Abstract
Endocytosis controls the perception of stimuli by modulating protein abundance at the plasma membrane. In plants, clathrin-mediated endocytosis is the most prominent internalization pathway and relies on two multimeric adaptor complexes, the AP-2 and the TPLATE complex (TPC). Ubiquitination is a well-established modification triggering endocytosis of cargo proteins, but how this modification is recognized to initiate the endocytic event remains elusive. Here, we show that TASH3, one of the large subunits of TPC, recognizes ubiquitinated cargo at the plasma membrane via its SH3 domain-containing appendage. TASH3 lacking this evolutionary specific appendage modification allows TPC formation, but the plants show severely reduced endocytic densities, which correlates with reduced endocytic flux. Moreover, comparative plasma membrane proteomics identified differential accumulation of multiple ubiquitinated cargo proteins for which we confirm altered trafficking. Our findings position TPC as a key player for ubiquitinated cargo internalization, allowing future identification of target proteins under specific stress conditions.
Introduction
Plant cells are outlined by their cell wall and plasma membrane (PM). The PM lipid bilayer houses surfeit of integral membrane proteins, which are essential for gathering information about the cell’s surroundings. To adapt to the constantly changing environment, cells need to rapidly refresh and adjust their PM proteome. Exocytosis deposits proteins at PM, whereas endocytosis is responsible for vesicle-mediated internalization of PM material and extracellular ligands (together termed cargo). Clathrin-mediated endocytosis (CME), characterized by the involvement of the scaffolding and cage-forming protein clathrin, is the best characterized and most prominent endocytic pathway1–3.
The initiation of endocytosis in plants is rather poorly understood, however it presumably starts with the arrival of adaptor protein complexes to cargo and lipid enriched spots at PM. In plants, two adaptor protein complexes function in CME, the heterotetrameric Adaptor Protein-2 complex (AP-2) and the octameric TPLATE complex (TPC). Six TPC components are evolutionary conserved: TPLATE, TML, TASH3, LOLITA, TWD40-1 and TWD40-2, and two subunits are plant-specific: AtEH1/Pan1 and AtEH2/Pan14,5. TPC is essential during pollen and somatic plant development4,6. This is also the case for the TWD40-2 subunit, yet besides null alleles causing male sterility (twd40-2-1 and twd40-2-2), a viable knockdown mutant (twd40-2-3) was also identified4,7.
During the initial steps of endocytosis, adaptor complexes recognize cargo via specific short linear motifs present in their cytoplasmic domains. In animals, the tyrosine motif (YXXΦ) and dileucine motif ([D/E]xxxL[L/I]) present in the PM cargo are well-studied AP-2 cargo recognition motifs8,9. Up to date, there is no direct evidence linking dileucine motifs with internalization of PM cargo via AP-2 in Arabidopsis10. Tyrosine motifs have been implicated in endocytosis of BRASSINOSTEROID INSENSITIVE 1 (BRI1), BORON TRANSPORTER 1 (BOR1) or PIN-FORMED 1 (PIN1)10–13. However, there are no examples in which tyrosine motifs were shown to be essential for internalization14,15. Recently, the double NPF motif of Secretory Carrier Membrane Protein 5 (SCAMP5) was reported to be recognized by the AtEH1/Pan1 TPC subunit and shown to be important for its internalization16.
Besides linear motifs, reversible post-translational modifications also function in endosomal trafficking of transmembrane proteins. Phosphorylation of NOD26-LIKE INTRINSIC PROTEIN 5;1 (NIP5;1), LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE 5 (LYK5) or FLAGELLIN-SENSITIVE 2 (FLS2) was shown to be a prerequisite for their internalization17–19. Within the last couple of years, ubiquitin became known as an essential mark for PM protein internalization from the cell surface and consequent degradation in the vacuole20,21. Mono-, di- or K63-linked poly-ubiquitin chains, covalently linked to the lysine residues of transmembrane proteins, drive the endosomal sorting of PM cargoes to the trans-Golgi network22. There, quality control enables vacuolar sorting of ubiquitinated PM proteins, while de-ubiquitinated cargo recycles back to PM21,23,24. A plethora of transmembrane proteins such as IRON-REGULATED TRANSPORTER 1 (IRT1), FLS2, LYK5, BRI1, PIN2 or BOR1 have been shown to undergo ubiquitin-dependent internalization14,25–31. Among the endocytic machinery, SH3P2, a member of the family of SH3 DOMAIN-CONTAINING PROTEIN (SH3P), and TARGET OF MYB 1-LIKE (TOL) proteins, of which some are located at PM and interact with TPC, bind ubiquitin23,32–34. Mechanistic insight as to how ubiquitinated cargo is recognized at PM and initiates CME remains however elusive.
Here we identified nosh (no SH3), a viable TPC mutant that produces a truncated variant of the large TASH3 subunit lacking the C-terminal SH3 domain. This truncation does not prevent complex formation, yet it severely compromises endocytosis. The SH3 domain of TASH3 binds ubiquitin, which consequently leads to an accumulation of ubiquitinated PM proteins in nosh, a feature which is less apparent in other endocytic mutants. Comparative PM proteomics identified multiple potential PM cargo proteins that differentially accumulate in nosh, including the TRANSMEMBRANE KINASE 1 (TMK1), for which we confirmed reduced internalization in nosh. Taken together, we provide evidence that TPC functions in recognizing and internalizing ubiquitinated cargo proteins via its evolutionary unique SH3-containing appendage domain on TASH3.
Results
A TASH3 mutant, lacking the SH3 domain, is viable
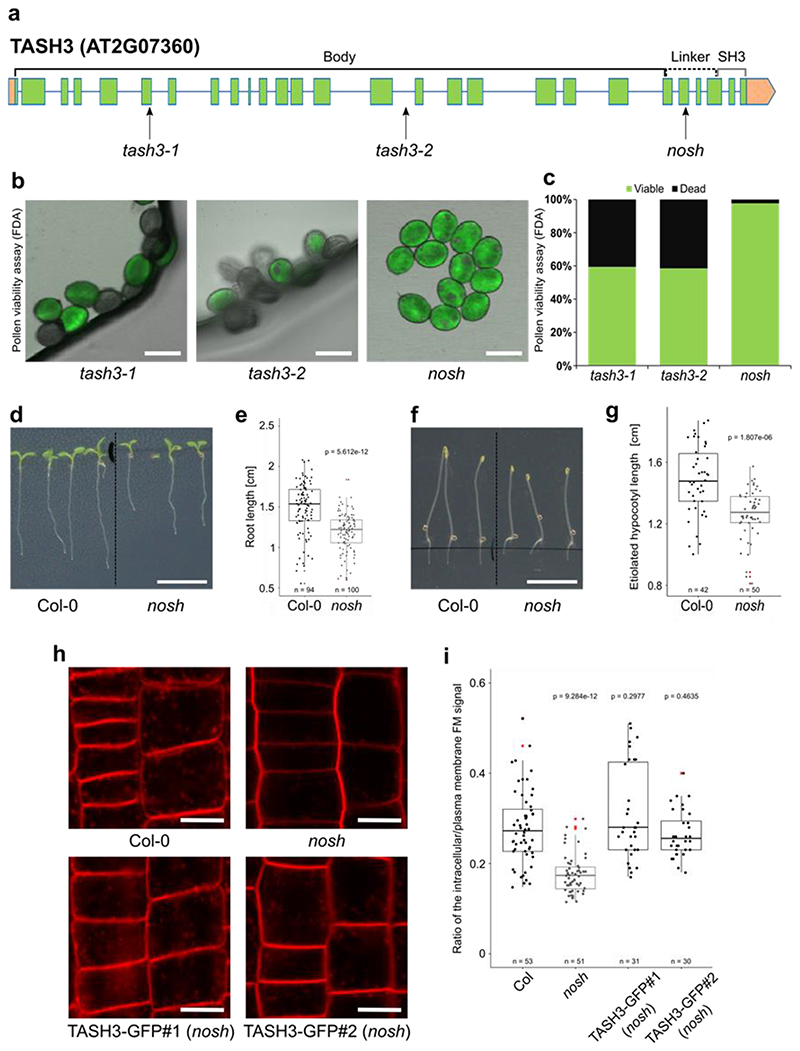
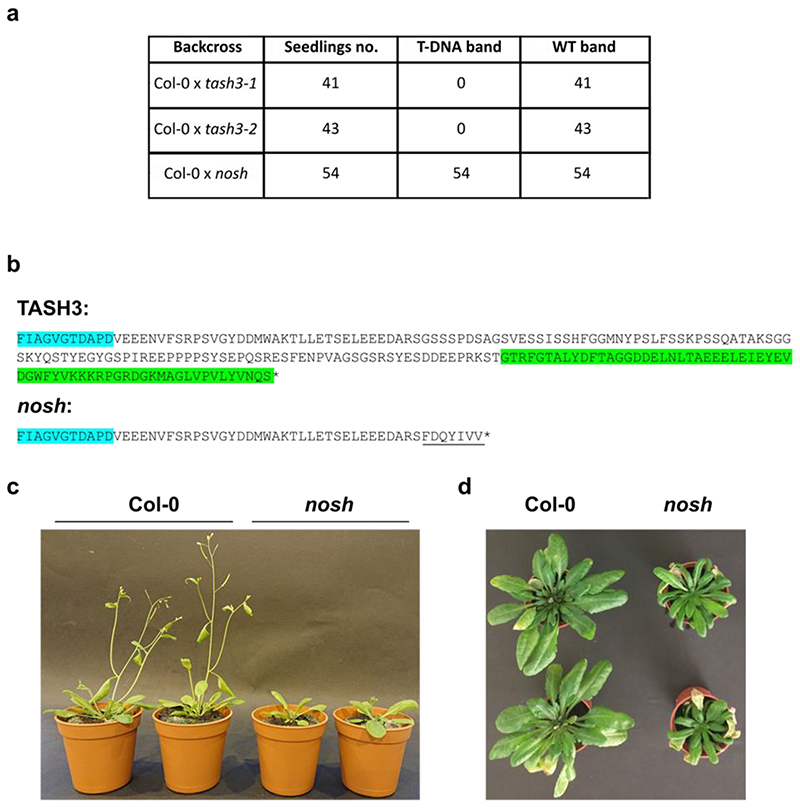
Knock-out mutants of any TPC subunit identified so far exhibit male gametophytic lethality, which is inhibiting the use of standard loss-of-function analysis4,6. Only two viable TPC subunit mutants were reported up to date: twd40-2-3, which reduces the amount of full-length protein present7 and WDXM2, a TPLATE isoform that reduces complex stability at room temperature and abolishes TPC function upon prolonged heat-shock35. TASH3 is one of the two large core TPC subunits whose role and function remains unexplored. In contrast to all large adaptor subunits of the heterotetrameric adaptor complexes, including TPLATE, which all carry a bilobal appendage domain composed of an N-terminal sandwich domain connected to a C-terminal platform domain, TASH3 has evolutionarily acquired an SH3 domain36. Similar to other TPC mutants4,6,37, available T-DNA insertion lines in TASH3, tash3-1 and tash3-2, harboring the T-DNA at the beginning or the middle part of the gene, respectively, caused gametophytic lethality. Both mutants exhibited more than 40% of fluorescein diacetate negative pollen and back-cross experiments failed to transfer the T-DNA to the next generation (Fig. 1a-c and Extended Data Fig. 1a).
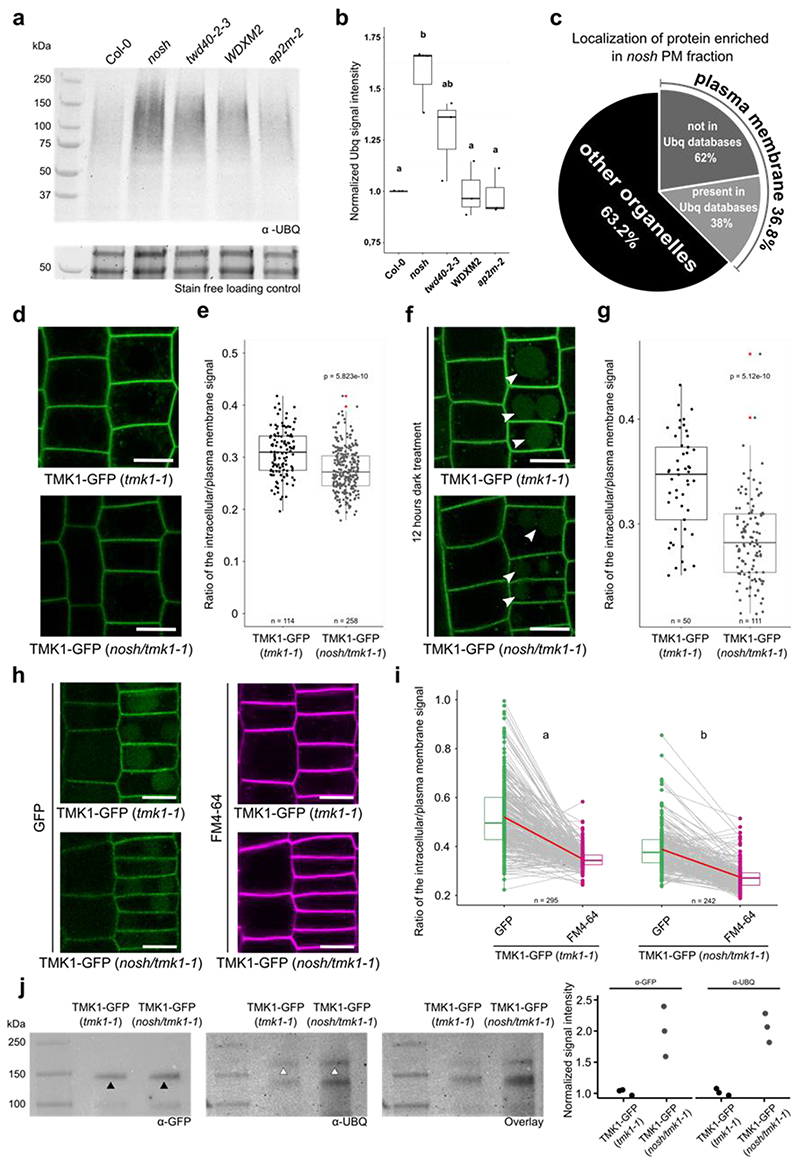
Figure 1. The viable nosh mutant in the TPC subunit TASH3 has reduced endocytic capacity.
a) TASH3 gene structure. Exons are represented by green squares, introns by blue lines and UTRs are in orange. Insertion sites of T-DNA alleles identified in this study (tash3-1, tash3-2 and nosh) are indicated by arrows. The structural domains of the protein (body, linker and SH3 domain) are depicted above the gene model. b) Pollen viability assay of tash3-1, tash3-2 and nosh. Pollen was stained with fluorescein diacetate (FDA) and visualized by confocal microscopy. Scale bar = 20 μm. c) Quantification of the percentage viable and dead pollen from tash3-1 (n=143), tash3-2 (n=41) and nosh (n=43) based on FDA staining. d-e) Representative images and corresponding root length box plot graph of 5-day-old Col-0 and nosh seedlings grown on ½ MS media without sucrose showing that nosh mutants exhibit slower root growth. Scale bar = 1 cm. f-g) Representative images and corresponding hypocotyl length box plot graph of 4-day-old seedlings of Col-0 and nosh grown on ½ MS media without sucrose in the dark showing that nosh mutant exhibit shorter etiolated hypocotyl length. Scale bar = 1 cm. h) Representative single confocal slices of FM4-64 stained root cells as a proxy for endocytic flux evaluation of Col-0, nosh and two independent complemented mutant lines (TASH3-GFP#1 and #2 in nosh). Scale bar = 10 μm. i) Box plot graph of the intracellular/PM intensity of FM4-64 in Col-0, nosh and two independent complemented nosh lines. Complementation with TASH3-GFP restores the endocytic capacity of the nosh mutant. e, g, i) The top and bottom lines of the box plot represent the 25th and 75th percentiles, the center line is the median and whiskers are the full data range. Red asterisks mark the outliers in the box plots. Numbers of quantified cells are indicated at the bottom of each graph. The indicated p-values were calculated using the two-sided Wilcoxon-signed rank test by comparing mutants to wild type.
In contrast, nosh allowed transfer of the T-DNA via the pollen (Fig. 1a-c and Extended Data Fig. 1a) and the isolation of homozygous mutant plants. The T-DNA in nosh is located C-terminally, in the linker before the SH3 domain (Extended Data Fig. 1b). nosh seedlings exhibited significantly reduced root and hypocotyl length compared to wild type (Fig. 1d-g). Adult nosh plants exhibited smaller rosette leaves, delayed flowering time and early senescence (Extended Data Fig. 1c,d). Uptake of the styryl dye FM4-64, a proxy for endocytic flux, revealed a significant reduction in internalization in nosh compared to wild type (Fig. 1h,i). Introducing full-length TASH3-GFP into nosh restored FM internalization to wild type levels for two independent complemented mutant lines (Fig. 1h,i), linking the T-DNA insertion with the reduced endocytic capacity. Given the gametophytic lethality of the other null alleles, nosh, therefore, likely produces a truncated variant of TASH3 lacking the C-terminal SH3 domain. Lack of this SH3 domain is not essential for plant life, but causes developmental delay and affects CME capacity.
