Summary
Gene transcription is a highly regulated process in all animals. In Drosophila, two major transcriptional programs, housekeeping and developmental, have promoters with distinct regulatory compatibilities and nucleosome organization. However, it remains unclear how the differences in chromatin structure relate to the distinct regulatory properties and which chromatin remodelers are required for these programs. Using rapid degradation of core remodeler subunits in Drosophila melanogaster S2 cells, we demonstrate that developmental gene transcription requires SWI/SNF-type complexes, primarily to maintain distal enhancer accessibility. In contrast, housekeeping gene transcription requires the Iswi and Ino80 remodelers to maintain nucleosome positioning and phasing at promoters. These differential remodeler dependencies relate to different DNA sequence-intrinsic nucleosome affinities, which favor a default ON state for housekeeping but a default OFF state for developmental gene transcription, respectively. Overall, our results demonstrate how different transcription-regulatory strategies are implemented by DNA sequence, chromatin structure and remodeler activity.
Introduction
The differential transcription of protein coding genes by RNA Polymerase II (Pol II) is a major determinant of cell types and state in multicellular organisms. In Drosophila, two major classes of genes, the ubiquitously expressed housekeeping genes and highly cell-type specific developmental genes, have characteristically distinct promoters. These not only differ in the occurrence of core-promoter sequence elements such as the Ohler 1 motifs or the TATA-box, respectively (FitzGerald et al., 2006; Haberle and Stark, 2018; Kadonaga, 2012; Lenhard et al., 2012; Ohler et al., 2002), but also display different regulatory properties and differentially respond to distinct enhancer types and cofactors (Haberle et al., 2019; Stampfel et al., 2015; Zabidi et al., 2015). In addition to these differences in sequence and function, the chromatin structure is also noticeably different (Figure 1A).Although promoters in general display characteristic nucleosome positioning and phasing (Mavrich et al., 2008), housekeeping promoters have a more stereotypical nucleosome arrangement with a well-defined nucleosome-free region and a strongly positioned +1 nucleosome (Haberle et al., 2019; Rach et al., 2011), whereas developmental promoters have a less pronounced nucleosome-free region and +1 nucleosome, and much weaker downstream nucleosome phasing. Nucleosomes prevent DNA access of regulatory proteins such as transcription factors or the transcription machinery (Groth et al., 2007; Radman-Livaja and Rando, 2010), but it remains unclear whether and how these differences in chromatin structure at promoters relate to the distinct regulatory properties of developmental versus housekeeping gene transcription.
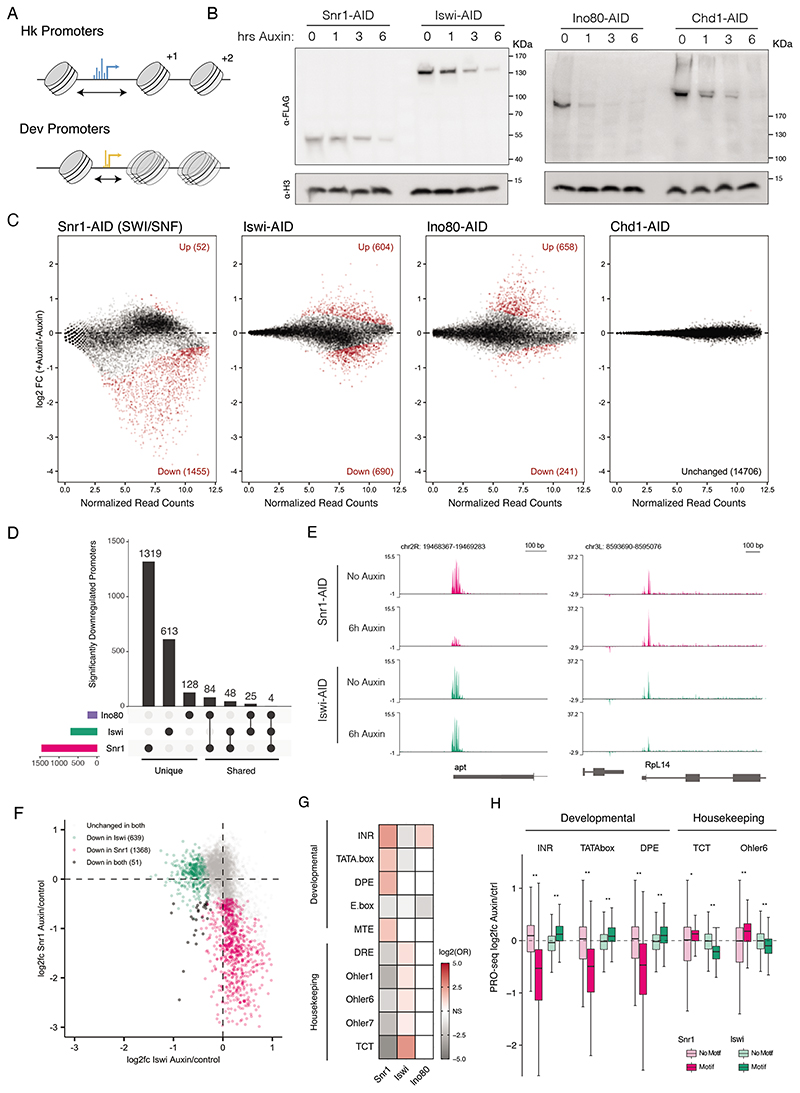
Figure 1. Chromatin remodeler depletion affects distinct gene sets.
A) Diagram of distinct nucleosome positioning at housekeeping (top) and developmental (bottom) promoter types. Housekeeping type promoters display wide NFRs (black arrow), dispersed initiation (blue) and precise downstream nucleosome positioning, while developmental type promoters have shorter NFRs, focused initiation (yellow) and fuzzy nucleosome positioning. B) Anti-FLAG (top) and anti-H3 (bottom) western blots against AID-tagged remodelers from engineered S2 cells after 0, 1, 3, and 6 hours of auxin treatment (molecular weights in KDa indicated by ticks).C) MA plots of differential transcription (PRO-seq at promoter regions of - 50bp to +150bp around the TSS) after 6 hours of auxin treatment vs. control (red points are padj < 0.05). D) Upset plot of significantly downregulated promoters after Ino80, Iswi, or Snr1 depletion. Horizontal bars indicate the total affected genes per perturbation (colors signify perturbed remodeler), vertical bars indicate number of promoters specific to a single perturbation or the indicated intersections. E)Genome browser screen shots of the apontic and Rpl14 loci showing normalized PRO-seq read coverage (library normalized counts per million) per condition. F) Comparison of gene expression changes after Snr1 depletion vs. Iswi depletion. PRO-seq promoter log2 fold-changes after Iswi/Snr1 depletion, non-affected promoters in gray, uniquely affected promoters in pink and green, and promoters affected in both conditions in black. G) Enrichment of housekeeping and developmental motifs in affected gene sets (OR: odds ratio; colors see legends; white for non-significant trends, i.e. Fisher test p-values > 0.05).H)Log2 fold-change of promoter groups split by motif presence after Snr1 or Iswi depletion (* p < 1x10-5, ** p < 2.2x10-16,Wilcoxon-test).
Chromatin remodelers facilitate nucleosome assembly, repositioning, and removal to shape chromatin structure, and are therefore likely key factors for establishing the nucleosome environment of developmental and housekeeping genes. Remodelers have been classified into four sub-families that share the same core ATPase domain but differ in domain organization and complex composition: SWI/SNF, Iswi, Ino80, and Chd (for a review and overview over complex composition and function see: Clapier et al., 2017; Clapier and Cairns, 2009).In Drosophila, there are two SWI/SNF-type complexes, BAP and PBAP,which are orthologous to yeast SWI/SNF and mammalian BAF/PBAF. Both Drosophila SWI/SNF-type complexes share the ATPase Brahma (also called Brm)and core subunits such as Snr1, and mediate nucleosome removal to grant accessibility of activators to DNA (Moshkin et al., 2007), but differ in some peripheral subunits (Tamkun et al., 1992, Mohrmann et al., 2004).The Iswi ATPase is found in three remodeling complexes: NURF, ACF and CHRAC (Ito et al., 1997; Tsukiyama et al., 1995; Tsukiyama and Wu, 1995; Varga-Weisz et al., 1997).Iswi complexes can assemble histones onto DNA and slide nucleosomes to create nucleosome free regions as well as regularly spaced nucleosome arrays (Corona et al., 2007; Fyodorov et al., 2004). Ino80 and Swr1 comprise the Ino80 family remodelers, and have been shown to be important for exchange of the Histone variant H2AZ at promoters (Mizuguchi et al., 2004; Shen et al., 2000; Yen et al., 2013). Finally, the Chd family remodelers are a family of at least 5 chromodomain containing remodelers that have diverse functions in transcription, replication, and DNA repair (Marfella and Imbalzano, 2007).
In yeast, specialized roles for these remodelers have been well characterized through reconstitution and deletion/depletion experiments: SWI/SNF establishes DNA accessibility at the promoter and enhances activator binding (Côté et al., 1994; Hartley and Madhani, 2009)whereas other remodelers such as Iswi and Chd1 set the appropriate downstream nucleosome positioning and spacing (Gkikopoulos et al., 2011; Krietenstein et al., 2016; Kubik et al., 2018; Parnell et al., 2015). However, the direct roles of individual remodelers in multicellular organisms with complex regulatory landscapes and distinct promoter types have received less attention. In Drosophila, loss of these remodelers is associated with early developmental lethality (Bhatia et al., 2010; Brizuela et al., 1994; Deuring et al., 2000) suggesting a critical role in early development or even basic cellular functions; however, the direct gene-regulatory changes underlying these defects cannot be determined from stable mutations. The potential for regulatory specialization of various essential regulators is supported by previous studies in human and flies which have shown that certain promoter and enhancer types can depend on different cofactors, including Mediator and/or P300 (Boija et al., 2017; Jaeger et al., 2020). Additionally, recent studies have found that DNA-binding of various factors, including CTCF and Oct4, differentially depend on the SWI/SNF and Iswi remodelers (Barisic et al., 2019; Liscovitch-Brauer et al., 2021). However, whether such dependencies on chromatin remodelers affect nucleosome organization and transcription at different promoter types and how this relates to the promoter’s regulatory properties have remained unclear.
Here, we combine rapid inducible protein depletion with nascent transcriptomics to uncover the genome-wide transcriptional dependencies on the four main classes of chromatin remodelers in Drosophila S2 cells. We demonstrate that the transcription of developmental genes specifically depends on the SWI/SNF-type complexes BAP/PBAP (hereafter referred to collectively as SWI/SNF), primarily because SWI/SNF is required to maintain the DNA accessibility of their enhancers. In contrast, housekeeping transcription is independent of SWI/SNF but modulated by Iswi and Ino80, which maintain these promoters’ highly stereotypical nucleosome arrangement. The differences in chromatin structure and remodeler dependencies relate to distinct DNA-sequence encoded nucleosome affinities, which are low at promoter-proximal housekeeping enhancers but high at promoter-distal developmental enhancers. Together, our results show how different chromatin structures and remodelers support the highly-cell-type specific versus broad nature of developmental versus housekeeping gene expression, respectively.
Results
Chromatin remodeler depletion affects different sets of genes
Given the differences in nucleosome positioning at the two major promoter types in Drosophila (Figure 1A), we hypothesized that they require different remodelers for transcriptional activation. To assess the direct regulatory targets of remodelers, we took advantage of auxin-inducible degradation (AID) (Nishimura et al., 2009) to rapidly deplete these proteins in Drosophila S2 cells. Specifically, we integrated a dual AID-FLAG tag and a selectable marker homozygously into the endogenous remodeler gene loci. For this, we used CRISPR/Cas9-based precise integration into target chromosome (PITCh) (Sakuma et al., 2016) in a parental cell line expressing the complementary E3 ligase from Oryza Sativa (Figure S1A). We confirmed correct homozygous integration of the tag with PCR and Sanger sequencing. Using this approach, we AID-FLAG-tagged selected core proteins from the four main sub-families of ATP dependent chromatin remodelers. To assess SWI/SNF transcriptional dependencies, we tagged Snr1, a core component of the SWI/SNF complex orthologous to human SMARCB1, which contacts the nucleosome core and is required for SWI/SNF assembly (He et al., 2020; Mashtalir et al., 2018) (we were unable to tag the core ATPase of SWI/SNF, brm). As Snr1 is a core SWI/SNF component found in both the BAP and PBAP complex types, its disruption should affect all SWI/SNF complexes.Indeed, Snr1 depletion releases Brm from the chromatin into the nucleoplasm (Figure S1C-E), confirming that Snr1 loss affects SWI/SNF complex integrity and/or localization to chromatin (Nakayama et al., 2017). We AID tagged ATP-translocase remodeler subunits of the remaining subfamilies, Iswi, Ino80, and Chd1 (we chose Chd1 as a representative of the diverse Chd family of remodelers, as it has been associated with maintenance of open chromatin and transcriptional elongation (Pray-Grant et. al., 2005 Sims et al., 2007)). Depletion of Iswi should affect the function of all Iswi complexes, including NURF, ACF and CHRAC. The tagged chromatin remodeler subunits were detected via the integrated FLAG-tag on a western blot at their expected sizes and showed close to complete degradation after 1 to 6 hours of treatment with 100 uM auxin (Figure 1B). After 24 hours, this depletion resulted in a strong proliferation defect for Snr1 and Ino80, a mild growth defect for Iswi, and no change for Chd1 (Figure S1B), suggesting that SWI/SNF, Iswi and Ino80, but not Chd1, are required in S2 cells, consistent with previous reports (Boutros et al., 2004). To confirm that the tagged remodelers, particularly Chd1, were indeed depleted, we performed quantitative mass-spectroscopy on nuclear lysates before and after auxin treatment. All targeted remodeler subunits were significantly depleted (Snr1, Iswi, and Chd1, Figure S1G) or undetectable (Ino80, Figure S1G-I) after auxin treatment, confirming tagging of all alleles and the functionality of the AID tag.
