Abstract
Solid cancers display dynamic balance between cell death and proliferation assuring continuous tumor maintenance and growth 1, 2. Increasing evidence links enhanced cancer cell apoptosis to paracrine activation of cells in the tumor microenvironment initiating tissue repair programs that support tumor growth3, 4, yet the direct effects of dying cancer cells on neighboring tumor epithelia and how this potentially contributes to therapy resistance are unclear. Here we demonstrate that chemotherapy-induced tumor cell death in patient-derived colorectal tumor organoids causes ATP release triggering a P2x4-mediated mTOR dependent pro-survival program in neighboring cancer cells, which renders surviving tumor epithelia sensitive to mTOR inhibition. The induced mTOR addiction in persisting epithelial cells is due to an elevated ROS production and subsequent increased DNA damage in response to death of neighboring cells. Accordingly, inhibition of the P2X4 receptor or direct mTOR blockade prevent induction of S6 phosphorylation and synergize with chemotherapy to cause massive ROS-induced cell death and marked tumor regression that is not seen when individually applied. Conversely, ROS scavenging prevents cancer cells from becoming reliant on mTOR activation. Collectively, we show that dying cancer cells establish a new dependency on anti-apoptotic programs in their surviving neighbors thereby creating a novel opportunity for combination therapy in P2X4 expressing epithelial tumors.
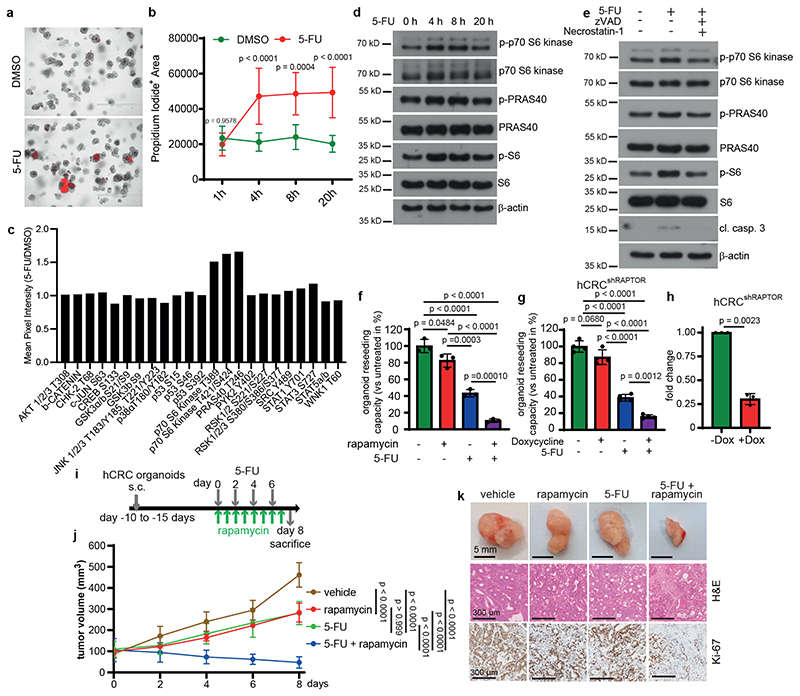
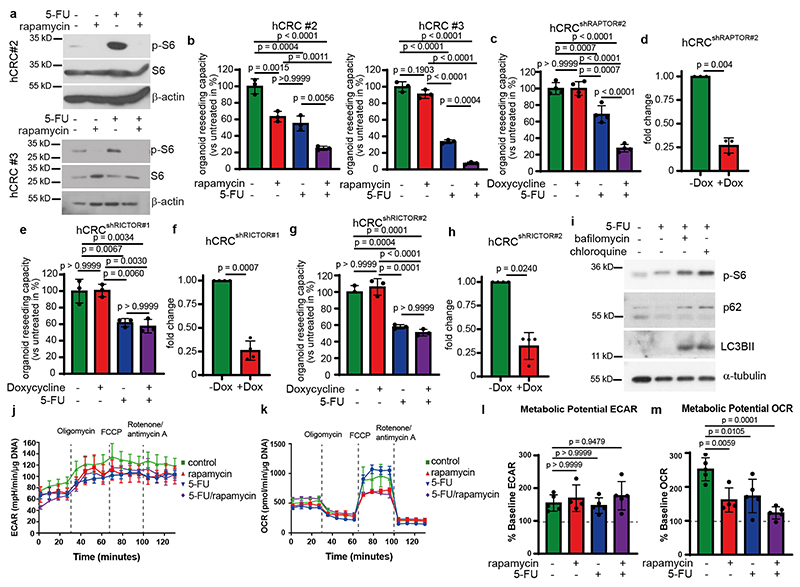
To examine a possible impact of tumor cell death on neighboring epithelia and to assess whether such paracrine effect may contribute to therapy resistance, we treated patient-derived colorectal tumor organoids (PDTO; hCRC) with the standard of care chemotherapeutic agent 5-Fluoruracil (5-FU) for up to 20 hours. Within 4 hours marked cell death could be observed (Fig. 1a-b). To determine which signaling pathways might be activated in surviving organoids in response to 5-FU induced cell death, we used a phospho-kinase array to profile kinase activity. Enhanced phosphorylation of p70 S6 kinase (T389 and T421/S424) as well as PRAS40 (T246) (Fig. 1c), two kinases involved in the regulation of mTORC15, 6, was detected 20 hours after 5-FU administration. Immunoblot analysis confirmed activation of p70 S6 Kinase, PRAS40 as well as mTOR determined by the elevated phosphorylation of S6 ribosomal protein starting from 4 hours after 5-FU administration when cell death started to be detected (Fig. 1d). These results could be also observed in two additional PDTO (Extended Data Fig. 1a). To confirm that activation of these kinases was a consequence of 5-FU-induced cell death, we added zVAD(carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone) and nec-1 (Necrostatin-1) to block both apoptosis and necroptosis7, which completely prevented induction of p-p70 S6 kinase, p-PRAS40 and p-S6 at 4 hours (Fig. 1e). We then investigated whether induced mTOR activation contributed to PDTO resistance to 5-FU and treated PDTO with 5-FU, rapamycin or a combination of both. While individual treatments had only a modest effect, the combined administration of 5-FU and rapamycin led to a marked suppression of PDTO reseeding capacity (Fig. 1f; Extended Data Fig.1b), confirming that mTOR signaling was key to 5-FU resistance in the surviving organoids. Such enhanced sensitivity of PDTO to 5-FU could also be observed by shRNA-mediated RAPTOR downregulation (Fig. 1g, h; Extended Data Fig. 1c, d) while loss of RICTOR expression had no effect (Extended Data Fig. 1e-h) confirming the dependence on mTORC1. These results were confirmed in vivo where co-administration of rapamycin and 5-FU led to a marked regression of subcutaneously (s.c.) transplanted PDTOs when compared to untreated tumors or each treatment alone (Fig. 1i-k). Recently, 5-FU has been shown to induce autophagy in colorectal cancer cells8, which could potentially increase the intracellular availability of nutrients and metabolites thereby activating mTORC1. To rule out involvement of a cell-autonomous activation mechanism, we treated tumor organoids with bafilomycin or chloroquine in the presence or absence of 5-FU. However, blocking autophagy did not prevent S6 phosphorylation (Extended Fig. 1i). Moreover, metabolic analysis revealed that tumor organoids were energetic and had significantly higher oxygen consumption rate (OCR) both under basal level and FCCP-uncoupled conditions (Extended Data Fig. 1j-m). The effect of 5-FU on the metabolic flexibility of organoids demonstrated a slight reduction in their capacity to increase their OCR and no change in extracellular acidification rate (ECAR), which is a proxy for their glycolytic rate, thus ruling out that this could trigger S6 phosphorylation.
Figure 1. Resistance of colorectal patient-derived tumor organoids to 5-FU depends on mTOR activation.
a, Propidium iodide (PI) staining of hCRC organoids treated with 5-FU for 4h (n = 3 biological replicates, scale bar = 1000 μm).
b, Quantification of PI+ area of 5-FU treated hCRC organoids (1h and 8h DMSO: n=6, 4h
DMSO: n=9, 20h DMSO: n=8h, 1h and 8h 5-FU: n=7, 4h and 20h 5-FU n=10, data were pooled from 3 biological replicates).
c, Kinase activation assay of 20h 5-FU treated hCRC organoids. Results represent fold increase of pixel intensity (5-FU/DMSO, n=1). For corresponding dot blot see SI.
d, Immunoblotting of hCRC organoids treated as indicated (n=2).
e, Immunoblotting of hCRC organoids, treated with zVAD and Necrostatin-1 2h prior to 4h 5-
FU treatment (n=2).
f, Reseeding capacity of hCRC organoids treated as indicated (n=3, one of four biological replicates).
g, Reseeding capacity of doxycycline-inducible hCRCshRAPTOR organoids treated as indicated (n=4, one of three biological replicates).
h, Raptor qRT-PCR analysis of doxycycline treated hCRCshRAPTOR tumor organoids (n=3 biological replicates).
i, Treatment regimen of subcutaneous hCRC tumors.
j, Growth rate of tumors in response to the treatment indicated in (i). n=6 mice for each condition. One set of mice was analysed.
k, Representative images of subcutaneous hCRC tumors (scale bar = 5mm) and H&E and Ki-67 staining (scale bar = 300 μm) of tumor tissues of mice treated with 5-FU +/- rapamycin as indicated in (j), (n=6, staining was performed on all tumors indicated in (j)).
