Summary
Ferns, and particularly homosporous ferns, have long been assumed to have experienced recurrent whole-genome duplication (WGD) events because of their substantially large genome sizes, surprisingly high chromosome numbers, and high degrees of polyploidy among many extant members. As the number of sequenced fern genomes is limited, recent studies have employed transcriptome data to find evidence for WGDs in ferns. However, they have reached conflicting results concerning the occurrence of ancient polyploidy, for instance, in the lineage of leptosporangiate ferns.
Because identifying WGDs in a phylogenetic context is the foremost step in studying the contribution of ancient polyploidy to evolution, we here revisited earlier identified WGDs in leptosporangiate ferns, mainly the core leptosporangiate ferns, by building Ks-age distributions and applying substitution rate corrections, and by conducting statistical gene tree – species tree reconciliation analyses.
Our integrative analyses confidently identified four ancient WGDs in the sampled core leptosporangiate ferns but also identified false positives and false negatives for WGDs that recent studies have reported earlier.
In conclusion, we underscore the significance of substitution rate corrections and uncertainties in gene tree – species tree reconciliations in calling WGD events and advance an exemplar workflow to overcome such often-overlooked issues.
Keywords: ferns, gene tree – species tree reconciliation, Ks-age distribution, phylogenomics, polyploidy, WGD
Introduction
Tracheophytes, or vascular plants, have shaped the diversity of the terrestrial ecosystem on Earth since their first appearance about 431 to 451 million years ago (Morris et al., 2018). Tracheophytes are composed of three major groups, spermatophytes (seed plants), Lycopodiopsida (lycophytes), and Polypodiopsida (ferns) (PPG I, 2016). Seed plants form a monophyletic group, including gymnosperms and angiosperms. Within the past two decades, strong evidence has accumulated for recurrent paleo-polyploidizations, or ancient whole-genome duplications (WGDs) in seed plants (Cui et al., 2006; Vekemans et al., 2012; Vanneste et al., 2014; Ming et al., 2015; Van de Peer et al., 2017; Stull et al., 2021) and their importance for the evolution of innovative traits and in facilitating the diversification of seed plant species are undisputed (Soltis & Soltis, 2016; Van de Peer et al., 2017; Landis et al., 2018; Fox et al., 2020; Van de Peer et al., 2021). Different from the lineage of seed plants, strong evidence for WGDs in lycophytes and ferns was lacking, although cytological evidence suggested that polyploidization may not be uncommon in ferns (Wood et al., 2009; Clark et al., 2016; Wang et al., 2022).
Ferns are the largest group of non-seed vascular plants and make up more than 90% of the extant diversity (PPG I, 2016). Compared to seed plants, very few have had their genome sequenced so far (Szövényi et al., 2021), because they, especially homosporous ferns, tend to have large genome sizes and large to huge chromosome numbers. For example, the modern fern or C-fern (Ceratopteris richardii) possesses 2n = 78 chromosomes with a genome of 11.25 Gbp (Marchant et al., 2019). More strikingly, Ophioglossum reticulatum is a fern species with the highest chromosome number known amongst eukaryotes, with 2n = 1,440 chromosomes (Khandelwal, 1990). The huge diversity as well as the high numbers of chromosomes of ferns are compelling mysteries that have fascinated evolutionary biologists for decades (Haufler & Soltis, 1986; Barker, 2009; Clark et al., 2016; Wang et al., 2022). Given that polyploidizations can increase both genome sizes and chromosome numbers directly, multiple rounds of polyploidizations, along with potential changes in chromosome compositions and/or processes of genome downsizing, have been hypothesized to explain the evolution of chromosomes and genomes (Clark et al., 2016; Wang et al., 2022) and further the species diversity in ferns (Fujiwara et al., 2021).
It is not until recently that genomic and transcriptomic data have begun to shed light on ancient polyploidies in ferns (Barker & Wolf, 2010; Szövényi et al., 2021). Analyses of the first two genomes of heterosporous ferns, Azolla filiculoides and Salvinia cucullata, have identified two WGDs, with one specific to the genus Azolla and the other shared by all core Leptosporangiates (Li et al., 2018). Furthermore, the flying spider-monkey tree fern (Cyathea spinulosa) genome has unveiled WGDs in Cyatheales (Huang et al., 2022), while the partial genome of the C-fern has provided some evidence for a WGD also in its lineage (Marchant et al., 2019). Also, analyses of Equisetum transcriptomes showed ancient polyploidy in the lineage (Vanneste et al., 2015; Clark et al., 2019). In addition, two recent studies have added valuable supplements of transcriptome data to the scarce genomic data of ferns and suggested several ancient WGDs during the evolutionary history of ferns (1KP initiative, 2019; Huang et al., 2020). However, conflicting results regarding the identified WGD events in ferns have been proposed in the previous studies.
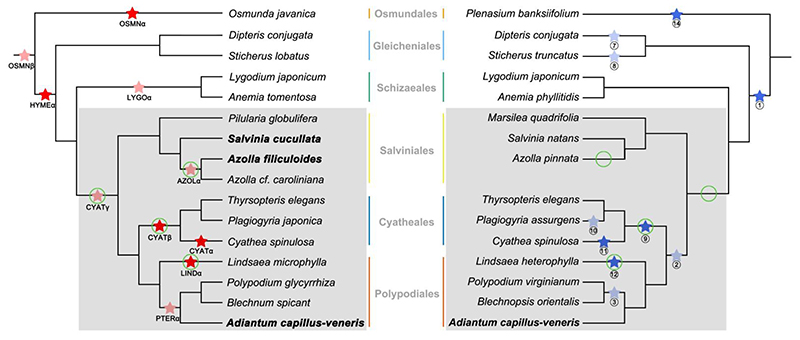
Here, we revisited the occurrence of WGDs in leptosporangiate ferns, or more specifically in the lineage of core leptosporangiates, for leptosporangiate ferns form most of the species in extant ferns (Pryer et al., 2004). Leptosporangiates are subdivided into seven orders, namely Osmundales (c. 18 species), Hymenophyllales (c. 434 species), Gleicheniales (c. 172 species), Schizaeales (c. 190 species), Salviniales (c. 82 species), Cyatheales (c. 713 species), and Polypodiales (c. 8,714 species), the last three of which include most species and constitute the lineage of core leptosporangiates (Smith et al., 2006; PPG I, 2016). To focus on those ferns that have been investigated in previous studies with a well-resolved phylogeny (Shen et al., 2018), we selected species in all three orders of the core leptosporangiates and used representatives of another three leptosporangiate orders as outgroups. This way, we could revisit ten out of 14 WGDs reported by the 1KP initiative (2019) and all ten WGDs reported by Huang et al. (2020) in leptosporangiates. For the ten WGD events retrieved from each study, only five are congruent and have been placed in the same phylogenetic position (Fig. 1).
Fig. 1. Identified WGD events in leptosporangiates, as reported by the 1KP initiative (2019) (left) and by Huang et al. (2020) (right).
Ten of 14 WGDs in leptosporangiates from the 1KP initiative (2019) are denoted as (light) red stars. Four WGDs are not included because they were placed in lineages not studied by Huang et al. (2020). Ten out of ten WGD events from Huang et al. (2020) are denoted as (light) blue stars. WGD events found in the same lineages by both studies are in solid red and solid blue. The grey background highlights the core leptosporangiates in the two phylogenetic trees. The names for species with fully sequenced genomes are in bold. The green circles denote the WGDs in core leptosporangiates identified in this study.
