Abstract
Background
Rapid tuberculosis (TB) drug susceptibility testing (DST) is crucial. Genotype MTBDRsl is a widely deployed World Health Organization (WHO)–endorsed assay. Programmatic performance data, including non-actionable results from smear-negative sputum, are scarce.
Methods
Sputa from Xpert MTB/RIF individuals (n = 951) were routinely-tested using Genotype MTBDRplus and MTBDRsl (both version 2). Phenotypic DST was the second-line drug reference standard. Discrepant results underwent Sanger sequencing.
Findings
89% (849 of 951) of individuals were culture-positive (56%, 476 of 849 smear-negative). MTBDRplus had at least 1 nonactionable result (control and/or TB-detection bands absent or invalid, precluding resistance reporting) in 19% (92 of 476) of smear-negatives; for MTBDRsl, 40% (171 of 427) were nonactionable (28%, 120 of 427 false-negative TB; 17%, 51 of 427 indeterminate). In smear-negatives, MTBDRsl sensitivity for fluoroquinolones was 84% (95% confidence interval, 67%–93), 81% (54%–95%) for second-line injectable drugs, and 57% (28%–82%) for both. Specificities were 93% (89%–98%), 88% (81%–93%), and 97% (91%–99%), respectively. Twenty-three percent (172 of 746) of Xpert rifampicin-resistant specimens were MTBDRplus isoniazid-susceptible. Days-to-second-line-susceptibility reporting with the programmatic advent of MTBDRsl improved (6 [5–7] vs 37 [35–46]; P < .001).
Conclusions
MTBDRsl did not generate a result in 4 of 10 smear-negatives, resulting in substantial missed resistance. However, if MTBDRsl generates an actionable result, that is accurate in ruling-in resistance. Isoniazid DST remains crucial. This study provides real-world, direct, second-line susceptibility testing performance data on non-actionable results (that, if unaccounted for, cause an overestimation of test utility), accuracy, and care cascade impact.
Keywords: Genotype MTBDRplus, Genotype MTBDRsl, smear-negative, TB, resistance
The MTBDRsl assay fails to generate a result in almost half of smear-negatives, resulting in substantial missed resistance, but has high rule-in value. Many rifampicin-resistant individuals would likely benefit from isoniazid. Programmatic direct testing was nevertheless associated with care cascade improvements.
Drug-resistant tuberculosis (DR-TB) is a leading cause of death. Globally, there were half a million rifampicin-resistant (RR) TB cases in 2019; 78% were estimated to be multidrug-resistant (MDR) [1]. Only 59% of RR-MDR individuals started on treatment in 2018 were treated successfully [2], partly due to the underdiagnosis of resistance to drugs other than rifampicin (RIF) such as isoniazid and the fluoroquinolones (FQs) [3, 4].
The Genotype MTBDRplus (Hain Lifesciences, Germany) and MTBDRsl (Hain Lifesciences, Germany) molecular line probe assays (LPAs) are globally used for rapid DR-TB detection. Both are World Health Organization (WHO)–endorsed and commercially available [5]. According to the Western Cape Province Department of Health TB guidelines [6], MTBDRplus is done after Xpert MTB/RIF (Xpert) to check for Xpert-detected false-positive rifampicin resistance and confirm MDR [7]. MTBDRsl is subsequently done to detect second-line resistance. One underappreciated yet important component of these workflows is that, even when an individual is confirmed as TB-positive using Xpert, the downstream reflex test must itself successfully amplify Mycobacterium tuberculosis complex (Mtb) DNA (LPAs Mtb detection is reported as TUB-band positivity). This applies to many reflex technologies and not just LPAs, including new drug susceptibility tests (DSTs) such as Xpert MTB/XDR [8, 9], which have yet to be available at scale.
As frontline TB test performance improves, it can outstrip reflex tests’ ability to detect TB and do DST (eg, Xpert MTB/RIF is almost always done before the LPAs, despite LPAs being an older technology) [10]. Both MTBDRplus and MTBDRsl can generate nonactionable results (indeterminate or invalid results) that are critical to report in order to quantify the overall number of drug-resistant cases missed (ie, not just due to imperfect sensitivity for resistance but also due to a failure of the test to detect TB). Such performance data that includes nonactionable results are scarce and a major limitation of the current literature. Despite increased demand for DST due to new oral regimens for RR-MDR TB (with the possibility of new FQ-based first-line regimens), MTBDRsl is 1 of only 2 WHO-endorsed rapid tests that can be used to confirm eligibility for these regimens.
The WHO recommends that MTBDRplus be used on smear-positive sputum (direct testing) and on culture isolates (indirect testing) for smear-negatives [11]. In contrast, MTBDRsl version 2 is recommended for direct smear-negative testing; however, evidence is of “low certainty” [5, 12], and meta-analyses have had insufficient data to create summary point estimates [13–16]. This uncertainty in performance is one reason why LPA uptake for the direct testing is suboptimal. In a global survey of 32 LPA-using laboratories, 66% and 50% tested smear-negative specimens with MTBDRplus and MTBDRsl, respectively [17], despite the positive WHO recommendation. Critically, more data are therefore needed.
Our overarching aim was to evaluate MTBDRplus (version 2) and MTBDRsl (version 2) performance, including in smear-negative specimens, and describe the nonactionable result rate. Importantly, we did this in a programmatic context that relies on affordable existing diagnostic tools to help guide therapeutic decisions. This approach enabled us to evaluate the association between the expansion of direct second-line DST and time to treatment and compare this to the period prior to the advent of direct second-line DST. Our intention was to provide data for laboratories and clinicians diagnosing and treating drug-resistant TB in resource-constrained settings where programmatic laboratory decisions and policies related to rapid diagnostic testing follow WHO guidance.
