Abstract
Asymmetric cell division gives rise to two daughter cells that inherit different determinants, thereby acquiring different fates. Polarized trafficking of endosomes containing fate determinants recently emerged as an evolutionary conserved feature of asymmetric cell division to enhance the robustness of asymmetric cell fate determination in flies, fish and mammals. In particular, polarized sorting of signalling endosomes by an asymmetric central spindle contributes to asymmetric cell division in Drosophila. However, how central spindle asymmetry arises remains elusive. Here, we identify a moonlighting function of the Elongator complex – an established protein acetylase and tRNA methylase involved in the fidelity of protein translation – as a key factor for central spindle asymmetry. Elongator controls spindle asymmetry by stabilizing microtubules differentially on the anterior side of the central spindle. Accordingly, lowering the activity of Elongator on the anterior side using nanobodies mistargets endosomes to the wrong cell. Molecularly, Elongator regulates microtubule dynamics independently of its acetylation and methylation enzymatic activities. Instead, Elongator directly binds to microtubules and increases their polymerisation speed while decreasing their catastrophe frequency. Our data establish a non-canonical role of Elongator at the core of cytoskeleton polarity and asymmetric signalling.
Introduction
Polarized trafficking of signalling endosomes is an evolutionary conserved feature of asymmetric cell division (ACD) to enhance the robustness of asymmetric cell fate determination in flies, fish and mammals1–7. In Drosophila, sensory organ precursor (SOP) cells in the notum divide asymmetrically to generate anterior pIIb and posterior pIIa cells8, with cell fate assignment controlled by differential Notch signalling via different endocytic mechanisms9. In particular, signalling endosomes labelled by the protein Sara (Smad anchor for receptor activation) are asymmetrically segregated to the pIIa cell during asymmetric cell division (ACD)2,4,10,11. Since these endosomes contain the cell fate determinant Notch and its ligand Delta, this contributes to asymmetric signalling and thereby to different cell fates in the SOP lineage2,4.
While the role of polarized trafficking in ACD is established, how this process is orchestrated remains unclear. In particular, how the cytoskeleton controls the asymmetric targeting of fate determinants is unknown. Recent work in SOPs showed that asymmetric endosome segregation is induced by symmetry breaking of the central spindle in anaphase. The central spindle is composed of microtubules that have their plus-ends facing the division plane where they are bundled in an antiparallel fashion (see12, and also Extended Data Fig. 1a). Specifically, in SOPs, the pIIb side of the central spindle has a higher density of microtubules than the pIIa side. During mitosis, Sara endosomes are transported along microtubules by the plus-end kinesin Klp98A, and therefore the asymmetry in microtubule density biases the motility of these vesicles towards the pIIa cell4. But how central spindle asymmetry arises remains poorly understood.
Results
Elongator controls central spindle asymmetry
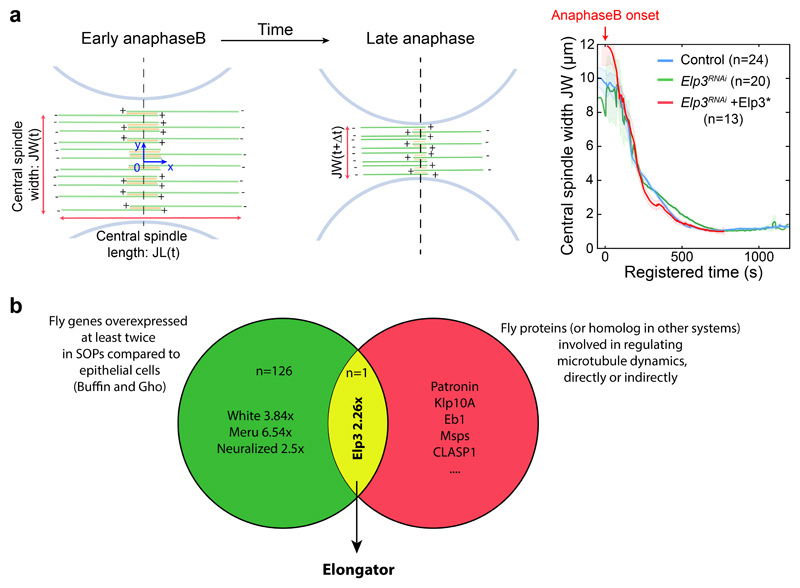
We thought to identify unknown members of the machinery controlling central spindle asymmetry using a candidate-based approach where we cross-referenced factors upregulated in SOPs13, which have at the same time been implicated in microtubule dynamics, even indirectly. This unexpectedly identified Elongator as the sole hit (see Extended Data Fig. 1b for details and Fig. 1).
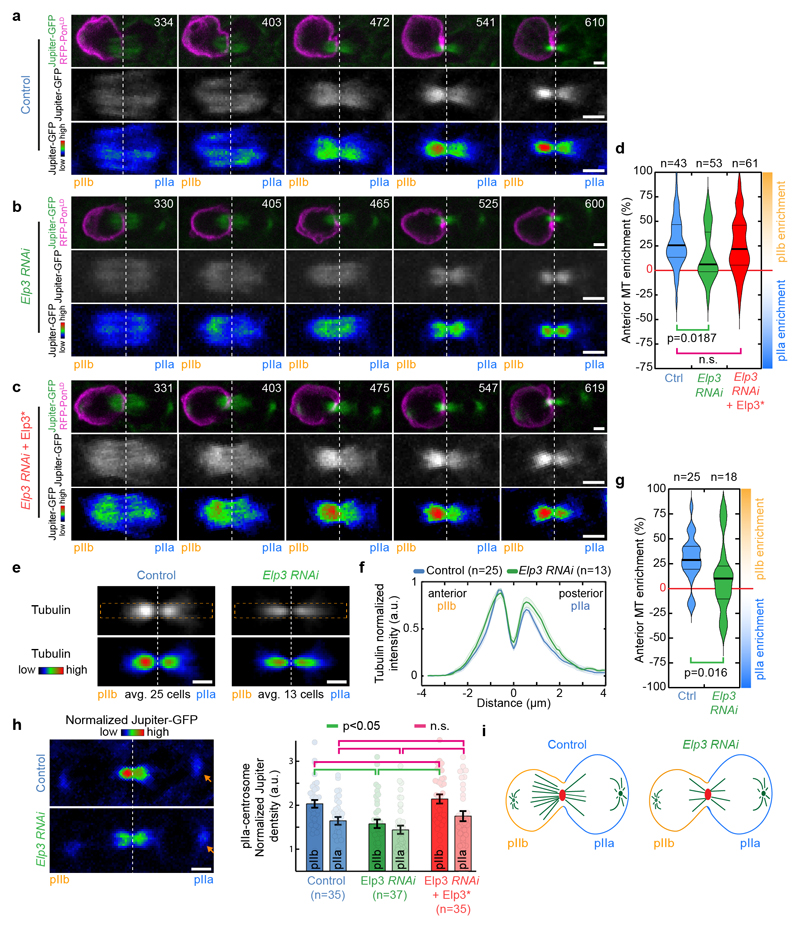
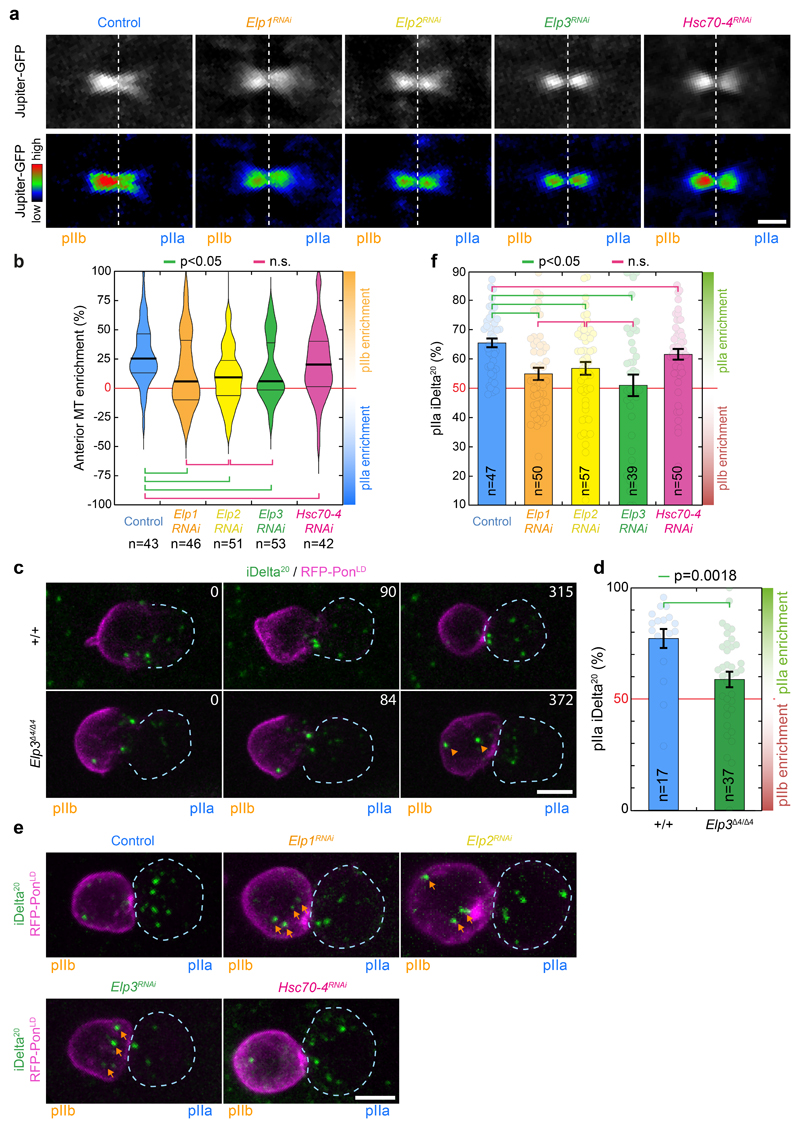
Figure 1. Elongator controls central spindle asymmetry.
(a-c) Timelapse of dividing SOP of indicated genotypes showing Jupiter-GFP/mRFP-PonLD (top rows); Jupiter-GFP (middle) or Jupiter-GFP with Rainbow RGB LUT (bottom rows). Movies were automatically registered in time (see Methods). Registered time=0s indicates anaphaseB onset. Anterior/pIIb orientation determined by the mRFP-PonLD signal. Images correspond to Spinning Disk Confocal Microscopy (SDCM) imaging (maximum intensity projection). (d) Enrichment in pIIb (see methods) for conditions shown in a-c. Thick line: median; thin line: quartile; n: number of SOPs analysed. Statistics: Kruskal-Wallis non-parametric ANOVA1 test (respective post-hoc test P-values indicated). Central spindles in WT SOPs are asymmetric, with an enrichment of Jupiter-GFP on the pIIb side of the spindle, while in Elp3RNAi they are symmetric. The phenotype can be rescued by expression of an RNAi-resistant version of Elp3, (Elp3*). (e-g) Tubulin immunostainings of central spindles in control and Elp3RNAi mutants. (e: top rows). Average central spindle images of SOPs after anti-β-tubulin immunolabeling, see methods. n: number of spindles. (f) Intensity profile of the β-tubulin signals across the spindle (dashed orange rectangle in e) displaying two peaks, one in pIIa, one in pIIb, due to the exclusion of antibodies from the core of the spindle. In control SOPs the pIIb peak is ≈25% higher than that of the pIIa, and this is abolished in Elp3RNAi mutants. Mean±SEM (shaded area). (g) Enrichment in pIIb. Thick line: median; thin line: quartile; n: spindles analysed. Statistics: two-sided Mann-Whitney test (P-values indicated). Central spindle asymmetry is abolished upon Elongator depletion. (h) Jupiter-GFP signal in dividing SOPs (left panel) and quantification (right panel; Mean±SEM) after normalization to pIIa centrosome (arrowhead, see methods). Statistics: ANOVA2 on square root transformed data, followed by a Tukey multiple comparison test (p value indicated). (i) Elp3 RNAi phenotype summary. Scale bars: 2 μm (a-c, h); 1 μm (e). Source numerical data available in source data.
Elongator is a conserved, 0.8 MDa protein complex composed of six subunits, Elp1-614,15, comprising two copies of each subunit arranged in two subcomplexes: Elp1-3 and Elp4-616,17. The catalytic subunit, Elp3, harbours a conserved lysine acetyl-transferase (KAT) and a S-adenosyl-L-methionine (SAM) domains that mediate acetylation and methylation, respectively. It was initially proposed that those domains would allow Elongator to directly modify a broad range of proteins, including histones, α-tubulin, or presynaptic proteins15,18–20. However, recent evidence suggests that Elongator’s main function is the regulation of translation through targeted tRNA modification21–23. As a result, defects in Elongator function are associated with severe neurodegenerative diseases and cancer24–26. As often with stable multiprotein complexes27, deletion of any Elongator subunit has the same phenotype in yeast, plants and mammals, suggesting that all subunits are essential to the integrity of the complex22,28–30. This is also true in flies, as whole-animal depletion of Elongator subunits for which tools exists (Elp1-5) leads to pupariation failure and lethality (Supplementary Table 1).
In SOPs, Elp3 depletion abolishes central spindle asymmetry (Fig.1). As described previously4, in controls, the central spindle gradually becomes asymmetric during anaphase B, culminating in a ≈25% enrichment of the density of the microtubule marker Jupiter-GFP on the pIIb side (Fig. 1a, d; Supplemental Movie S1). In contrast, upon Elp3 depletion, central spindle asymmetry is impaired (Fig. 1b, d; Supplemental Movie S2 for dynamics, Extended Data Fig. 2a-b, for controls; see also methods for automated temporal registration of movies). This effect is specific, as the phenotype can be rescued by a wild-type, RNAi-resistant Elp3 (termed Elp3*; Fig. 1c-d; see Extended Data Fig. 2a for validation). Similarly, depletion of other core subunits of Elongator also abolished central spindle asymmetry (Extended Data Fig. 3a, b). This phenotype on Jupiter-GFP was also confirmed by immunofluorescence against endogenous tubulin (Figs. 1e-g).
We then investigated whether spindles were symmetric in Elongator mutants because they lost microtubule density on the pIIb side of the spindle compared to wild type, or conversely, gained density on the pIIa side. To compare absolute microtubule densities between different SOPs, we considered the fluorescence intensity of the invariant pIIa centrosome (arrows in Fig. 1h left panel) as a reference to normalize fluorescence signals (see ref.4 and methods). This revealed that, in Elongator mutants, microtubule density has been lost on the pIIb side (Fig. 1h). Therefore, Elongator controls central spindle asymmetry by specifically increasing microtubule density on the pIIb side (Fig.1i).
Elongator controls signalling endosome polarized trafficking
We then wondered whether symmetric central spindles in Elongator mutants caused defects in the polarized trafficking of Sara signalling endosomes and thereby cell fate phenotypes (Fig. 2). Sara endosomes were monitored by following the Notch-ligand Delta 20 minutes after its internalization (iDelta20, see methods and ref.4). In controls, Sara endosomes are targeted to the spindle and segregate asymmetrically into the pIIa daughter cell during anaphase/telophase (Fig. 2a,b). Both central spindle targeting and the dynamics of departure are critical for polarized Sara endosome trafficking4,10, but this is not controlled by Elongator. Indeed, in Elp3RNAi SOPs, targeting is unaffected (first time point in Fig. 2c) and the dynamics of departure from the spindle and the speed of constriction of the central spindle are also unaffected (Fig. 2c and Extended Data Fig. 1a).
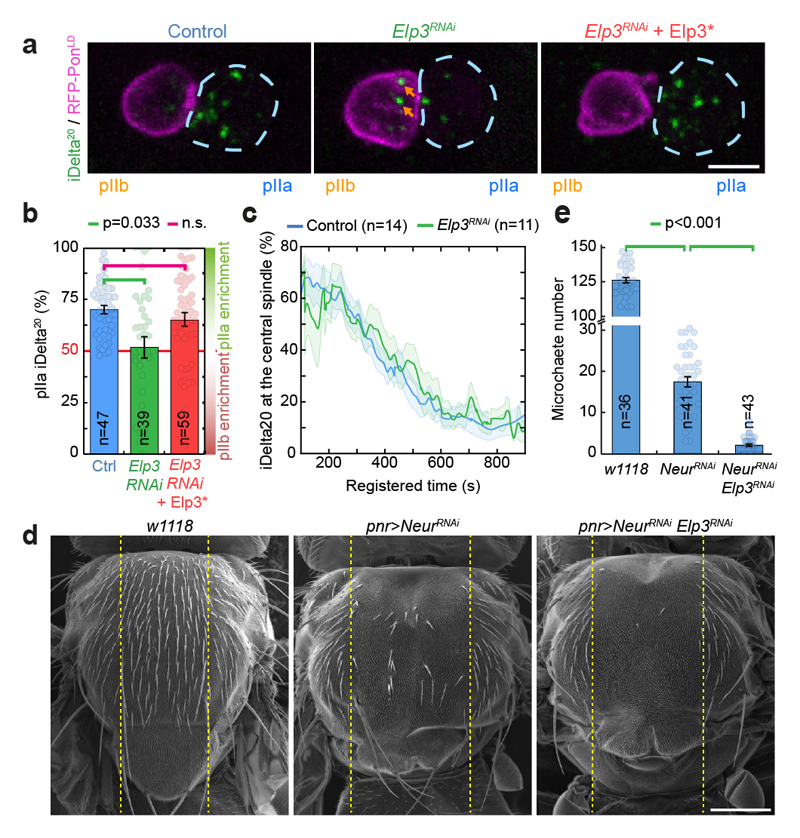
Figure 2. Elongator controls asymmetric Sara endosome segregation and thereby contributes to Notch signalling.