The conflicting results concerning the identified WGDs in previous studies could result from several pitfalls that are often overlooked in the two commonly used approaches to find evidence for ancient WGDs, namely Ks-age distributions of paralogs and gene tree – species tree reconciliation approaches. Although these two approaches have great power and have been widely applied to detect WGDs based on genomic and transcriptome data (Jiao et al., 2011; Vanneste et al., 2013; Li et al., 2015; McKain et al., 2016; Zhang et al., 2017; Ren et al., 2018), they must be used with caution (Tiley et al., 2018; Zwaenepoel et al., 2019; Zwaenepoel & Van de Peer, 2019). WGDs in so-called Ks-age distributions, where the number of duplicates is plotted against their age as inferred from the expected number of synonymous substitutions per synonymous site (Ks), can be identified as peaks in the distribution, which suggest that many genes have been duplicated at the same time (Vanneste et al., 2013). Such Ks peaks are often compared with speciation events characterized by Ks distributions of orthologs between species to infer the relative or absolute timing of the WGDs. However, such comparisons admit meaningful interpretation only if substitution rates of the species under consideration are similar, while substitution rates naturally vary across lineages. It has been gradually acknowledged that different substitution rates can affect the placement of WGDs (Barker et al., 2008; Chen et al., 2020; Sensalari et al., 2021). For instance, if two species sharing one WGD diverged and have evolved at different substitution rates afterward, given no correction for the difference in substitution rates, the WGD Ks peak identified in the species with a lower substitution rate may be incorrectly interpreted as a younger and lineage-specific WGD. In contrast, species with a higher substitution rate may still support a shared WGD. This could eventually lead to erroneous conclusions, especially when no genome is available to determine the inference of WGDs via collinear analysis.
A second approach to identify and date WGDs is to use gene tree – species tree reconciliation, where events underlying the evolutionary history of a gene, like gene duplication and loss, hybridization, introgression, horizontal gene transfer, and incomplete lineage sorting, are identified by mapping gene trees onto species trees. When many duplicated genes are reconciled on one specific branch of the species tree, this can be considered evidence for a WGD. Although the 1KP initiative (2019) and Huang et al. (2020) have implemented the reconciliation approaches differently, both have employed the least common ancestor (LCA) reconciliation to determine duplication events on a species phylogeny based on gene trees inferred by maximum likelihood (ML) inference. In LCA reconciliation, a duplication event involving genes from some species is placed on a species phylogeny at the node associated with the most recent common ancestor of these species (Zmasek & Eddy, 2001). Even if gene trees have been filtered based on their quality before reconciliations (based on bootstrap support values, for instance), the LCA reconciliation is still error-prone in placing gene duplication and loss events, and its accuracy depends on the correctness of inferred gene tree topologies (Hahn, 2007). Nevertheless, the true gene tree topology for a gene family is often one among many statistically equivalent gene trees (Wu et al., 2013), so only considering the one ‘best’ ML tree for each gene family may cause systematic bias when using LCA reconciliation to identify WGDs (Hahn, 2007; Zwaenepoel & Van de Peer 2019). In addition, a WGD and its phylogenetic position are often determined when the number of duplication events on a branch exceeds a certain cut-off, which is usually set somewhat arbitrarily without acknowledging the varying contribution of small-scale duplications (SSDs) along different branches of the species tree (Li et al., 2015; McKain et al., 2016; Ren et al., 2018), which may result in false positive WGD identification towards the tips of a species phylogeny (Zwaenepoel & Van de Peer, 2019).
To revisit ancient polyploidy in leptosporangiate ferns, we retrieved relevant transcriptome data from the 1KP initiative (2019), for its relatively high quality and reasonably high gene numbers (Fig. S1). Also, we added two publicly available genomes of heterosporous ferns and a newly sequenced homosporous fern, Adiantum capillus-veneris L. (Fang et al., 2022). We did not include the C-fern genome because of its partial and fragmented nature (Marchant et al., 2019). By considering differences in substitution rates and performing statistical gene tree – species tree reconciliations under a model integrating small-scale gene duplication and loss (DL) and WGDs, we confidently identified four WGDs in core leptosporangiate ferns (Fig. 1), fewer than the six WGDs as found by 1KP initiative (2019) and Huang et al. (2020), while some WGDs have also been predicted at different phylogenetic positions, suggesting that some WGDs identified by the two previous studies are likely false positives. Our study again highlights the importance of fully recognizing the caveats and limitations of commonly used approaches in calling WGD events.
Materials and Methods
Transcriptomes and Genomes of Leptosporangiates
We selected 16 and 15 species and their corresponding assembled transcriptomes from the 1KP initiative (2019) and Huang et al. (2020), respectively. Except for the order Hymenophyllales due to its uncertain phylogenetic position (PPG I, 2016;1KP initiative, 2019), the remaining six orders in leptosporangiate ferns were sampled (Fig. 1; Table S1). We removed unigenes with identical sequences by SeqKit (v0.7.1) (Shen et al., 2016) and filtered out coding sequences that were not divisible by three or had unknown nucleotides or premature stop codons. We retrieved the genomes of Azolla filiculoides and Salvinia cucullata from fernbase.org (Li et al., 2018) and used the Adiantum capillus-veneris genome (Fang et al., 2022). We then used BUSCO (v4.0.2) to assess gene space completeness in the three ferns with complete genomes and the 13 ferns with 1KP transcriptomes using embryophyta_odb10 (Simão et al., 2015; Kriventseva et al., 2019) (Fig. S2).
Constructing Ks-age distributions
Ks-age distributions for all paralogous genes (paranome) in transcriptomes and genomes were constructed by wgd (v1.1.1) (Zwaenepoel & Van de Peer, 2018). To detect peaks that could be signatures of WGD events in the Ks distributions, we performed mixture modeling using the R package mclust (v5.4.7) (Scrucca et al., 2016). We first transformed Ks distributions into log-scale, which were further fitted to a serial of Gaussian mixture models (GMM) (Rasmussen, 1999). We increasingly fitted one to eight components per mixture model and used the Bayesian Information Criterion (BIC) to select the optimal number of components. Although BIC strongly penalizes increases in the number of parameters, the GMM is still prone to overfitting, so we further performed SiZer (Significance of Zero Crossings of the Derivative) analysis using the R package feature (v1.2.15) (https://cran.r-project.org/web/packages/feature/index.html) to distinguish bona fide peaks in the Ks distributions from those that represent noises (Chaudhuri & Marron, 1999) (Fig. S3, S4).
Correcting differences in synonymous substitution rates
Orthologous Ks distributions were constructed by wgd (Zwaenepoel & Van de Peer, 2018). To circumscribe the phylogenetic placements of the identified WGDs in the Ks-age distributions for paranomes, we corrected the differences in synonymous substitution rates across species using two approaches. In the first approach, we used OrthoFinder (v2.3.3) (Emms & Kelly, 2019) with default settings, except “-M msa”, to identify gene families with the 16 species in Fig. 1. We used MUSCLE (v3.8.31) (Edgar, 2004) to perform multiple sequence alignment of the proteins for the 34 identified single-copy gene families, which were further concatenated after being trimmed and back-translated by trimal (v1.4.1) (Capella-Gutiérrez et al., 2009). PAML (v4.9j) with the free-ratio model (Yang, 2007) was then used to estimate branch lengths in Ks unit for the species phylogeny. To map all the identified Ks peaks onto the species phylogeny in Ks units, we halved Ks values of the identified peaks in the GMM analyses (Fig. S3) and placed each peak from the tip towards the root of the phylogeny to date when WGD events have occurred in the phylogeny, with the assumption that duplicate genes on average evolved at similar substitution rates after WGD events.
