Therapy-induced senescence renders cancer cells highly immunogenic, making them very efficient in triggering protective CD8-dependent antitumor immune responses.
Abstract
Cellular senescence is a stress response that activates innate immune cells, but little is known about its interplay with the adaptive immune system. Here, we show that senescent cells combine several features that render them highly efficient in activating dendritic cells (DC) and antigen-specific CD8 T cells. This includes the release of alarmins, activation of IFN signaling, enhanced MHC class I machinery, and presentation of senescence-associated self-peptides that can activate CD8 T cells. In the context of cancer, immunization with senescent cancer cells elicits strong antitumor protection mediated by DCs and CD8 T cells. Interestingly, this protection is superior to immunization with cancer cells undergoing immunogenic cell death. Finally, the induction of senescence in human primary cancer cells also augments their ability to activate autologous antigen-specific tumor-infiltrating CD8 lymphocytes. Our study indicates that senescent cancer cells can be exploited to develop efficient and protective CD8-dependent antitumor immune responses.
Significance:
Our study shows that senescent cells are endowed with a high immunogenic potential—superior to the gold standard of immunogenic cell death. We harness these properties of senescent cells to trigger efficient and protective CD8-dependent antitumor immune responses.
See related article by Chen et al., p. 432.
This article is highlighted in the In This Issue feature, p. 247
INTRODUCTION
Cellular senescence is a stress response aimed to eliminate unwanted, damaged, or aberrant cells (1, 2). This response consists of a stable proliferative arrest together with the development of a vigorous proinflammatory secretome (known as senescence-associated secretory phenotype, or SASP; refs. 3, 4). Through the SASP, senescent cells recruit immune cells to promote their own immune clearance, thereby restoring tissue homeostasis (5).
Cancer cells are usually exposed to a multitude of stressors known to trigger senescence, including oncogenic signaling, replicative stress, hypoxia, reactive oxygen species, nutrient deprivation, and exposure to cytokines present in the tumor microenvironment, such as TGFβ (1, 6, 7). Also, cytokines produced by immune cells induce intratumoral senescence, as is the case of IFNγ produced by Th1 cells (8, 9). Moreover, a variety of anticancer therapies can also induce senescence in cancer cells (1, 6). Consequently, tumors, both before and after therapy, generally contain a variable fraction of senescent cells, and this is now considered a “hallmark of cancer” (10, 11). Senescent cancer cells, due to their low or null proliferative capacity, do not contribute per se to tumor growth; however, they contribute to modify the tumor microenvironment through their SASP. The SASP produced by intratumoral senescent cells has complex and often opposite effects on tumor behavior depending on multiple factors that partly reflect the intrinsic heterogeneity of cancer and the response to cancer therapies (3, 6, 7). Notably, the SASP of tumor cells can recruit and activate CD4 and CD8 T cells eliciting antitumor protection (12–15).
The immune clearance of senescent cells is mediated by different populations of leukocytes, being better studied those belonging to the innate immune system (5, 16, 17). In particular, macrophages (18–21) and natural killer (NK) cells (12, 21–27) have been described as the main cell types responsible for the elimination of senescent cells. Other components of the innate immune system, such as invariant natural killer T cells (iNKT; ref. 28), neutrophils (29), and natural IgM antibodies (30), have also been shown to participate in the elimination of senescent cells.
However, sparse evidence is available regarding the adaptive immune system. Specifically, it has been reported that senescent hepatocytes expressing oncogenic mutant Nras activate CD4 T cells, and this triggers macrophage-mediated elimination of premalignant hepatocytes (18). In this experimental system, CD4 T activation was found to be mediated by antigen-presenting cells and senescent hepatocytes expressing MHC class II (MHC-II; ref. 18). MHC-II expression has also been observed in senescent melanocytes, but not in senescent keratinocytes or fibroblasts, suggesting that it is not a general feature of senescent cells (14). Senescent cells also upregulate the nonclassical MHC class I (MHC-I) molecule HLA-E, which is an inhibitory signal for NK and CD8 T cells, and depletion of HLA-E renders senescent cells susceptible to elimination by both NK and CD8 T cells (25). Finally, a particular type of lymphoma is characterized by containing a large fraction of tumor senescent cells, which are strongly immunogenic but escape immune control by expressing immunosuppressive ligands (31).
Here, we show that senescent cells combine multiple features that render them strongly immunogenic and very efficient in triggering antitumor adaptive immune responses.
RESULTS
Senescent Cells Upregulate MHC-I Antigen Presentation
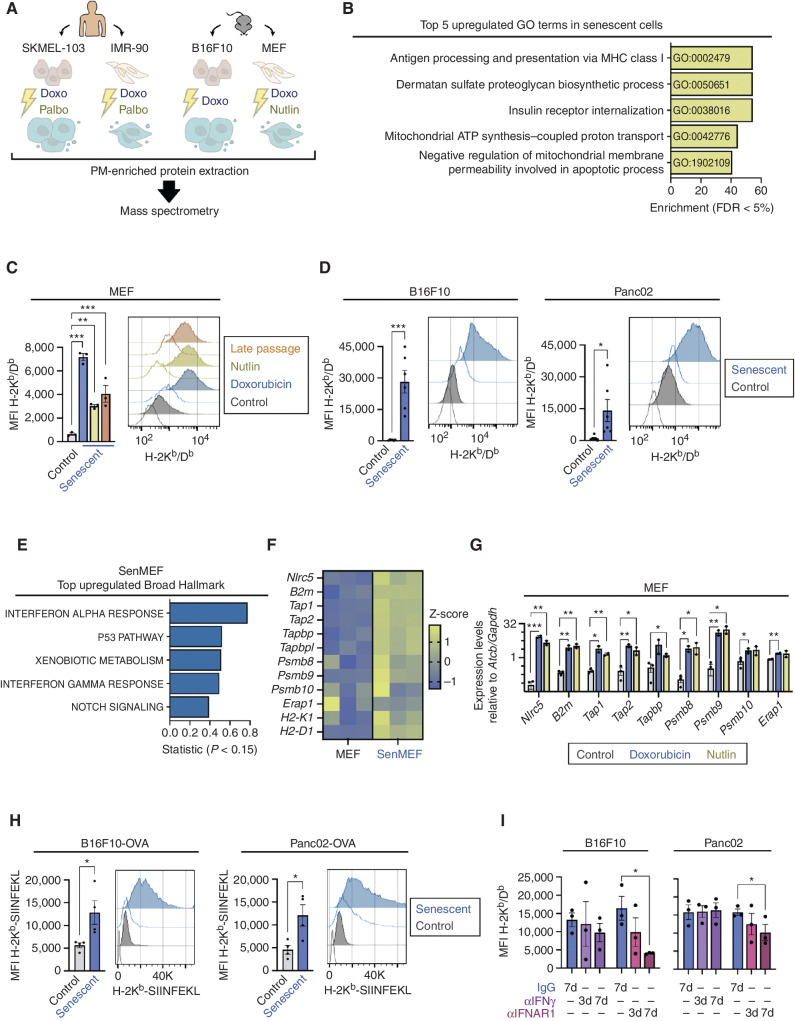
To identify proteins with potential immune regulatory activity specific to senescent cells, we performed a proteomic screen for plasma membrane–enriched fractions from senescent cells. Aiming to find proteins generally associated with senescence and ideally independent of the cell type and senescence trigger, we analyzed a total of four different cell types: two primary fibroblasts [human IMR-90 fibroblasts and mouse embryonic fibroblasts (MEF)] and two cancer cell lines [human melanoma SKMEL-103 and mouse melanoma B16-F10 (B16F10)]. Also, we used different types of senescence inducers, in particular, doxorubicin (genotoxic), palbociclib (CDK4/6 inhibitor), and nutlin-3A (p53 activator). Although doxorubicin is able to induce senescence in all the cell types used, the other targeted inhibitors are restricted in their ability to induce senescence depending on the cell type. Altogether, we tested seven different senescence conditions (Fig. 1A). Successful induction of senescence was confirmed by monitoring senescence-associated β-galactosidase (SAβG) activity and the mRNA expression of at least one senescence-associated gene CDKN1A/Cdkn1a or IL6 (Supplementary Fig. S1A and S1B). The “surfaceome” of the seven senescent and the four nonsenescent samples was obtained by mass spectrometry (each sample in biological triplicate; Supplementary Table S1). Gene ontology (GO) analysis of the genes encoding the proteins that were significantly increased in four or more senescence conditions as compared with their corresponding nonsenescent controls revealed that “Antigen processing and presentation of antigen via MHC class I” was the top enriched category (Fig. 1B; Supplementary Table S2). Increased expression of classical MHC-I molecules in senescent cells was confirmed by flow cytometry of H-2Kb/Db in MEFs and B16F10 (Fig. 1C and D), as well as HLA-A/B/C in IMR-90 and SKMEL-103 cells (Supplementary Fig. S1C). Moreover, we expanded our findings to other cancer cell lines, including mouse pancreatic adenocarcinoma Panc02 cells (Fig. 1D), and human squamous cell carcinoma UT-SCC-2 and UT-SCC-42B cells (Supplementary Fig. S1D). In addition, we observed augmented levels of the nonclassical MHC-I molecules H2-Qa1 and H2-Qa2 in senescent MEFs (Supplementary Fig. S1E).
Figure 1.
Senescent cells upregulate MHC-I antigen presentation. A, Schematics of the proteomic screen of the plasma membrane (PM)–enriched fraction of human (SKMEL-103 and IMR-90) and murine (B16F10 and MEF) cells, either untreated or exposed to various senescence-inducing stimuli (doxo, doxorubicin; nutlin, nutlin-3A; palbo, palbociclib). Three independent biological replicates per cell line were analyzed. B, Top five upregulated GO terms enriched in the proteins found upregulated in the plasma membrane fraction of senescent cells (in 4 or more conditions of senescence, with a linear fold change >1.5, FDR < 5%). C, Flow cytometry analysis of H-2Kb/Db expression in control versus senescent MEFs treated with doxorubicin or nutlin or late passaged. Representative histograms showing the fluorescence signal of each stained sample and its unstained control (uncolored histogram) and quantification after autofluorescence subtraction of n = 3 independent experiments are shown. ***, P < 0.001; **, P < 0.01; one-way ANOVA compared with control MEFs. MFI, mean fluorescence intensity. D, Flow cytometry analysis of H-2Kb/Db expression in control or senescent B16F10 and Panc02 cancer cells, treated with doxorubicin. Representative histograms showing the fluorescence signal of each stained sample and its unstained control (uncolored histogram) and quantification after autofluorescence subtraction of n = 5 independent experiments are shown. ***, P < 0.001; *, P < 0.05; unpaired Student t test compared with control cells. E, Top five upregulated “Broad Hallmarks” from the differential expression analysis (RNA-seq) of senescent MEFs (senMEF), in which senescence was induced by doxorubicin compared with control MEFs. n = 3 independent biological replicates were analyzed. F, Normalized expression levels of antigen presentation machinery– and immunoproteasome-related genes from the RNA-seq analysis of control versus senescent MEFs. G, mRNA expression levels of antigen presentation machinery– and immunoproteasome-related genes in control versus senescent MEFs treated with doxorubicin or nutlin, as measured by qRT-PCR (relative to the average expression of housekeeping genes Actb and Gapdh). n = 2 independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05, unpaired Student t test compared with control cells. H, Flow cytometry analysis of OVA-derived SIINFEKL peptide bound to H-2Kb presentation in control or senescent B16F10 and Panc02 cells stably expressing OVA. Representative histograms showing the fluorescence signal of each stained sample and its unstained control (uncolored histogram) and quantification after autofluorescence subtraction are shown. *, P < 0.05; unpaired Student t test compared with control cells. I, Flow cytometry analysis of H-2Kb/Db expression in senescent B16F10 or Panc02 cells, treated with doxorubicin, after treatment with blocking antibodies against IFNγ, IFNAR1, or their respective IgG isotype controls. Cells were treated with doxorubicin at day 0 and collected at day 7. The blocking antibodies were added to the culture medium for the indicated number of days (7d: from day 0 to 7; 3d: from day 4 to 7). Quantification after autofluorescence subtraction of n = 3 independent experiments is shown. *, P < 0.01; two-way ANOVA test compared with IgG-treated senescent cells.
Type I IFNs are generally present in the SASP, and IFN signaling is characteristically active in senescent cells (32). Considering the role of IFNs as key stimulators of MHC-I presentation (33), we hypothesized that this pathway could be responsible for the elevated levels of MHC-I in senescent cells. We performed RNA-sequencing (RNA-seq) of doxorubicin-senescent MEFs, doxorubicin-senescent B16F10, doxorubicin-senescent Panc02, palbociclib-senescent SKMEL-103, and doxorubicin-senescent IMR-90 and their nonsenescent counterparts. As expected, upregulation of IFN transcriptional signatures, including main gene components for MHC-I antigen processing and presentation, was found in all the tested senescent cells compared with their nonsenescent controls (Fig. 1E and F; Supplementary Fig. S1F–S1N and Supplementary Table S3). Validation by qRT-PCR analysis in separate biological replicates confirmed the upregulation of most of the genes tested, notably including the master transcriptional regulator of MHC-I–dependent immune responses Nlrc5 (ref. 34; Fig. 1G; Supplementary Fig. S1O). To test whether the transcriptional upregulation of the MHC-I–associated machinery was mirrored by an elevated presentation of actual antigens, we analyzed the expression of the H-2Kb restricted ovalbumin (OVA)-derived peptide SIINFEKL in cancer cell lines stably expressing OVA (B16F10 or Panc02), untreated or after senescent induction. We found that senescent cells presented more SIINFEKL bound to H-2Kb than their proliferating counterparts (Fig. 1H).
Finally, we studied the impact of type I and II IFN signaling as drivers for increased MHC-I expression in senescent cells. Using blocking antibodies at different time points from senescence induction, we found that blockade of type I IFNAR1 receptor, but not IFNγ, diminished MHC-I expression in senescent B16F10 and Panc02 cells (Fig. 1I; Supplementary Fig. S1P). This inhibition was more pronounced when the IFNAR1 blockade was initiated simultaneous to the addition of the senescence trigger and maintained for 7 days. As expected, chemical inhibition of JAK kinases, which are key mediators of IFN signaling, strongly reduced MHC-I expression in senescent cells (Supplementary Fig. S1Q–S1S).
Taken together, these data support the concept that enhanced MHC-I antigen processing and presentation are general features of senescent cells, and are explained by their hyperactivated type I IFN signaling.
