Abstract
Prime editing is a highly versatile CRISPR-based genome editing technology that works without DNA double-strand break formation. Despite rapid technological advances, in vivo application for the treatment of genetic diseases remains challenging. Here, we developed a size-reduced SpCas9 prime editor (PE) lacking the RNaseH domain (PE2ΔRnH) and an intein-split construct (PE2 p.1153) for adeno-associated virus (AAV)-mediated delivery into the liver. Editing efficiencies reached 15% at the Dnmt1 locus, and were further elevated to 58% by delivering unsplit PE2ΔRnH via human adenoviral vector 5 (AdV). To provide proof-of-concept for correcting a genetic liver disease, we next employed the AdV approach for repairing the disease-causing Pahenu2 mutation in a mouse model of phenylketonuria (PKU) via prime editing. Average correction efficiencies of 11.1% (up to 17.4%) in neonates led to therapeutic reduction of blood phenylalanine (L-Phe), without inducing detectable off-target mutations or prolonged liver inflammation. Although the current in vivo prime editing approach for PKU has limitations for clinical application due to the requirement of high vector doses (7×1014 vg/kg) and the induction of immune responses to the vector and the PE, further development of the technology may lead to curative therapies for PKU and other genetic liver diseases.
Introduction
More than 75,000 disease-associated genetic variants have been identified to date. The majority of them are caused by small insertion/deletion mutations or base substitutions (1). Programmable CRISPR-Cas9 nucleases have previously been applied to correct pathogenic alleles in vivo (2, 3). However, these systems rely on DNA double strand (dsDNA) break formation and homology-directed repair (HDR) to install precise edits (4–6), making them inefficient and error-prone in most somatic tissues (7, 8). Base editors (BEs) are CRISPR-based genome editing tools where nuclease-impaired Cas9 directs a deaminase to the locus of interest in order to install single nucleotide conversions. They enable precise genome editing independent of HDR (9, 10), and also operate with high efficiency and accuracy in somatic tissues in vivo (11–17). However, BEs can only install transition substitutions (A-T to G-C or C-T to T-A conversions), and are therefore not applicable to a large proportion of known pathogenic mutations.
Prime editors (PEs) are more recently developed genome editing tools that consist of an RNA-programmable SpCas9 nickase (H840A) fused to an engineered reverse transcriptase (RT). Additional to a guide-scaffold sequence, the prime editing guide RNA (pegRNA) contains a primer binding site (PBS) and a template sequence, which is reverse transcribed into DNA and copied into the targeted locus. Because PEs directly write new genetic information, they are extremely versatile, and allow the installation of all possible nucleotide conversions as well as short insertions and deletions up to 80 bp (18). To improve editing efficiency of PEs, mutations that enhance the activity of the RT have been introduced (PE2). Moreover, a second sgRNA can be designed to nick the non-edited DNA strand either in parallel (PE3) or after installing the edit (PE3b), further augmenting DNA repair of the edited DNA strand (18). Over the last two years prime editing has been applied in numerous cell types in vitro in cell lines and primary cells (18–21), and in vivo in rice, fruit flies and mice (22–27). Here, we develop in vivo prime editing approaches for the murine liver and describe the correction of a phenylketonuria (PKU) mouse model with therapeutic reduction of blood phenylalanine (L-Phe).
Results
Establishment of the size-reduced PE2ΔRnH variant
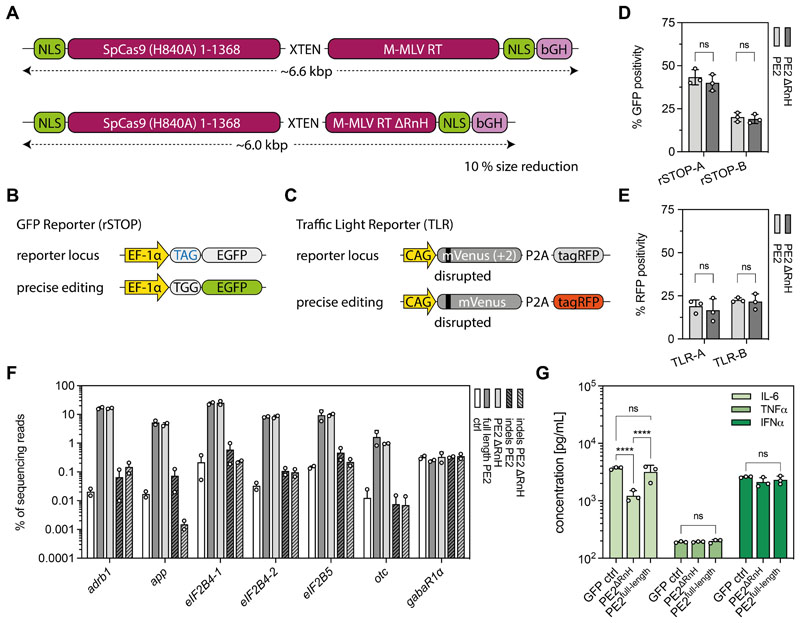
In vivo prime editing holds great potential for therapeutic applications, however the large size of PEs (~6.6 kb excluding non-coding regulatory sequences and the pegRNA; Fig. 1A) poses a challenge for in vivo delivery via several commonly used viral and non-viral vectors. Because previous studies have indicated that the polymerase and RNaseH domains of the M-MLV RT can function independently of each other (28, 29), we speculated that the RNaseH domain (~0.6 kb) could be negligible in the context of prime editing. Therefore, we compared editing efficiencies of PE2ΔRnH (RTΔ486-677) and PE2 in two different HEK293T fluorescent reporter cell lines: the rSTOP reporter, in which GFP expression is abolished by a premature TAG stop codon (Fig. 1B), and the traffic light reporter (TLR), where translation of a functional tagRFP is prevented by a 2 bp frameshift (Fig. 1C). Using multiple pegRNAs, we observed comparable editing efficiencies between PE2 and PE2ΔRnH by FACS analysis (Fig. 1D, 1E; fig. S1). To corroborate these results, we further targeted seven pathogenic mutations (transversions and transitions) at different loci, and analyzed them by deep sequencing. We again did not observe differences in editing rates or indel formation rates between PE2 and PE2ΔRnH (Fig. 1F). Because the lack of the RNaseH domain in the RT could increase DNA-RNA heteroduplexes (30), we speculated that PE2ΔRnH might trigger an elevated Toll-like receptor 9 (TLR9) response (31). However, IL-6 concentrations were lower in supernatants of PE2ΔRnH-treated RAW264.7 macrophages compared to full-length PE-treated macrophages and no differences in TNFα or IFNα concentrations were observed (Fig. 1G), suggesting comparable or even lower immunogenicity of size-reduced PE2ΔRnH compared to full length PE2.
Figure 1. Establishment of the size-reduced PEΔRnH variant.
(A) Schematic representation of the full-length PE2 and our size-reduced variant PE2ΔRnH lacking the RNaseH (RnH) domain (A486-677) of the RT. (B) rSTOP reporter: conversion of a TAG stop codon results in GFP expression. (C) TLR reporter: correction of a 2 bp frameshift results in tagRFP expression. (D, E) Performance of PE2 and PE2ΔRnH in rSTOP (D) and TLR (E) reporter cells using 2 different protospacers (labelled as A and B). Editing efficiency was scored by flow cytometry. (F) Comparative analyses of on-target editing efficiency and indel formation of PE2 and PE2ΔRnH at seven genomic sites. pegRNA plasmids were transfected as negative controls. (G) In vitro analysis of IL-6, TNFα, and IFNα production upon expression of PE2 and PE2ΔRnH in supernatants of RAW264.7 macrophages. A plasmid expressing GFP was used as an additional control (two-way ANOVA with Tukey’s multiple comparisons test; ****P<0.0001). Data from all experiments are represented as mean ± s.d. (three independent experiments; D, E, G) or range (two independent experiments; F). PE, prime editor; M-MLV, Moloney murine leukemia virus; RT, reverse transcriptase; NLS, nuclear localization signal; bGH, bovine growth hormone polyadenylation signal; EF-1α, eukaryotic translation elongation factor 1α; rSTOP, remove stop codon; ADRB1, β1-adrenergic receptor; APP, amyloid β-precursor protein; EIF2B, eukaryotic translation initiation factor 2B; OTC, ornithine carbamoyltransferase; GABAR1α, gamma-aminobutyric acid receptor subunit α-1.