In the second approach, we used ksrates (v1.0), which corrects synonymous substitution rates of other species to the rate of a focal species, i.e., the species desired to implement comparisons between the relative date of WGD and species divergence (Sensalari et al., 2021). To identify peaks representing WGDs, ksrates fits an exponential-lognormal mixture model to each Ks-age distribution, using the BIC to evaluate one exponential component for the L-shaped SSDs (Lynch & Conery, 2003) and one to five lognormal components for potential WGD peaks. Then, ksrates compared WGD peaks and corrected orthologous Ks peaks to infer the timing of WGDs (Fig. S5). Besides using default parameters, we set a maximum of 14 sets of trios to correct each divergence with multiple outgroups and used means to form consensus divergence peaks.
Statistical Gene Tree – Species Tree Reconciliation
To prepare data for the reconciliation analysis, we retrieved a species tree with divergence times from TimeTree (Kumar et al., 2017) (Fig. S6). Based on the 16,305 gene families identified above, we filtered 9,442 gene families for large family sizes or no common ancestor at the root by “orthofilter.py” (https://github.com/arzwa/Whale.jl). We then used PRANK (v150803) (Löytynoja, 2014) to perform multiple sequence alignment for each gene family and MrBayes (v.3.2.6) (Ronquist et al., 2012) to infer posterior probability distributions of gene trees under the LG+GAMMA model. MrBayes ran 110,000 generations and sampled at a frequency of 10 to get in total 11,000 posterior samples for each of the 6,863 gene families. Lastly, ALEobserve (Szöllõsi et al., 2013) constructed the conditional clade distribution containing marginal clade frequencies with a burn-in of 1,000.
Using Whale (v2.0.3) (Zwaenepoel & Van de Peer, 2019), we carried out statistical gene tree – species tree reconciliation and tested the occurrences of eight WGDs (Fig. S6) under the so-called DL+WGD model, which considers both small-scale gene duplication and loss (DL), and WGDs. Two DL+WGD models were adopted to incorporate various DL rates of SSDs across the species tree (Methods S1). In the critical branch-specific DL+WGD model, where we assumed the duplication (λ) and loss rates (μ) to be equal on each branch, a Beta(3,1) prior distribution was used for η, the parameter of the geometric prior distribution on the number of genes at the root. We used an improper flat prior for the mean branch rate r. The branch rates were assumed to follow a multivariate Gaussian prior with an Exponential prior with mean 0.1 for the standard deviation. For the more flexible DL+WGD model with branch-specific DL rates model, we assumed an independent bivariate normal prior with mean 0 and standard deviation 1 for the mean log-DL rate, assumed a Uniform(-1,1) prior for the correlation coefficient of DL rate for each individual branch, and assumed an exponential prior with mean 1 for the standard deviation of the branch rates. Lastly, because most of the analyzed species only have transcriptome data, missing genes were considered in the models by leveraging the BUSCO missing values of each species (Fig. S2). Generally, the effective sample sizes (ESSs) of all the inferred parameters were over 200, with a median over 500, indicating good approximations of the posteriors (Table S2, S3). We further checked the convergence of parameters with a subset of the 6,863 gene families by randomly selecting 1,000 gene families. We inferred their gene trees using MrBayes with 1,100,000 generations and a sampling frequency of 100. We filtered out 124 gene families (Fig. S7) and ran Whale under both models. The 95% uncertainty intervals for most of the estimates based on the subset overlapped with the ones based on the 6,863 gene families (Table S4, S5).
Collinear Analysis of Available Fern Genomes
We used i-ADHoRe (v.3.0.01) (Proost et al., 2011) to delineate both intra- and intergenomic collinearity with the three available fern genomes. For the intragenomic comparisons, i-ADHoRe in wgd identified 361, 414, and 375 anchor pairs – duplicate pairs retained in the collinear regions – in the genomes of A. capillus-veneris, A. filiculoides, and S. cucullata, respectively. Ks distributions for the anchor pairs show a larger fractions of anchor pairs with Ks values ranging from 0 to 0.1 in A. filiculoides and S. cucullata, compared to those in A. capillus-veneris (Fig. S8). Most of the anchor pairs with small Ks values are located on short scaffolds in the A. filiculoides and S. cucullata assemblies (Fig. S9), reflecting that they are still fragmented to a certain extent. We hence removed short scaffolds with fewer than ten genes and anchor pairs with Ks values less than 0.1 for intergenomic comparisons. To infer collinear ratios among the three fern genomes, we performed all-against-all BLASTP (v.2.6.0+) (Camacho et al., 2009) for all proteins from the three ferns with an E-value of 1 × 10-5 and ‘-max_target_seqs = 100000’. Homologous pairs were filtered using the c-score of 0.5 (Putnam et al., 2007) and were fed into i-ADHoRe to analyze intergenomic collinear ratio (Fig. S9).
Also, we used Whale without hypothetical WGD events to perform gene tree – species tree reconciliations for gene families having anchor pairs to estimate the expected number of duplication and loss on each branch (Methods S2). We fixed the η parameter to 0.75 based on the average number of observed genes in a family. An exponential prior with r for the expected number of duplication/loss events along a branch was assumed, where a noninformative prior was set for r. To reserve duplicates that were likely derived from WGD events, we kept 30, 75, and 36 anchor pairs in 137 gene families, which had a common ancestor at the root of the species tree and whose Ks values fell in the ranges of [1.8,2.5], [0.6,1.2] and [1.0,1.9] according to the paranome Ks-age distributions of A. capillus-veneris, A. filiculoides, and S. cucullata, respectively (Fig. S8).
Results
Different substitution rates among ferns in core leptospongiates
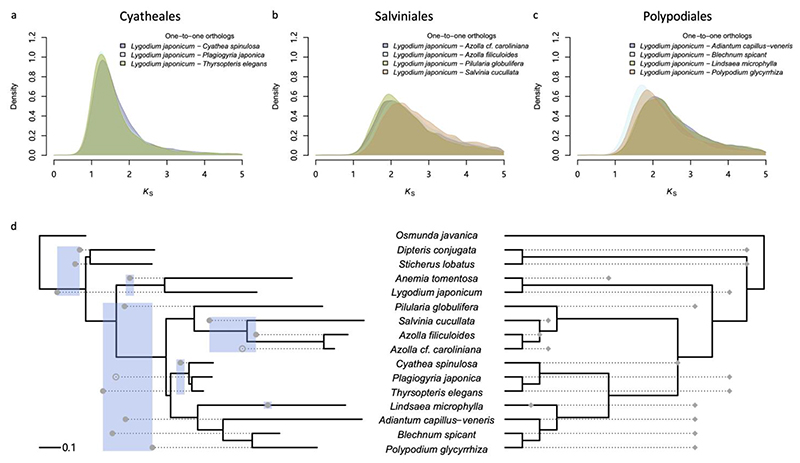
To compare synonymous substitution rates among core leptosporangiate ferns, we compared one-to-one orthologous Ks distributions between Lygodium japonicum from Schizaeales (or Dipteris conjugata from Gleicheniales in Fig. S10) and species from Cyatheales, Salviniales, and Polypodiales within core leptospongiates (Fig. 2a-c). Because peaks in the orthologous Ks distributions all represent the divergence between Schizaeales and the core leptospongiates, they should have identical or at least very similar Ks peak values if the selected species all have similar substitution rates. However, the orthologous Ks peak values are smaller for species from Cyatheales than species from Salviniales and Polypodiales, suggesting that nuclear genes in Cyathealean species, like chloroplast genes (Zhong et al., 2014), have slower substitution rates than their counterparts from the other two orders in the core leptosporangiates. In addition, the orthologous Ks peak values for the species belonging to Salviniales and Polypodiales show more variation than the ones in Cyatheales, indicating more variable substitution rates among Salvinialean and Polypodialean species (Fig. 2a-c).
