNs were enriched at 130 days and decreased over time. Excitatory DNs increased over time. See Fig. S20I and J for statistical analysis.
I GFAP identified early dysplastic regions at 25GW located in the white matter below the cortex. There was a focal enrichment of SCGN expressing CGE interneurons, while PV expressing MGE interneurons were rare (See. Fig. S21A and B).
J Quantification of MGE marker PV and CGE marker SCGN in ~25GW fetal brain. Unaffected cortical areas, unaffected white matter, as well as white matter lesions (WML) and adjacent cortex (perilesional cortex) quantified for density of cells expressing PV or SCGN (Fig. S21A and B). WMLs were significantly enriched in CGE interneurons compared to all other regions (One-way ANOVA, see Fig. S21C and D for statistical analysis).
K At 35GW Vimentin staining identified streams of cells in WMLs and affected cortex. SCGN expressing cells were abundant in the WMLs, and cortex, while PV expressing cells were mainly found in cortical regions.
L and M Quantification of MGE marker PV and CGE marker SCGN in 35GW TSC case. Density of all cells is shown in L, with increase of SCGN cells in tuber and WML areas. In M the density of DNs expressing PV and SCGN in tuber and WML areas is shown. SCGN DNs were significantly enriched in WMLs, while tubers contained similar density of PV and SCGN cells (all neurons: Tuber p = 0.0002, WML p = <0.0001; DNs: Tuber p = 0.248, WML p = <0.0001; pairwise Wilcoxon-test, see Fig. S21E for overview of samples).
(Scale bars: A, B, D, E, inset K: 50μm; G: 20μm; overview I and K: 500μm)
Dysmorphic neurons in cortical tubers have been shown to express excitatory and inhibitory neuron markers (4). In addition, early TSC lesions are populated by a high density of migrating neurons of unknown origin (4). To determine whether CGE interneurons produced by CLIP cells contribute to cortical tubers we evaluated expression of CGE (SCGN and COUP-TFII), MGE (Parvalbumin, PV), and excitatory neuron markers (SATB2) in organoids and patients. We found that at early stages most dysmorphic neurons in organoids were CGE interneurons (Fig. 5H and Fig. S20A to E). At later stages the contribution of excitatory neurons increased, while only few MGE interneurons are found (Fig. 5G and H; Fig. S20F and G).
To test whether CGE-interneurons are involved in early tuber lesions in patients we tested expression of SCGN and PV in a 25GW TSC case. At this stage tuber pathogenesis initiates with white matter lesions (WMLs). Similar to the organoid model, we found that WMLs were highly enriched in CGE-interneurons, whereas no MGE cells were detected (Fig. 5I and J; Fig. S21 A to D). This suggests that CGE interneurons are the migrating neurons previously described in TSC lesions.
Since excitatory dysmorphic neurons increased over time in organoids, we evaluated the contribution of different lineages during the development of TSC tubers. Around 35GW CGE interneurons were still increased (Fig. 5K and L). At the same time the first dysmorphic neurons (DNs) appeared, with CGE-DNs being more abundant in WMLs. (Fig. 5M and Fig. S21E). With progression of tuber lesions at post-natal stages, however, numbers of both excitatory and MGE neurons increased (Fig. S22A to E).
MGE and CGE markers identify distinct populations during normal brain development. In TSC patient organoids, we detected a sub-population of dysmorphic interneurons expressing the MGE marker PV together with the CGE marker SCGN (Fig. S20G and H). To test whether this mis-differentiated population is present in cortical tubers we tested expression of PV with the CGE markers SCGN and SP8. In a matched control case, no cells co-expressing these markers were found, whereas in a cortical tuber several triple positive cells were detected (Fig. S22F). Taken together, our data suggest that CGE lineages initiate cortical lesion development in TSC. Excitatory and MGE dysmorphic neurons appear over time and are frequent in post-natal lesions. Furthermore, our data show that a comprehensive analysis of different markers is necessary to study the contribution of different lineages to cortical tubers as mis-differentiated cells can be observed.
