Abstract
Adipose triglyceride lipase (ATGL) is the key-enzyme for the release of fatty acids (FAs) from triacylglycerol (TG) stores during intracellular lipolysis and produces FAs used for energy production. There is growing evidence that the products and intermediates from lipolytic breakdown during the FA mobilization process also have fundamental regulatory functions affecting cell signaling, gene expression, metabolism, cell growth, cell death and lipotoxicity. Regulation of ATGL is therefore vital for maintaining a defined balance between lipid storage and mobilization. This review addresses the regulation of ATGL activity at the post-translational level with special emphasis on protein-mediated interaction at the site of hydrolytic action, namely to the lipid droplet.
Keywords: Lipolysis, ATGL, adipose triglyceride lipase, PNPLA2, patatin-like phospholipase domain-containing protein 2, G0S2, G0/G1 switch gene 2, CGI-58, comparative gene identification 58, ABHD5, α/β hydrolase domain containing protein 5, perilipin, Plin, proteins cell death activator CIDE-3, CIDEC, fat-specific protein 27, FSP27, hypoxia-inducible lipid dropled-associated, HILPDA, pigment epithelium derived factor, PEDF, serpin family F member 1, SERPINF1, oleoyl-CoA, Atglistatin
1. Introduction
In times of nutrient abundance, fatty acids (FAs) are re-estrified into triacylglycerols (TGs) and are deposited in lipid droplets (LD) in vertebrate adipocytes or seeds in plants. This process represents the main form of energy storage in most organisms, including humans. Upon increasing energy demand, these FAs are released from TG depots in a physiological process termed intracellular lipolysis (Figure 1). The protein named adipose triglyceride lipase (ATGL) plays a prominent role in this reaction by catalyzing the initial step of cleaving TG to diacylglycerol (DG) and FA [1, 2]. Subsequently, hormone sensitive lipase (HSL) hydrolyzes DG to monoacylglycerol (MG) and FA [3, 4], whereas monoglyceride lipase (MGL) cleaves MG to glycerol and FA [5]. Generated FAs can either be used as substrates for energy production or they can serve as precursors for membrane lipids and signaling molecules [6, 7]. Dysregulated lipolysis leads to the excess of circulating FAs, which has destructive, lipotoxic effects on the organism, including insulin resistance, type 2 diabetes, fatty liver and inflammation [8–11].
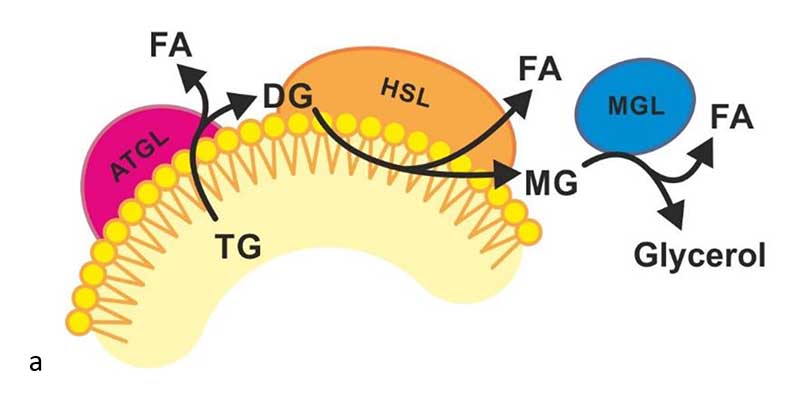
Figure 1.
Schematic overview of intracellular lipolysis, the sequential breakdown of TG to glycerol and three molecules of FAs. ATGL catalyzes the hydrolysis of TG stored in LDs to DG and FA, which is the first step of lipolysis. The next step of lipolysis is catalyzed by HSL to degrade DGs to MGs and FAs. The last step of lipolysis is catalyzed by MGL, which hydrolyzes MG to glycerol and FA.
As the tight regulation of lipolysis is of great physiological importance, ATGL - the key-enzyme of this process - is regulated on different levels [2]. Transcriptional control of ATGL is extensively reviewed elsewhere [12, 13]. Post-transcriptionally, ATGL is regulated by different proteins, which work in concert to ensure ATGL’s proper function (Figure 2). Within the last years, a lot of effort was put to identify these players in ATGL regulation and here we review the current literature (mainly protein-based) on regulation of the hydrolytic activity of ATGL and outline possible inter- dependencies. ATGL is directly delivered from the endoplasmic reticulum to LDs by the Golgi-ER transport protein complex GBF1-Arf1-COPI. This process involves the direct interaction of ATGL with the Arf1 exchange factor GBF1, which has been subject of detailed investigations within the last few years [14–18].
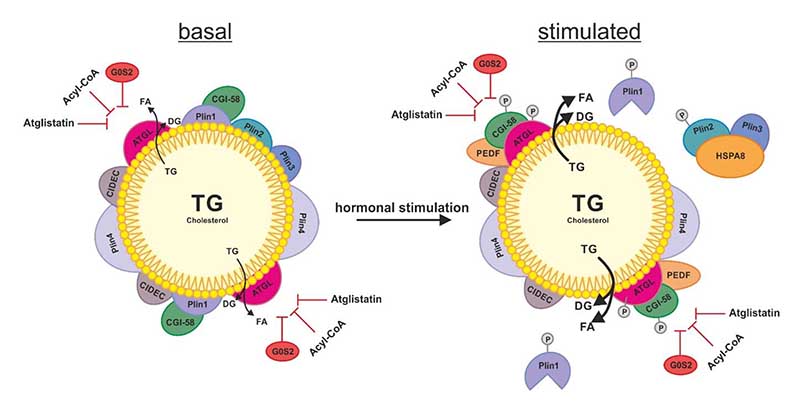
Figure 2.
Schematic overview of the regulation of ATGL on the LD. In basal condition of lipolysis, the LD is decorated with perilipins (Plins) and cell death activator CIDE-3 (CIDEC) to restrict the access of ATGL to the TG stores of the LD. The additional role of Plin1 is to sequester the co-activator protein comparative gene identification 58 (CGI-58) preventing its stimulating interaction with ATGL. ATGL hydrolyzes TG to DG and FA which also takes place to a small extent in basal conditions. Upon hormonal stimulation, the phosphorylation of Plin1, Plin2, CGI-58 and ATGL leads to a change on the surface of the LD: The chaperone heat shock protein HSPA8/hsc70 (HSPA8) shuttles Plin2 and Plin3 to the proteasome for degradation. The dissociation of CGI-58 from Plin1 enables its interaction with ATGL, which activates ATGL’s TG hydrolyzing activity. In both, basal and stimulated state of lipolysis, ATGL can be inhibited by the protein G0/G1 switch gene 2 (G0S2), acyl-CoA and synthetic inhibitor Atglistatin (which is effective in murine ATGL only).
The surface of LDs is decorated by five members of the perilipin (Plin) family, Plin1-Plin5, which form a barrier to prevent the access of ATGL to TG stores of the LD [19]. Chaperone-mediated autophagy mediates the degradation of Plin2 and Plin3 and thus aids in access of ATGL to LDs [20]. The direct regulation of ATGL activity on the protein level involves the interaction with its co-activator protein comparative gene identification 58 (CGI-58), also known as α/β hydrolase domain containing protein 5 (ABHD5), and inhibitor protein G0/G1 switch gene 2 (G0S2) [21–23].
Other protein interaction partners of ATGL have been described, but their exact mechanism is rather poorly understood. This includes the interaction of ATGL with two other LD associated proteins, namely cell death activator CIDE-3 (CIDEC) (also known as fat-specific protein 27, (FSP27)), hypoxia-inducible lipid dropled-associated (HILPDA), and with the adipocyte secreting protein pigment epithelium derived factor (PEDF), also known as serpin family F member 1 (SERPINF1) [24–31].
Small molecules have also been reported to influence ATGL activity, which includes inhibition by the naturally occurring activated long-chain FA intermediate oleoyl-CoA [7, 32]. A potent synthetic inhibitor, termed Atglistatin, was developed and is effective in inhibiting the activity of mouse ATGL [33, 34]. A similar effective inhibitor for human ATGL might have potential use in future therapeutic application.
1.1. Adipose triglyceride lipase (ATGL)
The gene for human ATGL is located on chromosome 11p15.5 and includes 10 exons encoding for a 504 amino acid protein (NCBI Reference Sequence: NG_023394.1, [35]). ATGL expression is found in virtually all tissues, but it is predominantly expressed in adipose tissue [2, 36–39]. ATGL is a triacylglycerol lipase (EC 3.1.1.3) and was first described in 2004 by three independent groups with different names (PNPLA2; desnutrin; iPLA2zeta; PEDF-R; TTS-2.2; TTS2 [2, 39, 40]; the recommended name (according to UniProt consortium) is patatin-like phospholipase domain-containing protein 2 (PNPLA2)).
Orthologs of ATGL have been described in a variety of other organisms, including mouse, rat, chicken, flies, plants and yeast [2, 41–47]. The mouse ortholog of ATGL consists of 486 amino acids and shares 86.8 % sequence identity to the human protein [2]. Sequence conservation is highest within residues 1-250 (92.4 %), whereas the C-terminal half of the protein is less conserved at amino acid level (80.8 %).
To date, no experimental three-dimensional structure of ATGL is available. Sequence based structure prediction and homology modelling groups ATGL to the patatin-like domain containing protein family (PNPLA) [48].
Crystal structures of members of the PNPLA family (Pat17 and Phospholipase A2, ExoU and VipD) are available and reveal that they contain a three-layer (α-β-α) sandwich architecture (Figure 3) and fulfill their hydrolytic function via a catalytic Ser-Asp dyad [49–54]. Ser47 and Asp166, located within the patatin-like domain, form the catalytic dyad in both, human and mouse ATGL. The nucleophilic Ser47 resides in a GXSXG motif generally found in α-β hydrolases [49, 52, 55] and the importance of this residue was confirmed via mutational studies [21, 38].