To assess the direct transcriptional consequences of depleting each remodeler, we combined acute degradation of these proteins with the nascent transcription assay precision run-on sequencing (PRO-seq) (Core et al., 2008),using spike-ins. For each AID cell line, we performed PRO-seq under auxin-treated (6 hours after treatment) and control conditions, each with two independent replicates, which showed an overall good agreement (pairwise Pearson-correlation coefficients (PCC)>0.95 between replicates; Figure S2A). We assessed differential protein-coding gene transcription between the auxin and control conditions by the PRO-seq signal at transcription-start-site (TSS) proximal positions (- 50,+150bp around the TSS; analysing PRO-seq signal at gene bodies showed consistent effects, ruling out any effects on pausing or elongation; Figure S2B-C) using DeSeq2 (Love et al., 2014). Auxin-treatment of the parental cell line containing no AID-FLAG-tagged protein showed no significant changes in transcription (at FDR<0.05), ruling out any off-target effects from OsTir1 in the presence of auxin or indirect effects from auxin treatment itself (Figure S1F). We also did not observe significant changes in transcription (at FDR<0.05) upon auxin-induced degradation of Chd1, consistent with previous results from RNAi-mediated depletion of Chd1 (Boutros et al., 2004). In contrast, depletion of Snr1 led to strong down-regulation of 1455 genes, depletion of Iswi had moderate effects in both directions (604 genes were up and 690 genes down-regulated), and Ino80 depletion led mainly to up-regulation (658 genes, only 241 genes were moderately down-regulated; Figure 1C).
We observed little or no overlap between the genes that were down-regulated upon depletion of the Snr1 and Iswi (Figure 1D). Of the 1455 and 690 genes down-regulated after Snr1 or Iswi depletion, respectively, only 48 were affected in both conditions. Ino80 caused only slight downregulation of 241 genes, which were partially overlapping with those affected after Snr1 depletion. These results did not depend on the significance threshold: genes affected by Snr1 depletion were generally unaffected by Iswi depletion and vice versa. For example, transcription of the gene encoding the transcription factor apontic (apt) was strongly dependent on Snr1 but independent of Iswi, whereas the opposite was true for the gene encoding ribosomal protein RpL14 (Figure 1E). These opposite trends were also true globally for essentially all affected genes (Figure 1F). Thus, different remodelers sharing the same core activity are required for transcription of distinct target genes.
To test whether the difference in regulatory behavior is explained by a requirement for specific remodelers at different promoter classes, we asked whether any of the canonical core promoter motifs were over-represented in the downregulated promoters. Indeed, promoters impaired by SWI/SNF depletion were strongly enriched for the initiator motif (Inr) and the TATA box, whereas promoters affected by Iswi depletion were enriched for the Ohler 1, 6 and 7 Motifs, and the TCT motif (Figure 1G). Given the characteristic occurrence of these motifs in developmental versus housekeeping type promoters, respectively, these analyses suggest a dependence of developmental gene transcription on SWI/SNF and of housekeeping transcription on Iswi. In contrast, Ino80-dependent promoters were only weakly enriched for the broadly occurring Inr motif, suggesting that Ino80 is not a general dependency of one of the main transcriptional programs in Drosophila. GO-term enrichment confirms these trends, including an enrichment of developmental GO terms for the SWI/SNF-dependent genes and of housekeeping GO terms, including ribosomal protein genes, among the Iswi-dependent genes (Figure S2D-E). These analyses reveal a general dependency of developmental gene transcription on SWI/SNF and of housekeeping transcription on Iswi. In addition, the data suggest that developmental and housekeeping genes can be transcribed independently of Iswi and SWI/SNF, respectively.
To test whether all active promoters of a certain type depend on distinct chromatin remodelers, we grouped all active promoters according to whether they contain developmental or housekeeping motifs and assessed their activity after SWI/SNF or Iswi depletion. Indeed, most promoters containing developmental motifs lost activity upon SWI/SNF depletion, whereas promoters that lack these motifs were largely unaffected (Figure 1H; compare dark pink with light pink). In contrast, housekeeping promoters containing TCT or Ohler 6 motifs lost activity upon Iswi depletion,whereas other promoters were unaffected or appeared even slightly upregulated (Figure 1H, compare dark green with light green). Although subtle on average, these changes were statistically significant and biologically relevant given the growth defect seen upon Iswi depletion (Figure S1B), indicating that wildtype transcription levels at these promoter types depend on the SWI/SNF or Iswi remodelers. Thus, the differential requirements for specific remodelers in developmental and housekeeping genes can be explained by differences in core-promoter structure.
SWI/SNF localization reflects transcriptional dependency
The distinct promoter dependencies on SWI/SNF and Iswi, respectively, might arise from differential localization of these remodelers. To address this hypothesis, we performed CUT&RUN for Snr1 and Iswi, using the integrated FLAG tag as an epitope (Skene and Henikoff, 2017) (Figure 2A).To control for general DNA accessibility, we performed anti-FLAG CUT&RUN in the parental cell line,which lacks the FLAG tag (no-tag control),and normalized all CUT&RUN signal to this background. We performed two independent replicates for each condition,which showed excellent correlation at genomic bins of 1kb (PCC>0.96, Figure S3A). CUT&RUN against the tagged proteins showed strong visible enrichment against the no-tag control, and we used this enrichment as a measure of protein localization (Figure 2B). Auxin treatment reduced the CUT&RUN signals to background levels, partly even slightly below the no-tag control (presumably due to subtle changes in DNA accessibility after remodeler depletion; Figure S3B-C).
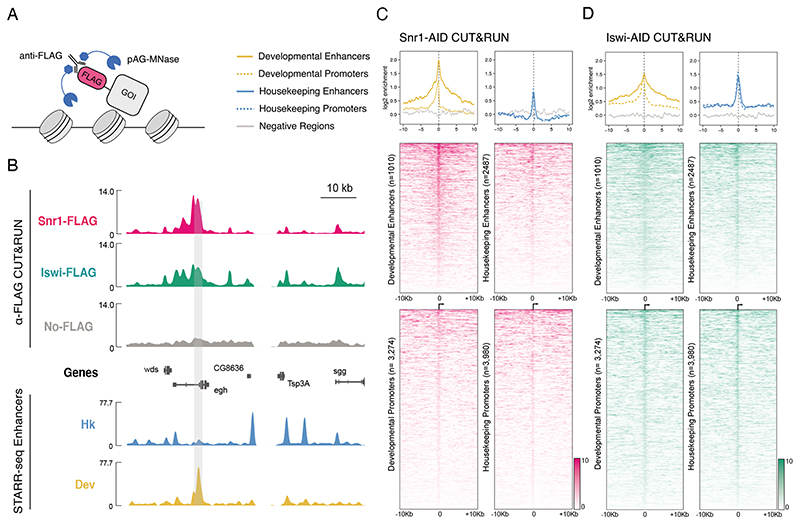
Figure 2. CUT&RUN profiling of Snr1 and Iswi genome occupancy.
A) Depiction of CUT&RUN assay: anti-FLAG antibody is bound to FLAG tagged gene of interest (GOI) and subsequently by AG-MNase fusion protein. B) Chromatin remodeler occupancy in Drosophila S2 cells. Top: genome browser tracks of anti-FLAG CUT&RUN coverage in Snr1-AID cells (pink), Iswi-AID cells (green), and No-FLAG parental cells as control (grey). Bottom: STARR-seq tracks of housekeeping and developmental enhancer activity. A developmental enhancer and the corresponding Snr1 peak is highlighted by grey shading; units: library normalized counts per million. C) Snr1 occupancy at extended genomic regulatory regions (+/- 10kb). CUT&RUN enrichments of FLAG-tagged Snr1 against parental control shown as metaplots (top; colors see legend) and heatmaps (bottom, enrichments as shades of pink, see legend) at developmental (left) and housekeeping (right) enhancers and promoters. D) as (C) but for Iswi (enrichments as shades of green, see legend).
We examined localization of the tested remodelers at promoters and enhancers selected to be accessible in S2 cells and specific to either the developmental or housekeeping classes, using previously established classifications (Zabidi et al., 2015;Haberle et al., 2019; see methods). The genomic distribution of Iswi and Snr1 revealed that both preferentially localized to promoter and enhancer elements in general (Figure 2C-D). Moreover, Iswi was enriched strongly at both developmental and housekeeping type enhancers and promoters (Figure 2D, S3D-E), suggesting that the preferential downregulation of housekeeping type genes is not explained by specific localization.In contrast, Snr1 showed much stronger enrichment specifically at developmental enhancers but not at promoters or at housekeeping enhancers (Figure 2C, S3F), in line with the developmental dependency. Snr1 and Iswi CUT&RUN signals remained enriched compared to the parental no-tag control for several kilobases from active regulatory regions (Figure 2C-D), but were at background level for randomly chosen genomic positions and appeared depleted at heterochromatic and Polycomb marked regions (Figure S3G).Overall, our data provides evidence that SWI/SNF is preferentially recruited to developmental regulatory regions; however, as there is also some localization to housekeeping regions and euchromatin in general, we considered if there are additional means by which SWI/SNF achieves specificity.
SWI/SNF dependency is enforced by intrinsic promoter compatibility
Transcriptional cofactors are known to exhibit specific intrinsic compatibilities towards certain promoter classes (Haberle et al., 2019), in addition to the distinct localization observed above. Such compatibilities may arise from interaction with specific complexes at different promoter types, or more optimal DNA substrates in the case of remodelers (van Arensbergen et al., 2014). Importantly, to characterize this property, activation must be disentangled from differential localization,for example, through forced recruitment. We therefore considered that SWI/SNF might be intrinsically compatible with developmental but not housekeeping promoters.