Establishment of an intein-split PE variant for dual AAV-mediated delivery
Due to their low immunogenicity and broad range of serotype specificity, AAVs are promising candidates for in vivo delivery of genome editing tools (32). To stay within the limited AAV packaging capacity (~4.7 kb) (33), the coding sequence of Cas9 nucleases or BEs has previously been split onto two separate AAVs using the intein-mediated protein trans-splicing system from cyanobacterium Nostoc punctiforme (Npu) (12–14, 34, 35). To optimize the intein-split approach for PE delivery, we assessed the activity of SpCas9-PE variants, which were split at different sites. Within the region where both generated PE segments would not exceed the packaging limit of an AAV, we identified eight surface-exposed positions with either a Cys, Ser or Thr at the N-terminal position of the C-intein PE moiety (36). When tested on our rSTOP reporter construct, PE2-p.1153 was the most efficient intein-split variant, maintaining 75% of the activity of unsplit PE2 (Fig. S2A, S2B). Further analysis of the intein-split architecture of PE2-p.1153 revealed the importance of flexible glycine-serine ([GGGGS]3)-linkers between the intein domains and PE segments (Fig. S2C, S3A), and showed that removal of the nuclear localization signal (NLS) from the N-intein splice-donor was beneficial for activity (Fig. S3B). Notably, we also evaluated potential split sites for circular permutant SpCas9-PE (Fig. S3C), and for PE variants based on orthogonal Cas9 from Staphylococcus aureus (SaCas), Staphylococcus auricularis (SauriCas), and Campylobacter jejuni (CjCas) (Fig. S4). However, none of the tested variants led to editing rates comparable to SpCas9-PE, prompting us to continue with linker- and NLS-optimized PE2-p.1153 for in vivo experiments.
AAV-mediated prime editing at the Dnmt1 locus in the mouse liver
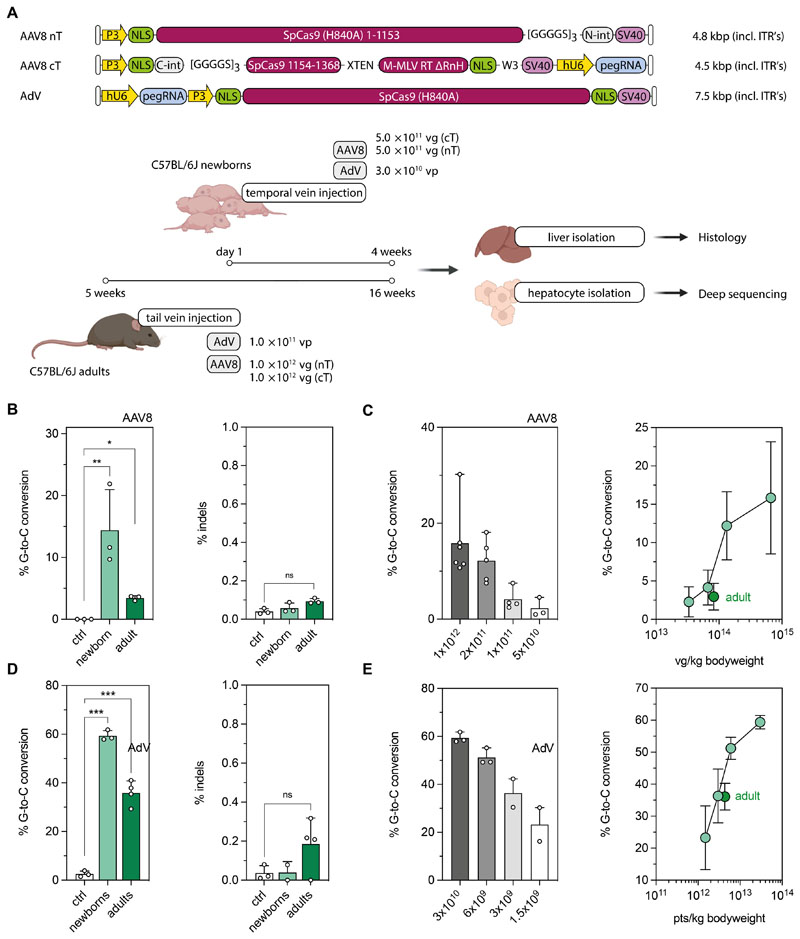
To establish in vivo prime editing approaches in the liver, we employed a pegRNA that has previously been demonstrated to install a G-to-C transversion mutation in the Dnmt1 gene with high efficiency in mouse embryos (25). We first confirmed on-target editing without significant induction of indels at the endogenous Dnmt1 locus when we co-expressed the pegRNA with PE2ΔRnH in vitro in Hepa1-6 cells (Fig. S5A, P>0.05). We next generated AAV constructs in which the intein-split PE2ΔRnH and the pegRNA are expressed under the synthetic liver-specific P3 (37–39) and U6 promoter, respectively, and packaged them into AAV2 serotype 8 (Fig. 2A; hereafter referred to as AAV8). AAV particles were systemically delivered into C57BL/6J pups on postnatal day 1 (P1) via the temporal vein at a dose of 5×1011 vector genomes (vg) per construct and animal (Fig. 2A), and primary hepatocytes were isolated after 4 weeks to analyze editing rates by targeted amplicon sequencing. The intended G-to-C conversion was installed with an efficiency of 14.4 ± 6.6%, with no indels above background detected at the target locus (Fig. 2B). Moreover, when mice were analyzed 12 weeks post injection (p.i.), comparable editing rates were observed (17.2 ± 4.7%; fig. S5C), suggesting that edited hepatocytes were maintained over time. Notably, co-expression of a second sgRNA that binds to the target locus upon introduction of a novel protospacer adjacent motif (PAM) that was generated by the G-to-C edit did not increase editing rates in vitro and in vivo (12.2 ± 1.5% and 15.9 ± 7.3%, fig. S5A, S5B). This was likely due to the fact that unlike for the PE3b approach (18), where the unedited strand is directly nicked after the initial edit is installed, introduction of a novel PAM site requires copying of the edit to the opposite DNA strand before the Cas9 complex can bind.
Figure 2. AAV8- and AdV-mediated prime editing at the Dnmt1 locus in the mouse liver.
(A) Schematic outline of the experimental setup with AAV8- or AdV-mediated prime editing in newborn and adult mice. Constructs used for in vivo prime editing at the Dnmt1 locus in the mouse liver are not depicted to scale. (B) Correction and indel rates in newborn and adult animals after AAV8-mediated delivery (injected dose per neonate and adult: 1 × 1012 and 2× 1012 vector genomes; vg). Untreated mice were used as negative controls. Percentage of sequencing reads with indels around the protospacer region were determined by deep sequencing. (C) Editing rates in neonates relative to AAV doses per animal (left panel) and per kg bodyweight (right panel). The respective values for adult mice were added (dark green) for comparison. (D) Correction and indel rates in newborn and adult mice at 4 weeks after AdV-mediated delivery (injected dose per neonate and adult: 3×1010 and 1×1011 viral particles, vp). (E) Editing rates in neonates relative to AdV doses per animal (left panel) and per kg bodyweight (right panel). The respective values for adult mice were added (dark green) for comparison. Untreated mice were used as negative controls. Data are represented as mean ± s.d. (n=3-6 mice per group) and were analyzed using a two-way ANOVA with Tukey’s multiple comparisons test (ns, not significant, P>0.05; *P<0.05; **P<0.005).
