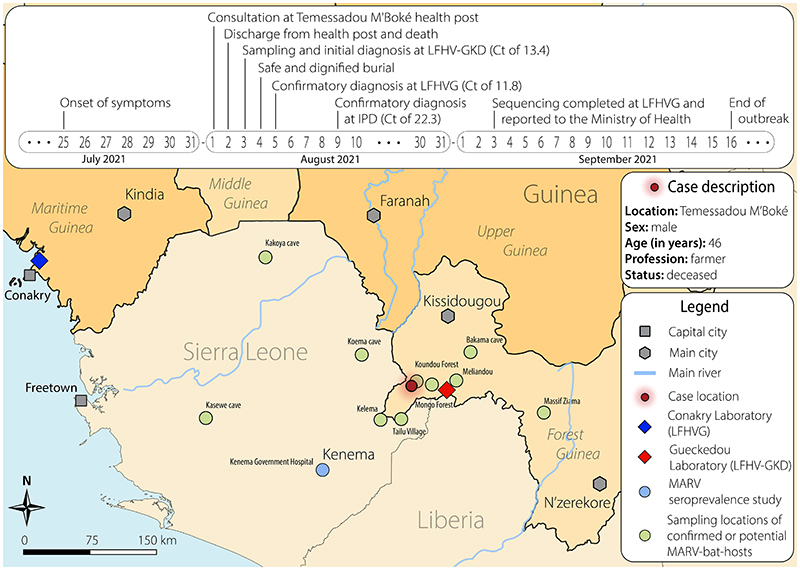
On August 2nd, 2021, a patient from Temessadou M’Boké town (Gueckédou prefecture) died with presentation of hemorrhage from several natural orifices (Figure 1A and Supplementary Appendix, available with the full text of this letter at NEJM.org). On August 3rd, the initial diagnosis of Marburg virus (MARV) was made on a post-mortem buccal swab sample by real time reverse transcription PCR with a cycle threshold (Ct) value of 13.4. Field investigation teams were deployed and validation of the diagnostic finding in two additional laboratories occurred within a few days (Figure 1A). In-country metagenomic next generation sequencing allowed for full-length MARV genome recovery (99.3%) and phylogenetic analysis indicated that the new Guinea MARV strain clusters with MARV strains isolated from bats in Sierra Leone and from humans in Angola (Figure 1B and Supplementary Appendix). Close monitoring for a period of 21 days identified that all contacts remained asymptomatic, and no additional cases were detected.
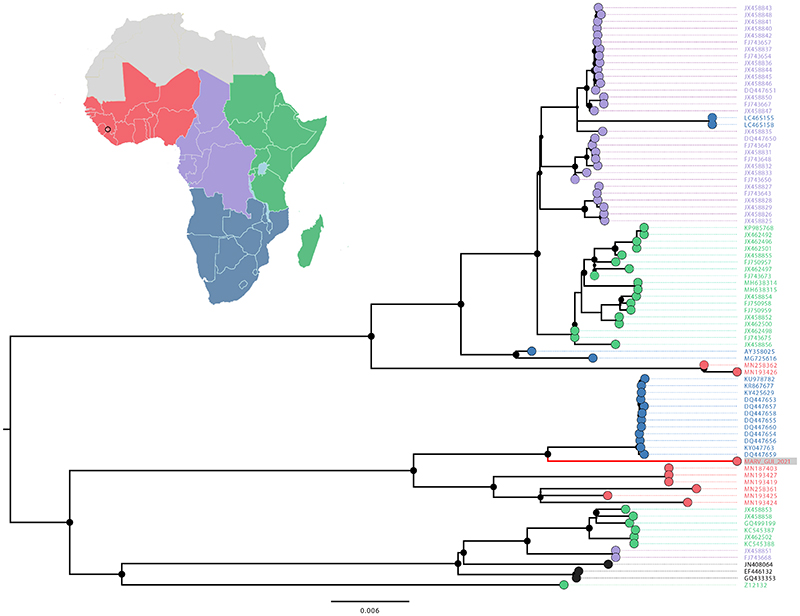
Figure 1. Map and timeline of events of the case occurrence (A) and phylogenetic analysis of MARV genomes (B).