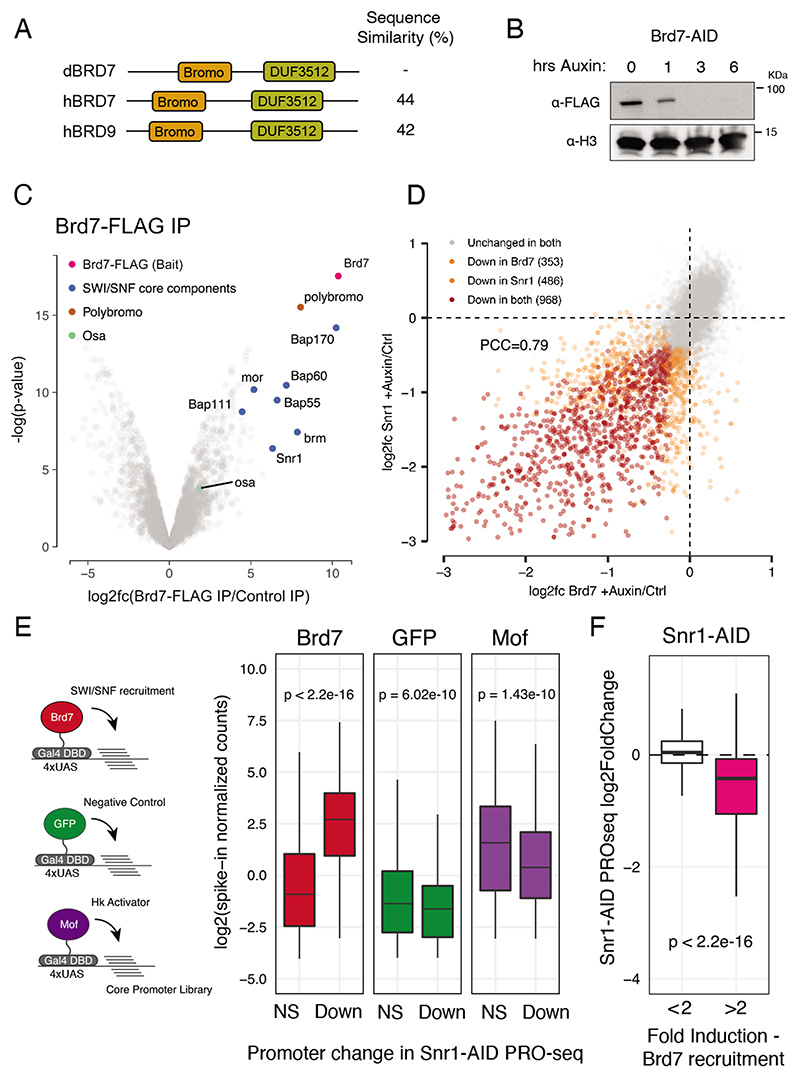
Previous studies showed that CG7154, a bromodomain-containing protein orthologous to the human SWI/SNF subunits Brd7 and Brd9 (thus referred to in this paper as Brd7) (Figure 3A), can activate transcription when recruited to minimal promoters via a Gal4 DNA binding domain and Upstream-Activating-Sequence sites (Haberle et al., 2019; Stampfel et al., 2015). In contrast, Snr1 and Iswi do not show any activating capability when tethered in the same setting (Stampfel et al., 2015), suggesting that these factors are not amenable to this approach. To confirm that Brd7 is indeed a SWI/SNF component, we tagged the endogenous Brd7 gene locus with an AID-FLAG tag as described above and found that it stably associated and co-purified with SWI/SNF (Figure 3B-C, Table S2). In particular, it co-purified with Polybromo, a PBAP-specific subunit, but not Osa, a BAP specific subunit, suggesting that it associates particularly strongly with PBAP-type SWI/SNF. Moreover, depletion of Brd7 led to strong and specific transcriptional changes that were similar to the ones observed after Snr1 depletion (PCC=0.79; Figure 3D). These results suggest that the transcriptional functions of BAP and PBAP are either interdependent or are – in this model system – largely mediated by PBAP.
Figure 3. SWI/SNF shows intrinsic activity towards developmental core-promoters.
A) Protein domains of dBrd7 and its human orthologs Brd7 and Brd9. B) anti-FLAG (top) and anti-H3 (bottom) western blot of Brd7-AID S2 cells after 0, 1, 3 and 6 hours of auxin treatment. C) Volcano plot of enrichment and Limma-computed p-value of proteins detected from Brd7-FLAG IP-MS. Highlighted are core components of SWI/SNF in blue and accessory components Polybromo (PBAP specific) and Osa (BAP specific) in in brown and green, respectively. D) Gene expression changes after Snr1 depletion vs. Brd7 depletion.Scatterplot of PRO-seq log2 fold-changes per promoter, with unaffected promoters in grey, uniquely affected promoters in orange, and promoters affected after both depletions in red. E) Assessment of intrinsic core-promoter preference of SWI/SNF. Left: schematic of cofactor-recruitment STAP-seq. Cofactors (colored circles) are recruited to a core-promoter library (lines) via Gal4-DBD. Activation of Snr1 independent (NS) and dependent (Down) promoters as measured by STAP-seq (spike-in normalized counts) after recruitment of Brd7, GFP (negative control) and Mof (housekeeping control). F) Snr1-dependency of promoters activatable by Brd7 recruitment in STAP-seq (pink) versus control (white) shown as PRO-seq changes (log2fc) after Snr1 depletion.
To determine whether SWI/SNF is intrinsically compatible with developmental promoters, we reanalysed genome-wide promoter activity data after recruitment of various cofactors, including Brd7(Haberle et al., 2019). Of the 10,064 promoters that were active in S2 cells according to PRO-seq, 6,990 of the corresponding 133bp core promoters were active or could be activated by cofactor recruitment. If the dependence of developmental promoters on Snr1 depends on specific compatibility, then Brd7 recruitment should result in greater activation at Snr1-dependent compared to Snr1-independent promoters. Indeed,although both promoter sets showed similar basal activities, based on GFP recruitment, Snr1-dependent developmental promoters were more than 8-fold more strongly activated by Brd7 recruitment compared to Snr1-independent promoters (Figure 3E, S3F). In contrast, Snr1-independent promoters, which largely consist of housekeeping type promoters, were activated readily and preferentially by males absent on the first (Mof), a histone acetyl transferase previously shown to be a housekeeping specific activator (Figure 3E, S3F) (Feller et al., 2011; Haberle et. al. 2019; Lam et al., 2012; Stampfel et al., 2015).Mof-mediated activation of the Snr1-independent promoters rules out that these promoters cannot be activated at all. The reverse analysis showed consistent results: promoters that were activated by Brd7 recruitment were also endogenously more dependent on Snr1 than promoters that could not be activated by Brd7 recruitment (Figure 3F). These data suggest that, in addition to the preferential localization of SWI/SNF to developmental promoters, SWI/SNF also exhibits intrinsic regulatory compatibility towards this promoter type, contributing to the observed functional dependency.
SWI/SNF maintains accessibility of developmental enhancers
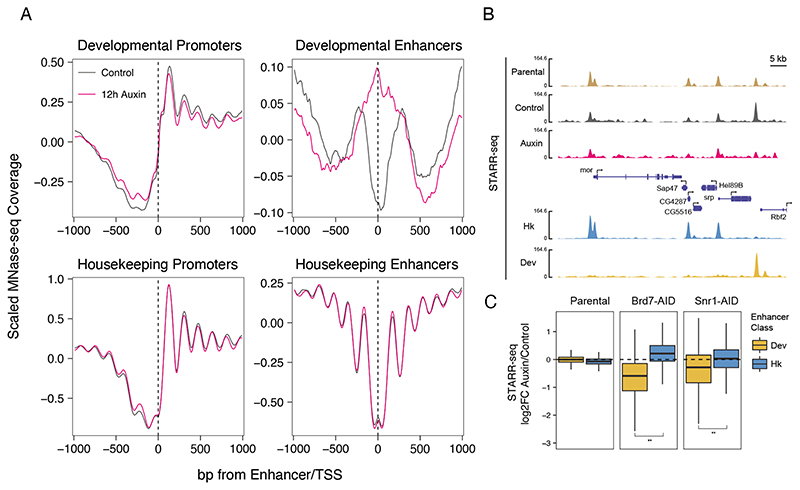
The failure of developmental transcription after SWI/SNF depletion might also arise from specific changes to the chromatin structure. To assess how SWI/SNF depletion affects chromatin structure, we performed MNase-seq after 12 hours of Snr1 depletion (Figure 4A).We did not observe changes in nucleosome occupancy and positioning at housekeeping promoters and enhancers, in line with the lack of change in transcription and the preferential localization of SWI/SNF to developmental genes. At developmental promoters, DNA accessibility and nucleosome positioning were also essentially unchanged. However, accessibility at developmental enhancers was almost completely lost, consistent with the strong localization of Snr1 to developmental enhancers. These results are in line with recent studies in mammalian cells that found chromatin accessibility of distal sites to be dependent on SWI/SNF (Iurlaro et al., 2021; Schick et al., 2021). Depletion of Brd7 also resulted in a loss of accessibility at developmental type enhancers (Figure S4F), similar to the depletion of Snr1 and consistent with the effects of depleting either protein on nascent transcription. Together these observations demonstrate the requirement of SWI/SNF for maintaining the DNA accessibility and function at developmental enhancers. Overall, SWI/SNF might therefore provide two complementary functions, to mediate DNA accessibility at enhancers and to directly transactivate transcription at promoters.
Figure 4. SWI/SNF maintains enhancer accessibility and activity.
A) Nucleosome occupancy changes after Snr1 depletion. Scaled MNase-seq coverage of Snr1-AID cell lines 12 hours after auxin (pink) or mock (black) treatment. Mean coverage of 2 replicates shown per sample, centered on developmental and housekeeping TSSs or enhancer centers. B) Genome browser tracks of mixed-library STARRseq coverage in Parental S2 cells (brown), and Snr1-AID cells before (grey) and after (pink) Snr1 depletion. Below: housekeeping and developmental enhancer activities in S2 cells, with a developmental-specific enhancer highlighted by grey shading.C) Changes in housekeeping and developmental enhancer activity after SWI/SNF depletion. Boxplots of log2 fold-change in enhancer activity as measured by STARR-seq after auxin treatment in parental, Brd7-AID, or Snr1-AID cells (** p < 2.2x10-16 Wilcoxon-test).
To determine whether enhancer activity, i.e. the enhancers’ ability to activate transcription, is indeed compromised after Snr1 or Brd7 depletion, we performed STARR-seq, a genome-wide enhancer-activity assay, on a mixed housekeeping (Rps12) and developmental (DSCP)STARR-seq library (Zabidi et al., 2015). We separately transfected Snr1-AID and Brd7-AID cells with the library, treated half of the cells in each case with auxin to induce Snr1 or Brd7 depletion, and harvested the auxin-treated and control cells 24 hours after transfection for reporter RNA processing, as described previously (Muerdter et al., 2015). We performed two independent replicates for each factor, which showed good overall correlation with PCC > 0.8 (Figure S4A). We observed a significant loss of developmental but not housekeeping enhancer activity after Snr1 or Brd7 depletion (Wilcoxon test, p<2.2E-16), confirming that SWI/SNF is specifically required for developmental enhancer activity (Figure 4B-C, S4B-E).
Housekeeping nucleosomes are positioned by Iswi and Ino80
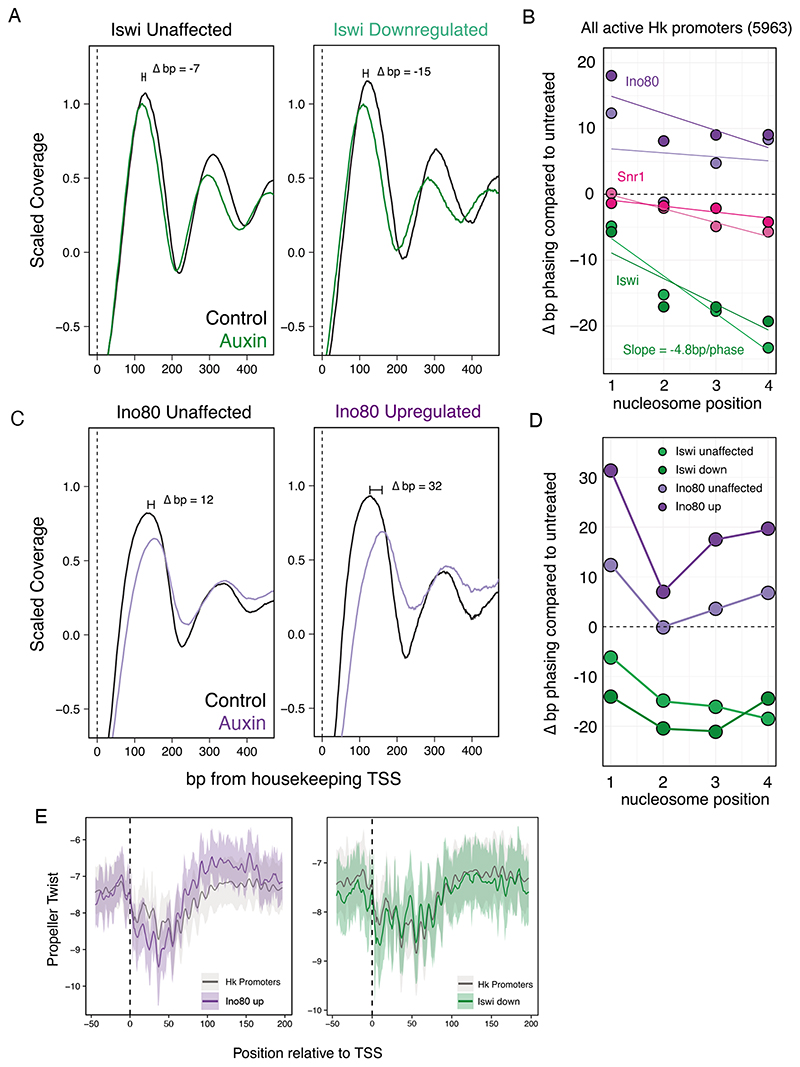
In contrast to Snr1, depletion of Iswi did not result in strong changes in DNA accessibility at any of the regulatory elements, despite the transcriptional downregulation of many housekeeping promoters (Figure S5A). Interestingly, however, Iswi depletion did affect the stereotypic and highly organized nucleosome positioning and phasing at housekeeping promoters. Iswi depletion led to an encroachment of the +1 nucleosome on the TSS by about 7bp, as well as a decrease in the nucleosome repeat length of about 4.8bp per phase (Figure 5A-B), while Snr1 depletion resulted in no change in +1 nucleosome positioning. Moreover, the promoters that showed significant downregulation after Iswi depletion also showed a larger +1 nucleosome shift of about 15bp towards to the TSS, suggesting that nucleosome positioning and phasing are directly linked to promoter activity (Figure 5A,D). We confirmed this dependency of housekeeping type transcription on Iswi by performing STARR-seq as described above, which showed a loss in housekeeping enhancer activity, while developmental enhancers appeared unaffected (Figure S5B).