To next assess editing efficiencies in adult animals, we injected 5 week-old C57BL/6J mice via the tail vein at a dose of 1×1012 vg per construct and animal (Fig. 2A). After 12 weeks, hepatocytes were isolated and on-target editing rates were analyzed by deep sequencing. Although we again observed editing of the target base without indel formation (Fig. 2B), editing rates were lower, with the intended G-to-C conversion present in 3.4 ± 0.4% of Dnmt1 alleles (Fig. 2B). One potential explanation for lower editing rates in adult animals is the lack of hepatocyte proliferation compared to neonates. However, when we induced ectopic hepatocyte proliferation at three or six weeks after PE administration via two-third partial hepatectomy (PHx, fig. S6A, S6B), no increase in editing rates was monitored (Fig. S6C). An alternative explanation for the reduced editing rates observed in adult animals is different dosing, as neonates were infused with higher viral doses per kg bodyweight. Indeed, when we injected P1 pups with 5-, 10-, or 20-fold lower concentrations of AAV8 we observed a dose-dependent decline of editing rates, with neonates that received a viral dose comparable to adults displaying similar C-to-G conversion rates (Fig. 2C).
AdV-mediated prime editing at the Dnmt1 locus in the mouse liver
Due to the relatively low reconstitution efficiency of full-length proteins from intein-split moieties (12) (Fig. S7) and the observed dose-dependent increase in editing rates (Fig. 2C), we hypothesized that delivery of unsplit PEs using a vector with higher packaging capacity could further enhance prime editing rates. We therefore next employed the human Adenovirus 5 (AdV) vector for PE delivery. Although AdV has the disadvantage of being more immunogenic than AAVs, raising safety concerns for its application in patients, it can package the unsplit PE2ΔRnH together with a pegRNA on a single vector. Targeting the Dnmt1 locus with unsplit PE2ΔRnH into C57BL/6J adult or neonatal mice indeed led to a substantial increase in editing compared to AAV-treated animals, with editing rates of 58.2 ± 0.4% and 35.9 ± 4.9% in neonates and adults, respectively (Fig. 2D). High on-target editing did not result in an increase of unintended edits at the target locus, as indel mutations were still within the range of untreated control animals (Fig. 2D). Prime editing efficiencies, moreover, were again strongly dependent on the vector dose, and declined when a 5-, 10- or 20-fold lower AdV concentration was injected (Fig. 2E).
Establishment of prime editing strategies to correct the Pahenu2 allele
For a number of genetic liver diseases, including PKU, correction rates below 10% could lead to therapeutic benefits (38). PKU is caused by inactivating mutations in the phenylalanine hydroxylase (Pah) gene, which lead to toxic accumulation of phenylalanine (L-Phe) and its byproducts in the blood. The Pahenu2 mouse model for PKU carries a homozygous point mutation on exon 7 (c.835T>C; p.F263S), resulting in abnormally high blood L-Phe concentrations of ~2000 μmol/L (40).
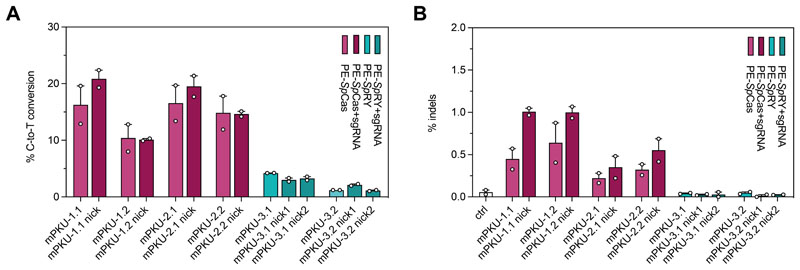
To assess whether the Pahenu2 locus could be corrected by prime editing, we first evaluated on-target editing rates and indel formation using various pegRNAs in vitro in a HEK293T cell line with stably integrated exon 7 of the Pahenu2 allele. We tested pegRNAs for two different SpCas9 protospacers and one SpRY protospacer. pegRNAs harbored a 13-nucleotide long PBS domain combined with 16- (mPKU-*.1) or 19- (mPKU-*.2) nucleotide long RT templates. Deep amplicon sequencing revealed highest editing rates with pegRNAs mPKU-1.1 (PE2: 19.6%) and mPKU-2.1 (PE2: 19.7%) (Fig. 3A), but due to the lower indel rates we chose mPKU-2.1 for in vivo experiments (Fig. 3B).
Figure 3. In vitro correction of the Pahenu2 allele using PE-SpCas and -SpRY variants.
(A, B). In vitro editing rates (A) and indel formation (B) of pegRNAs designed for SpCas-(light and dark purple) and SpRY-PEs (light and dark green) to target the disease-causing Pahenu2 mutation (c.835T>C; p.F263S) on exon 7. pegRNAs for the SpCas-PE were also combined with an additional PE3 nicking sgRNA (pegRNA+sgRNA, dark purple). Two nicking sgRNAs were designed for two PE3b approaches using the SpRY variant (dark green). Experiments were performed in reporter HEK293T cells in which the mutated exon 7 of the Pahenu2 gene was stably integrated. Data are represented as mean ± range of two independent experiments.
Restoring physiological L-Phe concentrations by AdV-mediated in vivo prime editing
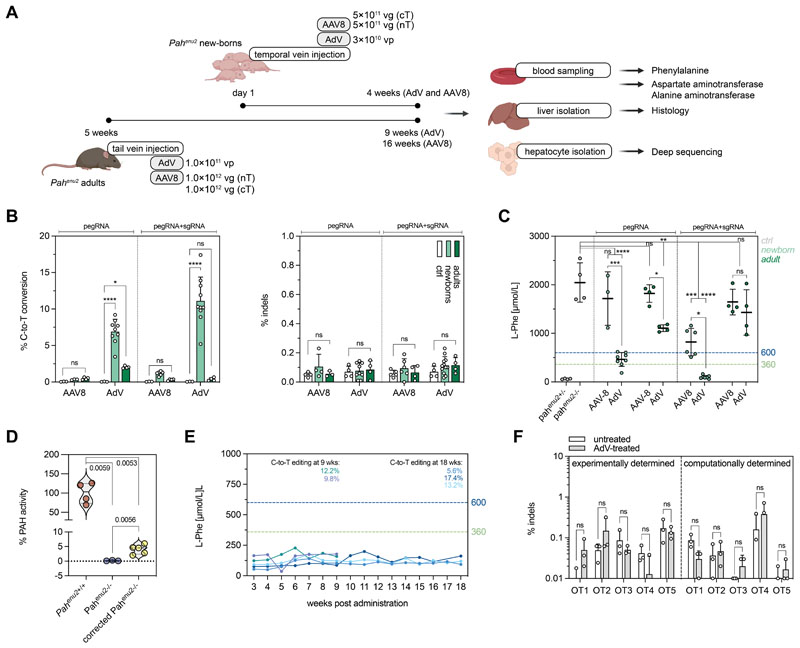
To determine whether we could reach therapeutic correction of the pathogenic Pahenu2 mutation by in vivo prime editing, we first generated AAV vectors co-expressing intein-split PE2ΔRnH and mPKU-2.1 pegRNA. Systemic delivery in a 1:1 ratio into newborn and adult PKU mice at a dose of 5×1011 (newborn) or 1×1012 (adult) vg per construct and animal (Fig. 4A), however, led to consistently lower editing rates as compared to the Dnmt1 locus (<2%, Fig. 4B). Because AAV transduction rates were comparable in Dnmt1 and Pahenu2 experiments (Fig. S8A), and both loci were edited with similar efficiencies in vitro when stably integrated into the genome of HEK293T cells (Fig. S8B), we speculate that the active DNA mismatch repair machinery in hepatocytes disfavored the C-to-T conversion at the Pahenu2 locus over the G-to-C conversion at the Dnmt1 locus (41–43).
Figure 4. Correction of the Pahenu2 allele in vivo in mice using prime editing.