Fig. 2. Orthologous Ks distributions and WGD events identified based on Ks distributions for the whole paranomes of different fern species.
(a-c) One-to-one orthologous Ks distributions between Lygodium japonicum and species from Cyatheales (a), Salviniales (b), and Polypodiales (c). (d) WGD events identified based on Ks distributions for the whole paranome of species in the phylogeny. On the left, a species phylogram is shown with branch lengths in Ks units, while WGD events are depicted as dots (calculated as half the Ks peak value of each species starting from the corresponding tip). Solid dots denote significant Ks peaks in the SiZer analysis, whereas hollow dots denote Ks peaks only identified by GMM but not SiZer (see Materials and Methods and Fig. S3, S4). On the right, a species cladogram is shown, where WGD events are depicted as rhombs according to the analyses of ksrates (Sensalari et al. 2021). Note that when a WGD and a speciation event overlap in the ksrates analysis, the WGD event is placed at the speciation event in the cladogram.
Inferring WGDs by applying substitution rate corrections
As substitution rates affect Ks distributions, peaks in the paralogous Ks distributions may differ for species with different substitution rates, even if they have experienced the same WGD event. We hence wondered if the differences in calling WGDs within core leptosporangiates in previous studies could be due to different substitution rates. Ks-age distributions for the whole paranomes of the 16 selected species, except for Plagiogyria japonica and Azolla cf. caroliniana, all show a peak verified by the SiZer analyses (Fig. S3, S4). The peaks show various Ks values and are largely in line with the paranome Ks distributions from the 1KP initiative (2019).
To correct for different synonymous substitution rates among species, we adopted two recently developed approaches. In the first approach, we inferred a species phylogeny in Ks units so that we could compare WGD peaks in one species with its divergence from other species (Chen et al., 2020). If we assume that both paralogs, on average, evolved at a similar rate after a WGD event, we could simply consider half the Ks values of all identified peaks in each species to position, starting from each tip, the WGDs on the phylogenetic tree (Fig. 2d). By applying this approach, we show that the Ks peak identified in Lindsaea microphylla supports one WGD in the lineage of Lindsaea (Fig. 2d), while the Ks peaks found in Blechnum spicant, Polypodium glycyrrhiza, and Adiantum capillus-veneris all support a more ancient WGD likely shared by the core leptospongiates (Fig. 2d), although they all have different Ks peak values (Fig. S3) for different substitution rates (Fig. 2c; Fig. S10).
In Cyatheales, the Ks peak in Cyathea spinulosa supports a WGD shared by Cyathales. If true, we would expect to observe clear Ks peaks in Thyrsopteris elegans and Plagiogyria japonica as well. Unexpectedly, only a Ks peak supporting a WGD before the divergence of core leptospongiates and Schizaeales has been observed in Thyrsopteris elegans (Fig. 2d). Similarly, a less perceptible Ks peak in Plagiogyria japonica, which is supported by the GMM but the SiZer analysis, also suggests an ancient WGD event. We argue that Thyrsopteris elegans and Plagiogyria japonica have even lower synonymous substitution rates than Cyathea spinulosa (Fig. 2a). If the Ks peak for the Cyathealean WGD is at Ks≈ 0.3 in the paranome Ksdistribution of Cyathea spinulosa, the expected Kspeaks in slower evolving Thyrsopteris elegans and Plagiogyria japonica must have even smaller values, which may be confounded by the background Ksdistribution from SSDs (Fig. S11).
In Salviniales, the Ks peak in Azolla filiculoides supports a WGD in the lineage leading to Azolla. However, the paranome Ks distribution of Salvinia cucullata has a peak supporting a WGD before the divergence of Salvinia and Azolla, instead of a WGD before the divergence of core leptosporangiates, as suggested earlier (Li et al., 2018). Also, the GMM has disentangled a peak in the paranome Ks distribution of Azolla cf. caroliniana, suggesting a WGD shared by Salvinia and Azolla, but the peak is not significant in the SiZer analysis (Fig. S3). Lastly, a WGD shared by core leptosporangiates is evidenced by the Ks peak found in Piluaria globulifera.
In the second approach, we used ksrates, which adjusts synonymous substitution rates to the rate of a focal species by relative rate tests (Sensalari et al., 2021). Therefore, peaks identified in the Ks-age distribution of the focal species can be directly compared with speciation events represented by orthologous Ks distributions (Fig. S5). By correcting for unequal substitution rates among species, the ksrates analysis confirms our previous results, except that the ksrates analysis provides extra support from Lindsaea microphylla for a WGD shared by the core leptospongiates (Fig. 2d).
Evaluating WGDs with two different DL+WGD models
The analyses of Ks-age distributions as described above suggest eight branches in the species phylogeny potentially associated with WGDs, but the occurrence of WGDs on some branches is ambiguous, because Ks peaks from different species fall onto adjacent branches (Fig. 2d). For example, the Ks peaks from Polypodium glycyrrhiza, Pilularia globulifera, and Adiantum capillus-veneris support a WGD shared by all core leptospongiates, whereas the ones from Thyrsopteris elegans, Blechnum spicant, and Plagiogyria japonica support a WGD before the divergence between core leptospongiates and Schizaeales. Similarly, the Ks peak from Azolla filiculoides supports a WGD specific to Azolla. Still, the Ks peak for Salvinia cucullata favors a shared WGD by Azolla and Salvinia. These results could point to two independent WGDs, one before and one after the speciation event, or alternatively, to one WGD event that is, however, represented by Ks peaks with different values for different species.
To determine the exact positions of these potential WGDs, we used the so-called DL+WGD model implemented in Whale to perform statistical gene tree – species tree reconciliation (Zwaenepoel & Van de Peer, 2019), with inferred posterior probability distributions of gene trees for 6,863 gene families. To this end, the eight hypothetical WGDs, according to the Ks analyses, were placed on the species tree (Fig. 3; Fig. S6), each with a uniform prior for the WGD retention rate (q). Whale then used amalgamated likelihood estimation (szöllõsi et al., 2013) to test WGD hypotheses in a phylogenetic context through estimating duplication (λ) and loss (μ) rates for SSDs, and q for WGDs (Zwaenepoel & Van de Peer, 2019). Because assuming constant DL rates of SSDs across the species tree could substantially affect WGD testing (Zwaenepoel & Van de Peer, 2019), we adopted two models to incorporate various DL rates of SSDs across the species tree: 1) the critical branch-specific model, where each branch in the species tree has an equal DL rate, i.e., λ = μ, but the rates vary across branches; and 2) the relaxed branch-specific model, where DL rates again vary across branches but not necessarily equal, i.e., λ ≠ μ. Comparing results from different models may aid in assessing the robustness of particular inferences to model violations, because the basic linear birth-death process in the DL model may not be an ideal model of gene family evolution (Zwaenepoel & Van de Peer, 2021).