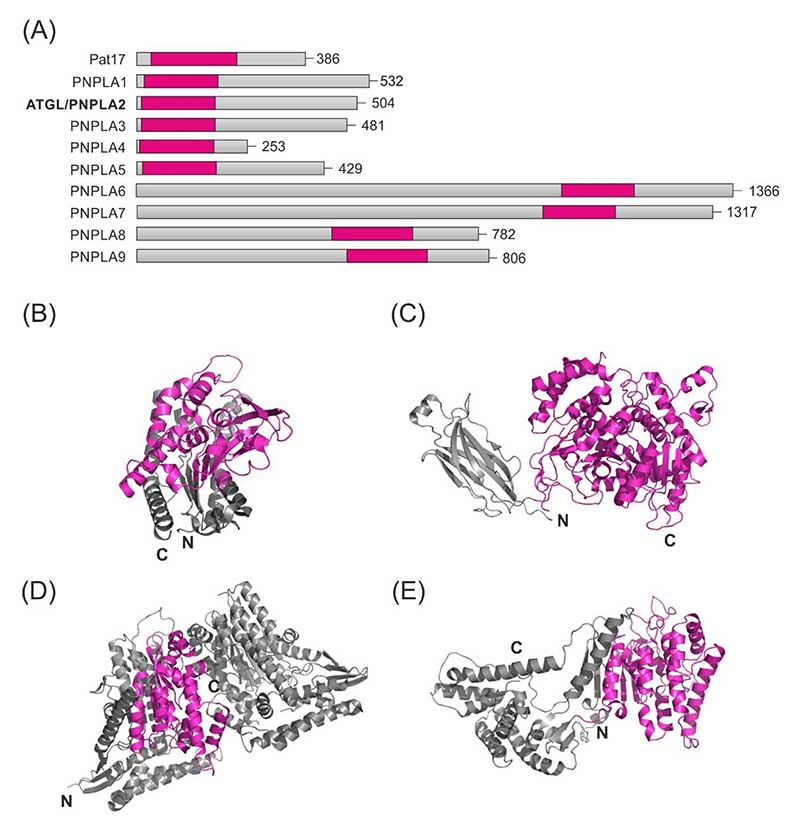
Figure 3. Overview of patatin-like domain containing proteins.
(A) Graphical representation of Pat17 and nine members of the human PNPLA family and the location of their patatin-like domains, which are depicted in magenta. Crystal structures of (B) Pat17, (C) cytosolic phospholipase A2, (D) ExoU and (E) VipD. Patatin-like domains of Pat17, ExoU and VipD and the catalytic phospholipase A2 (PLA2c) domain in cytosolic phospholipase A2 are depicted in magenta.
Other structural features of ATGL include an N-terminal amphipathic helix (Ile10-Gly24) potentially involved in TG binding, and a hydrophobic region (Pro315-Pro360), which is considered to be responsible for the localization to LDs [56, 57]. C-terminal truncated versions of human ATGL (Q289X, deletion of 215 residues) fail to localize to LDs, but surprisingly exhibit higher lipolytic activity in vitro, indicating that the C-terminus of ATGL possesses auto-regulatory function [57]. This goes in line with higher activities, which were determined for C-terminally truncated mouse ATGL (Met1-Leu254, deletion of 232 residues) in vitro [58].
Human patients with a loss of functional ATGL, suffer from neutral lipid storage disease with myopathy (NLSD-M) characterized by abnormal TG accumulation in multiple organs and tissues leading to cardiac and skeletal muscle myopathy [55, 59–61]. Besides massive TG accumulation, global inactivation of ATGL in mice results in reduced FA concentrations in the plasma, having beneficial effects on glucose tolerance and insulin sensitivity [1, 62].
Ser404 and Ser428 of human ATGL are targets for phosphorylation [63]. Phosphorylation of Ser404 (corresponds to Ser406 in mouse ATGL) increases the activity of ATGL, but it is still controversial if AMP-activated protein kinase (AMPK) or cAMP dependent protein kinase A (PKA) is mediating this phosphorylation process [2, 64–68]. Furthermore, phosphorylation of Thr372, which is located within the lipid binding region of ATGL, potentially contributes to the localization of ATGL to LDs, but the kinase involved in this process was not characterized yet [69].
The N-terminal region (residues 1-254) of ATGL directly interacts with its activator protein comparative gene identification 58 (CGI-58) and its inhibitor protein G0/G1 switch gene 2 (G0S2) [58]. Under basal conditions, ATGL catalyzes the hydrolysis of TGs preferable at the sn-2 position of the glycerol backbone generating sn-1,3 diacylglycerols and FAs [70]. Upon stimulation with CGI-58 however, ATGL expands its selectivity towards the sn-1 position [70]. The molecular mechanism behind the co-activation with CGI-58 and changed hydrolytic patterns are not understood.
Other interaction partners of ATGL include the LD associated proteins Plin5, CIDEC and PEDF [28, 30, 31, 71]. The protein HILPDA is discussed to be involved in ATGL regulation, but this still has to be further elucidated [29]. Recent findings concerning these interaction and the consequences on ATGL activity are discussed below.
1.2. Regulation of ATGL by proteins
The lipolytic activity of ATGL is regulated by the direct and indirect interaction with several proteins and individual interaction partners are described here in detail.
1.2.1. Perilipins (Plins)
The genes for the human Plin protein family are located on different chromosomes and are expressed in a tissue-specific manner to enable the precise management of lipid storage and breakdown in different tissues [19, 72]. The family consists of five structurally related proteins (Plin1-5), which share a conserved PAT domain (approx. 100 amino acids) in their N-terminal region [19, 73, 74] and a conserved 11-mer repeat region forming a helical motif C-terminal of the PAT domain, which is associated with LD localization [75]. Orthologs of Plins have been described in multiple organisms, including mouse, rat, fly, C. elegans and fungi [76–79]. Plins are suggested to serve as protective barrier to prevent the access of lipases to the surface of LDs. In response to hormonal stimulation, some Plins are phosphorylated and thus change from their protective state to a lipolysis-supporting state [19]. The gene for human Plin1 (also known as Plin or Peri) lies on chromosome 15q26.1 and comprises 10 exons transcribing for a 522 amino acid protein (NCBI Reference Sequence: NG_029172.1). Plin1 is predominantly expressed in adipose tissue and is the most abundant coating protein on the surface of LDs [72, 73, 80]. Mouse Plin1 consists of 517 amino acids and shows a sequence identity of 82.3 % to the human ortholog.
Compared to the structural features of Plin2-5, Plin1 has a unique, additional C-terminal region which is responsible for the interaction with CGI-58 [19, 81] thereby sequestering it on the surface of LDs to prevent its stimulating interaction with ATGL [82–84]. Upon hormonal (e.g. β-adrenergic) stimulation, Plin1 is phosphorylated by PKA, which triggers the release of CGI-58 from the LD surface. This facilitates the interaction of CGI-58 with ATGL and is considered to represent the switch from basal to stimulated lipolysis [72, 82–84].
The gene for human Plin2 (also known as ADFP or ADRP) is located on chromosome 9p22.1 and includes 9 exons, which encode for a 437 amino acid protein (NCBI Reference Sequence: NM_001122.3). The mouse ortholog of Plin2 consists of 425 amino acids and shares 82.6 % sequence identity compared to the human protein. The human gene for Plin3 (also known as TIP47 or M6PRBP1) lies on chromosome 19p13.3 and comprises 8 exons encoding for a 434 amino acid protein (NCBI Reference Sequence: NG_028080.1). The ortholog for mouse Plin3 consists of 437 residues and shows 75.8 % sequence identity to human Plin3. Plin2 and Plin3 are expressed in virtually all tissues and therefore contribute to the surface of LDs in all cells [19, 36, 72]. Both proteins, Plin2 and Plin3, miss a C-terminal part of Plin1 required for the interaction with CGI-58. Interestingly, chimeric proteins containing Plin2 or Plin3 and the C-terminus of Plin1 restore the ability to sequester CGI-58 [81]. The three-dimensional structure of the C-terminal domain of mouse Plin3 missing the N-terminal PAT domain and the 11-mer repeat region responsible for LD localization has been determined and suggests to contain an α/β domain of unusual topology and a four-helix bundle which forms the shape of an “L” ([85], PDB entry: 1szi). The four-helix bundle shows structural similarities to the N-terminal LDL-receptor binding domain of apolipoprotein E [86]. A deep hydrophobic cleft is formed between the α/β domain and the four-helix bundle. The shape, size and hydrophobicity of the cleft rather suggest the potential interaction with hydrophobic peptides, proteins or monomeric lipid species than with phospholipids from the surface of LDs [85]. Sequence alignments show that the hydrophobic cleft is present in all proteins of the Plin family suggesting an important but yet unidentified functional role [19, 85, 87].
Upon lipolytic stimulation, chaperone-mediated autophagy triggers Plin2 and Plin3 to undergo proteasomal degradation. The chaperone heat shock protein HSPA8/hsc70 interacts with Plin2 and Plin3. The subsequent phosphorylation of Plin2 by AMPK enables Plin2 and Plin3 in complex with HSPA8/hsc70 to enter the proteasome [88]. Depletion of Plin2 and Plin3 from the surface of LDs implements the association of cytosolic ATGL to the LD and enables the first step of lipolysis [20, 89].