METHODS
Study Design
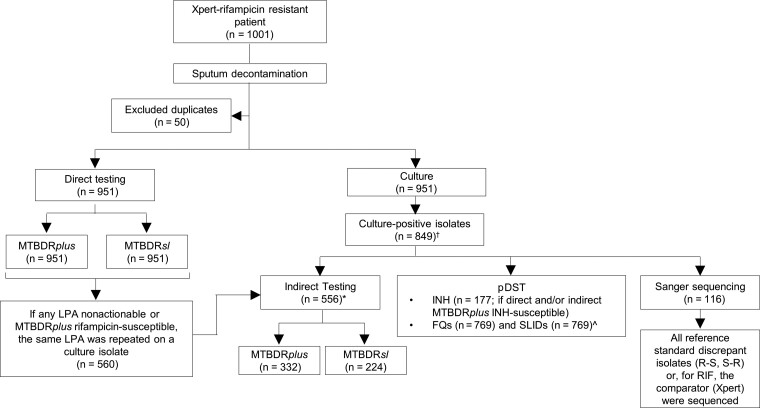
This study was performed in a programmatic context following the TB diagnostic algorithm in the Western Cape, South Africa (Figure 1). Direct testing was performed initially using MTBDRplus and MTBDRsl on sputum consecutively tested with no study-specific criteria between 1 June 2016 and 30 September 2019. MTBDRplus was performed on specimens of all smear status, defined below as the “after period.” All valid results were reported and reflexed for MTBDRsl testing. All TUB-band negative, indeterminate for 1 or both drugs were reported as invalid (MTBDRplus/MTBDRsl); rifampicin-susceptible results were reported as discrepant and reflexed for indirect testing using a confirmed culture-positive isolate. All culture isolate results, except Sanger sequencing, formed part of indirect diagnostic workflows including Genotype MTBDRplus, MTBDRsl, and phenotypic drug susceptibility testing (pDST), and all valid results were reported immediately. Phenotypic DST was done on specimens with valid direct and indirect LPA results. All discrepant results for MTBDRplus/MTBDRsl with reference standard pDST were resolved with repeat testing on the cultured isolate. For discrepancies that remained even after repeat testing, sequencing was performed (Figure 1).
Figure 1.
Testing flow diagram showing direct and indirect testing using MTBDRplus and MTBDRsl and the use of reference standard phenotypic testing for second-line drugs, irrespective of the LPA result. Prior to the study, the flow of tests were the same except MTBDRsl was not used and MTBDRplus was only done directly if the specimen was smear-positive. *4 direct nonactionables were culture-negative and unable to be tested indirectly. †102 Xpert-positives were not culture-positive and hence did not have an isolate available. ^80 isolates were contaminated upon regrowth for FQ and SLID pDST. Abbreviations: FQ, fluoroquinolones; INH, isoniazid; LPA, line probe assay; MDR, multidrug-resistant; pDST, phenotypic drug susceptibility testing; R, resistant; RIF, rifampicin; S, susceptible; SLID, second-line injectable drugs; Xpert, Xpert MTB/RIF.
Sputum Collection and Preparation
In the Western Cape Province, 2 sputum samples were collected upfront for screening of presumptive TB per local guidelines [6]. Sputum processing and testing was done at the National Health Laboratory Service Green Point reference laboratory in Cape Town, South Africa. Pretreatment individuals who were first tested using Xpert MTB/RIF (version 4.3; Xpert) formed part of the then standard-of-care algorithm [18]. A paired sputum specimen from Xpert–RR individuals (n = 1001) was decontaminated using n-acetyl-L-cysteine-sodium hydroxide (final concentration, 1%) and the sediment resuspended in 2 mL phosphate buffer [19]. Auramine microscopy was performed. From decontaminated sputum, 0.5 mL was inoculated into a mycobacteria growth indicator tube (MGIT; Becton Dickinson) and incubated in a BACTEC MGIT960 instrument for ≤35 days (our programmatic standard of care due to space limitations).
DNA Extraction and Line Probe Assay Testing
DNA extracted per manufacturer’s guidelines [20, 21] from resuspended sputum sediments was tested directly with MTBDRplus and MTBDRsl (version 2 of both) in parallel by a single operator irrespective of smear status. The GT blot (Hain Lifesciences) and Genoscan software (GS-001, Hain Lifesciences) were used to analyze results followed by operator visual confirmation. All invalid tests (direct testing) were repeated as recommended (the repeat result was reported in analyses). For specimens (direct testing) that were TB-negative per LPAs (ie, TUB-band negative), indeterminate for at least 1 locus, or with an LPA DST result discrepant with pDST, the corresponding isolate was tested using the same LPA (indirect testing). A total of 332 and 224 isolates were tested using MTBDRplus and MTBDRsl, respectively. The manufacturer-recommended 2.2°C/s ramp rate [17, 22] and ISO15189 standards were used. Results were interpreted per Supplementary Table 1.
TB and Phenotypic Drug Susceptibility Testing Reference Standards
MGIT960 culture positivity with MTBDRplus TUB-positivity was used for the detection of TB. Rifampicin pDST was not done. pDST was done programmatically for isoniazid, FQs, and second-line injectable drugs. Per the algorithm, only MTBDRplus RR, isoniazid-susceptible isolates received isoniazid pDST to ensure resistance was not excluded (we are hence unable to calculate MTBDRplus’s sensitivity, specificity, and positive predictive value (PPV) for isoniazid resistance). If direct MTBDRplus was nonactionable or isoniazid susceptible, indirect MTBDRplus testing was done and, only based on this result, was isoniazid pDST done (hence, only the negative predictive value (NPV) of indirect MTBDRplus for resistance was calculable). See the Supplementary Methods for more information.
Discrepant Analysis
Sanger sequencing was used as the composite reference standard to resolve discrepancies involving LPAs, pDST, and Xpert RR and MTBDRplus rifampicin-susceptible specimens (Supplementary Methods, Supplementary Table 6).
Implementation and Effect of Programmatic MTBDRplus and MTBDRsl Testing
We compared the diagnostic care cascade in the “before algorithm” (2 January 2012–30 December 2015) vs the “after algorithm” (1 June 2016- 30 September 2019) periods. In the before algorithm period, programmatic DST for isoniazid, FQs, and amikacin was done phenotypically. MTBDRplus (includes v1) was done routinely for both rifampicin and isoniazid directly in smear-positives or on culture isolates. In the after algorithm period, MTBDRplus and MTBDRsl (both version 2) were implemented programmatically and reported for potential patient management (see the Supplementary Methods for more detail on these periods).
Statistical Analyses
GraphPad Prism (version 6; GraphPad Software) and Stata (version 14.0; StataCorp; 2 sample proportion test and McNemar test) were used. P values ≤.05 were significant.
Ethics
This study was done in accordance with relevant guidelines and regulations approved by the Health Research Ethics Committee of Stellenbosch University and the Western Cape Province Department of Health. Permission was granted to access anonymized residual specimens collected as part of routine diagnostic practice, and informed consent waived.