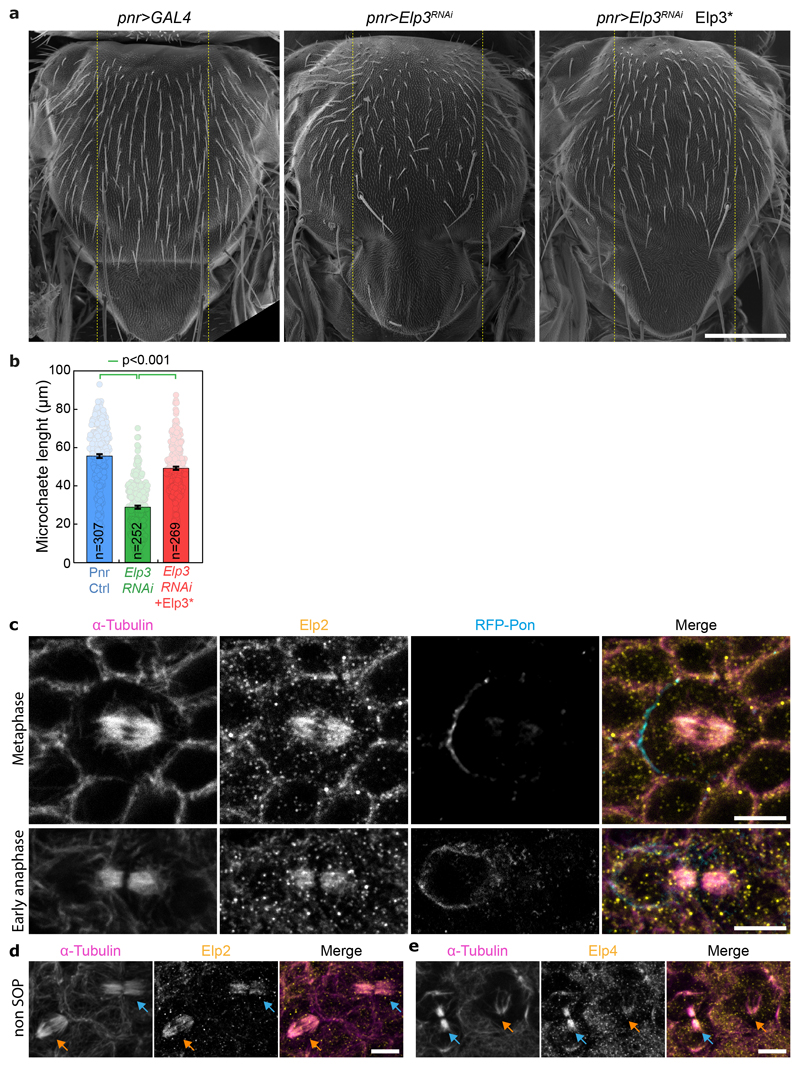
(a) Dividing SOP showing Sara endosomes (iDelta20) and mRFP-Pon (pIIa cortex; dashed blue) at the end of cytokinesis. Image corresponds to Maximum intensity z-projection of entire cells (SDCM). Note the endosomes in the pIIb daughter cell upon Elongator depletion (arrows). (b) Percentage of iDelta20 in pIIa after abscission in SOPs (mean ± SEM; Statistics: one-way ANOVA, followed by a Tukey post-hoc test (p value=0.033); n: number of SOPs analysed). (c) Dynamics of Sara endosomes at the central spindle, mean ± SEM (shaded area). Time is automatically registered between movies (for registration, see methods). Registered t=0 seconds: anaphase B onset. Note that the fraction of total endosomes at the spindle at t=100s is similar between control and Elp3RNAi, and that the kinetics of departure are the same. (d-e) Scanning EM (d) and microchaete numbers in the central of the notum (panier region, dashed lines in d) in Elp3RNAi/NeurRNAi mutants (e). Mean±SEM. Statistics: Kruskal-Wallis non-parametric ANOVA1 test. post-hoc test P-values indicated. Scale bars: 5 μm (a); 200 μm (d). Source numerical data available in source data.
Yet, Sara endosomes are symmetrical upon Elp3 depletion (Fig. 2a middle panel, see also Fig. 2b and methods for automated quantification). Symmetrical Sara endosome inheritance in Elp3RNAi SOPs can be rescued by the RNAi-resistant Elp3* (Fig. 2a right panel, b), and is also observed in MARCM clones for a loss-of-function lethal allele of Elp3, Elp3Δ4 (ref.18; Extended Data Fig. 3c,d and Supplemental Movie S3). Similarly, depletion of other Elongator subunits also abolished asymmetric endosome segregation, confirming the specificity of the phenotype to Elongator depletion (Extended Data Fig. 3e,f). Thus, Elongator is critical for polarized Sara endosome trafficking.
Sara-endosome motility contributes to cell fate assignation through asymmetric Notch signaling, but this activity is redundantly covered by Neuralized9. Neuralized is an E3 ubiquitin ligase that regulates asymmetric activation of Delta, the Notch ligand31. Consequently, strong synergistic cell fate phenotypes are observed in the bristle lineage of adult flies when both Sara endosome asymmetry and Neuralized activity are compromised4,10. We thus wondered if abolishing Sara-endosome polarized segregation in Elongator mutants also induced similar fate phenotypes. Indeed, pnr>neurRNAi pnr>Elp3RNAi double mutants show a synergistic fate phenotype where the notum is largely void of bristles (Fig. 2d,e ; Extended Data Fig. 4 for controls). Therefore, Elongator controls the formation of an asymmetric central spindle, which in turns mediates asymmetric Sara endosome trafficking, thereby contributing to asymmetric Notch signaling and cell fate assignation.
Elongator directly interacts with microtubules
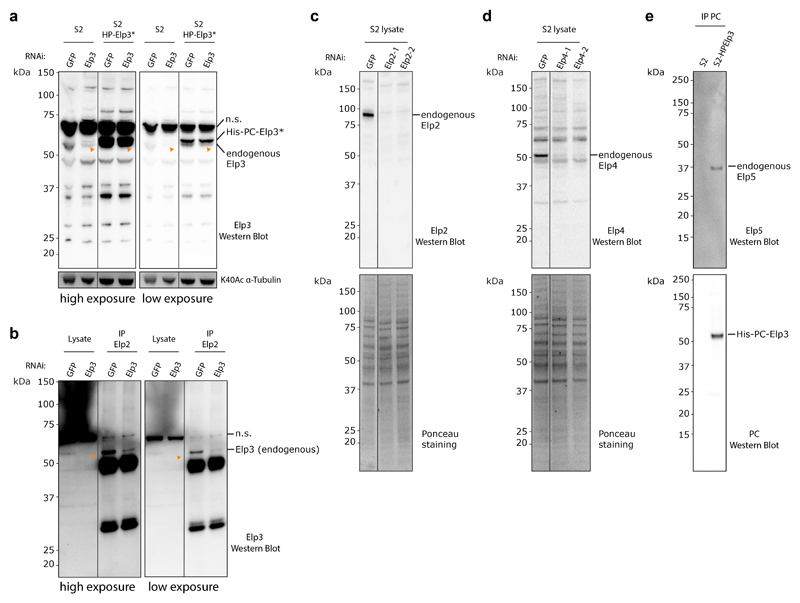
We next sought to shed light on how Elongator controls central spindle asymmetry by examining its distribution in dividing SOPs (Fig. 3). Using antibodies raised against Drosophila Elongator subunits (characterisation: Extended Data Fig. 2c,d), Elongator was found to localize to the spindle throughout mitosis (Endogenous Elp4 immunostainings in Fig. 3a,b, Extended Data Fig. 4c-e for endogenous Elp2 immunostaining and confirmation in non-SOP cells).
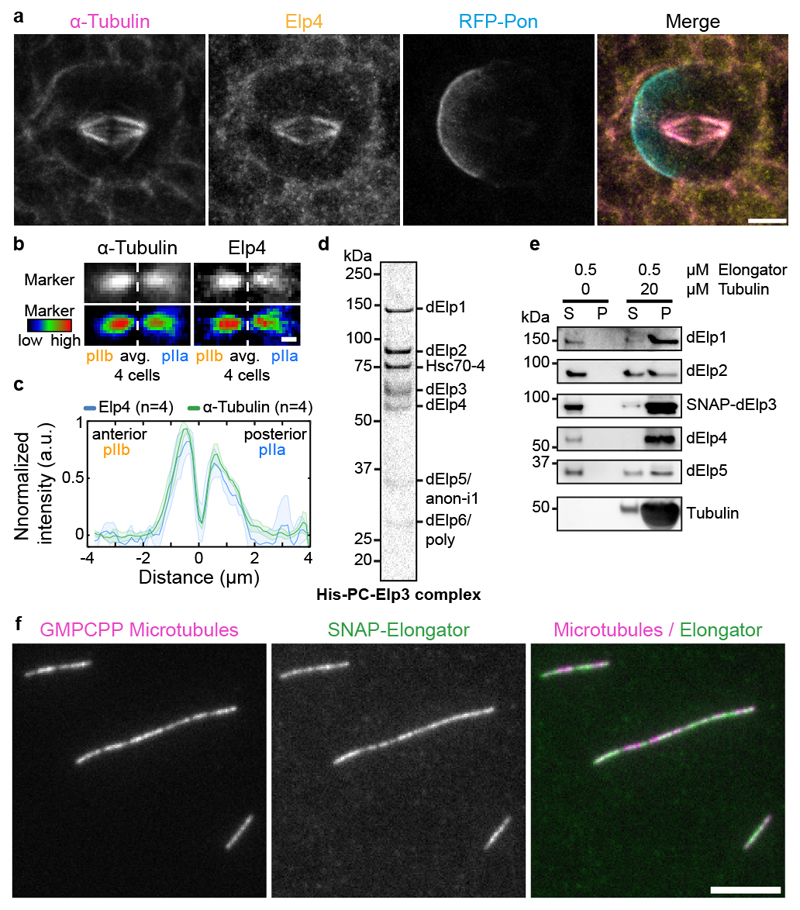
Figure 3. Elongator is recruited to the central spindle via its direct interaction with microtubules.
(a) Dividing SOP showing mRFP-Pon after immunolabelling with antibodies against endogenous dElp4 and α-tubulin. Image corresponds to a maximum intensity z-projection (SDCM). (b) Average central spindle of SOPs after immunolabelling using antibodies against endogenous dElp4 and α-tubulin (n=4 central spindles, SDCM). (c) Intensity profile of the β-tubulin signals across the average spindle shown in (b), mean ± SEM (shaded area). (d) Coomassie-stained gradient SDS-PAGE gel of Elongator complex purified using a His-PC-tagged Elp3 subunit after two steps of purification (PC affinity then Superose6 size exclusion). (e) Western blot analysis of in vitro co-sedimentation assays with dynamic microtubules and purified Elongator complex (P pellet; S, supernatant). In the presence of microtubules, all the subunits of Elongator identified by immunoblotting using specific antibodies for each subunit are found in the pellet. (f) Rhodamine-labelled GMPCPP-stabilized microtubules incubated with 25 nM Elongator complex labelled with BG-Alexa Fluor 488 dye and observed by TIRFM. Elongator directly binds GMPCPP microtubules. Scale bars: 5 μm (a,f); 1 μm (b). Source numerical data and unprocessed blots are available in source data.
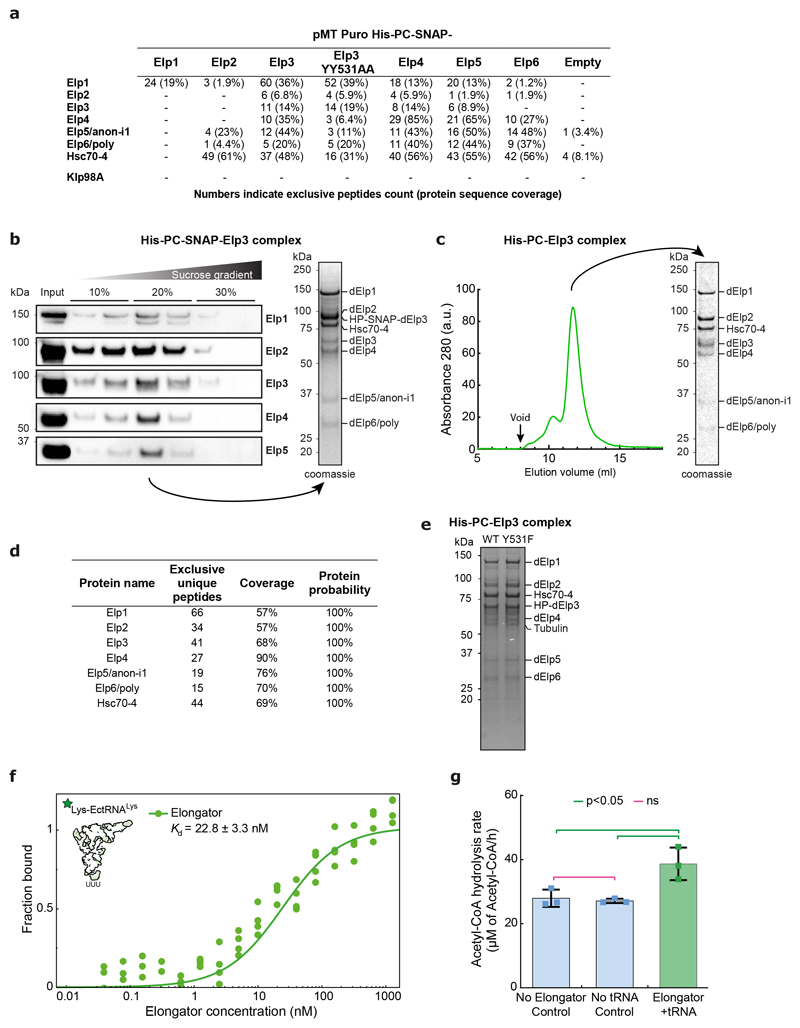
To test if Elongator directly interacts with microtubules, we established the purification of the full Drosophila Elongator complex (see methods). Briefly, a collection of stable Drosophila S2 cell lines was established, each one expressing a (His)6-PC-SNAP tagged version of each subunit. From these stable lines, the endogenous Elongator complex can be recovered by pulling on the exogenous subunit via the tag. Tagged Elp3 showed the highest incorporation into the endogenous complex (Extended Data Fig. 5a), and thus this subunit was selected for large-scale purification. Our method yielded milligrams of pure Elongator complex (Fig. 3d, Extended Data Fig. 5c, b for SNAP-tagged version). Importantly, the Drosophila Elongator complex purified by this method is functional and displays the expected activities reported in other species23,32, namely high affinity for tRNA (22.8 ±3.3nM, Extended Data Fig. 5f), and tRNA modification (Extended Data Fig. 5g).
Interestingly, Drosophila Elongator co-purified in seemingly stochiometric amounts with Hsc70-4, a protein previously implicated in clathrin-mediated endocytosis and neurotransmitter exocytosis33,34, even after size exclusion chromatography or sucrose density gradient ultracentrifugation (Fig.3d; Extended Data Fig.5a-d). This suggests that Hsc70-4 associates tightly with the complex. We refer to this [Elp1-6+Hsc70-4] complex simply as the “Elongator complex” in the following. Note that while ubiquitous depletion of Hsc70-4 is lethal, as is depletion of any Elongator subunit (Supplementary Table 1), SOP-specific depletion of Hsc70-4 does not affect central spindle asymmetry nor polarized segregation of Sara endosomes (Extended Data Fig. 3). This suggests that Hsc70-4 is required for some functions of Elongator, but not for regulating microtubule dynamics, which is the focus of this study.
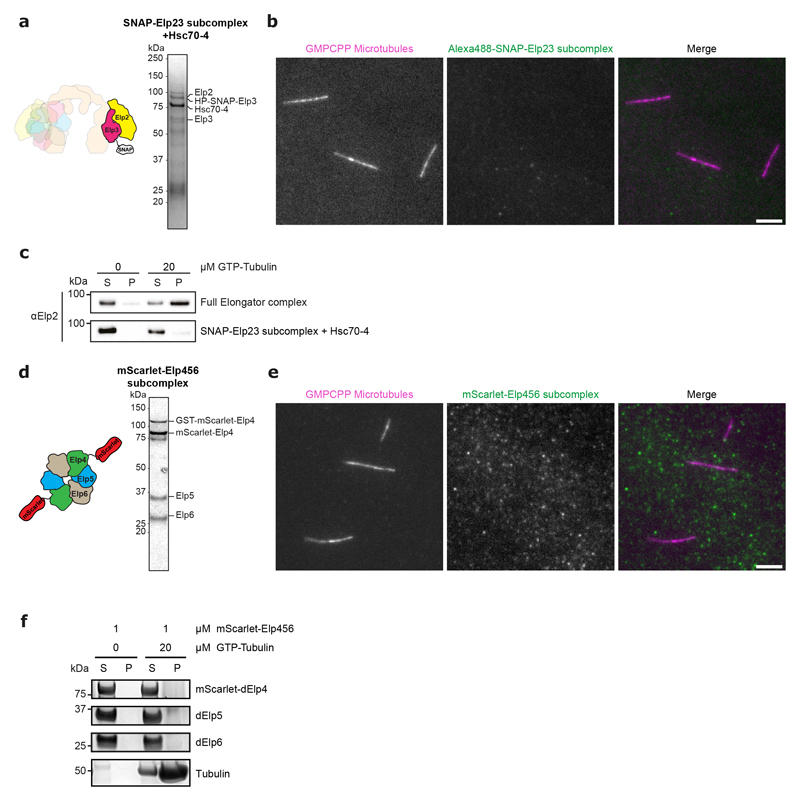
Using this purified complex in co-pelleting assays, we found that Elongator directly binds to dynamic microtubules in vitro (Fig. 3e). This was confirmed using total internal reflection fluorescence microscopy (TIRFM) using fluorescently labelled complex and stabilized microtubules (Fig. 3f). Interestingly, efficient microtubule binding requires the full complex: an Elp23+Hsc70-4 complex, a by-product of our purification, binds to microtubules much less than the full complex (Extended Data Fig. 6a-c), and an Elp456 subcomplex hexamer produced in E. coli was unable to bind to microtubules at all (Extended Data Fig. 6d-f). These data indicate that the Elongator complex binds to microtubules in vitro and in vivo.
