Abstract
Many DNA-binding factors, such as transcription factors, form oligomeric complexes with structural symmetry that bind to palindromic DNA sequences1. Palindromic consensus nucleotide sequences are also found at the genomic integration sites of retroviruses2–6 and other transposable elements7–9, and it has been suggested that this palindromic consensus arises as a consequence of the structural symmetry in the integrase complex2,3. However, we show here that the palindromic consensus sequence is not present in individual integration sites of human T-cell lymphotropic virus type 1 (HTLV-1) and human immunodeficiency virus type 1 (HIV-1), but arises in the population average as a consequence of the existence of a non-palindromic nucleotide motif that occurs in approximately equal proportions on the plus strand and the minus strand of the host genome. We develop a generally applicable algorithm to sort the individual integration site sequences into plus-strand and minus-strand subpopulations, and use this to identify the integration site nucleotide motifs of five retroviruses of different genera: HTLV-1, HIV-1, murine leukaemia virus (MLV), avian sarcoma leucosis virus (ASLV) and prototype foamy virus (PFV). The results reveal a non-palindromic motif that is shared between these retroviruses.
Integration of a cDNA copy of the viral RNA genome is essential to establish infection by retroviruses. This process (see, for example, ref. 10 for a review) is catalysed by the virus-encoded enzyme integrase (IN) and is composed of two steps: (1) the 3’ processing reaction and (2) strand transfer. During the 3’ processing reaction, a di- or tri-nucleotide is removed from the 3’ ends of the viral long terminal repeats (LTRs) to expose the nucleophilic 3’OH groups, which consequently attack the phosphodiester backbone of the target DNA during strand transfer. Strand transfer results in single-stranded DNA gaps that are filled in and repaired by host cellular enzymes. Depending on the retrovirus, the strand transfer reaction takes place with a four (for example, murine leukaemia virus (MLV) and prototype foamy virus (PFV)), five (for example, human immunodeficiency virus type 1 (HIV-1)) or six (for example, human T-cell lymphotropic virus (HTLV-1 and 2)) base pair stagger, giving rise to a duplication of the respective number of nucleotides at the integration site.
Integration is not random. Each retrovirus has characteristic preferences for the genomic integration site (InS) (for example, refs 11–15). These preferences are evident on at least three scales: chromatin conformation and intranuclear location; proximity to specific genomic features such as transcription start sites or transcription factor binding sites; and the primary DNA sequence at the InS itself. Certain host factors also play an active part, the best characterized of which is lens epithelium-derived growth factor16,17, which biases HIV-1 integration into genes in preference to intergenic regions18, and bromodomain and extraterminal proteins, which direct MLV integration into the 5’ end of genes10.
A nucleotide sequence is said to be palindromic if it is equal to its reverse complement (for example, GAATTC and its complement, CTTAAG). Previous studies have revealed a weak palindromic consensus sequence at the InS in several retroviral infections, including HTLV-1, avian sarcoma leucosis virus (ASLV), PFV, MLV, simian immunodeficiency virus (SIV) and HIV-1 (refs 2,3,19–23). The reason for the presence of a palindromic consensus sequence remains unknown, but authors have speculated that it reflects the binding to the DNA of the pre-integration complex (PIC) in symmetrical dimers or tetramers, so that each half-complex has a similar DNA target (that is, potential integration site) preference2. However, the consensus sequence is a population average, defined by taking the modal nucleotide at each position in a population of InS sequences. The question arises whether or not the consensus is truly representative of the population. It may be a poor representation of the population if, for example, the population is highly variable or is composed of two or more distinct subpopulations (and hence is bi- or multimodal). Retroviral InS sequences are known to be highly diverse, which immediately indicates the need for caution when interpreting the consensus. Here, we perform statistical analyses to determine whether or not the palindromic consensus sequences efficiently represent the populations of InS sequences from which they are calculated. We find strong evidence that this is not the case and investigate the possibility that these palindromic consensus sequences arise from the presence of motif sequences that appear in both ‘forward’ and ‘reverse complement’ orientations in the genome.