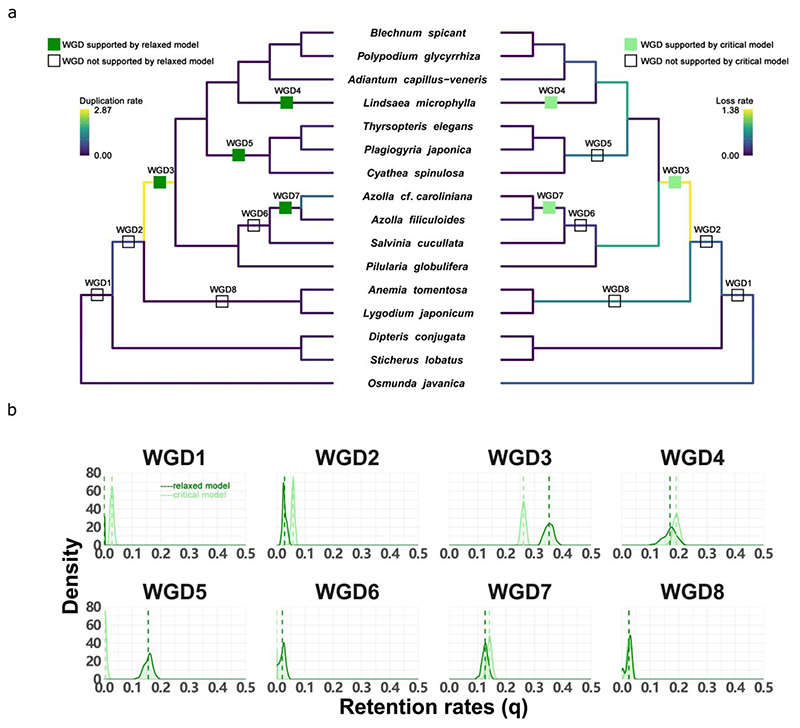
Fig. 3. Whale (gene tree – species tree reconciliation) analysis for eight hypothetical WGDs under the DL+WGD model.
(a) The species cladograms with the eight putative WGD events mentioned in the previous Ks-age analyses. The WGD bars in green on the left cladogram (for the relaxed branch-specific model) and in light green on the right cladogram (for the critical branch-specific model) are supported WGDs with retention rates significantly different from zero, while the hollow WGD bars in each cladogram are the ones with retention rates not different from zero (Table 1). Posterior mean of duplication (left) and loss (right) rates estimated under the relaxed DL+WGD model (see Materials and Methods) are colored on the cladograms. (b) The posterior distributions of the WGD retention rates (q) for the eight putative WGDs under the relaxed branch-specific model (green) and the critical branch-specific model (light green). The dotted lines show the posterior mean of each posterior distribution.
After obtaining posterior distributions of all the parameters under both DL+WGD models (Fig. 3), we estimated q for each putative WGD by its posterior mean. Further, we used the posterior distributions of q to estimate the Bayes Factor (K) to test if q is significantly different from zero using the Savage-Dickey density ratio (Zwaenepoel & Van de Peer, 2019). A putative WGD with q significantly larger than zero would hence indicate the occurrence of a WGD on a specific branch (Table 1). With the relaxed DL+WGD model, our results support four WGDs, i.e., WGD3, WGD4, WGD5, and WGD7, which all have over 0.05. Similarly, the results based on the critical branch-specific DL+WGD model support WGD3, WGD4, and WGD7 (Fig. 3). Our gene tree – species tree reconciliation analyses with both DL+WGD models raised our confidence in resolving the two ambiguous WGDs discussed higher.
Table 1.
Hypothetical WGDs, posterior mean of duplicate retention rate (q), and the Bayes Factor (K) to compare the likelihood of q = 0 (H0) to the likelihood of q > 0 (H1) using the Savage-Dickey density ratio.
| Relaxed branch-specific model | Critical branch-specific model | |||
|---|---|---|---|---|
| WGD1 | 0.001 | 1272.297 | 0.029 | 2.693 |
| WGD2 | 0.028 | 2.649 | 0.058 | 0.393 |
| WGD3 | 0.352 | 0.047** | 0.263 | 0.061** |
| WGD4 | 0.170 | 0.197* | 0.193 | 0.094** |
| WGD5 | 0.156 | 0.149* | 0.006 | 128.480 |
| WGD6 | 0.020 | 21.026 | 0.000 | 6919.539 |
| WGD7 | 0.127 | 0.166* | 0.142 | 0.135* |
| WGD8 | 0.025 | 12.864 | 0.027 | 6.007 |
K < 1/10 or K < 0.1, strong evidence against H0;
K < 1/100.5 or K < 0. 3162 substantial evidence against H0;
K < 1, H1 supported, not worth more than a bare mention; K > 1, H0 supported.
The Azolla WGD
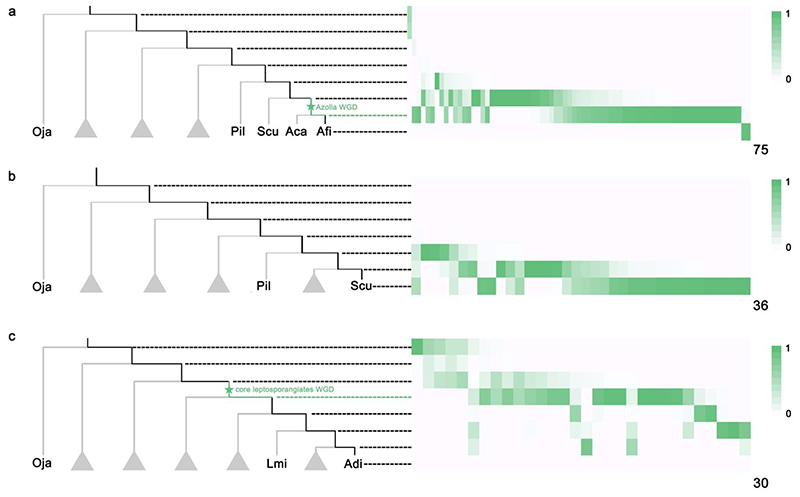
Both the relaxed and critical branch-specific models strongly support a WGD in the lineage leading to Azolla (WGD7) rather than a WGD shared by Azolla and Salvinia (WGD6) (Fig. 3; Table 1). Although the latter seems to have some support from the Ks analyses (Fig. 2d), the peak in Azolla cf. caroliniana is insignificant in the SiZer analysis (Fig. S3), and the peak in Salvinia cucullata may be artificial due to substitution saturation for its highest substitution rate among the analyzed species in Salviniales (Fig. 2b). Given the availability of Azolla filiculoides and Salvinia cucullata genomes (Li et al., 2018), we studied intra-genomic collinearity in each species and identified anchor pairs. Further, we examined the Ks distributions (Fig. S8) and the Whale reconciliation results for anchor pairs (Fig. 4). Except for some anchor pairs reconciled with high posterior probability to the species-specific branch, anchor pairs from the Azolla filiculoides genome tend to support the WGD specific to the Azolla genus (WGD7) rather than a WGD shared by Azolla and Salvinia (WGD6) (Fig. 4a). Also, the reconciliation results for anchor pairs from Salvinia cucullata only lend little support for a WGD shared by Azolla and Salvinia (Fig. 4b). In addition, a further inter-genomic collinearity comparison shows that the syntenic ratio of Azolla filiculoides : Salvinia cucullata : Adiantum capillus-veneris is 2 : 1 : 1, which again confirms our conclusion of one round of WGD experienced by Azolla, while no evidence for a WGD on the branch leading to Adiantum and Salvinia (Fig. S12).
Fig. 4. Gene tree - species tree reconciliation analyses for anchor pairs identified in the three fern genomes, Azolla filiculoides, Salvinia cucullata, and Adiantum capillus-veneris.
On the phylogenetic trees, branches highlighted in black are the ones to which an anchor pair in the genomes of (a) Azolla filiculoides, (b) Salvinia cucullata, and (c) Adiantum capillus-veneris can be reconciled in the gene tree – species tree reconciliation analyses. Note that the reconciliation result of a pair of paralogs in Whale is not a duplication event on a specific branch but a posterior distribution over the possible branches in the species phylogeny on which the duplicate may reconcile to. On the right, the total number of columns in each heatmap, denoted at the right bottom corner, is the number of anchor pairs in analyzed. The squares in white to green in each column show the posterior probability that an anchor pair is reconciled as a duplication event to the respective branch. The color code ranges from white (posterior probability equal to zero) to green (posterior probability equal to one).