Overall CME dynamics are delayed in nosh
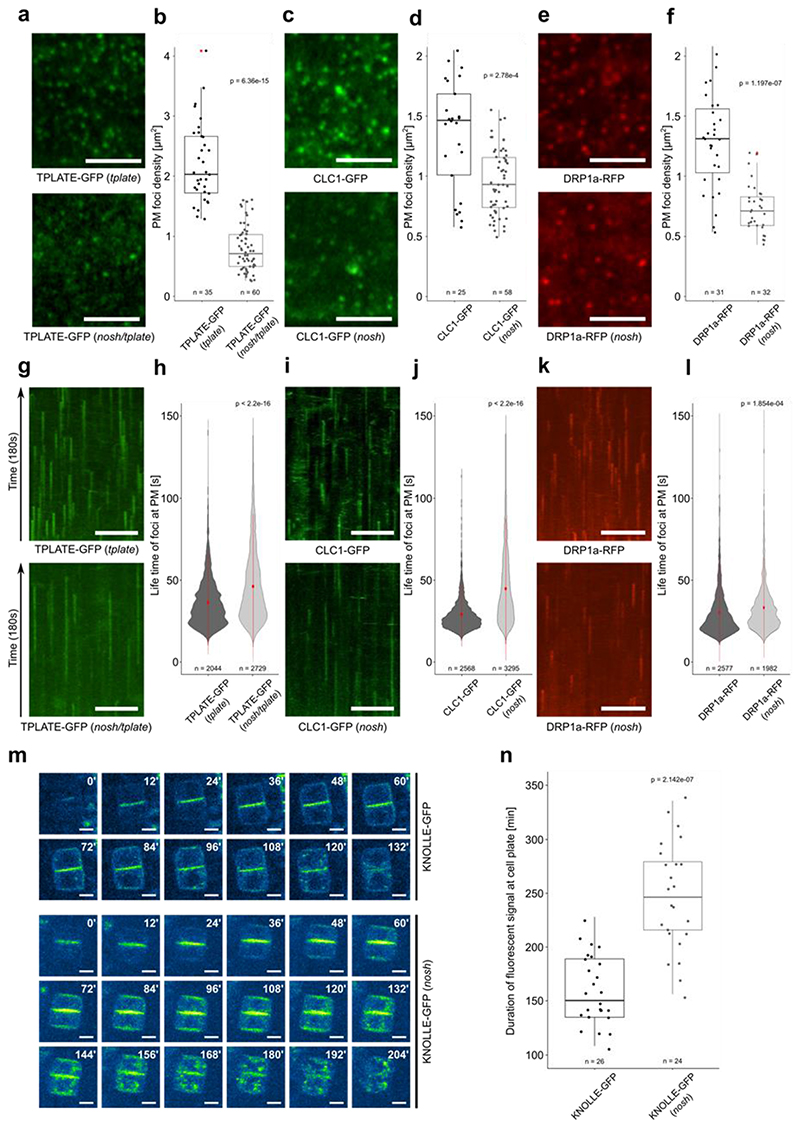
To properly assess CME in nosh, we crossed TPLATE-GFP, CLATHRIN LIGHT CHAIN 1 (CLC1)-GFP and DYNAMIN-RELATED PROTEIN 1A (DRP1a)-RFP into nosh and evaluated their dynamic behavior at PM. All three endocytic markers revealed significantly lower densities of endocytic foci in nosh compared to control etiolated hypocotyl cells (Fig. 2a-f). Kymograph analysis revealed significantly prolonged life-times of endocytic events at PM in nosh compared to control cells (Fig. 2 g-l). This was most pronounced for TPLATE and CLC1 (Fig. 2g-j), and to a lesser extent for DRP1a (Fig. 2k,l).
Figure 2. nosh is defective in endocytosis.
a-f) Representative single slice spinning disk images and box plot quantifications of TPLATE-GFP, CLC1-GFP and DRP1a-RFP densities in dark grown hypocotyl epidermal cells of control (top row) and nosh (bottom row) backgrounds. For each marker, the endocytic foci density is significantly reduced in nosh. Numbers of quantified cells (with 2 cells analyzed per individual seedling) are indicated. Red asterisks mark the outliers. Scale bar = 5 μm. g-l) Representative kymographs and violin plot graphs of spinning disk time lapses (from a, c, e) showing increased average life-time at PM for TPLATE-GFP, CLC1-GFP and DRP1a-RFP in nosh compared to the controls. The number of events analyzed for each independent line is indicated at the bottom of each graph. At least 12 movies from 6 seedlings were imaged and analyzed for each independent transgenic line. Red circles represent the mean and the red line represents the standard deviation. Scale bar = 50 μm. m) Single frame images of a root tracking time lapse of KNOLLE-GFP in Col-0 and nosh. Numbers in the upper right corner represent the relative time in minutes. KNOLLE-GFP displayed prolonged cell plate localization and overall cellular presence post cytokinesis in nosh. Scale bar = 5 μm. n) Box plot graph showing the duration of cell plate localization of KNOLLE-GFP. Numbers of quantified cells from independent seedlings (≥ 6 cells per seedling) are indicated. The top and bottom lines of the box plots represent the 25th and 75th percentiles, the center line is the median and whiskers are the full data range. The widest part of the violin plot represents the highest point density, whereas the top and bottom are the maximum and minimum data respectively. The indicated p-values (panels b, d, f, h, j, l and n) were calculated using the two-sided Wilcoxon-signed rank test by comparing mutant to control.
The cytokinetic syntaxin KNOLLE is specifically degraded by CME following cell plate maturation38,39. Time lapse imaging of growing root tips monitored the time that KNOLLE-GFP was present in the dividing cells. The presence of KNOLLE-GFP at the fully formed cell plate in nosh was strongly prolonged compared to wild type (Fig. 2m,n). The lower densities and prolonged life-times of endocytic foci in nosh therefore correlate with a significant delay in the removal and degradation of endocytic cargo.
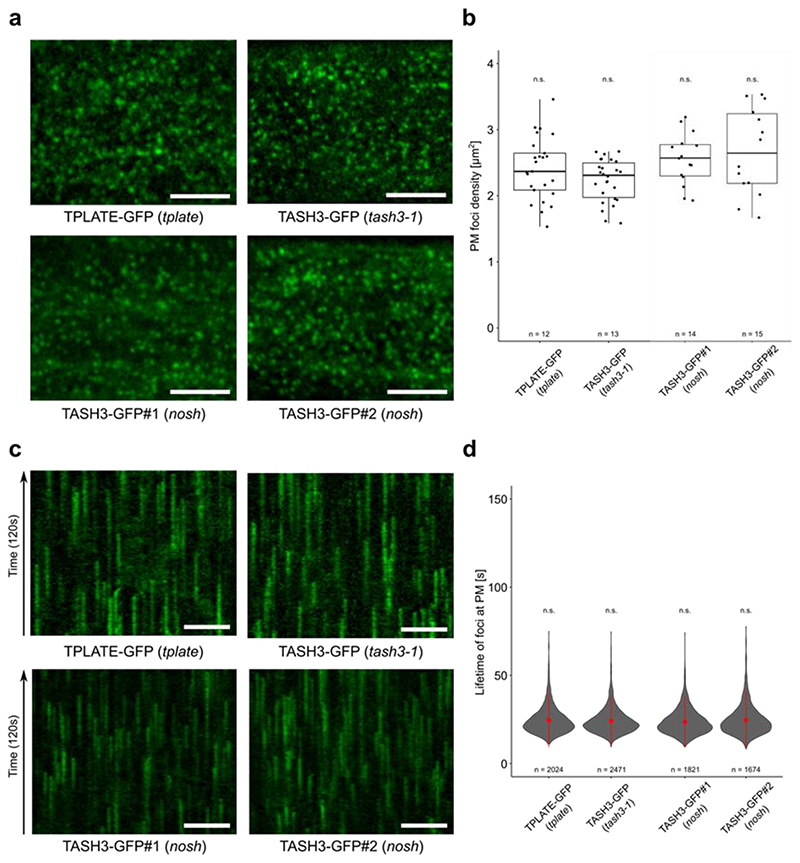
Introducing full length TASH3-GFP into nosh complemented the deviating density and life-times, indicating that the observed effects are caused by truncating TASH3 (Extended Data Fig. 2a-d). The foci density and endocytic life-time in the nosh complemented lines, visualized using TASH3-GFP, were analogous to those observed in the complemented tash3-1 mutant expressing TASH3-GFP, and were similar to those obtained using TPLATE-GFP in the complemented tplate mutant (Extended Data Fig. 2a-d). TASH3-GFP and TPLATE-GFP can therefore be interchangeably used as markers to visualize TPC dynamics and full-length TASH3-GFP complements the endocytic defects in nosh.
AtEH/Pan1 proteins specifically dissociate from TPC in nosh
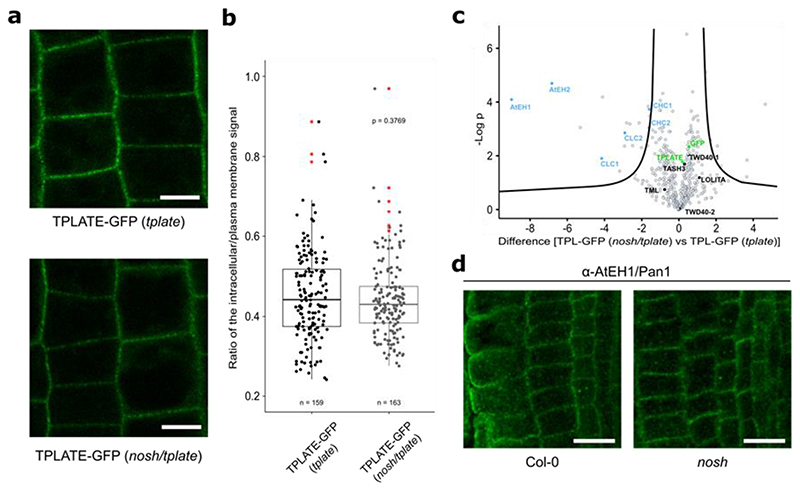
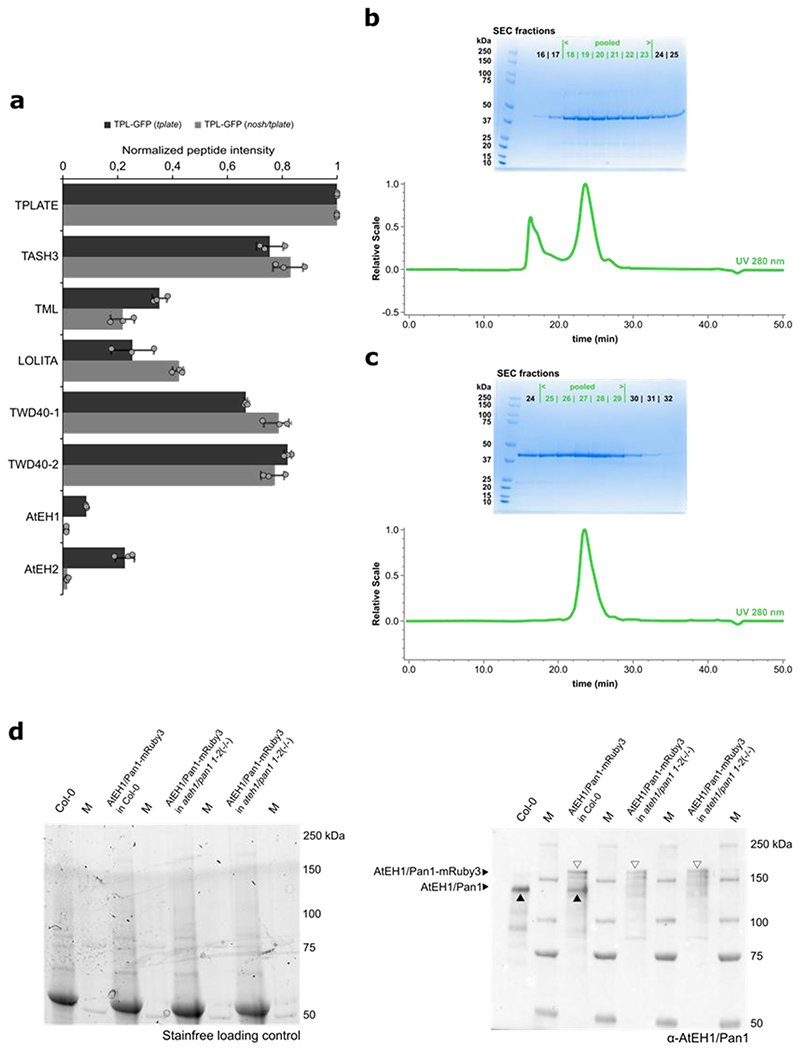
Despite the changes in endocytic density and dynamics, TPLATE-GFP localized preferentially to the PM in the nosh/tplate double mutant, with no markable differences compared to the control (TPLATE-GFP in tplate) (Fig. 3a,b). We evaluated TPC assembly in nosh, by affinity purification coupled with mass spectrometry (AP-MS) using TPLATE as bait, similarly to what we did for the destabilized TPLATE isoform WDXM235. We observed significant enrichment of AtEH1/Pan1 and AtEH2/Pan1 proteins alongside clathrin light and heavy chain proteins when we compared the complemented tplate mutant line to the nosh/tplate double mutant (Fig. 3c, Supplementary Dataset). However, relative to the bait, the average intensity of shared peptides for all other TPC subunits was similar between nosh/tplate compared to the original TPLATE-GFP complemented line (Extended Data Fig. 3a). These results imply that in nosh, a stable hexameric TPC forms, but that the AtEH/Pan1 proteins associate less strongly with the complex and fail to co-purify, similar to what was observed for the C-terminal truncation of TML4. To validate if the nosh mutation affects the localization of AtEH/Pan1, we performed immunolocalization using a self-raised antibody against the C-terminus of AtEH1/Pan1 (Extended Data Fig. 3b-d). Similarly to the control, AtEH1/Pan1 localized at PM in nosh (Fig. 3d).
Figure 3. nosh destabilizes the association between the hexameric core and the AtEH/Pan1 subunits.
a-b) Representative single-slice epidermal root cell confocal images of TPLATE-GFP and box plot graph representing the intracellular/PM intensity of TPLATE-GFP in the complemented tplate mutant and in the nosh/tplate double mutant. Numbers of quantified cells (approximately 10 cells per individual root) are indicated. Scale bar = 10 μm. No significant differences in signal intensities could be identified. The p-value was calculated using the two-sided Wilcoxon-signed rank test by comparing mutant to control. The top and bottom lines of the box plot represent the 25th and 75th percentiles, the center line is the median and whiskers are the full data range. Red asterisks mark outliers. c) Volcano plot of MS analysis following GFP pull-down from TPLATE-GFP (tplate) and TPLATE-GFP (nosh/tplate) lines. AtEH1/Pan1 and AtEH2/Pan1 subunits specifically dissociate from TPC during immunoprecipitation in the nosh/tplate double mutant. For pairwise comparison of the samples, a two-sided t-test was performed with correction for multiple testing being done by permutation based FDR, with FDR=0.01 and S0=1 thresholds applied. d) Representative single-slice confocal images of immunolocalized endogenous AtEH1/Pan1 detected by the self-raised α-AtEH1/Pan1 antibody showing that AtEH1/Pan1 remains localized at PM in nosh. Scale bar = 20 μm.
The coverage of TASH3 peptides identified by MS in complemented tplate mutants versus the nosh/tplate double mutant allowed us to characterize NOSH. Whereas peptides covered almost the entire length of TASH3, in the tplate background, peptides from the C-terminal part of TASH3 were absent in the nosh/tplate double mutant line (Extended Data Fig. 4a). This confirmed our prediction that a truncated TASH3 protein without the SH3 domain is being produced in nosh. Taken together, we confirmed that NOSH lacks the SH3 domain and that it is integrated into a TPC where the interactions between the hexameric complex and the AtEH/Pan1 proteins are weakened.