(A) The top panel highlights the timeline of events while the map depicts parts of Guinea, Sierra Leone and Liberia with a focus on Forest Guinea and the Gueckédou prefecture where the MVD emerged. Note that on 3 September, LFHVG further shared the Marburg virus sequence with the public (https://virological.org/t/marburg-virus-sequence-from-guinea-2021/755) to support the public health response as well as the development and evaluation of Marburg virus disease diagnostics and therapeutics. The map depicts locations of the laboratory based in Gueckédou (LFHV-GKD), that of Conakry (LFHVG), and other relevant locations of sites that have reported (i) evidence of MARV circulation in bats1 and in humans3 in Sierra Leone, and (ii) identification of bat species known to be potential reservoir hosts of MARV in Guinea, including Méliandou, the location of the 2014-2016 Ebola virus disease outbreak, as well as Mongo Forest, Koundou Forest, Bakama cave, and Massif Ziama. The names and GPS locations of study sites from published works were used (Supplementary Appendix). The map was drawn using QGIS software version 3.22.0 and Adobe Illustrator®. Ct, cycle threshold; LFHV-GKD, “Laboratoire des Fièvres Hémorragiques Virales de Gueckédou”; LFHVG, “Laboratoire des Fièvres Hémorragiques Virales de la Guinée” ; IPD, “Institut Pasteur de Dakar”.
(B) The map highlights four geographic regions of the African Union with known reports of MARV circulation (red, West Africa; green, East Africa; purple, Central Africa; dark blue, Southern Africa). This same color code is applied in the phylogenetic tree as per geographic origin of the sequence. Note that sequences in black refer to MARV cases that occurred outside Africa. The new Guinea MARV genome (MARV_GUI_2021, red and grey background, long branch) clusters between the Sierra Leone bat MARV clade (red) and the human Angola clade (blue). The internal node shapes are proportional to bootstrap support and accession numbers are provided. The accession numbers of the new MARV Guinea are OK665848 for LFHVG and OL702894 for IPD.
Forest Guinea, along with other areas of West Africa including Sierra Leone, is predicted to be environmentally suitable for zoonotic transmission of MVD by bats and particularly by Rousettus aegyptiacus (ERB) which has been identified as a natural MARV reservoir host.1–3 Most MARV bat reservoir hosts are present in the forested region of Guinea, particularly in Koundou, which is close to the case emergence location (Figure1A and Supplementary Appendix). The patient had limited social interactions and lived in a household of four people. There was no evidence of travel history to country outside Guinea for the patient or his close contacts, nor contacts to returning travelers. He was a farmer living in close contact with nature and wildlife and may therefore have experienced repeated exposure to an environment or food contaminated with excreta of MARV-infected bats. Community surveys showed that while he may have harvested wild fruits for personal consumption, there was no suggestion of visits to caves, nor hunting activities for bushmeat, including bats, or its consumption. Traditional practices of bushmeat consumption or preparation (i.e., direct exposure to bodily fluids) cannot be fully excluded as they would not be disclosed in the context of the national ban that was enforced following the 2021 EVD outbreak.
The new Guinea MARV and the Angola MARV clade share a common ancestor that probably existed in 1965 [95% confidence interval: 1944,1981, Bayesian molecular clock analysis]. This indicates that about 55 years ago, these lineages diverged from a common ancestor, and each evolved independently in its respective reservoir host with the Guinea MARV remaining undetected until this 2021 spillover event. This time scale of decades provided ample opportunity for the virus to be dispersed over large distances by bat migration. One could draw a parallel with the emergence of the West-African Ebola virus lineage (Makona) that diverged from a central African ancestor and independently evolved in its host until the spillover event happened.4 In the case of MARV, the basal clustering of bat MARV in Sierra Leone suggests that even the Angola outbreak may have had its eventual roots in West Africa (Figure 1B).
Both the epidemiology and phylogenetic history argue against the possibility the new MVD may be imported. Overall, it seems plausible that the MVD emergence in Guinea is due to a zoonotic transmission event from a bat reservoir at the end of July 2021.