All data are mean ±SD and analysed by two-tailed Student’s t-test (h) or 1-way ANOVA (f, g) and 2-way ANOVA (b, j), with Bonferroni’s multiple comparison.
Paracrine mTOR activation mediates tumor cell survival upon cell death induction
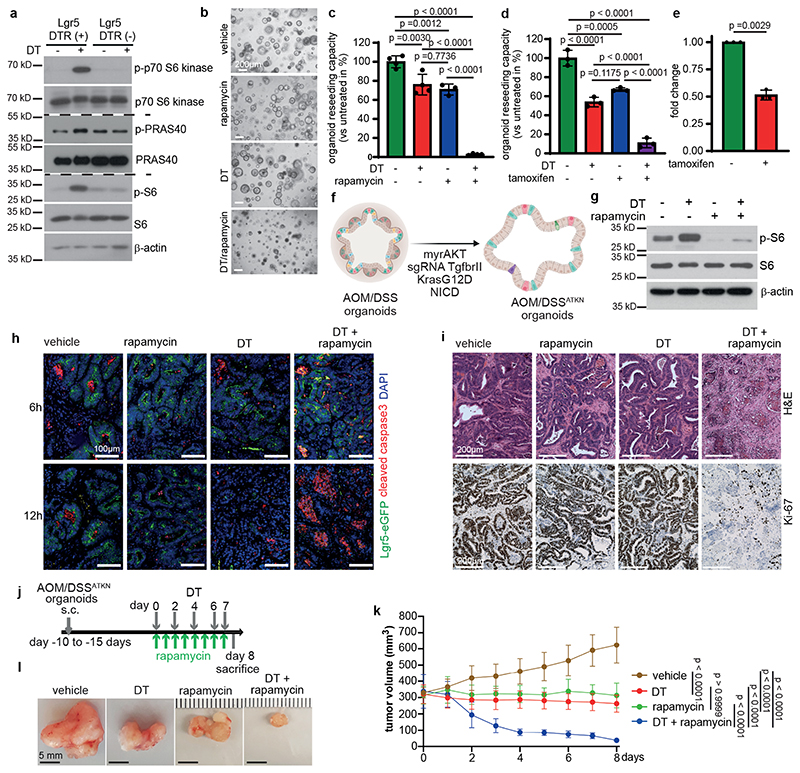
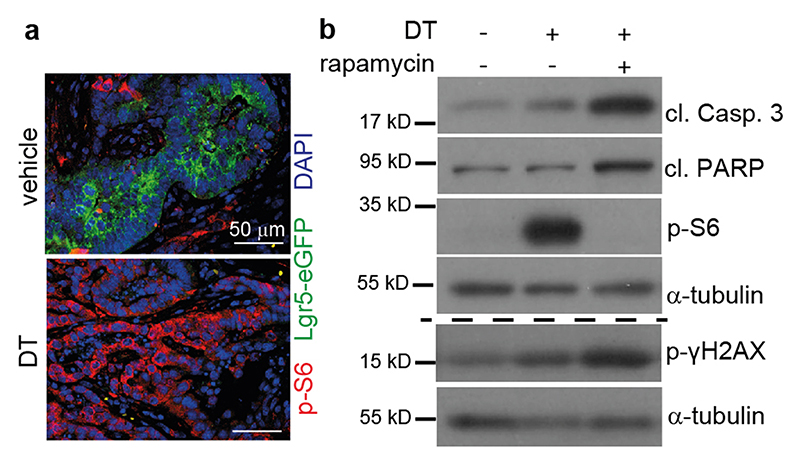
To investigate whether cell death-induced mTOR activation occurred in a paracrine manner, we took advantage of a genetic tool. Lgr5EGFP-DTR(+) mice9 allow targeted depletion of a well-defined epithelial tumor compartment by diphteria toxin (DT)-induced apoptosis thus providing the chance to study secondary effects on remaining neighboring cell populations that do not express the transgene. We established organoids from AOM/DSS-induced tumors derived from Lgr5EGFP-DTR(+) mice and confirmed enhanced phosphorylation of p70 S6 kinase, PRAS40 as well as S6 ribosomal protein upon a single DT administration (Fig. 2a). S6 phosphorylation could also be confirmed 24 hours after DT administration in Lgr5- AOM/DSS tumor cells in vivo (Extended Data Fig. 2a). In line with the notion that Lgr5+ cell loss causes tumor stasis10, Lgr5+ cell ablation alone in tumor organoids did not cause marked changes in gross morphology or growth rates of tumor organoids (Fig. 2b). However, mTOR inhibition by rapamycin combined with DT led to substantial cell death within 24 hours (Fig. 2b) and a significantly reduced capacity to form organoids upon re-seeding, which was not observed when either compound was applied individually (Fig. 2c), thus mimicking the phenotype of PDTO exposed to the combination of 5-FU and rapamycin. In agreement with a critical role for mTORC1 in survival upon Lgr5+ cell loss, tamoxifen induced deletion of Raptor in tumor organoids derived from AOM/DSS treated villin-Cre ERT2;RaptorF/F;Lgr5 ERFP-DTR(+) mice confirmed the greatly diminished reseeding capacity of DT-treated tumor organoids although raptor expression was reduced only by about 50% (Fig. 2d, e). To generate more aggressive tumor organoids that could be grown subcutaneously, we introduced either retrovirally or by lentiviral transduction a constitutively active form of AKT (myristoylated AKT), mutant K-RAS (K-RASG12D) as well as an active form of Notch1 (NICD) into AOM/DSS-induced tumor organoids from Lgr5EGFP-DTR(+) mice. Furthermore, Tgfbr2 was knocked out by CRISPR/Cas9 mediated gene editing resulting in AOM/DSSATKN tumor organoids (Fig. 2f). In AOM/DSSATKN tumor organoids, DT-induced Lgr5+ cell ablation also triggered S6 ribosomal protein phosphorylation (Fig. 2g), which could be prevented by rapamycin thereby strongly enhancing apoptosis (Extended Data Fig. 2b). Interestingly, apoptosis was also induced in Lgr5- cells in s.c. grown AOM/DSSATKN tumor organoids following combined DT/rapamycin treatment, an effect that was confirmed by immunohistochemical analysis of cleaved caspase 3 staining (Fig. 2h). Apoptosis peaked at 12 hours and markedly exceeded the levels observed in mice that received DT or rapamycin only (Fig. 2h). Consequently, the combination of DT and rapamycin resulted in a significant loss of viable, Ki-67+ tumor cells after 24 hours (Fig. 2i) and a nearly complete tumor regression within 8 days of treatment (Fig. 2j-l). In contrast, single agent rapamycin or DT administration treatment was less efficient in suppressing tumor growth and led to tumor stasis. Collectively, these data confirmed that tumor cell death induced a novel mTOR dependence in neighboring epithelial cells.
Figure 2. Dying tumor cells induce mTOR survival signaling in a paracrine manner.
a, Immunoblotting of DT treated Lgr5EGFP-DTR(+) and Lgr5EGFP-DTR(-) tumor organoids (n = 3). PRAS40 and phospo-PRAS40 were probed on a separate gel (see SI).
b, Representative Lgr5EGFP-DTR(+) tumor organoids treated as indicated (n=3, scale bar = 200μm).
c, Reseeding capacity of Lgr5EGFP-DTR(+) tumor organoids treated as indicated (n=4, one of three biological replicates).
d, Reseeding capacity of Villin-CreER Raptor ;Lgr5EGFP-DTR(+) tumor organoids treated as
indicated (n=3, one of three biological replicates).
e, Raptor qRT-PCR analysis of tamoxifen-induced Villin-Cre ERT2;RaptorF/F;Lgr5EGFP-DTR(+) tumor organoids (n=3 biological replicates).
f, Schematic representation of AOM/DSSATKN tumor organoid generation.
g, Immunoblotting of AOM/DSSATKN tumor organoids treated as indicated (n=3).
h, AOM/DSSATKN tumor organoids were injected subcutaneously. Once tumors reached 200 mm3, mice were treated as indicated and analysed after 6h or 12h for cleaved caspase-3 and Lgr5-GFP. Nuclei were counterstained with Dapi. Representative images from at least 2 mice/timepoints (scale bar = 100μm).
i, AOM/DSSATKN tumor organoids were injected subcutaneously. Once tumors reached 200 mm3, mice were treated as indicated for 24h and analysed by H&E and Ki-67 staining. Representative images are shown (n=3 mice, scale bar = 200μm).
j, Treatment regimen of subcutaneous AOM/DSSATKN tumors.
k, Growth rate of subcutaneous AOM/DSSATKN tumors in response to treatment as indicated in (j) (n=5 mice for vehicle and rapamycin, n=6 mice for DT and DT + rapamycin). One set of mice was analysed.
l, Representative images of subcutaneous AOM/DSSATKN tumors on day 8 treated as indicated in (k), (n= same as (k), scale bar = 5mm).
All data are mean ±SD and analysed by two-tailed Student’s t-test (e) or by 1-way ANOVA (c, d) and 2-way ANOVA (k) with Bonferroni’s multiple comparison.