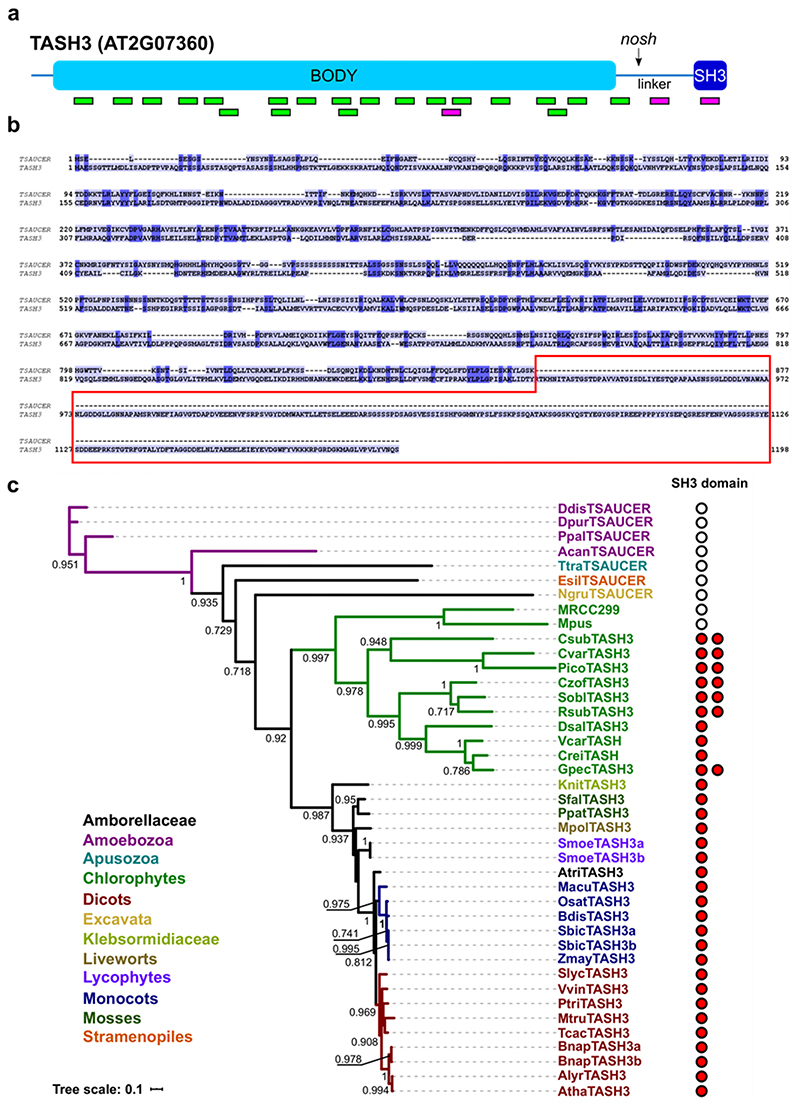
Loose association of AtEH/Pan1 subunits with the hexameric TPC resembles the TSET complex from Dictyostelium5. Moreover, the partial functionality of TPC lacking an SH3 domain is expected given that Dictyostelium TASH3 (TSAUCER) also lacks this domain (Extended Data Fig. 4b). Phylogenetic analysis of TASH3 from different eukaryotic species revealed that this domain firstly appeared in a common ancestor of Chlorophytes and Streptophytes, while in the other eukaryotic supergroups like Excavata, Apusozoa or Amoebozoa, only TSAUCER without SH3 domain is present (Extended Data Fig. 4c). Interestingly, in the TASH3 sequences of some Chlorophytes we identified TASH3 homologs possessing two SH3 domains (Extended Data Fig. 4c), pointing to potential divergence and specialized functions of these domains. To unravel why Archaeplastida did evolve a TPC containing an SH3 domain, we examined its function.
The SH3 domain of TASH3 binds ubiquitin
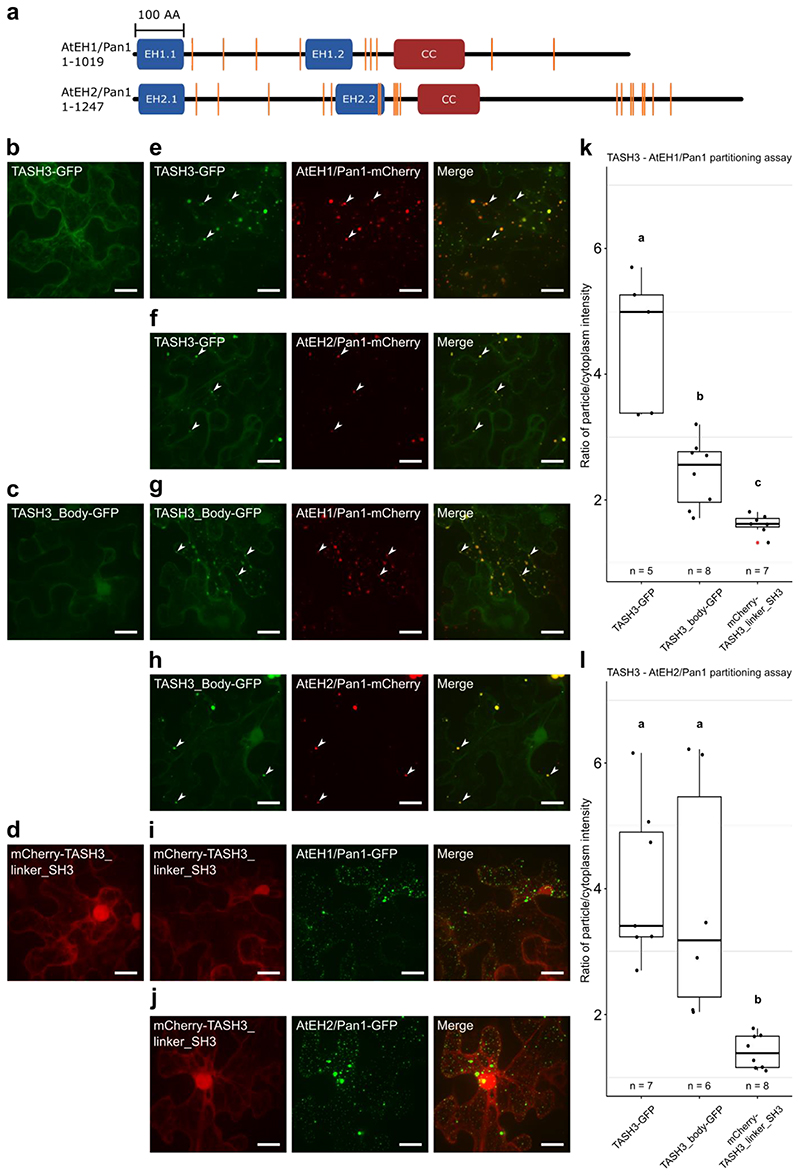
Based on the available literature, SH3 domains carry out a variety of activities and interactions. Among all, two well-characterized roles are recognizing proline-rich regions (PRR, defined as PXXP)40,41 and binding ubiquitin molecules42,43. TPC containing NOSH has reduced association with the AtEH/Pan1 subunits and these contain several PRR (Extended Data Fig. 5a). We therefore tested interactions between AtEH/Pan1 subunits and different TASH3 truncated variants: TASH3 full-length, the body part of TASH3 and the linker and SH3 domain of TASH3. Overexpression of AtEH/Pan1 in N. benthamiana can be used to perform protein-protein interaction analysis via partitioning16,37. In contrast to single infiltrations (Extended Data Fig. 5b-d), both full-length TASH3-GFP and TASH3_body-GFP partitioned together with AtEH1/Pan1-mCherry or AtEH2/Pan1-mCherry whereas mCherry-TASH3_linker_SH3 did not (Extended Data Fig. 5e-l). This strongly suggests that the SH3 domain of TASH3 has no binding capacity for the AtEH/Pan1 subunits. Thus, the detachment of AtEH/Pan1 subunits from the hexameric TPC observed in nosh is not caused by the absence of the SH3 domain.
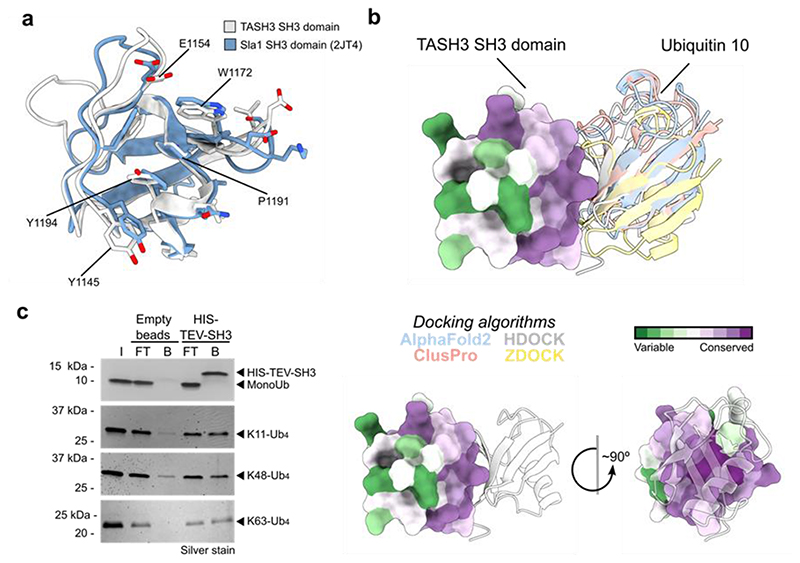
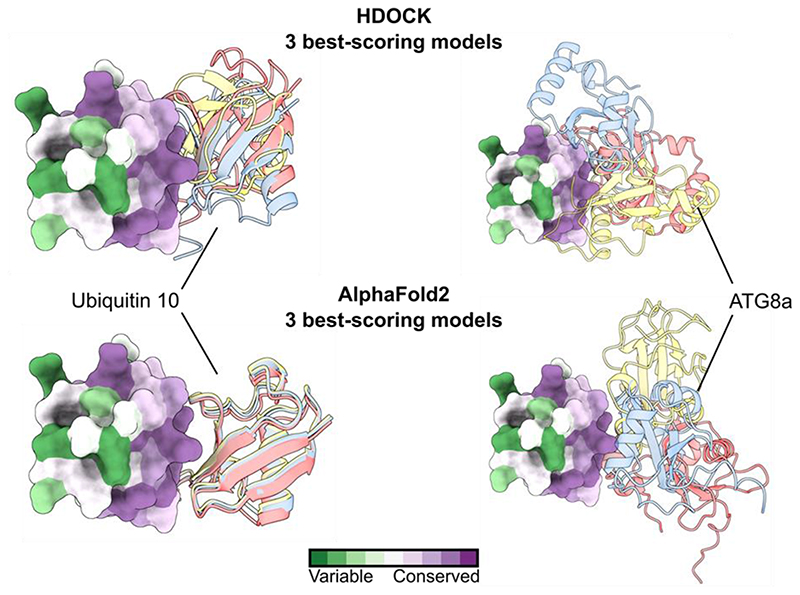
Besides recognizing PRR, some SH3 domains also bind ubiquitin42. To assess, if the SH3 domain of TASH3 binds ubiquitin, we structurally aligned our SH3 domain with that of yeast Sla1p, for which the ubiquitin binding residues have been experimentally determined42. We focused on residues forming the binding interface (defined as Sla1p residues within a distance of 0.3 nm from the ubiquitin molecule). Despite the F1194Y mutation in TASH3 SH3, the other ubiquitin-interacting residues (Y1145, E1154, W1172 and P1191) of Sla1p SH3 and TASH3 SH3 are well conserved. Moreover, the tyrosine in the TASH3 SH3 domain might mediate the interaction analogously as the phenylalanine in Sla1p (Fig. 4a). Further, we performed in silico docking and folding of the SH3 domain and a single Ubq10 molecule from Arabidopsis. Three different algorithms for protein-protein docking and one for protein folding, positioned the ubiquitin molecule and the SH3 domain almost identically (Fig. 4b). As a control, we performed docking of the TASH3-SH3 domain with ATG8a, which has a ubiquitin-like structure and size44. Using two different algorithms, docking of ATG8a did not result in a single orientation/binding with any of them (Extended Data Fig. 6). These results corroborate the specificity of the docking approach and indicate a preference of the TASH3 SH3 domain towards ubiquitin.
Figure 4. The SH3 domain of TASH3 binds poly-ubiquitin chains.
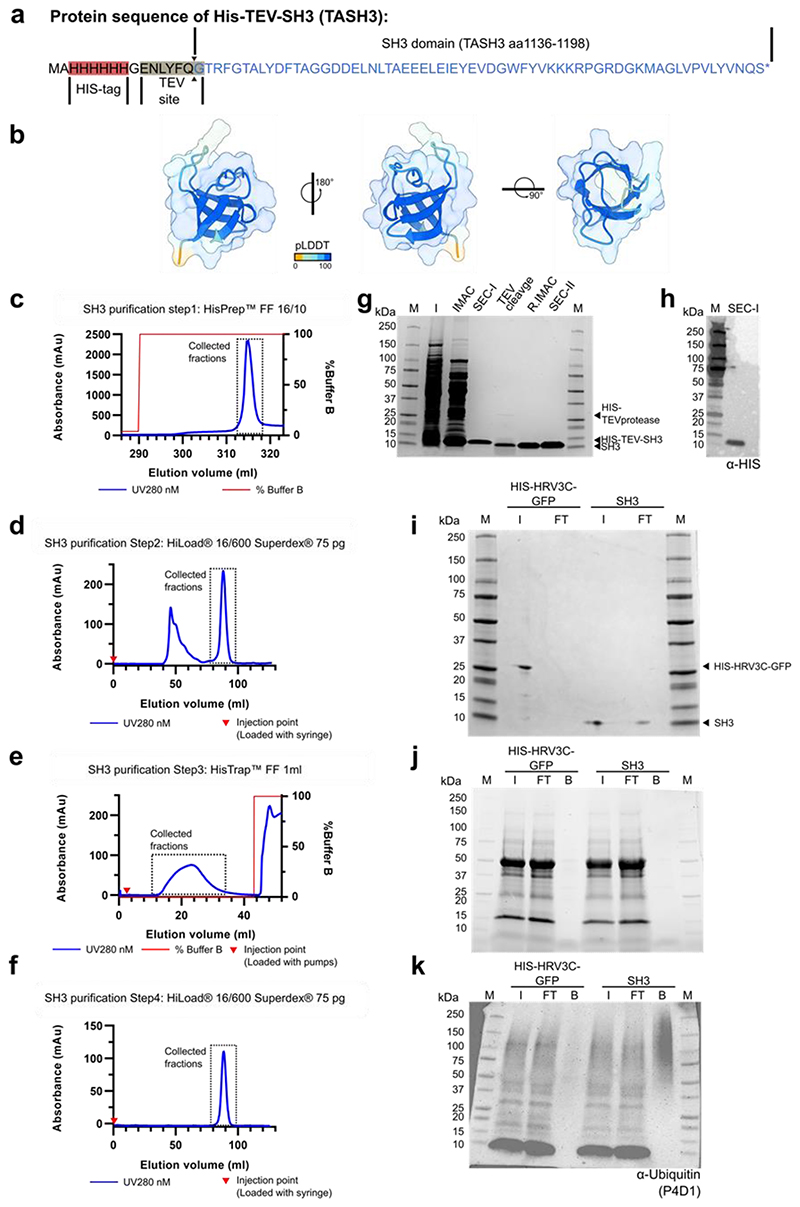
a) Comparison of the ubiquitin-binding interface of the yeast Sla1_SH3 domain (in blue, RCSB code 2JT4) and the TASH3-_SH3 domain (in white). Residues forming a binding interface (defined as residues in 0.3 nm distance from Ubq) are shown in the liquorice representation. Conserved (Y1145, E1154, W1172, P1191) or similar (Y1194) amino acid residues are highlighted for the TASH3_SH3 domain. b) Modelling of TASH3_SH3-Ubiquitin interaction reveals a highly conserved binding interface. Four different modelling algorithms resulted in an almost identical position of Arabidopsis Ubiquitin 10 with respect to the TASH3_SH3 domain. TASH3_SH3 is shown in the surface representation with mapped conserved amino acid residues (dark purple). Ubiquitin is shown in the ribbon representation. The different colours depicted for Ubiquitin 10 represent the best scoring model from the respective modelling algorithms. c) Co-immunoprecipitation assay using the purified recombinant HIS-TEV-SH3 domain from TASH3 or the empty beads control with different forms of ubiquitin (mono-ubiquitin, K11-linked tetra-ubiquitin, K48-linked tetra-ubiquitin and K63-linked tetra-ubiquitin). The assay was visualized via SDS-PAGE and subsequent silver-staining. Compared to the controls, the SH3 domain binds more K11-, K48- and K63-linked tetra-ubiquitin. I – input, FT – flow through, B – beads fraction.
To show direct ubiquitin binding, we heterologously expressed and purified the SH3 domain of TASH3 in E. coli (Extended Data Fig. 7a-j). Co-immunoprecipitation assays with the ubiquitin moiety from total cell extracts of Arabidopsis seedlings showed that the SH3 domain can bind ubiquitinated proteins (Extended Data Fig. 7k). Further, we analyzed the specificity of the SH3 domain to different ubiquitin molecules that are present in plants: mono-ubiquitin, K11-linked, K48-linked and K63-linked ubiquitin chains using co-immunoprecipitation. In contrast to empty beads, the SH3 domain clearly bound K11, K48 and K63-linked tetra-ubiquitin chains (Fig. 4c), while it did not bind mono-ubiquitin (Fig. 4c). These results suggest a preference for poly-ubiquitin, but in vitro, no clear specificity of the TASH3 SH3 domain to differently linked chains could be determined.