Senescent Cells Stimulate CD8 T Cells
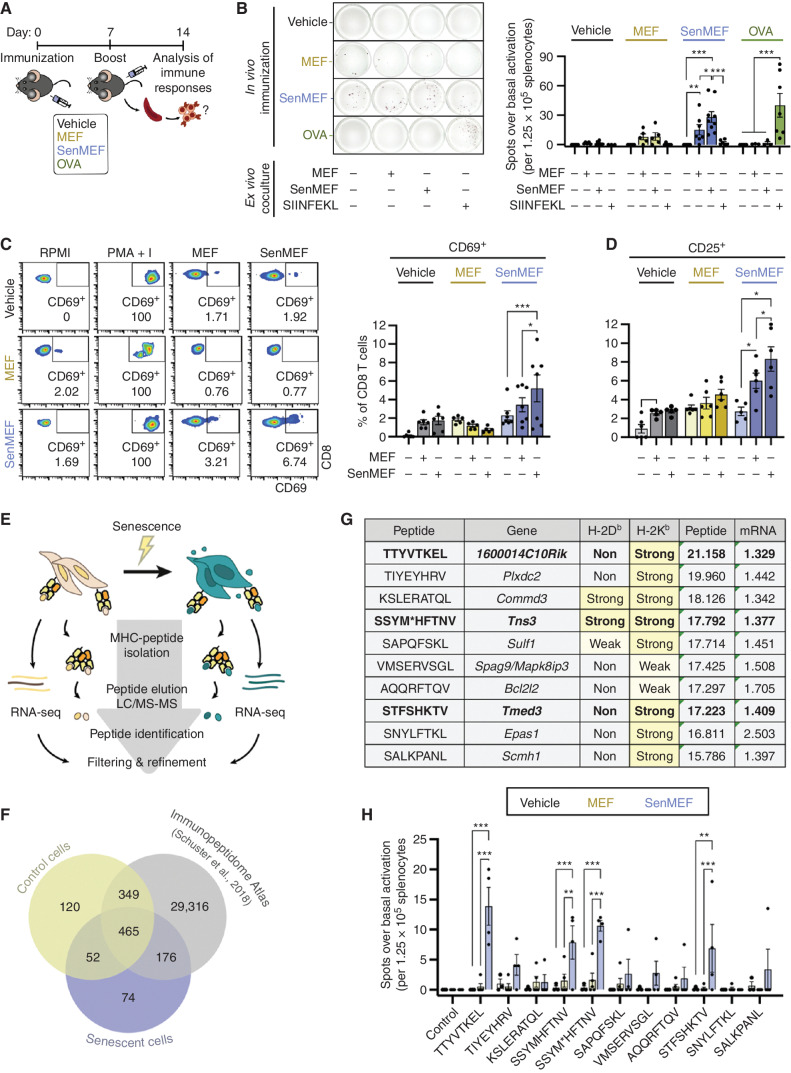
It is well established that cellular stress partially changes the repertoire of self-peptides presented by MHC-I (35–39). Considering that senescence is indeed a state of cellular stress, and in view of the enhanced capacity of senescent cells to process and present MHC-I antigens, we wondered whether senescent cells could induce antigen-dependent immune responses in vivo. For this, we used noncancer cells because, in contrast to cancer cells, they are expected to be poorly immunogenic. We immunized mice with syngeneic primary fibroblasts, either untreated (MEF) or senescent (senMEF). As a negative control, we used vehicle with no cells (PBS); as a positive control, we immunized animals with OVA to trigger T-cell activation against a known antigen (SIINFEKL; Fig. 2A). These immunizations were performed using CpG oligodeoxynucleotide as adjuvant. After immunization, splenocytes were isolated, and the number of activated T cells was measured by IFNγ enzyme-linked immunospot (ELISpot) in the absence of ex vivo stimuli (basal activation) or in the presence of MEF, senMEF, or SIINFEKL peptide. Notably, basal activation was significantly higher in mice immunized with senMEF than under all other conditions (Supplementary Fig. S2A). Splenocytes from senMEF-immunized mice responded strongly to ex vivo reexposure to senMEF, yielding a number of spots over basal activity comparable with that of OVA-immunized splenocytes exposed to SIINFEKL (Fig. 2B). Interestingly, splenocytes from mice immunized with senMEF also responded to nonsenescent MEF ex vivo, although not as strongly as to senMEF (Fig. 2B). Maximal possible activation of T cells was achieved by treatment with a phorbol ester and a calcium ionophore (PMA + I; Supplementary Fig. S2B). As additional evidence, CD69 and CD25 were both elevated in CD8 T cells from senMEF-immunized animals when exposed ex vivo to senMEF, and this activation was also detectable, albeit of lower magnitude, when CD8 T cells were exposed to nonsenescent MEF (Fig. 2C and D). In contrast to the upregulation of MHC-I, MHC-II (I-A/I-B) was not upregulated in senMEF (Supplementary Fig. S2C), and, accordingly, CD4 T cells from senMEF-immunized mice showed a weak activation when exposed ex vivo to senMEF (CD25, but not CD69, was elevated; Supplementary Fig. S2D and S2E).
Figure 2.
Senescent noncancer cells induce an adaptive immune response in vivo and present unique immunogenic peptides. A, Schematic outline of the immunization protocol used in this study. Briefly, immunocompetent C57BL/6 animals were subcutaneously immunized on days 0 and 7 with vehicle (no cells), control or senescent syngeneic fibroblasts (MEF or senMEF, respectively), or OVA, all done concomitantly with an immune adjuvant (CpG). One week later, animals were sacrificed, and immune responses were tested ex vivo.B, ELISpot assay to detect IFNγ production in splenocytes isolated from nonimmunized mice (vehicle) or animals immunized with control MEF, senMEF, or OVA (n = 3–7 mice per group). Splenocytes were cultured in RPMI either alone (control) or cocultured with control MEF (1:10 target-to-splenocyte ratio), senMEF (1:10 target-to-splenocyte ratio), or SIINFEKL OVA-derived peptide (400 nmol/L). The number of spots for each condition above the control condition (background) was quantified. Representative pictures (left) and quantification (right) are shown. ***, P < 0.001; **, P < 0.01; *, P < 0.05; two-way ANOVA test. C, Flow cytometry analysis of CD69 activation marker in CD8 T cells from naïve (vehicle) versus MEF- or senMEF-immunized animals after culture in RPMI medium either alone or with PMA + I, MEF, or senMEF ex vivo. Representative pseudocolor plots and quantification of n = 5–7 mice per group are shown. ***, P < 0.001; *, P < 0.05; two-way ANOVA test. D, Flow cytometry analysis of CD25 activation marker in CD8 T cells from naïve (vehicle) versus MEF- or senMEF-immunized animals after culture in RPMI medium either alone or with PMA + I, MEF, or senMEF ex vivo (n = 5–7 mice per group). *, P < 0.05; two-way ANOVA test. E, Layout of combined immunopeptidomics and RNA-seq analyses in control versus senescent MEFs. F, Venn diagram displaying peptides identified in control cells, senescent cells, and the Mouse Immunopeptidome Atlas dataset (40). G, List of selected peptides presented exclusively on senescent cell MHC-I together with their corresponding coding gene and their predicted binding affinity to H-2Kb and H-2Db (NetMHCpan v4.1; classified as non-, weak, or strong binders, with the latter two highlighted in yellow). “Peptide” indicates the normalized log2(area) of the signal obtained in the immunopeptidomic mass spectrometry analysis for each peptide. Similarly, “mRNA” indicates the linear fold-change expression of the corresponding gene (senescent vs. control MEF) from the RNA-seq transcriptomic analysis are indicated. M* indicates M(+15.99), oxidized methionine. Bold font indicates those peptides that were validated as immunogenic (see next panel). H, Selected peptides validated using ELISpot assay to detect IFNγ production in splenocytes isolated from nonimmunized mice (vehicle) or animals immunized with MEF or senMEF (n = 3–5 mice per group). Splenocytes were cultured in RPMI medium either alone as negative control (control) or supplemented with the different peptides selected from the immunopeptidome analysis, as indicated. For SSYM*HFTNV peptide, both SSYMHFTNV and the modified SSYM*HFTNV were tested. The number of spots for each condition above the control condition (background) was quantified. ***, P < 0.001; **, P < 0.01; two-way ANOVA test.
We conclude that senescence induction renders fibroblasts able to induce a CD8-dependent immune response.
Senescence Is Associated with an Altered Immunopeptidome
To test whether CD8 T-cell activation was directly linked to the recognition of epitopes presented by senescent cells, we profiled the immunopeptidome of control and senescent MEF, in which senescence was induced by doxorubicin treatment (Fig. 2E). Mass spectrometry analysis of peptides eluted from classical MHC-I molecules identified 767 bona fide peptides corresponding to annotated proteins (see Methods). Interestingly, about 10% of these peptides (74 peptides) were detected in senescent cells, but not in their nonsenescent counterparts or in the Mouse Immunopeptidome Atlas (Fig. 2F; Supplementary Table S4). The latter consists of more than 30,000 MHC-I (H-2Kb/Db) peptides obtained from 19 normal tissues and four murine cancer cell lines (40). As a criterion to prioritize the 74 senescence-associated MHC-I peptides, we selected those with upregulation (>1.3 linear fold-change) of the coding mRNA in senescent versus untreated MEF obtained by RNA-seq (10 of 74 peptides; Fig. 2G). To evaluate the immunogenic potential of these senescence-associated epitopes, we used individual peptides (of the 10 prioritized peptides; Fig. 2G) or pooled combinations (of the remaining 64 peptides; Supplementary Table S4) to restimulate splenocytes isolated from animals immunized with vehicle, MEF, or senMEF. As revealed by IFNγ ELISPOT assays, splenocytes isolated from senMEF-immunized animals were specifically activated by a subset of senescence-associated peptides (Fig. 2H; Supplementary Fig. S2F–S2H). In total, we identified three peptides from the set of 10 that were prioritized as immunogenic (Fig. 2H and marked in bold in Fig. 2G). [Note that one peptide has two versions with oxidized (M*) or nonoxidized (M) methionine, both giving similar results.] Additionally, three peptide pools of the remaining set of peptides (up to 64 peptides) were recognized by splenocytes from senMEF-immunized animals (Supplementary Fig. S2F).
These results suggest that noncancer senescent cells can elicit CD8 T-cell responses against senescence-associated self-antigens, which further reinforces the notion that induction of senescence renders cells immunogenic.
Senescent Cancer Cells Efficiently Activate Dendritic Cells
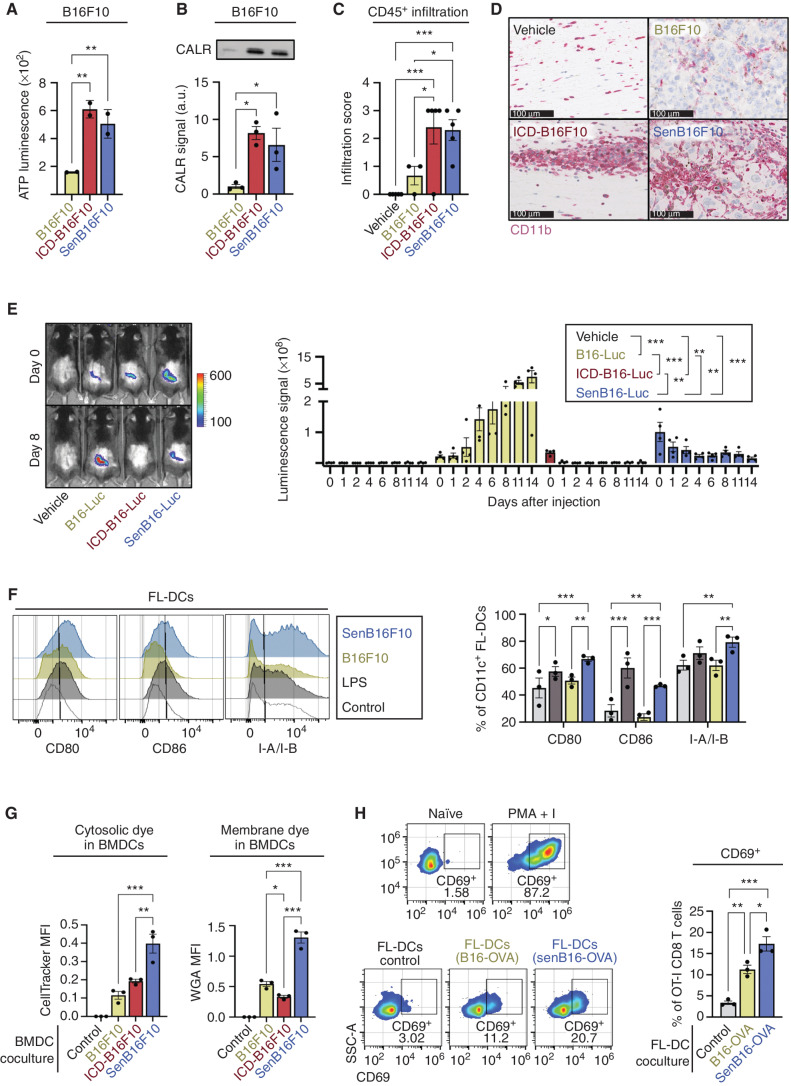
Having demonstrated the enhanced immunogenicity of noncancer senescent cells, we then moved to cancer settings with the final goal of triggering protective antitumor immune responses using senescent cancer cells. Senescent cells secrete a plethora of proinflammatory cytokines and chemokines that recruit and activate immune cells (3, 4), including damage-associated molecular patterns (DAMP), such as calreticulin (CALR; ref. 41), but direct measurement of their adjuvant properties has not been reported. Therefore, we compared the levels of DAMPs released by senescent cancer cells and by cells undergoing immunogenic cell death (ICD). We chose ICD because it is the best established method for efficient antitumor immunization (42, 43). Of note, we induced ICD and senescence using the same agent, doxorubicin, at different concentrations (5 μmol/L in the case of ICD, 0.1 μmol/L in the case of senescence). We observed that both ATP and CALR, prototypical DAMPs, were present in the conditioned media (CM) of senescent cells (B16F10, Panc02, and MEF) at similar levels to ICD cells (Fig. 3A and B; Supplementary Fig. S3A and S3B).
Figure 3.