RESULTS
Cohort Characteristics
Of 1001 Xpert RR sputa, 95% (951) were from unique patients, 89% (849) were confirmed culture-positive (93 were culture-negative and 10 culture-contaminated), and 81% (769) had a usable second-line pDST result (8%; 80 contaminated; Figure 1). Most individuals were male with smear-negative TB (Supplementary Table 2). In individuals with a known human immunodeficiency virus (HIV) status, 50% (203 of 404) were with HIV. Those with HIV were more likely to be sputum smear-negative than those not with HIV (59%, 120 of 203 vs 48%, 110 of 230; P = .018).
Smear Microscopy, Culture, and Phenotypic DST Results
Among the culture-positives, 44% (373 of 849) and 56% (476 of 849) were sputum smear-positive and smear-negative, respectively. Using MTBDRplus, 21% (177 of 849) and 60% (509 of 849) were classified as rifampicin-monoresistant and MDR (Figure 2). Using MTBDRsl, 5% (42 of 769), 1% (11 of 769), and 2% (19 of 769) were FQ-resistant, second- line injectable drug (SLID)-resistant, or both FQ- and SLID-resistant, respectively (Figure 3).
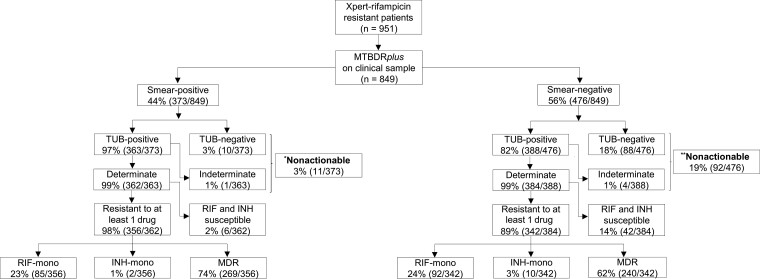
Figure 2.
Direct MTBDRplus testing of sputum is successful in almost all smear-positives and most smear-negatives; however, it fails to generate a susceptibility result in a significant minority of smear-negatives (1 in 5), indicating that a failure to detect tuberculosis is the primary cause of drug resistance being missed (ie, nonactionable results). Furthermore, a significant minority of Xpert RIF-resistant patients do not have MDR per MTBDRplus, suggesting a continued role for isoniazid drug susceptibility testing. Importantly, in patients with actionable MTBDRplus results, sensitivity and specificity for resistance did not differ by smear status. Resistance classifications on the bottom 2 rows of boxes are per direct MTBDRplus. Of the 951 Xpert rifampicin-resistant patients, only 849 were confirmed culture-positive. *Indirect smear-positive MTBDRplus results: MDR (n = 7), RIF-mono (n = 0), INH-mono (n = 1), fully susceptible (n = 3), and nonactionable (n = 0). **Indirect smear-negative MTBDRplus results: MDR (n = 69), RIF-mono (n = 0), INH-mono (n = 3), fully susceptible (n = 20), and nonactionable (n = 0). Abbreviations: INH, isoniazid; mono, monoresistant; MDR, multidrug-resistant; RIF, rifampicin; TUB, TUB-band; Xpert, Xpert MTB/RIF.
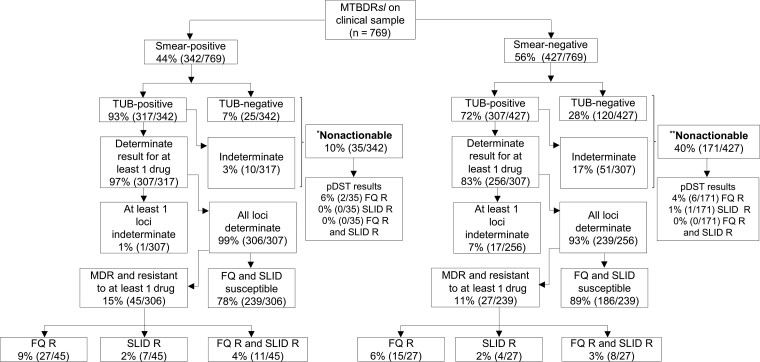
Figure 3.
Although direct MTBDRsl testing of sputum is successful in most patients, it results in relatively high proportions of nonactionable results in smear-positives and especially in smear-negatives. MTBDRsl failed in 4 of 10 smear-negative patients with Xpert-diagnosed rifampicin resistance. As seen for MTBDRplus, a failure to generate an actionable result on smear-negatives was the primarily cause of missed resistance (as opposed to a false-negative susceptible result). Resistance classifications on the bottom 2 rows of boxes are per direct MTBDRsl. Of the 849 culture-positive patients, only 769 had usable pDST (80-contaminated). *Indirect smear-positive MTBDRsl results: FQ-R (n = 3), SLID-R (n = 0), FQ-R and SLID-R (n = 0), fully susceptible (n = 33), and nonactionable (n = 0). **Indirect smear-negative MTBDRsl results: FQ-R (n = 7), SLID-R (n = 4), FQ-R and SLID-R (n = 2), fully susceptible (n = 175), and nonactionable (n = 0). Abbreviations: FQ, fluoroquinolones; MDR, multidrug-resistant; pDST, phenotypic drug susceptibility testing; R, resistant; SLID, second-line injectable drug; TUB, TUB-band.
MTBDRplus
Nonactionables
Three percent (11 of 373) and 19% (92 of 476) of sputum smear-positives and smear-negatives had nonactionable results, respectively; of those, 70% (521 of 746) were phenotypically isoniazid resistant (Figure 2). Of the sputum smear-negative nonactionables, 18% (88 of 476) were due to a false-negative TB result and 1% (4 of 476) were due to an indeterminate call (Figure 2). Nonactionable results from indirect testing are provided in Supplementary Figure 1. No MTBDRplus invalid results occurred.
MTB
The sensitivity of MTBDRplus was 97% (363 of 373) and 82% (388 of 476; P < .001) for sputum smear-positive and smear-negative TB, respectively (Table 1).
Table 1.