The gene for human Plin4 (also known as S3-12 or KIAA1881) is located on chromosome 19p13.3, comprises 9 exons and encodes for a 1,357 amino acid protein (NCBI Reference Sequence: NM_001080400.1). Plin4 is predominately expressed in adipose tissue but also in skeletal muscle and heart [36, 90]. The ortholog of mouse Plin4 consists of 1,403 amino acids and shares 62.5 % sequence identity to human Plin4. The extended size of Plin4 is a result of a 900 amino acid insert in the 11-mer repeat [19]. Plin4 localizes to nascent LDs [90, 91], however the inactivation of Plin4 in mice did not lead to any alterations in adipose tissue mass or adipocyte differentiation [92]. Interestingly, the knock-out of Plin4 was also associated with a reduction of Plin5 mRNA and protein content especially in the heart thereby decreasing cardiac lipid accumulation in mice [92]. The exact mechanism remains unclear but can also be an artifact by the experimental set-up since the genes encoding for Plin4 and Plin5 are located on the same chromosome in close proximity [92]. There is no evidence that Plin4 is target of phosphorylation, but the protein was not subject to extensive researched up to now [19].
The gene for human Plin5 (also known as LSDP5, MLDP, OXPAT or PAT1) is located on chromosome 19p13.3 and comprises 8 exons encoding for a 463 amino acid protein, which is predominantly expressed in oxidative tissue (NCBI Reference Sequence: NM_001013706.2, [36, 87, 93, 94]). The mouse ortholog of Plin5 consists of 463 amino acids and shows 73.2 % sequence identity compared to the human protein. Heart specific knock-out of Plin5 in a mouse model results in a reduction of TG content in the cardiac muscle as a result of a reduction in ATGL activity [95, 96]. Inactivation of Plin5 in mice also goes in line with a reduced TG content in the liver [97]. This is in accordance with heart specific overexpression of Plin5, which increases cardiac TG content and promotes cardiac steatosis [98].
Plin5 directly interacts with ATGL and CGI-58 [82–84], co-locates with ATGL and CGI-58 to the surface of LDs and increases their local concentration on LDs [71, 99, 100]. Plin5 does not bind ATGL and CGI-58 simultaneously; both proteins compete for the interaction with Plin5, which prevents the interaction of CGI-58 with ATGL. Residues Arg417-Phe463 of Plin5 are responsible for the interaction with ATGL as mutants covering this region were not able to bind ATGL but still interacted and co-localized with CGI-58 to LDs [100].
Ser155 of mouse Plin5 is target for PKA dependent phosphorylation. Plin5 regulates ATGL activity in oxidative tissue analogous to Plin1 in adipose tissue. After the phosphorylation event, Plin5 releases CGI-58, which in turn facilitates interaction of CGI-58 with ATGL leading to the induction of lipolysis [101–103].
1.2.2. Cell death activator CIDE-3 (CIDEC)
The gene for human CIDEC, also known as FSP27, is located on chromosome 3p25.3 and comprises 9 exons encoding for a 238 amino acid protein in the ‘canonical’ sequence (NCBI Reference Sequence: NG_042291.1). It is highly expressed in adipocytes and associated to LDs [36, 104]. Orthologs of CIDEC have been only described in higher organisms including mouse, rat and chicken. Mouse CIDEC consists of 239 amino acids and shares 79.8 % sequence identity to the human ortholog. Three alternative splicing variants are described for human CIDEC, which vary in their length. CIDEC promotes LD clustering and enlargement primarily mediated by residues Ser120-Gln210 [105]. Interaction of ATGL with the C-terminal region of CIDEC harboring residues Ser120-Pro220 [28], decreases lipolysis by restricting access of ATGL to TG stores of the LDs [31]. This C-terminal region of CIDEC is also responsible for the interaction with Plin1 promoting its co-localization and the enlargement of LD size [106]. In addition, CIDEC directly interacts with the repressor protein Erg1, leading to binding of Erg1 to the ATGL/PNPLA2 promotor region, which suppresses its transcription [107]. The crystal structure of the N-terminal domain of CIDEC (CIDE-N, Val32-Ser120, PDB entry: 1d4b) reveals a CIDE domain fold with an α/β roll fold with two α helices and five β strands which form a homo-dimer in solution [108].
1.2.3. Hypoxia-inducible lipid droplet-associated (HILPDA)
The gene for human HILPDA is located on chromosome 7q32.1, comprised 2 exons which encodes for a small, 63 amino acid protein (NCBI Reference Sequence: NM_001098786.1). As the name indicates, its expression is induced by hypoxic stress in virtually all tissues [36, 109]. Orthologs of HILPDA have been only described in higher organisms, including mouse and rat. The mouse ortholog of HILPDA consists of 64 amino acids and shares 77.8 % sequence identity to the human protein. HILPDA coats the surface of LDs and co-localizes with Plin 2 and Plin 3 [109]. Residues Met1-Gly37 of mouse HILPDA are associated with LD localization [109]. Hepatic overexpression of HILPDA was shown to promote TG accumulation, whereas downregulation reduces TG content [27, 29]. As HILPDA shows similarities to the hydrophobic region of G0S2 involved in the interaction with ATGL [23, 110], it is speculated that HILPDA has potential to inhibit ATGL activity. To date one study had addressed this question, but did not find any effect on the TG hydrolyzing activity of basal ATGL [29]. Consequently, this assumption still awaits unambiguous experimental evidence.
1.2.4. Comparative Gene Identification 58 (CGI-58)
The gene for human CGI-58, also known as ABHD5, is located on chromosome 3p21.33, it includes 8 exons and encodes for a 349 amino acid protein (NCBI Reference Sequence: NG_007090.3, [111]). CGI-58 is ubiquitously expressed in all tissues including adipose tissue, testes, liver, kidney, heart and skin [21, 36, 83, 112]. Orthologs of CGI-58 have been described in multiple organisms, including mouse, rat, drosophila, plants and yeast [21, 113–116]. Mouse CGI-58 consists of 351 amino acids and shows a high sequence identity of 94.3 % to the human ortholog. An alternative splicing isoform, missing exon 2 and 3, is described for mouse CGI-58. It encodes for a 202 amino acid protein that lacks the N-terminal region of CGI-58 [117].
To date no three-dimensional structure is known, but CGI-58 is predicted to harbor an α/β hydrolase fold spanning from residue Arg102-Cys345. Compared to functional α/β hydrolase hydrolases, CGI-58 harbors an impaired catalytic triad composed of Asp301, His327, with the nucleophilic serine usually found in the GXSXG motif being replaced by Asn153 [111]. However, reconstruction of the catalytic triad could not restore lipase activity of CGI-58 [12]. Weak lysophosphatic acid acyl transferase (LPAAT) activity mediated by a conserved HX4D motif had been reported for murine and plant CGI-58, yet was later dismissed [118, 119].
Interestingly, human patients with mutations (including single nucleotide-, deletion-, insertion- and splice site mutations leading to single-amino acid exchanges and truncated proteins) in the gene encoding for CGI-58 have been shown to cause NLSD with ichthyosis and show a phenotype different to patients with a loss of function of ATGL [55, 111]. They suffer from neutral lipid storage disease accompanied by ichthyosis (NLSD-I), which is characterized by the abnormal accumulation of TGs especially in the skin [55, 111]. Global inactivation of CGI-58 in mice leads to a drastic skin barrier defect also indicating a role different to ATGL activation. This indicates a function of CGI-58 in the skin and it has been speculated if CGI-58 may also activate a skin specific triacylglycerol lipase [120, 121].
CGI-58 directly interacts with ATGL and translocates it to LDs, which leads to an activation of the lipase up to 20 fold [21]. CGI-58 contains a hydrophobic Trp-rich stretch at its N-terminus, which forms an anchor essential for localization to LDs [122, 123]. LD binding mediated by this region appears to be a prerequisite for ATGL co-activation [123]. Plin1 sequesters CGI-58 at the surface of LDs and stimulation of ATGL by CGI-58 can thus take place only upon PKA mediated phosphorylation of Plin1 and subsequent release of CGI-58 [82–84]. CGI-58 is also target for phosphorylation and PKA dependent phosphorylation of Ser239 is additionally required for the release of CGI-58 from Plin1 [124].
Similarly, Plin5 interacts with CGI-58 in oxidative tissue thus preventing stimulatory interaction of CGI-58 with ATGL [71, 99, 100]. Interestingly, the presence of activated long-chain FA intermediates such as oleoyl-CoA triggers the interaction of CGI-58 with Plin1 and Plin5 suggesting a negative feedback mechanism to inhibit lipolysis by excluding ATGL from the LD [125]. Recent studies identified Gly328 and Arg299 within CGI-58 as essential residues for ATGL activation. ABHD4, a closely related family member of CGI-58 possesses hydrolase activity but fails to activate ATGL. When Gly328 and Arg299 were introduced in ABHD4 at equivalent positions as in CGI-58/ABHD5, ABHD4 partially gained ATGL activating function [126]. Conversely, the loss of function mutant of CGI-58/ABHD5 failed to activate ATGL but was still capable to interact with Plin1 and Plin5 and to translocate ATGL to the LD. Moreover, a negatively charged amino acid at position Asp334 of CGI-58/ABHD5 is required for ATGL activation, as the exchange to Ala or Asn but not Glu abolished the potency to activate ATGL. In a suggested mechanism, Arg299 and Asp334, which are in close proximity in the structural model, form a perturbation at the surface of LDs to enable the access of ATGL to the TG stores [126]. Additionally, CGI-58 was reported to bind to five out of nine members of the fatty acid-binding protein (FABP) family. In the presence of A-FABP, CGI-58 is even more potent to activate ATGL [127]. FABPs bind long-chain FAs with high affinity and are important for the uptake and transport of free FAs. The interaction with CGI-58 may facilitate the adsorption of generated FAs and therefore remove them from ATGL resulting in an increase in TG hydrolysis [127–129]. Additionally, this interaction provides a direct link of lipolysis to lipid signaling and induction of peroxisome proliferator-activated receptors [127].