Cell death associated ATP release engages P2X4 to induce mTOR activation
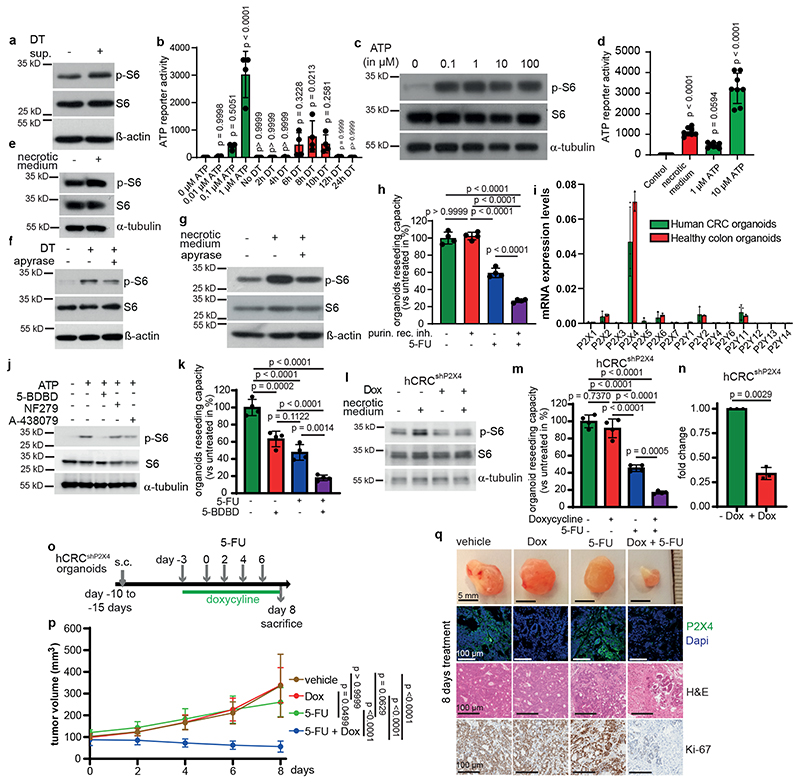
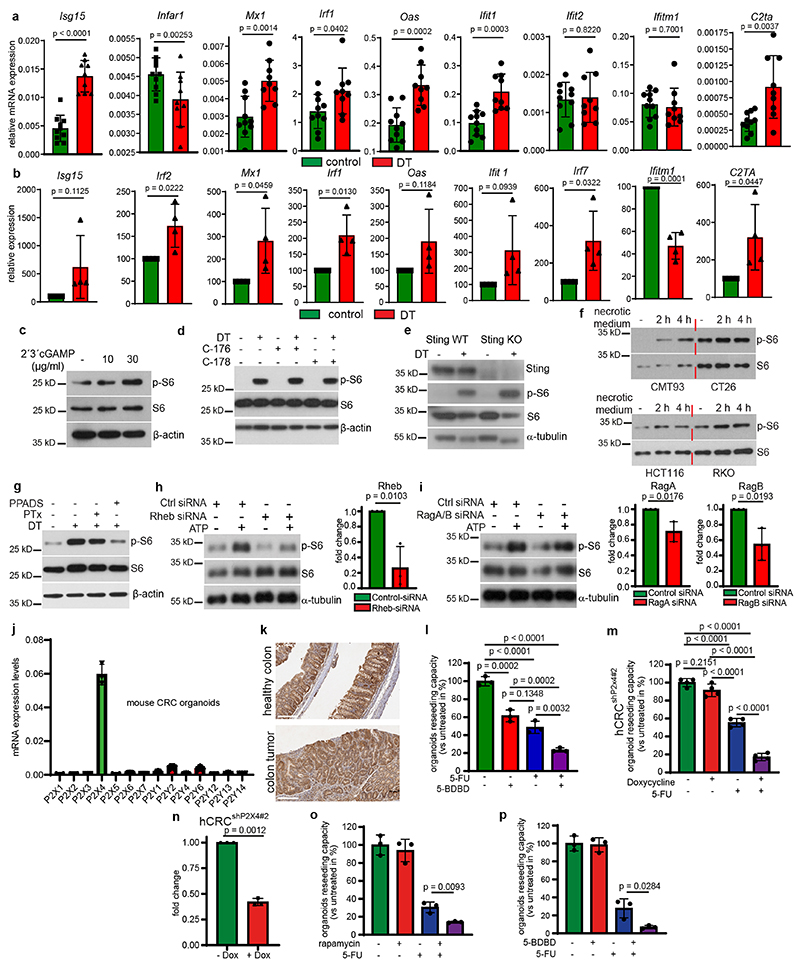
Cell death leads to the release of damage-associated molecular patterns (DAMPs) including various growth factors, inflammatory mediators, mitochondrial DNA (mtDNA), reactive oxygen species (ROS) and metabolites 11–13 that trigger inflammation and activate innate immunity to stop the damage and orchestrate tissue repair and wound healing14. To explore the mechanism causing paracrine mTORC1 activation, we treated Lgr5EGFP-DTR(-) tumor organoids for 2 hours with supernatants from either vehicle- or DT-treated Lgr5EGFP-DTR(+) tumor organoids and confirmed increased phosphorylation of S6 ribosomal protein in organoids exposed to supernatants from DT treated cells (Fig. 3a). Interestingly, we had observed an upregulated interferon response upon killing of Lgr5+ cells in AOM/DSS induced tumors (Extended Data Fig. 3a) similarly to s.c. transplanted tumors10 and which could also be observed in DT-treated tumor organoids (Extended Data Fig. 3b). Transcription of type-I interferons (IFNs) and interferon-stimulatory genes (ISGs) by the cGAS-STING pathway15, which had recently also been shown to confer pro-tumorigenic and pro-metastatic properties when activated in tumor cells16–19 can be triggered by mtDNA20. Therefore, we speculated that release of mtDNA by dying tumor epithelial cells could be involved in mTOR activation as well. Indeed, exogenous cGAMP induced phosphorylation of S6-ribosomal protein in a dose dependent manner (Extended Data Fig. 3c). However, neither selective STING inhibitors C176 and C-17821 nor CRISPR-mediated knockout of Sting in Lgr5EGFP-DTR(+) AOM/DSS-induced tumor organoids was able to prevent S6-ribosomal protein phosphorylation after Lgr5+ cell depletion (Extended Data Fig. 3d, e), ruling out an essential contribution of mtDNA to paracrine mTOR activation. Therefore, we focused on extracellular ATP that can be hydrolyzed by ATP-hydrolyzing enzymes present in the tumor microenvironment to generate ADP and adenosine that can act as ligands for purinergic receptors (P2R) and adenosine receptors (P1R)22. We confirmed a marked release of ATP (in the range of 0.1-1μM) into the supernatant of Lgr5+ containing organoids 6-10 hours after DT treatment initiation (Fig. 3b). Addition of extracellular ATP in a comparable concentration range induced phosphorylation of S6-ribosomal protein in Lgr5EGFP-DTR(-) tumors organoids (Fig. 3c). Similarly, supernatant of necrotic colorectal cancer cells, which also contains a substantial amount of ATP (1-10μM) (Fig. 3d), triggered S6 phosphorylation in tumor organoids and cancer cells of both murine and human origin (Fig. 3e, Extended Data Fig. 3f). Hydrolyzing ATP in supernatants from apoptotic or necrotic cells using apyrase prevented S6 phosphorylation (Fig. 3f, g). Reduced S6 phosphorylation could also be observed when applying non-selective purinergic receptor blockers or siRNA-mediated knockdown of Rheb, but not of Rag GTPases RagA and RagB (Extended Data Fig. 3g-i). Furthermore, blocking purinergic receptors using a combination of non-selective P2 receptor antagonists (50 μM theophylline; 20 ng/ml Pertussis toxin and 30 μM PPADS) in combination with 5-FU phenocopied the effect of the 5-FU/rapamycin double treatment on organoid survival (Fig. 3h). Collectively, these results support the notion that extracellular ATP release leads to paracrine mTORC1 activation. ATP signals via purinergic receptors of the P2 family, which comprises seven subtypes of P2X receptors and 8 (in mice 7) subtypes of P2Y receptors23. To examine whether one or several distinct receptor subtypes may be engaged in the observed signaling cascade, we determined gene expression of all known subtypes in both human untransformed and tumor organoids as well as in murine tumor organoids and detected prominent expression of P2X4 (Fig. 3i and Extended Data Fig. 3j). Immunohistochemistry confirmed expression of P2X4 in unchallenged colon and colonic tumors (Extended Data Fig. 3k), indicating that P2X4 may be the dominant purinergic receptor present on intestinal epithelial cells. Indeed, selective P2X4 inhibition using 5-(3-Bromophenyl)-1,3-dihydro-2H-Benzofuro[3,2-e]-1,4-diazepin-2-one (5-BDBD)24 reduced ATP-induced S6 phosphorylation in PDTOs, whereas selective P2X1 (NF279) and P2X7 inhibitors (A-438079) had no effect (Fig. 3j). Accordingly, combining 5-BDBD with 5-FU led to decreased survival of PDTO when compared to each treatment alone (Fig. 3k and Extended Data Fig. 3l). P2X4 knockdown in PDTO ablated necrotic medium-induced S6 phosphorylation and enhanced 5-FU mediated cytotoxicity significantly both in ex vivo cultured organoids (Fig. 3l-n and Extended Data Fig. 3m, n) and in s.c. tumors (Fig. 3o-q). PDTO from pancreatic ductal adenocarcinoma, which are known to express high levels of P2X425, were sensitive to the combination of rapamycin or 5-BDBD and 5-FU as well (Extended Data Fig. 3o, p).