Accuracy of Direct MTBDRplus and MTBDRsl Testing for Tuberculosis and Phenotypic Second-line Drug Resistance in Sputum of Xpert-Positive Rifampicin-Resistant Patients
| Assay | Overall | Smear-Positive | Smear-Negative | ||||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | ||
| MTBDRplus | Tuberculosis | 88 (751/849) 86–90 | 43 (40/93) 32–53 | 97 (363/373) 94–98 | 36 (4/11) 10–69 | 82 (388/476) 77–84 aP < .001 | 44 (36/82) 32–54 aP = .635 |
| MTBDRsl | Tuberculosis | 82 (696/849) 79–84 bP < .001 | 51 (47/93) 32–54 bP = .303 | 93 (347/373) 90–95 bP = .006 | 73 (8/11) 39–93 bP = .086 | 73 (349/476) 69–77 aP < .001 bP = .002 | 48 (39/82) 36–58 aP = .117 bP < .001 |
| Fluoroquinolones | 87 (71/82) 77–93 | 93 (297/321) 90–96 | 89 (40/45) 75–96 | 92 (180/195) 88–96 | 84 (31/37) 67–93 aP = .105 | 93 (117/126) 89–98 aP = .855 | |
| Second-line injectable drugs | 84 (32/38) 68–93 cP = .720 | 94 (317/339) 90–95 cP = .820 | 86 (19/22) 65–97 cP = .001 | 97 (205/212) 93–98 cP = .108 | 81 (13/16) 54–95 aP = .011 cP = .821 | 88 (112/127) 81–93 aP = .002 cP = .052 | |
| Fluoroquinolone and second-line injectable drugs | 70 (19/27) 69–98 | 97 (257/264) 94–98 | 85 (11/13) 54–98 | 97(165/169) 94–99 | 57 (8/14) 28–82 aP = .118 | 97 (92/95) 91–99 aP = .701 | |
Data are % (n/N), 95% confidence interval. All P values which were statistically significant appeared in bold.
Within-row comparisons between smear statuses.
Within-column comparisons for MTBDRsl vs MTBDRplus.
Within-column comparisons for second-line injectable drugs vs fluoroquinolones.
Rifampicin
Ninety-one percent (686 of 746) of the Xpert RR patients whose direct MTBDRplus was actionable were MTBDRplus RR (24% [177 of 746] had MTBDRplus-defined rifampicin monoresistance). In a head-to-head comparison of direct MTBDRplus and Xpert actionable results, 8% (60 of 746) were Xpert-resistant MTBDRplus-susceptible, with most discrepants in smear-negative TB rather than in smear-positive TB (Figure 2). Overall, of the discrepants successfully sequenced (9 culture-contaminated, 3 nonamplifiable), 85% (22 of 26) resolved in favor of Xpert (Table 2). Indirect MTBDRplus results are provided in Supplementary Figure 1.
Table 2.
Sequencing of MTBDRplus Targets (rpoB, katG, inhA Promoter Region) Done to Resolve Discrepant Results Either Between MTBDRplus and Xpert (Rifampicin) or MTBDRplus and Phenotype (Isoniazid)
| Sequencing | ||||||||
|---|---|---|---|---|---|---|---|---|
| Locus | MTBDRplus | Comparator Result | Mutation | No. of Isolates | No. With Heteroresistance | Susceptibility Result | Resolved in Favor of Line Probe Assay or Comparator | |
| Rifampicin | rpoB a (n = 29) | S | R | S531L | 8 | 1 | R | Xpert |
| … | … | H526Y | 2 | 0 | R | Xpert | ||
| … | … | D516V | 3 | 1 | R | Xpert | ||
| … | … | Q513P | 1 | 0 | R | Xpert | ||
| … | … | L511Pb | 8 (1 Double mutant with D485N) | 1 | R | Xpert | ||
| … | … | WT | 4 | 0 | S | MTBDRplus | ||
| … | … | … | NR | 3 | … | … | … | |
| Discrepant resolution by sequencing | 85% (22/26) resistant (resolved in favor of Xpert) 15% (4/26) susceptible (resolved in favor of MTBDRplus) |
|||||||
| Isoniazid | katG c (n = 24) | S | R | G312C | 1 | … | R | pDST |
| … | … | … | S315T | 3 | … | R | pDST | |
| … | … | … | WT | 19 | … | S | MTBDRplus | |
| … | NR 1 | |||||||
| inhA promoterc (n = 24) | S | R | −8 T/C WT | 1 23 | … | R, S | pDST MTBDRplus | |
| … | Discrepant resolution by sequencing | 21% (5/24) resistant (resolved in favor of pDST) 79% (19/24) susceptible (resolved in favor of MTBDRplus) |
||||||
Sequencing suggested Xpert is more sensitive for rifampicin resistance than MTBDRplus. MTBDRplus detected mutations known to cause isoniazid resistance better than pDST. See Supplementary Methods for how line probe assay results were categorized as discrepant.
Abbreviations: NR, not reportable (did not amplify for sequencing); pDST, phenotypic drug susceptibility testing; R, resistant; S, susceptible; WT, wild type; Xpert, Xpert MTB/RIF.
Only Xpert rifampicin-resistant and MTBDRplus rifampicin-susceptible discrepant sputa were sequenced from the isolate.
L511P is considered borderline by the World Health Organization, which recommends that people found with this mutation be classified as resistant [23].
Discrepant isolates sequenced included only MTBDRplus-susceptible that were phenotypic-resistant (due to contemporaneous programmatic algorithm).
Isoniazid
Sixty-eight percent (509 of 746) of Xpert RR patients whose direct MTBDRplus was actionable had, per MTBDRplus, MDR and 2% (12 of 746) isoniazid monoresistance (the remainder were rifampicin-monoresistant). A total of 328 received indirect MTBDRplus testing, and 53% (177 of 328) were MTBDRplus RR, isoniazid-susceptible (Supplementary Figure 1). There were 17% (30 of 177) that were phenotypically resistant. We could only calculate MTBDRplus’s NPV for isoniazid resistance when done indirectly, which was 83% (147 of 177). When discrepant isoniazid results (indirect MTBDRplus-susceptible, pDST-resistant, n = 30) were analyzed, 80% (24 of 30) had usable sequences. Seventy-nine percent (19 of 24), all of which were sequencing wild-type, resolved in favor of MTBDRplus (Table 2), resulting in NPV increasing to 97% (166 of 171).