Elongator enzymatic activities are dispensable for asymmetry
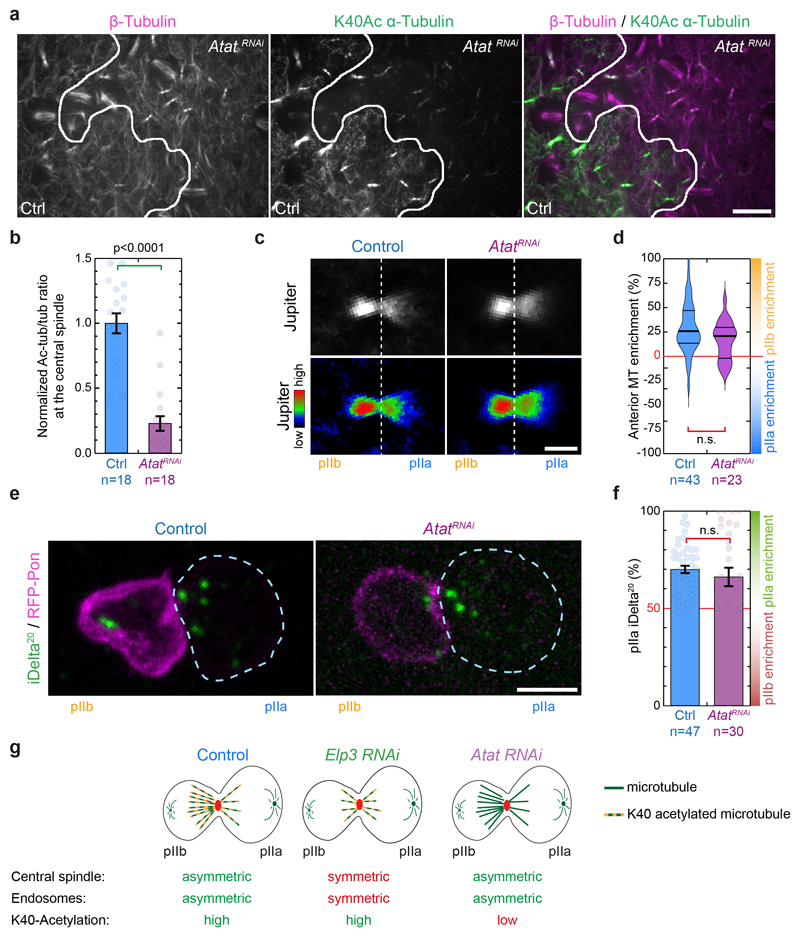
Next, we wondered whether Elongator effects on central spindle asymmetry required its enzymatic activities. Indeed, a tantalizing hypothesis to explain the Elongator phenotype is its previously reported acetyl-transferase activity for α-tubulin at position K4019, although this was later challenged35–38. First, we depleted a bona fide tubulin K40 acetyl-transferase39, Atat, and confirmed using ratiometric quantitative immunofluorescence that it dramatically decreases the acetylation levels of central spindle microtubules (Extended Data Fig. 7a,b). However, neither central spindle asymmetry nor endosome asymmetric segregation are affected in AtatRNAi mutants (Extended Data Fig. 7c-g), excluding that central spindle asymmetry is controlled by tubulin acetylation.
Importantly, in Elongator mutants, microtubule acetylation is still detected (Extended Data Fig. 8a,b), and the loss of microtubule asymmetry due to Elongator depletion (Fig. 1) is also seen for the pool of acetylated microtubules (Extended Data Fig. 8a-c). Furthermore, microtubules on either side of the central spindle are as acetylated in Elp3RNAi SOPs as in control (Extended Data Fig. 8d). This was confirmed by quantitative western blot in Elongator-depleted S2 cells (Extended Data Fig. 8e). Together with the Atat data above, this confirms that Elongator does not regulate microtubule acetylation, and that microtubule acetylation is anyway irrelevant for central spindle asymmetry and polarized endosome dynamics (Extended Data Fig. 7g).
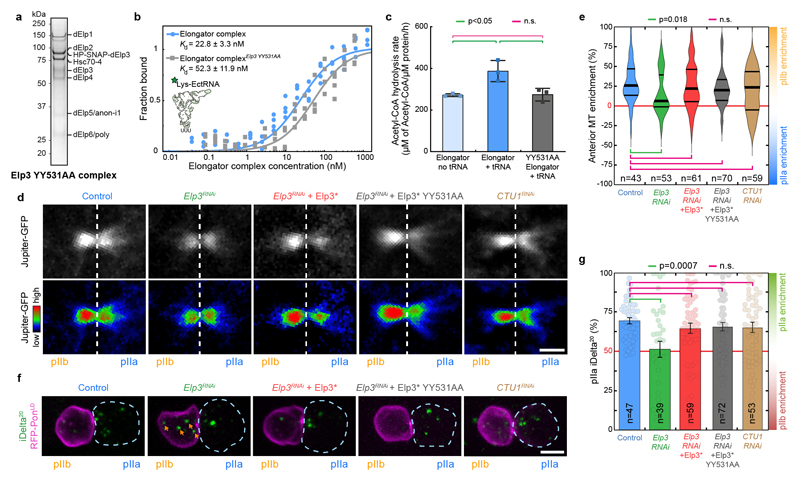
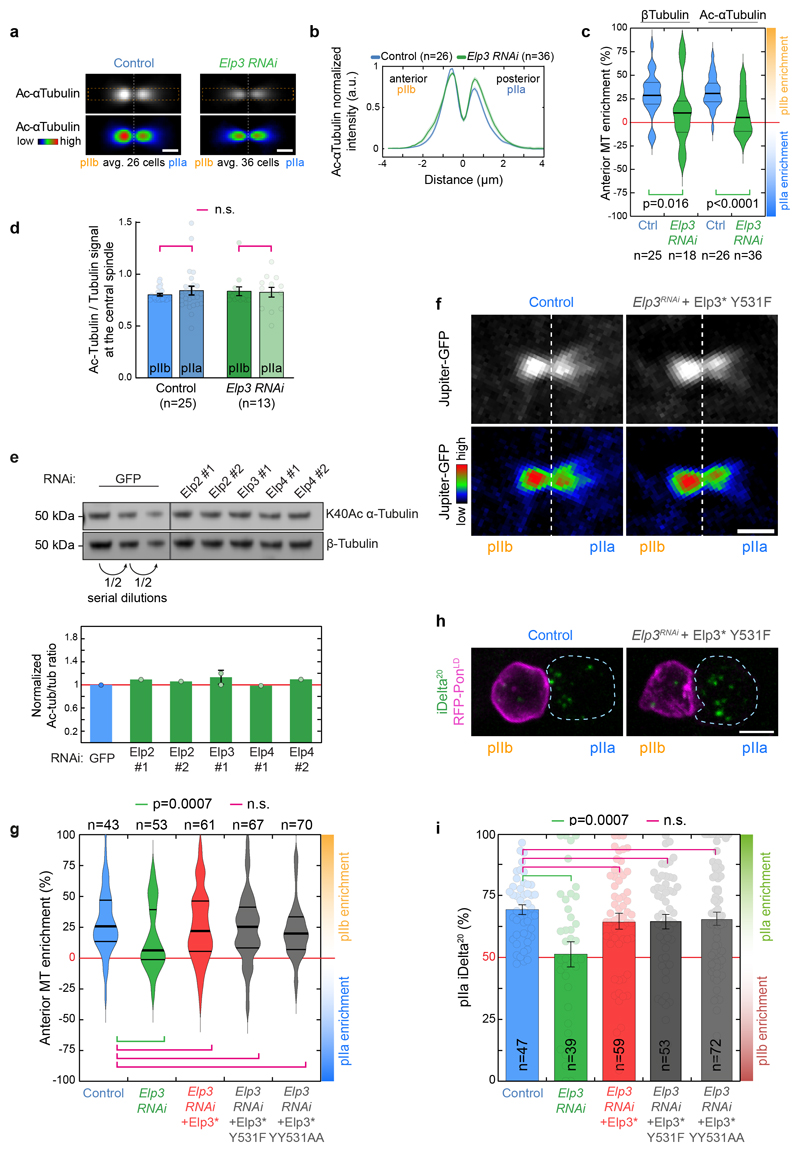
We still wondered whether the enzymatic activities of Elongator for targets other than tubulin were at all involved in its effects on the central spindle. We thus mutated two tyrosine residues known to be critical for the acetylase and methylase enzymatic activities of yeast Elp340 in our RNAi-resistant Elp3* construct. Importantly, this YY531AA mutant of Elp3* is not destabilized and still supports assembly of the full complex (Fig. 4a). Furthermore, while Elp3*-YY531AA Elongator complex is able to bind to tRNAs with similar affinity as the wild-type (Fig. 4b), it can no longer modify them (Fig. 4c) establishing that it is a bona fide enzymatic-dead mutant in flies. As expected since Elongator controls the translation of many genes via its tRNA modification activity, Elp3*-YY531AA mutant cannot rescue the characteristic pupariation failure phenotype of ubiquitous Elp3 depletion, while WT Elp3* can (Supplementary Table 2). But strikingly, Elp3*-YY5311AA rescues the phenotypes of SOP-specific depletion of Elp3 on central spindle asymmetry (Fig. 4d,e) and polarized trafficking of Sara endosomes (Fig. 4f,g). This was confirmed with another Elp3 mutant, Y531F, previously shown to be dominant negative in flies41 (Extended data Fig. 5e, Extended Data Fig. 8f-i., Supplementary Table 2).
Figure 4. The enzymatic activity of Elp3 is not required for Elongator effects on the central spindle.
(a) Coomassie-stained gradient SDS-PAGE gel of Elongator YY531AA mutant complex purified using a His-PC-SNAP-tagged Elp3 subunit after two steps of purification (PC affinity then sucrose gradient). (b,c) Biochemical characterization of the YY531AA Elongator complex. (b) Measurement of the affinity of the direct interaction between purified His-PC-Elp3 Elongator and tRNALys measured by MST (see methods). (c) Measurements of the tRNA modification activity of Elongator (mean ± SD, see methods). The YY531AA mutant of Elongator binds to tRNA with a similar affinity compared to WT Elongator, but is unable to modify tRNA. Note that the control curves are identical to the ones shown in Extended Data Fig. 5f,g. n=3 independent experiments. Statistics: one-way ANOVA followed by a Tukey multiple comparison test (p value indicated). (d) Dividing SOP showing Jupiter-GFP (top row) or Jupiter-GFP with the Rainbow RGB LUT applied (bottom row). Image corresponds to maximum intensity z-projection of the central spindle (SDCM). (e) Enrichment in pIIb. Thick line: median; thin line: quartile; n: number of central spindles analysed. Statistics: Kruskal-Wallis non-parametric ANOVA1 test. P-values of the respective post-hoc tests are indicated. Note that the control, Elp3 RNAi and Elp3 RNAi + Elp3* datasets are the same as the ones presented in Fig. 1d, shown here for convenience. (f) Dividing SOP showing Sara endosomes (iDelta20) and mRFP-Pon (pIIa cortex; dashed blue). Image corresponds to Maximum intensity z-projection of entire cells (SDCM). Note the endosomes in the pIIb daughter cell upon Elongator depletion (arrows). (g) Percentage of iDelta20 in pIIa after abscission in SOPs (mean ± SEM; Statistics: one-way ANOVA (P<0.0001), followed by a Tukey post-hoc test (p value indicated); n: number of SOPs analysed. Note that the control, Elp3 RNAi and Elp3 RNAi+Elp3* datasets are the same as the one presented in Fig. 2b, shown here for convenience). Expression of a RNAi-resistant version of Elp3 YY531AA is able to rescue the Elp3 RNAi phenotype on the central spindle and the asymmetric segregation of Sara endosomes. Scale bars: 2 μm (2); 5 μm (f). Source numerical data are available in source data.
Furthermore, it is well known that, in vivo, the tRNA modification activity of Elongator requires downstream accessory proteins such as CTU1 (NCS6 in yeast)42,43. This provided an orthogonal way to confirm that the Elongator effects on the central spindle can be untangled from its roles in translation. Importantly, while ubiquitous CTU1 depletion phenocopies Elongator depletion (Supplementary table 1), SOP-specific depletion of CTU1 did not affect spindle asymmetry (Fig. 4d,e) nor polarized endosome dynamics (Fig.4 g,f). This establishes that the KAT and SAM activities of Elp3 are not involved in the role of Elongator in establishing central spindle asymmetry.
Elongator directly stabilizes microtubules
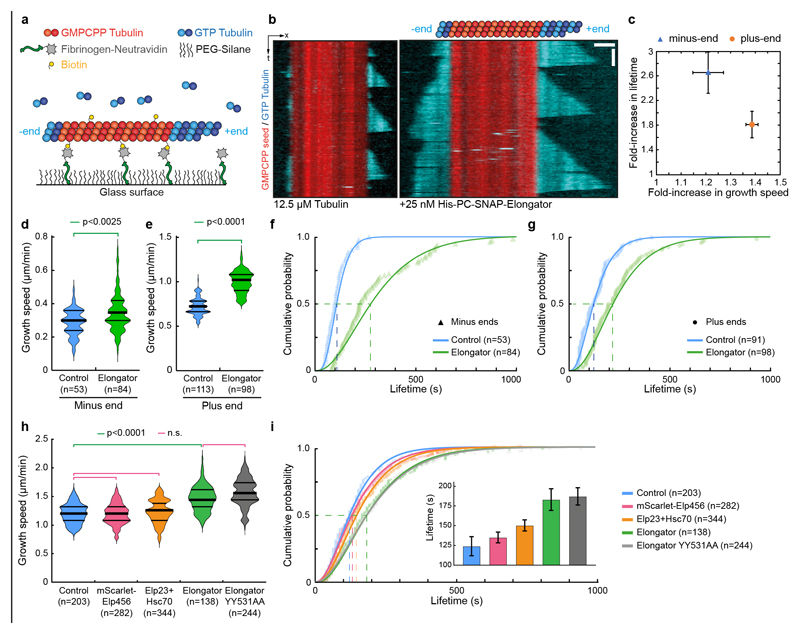
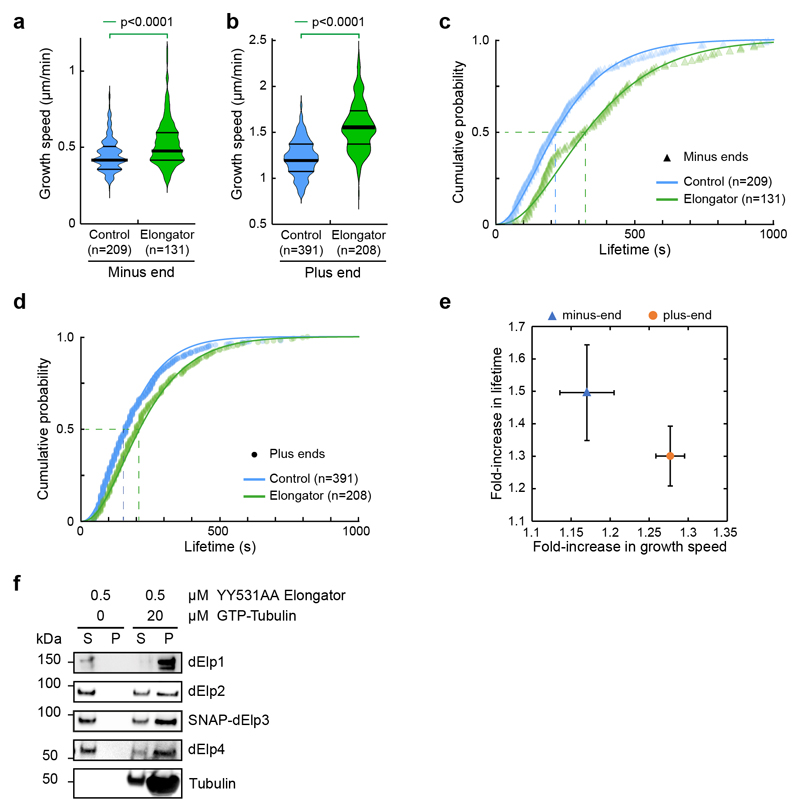
Because the enzymatic activities of Elongator are dispensable for central spindle asymmetry, we wondered if Elongator could directly affect microtubule dynamics. The effects of Elongator were monitored in a standard assay where cycles of growth/catastrophe of dynamic microtubules from stable seeds are observed by TIRFM44 (Fig. 5a,b, Supplemental Movie S4). Using physiological concentrations of Elongator and Tubulin45, Elongator increases the growth speed of microtubules by ≈l.4- and ≈l.2-fold at the plus and minus ends, respectively (Fig. 5b-e and Supplemental Movie S4).
Figure 5. Elongator directly controls microtubule dynamics.