Figure 5. Iswi and Ino80 position nucleosomes at housekeeping promoters.
A) MNase-seq scaled coverage in Iswi-AID cells in control (black) and auxin treated (green) conditions. Nucleosome positioning at expressed housekeeping promoters which were unaffected (left) or downregulated (right) after Iswi depletion. B) Quantification of nucleosome positioning shifts at housekeeping promoters before and after remodeler depletion. Two replicates and linear best fit per replicate are displayed per remodeler. For Iswi, the linear model coefficient, representing the change in phase per nucleosome is displayed. C) As A but for Ino80-AID cells and at Ino80 unaffected (left) versus upregulated (right) housekeeping promoters. D) Quantification of the change in nucleosome positioning (bp) after Iswi or Ino80 depletion in unaffected and affected gene sets for Iswi and Ino80. E) Average DNA shape profiles at TSSs and +1 nucleosome regions. Left: at all housekeeping promoters and Ino80 upregulated promoters. Right: at all housekeeping promoters and Iswi downregulated promoters. Solid line indicates mean propeller twist per nucleotide and shaded areas represent the 10th to 90th percentile interval.
Given the specific sensitivity of housekeeping promoters to Iswi depletion, we were intrigued that housekeeping promoters were also enriched among the 658 promoters that were upregulated after Ino80 depletion (Figure S5C). To determine whether nucleosome positioning could explain the upregulation of these genes, we performed MNase-seq after 12 hours of Ino80 depletion. Like the depletion of Iswi, Ino80 depletion also did not lead to changes in promoter accessibility (Figure S5D) but affected nucleosome positioning and phasing (Figure 5B-C; see Figure S5F-G for the analyses of all gene sets). Interestingly,however,unlike Iswi depletion, Ino80 depletion moved the +1 nucleosome downstream and further away from the TSS.In addition, the +1 nucleosome shifted even farther away from the TSS at the promoters that were upregulated after Ino80 depletion compared to those that were unaffected (Figure 5D) or downregulated (Figure S5G).
Overall, these results suggest that Iswi and Ino80 function at housekeeping promoters to create and/or maintain the stereotypical nucleosome positioning and phasing that is characteristic of these types of promoters. Similar to Iswi (Figure 2D), Ino80 seems to localize rather un-specifically and broadly to all regulatory elements (CUT&RUN; Figure S5G), suggesting that localization cannot explain the housekeeping-promoter specific requirements of either remodeler. Iswi seems to push nucleosomes away from the TSS to increase the width of the nucleosome-free region and the spacing between nucleosomes, whereas Ino80 seems to push nucleosomes towards the TSS and decreases the spacing between nucleosomes.
Interestingly, the effects of Iswi and Ino80 depletion are greater at the subset of housekeeping promoters that show transcriptional changes, suggesting that sequence features such as DNA shape may render promoters differentially sensitive to remodeler depletion. Previous in vitro and in vivo studies have suggested a role for DNA shape of the +1 nucleosome region in influencing Ino80 activity. In yeast, promoter regions with a lower degree of DNA propeller twist are more efficiently remodelled by Ino80, resulting in closer positioning of the +1 nucleosome to the TSS (Krietenstein et al., 2016; Oberbeckmann et al., 2021). We therefore wondered whether housekeeping promoters that are significantly affected by Ino80 depletion might have a such a DNA shape sequence signature, which could explain their preferential upregulation after Ino80 depletion. Indeed, around the dyad of the +1 nucleosome position, we found that Ino80 affected promoters had lower propeller twist than the average housekeeping promoter, at least partially explaining the selective effect of Ino80 on a subset of housekeeping promoters (Figure 5E).In contrast, Iswi transcriptionally affected promoters did not show any difference in DNA shape from the average housekeeping promoter, indicating that this feature is specific to Ino80 (Figure 5E).
Different nucleosome affinity profiles underlie remodeler dependencies
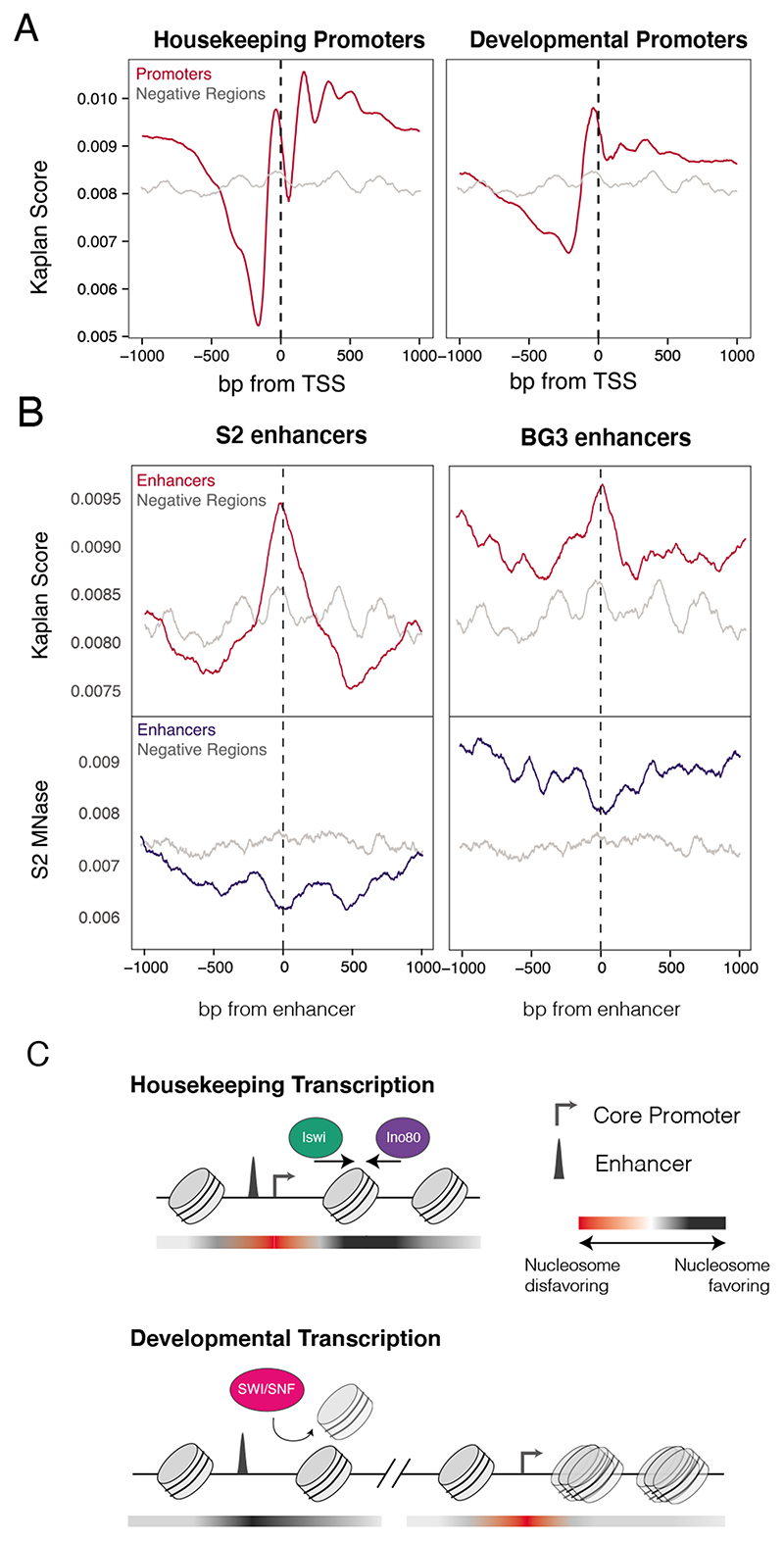
The striking differences in remodeler dependencies for the different transcriptional programs prompted us to examine whether these regions have different intrinsic affinities for nucleosomes. It has previously been described that different DNA sequences have a range of sequence intrinsic affinities that result from sequence composition as well as biophysical properties such as DNA bendability and shape. We made use of a previously described nucleosome occupancy prediction score that reflects the intrinsic affinity of DNA for nucleosomes based on a probabilistic model trained on in vitro nucleosome-to-DNA assembly (Kaplan et al., 2009).Promoters of both types showed a low affinity region upstream of the TSS, potentially explaining the lack of any changes in accessibility at these regions after remodeler depletion (Figure 6A). Closer examination revealed that housekeeping-type promoters have lower affinity sequences upstream of the TSS than developmental promoters, and conversely, stronger nucleosome affinity sequence at consistent and phased positions downstream of the TSS.
Figure 6. Nucleosome affinity profiles reflect remodeler dependencies.
A) Predicted nucleosome occupancy scores for housekeeping and developmental promoters (red), compared to negative regions (grey). B) Nucleosome affinity and accessibility for active vs. inactive cell-type-specific enhancers. Top: Nucleosome affinity scores for S2-cell specific (left) and BG3 specific (right) developmental enhancers (red) and negative regions (grey). Bottom: MNase-seq coverage profiles in S2 cells at the same regions (blue), compared to negative regions (grey). C) Model of remodeler activity and dependencies at enhancers and promoters. Red-to-black gradient depicts nucleosome favorability of DNA sequence, where red is less favorable and black is more favorable.
Unlike housekeeping enhancers, which are typically located proximal to the TSS of a gene and therefore overlap with the nucleosome low affinity region, developmental enhancers are often many kilobases away from their target gene (Zabidi et al., 2015). The sequences of enhancers that are active in S2 cells have a high intrinsic affinity for nucleosomes (Figure 6B, top). Although these would data suggest that the enhancers should be preferentially occupied by nucleosomes, our MNase data showed instead that they were less occupied by nucleosomes than non-enhancer control regions (Figure 6B, bottom). This result and the observation that developmental enhancers become nucleosome-occupied after depletion of Snr1 (Figure 4A) suggest that developmental enhancers are nucleosome-occupied by default and that establishing accessibility is an active process, requiring the joint action of cell-type specific transcription factors and SWI/SNF.This ‘default off’ model (Barozzi et al., 2014) predicts that enhancers should be nucleosome-occupied when they are inactive. Indeed, enhancer sequences that are active in neuronal BG3 cells derived from larvae (Ui et al., 1994.) but inactive in S2 cells have high intrinsic nucleosome affinity and were nucleosome-occupiedin S2 cells (Figure 6B). In addition, differences in nucleosome score across developmental enhancers resulted in different nucleosome occupancy profiles after Snr1 depletion, whereby the enhancers with the highest nucleosome affinities showed a centrally positioned nucleosome while the enhancers with the weakest affinity appeared relatively nucleosome depleted (Figure S6A-B). Overall, the intrinsic nucleosome affinities of the DNA sequences help explain the changes seen in accessibility at promoters and enhancers after Snr1 depletion.
Discussion
The two main transcriptional programs of Drosophila driving expression of housekeeping and developmental genes, respectively, have fundamentally different promoter and enhancer types that differ in regulatory properties, characteristic sequence motifs, and chromatin structure. In the present study, we demonstrate that these programs differentially depend on different ATPase chromatin remodelers. We show that developmental gene transcription depends on SWI/SNF whereas housekeeping gene transcription is generally remodeler independent but fine-tuned by Iswi and Ino80 (Figure 6C).