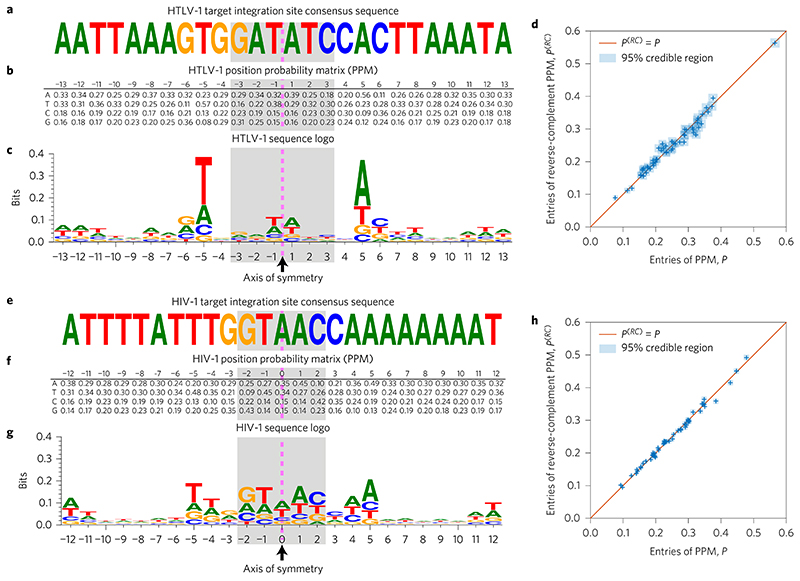
To depict the sequence of the consensus integration site motif, we calculated the frequency of each nucleotide at each respective position in the motif. The result, shown as a sequence logo (Fig. 1), shows a clear palindrome for each virus, as previously described2,3,19. However, on close inspection an anomaly becomes evident: the sequence is palindromic not only in the most frequent nucleotide, but also at the second, third and (therefore) fourth nucleotide at every position. Although it is plausible that the symmetry of the integrase complex should favour a palindromic motif in the nucleotides that make contacts with the integrase protein, it is not clear why the less frequent nucleotides across all positions in the motif should also be perfectly palindromic.
Figure 1. Palindromic HTLV-1 and HIV-1 target integration site consensus sequences and position probability matrices (PPMs), calculated from 4,521 HTLV-1 and 13,442 HIV-1 InS sequences.
a, In agreement with previous studies, we find the HTLV-1 consensus sequence to be a distinctive weak palindrome. The dashed pink line indicates the palindrome’s axis of symmetry, while the shaded area indicates the duplicated region. b, The PPM, P, for the target integration sites is also palindromic; that is, P1,–j ≈ P2,j, P2,–j ≈ P1,j, P3,–j ≈ P4,j and P4-j≈ P3J for j = 1,…,13. Sequence positions to the left of the symmetry line are labelled as negative, and those to the right as positive. c, The symmetry in the PPM may be conveniently visualized using a sequence logo, which also highlights that the palindrome is only weak (has low information content). d, We plot the entries in the first 13 columns of the PPM, P, against the corresponding entries in the reverse-complement PPM, P(RC) (that is, the PPM obtained after first taking the reverse complement of all of the sequences). Uncertainty in the PPM entries is indicated using blue squares showing the 95% credible interval (highest posterior density) range (see Methods). A perfectly palindromic PPM would be one for which P(RC = P, the entries of which would lie along the diagonal shown in the plot. e-h, As in a-d, but using the HIV-1 integration sites.
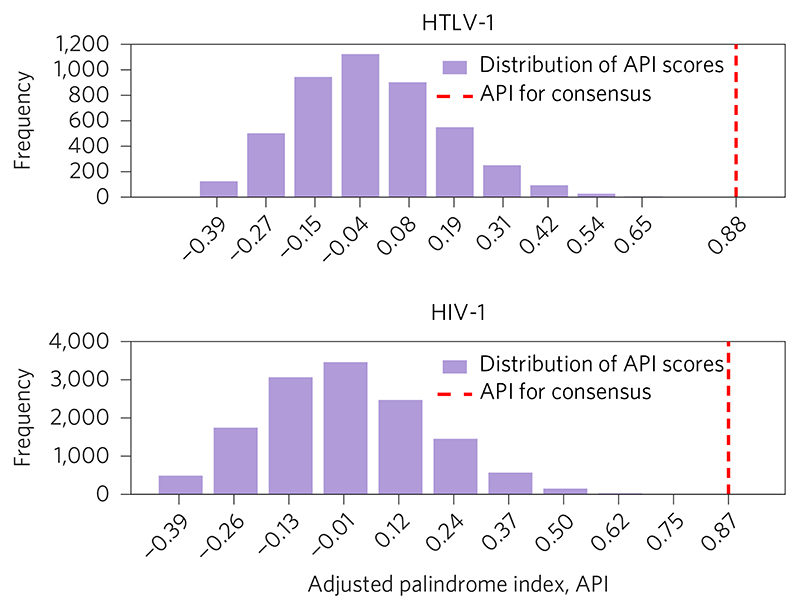
To quantify whether or not an individual sequence is palindromic, we defined the adjusted palindrome index (API), as described further in the Methods. The API is 1 if the sequence is perfectly palindromic, 0 if the sequence is as palindromic as expected by chance, and negative if the sequence is less palindromic than expected by chance. The APIs of the HTLV-1 and HIV-1 motifs confirmed the very high palindromicity of the consensus sequence in each case (Fig. 2). However, examination of the APIs of individual observed integration site sequences reveals a second anomaly: the mean values of the API across the populations of InS sequences are significantly less than zero, for both the HTLV-1 (Table 1) and HIV-1 (Table 2) InS sequences. Although the effect size is small (as might be expected given that the sequences are highly diverse), the key point is that, on average, the InS sequences are less palindromic than we would expect by random chance.