Figure 3. Dying tumor cells release ATP to trigger mTORC1 via P2X4.
a) Immunoblotting of Lgr5EGFP-DTR(-) tumor organoids treated with supernatant from DT-treated Lgr5EGFP-DTR(+) tumor organoids for 2h (n=2).
b, ATP content in supernatants of DT treated organoids (n=4 biological replicates).
c, Immunoblotting of Lgr5EGFP-DTR(-) tumor organoids treated with ATP for 15 min (n=3).
d, ATP content in control or necrotic medium (n=8 biological replicates).
e-g, Immunoblotting of Lgr5EGFP-DTR(+) tumor organoids treated as indicated (e, g were treated 2h, f for 6h) (n=3 for all immunoblots).
h, Reseeding capacity of Lgr5EGFP-DTR(+) tumor organoids treated as indicated (n=4, one of four biological replicates).
i, qRT-PCR analysis of indicated genes in hCRC or healthy colon organoids (n=3, one of two biological replicates).
j, Immunoblotting of hCRC organoids treated as indicated for 15 min. Inhibitors were added 15 min prior to treatment with 100 μM ATP (n=3).
k, Reseeding capacity of hCRC organoids treated as indicated (n=4, one of three biological replicates).
l, Immunoblotting of P2x4 knockdown hCRC organoids (hCRCshP2X4) treated as indicated for 2h (n=3).
m, Reseeding capacity of doxycycline-inducible hCRCshP2X4 tumor organoids treated as indicated (n=4, one of three biological replicates).
n, P2X4 qRT-PCR analysis of doxycycline treated hCRCshP2X4 tumor organoids (n=3 biological replicates).
o, Treatment regimen of subcutaneous hCRCshP2x4 tumors.
p, Growth rate of subcutaneous hCRCshP2X4 tumors in response to 5-FU +/- doxycycline treatment as indicated in (o). (n=6 mice for each condition). One set of mice was analysed.
q, Representative images of tumors and P2x4, Ki67 and H&E staining on tumor sections of mice indicated in (p), (n=6, scale bar = 100μm). Staining was performed on all tumors described in (p).
All data are mean ±SD and analysed by two-tailed Student’s t-test (n) or 1-way ANOVA (b, d, h, k, m) and 2-way ANOVA (p) with Bonferroni’s multiple comparison.
Cell death-induced mTOR counteracts ROS dependent apoptosis in neighboring tumor epithelia
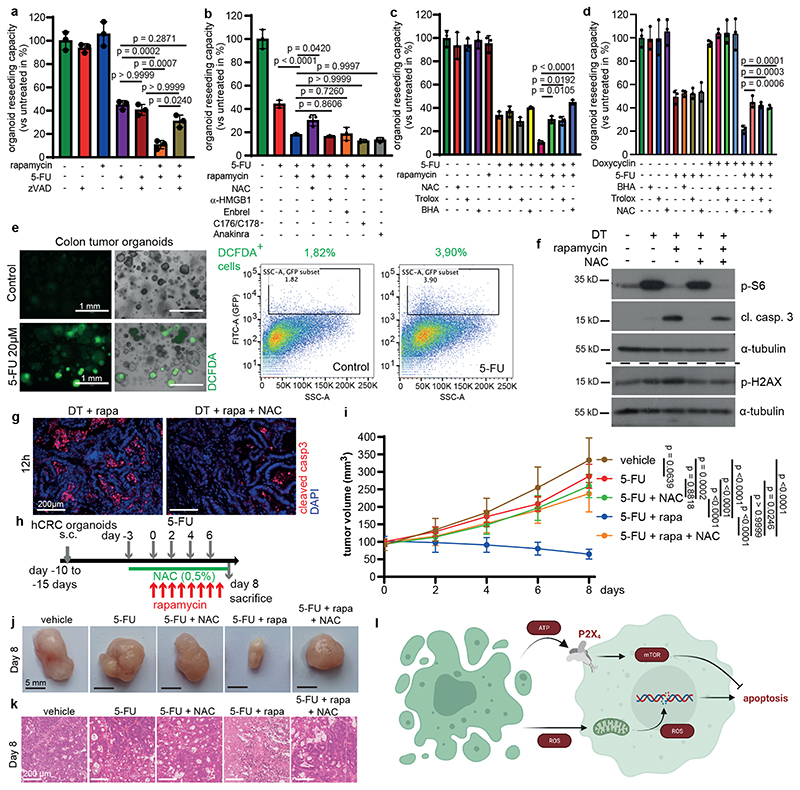
Our data clearly show that cell death induces paracrine ATP dependent P2X4-mediated mTORC1 activation in surviving tumor cells. Strikingly, surviving tumor cells were not dependent on mTOR prior to induction of apoptosis of neighboring cells as inhibition of mTOR alone did not cause a comparable tumor regression. Therefore, we reasoned that dying cells induce a second, pro-apoptotic signaling cascade in adjacent tumor epithelial cells that was counteracted by the simultaneous activation of P2X4/mTORC1. In line with this notion, inhibition of apoptosis using zVAD reverted the synergistic effect of combining 5-FU and rapamycin on organoid survival without having a significant impact on single 5-FU administration (Fig. 4a). To examine whether 5-FU-dependent release of other well-characterized DAMPS may be involved in the induction of a pro-apoptotic signal in adjacent tumor cells we blocked the action of oxygen radicals (N-acetylcysteine (NAC), mtDNA (STING inhibitors C176/C178), HMGB1 (neutralizing antibody), TNFα (Enbrel) or IL-1α (sIL-1ra; anakinra). Notably, only ROS scavenging by NAC was able to prevent decreased survival of 5-FU/rapamycin treated organoids (Fig. 4b), which could also be confirmed when ROS formation was antagonized by the vitamin E analogue 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) or butylated hydroxytoluene (BHA) (Fig. 4c). The same protective effects could be observed when these anti-oxidants were added to 5-FU treated P2X4 silenced PDTO (Fig. 4d). Furthermore, 2’,7’–dichlorofluorescein diacetate (DCFDA) staining indicated increased ROS production upon 5-FU administration (Fig. 4e). NAC treatment also prevented caspase 3 cleavage of rapamycin treated Lgr5+ cell ablated organoids (Fig. 4f), confirming that ROS induced cell death was indeed triggered in a paracrine manner and this was not caused by a cell autonomous 5-FU induced mechanism. This correlated with changes in H2AX phosphorylation, suggesting that elevated paracrine ROS levels induced DNA damage, which would possibly be involved in cell death induction (Fig. 4f). Importantly, NAC administration reduced apoptosis in tumor tissues of DT/rapamycin treated animals after 24 hours (Fig. 4g) and completely prevented inhibition of tumor growth in 5-FU/rapamycin treated mice (Fig. 4h-k).
Figure 4. ROS scavenging prevents induced mTOR addiction.
a, Reseeding capacity of AOM/DSSATKN tumor organoids treated as indicated (n=3, one of three biological replicates).
b, Reseeding capacity of AOM/DSSATKN tumor organoids treated as indicated (n=3, one of three biological replicates).
c, Reseeding capacity of AOM/DSSATKN tumor organoids treated as indicated (n=3, one of four biological replicates).
d, Reseeding capacity of hCRCshP2X4 tumor organoids treated as indicated (n=3, one of four biological replicates).
e, Fluorescence microscopic and flow cytometric ROS analysis using DCFDA in 5-FU (20h) treated colon tumor organoids. One representative of four biological replicates (scale bar = 1mm).
f, Immunoblot analysis of AOM/DSSATKN tumor organoids treated as indicated. p-γH2AX and the corresponding α-tubulin loading were probed on a separate gel (see SI) (n=3).
g, AOM/DSSATKN tumor organoids were injected subcutaneously. Once tumor reached 200 mm, mice were treated as indicated for 12h and analysed for cleaved caspase 3. Nuclei were counterstained with DAPI. (n=3 mice, scale bar = 200 μm).
h, Treatment regimen of subcutaneous hCRC tumors.
i, Growth rate of subcutaneous hCRC tumors in response to treatment as indicated in (h) (n=6 mice for vehicle treatment, n=7 mice for all other treatments. One set of mice was analysed.
j, Representative images of subcutaneous hCRC tumors on day 8 treated as indicated in (i), (n=6 for vehicle, n=7 for all other treatments, scale bar = 5 mm).
k, Representative H&E staining of subcutaneous hCRC tumors described in (i) (n=6 for vehicle, n=7 for all other treatments, scale bar = 200 μm). Staining was performed on all tumors indicated in (i).
l, Schematic representation of how dying cells activate mTOR in adjacent cells to counteract apoptosis induction by increased ROS levels.
All data are mean ±SD and analysed by 1-way ANOVA (a-d) or 2-way ANOVA (i) with Bonferroni’s multiple comparison.