1.2.5. G0/G1 Switch Protein 2 (G0S2)
The gene for human G0S2 is located at chromosome 1q32.2, comprises 2 exons and encodes a 103 amino acid protein (NCBI Reference Sequence: NM_015714.3, [130]). G0S2 is ubiquitously expressed with highest levels in adipose tissue, followed by bone marrow, skeletal muscle and liver [23, 36, 130–132]). Orthologs of G0S2 have only been identified in vertebrates including human, mouse, rat and chicken. Orthologs in plants or other organisms including yeast or flies have not been reported [23, 116, 133, 134]. Mouse G0S2 comprises 103 amino acids and shares 77.7 % sequence identity to the human protein. Highest conservation throughout all species is found at the N-terminal region of G0S2 while the C-terminus shows higher alterations.
To date, no experimental three-dimensional structure of G0S2 has been published and even homology modelling attempts fail to identify a suitable structural homolog. G0S2 harbors a highly conserved hydrophobic region predicted to be an α-helix within the N-terminal half of the protein, whereas the C-terminus of G0S2 is predicted to be unstructured.
G0S2 directly interacts with ATGL and thereby inhibits its TG hydrolase activity [23, 110]. N-terminal and C-terminal deletions of G0S2 retaining only the mainly hydrophobic stretch Lys20-Ala52 was shown to be sufficient for inhibiting ATGL [110]. The corresponding synthetic peptide containing Lys20-Ala52 of human G0S2 is sufficient for the effective and selective inhibition of ATGL activity and acts in a non-competitive mode with an inhibitory constant of 25 nM [110]. The interaction of G0S2 with ATGL is independent from the interaction with CGI-58 since G0S2 inhibits ATGL even in the presence of the activator CGI-58 [22, 58, 110]. Protein-protein interaction of G0S2 takes place at the N-terminal patatin like domain of ATGL [23, 58, 110]. Using different deletion variants the interaction region was delineated to the first 254 amino acids of ATGL [58]. Human G0S2 is capable to inhibit mouse ATGL, which is in agreement to the high level of conservation of the involved hydrophobic region essential for mediating this interaction [110]. Interestingly, G0S2 is also involved in the localization of ATGL to the surface of LD as C-terminally truncated ATGL, which fails to localize to LDs, could be recruited again to LDs by G0S2 [134].
Interestingly, no pathologies are described that are associated with defective G0S2. Inactivation of G0S2 in mice leads to enhanced lipolysis, reduced adipose tissue mass, improved glucose tolerance and insulin sensitivity, while overexpression of G0S2 results in reduced lipolysis, increased fat mass and fatty liver [135–139] which is very similar to the phenotype observed for knock-out of ATGL [1, 62]. Moreover, it was shown, that G0S2-knock-out mice are resistant to high-fat died induced body weight gain, are glucose tolerant and insulin sensitive, which renders the protein interesting in the treatment of obesity and different metabolic disorders [140]. To date no phosphorylation sites of G0S2 are described. G0S2 undergoes proteasomal degradation [141]. Lys25 of G0S2 represents a target for ubiquitination, which triggers it to the proteasome. Interestingly, G0S2 could be stabilized upon interaction with ATGL, and this increase in protein stability appears to be independent of CGI-58 [142].
G0S2 is also involved in multiple other important cellular functions, including apoptosis, oxidative phosphorylation and cell proliferation [131, 143–146]. The gene coding for G0S2 was reported to be hypermethylated in certain cancer types, thus indicating a potential role in the development of cancer [131, 147]. Direct interaction of G0S2 with nucleolin in the cytosol leads to an inhibition of cell proliferation. The hydrophobic region of G0S2 and the C-terminal stretch of nucleolin, which is rich in Arg-Gly-Gly repeats, are putatively involved in the interaction, as the generation of variants in which the hydrophobic region of G0S2 and the Arg-Gly-Gly repeats of nucleolin were deleted, resulted in proteins that were unable to interact [145, 146]. In mitochondria, G0S2 serves the role of promoting apoptosis as it specifically interacts with Bcl-2 to prevent its interaction with Bax and the formation of the anti-apoptotic Bcl-2/Bax heterodimer complex [131]. No Bcl-2 homology domain is known in G0S2, but it is suggested to interact with Bcl-2 through its hydrophobic region since residues Leu33-Gln67 of G0S2 are sufficient for the interaction with Bcl-2 [131]. Under hypoxic stress, G0S2 was observed to interact with F0/F1-ATP synthase to protect cells from a critical energy crisis. Interestingly, the binding of G0S2 to ATP5A subunit of F0/F1-ATP synthase leads to increased ATP production and enhanced oxidative phosphorylation [144, 148]. The exact mechanism underlying these upregulation is unknown.
1.2.6. Pigment epithelium derived factor (PEDF)
The gene for human PEDF, also known as SERPINF1 is located at chromosome 17p13.3 and comprises 9 exons encoding a 418 amino acid protein (NCBI Reference Sequence: NG_028180.1, [149]). PEDF is a non-inhibitory member of the serpin protein family and was first discovered as an extracellular protein derived from pigment epithelial cells [150–152]. Expression of PEDF is found in virtually all tissues as it is an important secretion factor of adipocytes with the ability to modulate insulin sensitivity [36, 153]. Orthologs of PEDF are described in higher organisms, including mouse and rat, as well as in bacteria. The ortholog of mouse PEDF consists of 417 amino acids and shows 86.1 % sequence identity compared to human PEDF.
The experimental three-dimensional structure of glycosylated human PEDF ([154], PDB entry: 1imv) reveals an α/β core serpin domain. Moreover, PEDF shows an asymmetric charge distribution unlike other members of the serpin protein family with potential physiological relevance. PEDF is a multifunctional protein with neurotrophic properties and residues Val58-Thr101 are potentially involved in binding to PEDF-receptors [151, 154].
Endogenous PEDF was shown to directly interact with ATGL and stimulate lipolysis in adipocytes, hepatocytes and in the retina [24, 25, 30]. Dai et al. reported an important role of residues Arg268-Lys504 in ATGL to be responsible for the binding to PEDF [26]. PEDF interacts with ATGL independently of ATGL’s interaction with G0S2, which is in full agreement of the completely different interaction regions [23, 26, 58, 110]. PEDF only activates basal, but not stimulated lipolysis and it was also reported that PEDF is able to translocate ATGL to LDs [25, 26]. Interestingly, residues Leu159-Met325 in ATGL exhibit same binding affinity to PEDF as full length ATGL, and truncated variants Ile193-Met232 and Thr210-Leu249 of ATGL selectively bind to PEDF [155]. A 44-mer sequence comprising Val78-Thr121 of PEDF was identified to be sufficient to directly interact with ATGL and to induce ATGL’s TG-hydrolytic activity [156]. Residues Thr210-Leu249 of ATGL selectively bind to the 44-mer sequence and even to a shorter 17-mer fragment (Gln98-Ser114) of PEDF. Within that 17-mer sequence, the exchange of Arg99 to Ala abolishes the binding, whereas mutating His105 to Ala increases binding affinity to Thr210-Leu249 of ATGL [157].
Elevated PEDF levels induce insulin resistance as a consequence of elevated levels of FAs in obese mice and humans [153, 158]. This lead to the speculation that PEDF mediated insulin resistance is a consequence of the upregulation of ATGL activity by PEDF [24]. Besides the effect of stimulating lipolysis, PEDF decreases ATGL protein levels in vivo and in vitro by triggering ATGL to undergo proteasome mediated degradation via the ubiquitin dependent pathway [159]. However, it remains ambiguous how enhanced proteasomal degradation of ATGL relates to the observed elevated levels of FAs.
1.3. Regulation of ATGL by small molecules
Naturally occurring and synthetically derived small molecules have been reported to influence ATGL’s TG hydrolyzing activity and are described in this chapter.
1.3.1. Long-chain acyl-CoAs inhibit ATGL
Long-chain acyl-CoAs are activated FAs, which can further serve for β-oxidation, protein acylation or as precursors for membrane lipids. Long-chain acyl-CoAs regulate a set of important cellular functions and metabolic enzymes including HSL [6, 7]. Long-chain acyl-CoAs non-competitively inhibit ATGL activity. Oleoyl-CoA has the most effect and inhibits ATGL dose-dependently with a half maximal inhibitory concentration (IC50) of approx. 25 µM [32]. This is in the similar range as HSL inhibition by Acyl-CoAs (IC50 of 25-40 µM [7]). As observed with other ATGL-interactions, the presence of the activator protein CGI-58 does not seem to influence acyl-CoA mediated inhibition. Physiologic concentrations of long-chain acyl-CoAs vary from 5-160 µM and strongly depend on the metabolic condition of the organism, such as fasting, feeding or insulin resistance [160]. According to this, inhibition of ATGL by oleoyl-CoAs is physiological relevant and could represent a product feedback mechanism in dysregulated lipolysis [32]. It is established that long-chain acyl-CoAs are predominantly bound to acyl-CoA binding proteins (ACBPs) and levels of free long-chain acyl-CoAs are rather low [161]. However, long-chain acyl-CoAs bound to ACBP are able to enter metabolic processes like β-oxidation [160, 162, 163], but it is currently unknown whether the ACBP-Acyl-CoA complex can also affect ATGL activity [160].