MTBDRsl
Nonactionable
When done directly, 10% (35 of 342) of sputum smear-positives and 40% (171 of 427) of smear-negatives were nonactionable (Figure 3). In addition, 4% (8 of 206), 0% (1 of 206), and 0% (0 of 206) of nonactionables were phenotypically resistant to FQs, SLIDs, or both FQs and SLIDs, respectively. Like MTBDRplus on sputum smear-negatives, most MTBDRsl smear-negative results were nonactionable due to a false-negative TB result (28%, 120 of 427) or an indeterminate result (17%, 51 of 427; Figure 3). A total of 28 MTBDRsl results were initially invalid prior to pDST (1%, 2 of 373 for sputum smear-positives vs 5%, 26 of 476 for sputum smear-negatives; P < .001; Supplementary Table 3), but all resolved upon retesting (and were hence ultimately not nonactionable). No indirect nonactionable results occurred (Supplementary Figure 2).
MTB
Sensitivity was 93% (347 of 373) and 73% (349 of 476; P < .001) for sputum smear-positive and smear-negative specimens, respectively (Table 1), and less than MTBDRplus in the same individuals (97%; 95% confidence interval [CI], 94%–98% vs 93%; 95% CI, 90%–95%; P < .001) for sputum smear-positives and (82%; 95% CI, 77%–84% vs 73%; 95% CI, 69%–77%; P < .001) for smear-negatives.
Fluoroquinolones
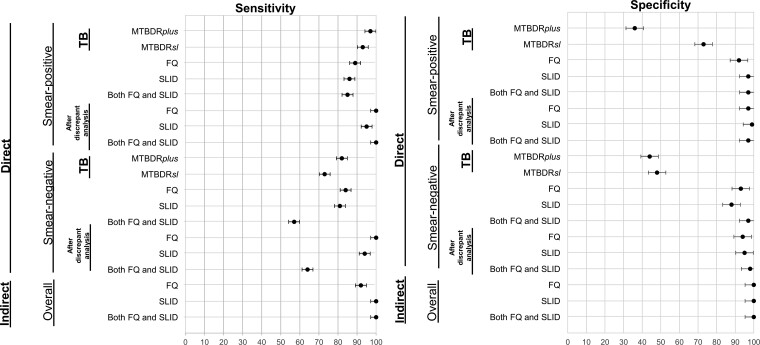
For direct sputum smear-positive and smear-negative testing, sensitivities were 89% (40 of 45) and 84% (31 of 37; P = .105) and specificities were 92% (180 of 195) and 93% (117 of 126; P = .855), respectively (Table 1, Figure 4). For indirect testing, sensitivity was 92% (12 of 13) and specificity was 100% (211 of 211; Supplementary Table 4). When discrepant FQ results from direct testing were analyzed (MTBDRsl-resistant pDST-susceptible, n = 24; MTBDRsl-susceptible pDST-resistant, n = 11), 83% (29 of 35) generated usable sequences. Sixty-nine percent (20 of 29) of discrepancies favored MTBDRsl and 31% (9 of 29) favored pDST (Table 3). MTBDRsl falsely reported 2 specimens with gyrA S95T natural polymorphisms [24] as resistant through the absence of a wild-type band (WT3, MUT3C). After following discrepant analysis reclassification, sensitivities and specificities increased (Figure 4, Supplementary Table 5).
Figure 4.
Selected summary forest plots showing sensitivity and specificity estimates (with 95% confidence intervals) for MTBDRplus and MTBDRsl. Importantly, only patients first detected as TB positive (top 2 rows) can generate an actionable line probe assay drug susceptibility testing result. Estimates for smear-negatives were lower than for smear-positives and, overall, estimates for SLIDs were lower than for FQs. All estimates improved in favor of LPAs after discrepant resolution. Abbreviations: FQ, fluoroquinolones; SLID, second-line injectable drugs; TB, tuberculosis.
Table 3.
Sequencing of MTBDRsl Targets (gyrA, rrs) to Resolve Results Discrepant With Phenotypic Drug Susceptibility Testing
| Sequencing | ||||||||
|---|---|---|---|---|---|---|---|---|
| Locus | MTBDRsl | pDST | Mutation | No. of Isolates | Susceptibility Result | Resolved in Favor of Line Probe Assay or pDST | ||
| Fluoroquinolones | gyrA (n = 11) | S | R | G81Ca | 1 | S | MTBDRsl | |
| … | … | … | A88T | 1 | R | pDST | ||
| … | … | … | WT | 9 | S | MTBDRsl | ||
| (n = 24) | R | S | A88T | 1 | R | MTBDRsl | ||
| … | … | C86T | 1 | R | MTBDRsl | |||
| … | … | D89N | 1 | R | MTBDRsl | |||
| … | … | A90V | 4 | R | MTBDRsl | |||
| … | … | S91P | 1 | R | MTBDRsl | |||
| … | … | D94G | 2 | R | MTBDRsl | |||
| … | … | S95Tb | 2 | S | pDST | |||
| … | … | WT | 6 | S | pDST | |||
| … | … | NR | 6 | … | … | |||
| Discrepant resolution by sequencing | 69% (20/29) in favor of MTBDRsl 31% (9/29) in favor of pDST |
|||||||
| Second-line injectable drugs | rrs (n = 6) | S | R | WT | 3 | … | S | MTBDRsl |
| … | … | … | NR | 3 | … | … | … | |
| (n = 22) | R | S | WT | 8 | … | S | pDST | |
| … | … | … | A1401G | 1 | … | R | MTBDRsl | |
| … | … | … | NR | 13 | … | … | … | |
| … | … | … | … | … | … | … | … | |
| Discrepant resolution by sequencing | 33% (4/12) in favor of MTBDRsl 67% (8/12) in favor of pDST |
|||||||
Most fluoroquinolone discrepants resolved in favor of MTBDRsl, whereas most second-line injectable drug discrepants resolved in favor of pDST.
Abbreviations: NR, number of specimens that did not amplify for sequencing; pDST, phenotypic drug susceptibility testing; R, resistant; S, susceptible; WT, wild-type.
G81C, silent mutation.
Second-Line Injectable Drugs
For direct testing in sputum smear-positives and smear-negatives, sensitivities were 86% (19 of 22) and 81% (13 of 16; P = .011), respectively, and specificities were 97% (205 of 212) and 88% (112 of 127; P = .002), respectively (Table 1, Figure 3). For indirect testing, sensitivity was 100% (6 of 6) and specificity was 100% (218 of 218; Supplementary Table 4, Figure 4). When direct MTBDRsl-pDST discrepant results (MTBDRsl-resistant pDST-susceptible, n = 22 and MTBDRsl-susceptible pDST-resistant, n = 6) were analyzed, 43% (12 of 28) had sequenceable isolate DNA. In contrast to FQs, most discrepancies (67%, 8 of 12) resolved in favor of pDST (Table 3, Supplementary Table 7). Following reclassification, sensitivity and specificity increased (Figure 4, Supplementary Table 5).