Senescent cancer cells efficiently activate dendritic cells (DC). A, Levels of extracellular ATP in the CM of 106 control (B16F10), ICD B16F10 (5 μmol/L doxorubicin; ICD-B16F10), and senescent B16F10 (0.1 μmol/L doxorubicin; senB16F10). n = 2 independent experiments. **, P < 0.01; one-way ANOVA test compared with control B16F10. B, Immunoblot detection of CALR in the CM of 106 B16F10, ICD-B16F10, and senB16F10. Representative images (left) and quantification (right) of n = 3 independent experiments are shown. *, P < 0.05; one-way ANOVA test. C, Semiquantitative infiltration score of CD45+ cells within or closely surrounding the melanoma foci in skin sections of animals 7 days after subcutaneous injection of vehicle (no cells), B16F10, ICD-B16F10, or senB16F10 (n = 5 animals per group). Analysis performed by a histopathologist (N. Prats). ***, P < 0.001; *, P < 0.05; one-way ANOVA test. D, Immunochemistry staining of CD11b+ cells (purple) in skin sections of animals 7 days after subcutaneous injection of vehicle (no cells), B16F10, ICD-B16F10, or senB16F10. Representative images selected by a histopathologist (N. Prats) of n = 5 animals per group are shown. Note that the brown pigmentation is due to melanin. Scale bars for each image are shown (100 μm). E,In vivo imaging detection of luciferase-expressing B16F10 (B16-Luc) in animals subcutaneously injected with vehicle, 106 control, ICD B16-Luc, or senescent B16-Luc (n = 4 per group) at different time points after injection (as indicated). Representative images (left) and quantification (right) are shown. ***, P < 0.001; **, P < 0.01; two-way ANOVA test. F, Flow cytometry analysis of the DC activation markers CD80, CD86, and MHC-II (I-A/I-B) in CD11c+ FL-DCs upon coculture with RPMI medium either alone or with LPS, B16F10, or senB16F10. Representative histograms and quantification of n = 3 biological replicates are shown. ***, P < 0.001, **, P < 0.01; *, P < 0.05; one-way ANOVA test. G, Flow cytometry analysis of the uptake of CFSE (cytosolic dye) or WGA-Alexa647 (membrane dye) by BMDCs from labeled B16F10, ICD-B16F10, or senB16F10. Quantification after subtraction of autofluorescence from unstained BMDCs of n = 3 biological replicates. ***, P< 0.001; **, P < 0.01; *, P < 0.05; one-way ANOVA test. MFI, mean fluorescence intensity. H, Flow cytometry analysis of OT-I CD8 T-cell activation, as measured by CD69 expression, upon coculture with RPMI medium either alone (naïve) or with PMA + I, control FL-DCs or FL-DCs previously cocultured with control (B16-OVA) or senescent (senB16-OVA) B16-OVA cells, as indicated. Representative histograms and quantification of n = 3 biological replicates are shown. SSC-A, side scatter area. ***, P < 0.001; **, P < 0.01; *, P < 0.05; one-way ANOVA test.
To test whether the secretion of alarmins by senescent cells reflects on an enhanced immune recruitment in vivo, we injected mice subcutaneously with vehicle (PBS) or with B16F10 cells untreated, undergoing ICD, or senescent, and performed histologic analysis of the site of injection. Interestingly, 1 week after injection, senescent melanoma cells were clearly visible in the subcutis as large pleomorphic and anaplastic cells, often containing granular melanin pigment, surrounded by abundant immune cells (CD45+), mainly of myeloid lineage (CD11b+) together with T cells (CD3+; Fig. 3C and D; Supplementary Fig. S3C–S3E). Abundant immune and myeloid cells were also present at the ICD injection sites, although, as expected, no evidence of viable tumor cells was found. Control B16F10 cells started to form visible tumors after 1 week, with lower relative immune infiltration. We then wondered for how long the senescent cells would persist at the injection site. To explore this, we injected luciferase-expressing B16F10 cells untreated, ICD, or senescent (106 cells for each condition) and monitored their bioluminescence over time (Fig. 3E). As expected, untreated B16F10 cells grew and formed tumors with strong luciferase activity. In the case of cells undergoing ICD, the initial luciferase signal detected at the time of injection (day 0) disappeared after 1 day. In contrast, the luciferase signal from senescent cells was clearly detected up to 11 days after injection (Fig. 3E). Because senescent cells are considerably larger than their nonsenescent counterparts, we also performed this experiment injecting equal amounts of protein mass of ICD and senescent cells (106 ICD vs. 2 × 105 senescent cells), obtaining similar results (Supplementary Fig. S3F). Therefore, although senescent and ICD cells produce similar levels of alarmins, this production is expected to be limited in time in the case of ICD cells (less than 1 day), while maintained over several days in the case of senescent cells.
Upon recruitment of immune cells, the next essential steps for triggering an adaptive immune response involve the capture of antigens by dendritic cells (DC), as well as DC activation and maturation (44, 45). To evaluate this, we cocultured DCs with untreated or senescent B16F10 cells and measured the surface expression of DC activation and maturation markers CD80, CD86, and MHC-II (I-A/I-B) on DCs. We performed these experiments using bone marrow–derived DCs stimulated ex vivo with GM-CSF and IL4 (abbreviated as BMDCs, as a model of myeloid DCs) or with FLT3L (abbreviated as FL-DCs, as a model of conventional DCs; ref. 46). Lipopolysaccharide (LPS) stimulation was used as a positive control for DC activation. We found that coculture of DCs with senescent, but not untreated, B16F10 cells, led to the activation of DCs to similar levels as LPS stimulation (Fig. 3F; Supplementary Fig. S3G). Interestingly, DC activation by senescent cells required direct cell-to-cell contact because upregulation of the CD80 activation marker was abolished when a transwell coculture system was used (Supplementary Fig. S3H).
Next, we evaluated the efficiency of DCs in capturing cytosolic and membrane antigens from senescent cells. Untreated, ICD, or senescent cancer cells were stained with membrane or cytosolic fluorescent dyes and cocultured with BMDCs. We found that BMDCs captured both cytosolic and membrane dyes from B16F10 or Panc02 senescent cells more efficiently than from untreated or ICD cells (Fig. 3G; Supplementary Fig. S3I). As previously reported, ICD cells were efficient in delivering cytosolic dyes, but inefficient in delivering membrane dyes compared with untreated cells (47). Finally, we tested if senescent cells result in a better acquisition and presentation of actual antigens by DCs. For this, we cocultured DCs (BMDCs and FL-DCs) with untreated or senescent B16F10 cells expressing OVA (B16-OVA). Then, we sorted the DCs (CD11c+) and cocultured them with OT-I CD8 T cells, whose T-cell receptor (TCR) specifically recognizes the OVA-derived SIINFEKL antigen, and measured CD8 activation (via CD69 expression). We found that DCs previously cocultured with OVA-expressing senescent cells were more efficient at activating OT-I CD8 T cells than those DCs cocultured with nonsenescent cells (Fig. 3H; Supplementary Fig. S3J).
Collectively, these data support the notion that senescent cells display a strong adjuvanticity, promoting the recruitment and activation of DCs, and this is accompanied by an efficient delivery of antigens to DCs.
Immunization with Senescent Cancer Cells Promotes Anticancer Immune Surveillance
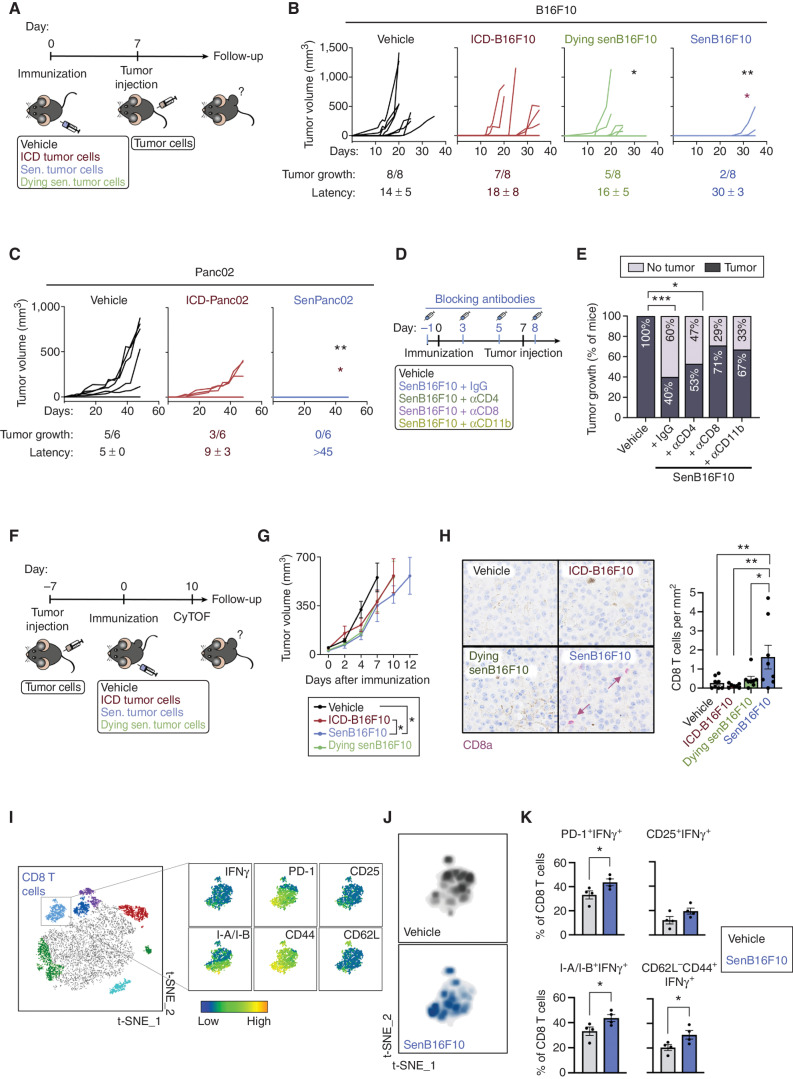
Given that senescent cells exhibit enhanced antigenicity and adjuvanticity, we postulated that senescent cancer cells could be used to promote an immune response against cancer. Previous pioneering work has shown that senescent cancer cells can trigger antitumor protection when used in a vaccination setting, and this property was attributed to their SASP (13). Here, we have extended these findings by first comparing the prophylactic effect of immunization with senescent versus ICD B16F10 melanoma cells against a subsequent rechallenge with proliferating B16F10 cells. Of note, we used the same agent, doxorubicin, to generate both immunogens (0.1 μmol/L for 7 days to induce senescence, and 5 μmol/L for 18 hours to induce ICD). To test the importance of cell viability in the case of senescent cells, we introduced another experimental condition that consisted of dying senescent cells, and, for this, we treated senescent B16F10 (senB16F10) cells with navitoclax, an agent known to induce apoptosis in senescent cells (i.e., senolysis; Fig. 4A). Immunizations were performed using equal amounts of protein for each condition (thus, animals received either 106 ICD or 2 × 105 senescent cells). One week after immunization by subcutaneous injection, all experimental groups were rechallenged subcutaneously with 3 × 104 untreated B16F10 cells in the opposite flank. We observed that immunization with senB16F10 elicited better anticancer protection than ICD-B16F10 or dying senB16F10 (Fig. 4B; Supplementary Fig. S4A). Similarly, senescent Panc02 (senPanc02) cells induced a stronger protective response than ICD-Panc02 cells (Fig. 4C; Supplementary Fig. S4B). The antitumor protection afforded by senescent cancer cells, B16F10 and Panc02, was reflected by a lower rate of tumor growth and by a longer latency of those tumors that escaped immune control (in the case of Panc02, none of the animals developed tumors; Fig. 4B and C).
Figure 4.
Immunization with senescent cancer cells promotes anticancer immune surveillance. A, Schematics of the cancer immunization protocol used in these studies. Sen., senescent. B, Individual tumor growth curves from vehicle-treated mice or mice immunized with ICD-B16F10, senB16F10, or senB16F10 cells dying by senolysis (induced by 10 μmol/L navitoclax, dying senB16F10; n = 8 mice per group). Tumor growth (number of animals developing tumors out of the total) and tumor latency (mean ± SD of the day on which the tumor appeared) are indicated for each group. **, P < 0.01; *, P < 0.05; two-way ANOVA test compared with vehicle-treated group (black) or ICD-B16F10 group (red). C, Individual tumor growth curves from vehicle-treated mice or mice immunized with Panc02 cells dying by ICD (induced by a high dose of doxorubicin, ICD-Panc02) or senescent Panc02 (low dose of doxorubicin, senPanc02; n = 6 mice per group). Tumor growth (number of animals developing tumors out of the total) and tumor latency (mean ± SD day of appearance of the tumor) are indicated for each group. **, P < 0.01; *, P < 0.05; two-way ANOVA test compared with vehicle-treated (black) group or ICD-B16F10 group (red). D, Schematics of the cancer immunization and immune depletion protocol used in this study. E, Tumor appearance after rechallenge in vehicle-treated mice (n = 14) or mice immunized with senB16F10 treated with IgG (n = 14) or the indicated blocking antibodies as described in D (n = 15 for aCD4, n = 14 for aCD8, or n = 6 for aCD11b). ***, P < 0.001; *, P < 0.05; Fisher exact test. F, Schematics of the therapeutic cancer immunization protocol used in these studies. CyTOF, cytometry by time of flight. G, Grouped tumor growth of B16F10 tumor–bearing animals immunized with vehicle, ICD-B16F10, dying senB16F10, or senB16F10. *, P < 0.05; two-way ANOVA test (n = 7–8) H, CD8a staining (purple) in B16F10 tumor sections from animals immunized with vehicle, ICD-B16F10, dying senB16F10, or senB16F10 and sacrificed at humane endpoint. Note that the brown pigmentation is due to melanin. Representative images and quantification of n = 7–8 mice per group. **, P < 0.01; *, P < 0.05; one-way ANOVA test. I, t-Distributed stochastic neighbor embedding (t-SNE) representation of tumor-infiltrating immune (CD45+) cells detected by CyTOF of B16F10 tumors from animals immunized with vehicle or senB16F10 and sacrificed 10 days after immunization. The cluster of CD8 T cells is amplified, and the expression pattern of T-cell markers of activation (IFNγ, PD-1, CD25, and I-A/I-B), and differentiation to effector (CD44) or naïve T cells (CD62L) are shown (left). J, Density plots of the distribution of infiltrating CD8 T cells from nonimmunized and senB16F10-immunized animals (n = 4 animals per group). K, Percentage of activated tumor-infiltrating CD8 T cells (PD-1+IFNγ+, CD25+IFNγ+, I-A/I-B+IFNγ+, CD62L−CD44+IFNγ+) from tumors of nonimmunized animals (vehicle) or immunized with senB16F10 (n = 4 mice per group). *, P < 0.05; unpaired Student test.
To gain a deeper understanding of the cancer-preventive response induced by live senescent cancer cells, we depleted the major immune populations involved in adaptive antitumor immunity, namely, CD4 and CD8 T cells and CD11b+ myeloid cells (Fig. 4D; Supplementary Fig. S4C). We found that depletion of CD8 T cells or CD11b+ myeloid cells significantly impaired the protective effect of vaccination with senescent cells, whereas depletion of CD4 T cells had a modest effect (Fig. 4E; Supplementary Fig. S4D and S4E).