Ubiquitinated PM proteins accumulate in nosh
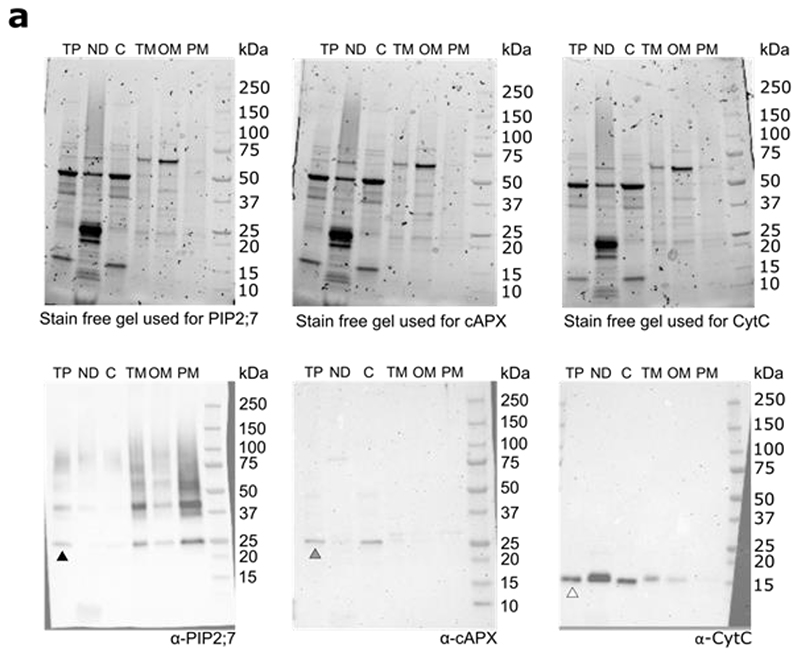
Ubiquitin is an anchoring molecule, which marks PM cargo for internalization45. The lack of an SH3 domain in TPC containing NOSH could prevent the internalization of ubiquitinated proteins at PM. To investigate this, we extracted PM fractions from wild type and several other viable endocytic mutants with documented defects in CME7,35,46. Western blot analysis of PM fractions from various endocytic mutants clearly revealed an increased amount of ubiquitin signal in the PM fraction of nosh compared to the wild type control (Col-0), but also compared to twd40-2-3, WDXM2 and ap2m-2 mutants (Fig. 5a,b). We evaluated the purity of the extracted PM fraction by probing all fractions with specific antibodies. We detected a clear enrichment of Aquaporin PIP2;7 (PIP2;7, a PM marker), while we could not detect the presence of cytosolic Ascorbate Peroxidase (cAPX, a cytosol marker) or Cytochrome C (CytC, a mitochondrial marker) in the PM fraction (Extended Data Fig. 8).
Figure 5. Ubiquitinated plasma membrane proteins accumulate in nosh seedlings.
a) Presence of ubiquitinated proteins in PM fraction of Col-0, nosh, twd40-2-3, WDXM2 and ap2m-2 mutants. Below the blot the stain free gel loading control is shown to verify that equal amounts of extracted PM proteins were loaded.. b) Box plot graph quantifying the normalized intensity of ubiquitinated PM proteins of the different endocytic mutants. c) SUBA depicted localization of the proteins enriched in the PM fraction of dark grown nosh seedlings compared to Col-0. d) Representative single slice confocal images of TMK1-GFP in tmk1-1 and nosh/tmk1-1. e) Box plot graph of the intracellular/PM intensity of TMK1-GFP signal in tmk1-1 and nosh/tmk1-1mutant f) Representative single slice confocal images of TMK1-GFP in tmk1-1 and nosh/tmk1-1mutant after 12h of dark treatment. White arrowheads mark GFP signal from TMK1 degradation in the vacuole. g) Box plot graph of the intracellular/PM signal intensity of TMK1-GFP in tmk1-1 and nosh/tmk1-1. h) Detection of ubiquitination levels of TMK1-GFP in the tmk1-1 and nosh/tmk1-1 backgrounds and quantification of band intensities (normalized to TMK1-GFP in tmk1-1) of three independent repetitions (GFP-band marked by black arrowheads and the higher MW band representing the ubiquitinated full-length TMK1-GFP marked by white arrowheads). i) Representative single slice confocal images of TMK1-GFP in tmk1-1 and in the nosh/tmk1-1mutant after 12h of dark treatment and 30 min of FM4-64 treatment. j) Paired slope graph of the inracellular/PM signal intensity of TMK1-GFP and FM4-64 measured from the same cells in tmk1-1 and nosh/tmk1-1. Letters represent a two-sided mixed linear model statistic used to determine the difference between GFP/FM4-64 signal in the tmk1-1 and in the nosh/tmk1-1 background. Numbers of quantified cells are indicated at the bottom of each graph. The top and bottom lines of box plots represent the 25th and 75th percentiles, the center line is the median and whiskers are the full data range. Red asterisks mark the outliers. For graphs in 5b, e, g the indicated p-values were calculated using the two-sided Wilcoxon-signed rank test by comparing mutant to control. No adjustment for multiple comparisons was performed. Scale bar = 10 μm.
To identify which PM proteins accumulate in nosh, we extracted the PM fraction from dark grown Col-0 and nosh seedlings and performed MS analysis. We chose the dark treatment as it was previously shown to induce ubiquitination and degradation of proteins47. From all identified and significantly enriched proteins in nosh compared to Col-0, almost 37% of them were annotated as PM proteins (Fig. 5c, Supplementary Dataset). Further, comparing our significantly enriched PM proteins, with published databases of ubiquitinated proteins48–54 we identified 38% of enriched PM proteins previously shown to be ubiquitinated (Fig. 5c). These results suggest that deletion of the SH3 domain of TASH3 causes accumulation of ubiquitinated PM proteins and that this is not a feature that correlates with an overall reduction of endocytic capacity.
TMK1 protein exhibits altered dynamics in nosh
To validate the obtained data from our MS analysis, we decided to focus on the receptor protein kinase TMK1, one of the PM proteins that was significantly over-accumulated in the PM fraction of nosh compared to the wild type control Col-0 (Supplementary Dataset). We chose TMK1, as it was previously shown to be ubiquitinated in etiolated seedlings52. We crossed pTMK1::TMK1-GFP (in tmk1-1) expressing plants with nosh and analyzed the localization of TMK1-GFP in the nosh/tmk1-1 double mutant background. The ratio between the endosomal and PM signal for TMK1-GFP in the nosh/tmk1-1 double mutant shifted towards PM compared to the complemented tmk1-1 mutant (Fig. 5d,e) and vacuolar accumulation of GFP, visualized after prolonged dark treatment, was significantly reduced in nosh/tmk1-1 compared to the complemented tmk1-1 mutant (Fig. 5f,g). Both the increased PM and reduced vacuolar accumulation support diminished internalization of TMK1 in nosh/tmk1-1, confirming our PM proteomics results.
Further, we evaluated whether TMK1 was differentially post-translationally modified by ubiquitination in nosh. Immunoprecipitated TMK1-GFP from the tmk1-1 complemented mutant and the nosh/tmk1-1 double mutant, probed with the general P4D1 anti-ubiquitin antibody, visualized substantially more ubiquitinated TMK1 in nosh/tmk1-1 compared to the control (Fig. 5h, white arrowheads; lower bands on the ubiquitin blot likely represent ubiquitinated degradation products of TMK1-GFP). These results confirmed that the absence of the SH3 domain of TASH3 reduces internalization and causes accumulation of ubiquitinated cargo at PM.
To evaluate whether nosh is more affected in ubiquitinated cargo internalization than in general endocytic flux, we combined prolonged dark treatment with FM4-64 staining. We measured the vacuole over PM ratio of GFP as well as the internalization of FM4-64 in the same cells for both the tmk1-1 and the nosh/tmk1-1 backgrounds. Plotting the internalization values obtained for GFP and FM4-64 per cell, revealed a steeper slope in tmk1-1 compared to the double mutant (Fig. 5i,j). Deletion of the SH3 domain in nosh therefore affects the internalization of ubiquitinated cargo more than it impairs general endocytic flux.
The TASH3 SH3 domain recognizes ubiquitinated cargo
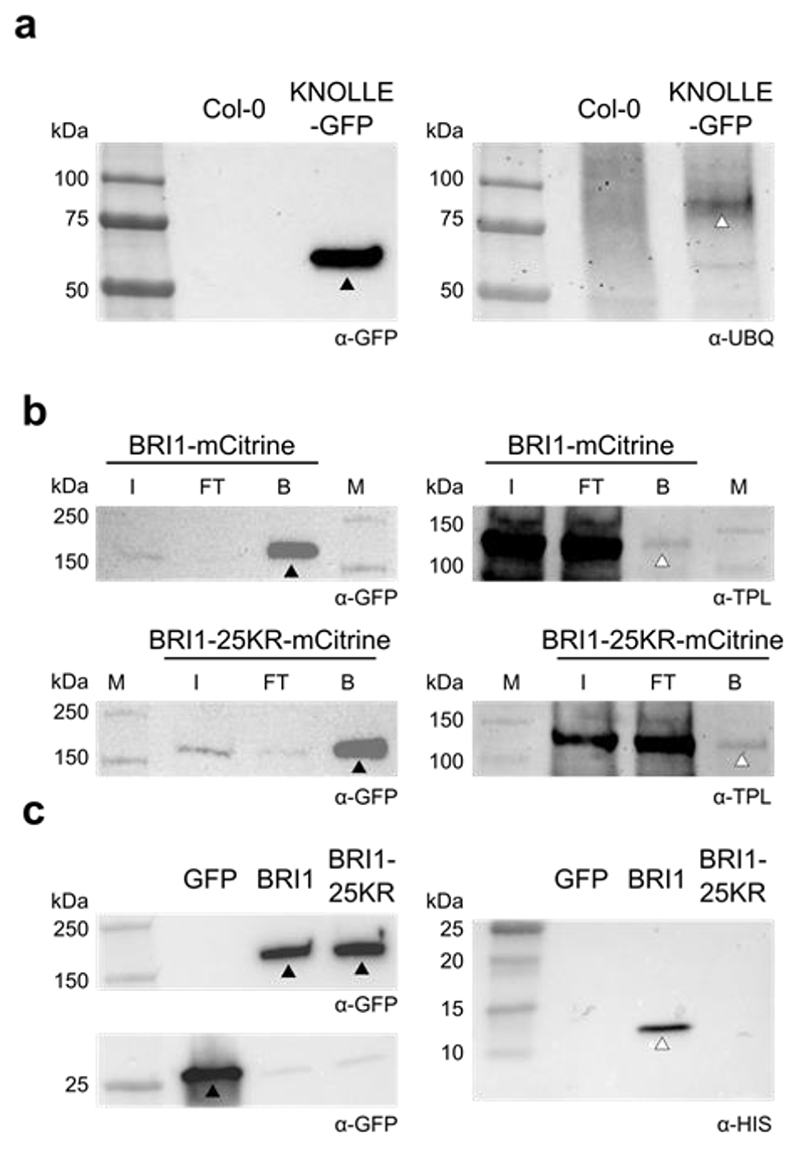
Degradation of KNOLLE from the cell plate in nosh is delayed compared to the control (Fig. 2m,n). To correlate the delay in KNOLLE degradation with the inability of TPC containing NOSH to recognize ubiquitinated cargo, we checked whether KNOLLE is ubiquitinated. We probed immunoprecipitated KNOLLE-GFP with a general anti-ubiquitin antibody (P4D1) and observed a specific band representing ubiquitinated KNOLLE-GFP (Fig. 6a). Delayed degradation and ubiquitination of KNOLLE are therefore in agreement with the inability of TPC in nosh to properly recognize ubiquitinated cargo.
Figure 6. The SH3 domain of TASH3 recognizes ubiquitinated cargo.
a) Detection of the ubiquitination status of KNOLLE following immunoprecipitation of KNOLLE-GFP with Col-0 as control. The black arrowhead indicates the band representing KNOLLE-GFP and the white arrowhead indicates Ubiquitinated- KNOLLE-GFP. b) Co-immunoprecipitation of TPLATE with pBRI1::BRI1-mCitrine and pBRI1::BRI1-25KR-mCitrine. TPLATE (marked by the white arrowhead) interacts similarly with both BRI1 variants (marked by the black arrowheads). I – input, FT – flow through, B – beads, M – marker. c) Co-immunoprecipitation experiment comparing the binding of the TASH3 HIS-TEV-SH3 domain to 35S::eGFP (GFP), pBRI1::BRI1-mCitrine (BRI1) and pBRI1::BRI1-25KR-mCitrine (BRI1-25KR) expressing lines. The SH3 domain specifically co-purifies with BRI1. Full-length bands of the proteins are marked with arrowheads (black arrowheads: eGFP, BRI1 and BRI-25KR; white arrowhead: SH3 domain). The antibodies used are depicted in the bottom right corner below the blots.
To investigate if TPC exclusively interacts with ubiquitinated cargo, we compared pBRI1::BRI1-mCitrine with the ubiquitin-dead pBRI1::BRI1-25KR-mCitrine line, which is functional, yet mutated in 25 lysine residues55. TPLATE co-purified similarly with BRI1-mCitrine and BRI1-25KR-mCitrine (Fig. 6b), indicating that ubiquitin is not the sole mechanism linking TPC to PM cargo. Next, we co-immunoprecipitated pBRI1::BRI1-mCitrine and pBRI1::BRI1-25KR-mCitrine line in the presence of recombinant HIS-TEV-SH3. The TASH3 SH3 domain specifically co-purified with BRI1-mCitrine (Fig. 6c), providing direct evidence that the TASH3 SH3 domain functions in ubiquitinated cargo recognition.
Discussion
T-DNA knockout mutants in TPC subunits do not allow to evaluate the somatic function of this complex. This has necessitated the search for alternative ways to inhibit TPC, including inducible amiRNA, nanobody-dependent inhibition, conditional inactivation4,35,37,38,56, or by identifying weak alleles7. Here, we introduce a unique partial loss of function TPC mutant, nosh, producing a TPC containing a truncated variant of the TASH3 subunit lacking its C-terminal SH3 domain. Overall, this mutant displays reduced endocytic flux, which is correlated with reduced density and increased life-time of endocytic foci. Both formerly described viable TPC mutants, twd40-2-3 and the destabilized TPLATE mutant WDXM2, displayed reduced endocytic flux and prolonged endocytic foci life-time, however, neither of them exhibited decreased endocytic densities7,35. On the other hand, ap2 σ manifested reduced density of CLC labelled foci and a decreased turnover rate although actual life-time comparisons were not performed57. It is tempting to speculate that the reduced density of endocytic foci in nosh represents a failure to recognize a distinct subset of cargo proteins due to the absence of the SH3 domain.
The delayed CME observed in nosh correlates with changes in TPC composition. AP-MS analysis revealed formation of the hexameric complex, while both AtEH/Pan1 subunits associated less strongly with the hexameric core in nosh compared to the control. Immunolocalization revealed no visible difference in AtEH1/Pan1 PM localization in nosh compared to wild type. Together with the observation that AtEH1/Pan1 remains at PM upon temperature-dependent inactivation of TPLATE35, our results indicate that AtEH1/Pan1 proteins do not need other TPC subunits to localize at PM, and they likely do so via their anionic phospholipid and protein binding EH domains16. Moreover, relative to the bait, we identified similar levels of all hexameric core subunits, indicating that comparable amounts of TPC are present in Col-0 and nosh. This differs from our previous results with WDXM2 as bait, where an imbalance between the amount of bait protein versus the other core TPC subunits strongly suggested a reduced amount of TPC units35.
TPC organization in nosh therefore resembles the situation in the slime mold Dictyostelium discoideum, which contains a similar, yet hexameric complex, called TSET, lacking AtEH/Pan1-like subunits4,5. Our previous work indicated the μHD of TML as a major factor for stabilizing the interactions between the hexameric core and the AtEH/Pan1 subunits. Dictyostelium TSET does not contain a μHD, which can explain the absence of robust interactions with AtEH/Pan14,5,36. Interactions between TASH3 and AtEH/Pan1 are independent from the SH3 domain, indicating that the reason why AtEH/Pan1 detaches from TPC containing NOSH is caused by another mechanism. This could include structural destabilization by NOSH or even the absence of an unknown stabilizing factor that connects to TPC via a binding motif in the linker region that is partially truncated in nosh.
Despite the SH3 domain of TASH3 does not have an obvious function to stabilize the different TPC subunits, it obviously has an important, albeit not essential, function that is underlined by all the observed phenotypes. The SH3-containin appendage domain is a modification which is absent in any other large core subunit from other heterotetrameric adaptor complexes containing a coat family members. The SH3 domain first appeared as a part of TASH3 in Archaeplastida. Moreover, several species of Chlorophytes possess TASH3 isoforms containing two SH3 domains. This might suggest that SH3 domains underwent specialization for different cellular functions. Establishment of a singular SH3 domain in TASH3 throughout evolution indicates the divergence of these domains and potentially roles for independent proteins possessing SH3 domains. Compared to animal genomes possessing more than hundred SH3 domain containing proteins, there are only five such proteins identified in Arabidopsis. Three members of the SH3P family, TASH3 and a SH3 domain-containing protein (AT4G39020). Expression of the last one has not been confirmed by any recent single cell data set, indicating that it likely has no major function58–60.