Collectively, we describe the initiation of a complex paracrine signaling rewiring in tumor cells in response to cell death of neighboring cells that is established to assure the maintenance of tumor integrity and is not a simple response to stress-induced cell autonomous mechanisms such as autophagy or glycolysis. Notably, such paracrine induced shift represents a general response to cells undergoing necrosis or apoptosis. Engagement of purinergic receptors upon cell death is critical for immune cell recruitment and balancing an adequate inflammatory response26. However, we demonstrate here that intestinal epithelial cells are readily equipped to mount an appropriate defense mechanism in response to danger signals that is mediated by a distinct purinergic receptor subtype. The newly installed balance of pro- and anti-apoptotic signaling cascades provides an adaptive mechanism for tumor cells to cope with pro-apoptotic therapies and offers a new opportunity for the treatment of colorectal cancer as it renders cells dependent on mTOR signaling (Fig. 4l), thus, providing a novel rationale for combination therapy of cytotoxic compounds with rapamycin or its analogs that goes beyond a cell-autonomous induction of autophagy27. Considering that mTOR blockade has a limited therapeutic index28, targeting P2X4 may represent alternative strategy that is better tolerated and may be applicable to CRC or other tumor types known to express high levels of P2X4, such as lung and pancreas.
Methods
No statistical methods were used to predetermine sample size. Investigators were blinded to outcome assessment of tissue staining and qRT-PCR but were not blinded to allocation during outcome assessment of other experiments.
Animal studies
All experiments involving animals were reviewed and approved by Regierungspräsidium Darmstadt, Darmstadt, Germany. Lgr5EGFP-DTR(+) mice9, 10 and Villin-CreERT2;RaptorF/F mice29 were described previously. Lgr5EGFP-DTR(+) mice were crossed with Villin-CreER Raptor mice in order to generate Villin-CreERT2;RaptorF/F;Lgr5EGFP-DTR(+) mice NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratory and colony was maintained at animal facility of Georg-Speyer-Haus, Frankfurt. The mice were housed under standard laboratory conditions in SPF cages in an animal room at constant temperature (19–23 °C) and regulated humidity under a 12 h/12 h light-dark cycle and received standard laboratory chow and water ad libitum. All mice entered the experiments at 8-12 weeks of age. Both, male and female mice were used for the experiments. No statistical method was used to predetermine the sample size. Sample size was based on experimental feasibility, sample availability and according to the published literature. For AOM/DSS induced colon tumorigenesis in Lgr5-DTR or Villin-CreERT2;RaptorF/F;Lgr5EGFP-DTR(+) mice, both, male and females were used to induce tumors and to generate organoids. For subcutaneous transplantation experiments in NSG mice, male and female mice were randomly distributed among treatment groups when tumor size reached a measurable volume.
AOM/DSS induced colon tumorigenesis and mini-endoscopy
Colon tumors were induced as previously described30. Mice were injected intraperitoneally (i.p.) with AOM (10 mg/kg; Sigma, #A5486) and after 5 days, 2% DSS (molecular weight, 36-50 kDA; MP Biochemicals, #SKU 0216011090) was given in the drinking water for 5 days, followed by 14 days for regular water. The DSS treatment was repeated for two additional cycles. Mini endoscopy was performed at around day 80 and after the treatments on anesthetized mice using a mouse mini endoscopy system (Karl Storz). Recorded videos from endoscopy system were opened and viewed in VLC Media Player (v2.2.4) and still images were captured from these videos.
Xenograft experiments
Organoids were collected using ice cold Cell Recovery Solution (Corning, #354253) and mechanically dissociated into small clusters. Dissociated organoid cells were resuspended in PBS, admixed with 50% Matrigel (Corning, #356231) in a final volume of 100 μl, and injected subcutaneously in the right flank of NSG mice (AOM/DSSATKN and human CRC organoids). Tumor size was monitored with a digital caliper and tumor volumes were calculated according to the formula (length × width × width)/2. The experimenters performing mouse experiments were not blinded as the mice were assigned to the treatment groups by a visible tailmark. This tailmark was known to the researchers, as the researchers performed both, injections for treatments and tumor measurements. The maximal permitted tumor size was 1.5 cm which was not exceeded in the described xenograft experiments. Other endpoints were weight loss of more than 20% or tumor ulceration which were not reached during the experiments. At the end of experiments mice were humanely euthanized to collect tumors.
Cell lines
All cell lines (HEK293, CMT93 and CT26, HCT116 and RKO were originally obtained from ATCC and were maintained in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS). The cell lines were not authenticated or tested for mycoplasma contamination.
Diphtheria toxin and other in vivo treatments
DT (Millipore, #322326) treatment was performed as previously described9, 10. Briefly, DT was injected intraperitonially at a dose of 50 μg/kg for the time indicated in individual experiments. mTOR inhibitor rapamycin (LC Labs, #-5000) was dissolved in 5% Ethanol, 5% PEG400 and 5% Tween 80 in PBS and injected i.p. at 10 mg/kg once a day for indicated time. 0.5% N-acetylcysteine (Sigma-Aldrich, #A91165) was applied ad libitum in drinking water 3 days before starting the treatment with 5-FU and rapamycin. 1 mg/ml Doxycycline (Sigma#D9891) was applied ad libitum in drinking water containing 3% Sucrose 3 days before treatment with 5-FU and rapamycin and remained in the drinking water until the end of the experiment. 5-FU (Sigma#F6627) was dissolved in sterile H2O and injected intraperitoneally for the times indicated in individual experiments at a concentration of 20 mg/kg.
Human derived research material
Human tumor organoids were collected as part of the interdisciplinary Biobank and Database Frankfurt (iBDF) after prior written informed consent the study was approved by the institutional review board of the UCT and the Ethical Committee at the University Hospital Frankfurt (Ethics vote: 4/09; project-numbers: SGI-06-2015, SGI-12-2018 and SGI-7-2018). hCRC line1 was established from a rectal adenocarcinoma tissue of 59 old male, hCRC line 2 from sigmoidal colon cancer tissue of a 69 year old male and hCRC line 3 from a caecal adenocarcinoma tissue of a 74 old female. Pancreatic adenocarcinoma (PDAC) organoids were established from tumor tissue of a 68 old male diagnosed with PDAC.
Tumor organoid culture
Colon tumor organoids were cultured as previously described31. AOM/DSS induced colon tumors were harvested by opening the colon longitudinally and dissociated enzymatically for 30-40 min in digestion media containing 1 mg/ml collagenase I (Sigma-Aldrich), 0.5 mg/ml Dispase (Roche), 50 μg/ml dNase (Sigma-Aldrich) in RPMI medium containing 2% FCS. Supernatant was washed and filtered through a 70 μM cell strainer. Cells were then embedded in Matrigel (Corning, #356231) and cultured in Advanced Dulbecco’s modified Eagle’s medium/F12 (Invitrogen, #12634-028) supplemented with Glutamax (Invitrogen, #35050038), Hepes (Invitrogen, #15630056), Pen/Strep (Invitrogen, #15140122), N2 supplement (Invitrogen, #17502048), B27supplement (Invitrogen, #17504044) and n-Acetylcysteine (Sigma-Aldrich, #A91165). EGF (Invitrogen, #PMG8045), RspoI and noggin were added as niche factor. To culture ATKN organoids, puromycin (2 μg/ml), Hygromycin (200 μg/ml) and blasticidin (5 μg/ml) was added to culture medium. Human CRC organoids were cultured in Advanced Dulbecco’s modified Eagle’s medium/F12 containing RspoI, noggin, B27, n-Acetylcysteine, hEGF (Peprotech, # AF-100-15, A83_01 (Tocris, #2939) and SB202190 (Sigma, #S7067). Human organoids were collected as part of the interdisciplinary Biobank and Database Frankfurt (iBDF) after prior written informed consent and the study was approved by the institutional review board of the UCT and the Ethical Committee at the University Hospital Frankfurt (Ethics vote: 4/09; project-numbers: SGI-06-2015 and SGI-12-2018).
Organoids treatments and organoid harvesting
In vitro Lgr5+ cells ablation experiments, organoids were treated with 20 ng/ml DT (Millipore, #322326) in the culture medium. Other treatments were performed as described in each figure legends or the main text. Rapamycin (10 μM, Selleckchem, #S1039), 5-BDBD (1μM and 10μM, Tocris#3579), NF279 (10 μM, Tocris, #1199), PPADS (60 μM, Santacruz, #sc-202770), PTx (40 ng/ml, Tocris, #3097), NAC (2 mM, Sigma-Aldrich, #A91165), Trolox (1mM, Tocris#6002), BHA (10 μM, Sigma#B1253), theophyllin (50 μM, Sigma, #T1633), A438079 (10 μM, Sigma, #A9736), apyrase (10U/ml, Sigma, #A6132, was applied to the medium 30 min prior to treatment), 2’3’ cGAMP (10 and 30 μg/ml, Invivogen, #tlrl-nacga23), Bafilomycin (10 nM, Selleck chemicals, #S1413), chloroquine (15 μM, Sigma#6628) ATP (100 μM, Sigma, #A6419), 5-Fluorouracil (20 μM, Sigma, #F6627-5G); C-176 and C-178 (500 nM, Ablasser Lab21), Doxycycline (Sigma#D9891, 1μg/ml), anti-HMGB1 antibody (5μg/ml), Enbrel (Etanercept, Pfizer, 10 μg/ml), Anakinra (Kineret, SOBI, 100 μg/ml), zVAD (Z-VAD-FMK, 50 μM, Selleck chemicals, #S7023) and Necrostatin-1 (Selleck chemicals, #S8037, 20 μM). Treatment with 4-hydroxytamoxifen (4-OHT; Sigma; 1uM) was performed for 24 hours after which Tamoxifen containing media was replaced by normal organoid culture medium. Tamoxifen treatment was performed 48h prior to DT treatment. For analyzing the organoid reseeding capacity, organoids were seeded in equivalent numbers for each treatment condition. After 48 hours the organoids were treated as indicated in the figure legends for additional 48 hours. The organoids were then dissociated by pipetting up and down 50 times using a P200 pipette-tip, re-seeded and newly generated organoids were counted 72 hours after re-seeding. The reseeding capacity of treated organoids was compared to the reseeding capacity of vehicle treated control organoids which was set at 100%. Human pancreatic organoids were cultured as previously described32. For the reseeding assay human pancreatic organoids were treated with 5-FU (20μM), rapamycin (10μM), 5-BDBD (1μM) or their combination for 48 hours in triplicate. After 48 hours organoids were dissociated by pipetting up and down through a 1ml serological pipette adapted with a P200 non-filtered tip 30 times to break them apart. Organoids were re-seeded and counted 72 after re-seeding. The reseeding capacity of treated organoids was compared to the reseeding capacity of vehicle treated control organoids which was set at 100%. For immunoblot analysis, organoids were collected 24h after treatment (if not indicated otherwise in the figure legend) in ice-cold Cell Recovery Solution (Corning, #354253) and pelleted by centrifugation. To prepare protein lysates for immunoblot analysis, organoid pellet was washed twice with cold PBS and pellet was snap frozen or lysed in lysis buffer. Survival in organoids were determined by treatment with propidium iodide (10 ug/mL) for 30 minutes and analyzed using ImageJ software.