Figure 2. Distribution of adjusted palindrome index (API) scores.
Scores over all 4,521 HTLV-1 integration site sequences (top, taking the sequence length to be 2n = 26, where n is the number of positions each side of the line of palindromic symmetry) and over all 13,442 HIV-1 integration sequences (bottom, with 2n +1 = 25). In both cases, the API for the corresponding consensus sequence (indicated by the red dashed line) is in the extreme positive tail of the distribution.
Table 1. API scores for HTLV-1 integration site sequences.
| 26 | 0.79 | –0.01 | 2.12 ×10–6 |
| 24 | 0.89 | –0.01 | 2.99 ×10–7 |
| 22 | 0.87 | –0.01 | 5.31 × 10–7 |
| 20 | 0.86 | –0.02 | 1.58 × 10–7 |
| 18 | 0.85 | –0.02 | 1.08 × 10–7 |
| 16 | 1 | –0.02 | 2.41 × 10–11 |
| 14 | 1 | –0.03 | 5.00 × 10–15 |
| 12 | 1 | –0.03 | 1.08 × 10–14 |
| 10 | 1 | –0.04 | 1.58 × 10–18 |
| 8 | 1 | –0.03 | 1.15 ×10–14 |
| 6 | 1 | –0.04 | 5.04 × 10–18 |
| 4 | 1 | –0.05 | 1.28 ×10–15 |
| 2 | 1 | –0.08 | 2.83 ×10–21 |
We consider a variety of possible sequence lengths, ranging from 2n = 26 to 2n = 2, where n is the number of positions each side of the line of palindromic symmetry. The mean API values were calculated by finding the API for each of the 4,521 individual InS sequences, and then taking the mean. The final column contains p values resulting from one-sample ř-tests assessing the null hypothesis (H0) that the population mean value is equal to zero.
Table 2. Adjusted palindrome index (API) scores for HIV-1 integration site sequences.
| 25 | 0.88 | –0.01 | 8.21 × 10–9 |
| 23 | 0.87 | –0.01 | 1.60 ×10–8 |
| 21 | 0.86 | –0.01 | 4.29 ×10–9 |
| 19 | 0.85 | –0.01 | 1.29 × 10–11 |
| 17 | 0.83 | –0.01 | 1.08 ×10–12 |
| 15 | 0.8 | –0.02 | 1.04 ×10–13 |
| 13 | 1 | –0.02 | 3.16 ×10–18 |
| 11 | 1 | –0.03 | 1.69 × 10–26 |
| 9 | 1 | –0.03 | 1.02 ×10–27 |
| 7 | 1 | –0.03 | 8.57 × 10–25 |
| 5 | 1 | –0.04 | 1.09 ×10–24 |
| 3 | 1 | –0.07 | 1.95 × 10–35 |
How can a population of individually non-palindromic sequences generate a palindromic consensus motif? We hypothesized that the retroviral integrase complex recognizes a non-palindromic motif present either on the plus strand (‘forward’ orientation) or the minus strand (‘reverse’ orientation) of the host genome: the reverse complement of the minus-strand motif appears as the mirror image of the plus-strand motif, so that when the two are combined in a population of sequences, the consensus appears as a palindrome.