The WGD shared by core leptosporangiates
With respect to WGD2 and WGD3, our Whale results support the WGD shared by core leptosporangiates (WGD3) but reject a WGD before the divergence between core leptosporangiates and Schizaeles (WGD2) (Fig. 3; Table 1). In the critical branch-specific model, the for WGD2 is over 0.05 and larger than that in the relaxed branch-specific model. Also, although the Bayes Factor of WGD2 slightly favors q over zero, it cannot provide strong evidence for the occurrence of a WGD. By further examining the anchor pairs in the A. capillus-veneris genome (Fig. S8), we found that the Whale reconciliation results for the anchor pairs also only support a WGD shared by the core leptosporangiates (Fig. 4c).
One WGD in Cyatheales and one WGD in Lindsaea
The support of a WGD shared by Cyatheales is not as decisive as the one in the lineage leading to Lindsaea. The latter is supported by both DL+WGD models, as well as by the Ks analysis of Lindsaea microphylla. The former is, however, only supported by the Ks peak in Cyathea spinulosa, but not in the other two Cyathealean species. The critical branch-specific model has a low estimate for ≈ 0, compared with = 0.16 in the relaxed branch-specific model. In the relaxed model, the duplication rate is low ( = 0.047), but the loss rate is high (= 0.551) on the branch leading to Cyatheales, so in the critical branch-specific model, the duplication rate is higher, whereas the loss rate is lower compared to the two rates estimated by the relaxed branch-specific model, respectively ( = = 0.313) (Fig. S6). Therefore, on the branch with WGD5, the assumption of equal DL rates in the critical branch-specific model appears to be strongly violated, although the rate differences in the branch-specific model appear to be unrealistic when interpreted as a model of gene family evolution. It seems prudent to conclude that support for WGD5 is not robust to model violations, and to abstain from further judgment based on these phylotranscriptomic analyses.
Discussion
Accurate identification of WGDs in a phylogenetic context is the first and vital step to studying genome and chromosome evolution and the consequences of ancient polyploidy during evolution. Lacking high-quality genome assemblies, identifying WGDs in seed-free vascular plants has been primarily based on paranome Ks-age distributions and gene tree – species tree reconciliations using transcriptome data. By revisiting both genomic and transcriptome data for leptosporangiate ferns, especially core leptosporangiates, we show that various synonymous substitution rates are present among the lineages of core leptosporangiates, suggesting that direct comparisons between WGD Ks peaks and speciation events under the assumption that lineages have similar substitution rates can be misleading. Therefore, considering various substitution rates across lineages is essential in correctly interpreting the identified Ks peaks.
In our analyses of the three species in Cyatheales, for instance, the Ks peak identified in Cyathea spinulosa supports a WGD shared by the three species, while the Ks peak identified in Thyrsopteris elegans supports a WGD shared by core leptosporangiates. In contrast, the 1KP initiative (2019), Huang et al. (2020), and Huang et al. (2022) all support a WGD in the lineage leading to Cyathea (‘CYATα’ and ‘11’ in Fig. 1) and a shared WGD for Cyatheales (‘CYATβ’ and ‘9’ in Fig. 1). In addition, according to Huang et al. (2020), there is another WGD in the lineage leading to Plagiogyria (‘10’ in Fig. 1). However, the Ks peaks identified in our analyses neither support a WGD in Cyathea (‘CYATα’ and ‘11’ in Fig. 1) or in Plagiogyria (‘10’). Evidently, the three species in Cyatheales have the lowest substitution rates among the core leptosporangiates (Fig. 2a). Therefore, the three studies above might have misinterpreted the peak in Cyathea spinulosa at Ks ≈ 0.3 as evidence for a recent WGD event in the lineage leading to Cyathea, while this peak is actually the result of a more ancient WGD with small Ks values due to the comparatively low substitution rates. Correspondingly, although Huang et al. (2022) specified consistent peaks (Ks > 1) in several Cyathealean species as the evidence for the Cyathealean WGD, those peaks are all in support of the WGD shared by core leptosporangiates.
Similarly, considering various substitution rates in Salviniales and Polypodiales also show placements of WGDs different from the 1KP initiative (2019) and Huang et al. (2020). In Salviniales, Huang et al. (2020) found no evidence for WGD, but our Ks analyses, along with the 1KP initiative (2019), and Li et al. (2018), identified one WGD in the lineage leading to Azolla. Within Polypodiales, both the 1KP initiative (2019) and Huang et al. (2020) identified two WGDs. One is placed in the lineage of Lindsaea and is supported by both studies (‘LINDα’ and ‘12’ in Fig. 1), as well as by our Ks analyses (Fig. 2d). Further, the 1KP initiative (2019) suggests that the other WGD (‘PTERα’ in Fig. 1) is shared by Polypodium, Blechnum, and Adiantum, whereas Huang et al. (2020) suggests that the WGD (‘3’ in Fig. 1) is only shared by Polypodium and Blechnum. However, the synonymous substitution rates in Blechnum spicant and Polypodium glycyrrhiza are lower than that in Lindsaea microphylla (Fig. 2c), suggesting that the Ks peaks identified in the two species may signal a more ancient WGD shared by core leptospongiates instead of WGD ‘PTERα’ or WGD ‘3’. Note that in each of the three orders of core leptosporangiates, there is at least one species that lends support for an ancient WGD shared by all the core leptosporangiates (or even before the divergence between Schizaeales and core leptosporangiates) as identified by the 1KP initiative (2019) (‘CYATγ’ in Fig. 1). In contrast, Huang et al. (2020) have indicated a WGD shared by Polypodiales and Cyatheales (‘2’ in Fig. 1), which has no support from the 1KP initiative (2019), nor from our results.
Although analyses of Ks-age distributions and considering different substitution rates across lineages could already reject some of the WGDs proposed in earlier studies (Fig. 2d), some Ks peaks from different species fall in competing branches adjacent to each other in the species phylogeny and remain therefore ambiguous. Two such cases are related to WGD2 and WGD3, and WGD6 and WGD7 (Fig. 3). As a signal to identify WGDs, the accuracy of estimating peak values in Ks-age distributions can be affected by the WGD age and substitution rates. Indeed, taking a species with two WGDs as an example, the Ks peak for the recent WGD is often clear, but the Ks peak from the ancient WGD tends to be hidden in a flattened distribution and sensitive to distribution curve fluctuations due to different rates between paralogs or gene loss (or missing). Moreover, for species with high substitution rates, identifying peaks representing an ancient WGD at large Ks value may be confounded by substitution saturation to different extents (Vanneste et al., 2013), leading to less accurate estimates of Ks peak values. Also, both our rate correction approaches keep corrected rates as constants, while substitution rates likely have changed over time during evolution, leading to potentially inaccurate comparisons between Ks peaks and (especially ancient) speciation events. Therefore, solely relying on Ks-age distributions is sometimes problematic, especially in species with high substitution rates.