Joint FQ and SLID Resistance
For sputum smear-positives and smear-negatives, direct sensitivities were 85% (11 of 13) and 57% (8 of 14; P = .118), respectively, and specificities were 97% (165 of 169) and 97% (92 of 95; P = .701), respectively (Table 1, Figure 3). Indirect testing sensitivity and specificity were very high (Supplementary Table 4, Figure 4). Like that observed for the individual drug classes, after discrepancy resolution, MTBDRsl sensitivity and specificity increased (Supplementary Table 5).
Diagnosis Care Cascade Gaps in Before and After Periods
We compared programmatic data from the period immediately preceding the study (before period when MTBDRplus was the only LPA done directly, only on sputum smear-positives, and the only second-line testing was pDST) to a similar period after the start of study testing (after period; both LPAs were done, at a minimum, directly and reported for routine patient management). With MTBDRsl implementation, the proportion of individuals on treatment without second-line DST results decreased from 23% (668 of 2938) to 5% (40 of 799; P < .001; Table 4), and second-line DST results were available more quickly (33 [29–38] to 16 [13–22] days for smear-positives and 42 [36–50] to 22 [18–27] days for smear-negatives), even after factoring in many smear-negatives with direct nonactionable results that required subculture for further testing compared with smear-positives (37%, 143 of 383 vs 9%, 36 of 416; P < .001; Table 4).
Table 4.
Comparison of Key Care Cascade Gaps for the Diagnosis of Drug Resistance Before and After the Implementation of Improved Molecular Diagnostics for Resistance Beyond Rifampicin
| … | Retrospective Period MTBDRplus Only on Smear-Positives Second-line DST by pDST Only | Prospective Period MTBDRplus and MTBDRsl Irrespective of Smear Status Second-line pDST Still Done | ||||
|---|---|---|---|---|---|---|
| Overall (n = 2938) | Smear-Positive (n = 1674) | Smear-Negative (n = 1264) | Overall (n = 799) | Smear-positive (n = 416) | Smear-negative (n = 383) | |
| On treatment without receiving any second-line DST | 23 (668/2938) | 21 (357/1674) | 25 (311/1264) aP = .358 | 5 (40/799) bP < .001 | 2 (7/416) bP < .001 | 9 (33/383) bP < .001 bP < .001 |
| MTBDRplus direct testing | N/A | 100 (1674/1674) | N/A | 100 (799/799) | 100 (416/416) | 100 (383/383) |
| With an actionable result | N/A | 79 (1317/1674) | N/A | 99 (797/799) | 100 (416/416) | 99 (381/383) aP = .140 |
| Without an actionable result | N/A | 21 (357/1674) | N/A | 0 (2/799) | 0 (0/416) | 1 (2/383) |
| MTBDRsl direct testing | N/A | N/A | N/A | 100 (799/799) | 100 (416/416) | 100 (383/383) |
| With an actionable result | N/A | N/A | N/A | 78 (622/799) | 91 (380/416) | 63 (242/383) aP < .001 |
| Without an actionable result | N/A | N/A | N/A | 22 (177/799) | 9 (36/416) | 37 (141/383) aP < .001 |
| Days to result (actionable or nonactionable) | N/A | N/A | N/A | 6 (5–7) | 6 (5–7) | 6 (5–7) aP < .001 |
| MTBDRsl indirect testing | N/A | N/A | N/A | 22 (177/177) | 9 (36/36) | 37 (141/141) |
| With an actionable result | N/A | N/A | N/A | 22 (177/177) | 9 (36/36) | 37 (141/141) |
| Without an actionable result | N/A | N/A | N/A | 0 | 0 | 0 |
| Days to result (actionable or nonactionable) | N/A | N/A | N/A | 22 (16–26) | 16 (13–22) | 22 (18–27) aP = .081 |
| pDST | 77 (2270/2938) | 79 (1317/1674) | 75 (953/1264) aP = .358 | 94 (750/799) bP < .001 | 96 (400/416) bP < .001 | 91 (350/383) aP = .500 bP < .001 |
| Days to result (interquartile range ) | 37 (35–46) | 33 (29–38) | 42 (36–50) aP < .001 | 30 (27–36) bP < .001 | 28 (25–35) bP < .001 | 34 (30–40) aP < .001 bP < .001 |
| Overall, second-line DST | … | … | … | … | … | … |
| Patients who required second-line DST on isolates (indirect MTBDRsl or pDST) when direct MTBDRsl was nonactionable | 0 | 0 | 0 | 22 (177/799) | 9 (36/416) | 37 (141/383) aP < .001 |
| Days to first actionable second-line DST result (direct MTBDRsl, indirect MTBDRsl, or pDST) | 37 (35–46) | 33 (29–38) | 42 (36–50) aP < .001 | 6 (5–7) | 6 (5–7) | 6 (5–7) aP < .001 |
Implementation of first-line MTBDRplus testing on Xpert rifampicin-resistant sputum to include smear-negatives and MTBDRsl testing on all sputum resulted in a greater proportion of patients receiving second-line DST, reduced reliance on culture, and reduced turnaround time. The Supplementary Methods section contains more information on these periods. Data are median (interquartile range) or % (n/N). All P values which were statistically significant appeared in bold.
Abbreviations: DST, drug susceptibility testing; N/A, nonapplicable; pDST, phenotypic drug susceptibility testing.
Comparisons within rows and between columns by same smear status.
Comparisons within rows in retrospective vs prospective periods.
DISCUSSION
There are limited data on nonactionable results, accuracy, and effect of rapid molecular assays for the diagnosis of resistance beyond rifampicin, especially on smear-negative sputum. To address this, we performed a large-scale evaluation of the newest-generation LPAs in a routine programmatic setting, did comprehensive reference standard testing, and compared care cascade data before and after. Definitive data on MTBDRsl’s performance on smear-negative specimens is essential as the need for FQ susceptibility testing increases and new tools such as Xpert MTB/XDR remain expensive (cost per cartridge $19.80, at least $3860 to upgrade existing modules [25]).