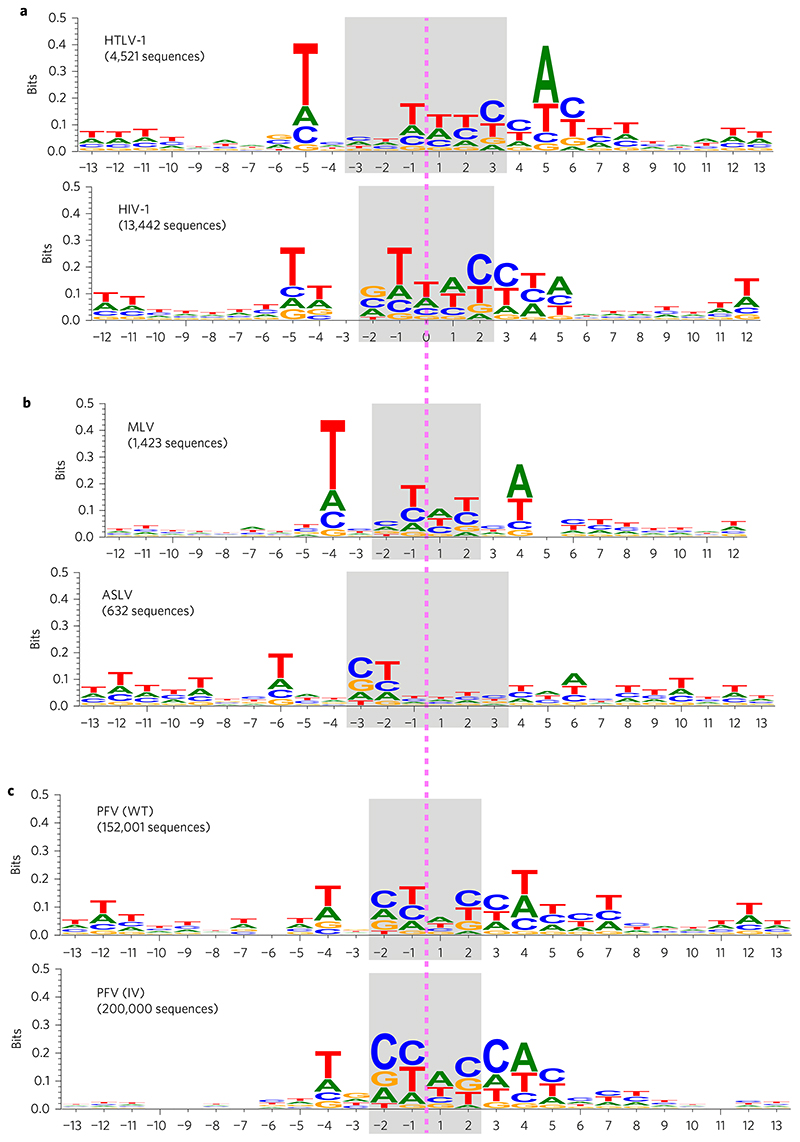
To test this hypothesis, we fitted a model to resolve the population of observed integration sites into two components, one component corresponding to the subpopulation of sequences in the forward orientation and the other corresponding to those in the reverse orientation. We fitted the model by maximum likelihood (see Methods for details of the model and fitting procedure, and the ‘Code availability’ section for an implementation). We additionally considered a number of alternative algorithms for fitting the models (maximum profile likelihood and Gibbs sampling approaches), which provided qualitatively identical results (Supplementary Fig. 1). For both HTLV-1 and HIV-1, the algorithms identified complementary subpopulations within the collections of InS sequences (Fig. 3a), with the subpopulations appearing in approximately equal proportions (λHTLV = 0.47 and λHIV = 0.49, where λ denotes the proportion of sequences in the ‘forward orientation’). As a further check, we also considered an unconstrained clustering of the sequences, which also identified complementary clusters among the InS sequences (Supplementary Figs 2 and 3).
Figure 3. Summary of results from fitting the two-component mixture model by maximum likelihood.
a, Sequence logo summaries of one of the two subpopulations of integration site sequences in the HTLV-1 and HIV-1 data sets (in each case, the other subpopulation is characterized by the reverse complement of the sequence logo shown). b, As in a, but for the MLV and ASLV data sets. c, As in a, but for the PFV (WT) and PFV (IV) data sets.