(A) Schematic outline of the experimental setup for AAV8- and AdV-mediated treatment in newborn and adult PKU mice (vg, viral genomes; vp, viral particles). (B) In vivo correction and indel rates in newborn and adult animals after AAV8- or AdV-mediated delivery of SpCas-PEs (injected dose of AAV8 in neonates and adults: 1×1012 and 2×1012 vg; injected dose of AdV in neonates and adults: 3.0×1010 and 1.0×1011 vp). Untreated mice were used as negative controls. Indels are calculated as percentage of sequencing reads with indels at the protospacer region. (C) Blood L-Phe concentrations after in vivo prime editing compared to untreated, heterozygous, and homozygous control animals. L-Phe concentrations below 600 μmol/L (U.S.) and 360 μmol/L (Europe) are considered therapeutically satisfactory (61, 62). (D) Enzymatic activity of PAH at experimental endpoints (4 weeks, AdV). (E) Blood L-Phe concentrations in newborn AdV-treated mice over time. Editing rates of the corresponding mice are color-coded and indicated at 9 and 18 weeks. (F) Deep amplicon sequencing of the top 5 experimentally determined off-target (OT) sites for the protospacer of the pegRNA mPKU-2.1 and of the top 5 computationally predicted off-target sites in untreated and AdV-treated mice (>20’000 reads per site). Data are represented as mean ± s.d. (n=3-8 mice per group) and were analyzed using a two-way ANOVA with Tukey’s multiple comparisons test (ns, not significant, P>0.05; *P<0.05; **P<0.005; ***P<0.0005; ****P<0.0001).
Based on our observations at the Dnmt1 locus we considered that delivery of unsplit PE might also enhance editing at the Pahenu2 locus. Indeed, systemic intravenous (i.v.) delivery of an AdV vector encoding for PE2ΔRnH and mPKU-2.1 into adult (tail vein) and neonatal (temporal vein) PKU mice (Fig. 4A) led to a significant increase in editing (P=0.0265 in adults and P<0.0001 in neonates). We obtained correction rates of 2.0 ± 0.2% in treated adults and 6.9 ±1.7% in treated neonates without observing indel mutations above background (Fig. 4B). Co-expression of a PE3 sgRNA targeting a non-canonical NAG PAM (44) 4 bp upstream of the disease-causing mutation, moreover, led to slightly increased editing rates in vitro (19.6 ± 2.6%; fig. 3A), and in injected newborn animals (11.1 ± 3.3%; fig. 4B).
To next assess therapeutic efficacy of in vivo prime editing on the Pahenu2 locus, we quantified blood L-Phe concentrations in treated PKU mice. Compared to L-Phe concentrations of untreated homozygous animals (2046±404 μmol/L), we observed a significant, though not therapeutic (45) reduction of L-Phe in PKU mice treated as adults with AdV (1106±70 μmol/L; P<0.0038) (Fig. 4C, pegRNA). However, in animals injected with AdV as newborns, L-Phe concentrations were reduced to therapeutically satisfactory levels of 463±143 μmol/L (mPKU-2.1 pegRNA) and 100±34 μmol/L (mPKU-2.1 pegRNAs+PE3 sgRNA) (Fig. 4C), with the latter group of animals displaying 2.0%-6.0% of the wild-type PAH enzyme activity (Fig. 4D). L-Phe concentrations in these animals, moreover, maintained stable below the therapeutic threshold of 360 μmol/L over a period of 18 weeks after injection (Fig. 4E).
In vivo prime editing does not induce detectable off-target mutations
For clinical application of in vivo prime editing, extensive off-target editing or persistent liver damage triggered by an immune response to the PE or the shuttling vector could be a critical limitation. To assess potential off-target editing effects, we first analyzed whether prime editing of the Pahenu2 locus also occurred in other tissues than the liver. However, as expected based on the hepatotropism of the AdV and P3 promoter, deep sequencing of various tissues in AdV-treated mice revealed that editing was mainly limited to the liver (Fig. S9A). Next, we used CHANGE-seq (46) to experimentally assess potential off-target binding sites of the mPKU-2.1 pegRNA (Fig. S9B) and performed deep sequencing of identified sites in the liver of treated animals. In line with previous studies reporting higher specificities of PEs compared to CRISPR-Cas9 nucleases (18, 47), no editing above background was observed in the top 5 experimentally identified off-target sites or in the top 5 computationally predicted off-target sites (Fig 4F; n=3 mice per group). Similarly, treatment with the Dnmt1-targeting AdV did not induce editing at the top 5 experimentally identified off-target binding sites of the Dnmt1 pegRNA (Fig. S9C, S9D).
In vivo prime editing does not induce prolonged liver damage but induces a humoral immune response in adult animals
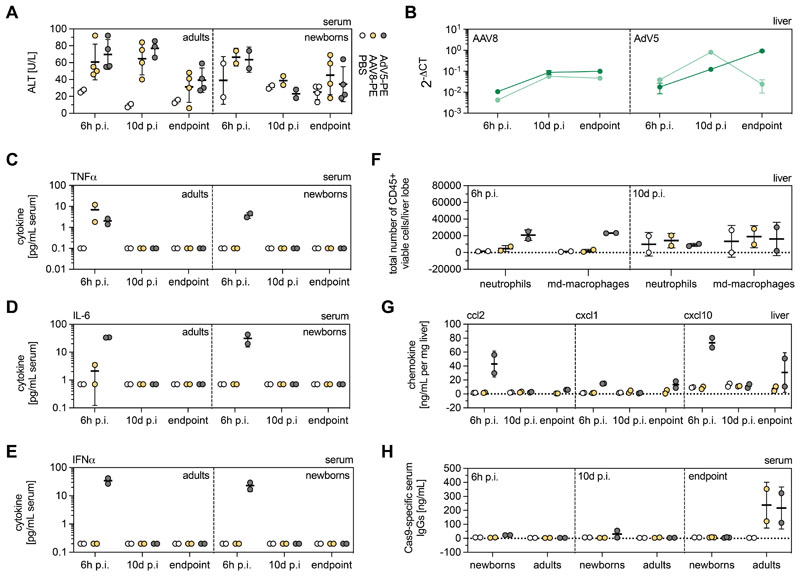
To determine vector- or PE-mediated hepatotoxicity, we analyzed transaminase alanine aminotransferase (ALT) concentrations in PBS-, AAV8- and AdV-treated animals at different time points after vector administration (6h p.i., 10d p.i. and experimental endpoints). Although we observed a slight elevation of ALT levels in all vector-treated animals at 6h post treatment, the concentrations decreased to background at 10d p.i. in newborns and at experimental endpoints in adults (AdV: 4 weeks for newborns and adults; AAV8: 4 weeks for newborns, 8 weeks for adults) (Fig. 5A). Notably, in adult animals treated with AAV8 and AdV, the PE was still highly expressed at the experimental endpoints (Fig. 5B), suggesting that ALT elevation was not triggered by PE expression but by viral vector components. Histological examination of the liver, moreover, did not reveal obvious signs of tissue necrosis in treated animals at any of the analyzed timepoints (Fig. S10), indicating that the transient ALT upregulation was triggered by mild hepatotoxicity.
Figure 5. Innate and adaptive immunity to AAV8 and AdV vectors.
(A) ALT concentrations in PBS-, AAV8- and AdV-treated mice at 6h p.i., 10d p.i., and experimental endpoints. (B) Cas9 transcript abundance in AAV8- and AdV-treated newborns (light green) and adults (dark green) at 6h p.i., 10d p.i., and endpoints. Transcript counts were normalized to the housekeeping gene Rplp0. (C-E) TNFα (C), IL-6 (D), and IFNα (E) concentrations in the serum of PBS-, AAV8- and AdV-treated adults and neonates at 6h, 10d p.i. and experimental endpoints. (F, G) Presence of neutrophils and monocyte-derived (md) macrophages (F) and pro-inflammatory chemokines ccl2, cxcl1, and cxcl10 (G) in the livers of adult mice at 6h p.i. and 10d p.i. (H) Cas9-specific adaptive immune response in control and treated mice 6h, 10d p.i. and endpoints. Data are represented as mean ± s.d (n=2-4 animals per group). Experimental endpoints for AdV, 4 weeks (newborns and adults); for AAV8, 4 weeks (newborns), 8 weeks (adults).