(a) Assay design: Biotinylated, Rhodamine-labelled, GMPCPP-stabilized seeds (red) are anchored via Fibrinogen-neutravidin. Free tubulin (10% HiLyte647-labelled, cyan) is added, and microtubule dynamics are observed by TIRFM. (b) Representative kymographs (12.5 μM GTP-tubulin, 10% HiLyte647-tubulin) in the absence (left) or in the presence (right panel) of 25 nM (His)6-PC-SNAP-Elongator (note that the SNAP-tagged Elongator is not conjugated to any dye). The control was performed with the same buffer used for the experiment in the presence of Elongator. Microtubules grow faster and undergo less catastrophe in the presence of Elongator. (c) Increase of the polymerisation speed versus increase of the mean microtubule lifetime (± error) at both ends, see d-g. (d,e) Microtubule growth rate at both the minus- (d) and plus-ends (e) under the conditions described for (b). Data points represent individual growth events quantified from at least two independent experiments. Thick line: median; thin line: quartile. Statistics: two-sided Mann-Whitney test (p-value as indicated; n: microtubule number). (f,g) Cumulative lifetime distribution of microtubules grown from each end under the conditions described for (b). Elongator complex increases the microtubule lifetime at both the minus- (f) and the plus-ends (g). Lines: gamma distribution fits. Mean lifetime estimate ±error (i.e. lifetime at half cumulative distribution, dashed lines). Minus-end control: 102± 9s (n=53; blue) and 270 ± 25s for Elongator (n=84; green). Plus-end control: 121 ± 10s (n=91; blue) and 219 ± 18s for Elongator (n=98; green). Estimate of microtubule catastrophe rate by fitting lifetime histograms to a mono-exponential decay model: Minus end: Control 0.0134±0.0030 sec-1 / Elongator 0.0040±0.0013 sec-1 ; Plus end: Control 0.0084±0.0021 sec-1 / Elongator 0.0044±0.0020 sec-1. (h,i) Effect of indicated conditions on microtubule growth speed (h) or lifetime (i) at the plus end. The inset represents the lifetime estimate±error from the bootstrapped mean lifetimes (see Methods). Statistics: Mann-Whitney test followed by Dunn’s multiple comparison test (p-value indicated). n: microtubule growing events analysed. Microtubule catastrophe rate with a mono-exponential decay model: Control: 0.0074±0.0040 sec-1/Elp23: 0.0062±0.0018 sec-1/Elp456: 0.068±0.0019 sec-1 / Elongator WT: 0.0049±0.0015 sec-1/Elongator YY531AA: 0.0040±0.0018 sec-1. Source numerical data are available in source data.
In addition, microtubules grew significantly longer before undergoing catastrophe in the presence of Elongator (Fig. 5b and Supplemental Movie S4; 0.52±0.03 to 1.62±0.13 μm at the minus-end; 1.62±0.09 to 4.21±0.32 μm at the plus-end; mean±SEM; n=53/84 and 113/98 microtubules for the minus and plus ends, respectively). Consistent with these findings, in the presence of Elongator, the lifetime of microtubules is increased (Fig. 5c, f, g and Supplemental Movie S4). The effect of Elongator on enhancing the growth rate is stronger at the plus-end, while the effect on decreasing the catastrophe rate is stronger at the minus-end (Fig. 5c). All the results were confirmed using a version of Elongator devoid of the SNAP tag and at different tubulin/Elongator concentrations (Extended Data Fig. 9), confirming that Elongator tagging on Elp3 does not affect its activity. Importantly, these effects require the full complex, as partial Elongator sub-complexes can neither bind (Extended Data Fig. 6) nor stabilize microtubule or affect their growth rate (Fig. 5h,i). Interestingly, one of these inactive partial complexes contained Hsc70-4, thereby confirms our in vivo results that Hsc70-4 is not sufficient to explain the effects of Elongator on microtubules. Last, the effect on microtubule dynamics of the Elp3-YY531AA Elongator complex are indistinguishable from wild-type Elongator (Fig. 5h,i), confirming that the KAT and SAM activities are indeed dispensable for the role of Elongator on microtubules. This demonstrates that Elongator directly tunes microtubule dynamics to favour growth.
Elongator asymmetry controls asymmetric endosome trafficking
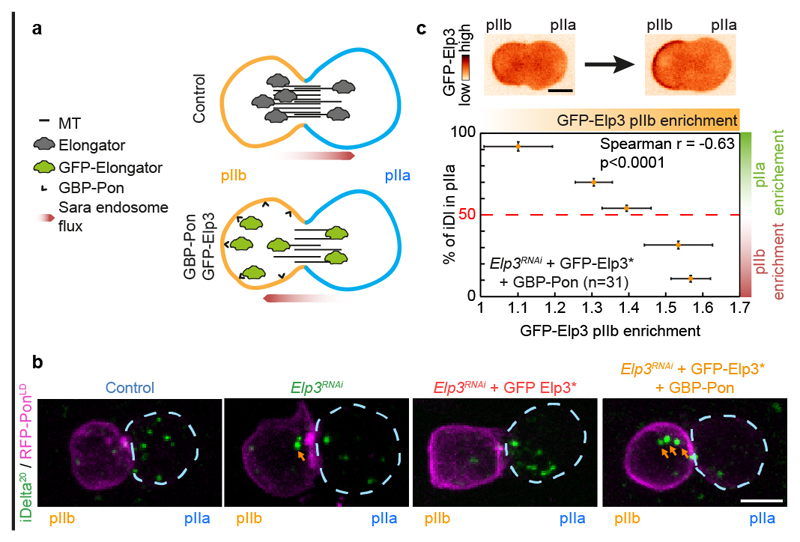
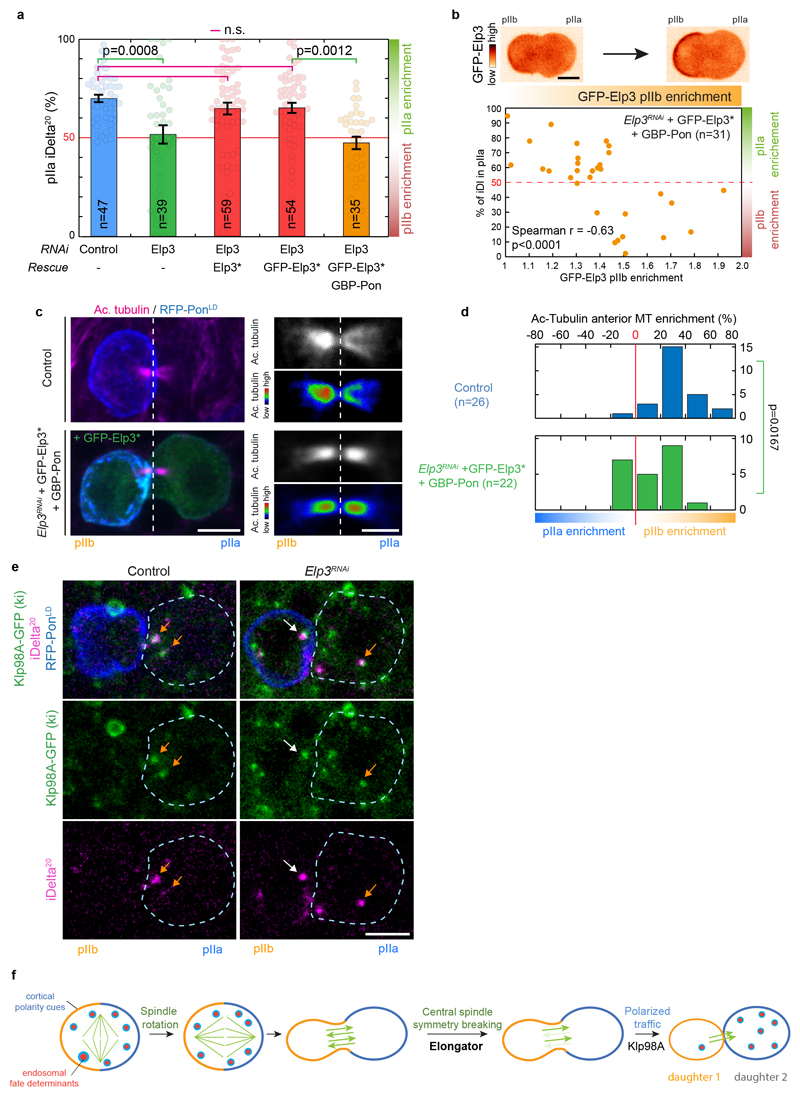
Our data shows that Elongator stabilises microtubules in vitro (Fig. 5a-i), and differentially stabilizes microtubules on the pIIb side of the central spindle in vivo (Fig. 1). In this context, it is worth noting that microtubule asymmetry matches Elongator asymmetry at the central spindle (pIIb enrichment: 43±8% for endogenous Elp4 versus 43±15% for microtubules, mean±SEM; n=4; Fig. 3b,c). Thus, we wondered if the asymmetry of Elongator activity could induce the asymmetric stabilization of microtubules in vivo. To test this, we specifically depleted Elongator from the pIIb side of the central spindle and investigated if this affects central spindle asymmetry and polarized trafficking.
To spatially alter the distribution of Elongator complex in cells, we capitalized on our previously established nanobody assay4, where GFP-tagged proteins of interest are asymmetrically mistargeted to the anterior cortex via an anti-GFP nanobody (GBP) fused to the polarity marker PonLD (GBP-PonLD), which itself localizes at the anterior cortex. This lowers the distribution of the GFP-tagged target at the pIIb side of the spindle in anaphase due to the establishment of a diffusion barrier (Fig. 6a and ref.4). Note that here we relocalized GFP-Elongator in a background where the endogenous Elongator was absent, capitalizing on a functional, RNAi-resistant GFP-Elp3* (Fig. 6b, Extended Data Fig. 10a and Supplementary Table 2 for validation).
Figure 6. Asymmetric control of microtubule dynamics by Elongator governs the asymmetric trafficking of Sara endosomes.
(a) Schematic of the nanobody assay. In control conditions (upper panel), the Elongator complex is enriched on the pIIb side of the central spindle (Fig 1), hence the central spindle is asymmetric with a higher microtubule density in the pIIb side, thereby delivering Sara endosomes preferentially to the pIIa cell. In contrast, upon expression of GBP-PonLD, a fusion between the localisation domain of the anterior cortical polarity marker Pon with an anti-GFP nanobody, in a background where endogenous Elp3 has been replaced by GFP-Elp3*, the whole Elongator complex becomes targeted to the anterior cortex. Due to the diffusion barrier at the central spindle in late anaphase, the pIIb and pIIa pools of Elongator are uncoupled, and the additional sink in pIIb provided by the anterior cortex effectively modifies the amount of Elongator at the spindle4. As Elongator directly stabilizes the microtubules, this situation leads to a higher microtubule density in the pIIa cell and, as a consequence, Sara endosomes can be preferentially delivered to the pIIb cell. (b) Dividing SOPs showing Sara endosomes (iDelta20) and mRFP-PonLD (pIIa cortex; dashed blue). Image corresponds to Maximum intensity z-projection of entire cells (SDCM). Note that while the Elp3RNAi phenotype can be rescued by RNAi-resistant GFP-Elp3*, this is not the case upon additional expression of GBP-Pon where most Sara endosomes can be targeted in the wrong cell (arrows). See also Extended Data Fig.10a for quantifications. (c) Percentage of Sara endosome in pIIa cell as a function of the extent of relocalisation of Elongator to the anterior cortex. Note that in this experiment, a phenotypic series is generated. The extent of relocalisation of Elongator to the anterior cortex positively correlates with the extent of Sara endosomes being targeted to the wrong, pIIb cell. Data were binned (mean ± SEM) according to their percentage of Sara in the pIIa cell (see Extended Data Fig. 10d for unbinned data). Statistics: Spearman’s correlation coefficient on unbinned data (p value<0.0001). n=31, number of SOPs analysed Scale bar: (b, c), 5 μm. Source numerical data are available in source data.
Mistargeting GFP-Elp3* away from the spindle with GBP-Pon induced a statistically significant reversal of central spindle asymmetry towards the wrong, pIIa, cell (Extended Data Fig. 10b,c), and thereby affected the asymmetric segregation of Sara endosomes (Extended Data Fig. 10a). In extreme cases, re-localization of GFP-Elp3* to the anterior cortex causes reversal of the polarity of endosomal trafficking, thereby directed towards the pIIb cell (Fig. 6b). Interestingly, due to the uncoupled variability of expression levels of Elp3RNAi, GFP-Elp3* and GBP-Pon in this experiment, we observed a phenotypic series among SOPs: different levels of cortical re-localisation of Elongator induce different levels of endosome segregation reversal. We could thus use this variability to test the correlation between the degree of cortical re-localisation and the degree of Sara endosome asymmetry. Indeed, there is a strong statistically significant correlation (p<0.0001) between the level of Elp3 re-localized to the cortex and the fraction of endosomes mistargeted to the wrong, pIIb cell (strong relocalisation: endosome reversal; mild relocalisation: symmetric endosomes; weak relocalisation: normal targeting phenotype; Fig. 6c and Extended Data Fig. 10d for unbinned data). Thus, differential Elongator activity between the two poles of the SOP can drive central spindle symmetry breaking and thereby polarized endosome trafficking.
To independently establish that Sara endosomes are symmetric upon Elongator depletion because the landscape of tracks they move on has lost its polarity, rather than because Elongator depletion affected the molecular motors at their surface, we studied the relationship between Elongator and Klp98A, the sole motor responsible for the motility of Sara endosomes4. Importantly, no interaction between Elongator and Klp98A could be detected by quantitative proteomics (Extended Data Fig. 5a), and, similarly, Elongator depletion did not affect the recruitment of endogenous Klp98A onto Sara endosomes (Extended Data Fig. 10e). This suggests a paradigm whereby Elongator first induces central spindle symmetry breaking, then Klp98A targets endosomes to the central spindle, but due to the local asymmetry in microtubule density, motility becomes polarized, leading to polarized trafficking. In other words, Elongator “writes” central spindle asymmetry, while Klp98A “reads” it (Extended Data Fig.10f).
Discussion
A link between cytoskeleton and transcription/translation?
While the role of Elongator on the regulation of translation during interphase is well established, here we identified an additional role for this complex during mitosis: the control of central spindle asymmetry and thereby of the trafficking of fate-determinants (Figs. 1,2). Five lines of evidence suggest that this Elongator activity is not an indirect effect through translational control of unknown factors. First, Elongator localizes to the central spindle in vivo, and directly stabilizes microtubules in vitro (Figs. 3,5). Second, Elongator phenotypes in the SOP can be rescued with mutants abolishing its KAT/SAM activities, known to mediate its effects on translation/transcription40,41 (Figs. 4). Third, abolishing these KAT/SAM activities does not affect Elongator’s ability to bind to microtubules or tune their dynamics (Fig.5). Fourth, depletion of CTU1, an Elongator partner critical for its effects on transcription, has no effect on central spindle asymmetry/endosome motility (Fig. 4d,e). Last, the long timescale of transcription/translation (hours) is not compatible with the fast effects we observed on central spindle asymmetry upon cortical re-localization of Elongator during mitosis (minutes; Fig.6).
Rather, our data suggest that Elongator is a bona fide Microtubule Associated Protein, which achieves control of central spindle asymmetry by directly stabilizing microtubules on the anterior side of the spindle in vivo (Figs.1,5,6). Importantly, this activity of Elongator likely requires the whole complex. Indeed: i) multiple Elongator subunits phenocopy each other in SOPs (Extended Data Fig.2) ; ii) Elongator always directly binds to microtubules as a whole entity (Fig.3e, Extended Data 9f); iii) multiple Elongator subunits can be detected at the spindle (Fig.3a,b, Extended Data Fig.4c); iv) partial Elongator subcomplexes do not bind to microtubules, nor can affect their dynamics (Fig.5 h,i, Extended Data Fig. 6,). While this “moonlighting” function of Elongator we identified is specific of mitosis, Elongator could well bind to microtubules during interphase. Future research is needed to investigate if Elongator can facilitate rapid changes in the local regulation of translation by linking the cytoskeleton, and therefore cortical signalling, to the translation machinery in large, polarized cells like neurons, where translation is controlled differentially in the cell body and synapses46.
Local control of microtubule dynamics in polarized cells
Having dedicated regulators of microtubule dynamics confers opportunities for local cytoskeleton remodelling in polarized systems, such as neurons and asymmetrically dividing cells. During SOP asymmetric division, the concentration of tubulin is likely the same in both daughter cells, but by locally controlling the activity of microtubule stabilizers like Elongator, which is upregulated in these cells13, an asymmetric structure of polarized microtubules can be generated to serve as tracks for the polarized segregation of fate determinants. Other MAPs, such as Patronin/CAMSAP, have also been reported to have polarized activity in polarized cells and during ACD4,47,48 by locally controlling the depolymerisation rate of microtubules minus ends, offering yet another knob to tweak in order to generate the right microtubule landscape.
The microtubule network is deployed in the cytosol, distant from the polarity cues which reside at the cortex of the cell. For instance, in the SOP, the main regulator of polarity, the Par complex, is found at the posterior cortex, which is several microns away from the central in the centre of the cell. A fascinating question for future research is how information flows from the Par complex to the microtubules, including the role of Elongator in this process. A number of PTMs have been described for Elongator subunits49,50, which might be targets for the kinase subunit of the Par complex.