We next assessed whether the hypothesis of two complementary subpopulations provided a significantly better description of the data than the hypothesis of a single population characterized by a palindromic motif. A likelihood ratio test (see Methods) decisively rejected the single-population hypothesis (p < 0.001). We also calculated for each model the Bayesian information criterion24 (BIC), which provides a measure of the ability of a model to explain the observed data. The results again showed that for both HIV-1 and HTLV-1 there was very strong evidence against the one-population (palindromic) model (ΔBICHIV = 2.86 × 103 and ΔBICHTLV = 1.48 × 103).
We fitted our two-component mixture model to smaller data sets on HTLV-1, HIV-1, MLV and ASLV taken from the literature19. The results on MLV and ASLV are given in Fig. 3b. The results for HTLV-1 and HIV-1 are qualitatively identical to those obtained from the larger data sets and are given in Supplementary Fig. 4. We also considered two large PFV data sets from Maskell et al.25: (1) the PFV (WT) data set, which comprises integration sites for 153,447 unique integration events in HT1080 cells and (2) the PFV (IV) data set, comprising ~2 × 106 integration sites determined using purified PFV intasomes and deproteinized human DNA.
After pre-processing to remove duplicates and sequences containing indeterminate nucleotides (Ns), 152,001 integration sites remained in the PFV (WT) data set and 2,197,613 in the PFV (IV) data set. To reduce computation time, we randomly sampled 200,000 integration site sequences from the PFV (IV) data set to use for analysis. The results on PFV (WT) and PFV (IV) are given in Fig. 3c. The results obtained for all retroviruses reveal similarities between the non-palindromic motifs.
The factors that influence the pattern of integration of retroviruses and transposable elements operate at different physical scales. The strength of association between specific genomic features and retroviral integration frequency depends on the genomic scale on which the data are analysed20,26. Broadly, three scales have been studied: chromosome domains and euchromatin/heterochromatin; genomic features such as histone modifications and transcription factor binding sites; and primary DNA sequence.
The primary DNA sequence of the host genome is thought to influence the site of retroviral integration by determining both the binding affinity of the intasome and the physical characteristics of the target DNA, especially the ability of the double helix to bend7,27, which depends in turn on the presence of specific dinucleotides and trinucleotides. Müller and Varmus28 concluded that the bendability of DNA could explain the preferential integration of certain retroviruses in DNA associated with nucleosomes. The requirement for DNA bending during retroviral integration has been explained by the discovery of the crystal structure of the foamy viral intasome complexed with target DNA29,30. Complete unstacking of the central dinucleotide at the site of integration allows the scissile phosphodiester backbone to reach the active sites of the IN protomers. Although the bending of the tDNA observed in the crystal structure does not correspond with the bend described in nucleosomal DNA31, the cryo-electron microscopy structure of the foamy viral intasome in complex with mononucleosomes25 showed that the nucleosomal DNA is lifted from the histone octamer to allow proper accommodation within the active sites of the IN protomers. Given that integration catalysed by different retroviral INs gives rise to a different target duplication size, it is expected that DNA bending at the site of integration will be more severe for integrations with a 4 bp target duplication compared to those with a 6 bp target duplication29.
Whereas some retroviruses preferentially integrate into regions of dense nucleosome packing (for example, PFV and MLV)25, others prefer regions of sparse nucleosome packing (for example, HIV and ASV32). However, even in cases where nucleosome sparseness is preferred, a nucleosome at the integration site itself contributes to efficient integration.
In addition to the impact of specific dinucleotides and trinucleotides on DNA bendability, the other chief impact of primary DNA sequence on retroviral integration is the presence of a primary DNA motif, that is, preferred nucleotides at specific positions in relation to the integration site. Palindromic DNA sequences have been reported at the insertion site of transposable elements in Drosophila7, yeast8,9 and retroviruses2–6,19. The presence of the palindrome has been attributed by several workers to the symmetry of the multimeric viral pre-integration complex2,3. However, Liao et al.7 noted that, although the palindromic pattern that they observed at the insertion site of a P transposable element in Drosophila could be discerned when as few as fifty insertion sites were aligned and averaged, the palindrome was not evident at the level of a single insertion site.