The apparent differences in remodeler dependencies of the developmental and housekeeping regulatory regions could result from these regions’ different intrinsic nucleosome favorability (Figure 6C). Distal developmental enhancers have a high affinity for nucleosomes, and therefore require the action of SWI/SNF to create accessible DNA. Given that these regulatory regions control genes that are meant to be expressed in a highly cell type-specific manner, this intrinsically inhibited,or ‘default-off’,state is appropriate (Barozzi et al., 2014). In contrast, housekeeping gene promoters and their enhancers, which often overlap the core promoter,have a low affinity for nucleosomes, which might explain their lack of dependence on SWI/SNF.We only observed moderate effects on housekeeping transcription, and many genes were not significantly changed after depletion of the tested remodelers. The ‘default-on’ state of these promoters is appropriate given that the downstream genes are meant to be broadly expressed. This limited effect may be due to remodeler redundancy or requirement of another chromatin remodeler that was not tested in this study. We cannot rule out that these sequences require remodeler activity to be opened initially but then are resistant to closing after acute depletion of a given remodeler. Alternatively, accessibility may be maintained independently of chromatin remodeler activity through a combination of the intrinsically low nucleosome affinity and the presence of many binding sites for sequence-specific DNA binders such as BEAF-32, Dref, and M1BP.
In addition to nucleosome affinity based on DNA sequence, our results also reveal two mechanisms that explain the dependency of developmental genes on SWI/SNF. First, SWI/SNF preferentially localizes to developmental regulatory regions, particularly distal enhancers, consistent with the strong loss of enhancer accessibility after SWI/SNF depletion. Second, the observation that SWI/SNF is compatible with developmental but not housekeeping promoters also suggests a role of this remodeler at the promoter, consistent with its described in vitro function (Agalioti et al., 2000). The specific loss of accessibility at enhancers, but not promoters, may arise from greater robustness of promoters due to their low nucleosome affinity as well as the many factors that strongly bind this region including TFs, GTFs and Pol II. The enhancer-specific loss of DNA accessibility is consistent with SWI/SNF’s requirement in mammals: Loss of core SWI/SNF subunits results in loss of enhancer accessibility and of active marks such as H3K27ac, but few changes at promoters (Alver et al., 2017; Iurlaro et al., 2021; Nakayama et al., 2017; Schick et al., 2021). The requirement of SWI/SNF to maintain accessibility of such distal regulatory regions and transcription factor binding sites is consistent with their high sequence intrinsic nucleosome favorability, pointing to a conserved regulatory role in metazoans (Barozzi et al., 2014; Tillo et al., 2010). Interestingly, some promoter-distal regions such as CTCF-binding sites have been shown to be SWI/SNF independent but Iswi dependent (Barisic et al., 2019), suggesting that remodelers are also used distinctly at different types of regulatory elements in mammals.
Although the accessibility of housekeeping type promoters was generally unperturbed after depletion of any of the remodelers, we did observe changes in downstream nucleosome positioning that could explain the observed changes in transcription. Loss of Iswi caused +1 nucleosome encroachment on the promoter whereas Ino80 had the opposite effect and these effects correlated with changes in transcription. While the effects on nucleosome positioning could be a consequence of the transcriptional changes (for example, in the absence of Ino80 increased passage of Pol II may shift the +1 nucleosome further downstream), the established roles of Iswi and Ino80 as chromatin remodelers make it more likely that Iswi and Ino80 depletion leads to changes in nucleosome positioning, which in turn affects transcription. Consistent with previous studies, our data support a model in which encroachment of the +1 nucleosome of the promoter may interfere with Pre-Initiation Complex binding through steric occlusion (Reja et al., 2015). In the same vein, movement of the +1 nucleosome away from the promoter may augment transcriptional output of the promoter by increasing accessibility and/or revealing previously inaccessible TSSs, a phenomenon also seen in yeast (Kubik et al., 2019a). Interestingly, some of the TSSs most affected by Iswi depletion were ribosomal protein genes, which have focused initiation patterns, an atypical feature of housekeeping promoters. We speculate that the focused nature of these promoters makes them susceptible to +1 nucleosome encroachment by preventing initiation at the native site; in contrast, most housekeeping promoters have dispersed initiation patterns which could provide robustness against nucleosome encroachment. Similarly, Inr-containing promoters were downregulated after Ino80 depletion suggesting that they might be sensitive to precise TSS to +1 nucleosome spacing for PIC assembly.
In contrast to its role in flies, Iswi has been shown in yeast and mammals to act as a ‘puller’, moving nucleosomes towards promoters and other boundaries; for example, in S. cerevisiae, Iswi counteracts opening of the promoter by RSC, and when depleted, causes expansion of the nucleosome-free region at the promoter (Krietenstein et al., 2016; Kubik et al., 2019b; Whitehouse et al., 2007; Yen et al., 2012). In addition, Iswi has been shown to be directly recruited by sequence-specific factors, such as RSC, in yeast. A recent study noted that Drosophila Iswi has lost critical acidic residues required for activator interaction (Donovan et al., 2021), which may help explain the species-specific differences in chromatin positioning and accessibility after Iswi loss. The broad localization of Iswi seen in our study shows that it indeed may lack mechanisms of direct recruitment to specific cis-regulatory elements.
Overall, our demonstration that distinct transcription regulatory programs depend on different chromatin remodelers and are affected distinctly after remodeler depletion is intriguing. It illustrates how promoter DNA sequences and chromatin structure can evolve to adopt distinct default regulatory states that reflect the promoters’ functions; and shows that chromatin remodelers are both functionally specialized and differentially employed during gene regulation.Our results also assign regulatory significance to the stereotypical nucleosome arrangement at housekeeping promoters, a decade-old observation whose functional implication has remained unclear. Together, these insights have important implications for our understanding of broad and cell type-specific gene regulatory programs and their implementation at the DNA sequence and chromatin level in animals.
Limitations of this study
This study has tested the hypothesis that the characteristically different chromatin structure at different types of promoters, particularly developmental versus housekeeping promoters, implies promoter-class specific remodeler functions and dependencies. To achieve this, we have selected representative remodelers for rapid auxin-inducible degradation and assessed direct transcriptional changes. Therefore, our study has several limitations inherent to the experimental design:
As expected for an approach based on protein depletion, depleting remodelers with partially or entirely redundant functions may not result in transcriptional changes. This could be redundancies within a remodeler family, which is presumably the case for Chd1, or due to compensation from other remodelers, as might be partially the case for Iswi at housekeeping promoters. These limitations stemming from remodeler redundancies could be overcome by the combined depletion of multiple redundantly acting proteins.
Certain remodelers may also have other context-specific functions in other cell types where different transcription factors are expressed that we could not assess here. For example, Ino80 is known to act in concert with Polycomb during development, which may extend its regulatory role from the housekeeping role we observe in our differentiated cell type.
Given our aim and to avoid redundancies between different subcomplexes as much as possible, we targeted core components (typically the catalytic subunits) shared between entire remodeler families rather than accessory subunits that are unique to one or some family members. We therefore cannot assess differences between family members and future work will be needed to determine the potentially more fine-grained regulatory roles within each remodeler family. For example, differences between the distinct Iswi containing complexes NURF, ACF and CHRAC might explain how depletion of the Iswi ATPase resulted in downregulation of housekeeping promoters and upregulation of developmental promoters. Moreover, as we wanted to inactivate remodelers to study the direct transcriptional consequences, our approach could not assess more detailed aspects of remodeler complex assembly or disassembly beyond the stability of individual subunits.
STAR methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alexander Stark (stark@starklab.org).
Materials availability
All cell lines and plasmids generated in this study are available upon request.
Experimental model and subject details
S2 cells were maintained in Schneider’s Medium supplemented with 10% heat-inactivated Fetal Bovine Serum, and grown at 27°C, in 0.4% CO2. Live cell counts were obtained using Countess automated cell counter (Invitrogen) with Trypan Blue staining. Conditioned media for single cell sorting and serial dilution was prepared as follows: ~70% confluent passages of S2 cells were split 1:10 and grown in T175 flasks to 60-70% confluency in 25mL media. Media was then removed and spun down at 1000g for 5 minutes to pellet cells. Supernatant was then filtered through 0.2um sterile filter and diluted 1:1 with regular media, and used within 1-2 weeks.
Method details
Plasmids and vectors used in this study
SpCas9 and and sgRNAs were expressed in S2 cells from the plasmid pAc-sgRNA-Cas9 (Addgene plasmid #49330). sgRNA sequences used for targeted integration can be found in Table S1. Donors for AID tagging were cloned into pCRIS-PITChv2-FBL (Addgene Plasmid #63672) with 20bp microhomology arms as described in (Sakuma et al., 2016). For 5’ tagging we generated cassette containing the following construct (5’ BlastRes-P2A-FLAG-AID-3xGGGS 3’) and for 3’ tagging the following (5’ 3xGGGS – AID – FLAG – P2A – BlastREs 3’). AID sequence was ordered as a GeneBlock from IDT, using the mini-degron AID* sequence (IAA17_71-114) from (Morawska and Ulrich, 2013). Vector maps and plasmids are available upon request.
OsTir1 Parental cell line generation
A parental cell line expressing the E3 ligase for AID was generated using an mCherry-P2A-OsTir1-MYC cassette with 50bp homology arms for the C terminus of the essential Actin 5C locus (see Table S1 for sequences). The OsTir1-MYC sequence was derived from pBabe Puro osTIR1-9Myc (Addgene plasmid #80074). Cells were transfected using MaxCyte STX Electroporation system with the Tir1 recombination cassette and gRNA against the 3’ end of Actin5c, and after 4 days single mCherry positive cells were sorted into 96 well plates with conditioned media. Cells were genotyped using primers flanking the 3’ end of Actin 5C locus and a heterozygous clone was selected to avoid potential silencing of one allele.
AID cell line generation
Parental cells were transfected with 5ug of gene specific pAc-sgRNA-Cas9, 3ug PITCh gRNA pAc-sgRNA-Cas9, and 1ug of pCRIS-PITChv2-FBL containing AID donor and gene specific microhomology. 5x106 cells were transfected in an OC-100 using the Maxcyte electroporation system. After 2 days of recovery cells were selected for integration of the Blasticidin cassette with 5ug/mL Blasticidin. After patches of cells grow out (approximately 1-2 weeks), cells were seeded for single cell colonies using serial dilution in conditioned media. Correct integration was confirmed by PCR-genotyping on cell lysates obtained with QuickExtract, and using primers surrounding integration site (for primers see table S1) and confirmed with sanger sequencing.
Competition Assays
For competition assays, tagged cell lines were seeded at 1:1 ratio with WT cells at a density of 1 x 106 cells/mL in a T-75 with volume 10mL, and split into control and treatment groups. WT to tagged cell lines ratios were quantified every 24hrs by Flow Cytometry using the mCherry tag present only in the tagged cell lines expressing Tir1.
Western blotting
Cells were lysed in RIPA buffer and the chromatin fraction solubilized by sonication with Diagenode Bioruptor for 10 minutes on low setting with 30 seconds sonication followed by 30 seconds break. Laemmeli Sample Buffer was added with 2-mercaptoethanol, and denatured at 95C for 10 minutes. Samples were ran on 4-20% Mini-PROTEAN TGX Precast Protein gels, and transferred onto PVDF membrane using the ThermoFischer Power Blotter kit. Blocking was performed with 5% milk in TBST for 1 hour followed by overnight incubation with primary antibody at 4°C. Membrane was washed 3 times with TBST for 5 minutes and incubated with appropriate HRP conjugated secondary antibody for 1 hour. Blots were visualized using Clarity and Calrity Max ECL substrate (Biorad) and ChemicDoc (Biorad).
Nuclear Fractionation
For separation of soluble and chromatin fractions of nuclei we isolated nuclei through incubation of S2 cells in nuclei isolation buffer (25mM HEPES pH 7.5, 300mM Sucrose, 10mM CaCl2, 5mM MgCl2, .1% IGEPAL-CA630) for 5 minutes followed by 2 washes. Nuclei were then lysed with 25mM HEPES pH 7.5, 300mM sucrose, 50mM NaCl, 3mM MgCl2, 0.5% Triton X-100 for 5 minutes on ice. The chromatin fraction was pelleted at 2500g for 5 minutes and resuspended in RIPA buffer and subject to sonication as described above.
PRO-seq
We performed PRO-seq following the published protocol from (Mahat et al., 2016), with several customizations to increase scalability and accuracy. 20 x 106 S2 cells were harvested per replicate and 1 x 106 HCT-116 cells were spiked in. We performed a run on using Biotin-11-CTP. Following RNA purification, Base Hydrolysis, and Streptavidin enrichment as described in Mahat et al, we extracted biotin labelled RNA off streptavidin beads using Zymo Direct-zol micro columns according to kit specifications. For elution, we used 5.5 uL 1:8 diluted 3’ RNA linker and proceeded directly to 3’ RNA ligation. Following ligation and bead washing, we performed TAP and PNK reactions directly on beads, and following bead washing, 5’ adapter ligation was also performed on beads. The 3’ RNA adapter was modified to include an 8bp UMI with the following final sequence and modifications : 5’ - /5Phos/rNrNrNrN rNrNrNrN rGrArUrC rGrUrCrG rGrArCrU rGrUrArG rArArCrU rCrUrGrA rArC/3InvdT/ - 3’.