1.3.2. Atglistatin, the first specifically developed synthetic ATGL inhibitor
As ATGL is a promising target to affect lipolysis and as such many physiological processes downstream of lipolysis, considerable effort was put into the development of a small synthetic ATGL inhibitor. Starting with a high-throughput screen originally established to identify HSL inhibitors [164, 165] a potent molecule, termed Atglistatin was discovered [33]. Atglistatin selectively inhibits the activity of mouse ATGL in vitro and in vivo in a competitive mode with an IC50 of 0.7 µM. Oral gavage of Atglistatin to mice also leads to the inhibition of ATGL as evidenced by reduced plasma TG and FA levels [33]. Administration of Atglistatin is associated with different positive effects, e.g. suppression of tumor cell growths [166] and effectively reduced high-fat diet induced weight gain, insulin resistance and fatty liver [34]. Notably, the long-term treatment of mice with Atglistatin was not accompanied with TG accumulation in the skeletal muscle or the heart as observed for ATGL knock-out mice. These observations render small molecule inhibition of ATGL a very interesting target for the treatment of obesity-associated diseases and the concomitant metabolic syndrome comprising of multiple pathologies. Especially pathologies associated with the concept of lipotoxicity - where adipose tissue-derived FAs and FA-derivatives mediate toxic effects in non-adipose tissues and cells including dysregulation of metabolic and signaling (insulin-, inflammatory-, nitric oxide-signaling) pathways, cell- and organelle-dysfunction and cell death - might benefit greatly from pharmacological inhibition of intracellular lipolysis [33, 34, 167, 168].
1.4. Unspecific inhibition of ATGL
Many experimental studies on ATGL crucially depend on enzymatic assays testing for the hydrolytic activity of ATGL in tissues and in vitro. ATGL’s TG hydrolyzing activity is very sensitive to pH, as maximal in vitro ATGL activity was reported at a pH value of 7.0, with changes of 1 unit in both directions leading to the reduction of ATGL activity to 50 %. ATGL exerts highest activity using a phosphatidylcholine/phosphatidylinositol (PC/PI) or Triton-X emulsion as LD mimic. ATGL shows a reduction of 35 % in activity when mimicking the LD with cholate. Interestingly, the interaction of ATGL with its activator protein CGI-58 highly depends on the nature of the detergent used as LD mimic since the stimulation with CGI-58 is only facilitated using a PC/PI LD mimic. These findings are based on empirical reports and might be due to unspecific interactions or conformational changes induced in different assay conditions [169].
Conclusion
For more than a decade, ATGL has been considered to be one of the most important enzymes in intracellular lipolysis for very good reasons: The lipase exerts the first step in the breakdown of TGs [1, 2] and dysregulated lipolysis is associated with metabolic diseases including insulin resistance, diabetes, inflammation and non-alcoholic fatty liver. Therefore, it is of great importance to understand the regulation of this crucial metabolic process [8–10].
To date, a number of interaction partners involved in the regulation of ATGL’s TG hydrolyzing activity have been identified. Direct protein-protein interactions have been established between ATGL and its activator protein CGI-58, or inhibitor protein G0S2 [21–23]. Additional regulation is achieved by direct interactions with the LD associated proteins Plin5 in oxidative tissue [100, 103] and CIDEC [28, 31], which restrict the access of ATGL to LDs. ATGL also interacts with the adipocyte secretion protein PEDF leading to a stimulation of ATGL activity [24, 25, 30]. However, questions about their exact interplay to collaboratively regulate ATGL still need to be addressed.
Although mutational analysis narrows down interaction regions of the single players, experimental three dimensional structures of the involved proteins, especially of ATGL, CGI-58 and G0S2 are missing to map a precise interaction surface and more important to investigate their mode of action. Clearly, further research on the regulation of ATGL has to be performed to finally answer this important questions and to identify other potential interaction partners crucial for the regulation of ATGL.
Acknowledgements
This work was supported by the grant SFB LIPOTOX F30, the grant P22170 and the DK Molecular Enzymology W09, all funded by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung.
List of Abbreviations
- ATGL
adipose triglyceride lipase
- ABDH5
α/β hydrolase domain containing protein 5
- ACBP
acyl-CoA binding protein
- CGI-58
comparative gene identification 58
- CIDEC
cell death activator CIDE-3
- FA
fatty acid
- FABP
fatty acid binding protein
- FSP27
fat-specific protein 27
- G0S2
G0/G1 switch gene 2,
- HILPDA
hypoxia-inducible lipid dropled-associated
- HSL
hormone sensitive lipase
- IC50
half maximal inhibitory concentration
- LD
lipid droplet
- PC
phosphatidylcholine
- PEDF
pigment epithelium derived factor
- PI
phosphatidylinositol
- Plin
perilipin
- PNPLA
patatin domain containing protein family
- TG
triacylglycerol
References
- [1].Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–7. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- [2].Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–6. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- [3].Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–15. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- [4].Osuga J, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujimoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O, Yamada N. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci U S A. 2000;97:787–92. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem. 1997;272:27218-23. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- [6].Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Severson DL, Hurley B. Inhibition of the hormone-sensitive lipase in adipose tissue by long-chain fatty acyl coenzyme A. Lipids. 1984;19:134–8. doi: 10.1007/BF02534504. [DOI] [PubMed] [Google Scholar]
- [8].Li LO, Klett EL, Coleman RA. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim Biophys Acta. 2010;1801:246–51. doi: 10.1016/j.bbalip.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–77. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim Biophys Acta. 2010;1801:209–14. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- [11].Unger RH, Scherer PE. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol Metab. 2010;21:345–52. doi: 10.1016/j.tem.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Young SG, Zechner R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 2013;27:459–84. doi: 10.1101/gad.209296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ellong EN, Soni KG, Bui QT, Sougrat R, Golinelli-Cohen MP, Jackson CL. Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1. PLoS One. 2011;6:e21889. doi: 10.1371/journal.pone.0021889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–61. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci. 2009;122:1834–41. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV, Jr, Walther TC. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kimmel AR, Sztalryd C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu Rev Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- [20].Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–70. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–19. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- [22].Lu X, Yang X, Liu J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle. 2010;9:2719–25. doi: 10.4161/cc.9.14.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Borg ML, Andrews ZB, Duh EJ, Zechner R, Meikle PJ, Watt MJ. Pigment epithelium-derived factor regulates lipid metabolism via adipose triglyceride lipase. Diabetes. 2011;60:1458–66. doi: 10.2337/db10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chung C, Doll JA, Gattu AK, Shugrue C, Cornwell M, Fitchev P, Crawford SE. Anti-angiogenic pigment epithelium-derived factor regulates hepatocyte triglyceride content through adipose triglyceride lipase (ATGL) J Hepatol. 2008;48:471–8. doi: 10.1016/j.jhep.2007.10.012. [DOI] [PubMed] [Google Scholar]
- [26].Dai Z, Zhou T, Li C, Qi W, Mao Y, Lu J, Yao Y, Li L, Zhang T, Hong H, Li S, et al. Intracellular pigment epithelium-derived factor contributes to triglyceride degradation. Int J Biochem Cell Biol. 2013;45:2076–86. doi: 10.1016/j.biocel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- [27].DiStefano MT, Danai LV, Roth Flach RJ, Chawla A, Pedersen DJ, Guilherme A, Czech MP. The Lipid Droplet Protein Hypoxia-inducible Gene 2 Promotes Hepatic Triglyceride Deposition by Inhibiting Lipolysis. J Biol Chem. 2015;290:15175–84. doi: 10.1074/jbc.M115.650184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Grahn TH, Kaur R, Yin J, Schweiger M, Sharma VM, Lee MJ, Ido Y, Smas CM, Zechner R, Lass A, Puri V. Fat-specific protein 27 (FSP27) interacts with adipose triglyceride lipase (ATGL) to regulate lipolysis and insulin sensitivity in human adipocytes. J Biol Chem. 2014;289:12029–39. doi: 10.1074/jbc.M113.539890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mattijssen F, Georgiadi A, Andasarie T, Szalowska E, Zota A, Krones-Herzig A, Heier C, Ratman D, De Bosscher K, Qi L, Zechner R, et al. Hypoxia-inducible lipid droplet-associated (HILPDA) is a novel peroxisome proliferator-activated receptor (PPAR) target involved in hepatic triglyceride secretion. J Biol Chem. 2014;289:19279–93. doi: 10.1074/jbc.M114.570044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, Notario PM, Saravanamuthu S, Nueda ML, Sanchez-Sanchez F, Escribano J, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–37. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- [31].Yang X, Heckmann BL, Zhang X, Smas CM, Liu J. Distinct mechanisms regulate ATGL-mediated adipocyte lipolysis by lipid droplet coat proteins. Mol Endocrinol. 2013;27:116–26. doi: 10.1210/me.2012-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nagy HM, Paar M, Heier C, Moustafa T, Hofer P, Haemmerle G, Lass A, Zechner R, Oberer M, Zimmermann R. Adipose triglyceride lipase activity is inhibited by long-chain acyl-coenzyme A. Biochim Biophys Acta. 2014;1841:588–94. doi: 10.1016/j.bbalip.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mayer N, Schweiger M, Romauch M, Grabner GF, Eichmann TO, Fuchs E, Ivkovic J, Heier C, Mrak I, Lass A, Hofler G, et al. Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat Chem Biol. 2013;9:785–7. doi: 10.1038/nchembio.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schweiger M, Romauch M, Schreiber R, Grabner GF, Hutter S, Kotzbeck P, Benedikt P, Eichmann TO, Yamada S, Knittelfelder O, Diwoky C, et al. Pharmacological inhibition of adipose triglyceride lipase corrects high-fat diet-induced insulin resistance and hepatosteatosis in mice. Nat Commun. 2017;8:14859. doi: 10.1038/ncomms14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fischer J, Lefevre C, Morava E, Mussini JM, Laforet P, Negre-Salvayre A, Lathrop M, Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- [36]. http://www.proteinatlas.org .