It was previously assumed that the non-appearance of the palindromic nucleotide sequence in individual retroviral integration sites was due to the fact that the palindrome was weak, that is, poorly conserved. However, in the present study we found evidence that the palindrome was statistically significantly disfavoured at the level of individual sites: the palindrome is evident only as an average—a consensus—of the population of integration sites. We propose that the most likely explanation is that the palindrome results from a mixture of sequences that contain a non-palindromic nucleotide motif in approximately equal proportions on the plus strand and minus strand of the genome. In fact, while the integrase components of the in vitro purified intasome form a highly symmetrical structure, within the in vivo pre-integration complex, which also includes other viral and host proteins, a degree of asymmetry is imposed by the presence of the retroviral DNA. This asymmetry may be sufficient to favour a non-palindromic sequence at the integration site.
On the hypothesis of a non-palindromic nucleotide motif in approximately equal proportions on the plus strand and minus strand of the genome, we sorted the populations of sequences of several different retroviral integration sites into those with a conserved motif respectively on the plus and minus strand of the genome. The resulting alignment revealed the putative true nucleotide motif that is recognized by the intasome in each case. Comparison of these motifs among the respective viruses showed certain similarities between the sequences (Fig. 3), including two T residues upstream of the integration site and an A residue two or three nucleotides downstream. There is a shared motif 5’-T(N1/2) [C(N0/1)T|(W1/2)C]CW-3’, where the square brackets ([and]) represent the start and end of the duplicated region, W denotes A or T, and | represents the axis of symmetry. The preference for an A (T) two or three nucleotides downstream (upstream) of the integration site was previously observed and was explained by a direct contact between A and the residue at the PFV IN Ala188 equivalent position29,30,33. Indeed, the recent X-ray structure of the poststrand-transfer complex of the alpharetrovirus Rous sarcoma virus (RSV) IN illustrates a direct contact with an A (T) three nucleotides downstream (upstream) of the integration site and the homologous Ser124 residue site34. Using the same algorithm on InS sequences generated with HIV-1 IN Ser119Thr (equivalent to PFV IN Ala188)33 the shared motif is preserved (Supplementary Fig. 5), with a stronger preference for an A(T) three nucleotides downstream (upstream) of the InS. It remains to be seen whether the nucleotide composition of the remainder of the shared motif, in particular the central T-rich region, is preferred because of the flexibility of the DNA at such sequences, or is due to direct contact between IN and the bases. Further structural information on lenti-, gamma- and delta-retroviral synaptic complexes is needed to answer this question.
To summarize, we conclude that, in contrast to the palindromic sequence motifs that are bound by many transcription factors, the primary DNA motif recognized by the retroviral intasome is non-palindromic.
Methods
Mapped integration sites
To focus on the initial integration targeting profile of HTLV-1 and HIV-1, integration sites were identified in DNA purified from cells infected experimentally in vitro. Jurkat T cells were infected either by short co-culture with HTLV-1-producing cell line MT2 (ref. 35) or by VSV-G pseudotyped HIV-1 (gift from A. Fassati, UCL). The identification of4,521 HTLV-1 integration sites from in vitro infected Jurkat T cells has been described before15,36. The identification of 13,442 HIV-1 integration sites was carried out using a similar approach, using the following HIV-specific PCR forward primers: HIVB3 5’-GCTTGCCTTGAGTGCTTCAAGTAGTGTG-3’, HIVP5B5 5’-AATGATACGGCGACCACCGAGATCTACACGTGCCCGTCTGTTGTGTGAC TCTGG-3’ and HIV-specific sequencing primer 5’-ATCCCTCAGACCCTTTT AGTCAGTGTGGAAAATCTC-3’.
Credible intervals for entries of the position probability matrices (PPMs)
To obtain the credible intervals given in Fig. 1d,h, we regard the elements of the PPM as parameters, which we then infer using Bayesian methods. Let pX,k denote the probability that nucleotide X ∈ {A, T, C, G} is observed in position k, and define nx,k to be the number of times X was observed in position k. For column k of the PPM, which we denote pk = [pA,k pT,k PC,k pG,k], we know that each pX,k ≥ 0 and that Σx∈{A,T,C,G}pXk = 1, so a Dirichlet prior is appropriate. We take a symmetric Dirichlet prior with a = 1 (which is equivalent to a uniform prior). Assuming [nA,k nT,k nC,k nG,k] are jointly distributed according to a multinomial distribution with nTOTAL = ΣX∈ {A,T,C,G} nX,k trials and probabilities [pA,k pT,k pC,k pG,k], it can be shown that the marginal posterior distributions for the entries of column k of the PPM are pX,k ~ Beta(1 + nX,k, 4 + nTOTAL – (1 + nX,k)). Using these, we find 95% highest posterior density (HPD) regions using the betaHPD function from the pscl package37 in the R statistical programming language38.