Our key findings include that 19% and 40% of smear-negative individuals tested by MTBDRplus and MTBDRsl were nonactionable, respectively, resulting in many individuals with resistance missed; about 25% of Xpert RR patients have MTBDRplus-defined isoniazid susceptibility; and deployment of direct LPA testing was associated with improvements in days to diagnosis, more individuals receiving DST, and reduced culture reliance.
MTBDRsl had almost double the nonactionable result rate of MTBDRplus in smear-negatives for TB detection, causing diagnostic and treatment delays. Our data highlight the suboptimal ability of reflex DSTs to detect TB even in individuals already identified as TB-positive by frontline tests. This information loss will persist as the limit of detection of new frontline tests outstrips that of reflex tests (Xpert MTB/RIF Ultra vs Xpert MTB/XDR). We recommend that all studies that evaluate reflex test report this key metric (nonactionable results).
In Xpert RR specimens that were MTBDRplus rifampicin-susceptible, Xpert was correct more frequently than MTBDRplus [26, 27]. Possible reasons include heteroresistance and variants not included in MTBDRplus. These findings question diagnostic algorithms that use MTBDRplus to confirm Xpert-detected rifampicin resistance [7, 27, 28].
Importantly, MTBDRplus has value for isoniazid-susceptibility detection. Our data suggest that isoniazid is likely effective in 25% of Xpert RR individuals. In agreement with that observed in the Democratic Republic of the Congo [29] and Iran [30], we recommend that RR TB not be automatically assumed to be MDR and all Xpert RR individuals receive isoniazid DST (which should always be done as isoniazid resistance prevalence is globally in excess of that of rifampicin [31].
Fluoroquinolones are key components of new regimens, and SLIDs such as amikacin remain important. Although important new tools such as Xpert MTB/XDR are emerging [32], MTBDRsl is already established in many laboratories worldwide. The sensitivity and specificity for FQ on smear-negatives were 84% and 93%, respectively. High MTBDRsl sensitivity (81%) was observed on smear-negatives for SLID; however, specificity was less (88%); both improved after discrepant analysis. Importantly, in contrast to FQs, most SLID MTBDRsl-pDST discrepant results resolved in favor of pDST-confirmed susceptibility.
In the after period, we found significant improvements in the proportion of people who had any second-line DST results (such individuals are thus more likely to start effective treatment) and time to result. Such real-world data regarding the programmatic impact of TB diagnostics are scarce but important. With the scale-up of second-line LPAs, individual with smear-negative TB still suffered from unacceptably long times to diagnosis. This subset of individuals should be targeted for interventions in order to accelerate treatment initiation, such as new expensive assays such as Xpert MTB/XDR or Deeplex Myc-TB (Genoscreen) [8, 33].
A strength and limitation of our study is the programmatic context of the study, permitting it to be large and the results reported for potential patient management within the South African care cascade. However, this meant that the study was constrained by contemporary diagnostic algorithms, which affected specimen and meta-data availability given the suboptimal quality of care common in high-volume resource-scarce settings.
Time to DST results associated with LPA scale-up may vary across other provinces within South Africa as, unlike in the Western Cape, only 1 specimen is collected initially for presumptive TB and a second sputum specimen is dependent on an individual returning to a clinic (this may affect generalizability). We were unable to do pDST for rifampicin and isoniazid; however, our primary objective was to evaluate LPA performance for second-line drugs. We also did targeted sequencing rather than whole-genome sequencing, and discrepant analyses may have missed rare noncanonical variants; however, WHO-recommended second-line pDST was done in all isolates [34].
LPA use in our programmatic laboratory was associated with improvements in the care cascade, and patient-important outcomes remained suboptimal. Until next-generation reflex DSTs are widely available, expanded LPA testing remains key to the successful scale-up of new regimens, despite important paucibacillary specimen performance caveats.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Samantha Pillay, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa; National Health Laboratory Services, Green Point, Cape Town, South Africa.
Margaretha de Vos, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Brigitta Derendinger, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Elizabeth Maria Streicher, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Tania Dolby, National Health Laboratory Services, Green Point, Cape Town, South Africa.
Leeré Ann Scott, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Amy Debra Steinhobel, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Rob Mark Warren, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Grant Theron, DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa.
Notes
Author Contributions. S. P., M. dV., G. T., and R.W. conceptualized the experiments. T. D., S. P., B. D., and R.V. assisted with data curation. S.P. performed formal analysis and methodology and wrote the original draft. A. D. S., L. A. S., and E. S. assisted S.P. with the investigation. All authors reviewed and edited the manuscript.
Acknowledgments. The authors thank the National Health Laboratory and Hain Lifesciences. The authors thank Dr Rouxjeane Venter for help with patient data collection.
Disclaimer. Hain Lifesciences donated MTBDRsl kits but had no role in study design or results interpretation.
Financial support. This work was supported by the Stellenbosch University Faculty of Health Sciences, the National Research Foundation, and Harry Crossley. R. M. W. acknowledges funding from the South African Medical Research Council. G. T. acknowledges funding from the EDCTP2 Programme supported by the European Union (SF1401, OPTIMAL DIAGNOSIS), and the National Institute of Allergy and Infection Diseases of the National Institutes of Health (U01AI152087).
References
- 1. World Health Organization . Global Tuberculosis Report. Geneva, Switzerland: 2020. [Google Scholar]
- 2. World Health Organization . Global Tuberculosis Report. Geneva, Switzerland: 2021. [Google Scholar]
- 3. Naidoo P, Theron G, Rangaka MX, et al. The South African tuberculosis care cascade: estimated losses and methodological challenges. J Infect Dis 2017; 216:S702–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox H, Dickson-Hall L, Ndjeka N, et al. Delays and loss to follow-up before treatment of drug-resistant tuberculosis following implementation of Xpert MTB/RIF in South Africa: a retrospective cohort study. PLoS Medicine 2017; 14:e1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization . The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: policy guidance. Geneva, Switzerland: 2016. [Google Scholar]
- 6. Director AC, Kruger MJ. Clinical Guidelines & Standard Operating Procedure for the Implementation of the Short & Long DR-TB regimens for Adults, Adolescents and Children.Published by National Department of Health. Available from: https://www.westerncape.gov.za/assets/departments/health/tuberculosis_-_dr-tb_clinical_guidelines_2018.pdf.