Apoptosis assay
Human PDOs were treated with DMSO or 5-FU (20μM) for the indicated timepoints. Organoids were then stained with propidium iodide to visualize dying cells and analyzed by fluorescence microscopy. The propidium iodide positive area was then quantified using ImageJ and compared between DMSO and 5-FU treated organoids.
Measurement of ROS and ATP
ATP level in organoid supernatant or necrotic medium was measured using the CellTiter-Glo 2.0 Cell Viability Assay (Promega#G9242) according to the manufacturer’s instruction. ROS levels were measured using H2DCFDA (ThermoFisher #D339). Briefly, organoids were treated as indicated with 5-FU, stained with H2DCFDA (10μM), analysed by flurorescence microscopy then collected in ice-cold Cell Recovery Solution (Corning, #354253), pelleted by centrifugation and dissociated to single cells using Accutase (Sigma, #A6964) at RT for 5 min. Single cells were then analysed for H2DCFDA positivity via FACS analysis. The following gating strategy was used for the identification of Reactive oxygen species (ROS) positive cells: Dead cells were excluded from the analysis on the basis of forward scatter area (FSC-A) and side scatter area (SSC-A). The selected live cells were used to quantify the frequency of Ros positive cells (H2DCFA subset) based on the signal of the fluorescent H2DCFA dye in the Fitc-GFP and the SSC-A channel. Positive cells were determined by comparing H2DCFA labelled cells with non labelled cells. The threshold of positive cells was set upon which no unstained cells were detected.
Necrotic and DT Supernatant transfer experiments
Murine colorectal cancer cells (CMT93 and CT26) and human colorectal cancer cells (HCT116 and RKO) were cultured overnight in DMEM medium containing 2% FCS and culture supernatant was collected with or without cells and snap frozen in liquid nitrogen for 5 minutes, thaw at 37°C and filtered with 0.45 μM filters. Filtered control supernatant (without cells) and necroptotic supernatant (filtered supernatant of culture medium collected with cells) was transferred to 60-70% confluent cells (cultured overnight in DMEM medium containing 2% FCS). Cells were collected in lysis buffer after 2 hours or 4 hours for immunoblot analysis. For necroptotic supernatant transfer experiment in organoids control and necroptotic supernatant was prepared from CT26 cells cultured in Advanced DMEM/F12 without serum overnight and transferred to Lgr5EGFP-DTR(+) tumor organoids. If not indicated otherwise organoids were treated for 2 hours with necrotic medium and lysed in lysis buffer for immunoblot analysis. For treatment of Lgr5EGFP-DTR(-) organoids with supernatant from DT treated Lgr5EGFP-DTR(+) organoids, Lgr5EGFP-DTR(+) were treated either with vehicle or DT for 10h. Then supernatant was used to treat Lgr5EGFP-DTR(-) tumor organoids for 2 hours before lysis for immunoblot analysis.
Immunoblot analysis
Collected organoids were lysed in the lysis buffer as described in30. Protein content was quantified using protein assay dye reagent (Bio-Rad, #5000006) and 40 μg of total protein lysates in Laemmli buffer (Bio-Rad, #161-0747) were separated on SDS-PAGE, transferred to 0.45 μm PVDF membrane (Millipore, #IPVH00010) and blocked with 5% non-fat milk and 1% BSA in PBS at RT for 1 hour. After blocking membranes were incubated at 4°C overnight with primary antibodies. The following antibodies were used: cleaved caspase 3(#9661) diluted 1:500, cleaved PARP (#9548) diluted 1:1000, phospho-histone H2a.X (#2577) diluted 1:1000, phospho-p70 S6 kinase (#9205) diluted 1:1000, p70 S6 kinase (#9202) diluted 1:1000, phospho-S6 (#2211) diluted 1:2000, S6 (#2217) diluted 1:2000, phospho-Pras40 (#2997) diluted 1:1000, Pras40 (#2691) diluted 1:1000, LC3B (#2775) diluted 1:500 from Cell Signaling; Sting (Abcam, #ab92605) diluted 1:500, p62 (Santa Cruz, #sc28359) diluted 1:500. β-Actin (Sigma, #A4700) diluted 1:1000 or α-Tubulin (Santa-Cruz, #sc32293) diluted 1:1000, was used as loading control. After washing with PBS, membranes were incubated with appropriate HRP-conjugated secondary antibodies (mouse anti-rabbit IgG-HRP (Santa Cruz, Cat. No. #sc-2357) and Rabbit Anti-Mouse IgG H&L (HRP) (Abcam, Cat. No. #ab6728)) at 1:5000 dilution for 1h at RT and membranes were developed using SuperSignal West Pico Chemoluminescence Substrate (Thermo Fisher Scientific, #34580) and the ChemiDocTM MP Imaging System or Foto films. For the kinase activation assay human PDOs were treated either with DMSO or 5-FU (20μM) for 20h and lysed for protein extraction. Lysates were analysed using the Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems, #ARY003C) according to the manufacturer’s instructions. Immunoblots in the manuscript are representative experiments, the number of successful biological replicates is indicated as n in the figure legends).
Flow cytometry
For flow cytometric analysis, organoids were collected in ice-cold Cell Recovery Solution, washed with cold PBS and dissociated to single cells using Accutase (Sigma, #A6964) at RT for 5 min, filtered through a 40 μM cell strainer and resuspended in FACS buffer (1 mM EDTA and 1% FCS in cold PBS). Samples were acquired using BD LSRFortessa™ cell analyzer (BD Bioscience, with BD FACS Diva software (version 8.0) and analyzed using FlowJo v9 (FlowJO LLC).
Seahorse analysis
To perform metabolic measurements using the Seahorse XFe24 Metabolic Analyzer, ATKN organoids were collected 2 days after passaging. Organoids were first harvested from matrigel in 10 ml Hank’s Balanced salt solution (HBSS, Sigma) containing 1% BSA. Then they were spun down at 400g for 5 minutes, resuspended in 6 ml of cell recovery solution (Corning) and incubated for 10 minutes on ice. Organoids were pelleted at 400g for 5 minutes, supernatant was discarded, the pellet was resuspended in 1 ml of TrypLE (Life technologies) with 30μl Dnase (5000U/ml, Roche) and incubated at 37°C for 2 minutes. 9 ml of HBSS were added and organoids were pipetted with a serological 10 ml pipette for 10 times in order to shear them and pelleted at 400g for 5 minutes. The pellet was resuspended in 1 ml HBSS and cells were counted in a Kova Glasstic (KOVA international). 50.000 cells were plated per well in 10 μl Matrigel (Corning) on a Seahorse XF24 V7 PS Cell Culture Microplate. The plate was incubated for 12 minutes at 37°C to let Matrigel solidify. 500 μl of pre-warmed ATKN feeding media with 0,1% y27632 inhibitor (Selleck) were overlayed. Two days after plating 5-FU (20μM) and/or rapamycin (10μM) were applied to organoids. 24h later the Seahorse run was performed according to instructions in Agilent Seahorse XF Cell Mito Stress Test protocol with concentrations of 1 μM oligomycin, 1.5 μM FCCP and 0.5 μM rotenone/antimycin.
siRNA mediated knockdown
60-70% confluent CT26 cells were transfected with 25 nM siRNA smartpool (Dharmacon) against mouse Rheb (#L-057044-00), RagA(#L-057667-01), RagB (#L-066440-01) or a pool of non-targeting control siRNA(#D-001810-10) using DharmaFECT (Dharmacon) and HiPerFect (Qiagen#301704) transfection reagents. The siRNA sequences used are listed in the Supplementary Table 3 in the Supplementary information file. RNA was purified after 48 hours to confirm the knockdown efficiency. For supernatant transfer experiments, culture medium was changed after 16 hours of transfection, and supernatant was collected 72 hours later, filtered and stored at 4°C.