API
We define the palindrome index (PI) for a sequence to be the proportion of positions at which it is equal to its reverse complement. For example, the PI for the sequence s = ATCCGGTT is 0.75, because the reverse complement sequence is s’ = AACCGGAT, and s and s’ are identical at six of the eight positions (6/8 = 0.75). For sequences of odd length, we first remove the central letter. Hence sequences may be assumed to be of even length. The API is a ‘corrected for chance’ version of the PI, which controls for the fact that the expected value of the PI depends on the length of the sequence. Such adjusted indexes are common (for example, ref. 39) and are calculated as Adjusted Index = (Observed Index – Expected Index)/(Maximum Index – Expected Index). For the PI, the maximum value is 1 (when a sequence is perfectly palindromic). Given sequence s = σ-n...σ-1σ+1...σ+n, the expected value for the PI is the expectation when σ+j and σ- are independent, which is given by
Here c(X) denotes the complement of X and p(σ±j = X) are the empirical marginal probabilities, which may be taken from the entries of the PPM.
Two-component mixture model
We model the InS sequences as being drawn from a two-component mixture model, p(s|P,λ) = λf(s|P) + (1 – λ)f(s|P(RC)), where f(s|P) is the likelihood of sequence s given PPM P, and P(RC) denotes the reverse complement of PPM P (which follows automatically from P by reversing the order of the columns, and swapping the A and T rows with one another and the C and G rows with one another). We define the likelihood straightforwardly as the product of probabilities of each of the elements of s, where the individual probabilities are given by the entries of the PPM. To fit the model, we must estimate parameters λ and P. We find the maximum likelihood estimates of these parameters using the expectation maximization algorithm.
Expectation-maximization (EM) algorithm for our model
We refer the reader to ref. 40 for general information about the EM algorithm, and here provide the update equations for the model parameters, λ and P. Suppose we have a collection of N InS sequences, s(1),...,s(N). At iteration t, define to be the posterior probability of sequence s(i) belonging to the subpopulation with PPM P, given λt–1 and Pt–1 (the parameter estimates at iteration t–1). That is, . Also, for X ∈ {A, T, C, G} and k = 1,.,.,n (or k = 0,.,.,n in the odd palindrome case), we define . Then , and defining the element of Pt in column k and row labelled by nucleotide X to be Pt,(k,X), we have Pt(k, X) = (Qt(k, X))/(Σx∈ {ATCG} Qt(k, X)).
Initialization and stopping criteria for the EM algorithm
We initialize the EM algorithm by setting the initial PPM, P0, to be the original (palindromic) PPM and setting the initial mixture weight, λ0, to be 0.5. At iteration t, we calculate the loglikelihood associated with the full data set using the current parameter estimates, . We terminate the algorithm when lt+1 – lt < τ, for some preset threshold value τ. To obtain the results shown in Fig. 3, we set τ = 1 × 10–10. To reduce run times when finding the null distribution of the likelihood ratio test (LRT) statistic, we set τ = 0.1, because it was necessary to run the algorithm a large number of times.
Likelihood ratio tests for quality of fit
Although it is tempting to apply a simple LRT to determine if the unconstrained two-component mixture model provides a significantly better fit to the data than the constrained, single-component palindromic model (in which P = P(RC), it is well known that for mixture models the LRT statistic does not in general follow standard χ2 distributions41. We therefore adopted McLachlan’s approach42 to construct an empirical null distribution for the LRT statistic, D. Note that the null model here is a single component with PPM equal to the empirical PPM (given in Fig. 1b for HTLV-1 and Fig. 1f for HIV-1), while the alternative is the fitted two-component mixture model. Briefly, we simulated 1,000 new data sets using the null model, fitted both the null and alternative models to each simulated data set and calculated the LRT statistic each time. In this way, we obtained empirical null distributions for the LRT statistic, which we then used to assess the significance of the observed LRT statistic. For the HTLV-1 InS sequences, the 1,000 values sampled from the null distribution of the LRT statistic all fell between –28.64 and 18.79, while the observed LRT statistic was 1.49 × 103. For the HIV-1 InS sequences, the sampled LRT statistics all fell between –32.37 and 29.24, while the observed LRT statistic was 2.86 × 103. For both the HTLV-1 and HIV-1 data sets we may clearly reject the null model in favour of the alternative model (p <0.001).