To assess if viral vector-mediated PE delivery induced an innate immune response, we analyzed concentrations of the inflammatory cytokines IL-6, TNFα, and IFNα in the sera of treated mice. As expected, systemic administration of AdV into neonates and adult animals led to a transient increase of all three cytokines at 6h p.i., with concentrations returning to background values at 10d p.i. (Fig. 5C-E). To further analyze whether administration of PE vectors triggered immune cell infiltration in the liver, we performed FACS analysis on liver samples of vector- and control PBS-treated adult mice (Fig. S11). We identified an increase in liver-resident neutrophils and monocyte-derived macrophages in AdV-treated adult mice at 6h p.i., but not at later time points or in AAV8-treated animals (Fig. 5F). Moreover, only liver samples of AdV-treated adult mice at 6h p.i showed elevated concentrations of the pro-inflammatory chemokines ccl2, cxcl1, consistent with increased recruitment of monocytes and neutrophils, respectively, and the interferon-inducible chemokine cxcl10 (Fig. 5G).
Activation of an innate immune response together with the expression of bacterial- and viral- derived proteins (Cas9 and RT) suggests that an adaptive immune response to the PE could have been established during treatment. Indeed, using ELISA we detected Cas9-specific antibodies in adult mice treated with AAV8 or AdV (Fig. 5H). By contrast, no humoral immune response against Cas9 was observed when mice were treated as newborns, in line with previous studies demonstrating that neonatal mice have an undeveloped immune system (48, 49). For clinical application of prime editing, nevertheless, the establishment of adaptive immunity raises concerns, as Cas9-specific cytotoxic T cells could lead to the elimination of PE-expressing cells during states of acute inflammation. Thus, in the future it would be important to establish transient prime editing systems, which could for example be achieved by delivery of PE-encoding RNA into the liver via lipid nanoparticles (LNPs).
Discussion
In vivo prime editing in somatic tissues holds great promise for therapeutic application in patients with genetic diseases. In our study we demonstrate the installation of transition and transversion edits in vivo in the liver of mice. Unlike CRISPR-Cas9 nucleases PEs work independent of dsDNA break formation. This is an important aspect for safety considerations as dsDNA breaks can trigger excessive genetic damage, including translocations, inversions and large deletions (50–52). Another genome editing tool that works without inducing dsDNA breaks is base editing, which has previously been applied to correct disease-causing mutations in vivo at a number of genomic sites (11–14), including the Pahenu2 locus (12, 15). Although cytidine base editing was more efficient than prime editing in correcting the Pahenu2 mutation, substantial amounts of bystander mutations and indel mutations were observed. These data suggest that PEs are not only more versatile than BEs, but also more precise.
In our study, systemic administration of adenoviral vectors encoding for prime editing components into newborn mice resulted in 58% editing at the Dnmt1 locus and 11.1% editing at the Pahenu2 locus. These editing rates are substantially higher than in previously reported in vivo prime editing studies (24, 27), and in theory should be sufficient to treat a variety of genetic liver diseases other than PKU, including urea cycle disorders (53, 54). The presented approach, nevertheless, has limitations in terms of clinical translation. First, reaching therapeutic editing rates required administration of 3×1013 of AdV vg/kg bodyweight, which initiated a potent innate immune response and exceeds the acceptable vector doses for human trials. Second, prolonged PE expression after viral vector mediated delivery and the formation of an adaptive immunity to the PE raises safety concerns, because corrected cells could be rejected later in life, for example if triggered by acute hepatitis. Third, we observed a large variation in in vivo prime editing rates at the two tested loci, despite obtaining similar editing in vitro and administering comparable vector doses. While we speculate that the active DNA mismatch repair machinery in hepatocytes might have favored the G-to-C conversion at the Dnmt1 locus over the C-to-T conversion at the Pah locus (41–43), an in-depth evaluation at multiple loci would be required to better understand and predict in vivo prime editing efficiencies across various genomic loci.
Additional work is therefore needed to translate in vivo prime editing to the clinic, and should focus on enhancing the PE activity to reduce required vector doses and developing transient delivery approaches, such as LNP-mediated RNA delivery. Examples for strategies to increase the efficiency of PEs could be increasing the nuclear import efficiency of PEs through optimizing NLS designs (20, 24), or enhancing the processivity of the RT domain via directed protein evolution (55). Recent efforts achieved improved prime editing efficiencies in vitro in cell lines by enhancing the stability of the pegRNA and by inhibiting the DNA mismatch repair machinery (20, 56), and it will be interesting to assess whether these approaches also increase prime editing rates in vivo.
Materials and Methods
Study design
The aim of this study was to establish in vivo prime editing approaches for preclinical treatment of metabolic liver disease. Optimized intein-split PEs and pegRNA designs for various loci were determined in vitro in reporter cell lines in at least two independent experiments using flow cytometry and targeted amplicon sequencing. To determine sample sizes for animal experiments, a priori power calculations were performed using the R ‘pwr’ package. Treatment groups (AAV or AdV) were randomly assigned without gender bias. For viral vectors targeting the Dnmt1 locus, injected animals were used for targeted amplicon sequencing, RT-qPCR, Western blot, flow cytometry, histological analysis and determination of off-target editing. For viral vectors targeting the Pahenu2 locus, injected adults were used for targeted amplicon sequencing, RT-qPCR, Western blot, histological analysis, determination of blood L-Phe concentrations, ALT values, and off-target editing. For newborns, all animals of a litter were treated, but only homozygous animals were used for analyses. No data was excluded from the study. Blinding was not performed in this study. Biological replicates, animal numbers and group sizes are indicated for each experiment in the respective figure legends. A detailed table containing the primary data and animal genders is reported in data file S1.
Generation of plasmids
To generate pegRNA plasmids, annealed spacer, scaffold, and 3’ extension oligos were cloned into pU6-pegRNA-GG-acceptor by Golden Gate assembly as previously described (18). To generate nicking sgRNA plasmids, annealed and phosphorylated oligos were ligated into BsmBI-digested lentiGuide-Puro backbone. Sequences of all pegRNAs and nicking sgRNAs are listed in Table S1. For the generation of split-intein and orthogonal PEs, inserts were ordered as gBlocks from Integrated DNA Technologies (IDT) and cloned into pCMV-PE2 backbone using HiFi DNA assembly MasterMix (NEB). To generate piggyBac disease reporter plasmids, inserts with homology overhangs for cloning were ordered from IDT and cloned into the pPB-Zeocin backbone using HiFi DNA assembly MasterMix (NEB). To engineer plasmids for virus production, inserts were ordered as gBlocks (IDT) and cloned into AAV backbones using HiFi DNA assembly MasterMix (NEB). All PCR reactions were performed using Q5 High-Fidelity DNA polymerase (New England Biolabs).
Cell culture transfection and genomic DNA preparation
HEK293T (ATCC CRL-3216), Hepa1-6 (ATCC CRL-1830) and RAW264.7 (ATCC TIB-71) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) plus GlutaMax (Thermo Fisher), supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin/streptomycin (Thermo Fisher) at 37°C and 5% CO2. Cells were maintained at confluency below 90% and seeded on 48-well cell culture plates (Greiner). Cells were transfected at 70% confluency using 1.5 μL of Lipofectamine 2000 (Thermo Fisher) with 375 ng PE, 125 ng pegRNA, and 40 ng sgRNA according to the manufacturer’s instructions. When intein-split PEs were transfected, 375 ng of each PE half was used for transfection. Unless otherwise noted, cells were incubated for 3 days and genomic DNA was isolated by direct lysis.
Generation of reporter cells by lentivirus transduction or PiggyBac transposon
For Lentivirus production, HEK293T cells were seeded into T75 flask (Greiner) with Opti-MEM (Thermo Fisher) and transfected at 70% confluency using polyethylenimine (PEI). Briefly, 60 μL PEI (0.1 mg/mL) was mixed with 370 μL Opti-MEM, incubated at room temperature for 5 min, and added to 4.4 μg PAX2, 1.5 μg VSV-G, and 5.9 μg lentiviral vector plasmid (filled to 430 μL Opti-MEM). Following 20 min incubation at room temperature, cells were transfected. The culture medium was changed one day after transfection. After two days, the cell culture supernatant was harvested and lentiviral particles were purified by filtration (0.20 μm, Sarstedt). Fresh HEK293T cells were subsequently transduced with lentiviral particles in a 24-well cell culture plate (Greiner). Two days after transduction, cells were enriched for 7 days using 2.5 μg/mL puromycin.