EGFR inhibition reduces tumor burden
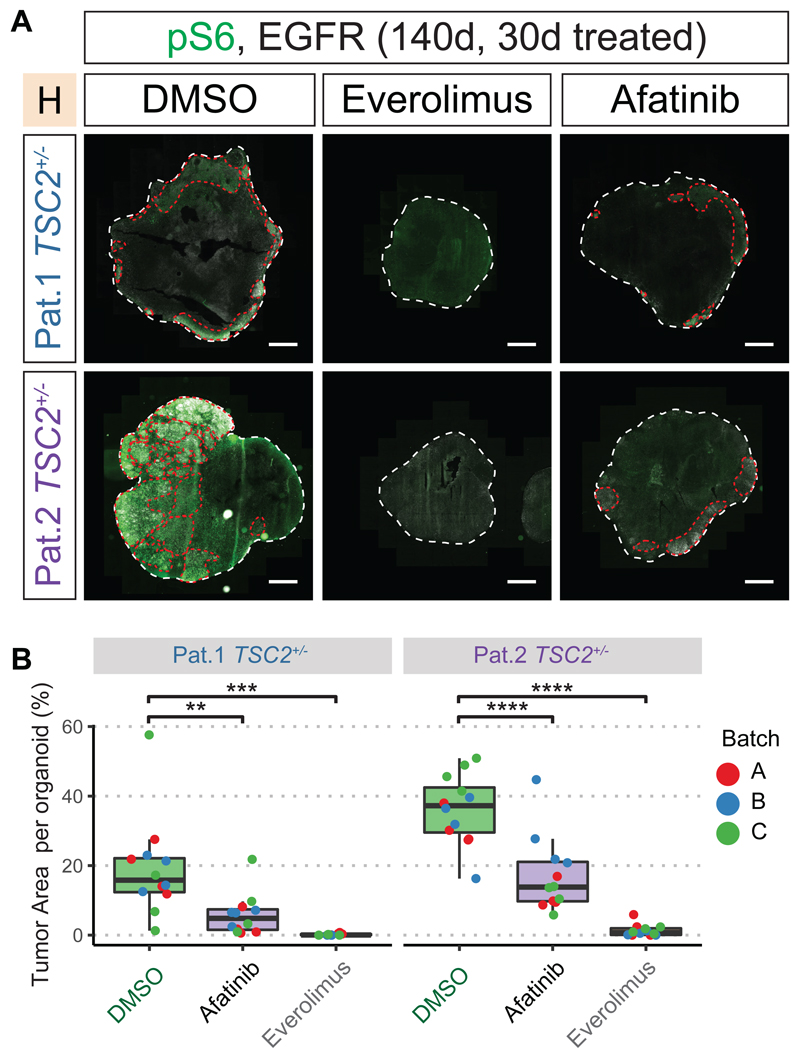
mTOR inhibition has been clinically used to treat SEN/SEGAs in TSC patients. However, known side-effects and limitations like tumor re-growth after drug discontinuation, necessitate exploring alternative therapeutic strategies (37–39). Both CLIP cells and proliferating cells in TSC tumors express EGFR. To assess the role of the EGFR pathway in tumor growth, we performed a drug testing assay in TSC2+/- H-organoids at 110 days, when tumors were already apparent. We used the EGFR receptor tyrosine kinase inhibitor (RTKI) Afatinib and Everolimus, an mTOR Complex 1 inhibitor. Organoids were treated for 30 days with Everolimus, Afatinib, or DMSO (Fig. S23A). Tumor reduction was determined by measuring areas co-expressing pS6 and EGFR. Everolimus treatment almost completely abolished tumors in 140-day-old organoids (Fig. 6A, B; Fig. S23B). After Afatinib treatment, both tumor load and mean tumor size were significantly reduced when compared to untreated organoids (Fig. 6A and B, Fig. S23B to D). Thus, targeting the EGFR pathway could be an alternative strategy for the treatment of TSC brain lesions.
Fig 6. EGFR-inhibition reduces tumor burden.
A 30 days of treatment started at 110 days after EB formation (see Fig. S23A). Immunostaining for pS6 and EGFR identified tumors (red lines) in the control group (DMSO) in both patients. Tumors were reduced by Afatinib and Everolimus treatment.