- 7. Beylis N, Ghebrekristos Y, Nicol M, et al. Management of false-positive rifampicin resistant Xpert MTB/RIF. Lancet Microbe 2020; 1:e238. [DOI] [PubMed] [Google Scholar]
- 8. World Health Organization. World Health Organization consolidated guidelines on tuberculosis: module 3: diagnosis-rapid diagnostics for tuberculosis detection: web annex 4: evidence synthesis and analysis. Geneva, Switzerland: 2021. [Google Scholar]
- 9. Penn-Nicholson A, Georghiou SB, Ciobanu N, et al. Detection of isoniazid, fluoroquinolone, ethionamide, amikacin, kanamycin, and capreomycin resistance by the Xpert MTB/XDR assay: a cross-sectional multicentre diagnostic accuracy study. Lancet Infect Dis 2022:22:242–9. [DOI] [PubMed] [Google Scholar]
- 10. García-Basteiro AL, DiNardo A, Saavedra B, et al. Point of care diagnostics for tuberculosis. Pulmonology 2018; 24:73–85. [DOI] [PubMed] [Google Scholar]
- 11. Samra Z, Kaufman L, Bechor Jet al. Comparative study of three culture systems for optimal recovery of mycobacteria from different clinical specimens. Eur J Clin Microbiol Infect Dis 2000; 19:750–4. [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization . WHO consolidated guidelines on drug-resistant tuberculosis treatment. Geneva, Switzerland: World Health Organization; 2016. [Google Scholar]
- 13. Theron G, Peter J, Richardson M, et al. The diagnostic accuracy of the GenoType® MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev 2014; CD010705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bai Y, Wang Y, Shao Cet al. Genotype MTBDRplus assay for rapid detection of multidrug resistance in Mycobacterium tuberculosis: a meta-analysis. PLoS One 2016; 11:e0150321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drobniewski F CM, Jordan J. , et al. Systematic review, meta-analysis and economic modelling of molecular diagnostic tests for antibiotic resistance in tuberculosis. Southampton (UK): NIHR Journals Library. (Health Technology Assessment, No. 19.34.) Chapter 3, Systematic review. Available from: https://www.ncbi.nlm.nih.gov/books/. NBK293793. 2015.
- 16. Theron G, Peter J, Richardson Met al. Genotype® MTBDRsl assay for resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev 2016; 9:CD010705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derendinger B, De Vos M, Nathavitharana R, et al. Widespread use of incorrect PCR ramp rate negatively impacts multidrug-resistant tuberculosis diagnosis (MTBDR plus). Sci Rep 2018; 8:3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. National Institute for Communicable Diseases Division of National Health Laboratory Service . South African Tuberculosis Drug Resistant Survey. 2015 [cited 2022 July 14]. Available from: http://www.nicd.ac.za/assets/files/K-12750%20NICD%20National %20Survey%20Report_Dev_V11-LR.pdf.
- 19. Kent PT, Kubica GP. , et al. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, GA: Centers for Disease Control and Prevention; 1985. [Google Scholar]
- 20. Hain Lifescience GmbH . Genotype MTBDRplus version 2.0: instruction manual. Germany: Hain Lifescience GmbH, Nehren, 2012. [Google Scholar]
- 21. Hain Lifescience GmbH . Genotype MTBDRsl version 2.0: instruction manual. Germany: Hain Lifescience GmbH, Nehren; 2015. [Google Scholar]
- 22. Derendinger B, de Vos M, Pillay S, et al. Frequent suboptimal thermocycler ramp rate usage negatively impacts MTBDRsl performance for second-line drug resistant tuberculosis diagnosis. J Mol Diagn 2022; 24:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. World Health Organization, Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. World Health Organization Geneva. 2021. [Google Scholar]
- 24. Miotto P, Tessema B, Tagliani E, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur J Respir J 2017; 50:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naidoo K, Dookie N. , et al. Can the GeneXpert MTB/XDR deliver on the promise of expanded, near-patient tuberculosis drug-susceptibility testing? Lancet Infect Dis 2022; 22:e121–7. [DOI] [PubMed] [Google Scholar]
- 26. Van Rie A, Whitfield MG, De Vos E, et al. Discordances between molecular assays for rifampicin resistance in Mycobacterium tuberculosis: frequency, mechanisms and clinical impact. J Antimicrob Chemother 2020; 75:1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ngabonziza JCS, Decroo T, Migambi P, et al. Prevalence and drivers of false-positive rifampicin-resistant Xpert MTB/RIF results: a prospective observational study in Rwanda. Lancet Microbe 2020; 1:e74–83. [DOI] [PubMed] [Google Scholar]
- 28. Ghebrekristos Y. Characterization of Mycobacterium tuberculosis isolates with discordant rifampicin susceptibility test results. University of Cape Town, South Africa. 2018. Available at: https://open.uct.ac.za/handle/11427/29248. [Google Scholar]
- 29. Bisimwa BC, Nachega JB, Warren RM. , et al. Xpert MTB/RIF-detected Rifampicin Resistance is a Sub-Optimal Surrogate for Multidrug Resistant Tuberculosis in Eastern Democratic Republic of the Congo: Diagnostic and Clinical Implications 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nasiri M, Zamani S, Pormohammad A. , et al. The reliability of rifampicin resistance as a proxy for multidrug-resistant tuberculosis: a systematic review of studies from Iran 2018; 37:9–14. [DOI] [PubMed] [Google Scholar]
- 31. Dean Anna S, Zignol Matteo, Cabibbe Andrea Maurizioet al. Prevalence and genetic profiles of isoniazid resistance in tuberculosis patients: A multicountry analysis of cross-sectional data. PLOS Medicine 2020; 17(1):e1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bainomugisa A, Gilpin C, Coulter Cet al. New Xpert MTB/XDR: added value and future in the field. European Respiratory Journal 2020;. 56. [DOI] [PubMed] [Google Scholar]
- 33. Feuerriegel S, Kohl TA, Utpatel C. , et al. Rapid genomic first- and second-line drug resistance prediction from clinical Mycobacterium tuberculosis specimens using deeplex-MycTB. Eur Respir J 2021.57. [DOI] [PubMed] [Google Scholar]
- 34. Georghiou S B, Schumacher S G, Rodwell T C. , et al. Guidance for studies evaluating the accuracy of rapid tuberculosis drug-susceptibility tests. J Infect Dis 2019:220):S126–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.