CUT&RUN
0.5 x 106 cells were harvested per replicate and washed 1x with PBS, and resuspended in 1.5 mL Wash Buffer (20mM HEPES pH 7.3, 150mM NaCl, 0.5mM Spermidine, Roche complete Inhibitor, EDTA-free). Cells were washed 3x with Wash buffer, and resuspended in 10uL ConcavadinA magnetic beads equilibrated in Binding buffer (20mM HEPES pH 7.3, 10mM KCl, 1mM CaCl2, 1mM MnCl2, Roche complete Inhibitor, EDTA-free). After 10 minutes of binding to beads, cells were resuspended in 50uL Antibody (20mM HEPES, 150mM NaCl, 0.5mM Spermidine, 0.025% Digitonin, 2mM EDTA, Roche Complete Inhibitor). M2 FLAG antibody was added to a final dilution of 1:100, and incubated overnight at 4ºC. Beads were washed 3x and resuspended in 150uL Digitonin Buffer (20mM HEPES, 150mM NaCl, 0.5mM Spermidine, 0.025% Digitonin, Roche Complete Inhibitor). pAG-MNase was added to a final concentration of 700 ng/mL and incubated rotating for an hour at 4ºC. Cells were washed 3 times with Digitonin Buffer, CaCl2 added to final concentration of 2mM, and rotated for 2 hours at 4ºC. Digestion was stopped with addition of 100uL STOP buffer (340 mM NaCl, 20mM EDTA, 4mM EGTA, 0.02% Digitonin, 50ug/mL RNase A, 50 ug/mL Glycogen). Samples were incubated for 10 minutes at 37ºC at 500rpm, and centrifuged for 5 minutes at 16000g. The supernatant was taken and digested with Proteinase K and DNA extracted using an in-house PCR purification kit. The resulting DNA was cloned into a library using the NEBNext Ultra II DNA Library Prep Kit for Illumina.
STARRseq Library Transfection and Processing
A genome wide STARR-seq library containing a DSCP synthetic core promoter library was mixed at a 1:1 molar ratio with genome wide library containing Rps12 core promoter to achieve a mixed library. Per replicate, we transfected 80 x 106 S2 cells at 70-80% confluence using the MaxCyte STX Scalable Transfection System. Per OC-400, we transfected 20 x 106 cells with 20ug of mixed library. After Electroporation and recovery as described previously, we resuspended and mixed all cells together in a total of 100mL of media. Cells were then split into a control and treatment group where the latter was treated with 100uM Auxin. 24 hours after transfection, total RNA was isolated from cells and the STARR-seq library processed and sequenced as described previously (Neumayr et al., 2019).
MNase Seq
MNase Seq was performed as described in (Ramachandran et al., 2017) with modifications. In detail, cells from a 70% confluent 75mm2 flask were harvested per sample, and washed once with PBS. After resuspension in buffer HM2 (10mM HEPES pH 7.4, 2mM MgCl2, 0.5mM PMSF), IGEPAL-40 was added to a final concentration of 0.75%. Cells were incubated for 5 minutes on ice and then spun down and washed once with buffer HM2. The resulting nuclei were resuspended in buffer HM2 and CaCl2 added to a final concentration of 1mM and incubated for 5 minutes at 37°C. The MNase reaction was caμried out for 25 minutes at 37°C and stopped by addition of EGTA to a final concentration of 1.5mM and SDS to a final concentration of 1%. Sample was digested with ProteinaseK and DNA was Isolated with Phenol Chloroform extraction and precipitation. The resulting DNA was size selected on gel for mono-nucleosomal fragments and the purified DNA was cloned into a library using the NEBNext Ultra II DNA Library Prep Kit for Illumina. Samples were sequenced PE36 on an Illumina NextSeq2500 to a depth of 15-30 million reads.
LC-MS/MS on S2 Nuclei
Samples for mass spectrometry analysis were prepared using the iST kit (PreOmics GmbH #P.O.00027), according to the manufacturer’s instructions. Nuclei were frozen and then protein extraction was performed for 10 min in 50 μl of lysis buffer at 95°C. Nuclei were sonicated as described above using the Diagenode Bioruptor. An UltiMate 3000 RSLC nano system coupled to a Q Exactive HF-X mass spectrometer, equipped with an EASY-spray ion source was used (Thermo Fisher Scientific) with a JailBreak 1.0 adaptor insert for a spray emitter (Phoenix S&T, Inc., USA). Peptides were loaded onto a trap column (Thermo Fisher Scientific, PepMap C18, 5 mm × 300 μm ID, 5 μm particles, 100 Å pore size) at a flow rate of 25 μL/min using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with the analytical column (Thermo Fisher Scientific, PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å). Peptides were eluted using a flow rate of 230 nl/min, and a binary 3h gradient, respectively 220 min. The gradient starts with the mobile phases: 98% A (water/formic acid, 99.9/0.1, v/v) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, v/v/v), increases to 35%B over the next 180 min, followed by a gradient in 5 min to 90%B, stays there for 5 min and decreases in 2 min back to the gradient 98%A and 2%B for equilibration at 30°C.
FLAG-Immunoprecipitation
Drosophila S2 cells expressing endogenously AID-3xFLAG-tagged Brd7 protein were used for immunoprecipitation. Parental cells without the AID-3xFLAG tag were used as a background. Two hundred million Drosophila S2 cells were collected through centrifugation at 300g, washed in PBS, resuspended in 10mL of hypotonic swelling buffer (10mM Tris pH7.5, 2mM MgCl2, 3mM CaCl2, protease inhibitors) and incubated for 15 minutes at 4°C. Cells were pelleted at 700g for 10 minutes at 4°C, resuspended in 10mL of GRO lysis buffer (10mM Tris pH7.5, 2mM MgCl2, 3mM CaCl2, 0.5% NP-40, 10% glycerol, 1mM DTT, protease inhibitors) and incubated for 30 minutes with rotation at 4°C. Samples were pelleted at 700g for 10 minutes at 4°C, resuspended in 1mL of IP lysis buffer (100mM NaCl, 20mM HEPES pH7.6, 2mM MgCl2, 0.25% NP-40, 0.3% Triton X-100, 10% glycerol, 1 mM DTT, protease inhibitors) and lysed with rotation for 30 minutes at 4°C. Lysed samples were pelleted for 5 minutes at 20000g at 4°C and supernatant (soluble nucleoplasm) and pellet (chromatin) were separated. The pellet was resuspended in 300uL of IP lysis buffer supplemented with 300mM NaCl, sonicated for 10 minutes (30 sec on/30 sec off, low intensity, 4°C) on Diagenode Bioruptor sonicator, pelleted for 5 minutes at 20000g at 4°C and soluble fraction mixed with the supernatant from earlier. The resulting mixture was then cleared by centrifugation for 5 minutes at 20000g at 4°C and protein concentrations measured using Qubit Protein Assay Kit. For each immunoprecipitation, 1mg of total protein was incubated with 50ul previously buffer-equilibrated Anti-FLAG M2 magnetic beads for 6 hours at 4°C with rotation. The beads were then washed three times for 10 minutes in IP lysis buffer supplemented with 150mM NaCl, and four times for 10 minutes in a no-detergent buffer (130mM NaCl, 20mM Tris pH7.5). The beads were resuspended in 50ul of 50mM ammonium bicarbonate and subject to tryptic digest and mass-spectrometry.
LC-MS/MS analysis for immunoprecipitations
The Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific), was operated in data-dependent mode, performing a full scan (m/z range 380-1200, resolution 60,000, target value 3E6) at 2 different CVs (-50, -70), followed each by MS/MS scans of the 10 most abundant ions. MS/MS spectra were acquired using a collision energy of 30, isolation width of 1.0 m/z, resolution of 45.000, the target value of 1E5 and intensity threshold of 2E4 and fixed first mass of m/z=120. Precursor ions selected for fragmentation (include charge state 2-5) were excluded for 30 s. The peptide match feature was set to preferred and the exclude isotopes feature was enabled.
Quantification and statistical analysis
Promoter and Enhancer Annotation
A set of ~17,000 transcripts from Dmel 5.57, containing unique 5’ ends was used as a starting set of unique TSSs. TSSs were corrected by CAGE signal from S2 cells downloaded from modENCODE 5331 that lie within a window of ±250 bps, and motifs were annotated as described in (Haberle et al., 2019). Promoters were classified as housekeeping or developmental based on their motif content, considering TATAbox, Inr, DPE, MTE, E-box, Ohler1, Ohler6, Ohler7, Ohler8, DRE, and TCT motifs. The classification is available on GEO at GSE184187. For scoring motif presence, thresholds for motif matching were as described in (Haberle et al., 2019) with small changes regarding match thresholds for TATAbox to 90%, DPE to 98% DRE to 98% and Ohler6 to 97%.
Housekeeping and developmental enhancer annotation was done based on differential STARR-seq signals as described previously (Zabidi et al., 2015). Briefly, STARR-seq peaks and enrichments from previously published housekeeping and developmental screens were compared to determine enhancer-core promoter preference as the ratio of both enrichments. For enhancers with peak summits in both screens within 250bp of each other, the enhancer summit in the screen with the higher enrichment was chosen. As STARR-seq can detect enhancer activity from regions that are endogenously inaccessible, we filtered enhancers for being in DHS peaks in S2 cells. Enhancers were then filtered for having greater than four-fold enrichment compared to input in STARR-seq, resulting in a dataset of 4643 enhancers, with 1713 developmental and 2930 housekeeping.
NGS processing
All NGS sequencing data was mapped to the dm3 genome using bowtie 1.2.2 with the following settings: -v 1 --best --strata unless otherwise indictaed. Cutadapt 1.18 was used with default options to trim adapter sequences. For PRO-seq, reads were collapsed based on position and on UMI, allowing for 1bp mismatch to avoid UMI overestimation from sequencing errors. All other methods were collapsed based on position after mapping.
PRO-seq Analysis
Mappled PRO-seq reads were resized to 1bp fixing the start nucleotide and strand switched. For gene level analysis, tag counts along the entire length of the longest transcript isoforms per gene were quantified and used for differential analysis. Unexpressed genes were removed from the count table and total dm3 mapped reads were used for size factors in Deseq2, as no significant changes in spike-in ratio were observed. TSS reads from the CAGE TSS to 150bp downstream were counted and used for differential analysis with DeSeq2, calling significant changes at 5% FDR. For promoter motif enrichment, Two-sided Fisher’s exact test was used, which accounts for the different set sizes and compares the proportions of motif found, with significance cutoff of 0.05. Differences in Fold Change split by motif presence was tested using Wilcoxon rank-sum test.
Gene Ontology analysis
We assessed whether genes significantly downregulated or upregulated after Snr1, Iswi, or Ino80 depletion were enriched for a particular gene ontology (GO) term by calculating hypergeometric p-values for every GO term with R/Bioconductor package ClusterProfiler (Yu et al., 2012). Gene universe or background set was defined as all expressed genes not in the affected set. Significance cutoff set at p<0.01
CUT&RUN analysis
CUT&RUN bedGraph coverage was obtained from mapped reads using Bedtools. For each replicate, mean coverage (counts per million) at enhancer and promoter regions was used to calculate enrichment by normalizing per sample to input coverage. To assess replicate quality, we calculated cpm in 1Kb bins across the Drosophila genome and used Pearson Correlation Coefficient. Heatmaps were generated using the image function in R with an enrichment cutoff of 10.
MNase seq analysis
MNase was mapped coverage was obtained from mapped reads using Bedtools. Median coverage across region was calculated per region, and profiles were smoothed using the frollmean function from R data.table, with a sliding window of 20 fixed on the center position. At housekeeping promoters, downstream nucleosome positions were called by looking at the maximum profile position on smoothed metaprofiles, and mean differences in nucleosome positioning were calculated from the two replicates. For changes in phasing, a linear regression was performed on the nucleosome positioning differences at the first 4 downstream nucleosomes, and the mean coefficient from both replicates used. DNA shape analysis was performed using the DNAshapeR package (Chiu et al., 2016).