We also tested whether therapeutic immunization with senescent cells could inhibit tumor growth in mice already bearing tumors at the time of immunization (Fig. 4F). We found that animals immunized with senescent cells showed a moderate but significantly reduced tumor growth rate compared with animals immunized with ICD cells (Fig. 4G; Supplementary Fig. S4F). We examined the infiltration of CD8 T cells within B16F10 tumors, which are considered highly immunosuppressive and poorly permissive to immune infiltration (48). Interestingly, immunization with senB16F10 cells enhanced the infiltration of CD8 T cells into B16F10 tumors compared with all other experimental groups (Fig. 4H). To further understand the effects of therapeutic immunization within the tumor infiltrate, we simultaneously immunophenotyped 19 markers using mass cytometry [cytometry by time of flight (CyTOF)] at day 10 after immunization (Fig. 4F). Interestingly, the phenotype of the tumor-infiltrating CD8 T cells from animals immunized with senB16F10 was skewed toward a more activated state (Fig. 4I and J). Accordingly, the percentage of CD8 T cells expressing T-cell activation markers (PD-1+IFNγ+, CD25+IFNγ+, and I-A/I-B+IFNγ+) was increased in infiltrating CD8 T cells from immunized animals compared with nonimmunized ones (Fig. 4K). Regarding other immune cell populations, the percentage (relative to the total number of CD45+ cells) of tumor-infiltrating DCs (CD11b+ CD11c+) was increased in immunized animals compared with nonimmunized ones (Supplementary Fig. S4G). The percentages of the rest of the main immune populations were not significantly changed (Supplementary Fig. S4G).
Taken together, our observations suggest that immunization with viable senescent cancer cells can promote a tumor prophylactic and therapeutic CD8 T cell–dependent antitumor immune response.
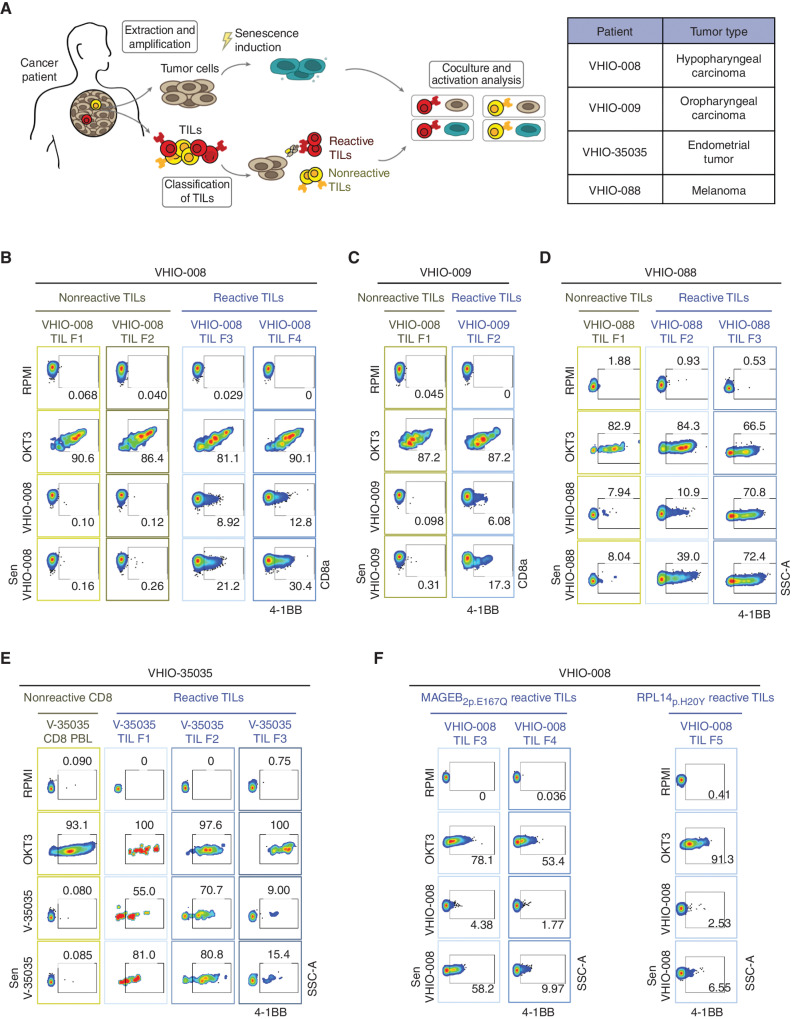
Senescent Cancer Cells from Human Patients Hyperstimulate Autologous Reactive Tumor-Infiltrating Lymphocytes
Finally, we extended our findings to a clinically relevant experimental setting. In particular, we investigated whether the induction of senescence in patient-derived primary cancer cells enhances the activation of autologous tumor-infiltrating lymphocytes (TIL). To address this, TILs from different fragments of primary tumors (labeled F1, F2, and so on) were expanded ex vivo and tested for recognition of their autologous cancer cells. In this way, TILs from different fragments were classified as tumor-reactive or nonreactive (Fig. 5A). A total of four primary cancers of different types were analyzed in this way (listed in Fig. 5A). [Note that for the VHIO-35035 patient, all TIL fractions reacted against tumor cells, so CD8 T cells from peripheral blood (PBL) of the same patient were used as a nonreactive control population.] Upon treatment with anti-CD3–coated beads (OKT3), both reactive and nonreactive TILs showed robust upregulation of the activation marker 4-1BB (also known as CD137), indicating that both types of TILs were functional (Fig. 5B–E; Supplementary Fig. S5A). As expected, all cancer cells activated their autologous reactive TILs, but not their nonreactive TILs (Fig. 5B–F; Supplementary Fig. S5A). In the case of VHIO-088 nonreactive TILs, a modest reactivity was detected in the fraction of TILs originally classified as nonreactive (Fig. 5D). The four primary cancer cells were treated with bleomycin to induce senescence, which manifested in three of the cancer cell isolates (VHIO-008, -35035, and -088) as a flat and large cellular morphology, high levels of SAβG, and upregulation of CDKN1A mRNA (Supplementary Fig. S5B and S5C). In the case of bleomycin-treated VHIO-009 cells, their morphology was also flat and enlarged and upregulated CDKN1A; however, they remained negative for SAβG; then, we also tested IL8, as a SASP marker, observing a notable transcriptional upregulation (Supplementary Fig. S5B and S5C). Importantly, the four senescent patient-derived cancer cells activated their corresponding reactive TILs with higher efficiency than nonsenescent cells (Fig. 5B–E). Altogether, a total of six of eight reactive TIL fractions presented a higher level of activation by senescent cells compared with nonsenescent cells (ranging from 1.5- to 4-fold higher). Two TIL fractions (VHIO-35035 TIL F2 and VHIO-088 TIL F3) were strongly activated by nonsenescent cells (>70%), and these levels were only marginally increased by senescent cells. It is important to emphasize that senescent cancer cells did not stimulate nonreactive TILs, further indicating that TIL stimulation by senescent cells requires antigen recognition.
Figure 5.
Senescent cancer cells from human patients hyperstimulate autologous reactive TILs. A, Schematics of the procedure for isolating, amplifying, classifying, and coculturing patient-derived tumor cells with autologous reactive and nonreactive TILs (left). Table indicating patients and corresponding tumor type used in this study (right). B, Flow cytometry analysis of 4-1BB activation marker in CD8 cells from nonreactive (F1 and F2 fragments) and reactive (F3 and F4 fragments) autologous TILs from VHIO-008 patient after culture in RPMI medium either alone or with anti-CD3 (OKT3), control VHIO-008 cells, or bleomycin-treated senescent VHIO-008 cells (as indicated). C, Flow cytometry analysis of 4-1BB activation marker in CD8 cells from nonreactive (F1 fragment) and reactive (F3 fragment) autologous TILs from patient VHIO-009 after culture in RPMI medium either alone or with anti-CD3 (OKT3), control VHIO-009 cells, or bleomycin-treated senescent VHIO-009 cells (as indicated). D, Flow cytometry analysis of the 4-1BB activation marker in CD8 cells from nonreactive (F1 fragment) and reactive (F2 and F3 fragment) autologous TILs from patient VHIO-088 after culture in RPMI medium either alone or with anti-CD3 (OKT3), control VHIO-088 cells, or bleomycin-treated senescent VHIO-088 cells (as indicated). SSC-A, side scatter area. E, Flow cytometry analysis of the 4-1BB activation marker in nonreactive PBL CD8 T cells and CD8 cells from reactive (F1, F2, and F3 fragments) autologous TILs from patient VHIO-35035 (abbreviated as V-35035) after culture in RPMI medium either alone or with anti-CD3 (OKT3), control V-35035 cells, or bleomycin-treated senescent V-35035 cells (as indicated). F, Flow cytometry analysis of 4-1BB activation marker in CD8 cells from reactive autologous TILs (reactive F3, F4 and F5 fragments) enriched to be reactive against MAGEB2p.E167Q and RPL14p.H20Y (two neoantigens previously identified by whole-exome sequencing of the autologous tumor cell line) after culture in RPMI medium either alone or with anti-CD3 (OKT3), control VHIO-008 cells, or bleomycin-treated senescent VHIO-008 cells (as indicated).
The above observations were validated in an antigen-specific setting. For this, we used enriched populations of TILs from patient VHIO-008 specific to MAGEB2p.E167Q and RPL14p.H20Y, two neoantigens previously identified by whole-exome sequencing of the autologous tumor cell line (49). Neoantigen-specific TILs were cocultured with parental VHIO-008 cancer cells, either untreated or senescent, and their activation was assessed. In this case, TIL activation upon coculture with senescent VHIO-008 was more potent (2–6-fold higher) than when using the nonsenescent parental cells (Fig. 5F). To test whether senescence induction with other agents promoted equivalent responses, we also tested doxorubicin-induced and palbociclib-induced senescent VHIO-008 cells, observing similar results regarding the stimulation of nonreactive TILs and antigen-specific TILs (Supplementary Fig. S5D–S5E).
Senescence induction was accompanied by different degrees of upregulation of MHC-I (HLA-I), whereas MHC-II (HLA-II) levels remained largely unchanged (Supplementary Fig. S5F and S5G). Moreover, although CD8 T cells from reactive TILs were activated upon coculture with cancer cells, CD4 T cells remained unaffected, further supporting the main role of the CD8 T-cell population in the recognition of senescent cancer cells (Supplementary Fig. S5H–S5K).
Together, these data indicate that the induction of senescence in human cancer cells potentiates antigen-dependent CD8 T-cell activation.
DISCUSSION
The activation of adaptive immunity is strictly associated with two factors, adjuvanticity and antigenicity (42). Danger signals acting as adjuvants are essential to activate adaptive immune cells and, if absent, presentation of antigenic peptides to T cells drives peripheral tolerance rather than immune activation (43–45). Conversely, adjuvant signals in the absence of antigenic determinants drive inflammation, but not adaptive immune responses (50). In this study, we show that senescent cells combine an enhanced antigenicity with a strong adjuvanticity. These properties, together with the long-term viability of injected senescent cells over several days, make senescent cells ideal agents to trigger CD8 T cell–dependent adaptive immunity.
We compared a number of adjuvant-related features in parallel between senescent cells and cells undergoing ICD induced by the same agent, with the latter constituting the current gold standard for eliciting cell-based antitumor immune responses (42, 43). The levels of alarmins released into the extracellular milieu, specifically ATP and CALR, were similar between senescent and ICD cells, and this was reflected by the abundant recruitment of myeloid cells when injected subcutaneously. Notably, senescent cells outperformed ICD cells in the following two aspects. First, senescent cancer cells had a higher persistence in the skin for up to 11 days after injection, whereas ICD cells were essentially undetectable 1 day after injection. Second, senescent cells were superior to ICD cells in transferring cellular material to DCs, both from the cytosol and cytoplasmic membrane. Moreover, senescent cancer cells promoted a better activation and maturation of DCs and were more efficient in mediating CD8 T-cell activation against a specific experimental antigen (OVA). As expected, the combination of these features resulted in a stronger activation of antigen-specific CD8 T cells by senescent cells compared with ICD or nonsenescent cells.
Regarding antigenicity, we initially focused on the expression of MHC-I, which we identified upregulated in an unbiased proteomic screening comparing the plasma membrane proteins of senescent and nonsenescent cells. Remarkably, senescent cells also upregulated the MHC-I–associated presentation machinery, including its master transcriptional regulator, Nlrc5 (34). In agreement with previous observations (32), we also found that the IFN transcriptomic signature was elevated in senescent cells. Interferons are considered main inducers of MHC-I presentation (33), and we show that IFN signaling, specifically type I, is a key mediator of MHC-I upregulation in senescent cells in vitro. Of note, while type I IFNs are produced by all cell types in response to damage and senescence, IFNγ is produced almost exclusively by immune cells. Therefore, in an in vivo context, IFNγ may also contribute to the MHC-I upregulation observed in senescent cells, as recently shown (51).
Importantly, we show that immunization with noncancer syngeneic senescent fibroblasts triggers CD8 T-cell responses against both senescent and nonsenescent fibroblasts. To pinpoint senescence-specific responses, we wondered if we could identify senescence-associated MHC-I peptides. The immunopeptidome presented by the MHC-I of a given individual cell reflects a minority (at most 1%) of all possible peptides from the proteome (50), and it is highly dynamic depending on the cellular context, particularly under conditions of stress (43, 52). We speculated that senescent cells, even if derived from noncancer cells, could be enriched in senescence-associated self-peptides. Indeed, by isolating and sequencing the immunopeptidome of senescent cells, we demonstrate the presence of senescence-associated self-peptides absent in their parental nonsenescent cells or in the Mouse Immunopeptidome Atlas (which includes >30,000 peptides normally present in a large collection of murine tissues and cancer cell lines; ref. 40). Moreover, CD8 T cells from mice immunized with senescent cells were activated ex vivo when exposed to senescence-associated self-peptides. This was not the case when mice were immunized with nonsenescent parental cells. These observations indicate that senescent cells have the capacity to elicit CD8 T-cell responses partly evoked against senescence-associated self-antigens.
Having demonstrated that the induction of senescence increases the immunogenicity of noncancer cells, we extended this concept to cancer cells. Indeed, previous work has shown that senescent cancer cells can trigger antitumor protection when used in a vaccination setting (13). We reinforce this concept by showing that senescent cancer cells are superior to ICD cancer cells in triggering protection against a subsequent rechallenge with untreated cancer cells. As expected, antitumor protection involved a classic adaptive immune response mediated by antigen-presenting cells (APC) and CD8 T cells. Moreover, we compared the immunogenic potential of live versus dying senescent cancer cells and observed that live senescent cells were more efficientin eliciting antitumor protection. This finding further reinforces the idea that senescent cells play an active role in triggering an efficient immune response. We also applied this strategy in a therapeutic setting by immunizing tumor-bearing mice. In this case, immunization significantly delayed B16F10 melanoma growth, which is considered a highly immunosuppressive tumor model (48). This delay in tumor growth was accompanied by an increased infiltration of CD8 T cells with a superior activation phenotype.