Ubiquitination is involved in internalization and subsequent degradation of PM proteins in plants21. Different types of ubiquitin attachments can induce different fates for the cargo proteins. Mono-ubiquitination of IRT1 triggers its internalization from PM27, while degradation of BOR1, IRT1, BRI1 or PIN2 requires K63-linked poly-ubiquitination26,61–64. Degradation of ubiquitinated PM cargo is negatively regulated by deubiquitinating proteins (DUB), as shown recently for BRI1, thus negatively modulate its vacuolar targeting65. However, the evidence of ubiquitinated cargo recognition at PM by endocytic machinery is still missing. In the animal field, a subfamily of clathrin-associated sorting proteins (CLASPs) epsin 1, epidermal growth factor pathway substrate clone 15 (EPS15) and Eps15-related protein (EPS15-R), cargo-specific monomeric endocytic adaptor proteins, contain ubiquitin-interacting motifs that have been implicated in the recognition of ubiquitinated cargo66–71. Plant homologues of these proteins, however do not contain these conserved ubiquitin binding domains, suggesting that ubiquitinated proteins need to be recognized prior to internalization by other mechanisms.
Here we suggest that this role can be performed by the SH3 domain of TASH3. We show that it binds different ubiquitin-linked chains and PM fractions of nosh contain the highest levels of ubiquitinated proteins compared to other endocytic mutants. Moreover, the SH3 domain of TASH3 specifically co-purifies with BRI1 and not with the ubiquitin-dead BRI1-25KR isoform. Together with the in vitro data, this strongly favors the SH3 domain of TASH3 as a poly-ubiquitin binding domain, recognizing cargo at PM. However, it is likely not the only one. Proteins associated with the endocytic machinery such as TARGET OF MYB1 (TOM1)-LIKE (TOL) or SH3P2 are also capable of binding ubiquitin23,32,33. A role of SH3P in recognizing ubiquitinated cargo would be in agreement with the recent finding that nosh enhanced the plant developmental defects of a triple sh3p mutant, indicating that these proteins have a non-redundant role in plant development72. In mammalian cells, WD40-repeat β-propellers also bind ubiquitin73, which suggests that two TWD40 subunits of TPC might potentially also play a role in this. Another group of ubiquitin recognizing proteins is a family of AMSH-like proteins, which can bind K63- and K48-linked poly-ubiquitin chains. AMSH proteins were shown to participate in autophagic degradation as well as vacuolar degradation of endocytic cargo74–77. AMSH3 protein can also associates with SH3P2, which was shown to also participate in autophagic degradation23. Recently, TPC and especially AtEH/Pan1 subunits have been shown to participate in autophagosome formation at ER-PM contact sites37, therefore the role of the TASH3 SH3 domain in autophagic degradation cannot be completely excluded.
In conclusion, the addition of an SH3 domain to the large TPC subunit TASH3 represents a unique evolutionary plant synapomorphy. Our study reveals that this adaptation modulates endocytosis in plants by recognizing poly-ubiquitinated cargo at PM destined for degradation. As such, it provides the first mechanistic insight into ubiquitin-mediated cargo recognition leading to endocytic initiation.
Materials and Methods
Molecular cloning
Primers used for constructing all the constructs in this manuscript are present in the Supplementary Dataset. To yield the expression construct for full length TASH3-GFP, entry clones of TASH3 without a stop codon in pDONR2214, pDONRP4-P1R-Histone3p78 and pDONRP2-P3R-GFP79 were combined with pB7m34GW79 in a triple gateway reaction.
To create the TASH3_body-GFP construct, TASH3_body without stop codon was amplified from the full length TASH3 entry clone in pDONR221 using AtTASH3_Body_221_Fw and AtTASH3_Body_221_Rv primers, cloned in pDONR221 and combined with pDONRP4-P1R-Histone3p78, pDONRP2-P3R-GFP79 and pB7m34GW79 in a triple gateway reaction.
The mCherry-TASH3_linker_SH3 construct was created by combining the Golden Gate entry clones pGGC-TASH3_linker_SH3 without a stop codon, pGG-A-pH3.3-B, pGG-B-mCherry-C80, pGG-D-Decoy_v2-E81, pGG-E-tHSP18.2M-F82 and pGG-F-linkerII-G83 with the pGGB A-G (Basta)83 destination vector in a Golden Gate reaction.
The SH3 domain of TASH3 was amplified from TASH3 without a stop codon in pDONR2214 using SH3domain_Fwd and SH3domain_Rv primers. The PCR fragment was cloned into the pET22b plasmid (Novogen) by restriction digestion (Nde/Xho). The final construct has an N-terminal HIS-tag followed by a TEV-protease cleavage site and contains amino acids 1136-1198 of TASH3.
UBI10::AtEH1-mCherry was created using pGG-A-pUBI10-C, pGG-C-AtEH1-D, pGG-D-mCherry-E80, pGG-E-tHSP18.2M-F82 and pGG-F-linkerII-G83 and combining them with the pGGH A-G (Hygro)83 destination vector in a Golden Gate reaction.
To create the UBI10::AtEH2/Pan1-mCherry construct, entry clones of pGG-A-pUBI10-C, pGG-C-AtEH2/Pan1-D, pGG-D-mCherry-E80, pGG-E-tHSP18.2M-F82 and pGG-F-linkerII-G83 were combined with the pGGH A-G (Hygro)83 destination vector in a Golden Gate reaction. To yield the vector for antibody production, the expression plasmid peAtEH1-Pan1c-term for heterologous expression of the C-terminal part of AtEH1/Pan1 with a 6 times HIS tag in E. coli was generated via restriction digest-mediated (NdeI/BamHI) cloning into the pLT32 expression vector84. The pDONR221-AtEH1_no_stop4 plasmid was used as a template for PCR with primers EH_atb_Fw and EH_atb_Rv.
Plant material
The Arabidopsis mutant lines tash3-1 (SALKseq_122269), tash3-2 (SALK_020985) and nosh (SALK_011079) were obtained from NASC. twd40-2-37, WDXM235 and ap2m-285 were described previously. The Arabidopsis lines expressing pLAT52::TPLATE-GFP4,6, p35S::DRP1a-mRFP86, pKNOLLE::KNOLLE-GFP87, pCLC1::CLC1-GFP88, pTMK1::TMK1-GFP89, pH3.3::AtEH1/Pan1-mRuby337, p35S::eGFP90, pBRI1::BRI1-mCitrine91 and pBRI1::BRI1-25KR-mCitrine55 were described previously.
For backcross experiments, heterozygous mutant plants of tash3-1, tash3-2 and nosh were used as male to cross with Col-0 as female. The transfer of the T-DNA, requiring the functionality of the complementing fusion construct, was analyzed by genotyping PCR on F1 plants.
Homozygous tplate mutant plants carrying pLat52::TPLATE-GFP were crossed with nosh. F2 plants in the double homozygous nosh/tplate mutant background were identified by genotyping PCR in the following generations. Homozygous tmk1-1 mutant plants carrying pTMK1::TMK1-GFP were crossed with nosh. F2 nosh/tmk1-1 double homozygous mutant plants were identified by genotyping PCR. Lines expressing p35S::DRP1a-mRFP, pKNOLLE::KNOLLE-GFP and pCLC1::CLC1-GFP were crossed with nosh and F2 plants in the nosh homozygous background were identified by genotyping PCR. Genotyping primers are in provided Supplementary Dataset.
The pH3.3::TASH3-GFP construct was used for complementing the nosh and tash3-1 mutant backgrounds. F2 homozygous mutant plants were identified by genotyping PCR.
Nicotiana benthamiana plants were grown in a greenhouse under long-day conditions (06–22 h light, 100 PAR, 21°C) in soil (Saniflo Osmocote pro NPK: 16-11-10+ magnesium and trace elements). Transient expression was performed by leaf infiltration according to92. An optical density of 0.5 of Agrobacterium strains was used for all constructs during co-expression. Transiently transformed N. benthamiana epidermal leaf cells were imaged two to three days after infiltration.
Root and hypocotyl growth
Arabidopsis seedlings were grown on ½ strength MS medium without sucrose. Plates with seeds were stratified for 48 h at 4 °C, and then placed at 21 °C in continuous light. For root growth analysis, seedlings were grown in continuous light for 5 days. Two days after transfer to the light, seeds which did not germinate or which grew into the agar were marked and excluded from further analysis. Root length measurements were carried out with the Fiji93 software package. For hypocotyl analysis, following stratification, plates were transferred to continuous light for at least 3 hours and afterwards covered in aluminum foil. Plates were kept covered for another 4 days. Hypocotyl length measurements were carried out with the Fiji93 software package.
Phylogenic analysis
To identify TASH3 homologs, predicted proteins of selected genomes from the Joint Genome Institute database (https://genome.jgi.doe.gov/portal/) were searched using the BLASTP algorithm94 with Arabidopsis TASH3 as input sequence. TSAUCER sequences were obtained from5. The SMART database95 was used to decipher the presence of the SH3 domain. A multiple sequence alignment was constructed with the MAFFT algorithm in the einsi mode96 (Supplementary alignment file, a Jalview compatible multiple alignment file). The phylogenetic analysis was carried out utilizing PhyML v3.097 with the smart model selection98. The phylogenetic tree was visualized using iTOL v699.
Docking
Structures of the Arabidopsis TASH3_SH3 domain (Uniprot code F4IL68, amino acids 1137-1198), the Arabidopsis UBIQUITIN10 (Uniprot code Q8H159) and the Arabidopsis ATG8a (Uniprot code Q8LEM4) were downloaded from the AlphaFold2 structure database100. Four different modelling algorithms were used to position the TASH3_SH3 domain to either UBIQUITIN10 or ATG8a. Namely, we used the AlphaFold2 algorithm as implemented in ColabFold101, ClusPro102, HDOCK103 and ZDOCK104. The best scoring models were analyzed and visualized in the ChimeraX program105. To analyze amino acid conservation, we employed the Consurf server106.
SH3 domain purification
The pET22b-HIS-TEV-SH3 construct was transformed into BL21(DE3) (#C2527H, NEB). Cells were grown at 37 °C in LB+ medium and induced by adding 0.4 mM IPTG at OD 0.6 for 5h. Proteins were extracted using sonication in 20 mM HEPES pH 7.4, 150 mM NaCl, 10mM imidazole and Protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail [Roche]). Purification was performed on an ÄKTA purifier (GE Healthcare) system by subsequent purification using immobilized metal affinity chromatography (IMAC) via a HisPrep™ FF 16/10 column (GE Healthcare) using 20 mM HEPES pH 7.4, 150 mM NaCl, 10mM imidazole as binding buffer and 20 mM HEPES pH 7.4, 150 mM NaCl, 500mM imidazole as elution buffer. The first size exclusion chromatography (SEC) step was performed using a HiLoad® 16/600 Superdex® 75 pg column (GE Healthcare) using 20 mM HEPES pH 7.4, 150 mM NaCl as elution buffer. When no HIS-tag was required, the protein was incubated overnight with 1/40 protein:HIS-TEV-protease (own production)107 at room temperature without shaking. Uncleaved protein and protease were removed via reverse IMAC using a 1ml HisTrap™ FF column (GE Healthcare) using the same buffers as in the first IMAC step followed by SEC using a HiLoad® 16/600 Superdex® 75 pg column (GE Healthcare) using the same buffer as in the first SEC step. The yield was approximately 0.250mg/L culture after all purification steps. The protein sequence of the SH3 domain along with its native molecular weight was verified by MS analysis. The HIS-HRV3C-GFP control (Ray Owens, OPPF, UK) on an OPINF backbone16 was produced and purified analogously to the TASH3 SH3 domain.
SH3 ubiquitin binding assays
For the SH3 - ubiquitin binding assays, 100 μg HIS-TEV-SH3 and 326 μg HIS-HRV3C-GFP were covalently coupled via their primary amines to 3 mg Pierce™ NHS-Activated Agarose (Thermo Scientific™). This was done by incubating the recombinant proteins with the beads in purification buffer: 20 mM HEPES pH 7.4, 150 mM NaCl and Protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail [Roche]) rotating at room temperature for 1 hour. For the empty beads control, recombinant protein was omitted. The coupling efficiency of HIS-TEV-SH3 and HIS-HRV3C-GFP was determined via a Coomassie-stained SDS-PAGE gel by comparing input and flow through. The beads were washed 3 times with purification buffer. Subsequently, free binding spots on the beads were quenched with 1 M TRIS-HCl pH 7.4 for 20 min rotating at room temperature. The beads were washed with binding buffer: 20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween-20 and then incubated with 10 μg/ml tetra-ubiquitin (K11-, K48- and K63-linked ubiquitin, Biotechne/R&Dsytems) or 40 μg/ml mono-ubiquitin (Biotechne-R&Dsystems) in binding buffer, rotating at 4 °C for 2 hours. The bound fraction was eluted by incubating at 70°C for 10 min in 1x Laemmli buffer (BioRad) and 1x NuPage ™ reducing agent (Invitrogen). Input, flow-through and bound fractions were assessed via SDS-PAGE using a 4-20% SDS-PAGE TGX gel (BioRad) and subsequent silver-staining using the Pierce™ Silver Stain Kit (ThermoFisher Scientific).
For the SH3 – Ubiquitinated proteins pulldown from Arabidopsis thaliana Col-0 extracts, 100 μg SH3 (TEV-cleaved) and 100 μg HIS-HRV3C-GFP were coupled as described above. Six extracts from Arabidopsis thaliana Col-0 were prepared by flash-freezing and grinding the material in liquid nitrogen. The material was then incubated in a 1:1 ratio with protein extraction buffer: 50 mM Tris pH 7.6, 150 mM NaCl, 5 mM DTT, 1 mM PMSF and protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail [Roche]) and allowed to rotate for 30 min at 4 °C. Subsequently, the supernatant was cleared by centrifugation at 20,000 g at 4 °C for 20 min. Protein concentrations from the extracts were measured using the Qubit™ Protein assay kit (Thermo Fisher). Equal concentrations of the extracts were incubated with the beads rotating at 4°C for 2 hours. The beads were washed 3 times with protein extraction buffer. The bound fraction was eluted by incubating at 70 °C for 10 min in 1x Laemmli buffer (BioRad) and 1x NuPage ™ reducing agent (Invitrogen). Input, flow through and bound fractions from different replicates were pooled and assessed via Western blot.
KNOLLE and TMK1 Ubiquitination assays
For immunoprecipitation of KNOLLE-GFP, 3-day-old light grown Col-0 seedlings and seedlings expressing pKNOLLE::KNOLLE-GFP (in the Col-0 background) were used. For immunoprecipitation of TMK1-GFP, 7-day-old dark grown seedlings expressing pTMK1::TMK1-GFP in the tmk1-1 and the nosh/tmk1-1 background were used. Seedlings were harvested and grinded using a mortar and a pestle. Tissue was lysed using 1:1 w/v extraction buffer (50 mM Tris-HCl pH 7.5, 5 mM DTT, 100 mM NaCl, 10% [v/v] glycerol, 2% [v/v] NP-40, 20 mM N-ethylmaleimide, 1x cOmplete Mini Protease Inhibitor Cocktail [Roche]). Cell debris was removed by centrifuging two times at 14,000g at 4 °C for 15 min. Anti-GFP immunoprecipitation was carried out following the manufacturer’s instructions (Miltenyi Biotec) with slight modifications. Columns were equilibrated using 200 μL of extraction buffer and washed 2 times with washing buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% [v/v] glycerol, 20 mM N-ethylmaleimide, 1x cOmplete Mini Protease Inhibitor Cocktail [Roche]). Samples were subsequently analyzed using Western blot.
SH3 domain cargo recognition assays
6-day-old light grown seedlings expressing pBRI1::BRI1-mCitrine86 and pBRI1::BRI1-25KR-mCitrine87 were used. Seedlings were harvested and grinded using a mortar and a pestle. Tissue was lysed using 1:1 w/v extraction buffer (50 mM Tris-HCl pH 7.5, 5 mM DTT, 100 mM NaCl, 10% [v/v] glycerol, 2% [v/v] NP-40, 20 mM N-ethylmaleimide, 1x cOmplete Mini Protease Inhibitor Cocktail [Roche]). Cell debris was removed by centrifuging two times at 14,000g at 4 °C for 15 min. Supernatant was incubated with purified TASH3_SH3 [10 μM] for 30 min at 4 °C. Anti-GFP immunoprecipitation was carried out following the manufacturer’s instructions (Miltenyi Biotec) with slight modifications. Columns were equilibrated using 200 μL of extraction buffer and washed 2 times with washing buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% [v/v] glycerol, 20 mM N-ethylmaleimide, 1x cOmplete Mini Protease Inhibitor Cocktail [Roche]). Samples were subsequently analyzed using Western blot.