Mass-Spectrometry Data Processing
For peptide identification, the RAW-files were loaded into Proteome Discoverer (version 2.5.0.400, Thermo Fisher Scientific). All hereby created MS/MS spectra were searched using MSAmanda v2.0.0.16129 (Dorfer et al., 2014). RAW-files were against the drosophila database called dmel-all-translation-r6.34.fasta (Flybase.org, 22,226 sequences; 20,310,919 residues), each case supplemented with common contaminants, using the following search parameters: The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±15 ppm (HF-X) or to ±6 ppm (Exploris). The maximal number of missed cleavages was set to 2, using tryptic specificity with no proline restriction. Beta-methylthiolation on cysteine was set as a fixed modification, oxidation on methionine was set as a variable modification, the minimum peptide length was set to 7 amino acids. The result was filtered to 1 % FDR on protein level using the Percolator algorithm (Käll et al., 2007) integrated in Thermo Proteome Discoverer and was used to generate a smaller sub-database for further processing. As a second step, the RAW-files were searched against the created sub-database using the same settings as above plus the following search parameters: Deamidation on asparagine and glutamine were set as variable modifications. In some data sets acetylation on lysine, phosphorylation on serine, threonine and tyrosine, methylation on lysine and arginine, di-methylation on lysine and arginine, tri-methylation on lysine, ubiquitinylation residue on lysine, biotinylation on lysine, formylation on lysine were set as additional variable modifications. The localization of the post-translational modification sites within the peptides was performed with the tool ptmRS, based on the tool phosphoRS (Taus et al., 2011). Peptide areas were quantified using the in-house-developed tool apQuant (Doblmann et al., 2018). Proteins were quantified by summing unique and razor peptides. Protein-abundances-normalization was done using sum normalization. Statistical significance of differentially expressed proteins was determined using limma (Smyth, 2005). For processed data see Table S2 (Brd7 IP-MS) and Table S3 (Nuclear Lysate MS).
Supplementary Material
Acknowledgements
We thank Ursula Schoeberl (IMP) and Maja Gehre (IMBA) for advice and help establishing PRO-seq and CUT&RUN, respectively, Vanja Haberle, Bernardo Almeida, Anna Vlasova and Lisa-Marie Pleyer (all IMP) for advice and help with data analysis, Martina Rath and Michaela Pagani (IMP) for technical assistance, Steve Henikoff (Hutch), Philip Korber (LMU) for discussions, Vincent Loubiere, Clemens Plaschka (both IMP), Ian Patten, and Life Science Editors for comments on the manuscript. We thank Andrew Dingwall (Loyola) for sharing the anti-Brm serum. We thank the IMP/IMBA/GMI in-house FACS and Mass Spectrometry Facility, and Molecular Biology Service. Deep sequencing was performed at the Vienna Biocenter Core Facilities GmbH. OH and LS are supported by DOC-fellowship/OEAW. F.M. was supported by an EMBO long-term fellowship (EMBO ALTF 491–2014). FN is supported by BIF. Research in the Stark group is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 647320) and by the Austrian Science Fund (FWF, P29613–B28 and P33157–B). Basic research at the IMP is supported by Boehringer Ingelheim GmbH and the Austrian Research Promotion Agency (FFG).
Footnotes
Author Contributions
O.H. and A.S. conceived the project. O.H., F.M., L.H., L.S., and K.B., generated CRISPR/Cas9 edited cell lines. O.H performed all experiments with the help of L.S. and analyzed the data. F.N. performed IP-MS experiments. O.H. and A.S. interpreted the data and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Data and code availability
All sequencing data have been deposited at GEO repository GSE184187 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.Imaging data has been deposited and published through Mendeley Data: https://data.mendeley.com/datasets/zj2f34rwff/draft?a=af5adcbd-3cd8-4b91-9fcd-bd900684f1bc. Publicly available datasets re-analyzed in this study are available in the GEO repositories listed in the Key Resources Table.
This paper does not report original code. All custom code used for the analyses was written with existing software as detailed in the STAR Methods and Key Resources table and is available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal Anti-FLAG M2 Antibody | Merck | F3165 |
| Histone-H3 | Abcam | ab1791 |
| Secondary anti mouse HRP | Sigma-Aldrich | 12-349 |
| Secondary anti rabbit HRP | Sigma-Aldrich | 12-348 |
| Bacterial and virus strains | ||
| MegaX DH10B T1R Electrocomp™ Cells | Thermo Fisher | C640003 |
| Experimental models: Cell lines | ||
| D. melanogaster Schneider S2 cells | Thermo Fisher | R69007 |
| HCT-116 | ATCC | CCL-247 |
| S2OsTir1 | This Paper | N/A |
| S2OsTir1-Snr1-AID | This Paper | N/A |
| S2OsTir1-Iswi-AID | This Paper | N/A |
| S2OsTir1-Ino80-AID | This Paper | N/A |
| S2OsTir1-Chd1-AID | This Paper | N/A |
| S2OsTir1-Brd7-AID | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Puromycin | Invivogen | ant-pr-1 |
| FastDigest MluI | ThermoFisher | FD0564 |
| BspQI | NEB | R0712S |
| Blasticidin S HCl | ThermoFisher | R21001 |
| 3-Indoleacetic acid | Merck | I3750 |
| QuickExtract™ DNA Extraction Solution | Lucigen | QE9059 |
| 2x Laemmli Sample Buffer | BioRad | 1610737 |
| EGTA | Merck | E4378 |
| Biotin-11-CTP | PerkinElmer | NEL542001EA |
| Trizol | ThermoFisher | 15596026 |
| Trizol-LS | ThermoFisher | 10296010 |
| GlycoBlue™ Coprecipitant | ThermoFisher | AM9515 |
| NTP Set, 100 mM Solution | ThermoFisher | R0481 |
| N-Lauroylsarcosine sodium salt | Merck | L5125 |
| Dynabeads™ M-280 Streptavidin | ThermoFisher | 11205D |
| Cap-CLIP | BioZym | C-CC15011H |
| T4 Polynucleotide Kinase | NEB | M0201S |
| Murine RNAse Inhibitor | NEB | M0314L |
| T4 RNA Ligase | NEB | M0204L |
| SuperScript™ III Reverse Transcriptase | ThermoFisher | 18080093 |
| KAPA HiFi HotStart Real-Time Library Amp Kit | Roche | 7959028001 |
| AMPure XP beads | Beckman Coulter | A63882 |
| Anti-FLAG® M2 Magnetic Beads | Merck | M8823 |
| Nuclease micrococcal from Staphylococcus aureus | Merck | N3755 |
| Proteinase K, recombinant, PCR Grade | Merck | 3115828001 |
| TissueRuptor Disposable Probes | Qiagen | 990890 |
| ROTI® Phenol/ Chloroform/ Isoamylalkohol, 500 ml | Roth | A156.2 |
| Turbo DNase | Thermo | AM2239 |
| RNase A (17,500 U) | Qiagen | 19101 |
| Digitonin | Merck | D141 |
| Spermidine trihydrochloride | Merck | 85580 |
| Lysyl endopeptidase | Wako Chemicals | Cat#7041 |
| Ammoniumbicarbonate | Sigma-Aldrich | Cat#09830 |
| Tris-(2-carboxyethyl)-phosphin-hydrochloride (TCEP) | Sigma-Aldrich | Cat#646547 |
| S-Methyl-thiomethanesulfonate (MMTS) | Sigma-Aldrich | Cat#64306 |
| Trifluoroacetic acid | Sigma-Aldrich | Cat#T6508 |
| oComplete mini protease inhibitors | Sigma-Aldrich | Cat# 11836170001 |
| Axygen 1.5mL MaxyClear tube | Corning | Cat#MCT-150-A |
| Critical commercial assays | ||
| Direct-zol RNA Microprep | Zymo | R2061 |
| Micro Bio-spin P-30 gel columns | Bio-rad | 7326251 |
| NEBNext® Ultra™ II DNA Library Prep Kit for Illumina | NEB | E7645 |
| CUTANA pAG-MNAse | Epicypher | 15-1016 |
| CUTANA Concanavalin A Conjugated Paramagnetic Beads | Epicypher | 21-1401 |
| Power Blotter Station | ThermoFisher | PB0010 |
| MaxCyte STX Scalable Transfection System | Maxcyte | NA |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 15-well, 15 μl | Bio-Rad | #34561096 |
| Mini-PROTEAN Tetra Vertical Electrophoresis Cell | Bio-Rad | 1658004 |
| Monarch Gel Extraction | NEB | T1020L |
| Deposited data | ||
| Raw and analyzed sequencing data | This paper | GEO: GSE184187 |
| Brd7 IP-MS | This paper | PRIDE: PXD036007 |
| Mendeley Dataset | This paper | DOI: 10.17632/zj2f34rwff.1 |
| STAP-seq cofactor recruitment data | Haberle et al. 2019 | GEO: GSE116197 |
| S2 dCP and hkCP STARR-seq | Zabidi et al., 2015 | GEO: GSE57876 |
| BG3 STARRseq | Yanez Cuna et al., 2014 | GEO: GSE49809 |
| Drosophila CAGE data | modEncode | http://www.modencode.org/publications/fly_2010pubs/index.shtml |
| Nucleosome Occupancy Prediction score | Segal Lab | https://genie.weizmann.ac.il/pubs/nucleosomes08/. |
| Oligonucleotides | ||
| Primers for AID tagging | This Paper | Table S1 |
| 5’- /5Phos/rNrNrN rNrNrN rNrNrG rArUrC rGrUrC rGrGrA rCrUrG rUrArG rArArC rUrCrU rGrArA rC/3InvdT/ -3’ | IDT | N/A |
| 5- rCrCrU rUrGrG rCrArC rCrCrG rArGrA rArUrU rCrCrA rNrNrN rN -3 (5’ RNA linker) | IDT | N/A |
| Recombinant DNA | ||
| pBabe Puro osTIR1-9Myc | Addgene | plasmid #80074 |
| pAc-sgRNA-Cas9 | Addgene | plasmid #49330 |
| pCRIS-PITChv2-FBL | Addgene | plasmid #63672 |
| Software and algorithms | ||
| Benchling | N/A | https://benchling.com |
| R version 3.5.3 | R Development Core Team, 2019 | https://www.r-project.org |
| Cutadapt | Martin et al. 2011 | https://bioweb.pasteur.fr/packages/pack@cutadapt@1.18 |
| Samtools version 1.9 | Li et al. 2009 | http://www.htslib.org/ |
| bowtie version 1.2.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.2.2/ |
| GenomicRanges 1.34.0 | Lawrence et al. 2013 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| bigBedtoBed | Kent et al. 2010 | https://github.com/ENCODE-DCC/kentUtils/blob/master/src/utils/bigBedToBed/bigBedToBed.c |
| bedtools 2.27.1 | Quinlan & Hall 2010 | https://github.com/arq5x/bedtools2/releases/tag/v2.30.0 |
| DESeq2 package v.1.30.1 | Love et al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CWM. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648–10. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic D, Stadler MB, Iurlaro M, Schübeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019;569:136–140. doi: 10.1038/s41586-019-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Pawar H, Dasari V, Mishra RK, Chandrashekaran S, Brahmachari V. Chromatin remodeling protein INO80 has a role in regulation of homeotic gene expression in Drosophila. Genes Cells. 2010;15:725–735. doi: 10.1111/j.1365-2443.2010.01416.x. [DOI] [PubMed] [Google Scholar]
- Boija A, Mahat DB, Zare A, Holmqvist P-H, Philip P, Meyers DJ, Cole PA, Lis JT, Stenberg P, Mannervik M. CBP Regulates Recruitment and Release of Promoter-Proximal RNA Polymerase II Mol. Cell. 2017;68:491–503.:e5. doi: 10.1016/j.molcel.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N Heidelberg Fly Array Consortium. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Brahma S, Henikoff S. RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Mol Cell. 2019;73:238–249.:e3. doi: 10.1016/j.molcel.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T-P, Comoglio F, Zhou T, Yang L, Paro R, Rohs R. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics. 2016;32:1211–1213. doi: 10.1093/bioinformatics/btv735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78(1):273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DFV, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS biology. 2007;5(9):e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Doblmann J, Dusberger F, Imre R, Hudecz O, Stanek F, Mechtler K, Dürnberger G. apQuant: Accurate Label-Free Quantification by Quality Filtering. J Proteome Res. 2018 doi: 10.1021/acs.jproteome.8b00113. [DOI] [PubMed] [Google Scholar]