Methods
Fly strains
Transgenes used in this study included UAS>GFP-Elp3* (this study), UAS>Elp3* (this study), UAS>Elp3* Y531F (this study), UAS>Elp3* YY531AA (this study), Klp98A-GFP knock-in (this study), UAS>Jupiter-mCherry 4, UAS-GBP-PonLD (ref.4), Jupiter-GFP knock-in at the endogenous locus (ref. 55, Bloomington #6836), UAS-mRFP-PonLD (ref. 56), Neur>Gal4 57, pnr>Gal4, phyllopod>GFP-PonLD (ref. 58), pnr>Gal4 (Bloomington #3039), UAS>DsRed (kind gift from François Karch), Ase>GFP-PonLD (ref. 4), UAS>Elp1RNAi (VDRC #109402), UAS>Elp2RNAi (VDRC #105393), UAS>Elp3RNAi (NIG #15433R-3), UAS>Elp4RNAi (VDRC #22460), UAS>Elp5RNAi (VDRC #20785), UAS>Hsc70-4RNAi (Bloomington #35684), UAS>CTU1RNAi (VDRC #104409), UAS>AtatRNAi (VDRC #26338); Elp3Δ4 (null allele of Elp3, see ref.18, kind gift from Patrik Verstreken. This mutant lacks most of the protein, including the SAM and KAT domains); P{neoFRT}40A (Bloomington); UAS>NeuralizedRNAi (VDRC #108239). The genotypes of recombined stocks were always verified by PCR and sequencing. To generate the desired genotypes and remove balancers, all experiments were performed on F1 of crosses at 25°C. Embryos were laid at 25°C. For SOP imaging, larvae were then shifted to 16°C until puparium formation (or at room temperature for MARCM experiments) and were shifted to 25°C or 29°C 16h prior to imaging as indicated. For MARCM experiments (Extended Data Fig. 3e,f), L3 larvae were heat-shocked for 1 hour at 37°C in a waterbath to induce clones.
Detailed genotypes and temperatures
In the main text
Fig. 1a: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Fig. 1b: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Fig. 1c: w1118; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Fig. 1d :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4/UAS>Elp3RNAi (29°C)
Fig. 1h :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3’RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118; UAS>mRFP-Pon / UAS>Elp3*; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Fig. 2c :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
w1118 (25°C)
pnr>NeurRNAi: w1118; Ase>GFP-PonLD /UAS>NeurRNAi; pnr>Gal4 UAS>DsRed/TM6B (29°C)
pnr>NeurRNAi Elp3RNAi: w1118; Ase>GFP-PonLD /UAS>NeurRNAi ; pnr>Gal4 UAS>DsRed / UAS>Elp3RNAi (29°C sibling of fly above)
Fig. 3a-c: w1118; Neur>Gal4, UAS>mRFP-PonLD / + (25°C)
Fig.4d-f :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* YY531AA: w1118; UAS>mRFP-PonLD/UAS>Elp3* YY531AA; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
CTU1 RNAi: w1118; UAS>mRFP-PonLD / UAS>CTU1RNAi; Neur>Gal4, Jupiter-GFP/+ (29°C)
Fig.4g :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* YY531AA: w1118; UAS>mRFP-PonLD/UAS>Elp3* YY531AA; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
CTU1 RNAi: w1118; UAS>mRFP-PonLD / UAS>CTU1RNAi; Neur>Gal4, Jupiter-GFP/+ (29°C)
Fig. 6b :
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4/UAS>Elp3RNAi (29°C)
Elp3 RNAi + GFP-Elp3*: w1118; UAS>mRFP-PonLD / UAS>GFP-Elp3*; Neur>Gal4/UAS>Elp3RNAi (29°C)
Elp3 RNAi + GFP-Elp3*+GBP-Pon: w1118; UAS>GBP-PonLD / UAS>GFP-Elp3*; Neur>Gal4 UAS>mRFP-PonLD/ UAS>Elp3RNAi (29°C)
Fig. 6c :
Elp3 RNAi + GFP-Elp3*+GBP-Pon: w1118; UAS>GBP-PonLD / UAS>GFP-Elp3*; Neur>Gal4 UAS>mRFP-PonLD/UAS>Elp3RNAi (29°C), or w1118 ; UAS>GBP-PonLD / +; Neur>Gal4 UAS>mRFP-PonLD/UAS>GFP-Elp3* (29°C).
In the supplementary materials
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp1 RNAi: w1118; UAS>mRFP-PonLD / UAS>Elp1RNAi; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp1 RNAi: w1118; UAS>mRFP-PonLD / UAS>Elp2RNAi; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PoLDn /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Hsc70-4 RNAi: w1118; UAS>mRFP-PoLDn /+; Neur>Gal4, Jupiter-GFP/UAS>Hsc70-4RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp1 RNAi: w1118; UAS>mRFP-PonLD / UAS>Elp1RNAi; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp1 RNAi: w1118; UAS>mRFP-PonLD / UAS>Elp2RNAi; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Hsc70-4 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Hsc70-4RNAi (29°C)
+/+: w1118 hsFlp12 /X; FRT40A / tub>Gal80 FRT40A ; Neur>Gal4, UAS>mRFP-PonLD / + (25°C)
Elp3Δ4/Δ4: w1118 hsFlp12 / X; Elp3Δ4 FRT40A / tub>Gal80 FRT40A ; Neur>Gal4, UAS>mRFP-PonLD/ + (25°C)
pnr>Gal4: w1118 ; pnr>Gal4 phyllopod>GFP-PonLD/ + (29°C)
pnr>Elp3RNAi: w1118; Ase>GFP-PonLD /+ ; pnr>Gal4 UAS>DsRed/UAS>Elp3RNAi (29°C)
pnr>Elp3RNAiElp3*: w1118; Ase>GFP-PonLD / UAS>Elp3* ; pnr>Gal4 UAS>DsRed/UAS>Elp3RNAi (29°C)
Extended Data Fig. 7a,b: w1118 ; pnr>Gal4 phyllopod>GFP-PonLD/UAS>AtatRNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
AtatRNAi: w1118 ; UAS>Jupiter-mCherry /+ ; pnr>Gal4 phyllopod>GFP-PonLD/UAS>AtatRNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
AtatRNAi: w1118 ; pnr>Gal4 phyllopod>GFP-PonLD/UAS>AtatRNAi (29°C)
or w1118 ; UAS>Jupiter-mCherry /+ ; pnr>Gal4 phyllopod>GFP-PonLD/UAS>AtatRNAi (29°C)
Control: w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi: w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi + Elp3* Y531F: w1118 ; UAS>mRFP-PonLD / UAS>Elp3* Y531F; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* Y531F: w1118 ; UAS>mRFP-PonLD / UAS>Elp3* Y531F; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* YY531AA: w1118; UAS>mRFP-PonLD/UAS>Elp3* YY531AA; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
Elp3 RNAi + Elp3* Y531F: w1118 ; UAS>mRFP-PonLD / UAS>Elp3* Y531F; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118 ; UAS>mRFP-PonLD / UAS>Elp3*; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* Y531F: w1118 ; UAS>mRFP-PonLD / UAS>Elp3* Y531F; Neur>Gal4, Jupiter-GFP / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3* YY531AA: w1118; UAS>mRFP-PonLD/UAS>Elp3* YY531AA; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Control: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/ + (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi: w1118; UAS>mRFP-PonLD /+; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
or w1118; Neur>Gal4, UAS>mRFP-PonLD / UAS>Elp3RNAi (29°C)
Elp3 RNAi + Elp3*: w1118; UAS>mRFP-Pon / UAS>Elp3*; Neur>Gal4, Jupiter-GFP/UAS>Elp3RNAi (29°C)
Elp3 RNAi + GFP-Elp3*: w1118; UAS>mRFP-PonLD / UAS>GFP-Elp3*; Neur>Gal4/UAS>Elp3RNAi (29°C)
Elp3 RNAi + GFP-Elp3*+GBP-Pon: w1118; UAS>GBP-PonLD / UAS>GFP-Elp3*; Neur>Gal4 UAS>mRFP-PonLD/UAS>Elp3RNAi (29°C)
Control: w1118; Neur>Gal4, UAS>mRFP-PonLD / + (29°C)
Elp3 RNAi + GFP-Elp3*+GBP-Pon: w1118; UAS>GBP-PonLD / UAS>GFP-Elp3*; Neur>Gal4 UAS>mRFP-PonLD/UAS>Elp3RNAi (29°C).
Elp3 RNAi + GFP-Elp3*+GBP-Pon: w1118; UAS>GBP-PonLD / UAS>GFP-Elp3*; Neur>Gal4 UAS>mRFP-PonLD/UAS>Elp3RNAi (29°C), or w1118 ; UAS>GBP-PonLD / +; Neur>Gal4 UAS>mRFP-PonLD/UAS>GFP-Elp3* (29°C).
control: w1118;; Neur>Gal4 UAS>mRFP-PonLD Klp98A-GFP (29°C)
Elp3 RNAi: w1118;; Neur>Gal4 UAS>mRFP-PonLD Klp98A-GFP/UAS>Elp3RNAi (29°C)
Elp1: w1118 ; (If,CyO)/UAS>Elp1RNAi; Da>Gal4 / + (29°C)
Elp2: w1118 ; (If,CyO)/UAS>Elp2RNAi; Da>Gal4 / + (29°C)
Elp3: w1118; (If,CyO)/+; Da>Gal4 / UAS>Elp3RNAi (29°C)
Elp4: w1118; (If,CyO)/+; Da>Gal4 / UAS>Elp4RNAi (29°C)
Elp5: w1118; (If,CyO)/UAS>Elp5RNAi; Da>Gal4 / + (29°C)
Hsc70-4: w1118; (If,CyO)/+; Da>Gal4 /UAS>Hsc70-4RNAi (29°C)
CTU1: w1118; (If,CyO)/UAS>CTU1RNAi; Da>Gal4 / + (29°C)
CTU2#1: w1118; (If,CyO)/UAS>CTU2RNAi #1; Da>Gal4 / + (29°C)
CTU2#2: w1118; (If,CyO)/UAS>CTU2RNAi #2; Da>Gal4 / + (29°C)
Kti11/DPH3: w1118; (If,CyO)/UAS> Kti11RNAi; Da>Gal4 / + (29°C)
none: Elp3: w1118; (If,CyO)/+; Da>Gal4 / UAS>Elp3RNAi (29°C)
Elp3*: Elp3: w1118; (If,CyO)/UAS>Elp3*; Da>Gal4 /UAS>Elp3RNAi (29°C)
GFP-Elp3*: Elp3: w1118; (If,CyO)/UAS>GFP-Elp3*; Da>Gal4 / UAS>Elp3RNAi (29°C)
Elp3*-Y531F: Elp3: w1118; (If,CyO)/UAS>Elp3*-Y531F; Da>Gal4 / UAS>Elp3RNAi (29°C)
Elp3*-YY531AA: Elp3: w1118; (If,CyO)/UAS>Elp3*-YY531AA; Da>Gal4 / UAS>Elp3RNAi (29°C)
Generation of Klp98A GFP Knock In
Klp98A-GFP is a GFP knock in allele generated by CRISPR gene editing. Briefly, we modified our previously published Klp98A pW25 repair plasmid4 to replace the 5’ UTR region of Klp98A by a 3145bp homology fragment upstream of the stop codon in Klp98A (this encompasses exons 6-12 and associated introns). The 5’ homology region is flanked by the coding sequence of GFP. Upon repair of a CRISPR-mediated DNA cut, this construct replaces the stop codon of Klp98A by GFP followed by a stop codon, then by the whs gene flanked by two LoxP sites followed by an AttP ΦC31 site. To induce CRISPR-mediated DNA cut, a Chi RNA for Klp98 was cloned into the BbsI sites of pU6-BbsI-chiRNA plasmid (Addgene #45946) as described59 using the oligos: chiRNA klp98BbsI up: CTT CGC AGG AGA TAA TTC AGC AGT and chiRNA klp98BbsI low: AAA CAC TGC TGA ATT ATC TCC TGC. Both plasmids were injected by BestGene Inc at a concentration of 125 ng/μl for the pU6 chiKlp98 and 500 ng/μl for the donor plasmid. The whs gene was subsequently floxed to generate Klp98A-GFP which corresponds to a GFP knock-In. Gene editing was confirmed by PCR. The GFP signal Klp98A-GFP displays the characteristics endosomal signal we previously reported for endogenous Klp98A4, and, furthermore, it colocalizes with iDetla20 as expected4 (Extended Data Figure 10e), suggesting it is a bona fide Klp98A live reporter.
S2 and Dmel-2 stable cell culture and cell line establishment
S2 cells (UCSF, sold by ThermoFisher #R69007, mycoplasm-free judged by Dapi staining) were grown at 25°C in Schneider Medium (Lonza #04-351Q) supplemented with 1% Pen/Strep (Gibco #15140122) and 10 % (vol/vol) Foetal Bovine Serum (Gibco; #10270106; Heat-inactivated for 1h at 70°C). Stable cell lines were obtained by transfection with pMT puro vectors (see above) using Effectene (Qiagen #301425) according to the manufacturer’s instructions, followed by selection in 5 μg/mL Puromycin (ThermoFisher #A1113803). Similarly, D.mel-2 cells (ATCC #CRL-1963) were grown in a shaking incubator 27°C in protein-free Insect Express medium (Lonza #BE12-730Q) supplemented with 1% vol/vol Pen/Strep (Gibco) and 1% vol/vol Pluronic F-68 (Gibco # 24040032). Stable cell lines were obtained by transfection with pMT puro vectors (see below) using ExpiFectamine (ThermoFisher #A38915) according to the manufacturer’s instructions, followed by selection in 5 μg/mL Puromycin.
Plasmids
All the Open Reading Frames (ORFs) cloned by PCR for this study were flanked by FseI and AscI sites for convenient shuttling between compatible plasmids. Elp3 (CG15433-PA) was cloned by w1118 fly cDNA. In addition, we also designed Elp3*, a codon-optimized version of Elp3 (CG15433-PA) which has been specifically engineered to be resistant to the Elp3 RNAi sequence used in this study via numerous silent mutations. Elp3* was synthetized by GeneArt. Elp3* Y531F and Elp3* YY531AA were generated by site directed mutagenesis (GeneArt). Similarly, Drosophila codon-optimized versions of Elp1 (CG10535-PA), Elp2 (CG11887-PA), Elp4 (CG6907), Elp5 (CG2034-PA), Elp6 (CG9829-PA) were synthetized by IDT and are referred to as Elp1*, Elp2*, Elp4*, Elp5*, Elp6*, respectively. Note that we experimentally confirmed by Mass-Spectrometry in Extended Data Fig. 5 that the Drosophila homologs Elp5 and Elp6 are indeed CG2034 (Anon-i1) and CG9829 (Poly), as previously proposed by distant phylogenetic inference53,54.
For coexpression of mCherry-Elp4*, Elp5* and Elp6* in bacteria, we first cloned Elp4* into a modified pGEX vector that tags Elp4* with an N-terminal GST tag followed by a TEV protease cleavage and an mScarlet tag. We then synthetized a fragment comprising both Elp5* and Elp6* ORFs, with their respective stop codons and a 5’ Ribosome Binding sequence (RBS): [RBS-Elp5*-STOP-RBS-Elp6*-STOP], which we cloned in 3’ of Elp4*, after the stop codon and before the transcription termination side. Thus, the single transcription unit in this vector is Promoter-GST-TEV-mCherry-Elp4*-STOP-RBS-Elp5*-STOP-RBS-Elp6*-STOP-terminator, leading to all tree proteins, GST-TEV-mCherry-Elp4*, Elp5* and Elp6 being expressed. This vector is referred to as pGEX-mScarletElp4*/5*/6* in the following.
For fast generation of stable cell lines in S2 cells, ORFs were cloned into a modified pMT vector (Life Technologies), bringing a Puromycin selection gene and either a (His)6-PC or a (His)6-PC-SNAP tandem N-terminal tags. PC stands for the Protein C epitope tag: EDQVDPRLIDG. (His)6, the His tag, is abbreviated “His” in the following for simplicity. N-terminal tagging of Elp3 with SNAP is functional, as the SNAP-Elp3 complex has the same activity as the Elp3-complex in vitro (Fig. 5 and Extended Data Fig. 9).
For expression in flies with the UAS/Gal4 system, the various versions of the Elp3* ORF described above were subcloned into modified pUAST4 vectors tagging the ORF with either N-terminal PC-eGFP (UAS>GFP-Elp3*) or leaving it untagged (for UAS>Elp3*, UAS>Elp3* Y531F, UAS>Elp3* YY531AA). Note that N-terminal GFP tagging of Elp3 is functional as it can rescue the Elp3RNAi phenotype in vivo (Fig. 6, Extended Data Fig. 10a, Supplementary Table 2), confirming previous results with GFP-Elp318. Injection of plasmids into Drosophila embryos to generate transgenics was performed by BestGene Inc.
Antibodies
All antibodies were used at 1μg /mL for western blot and immunofluorescence.
Previously characterized Oregon Green 514-labelled mouse anti-β-tubulin (E7, Developmental studies hybridoma bank), Atto-647N-labelled anti-α K40 acetylated tubulin (C3B9, HPA Cultures) and ATTO-647N-labelled anti-Delta (C594.9B, Developmental studies hybridoma bank), were prepared as previously described 4. DM1A antibody anti alpha tubulin was purchased from Sigma (#T6199).