Gene tree – species tree reconciliation is a complementary approach that, in some cases, can provide support for some very ancient WGDs (Jiao et al., 2011; Zwaenepoel & Van de Peer, 2019). Here, we adopted a statistical gene tree – species tree reconciliation method that considers gene tree uncertainties and other pitfalls known in LCA reconciliation. Unlike the DL+WGD model with an ML scheme, in which a series of likelihood ratio tests are performed by removing only one WGD at a time to test its likelihood of occurrence (Tiley et al., 2016), Whale adopts a Bayesian scheme to formally test the eight hypothetic WGDs, obtained by the Ks analyses (Fig. 3). For the four hypothetical WGDs on adjacent branches that could not be fully resolved by Ks-age distributions, the Whale analyses with both DL+WGD models raised our confidence in WGD7 over WGD6, and WGD3 over WGD2. Also, the support of a WGD in the lineage leading to Lindsaea (WGD4) is clear. Nevertheless, the WGD shared by Cyatheales (WGD5) shows the necessity to assess the performance of the critical and relaxed branch-specific DL+WGD models, suggesting the need for more realistic models for genome evolutionary processes.
Here, we focused on the core leptosporangiates, because both our Ks and reconciliation analyses required suitable outgroups. Beyond the core leptosporangiates, the Ks peak identified in Anemia tomentosa indicates a shared WGD with Lygodium japonicum. However, the Ks peak in Lygodium japonicum goes against a shared WGD with Anemia tomentosa but suggests a more ancient WGD, which is also supported by the Ks peaks in Dipteris conjugata and Sticherus lobatus (Fig. 2d). In general, our results seem to largely agree with the 1KP initiative (2019), which identified a WGD in Anemia tomentosa (‘LYGOα’ in Fig. 1), but no WGDs in Dipteris and Sticherus, in contrast to those in Huang et al. (2020) (‘7’ and ‘8’ in Fig. 1). For the more ancient WGDs identified by both previous studies (‘OSMNβ’, ‘HYMEα’, and ‘1’ in Fig. 1), our analyses could not resolve whether a WGD occurred before the divergence of leptosporangiates (‘OSMNβ’ in Fig. 1) and/or a WGD occurred after Osmunda javanica diverged from the rest of leptosporangiates (‘HYMEα’ and ‘1’ in Fig. 1), because Osmunda javanica is an outgroup in the phylogenetic tree (Fig. 2d). Without extra information we cannot determine when Osmunda javanica diverged from other leptosporangiates. This is also why, although there is a Ks peak in the distribution for Osmunda javanica (Fig. S3), we were uncertain about assuming a WGD either shared with other leptosporangiates or a species-specific WGD (‘OSMNα’ and ‘14’ in Fig. 1). Likewise, our Whale analyses show no support for WGD1 and WGD8 (Fig. 3), as they were placed in the outgroups of core leptosporangiates, so the species sampling may be less suitable to resolve these WGDs. For example, WGD1 may be the result of two WGDs that have occurred on two consecutive branches, if we accept the results from the 1KP initiative (2019). Without species that can further break down the branch in the species phylogeny where WGD1 is located, it is difficult to neatly solve the problem with either the Ks-age or the reconciliation approaches (Zwaenepoel & Van de Peer, 2019). Although we could add extra species to determine the root or further break down long branches, this may introduce another species with a WGD event that cannot be resolved with certainty, so here we decided to only focus on the analyses in the core leptosporangiates.
In conclusion, neglecting differences in substitution rates and performing LCA reconciliations could lead to both false positives and false negatives in calling WGDs. Therefore, we underscore the importance of careful analysis, including the consideration of differences in substitution rates and appreciation of gene tree – species tree reconciliation uncertainties, prompting that failure to do so is likely to lead to unreliable or incorrect conclusions. In addition, we highlight the importance of developing better and more robust statistical models for genome evolutionary processes if we are to reliably characterize the evolutionary history of species at the genomic level.
Supplementary Material
Acknowledgements
Y. V.d.P. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (No. 833522) and from Ghent University (Methusalem funding, BOF.MET.2021.0005.01). H.C. acknowledges funding from the Research Foundation – Flanders (FWO) (No. 3G032219). A.Z. acknowledges the PhD Fellowship of FWO.
Footnotes
Author contributions
Z. L. and Y.V.d.P. conceived and managed the project. H.C., Z.L., and A.Z. conducted analyses. Z.L., H.C., Y.V.d.P., Y.F., and S.H. wrote the manuscript. All authors read and approved the manuscript.
Competing interests
None declared.
Data availability
The data that support the findings of this study are openly available as summarized in Table S1.
References
- 1KP initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574(7780):679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. Multiple Paleopolyploidizations during the Evolution of the Compositae Reveal Parallel Patterns of Duplicate Gene Retention after Millions of Years. Molecular Biology and Evolution. 2008;25(11):2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS. Evolutionary Genomic Analyses of Ferns Reveal that High Chromosome Numbers are a Product of High Retention and Fewer Rounds of Polyploidy Relative to Angiosperms. American Fern Journal. 2009;99(2):136–141. [Google Scholar]
- Barker MS, Wolf PG. Unfurling Fern Biology in the Genomics Age. BioScience. 2010;60(3):177–185. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri P, Marron JS. SiZer for Exploration of Structures in Curves. Journal of the American Statistical Association. 1999;94(447):807–823. [Google Scholar]
- Chen Y-C, Li Z, Zhao Y-X, Gao M, Wang J-Y, Liu K-W, Wang X, Wu L-W, Jiao Y-L, Xu Z-L, et al. The Litsea genome and the evolution of the laurel family. Nature Communications. 2020;11(1):1675. doi: 10.1038/s41467-020-15493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Hidalgo O, Pellicer J, Liu H, Marquardt J, Robert Y, Christenhusz M, Zhang S, Gibby M, Leitch IJ, et al. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytologist. 2016;210(3):1072–1082. doi: 10.1111/nph.13833. [DOI] [PubMed] [Google Scholar]
- Clark JW, Puttick MN, Donoghue PCJ. Origin of horsetails and the role of whole-genome duplication in plant macroevolution. Proceedings of the Royal Society B: Biological Sciences. 2019;286(1914):20191662. doi: 10.1098/rspb.2019.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16(6):738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Qin X, Liao Q, Du R, Luo X, Zhou Q, Li Z, Chen H, Jin W, Yuan Y, et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nature Plants. 2022 doi: 10.1038/s41477-022-01222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Soltis DE, Soltis PS, Ashman T-L, Van de Peer Y. Polyploidy: A biological force from cells to ecosystems. Trends in Cell Biology. 2020;30(9):688–694. doi: 10.1016/j.tcb.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Liu H, Meza-Torres EI, Morero RE, Vega AJ, Liang Z, Ebihara A, Leitch IJ, Schneider H. Evolution of genome space occupation in ferns: linking genome diversity and species richness. Annals of Botany. 2021:mcab094. doi: 10.1093/aob/mcab094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biology. 2007;8(7):R141. doi: 10.1186/gb-2007-8-7-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haufler CH, Soltis DE. Genetic evidence suggests that homosporous ferns with high chromosome numbers are diploid. Proceedings of the National Academy of Sciences. 1986;83(12):4389–4393. doi: 10.1073/pnas.83.12.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, Qi X, Chen D, Qi J, Ma H. Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. Journal of Integrative Plant Biology. 2020;62(4):433–455. doi: 10.1111/jipb.12877. [DOI] [PubMed] [Google Scholar]
- Huang X, Wang W, Gong T, Wickell D, Kuo L-Y, Zhang X, Wen J, Kim H, Lu F, Zhao H, et al. The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nature Plants. 2022;8(5):500–512. doi: 10.1038/s41477-022-01146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Khandelwal S. Chromosome evolution in the genus Ophioglossum L. Botanical Journal of the Linnean Society. 1990;102(3):205–217. [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, Zdobnov EM. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Research. 2018;47(D1):D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Molecular Biology and Evolution. 2017;34(7):1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Landis JB, Soltis DE, Li Z, Marx HE, Barker MS, Tank DC, Soltis PS. Impact of whole-genome duplication events on diversification rates in angiosperms. American Journal of Botany. 2018;105(3):348–363. doi: 10.1002/ajb2.1060. [DOI] [PubMed] [Google Scholar]