B Quantification of tumor area per organoid. All sections of each organoid stained on one slide were used for quantification. Tumors were identified as regions of overlapping pS6 and EGFR staining. While tumors were detected in DMSO-control for both patients, Afatinib or Everolimus treatment both reduce tumor burden. (two-way ANOVA for treatment condition controlling for batches, Tukey's multiple comparisons test, see Fig. S23B)
(Scale bars: A.: 1mm)
Here, we show that the neurodevelopmental disorder TSC is initiated by a caudal late interneuron progenitor, the CLIP cells (Fig. S24). Early lesions consisted almost exclusively of CLIP cell lineages, whereas other cell types appeared during disease progression. Although our scRNA sequencing analysis is descriptive and we only analyzed organoids from one patient, our extensive validation in organoids from 3 TSC patients plus tissues from more than 10 additional TSC cases demonstrates that the TSC organoid model recapitulated fetal disease dynamics. However, the organoid model was limited in modeling post-natal processes, possibly due to the absence of environmental factors that are present in vivo.
The cell-of-origin for many human brain tumors remains elusive; however, the idea that cancer stems from the reactivation of a remnant of developmental tissue was proposed more than a century ago (40, 41). Studies in mice have revealed sensitivity of adult neural stem cells to cancer-initiating mutations, resulting in the formation of glioblastoma, a high-grade brain tumor. Transcriptional similarities between CLIP cells and mouse adult neural stem cells suggest that CLIP cells could be involved more generally in brain cancers. Our data suggest that a sensitivity to increased mTOR signaling makes CLIP cells vulnerable to mutations in TSC2. We hypothesize that a similar mechanism could explain other malformations of cortical development caused by mTOR dysregulation, e.g., FCD type II.
Extensive migration of interneurons into the cortex continues in humans even after birth (42). As these late migrating neurons also arise from the CGE and share markers with CLIP cells, we speculate that CLIP cells give rise to late migrating interneurons in the healthy human brain. This is consistent with previous results showing that CGE-derived interneurons contribute to the human brain in much higher percentages (43, 44) and with the observation that late-migrating SCGN interneurons from the CGE are found in humans, but not in mice (45). The protracted brain development seen in large, gyrated cortices was accompanied by the generation or expansion of cell types. These are not or less present in small lissencephalic brains like the mouse brain, necessitating human disease models. Our data suggest that CLIP cells are among the cell types specific for or amplified in the human brain, which would make TSC a disease unique to large, gyrated brains.
Materials and Methods summary
Detailed information on all materials and methods performed are provided in the supplementary materials.
IPS cell generation and culture
In brief, patients were selected from the TSC data registry of the MUV. Blood was collected from patients and PBMCs were isolated. Reprogramming was perfomed using Sendai Vectors. iPSCs cells were cultured using the Cellartis DEF-CS 500 culture system (Takara). Isogenic control cell lines were directly isolated (mosaic patient 1) or generated using scarless CRISPR repair (patient 2).
Organoid generation
Organoids were generated using either a high-nutrient (H-organoids) or low nutrient (L-organoids) medium to favor proliferation or neuronal maturation respectively. In addition, organoids were patterned towards dorsal or ventral brain regions as described (35, 36).
Single cell transcriptomics and analysis
Organoids were dissociated. Library preparation was performed with the Chromium Single Cell 3' Library & Gel Bead Kit v.3 (10x Genomics, PN-1000075). Libraries were sequenced on a NextSeq 550 (Illumina) or on a NovaSeq SP lane (Illumina). Quality control and pre-processing was performed using Seurat R package v.3. Visualization and pseudotime analysis were performed using monocle3.
Immunohistochemistry
Immunohistochemistry on frozen organoid samples was performed as described with slight modifications (18, 35). Human brain tissue samples were collected in strict observance of the local legal and institutional ethical regulations. Tissue was processed for cryosections or paraffin sections. Antigen retrieval was performed followed by immunohistochemistry as described in Table S6.
Supplementary Material
One Sentence Summary.
Caudal late interneuron progenitors initiate tumors and cortical tubers in Tuberous Sclerosis Complex.