The above concepts were corroborated in a human setting based on cancer cells and autologous TILs from four patients (oropharyngeal, hypopharyngeal, endometrial, and melanoma). We found that the coculture of TILs with senescent cancer cells evokes a stronger antigen-dependent activation of CD8 T cells than their nonsenescent parental cancer cells. This was also demonstrated in the case of TILs that recognize a single cancer-derived mutated antigen. In parallel to our work, similar findings have been reported in human acute myeloid leukemia, where therapy-induced senescence also triggers antigen presentation and antitumor immunity (53).
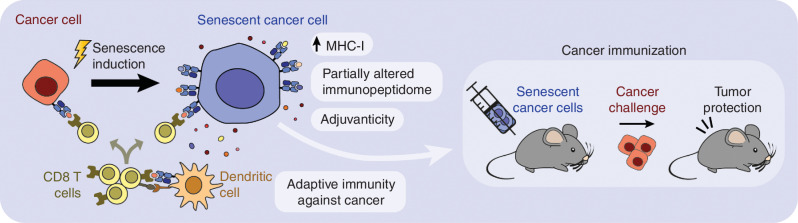
In summary, our findings demonstrate that senescent cells are strongly immunogenic owing to the combination of several features: their long-term persistence in vivo, release of adjuvant factors, activation of IFN signaling, efficient antigen transfer and activation of APCs, upregulated antigen presentation, and altered immunopeptidome (Fig. 6). The upregulation of MHC-I associated with senescence can be a strategy to overcome immune evasion linked to MHC-I downregulation, which remains the primary mechanism through which cancer cells escape CD8 T-cell killing (54, 55). Our results suggest the possibility of improving anticancer vaccination strategies based on senescent cancer cells. Also, in view of the capacity of senescent cells to deliver antigens and activate DCs, senescent cancer cells could be used to generate improved DC-based vaccines. Beyond cancer, many human diseases and aging-associated frailty are partly driven by the presence of senescent cells (1), and it is tempting to speculate that unleashing adaptive immune responses or engineered T cells against senescent cells could have therapeutic and health benefits, as recently shown (56, 57).
Figure 6.
Graphical summary.
METHODS
Cell Culture
SKMEL-103 (human melanoma), IMR-90 (human lung fibroblasts), B16-F10 (B16F10, mouse melanoma), and 293T (human embryonic kidney) cells were obtained from the ATCC. Panc02 (mouse pancreatic adenocarcinoma) cells were kindly provided by Dr. Abdul Azad (Oxford University, United Kingdom). UT-SCC-2 and UT-SCC-42B (human head and neck squamous cell carcinoma) were provided by Dr. Reidar Grenman (University of Turku, Finland). OVA-expressing B16F10 and Panc02 cells were kindly provided by the Kroemer Lab (CRC, Paris, France). Luciferase-expressing B16F10 cells were kindly provided by the Soengas Lab (CNIO, Madrid, Spain). Primary MEFs were obtained from C57BL/6 embryos at E13.5 as previously described (58). Panc02 cells and all primary immune cells were maintained in a standard RPMI medium. All the other cell lines were maintained in standard DMEM. All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco) and 1% antibiotics (penicillin/streptomycin 100 U/mL; Gibco). Cells were maintained in a humidified incubator at 37°C and 5% CO2. Cells were tested monthly for Mycoplasma contamination using standard PCR, and only negative cells were used.
Senescence was induced by treatment with the DNA-damaging agents doxorubicin (100 or 200 nmol/L; Sigma, #D1515) or bleomycin (3 or 6 mU/L; Sigma, #B8416) for 48 hours, the CDK4/6 inhibitor palbociclib (1 μmol/L; PD033299, Absource Diagnostic, #S1116), or the p53 activator nutlin-3A (5 μmol/L; Sigma, #SML0580), as indicated. Seven to 10 days after the beginning of the treatment, senescent cells were collected and used for experiments. Late passage senescence was triggered by the repeated subculture of nontransformed primary cells until proliferation stopped. ICD was induced by treatment with a high dose of doxorubicin (5 μmol/L) for 18 hours. Senescent cell death was induced by treatment with the senolytic agent navitoclax (10 μmol/L; Quimigen, #HY-10087) for 18 hours.
For experiments blocking IFNγ or IFNAR1, blocking antibodies against IFNy (InVivoMAb, #BE0055), IFNRA-1 (InVivoMAb, #BE0241), or their respective IgG1 isotypes control (InVivoMAb, #BE0088 and #BE0083) were added at 50 μg/mL to the cultured media at the indicated day and refreshed every 3 days. For chemical inhibition of JAK signaling in MEFs, cells were treated with the JAK inhibitor SAR-20347 (Selleckchem, #S0437), which was added at a concentration of 2 μmol/L to the culture media on the indicated day; for inhibition in B16F10 or Panc02 cells, JAK inhibitor I (Calbiochem; Sigma, #42099) was added to the culture media at a concentration of 0.6 μmol/L (B16F10) or 1.2 μmol/L (Panc02) on the indicated day and refreshed every 3 days.
SAβG Assay
Cells were washed with PBS, fixed with 0.2% glutaraldehyde for 10 minutes, washed with PBS, and incubated overnight at 37°C with a staining solution containing 1 mg/mL X-Gal (Melford BioLaboratories, #MB1001) prepared in dimethylformamide (DMF; Sigma, D4551) at pH 6. Cells were then washed in PBS and visualized using a Nikon Eclipse TS2 brightfield microscope.
Plasma Membrane Proteomic Screening
Up to 5 × 106 cells per condition were collected in cold PBS by scraping and pelleted by centrifugation. Plasma membrane proteins were extracted using a plasma membrane protein extraction kit (Abcam, #ab65400) following the manufacturer's instructions.
Proteins were dissolved in UT buffer (8 mol/L urea, 2 mol/L thiourea, 100 mmol/L Tris–HCl pH 8) and digested by means of the standard FASP protocol. Briefly, proteins were reduced [15 mmol/L TCEP for 30 minutes at room temperature (RT)], alkylated (50 mmol/L CAA for 20 minutes in the dark at RT), and sequentially digested with Lys-C (Wako; protein:enzyme ratio 1:50, overnight at RT) and trypsin (Promega; protein:enzyme ratio 1:100 for 6 hours at 37°C). Resulting peptides were desalted using Sep-Pak C18 cartridges (Waters). Liquid chromatography with tandem mass spectrometry (LC-MS/MS) was done by coupling an UltiMate 3000 RSLCnano LC system to either a Q Exactive HF or Q Exactive HF-X-mass spectrometer (Thermo Fisher). In both cases, peptides were loaded into a trap column (Acclaim PepMap 100 C18 LC Columns 5 μm, 20 mm length) for 3 minutes at a flow rate of 10 μL/minute in 0.1% formic acid (FA). Then, peptides were transferred to an EASY-Spray PepMap RSLC C18 column (Thermo Fisher; 2 μm, 75 μm × 50 cm) operated at 45°C and separated using a 90-minute effective gradient (buffer A: 0.1% FA; buffer B: 100% ACN, 0.1% FA) at a flow rate of 250 nL/minute. The gradient used was from 4% to 6% of buffer B in 2.5 minutes, from 6% to 25% B in 72.5 minutes, and from 25% to 42.5% B in 14 minutes plus 6 additional minutes at 98% B. Peptides were sprayed at 1.8 kV into the mass spectrometer via the EASY-Spray source, and the capillary temperature was set to 300°C. The Q Exactive HF was operated in a data-dependent mode with an automatic switch between MS and MS/MS scans using a top 15 method (intensity threshold ≥ 6.7e4, dynamic exclusion of 26.25 seconds and excluding charges +1 and > +6). MS spectra were acquired from 350 to 1,400 m/z with a resolution of 60,000 FWHM (200 m/z). Ion peptides were isolated using a 2.0 Th window and fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 27. MS/MS spectra resolution was set to 15,000 or 30,000 (200 m/z). The ion target values were 3e6 for MS (maximum IT of 25 ms) and 1e5 for MS/MS (maximum IT of 15 or 45 ms). The Q Exactive HF-X was operated in a data-dependent mode with an automatic switch between MS and MS/MS scans using a top 12 method (intensity threshold ≥ 3.6e5, dynamic exclusion of 34 seconds and excluding charges +1 and > +6). MS spectra were acquired from 350 to 1,400 m/z with a resolution of 60,000 FWHM (200 m/z). Ion peptides were isolated using a 1.6 Th window and fragmented using HCD with a normalized collision energy of 27. MS/MS spectra resolution was set to 15,000 (200 m/z). The ion target values were 3e6 for MS (maximum IT of 25 ms) and 1 × 105 for MS/MS (maximum IT of 22 ms). Raw files were processed with MaxQuant using the standard settings against either a human protein database (UniProtKB/Swiss-Prot, 20,373 sequences) or a mouse database (UniProtKB/TrEMBL, 53,449 sequences). Carbamidomethylation of cysteines was set as a fixed modification, whereas oxidation of methionines and protein N-term acetylation were set as variable modifications. Minimal peptide length was set to 7 amino acids, and a maximum of two tryptic missed-cleavages were allowed. Results were filtered at 0.01 FDR (peptide and protein level). Afterward, the “proteinGroups.txt” file was loaded in Prostar (59) using the label-free quantitation intensity values for further statistical analysis. Briefly, proteins with less than 75% valid values in at least one experimental condition were filtered out. When needed, a global normalization of log2-transformed intensities across samples was performed using the LOESS function. Missing values were imputed using the algorithms SLSA (60) for partially observed values and DetQuantile for values missing on an entire condition. Differential analysis was performed using the empirical Bayes statistics Limma. Proteins with a P < 0.05 and a log2 ratio >0.58 (1.5 in nonlog scale) were defined as upregulated. The FDR was estimated to be below 5%. Upregulated proteins in senescent cells following the indicated criteria in four or more conditions of senescence were selected, and GO analysis was performed using the GOrilla Gene Ontology tool (61).
Flow Cytometry Analysis
For analysis of cultured cell lines, single cells were digested into single cells by trypsinization (0.25% trypsin-EDTA, Invitrogen). Nonadherent primary immune cells maintained in suspension were collected from the culture by pipetting. Blood was collected in EDTA-coated tubes (Microvette, #16.444) to assess immune cell depletion and incubated with red blood cells (RBC) lysis buffer (BioLegend, #42031) for 5 minutes at RT. Cells were resuspended in fluorescence-activated cell sorting (FACS) buffer (5 mmol/L EDTA and 0.5% BSA in PBS). Cell viability was assessed using DAPI (0.1 μmol/L, Molecular Probes, #D1306) or Live/Dead Fixable Yellow dye (Invitrogen, #L34967) following the manufacturer's instructions. Dead cells were excluded from the analysis. Mouse cells were incubated with mouse BD Fc Block containing purified antimouse CD16/CD32 mAb 2.4G2 (1:400, BD Biosciences, #553142) for 10 minutes at 4°C. Cells were washed and then stained with the appropriate antibody (Supplementary Table S5) for 40 minutes at 4°C. Cell suspensions were run on a Gallios Beckman Coulter flow cytometer (BD Biosciences). Autofluorescence signal from the unstained samples was obtained and subtracted from each sample in all experiments. Data were analyzed using FlowJo v10 software.
RNA Extraction, RNA-seq Library Preparation, and Sequencing
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, #QIA74106) following the manufacturer's instructions.
For MEF, Panc02, and SKMEL-103 RNA-seq analysis, library preparation and quality control were performed at the Genomics Facility of IRB Barcelona. The total RNA concentration was quantified using the Qubit RNA HS Assay kit (Invitrogen, #Q32852), and RNA integrity was assessed using the Bioanalyzer 2100 RNA Nano assay (Agilent). RNA-seq libraries were prepared at the IRB Barcelona Functional Genomics Core Facility. Briefly, mRNA was isolated from 1.1 μg total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). Isolated mRNA was used to generate dual-indexed cDNA libraries using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Eight cycles of PCR amplification were applied to all the libraries. The final libraries were quantified using the Qubit dsDNA HS assay (Invitrogen), and the quality was controlled using the Bioanalyzer 2100 DNA HS assay (Agilent). An equimolar pool was prepared using the 13 libraries and submitted for sequencing at the National Centre for Genomic Analysis (CRG-CNAG). Final quality control by qPCR was performed by the sequencing provider before paired-end 150 nt sequencing on a NovaSeq6000 S4 (Illumina). The sequencing results exceeded 312 Gbp, with a minimum of 53.97 million paired-end reads sequenced for each sample. Adapters were trimmed from the initial sequences using Cutadapt (62), version 1.18. Trimmed paired-end reads were then aligned to the mm10 version of the mouse genome using STAR (63) version 2.3.0e under default parameters. SAM files were sorted and indexed using Sambamba (64) version 0.5.9. The R package Casper (ref. 65; version 2.16.1; https://www.r-project.org/) was used to quantify the intensities at the transcript and gene levels. The Ensembl database was used for the transcript annotation. Differential expression between senescence conditions and control was performed using the limma R package (66), version 3.38.2, on the gene-level intensities using the replicate Id as an adjusting covariate to account for paired samples. Genes were annotated to hallmark terms (67) using the R package (version 3.7.0, org.Hs.eg.db). Gene set analysis was then performed to filter out low-expression genes (genes with less than an average count of 5 reads). The rotation-based approach for enrichment (68) implemented in the R package limma was used to represent null distribution. The maxmean enrichment statistic proposed in ref. 69 under restandardization was considered for competitive testing.
For B16F10 and IMR-90 cell RNA-seq analysis, total RNA was subjected to quality control with Agilent Tapestation according to the manufacturer's instructions. To construct libraries suitable for Illumina sequencing the Illumina Stranded mRNA Prep, ligation preparation protocol was used, which includes mRNA isolation, cDNA synthesis, ligation of anchors, and amplification and indexing of the libraries. The yield and quality of the amplified libraries were analyzed using Qubit by Thermo Fisher and the Agilent Tapestation. The indexed cDNA libraries were normalized and combined, and the pools were sequenced on the Illumina Nextseq 2000 P2 flowcell for a 120-cycle paired-end sequencing run generating 60 bp paired-end reads. For analysis, Bcl files were converted and demultiplexed to fastq using the bcl2fastq v2.20.0.422 program. STAR 2.7.9a was used to index the mouse (mm10/GRCm38) and human (hg38/GRCh38) reference genomes and align the resulting fastq files. Mapped reads were then counted in annotated exons using featureCounts v1.5.1. The gene annotations (Mus_musculus.GRCm38.99.gtf; Homo_sapiens.GRCh38.101.gtf) and reference genome were obtained from Ensembl. The count table from featureCounts was imported into R/Bioconductor, and differential gene expression was performed using the EdgeR package and its general linear models’ pipeline and DESeq2. For the gene expression analysis, genes were filtered to remove non- or very low expressing genes and normalized using the default normalization for each analysis package. Genes with an FDR < 0.05 were termed significantly regulated. Gene set enrichment analysis was performed using GSEA 4.2.3.