- [37].Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–57. [PMC free article] [PubMed] [Google Scholar]
- [38].Lake AC, Sun Y, Li JL, Kim JE, Johnson JW, Li D, Revett T, Shih HH, Liu W, Paulsen JE, Gimeno RE. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005;46:2477–87. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- [39].Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–75. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- [40].Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–75. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- [41].Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–9. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- [42].Chen JF, Dai LH, Xu NY, Xiong YZ, Jiang SW. Assignment of the patatin-like phospholipase domain containing 2 gene (PNPLA2) to porcine chromosome 2p17 with radiation hybrids. Cytogenet Genome Res. 2006;112:342G. doi: 10.1159/000089897. [DOI] [PubMed] [Google Scholar]
- [43].Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–75. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Festuccia WT, Laplante M, Berthiaume M, Gelinas Y, Deshaies Y. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 2006;49:2427–36. doi: 10.1007/s00125-006-0336-y. [DOI] [PubMed] [Google Scholar]
- [45].Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–30. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- [46].Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- [47].Saarela J, Jung G, Hermann M, Nimpf J, Schneider WJ. The patatin-like lipase family in Gallus gallus. BMC Genomics. 2008;9:281. doi: 10.1186/1471-2164-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kienesberger PC, Oberer M, Lass A, Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009;50:S63–8. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–60. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- [50].Gendrin C, Contreras-Martel C, Bouillot S, Elsen S, Lemaire D, Skoufias DA, Huber P, Attree I, Dessen A. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 2012;8:e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ku B, Lee KH, Park WS, Yang CS, Ge J, Lee SG, Cha SS, Shao F, Heo WD, Jung JU, Oh BH. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 2012;8:e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- [53].Tyson GH, Halavaty AS, Kim H, Geissler B, Agard M, Satchell KJ, Cho W, Anderson WF, Hauser AR. A novel phosphatidylinositol 4,5-bisphosphate binding domain mediates plasma membrane localization of ExoU and other patatin-like phospholipases. J Biol Chem. 2015;290:2919–37. doi: 10.1074/jbc.M114.611251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wijeyesakere SJ, Richardson RJ, Stuckey JA. Crystal structure of patatin-17 in complex with aged and non-aged organophosphorus compounds. PLoS One. 2014;9:e108245. doi: 10.1371/journal.pone.0108245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–96. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- [56].Kobayashi K, Inoguchi T, Maeda Y, Nakashima N, Kuwano A, Eto E, Ueno N, Sasaki S, Sawada F, Fujii M, Matoba Y, et al. The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J Clin Endocrinol Metab. 2008;93:2877–84. doi: 10.1210/jc.2007-2247. [DOI] [PubMed] [Google Scholar]
- [57].Schweiger M, Schoiswohl G, Lass A, Radner FP, Haemmerle G, Malli R, Graier W, Cornaciu I, Oberer M, Salvayre R, Fischer J, et al. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J Biol Chem. 2008;283:17211–20. doi: 10.1074/jbc.M710566200. [DOI] [PubMed] [Google Scholar]
- [58].Cornaciu I, Boeszoermenyi A, Lindermuth H, Nagy HM, Cerk IK, Ebner C, Salzburger B, Gruber A, Schweiger M, Zechner R, Lass A, et al. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011;6:e26349. doi: 10.1371/journal.pone.0026349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Coassin S, Schweiger M, Kloss-Brandstatter A, Lamina C, Haun M, Erhart G, Paulweber B, Rahman Y, Olpin S, Wolinski H, Cornaciu I, et al. Investigation and functional characterization of rare genetic variants in the adipose triglyceride lipase in a large healthy working population. PLoS Genet. 2010;6:e1001239. doi: 10.1371/journal.pgen.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Missaglia S, Tasca E, Angelini C, Moro L, Tavian D. Novel missense mutations in PNPLA2 causing late onset and clinical heterogeneity of neutral lipid storage disease with myopathy in three siblings. Mol Genet Metab. 2015;115:110–7. doi: 10.1016/j.ymgme.2015.05.001. [DOI] [PubMed] [Google Scholar]
- [61].Pennisi EM, Missaglia S, Dimauro S, Bernardi C, Akman HO, Tavian D. A myopathy with unusual features caused by PNPLA2 gene mutations. Muscle Nerve. 2015;51:609–13. doi: 10.1002/mus.24477. [DOI] [PubMed] [Google Scholar]
- [62].Hoy AJ, Bruce CR, Turpin SM, Morris AJ, Febbraio MA, Watt MJ. Adipose triglyceride lipase-null mice are resistant to high-fat diet-induced insulin resistance despite reduced energy expenditure and ectopic lipid accumulation. Endocrinology. 2011;152:48–58. doi: 10.1210/en.2010-0661. [DOI] [PubMed] [Google Scholar]
- [63].Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–65. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- [64].Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, Wang Y, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13:739–48. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Duncan RE, Wang Y, Ahmadian M, Lu J, Sarkadi-Nagy E, Sul HS. Characterization of desnutrin functional domains: critical residues for triacylglycerol hydrolysis in cultured cells. J Lipid Res. 2010;51:309–17. doi: 10.1194/jlr.M000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kim SJ, Tang T, Abbott M, Viscarra JA, Wang Y, Sul HS. AMPK Phosphorylates Desnutrin/ATGL and Hormone-Sensitive Lipase To Regulate Lipolysis and Fatty Acid Oxidation within Adipose Tissue. Mol Cell Biol. 2016;36:1961–76. doi: 10.1128/MCB.00244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mason RR, Meex RC, Lee-Young R, Canny BJ, Watt MJ. Phosphorylation of adipose triglyceride lipase Ser(404) is not related to 5’-AMPK activation during moderate-intensity exercise in humans. Am J Physiol Endocrinol Metab. 2012;303:E534–41. doi: 10.1152/ajpendo.00082.2012. [DOI] [PubMed] [Google Scholar]
- [68].Pagnon J, Matzaris M, Stark R, Meex RC, Macaulay SL, Brown W, O’Brien PE, Tiganis T, Watt MJ. Identification and functional characterization of protein kinase A phosphorylation sites in the major lipolytic protein, adipose triglyceride lipase. Endocrinology. 2012;153:4278–89. doi: 10.1210/en.2012-1127. [DOI] [PubMed] [Google Scholar]
- [69].Xie X, Langlais P, Zhang X, Heckmann BL, Saarinen AM, Mandarino LJ, Liu J. Identification of a novel phosphorylation site in adipose triglyceride lipase as a regulator of lipid droplet localization. Am J Physiol Endocrinol Metab. 2014;306:E1449–59. doi: 10.1152/ajpendo.00663.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Eichmann TO, Kumari M, Haas JT, Farese RV, Jr, Zimmermann R, Lass A, Zechner R. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J Biol Chem. 2012;287:41446–57. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wang H, Bell M, Sreenivasan U, Hu H, Liu J, Dalen K, Londos C, Yamaguchi T, Rizzo MA, Coleman R, Gong D, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286:15707–15. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. 2012;287:2273–9. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–59. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- [74].Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–71. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Rowe ER, Mimmack ML, Barbosa AD, Haider A, Isaac I, Ouberai MM, Thiam AR, Patel S, Saudek V, Siniossoglou S, Savage DB. Conserved Amphipathic Helices Mediate Lipid Droplet Targeting of Perilipins 1-3. J Biol Chem. 2016;291:6664–78. doi: 10.1074/jbc.M115.691048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chughtai AA, Kassak F, Kostrouchova M, Novotny JP, Krause MW, Saudek V, Kostrouch Z, Kostrouchova M. Perilipin-related protein regulates lipid metabolism in C. elegans. PeerJ. 2015;3:e1213. doi: 10.7717/peerj.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm Genome. 2001;12:741–9. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- [78].Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–7. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- [79].Wang C, St Leger RJ. The Metarhizium anisopliae Perilipin Homolog MPL1 Regulates Lipid Metabolism, Appressorial Turgor Pressure, and Virulence. J Biol Chem. 2007;282:21110–5. doi: 10.1074/jbc.M609592200. [DOI] [PubMed] [Google Scholar]
- [80].Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–91. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- [81].Patel S, Yang W, Kozusko K, Saudek V, Savage DB. Perilipins 2 and 3 lack a carboxy-terminal domain present in perilipin 1 involved in sequestering ABHD5 and suppressing basal lipolysis. Proc Natl Acad Sci U S A. 2014;111:9163–8. doi: 10.1073/pnas.1318791111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J Biol Chem. 2009;284:34538–44. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–71. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- [84].Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem. 2004;279:30490–7. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- [85].Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH. Structure of a lipid droplet protein; the PAT family member TIP47. Structure. 2004;12:1199–207. doi: 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- [86].Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–22. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- [87].Dalen KT, Dahl T, Holter E, Arntsen B, Londos C, Sztalryd C, Nebb HI. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim Biophys Acta. 2007;1771:210–27. doi: 10.1016/j.bbalip.2006.11.011. [DOI] [PubMed] [Google Scholar]