RNA extraction and qRT-PCR
For RNA extraction organoids were harvested directly in RLT buffer, RNA extraction was performed using rNeasy Mini kit (Qiagen, #74106) according to the manufacturer’s instructions. Total RNA (1-2 μg) was reverse transcribed into cDNA using a Super script II reverse transcriptase (Invitrogen, #18064-071) and OligodT (Invitrogen, #18418020). Real-Time quantitative PCR was performed using FastStart Universal SYBR Master Mix (Roche, # 4913914001) in 20 μl total volume on a StepOne Plus Real-Time PCR system (Applied Biosystems, StepOne software v2.2). Relative gene expression levels were quantified by using cyclophilin as a housekeeping gene (2[Ct cyclophilin – Ct target gene]). The primers used are listed in the Supplementary Table 1 in the Supplementary information file.
Immunostaining
Immunostaining was performed as previously described33. Briefly, tissues were fixed with 4% paraformaldehyde. 6 μm OCT frozen tissue sections or 5 μm paraffin-embedded tissue sections were processed for immunostaining using a standard histological protocol. Following primary antibodies were used: EGFP (Abcam, #ab6673) dilution 1:400, cleaved caspase 3 (Cell Signaling, #9579) diluted 1:500, Ki67 (Leica Biosystems, #NCL-L-Ki67-MM1) diluted 1:200, phospho-S6 (Cell Signaling, #2211) diluted 1:500, P2x4 (Abcam#134559) diluted 1:100. Sections were then incubated with the corresponding secondary antibodies (Horse Anti-Mouse IgG Antibody (H+L), Biotinylated (Vectorlaboratories, #BA-2000, diluted 1:200) and Rabbit Anti-Goat IgG Antibody (H+L), Biotinylated (Vectorlaboratoriess, #BA-5000, diluted 1:200)). The signal was detected with the VECTASTAIN® ABC-HRP Kit, Peroxidase (Vectorlaboratories, #PK-4000) according to the manufacturer’s instructions. For immunofluorescence staining after primary antibody incubation, sections were incubated with fluorochrome-labelled secondary antibodies (Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 (Invitrogen, Cat. No. #A-11055, diluted 1:500) and Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 594 (Invitrogen, Cat. No. #A-21207, diluted 1:500) and sections were mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen, #P36931). Images were captured on a Zeiss microscope (Axio imager.M2) and analysed using ImageJ (v1.53e, National Institutes of Health).
Plasmids, Lentivirus and retrovirus production
shRNA sequences were obtained from and cloned into mir-E based retroviral backbone as previously described34. The following shRNAs were used: P2x4_sh849 (TCTCATAGGTGAAGAAGTCGGA), P2x4_sh877 (TTCCATTTTGTAGTACTTGGCA), Rictor_sh4860 (TAATCTTAGAACTTCTTTGCGA), Rictor_sh5055 (TTGTAGAAACTGTACATCTTGA) Raptor_sh973 (TAAGGCAACACTGACTGTCTTC) Raptor_sh4182 (TTGTTGATGAGCTCTCCGCTGC). K-rasG12D was cloned in pBABE-puro (Addgene plasmid #1764). An IRIS-AKT (myristoylated) cDNA was cloned downstream of hygromycin cassette in pMSCV-rtTA3-PGK-Hygro (kindly provided by Lars Zender, Tübingen). Retroviral particles were produced by co-transfection of retroviral plasmids with packaging plasmid pCL-ECO (Addgene plasmid #12371) to HEK293T cells using the calcium phosphate method. Notch intracellular domain (NICD) was cloned in LentiCas9-BSD (Addgene plasmid #52962) by replacing Cas9. Lentiviral supernatants were prepared by co-transfection of Lenti-NICD-BSD plasmid, the packaging vector psPAX2 (Addgene plasmid #12260) and the envelope vector pMD2.G (Addgene, #12259) using Lipofectamine 2000. The culture medium was replaced 12 hours after transfection and viral supernatant was collected 48 hours later, filtered through a 0.45 μm PVDF filter (Millipore). Viral supernatants were concentrated by overnight centrifugation at 12,000 rpm at 4°C and viral pellets were resuspended in organoid culture medium. Single cells from organoids were infected with polybrene-supplemented viral supernatant (8 μg/ml polybrene, Sigma). Cells were embedded in matrigel 4 hours post infection and cultured in organoid culture medium supplemented with 10 μM Y-27632 (Sigma). 48 hours post infection organoids were selected in 2 μg/ml puromycin, 200 μg/ml Hygromycin or 5 μg/ml Blasticidin for 5-7 days. Tgfr2 gRNA (GGCCGCTGCATATCGTCCTG) was cloned into gRNA cloning vector (Addgene plasmid #41824) as previously described35. Single cells from organoids were co-transfected with gRNA plasmid and Cas9 expressing plasmid (Addgene plasmid #41815) using Lipofectamine 2000 as previously described36. Tgfbr2 mutant organoids were selected in the absence of Noggin and presence of 50 ng/ml TGF-β (R&D Systems, # 7666-MB-005). The guide sequences for shRNAs are listed in the Supplementary Table 2 in the Supplementary information file.
Statistical analysis
No statistical method was used to predetermine the sample size for in vivo experiments. For subcutaneous transplantation experiments, mice were distributed among treatment groups when tumor size reached a measurable size. Tables and graphs for statistical analysis were created using GraphPad Prism 4.03 (GraphPad Software, Inc.) or Microsoft® Excel® 2016 MSO (Version 2207 Build 16.0.15427.20166) 32 Bit). Statistical significance between two groups was determined by two-tailed Student’s /-test and for more than two groups 1-way ANOVA or 2-way ANOVA with Bonferroni’s multiple comparison test was performed (GraphPad Prism 4.03, GraphPad Software, Inc.). All the data in the graphs are shown as mean ± SD, unless stated otherwise.
Extended Data
Extended Data Fig. 1. (Related to Figure 1): CRC resistance to 5-FU depends on mTORC1 but not mTORC2.
a, Immunoblot analysis of two different human tumor organoid lines derived from sporadic CRC treated as indicated (n=3).
b, Reseeding capacity of hCRC organoid lines treated with 5-FU, rapamycin or 5-FU/rapamycin (n=3, one of three biological replicates).
c, Reseeding capacity of doxycycline-inducible hCRCshRAPTORr#2 tumor organoids treated as indicated (n=4, one of three biological replicates).
d, Raptor qRT-PCR analysis of doxycycline treated hCRCshRAPTOR#2 tumor organoids (n=3 biological replicates).
e, Reseeding capacity of doxycycline-inducible hCRCshRICTOR tumor organoids treated as indicated (n=4, one of three biological replicates).
f, Rictor qRT-PCR analysis of doxycycline treated hCRCshRictor#1 tumor organoids (n=3 biological replicates).
g, Reseeding capacity of doxycycline inducible hCRCshRICTOR#2 tumor organoids treated as indicated (n=4, one of three biological replicates).
h, Rictor qRT-PCR analysis of doxycycline treated hCRCshRictor#2 tumor organoids (n=3 biological replicates).
i, Immunoblot analysis of Lgr5 - tumor organoids treated as indicated (n=3).
j) Profiles for Extracellular Acidification Rate (ECAR) of AOM/DSSATKN tumor organoids treated as indicated for 24h (n=4 for rapamycin treated organoids in all other conditions n=5 biological replicates of one experiment).
k) Profiles for Oxygen Consumption Rate (OCR) of AOM/DSSATKN tumor organoids described in (j) (n=4 for rapamycin treated organoids in all other conditions n=5 biological replicates of one experiment).
l) Percentage of metabolic potential over baseline in ECAR in organoids described in (j) (n=4 for rapamycin treated organoids in all other conditions n=5 biological replicates of one experiment).
m) Percentage of metabolic potential over baseline in OCR in organoids treated described in (j) (n=4 for rapamycin treated organoids in all other conditions n=5 biological replicates of one experiment).
All data are mean ±SD (except for j, k where data are mean ±SEM) and analysed by twotailed Student’s t-test (d, f, h) or 1-way ANOVA with Bonferroni’s multiple comparison (b, c, e, g, l, m).
Extended Data Fig. 2. (related to Figure 2): Dying tumor cells activate mTORC1 in surrounding tumor cells in vivo.
a, Immunofluorescence analysis of p-S6 (red) and EGFP (Lgr5, green) expression in tumor tissues of vehicle or DT treated AOM/DSS mice 24h after injection. Nuclei were counterstained with DAPI. Representative images are shown (n=3 biological replicates, scale bar = 50 μm).
b, Immunoblot analysis of AOM/DSSATKN tumor organoids treated as indicated. For p-γH2AX and the corresponding α-tubulin loading samples were run on a separate gel (see SI). Representative results are shown (n=3).