For generation of disease reporter cell lines with the PiggyBac transposon, 30’000 HEK293T cells were seeded into a 24-well culture plate (Greiner) and transfected at 70% confluency using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions. Briefly, 1.5 μL Lipofectamine was mixed with 23.5 μL Opti-MEM, incubated at room temperature for 10 min, and added to 225 ng transposon plasmid and 25 ng transposon helper plasmid (filled up to 25 μL Opti-MEM). Following 30 min incubation at RT, cells were transfected. Three days after transfection, cells were enriched for 10 days using 150 μg/mL Zeocin.
In vitro assessment of DNA-RNA heteroduplexes
RAW264.7 cells were seeded in a 48-well cell culture plate (Greiner) and transfected at 50% confluency with 750 ng PE2, PE2ΔRnH and 250 ng pegRNA plasmid or with 1000 ng GFP control plasmid using 1.5 μL of Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions. Cells were incubated for 48h before supernatants were collected for analyses of cytokine production. IL-6, TNFα and IFNα in the cell culture supernatant were assessed via ELISA by Cytolab (Regensdorf).
Fluorescence reporter assays and fluorescence-activated cell sorting
Reporter cells were transfected with prime editing tools that are programmed to restore the expression of a fluorescent protein. Cells were incubated for 3 days post-transfection and trypsinized with TrypLE (Gibco). Cells were washed twice with phosphate-buffered saline (PBS) and resuspended in FACS Buffer (PBS supplemented with 2% FBS and 2 mM EDTA). Cell suspensions were filtered through 35 μm nylon mesh cell strainer snap caps (Corning) and kept on ice until analysis. For each sample, 100’000 events were counted on an LSR Fortessa (BD Biosciences) using the FACSDiva software version 8.0.1 (BD Biosciences). Experiments were performed in up to four replicates on different days. Data are reported as mean values ± standard deviation (s.d.). The gating strategy is shown in Fig. S1.
AAV and AdV5 production
All pseudo-typed vectors (AAV2 serotype 8) were produced by the Viral Vector Facility of the Neuroscience Center Zurich. Briefly, AAV vectors were ultracentrifuged and diafiltered. Physical titers (vector genome per mL) were determined using a Qubit 3.0 fluorometer as previously published (57). Identity of the packaged genomes of each AAV vector was confirmed by Sanger sequencing. AdV vectors were produced by ViraQuest, Inc.
Animal studies
Animal experiments were performed in accordance with protocols approved by the Kantonales Veterinäramt Zürich and in compliance with all relevant ethical regulations. Pahenu2 and C57BL/6J mice were housed in a pathogen-free animal facility at the Institute of Pharmacology and Toxicology of the University of Zurich. Mice were kept in a temperature- and humidity-controlled room on a 12-h light-dark cycle. Mice were fed a standard laboratory chow (Kliba Nafag no. 3437 with 18.5% crude protein) and genotyped at weaning. Heterozygous Pahenu2 littermates were used as controls for physiological L-Phe concentrations in the blood (<120 μM). For sampling of blood for L-Phe determination, mice were fasted for 3-4 h and blood was collected from the tail vein. Unless otherwise noted, newborn animals (P1) received 3.0×1010 (AdV, 30 μL total) viral particles (vp) per animal via the temporal vein, or 5×1011 (AAV, 30 μL total) vg per animal and construct via the temporal vein. Adult mice were injected with 1.0×1011 vp (AdV, 150 μL total) per animal or 1×1012 (AAV, 150 μL total) vg per animal and construct via the tail vein.
The selected AdV and AAV doses were based on the maximum injection volume for neonates (30 μL undiluted viral vectors via the temporal vein) and adults (150 μL undiluted viral vectors via the tail vein). The average weight of neonatal and adult mice (5 weeks) was 1.5 g and 20 g, respectively. Newborn mice were euthanized 4 weeks after injection. Adult mice were euthanized 4 weeks (AdV) or 12 weeks (AAV) after injection if not stated otherwise.
Primary hepatocyte isolation
Primary hepatocytes were isolated using a two-step perfusion method. Briefly, pre-perfusion with HANKS’s buffer (HBSS supplemented with 0.5 mM EDTA, 25 mM HEPES) was performed by inserting the cannula through the superior vena cava and cutting the portal vein. Next, livers were perfused at low flow for approximately 10 min with Digestion Buffer (low glucose DMEM supplemented with 1 mM HEPES) containing freshly added Liberase TM (32 μg/mL; Roche). Digestion was stopped using Isolation Buffer (low glucose DMEM supplemented with 10% FBS) and cells were separated from the matrix by gently pushing with a cell scraper. The cell suspension was filtered through a 100 μm filter (Corning) and hepatocytes were purified by two low speed centrifugation steps (50×g for 2 min).
RNA isolation and RT-qPCR
RNA was isolated from shock frozen liver samples using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. RNA was reverse transcribed to cDNA using random primers and GoScript Reverse Transcriptase kit (Promega). RT-qPCR was performed using Firepol qPCR Master Mix (Solis BioDyne) and analyzed by 7900HT Fast Real-Time PCR System (Applied Biosystems). Fold changes were calculated using the delta Ct method. Used primers are listed in Table S4.
Western blot
Proteins were isolated from transfected HEK293T cells or transduced primary hepatocytes of treated and untreated animals. Briefly, cells were lysed in RIPA buffer, supplemented with protease inhibitors (Sigma-Aldrich). Protein amounts were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher). Equal amounts of protein (in vitro samples: 3 μg; in vivo samples: 80 μg) were separated by SDS-PAGE (Thermo Fisher) and transferred to a 0.45 μm nitrocellulose membrane (Amersham). Membranes were incubated with mouse anti-Cas9 (1:1’000; Cat. No. #14697T; Cell Signaling) and rabbit anti-GAPDH (1:10’000; Cat. No. ab181602; abcam). Signals were detected by fluorescence using IRDye-conjugated secondary antibodies (Licor).
Liver immune cell infiltration panels and flow cytometry
Same liver lobes from treated animals were collected 6h and 10d post injection. Single-cell suspensions from the livers were prepared by digestion of the organs with collagenase IV (300U/mL) and DNaseI (200U/mL, Worthington) before dissociation with gentleMACS (Miltenyi) and filtration through a 70μm cell strainer. Leukocytes from the liver were isolated using 40% and 70% Percoll gradient centrifugation (GE Healthcare). Viability staining was performed using the fixable viability dye eFluor780 (Thermo Fisher). Staining with fluorescent antibodies was carried out using fluorescently labelled antibodies summarized in (Table S6). After washing in FACS buffer (2 mM EDTA, 2% FBS), cell populations were analyzed using BD Fortessa and FlowJo software (version 10). The gating strategy is shown in figure S12.
Chemokine and cytokine panels
Each liver piece was weighed before homogenization in 250μL PBS at 30 Hz for 4 min, shock frozen and stored at -80°C. Analysis was performed simultaneously with all samples using LEGENDplex Mouse Proinflammatory Chemokine Panel (13-plex) and LEGENDplex Mouse Inflammation Panel (13-plex) (Cat. No. 740007 and 740446; BioLegend). Serum TNFα, IL-6 and IFNα concentrations were determined via ELISA by Cytolab (Regendsdorf).
Cas9-specific IgG ELISA
Cas9-specific antibodies were determined by an in-house set-up direct ELISA. In short, 10 ng of Cas9 were immobilized on 96-well polystyrene MaxiSorp plates (Thermo Fisher Scientific, cat. no. 439454) diluted in 1×ELISA Coating Buffer (BioRad, cat. no. BUF030A) for 2h at room temperature. After washing in 1 ×ELISA Wash Buffer (Bio-Rad, cat. no. BUF031C), the wells were blocked for 30min in ELISA BSA blocking solution (Bio-Rad, cat. no. BUF032C). For Cas9 detection, mouse-anti-Cas9 mAB (7A9-3A3; clone no. 14697T, Cell Signaling, cat. no. 14697) was used as a positive control and standard curve. Plasma samples were diluted 100-fold for mouse plasma in Tris-buffered saline with Tween (TBS-T) and incubated for 2h at room temperature. Goat-anti-mouse HRP-linked secondary antibodies (SouthernBiotech, cat. no. 1030-05) were used to detected protein-binding antibodies and developed using 1-Step Turbo TMB-ELISA Substrate Solution (Thermo Fisher Scientific, cat. no. 34022) and stopped after 20min with Stop Solution for TMB Substrates (Thermo Fisher Scientific, cat. no. N301). Absorbance was measured at 450nm and background at 540nm; the latter was subtracted for quantification.