Acknowledgments
We thank C. da Cunha E Silva Martins Costa, P. Möseneder, H. Eleanor Gustafson and S. Wolfinger for help with experiments and analyses; the IMBA Stem Cell Core Facility and C. Allison Agu for generation of IPS Cell Lines; B. Gebarski and A. Vogt for library preparation and sequencing performed at the VBCF NGS Unit (https://www.viennabiocenter.org/facilities/next-generation-sequencing/); the Genome Engineering Unit of VBCF ProTech facility (https://www.viennabiocenter.org/facilities/protein-technologies/) for assistance with isogenic control line preparation; K. Stejskal and E. Roitinger for Mass spectrometry performed at the IMBA/IMP Mass spectrometry facility; the IMBA/IMP Biooptics facility; A. Mancebo Gimenez and M. Zeba of the VBCF HistoPathology facility for immunohistochemistry; A. Meixner for coordinating ethical approvals; R. Diehm; G. Kasprian for providing MRIs; the KIN Biobank of the MUV; K. Auguste and the UCSF Brain Tumor SPORE Biorepository (NIH/NCI 5P50CA097257) for their coordination for surgical tissue collection; V. Elorriaga Benavides for work on primary material, O. Wüseke for establishing contact with the MUV. We thank G. Riddihough/Life Science Editors for help with editing the manuscript. We especially thank all patients and their families for participating in this study or donating tissue.
Funding
A.V. was supported by an EMBO Fellowship (ALTF-1112-2019). Work in J.A.K.'s laboratory is supported by the Austrian Federal Ministry of Education, Science and Research, the Austrian Academy of Sciences, the City of Vienna and the SFB F78 Stem Cell (F 7803-B). This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation (695642).
Footnotes
Author contributions: O.L.E, N.S.C., M.F. and J.A.K. designed the study and analysis. Experiments were performed by O.L.E., I.M., N.S.C., T.S., V.E.G., A.M.P., and J.C.. Data analysis was performed by O.L.E., N.S.C., J.A.H., A.V., M.N. and M.F.P.. The study was supervised by N.S.C. and J.A.K. The manuscript was prepared by O.L.E, N.S.C. and J.A.K. with input from all authors.
Competing interests: J.A.K. is on the supervisory and scientific advisory board of a:head bio AG (https://aheadbio.com) and is an inventor on several patents relating to cerebral organoids.
Ethics statement:
The study was approved by the local ethics committee of the Medical University of Vienna (MUV).
Data and materials availability
WGS and scRNA data are available through controlled access at the European genome-phenome Archive (EGA). Study Number: EGAS00001004586 Datasets: Single Cell RNA seq: EGAD00001006332, Whole Genome Sequencing: EGAD00001006333. All code used in this study is available on GitHub (https://github.com/OliverEichmueller/TSC_Science2021, DOI: 10.5281/zenodo.5741170) and upon request.
TSC patient IPS cell lines will be made available upon request after obtaining ethical approval from the Ethics Committee of the Medical University of Vienna under a material transfer agreement with the Institute of Molecular Biotechnology of the Austrian Academy of Sciences.
References and Notes
- 1.Kuzniecky RI. MRI in cerebral developmental malformations and epilepsy. Magnetic Resonance Imaging. 1995;13:1137–1145. doi: 10.1016/0730-725x(95)02024-n. [DOI] [PubMed] [Google Scholar]
- 2.Thiele EA. Managing and understanding epilepsy in tuberous sclerosis complex. Epilepsia. 2010;51(Suppl 1):90–91. doi: 10.1111/j.1528-1167.2009.02458.x. [DOI] [PubMed] [Google Scholar]
- 3.Ruppe V, et al. Developmental brain abnormalities in tuberous sclerosis complex: a comparative tissue analysis of cortical tubers and perituberal cortex. Epilepsia. 2014;55:539–550. doi: 10.1111/epi.12545. [DOI] [PubMed] [Google Scholar]
- 4.Gelot AB, Represa A. Progression of Fetal Brain Lesions in Tuberous Sclerosis Complex. Front Neurosci. 2020;14:899. doi: 10.3389/fnins.2020.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henske EP, Jozwiak S, Kingswood JC, Sampson JR, Thiele EA. Tuberous sclerosis complex. Nat Rev Dis Primers. 2016;2:16035. doi: 10.1038/nrdp.2016.35. [DOI] [PubMed] [Google Scholar]
- 6.Martin KR, et al. The genomic landscape of tuberous sclerosis complex. Nat Commun. 2017;8:15816. doi: 10.1038/ncomms15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crino PB. Evolving neurobiology of tuberous sclerosis complex. Acta Neuropathol. 2013;125:317–332. doi: 10.1007/s00401-013-1085-x. [DOI] [PubMed] [Google Scholar]