- Li F-W, Brouwer P, Carretero-Paulet L, Cheng S, de Vries J, Delaux P-M, Eily A, Koppers N, Kuo L-Y, Li Z, et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature Plants. 2018;4(7):460–472. doi: 10.1038/s41477-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, Scascitelli M, Graham SW, Rieseberg LH, Barker MS. Early genome duplications in conifers and other seed plants. Science Advances. 2015;1(10):e1501084. doi: 10.1126/sciadv.1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A. In: Multiple Sequence Alignment Methods. Russell DJ, editor. Totowa, NJ: Humana Press; 2014. Phylogeny-aware alignment with PRANK; pp. 155–170. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. Journal of Structural and Functional Genomics. 2003;3(1):35–44. [PubMed] [Google Scholar]
- Marchant DB, Sessa EB, Wolf PG, Heo K, Barbazuk WB, Soltis PS, Soltis DE. The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly. Scientific Reports. 2019;9(1):18181. doi: 10.1038/s41598-019-53968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain MR, Tang H, McNeal JR, Ayyampalayam S, Davis JI, dePamphilis CW, Givnish TJ, Pires JC, Stevenson DW, Leebens-Mack JH. A Phylogenomic Assessment of Ancient Polyploidy and Genome Evolution across the Poales. Genome Biology and Evolution. 2016;8(4):1150–1164. doi: 10.1093/gbe/evw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang M-L, Chen J, Biggers E, et al. The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics. 2015;47(12):1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ. The timescale of early land plant evolution. Proceedings of the National Academy of Sciences. 2018;115(10):E2274–E2283. doi: 10.1073/pnas.1719588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PPG I. A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution. 2016;54(6):563–603. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, Vandepoele K. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Research. 2011;40(2):e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. American Journal of Botany. 2004;91(10):1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rabier C-E, Ta T, Ané C. Detecting and Locating Whole Genome Duplications on a Phylogeny: A Probabilistic Approach. Molecular Biology and Evolution. 2013;31(3):750–762. doi: 10.1093/molbev/mst263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen CE. The infinite Gaussian mixture model; Proceedings of the 12th International Conference on Neural Information Processing Systems; Denver, CO. 1999. pp. 554–560. [Google Scholar]
- Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, Ma H, Qi J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Molecular Plant. 2018;11(3):414–428. doi: 10.1016/j.molp.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. The R journal. 2016;8(1):289–317. [PMC free article] [PubMed] [Google Scholar]
- Sensalari C, Maere S, Lohaus R. ksrates: positioning whole-genome duplications relative to speciation events in Ks distributions. Bioinformatics. 2021;38(2):530–532. doi: 10.1093/bioinformatics/btab602. [DOI] [PubMed] [Google Scholar]
- Shen H, Jin D, Shu J-P, Zhou X-L, Lei M, Wei R, Shang H, Wei H-J, Zhang R, Liu L, et al. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience. 2017;7(2) doi: 10.1093/gigascience/gix116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLOS ONE. 2016;11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG. A classification for extant ferns. TAXON. 2006;55(3):705–731. [Google Scholar]
- Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology. 2016;30:159–165. doi: 10.1016/j.pbi.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Stull GW, Qu X-J, Parins-Fukuchi C, Yang Y-Y, Yang J-B, Yang Z-Y, Hu Y, Ma H, Soltis PS, Soltis DE, et al. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nature Plants. 2021;7(8):1015–1025. doi: 10.1038/s41477-021-00964-4. [DOI] [PubMed] [Google Scholar]
- Szöllősi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. Efficient Exploration of the Space of Reconciled Gene Trees. Systematic Biology. 2013;62(6):901–912. doi: 10.1093/sysbio/syt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szövényi P, Gunadi A, Li F-W. Charting the genomic landscape of seed-free plants. Nature Plants. 2021;7(5):554–565. doi: 10.1038/s41477-021-00888-z. [DOI] [PubMed] [Google Scholar]
- Tiley GP, Ané C, Burleigh JG. Evaluating and Characterizing Ancient Whole-Genome Duplications in Plants with Gene Count Data. Genome Biology and Evolution. 2016;8(4):1023–1037. doi: 10.1093/gbe/evw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley GP, Barker MS, Burleigh JG. Assessing the Performance of Ks Plots for Detecting Ancient Whole Genome Duplications. Genome Biology and Evolution. 2018;10(11):2882–2898. doi: 10.1093/gbe/evy200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Ashman T-L, Soltis PS, Soltis DE. Polyploidy: an evolutionary and ecological force in stressful times. The Plant Cell. 2020;33(1):11–26. doi: 10.1093/plcell/koaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nature Reviews Genetics. 2017;18(7):411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Van Dongen SM. Graph clustering by flow simulation. PhD thesis, University of Utrecht; Netherlands: 2000. https://dspace.library.uu.nl/handle/1874/848 . [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Research. 2014;24(8):1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E. Horsetails Are Ancient Polyploids: Evidence from Equisetum giganteum. The Plant Cell. 2015;27(6):1567–1578. doi: 10.1105/tpc.15.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Van de Peer Y, Maere S. Inference of Genome Duplications from Age Distributions Revisited. Molecular Biology and Evolution. 2013;30(1):177–190. doi: 10.1093/molbev/mss214. [DOI] [PubMed] [Google Scholar]
- Vekemans D, Proost S, Vanneste K, Coenen H, Viaene T, Ruelens P, Maere S, Van de Peer Y, Geuten K. Gamma Paleohexaploidy in the Stem Lineage of Core Eudicots: Significance for MADS-Box Gene and Species Diversification. Molecular Biology and Evolution. 2012;29(12):3793–3806. doi: 10.1093/molbev/mss183. [DOI] [PubMed] [Google Scholar]
- Wang F-G, Wang A-H, Bai C-K, Jin D-M, Nie L-Y, Harris AJ, Che L, Wang J-J, Li S-Y, Xu L, et al. Genome size evolution of the extant lycophytes and ferns. Plant Diversity. 2022;44(2):141–152. doi: 10.1016/j.pld.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences. 2009;106(33):13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-C, Rasmussen MD, Bansal MS, Kellis M. TreeFix: Statistically Informed Gene Tree Error Correction Using Species Trees. Systematic Biology. 2012;62(1):110–120. doi: 10.1093/sysbio/sys076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang G-Q, Liu K-W, Li Z, Lohaus R, Hsiao Y-Y, Niu S-C, Wang J-Y, Lin Y-C, Xu Q, Chen L-J, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549(7672):379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Fong R, Collins LJ, McLenachan PA, Penny D. Two New Fern Chloroplasts and Decelerated Evolution Linked to the Long Generation Time in Tree Ferns. Genome Biology and Evolution. 2014;6(5):1166–1173. doi: 10.1093/gbe/evu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17(9):821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Li Z, Lohaus R, Van de Peer Y. Finding Evidence for Whole Genome Duplications: A Reappraisal. Molecular Plant. 2019;12(2):133–136. doi: 10.1016/j.molp.2018.12.019. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. wgd—simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2018;35(12):2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. Inference of Ancient Whole-Genome Duplications and the Evolution of Gene Duplication and Loss Rates. Molecular Biology and Evolution. 2019;36(7):1384–1404. doi: 10.1093/molbev/msz088. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. A two-type branching process model of gene family evolution. bioRxiv. 2021 doi: 10.1101/2021.03.18.435925. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available as summarized in Table S1.