Amplification for deep sequencing
Genomic DNA from mouse liver tissues were isolated from whole liver lysate by direct lysis. Locus-specific primers were used to generate targeted amplicons for deep sequencing. First, input genomic DNA was amplified in a 20 μL reaction for 25 cycles using NEBNext High-Fidelity 2× PCR Master Mix (NEB). PCR products were purified using AMPure XP beads (Beckman Coulter) and subsequently amplified for 8 cycles using primers with sequencing adapters. Approximately equal amounts of PCR products from each sample were pooled, gel-purified and quantified using a Qubit 3.0 fluorometer and the dsDNA HS assay kit (Thermo Fisher). Paired-end sequencing of purified libraries was performed on an Illumina Miseq.
HTS data analysis
Sequencing reads were demultiplexed using MiSeq Reporter (Illumina). Amplicon sequences were aligned to their reference sequences using CRISPResso2 (58). Prime editing efficiencies were calculated as percentage of (number of reads containing only the desired edit)/(number of total reads). Indel yields were calculated as percentage of (number of indel-containing reads)/(total reads).
Guide-dependent off-target prediction and analysis using CHANGE-seq
For CHANGE-seq, the protospacer of the pegRNA was first tested for functionality: a 599 bp long construct was PCR amplified from genomic DNA isolated from the tail of a Pahenu2 homozygous animal using GoTaq G2 HotStart Green Master Mix (Promega). Primers are listed in Suppl. Table 2. The library was prepared as previously described (46). Data was processed using v1.1 of the CHANGE-Seq analysis pipeline (https://github.com/tsailabSJ/changeseq) with parameters: “window_size: 3; mapq_threshold: 50; start_threshold: 1; gap_threshold: 3; mismatch_threshold: 15; merged_analysis: True; variant_analysis: True”. The top 5 off-target sites for the protospacer of the pegRNA were selected for targeted amplicon deep sequencing and covered by at least 20’000 reads per site. sgRNA functionality of the DNMT1 protospacer was assessed on PCR-amplified DNA isolated from wildtype animals. The mismatch threshold parameter was reduced to 6.
PAH enzyme activity assay
Whole liver extracts were analyzed using isotope-dilution liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) according to a previously published method (59).
Quantification of phenylalanine in blood
Amino acids were extracted from a 3.2 mm dried blood sample using the Neomass AAAC Plus newborn screening kit (Labsystems Diagnostics). A UHPLC Nexera X2 coupled to an LCMS-8060 Triple Quadrupole mass spectrometer with electrospray ionization (Shimdazu) was used for the quantitative analysis of Phenylalanine. Labsolutions and Neonatal Solutions software (Shimdazu) were used for data acquisition and data analysis.
Partial (70%) hepatectomy
0.1 mg/kg bodyweight of buprenorphine was administered into the mice subcutaneously 30 min before starting the operative procedure. Animals were anesthetized with isoflurane: 5% isoflurane with 1000 mL/min in 100% O2. During the surgical procedure, anesthesia was maintained at 1.5-2.5% isoflurane with 400 mL/min in 100% O2. Correct administration of anesthesia was verified by toe-pinch reflex. The animals were placed on a warming surface for maintenance of body temperature. The incision site was disinfected with antiseptic, povidone-iodine solution (Braunol, Braun). A midline abdominal skin and muscle incision (about 3 cm long) was performed to expose the abdominal cavity. The liver was freed from ligaments. After cholecystectomy (Prolene, 8/0), the left lateral lobe was ligated with a 6/0 silk thread (Fine Science Tools) and resected. Successful ligation was confirmed by observing a color change of the lobe. The median lobe was ligated in two steps (silk 6/0) and resected. For closing the peritoneum and the abdomen, VICRYL 5-0 Suture (Ethicon) was used.
Histology
Livers were fixed using 4% paraformaldehyde (PFA, Sigma-Aldrich), followed by ethanol dehydration and paraffinization. Paraffin blocks were cut into 5 μm thick sections, deparaffinized with xylene, and rehydrated. Sections were HE-stained and examined for histopathological changes.
Immunofluorescence
Antigen retrieval was performed on deparaffinized and rehydrated 5 μm-thick liver sections using HistosPro Microwave histoprocessor. Slides were blocked using PBS supplemented with 2% Normal-donkey serum (Abcam, ab7475) and 0.1% Triton-X for 30 min. Slides were incubated with primary antibody at 4°C overnight using rabbit-anti-CK19 (Abcam, ab52625, dilution 1:500) and mouse-anti-Ki67 (Agilent, M7248, dilution 1:100). Goat-anti-mouse-488 and Goat-anti-rabbit-568 were used as secondary antibodies and sections were counterstained with DAPI. Mounting was performed using Fluorescence Mounting Medium (Agilent, S302380-2). Confocal images were taken with a Zeiss LSM 700. Ki67-positive cells per area were quantified using the cell counter plugin of FIJI (60).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9.0.0. for MacOS. Data are represented as biological replicates and are depicted as mean ± s.d. or range as indicated in the corresponding figure legends. Likewise, sample sizes and the statistical tests used are described in detail in the respective figure legends. The data was tested for normality using the Shapiro-Wilk test as described in detailed in data file S1. Unpaired Student’s t tests and one/two-way analysis of variants (ANOVA) tests were performed followed by the appropriate post-hoc test when more than two groups were compared. For all analyses, P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dario Hüsler for providing macrophage cultures, the Functional Genomics Center Zurich and ZMB for technical support and access to instruments at the University of Zurich and ETH Zürich, and members of the Kopf, Thöny and Schwank labs for discussions. pU6-pegRNA-GG-acceptor (Addgene plasmid no. 132777) and pCMV-PE2 (Addgene plasmid no. 132775) were gifts from David Liu. lentiGuide-Puro and PX404 Campylobacter jejuni Cas9 were gifts from Feng Zhang (Addgene plasmid no. 52963 and 68338). pCMV-VSV-G was a gift from B. Weinberg (Addgene plasmid no. 8454) and psPAX2 was a gift from D. Trono (Addgene plasmid no. 12260). SauriABEmax was a gift from Yongming Wang (Addgene plasmid no. 135968).
Funding
This study was supported by Swiss National Science Foundation (SNSF) grant no. 310030_185293 (to G.S.), SNSF Sinergia grant no. 180257 (to B.T.), Novartis Foundation for Medical-Biological Research no. FN20-0000000203 (to D.B.), SNSF Spark fellowship no. 196287 (to D.B.), PHRT grant no. 528 (to G.S. and B.T.), the URPP Itinerare (G.S.) and the Helmut Horten Foundation (to G.S.).
Footnotes
Author contributions: D.B., T.R., L.V. and G.S. designed the study. D.B., T.R., L.V. and H.M.G.C. performed and analyzed in vivo experiments. D.B., T.R., L.V., M.M., L.S., N.M. and E.I. performed and analyzed in vitro experiments. N.R. performed blood L-Phe and ALT measurements and measured PAH enzyme activity. D.B., T.R. and L.V. prepared figures. D.B., T.R., L.V. and G.S. wrote the manuscript. All authors reviewed the manuscript.
Competing interests: D.B, L.V., and G.S are named on patent applications related to CRISPR-Cas technologies (“Split Prime Editing Enzyme”, EP21172542.9).
Data and materials availability
All data related to this study is present in the paper or supplementary materials. Illumina sequencing data is available at the Gene Expression Omnibus (GEO) database under accession number GSE174757. The authors declare that all other data supporting the findings of this study are available within the paper and its extended data files.