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust UK (Senior Investigator Award 100291 to C.R.M.B.; Investigator Award 107005 to G.N.M.) and the MRC (project reference MC_UP_0801/1). The authors thank the following individuals for providing materials: A. Zhyvoloup and A. Fassati (Division of Infection and Immunity, University College London) and H. Niederer (Division of Infectious Diseases, Imperial College London). The authors also thank L. Game and M. Dore at the Medical Research Council Clinical Sciences Centre Genomics Laboratory at Hammersmith Hospital, London, UK.
Footnotes
Author contributions
P.K. and C.B. conceived the project. A.M. and G.M. performed the experiments. P.K. and M.H. performed the statistical analysis and modelling. P.K. and C.B. co-wrote the paper.
Competing interests
The authors declare no competing financial interests.
Supplementary information is available for this paper.
Reprints and permissions information is available atwww.nature.com/reprints.
Data availability
The data used to reproduce the results on HTLV-1 presented in this study are included with the code (see section ‘Code availability’). All other data that support the findings of this study are available from the corresponding author upon request.
Code availability
Code is available from http://www.mrc-bsu.cam.ac.uk/software/bioinformatics-and-statistical-genomics/.
References
- 1.Pabo CO, Sauer RT. Protein–DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grandgenett DP. Symmetrical recognition of cellular DNA target sequences during retroviral integration. Proc Natl Acad Sci USA. 2005;102:5903–5904. doi: 10.1073/pnas.0502045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowrouzi A, et al. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- 6.Meekings KN, Leipzig J, Bushman FD, Taylor GP, Bangham CRM. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PLoS Pathog. 2008;4:e1000027. doi: 10.1371/journal.ppat.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao G-c, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ, Craig NL. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc Natl Acad Sci USA. 2010;107:21966–21972. doi: 10.1073/pnas.1016382107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee AG, et al. Serial number tagging reveals a prominent sequence preference of retrotransposon integration. Nucleic Acids Res. 2014;42:8449–8460. doi: 10.1093/nar/gku534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA integration. Chem Rev. 2016;116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schröder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narezkina A, et al. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melamed A, et al. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013;9:e1003271. doi: 10.1371/journal.ppat.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherepanov P, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 17.Maertens G, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting ofHIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 18.Shun M-C, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derse D, et al. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carteau S, Hoffmann C, Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens SW, Griffith JD. Sequence analysis of the human DNA flankingsites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 25.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong J, et al. Chromatin landscapes of retroviral and transposon integration profiles. PLoS Genet. 2014;10:e1004250. doi: 10.1371/journal.pgen.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 28.Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrao E, Ballandras-Colas A, Cherepanov P, Maertens GN, Engelman AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tachiwana H, et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci USA. 2010;107:10454–10459. doi: 10.1073/pnas.1003064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benleulmi MS, et al. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology. 2015;12:13. doi: 10.1186/s12977-015-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrao E, et al. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014;42:5164–5176. doi: 10.1093/nar/gku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Z, et al. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362–366. doi: 10.1038/nature16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyoshi I, et al. A novel T-cell line derived from adult T-cell leukemia. Gan. 1980;71:155–156. [PubMed] [Google Scholar]
- 36.Gillet NA, et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackman S. pscl: Classes and Methods for R Developed in the Political Science Computational Laboratory. Stanford Univ; 2015. [Google Scholar]
- 38.R Core Team. R A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2014. http://www.R-project.org/ [Google Scholar]
- 39.Kuncheva L. A stability index for feature selection; Proceedings of the 25th International Multi-Conference on Artificial Intelligence and Applications; 2007. pp. 390–395. [Google Scholar]
- 40.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J Roy Stat Soc B Met. 1977;39:1–38. [Google Scholar]
- 41.Aitkin M, Rubin DB. Estimation and hypothesis testing in finite mixture models. J Roy Stat Soc B Met. 1985;47:67–75. [Google Scholar]
- 42.McLachlan GJ. On bootstrapping the likelihood ratio test stastistic for the number of components in a normal mixture. J Roy Stat Soc C Appl Stat. 1987;36:318–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to reproduce the results on HTLV-1 presented in this study are included with the code (see section ‘Code availability’). All other data that support the findings of this study are available from the corresponding author upon request.
Code is available from http://www.mrc-bsu.cam.ac.uk/software/bioinformatics-and-statistical-genomics/.