Polyclonal rabbit raised against the Elp1 to Elp5 subunits were generated by injecting rabbits (Eurogentec Speedy program) with synthetized peptides (Eurogentec) spanning flexible regions of each subunit (Elp1: H2N-CSNYDASKQTYPSDYR-CONH2; Elp2: H2N-FRHISGIENDDAGDV-CONH2; Elp3: H2N-CQLDGPYMSKSIEEN-CONH2; Elp4: H2N-MTSFRKRTVQKPIRGC-CONH2; Elp5: H2N-CASPKQPSPEAEQTTE-CONH2). We attempted but could not obtain functional Elp6 antibodies using the same method. Immunized sera were affinity-purified with a sepharose resin coupled with the respective peptide using standard glycine elution (0.1M glycine pH 3.0). Eluted antibodies were subsequently dialyzed against PBS supplemented with 50% glycerol and stored at -20°C. The characterization of these antibodies is presented in Extended Data Fig. 2.
SDS-PAGE and Western blot
SDS-PAGE was performed using NuPAGE 4-12% Bis-Tris gels (Life Technologies) according to the manufacturer's instructions. InstantBlue (Sigma) was used for total protein staining of gels.
For Western Blot, gels were transferred on nitrocellulose membranes using iBLOT (Life Technologies) following manufacturer’s instructions. Following Ponceau staining (Sigma), membranes were washed in TBS and blocked in TBS enriched with 5% milk powder for 20 min at RT. Primary antibodies were incubated overnight at 4°C using a solution made of 1 μg/ml antibody in TBS supplemented with 0.2% BSA and 0.02% Thymerosal. The membranes were revealed using HRP-conjugated antibodies (Jackson immunoResearch, #AB_2313567, dilution 1:10,000 in TBS-5% milk 0.1% Tween-20 for 1h at RT), Western Bright Quantum (Advansta) or SuperSignal West Pico (Pierce) chemiluminescence reagents and a ChemiDoc (Bio-Rad) or a Fusion (Vilber Lourmat) imagers using manufacturer’s instructions.
Alternatively, for fluorescent western blot with Oregon Green 514-labelled mouse anti-β-tubulin and Atto-647N-labelled anti-α K40 acetylated tubulin antibodies (Extended Data Figs. 1a and 8e), membranes were incubated overnight at 4°C using a solution containing both antibodies (1 μg/ml each) in TBS supplemented with 0.2 % BSA and 0.02 % Thymerosal. The membranes were then imaged using a Typhoon or an Ettan DIGE fluorescence scanner (GE Healthcare).
RNAi-treatment of S2 cells and immunoprecipitation
For RNAi-treated S2 cell extracts (Extended Data Figs. 1 and 8e), cells were cultured and incubated with 5 μg dsRNA for 4 days as previously described 60. The Elp3 dsRNA sequence we used corresponds to the sequence in the UAS>Elp3RNAi fly stock used through this paper (NIG-15433R-3). Elp2 and Elp4-depleted S2 extracts were similarly derived using the RNAi sequences found in the following fly stocks: Elp2-1: VDRC #7560; Elp2-2: VDRC #105393; Elp4-1: VDRC #22460 ; Elp4:2: VDRC #110450. Note that the Elp2-2 and Elp4-1 RNAi sequences are the ones we used in vivo in Supplementary Table 1 and Extended Data Fig. 3). As a control, we used dsRNA against GFP. Specific RNAi sequences were amplified by PCR from genomic DNA using the following primers:
rnai_elp2_1f TAATACGACTCACTATAGGG TGGAGCACACAAAGCGAGTAAACA
rnai_elp2_1r TAATACGACTCACTATAGGG GCCACTGAAGGCGAGCAGAA
rnai_elp2_2f TAATACGACTCACTATAGGG GGTTGGATTGCGACAAACTT
rnai_elp2_2r TAATACGACTCACTATAGGG AGCTCACTGCATACCAGGCT
rnai_Elp3f TAATACGACTCACTATAGGG TCTTCAGACCCGACACCGCGT
rnai_Elp3r TAATACGACTCACTATAGGG AGCGGAAGGCAGGGTTCTCAA
rnai_elp4_1f TAATACGACTCACTATAGGG CAGCAGCGTGACCAGTAGCACAAA
rnai_elp4_1r TAATACGACTCACTATAGGG CGAACCGGCAAACGACTCCA
rnai_elp4_2f TAATACGACTCACTATAGGG ACGAAGATGACCAGTTTCCG
rnai_elp4_2r TAATACGACTCACTATAGGG GGCTATCGTCCCACAAGGTA
rnai_GFP_f TAATACGACTCACTATAGGG ATGGTGAGCAAGGGCGAGGA
rnaiGFP_r TAATACGACTCACTATAGGG CTTGTACAGCTCGTCCATGC
DsRNA-treated cells were washed in XB (20 mM Hepes, 150 mM KCl, pH 7.7), then lysed in lysis buffer (20 mM Hepes, 150 mM KCl, 1 mM CaCl2, 1 % Tx100, 5 % Glycerol, pH 7.7 enriched with protease inhibitors: 1mM Benzamidine, 40 ug/ml Chymostatine, 40 μg/ml Antipain, 1 μM Leupeptine, 1 mM Pefabloc and 0.5 mM PMSF. After a 10 min incubation at 4°C with rocking, cellular debris were cleared by centrifugation at 16'000 x g for 10 min at 4°C. Extracts were then diluted in LDS sample buffer enriched with 2.5% β-mercaptoethanol and analyzed by SDS-PAGE and Western blot as above.
For immunoprecipitations, cell extracts were incubated with 2 μg of purified anti-Elp2 antibodies (see above), or non-specific Rabbit IgG control (Thermo Fisher), and 20 μL of protein G beads (Pierce). After 2 hours incubation at 4°C with rocking, the beads were washed three times in lysis buffer and the samples analysed by SDS-PAGE and Western blot as above.
Protein purification
Unless stated otherwise, all purification steps were performed at 4 °C. An aliquot of the last buffer exchange step of each purification was always kept to serve as a true negative control in all tubulin dynamics experiments. Protein concentrations were determined using a BCA protein assay kit (Pierce) or by densitometry on Coomassie-stained SDS-PAGE gels against a BSA dilution series. Protein identity was always verified by Mass Spectrometry (see below). Controls of biochemical experiments were always performed using an identical volume of the specific buffer of the protein of interest (usually the last dialysis or gel filtration buffer) to ensure that buffer variations are not a variable in our experiments.
The SNAP-tagged Elongator complex was purified from a puromycin-resistant D.mel-2 cell stable cell line expressing His-PC-SNAP-dElp3 under the inducible metallothionein promoter. Cells were grown in Insect-Xpress medium (Lonza) supplemented with 5 μg/ml puromycin (Gibco) and 0.6 mM CuSO4. After amplifying this cell line to litre-scale over several weeks, cells were collected by centrifugation, washed in 20 mM HEPES pH 7.6, 150 mM KCl, 1 mM CaCl2 buffer, frozen in liquid N2 and stored at -80 °C. Frozen cells were thawed, diluted in lysis buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 1% Triton X-100, 1 mM CaCl2, 5 mM MgCl2, 20% glycerol, 0.12 mg/ml Benzamidine, 20 μg/ml chymostatin, 20 μg/ml antipain, 0.5 μg/ml leupeptin, 0.24 mM Pefabloc SC and 0.5 mM PMSF), lysed by extrusion using a teflon dounce homogenizer and clarified by centrifugation at 25,000 x g for 30 minutes using a JA 25.50 rotor (Beckman). The clarified lysate was incubated with 2 ml of pre-equilibrated Protein C affinity resin (Roche) for 3h at 4°C. The resin was then packed into an empty column (Bio-Rad), washed with 50 ml of wash buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5 mM MgCl2, 20% glycerol) supplemented with 1 mM CaCl2, followed by 50 ml of wash buffer without calcium. The Elongator complex was then eluted by incubating the resin with sequential incubation with 2 ml of elution buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5 mM MgCl2, 20% glycerol, 5 mM EGTA). All elution steps were analyzed by SDS-PAGE, and Elongator-containing fractions were concentrated (Amicon Ultra,-4 3kD MWCO, Millipore) and further purified by size-based separation on a sucrose gradient by layering concentrated fraction atop a manually prepared 10-30% discontinuous sucrose gradient (in 5% steps) in elution buffer without glycerol. Following ultracentrifugation for 1h at 258,488 x g at 4°C using a TLS-55 rotor (Beckman) with the slowest acceleration and deceleration settings, the fractions were analyzed by SDS-PAGE. Fractions containing the full complex (Elp1-6+Hsc70-4) were then buffer-exchanged against Elongator buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5 mM MgCl2, 20% sucrose, 5 mM EGTA) and either concentrated, flash-frozen in liquid N2 and stored in small aliquots at -80°C or labelled using SNAP-Surface Alexa Fluor 488 (NEB). Labelling of the SNAP Elongator complex with SNAP-Surface Alexa Fluor 488 (NEB) was performed following manufacturer’s instructions. The labelled protein was then desalted into Elongator buffer using Zeba Spin columns (Thermo Fisher) to remove the non-bound dye, then concentrated, flash-frozen in liquid N2 and stored at -80°C. The purification of the Elongator complex devoid of the SNAP tag (Fig. 3d and Extended Data Figure 5c) was performed in a similar fashion, except that cells were transfected with a pMT Puro His-PC-Elp3* plasmid, and that the sucrose gradient step was replaced by a gel filtration step on a superose 6 increase column (see Extended Data. Fig. 5c). The complex isolated by this method is functional for previously reported Elongator activity, in particular tRNA binding (Extended Data Fig.5 f) and tRNA modification (Extended Data Fig.5 g).
The Elongator Elp3 YY531AA complex (Fig. 4) was purified as the WT complex, albeit expression of Elp3 YY531AA was found to strongly affect cell growth and viability, likely because of a dominant negative effect (Elp3 YY531AA is unable to rescue WT Elp3 in flies, see Supplementary Table 2). We thus capitalised on the inducible nature of our expression system and grew large quantities of cells (several litters) in the absence of CuSO4. Only then did we induce them, and harvested the cells when viability started to decrease (usually after 3-5 days). As expected, the yield was markedly lower compared to constitutive expression due to the poor incorporation of the exogenous Elp3 YY531AA into the endogenous complex, but by scaling up the culture, we could get enough for functional assays. As expected from previous work on yeast Elongator, the YY531AA complex isolated retains tRNA binding but does not exhibit tRNA modification activity (Fig. 4). As a final purification step, the YY531AA complex was buffer-exchanged against a buffer aliquot saved from the WT complex purification, resulting in both complexes being in the same exact buffer for its use in in vitro experiments.
Note that we found that the Drosophila Elongator complex copurifies with the protein Hsc70-4 as a stochiometric partner after size-exclusion chromatography (both gel filtration and sucrose gradient, see Extended Data Fig. 5c,d). To our knowledge, this was not previously reported for the yeast and human complexes, for which most of the biochemical characterization has been done. While this could reflect some Drosophila specificity, it could also reflect some transient interactions that are lost during the purification and/or regulation by post-translational modifications. Note that global depletion of Hsc70-4 at the level of the fly is lethal, as is depletion of any Elongator subunit (Supplementary Table 1). However, SOP-specific depletion of Hsc70-4 does not affect central spindle asymmetry nor polarized segregation of Sara endosomes (Extended data Fig. 3), and, in agreement, the Elp23-Hsc70-4 does not bind to microtubules nor markedly affect their dynamics in vitro (Fig. 5h,I and Extended Data Fig.6 a-c). This likely reflect that the interaction of Hsc70-4 with Elongator is important for its roles in translation, but not for its roles in regulating microtubule dynamics.
A by-product of our Elongator purification method is the SNAP-Elp23 heterodimer with Hsc70-4, which comes as an isolated fraction in the sucrose gradient (Extended Data Fig 6). This is not an artefact of the SNAP-tag, as a similar fraction can be isolated from cells expressing His-PC-Elp3*.
The mScarlet-Elp456 subcomplex was purified from BL21 (DE3) Rosetta2 (Invitrogen) Escherichia coli transformed with the pGEX-mScarletElp4*/5*/6* vector (see above). Bacteria were grown at 37°C in 2xYT medium to OD600=0.8 and then induced using 0.5 mM IPTG at 20°C for at least 16 hours. For the mScarlet-Elp456, the cells were collected by centrifugation, resuspended in lysis buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5% glycerol, 1 mM DTT, 10 mM MgCl2, 1% Triton X-100 and complete™ Protease Inhibitor Cocktail Tablets (Roche)) and lysed by sonication. After clarification (40,000 x g, Beckman JA 25.5, 30 min, 4°C), the soluble fraction of the lysate was applied to a Glutathione Sepharose 4B column (GE Healthcare), washed with wash buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5% glycerol, 1 mM DTT) and eluted in wash buffer supplemented with 10 mM GSH. The eluted complex was then incubated with rTEV protease to remove the N-terminal GST-tag, dialyzed against fresh wash buffer and further purified by gel filtration on a Superdex 200 column (GE Healthcare) in a final buffer containing 50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5% glycerol, 1 mM DTT, 5 mM EGTA and 5 mM MgCl2. Fractions containing the complex were pooled, concentrated, flash-frozen in liquid N2 and stored in small aliquots at -80 °C. Note that this purification contains some traces of uncleaved GST-mScarlet-Elp4, but since no mScarlet signal is observed on microtubules, this implies that a potentially dimeric GST-tagged Elp456 also does not bind to microtubules.
Unlabelled porcine tubulin, or HiLyte647- and rhodamine-labelled porcine tubulin were purchased from Cytoskeleton (#T240, #TL590M and #TL670M), reconstituted at 5 mg/ml in BRB80 buffer (80 mM K-Pipes, 1 mM MgCl2 pH 6.9) supplemented with 1 mM GTP (Roche) or 1 mM GMPPCP (Jena Bioscience # NU-405S), flash frozen and kept in liquid N2.
Microtubules seeds stocks were prepared at 50 μM tubulin concentration (20% rhodamine-tubulin, 20% biotinylated tubulin) in BRB80 buffer supplemented with GMPCPP-Tubulin buffer (1 mM GMPCPP, 2 mM MgCl2 and 0.5 mM EGTA), aliquoted and stored in liquid N2. Before use, each aliquot of microtubule seed stock was incubated at 37°C for 30 minutes, sedimented on a table-top centrifuge by centrifugation (8 minutes at 14000 x g), and resuspended in GMPCPP-Tubulin buffer buffer.
Fibrinogen-neutravidin was prepared as described 61 by mixing four-fold molar excess of Maleimide-Neutravidin with thiolated-Fibrinogen. The resulting solution was incubated overnight at room temperature under gentle agitation. The reaction was quenched by the addition of 50 mM cysteine. The fibrinogen-neutravidin was then precipitated using a 25% ammonium sulfate solution and resuspended in carbonate buffer. This process was repeated twice to improve the purity of the mix. The final solution was aliquoted, flash frozen in liquid N2 and stored at -80°C. The thiolated-Fibrinogen was prepared by mixing 25-fold molar excess of Traut’s Reagent (Pierce #26101) with fibrinogen (MP Biomedicals #08820224) for 45 minutes at room temperature while gently rocking. The reaction was quenched by the addition of 10 mM DTT (incubated 1h at room temperature) and desalted using Zeba Spin columns equilibrated and eluted using carbonate buffer supplemented with 1 mM EDTA. Maleimide-Neutravidin was prepared by incubating 6X molar excess of NHS-Maleimide-PEG8-succinimidyl ester (Sigma #746207) with Neutravidin (ThermoFisher Scientific #31000) for 1 hour at room temperature, then desalted using the same procedure as for the thiolated-Fibrinogen.
Microtubule dynamics assays
Unless stated otherwise, all proteins were centrifuged for 10 minutes at 20000 x g at 4°C before use in biochemical assays. Glass 22x22 mm coverslips (NEXTERION, Schott) were cleaned using a plasma cleaner (PlasmaPrep2, GaLa instruments) and incubated overnight at room temperature with gentle agitation in a solution of 1 mg/ml mPEG-Silane (30 kDa, PSB-2014, Creative PEGWorks) in 96 % (v/v) ethanol and 0.2 % (v/v) HCl. The coverslips were then washed in ethanol and ultrapure water, dried with nitrogen gas and assembled into an array of flow cells on a mPEG-Silane passivated slide using double-sided tape (Adhesive Research AR-90880 precisely cut with a Graphtec CE6000 cutting plotter). The chamber was first perfused with one chamber volume of 0.04 mg/ml Fibrinogen-Neutravidin (see above) solution in carbonate buffer (0.1M sodium bicarbonate, 0.5 mM EDTA, pH 8.3), washed with 10 volumes of carbonate buffer, quenched for 10 minutes with a 0.2 mg/ml solution of PLL-g-PEG (Susos) and finally washed with 10 volumes of BRB80 buffer. Biotin-GMPCPP stabilized microtubule seeds (20% with rhodamine-tubulin, 20% biotin-tubulin) were then injected and let to adhere to the neutravidin for 5 minutes. The chamber was then washed with 10 chamber volumes of imaging buffer (BRB80 enriched with 0.1 mg/ml K-Casein, 0.1 mg/ml BSA, 40 μM DTT, 64 mM D-glucose, 160 μg/ml glucose oxidase, 20 μg/ml catalase and 0.2% methylcellulose). Seeds were elongated with a mixture containing 12.5 μM of 10% labeled HiLyte647-GTP Tubulin and Elongator complex (or an equivalent volume of Elongator buffer as a control) and microtubule dynamics were monitored by TIRF microscopy. The microscope stage was kept at 37°C. Alternatively, for Extended Data Fig. 9a-d, anchoring of the biotinylated seeds was performed by doping the mPEG-Silane coverslip treatment with 10% biotin-mPEG-Silane (Laysan Bio #Biotin-PEG-SIL-3400) instead of using fibrinogen-neutravidin. Note that all Elongator complex used for dynamic assays was purified by size exclusion (sucrose gradient or gel filtration) to ensure the activity of the full complex is assayed. Note that an aliquot of the last buffer exchange step during the purification of each protein was always kept to serve as a true negative control in all tubulin dynamics experiments.