- 8.Feliciano DM, Su T, Lopez J, Platel JC, Bordey A. Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J Clin Invest. 2011;121:1596–1607. doi: 10.1172/JCI44909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feliciano DM, Quon JL, Su T, Taylor MM, Bordey A. Postnatal neurogenesis generates heterotopias, olfactory micronodules and cortical infiltration following single-cell Tsc1 deletion. Hum Mol Genet. 2012;21:799–810. doi: 10.1093/hmg/ddr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Way SW, et al. Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum Mol Genet. 2009;18:1252–1265. doi: 10.1093/hmg/ddp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson RP, Van Nielen DL, Winzenburger PA, Ess KC. Neuronal and glia abnormalities in Tsc1-deficient forebrain and partial rescue by rapamycin. Neurobiol Dis. 2012;45:369–380. doi: 10.1016/j.nbd.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto J, et al. Regulable neural progenitor-specific Tsc1 loss yields giant cells with organellar dysfunction in a model of tuberous sclerosis complex. Proc Natl Acad Sci U S A. 2011;108:E1070–1079. doi: 10.1073/pnas.1106454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blair JD, Hockemeyer D, Bateup HS. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med. 2018;24:1568–1578. doi: 10.1038/s41591-018-0139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin W, et al. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol. 2010;20:1096–1105. doi: 10.1111/j.1750-3639.2010.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henske EP, et al. Allelic Loss Is Frequent in Tuberous Sclerosis Kidney Lesions but Rare in Brain Lesions. Am J Hum Genet. 1996:400–406. [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JA, et al. Pathogenesis of Tuberous Sclerosis Subependymal Giant Cell Astrocytomas: Biallelic Inactivation of TSC1 or TSC2 Leads to mTOR Activation. Journal of Neuropathology and Experimental Neurology. 2004 doi: 10.1093/jnen/63.12.1236. [DOI] [PubMed] [Google Scholar]
- 17.Knudson AG., JR Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Nat Acad Sci USA. 68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardy C, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A. 2015;112:E3312. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SH, et al. Tuberous sclerosis in a 20-week gestation fetus: immunohistochemical study. Acta Neuropathol. 1997;94:180–186. doi: 10.1007/s004010050691. [DOI] [PubMed] [Google Scholar]
- 21.Mizuguchi M, Takashima S. Neuropathology of tuberous sclerosis. Brain & Development. 2001:508–515. doi: 10.1016/s0387-7604(01)00304-7. [DOI] [PubMed] [Google Scholar]
- 22.Buccoliero AM, et al. Subependymal giant cell astrocytoma: a lesion with activated mTOR pathway and constant expression of glutamine synthetase. Clin Neuropathol. 2016;35:295–301. doi: 10.5414/NP300936. [DOI] [PubMed] [Google Scholar]
- 23.Buccoliero AM, et al. Subependymal giant cell astrocytoma (SEGA): Is it an astrocytoma? Morphological, immunohistochemical and ultrastructural study. Neuropathology. 2009;29:25–30. doi: 10.1111/j.1440-1789.2008.00934.x. [DOI] [PubMed] [Google Scholar]
- 24.Cotter JA. An update on the central nervous system manifestations of tuberous sclerosis complex. Acta Neuropathol. 2020;139:613–624. doi: 10.1007/s00401-019-02003-1. [DOI] [PubMed] [Google Scholar]
- 25.McGinnis CS, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019;16:619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinters HV, et al. Tuberous sclerosis-related gene expression in normal and dysplastic brain. Epilepsy Res. 1998;32:12–23. doi: 10.1016/s0920-1211(98)00036-9. [DOI] [PubMed] [Google Scholar]