Western blotting
Samples with 1x Laemmli loading buffer (BioRad) and 1x NuPage ™ reducing agent (Invitrogen) were heated for 10 min at 70 °C. Equal protein amounts were loaded on a 4-20% SDS-PAGE TGX gel (BioRad). SDS-PAGE separated proteins were blotted on PVDF (BioRad) or nitrocellulose membranes (BioRad) depending on the protein. Membranes were blocked overnight at room temperature in 5% skimmed milk. The blots were then incubated at room temperature with the primary antibodies (α-GFP [Miltenyi Biotec, 1:2000, PVDF], α-ubiquitin P4D1 [Santa Cruz Biotechnology, PVDF, 1:500; Millipore, 1:1000, PVDF], α-Penta-HIS HRP Conjugate [Qiagen, 1:2000, PVDF]), α-PIP2;7 [Agrisera, 1:2000, nitrocellulose], α-cAPX [Agrisera, 1:1000, PVDF], α-CytC [Agrisera, 1:1000, PVDF] and secondary antibodies (α-mouse-HRP, from sheep [Amersham/Cytiva, 1:10 000], α-rabbit-HRP, from donkey [Amersham/Cytiva, 1:10 000]) in 5% skim milk for 1h. Antigen-antibody complexes were detected using chemiluminescence (Perkin-Elmer).
Protein expression for antibody production
Protein expression and purification was performed by the VIB protein core facility (https://vib.be/labs/vib-protein-core/research) in two independent runs. One batch was used for rabbit immunization (Extended Data Fig. 3b) and one batch was used for antibody purification from the obtained serum (Extended Data Fig. 3c). Protein expression was induced via IPTG, afterwards the cells were pelleted and resuspended in lysis buffer (50 mM HEPES pH 7,4; NaCl; 20 mM Imidazole; 1 tablet/50 ml cOmplete Mini Protease Inhibitor Cocktail [Roche]; 1 mg/100 ml DNase1) followed by sonication. The sonicated solution was pelleted twice for 30 min at 10,000 and 18,000 rpm at 4 °C to separate the cell lysate from the debris. The lysate was subsequently filtered with a 0.22 μm filter and the protein was purified on an Äkta purifier (Ge Healthcare) in three independent steps. First the protein was purified via IMAC using a HisPrep™ FF 16/10 GE 20ml column. After applying the lysate, the column was washed with 20 column volumes (CV) IMAC buffer (50mM HEPES pH 7,4; 20 mM NaCl; 20 mM Imidazole; 0,1% empigen detergent) and then with 5 column volumes IMAC buffer containing 50 mM Imidazole. The protein was then eluted with IMAC elution buffer (50 mM HEPES pH 7,4; 20 mM NaCl; 400 mM Imidazole). The protein was further purified via ion exchange chromatography using Source Q15 resin. Therefore, the IMAC eluate was diluted 1:100 to guarantee binding to the resin. After washing the column with 20 column volumes of 20 mM Tris-HCl pH 8.0 the protein was eluted via a NaCl gradient increasing the concentration from 0 M to 1 M over 5 column volumes. Finally, the protein was purified by size exclusion chromatography (SEC). SEC was performed with a HiLoad Superdex 200 26/600 prep grade column using PBS buffer. Coomassie stained gels of the obtained SEC fractions for each run are depicted in Extended Data Fig. 3b,c. For the batch used for rabbit immunization the fractions 18 - 23 were pooled (Extended Data Fig. 3b), for the batch used for antibody purification the fractions 25 - 29 were pooled (Extended Data Fig. 3c). Both pools were analyzed via HPLC using a Superdex 200 increase 10/300 as a quality control (chromatograms in Extended Data Fig. 3b,c).
Polyclonal AtEH1/Pan1 antibody generation
The Biotem antibody service (https://www.biotem-antibody.com/) was used to generate a polyclonal rabbit antibody against the C-terminus of AtEH1/Pan1. Rabbits for immunization were selected based on their blood serum responsiveness to the Arabidopsis proteome. Selected rabbits were injected on day 0, 7, 14 and 34 with purified protein concentrated to 0,724 mg/ml. After 42 days, the rabbits were exsanguinated and their sera were tested against the antigen. The antibody of the most responsive serum was purified by Biotem against the C-terminus of AtEH1/Pan1. The purification (Biotem) was performed using a second batch of recombinant protein concentrated at 3 mg/ml.
To test the purified antibody for specificity, 5-day-old seedlings expressing AtEH1/Pan1-mRuby3 in a Col-0 or ateh1/pan1 mutant background35 as well as Col-0 control seedlings were shock frozen and ground in liquid nitrogen. Subsequently, heated 1.2x Laemmli buffer was added to the plant material followed by thorough vortexing. After 8 min incubation at 75 °C, samples were centrifuged two times at 20,000 g for 1 min at 4 °C. Samples were separated on a 4-20% Gradient TGX SDS stain free gel (BioRad) and comparable protein amounts between samples were confirmed via stain-free imaging (Extended Data Fig. 3d). The gel was blotted on a nitrocellulose membrane (BioRad) and developed with the purified polyclonal antibody diluted 1:2000 in PBS-T containing 5% milk powder. The specificity could be confirmed, as Col-0 and the AtEH1/Pan1-mRuby3 (Col-0) lines show the native AtEH1/Pan1 band roughly at 140 kDa, which is not present in the complemented mutant lines (Extended Data Fig. 3d, marked by black arrowheads). Transgenic lines expressing AtEH1/Pan1-mRuby3 in (Col-0) and (ateh1/pan1 1-2 -/-) backgrounds also exhibited a band for AtEH1/Pan1-mRuby3 at roughly 175 kDa (Extended Data Fig. 3d, marked by white arrowheads).
Plasma membrane protein extraction
Sample preparation
Arabidopsis seedlings were grown on ½ strength MS medium without sucrose at 21 °C in dark for 4 days. We chose dark germination to induce ubiquitination and degradation of proteins. The tissue samples were flash-frozen and crushed using a liquid cooled mortar and pestle and the crushed material was used to extract the plasma membrane fraction using the Minute™ Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent Biotechnologies) based on the manufacturer’s manual. For LC-MS/MS analysis of the PM fractions, samples were prepared based on the Mini Urea protocol, using 90 μg of plasma membrane proteins, as described in108. For the analysis of the fractions by Western blot, samples were prepared based on the manufacturer’s manual with a slight modification. The pellet of the total membrane fraction and the pellet of the plasma membrane fraction were each washed once in the respective buffers that were used to acquire the pellet during that step (Buffer A for total membrane; Buffer B + PBS for plasma membrane). This was followed by an identical centrifugation step. For Western blot analysis, samples were prepared as described in the Western blot section.
LC-MS/MS analysis
Purified peptides were re-dissolved in 25 μl loading solvent A (0.1% TFA in water/ACN (98:2, v/v)) and 5 μl was injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano LC (Thermo Fisher Scientific, Bremen, Germany) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Trapping was performed at 10 μl/min for 4 min in loading solvent A on a 20 mm trapping column (made in-house, 100 μm internal diameter (I.D.), 5 μm beads, C18 Reprosil-HD, Dr. Maisch, Germany) and the sample was loaded on a 200 mm analytical column (made in-house, 75 μm I.D., 1.9 μm beads C18 Reprosil-HD, Dr. Maisch). Peptides were eluted by a non-linear gradient from 5 to 55% MS solvent B (0.1% FA in water/acetonitrile (2:8, v/v)) over 145 min at a constant flow rate of 300 nl/min, reaching 99% MS solvent B after 150 min, followed by a 10 minute wash with 99% MS solvent B and re-equilibration with MS solvent A (0.1% FA in water). The column temperature was kept constant at 45 °C in a column oven (Butterfly, Phoenix S&T). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 16 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375-1500 m/z) were acquired at a resolution of 60,000 in the orbitrap analyzer after accumulation to a target value of 3E6. The 16 most intense ions above a threshold value of 2.0E4 were isolated for fragmentation at a normalized collision energy of 28% after filling the trap at a target value of 1E5 for maximum 50 ms. MS/MS spectra (200-2000 m/z) were acquired at a resolution of 15,000 in the orbitrap analyzer.
MS data-analysis
Data analysis was performed by MaxQuant (version 1.6.10.43) using the Andromeda search engine with default settings, including a false discovery rate set at 1% on both the peptide and protein level. Spectra were searched against the Araport11plus database, consisting of the Araport11_genes.2016.06.pep.fasta downloaded from arabidopsis.org, extended with sequences of all types of possible contaminants in proteomics experiments in general. These contaminants include the cRAP protein sequences, a list of proteins commonly found in proteomics experiments, which are present either by accident or by unavoidable contamination of protein samples (The Global Proteome Machine, http://www.thegpm.org/crap/). In addition, commonly used tag sequences and typical contaminants, such as sequences derived from the resins or the proteases used, were added. The Araport11plus database contains in total 49,057 sequence entries. The mass tolerance for precursor and fragment ions was set to 4.5 and 20 ppm, respectively, including matching between runs. Enzyme specificity was set as C-terminal to arginine and lysine (trypsin), also allowing cleavage at arginine/lysine-proline bonds with a maximum of two missed cleavages. Fixed modification was set to carbamidomethylation of cysteines. Variable modifications were set to oxidation of methionine residues and acetylation of protein N-termini. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software. Only proteins with at least one unique peptide were retained for identification.
Differential analysis was performed with the Perseus software (version 1.6.7.0) after loading the proteingroups.txt file from MaxQuant, with reverse and contaminant hits removed. LFQ intensities were log2 transformed and biological replicate samples were grouped together. Proteins with less than two valid values in at least one group were removed and missing values were imputed from a normal distribution around the detection limit. On the quantified proteins, a t-test was performed for pairwise comparison of the samples. Correction for multiple testing was done by permutation based FDR, with thresholds FDR=0.05, S0=0.5 or S0=0.1. The result is listed in Supplementary Dataset.
GFP pull-down
Sample Preparation
Samples were prepared as described in109. Briefly: samples were lysed in extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP-40, 1x cOmplete Mini Protease Inhibitor Cocktail [Roche]) and sonicated. After centrifugation (15 min at 4 °C at 18,000 rpm/39.000 g) 100 μL of anti-GFP μBeads (μMACS, Miltenyi) and samples were incubated for 2 h at 4 °C. Afterwards, the extracts were run through μMACS Separator columns, washed 2 times with extraction buffer, washed 2 times with ABC buffer (50 mM NH4HCO3 in H2O) and eluted with 95 °C preheated ABC buffer. For LC-MS/MS analysis samples were prepared as described in108.
LC-MS/MS analysis
Peptides were re-dissolved in 20 μl loading solvent A (0.1% TFA in water/ACN (98:2, v/v)) of which 5 μl was injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano LC (Thermo Fisher Scientific, Bremen, Germany) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). The peptides were first loaded on a trapping column made in-house, 100 μm internal diameter (I.D.) × 20 mm, 5 μm beads C18 Reprosil-HD, Dr. Maisch, Ammerbuch-Entringen, Germany) and after flushing from the trapping column the peptides were separated on a 50 cm μPAC™ column with C18-endcapped functionality (Pharmafluidics, Belgium) kept at a constant temperature of 50 °C. Peptides were eluted by a linear gradient from 99% solvent A’ (0.1% formic acid in water) to 55% solvent B’ (0.1% formic acid in water/acetonitrile, 20/80 (v/v)) in 30 min at a flow rate of 300 nL/min, followed by a 5 min wash reaching 95% solvent B’. The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and MS/MS acquisition for the 5 most abundant peaks in a given MS spectrum. One MS1 scan (m/z 400–2,000, AGC target 3 × 106 ions, maximum ion injection time 80 ms), acquired at a resolution of 70,000 (at 200 m/z), was followed by up to 5 tandem MS scans (resolution 17,500 at 200 m/z) of the most intense ions fulfilling predefined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 80 ms, isolation window 2 m/z, fixed first mass 140 m/z, spectrum data type: centroid, intensity threshold 1.3xE4, exclusion of unassigned, 1, 5-8, >8 positively charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time 12 s).
MS Data analysis
The raw data was searched with MaxQuant (version 1.6.10.43) as described for the plasma membrane proteins. Differential analysis was performed with the Perseus software (version 1.6.15.0) with the largely the same workflow as described for the plasma membrane proteins. Except here, proteins with less than three valid values in at least one group were removed, and thresholds FDR=0.01 and S0=1 were applied. The results are shown in the volcano plot in Fig. 3c, and listed in Supplementary Dataset.
Whole-mount immunofluorescence imaging
4-day-old Arabidopsis seedlings were analyzed by immunofluorescence using INTAVIS In situ robot as described before110 with minor modifications. Samples were fixed with 4% paraformaldehyde, incubated with 1,5% Driselase for 30 min, 3% IGEPAL CA-630 for 30 min and blocked with 3% BSA for 60 min. Afterwards the samples were incubated with the α-AtEH1/Pan1 antibody [1:600] for 4 hours and with the fluorochrome-conjugated secondary anti-rabbit-Alexa488 antibody (from donkey, Invitrogen [1:600]) for 4 hours.
Live-cell imaging
FM4-64 imaging
Prior to imaging, whole 5-day-old seedlings were incubated with 2 μM FM4-64 (Invitrogen) solution in ½ strength MS liquid medium without sucrose at room temperature for 30 min. For the FM4-64 uptake in Figure 1, confocal images were taken using a FluoView FV1000 (Olympus) confocal, equipped with a 60× water-corrected objective (NA = 1.2). Fluorescence was imaged in a single channel setting with 559 nm excitation light and emission fluorescence was captured in the frame-scanning mode via a 570- to 670 nm band pass emission window. For the FM4-64 uptake in Figure 5, FM4-64 was visualized on a Leica SP8X confocal with settings as outlined below.
Fluorescein diacetate (FDA) staining
FDA staining was performed on mature pollen stained on a glass slide in a solution containing 50 μM FDA in 10% sucrose for 5 min in the dark. Afterwards, confocal images were taken using a FluoView FV1000 confocal microscope (Olympus), equipped with a 60× water-immersion corrected objective (NA = 1.2). Fluorescence was imaged in a single channel setting with 488 nm excitation light and emission fluorescence was captured in the frame-scanning mode via a 515-565 nm band pass emission window.
Protein localization in Arabidopsis roots
Arabidopsis root images for PM signal quantification (TPLATE and TMK1) were obtained using a Leica SP8X confocal microscope equipped with a White Light Laser and using the LASX software package (Leica). Images were acquired on Hybrid detectors (HyD, gating 0.3-10 ns) using bidirectional imaging with a 40x water-immersion corrected objective (NA = 1.10), frame or line signal averaging and with a 3x digital zoom. A single excitation (488nm laser) line and emission windows ranging between 500-550 nm for GFP; and 600-740 nm for propidium iodide (PI; Invitrogen) were used. The PI served to generate the mask for quantification of the cytoplasm/PM ratio in Fig. 3a, b and Fig. 5d-g. For the combined imaging of GFP and FM4-64 in Figure 5, the GFP settings from above were combined with 561 nm laser excitation and a 570-630 nm detection window for FM4-64 in a line sequential imaging mode.
Root tracking
Arabidopsis seedlings expressing the pKNOLLE::KNOLLE-GFP marker were prepared as described in111. In Brief, 3 days old seedlings were transferred into an in house-made two-part chambered coverglass (adapted from https://cellgrowth-lab.weebly.com/3d-prints.html), covered with a slice of ½ MS agar and moved back to the growth chamber for 1h to recover. A Zeiss LSM 900 KMAT confocal microscope equipped with a Zeiss Plan-Apochromat 20x/0.8 dry objective, 488 nm Diode laser and a GaASP-PMT detector (detection window of 490-565 nm) was used to acquire the images. For several roots 3μm step size Z-stacks were taken over 120 timepoints with a time interval of 720s with 8 bit depth, 1024x1024 pixels, pixel time of 0.52 μs, bidirectional scanning and 4 times line averaging. A tip track Matlab script111 was used for tracking the root tips. Afterwards, assembly of the obtained videos was performed using the provided Fiji/Image script described in111.
N. benthamiana transient expression-based imaging
Imaging the interactions between TASH3 and AtEH/Pan1 in N. benthamiana was performed on a PerkinElmer UltraView spinning-disk system, attached to a Nikon Ti inverted microscope and operated using the Volocity software package (Quorum Technologies Inc.). Images were acquired on an 512x512 pixel ImagEM CCD camera (Hamamatsu C9100-13) using frame-sequential imaging with a 60x water immersion objective (NA = 1.40). Specific excitation and emission was achieved using a 488 nm laser combined with a single band pass filter (500-550 nm) for GFP. RFP was visualized using 561 nm laser excitation and a 410-480/580–650 nm detection window.