Dynamic microtubule co-sedimentation assay
All proteins were centrifuged for 10 minutes at 20000g at 4°C before use in biochemical assays. The protocol for microtubule pelleting assays using dynamic microtubules (Fig. 3e) was adapted from ref62. Briefly, 30 μl protein samples in BRB80+ buffer (BRB80 supplemented with 10% DMSO, 1 mM GTP and 1 mM MgCl2) were incubated for 20 minutes at 30°C to initiate microtubule polymerization. Protein samples include 0.4 μM purified Elongator complex (WT or YY531AA), 0.4 μM Elp23+Hsc70-4 or 1 μM mScarlet-Elp456 with or without 40 μM tubulin. After incubation, the samples were transferred into warm 20 mm polycarbonate ultracentrifuge tubes (Beckman) pre-filled with 150 μl of warm GTP-glycerol cushion solution (BRB80 supplemented with 60% glycerol, 1 mM GTP and 1 mM MgCl2). The samples were centrifuged at 100,000 x g for 30 minutes at 37°C using a warmed TLA100 rotor. After centrifugation, the top 20 μl were recovered and saved for SDS-PAGE/Western blot analysis, designated as “supernatant”. The remaining 10 μl atop the glycerol cushion were removed and discarded, and the interfaces were washed three times with 100 μl of warm BRB80+ buffer. The glycerol cushion solutions were then removed and the pellets gently washed with 3x100 μl of warm BRB80+ buffer. Finally, the pellets were resuspended in 30 μl of BRB80+ buffer, saved and designated as “pellet” for further analysis. Note that all Elongator complex used for co-sedimentation assays was purified by size exclusion (sucrose gradient or gel filtration) to ensure the activity of the full complex is assayed.
Scanning Electron Microscopy (SEM)
Flies were euthanized by exposure to diethyl ether for 20 minutes, then mounted on SEM holders using double-sided carbon tape (Electron Microscopy Sciences) and subsequently treated with a gold sputter coater (JFC-1200, JEOL). Imaging was performed using a JEOL JSM-6510LV scanning electron microscope operating in high vacuum mode using a working distance of 10 mm and an acceleration of 10 kV.
Affinity measurements
To measure the tRNA binding affinity to Elongator, microscale thermophoresis (MST) assays were performed following previously stablished protocols32,63. The assays were performed on a Monolith NT.115 (Nanotemper Technologies) using standard capillaries (MO-K022) at 25°C. Two-fold dilutions series of the non-fluorescent protein were performed in Elongator buffer (50 mM K-HEPES pH 7.5, 100 mM K-Acetate, 5 mM MgCl2, 20% sucrose, 5 mM EGTA, 1 mM DTT) supplemented with 0.05% (v/v) Tween-20. The fluorescent EctRNAUUULys fused to BODIPY-FL (Promega) was used at a constant concentration of 20 nM, and the was assessed using 10% LED power and heating for 20s at 20% Laser power. The dissociation constant values were obtained by analysing the change in thermophoresis using the MO.Affinity software provided by the manufacturer.
Acetyl-CoA hydrolysis assays
Acetyl-CoA hydrolysis rates were measured using the Acetyl-Coenzyme A Assay Kit (MAK039, Sigma-Aldrich) and following a previously stablished protocol23,63. In brief, WT and YY531AA Elongator complexes (100 nM), or equivalent quantities of Elongator buffer, were mixed with 2 μM bulk tRNA from baker’s yeast (TRNABAK-RO, Roche) and 100 μM acetyl-CoA in acetyl-CoA assay buffer. Following incubation at 25°C for 150 min, proteins and tRNA were removed using Amicon Ultra-0.5 columns. The acetyl-CoA concentration in the flow-through was measured following manufacturer’s instructions (λex = 535, λem = 587 nm) using a Tekan Spark 10M plate reader. Hydrolysis rates were calculated from three independent experiments.
Mass spectrometry
Polyacrylamide gel slices (1-2 mm) containing the purified proteins were prepared for mass spectrometric analysis by manual in situ enzymatic digestion. Briefly, the excised protein gel pieces were placed in a well of a 96-well microtitre plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, proteins were digested with 6 ng/μL Trypsin (Promega, UK) overnight at 37 °C. The resulting peptides were extracted in 2% v/v formic acid, 2% v/v acetonitrile. The digest was analysed by nano-scale capillary LC-MS/MS using a Ultimate U3000 HPLC (ThermoScientific Dionex, San Jose, USA) to deliver a flow of approximately 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm x 20 mm nanoViper (ThermoScientific Dionex, San Jose, USA), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 μm, 75 μm x 150 mm nanoViper (ThermoScientific Dionex, San Jose, USA). Peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a modified nano-flow electrospray ionisation source, with a hybrid dual pressure linear ion trap mass spectrometer (Orbitrap Velos, ThermoScientific, San Jose, USA). Data dependent analysis was carried out, using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision induced dissociation. LC-MS/MS data were then searched against an in house protein sequence database, containing Swiss-Prot and the protein constructs specific to the experiment, using the Mascot search engine programme (Matrix Science, UK)64. Database search parameters were set with a precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 Da. Two missed enzyme cleavages were allowed and variable modifications for oxidized methionine, carbamidomethyl cysteine, pyroglutamic acid, phosphorylated serine, threonine, tyrosine, tert-butyloxycarbonyl-lysine, norbornene-lysine and prop-2-yn-1-yloxycarbonyl-lysine were included. MS/MS data were validated using the Scaffold programme (Proteome Software Inc., USA)65. All data were additionally interrogated manually.
Fly Notum live imaging, Delta antibody uptake and immunofluorescence
Fly notum dissection and SOP imaging was performed in Clone 8 medium after embedding into a fibrinogen clot in order to diminish tissue movements during fast 3D image acquisition as described 66. Fluorescent Delta antibody uptake to label the Sara endosomes was performed using our previously described improved assay using anti Delta antibodies directly labelled with the photostable dye ATTO-647N (see above and also ref.4 for more details). Briefly, Sara endosomes were labelled with a 5-minute pulse (3.4 μg/ml ATTO-647N-anti Delta antibody in Clone 8) and a 20-minute chase (referred to as iDelta20). All notum live-imaging was performed on the Spinning disk confocal microscope described below.
For notum immunofluorescence, dissected fly nota were fixed according to a method designed to preserve the microtubule cytoskeleton67. Briefly, nota were first incubated in Hank’s balanced salt solution (Gibco) enriched with 1 mM DSP (Pierce) for 10 min at RT followed by a 10 min incubation in MTSB (microtubule stabilization buffer: 0.1 M PIPES, 1 mM EGTA, 4 % PEG 8000, pH 6.9) enriched with 1 mM DSP, then finally in MTSB enriched with 4% PFA (Electron Microsopy Science). Nota were then permeabilised in MTSB enriched with 4% PFA and 0,2% Triton X100 then processed for immunofluorescence as described4 and mounted in Prolong Gold antifade reagent (Molecular Probes). Unlabelled and fluorescently labelled (see Antibodies) primary antibodies were used at 1 μg/ml. When non-labelled primary antibodies were used (Fig. 3a), we added Alexa647- or Alexa488-coupled secondary antibodies (Life Technologies) at a 1/500 dilution. Imaging was performed on the Spinning disk or laser scanning confocal microscopes described below.
Microscopy
Most imaging presented in this paper was performed onto a custom TIRF/spinning disk confocal microscope composed of a Nikon Ti stand equipped with perfect focus, a fast piezo z-stage (ASI) and a Plan Apochromat lambda 100X NA 1.45 objective (for TIRF) or a 60X NA 1.49 Apochromat TIRF objective (for Spinning disk). The use of a high-NA TIRF objective for SDC imaging improves the light-throughput, thus allowing faster imaging framerates, at the expense of the depth of field. The confocal imaging arm is composed of a Yokogawa CSU-X1 spinning disk head and a Photometrics 95B back-illuminated sCMOS camera operating in global shutter mode and synchronised with the spinning disk rotation. The TIRF imaging arm is composed of an azimuthal TIRF illuminator (iLas2, Roper France) modified to have an extended field of view to match the size of the camera (Cairn). Images were recorded with a Photometrics Prime 95B back-illuminated sCMOS camera run in pseudo-global shutter mode and synchronised with the rotation of the azimuthal illumination. TIRF angle was set independently for all channels so that the depth of the TIRF field was identical for all channels. Excitation in both arms was performed using 488 nm (150 mW OBIS LX), 561 nm (100 mW OBIS LS) and 637 nm (140 mW OBIS LX) lasers fibered within a Cairn laser launch. To minimize bleedthrough, single band emission filters were used (Chroma 525/50 for Alexa 488/GFP/Alexa514; Chroma 595/50 for mScarlet/rhodamine/mRFP/mCherry and Chroma 680/40 (SDCM) or 655LP (TIRFM) for HiLyte647/Alexa647/ATTO647N) and acquisition of each channel was performed sequentially using a fast filter wheel in each arm (Cairn Optospin). To enable fast acquisition, the entire setup is synchronized at the hardware level by a Zynq-7020 Field Programmable Gate Array (FPGA) stand-alone card (National Instrument sbrio 9637) running custom code. In particular fast z-stacks are obtained by synchronizing the motion of the piezo z-stage during the readout time of the cameras. Sample temperature was maintained at 25°C (or 37°C for microtubule dynamics) using a heating enclosure (MicroscopeHeaters.com, Brighton, UK). Acquisition was controlled by Metamorph software (v7.10.1.161).
Alternatively, spinning disk imaging was performed on a 3i Marianas spinning disk confocal setup based on a Zeiss Z1 stand, a 63X PLAN APO NA 1.4 objective and a Yokogawa X1 spinning disk head followed by a 1.2X magnification lens and an Evolve EMCCD camera (Photometrics). Fast Z-stack acquisition was obtained using a piezo stage (Mad city labs). Single emitter emission filters were always used to avoid bleed-through and each channel was acquired sequentially. Acquisition was controlled by 3i Slidebook 6. Fig. 3a was obtained Leica SP5 confocal microscope equipped with a 63X Oil NA 1.4 objective and HyD detectors (Leica).
Image processing
Images were processed using Fiji 68 (ImageJ version: 1.53f) and Matlab 2020b (Mathworks) using custom codes available on our github page (https://github.com/deriverylab/2022_Planelles-Herrero). Figures were assembled in Adobe Illustrator 2021. Movies were edited in Adobe Premiere 2021. All lookup tables applied to images in this paper come from the collection from James Manton (https://github.com/jdmanton/ImageJ_LUTs).
Spatial drift during acquisition for TIRF movies was corrected using a custom GPU-accelerated registration code based on cross correlation between successive frames. Drift was measured on one channel and applied to all the channels in multichannel acquisitions. Code is available on our github page (https://github.com/deriverylab/GPU_registration).
For representation purposes, the Pon panel in Fig.3a, panel Pon was denoised using a safir69 filter (code availability: https://team.inria.fr/serpico/software/nd-safir). Similarly, a Wavelet “à trous” filter was applied to movie S3 (custom GPU-accelerated MATLAB port of a code originally developed by Fabrice Cordeliere for the “Improve Kymo” ImageJ plugin70. Code is available on our Github page (https://github.com/deriverylab/GPU_wavelet_a_trous). In the latter case, the raw movie was averaged with the filtered movie.
1). Temporal registration of movies
In this report, we used average data coming from several movie datasets in an absolute temporal reference frame (see for instance Figs. 1a-c, 2c and Extended Data Fig 3a). This relies on the fact that since the shrinking of the central spindle is stereotypic from cell to cell 4, we can use it to register movies in time. For each cell, we thus plotted the temporal dynamics of the Jupiter signal across a line perpendicular to the spindle JWcell(t) (see Extended Data Fig 3a left panel for conventions) together with that of a reference cell (JWref(t)).JWcell(t) essentially follows the actin ring constriction4. This reference cell movie was arbitrarily chosen as one that span from anaphase to cytokinesis, the relevant phases for this work. We then computed by least square fitting the time delay τcell that needs to be applied to the cell of interest to minimize the difference between the two Jupiter constriction temporal profiles (ΔJupi), i.e. find the τcell for which is minimum. We then set the initial time of each movie to be equal to τcell, thereby registering all the movies into an absolute time frame, we name “registered time”. In this reference frame, t=0 corresponds to the onset of anaphaseB. Importantly, the contraction of Jupiter is unaffected by Elp3 depletion (Extended Data Fig 3a), thus enabling automated temporal registration of movies acquired in these genetic backgrounds relatively to the control (Fig. 2c).
2). Quantification of endosome recruitment to the central spindle
To measure endosome recruitment to the central spindle (Fig. 2c), we first used the Jupiter-GFP signal to define a central spindle region of interest (ROI) over time. The central spindle ROI was operationally defined as a rectangle centred on the centroid of the central spindle, with the x-axis dimension set at 2 μm and for y-axis dimension the width of the spindle at that time JWcell(t). By convention, we consider the y-axis is parallel to the division plane and the x-axis perpendicular to it (see Extended Data Fig 3a left panel). We then measured the iDelta20 signal in the central spindle ROI over time by integrating the fluorescence signal in 3D using a constant manual threshold to segment the endosomes after homogeneous background subtraction. The iDelta20 intensity signal was similarly measured in the entire cell, including the central spindle. The central spindle-associated signal was then expressed as a percentage of the total signal present in the cell. As before, the Jupiter tracking data was also used for precise time registration of these movies to average data between cells.
3). iDelta20 asymmetry measurements
iDelta20 asymmetry was measured at late cytokinesis when all endosomes had departed from the central spindle (≈600 s in Registered time, see Fig. 2c). Asymmetry was automatically measured as follows: Endosomes were first detected by using 2D gaussian fitting (using the algorithm of Holden and colleagues71), then total intensity was then integrated for each endosome, with the local background (determined by Gaussian fitting) subtracted. The pIIa and the pIIb cells were then segmented manually using the PonLD channel as a reference. Finally, endosomes were assigned based on their coordinates to the segmented pIIa or the pIIb regions. The total endosomal signal for each daughter cell was subsequently computed. The percentage of iDelta20 in the pIIa daughter cell was then computed as:
We measured the percentage of iDelta20 signal in the pIIa daughter cell rather than the ratio of signal between the two cells (pIIa/pIIb) since our automatic detection method sometimes does not detect any particles in either daughter cell, leading to a pIIa/pIIb ratio of 0 or infinity.
4). Central spindle averaging
To generate average central spindle images (Figs. 1e,h and 3b as well as Extended Data Fig. 8a), images of fixed samples were obtained shortly before abscission. At this stage, the tubulin (or Ac-tubulin) signals have the characteristic hourglass shape of late mitotic spindles. We can thus use this characteristic shape as a registration cue to align spindles from different images and average them, as we previously established 4. Antero-posterior polarity was provided by the PonLD signal. All these operations were performed on z-projected images (max projection, 6 μm total, Δz = 0.27 μm).
5). Measurement of spindle asymmetry in anaphase
In this report, central spindle asymmetry was always measured by computing the difference of microtubule density between the two sides of the central spindle, rather than the absolute amounts of tubulin. This is because we previously established that what matters for polarized trafficking of Sara endosomes is the ratio of tubulin densities between the two sides of the spindle, rather than the absolute amount of tubulin 4. Briefly, we first projected z-stacks containing the entire central spindle (6 μm depth, Δz = 0.5 μm) using Sum-intensity projection. We then segmented the Jupiter (or tubulin) signal to define the x/y-axes of the spindle (see above). The intensity of the Jupiter-GFP signal was then measured along the x-axis upon signal integration over the y-axis within a region of interest (ROI) centered on the central spindle. This measurement thus conceptually resembles a linescan along the x-axis of the spindle, but where not a line, but a rectangular ROI is considered (ROI dimensions: 10 μm on the x-axis and JW on the y-axis). For this reason, we previously named it “pseudo-linescan” method 4. The signal intensity over the x-axis determined this way displays two peaks: one in pIIa, one in pIIb, see Fig. 1f for an example. This reflects the facts that most markers are excluded (at least in part) from the core of the central spindle. We then measured the value of each peak and subtracted the local background (average background was determined from 5 pixels adjacent to the spindle). Central spindle asymmetry was computed as the enrichment of the density of the marker in the pIIb relative to the pIIa according to:
We used the same procedure to compare the Tubulin, Elp4 and/or Ac-Tubulin enrichments at the central spindle in immunostainings (Figs. 1e and 3b).
6). Ratiometric measurement of the degree of microtubule acetylation
For measurements of the Acetyl-α-tubulin over Pan-β-tubulin ratio on quantitative fluorescent Western Blot (Extended Data Fig. 8e), the respective signals were measured after homogenous background subtraction, and the ratio computed.
For determination of the degree of microtubule acetylation across the central spindle in SOPs (Extended Data Fig 8d), pseudo-linescans were measured on both Acetyl-α-tubulin and Pan-β-tubulin channels as above. They were then normalized, and the Acetyl-α-tubulin signal was divided by the Pan-β-tubulin signal to obtain the acetyl-tubulin/total tubulin ratio along the length of the central spindle. The value of this normalized ratio was then measured in both the pIIb and pIIa side (determined from the Pan-β-tubulin signal), and averaged amongst different cells.