References
- 1.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 3.Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, Hu YJ, Hu JH, Thompson DB, Shu Y, Li Y, et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Sciences. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman JR, Taylor MRG, Boulton SJ. Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–71. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–4. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of AT to GC in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 12.Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, Fingerhut R, Häberle J, Matos J, Robinson MD, Thöny B, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 13.Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao X, Newby GA, et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villiger L, Rothgangl T, Witzigmann D, Oka R, Lin PJC, Qi W, Janjuha S, Berk C, Ringnalda F, Beattie MB, Stoffel M, et al. In vivo cytidine base editing of hepatocytes without detectable off-target mutations in RNA and DNA. Nat Biomed Eng. 2021;5:179–189. doi: 10.1038/s41551-020-00671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, DeNizio JE, Reiss CW, Wang K, Iyer S, Dutta C, Clendaniel V, Amaonye M, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 17.Rothgangl T, Dennis MK, Lin PJC, Oka R, Witzigmann D, Villiger L, Qi W, Hruzova M, Kissling L, Lenggenhager D, Borrelli C, et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat Biotechnol. 2021;39:949–957. doi: 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schene IF, Joore IP, Oka R, Mokry M, van Vugt AHM, van Boxtel R, van der Doef HPJ, van der Laan LJW, Verstegen MMA, van Hasselt PM, Nieuwenhuis EES, et al. Prime editing for functional repair in patient-derived disease models. Nat Commun. 2020;11:5352. doi: 10.1038/s41467-020-19136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen P-F, Chen C, Nelson JW, Newby GA, Sahin M, Osborn MJ, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geurts MH, de Poel E, Pleguezuelos-Manzano C, Oka R, Carrillo L, Andersson-Rolf A, Boretto M, Brunsveld JE, van Boxtel R, Beekman JM, Clevers H. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci Alliance. 2021;4:e202000940. doi: 10.26508/lsa.202000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Li X, He S, Huang S, Li C, Chen Y, Liu Z, Huang X, Wang X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020;6:27. doi: 10.1038/s41421-020-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch JA, Birchak G, Perrimon N. Precise genome engineering in Drosophila using prime editing. Proc Natl Acad Sci U S A. 2020;118:e2021996118. doi: 10.1073/pnas.2021996118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P, Liang S-Q, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K, Mir A, Sontheimer EJ, Gao G, Flotte TR, Wolfe SA, et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun. 2021;12:2121. doi: 10.1038/s41467-021-22295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aida T, Wilde JJ, Yang L, Hou Y, Li M, Xu D, Lin J, Qi P, Lu Z, Feng G. Prime editing primarily induces undesired outcomes in mice. bioRxiv. 2020:2020.08.06.239723 [Google Scholar]
- 26.Lin Q, Jin S, Zong Y, Yu H, Zhu Z, Liu G, Kou L, Wang Y, Qiu JL, Li J, Gao C. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat Biotechnol. 2021;39:923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- 27.Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G, Gopalappa R, Kim D, Cho SR, Kim JH, Kim HH. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng. 2021 doi: 10.1038/s41551-021-00788-9. [DOI] [PubMed] [Google Scholar]
- 28.Tanese N, Goff SP. Domain structure of the Moloney murine leukemia virus reverse transcriptase: Mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988;85:1777–81. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post K, Guo J, Kalman E, Crouch RJ, Levin JG, Uchida T. A Large Deletion in the Connection Subdomain of Murine Leukemia Virus Reverse Transcriptase or Replacement of the RNase H Domain with Escherichia coli RNase H Results in Altered Polymerase and RNase H Activities. Biochemistry. 1993;32:5508–17. doi: 10.1021/bi00072a004. [DOI] [PubMed] [Google Scholar]
- 30.Keller W, Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972;69:3360–4. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MAM, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, Modis Y, MacDonald AS, et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 2014;33:542–58. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat Rev Genet. 2011;12:341–55. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 33.Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–42. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truong DJJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–8. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Sun W, Wang B, Xiao X, Liu XQ. Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Hum Gene Ther. 2008;19:958–64. doi: 10.1089/hum.2008.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair N, Rincon MY, Evens H, Sarcar S, Dastidar S, Samara-Kuko E, Ghandeharian O, Viecelli HM, Thöny B, De Bleser P, VandenDriessche T, et al. Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood. 2014;123:3195–9. doi: 10.1182/blood-2013-10-534032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viecelli HM, Harbottle RP, Wong SP, Schlegel A, Chuah MK, Vandendriessche T, Harding CO, Thöny B. Treatment of phenylketonuria using minicircle-based naked-DNA gene transfer to murine liver. Hepatology. 2014;60:1035–43. doi: 10.1002/hep.27104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grisch-Chan HM, Schlegel A, Scherer T, Allegri G, Heidelberger R, Tsikrika P, Schmeer M, Schleef M, Harding CO, Häberle J, Thöny B. Low-Dose Gene Therapy for Murine PKU Using Episomal Naked DNA Vectors Expressing PAH from Its Endogenous Liver Promoter. Mol Ther Nucleic Acids. 2017;7:339–349. doi: 10.1016/j.omtn.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shedlovsky A, McDonald JD, Symula D, Dove WF. Mouse models of human phenylketonuria. Genetics. 1993;134:1205–10. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–4. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 42.Su SS, Lahue RS, Au KG, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–35. [PubMed] [Google Scholar]
- 43.Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–51. [PubMed] [Google Scholar]
- 44.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh JRJ, Aryee MJ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med. 2011;13:697–707. doi: 10.1097/GIM.0b013e3182141b48. [DOI] [PubMed] [Google Scholar]
- 46.Lazzarotto CR, Malinin NL, Li Y, Zhang R, Yang Y, Lee GH, Cowley E, He Y, Lan X, Jividen K, Katta V, et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat Biotechnol. 2020;38:1317–1327. doi: 10.1038/s41587-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DY, Bin Moon S, Ko JH, Kim YS, Kim D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 2020;48:10576–10589. doi: 10.1093/nar/gkaa764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adkins B, Leclerc C, Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat Rev Immunol. 2004;4:553–64. doi: 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- 49.Siegrist CA, Aspinall R. B-cell responses to vaccination at the extremes of age. Nat Rev Immunol. 2009;9:185–94. doi: 10.1038/nri2508. [DOI] [PubMed] [Google Scholar]
- 50.Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ. Large deletions induced by Cas9 cleavage. Nature. 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 51.Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–8. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meijer AJ, Lamers WH, Chamuleau RAFM. Nitrogen metabolism and ornithine cycle function. Physiol Rev. 1990;70:701–48. doi: 10.1152/physrev.1990.70.3.701. [DOI] [PubMed] [Google Scholar]
- 55.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 56.Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An M, Newby GA, Chen JC, Hsu A, Liu DR. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Düring DN, Dittrich F, Rocha MD, Tachibana RO, Mori C, Okanoya K, Boehringer R, Ehret B, Grewe BF, Gerber S, Ma S, et al. Fast Retrograde Access to Projection Neuron Circuits Underlying Vocal Learning in Songbirds. Cell Rep. 2020;33:108364. doi: 10.1016/j.celrep.2020.108364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, Pinello L. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heintz C, Troxler H, Martinez A, Thöny B, Blau N. Quantification of phenylalanine hydroxylase activity by isotope-dilution liquid chromatography-electrospray ionization tandem mass spectrometry. Mol Genet Metab. 2012;105:559–65. doi: 10.1016/j.ymgme.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 60.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Wegberg AMJ, MacDonald A, Ahring K, Bélanger-Quintana A, Blau N, Bosch AM, Burlina A, Campistol J, Feillet F, Gizewska M, Huijbregts SC, et al. The complete European guidelines on phenylketonuria: diagnosis and treatment. Orphanet J Rare Dis. 2017;12:162. doi: 10.1186/s13023-017-0685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vockley J, Andersson HC, Antshel KM, Braverman NE, Burton BK, Frazier DM, Mitchell J, Smith WE, Thompson BH, Berry SA. Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet Med. 2013;16:188–200. doi: 10.1038/gim.2013.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data related to this study is present in the paper or supplementary materials. Illumina sequencing data is available at the Gene Expression Omnibus (GEO) database under accession number GSE174757. The authors declare that all other data supporting the findings of this study are available within the paper and its extended data files.