- [88].Schweiger M, Zechner R. Breaking the Barrier--Chaperone-Mediated Autophagy of Perilipins Regulates the Lipolytic Degradation of Fat. Cell Metab. 2015;22:60–1. doi: 10.1016/j.cmet.2015.06.017. [DOI] [PubMed] [Google Scholar]
- [89].Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–8. doi: 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–21. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- [91].Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280:19146–55. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- [92].Chen W, Chang B, Wu X, Li L, Sleeman M, Chan L. Inactivation of Plin4 downregulates Plin5 and reduces cardiac lipid accumulation in mice. Am J Physiol Endocrinol Metab. 2013;304:E770–9. doi: 10.1152/ajpendo.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao-Borengasser A, Rasouli N, Kern PA, Finck BN, et al. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 2006;55:3418–28. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- [94].Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem. 2006;281:14232–40. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- [95].Kuramoto K, Okamura T, Yamaguchi T, Nakamura TY, Wakabayashi S, Morinaga H, Nomura M, Yanase T, Otsu K, Usuda N, Matsumura S, et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J Biol Chem. 2012;287:23852–63. doi: 10.1074/jbc.M111.328708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Pollak NM, Schweiger M, Jaeger D, Kolb D, Kumari M, Schreiber R, Kolleritsch S, Markolin P, Grabner GF, Heier C, Zierler KA, et al. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J Lipid Res. 2013;54:1092–102. doi: 10.1194/jlr.M034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang C, Zhao Y, Gao X, Li L, Yuan Y, Liu F, Zhang L, Wu J, Hu P, Zhang X, Gu Y, et al. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology. 2015;61:870–82. doi: 10.1002/hep.27409. [DOI] [PubMed] [Google Scholar]
- [98].Wang H, Sreenivasan U, Gong DW, O’Connell KA, Dabkowski ER, Hecker PA, Ionica N, Konig M, Mahurkar A, Sun Y, Stanley WC, et al. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J Lipid Res. 2013;54:953–65. doi: 10.1194/jlr.M032466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Granneman JG, Moore HP, Mottillo EP, Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem. 2009;284:3049–57. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J Biol Chem. 2011;286:5126–35. doi: 10.1074/jbc.M110.180711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Macpherson RE, Vandenboom R, Roy BD, Peters SJ. Skeletal muscle PLIN3 and PLIN5 are serine phosphorylated at rest and following lipolysis during adrenergic or contractile stimulation. Physiol Rep. 2013;1:e00084. doi: 10.1002/phy2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Pollak NM, Jaeger D, Kolleritsch S, Zimmermann R, Zechner R, Lass A, Haemmerle G. The interplay of protein kinase A and perilipin 5 regulates cardiac lipolysis. J Biol Chem. 2015;290:1295–306. doi: 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong DW, Stanley WC, Sztalryd C. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. 2011;52:2159–68. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Liang L, Zhao M, Xu Z, Yokoyama KK, Li T. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem J. 2003;370:195–203. doi: 10.1042/BJ20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jambunathan S, Yin J, Khan W, Tamori Y, Puri V. FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One. 2011;6:e28614. doi: 10.1371/journal.pone.0028614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Grahn TH, Zhang Y, Lee MJ, Sommer AG, Mostoslavsky G, Fried SK, Greenberg AS, Puri V. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem Biophys Res Commun. 2013;432:296–301. doi: 10.1016/j.bbrc.2013.01.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Singh M, Kaur R, Lee MJ, Pickering RT, Sharma VM, Puri V, Kandror KV. Fat-specific protein 27 inhibits lipolysis by facilitating the inhibitory effect of transcription factor Egr1 on transcription of adipose triglyceride lipase. J Biol Chem. 2014;289:14481–7. doi: 10.1074/jbc.C114.563080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lee SM, Jang TH, Park HH. Molecular basis for homo-dimerization of the CIDE domain revealed by the crystal structure of the CIDE-N domain of FSP27. Biochem Biophys Res Commun. 2013;439:564–9. doi: 10.1016/j.bbrc.2013.09.018. [DOI] [PubMed] [Google Scholar]
- [109].Gimm T, Wiese M, Teschemacher B, Deggerich A, Schodel J, Knaup KX, Hackenbeck T, Hellerbrand C, Amann K, Wiesener MS, Honing S, et al. Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J. 2010;24:4443–58. doi: 10.1096/fj.10-159806. [DOI] [PubMed] [Google Scholar]
- [110].Cerk IK, Salzburger B, Boeszoermenyi A, Heier C, Pillip C, Romauch M, Schweiger M, Cornaciu I, Lass A, Zimmermann R, Zechner R, et al. A peptide derived from G0/G1 switch gene 2 acts as noncompetitive inhibitor of adipose triglyceride lipase. J Biol Chem. 2014;289:32559–70. doi: 10.1074/jbc.M114.602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Lefevre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, Ozguc M, et al. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–12. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Akiyama M, Sakai K, Takayama C, Yanagi T, Yamanaka Y, McMillan JR, Shimizu H. CGI-58 is an alpha/beta-hydrolase within lipid transporting lamellar granules of differentiated keratinocytes. Am J Pathol. 2008;173:1349–60. doi: 10.2353/ajpath.2008.080005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ghosh AK, Chauhan N, Rajakumari S, Daum G, Rajasekharan R. At4g24160, a soluble acyl-coenzyme A-dependent lysophosphatidic acid acyltransferase. Plant Physiol. 2009;151:869–81. doi: 10.1104/pp.109.144261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ghosh AK, Ramakrishnan G, Rajasekharan R. YLR099C (ICT1) encodes a soluble Acyl-CoA-dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae. J Biol Chem. 2008;283:9768–75. doi: 10.1074/jbc.M708418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12:1115–26. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Turnbull PC, Ramos SV, MacPherson RE, Roy BD, Peters SJ. Characterization of lipolytic inhibitor G(0)/G(1) switch gene-2 protein (G0S2) expression in male Sprague-Dawley rat skeletal muscle compared to relative content of adipose triglyceride lipase (ATGL) and comparitive gene identification-58 (CGI-58) PLoS One. 2015;10:e0120136. doi: 10.1371/journal.pone.0120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Yang X, Lu X, Liu J. Identification of a novel splicing isoform of murine CGI-58. FEBS Lett. 2010;584:903–10. doi: 10.1016/j.febslet.2009.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Khatib A, Arhab Y, Bentebibel A, Abousalham A, Noiriel A. Reassessing the Potential Activities of Plant CGI-58 Protein. PLoS One. 2016;11:e0145806. doi: 10.1371/journal.pone.0145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].McMahon D, Dinh A, Kurz D, Shah D, Han GS, Carman GM, Brasaemle DL. Comparative gene identification 58/alpha/beta hydrolase domain 5 lacks lysophosphatidic acid acyltransferase activity. J Lipid Res. 2014;55:1750–61. doi: 10.1194/jlr.M051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Grond S, Radner FP, Eichmann TO, Kolb D, Grabner GF, Wolinski H, Gruber R, Hofer P, Heier C, Schauer S, Rulicke T, et al. Skin Barrier Development Depends on CGI-58 Protein Expression during Late-Stage Keratinocyte Differentiation. J Invest Dermatol. 2017;137:403–413. doi: 10.1016/j.jid.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, Theussl HC, et al. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J Biol Chem. 2010;285:7300–11. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Boeszoermenyi A, Nagy HM, Arthanari H, Pillip CJ, Lindermuth H, Luna RE, Wagner G, Zechner R, Zangger K, Oberer M. Structure of a CGI-58 motif provides the molecular basis of lipid droplet anchoring. J Biol Chem. 2015;290:26361–72. doi: 10.1074/jbc.M115.682203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Gruber A, Cornaciu I, Lass A, Schweiger M, Poeschl M, Eder C, Kumari M, Schoiswohl G, Wolinski H, Kohlwein SD, Zechner R, et al. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem. 2010;285:12289–98. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sahu-Osen A, Montero-Moran G, Schittmayer M, Fritz K, Dinh A, Chang YF, McMahon D, Boeszoermenyi A, Cornaciu I, Russell D, Oberer M, et al. CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: control of subcellular localization. J Lipid Res. 2015;56:109–21. doi: 10.1194/jlr.M055004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Sanders MA, Madoux F, Mladenovic L, Zhang H, Ye X, Angrish M, Mottillo EP, Caruso JA, Halvorsen G, Roush WR, Chase P, et al. Endogenous and Synthetic ABHD5 Ligands Regulate ABHD5-Perilipin Interactions and Lipolysis in Fat and Muscle. Cell Metab. 2015;22:851–60. doi: 10.1016/j.cmet.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Sanders MA, Zhang H, Mladenovic L, Tseng YY, Granneman JG. Molecular Basis of ABHD5 Lipolysis Activation. Sci Rep. 2017;7:42589. doi: 10.1038/srep42589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Hofer P, Boeszoermenyi A, Jaeger D, Feiler U, Arthanari H, Mayer N, Zehender F, Rechberger G, Oberer M, Zimmermann R, Lass A, et al. Fatty Acid-binding Proteins Interact with Comparative Gene Identification-58 Linking Lipolysis with Lipid Ligand Shuttling. J Biol Chem. 2015;290:18438–53. doi: 10.1074/jbc.M114.628958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Haunerland NH, Spener F. Fatty acid-binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43:328–49. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- [129].Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- [130].Russell L, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–91. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- [131].Welch C, Santra MK, El-Assaad W, Zhu X, Huber WE, Keys RA, Teodoro JG, Green MR. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782–9. doi: 10.1158/0008-5472.CAN-09-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S, Muller M, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–24. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Oh SA, Suh Y, Pang MG, Lee K. Cloning of avian G(0)/G(1) switch gene 2 genes and developmental and nutritional regulation of G(0)/G(1) switch gene 2 in chicken adipose tissue. J Anim Sci. 2011;89:367–75. doi: 10.2527/jas.2010-3339. [DOI] [PubMed] [Google Scholar]