For determination of the degree of microtubule acetylation in control versus AtatRNAi central spindles (Extended Data Fig 7b), pseudo-linescans were measured on both Acetyl-α-tubulin and Pan-β-tubulin channels as above for central spindles inside or outside the Pnr expression region. The peak value on both sides of the central spindle were then averaged to compute an estimate of the density of Acetyl-α-tubulin and Pan-β-tubulin at the central spindle per cell. The density ratio of Acetyl-α-tubulin over Pan-β-tubulin was then computed per cell, and normalized by the average ratio in cells outside the Pnr expression region in the same image. To avoid variability due to decreased fluorescence signals in deep imaging z-planes, we imaged central spindles around the frontier of the Pnr region so we could image central spindles in similar z-planes and thus accurately compare their absolute Acetyl-α-tubulin and Pan-β-tubulin fluorescence intensities. The normalized acetyl-tubulin/tubulin ratio was then averaged amongst different cells. Note that in this analysis, we are not considering specifically considering SOPs, but also epithelial cells, given the fact the expression domain of Pnr encompasses the entire epithelium between the dorsocentral machrochaete (i.e. SOP and epithelial cells).
7). Measurement of cortical relocalisation of Elongator
To evaluate the extent of cortical relocalisation of GFP Elongator in the nanobody experiment (Fig.6), we first selected the most equatorial zplane within the 3D z-stack, where the membrane is best in focus, using the PonLD signal as a reference. We then drew a 3px wide line perpendicular to the cell’s division plane, along which we measured the GFP-Elongator signal. This displays a peak of intensity corresponding to the membrane, followed by a lower intensity region corresponding to the cytosol. We then evaluated the ratio between these two values after background subtraction. This measurement was done at the onset of anaphase.
8). Measurement of microtubule dynamics in vitro
Microtubule polarity was established by measuring growth rates, with fast-growing extensions indicating plus ends and slow-growing extensions indicating minus ends. Microtubule dynamics were analyzed manually on kymographs using the ImageJ plugin ‘KymoToolBox’ developed by Fabrice Corderlières 70 after homogenous background subtraction. To account for the fact that microtubule sometimes show irregularities in their growth dynamics, and/or can leave briefly the TIRFM field, growth speeds were determined from the slope of microtubule growth on kymographs. In other words, determined an average growth rate per microtubule rather than the instantaneous growth.
Microtubule lifetime were computed for each microtubule as the elapsed time between the onset of growth and the last time frame before a catastrophe event. Following the method established by Gardner and colleagues72, the cumulative distribution of microtubule lifetimes was then plotted and fitted to a gamma distribution in Matlab using the gamfit function (Figs. 5f,g,i and Extended Data Fig 9c,d). As an estimate of the mean lifetime, we computed the lifetime at the half cumulative distribution from the fitted data (i.e. when the cumulative distribution is equal to 0.5). We then used boostrapping to estimate the error of this mean lifetime value. Briefly, we drew 10000 bootstrap data samples from the observed lifetime distribution and computed the lifetime at the half cumulative distribution as above for each of them. We then computed the interquartile range of the distribution of the boostrapped mean lifetimes (i.e. the difference between the 75th and the 25th percentiles of the boostrapped mean lifetime distribution), and used it as an estimate of the error.
When computing the increase in growth rate or lifetime due to the addition of Elongator (Fig. 5c and Extended Data Fig 9e), we evaluated the error of this increase by error propagation. For instance, the error of the fold increase in growth rate, δincrease in growth rate was computed as follows:
With and the mean growth speed in the absence of Elongator and its associated error, respectively, and and the mean growth speed in the presence of Elongator and its associated error, respectively. Error associated with lifetime ratios were similarly computed.
9). Absolute measurements of MT density
To compare Jupiter-GFP intensity between different movies (Fig. 1h), we needed to account for the variations of the Jupiter-GFP signal, which occurs even in identical imaging conditions at endogenous levels of Jupiter-GFP expression, probably due to different imaging depth into the tissue. As established previously4, we used the intensity of the centrosome of the pIIa daughter cell, which Jupiter labels throughout the cell cycle as a reference (orange arrow in Fig. 1h). The intensity of the pIIa centrosome was measured in a circular ROI centered on the centrosome (1.2 μm of diameter) and integration of the signal intensity within the ROI on 10 z-planes (5 μm total depth). Local background was subtracted by considering an adjacent ROI of identical diameter for each plane. We then measured the Jupiter-GFP signal at the central spindle in both the pIIa and the pIIb daughter cells by using the same circular ROI dimensions and background subtraction as above. We then normalized the obtained signal intensity by the pIIa centrosome value.
Statistics & Reproducibility
Unless stated otherwise, measurements are given in mean ± SEM. Statistical analyses were performed using GraphPad Prism 8 or SigmaStat 3.5 with an alpha of 0.05. Normality of variables was verified with Kolmogorov-Smirnov tests. Homoscedasticity of variables was always verified when conducting parametric tests. Post-hoc tests are indicated in their respective figure legends. No statistical method was used to predetermine sample size. The experiments were not randomized. No blind experiments were conducted in this study. No data were excluded from the analyses. The Investigators were not blinded to allocation during experiments and outcome assessment.
All representative results shown for tissue-immunolabelings, western-blots, protein purifications, microtubule pelleting assays and microtubule TIRFM experiments (including microtubule binding and microtubule dynamics assay) were performed at least three times independently with similar results. Note that the large scale purification of the endogenous Elongator complex carrying the dominant-negative, lethal YY531AA mutation was only performed once due to the high complexity of the experiment (see Methods).
Extended Data
Supplementary Material
Acknowledgments
This work has been supported by the Medical Research Council, as part of United Kingdom Research and Innovation (also known as UK Research and Innovation)(MC_UP_1201/13 to E.D), the Human Frontier Science Program (Career Development Award CDA00034/2017 to E.D.), as well as by grants from the SNSF, by the ERC (Sara and Morphogen), the NCCR Chemical Biology program, the DIP of the Canton of Geneva and the SystemsX EpiPhysX (SNSF) granted to M.G.G. V.P.H. is supported by an EMBO postdoctoral fellowship. We thank the electronics workshop of the LMB, in particular Martin Kyte for his help with the FPGA hardware required for precise synchronization of our microscope. We also thank the LMB Biophysics and Mass Spectrometry facilities, in particular Mark Skehel and Farida Begum for performing and analysing Mass Spectrometry data. We thank Irene Diaz (Venki Ramakrishnan lab) for sharing tRNA. We thank Tim Stevens and John O’Neill for expert advices on statistics and François Karch and Patrik Verstreken for flies. We thank Alicia Daeden and Sylvain Loubéry for help with microscopy. We thank members of the Derivery and Gonzalez-Gaitan labs, as well as Charlotte Aumeier, Buzz Baum, Simon Bullock and Karsten Kruse for critical reading of the manuscript.
Abbreviations
- SDCM
Spinning-Disk Confocal Microscopy
- TIRFM
Total Internal Reflection Microscopy
- EM
Electron Microscopy
- SOP
Sensory Organ Precursor
- MS
Mass Spectrometry
Footnotes
Author Contributions Statement
V.P.H conducted all biochemistry experiments, as well as all microtubule assays and associated analysis. E.D and A.B. conducted all fly imaging experiments and associated quantification. E.D. wrote the codes for automated image processing. C.S. generated the Klp98A-GFP knock-in, and conducted genetic interaction experiments. V.P.H. M.G.G and E.D. designed the project. All authors contributed to writing the paper.
Competing Interests Statement
The authors declare no competing interests
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE73 partner repository with the dataset identifier PXD036556 and 10.6019/PXD036556. Numerical data used to generate every panel of this study can be found in the Source data. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
Custom image processing code specific to this paper have been deposited on our github page in a dedicated repository (https://github.com/deriverylab/2022_Planelles-Herrero). Similarly, our general image registration and wavelet filtering codes can also be found on our Github page (https://github.com/deriverylab/GPU_registration and https://github.com/deriverylab/GPU_wavelet_a_trous). Safir denoising code can be found on the Inria website (https://team.inria.fr/serpico/software/nd-safir). All lookup tables applied to images in this paper come from the collection from James Manton (https://github.com/jdmanton/ImageJ_LUTs).
References
- 1.Kressmann S, et al. Directional Notch trafficking in Sara endosomes during asymmetric cell division in the spinal cord. Nat Cell Biol. 2015;17:333–339. doi: 10.1038/ncb3119. [DOI] [PubMed] [Google Scholar]
- 2.Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]
- 3.Montagne C, Gonzalez-Gaitan M. Sara endosomes and the asymmetric division of intestinal stem cells. Development. 2014;141:2014–2023. doi: 10.1242/dev.104240. [DOI] [PubMed] [Google Scholar]
- 4.Derivery E, et al. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature. 2015;528:280–285. doi: 10.1038/nature16443. [DOI] [PubMed] [Google Scholar]
- 5.Kawaguchi D, Furutachi S, Kawai H, Hozumi K, Gotoh Y. Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat Commun. 2013 doi: 10.1038/ncomms2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeffler D, et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature. 2019 doi: 10.1038/s41586-019-1531-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X, et al. Polarized endosome dynamics engage cytosolic Par-3 and dynein during asymmetric division. bioRxiv. 2020:2020.05.15.098012. doi: 10.1101/2020.05.15.098012. [DOI] [Google Scholar]
- 8.Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573 LP–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- 9.Furthauer M, Gonzalez-Gaitan M. Endocytic regulation of notch signalling during development. Traffic. 2009;10:792–802. doi: 10.1111/j.1600-0854.2009.00914.x. [DOI] [PubMed] [Google Scholar]
- 10.Loubéry S, et al. Sara phosphorylation state controls the dispatch of endosomes from the central spindle during asymmetric division. Nat Commun. 2017 doi: 10.1038/ncomms15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loubéry S, et al. Uninflatable and Notch Control the Targeting of Sara Endosomes during Asymmetric Division. Curr Biol. 2014:S0960-9822(14)00917-8. doi: 10.1016/j.cub.2014.07.054. [pii] [DOI] [PubMed] [Google Scholar]
- 12.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffin E, Gho M. Laser microdissection of sensory organ precursor cells of Drosophila microchaetes. PLoS One. 2010 doi: 10.1371/journal.pone.0009285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogan NJ, Greenblatt JF. Characterization of a Six-Subunit Holo-Elongator Complex Required for the Regulated Expression of a Group of Genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkes NA, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 16.Dauden MI, et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017;18:264–279. doi: 10.15252/embr.201643353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setiaputra DT, et al. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017;18:280–291. doi: 10.15252/embr.201642548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miśkiewicz K, et al. ELP3 controls active zone morphology by acetylating the ELKS family member bruchpilot. Neuron. 2011 doi: 10.1016/j.neuron.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Creppe C, et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of α-Tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 20.Solinger JA, et al. The Caenorhabditis elegans Elongator Complex Regulates Neuronal α-tubulin Acetylation. PLoS Genet. 2010;6:e1000820. doi: 10.1371/journal.pgen.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer F, et al. Translational Control of Cell Division by Elongator. Cell Rep. 2012;1:424–433. doi: 10.1016/j.celrep.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Tuck S, Byström AS. Defects in tRNA Modification Associated with Neurological and Developmental Dysfunctions in Caenorhabditis elegans Elongator Mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dauden MI, et al. Molecular basis of tRNA recognition by the Elongator complex. Sci Adv. 2019;5:eaaw2326. doi: 10.1126/sciadv.aaw2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaugenhaupt SA, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapino F, et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018;558:605–609. doi: 10.1038/s41586-018-0243-7. [DOI] [PubMed] [Google Scholar]
- 26.Bento-Abreu A, et al. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum Mol Genet. 2018;27:1276–1289. doi: 10.1093/hmg/ddy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derivery E, Gautreau A. Generation of branched actin networks: Assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays. 2010;32:119–131. doi: 10.1002/bies.200900123. [DOI] [PubMed] [Google Scholar]
- 28.Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelissen H, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci U S A. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehlgarten C, et al. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 32.Kojic M, et al. Elp2 mutations perturb the epitranscriptome and lead to a complex neurodevelopmental phenotype. Nat Commun. 2021;12:1–18. doi: 10.1038/s41467-021-22888-5. 2021 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang HC, et al. Hsc70 is required for endocytosis and clathrin function in Drosophila. J Cell Biol. 2002;159:477–487. doi: 10.1083/jcb.200205086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bronk P, et al. Drosophila Hsc70-4 Is Critical for Neurotransmitter Exocytosis In Vivo. Neuron. 2001;30:475–488. doi: 10.1016/s0896-6273(01)00292-6. [DOI] [PubMed] [Google Scholar]
- 35.Cheishvili D, et al. IKAP/Elp1 involvement in cytoskeleton regulation and implication for familial dysautonomia. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr036. [DOI] [PubMed] [Google Scholar]
- 36.Hunnicutt BJ, Chaverra M, George L, Lefcort F. Ikap/elp1 is required in vivo for neurogenesis and neuronal survival, but not for neural crest migration. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George L, et al. Familial dysautonomia model reveals Ikbkap deletion causes apoptosis of Pax3+ progenitors and peripheral neurons. Proc Natl Acad Sci U S A. 2013;110:18698–18703. doi: 10.1073/pnas.1308596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MZ, Gruner KA, Qin C, Tourtellotte WG. A neuron autonomous role for the familial dysautonomia gene ELP1 in sympathetic and sensory target tissue innervation. Dev. 2014;141:2452–2461. doi: 10.1242/dev.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akella JS, et al. MEC-17 is an α-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittschieben B, Fellows J, Wendy D, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and Elongator in vivo. EMBO J. 2000;19:3060–3068. doi: 10.1093/emboj/19.12.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker J, et al. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics. 2011;187:1067–1075. doi: 10.1534/genetics.110.123893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawer H, et al. Roles of Elongator Dependent tRNA Modification Pathways in Neurodegeneration and Cancer. Genes (Basel) 2019;10 doi: 10.3390/genes10010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gell C, et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 45.Hein MY, et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 2015 doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 46.Holt CE, Martin KC, Schuman EM. Local translation in neurons: visualization and function. Nature Structural and Molecular Biology. 2019;26:557–566. doi: 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 47.Feng C, et al. Patronin-mediated minus end growth is required for dendritic microtubule polarity. J Cell Biol. 2019;218:2309–2328. doi: 10.1083/jcb.201810155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noordstra I, et al. Control of apico-basal epithelial polarity by the microtubule minus-endbinding protein CAMSAP3 and spectraplakin ACF7. J Cell Sci. 2016;129:4278–4288. doi: 10.1242/jcs.194878. [DOI] [PubMed] [Google Scholar]
- 49.Abdel-Fattah W, et al. Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast. PLoS Genet. 2015;11:e1004931. doi: 10.1371/journal.pgen.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehlgarten C, Jablonowski D, Breunig KD, Stark MJR, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- 51.Yang K, Inoue A, Lee Y, Jiang C, Lin F. Loss of Ikbkap / Elp1 in mouse oocytes causes spindle disorganization, developmental defects in preimplantation embryos and impaired female fertility. Sci Rep. 2019;9 doi: 10.1038/s41598-019-55090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee JJ, et al. Meru couples planar cell polarity with apical-basal polarity during asymmetric cell division. Elife. 2017;6 doi: 10.7554/eLife.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002 doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolukbasi E, et al. Drosophila poly suggests a novel role for the Elongator complex in insulin receptor-target of rapamycin signalling. Open Biol. 2012 doi: 10.1098/rsob.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskelet. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- 56.Emery G, et al. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 57.Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol. 2001;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- 58.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson JL, et al. High-efficacy subcellular micropatterning of proteins using fibrinogen anchors. J Cell Biol. 2021 doi: 10.1083/jcb.202009063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell JN, Slep KC. αβ-tubulin and microtubule-binding assays. Methods Mol Biol. 2011 doi: 10.1007/978-1-61779-252-6_6. [DOI] [PubMed] [Google Scholar]
- 63.Lin TY, et al. The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-08579-2. 2019 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Electrophoresis. Vol. 20. Wiley-VCH Verlag; 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data; pp. 3551–3567. [DOI] [PubMed] [Google Scholar]
- 65.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 66.Loubery S, Gonzalez-Gaitan M. Monitoring notch/delta endosomal trafficking and signaling in Drosophila. Methods Enzym. 2014;534:301–321. doi: 10.1016/B978-0-12-397926-1.00017-2. [DOI] [PubMed] [Google Scholar]
- 67.Bell PB, Jr, Safiejko-Mroczka B. Improved methods for preserving macromolecular structures and visualizing them by fluorescence and scanning electron microscopy. Scanning Microsc. 1995;9:843–860. [PubMed] [Google Scholar]
- 68.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulanger J, et al. Patch-based nonlocal functional for denoising fluorescence microscopy image sequences. IEEE Trans Med Imaging. 2010 doi: 10.1109/TMI.2009.2033991. [DOI] [PubMed] [Google Scholar]
- 70.Zala D, et al. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013 doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 71.Holden SJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J. 2010;99:3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011 doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 73.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE73 partner repository with the dataset identifier PXD036556 and 10.6019/PXD036556. Numerical data used to generate every panel of this study can be found in the Source data. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Custom image processing code specific to this paper have been deposited on our github page in a dedicated repository (https://github.com/deriverylab/2022_Planelles-Herrero). Similarly, our general image registration and wavelet filtering codes can also be found on our Github page (https://github.com/deriverylab/GPU_registration and https://github.com/deriverylab/GPU_wavelet_a_trous). Safir denoising code can be found on the Inria website (https://team.inria.fr/serpico/software/nd-safir). All lookup tables applied to images in this paper come from the collection from James Manton (https://github.com/jdmanton/ImageJ_LUTs).