Extended Data Fig. 3. (related to Figure 3): Dying tumor cells activate mTORC1 via ATP/P2X4 in human CRC and PDAC.
a, Relative mRNA expression levels of the indicated genes in the colon tumors from Lgr5EGFP-DTR(+) mice 24h after injection of DT injection or vehicle determined by qRT-PCR. (n=10 tumors for controls and n=9 tumors for DT).
b, Relative mRNA expression levels of the indicated genes in Lgr5EGFP-DTR(+) tumor organoids at 24h after DT treatment determined by qRT-PCR. Expression of vehicle treated organoids was set to 100 arbitrary units (n=4 biological replicates).
c, Immunoblot analysis of Lgr5EGFP-DTR(+) tumor organoids treated as indicated for 30 min (n=3).
d, Immunoblot analysis of Lgr5EGFP-DTR(+) tumor organoids treated as indicated for 16h (n=3).
e, Immunoblot analysis of Sting WT and Sting KO Lgr5EGFP-DTR(+) tumor organoids treated as indicated (n=4).
f, Immunoblot analysis of murine (CMT93 and CT26, left panel) and human CRC cells (HCT116 and RKO, right panel) treated as indicated (n=2).
g, Immunoblot analysis of Lgr5EGFP-DTR(+) tumor organoids treated as indicated for 12h (n=3).
h, Left: Immunoblot analysis of ATP (100 μM) treated CT26 cells transfected with control or Rheb siRNA (n=3). Right: Relative mRNA expression levels of Rheb in siRNA transfected cells determined by qRT-PCR (n=3 biological replicates).
i, Left: Immunoblot analysis of ATP (100 μM) treated CT26 cells transfected with control or a combination of RagA and RagB siRNA (n=3). Right: Relative mRNA expression levels RagA and RagB in siRNA transfected cells determined by qRT-PCR (n=3 biological replicates).
j, Relative mRNA expression levels of P2x and P2y receptors in mouse colon tumor organoids tissues determined by qRT-PCR (n=3 biological replicates).
k, Immunohistochemistry analysis of P2x4 expression in healthy murine colon or colon tumors of AOM/DSS treated mice. Images from one representative staining (n=3, scale bar = 300 μm).
l, Reseeding capacity of hCRC tumor organoid line 2 treated as indicated (n=3, one of three biological replicates).
m, Reseeding capacity of doxycycline-inducible hCRCshP2x4#2 tumor organoids treated as indicated (n=4, one of three biological replicates).
n, P2x4 qRT-PCR analysis of doxycycline treated hCRCshP2x4#2 tumor organoids (n=3 biological replicates).
o, Reseeding capacity of human pancreatic tumor organoids treated as indicated (n=3, one of three biological replicates).
p, Reseeding capacity of human pancreatic tumor organoids treated as indicated (n=3, one of three biological replicates)
All data are mean ±SD and analysed by two-tailed Student’s t-test (a, b, h, i, n, o, p) or by 1way ANOVA with Bonferroni’s multiple comparison (l, m).
Supplementary Material
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Footnotes
Author Contributions
M.S., F.C., J.G and F.R.G. designed the experiments. M.S., F.C., J.G., E.E., K.B.K., Y.D. and M.Sche. performed and analyzed animal and organoid experiments. A.N. performed immunostainings and RT-PCR. M.P. generated mutant organoids, shRNA plasmids and contributed to animal experiments. A.K. performed and M.C.A. analyzed seahorse experiments. J.V. initiated project. M.R. helped in FACS experiments. M.K. helped in generation of Sting KO organoids. T.W.B. performed siRNA experiments. V.P. performed ROS analysis and ATP measurements. A.A., T.S. M.C.A. and F.J.d.S. provided essential material. F.R.G. conceptualized and conceived the project. M.S., J.G. and F.R.G. wrote the manuscript. All the authors commented on the manuscript draft.
Competing interests
M.S., J.G. and F.R.G. filed a patent regarding the use of P2X4 inhibitors in combination with cytotoxic compounds. F.J.d.S is employee of Genentech and own Roche shares. F.R.G. is a consultant for Amazentis not related to this study. All other authors declare no competing financial interests.
Ethics statement
Human tumour organoids were established at Georg-Speyer-Haus, Frankfurt, Germany from fresh human tumour tissue according to regional regulations and were approved by the regional ethics committee (Ethikkommission Universitätsklinikum Frankfurt/Main). Informed consent was obtained from all donors of tissue.
Data representation
For the presentation of our data, figures were created using Adobe Illustrator CS6 v16.0 _64-bit and Adode Photoshop CS6 v13.0*64. Illustrations were created with BioRender.com.
Acknowledgements
We thank Hana Kunkel, Susanne Bösser, Kathleen Mohs, Preeti Gupta, Eva Rudolf and Christin Danneil for expert technical assistance as well as the staff at the Animal Facility and the Histology and Flow Cytometry Core Facilities at the Georg-Speyer-Haus. Work in the lab of F.R.G. is supported by institutional funds from the Georg-Speyer-Haus, by the LOEWE Center Frankfurt Cancer Institute (FCI) funded by the Hessen State Ministry for Higher Education, Research and the Arts [III L 5 - 519/03/03.001 - (0015)], Deutsche Forschungsgemeinschaft (FOR2438: Gr1916/11-1; SFB1292-Project ID: 318346496-TP16; SFB1479-Project ID: 441891347-P02; GRK2336) and the ERC (Advanced Grant PLASTICAN-101021078). The Institute for Tumor Biology and Experimental Therapy, Georg-Speyer-Haus is funded jointly by the German Federal Ministry of Health and the Ministry of Higher Education, Research and the Arts of the State of Hessen (HMWK).
Data availability
Source data are provided with this paper. Reagents and any further information are available on request to the corresponding authors. Source data are provided with this paper.
References
- 1.Endo H, Inoue M. Dormancy in cancer. Cancer Sci. 2019;110:474–480. doi: 10.1111/cas.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudipaty SA, Conner CM, Rosenblatt J, Montell DJ. Unconventional Ways to Live and Die: Cell Death and Survival in Development, Homeostasis, and Disease. Annu Rev Cell Dev Biol. 2018;34:311–332. doi: 10.1146/annurev-cellbio-100616-060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Schaik TA, Chen KS, Shah K. Therapy-Induced Tumor Cell Death: Friend or Foe of Immunotherapy? Front Oncol. 2021;11:678562. doi: 10.3389/fonc.2021.678562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diwanji N, Bergmann A. Two Sides of the Same Coin-Compensatory Proliferation in Regeneration and Cancer. Adv Exp Med Biol. 2019;1167:65–85. doi: 10.1007/978-3-030-23629-8_4. [DOI] [PubMed] [Google Scholar]
- 5.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 6.Price DJ, Grove JR, Calvo V, Avruch J, Bierer BE. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 7.Oliver Metzig M, et al. Inhibition of caspases primes colon cancer cells for 5-fluorouracil-induced TNF-alpha-dependent necroptosis driven by RIP1 kinase and NF-kappaB. Oncogene. 2016;35:3399–3409. doi: 10.1038/onc.2015.398. [DOI] [PubMed] [Google Scholar]
- 8.Sui X, et al. JNK confers 5-fluorouracil resistance in p53-deficient and mutant p53-expressing colon cancer cells by inducing survival autophagy. Sci Rep. 2014;4:4694. doi: 10.1038/srep04694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Sousa e Melo F, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, et al. Inosine Released from Dying or Dead Cells Stimulates Cell Proliferation via Adenosine Receptors. Front Immunol. 2017;8:504. doi: 10.3389/fimmu.2017.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory CD, Pound JD. Cell death in the neighbourhood: direct microenvironmental effects of apoptosis in normal and neoplastic tissues. J Pathol. 2011;223:177–194. doi: 10.1002/path.2792. [DOI] [PubMed] [Google Scholar]
- 13.Rock KL, Kono H. The inflammatory response to cell death. Annu Rev Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 16.Ahn J, et al. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemos H, et al. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016;76:2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paludan SR, Reinert LS, Hornung V. DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer. Nat Rev Immunol. 2019;19:141–153. doi: 10.1038/s41577-018-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haag SM, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 22.Di Virgilio F, Adinolfi E. Extracellular purines, purinergic receptors and tumor growth. Oncogene. 2017;36:293–303. doi: 10.1038/onc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnstock G. Purinergic Signalling: Therapeutic Developments. Front Pharmacol. 2017;8:661. doi: 10.3389/fphar.2017.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balazs B, et al. Investigation of the inhibitory effects of the benzodiazepine derivative, 5-BDBD on P2X4 purinergic receptors by two complementary methods. Cell Physiol Biochem. 2013;32:11–24. doi: 10.1159/000350119. [DOI] [PubMed] [Google Scholar]
- 25.Hansen MR, Krabbe S, Novak I. Purinergic receptors and calcium signalling in human pancreatic duct cell lines. Cell Physiol Biochem. 2008;22:157–168. doi: 10.1159/000149793. [DOI] [PubMed] [Google Scholar]
- 26.Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol. 2016;16:177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- 27.Yang JW, Zhang QH, Liu T. Autophagy facilitates anticancer effect of 5-fluorouracil in HCT-116 cells. J Cancer Res Ther. 2018;14:S1141–S1147. doi: 10.4103/0973-1482.204898. [DOI] [PubMed] [Google Scholar]
- 28.Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faller WJ, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta J, et al. Dual function of p38alpha MAPK in colon cancer: suppression of colitis-associated tumor initiation but requirement for cancer cell survival. Cancer Cell. 2014;25:484–500. doi: 10.1016/j.ccr.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Canli O, et al. Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer Cell. 2017;32:869–883.:e865. doi: 10.1016/j.ccell.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallangyo CK, Ziegler PK, Greten FR. IKKbeta acts as a tumor suppressor in cancer-associated fibroblasts during intestinal tumorigenesis. J Exp Med. 2015;212:2253–2266. doi: 10.1084/jem.20150576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. Reagents and any further information are available on request to the corresponding authors. Source data are provided with this paper.