- [134].Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I, Oberer M, et al. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307–17. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Heckmann BL, Zhang X, Xie X, Saarinen A, Lu X, Yang X, Liu J. Defective adipose lipolysis and altered global energy metabolism in mice with adipose overexpression of the lipolytic inhibitor G0/G1 switch gene 2 (G0S2) J Biol Chem. 2014;289:1905–16. doi: 10.1074/jbc.M113.522011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Heier C, Radner FP, Moustafa T, Schreiber R, Grond S, Eichmann TO, Schweiger M, Schmidt A, Cerk IK, Oberer M, Theussl HC, et al. G0/G1 Switch Gene 2 Regulates Cardiac Lipolysis. J Biol Chem. 2015;290:26141–50. doi: 10.1074/jbc.M115.671842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Ma T, Lopez-Aguiar AG, Li A, Lu Y, Sekula D, Nattie EE, Freemantle S, Dmitrovsky E. Mice lacking G0S2 are lean and cold-tolerant. Cancer Biol Ther. 2014;15:643–50. doi: 10.4161/cbt.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wang Y, Zhang Y, Qian H, Lu J, Zhang Z, Min X, Lang M, Yang H, Wang N, Zhang P. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One. 2013;8:e72315. doi: 10.1371/journal.pone.0072315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63:934–46. doi: 10.2337/db13-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].El-Assaad W, El-Kouhen K, Mohammad AH, Yang J, Morita M, Gamache I, Mamer O, Avizonis D, Hermance N, Kersten S, Tremblay ML, et al. Deletion of the gene encoding G0/G 1 switch protein 2 (G0s2) alleviates high-fat-diet-induced weight gain and insulin resistance, and promotes browning of white adipose tissue in mice. Diabetologia. 2015;58:149–57. doi: 10.1007/s00125-014-3429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yang X, Zhang X, Heckmann BL, Lu X, Liu J. Relative contribution of adipose triglyceride lipase and hormone-sensitive lipase to tumor necrosis factor-alpha (TNF-alpha)-induced lipolysis in adipocytes. J Biol Chem. 2011;286:40477–85. doi: 10.1074/jbc.M111.257923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Heckmann BL, Zhang X, Saarinen AM, Liu J. Regulation of G0/G1 Switch Gene 2 (G0S2) Protein Ubiquitination and Stability by Triglyceride Accumulation and ATGL Interaction. PLoS One. 2016;11:e0156742. doi: 10.1371/journal.pone.0156742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lee PH, Yamada T, Park CS, Shen Y, Puppi M, Lacorazza HD. G0S2 modulates homeostatic proliferation of naive CD8(+) T cells and inhibits oxidative phosphorylation in mitochondria. Immunol Cell Biol. 2015;93:605–15. doi: 10.1038/icb.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Wang Y, Zhang Y, Zhu Y, Zhang P. Lipolytic inhibitor G0/G1 switch gene 2 inhibits reactive oxygen species production and apoptosis in endothelial cells. Am J Physiol Cell Physiol. 2015;308:C496–504. doi: 10.1152/ajpcell.00317.2014. [DOI] [PubMed] [Google Scholar]
- [145].Yamada T, Park CS, Burns A, Nakada D, Lacorazza HD. The cytosolic protein G0S2 maintains quiescence in hematopoietic stem cells. PLoS One. 2012;7:e38280. doi: 10.1371/journal.pone.0038280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Yamada T, Park CS, Shen Y, Rabin KR, Lacorazza HD. G0S2 inhibits the proliferation of K562 cells by interacting with nucleolin in the cytosol. Leuk Res. 2014;38:210–7. doi: 10.1016/j.leukres.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Kusakabe M, Watanabe K, Emoto N, Aki N, Kage H, Nagase T, Nakajima J, Yatomi Y, Ohishi N, Takai D. Impact of DNA demethylation of the G0S2 gene on the transcription of G0S2 in squamous lung cancer cell lines with or without nuclear receptor agonists. Biochem Biophys Res Commun. 2009;390:1283–7. doi: 10.1016/j.bbrc.2009.10.137. [DOI] [PubMed] [Google Scholar]
- [148].Kioka H, Kato H, Fujikawa M, Tsukamoto O, Suzuki T, Imamura H, Nakano A, Higo S, Yamazaki S, Matsuzaki T, Takafuji K, et al. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc Natl Acad Sci U S A. 2014;111:273–8. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Tombran-Tink J, Pawar H, Swaroop A, Rodriguez I, Chader GJ. Localization of the gene for pigment epithelium-derived factor (PEDF) to chromosome 17p13.1 and expression in cultured human retinoblastoma cells. Genomics. 1994;19:266–72. doi: 10.1006/geno.1994.1057. [DOI] [PubMed] [Google Scholar]
- [150].Becerra SP. Structure-function studies on PEDF. A noninhibitory serpin with neurotrophic activity. Adv Exp Med Biol. 1997;425:223–37. [PubMed] [Google Scholar]
- [151].Kawaguchi T, Yamagishi SI, Sata M. Structure-function relationships of PEDF. Curr Mol Med. 2010;10:302–11. doi: 10.2174/156652410791065255. [DOI] [PubMed] [Google Scholar]
- [152].Tombran-Tink J, Chader GG, Johnson LV. PEDF: a pigment epithelium-derived factor with potent neuronal differentiative activity. Exp Eye Res. 1991;53:411–4. doi: 10.1016/0014-4835(91)90248-d. [DOI] [PubMed] [Google Scholar]
- [153].Famulla S, Lamers D, Hartwig S, Passlack W, Horrighs A, Cramer A, Lehr S, Sell H, Eckel J. Pigment epithelium-derived factor (PEDF) is one of the most abundant proteins secreted by human adipocytes and induces insulin resistance and inflammatory signaling in muscle and fat cells. Int J Obes (Lond) 2011;35:762–72. doi: 10.1038/ijo.2010.212. [DOI] [PubMed] [Google Scholar]
- [154].Simonovic M, Gettins PG, Volz K. Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc Natl Acad Sci U S A. 2001;98:11131-5. doi: 10.1073/pnas.211268598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Subramanian P, Locatelli-Hoops S, Kenealey J, DesJardin J, Notari L, Becerra SP. Pigment epithelium-derived factor (PEDF) prevents retinal cell death via PEDF Receptor (PEDF-R): identification of a functional ligand binding site. J Biol Chem. 2013;288:23928–42. doi: 10.1074/jbc.M113.487884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zhang H, Sun T, Jiang X, Yu H, Wang M, Wei T, Cui H, Zhuang W, Liu Z, Zhang Z, Dong H. PEDF and PEDF-derived peptide 44mer stimulate cardiac triglyceride degradation via ATGL. J Transl Med. 2015;13:68. doi: 10.1186/s12967-015-0432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kenealey J, Subramanian P, Comitato A, Bullock J, Keehan L, Polato F, Hoover D, Marigo V, Becerra SP. Small Retinoprotective Peptides Reveal a Receptor-binding Region on Pigment Epithelium-derived Factor. J Biol Chem. 2015;290:25241–53. doi: 10.1074/jbc.M115.645846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Crowe S, Wu LE, Economou C, Turpin SM, Matzaris M, Hoehn KL, Hevener AL, James DE, Duh EJ, Watt MJ. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab. 2009;10:40–7. doi: 10.1016/j.cmet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- [159].Dai Z, Qi W, Li C, Lu J, Mao Y, Yao Y, Li L, Zhang T, Hong H, Li S, Zhou T, et al. Dual regulation of adipose triglyceride lipase by pigment epithelium-derived factor: a novel mechanistic insight into progressive obesity. Mol Cell Endocrinol. 2013;377:123–34. doi: 10.1016/j.mce.2013.07.001. [DOI] [PubMed] [Google Scholar]
- [160].Knudsen J, Neergaard TB, Gaigg B, Jensen MV, Hansen JK. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J Nutr. 2000;130:294S–298S. doi: 10.1093/jn/130.2.294S. [DOI] [PubMed] [Google Scholar]
- [161].Kragelund BB, Knudsen J, Poulsen FM. Acyl-coenzyme A binding protein (ACBP) Biochim Biophys Acta. 1999;1441:150–61. doi: 10.1016/s1388-1981(99)00151-1. [DOI] [PubMed] [Google Scholar]
- [162].Bouyakdan K, Taib B, Budry L, Zhao S, Rodaros D, Neess D, Mandrup S, Faergeman NJ, Alquier T. A novel role for central ACBP/DBI as a regulator of long-chain fatty acid metabolism in astrocytes. J Neurochem. 2015;133:253–65. doi: 10.1111/jnc.13035. [DOI] [PubMed] [Google Scholar]
- [163].Huang H, Atshaves BP, Frolov A, Kier AB, Schroeder F. Acyl-coenzyme A binding protein expression alters liver fatty acyl-coenzyme A metabolism. Biochemistry. 2005;44:10282–97. doi: 10.1021/bi0477891. [DOI] [PubMed] [Google Scholar]
- [164].Ebdrup S, Refsgaard HH, Fledelius C, Jacobsen P. Synthesis and structure-activity relationship for a novel class of potent and selective carbamate-based inhibitors of hormone selective lipase with acute in vivo antilipolytic effects. J Med Chem. 2007;50:5449–56. doi: 10.1021/jm0607653. [DOI] [PubMed] [Google Scholar]
- [165].Ebdrup S, Sorensen LG, Olsen OH, Jacobsen P. Synthesis and structure-activity relationship for a novel class of potent and selective carbamoyl-triazole based inhibitors of hormone sensitive lipase. J Med Chem. 2004;47:400–10. doi: 10.1021/jm031004s. [DOI] [PubMed] [Google Scholar]
- [166].Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6:28282–95. doi: 10.18632/oncotarget.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:319–36. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- [168].Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- [169].Schweiger M, Eichmann TO, Taschler U, Zimmermann R, Zechner R, Lass A. Measurement of lipolysis. Methods Enzymol. 2014;538:171–93. doi: 10.1016/B978-0-12-800280-3.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]