Gene Expression Analysis by RT-qPCR
Total RNA was extracted from cell samples using TRIzol reagent (Invitrogen, #15596018) according to the manufacturer's instructions. Up to 1 μg of total RNA was reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, #1725038) or gDNA Clear iScript cDNA Synthesis Kit for patient-derived tumor cells (Bio-Rad, #1725034) for RT-qPCR. Quantitative real-time PCR was performed using GoTaq PCR Master Mix (Promega, #A6002) or PowerUp SYBR Green Master Mix (Thermo Fisher, #A25741) for patient-derived tumor cells in a QuantStudio 6 Flex thermicycler (Applied Biosystems) 7900HT Fast Real-Time PCR System (Applied Biosystems). The average expression of both endogenous Actb and Gapdh in mouse and ACTB and GAPDH in human cell lines served as endogenous normalization controls. For patient-derived tumor cells, expression of the endogenous GAPDH gene served as an endogenous normalization control. Primers used in this study are listed in Supplementary Table S5.
Immunopeptidome
Up to 108 cells per condition were collected by trypsinization and pelleted by centrifugation. Cells were washed with PBS, thawed at −80°C, and shipped to the CHU Sainte-Justine Research Center (Montréal, Canada) for immunopeptidomics analysis using mass spectrometry (70).
For sample preparation, 1.5-mL and 2-mL microcentrifuge tubes (Protein LoBind Eppendorf, #022431081 and #02243100), low retention tips (Eppendorf; 10 μL #2717349, 20 μL #2717351, and 200 μL #2717352), acetonitrile (#A9964), trifluoroacetic acid (TFA, #AA446305Y), FA (#AC147930010), CNBr-activated sepharose 4 B (#45000066), and ammonium bicarbonate (#A643-500) were purchased from Fisher.
To isolate MHC-I peptides, a frozen pellet of cells was thawed by warming the bottom of the tube with a palm. The cell pellet was resuspended in 500 μL of PBS (Buph, #28372) by pipetting up and down until homogenization. An equivalent volume to the cell pellet suspension of cell lysis buffer [1% CHAPS (#22020110GM) in PBS containing protease inhibitors (Bio-Rad, #A32961), 1 pellet/10 mL] was added to the cell suspension, followed by incubation for 60 minutes at 4°C using a rotator device (10 RPM) and centrifugation at 18,000 × g for 20 minutes at 4°C. The cell lysis supernatant containing the MHC–peptide complexes was transferred to a new 2-mL microcentrifuge tube and kept on ice until use for immunopurification.
Bead coupling and immunopurification of MHC-I peptides were performed as previously described (71). Briefly, 80 mg of sepharose CNBr-activated beads were coupled with 2 mg of antimouse H2 antibody (M1/42.3.9.8; Bio X Cell, #BE0077). Sepharose antibody-coupled beads were incubated with the cell lysate supernatant in a 2-mL low binding microcentrifuge tube overnight at 4°C with rotation (22 RPM). The next day, a Bio-Rad polyprep chromatography column(Bio-Rad, #7311553) was installed on a rack and prerinsed with 10 mL of buffer A (150 mmol/L NaCl and 20 mmol/L Tris–HCl pH 8). The bead–lysate mixture was transferred into a Bio-Rad column, and the bottom cap was removed to discard the unbound cell lysate. The beads retained in the Bio-Rad column were washed sequentially with 10 mL of buffer A (150 mmol/L NaCl and 20 mmol/L Tris–HCl pH 8), 10 mL of buffer B (400 mmol/L NaCl and 20 mmol/L Tris–HCl pH 8), 10 mL of buffer A, and 10 mL of buffer C (20 mmol/L Tris–HCl pH 8). MHC–peptide complexes were eluted from the beads by adding 300 μL of 1% TFA, pipetting up and down 4 to 5 times, and collecting the flow-through. This step was repeated once, and the flow-throughs were collected and combined.
MHC-I peptides were desalted and eluted on a C18 solid phase extraction disk ultramicrospin C18 column (#SEMSS18V, 5–200 μL). MHC-I peptides were desalted and eluted on a C18 column preconditioned with 200 μL of (i) methanol, (ii) 80% acetonitrile/0.1% TFA, and (iii) 0.1% TFA, and spun at 1,545 × g in a fixed rotor to collect and discard the flow-throughs. Then, the MHC–peptide complexes previously collected in 600 μL of 1% TFA were loaded (3 × 200 μL) into the preconditioned C18 column and spun, and the flow-throughs were discarded. The final wash was performed with 200 μL of 0.1% TFA and spun again. Finally, the C18 column was transferred onto a 2-mL Eppendorf tube, and MHC-I peptides were eluted with 3 × 200 μL of 28% ACN, 0.1% TFA. The flow-through containing the eluted peptides was stored at −20°C for MS analysis. Prior to LC-MS/MS analysis, the purified MHC-I peptides were evaporated to dryness using a vacuum concentrator with presets of temperature 45°C for 2 hours, vacuum level: 100 mTorr, and vacuum ramp: 5.
Vacuum samples were resuspended in 52 μL of 4% FA, and each biological replicate was divided into 3 technical replicates of 16 μL each. Each replicate was loaded and separated on a homemade reversed-phase column (150 μm i.d. × 250 mm length, Jupiter 3 μm C18 300 Å) with a gradient from 10% to 30% ACN, 0.2% FA and a 600-nL/minute flow rate on an Easy nLC-1000 connected to an Orbitrap Eclipse (Thermo Fisher). Each full MS spectrum was acquired at a resolution of 240,000, an AGC of 4E5, and an injection time of 50 ms, followed by tandem-MS (MS-MS) spectra acquisition on the most abundant multiply charged precursor ions for a maximum of 3 seconds. Tandem-MS experiments were performed using HCD at a collision energy of 34%, a resolution of 30,000, an AGC of 1.5E4, and an injection time of 300 ms. Data files were processed using PEAKS X software (Peaks Pro V10.6, Bioinformatics Solutions) using the mouse database UniProtKB/Swiss-Prot (2019_09). “Unspecified enzyme digestion” was selected for the enzyme parameter, and mass tolerances on precursor and fragment ions were 10 ppm and 0.01 Da, respectively. All other search parameters were the default values. Final peptide lists were filtered using ALC of 80% and with an FDR of 1% using the Peaks software.
Only captured peptides between 8- and 12-mers (both included) and those predicted to be weak or strong binders to either H-2Kb or H-2Kd (NetMHCPan v4.0) were considered for the analysis (bona fide peptides). We combined all bona fide peptides detected as senescent MEFs and removed all peptides detected in nonsenescent cells. Peptides present in other tissues and cancer cell lines of C57BL/6 mice were obtained from the Mouse Immunopeptidome Atlas (40) and subtracted to filter our list of candidate peptides. Cross-analysis to identify unique peptides was performed using Venn diagrams with VENNY tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html). The values indicated for each peptide are the log2(area) obtained from the mass spectrometry. For the list of candidate peptides, we explored the senescent MEF versus MEF differences in mRNA for their predicted genes (under reverse translation) using RNA-seq data, giving extra priority to those peptides whose underlying genes were upregulated in senescent cells from the RNA-seq analysis.
Mice
All mice were maintained at the animal facility of the Scientific Parc of Barcelona (PCB) in strict adherence to Spanish and European Union regulations. Animal experiments were approved by the Animal Care and Use Ethical Committee of the PCB and the Catalan Government. Mice were maintained under specific pathogen–free conditions. Food and water were provided ad libitum. All in vivo experiments were performed using female C57BL/6 or C57BL/6-Tg(TcraTcrb)1100Mjb/Crl (OT-I) mice of 8 to 16 weeks of age, which were randomly allocated in the different groups of study. All mice were purchased from Charles River.
Mouse Immunizations
For immunization with noncancer cells, 8- to 12-week-old C57BL/6 mice were subcutaneously injected with 106 untreated or senescent MEFs resuspended in 100 μL PBS together with 25 μg of CpG immune adjuvant [synthetic oligodeoxynucleotide (ODT) containing unmethylated CpG motifs; ODT1825; Vaccigrade, #ac-1826-1] on days 0 and 7 on the left and right dorsal flanks, respectively. As negative controls, animals received only the vehicle plus adjuvant, and as a positive control, animals were immunized with 100 μg of OVA (Merck Life Sciences, #A5503) in a total volume of 100 μL PBS. One week after the last immunization, the animals were euthanized by CO2, and their spleens were used for the analysis of immune responses.
To analyze the adjuvant properties of cancer cells, 8- to 12-week C57BL/6 mice were subcutaneously injected with vehicle (PBS), 106 ICD cells, or 2 × 105 senescent cells resuspended in 100 μL PBS on day 0 on the left flank. On day 7, the subcutaneous inoculation site was dissected and subjected to histologic analysis.
For prophylactic immunization with cancer cells, 8- to 12-week-old C57BL/6 mice were subcutaneously injected with 106 ICD cells or 2 × 105 senescent cells (either live or dead, as indicated) resuspended in 100 μL PBS on day 0 on the left flank. On day 7, the animals were subcutaneously rechallenged with 3 × 105 live cancer cells in their right flank. Tumor appearance and growth on the right flank were monitored afterward.
For the depletion of immune populations, animals received 100 μg of blocking antibodies [anti-CD4 (clone GK1.5; Bio X Cell, #BP0003), anti-CD8 (clone 2.43; Bio X Cell, #BE0061), anti-CD11b (clone M1/70, Bio X Cell; #BE0007)] or isotype control IgG2b (clone LTF-2; Bio X Cell, #BP0090) via intraperitoneal injection of a total volume of 100 μL on days −1, 3, and 8 of the immunization experiments.
For therapeutic vaccination, the animals were subcutaneously injected with 5 × 105 live cancer cells in the right flank. When tumors were visible and palpable (from 40 mm3), animals were immunized subcutaneously with 106 ICD cells or 2 × 105 senescent cells (either live or dead, as indicated) resuspended in 100 μL PBS on the left flank. Tumor growth on the right flank was monitored afterward.
Tumors were measured using a caliper, and their volumes (v) were calculated using the formula v = (l × w2)/2 (where l is tumor length and w is tumor width). The measurements are shown in mm3. When tumors reached a size of 1,000 mm3 or became ulcerated, mice were euthanized by CO2 and samples were collected for further analysis.
For studying the persistence of cells after injection, 8- to 12-week-old C57BL/6 mice were subcutaneously injected with 106 luciferase-expressing untreated, ICD, cells or senescent cells resuspended in 100 μL PBS on the back. In a second experiment, the number of injected cells was adjusted in terms of protein, and 8- to 12-week-old C57BL/6 mice were subcutaneously injected with 106 luciferase-expressing ICD cells or 2 × 105 senescent cells resuspended in 100 μL PBS on the back. On the indicated day, mice received an intraperitoneal injection of 75 mg/kg luciferin. Bioluminescence was recorded 10 minutes after injection using an IVIS Spectrum Imaging System (PerkinElmer). Quantification was performed using Living Image 3.2 software (PerkinElmer).
Histologic Analysis
For formalin-fixed, paraffin-embedded samples, tissues were fixed overnight at 4°C with neutral buffered formalin (Sigma-Aldrich, #HT501128). Paraffin-embedded tissue sections (2–3 μm) were air-dried and dried overnight at 60°C.
For hematoxylin–eosin (H&E) staining, paraffin-embedded tissue sections were dewaxed and stained according to the H&E standard protocol using a CoverStainer (Dako, Agilent).
Immunohistochemistry for CD45 (30-F11; Thermo Fisher, #14-0451-82) at 1:100 for 60 minutes was performed using a Ventana Discovery XT, CD3 (Dako-Agilent, #IR503) at 1:10 for 120 minutes, CD11b (EPR1344; Abcam, # ab133357) at 1:6,000 for 120 minutes, and CD8α (EPR20305; Abcam, #ab209775) at 1:1,000 for 120 minutes with the Leica BOND RX. For CD45, antigen retrieval was performed using cell conditioning 1 (CC1) buffer (Roche, #950-124), followed by rabbit anti-Rat (Vector, #AI-4001) at 1:500 for 32 minutes and OmniMap anti-Rb HRP (Roche, #760-4311). Blocking was performed using casein (Roche, #760-219). Antigen–antibody complexes were revealed using the Discovery Purple Kit (Roche, #760-229). For CD3, CD11b, and CD8α, antigen retrieval was performed with BOND Epitope Retrieval 2 (ER2; Leica, #AR9640) for 40, 20, and 20 minutes, respectively, followed by Bond Polymer Refine Red Detection (Leica, #DS9390) without the post-primary for 30 minutes. The sections were mounted with mounting medium and toluene-free (Agilent, #CS705) using a Dako CoverStainer. The specificity of staining was confirmed by staining with rat IgG (R&D Systems, Biotechne, #6-001-F) or rabbit IgG (Abcam, #ab27478) as an isotype control. Brightfield images were acquired using a NanoZoomer-2.0 HT C9600 digital scanner (Hamamatsu) equipped with a 20× objective. All images were visualized with a gamma correction set at 1.8 in the image control panel of the NDP.view 2 U12388–01 software (Hamamatsu, Photonics). Representative images were selected by an independent histopathologist.
The semiquantitative analysis of CD45 infiltration (infiltration score) was performed by an independent histopathologist. Briefly, two halves were assessed for each sample, and the half used to perform the analyses was the one that presented a greater foci/number of tumor cells. The ratio of positive cells was performed considering total inflammatory cells within or closely surrounding the tumoral foci. The semiquantitative score was established as follows: 0 to 10% (0), 11% to 30% (1), 31% to 60% (2), or 61% to 100% (3). For the quantification of infiltrating CD8 T cells, a representative area of determined size was blindly selected for every sample and automatically quantified using QuPath Software o.1.2 (72).
Identification of Immune Cell Populations by Mass Cytometry (CyTOF)
Single-cell suspensions were prepared from B16F10 tumors. The tumors were briefly kept in DMEM on ice and manually minced in DMEM. The tumor fragments were incubated in 1 mg/mL collagenase I (Thermo Fisher, #17100017), 1 mg/mL collagenase IV (Thermo Fisher, #LS004188), and 20 U/mL DNase I (Merck, #D4513) in DMEM for 30 minutes, at 37°C, with gentle shaking, in gentleMACS C tubes, with several steps of processing in a gentleMACS Dissociator before and after the incubation following the manufacturer's instructions. The resulting cell suspension was passed through a 70-μm strainer, washed in PBS, incubated in RBC lysis buffer (BioLegend, #420301) for 5 minutes at RT, and then washed with DMEM a second time. Then, cells were stained following the Maxpar Nuclear Antigen Staining with Fresh Fix protocol (Fluidigm, #400277). Briefly, cells were stained for viability using Cell-ID Cisplatin-185Pt (Fluidigm, #201195) following the manufacturer's instructions. Then, cells were incubated with anti-mouse CD16/CD32 at a 1:400 ratio. Cells were stained using the indicated surface and intracellular antibodies (Supplementary Table S5) following the manufacturer's instructions. The cell suspension was fixed in 1.6% paraformaldehyde (Aname, #15710) in PBS for 10 minutes at RT. The samples were then resuspended in 1 mL of 125 nmol/L Cell-ID Intercalator-Ir (Fluidigm, #201192A) in Maxpar Fix and Perm Buffer (Fluidigm, #201067), incubated for 1 hour at RT, and cryopreserved at −80°C following the manufacturer's instructions. The acquisition was performed following the manufacturer's instructions in a Helios detector (Fluidigm). The gating strategy is fully standardized and described in detail by the manufacturer. Briefly, Intercalator-Ir–positive cells were selected, and only viable immune cells (negative for cisplatin and positive for CD45+) were analyzed. B cells were defined as CD19+B220+ cells. Next, TCRb+CD3+ T cells were selected, and, within this population, CD4+ and CD8+ T cells were detected. From the TCRb−CD3− population, we identified by sequential exclusion NK1.1+ NK cells and CD11b+CD11c+ DCs. Finally, among CD11b+ cells, macrophages and myeloid-derived suppressor cells were identified based on Gr1+ expression.
ELISpot Assay
To test immune responses in animals immunized with noncancer senescent cells (MEF), splenocytes from naïve [injected with vehicle (PBS) plus adjuvant] and immunized animals (injected with untreated, senescent cells, or OVA plus adjuvant) were collected 7 to 10 days after the last immunization. Briefly, spleens were harvested and mechanically digested into single-cell suspensions. RBCs were lysed using RBC lysis buffer for 5 minutes at RT. Cells were filtered sequencially through 100-μm and 70-μm strainers. After filtration, splenocytes were seeded as the effector cell population at 1.5 × 105 cells per well into mouse IFNγ ELISpot plates (Mabtech, #3321-4APT). Target cells (control or senescent cells) were added and cocultured at a 1:10 ratio (target:splenocytes). Peptides were added at concentrations of 400 nmol/L (when added alone) and 200 nmol/L (when added in pools) to the cultures. The OVA-derived peptide SIINFEKL (Sigma, #S795) was used as the positive control (400 nmol/L). Peptides obtained from immunopeptidomes were synthesized by Pepscan. Stimulation was maintained for 20 hours. Afterward, plates were developed according to the manufacturer's protocol. They were scanned and quantified using Elispot 7.0 iSpot software in an ImmunoSpot Plate Reader.
Measurement of ATP Levels
An equal volume of fresh medium was added to 106 cells per condition. After 24 hours, the culture medium was collected, centrifuged, and filtered through a 0.2-μm filter to eliminate cell debris. The medium was analyzed using CellTiter-Glo (Promega, #G7571) according to the manufacturer's protocol. The medium alone was used for background control of luminescence, and the signal was subtracted from the samples.
Measurement of CALR Secretion
A total of 106 cells per condition were washed 3 times with PBS, and FBS-free culture medium was added. After 24 hours, the CM was collected and concentrated using AMICON Ultra-15 tubes (Merck, #UFC900324) by centrifugation for 1 hour at 4°C. Up to 20 μL of concentrated CM per sample was loaded per lane and hybridized using antibodies against CALR (Abcam, #ab2907). Secondary fluorescent reagents (Goat anti-mouse or anti-rabbit IRDye 680RD and 800CW; LI-COR, #926-68070 and #926-32211) were used according to the manufacturer's instructions.
DC Generation
Bone marrow cells were harvested from the femurs and tibias of the C57BL/6 mice. The bone marrow was flushed and centrifuged at 350 × g for 5 minutes. Blood cells were lysed by incubation with RBC lysis buffer for 5 minutes at RT. Cells were filtered sequentially through 100-μm and 70-μm filters to remove aggregates. Cells were resuspended and cultured on 10-cm2 bacterial Petri dishes in RPMI with 20 ng/mL murine GM-CSF (PeproTech, #315-03) and 10 ng/mL murine IL4 (PeproTech, #214-14; for myeloid BMDC generation) or with 200 ng/mL human FLT3L (Miltenyi, #130-190-477; for cross-presenting conventional DC generation; FL-DC, as previously described; ref. 46). On day 3, a fresh medium containing fresh GM-CSF was added to the BMDC culture. The medium was replaced on day 6 with fresh cytokines. DCs were harvested on day 7 (for BMDC) or day 10 (for FL-DC) by gentle pipetting and used for the assays.
Assessment of DC Activation
Untreated, ICD, and senescent cancer cells were cocultured with DCs at a 1:1 ratio for 24 hours. Next, DCs were analyzed by flow cytometry for expression of DC activation and maturation markers. For transwell experiments, BMDCs were seeded in the lower chamber of 0.4-μm transwells (Corning Life Sciences, #3413) and cancer cells in the upper one in the same conditions as above. As a positive control for DC activation, DCs were treated with 100 ng/mL LPS (Sigma, #L2630).
Assessment of BMDC Antigen Capture
Untreated, ICD, and senescent cancer cells were stained using the fluorescent cytosolic dye CellTrace CFSE (Thermo Fisher, #C34554) or the fluorescent membrane-dye wheat germagglutinin (WGA)-Alexa Fluor 647 (Thermo Fisher, #W32466), following the manufacturer's instructions. Fluorescently labeled cancer cells were then cocultured with BMDCs at a 1:1 ratio for 18 hours, and BMDCs were analyzed by flow cytometry for the acquisition of CFSE or WGA-Alexa-647.
Assessment of OT-I CD8 T-Cell Activation by DCs
Splenocytes from OT-I mice were obtained as previously explained. CD8 T cells were purified using a CD8 T-cell isolation kit (Miltenyi, #130-104-075) following the manufacturer's instructions.
Untreated, ICD, and senescent cancer cells expressing OVA were cocultured with DCs at a 1:1 ratio for 24 hours. Then, DCs were sorted by CD45 and CD11c expression and cocultured at a 1:10 (DC:T-cell) ratio for 48 hours. CD8 T-cell activation was assessed by measurement of CD69 T-cell activation marker in CD3+CD8+ cells by flow cytometry.
Patient Characteristics and Patient-Derived Tumor Samples
Tumor specimens were obtained from two patients with metastatic head and neck cancer (VHIO-008, who had metastatic hypopharyngeal carcinoma, and VHIO-009, who had metastatic oropharyngeal carcinoma), one patient with endometrial cancer (VHIO-35035, classified as POLE molecular subtype), and another one with metastatic melanoma (VHIO-088). VHIO-008, -009, and -088 patients were refractory to standard lines of therapy prior to sample procurement. VHIO-35035 had not received any prior treatments. All samples were obtained from patients enrolled in two studies approved by the Vall d'Hebron Hospital ethical committee [PR(AG)252/2016, PR(AG)537/2019]. All patients provided written informed consent. Tumor biopsies were cut into small 2- to 4-mm3 fragments to expand independent TIL lines in the presence of 6,000 IU/mL IL2. Tumor cell lines were established by culturing one tumor fragment in RPMI 1640 supplemented with 20% human AB serum (Biowest), 100 U/mL penicillin, 100 μg/mL streptomycin, 25 mmol/L HEPES (Thermo Fisher), 10 μg/mL gentamicin, and 1.25 μg/mL amphotericin B at 37°C and 5% CO2. After 1 month, adherent and nonadherent cells were cultured in RPMI 1640 supplemented with 20% FBS (HyClone), 100 U/mL penicillin, 100 μg/mL streptomycin, 25 mmol/L HEPES (Thermo Fisher), 10 μg/mL gentamicin (Lonza), and 1.25 μg/mL amphotericin B (Gibco). Occasionally, we used differential trypsinization to enrich for epithelial cells. Media were changed once a month until the tumor cell line was established. Tumor cell lines were authenticated through whole-exome sequencing. Once established, cell lines were cultured in RPMI supplemented with 10% FBS and 1% antibiotics and maintained as explained above.
Assessment of Autologous Tumor Recognition by Upregulation of 4-1BB
To evaluate tumor recognition by TILs, autologous tumor cells were used as tumor targets in coculture assays. A total of 2 × 104ex vivo expanded TILs or TILs previously enriched for recognition of a specific neoantigen (i.e., MAGEB2p.E167Q and RPL14p.H20Y) identified as previously described (73) were cocultured with 5 × 104 autologous tumor cells. After 20 hours, T-cell recognition was assessed by measuring the upregulation of the activation marker 4-1BB on the surface of T cells by flow cytometry. Briefly, cocultured cells were pelleted, resuspended in a staining buffer, and incubated with the indicated antibodies for 30 minutes (Supplementary Table S5). Cells were washed, resuspended in staining buffer containing propidium iodide, and acquired on a BD LSRFortessa or BD FACSCanto. T-cell reactivities were considered positive when the frequency of 4-1BB by flow cytometry was higher than the control. A positive control was included by stimulating the T cells with the anti-CD3 antibody OKT3 (BioLegend).
Statistical Analysis
Data were analyzed using GraphPad Prism v.9.3.0 software and are represented as mean ± SEM of independent biological replicates. Statistical analyses were performed as described in the figures. Differences were considered significant based on P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Data Availability
Research data supporting this publication are deposited in public repositories. The RNA-sequencing data are available in the Gene Expression Omnibus located at https://www.ncbi.nlm.nih.gov/geo/ with the accession numbers GSE202032, GSE210334, GSE208048, GSE212085, and GSE212112. The mass spectrometry proteomic screen data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD033714. The mass spectrometry immunopeptidomic data are also available at ProteomeXchange with project accession PXD034059. Derived data supporting the findings of this study are available in the supplementary tables as indicated.
Supplementary Material
Proteomic screen of PM-enriched proteins in senescent cells
GO terms enrichment analysis of membrane proteins upregulated in 4 or more senescence conditions.
RNA-seq Differential Expression Analyses
Peptides presented exclusively on senescent cell MHC-I.
Supplementary materials
Senescent cells upregulate MHC-I antigen presentation
Noncancer senescent cells induce an adaptive immune response in vivo and present unique immunogenic peptides
Senescent cancer cells activate dendritic cells
Immunization with senescent cancer cells promotes anticancer immune surveillance
Senescent cancer cells from human patients hyperstimulate autologous reactive TILs
Acknowledgments
We are grateful to Maria Isabel Muñoz for assistance with the animal protocols; to Kevin Kovalchik for help with data sharing; to Francesca Castoldi for help in total RNA extraction for B16F10 and IMR-90 cells; to Fredrik Fagerstrom-Billai, Susann Fält, Anastasios Damdimopoulos, and David Brodin at Bioinformatics and Expression Analysis Core Facility, Karolinska Institute (KI), for assistance in RNA-seq and analysis; to the IRB core facilities (Functional Genomics, Biostatistics/Bioinformatics and Histopathology); and to the PCB (Animal House) for general research support. I. Marin was the recipient of an FPI fellowship from the Spanish Ministry of Science (PRE2018-083381). O. Boix was the recipient of an FPI-AGAUR fellowship from the Generalitat de Catalunya. A. Garcia-Garijo was supported by a PERIS grant (SLT017/20/000131) from the Generalitat de Catalunya. J.A. López-Domínguez and M. Kovatcheva were supported by a fellowship from the Spanish Association Against Cancer (AECC). Work in the laboratory of E. Caron was funded by the Fonds de recherche du Québec – Santé (FRQS), the Cole Foundation, CHU Sainte-Justine, the Charles-Bruneau Foundation, the Canada Foundation for Innovation, the National Sciences and Engineering Research Council (#RGPIN-2020-05232), and the Canadian Institutes of Health Research (#174924). E. Garralda received funding from the Comprehensive Program of Cancer Immunotherapy and Immunology II (CAIMI-II) supported by the BBVA Foundation (grant 53/2021). The M. Abad lab received funding from the Spanish Ministry of Science and Innovation (RTI2018-102046-B-I00A and RTC-2017-6123-1) and the AECC (PRYCO211023SERR). M. Abad was the recipient of a Ramón y Cajal contract from the Spanish Ministry of Science and Innovation (RYC-2013-14747). A. Gros received funding from the Spanish Ministry of Science cofunded by the European Regional Development Fund (ERDF; RTC-2017-6123-1), from the Instituto de Salud Carlos III (MS15/00058), and from CAIMI-II (grant 53/2021) supported by the BBVA Foundation. The work in the laboratory of F. Pietrocola is supported by a KI Starting Grant, a Starting Grant from the Swedish Research Council (2019_02050_3), and grants from the Harald Jeanssons Foundation, the Loo and Hans Osterman Foundation, and Cancerfonden (21 1637 Pj). Work in the laboratory of M. Serrano was funded by the IRB and La Caixa Foundation, and by grants from the Spanish Ministry of Science cofunded by the European Regional Development Fund (SAF-2017-82613-R, RTC-2017-6123-1), the European Research Council (ERC-2014-AdG/669622), Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement of Catalonia (Grup de Recerca consolidat 2017 SGR 282), and the AECC (PRYCO211023SERR).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
N. Prats reports grants from IRB Barcelona during the conduct of the study. J.A. López-Domínguez reports grants from the Spanish Association Against Cancer (AECC) during the conduct of the study. M. Kovatcheva reports grants from Asociación Española Contra el Cáncer (AECC), the European Research Council, and IRB BioMedTec during the conduct of the study, as well as other support from Galapagos NV outside the submitted work. E. Garralda reports personal fees (consulting) from Roche/Genentech, F. Hoffmann-La Roche, Ellipses Pharma, Neomed Therapeutics 1 Inc., Boehringer Ingelheim, Janssen Global Services, Seagen, TFS, Alkermes, Thermo Fisher, Bristol Myers Squibb, MabDiscovery, Anaveon, F-Star Therapeutics, and Hengrui, personal fees (speakers bureau) from Merck Sharp & Dohme, Roche, Thermo Fisher, Lilly, and Novartis, grants from Novartis, Roche, Thermo Fisher, AstraZeneca, Taiho, and BeiGene, and other support from Agios Pharmaceuticals, Amgen, Bayer, BeiGene USA, Blueprint Medicines, Bristol Myers Squibb, Cellestia Biotech, Debiopharm, F. Hoffmann-La Roche, Forma Therapeutics, Genentech, Genmab B.V., GSK, Glycotope Gmbh, Incyte Biosciences, Incyte Corporation, ICO, Kura Oncology, Lilly, S.A., Loxo Oncology, Macrogenics, Menarini Ricerche Spa, Merck Sharp & Dohme de Española, S.A, Nanobiotix, S.A, Novartis Farmaceutica, S.A, Pfizer, SLU, PharmaMar, S.A.U., Pierre Fabre Medicament, Principia Biopharma, Psioxus Therapeutics Ltd., Sanofi, Sierra Oncology, Sotio A.S., and Symphogen A/S outside the submitted work. M. Abad reports grants from the Spanish Ministry of Science and Innovation during the conduct of the study, as well as grants from the Spanish Association of Cancer Research (AECC), La Caixa Foundation, Fundación Mutua Madrileña, the Spanish Ministry of Science and Innovation (RTI2018-102046-B-I00 and SAF2015-69413-R), and the Fero Foundation outside the submitted work. A. Gros reports grants from the Spanish Ministry Science and Innovation, Instituto de Salud Carlos III, and Comprehensive Program of Cancer Immunotherapy and Immunology II (CAIMI-II) supported by the BBVA Foundation during the conduct of the study; personal fees from Achilles Therapeutics, Singula Bio, Genentech, Instil Bio, Pact Pharma, Roche, BioNTech SE, and Vall d'Hebron Institute of Oncology, and grants from Merck KGaA outside the submitted work; and a patent for E-059-2013/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., Tailored Therapeutics, LLC, Cellular Biomedicine Group, Inc., and Geneius Biotechnology, Inc., a patent for E-085-2013/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., and Geneius Biotechnology, Inc., and a patent for E-149-2015/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., and Tailored Therapeutics, LLC. F. Pietrocola reports grants from Karolinska Institute, Vetenskapsrådet (VR MH 2019-02050), Cancerfonden [Cancerfonden (21 1637 Pj)], Harald Jeanssons Stiftelse, and Loo och Hans Ostermans stiftelse for medicinsk forskning during the conduct of the study. M. Serrano reports grants from the Spanish Ministry of Science, the European Research Council, La Caixa Foundation, the Spanish Association for Cancer Research (AECC), and Departament d'Empresa i Coneixement of Catalonia during the conduct of the study, as well as other support from Senolytic Therapeutics, Inc., Rejuveron Senescence Therapeutics, AG, Life Biosciences, Inc., Altos Labs, Inc., and Galapagos NV, and grants from the Milky Way Research Foundation outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
I. Marin: Conceptualization, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. O. Boix: Investigation, methodology. A. Garcia-Garijo: Investigation, methodology. I. Sirois: Investigation, methodology. A. Caballe: Software, formal analysis, methodology. E. Zarzuela: Software, formal analysis, methodology. I. Ruano: Formal analysis, methodology. C. Stephan-Otto Attolini: Visualization. N. Prats: Formal analysis, supervision, methodology. J.A. López-Domínguez: Investigation. M. Kovatcheva: Investigation. E. Garralda: Resources. J. Muñoz: Software, formal analysis, supervision, methodology. E. Caron: Supervision, methodology. M. Abad: Conceptualization, resources, supervision. A. Gros: Conceptualization, resources, supervision. F. Pietrocola: Conceptualization, data curation, formal analysis, supervision, writing–original draft, writing–review and editing. M. Serrano: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014;15:482–96. [DOI] [PubMed] [Google Scholar]
- 2. Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, et al. Cellular senescence: defining a path forward. Cell 2019;179:813–27. [DOI] [PubMed] [Google Scholar]
- 3. Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol Mech Dis 2010;5:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birch J, Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev 2020;34:1565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prata LGPL, Ovsyannikova IG, Tchkonia T, Kirkland JL. Senescent cell clearance by the immune system: emerging therapeutic opportunities. Semin Immunol 2018;40:101275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang L, Lankhorst L, Bernards R. Exploiting senescence for the treatment of cancer. Nat Rev Cancer 2022;22:340–55. [DOI] [PubMed] [Google Scholar]
- 7. Calcinotto A, Kohli J, Zagato E, Pellegrini L, Demaria M, Alimonti A. Cellular senescence: aging, cancer, and injury. Physiol Rev 2019;99:1047–78. [DOI] [PubMed] [Google Scholar]
- 8. Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 2010;18:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Braumüller H, Wieder T, Brenner E, Aßmann S, Hahn M, Alkhaled M, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 2013;494:361–5. [DOI] [PubMed] [Google Scholar]
- 10. Park SS, Choi YW, Kim JH, Kim HS, Park TJ. Senescent tumor cells: an overlooked adversary in the battle against cancer. Exp Mol Med 2021;53:1834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 12. Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell 2008;134:657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meng Y, Efimova EV, Hamzeh KW, Darga TE, Mauceri HJ, Fu YX, et al. Radiation-inducible immunotherapy for cancer: senescent tumor cells as a cancer vaccine. Mol Ther 2012;20:1046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Tuyn J, Jaber-Hijazi F, MacKenzie D, Cole JJ, Mann E, Pawlikowski JS, et al. Oncogene-expressing senescent melanocytes up-regulate MHC class II, a candidate melanoma suppressor function. J Invest Dermatol 2017;137:2197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruscetti M, Morris JP 4th, Mezzadra R, Russell J, Leibold J, Romesser PB, et al. Senescence-induced vascular remodeling creates therapeutic vulnerabilities in pancreas cancer. Cell 2020;181:424–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song P, An J, Zou MH. Immune clearance of senescent cells to combat ageing and chronic diseases. Cells. 2020;9:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kale A, Sharma A, Stolzing A, Stolzing A, Desprez PY, Desprez PY, et al. Role of immune cells in the removal of deleterious senescent cells. Immun Ageing 2020;17:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011;479:547–51. [DOI] [PubMed] [Google Scholar]
- 19. Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell 2013;155:1104. [DOI] [PubMed] [Google Scholar]
- 20. Egashira M, Hirota Y, Shimizu-Hirota R, Saito-Fujita T, Haraguchi H, Matsumoto L, et al. F4/80+ macrophages contribute to clearance of senescent cells in the mouse postpartum uterus. Endocrinology 2017;158:2344–53. [DOI] [PubMed] [Google Scholar]
- 21. Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sagiv A, Burton DGA, Moshayev Z, Vadai E, Wensveen F, Ben-Dor S, et al. NKG2D ligands mediate immunosurveillance of senescent cells. Aging (Albany NY) 2016;8:328–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brighton PJ, Maruyama Y, Fishwick K, Vrljicak P, Tewary S, Fujihara R, et al. Clearance of senescent decidual cells by uterine natural killer cells in cycling human endometrium. Elife 2017;6:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ovadya Y, Landsberger T, Leins H, Vadai E, Gal H, Biran A, et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat Commun 2018;9:5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pereira BI, Devine OP, Vukmanovic-Stejic M, Chambers ES, Subramanian P, Patel N, et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat Commun 2019;10:2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med 2013;210:2057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruscetti M, Leibold J, Bott MJ, Fennell M, Kulick A, Salgado NR, et al. NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 2018;362:1416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arora S, Thompson PJ, Wang Y, Bhattacharyya A, Apostolopoulou H, Hatano R, et al. Invariant natural killer T cells coordinate removal of senescent cells. Med 2021;2:938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Binet F, Cagnone G, Crespo-Garcia S, Hata M, Neault M, Dejda A, et al. Neutrophil extracellular traps target senescent vasculature for tissue remodeling in retinopathy. Science 2020;369. [DOI] [PubMed] [Google Scholar]
- 30. Frescas D, Roux CM, Aygun-Sunar S, Gleiberman AS, Krasnov P, Kurnasov OV, et al. Senescent cells expose and secrete an oxidized form of membrane-bound vimentin as revealed by a natural polyreactive antibody. Proc Natl Acad Sci U S A 2017;114:E1668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reimann M, Schrezenmeier J, Richter-Pechanska P, Dolnik A, Hick TP, Schleich K, et al. Adaptive T-cell immunity controls senescence-prone MyD88- or CARD11-mutant B-cell lymphomas. Blood 2021;137:2785–99. [DOI] [PubMed] [Google Scholar]
- 32. Frisch SM, MacFawn IP. Type I interferons and related pathways in cell senescence. Aging Cell 2020;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raval A, Puri N, Rath PC, Saxena RK. Cytokine regulation of expression of class I MHC antigens. Exp Mol Med 1998;30:1–13. [DOI] [PubMed] [Google Scholar]
- 34. Kobayashi KS, Van Den Elsen PJ. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat Rev Immunol 2012;12:813–20. [DOI] [PubMed] [Google Scholar]
- 35. Reits EA, Hodge JW, Herberts CA, Groothuis TA, Chakraborty M, Wansley EK, et al. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J Exp Med 2006;203:1259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gravett AM, Trautwein N, Stevanović S, Dalgleish AG, Copier J. Gemcitabine alters the proteasome composition and immunopeptidome of tumour cells. Oncoimmunology 2018;7:e1438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oh CY, Klatt MG, Bourne C, Dao T, Dacek MM, Brea EJ, et al. ALK and RET inhibitors promote HLA Class i antigen presentation and unmask new antigens within the tumor immunopeptidome. Cancer Immunol Res 2019;7:1984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stopfer LE, Mesfin JM, Joughin BA, Lauffenburger DA, White FM. Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat Commun 2020;11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, et al. Translation from the 5’ untranslated region shapes the integrated stress response. Science 2016;351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuster H, Shao W, Weiss T, Pedrioli PGA, Roth P, Weller M, et al. A tissue-based draft map of the murine MHC class I immunopeptidome. Sci Data 2018;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Basisty N, Kale A, Jeon O, Kuehnemann C, Payne T, Rao C, et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. SSRN Electron J 2019;1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galluzzi L, Vitale I, Warren S, Adjemian S, Agostinis P, Martinez AB, et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J Immunother Cancer 2020;8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kroemer G, Galassi C, Zitvogel L, Galluzzi L. Immunogenic cell stress and death. Nat Immunol 2022;23:487–500. [DOI] [PubMed] [Google Scholar]
- 44. Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell 2001;106:255–8. [DOI] [PubMed] [Google Scholar]
- 45. Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 2021;21:298–312. [DOI] [PubMed] [Google Scholar]
- 46. Brasel K, De Smedt T, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood 2000;96:3029–39. [PubMed] [Google Scholar]
- 47. Mohapatra ADas TI, Benechet AP, Pattnayak S, Khanna KM, Srivastava PK. Cross-dressing of CD8a + dendritic cells with antigens from live mouse tumor cells is a major mechanism of cross-priming. Cancer Immunol Res 2020;8:1287–99. [DOI] [PubMed] [Google Scholar]
- 48. Wang J, Saffold S, Cao X, Krauss J, Chen W. Eliciting T cell immunity against poorly immunogenic tumors by immunization with dendritic cell-tumor fusion vaccines. J Immunol 1998;161:5516–24. [PubMed] [Google Scholar]
- 49. Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, et al. Isolation of T-cell receptors specifically reactive with mutated tumor-associated antigens from tumor-infiltrating lymphocytes based on CD137 expression. Clin Cancer Res 2017;23:2491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zitvogel L, Perreault C, Finn OJ, Kroemer G. Beneficial autoimmunity improves cancer prognosis. Nat Rev Clin Oncol 2021;18:591–602. [DOI] [PubMed] [Google Scholar]
- 51. Chen HA, Ho YJ, Mezzadra R, Adrover JM, Smolkin R, Zhu C, et al. Senescence rewires microenvironment sensing to facilitate anti-tumor immunity. Cancer Discov 2023;13:432–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Caron E, Vincent K, Fortier MH, Laverdure JP, Bramoullé A, Hardy MP, et al. The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol Syst Biol 2011;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gilioli D, Fusco S, Tavella T, Giannetti K, Conti A, Santoro A, et al. Therapy-induced senescence upregulates antigen presentation machinery and triggers anti-tumor immunity in acute myeloid leukemia. bioRxiv 2022.11.17.515658 [Preprint]. 2022. Available from: 10.1101/2022.11.17.515658. [DOI]
- 54. Cornel AM, Mimpen IL, Nierkens S. MHC class I downregulation in cancer: underlying mechanisms and potential targets for cancer immunotherapy. Cancers (Basel) 2020;12:1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Amor C, Feucht J, Leibold J, Ho YJ, Zhu C, Alonso-Curbelo D, et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 2020;583:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang TW, Johmura Y, Suzuki N, Omori S, Migita T, Yamaguchi K, et al. Blocking PD-L1-PD-1 improves senescence surveillance and ageing phenotypes. Nature 2022;611:358–64. [DOI] [PubMed] [Google Scholar]
- 58. Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16(INK4a). Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 59. Wieczorek S, Combes F, Lazar C, Giai Gianetto Q, Gatto L, Dorffer A, et al. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics 2017;33:135–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bø TH, Dysvik B, Jonassen I. LSimpute: accurate estimation of missing values in microarray data with least squares methods. Nucleic Acids Res 2004;32:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf 2009;10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–2. [Google Scholar]
- 63. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rossell D, Stephan-Otto Attolini C, Kroiss M, Stöcker A. Quantifying alternative splicing from paired-end rna-sequencing data. Ann Appl Stat 2014;8:309–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 2010;26:2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Efron BRT. On testing the significance of sets of genes. Ann Appl Stat 2007;1:107–29. [Google Scholar]
- 70. Caron E, Kowalewski DJ, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry. Mol Cell Proteomics 2015;14:3105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sirois I, Isabelle M, Duquette JD, Saab F, Caron E. Immunopeptidomics: isolation of mouse and human MHC class I- and II-associated peptides for mass spectrometry analysis. J Vis Exp 2021;176:e63052. [DOI] [PubMed] [Google Scholar]
- 72. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022;50:D543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic screen of PM-enriched proteins in senescent cells
GO terms enrichment analysis of membrane proteins upregulated in 4 or more senescence conditions.
RNA-seq Differential Expression Analyses
Peptides presented exclusively on senescent cell MHC-I.
Supplementary materials
Senescent cells upregulate MHC-I antigen presentation
Noncancer senescent cells induce an adaptive immune response in vivo and present unique immunogenic peptides
Senescent cancer cells activate dendritic cells
Immunization with senescent cancer cells promotes anticancer immune surveillance
Senescent cancer cells from human patients hyperstimulate autologous reactive TILs
Data Availability Statement
Research data supporting this publication are deposited in public repositories. The RNA-sequencing data are available in the Gene Expression Omnibus located at https://www.ncbi.nlm.nih.gov/geo/ with the accession numbers GSE202032, GSE210334, GSE208048, GSE212085, and GSE212112. The mass spectrometry proteomic screen data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD033714. The mass spectrometry immunopeptidomic data are also available at ProteomeXchange with project accession PXD034059. Derived data supporting the findings of this study are available in the supplementary tables as indicated.