- Donovan DA, Crandall JG, Truong VN, Vaaler AL, Bailey TB, Dinwiddie D, Banks OG, McKnight LE, McKnight JN. Basis of specificity for a conserved and promiscuous chromatin remodeling protein. Elife. 2021;10 doi: 10.7554/eLife.64061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfer V, Pichler P, Stranzl T, Stadlmann J, Taus T, Winkler S, Mechtler K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller C, Prestel M, Hartmann H, Straub T, Söding J, Becker PB. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res. 2012;40:1509–1522. doi: 10.1093/nar/gkr869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53–22. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes development. 2004;18(2):170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Haberle V, Arnold CD, Pagani M, Rath M, Schernhuber K, Stark A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–126. doi: 10.1038/s41586-019-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y. Structure of nucleosome-bound human BAF complex. Science. 2020;367:875–881. doi: 10.1126/science.aaz9761. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Iurlaro M, Stadler MB, Masoni F, Jagani Z, Galli GG, Schübeler D. Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet. 2021:1–9. doi: 10.1038/s41588-020-00768-w. [DOI] [PubMed] [Google Scholar]
- Jaeger MG, Schwalb B, Mackowiak SD, Velychko T, Hanzl A, Imrichova H, Brand M, Agerer B, Chorn S, Nabet B, Ferguson FM, et al. Selective Mediator dependence of cell-type-specifying transcription. Nat Genet. 2020;52:719–727. doi: 10.1038/s41588-020-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1:40–51. doi: 10.1002/wdev.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–721.:e12. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik S, Bruzzone MJ, Challal D, Dreos R, Mattarocci S, Bucher P, Libri D, Shore D. Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat Struct Mol Biol. 2019a;26:744–754. doi: 10.1038/s41594-019-0273-3. [DOI] [PubMed] [Google Scholar]
- Kubik S, Bruzzone MJ, Challal D, Dreos R, Mattarocci S, Bucher P, Libri D, Shore D. Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat Struct Mol Biol. 2019b;26:744–754. doi: 10.1038/s41594-019-0273-3. [DOI] [PubMed] [Google Scholar]
- Kubik S, O'Duibhir E, de Jonge WJ, Mattarocci S, Albert B, Falcone J-L, Bruzzone MJ, Holstege FCP, Shore D. Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Mol Cell. 2018;71:89–102.:e5. doi: 10.1016/j.molcel.2018.05.030. [DOI] [PubMed] [Google Scholar]
- Lam KC, Mühlpfordt F, Vaquerizas JM, Raja SJ, Holz H, Luscombe NM, Manke T, Akhtar A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- Liscovitch-Brauer N, Montalbano A, Deng J, Méndez-Mancilla A, Wessels H-H, Moss NG, Kung C-Y, Sookdeo A, Guo X, Geller E, Jaini S, et al. Profiling the genetic determinants of chromatin accessibility with scalable single cell CRISPR screens. Nat Biotechnol. 2021:1–8. doi: 10.1038/s41587-021-00902-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CGA, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashtalir N, D'Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, Valencia AM, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. 2018;175:1272–1288.:e20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Molecular and cellular biology. 2004;24(8):3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Mohrmann L, van Ijcken WFJ, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–661. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń ŁM, Arnold CD. STARR-seq-principles and applications. Genomics. 2015;106:145–150. doi: 10.1016/j.ygeno.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, Cassel SH, et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet. 2017;49:1613–1623. doi: 10.1038/ng.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayr C, Pagani M, Stark A, Arnold CD. STARR-seq and UMI-STARR-seq: Assessing Enhancer Activities for Genome-Wide-, High-, and Low-Complexity Candidate Libraries. Curr Protoc Mol Biol. 2019;128:e105. doi: 10.1002/cpmb.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann E, Krietenstein N, Niebauer V, Wang Y, Schall K, Moldt M, Straub T, Rohs R, Hopfner K-P, Korber P, Eustermann S. Genome information processing by the INO80 chromatin remodeler positions nucleosomes. Nat Commun. 2021;12:3231–19. doi: 10.1038/s41467-021-23016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao G-C, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087-12. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Schlichter A, Wilson BG, Cairns BR. The chromatin remodelers RSC and ISW1 display functional and chromatin-based promoter antagonism. Elife. 2015;4:e06073. doi: 10.7554/eLife.06073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA-and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rach EA, Winter DR, Benjamin AM, Corcoran DL, Ni T, Zhu J, Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011;7:e1001274. doi: 10.1371/journal.pgen.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Ahmad K, Henikoff S. Transcription and Remodeling Produce Asymmetrically Unwrapped Nucleosomal Intermediates. Mol Cell. 2017;68:1038–1053.:e4. doi: 10.1016/j.molcel.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reja R, Vinayachandran V, Ghosh S, Pugh BF. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015;29:1942–1954. doi: 10.1101/gad.268896.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Nakade S, Sakane Y, Suzuki K-IT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- Schick S, Grosche S, Kohl KE, Drpic D, Jaeger MG, Marella NC, Imrichova H, Lin J-MG, Hofstätter G, Schuster M, Rendeiro AF, et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat Genet. 2021;53:1–10. doi: 10.1038/s41588-021-00777-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen C-F, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York, NY: 2005. limma: Linear Models for Microarray Data; pp. 397–420. [DOI] [Google Scholar]
- Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, Stark A. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528:147–151. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and Confident Phosphorylation Site Localization Using phosphoRS. 2011 doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High Nucleosome Occupancy Is Encoded at Human Regulatory Sequences. PLoS ONE. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Ui K, Nishihara S, Sakuma M, Cellular STIV. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. Springer; 1994. [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, van Steensel B, Bussemaker HJ. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014;24:695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. 2012 doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at GEO repository GSE184187 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.Imaging data has been deposited and published through Mendeley Data: https://data.mendeley.com/datasets/zj2f34rwff/draft?a=af5adcbd-3cd8-4b91-9fcd-bd900684f1bc. Publicly available datasets re-analyzed in this study are available in the GEO repositories listed in the Key Resources Table.
This paper does not report original code. All custom code used for the analyses was written with existing software as detailed in the STAR Methods and Key Resources table and is available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal Anti-FLAG M2 Antibody | Merck | F3165 |
| Histone-H3 | Abcam | ab1791 |
| Secondary anti mouse HRP | Sigma-Aldrich | 12-349 |
| Secondary anti rabbit HRP | Sigma-Aldrich | 12-348 |
| Bacterial and virus strains | ||
| MegaX DH10B T1R Electrocomp™ Cells | Thermo Fisher | C640003 |
| Experimental models: Cell lines | ||
| D. melanogaster Schneider S2 cells | Thermo Fisher | R69007 |
| HCT-116 | ATCC | CCL-247 |
| S2OsTir1 | This Paper | N/A |
| S2OsTir1-Snr1-AID | This Paper | N/A |
| S2OsTir1-Iswi-AID | This Paper | N/A |
| S2OsTir1-Ino80-AID | This Paper | N/A |
| S2OsTir1-Chd1-AID | This Paper | N/A |
| S2OsTir1-Brd7-AID | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Puromycin | Invivogen | ant-pr-1 |
| FastDigest MluI | ThermoFisher | FD0564 |
| BspQI | NEB | R0712S |
| Blasticidin S HCl | ThermoFisher | R21001 |
| 3-Indoleacetic acid | Merck | I3750 |
| QuickExtract™ DNA Extraction Solution | Lucigen | QE9059 |
| 2x Laemmli Sample Buffer | BioRad | 1610737 |
| EGTA | Merck | E4378 |
| Biotin-11-CTP | PerkinElmer | NEL542001EA |
| Trizol | ThermoFisher | 15596026 |
| Trizol-LS | ThermoFisher | 10296010 |
| GlycoBlue™ Coprecipitant | ThermoFisher | AM9515 |
| NTP Set, 100 mM Solution | ThermoFisher | R0481 |
| N-Lauroylsarcosine sodium salt | Merck | L5125 |
| Dynabeads™ M-280 Streptavidin | ThermoFisher | 11205D |
| Cap-CLIP | BioZym | C-CC15011H |
| T4 Polynucleotide Kinase | NEB | M0201S |
| Murine RNAse Inhibitor | NEB | M0314L |
| T4 RNA Ligase | NEB | M0204L |
| SuperScript™ III Reverse Transcriptase | ThermoFisher | 18080093 |
| KAPA HiFi HotStart Real-Time Library Amp Kit | Roche | 7959028001 |
| AMPure XP beads | Beckman Coulter | A63882 |
| Anti-FLAG® M2 Magnetic Beads | Merck | M8823 |
| Nuclease micrococcal from Staphylococcus aureus | Merck | N3755 |
| Proteinase K, recombinant, PCR Grade | Merck | 3115828001 |
| TissueRuptor Disposable Probes | Qiagen | 990890 |
| ROTI® Phenol/ Chloroform/ Isoamylalkohol, 500 ml | Roth | A156.2 |
| Turbo DNase | Thermo | AM2239 |
| RNase A (17,500 U) | Qiagen | 19101 |
| Digitonin | Merck | D141 |
| Spermidine trihydrochloride | Merck | 85580 |
| Lysyl endopeptidase | Wako Chemicals | Cat#7041 |
| Ammoniumbicarbonate | Sigma-Aldrich | Cat#09830 |
| Tris-(2-carboxyethyl)-phosphin-hydrochloride (TCEP) | Sigma-Aldrich | Cat#646547 |
| S-Methyl-thiomethanesulfonate (MMTS) | Sigma-Aldrich | Cat#64306 |
| Trifluoroacetic acid | Sigma-Aldrich | Cat#T6508 |
| oComplete mini protease inhibitors | Sigma-Aldrich | Cat# 11836170001 |
| Axygen 1.5mL MaxyClear tube | Corning | Cat#MCT-150-A |
| Critical commercial assays | ||
| Direct-zol RNA Microprep | Zymo | R2061 |
| Micro Bio-spin P-30 gel columns | Bio-rad | 7326251 |
| NEBNext® Ultra™ II DNA Library Prep Kit for Illumina | NEB | E7645 |
| CUTANA pAG-MNAse | Epicypher | 15-1016 |
| CUTANA Concanavalin A Conjugated Paramagnetic Beads | Epicypher | 21-1401 |
| Power Blotter Station | ThermoFisher | PB0010 |
| MaxCyte STX Scalable Transfection System | Maxcyte | NA |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 15-well, 15 μl | Bio-Rad | #34561096 |
| Mini-PROTEAN Tetra Vertical Electrophoresis Cell | Bio-Rad | 1658004 |
| Monarch Gel Extraction | NEB | T1020L |
| Deposited data | ||
| Raw and analyzed sequencing data | This paper | GEO: GSE184187 |
| Brd7 IP-MS | This paper | PRIDE: PXD036007 |
| Mendeley Dataset | This paper | DOI: 10.17632/zj2f34rwff.1 |
| STAP-seq cofactor recruitment data | Haberle et al. 2019 | GEO: GSE116197 |
| S2 dCP and hkCP STARR-seq | Zabidi et al., 2015 | GEO: GSE57876 |
| BG3 STARRseq | Yanez Cuna et al., 2014 | GEO: GSE49809 |
| Drosophila CAGE data | modEncode | http://www.modencode.org/publications/fly_2010pubs/index.shtml |
| Nucleosome Occupancy Prediction score | Segal Lab | https://genie.weizmann.ac.il/pubs/nucleosomes08/. |
| Oligonucleotides | ||
| Primers for AID tagging | This Paper | Table S1 |
| 5’- /5Phos/rNrNrN rNrNrN rNrNrG rArUrC rGrUrC rGrGrA rCrUrG rUrArG rArArC rUrCrU rGrArA rC/3InvdT/ -3’ | IDT | N/A |
| 5- rCrCrU rUrGrG rCrArC rCrCrG rArGrA rArUrU rCrCrA rNrNrN rN -3 (5’ RNA linker) | IDT | N/A |
| Recombinant DNA | ||
| pBabe Puro osTIR1-9Myc | Addgene | plasmid #80074 |
| pAc-sgRNA-Cas9 | Addgene | plasmid #49330 |
| pCRIS-PITChv2-FBL | Addgene | plasmid #63672 |
| Software and algorithms | ||
| Benchling | N/A | https://benchling.com |
| R version 3.5.3 | R Development Core Team, 2019 | https://www.r-project.org |
| Cutadapt | Martin et al. 2011 | https://bioweb.pasteur.fr/packages/pack@cutadapt@1.18 |
| Samtools version 1.9 | Li et al. 2009 | http://www.htslib.org/ |
| bowtie version 1.2.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.2.2/ |
| GenomicRanges 1.34.0 | Lawrence et al. 2013 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| bigBedtoBed | Kent et al. 2010 | https://github.com/ENCODE-DCC/kentUtils/blob/master/src/utils/bigBedToBed/bigBedToBed.c |
| bedtools 2.27.1 | Quinlan & Hall 2010 | https://github.com/arq5x/bedtools2/releases/tag/v2.30.0 |
| DESeq2 package v.1.30.1 | Love et al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |