Abstract
Metabolic processes can influence disease risk and provide therapeutic targets. By conducting genome-wide association studies of 1,091 blood metabolites and 309 metabolite ratios, we identified associations with 690 metabolites at 248 loci; and associations with 143 metabolite ratios at 69 loci. Integrating metabolite-gene and gene expression information identified 94 effector genes for 109 metabolites and 48 metabolite ratios. Using Mendelian Randomization (MR), we identified 22 metabolites and 20 metabolite ratios having estimated causal effect on 12 traits and diseases, including orotate for estimated bone mineral density, alpha-hydroxyisovalerate for body mass index and ergothioneine for inflammatory bowel disease and asthma. We further measured orotate level in a separate cohort and demonstrated that, consistent with MR, orotate levels were positively associated with incident hip fractures. This study provides a valuable resource describing the genetic architecture of metabolites and delivers insights into their roles in common diseases, thereby offering opportunities for therapeutic targets.
Introduction
Metabolites are small molecules that are the intermediate or end products of metabolic reactions. Their levels are influenced by many factors, including genetics, diet and lifestyle, gut microbiota, and diseases1–3. They can also influence disease risk and are the target of therapeutic interventions4. Understanding the causal role of metabolites in disease etiology could provide tractable intervention points for therapies. One way to assess the role of metabolites in disease outcomes is through human genetics. The heritability of many metabolite levels is high5,6. This provides the opportunity to undertake Mendelian randomization (MR), which is a method of causal inference that uses genetic variants as instrumental variables to test the role of an exposure (in this case metabolites) in disease outcomes7. Since alleles are randomly assigned at conception, this randomization process generally breaks confounding with most risk factors, decreasing the propensity of confounding to bias results.
Previous genome-wide association studies (GWAS), whole-genome sequencing studies, and whole-exome sequencing studies have characterized the underlying genetic architecture of metabolite levels and implicated target genes1,5,8–22. Sequencing studies, in particular, can identify functional variants, yet have often been limited by sample size or the number of metabolites tested. Thus, larger studies, providing a more thorough set genetic determinants of more metabolites, could identify causal influences of metabolites upon diseases.
Here, we undertook a series of large GWASs, comprising 1,091 metabolites and 309 metabolite ratios in 8,299 individuals from the Canadian Longitudinal Study on Aging (CLSA) cohort. Using genetic signals that were identified to have strong biological plausibility to influence metabolites through known genes, we inferred the causal effect of metabolite levels and ratios on twelve traits and diseases that are predominantly influenced by different mechanisms (aging, metabolism, and immune response, respectively). We selected estimated bone mineral density (eBMD) from ultrasound measurements, Alzheimer’s disease, Parkinson’s disease, and osteoarthritis as outcomes influenced by aging; body mass index (BMI), coronary artery disease (CAD), ischemic stroke, and type 2 diabetes (T2D) as outcomes influenced by metabolism; and type 1 diabetes (T1D), inflammatory bowel disease (IBD), multiple sclerosis and asthma as outcomes influenced by immune responses. We also directly measured a lead candidate metabolite from our eBMD MR studies in a separate prospective nested case-control study on hip fractures.
Results
Genome-wide associations of blood metabolites
Of the 1,091 plasma metabolites tested, 850 had known identities across eight super pathways (i.e., lipid, amino acid, xenobiotics, nucleotide, cofactor and vitamins, carbohydrate, peptide, and energy). The remaining 241 were categorized as unknown or “partially” characterized molecules. The current study included 81 metabolites that were not tested in previous representative large metabolomics GWASs5,8,10,11,21, although these metabolites may include previously unnamed metabolites. Detailed cohort characteristics can be found in the published CLSA cohort profile23 and Supplementary Table 1, while metabolite information and their comparison with prior large-scale metabolomics GWAS is shown in Supplementary Tables 2, 3 and 4.
Undertaking a GWAS of metabolite levels identified 1,509 associations for 647 metabolites that passed a stringent Bonferroni correction, adjusted for the total number of tested metabolites (p < 5x10-8/1091 = 4.58x10-11). These included 85 associations for 46 metabolites that were tested uniquely in the current study. Associations passing this stringent Bonferroni correction were used to assess for novel findings (Supplementary Table 5). However, we note that such a multiple-testing correction is overly conservative, given the non-independence of the metabolites (Supplementary Figure 1 and Table 6). We therefore chose to use a more appropriate p-value threshold of 5x10-8 divided by the effective number of independent metabolites (N = 73), leading to a p-value threshold of 6.85x10-10 for subsequent analyses. This relaxed threshold identified 1,702 independent variant-metabolite associations from 690 metabolites that were used to explore the genetic architecture of metabolites and identify effector genes. (for details see Methods, Figure 1, Supplementary Table 5). No sign of excessive test statistic inflation or population stratification was detected by assessment of genomic inflation factors (max lambda=1.03) (Supplementary Table 7).
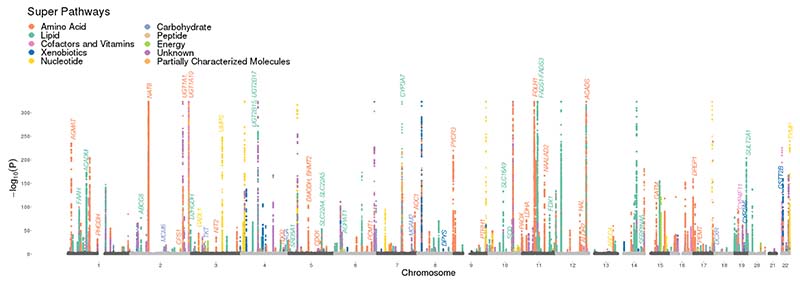
Figure 1. Summary of associations of metabolite levels and genetic loci.
A Manhattan plot displaying chromosomal positions (x axis) of significant associations (p < 6.85x10-10, accounting for multiple testing, y axis). Colors indicate metabolite super pathways. P values were obtained from genome-wide summary statistics from linear regression models using genetic variants as predictors and metabolite levels as outcomes. Effector genes identified for corresponding loci are annotated.
To assess novel associations, we compared our findings to variant-metabolite associations documented in four sources (PhenoScanner database24, previous leading associations from 26 metabolites GWAS studies (summarized by Yin et al.11, shown in Supplementary Table 8) and two recent metabolomics GWASs using a similar metabolomics platform11,21). We found 771 of 1,509 significant variant-metabolite associations are in linkage disequilibrium (LD; r2 > 0.8) with previously reported variants associated at p < 5x10-8 or stronger study-specific p-value thresholds for the same metabolites. Another 257 significant variant-metabolite associations were considered as potentially novel since these variants and/or their proxies were associated with previously unnamed metabolites that may be the same as those named in CLSA. The remaining 481 associations arising from 313 metabolites were considered as novel where some of these were conditionally independent and resided in the same locus. Additionally, for the shared variant-metabolite associations with Hysi et al.21, and Yin et al.11, 90% of these have the same direction of effect on the corresponding metabolites (Supplementary Table 5). Using all 1,702 independent variant-metabolite associations, we identified 248 loci, 216 of which contained genetic variants with previously reported genome-wide significant variants for the same or unnamed metabolites (Supplementary Table 5). Overall, 31.9% of significant genetic variant-metabolite associations and 12.9% of the loci reported in our study appear to be novel.
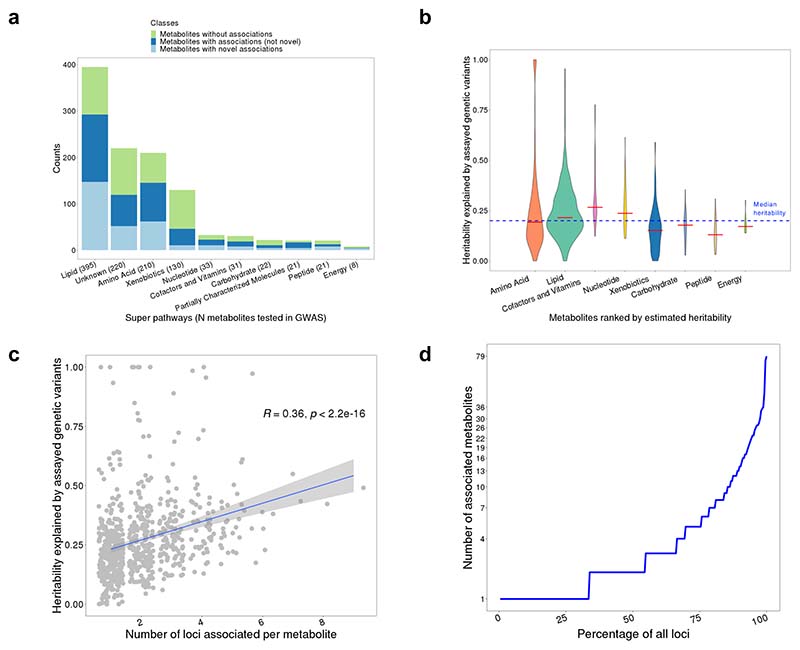
Our GWAS results provide insights into the genetic architecture of metabolite levels. Over 50% of tested metabolites within each super pathway had independent variant-metabolite associations, with the exception of xenobiotics and carbohydrate (Figure 2a). The median estimated SNP-based heritability of all tested metabolites was 19.7% while it was higher for cofactors/vitamins and nucleotides but lower for xenobiotics and peptides (Figure 2b; Supplementary Table 9).
Figure 2. Genetic architecture of metabolite levels.
a, Classification of tested metabolites with or without genetic associations in each metabolite super pathway. “Metabolites with novel associations” include metabolites that have at least one novel association. “Metabolites with associations (not novel)” include metabolites that only have independent variant-metabolite associations that are known. b, Distribution of heritability explained by assayed genotypes for metabolites in each super pathway (red lines indicate the median heritability of metabolites in each super pathway and blue dashed line indicates the median heritability for all tested metabolites). c, Distribution of variant-based heritability of metabolites, compared to the number of associated loci. Each point represents a different metabolite. The Spearman’s correlation coefficient is shown. The exact p value (two-sided) for the correlation coefficient is 2.4x10-22. 95% confidence interval around linear regression line were plotted. d, Distribution of number of associated metabolites per locus, demonstrating the pleiotropy of genetic effects on metabolites.
These genetic associations can also be characterized by their polygenicity and pleiotropy, which was also observed in previous studies8,25. The median number of loci associated with each metabolite was 2 and some metabolites (e.g., glutarylcarnitine (C5-DC)) were influenced by up to 9 loci (Figure 2c). We also found that the number of associated loci was overall positively correlated with the heritability of metabolites, but this relationship seems to be driven by lipids and the unknown metabolites (Figure 2c and Supplementary Figure 2). Assessing pleiotropy, we found a median of 2 metabolites per GWAS associated locus (range 1-79; Figure 2d). In particular, the locus on chromosome 11 containing the fatty acid desaturase (FADS) gene family is associated with 79 metabolites including 75 lipids (mostly fatty acids and glycerolipids), 3 unknown metabolites, and 1 amino acid (i.e., asparagine) (Supplementary Table 5). This suggests that such loci are highly pleiotropic. A similar genetic region has been reported by Lotta et al.,8 where they found associations with 66 lipids and asparagine.
Genetic determinants of metabolite ratios
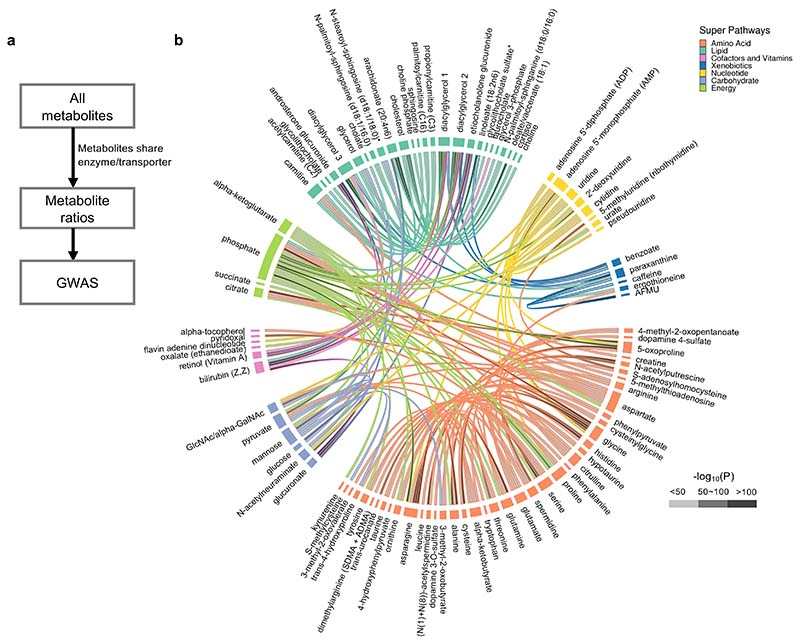
Since many metabolites are substrates and products of enzymatic reactions, identifying genetic determinants of the ratio of substrate to product may provide information on biological processes that cannot be discerned when studying only single metabolites. Similarly, knowledge of enzymes and transporters can also pinpoint genetic control points. To identify the genetic determinants of metabolic flux, we calculated the metabolite level ratios for metabolite pairs sharing an enzyme or transporter using metabolite-protein associations recorded in the Human Metabolome Database (HMDB)26 (Figure 3a, Supplementary Table 10 and Supplementary Figure 3). Compared to using a hypothesis-free approach to pair metabolites into ratios27, our evidence-based approach retains more statistical power by only testing a subset of metabolite ratios that have higher biological plausibility. Undertaking a GWAS for 309 metabolite ratios, we identified 247 associations for 143 metabolite ratios across 69 loci at a multiple testing-adjusted genome-wide significance threshold of p < 1.62x10-10 (using a conservative Bonferroni correction that does not account for non-independence of ratios: 5x10-8/309). This included 242 associations that have not been reported before while 63 of 69 loci only contain metabolite ratio associations that appear to be novel (Supplementary Table 11).
Figure 3. Summary of metabolite ratio GWAS results.
a, Construction of metabolite ratios for GWAS. b, Super pathway membership of metabolite ratio pairs with GWAS associations. The color of the connection line indicates the super pathway of the first metabolite (numerator of the ratio) of the metabolite pair that constructs the metabolite ratios. The grey scale gradient filling the connection line indicates the strength of the genetic association with darker color indicating stronger significance. For figure generation, five metabolite names were shortened. N-acetylglucosamine/N-acetylgalactosamine, GlcNAc/alpha-GalNAc; linoleoyl-arachidonoyl-glycerol (18:2/20:4) [1]*, diacylglycerol 1; linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2]*, diacylglycerol 2; oleoyl-linoleoylglycerol (18:1/18:2) [2], diacylglycerol 3; 5-acetylamino-6-formylamino-3-methyluracil, AFMU.
A limited number of studies have previously assessed the genetic determinants of metabolites ratios, and our findings replicated all 5 associations for 4 metabolite ratios that were reported in previous studies. For example, our results replicated the negative effect of the C allele at rs1260326 on glucose/mannose ratio which was reported by Suhre et al., (2011)28 and Shin et al. (2014)10. The variant rs2657879 is in LD with the rs2694917 (r2 = 0.83), where the latter was found to be associated with glutamine/histidine ratio in Shin et al. (2014)10. Furthermore, 16 genome-wide significant genetic variants were identified for 13 ratios where these variants were not significantly associated with either of the two metabolites forming the ratio, suggesting that the formation of ratios could help identify novel genetic determinants by focusing on specific metabolic reactions (Supplementary Table 12). For instance, we identified 5 genetic variants for caffeine/paraxanthine ratio with a minimum p-value of 8.8x10-35 while only one of these variants was found to be significantly associated with paraxanthine (p = 3x10-12).
To explore patterns among the metabolic ratios with genetic determinants, we annotated the super pathways of the involved metabolites. As shown in Figure 3b, most of the genetically associated metabolite ratios are from the amino acid and lipid super pathways and they tend to connect within their own super pathways. Metabolites from the energy super pathway show more connections to metabolites in other pathways. For example, genetic determinants were found for the ratios between phosphate and 21 metabolites from 4 different super pathways (Figure 3b). We also calculated the heritability of the metabolite ratios (Supplementary Figure 4a and Supplementary Table 13). We observed that the conversion from paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil, which is part of caffeine catabolism, has an estimated heritability of 83.6%, which is higher than the heritability of all other metabolite ratios and metabolites constructing this ratio.
Assessing the polygenicity of metabolite ratios, we found that most were influenced by fewer than 4 different loci (range 1-4) and there was low correlation between the number of associated loci and heritability (Supplementary Figure 4a). For pleiotropy, we found a median of 2 metabolite ratios per locus (range 1-21) and again the FADS locus demonstrated the highest count of associated metabolites ratios (i.e., 21) (Supplementary Figure 4b). Thus, through a genome-wide scan, we identified novel associations between genetic variants and metabolite ratios and demonstrated the genetic influences on metabolic flux.
Prioritization of effector genes for metabolites
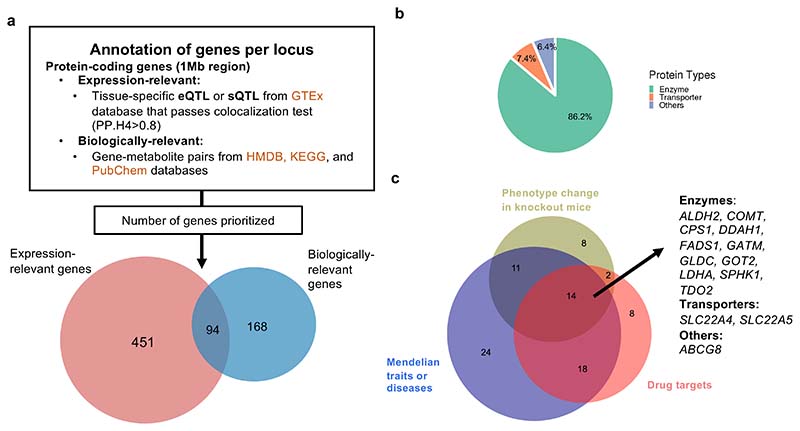
Identifying the genes, rather than genomic loci, that control metabolites and their ratios can help to pinpoint intervention points for therapeutic interventions. We applied two complementary approaches to identify the effector protein-coding genes overlapping a one megabase (Mb) region around associated genetic variants: (1) a gene expression-based approach colocalizing the identified metabolites or ratios in this study with expression quantitative trait loci (eQTL) or splicing quantitative trait loci (sQTL) from up to 49 human tissues in individuals of European ancestry29 (2) a biological knowledge-based approach integrating existing biological evidence with metabolite-gene associations using data from three databases: the HMDB26, KEGG pathway database30, and the PubChem Chemical Co-Occurrences in Literature database31 (Figure 4a). The gene expression-based approach tends to highlight genes mediating the genetic influence on metabolite levels through effects on genetic expression, while the biological knowledge-based approach prioritizes metabolism-related genes that are also near the genetic variants.
Figure 4. Assignment of effector genes by using evidence from gene expression and biological knowledge.
a, Identification of effector genes. b, Classification of the 94 effector genes with strong expression and biological evidence by protein types. c, Evidence from drug targets, phenotypic changes observed in murine knockouts, and associated Mendelian traits or diseases for the 94 effector genes.
The gene expression-based approach identified 545 expression-relevant genes for 625 genetic variants while the biological knowledge-based approach identified 262 biologically relevant genes for 321 variants (Figure 4a). By comparing the genes prioritized by these two approaches, we found 94 effector genes for 189 variant-metabolite associations (including 109 metabolites and 48 metabolite ratios) that have converging gene expression and biological evidence (Supplementary Table 14). Of these 94 genes, over 90% encode enzymes or transporters (Figure 4b). For the 35 effector genes identified for metabolic ratios, 9 also encode enzymes or transporters that were used to construct the metabolite pairs (Supplementary Table 14). For example, the effector genes identified for the genetic variants that influence ratios between bilirubin and glucuronide conjugates (e.g., etiocholanolone glucuronide and androsterone glucuronide) belong to the UDP-glucuronosyltransferase family. The genes in this family encode proteins catalyzing the glucuronidation reaction. The remaining 26 effector genes for metabolite ratios do not encode the enzymes or transporters that were used to construct ratios. They mainly encode proteins metabolizing one of the metabolites in the ratios, such as Arginase 1 (ARG1) for ratios involving arginine and Cytochrome P450 Family 2 Subfamily A Member 6 (CYP2A6) for ratios involving paraxanthine.
Integration of metabolic associations with disease and pharmacological information could help to better understand the mechanism underlying disease development. We therefore explored the drug information and diseases associated with the 94 effector genes. By surveying the DrugBank database32, we found 580 drugs at various stages of development which are antagonists, agonists, substrates, inhibitors, or inducers of the proteins encoded by 42 of the 94 effector genes. The International Mouse Phenotyping Consortium (IMPC)33 has described 35 of these 94 genes which when knocked out generate phenotypic changes in mice. Further, 67 of these 94 effector genes have associated Mendelian diseases, as described in the Online Mendelian Inheritance in Man (OMIM)34 (Figure 4c; details in Supplementary Tables 14 and 15). Integrating this information across all three external sources identified 14 effector genes whose associated metabolites have been described in murine knockouts, associated with Mendelian disease and have medicines which target them. Thus, these 14 genes could be explored for potential drug targets to modulate metabolite levels in disease management.
Causal metabolites and ratios implicated in human complex traits
Next, we explored the utility of the newly identified gene-metabolite associations by applying them in two-sample MR. A large available GWAS for the twelve representative traits and diseases that are predominantly influenced by three distinct processes (aging, metabolism, and immune response) were used to assess the effect of the metabolites on these outcomes. Specifically, we selected, eBMD35, Parkinson’s disease36, Alzheimer’s disease37, and osteoarthritis38 for aging; BMI39, T2D40, ischemic stroke41, and CAD42 for metabolism; asthma43, T1D44, IBD45 and multiple sclerosis46 for immune responses (Supplementary Table 16).
We selected 171 genetic variant-metabolite and variant-metabolite ratio associations with known effector genes, which helps strengthen the MR relevance assumption and could reduce the risk of horizontal pleiotropy (further details can be found in the Methods and Supplementary Table 17). Inverse variance weighted or Wald tests were then performed depending on the number of instrumental variables available for MR analyses. For the metabolites and ratios prioritized by MR, we further checked whether their instrumental variables had associations with other metabolites identified in this study or traits related to the outcomes in a compendium of GWAS results (PhenoScanner24). Then, to assess the possibility of reverse causality between these MR prioritized metabolites and ratios and traits, bidirectional MR analyses were performed using the twelve traits and diseases as the exposures and the circulating metabolites and ratios as outcomes.
We identified 36 metabolites and 26 ratios for these twelve traits and diseases using MR and applying Bonferroni multiple testing correction (Supplementary Tables 18, 19, and 20). We further pruned the likely causal metabolites and ratios by removing metabolites and ratios with potentially metabolically pleiotropic genetic instruments (as described in Methods). This step retained 25 metabolites and 20 ratios (Supplementary Table 21). By assessing PhenoScanner24, we found multiple associations for some variants (Supplementary Table 22). However, since we only used instrumental variables that colocalize with cis-eQTLs or cis-sQTLs of metabolite-related genes, these associations are most likely a result of vertical pleiotropy, which does not violate MR assumptions. In bidirectional MR analyses, we found revere causation was evident for three metabolism-related traits, BMI, T2D, and CAD where these traits had estimated causal effects on 6 metabolites and 3 metabolite ratios which were subsequently removed from further analyses (Supplementary Table 23). After evaluating for pleiotropy and reverse causation, 33 metabolite-outcome pairs (of 22 metabolites) and 30 ratio-outcome pairs (of 20 metabolite ratios) were retained (Figure 5).
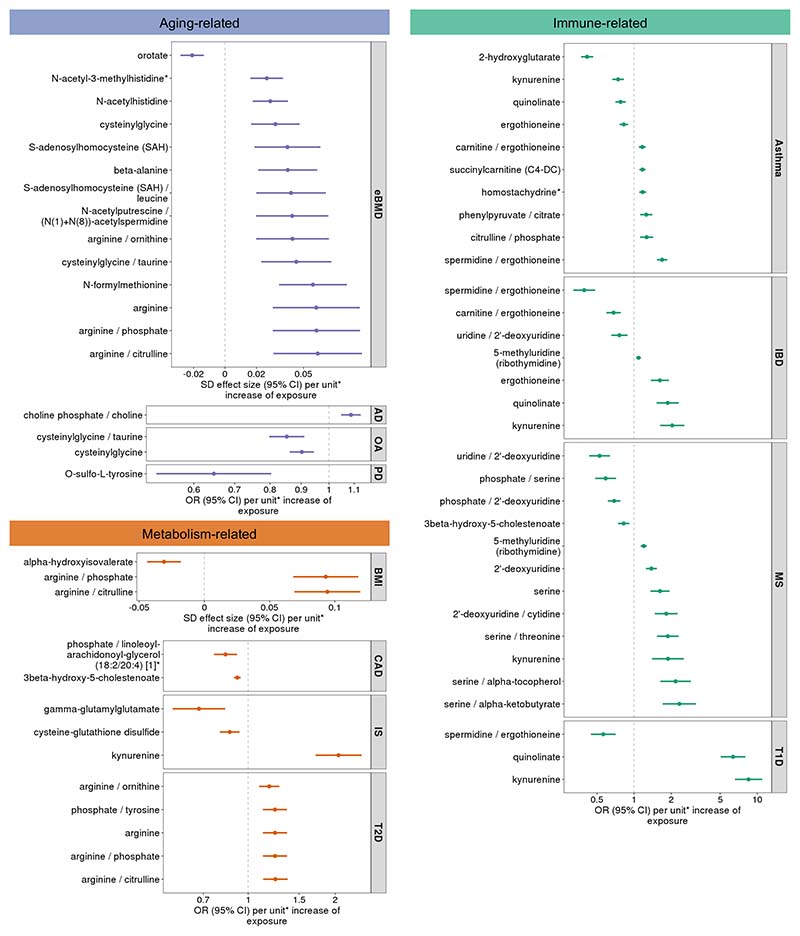
Figure 5. Forest plots showing effects (beta or OR estimates) and 95% confidence intervals from two-sample MR analyses.
Metabolites and metabolite ratios that have an estimated causal effect (with Bonferroni-corrected p < 0.05) and pass pleiotropy and reverse causation evaluations for twelve traits and diseases. MR estimates and p-values were calculated using inverse-variance weighted random effects test for instruments that contained more than one variant and Wald ratio test for instruments with one variant. *Metabolite unit: 1 standard deviation (SD) of log-normalized values. Metabolite ratio unit: 1 SD of inverse rank normalized values. Abbreviations: estimated bone mineral density (eBMD), Alzheimer’s disease (AD), osteoarthritis (OA), Parkinson’s disease (PD), body mass index (BMI), coronary artery disease (CAD), ischemic stroke (IS), type 2 diabetes (T2D), inflammatory bowel disease (IBD), multiple sclerosis (MS), type 1 diabetes (T1D). Specific sample sizes for each metabolite and trait can be found in Supplementary Tables 5, 11 and 16.
To check for possible bias due to LD, we tested the probability that the tested metabolites or metabolite ratios had the same genetic determinants as the outcomes using a Bayesian colocalization method as implemented in the coloc package47. We considered the exposure and outcome to have a shared single causal signal if the estimated probability of colocalization, i.e., coloc (PP.H4) was greater than 0.8. We found 10 metabolites and 12 ratios had their genetic associations colocalized with their target traits (Supplementary table 24). For aging-related traits, we found orotate for eBMD, choline phosphate/choline for Alzheimer’s disease, and O-sulfo-L-tyrosine for Parkinson’s disease. For metabolism-related traits, we identified the genetic determinants of arginine/phosphate, arginine/citrulline, and alpha-hydroxyisovalerate colocalized with BMI, phosphate/linoleoyl-arachidonoyl-glycerol ratio with CAD, kynurenine with ischemic stroke, and arginine, arginine/citrulline, arginine/phosphate, and phosphate/tyrosine with T2D. For immune response-related traits, 6 metabolites and 7 ratios were found to be colocalized with their target traits, such as serine with multiple sclerosis and spermidine/ergothioneine with T1D. Further, there are observational epidemiology descriptions of some metabolite-outcome associations such as serine and deoxyuridine with subtypes of multiple sclerosis48, arginine with T2D49, and alpha-hydroxyisovalerate with BMI50.
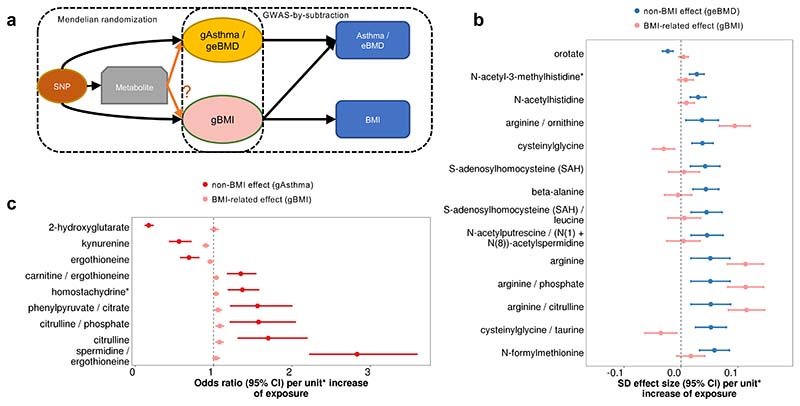
Since previous MR studies have found estimated causal effects of increased BMI on increased BMD and asthma risk51–53, we applied a GWAS-by-subtraction approach54–56 to further dissect BMI-related effects of the MR prioritized metabolites and ratios. As shown in Figure 6a, GWAS-by-subtraction models were used to obtain non-BMI (geBMD or gAsthma) and BMI-related (gBMI) latent genetic effects of the variants on eBMD and asthma risk, respectively, and then we used these latent genetic effects as outcomes in MR (detail see Method section). We found that 13 out of the 14 MR prioritized metabolites and ratios for eBMD had clear effects upon eBMD that were independent of BMI (p < 0.05/14 = 0.0036) (Figure 6b; Supplementary Table 25). In particular, 8 out of 13 had estimated BMI-related effects that largely overlapped with the null (p ≥ 0.1), implying that their effect was likely directly upon eBMD, rather than through BMI. For asthma, all tested metabolites and ratios had a significant non-BMI component in their effect upon asthma risk (p < 0.05/8 = 0.0063) (Figure 6c; Supplementary Table 26). For 2 out of these 8 metabolites and ratios (i.e., 2-hydroxyglutarate and spermidine/ergothioneine), no evidence was found for BMI-related effects (p ≥ 0.1), supporting their direct causal roles in modifying asthma risk. The remaining metabolites or ratios simultaneously exhibited suggestive or significant BMI-related effects. Therefore, for metabolites with BMI-related effects, the validity of their direct causal effects on eBMD or asthma risk, independent of BMI, could not be assessed under the current MR framework.
Figure 6. Comparison of estimated BMI-related and non-BMI effects on eBMD and Asthma.
a, Illustration of GWAS-by-subtraction models. b, Two-sample MR results showing the non-BMI (geBMD) and BMI-related (gBMI) effects (beta estimates) of MR prioritized metabolites and ratios for eBMD c, Two-sample MR results showing the non-BMI (gAsthma) and BMI-related (gBMI) effects (OR and beta estimates, respectively) of MR prioritized metabolites and ratios for asthma risk. *Metabolite unit: 1 SD of log-normalized values. Metabolite ratio unit: 1 SD of inverse normalized values. Specific sample sizes for each metabolite and trait can be found in Supplementary Tables 5, 11 and 16.
Orotate and hip fracture
Since no previous publication has reported the relationship between orotate with bone traits, we validated our MR findings of the relationship between orotate and eBMD using the Umeå Fracture and Osteoporosis (UFO) study, a population-based study focusing on osteoporotic fractures in Northern Sweden57. The nested case-control cohort used in the current study included 2,225 cases with hip fracture and 2,225 controls matched for sex, age, and fasting status (cohort characteristics in Supplementary table 26). The mean age of the nested case-control cohort was 58.8 and 71.6% of the participants were women. Logistic regression analysis adjusted for sex, age, and fasting status revealed that circulating orotate was directly associated with risk of hip fractures (OR of 1.15, 95% CI 1.08-1.22 per SD increase in orotate, p = 1.3x10-5), which is concordant with our MR results suggesting that orotate has a negative effect on eBMD since lowered eBMD is strongly associated with increased risk of fracture.
Discussion
In this study, we identified genome-wide significant associations with 690 metabolites and 143 metabolite ratios across 248 loci (32 novel) and 69 (63 novel) genetic loci, respectively. We assigned 94 effector genes for many of the identified associations by combining gene expression and gene-metabolism information. Using 171 identified variant-metabolite and variant-metabolite ratio associations with high biological plausibility in two-sample MR, we found 22 metabolites and 20 metabolite ratios to have estimated causal effects on one or more traits and diseases that are influenced by aging-, metabolism-, and immune-related mechanisms. Measuring one of the lead metabolites, orotate, in a separate nested case-control cohort, we found that increased orotate was associated with increased risk of fracture, a different but highly correlated outcome of eBMD. This result was consistent with our MR findings. Taken together, these results provide insights into the genetic architecture of metabolites, but also show how these data can be used to identify metabolites involved in risk of disease, thereby providing targets for interventions.
By triangulating MR and other genetic findings, we highlighted several metabolites and their corresponding genes that can be explored as targets for intervention. For example, MR and colocalization analyses for eBMD showed that genetically predicted higher plasma levels of orotate have an estimated negative effect on eBMD and this effect is likely independent from BMI. Since eBMD is a strong risk predictor for fracture35, we further checked the orotate-fracture risk association using the UFO cohort where we validated the detrimental effect of higher orotate levels on fracture risk. These results provide a road-map whereby GWAS of metabolite levels can be used to identify high-value MR associations, which when colocalized show the anticipated effect on a disease outcome in an independent cohort. A monogenic disease leading to orotic aciduria, due to mutations in UMPS leads to an increase in orotate (OMIM #258900) and to reduced life expectancy. UMPS encodes the uridine monophosphate synthase gene which uses orotate as substrate for uridine monophosphate synthesis. To our knowledge such patients have not usually been assessed for osteoporosis. More functional studies are needed to elucidate the potential causal link between orotate levels and eBMD.
These insights were not limited to eBMD. We showed that alpha-hydroxyisovalerate has an estimated negative causal effect on BMI. The implicated effector gene for this variant-metabolite association is lactate dehydrogenase A (LDHA) which can convert the product of degraded branched-chain amino acids, like 3-methyl-2-oxobutanoate, to alpha-hydroxyisovalerate58. In OMIM, rare mutations in LDHA have been reported to cause lactate dehydrogenase deficiency (OMIM #612933) which was linked to muscle degeneration, elevated blood pyruvate levels, and glycogen storage disease in humans59. Interestingly, from the IMPC database, heterozygous knockouts of Ldha have increased lean mass and decreased fat mass in mice, suggesting the involvement of this gene in energy metabolism and body composition.
The current study has several limitations. First, non-fasting plasma samples were used for metabolomics profiling. Although we adjusted the metabolomics measurements by including the number of hours since last meal or drink, additional variability may not be fully accounted for. Second, this study only focused on the most possible gene-metabolite pairs supported by the expression and biological knowledge available (i.e., with effector genes). This does not mean that other highly heritable metabolites or ratios are not disease-related. Future studies on identifying the effector genes for these metabolites and ratios are needed when more expression data or knowledge in the metabolism are available. Third, in the MR analyses, most of the metabolites and metabolite ratios only have one instrumental variable, limiting the application of common MR sensitivity tests such as MR-Egger60 which require multiple instrumental variables. Nevertheless, by using instrumental variables that are cis to effector genes influencing metabolite levels, and manually assessing for metabolic pleiotropy by removing instrumental variables that are associated with multiple metabolites that are not featured in the same metabolic process, our study design helps to guard against horizontal pleiotropy. Although these analyses help to reduce potential bias, we recognize that this potential source of bias may not be fully eliminated due to incomplete metabolome profiling and metabolite-protein connection databases. More comprehensive measurement of the metabolome will provide more accurate information regarding the genetic pleiotropy of metabolites in future studies. Last, this work mainly focused on older individuals of European ancestry, leaving the effect of the identified genetic variants on metabolites and ratios to be assessed in other populations.
In summary, in these large GWASs of metabolites, we identified novel gene-metabolite associations and showcased the use of these associations to highlight potential targets for various traits and diseases. The findings may assist in understanding the genetic regulation of human metabolism, allow future prospectively planned meta-analysis, and provide a valuable resource for the identification of targets for behavioral and pharmaceutical interventions.
Methods
Study cohort
The Canadian Longitudinal Study of Aging (CLSA) follows over 50,000 Canadians who were between the ages of 45 and 85 when recruited for biological, medical, physiological, social, lifestyle, and economic status information23. This metabolomics study focuses on 8,299 unrelated European subjects in CLSA who have been genome-wide genotyped and have had circulating plasma metabolites measured. We focused our study on individuals of European ancestries to reduce potential bias from population stratification (Supplementary Figure 5). We removed 203 European individuals with first- and second-degree relatives using kinship-based inference from KING package (v2.2.5)61.
pre-GWAS genotype quality control
The genome-wide genotyping has been done using the Affymetrix Axiom genotyping platform, which was followed with imputation using the Trans-Omics for Precision Medicine (TOPMed) programme62 , and genetic ancestry determination by the CLSA group63. We then removed low-quality imputed genetic variants by retaining only those single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) higher than 0.1%, imputation quality score > 0.3, and missing rate < 0.1, leading to approximately 15.4 million SNPs (in reference build 38) for GWAS testing.
Metabolite and metabolite ratio data processing
The levels of 1,458 metabolites were quantified in plasma samples by Metabolon, Inc. (Durham, NC, USA) using the Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) platform which is also known as Metabolon HD4 platform. Strict QC and curation of the metabolomics data were applied to ensure accurate and consistent identification of true chemical entities, and to remove those representing systemic artifacts, misassignments, and background noise. We then used batch normalized levels of metabolites generated by the Metabolon and only retained metabolites that have missing measurements in fewer than 50% of samples (N = 1,091). Novel metabolites tested in current study was also determined by comparing to five representative large-scale metabolomic GWAS studies5,8,10,11,21 (details see Supplementary Notes). For GWAS, metabolite levels were then natural log-transformed, trimmed to remove outliers that are 3 standard deviations away, and then standardized to have a mean of 0 and a standard deviation of 1.
For metabolite ratios, we first identified 309 metabolite pairs that share enzymes or transporters using the Human Metabolome Database (HMDB)26 (Supplementary Table 10 provides these pairs of metabolites and the HMDB evidence to support their pairing). Then the metabolite ratio was calculated for each pair of metabolites by dividing the batch-normalized measurement value of one metabolite by the measurement of the other metabolite in the same individual. The metabolite ratios were then trimmed (retaining those within 3 standard deviations), and inverse-rank normal transformed.
GWAS
After data processing and quality control, GWASs using linear regression of the metabolites and metabolite ratios were done using the fastGWA tool from GCTA version 1.93.2 beta64,65, adjusting for age, sex, hour since last meal or drink, genotyping batch, and the first 10 genetic principal components (PC). To improve diversity in genomic research, GWAS for South-Asian (n=108), East-Asian (n=104), and African (n=60) individuals were also performed (more details can be found in the Supplementary Notes). Although no further analyses were conducted, summary statistics of non-European groups were made available along with European GWAS results.
Calculation of effective number of independent metabolites
Since many metabolites were correlated, we used an eigendecomposition method to estimate the effective number of independent metabolites66 (more details can be found in Supplementary Notes). The estimated effective number of independent metabolites was 73 and it was used to adjust the p-value for multiple testing of metabolites GWAS, using a Bonferroni correction (5x10-8/73=6.85x10-10), to detect independent variant-metabolite associations.
Multi-SNP-based conditional & joint association analysis
To identify conditionally independent SNPs from the GWAS, we used GCTA-COJO67, which leverages correlation estimates (LD) between SNPs and summary statistics. We used the genotypes of the same 8,299 unrelated European individuals included in GWAS to compute the LD reference panel. The following parameters were used for COJO analyses: --maf 0.01, --cojo-p 5e-8, --cojo-wind 5000, --cojo-collinear 0.9.
Independent genome-wide significant associations
The SNPs for metabolites with an original p-value and COJO-adjusted p-value smaller than 6.85x10-10 and SNPs for metabolite ratios with an original p-value and COJO-adjusted p-value smaller than 1.62x10-10 (5x10-8/309 tested biologically plausible metabolite ratios) were considered as genome-wide significant.
Genomic inflation
The genomic inflation factor for each GWAS result was calculated as the median of the observed chi-squared test statistics divided by the expected chi-squared test statistics for each metabolite and metabolite ratios (Supplementary Table 7).
p-gain
To evaluation whether a metabolite ratio carries more information than the two corresponding metabolites alone, we calculated p-gain statistics using the universal p-gain equation68 where M1 and M2 are two metabolites, M1/M2 represents the metabolite ratio, and X is a genetic variant being tested (Supplementary Table 12).
Identification of novel associations and novel loci
To check if the associations have been identified before, we queried significant genetic variant-metabolite associations (p < 5x10-8 or stronger study-specific p-value threshold) from four sources. Source 1, PhenoScanner database24; Source 2, 26 previous studies in European ancestry (1394 associations for 622 traits) that have metabolite-variant associations summarized by Yin et al11 (Supplementary Table 8). The sources 3 and 4 are two recent metabolites GWAS, published by Hysi et al21 and Yin et al.11, that measured plasma metabolites in European ancestry cohorts. The significant variant-metabolite pairs we identified (p < 4.58x10-11) were classified into “Known associations” and “Potentially novel”, based on whether these genetic variants are the same or in LD (r2 > 0.8 using the 1000 Genomes European subset69) with previously reported variants that were associated with the same metabolites, or only unnamed metabolites, respectively. The remaining associations were considered as “Novel”. Since Hysi et al., and Yin et al., used a similar UPLC-MS/MS metabolomics platform, the direction of effect on the corresponding metabolites for the “Known associations” shared with CLSA was also checked (Supplementary Table 5). The novelty of variant-metabolite ratio associations was also assessed by checking the PhenoScanner database using the similar approach described above. More details on previous association assessments, and novelty checking can be found in Supplementary Notes.
Locus definitions can be found in Supplementary Notes. The novelty of loci was assessed by checking whether any of the genetic variants in the locus region were associated with the same metabolites named in CLSA or any unnamed metabolites. The related associations documented in PhenoScanner were searched using the genetic region of each locus in with the same parameters described above (Source 1). The Source 2, 3, and 4 mentioned above were also used. The labels “Known locus”, “Potentially novel locus”, and “Novel locus” were defined using the similar approach described above.
SNP-based heritability
We estimated the proportion of metabolites variance tagged by all SNPs on the genotyping array (i.e., the SNP-based heritability) using the GCTA-GREML program70. Specifically, all genotyped variants on autosomes that have MAF>0.01 were included in genetic relatedness matrix (grm) calculation which was then used in the estimation of variance with following flags (-grm, --reml, --pheno) in default setting. Overall, 655,452 out of 794,409 genotyped variants were used for heritability estimation. The power to detect the corresponding heritability was calculated using a method described before71 (Supplementary Table 9 and 13). The default values for the parameters were used: 2x10-5 for the variance of the SNP-derived genetic relationships and 0.05 for Type 1 error rate.
Identification of effector genes
To identify the effector genes influenced by GCTA-COJO independent genome-wide significant variants, we first retrieved protein-coding genes, from the human GENCODE resource (https://www.gencodegenes.org/), within or overlapping the metabolite-associated loci of the variants (1Mb region) using bedtools72. We found 4,404 unique protein-coding genes for 1,066 unique genetic variants. We next checked if the genes were:
Involved in the biological processes of the associated metabolites of the genetic variants. To do so, we checked if the protein-coding gene within or overlapping the 1Mb region of the variants were involved in the enzymatic reaction, transportation, or biological processes of the SNP-associated metabolites using three databases: the HMDB26, the KEGG pathway database30, and the PubChem Chemical Co-Occurrences in Literature database31.
Influenced by variants in terms of transcription and splicing. Specifically, we investigated whether the independent genome-wide significant genetic variants were also expression quantitative trait loci (eQTL) or splicing quantitative trait loci (sQTL) of the genes within or overlapping the 1Mb region of them, namely the cis-eQTL and cis-sQTL of the variants, by querying the multi-tissue gene expression data from the GTEx project29. We first included all variant-gene pairs that passed the statistical thresholds determined using permutation approach by the GTEx group in any tissue with European ancestries (data source: *.v8.EUR.signif_pairs.txt files from GTEx V8 release). To check if the same variant influencing both metabolites and gene expression, we conducted colocalization analyses (with the priors recommended by the original study47: p1 = 1x10-4, p2 = 1x10-4, p12 = 1x10-5) using full summary statistics of tissue-specific eQTL and sQTL from V8 release (European) with coloc R package (5.1.0)47. The SNPs in the 1Mb range of the tested genetic variants that have MAF > 0.05 were used for analysis. The metabolites or metabolite ratios that have PP.H4 > 0.8 (posterior probabilities of two traits share one causal SNP) with eQTL or sQTL were considered to pass colocalization test.
Next, genes that fit the following three criteria were highlighted as the effector genes:
were within or overlapping the 1Mb region of the independent genome-wide significant variants, and
are involved in the biological processes of the associated metabolites (defined as biologically-relevant genes) and
colocalized with significant cis-eQTLs or cis-sQTLs in at least one of the GTEx tested tissues (defined as expression-relevant genes).
For metabolite ratios, the gene that satisfied all three criteria for either of the two metabolites were identified as the effector gene for the ratio. A total of 94 effector genes were assigned to 113 unique genetic variants for 109 metabolites and 43 variants for 48 metabolite ratios. The protein types of the effector genes were checked using the UniProt database (https://www.uniprot.org/uniprot/).
Since not all the genetic variants have colocalized cis-eQTL, cis-sQTL or are in proximity to biologically relevant genes, we also annotated the variants with their closest protein-coding genes as they usually enrich for molecular QTL73 (Supplementary Table 5 and 11). The closest protein-coding gene for each genetic variant was retrieved by comparing the distance from the variants to the start and the end of the protein-coding genes within or overlapping the 1 Mb region.
Medical and pharmacological annotation the effector genes
To map the effector genes to associated Mendelian traits, murine knockout and pharmacological information, we retrieved the information from the Online Mendelian Inheritance in Man (OMIM) database34, the International Mouse Phenotyping Consortium (IMPC) database 33, and DrugBank database32 for these genes. More details on effector gene mapping can be found in Supplementary Notes.
Two-sample Mendelian randomization
Mendelian randomization (MR) studies use genetic variants that are associated with modifiable exposures to assess the causal effect of the exposure on outcomes and aim to reduce bias from confounding and reverse causation74. We applied two-sample MR to screen for potentially causal circulating metabolites and ratios as exposures for their role in influencing the selected outcomes. We followed the STROBE-MR (Strengthening the reporting of observational studies in epidemiology using mendelian randomization) guidelines for MR results reporting75,76.
Exposure definition
To satisfy the relevance assumption and mitigate partially against the possibility horizontal pleiotropy, we only included the independent genome-wide significant SNPs that were assigned to effector genes as instrumental variables for the exposure (number of instrumental variables = 188). We removed all SNPs on FADS locus (number of instrumental variables = 16) from MR analyses since they demonstrated extremely high pleiotropy (wherein they were associated with 79 metabolites). We also removed SNPs on extended Major histocompatibility complex region on chromosome 6 (+/- 500 Kb, number of instrumental variables removed = 1) as this region demonstrates strong pleiotropic cross-phenotype associations77. In total, 121 variant-metabolite pairs for 99 metabolites and 50 variant-metabolite ratio pairs for 43 metabolite ratios passed this selection criteria (Supplementary Table 17).
Outcome definition
Large GWASs for eBMD35, Parkinson’s disease36, Alzheimer’s disease37, osteoarthritis38, BMI39, T2D40, ischemic stroke41, CAD42, asthma43, T1D44, IBD45 and multiple sclerosis46 from European cohorts were used. The specific traits, sample size, and GWAS data location can be found in Supplementary Table 16. There was no overlap of participants between the metabolomics GWAS cohorts and the outcome GWAS cohorts.
SNP filtering
First, we harmonized SNPs associated with the exposures (in this case, metabolites and ratios) with SNPs associated with the outcomes (in this case, 12 traits and diseases) using “harmonise_data” function in TwoSampleMR package (version 0.5.6) in R (R version 4.0.5). When matching SNPs were not found, we used LD-proxy SNPs that were identified using Snappy78 using European 1000 Genomes phase 369 as the reference genome for LD structure and r2 > 0.8 as the LD threshold. The specific numbers of matching and LD-proxy SNPs used for MR for each traits can be found in Supplementary Table 17.
MR
MR analyses were performed using the “mr” function in TwoSampleMR package in R. Wald ratios were used to estimate the effect of the exposures on the outcome when there was only one SNP available as an instrumental variable. For the exposures that had multiple SNPs that qualified as instrumental variables, the inverse variance weighted (IVW) method was used to meta-analyze their combined effects. The metabolites and metabolite ratios that passed the corresponding multiple testing correction thresholds (Bonferroni corrected p-value: 4.24x10-4 for MS; 4.03x10-4 for eBMD, Parkinson’s disease, BMI, CAD, ischemic stroke, and T2D; 4.0x10-4 for IBD; 3.94x10-4 for AD; 3.85x10-4 T1D; 3.65x10-4 for asthma and osteoarthritis) were retained for pleiotropy risk evaluation and colocalization analyses.
Pleiotropy evaluation, colocalization, and reverse association check
Pleiotropy evaluation
The horizontal pleiotropy of each SNP was first assessed by checking the number of associated metabolites identified in current study. Specifically, we prioritized the metabolites and ratios with genetic variants that were
Only associated with one metabolite in the current study, or
Associated with more than one metabolite with known identity where these metabolites are in the same metabolic process
We repeated the MR test after removing the genetic variants at risk of horizontal pleiotropy. We further checked horizontal pleiotropy of genetic variants of the metabolites and ratios that were prioritized by MR test using PhenoScanner database24. The diseases and traits that were associated with these SNPs were extracted with specific searching criteria (p-value: 5x10-8, Proxies: EUR, r2: 0.8, reference Build: 38), see Supplementary Table 22.
Colocalization
To check whether the same genetic variants are driving the associations with metabolites and outcome traits, we undertook colocalization analysis. Specifically, a stringent Bayesian analysis implemented in the coloc R package (5.1.0) was performed (with the priors recommended by the original study47: p1 = 1x10-4, p2=1x10-4, p12 = 1x10-5) to estimate the posterior probability (PP) that the exposure and the outcome share a single causal SNP at the locus47. The SNPs in the 1Mb range of the tested instrumental variable that have MAF over 0.05 were used for analysis. The metabolites or metabolite ratios with PP.H4>0.8 (posterior probabilities of two traits sharing one causal SNP) were considered to be colocalized.
Bidirectional MR
To assess the possibility of reverse causality, bidirectional MR analyses were performed on the primary MR prioritized metabolites and ratios using the 12 traits and diseases as exposures and the circulating metabolites and metabolite ratios as outcomes. LD-independent SNPs from corresponding GWAS that passed the same SNP filtering and proxy search steps mentioned in the MR method section were used as instrument variables. We then undertook IVW to meta-analyze their effects on metabolite levels. LD-independent SNPs were selected using PLINK 1.979 clumping function (--clump-kb 1000 --clump-r2 0.001 --clump-p1 5x10-8) and the European 1000 Genomes phase 3 dataset was used as the reference genome. The metabolites and metabolite ratios that passed the specific Bonferroni multiple-testing correction threshold (Supplementary Table 23) were considered as statistically significant.
BMI-related genetic effect on eBMD and asthma
Since most of the tested metabolites and ratios have only one instrumental variable, we adapted a new structural equation modelling method, called “GWAS-by-subtraction”54,55 to disentangle the interplay between genetic determinants of these complex traits. Specifically, using GWAS-by-subtraction, we partitioned the genetic predisposition to eBMD onto two latent pathways: one latent pathway acting on both eBMD and BMI (gBMI), and the other latent pathway acting only on eBMD (geBMD). SNP effects on these two latent genetic components were estimated based on GWAS summary statistics, the LD score regression-estimated heritability of each trait, and the LD score regression-estimated genetic correlation between the two traits. Subsequently, we performed two-sample MR with the target metabolite as exposure and the two latent pathways separately as outcomes. We repeated the analyses for asthma and obtained the latent pathway shared by asthma and BMI (gBMI) as well as the latent pathway acting only on asthma (gAsthma).
Based on these MR results, we classified the metabolites’ effects on eBMD or asthma risk into two categories using the following criteria:
-
(1)
If a metabolite had a significant effect on geBMD or gAsthma passing the Bonferroni threshold (p < 0.05/14 = 0.0036 for eBMD and p < 0.05/8 = 0.0063 for asthma), and if its effect on gBMI had an uncorrected p-value ≥ 0.1, we considered this metabolite to have a direct causal effect on eBMD or asthma risk;
-
(2)
If a metabolite’s effect on geBMD or gAsthma did not pass the Bonferroni threshold, or if it also demonstrated at least a suggestive effect on gBMI (uncorrected p-value < 0.1), we would consider this metabolite’s causal effect to either have an indirect causal effect acting through BMI, or that the genetic instruments had horizontal pleiotropic effects.
UFO cohort and logistic regression analysis of orotate levels and fracture risk
The Umeå Fracture and Osteoporosis (UFO) study is a population-based, nested case-control study sampled from the Northern Sweden Health and Disease (NSHDS) study cohort57. We identified 2225 hip fracture cases (defined with ICD10-codes S72.0, S72.1 and S72.2 and ICD-9 code 820 in hospital records) that had also left a previous blood sample in the biobank. For each case, one control was selected from the NSHDS-cohort, matched for gender, age at baseline and fasting state, making a total number of 4,450 subjects. Orotate levels were quantified (using the Metabolon HD4 platform) and batch normalized along with other metabolites in plasma samples by Metabolon, Inc. (Durham, NC, USA). The association between standardized plasma orotate levels and hip fracture risk was evaluated using logistic regression models, adjusted for sex, age, and fasting status (more details can be found in Supplementary Notes).
Supplementary Material
Acknowledgements
This research was made possible using the data/biospecimens collected by the Canadian Longitudinal Study on Aging (CLSA). Funding for the Canadian Longitudinal Study on Aging (CLSA) is provided by the Government of Canada through the Canadian Institutes of Health Research (CIHR) under grant reference: LSA 94473 and the Canada Foundation for Innovation, as well as the following provinces, Newfoundland, Nova Scotia, Quebec, Ontario, Manitoba, Alberta, and British Columbia. This research has been conducted using the CLSA Metabolomics data (v1), CLSA Baseline dataset (v5), and Genomics data (v3), under Application Number 2006016. The CLSA is led by Drs. Parminder Raina, Christina Wolfson and Susan Kirkland. The authors also acknowledge the effort of the IPDGC (International Parkinson’s Disease Genomics Consortium), the IMSGC (International Multiple Sclerosis Genetics Consortium), the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) consortium, and the MEGASTROKE consortium in providing high quality GWASs data for other researchers.
The Richards research group is supported by the Canadian Institutes of Health Research (CIHR: 365825, 409511, 100558), the Lady Davis Institute of the Jewish General Hospital, the Canadian Foundation for Innovation, the NIH Foundation, Cancer Research UK (grant number C18281/A29019), Genome Québec, the Public Health Agency of Canada, Genome Québec, McGill University and the Fonds de Recherche Québec Santé (FRQS). JBR is supported by a FRQS Clinical Research Scholarship. Support from Calcul Québec and Compute Canada is acknowledged. TwinsUK is funded by the Welcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. These funding agencies had no role in the design, implementation or interpretation of this study. YC has been supported by an FRQS doctoral training fellowship and the Lady Davis Institute/TD Bank Studentship Award. TL has been supported by a Vanier Canada Graduate Scholarship, an FRQS doctoral training fellowship and a McGill University Faculty of Medicine studentship. TN is supported by a research fellowship of the Japan Society for the Promotion of Science for Young Scientists. CYS has been supported by the Lady Davis Institute/TD Bank Studentship Award. SZ has been supported by a CIHR fellowship and a FRQS fellowship. PR holds the Raymond and Margaret Labarge Chair in Optimal Aging and Knowledge Application for Optimal Aging, is the Director of the McMaster Institute for Research on Aging and the Labarge Centre for Mobility in Aging, and holds a Tier 1 Canada Research Chair in Geroscience.
Footnotes
Author Contributions
Conception and design: Y.C., V.F., S.Z., J.B.R. Methodology: Y.C., T.L., Y.F., V.F., C.O., S.Z., J.B.R. Data curation: Y.C. Data Analysis: Y.C., T.L. U.P.K. Visualization: Y.C., T.L. Writing – Original Draft: Y.C. Writing – Review & Editing: Y.C., T.L., U.P.K., I.D.S, G.B.L., T.N., A.C., K.Y.H.L., S.Y., J.D.S.W., C.Y.S., P.R., C.O., C.M.T.G., Y.F., V.F., C.L., S.Z., J.B.R. Supervision: J.B.R. Project administration: J.B.R. Funding acquisition: J.B.R.
Ethics declarations
Ethics declarations
This study was approved by the research ethics boards of the Jewish General Hospital, protocol number 2021-2762.
Competing Interests JBR has served as an advisor to GlaxoSmithKline and Deerfield Capital. JBR’s institution has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline and Biogen for projects unrelated to this research. JBR is the CEO of 5 Prime Sciences (http://www.5primesciences.com). TL and VF are employees of 5 Prime Sciences. YF consults for Fulcrum Genomics, 5 Prime Sciences, and Demetria. TN has received speaking fees from Boehringer Ingelheim and AstraZeneca. All other authors declare that there are no conflicts of interest. The opinions expressed in this manuscript are the authors’ own and do not reflect the views of the Canadian Longitudinal Study on Aging.
Data availability
The GWAS summary statistics were deposited to GWAS Catalog (https://www.ebi.ac.uk/gwas/). Accession numbers for European GWASs: GCST90199621-90201020; accession numbers for non-European GWASs: GCST90201021-90204063. Individual-level data are available from the Canadian Longitudinal Study on Aging (www.clsa-elcv.ca) for researchers who meet the criteria for access to de-identified CLSA data. HMDB database (http://www.hmdb.ca/system/downloads/current/serum_metabolites.zip). KEGG pathway database (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/database/keggPathway.txt.gz). PubChem database (https://pubchem.ncbi.nlm.nih.gov/). GTEx V8 release data can be found in https://www.gtexportal.org/home/datasets. UniProt database (https://www.uniprot.org/uniprot/). GENCODE resource (https://www.gencodegenes.org/). OMIM database (https://www.omim.org/downloads). IMPC database (https://www.mousephenotype.org/data/release). DrugBank database (https://go.drugbank.com)
Code availability
The GWAS was performed using GCTA-fastGWA (v1.93.2 beta). Multi-SNP-based conditional & joint association analysis was performed using GCTA-COJO (v1.93.2 beta). bedtools version v2.29.2 was used. KING package (v2.2.5) was used to remove individuals with first- and second-degree relatives. Snappy (available through Zenodo (https://doi.org/10.5281/zenodo.7328428, ref.78)) was used to identify LD-proxy SNPs. PLINK 1.9 was used to identify LD-independent SNPs from the trait and disease GWAS. All other data analyses were performed using R (version 4.0.5). R packages including dplyr (1.0.7), data.table (1.14.2), tidyverse (1.2.0), stringr (1.4.1), LDlinkR (1.1.2), TwoSampleMR (0.5.6), coloc (5.1.0), circlize (0.4.13), ComplexHeatmap (2.13.1), RcolorBrewer (1.1-3), ggpubr (0.4.0) and ggplot2 (3.3.5) were used for analysis and plotting. Other analyses and plotting scripts were made available through GitHub repository (https://github.com/richardslab/metabolomics_GWAS_CLSA) and also through Zenodo (https://doi.org/10.5281/zenodo.7331471) (ref.80).
References
- 1.Bar N, et al. A reference map of potential determinants for the human serum metabolome. Nature. 2020;588 doi: 10.1038/s41586-020-2896-2. [DOI] [PubMed] [Google Scholar]
- 2.Lee W-J, Hase K. Gut microbiota–generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 3.Pietzner M, et al. Plasma metabolites to profile pathways in noncommunicable disease multimorbidity. Nat Med. 2021;27:471–479. doi: 10.1038/s41591-021-01266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15 doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 5.Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49 doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 6.Hagenbeek FA, et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun. 2020;11 doi: 10.1038/s41467-019-13770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith GD. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33 doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 8.Lotta LA, et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet. 2021;53 doi: 10.1038/s41588-020-00751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feofanova Ev, et al. A Genome-wide Association Study Discovers 46 Loci of the Human Metabolome in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2020;107:849–863. doi: 10.1016/j.ajhg.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46 doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X, et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat Commun. 2022;13:1644. doi: 10.1038/s41467-022-29143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettunen J, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draisma HHM, et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kastenmüller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: an update. Hum Mol Genet. 2015;24:R93–R101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yazdani A, Yazdani A, Liu X, Boerwinkle E. Identification of Rare Variants in Metabolites of the Carnitine Pathway by Whole Genome Sequencing Analysis. Genet Epidemiol. 2016;40:486–91. doi: 10.1002/gepi.21980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B, et al. Loss-of-function variants influence the human serum metabolome. Sci Adv. 2016;2:e1600800. doi: 10.1126/sciadv.1600800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousri NA, et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat Commun. 2018;9:333. doi: 10.1038/s41467-017-01972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee EP, et al. An exome array study of the plasma metabolome. Nat Commun. 2016;7:12360. doi: 10.1038/ncomms12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gieger C, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hysi PG, et al. Metabolome Genome-Wide Association Study Identifies 74 Novel Genomic Regions Influencing Plasma Metabolites Levels. Metabolites. 2022;12:61. doi: 10.3390/metabo12010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bomba L, et al. Whole-exome sequencing identifies rare genetic variants associated with human plasma metabolites. Am J Hum Genet. 2022 doi: 10.1016/j.ajhg.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raina P, et al. Cohort Profile: The Canadian Longitudinal Study on Aging (CLSA) Int J Epidemiol. 2019;48:1752–1753j. doi: 10.1093/ije/dyz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamat MA, et al. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations. Bioinformatics. 2019;35 doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallois A, et al. A comprehensive study of metabolite genetics reveals strong pleiotropy and heterogeneity across time and context. Nat Commun. 2019;10:4788. doi: 10.1038/s41467-019-12703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen A-K, et al. On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinformatics. 2012;13 doi: 10.1186/1471-2105-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477 doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 2019;47:D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris JA, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51 doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tachmazidou I, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019;51:230–236. doi: 10.1038/s41588-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulit SL, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28 doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik R, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Harst P, Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Y, et al. Genome-wide analysis highlights contribution of immune system pathways to the genetic architecture of asthma. Nat Commun. 2020;11:1776. doi: 10.1038/s41467-020-15649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiou J, et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature. 2021;594:398–402. doi: 10.1038/s41586-021-03552-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365 doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giambartolomei C, et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahoor I, Rui B, Khan J, Datta I, Giri S. An emerging potential of metabolomics in multiple sclerosis: a comprehensive overview. Cell Mol Life Sci. 2021;78:3181–3203. doi: 10.1007/s00018-020-03733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganz T, et al. Serum asymmetric dimethylarginine and arginine levels predict microvascular and macrovascular complications in type 2 diabetes mellitus. Diabetes Metab Res Rev. 2017;33 doi: 10.1002/dmrr.2836. [DOI] [PubMed] [Google Scholar]
- 50.Moore SC, et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10:259–269. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song J, et al. The Relationship Between Body Mass Index and Bone Mineral Density: A Mendelian Randomization Study. Calcif Tissue Int. 2020;107:440–445. doi: 10.1007/s00223-020-00736-w. [DOI] [PubMed] [Google Scholar]
- 52.Skaaby T, et al. Estimating the causal effect of body mass index on hay fever, asthma and lung function using Mendelian randomization. Allergy. 2018;73:153–164. doi: 10.1111/all.13242. [DOI] [PubMed] [Google Scholar]
- 53.Yang X-L, et al. Causal link between lipid profile and bone mineral density: A Mendelian randomization study. Bone. 2019;127:37–43. doi: 10.1016/j.bone.2019.05.037. [DOI] [PubMed] [Google Scholar]
- 54.Grotzinger AD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demange PA, et al. Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction. Nat Genet. 2021;53:35–44. doi: 10.1038/s41588-020-00754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu T, Forgetta V, Greenwood CMT, Richards JB. Identifying Causes of Fracture Beyond Bone Mineral Density: Evidence From Human Genetics. J Bone Miner Res. 2022 doi: 10.1002/jbmr.4632. [DOI] [PubMed] [Google Scholar]
- 57.Nethander M, et al. BMD-Related Genetic Risk Scores Predict Site-Specific Fractures as Well as Trabecular and Cortical Bone Microstructure. J Clin Endocrinol Metab. 2020;105:e1344–e1357. doi: 10.1210/clinem/dgaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heemskerk MM, van Harmelen VJ, van Dijk KW, van Klinken JB. Reanalysis of mGWAS results and in vitro validation show that lactate dehydrogenase interacts with branched-chain amino acid metabolism. European Journal of Human Genetics. 2016;24:142–145. doi: 10.1038/ejhg.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanno T, et al. Lactate dehydrogenase M-subunit deficiency: a new type of hereditary exertional myopathy. Clinica Chimica Acta. 1988;173:89–98. doi: 10.1016/0009-8981(88)90359-2. [DOI] [PubMed] [Google Scholar]
- 60.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forgetta V, et al. Cohort profile: genomic data for 26 622 individuals from the Canadian Longitudinal Study on Aging (CLSA) BMJ Open. 2022;12:e059021. doi: 10.1136/bmjopen-2021-059021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics. 2011;88 doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang L, et al. A resource-efficient tool for mixed model association analysis of large-scale data. Nat Genet. 2019;51 doi: 10.1038/s41588-019-0530-8. [DOI] [PubMed] [Google Scholar]
- 66.Wang H, et al. Genotype-by-environment interactions inferred from genetic effects on phenotypic variability in the UK Biobank. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44 doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen A-K, et al. On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinformatics. 2012;13:120. doi: 10.1186/1471-2105-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43 doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visscher PM, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stacey D, et al. ProGeM: a framework for the prioritization of candidate causal genes at molecular quantitative trait loci. Nucleic Acids Res. 2019;47:e3. doi: 10.1093/nar/gky837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emdin CA, Khera Av, Kathiresan S. Mendelian Randomization. JAMA. 2017;318:1925. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 75.Skrivankova VW, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021:n2233. doi: 10.1136/bmj.n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skrivankova VW, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization. JAMA. 2021;326:1614. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- 77.Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 78.Forgetta Vincenzo. Snappy: A flexible SNP proxy finder. 2022 doi: 10.5281/zenodo.7328428. [DOI] [Google Scholar]
- 79.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y. Nature Genetics. Zenodo; 2022. Code for the analyses described in Chen et al. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS summary statistics were deposited to GWAS Catalog (https://www.ebi.ac.uk/gwas/). Accession numbers for European GWASs: GCST90199621-90201020; accession numbers for non-European GWASs: GCST90201021-90204063. Individual-level data are available from the Canadian Longitudinal Study on Aging (www.clsa-elcv.ca) for researchers who meet the criteria for access to de-identified CLSA data. HMDB database (http://www.hmdb.ca/system/downloads/current/serum_metabolites.zip). KEGG pathway database (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/database/keggPathway.txt.gz). PubChem database (https://pubchem.ncbi.nlm.nih.gov/). GTEx V8 release data can be found in https://www.gtexportal.org/home/datasets. UniProt database (https://www.uniprot.org/uniprot/). GENCODE resource (https://www.gencodegenes.org/). OMIM database (https://www.omim.org/downloads). IMPC database (https://www.mousephenotype.org/data/release). DrugBank database (https://go.drugbank.com)
The GWAS was performed using GCTA-fastGWA (v1.93.2 beta). Multi-SNP-based conditional & joint association analysis was performed using GCTA-COJO (v1.93.2 beta). bedtools version v2.29.2 was used. KING package (v2.2.5) was used to remove individuals with first- and second-degree relatives. Snappy (available through Zenodo (https://doi.org/10.5281/zenodo.7328428, ref.78)) was used to identify LD-proxy SNPs. PLINK 1.9 was used to identify LD-independent SNPs from the trait and disease GWAS. All other data analyses were performed using R (version 4.0.5). R packages including dplyr (1.0.7), data.table (1.14.2), tidyverse (1.2.0), stringr (1.4.1), LDlinkR (1.1.2), TwoSampleMR (0.5.6), coloc (5.1.0), circlize (0.4.13), ComplexHeatmap (2.13.1), RcolorBrewer (1.1-3), ggpubr (0.4.0) and ggplot2 (3.3.5) were used for analysis and plotting. Other analyses and plotting scripts were made available through GitHub repository (https://github.com/richardslab/metabolomics_GWAS_CLSA) and also through Zenodo (https://doi.org/10.5281/zenodo.7331471) (ref.80).