Endocytic dynamic imaging
Endocytic dynamics were imaged on the UltraView spinning-disk system (PerkinElmer) described above with similar laser and filter settings and using the Nikon Perfect Focus System (PFSIII) for Z-drift compensation. Images of hypocotyl epidermal cells of 3-day-old etiolated seedlings expressing single fluorescent markers were acquired with a 100x oil-immersion corrected objective (Plan Apo, NA = 1.45). Single-marker line movies were acquired with an exposure time of 1 s/frame. Movies were acquired with a duration of 2 or 3 min. Specific excitation and emission was achieved using a 488 nm laser combined with a single band pass filter (500-550 nm) for GFP. RFP was visualized using 561 nm laser excitation and a 410-480/580-650 nm dual band pass filter.
Image quantification
FM4-64 Uptake assays
Acquired FM4-64 labeled Arabidopsis root confocal images were analyzed using the Fiji software package. PM and cytosol regions of interest (ROIs) of individual epidermal cells were outlined using the Select Brush and Freehand selections tool, respectively and histograms of pixel intensities were generated for the indicated ROIs. Pictures which contained more than 1% saturated pixels were excluded from the quantifications. Cytoplasm/PM ratios were calculated from average intensities of the top 1% highest intensity pixels based on the histograms.
Quantification of endocytic dynamics
Densities of endocytic foci were measured using the Find Maxima function of the Fiji software package. In a single slice of the obtained videos, a ROI was selected in the middle of the image. The background was subtracted from this ROI and the number of endocytic foci was assessed using the Find Maxima function. We used the pixel size and the area to convert this to spots per μm2. For each of the analyzed sample sets, at least twelve cells from six different seedlings were analyzed.
Lifetimes of individual endocytic events were measured from kymographs generated by the Volocity software package (PerkinElmer). Lifetimes had to be measured manually using the Fiji software package. The cmeAnalysis package previously described in112,113 unfortunately could not be used with our samples due to the high fluorescent background present in nosh. For each of the analyzed sample sets, at least twelve cells from six different seedlings were analyzed.
Quantification PM versus cytoplasm fluorescence
Quantifications of PM over cytoplasm ratios were performed with a custom made Fiji-based script (available via GitHub - https://github.com/pegro-psb/Cyto-PM-signal-quantification).
The script uses the PI staining in the red channel as a mask to allow automatic detection of PM and cytoplasm regions in the GFP channel. GFP and PI images are merged together in Fiji. Cells are manually annotated and stored in the Fiji ROI Manager. The script automatically measures the top 5% mean intensity pixels of the channel 2 (GFP) in the detected PM and cytoplasm ROI of each annotated cell based on the PI signal in channel 1 (PI).
For quantification of GFP vs FM4-64 ratios within one cell, FM4-64 staining was used as a mask to detect plasma membrane and cytoplasm regions. GFP and FM4-64 images were merged together in Fiji and cells were manually annotated and stored in the Fiji ROI Manager. For quantification of the FM4-64 signal, acquired FM4-64 images were duplicated in Fiji and for one of them the LUT was changed to green. FM4-64-green and FM4-64-red images were merged together in Fiji and the ROIs stored in ROI Manager from GFP signal quantification were also used for FM4-64 internalization. The script automatically measured the top 5% mean intensity pixels of channel 2 (GFP or FM4-64-green) in the detected membrane and the cytoplasm ROI of each annotated cell based on the FM4-64 signal in channel 1 (FM4-64-red).
Partitioning assay quantification
Quantification of the partitioning assays in N. benthamiana between the TASH3 constructs and both AtEH/Pan1 proteins was performed using the MitoTally script114. Particle regions of interest were determined based on AtEH1/Pan1 or AtEH2/Pan1 positive foci.
Statistics and Reproducibility
All the statistical analyses were performed using RStudio (Version 1.2.5033) with R (RStudio, 2015). For all statistical analyses, a Mann-Whitney-Wilcoxon test was used except for Fig. 5i, where a linear mixed effect model was used to quantify the difference between GFP/FM4-64. Group characterization in Fig. 5 and Extended Data Fig. 5 was done via transformation of the p-values with the multicompView package.
Experiments presented in Figure 1d, 1f, 1h, 2a, 2c, 2e, 2g, 2i, 2k, 2m, 3a, 5a, 5d, 5f, 6a, 6c and in Extended Data Figure 2b, 2c and 7g were performed three times. Experiment presented in Figure 1b, 3d, 5i, Extended Data Figure 5b-5j and 8 were performed two times. Experiments presented in Figure 4c, 6b and in Extended Data Figure 7i-k were performed once during the revision period.
Extended Data
Extended Data Fig. 1. Phenotypical assessment of the nosh truncation.
a) Quantification of genotyping PCR reactions on the F1 progeny of tash3 1 (♂), tash3 2 (♂) and nosh (♂) backcrossed into Col-0 (♀) to evaluate the transfer of the T-DNA allele via the pollen. The male sterility of tash3-1 and tash3-2 mutants prevents transfer of the T-DNA insertion to the next generation (only WT band amplified). nosh mutants produce viable pollen and can transfer the T-DNA insertion to the next generation (WT band and T-DNA band amplified). b) Amino acid sequence of the TASH3 C-terminus and predicted amino acid sequence of the nosh C-terminus, based on the sequencing of the T-DNA insertion site. The body part sequence of TASH3 is marked in blue and the SH3 domain sequence is depicted in green. The sequence that is altered in nosh is underlined. c) Representative examples of 5-week-old Col-0 and nosh plants grown under long-day conditions (16h light/8h dark). Under these conditions, nosh exhibits predominantly delayed flowering. d) Representative examples of 8-week-old Col-0 and nosh plants grown at 12h light/12h dark conditions. Under these conditions, nosh exhibits reduced rosette growth and early senescence.
Extended Data Fig. 2. Complementation of nosh with full length TASH3-GFP restores its endocytic defects.
a-b) Representative single slice spinning disk images and box plot graphs of endocytic foci densities in epidermal dark grown hypocotyl cells of TPLATE-GFP (tplate), TASH3-GFP (tash3-1) and two independent TASH3-GFP expressing lines in the nosh background. The densities of endocytic foci in both complemented nosh mutants are similar to the values in TPLATE-GFP (tplate) and to those in the complemented tash3-1 mutant allele (TASH3-GFP in tash3-1). Numbers of quantified cells (2 cells per seedling) are indicated. The top and bottom lines of box plots represent 25th and 75th percentiles, the center line is the median and whiskers are the full data range. The statistical test used was a two-sided Wilcoxon-signed rank test by comparing mutants to wild type. No adjustment for multiple comparisons was performed. Scale bar = 5 μm. c-d) Representative kymographs and violin plot graphs of the life-time measurements from the spinning disk time lapses from panel a. Analogous life-time distributions of endocytic events were observed for TASH3-GFP in nosh as for TPLATE-GFP (tplate) and TASH3-GFP (tash3-1). The number of events analyzed for each independent line is indicated at the bottom of each graph. At least 12 movies from 6 seedlings were imaged and analyzed for each independent transgenic line. The widest part of the violin plot represents the highest point density, whereas the top and bottom are the maximum and minimum data respectively. Red circles represent the mean and the red line represents the standard deviation. The statistical test used was a two-sided Wilcoxon-signed rank test by comparing mutants to wild type. No adjustment for multiple comparisons was performed. Scale bar = 50 μm. n.s. = not significant.
Extended Data Fig. 3. Comparative interactomics and AtEH1/Pan1 antibody specification.
a) Graph depicting the normalized peptide intensities of TPC subunits obtained from MS analysis. For each TPC subunit, the intensities of only those peptides that were present in all experiments (for both baits and in all replicas) were averaged and normalized to the values of the corresponding bait protein. Error bars correspond to ± SD and are based on three technical repeats. The results show that nosh does not affect the hexameric TPC formation, but that the association with the AtEH/Pan1 proteins is weakened. b) Coomassie stained gel of the obtained SEC fractions and a quality control HPLC analysis performed using a Superdex 200 increase 10/300 for the batch of recombinant AtEH1 C-term fragment used for rabbit immunization. Fractions 18 - 23 were pooled to immunize (marked in green). c) Coomassie stained gel of the obtained SEC fractions and a quality control HPLC analysis performed using a Superdex 200 increase 10/300 for the batch used for antibody purification. Fractions 25 - 29 were pooled for purification (marked in green). d) Stain free gel and blot for testing AtEH1/Pan1 antibody specificity. In the Col-0 sample only the native AtEH1/Pan1 band is prominently observed (marked with a black arrowhead), while in the pH3.3::AtEH1/Pan1-mRuby3 (Col-0) sample, both the native and the transgene fusion protein (marked with a white arrowhead) can be observed. Two homozygous pH3.3::AtEH1/Pan1-mRuby3 (ateh1/pan1 1-2 -/-) lines show only the transgene fusion protein (marked with white arrowhead).
Extended Data Fig. 4. A TSAUCER-like variant is produced in nosh.
a) TASH3 protein structure with annotated peptides identified by MS analysis. Peptides marked in green were present in both samples, while peptides in magenta were missing in the TPLATE-GFP (nosh/tplate) sample. b) Amino acid sequence alignment of TASH3 (Arabidopsis) and TSAUCER (Dictyostelium) showing that the linker and SH3 domain are missing at the C-terminus of the TSAUCER protein (outlined in red). c) Phylogenetic tree representing the maximum likelihood phylogeny of TASH3. Numbers at nodes correspond to the approximate likelihood ratio test with Shimodaira–Hasegawa-like support. Red circles mark the presence of SH3 domain(s).
Extended Data Fig. 5. TASH3 interacts with AtEH/Pan1 subunits through its body and not via its SH3 domain.
a) Schematic representation of AtEH1/Pan1 and AtEH2/Pan1 proteins with PxxP motifs indicated by orange lines. b-d) Maximal intensity projection of representative Z-stacks of transiently expressed TASH3-GFP, TASH3_body-GFP and mCherry-TASH3_linker_SH3 in epidermal N. benthamiana cells, respectively. e-f) Maximal intensity projection of representative Z-stacks of TASH3-GFP recruitment to AtEH1/Pan1-mCherry and AtEH2/Pan1-mCherry positive foci upon transient co-expression in epidermal N. benthamiana cells. g-h) Maximal intensity projection of representative Z-stacks of TASH3_body-GFP recruitment to AtEH1/Pan1-mCherry and AtEH2/Pan1-mCherry positive foci upon transient co-expression in epidermal N. benthamiana cells. i-j) Representative Z-stacks showing that mCherry-TASH3_linker_SH3 is not recruited to AtEH1/Pan1-GFP or AtEH2/Pan1-GFP positive foci upon transient co-expression in epidermal N. benthamiana cells. White arrowheads indicate colocalization. Scale bar = 20 μm. k-l) Box plot graphs of the particle/cytoplasm intensity of TASH3, TASH3_body and TASH3_linker_SH3 upon transient co-expression of AtEH1/Pan1 and AtEH2/Pan1 in N. benthamiana. The number of Z-stacks analyzed for each combination is indicated at the bottom of each graph. The top and bottom lines of box plots represent 25th and 75th percentiles, the center line is the median and whiskers are the full data range. Letters represent a two-sided mixed linear model statistic used to determine the difference between samples. No adjustment for multiple comparisons was performed. One outlier is marked with a red asterix. TASH3 and TASH3_body are recruited to AtEH/Pan1, whereas TASH3_linker_SH3 is not.
Extended Data Fig. 6. Modelling of protein-protein interaction between TASH3_SH3 and UBIQUITIN10 or ATG8.
Three best-scoring models from two different modelling algorithms (HDOCK and AlphaFold2) show almost identical binding interfaces between TASH3_SH3 and UBIQUITIN10. In contrast, modelling ATG8a-TASH3_SH3 did not result in a single orientation of ATG8a towards the TASH3_SH3 domain as shown for three best-scoring models calculated by AlphaFold2 or HDOCK.
Extended Data Fig. 7. Recombinant TASH3_SH3 domain purification.
a) Protein sequence of the construct used for recombinant TASH3_SH3 production. b) Predicted structural model of amino acids 1136-1198 of TASH3 by Alphafold2. c-f) Purification of recombinant HIS-TEV-SH3 domain (TASH3). The collected fractions are indicated by a dotted line. c) Immobilized metal affinity chromatogram of the first step in the purification. d) Size exclusion chromatogram of the second step in the purification. The fractions collected in the first step were loaded on a HiLoad® 16/600 superdex®75pg column. e) Reverse Immobilized metal affinity chromatogram of the third step in the purification. HIS-TEV-SH3 collected in the second step was subjected to overnight TEV-cleavage with HIS-TEV protease and loaded on a HisprepTM Fast Flow 1 ml column (HIS-TEV was removed). f) Size exclusion chromatogram of the final step in the purification. The fractions collected in the third step were loaded on a HiLoad® 16/600 superdex®75pg column. g) Coomassie-stained SDS-PAGE gel of the different steps of the TASH3_SH3 domain purification process. IMAC – collected fractions from c, SEC-I – collected fractions from d, TEV cleavage - the cleavage of the HIS-TEV from the recombinant protein using a HIS-TEV protease, R.IMAC - the collected fractions from e, SEC-II – the collected fractions from f. HIS-TEV-SH3: 8.8kDa, SH3: 7.0kDa, HIS-TEV protease: 28kDa. h) Western blot detection using an anti-5xHIS antibody on the collected fractions of the second step of purification (SEC-II), showing that the collected fractions contain HIS-TEV-SH3. i) Coomassie-stained SDS-PAGE gel showing the coupling efficiency of HIS-HRV3C-GFP and TASH3_SH3 on PierceTM NHS-Activated agarose beads. Covalent coupling of the recombinant protein to the beads can be observed as a reduction in the intensity in the flow-through. j) Stain-freeTM SDS-PAGE gel showing Col-0 extracts which were incubated with the HIS-HRV3C-GFP or TASH3_SH3 coupled beads. k) Western blot detection using a general anti-Ubiquitin antibody (P4D1) showing Col-0 extracts which were incubated with the HIS-HRV3C-GFP or TASH3_SH3 coupled beads. The smear in the bound fraction indicates that the SH3 domain from TASH3 can bind ubiquitinated proteins as opposed to the HIS-HRV3C-GFP control. I – Input, FT - flow-through, B – bound fractions.
Extended Data Fig. 8. Analysis of PM fraction purity.
a) Immunoblot analysis of different fractions acquired using the Minute™ Plant Plasma Membrane Protein Isolation Kit. Collected fractions: total protein fraction (TP), nuclei and debris fraction (ND), cytosolic fraction (C), total membrane fraction (TM), organelle membrane fraction (OM) and plasma membrane fraction (PM). The obtained fractions were separated by SDS-PAGE, visualized using stain-free gel, blotted and probed with antibodies against the Aquaporin PIP2;7 (PIP2;7, a PM marker), cytosolic Ascorbate Peroxidase (cAPX, a cytosol marker) and Cytochrome C (CytC, a mitochondrial marker). Full length bands of the proteins are marked with arrowheads (PIP2;7 – black arrowhead; cAPX – gray arrowhead; CytC – white arrowhead). The antibodies used are listed at the right bottom corner beneath the blots.
Supplementary Material
Acknowledgements
We would like to thank Prof. Tongda Xu (FAFU-UCR, Fuzhou) and Prof. Jiří Friml (IST Austria) for sharing TMK1 seeds, Prof. Gregory Vert (CNRS, Toulouse) for sharing BRI1 and BRI1-25KR seeds, Prof. Eugenia Russinova for providing 35S::eGFP seeds and Prof. Ray Owens (OPPF, Research Complex, Harwell) for sharing an aliquot of the HIS-GFP vector. We express our gratitude to the VIB proteomics core facility for the help and expertise with running all MS experiments and the VIB protein core facility for the help and expertise with protein purifications. This work was supported by the European Research Council Grant T-REX 682436 (D.V.D.); The Research Foundation–Flanders (FWO) 1226420N (P.G.), 12S7222N (J.M.D.), 1124621N (A.D.M.), G017919N (M.K.); The Czech Science Foundation 22-35680M (R.P.) and the China Scholarship Council Grant 201906760018 (Q.J).
Footnotes
Author contributions
P.G. and D.V.D. designed the research and wrote the manuscript. P.G. performed most of the experiments. A.D.M. and K.Y. cloned and purified SH3 domain. A.D.M. performed SH3-ubiquitin binding assay, partitioning assay and PM fraction analysis. J.D.M. and M.V. helped with cloning. R.P. performed in silico docking and phylogenetic analysis. D.E. performed MS analysis. M.K. raised and characterized the AtEH1/Pan1 antibody. J.N. prepared samples for APMS analysis. Q.J. and B.P. created the script for measuring fluorescent signal on confocal images. E.M. performed Tip tracking experiments. B.D.R., G.D.J., D.V.D. and P.G. were responsible for experimental design and research supervision. All authors contributed to finalizing the text.
Competing interests
The authors declare no competing interests.