The isolated living habit of the patient likely played a role in minimizing the risk of secondary infections. Notably, capacity building programs, long-term collaborative partnerships, and establishment of decentralized laboratory capacities with well-trained staff have been valuable for a timely laboratory diagnosis. These same capacities proved key during the recent reemergence of EVD in Guinea.5
Supplementary Material
Funding
Supported by the German Federal Ministry of Health through support of the WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Viruses at the Bernhard-Nocht-Institute for Tropical Medicine (agreement ZMV I1-2517WHO005), the Global Health Protection Program (GHPP, agreements ZMV I1-2517GHP-704, ZMVI1-2519GHP704 and ZMI1-2521GHP921), the German Research Foundation (DFG, grants GU883/5-1 and GU883/5-2), the Coalition for Epidemic Preparedness Innovations (CEPI) and the Research and Innovation Programme of the European Union under H2020 grant agreement n°871029-EVA-GLOBAL. The BNITM is a member of the German Center for Infection Research (DZIF, partner site Hamburg–Lübeck–Borstel–Riems, Hamburg, Germany) and all works performed in this study have been supported by DZIF.. The European Mobile Laboratory (EMLab), coordinated by the BNITM, is a technical partner of the WHO Global Outbreak Alert and Response Network (GOARN) and the deployment of EMLab to Guinea has been coordinated and supported by the GOARN Operational Support Team at WHO/HQ. P.L. acknowledges support from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 725422 - ReservoirDOCS) and from the Research Foundation -- Flanders (“Fonds voor Wetenschappelijk Onderzoek – Vlaanderen”, G066215N, G0D5117N and G0B9317N). The Artic Network receives funding from the Wellcome Trust through project 206298/Z/17/Z. The work of IPD was supported in part by PraesensBio (Lincoln, NE, USA) and the University Nebraska Medical Center (UNMC). F.R.K., K.I., S.L.M., K.K., T.E.M., and F.M.B.T. operate the “Laboratoire des fièvres hémorragiques virales de Guéckédou”, Guinea, and acknowledge the support by the “Ministère de la Santé et de l’Hygiène Publique”, Guinea, and the “Direction Préfectorale de la Santé” of Guéckédou”.
Contributor Information
Fara R. Koundouno, Bernhard Nocht Institute for Tropical Medicine (BNITM) Hamburg, Germany
Sophie Duraffour, Email: duraffour@bnitm.de, Bernhard Nocht Institute for Tropical Medicine (BNITM) Hamburg, Germany.
N’Faly Magassouba, Laboratoire des Fièvres Hémorragiques Virales en Guinée (LFHVG) Conakry, Guinea.
A complete list of authors is available with the full text of this letter at NEJM.org.:
Fara R. Koundouno, Liana E. Kafetzopoulou, Martin Faye, Annick Renevey, Barré Soropogui, Kékoura Ifono, Emily V. Nelson, Aly A. Kamano, Charles Tolno, Giuditta Annibaldis, Saa L. Millimono, Jacob Camara, Karifa Kourouma, Ahmadou Doré, Tamba E. Millimouno, Fernand M.B. Tolno, Julia Hinzmann, Hugo Soubrier, Mette Hinrichs, Anke Thielebein, Glaucia Herzer, Meike Pahlmann, Georges A. Ki-Zerbo, Pierre Formenty, Anaïs Legand, Michael R. Wiley, Ousmane Faye, Moussa M. Diagne, Amadou A. Sall, Philippe Lemey, Aïssatou Bah, Stephan Günther, Sophie Duraffour, Sakoba Keita, and N’Faly Magassouba
References
- 1.Amman BR, Bird BH, Bakarr IA, et al. Isolation of Angola-like Marburg virus from Egyptian rousette bats from West Africa. Nat Commun. 2020;11:510. doi: 10.1038/s41467-020-14327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brauburger K, Hume AJ, Muhlberger E, Olejnik J. Forty-five years of Marburg virus research. Viruses. 2012;4:1878–927. doi: 10.3390/v4101878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hearn AE, Voorhees MA, Fetterer DP, et al. Serosurveillance of viral pathogens circulating in West Africa. Virol J. 2016;13:163. doi: 10.1186/s12985-016-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baize S, Pannetier D, Oestereich L, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–25. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 5.Keita AK, Koundouno FR, Faye M, et al. Resurgence of Ebola virus in 2021 in Guinea suggests a new paradigm for outbreaks. Nature. 2021 doi: 10.1038/s41586-021-03901-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.