- 27.Johnson MW, Emelin JK, Park SH, Vinters HV. Co-Localization of TSC1 and TSC2 Gene Products in Tubers of Patients with Tuberous Sclerosis. Brain Pathol. 1999:45–54. doi: 10.1111/j.1750-3639.1999.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, et al. Tsc1 mutant neural stem/progenitor cells exhibit migration deficits and give rise to subependymal lesions in the lateral ventricle. Genes Dev. 2011;25:1595–1600. doi: 10.1101/gad.16750211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim JS, et al. Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. Am J Hum Genet. 2017;100:454–472. doi: 10.1016/j.ajhg.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerfoot C, et al. Localization of Tuberous Sclerosis 2 mRNA and its Protein Product Tuberin in Normal Human Brain and in Cerebral Lesions of Patients with Tuberous Sclerosis. Brain Pathol. 1996:367–377. doi: 10.1111/j.1750-3639.1996.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 31.Huang W, et al. Origins and Proliferative States of Human Oligodendrocyte Precursor Cells. Cell. 2020;182:594–608.:e511. doi: 10.1016/j.cell.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhaduri A, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020 doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hang JF, et al. Thyroid transcription factor-1 distinguishes subependymal giant cell astrocytoma from its mimics and supports its cell origin from the progenitor cells in the medial ganglionic eminence. Mod Pathol. 2017;30:318–328. doi: 10.1038/modpathol.2016.205. [DOI] [PubMed] [Google Scholar]
- 35.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franz DN, et al. Everolimus for subependymal giant cell astrocytoma in patients with tuberous sclerosis complex: 2-year open-label extension of the randomised EXIST-1 study. The Lancet Oncology. 2014;15:1513–1520. doi: 10.1016/S1470-2045(14)70489-9. [DOI] [PubMed] [Google Scholar]
- 38.Krueger DA, et al. Long-term treatment of epilepsy with everolimus in tuberous sclerosis. Neurology. 2016;87:2408–2415. doi: 10.1212/WNL.0000000000003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martins F, et al. A review of oral toxicity associated with mTOR inhibitor therapy in cancer patients. Oral Oncol. 2013;49:293–298. doi: 10.1016/j.oraloncology.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Bhaduri A, et al. Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma. Cell Stem Cell. 2020;26:48–63.:e46. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Paredes MF, et al. Extensive migration of young neurons into the infant human frontal lobe. Science. 2016;354 doi: 10.1126/science.aaf7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodge RD, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raju CS, et al. Secretagogin is Expressed by Developing Neocortical GABAergic Neurons in Humans but not Mice and Increases Neurite Arbor Size and Complexity. Cereb Cortex. 2018;28:1946–1958. doi: 10.1093/cercor/bhx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agu CA, et al. Successful Generation of Human Induced Pluripotent Stem Cell Lines from Blood Samples Held at Room Temperature for up to 48 hr. Stem Cell Reports. 2015;5:660–671. doi: 10.1016/j.stemcr.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danecek P, McCarthy SA, HipSci C, Durbin R. A Method for Checking Genomic Integrity in Cultured Cell Lines from SNP Genotyping Data. PLoS One. 2016;11:e0155014. doi: 10.1371/journal.pone.0155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velasco S, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McInnes L, Healy J, Saul N, GroGberger L. UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software. 2018;3 [Google Scholar]
- 55.Garcia M, et al. Sarek: A portable workflow for whole-genome sequencing analysis of germline and somatic variants. F1000Research. 2020;9 doi: 10.12688/f1000research.16665.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milkereit R, et al. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Commun. 2015;6:7250. doi: 10.1038/ncomms8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hui KK, et al. GABARAPs dysfunction by autophagy deficiency in adolescent brain impairs GABAA receptor trafficking and social behavior. Sci Adv. 2019;5:eaau8237. doi: 10.1126/sciadv.aau8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS and scRNA data are available through controlled access at the European genome-phenome Archive (EGA). Study Number: EGAS00001004586 Datasets: Single Cell RNA seq: EGAD00001006332, Whole Genome Sequencing: EGAD00001006333. All code used in this study is available on GitHub (https://github.com/OliverEichmueller/TSC_Science2021, DOI: 10.5281/zenodo.5741170) and upon request.
TSC patient IPS cell lines will be made available upon request after obtaining ethical approval from the Ethics Committee of the Medical University of Vienna under a material transfer agreement with the Institute of Molecular Biotechnology of the Austrian Academy of Sciences.