Data availability
All materials are available from the corresponding authors upon request. All data generated or analyzed during this study are included in this published article (and its Extended Data information files) and/or in public repositories. The raw mass spectrometry data and MaxQuant result files have been deposited to the ProteomeXchange Consortium via PRIDE (PXD035444). The script for comparative quantification of fluorescent signal at PM versus cytoplasm is available for download (https://github.com/pegro-psb/Cyto-PM-signal-quantification). Araport11plus database consisting of the Araport11_genes.2016.06.pep.fasta downloaded from arabidopsis.org was used for MS analysis.
References
- 1.Bitsikas V, Corrêa IR, Nichols BJ. Clathrin-independent pathways do not contribute significantly to endocytic flux. Elife. 2014;3:1–26. doi: 10.7554/eLife.03970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19(5):313–326. doi: 10.1038/nrm.2017.132. 2018. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds GD, Wang C, Pan J, Bednarek SY. Inroads into Internalization: Five Years of Endocytic Exploration. Plant Physiol. 2018;176:208–218. doi: 10.1104/pp.17.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadeyne A, et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Hirst J, et al. Characterization of TSET, an ancient and widespread membrane trafficking complex. Elife. 2014;2014 doi: 10.7554/eLife.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme D, et al. Somatic Cytokinesis and Pollen Maturation in Arabidopsis Depend on TPLATE, Which Has Domains Similar to Coat Proteins. Plant Cell. 2006;18:3502–3518. doi: 10.1105/tpc.106.040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashline L, Li S, Zhu X, Gu Y. The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc Natl Acad Sci U S A. 2015;112:12870–12875. doi: 10.1073/pnas.1509292112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno H, et al. The Medium Subunits of Adaptor Complexes Recognize Distinct but Overlapping Sets of Tyrosine-based Sorting Signals. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- 9.Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. Conservation and Diversification of Dileucine Signal Recognition by Adaptor Protein (AP) Complex Variants. J Biol Chem. 2011;286:2022. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora D, van Damme D. Motif-based endomembrane trafficking. Plant Physiol. 2021;186:221–238. doi: 10.1093/plphys/kiab077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happel N, et al. ArabidopsisμA-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 2004;37:678–693. doi: 10.1111/j.1365-313x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 12.Takano J, et al. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci U S A. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mravec J, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–40. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 14.Yoshinari A, et al. Polar Localization of the Borate Exporter BOR1 Requires AP2-Dependent Endocytosis. Plant Physiol. 2019;179:1569–1580. doi: 10.1104/pp.18.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, et al. Endocytosis of BRASSINOSTEROID INSENSITIVE Is Partly Driven by a Canonical Tyr-Based Motif. Plant Cell. 2020;32:3598–3612. doi: 10.1105/tpc.20.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yperman K, et al. Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding. Nat Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-23314-6. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 19.Erwig J, et al. Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5) New Phytol. 2017;215:382–396. doi: 10.1111/nph.14592. [DOI] [PubMed] [Google Scholar]
- 20.Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10(9):583–596. doi: 10.1038/nrm2751. 2009. [DOI] [PubMed] [Google Scholar]
- 21.Dubeaux G, Vert G. Zooming into plant ubiquitin-mediated endocytosis. Curr Opin Plant Biol. 2017;40:56–62. doi: 10.1016/j.pbi.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Fan L, Li R, Pan J, Ding Z, Lin J. Endocytosis and its regulation in plants. Trends Plant Sci. 2015;20:388–397. doi: 10.1016/j.tplants.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Nagel MK, et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci U S A. 2017;114:E7197–E7204. doi: 10.1073/pnas.1710866114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg JS, Drubin DG. Regulation of clathrin-mediated endocytosis by dynamic ubiquitination and deubiquitination. Curr Biol. 2014;24:951–959. doi: 10.1016/j.cub.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinari A, et al. DRP1-Dependent Endocytosis is Essential for Polar Localization and Boron-Induced Degradation of the Borate Transporter BOR1 in Arabidopsis thaliana. Plant Cell Physiol. 2016;57:1985–2000. doi: 10.1093/pcp/pcw121. [DOI] [PubMed] [Google Scholar]
- 26.Leitner J, et al. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci U S A. 2012;109:8322–8327. doi: 10.1073/pnas.1200824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barberon M, et al. Monoubiquitin-dependent endocytosis of the Iron-Regulated Transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci U S A. 2011;108 doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vert G, et al. IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and for Plant Growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao D, et al. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017;214:1646–1656. doi: 10.1111/nph.14472. [DOI] [PubMed] [Google Scholar]
- 30.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci U S A. 2018;115:E1906–E1915. doi: 10.1073/pnas.1712251115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korbei B, et al. Arabidopsis TOL Proteins Act as Gatekeepers for Vacuolar Sorting of PIN2 Plasma Membrane Protein. Curr Biol. 2013;23:2500–2505. doi: 10.1016/j.cub.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 33.Moulinier-Anzola J, et al. TOLs Function as Ubiquitin Receptors in the Early Steps of the ESCRT Pathway in Higher Plants. Mol Plant. 2020;13:717–731. doi: 10.1016/j.molp.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Arora D, et al. Establishment of Proximity-Dependent Biotinylation Approaches in Different Plant Model Systems. Plant Cell. 2020;32:3388–3407. doi: 10.1105/tpc.20.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Conditional destabilization of the TPLATE complex impairs endocytic internalization. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2023456118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yperman K, et al. Molecular architecture of the endocytic TPLATE complex. Sci Adv. 2021;7 doi: 10.1126/sciadv.abe7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun. 2019;10 doi: 10.1038/s41467-019-12782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Damme D, et al. Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci U S A. 2011;108:615–620. doi: 10.1073/pnas.1017890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutté Y, et al. Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 2010;29:546–558. doi: 10.1038/emboj.2009.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nat. 1988;332(6161):272–275. doi: 10.1038/332272a0. 1988. [DOI] [PubMed] [Google Scholar]
- 41.Kurochkina N, Guha U. SH3 domains: modules of protein-protein interactions. Biophys Rev. 2013;5:29–39. doi: 10.1007/s12551-012-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamenova SD, et al. Ubiquitin binds to and regulates a subset of SH3 domains. Mol Cell. 2007;25:273. doi: 10.1016/j.molcel.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang J, Kang S, Hyuk NK, He W, Park S. Distinct interactions between ubiquitin and the SH3 domains involved in immune signaling. Biochim Biophys Acta. 2008;1784:1335–1341. doi: 10.1016/j.bbapap.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 44.Bu F, Yang M, Guo X, Huang W, Chen L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front Cell Dev Biol. 2020;8:466. doi: 10.3389/fcell.2020.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwihla M, Korbei B. The Beginning of the End: Initial Steps in the Degradation of Plasma Membrane Proteins. Front Plant Sci. 2020;11:680. doi: 10.3389/fpls.2020.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bashline L, Li S, Anderson CT, Lei L, Gu Y. The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 2013;163:150–160. doi: 10.1104/pp.113.221234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lian N, et al. COP1 mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation. Proc Natl Acad Sci U S A. 2017;114:12321–12326. doi: 10.1073/pnas.1708087114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JH, Kim WT. The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 2013;162:1733–1749. doi: 10.1104/pp.113.220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svozil J, Hirsch-Hoffmann M, Dudler R, Gruissem W, Baerenfaller K. Protein abundance changes and ubiquitylation targets identified after inhibition of the proteasome with syringolin A. Mol Cell Proteomics. 2014;13:1523–1536. doi: 10.1074/mcp.M113.036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walton A, et al. It’s Time for Some “Site”-Seeing: Novel Tools to Monitor the Ubiquitin Landscape in Arabidopsis thaliana. Plant Cell. 2016;28:6–16. doi: 10.1105/tpc.15.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson A, Vert G. Unraveling K63 Polyubiquitination Networks by Sensor-Based Proteomics. Plant Physiol. 2016;171:1808–1820. doi: 10.1104/pp.16.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguilar-Hernández V, et al. Mass Spectrometric Analyses Reveal a Central Role for Ubiquitylation in Remodeling the Arabidopsis Proteome during Photomorphogenesis. Mol Plant. 2017;10:846–865. doi: 10.1016/j.molp.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero-Barrios N, et al. Advanced Cataloging of Lysine-63 Polyubiquitin Networks by Genomic, Interactome, and Sensor-Based Proteomic Analyses. Plant Cell. 2020;32:123–138. doi: 10.1105/tpc.19.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grubb LE, et al. Large-scale identification of ubiquitination sites on membrane-associated proteins in Arabidopsis thaliana seedlings. Plant Physiol. 2021;185:1483–1488. doi: 10.1093/plphys/kiab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martins S, et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun. 2015;6 doi: 10.1038/ncomms7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler J, et al. Nanobody-Dependent Delocalization of Endocytic Machinery in Arabidopsis Root Cells Dampens Their Internalization Capacity. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.538580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan L, et al. Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Dev. 2013;140:3826–3837. doi: 10.1242/dev.095711. [DOI] [PubMed] [Google Scholar]
- 58.Wendrich JR, et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020;370 doi: 10.1126/science.aay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berrío RT, et al. Single-cell transcriptomics sheds light on the identity and metabolism of developing leaf cells. Plant Physiol. 2022;188:898–918. doi: 10.1093/plphys/kiab489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song Q, Ando A, Jiang N, Ikeda Y, Chen ZJ. Single-cell RNA-seq analysis reveals ploidy-dependent and cell-specific transcriptome changes in Arabidopsis female gametophytes. Genome Biol. 2020;21 doi: 10.1186/s13059-020-02094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin D, et al. Rho GTPase Signaling Activates Microtubule Severing to Promote Microtubule Ordering in Arabidopsis. Curr Biol. 2013;23:290–297. doi: 10.1016/j.cub.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Abas L, et al. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol. 2006;8(3):249–256. doi: 10.1038/ncb1369. 2006. [DOI] [PubMed] [Google Scholar]
- 63.Leitner J, Retzer K, Korbei B, Luschnig C. Dynamics in PIN2 auxin carrier ubiquitylation in gravity-responding Arabidopsis roots. 2012;7:1271–1273. doi: 10.4161/psb.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshinari A, et al. Transport-coupled ubiquitination of the borate transporter BOR1 for its boron-dependent degradation. Plant Cell. 2021;33:420–438. doi: 10.1093/plcell/koaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo Y, et al. Deubiquitinating enzymes UBP12 and UBP13 stabilize the brassinosteroid receptor BRI1. EMBO Rep. 2022 doi: 10.15252/embr.202153354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6(8):610–621. doi: 10.1038/nrm1701. 2005. [DOI] [PubMed] [Google Scholar]
- 67.Hawryluk MJ, et al. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;7:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 68.Sorkina T, et al. RNA interference screen reveals an essential role of Nedd4-2 in dopamine transporter ubiquitination and endocytosis. J Neurosci. 2006;26:8195–8205. doi: 10.1523/JNEUROSCI.1301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 70.Mayers JR, et al. Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc Natl Acad Sci U S A. 2013;110:11857–11862. doi: 10.1073/pnas.1302918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schuh AL, Audhya A. The ESCRT machinery: From the plasma membrane to endosomes and back again. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adamowski M, Matijević I, Narasimhan M, Friml J. SH3Ps recruit auxilin-like vesicle uncoating factors into clathrin-mediated endocytosis. doi: 10.1101/2022.01.07.475403. [DOI] [PubMed] [Google Scholar]
- 73.Pashkova N, et al. Article WD40 Repeat Propellers Define a Ubiquitin-Binding Domain that Regulates Turnover of F Box Proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Isono E, et al. The Deubiquitinating Enzyme AMSH3 Is Required for Intracellular Trafficking and Vacuole Biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22:1826–1837. doi: 10.1105/tpc.110.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katsiarimpa A, et al. The Arabidopsis Deubiquitinating Enzyme AMSH3 Interacts with ESCRT-III Subunits and Regulates Their Localization. Plant Cell. 2011;23:3026–3040. doi: 10.1105/tpc.111.087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katsiarimpa A, et al. The Deubiquitinating Enzyme AMSH1 and the ESCRT-III Subunit VPS2.1 Are Required for Autophagic Degradation in Arabidopsis. Plant Cell. 2013;25:2236. doi: 10.1105/tpc.113.113399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katsiarimpa A, et al. The ESCRT-III-Interacting Deubiquitinating Enzyme AMSH3 is Essential for Degradation of Ubiquitinated Membrane Proteins in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:727–736. doi: 10.1093/pcp/pcu019. [DOI] [PubMed] [Google Scholar]
- 78.Ingouff M, et al. Live-cell analysis of DNA methylation during sexual reproduction in arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017;31:72–83. doi: 10.1101/gad.289397.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karimi M, Depicker A, Hilson P. Recombinational Cloning with Plant Gateway Vectors. Plant Physiol. 2007;145:1144. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lampropoulos A, et al. GreenGate-A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. PLoS One. 2013;8:e83043. doi: 10.1371/journal.pone.0083043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waadt R, et al. Dual-Reporting Transcriptionally Linked Genetically Encoded Fluorescent Indicators Resolve the Spatiotemporal Coordination of Cytosolic Abscisic Acid and Second Messenger Dynamics in Arabidopsis. Plant Cell. 2020;32:2582–2601. doi: 10.1105/tpc.19.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waadt R, Krebs M, Kudla J, Schumacher K. Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis. New Phytol. 2017;216:303–320. doi: 10.1111/nph.14706. [DOI] [PubMed] [Google Scholar]
- 83.Decaestecker W, et al. CRISPR-TSKO: A Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis. Plant Cell. 2019;31:2868–2887. doi: 10.1105/tpc.19.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mertens N, Remaut E, Fiers W Versatile. multi-featured plasmids for high-level expression of heterologous genes in Escherichia coli: overproduction of human and murine cytokines. Gene. 1995;164:9–15. doi: 10.1016/0378-1119(95)00505-z. [DOI] [PubMed] [Google Scholar]
- 85.Kim SY, et al. Adaptor Protein Complex 2-Mediated Endocytosis Is Crucial for Male Reproductive Organ Development in Arabidopsis. Plant Cell. 2013;25:2970–2985. doi: 10.1105/tpc.113.114264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mravec J, et al. Report Cell Plate Restricted Association of DRP1A and PIN Proteins Is Required for Cell Polarity Establishment in Arabidopsis. Curr Biol. 2011;21:1055–1060. doi: 10.1016/j.cub.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 87.Reichardt I, et al. Plant Cytokinesis Requires De Novo Secretory Trafficking but Not Endocytosis. Curr Biol. 2007;17:2047–2053. doi: 10.1016/j.cub.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 88.Dejonghe W, et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat Chem Biol. 2019;15:641–649. doi: 10.1038/s41589-019-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu T, et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343:1025–8. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Di Rubbo S, et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell. 2013;25:2986–2997. doi: 10.1105/tpc.113.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belkhadir Y, et al. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci U S A. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1(4):2019–2025. doi: 10.1038/nprot.2006.286. 2006. [DOI] [PubMed] [Google Scholar]
- 93.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guindon S, et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 98.Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varadi M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mirdita M, et al. ColabFold-Making protein folding accessible to all. 2021 doi: 10.21203/rs.3.rs-1032816/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure. 2020;28:1071–1081.:e3. doi: 10.1016/j.str.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan Y, Tao H, He J, Huang SY. The HDOCK server for integrated proteinprotein docking. Nat Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 104.Pierce BG, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pettersen EF, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng Des Sel. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 108.Dahhan DA, et al. Proteomic characterization of isolated Arabidopsis clathrin-coated vesicles reveals evolutionarily conserved and plant-specific components. Plant Cell. 2022 doi: 10.1093/PLCELL/KOAC071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wendrich JR, Boeren S, Möller BK, Weijers D, De Rybel B. In Vivo Identifi cation of Plant Protein Complexes Using IP-MS/MS. Methods Mol Biol. 1497 doi: 10.1007/978-1-4939-6469-7_14. [DOI] [PubMed] [Google Scholar]
- 110.Sauer M, Paciorek T, Benkovó E, Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
- 111.von Wangenheim D, et al. Live tracking of moving samples in confocal microscopy for vertically grown roots. Elife. 2017;6 doi: 10.7554/eLife.26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson A, Vert G. Single Event Resolution of Plant Plasma Membrane Protein Endocytosis by TIRF Microscopy. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Narasimhan M, et al. Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. Elife. 2020;9 doi: 10.7554/eLife.52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Winkler J, et al. Visualizing protein-protein interactions in plants by rapamycin-dependent delocalization. Plant Cell. 2021;33:1101–1117. doi: 10.1093/plcell/koab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials are available from the corresponding authors upon request. All data generated or analyzed during this study are included in this published article (and its Extended Data information files) and/or in public repositories. The raw mass spectrometry data and MaxQuant result files have been deposited to the ProteomeXchange Consortium via PRIDE (PXD035444). The script for comparative quantification of fluorescent signal at PM versus cytoplasm is available for download (https://github.com/pegro-psb/Cyto-PM-signal-quantification). Araport11plus database consisting of the Araport11_genes.2016.06.pep.fasta downloaded from arabidopsis.org was used for MS analysis.