Abstract
Ensembl Plants (http://plants.ensembl.org) offers genome-scale information for plants, with four releases per year. As of release 47 (April 2020) it features 79 species and includes genome sequence, gene models, and functional annotation. Comparative analyses help reconstruct the evolutionary history of gene families, genomes, and components of polyploid genomes. Some species have gene expression baseline reports or variation across genotypes. While the data can be accessed through the Ensembl genome browser, here we review specifically how our plant genomes can be interrogated programmatically and the data downloaded in bulk. These access routes are generally consistent across Ensembl for other non-plant species, including plant pathogens, pests, and pollinators.
Keywords: Database, Genomics, Comparative genomics, Genetic variation, Crops, Model plants, Polyploids, Scripting, API
1. Introduction
Plants play a central role in the ecology and economy of our planet and are essential to our food security. As the world population increased by 145% in the last 60 years, the yields of cereals increased even more, while not needing much more land [1]. This has been possible as a result of improved agricultural practices and crops. Currently, breeding programs take advantage of inexpensive genomic and phenotypic data. The next steps towards what is being called Breeding 4.0 [2] include adapting crops to changing environments and broadening the diversity pool to compensate for the losses occurred during domestication. For this reason wild relatives of crops are being sequenced increasingly and added to pre-breeding programs [3]. In addition, natural plant populations and model plants are being studied to understand their ecology and the genetic basis of their adaptation mechanisms, which can then be applied to in crops. In this context, genomics is a foundation of plant sciences, as standard approaches such as marker-assisted breeding, QTL analysis, and genome-wide association studies, as well as genomic selection, induced variation experiments, and genome editing, all depend on genomic technologies and databases. These tools are accelerating breeding and helping to untangle complex polyploid genomes, such as that of bread wheat [4].
Ensembl Plants (http://plants.ensembl.org) is the Ensembl portal for plants and red algae [5] and provides a consistent set of interfaces to genomic data, including reference genome sequences, gene and transcript models, genetic variation, gene expression, markers, and comparative genomics. There are up to four releases per year. At the time of writing, the latest release of Ensembl Plants is version 47 (April 2020), which corresponds to Ensembl version 100. This release comprises 79 genomes, containing several cultivars and ecotypes for some species. Ensembl Plants is developed with our long-term partners Gramene [6] and with individual groups that publish plant genomes around the world. This chapter documents how the data at Ensembl Plants can be downloaded in bulk and interrogated programmatically using a variety of approaches. It provides a series of recipes, available as source code at https://github.com/Ensembl/plant-scripts, that can be modified to carry out more complex analyses of plant genomes.
2. Materials
2.1. Database Structure and Data Access
Ensembl Plants is implemented primarily as a collection of MySQL relational databases. The overall data structure is modular, with different data (e.g., core annotation, comparative genomics, functional genomics, variation data) modeled by distinct schemas. The core schema is modeled on the central dogma of molecular biology, linking genome sequence to genes, transcripts, and their translations, each of which can be decorated with functional annotation (see Note 1). Much annotation takes the form of cross-references, which are web links to entries in other resources, such as InterPro [7] or Gene Ontology [8], that either represent the primary source of the biological entity or provide additional information. Cross-references describe functional entities such as domains, reactions, and processes. Some also serve as controlled vocabularies for functional annotation.
The databases can be downloaded for local installation or alternatively accessed via a public MySQL server. Local MySQL databases are an efficient alternative to the public MySQL server, particularly if heavy use is anticipated (see Note 2). Programmatic access is supported by two APIs, which allow data discovery and access through an abstraction layer that hides the detailed structure of the underlying data store. One is a Perl API, while the other uses a language-agnostic REST interface [9]. The REST service allows up to 15 requests per second.
In addition to the primary databases, Ensembl Plants also provides access to denormalized data warehouses, constructed using the BioMart tool kit [10]. These are specialized databases that support efficient gene- and variant-centric queries. Finally, a variety of data selections are exported from the databases in common file formats and made available for download via an FTP site.
These resources are summarized in Table 1. Recipes to query each of them are listed in Table 7.
Table 1.
Programming interfaces and data sources in Ensembl Plants. The public MySQL server contains databases from the most recent ten releases
| Resource | Description |
|---|---|
| Perl API | A comprehensive Perl-based API for accessing all types of data available: http://plants.ensembl.org/info/docs/api/index.html |
| REST service |
A language-independent API for retrieving selected data: http://plants.ensembl.org/info/data/rest.html |
| BioMart | A data mining tool for batch retrieval of gene-related data. Accessible via web interface and a Bioconductor package: http://plants.ensembl.org/info/data/biomart/index.html |
| FTP server | Pre-generated genome-scale data files in a variety of commonly used formats: http://plants.ensembl.org/info/data/ftp/index.html |
| MySQL server |
Public access to Ensembl Genomes MySQL databases: http://plants.ensembl.org/info/data/mysql.html |
Table 7.
Programming recipes to analyze data in Ensembl Plants, including perl API (A), R BiomaRt (B), FTP (F), SQL (S), REST (R), and Ensembl VEP (V) examples. These recipes and their software dependencies, together with a few more scripts for phylogenomic analyses, are updated at https://github.com/Ensembl/plant-scripts
| Recipe | Description |
|---|---|
| A1 | Load the Registry object with details of genomes available |
| A2 | Check which analyses are available for a species |
| A3 | Get soft-masked sequences from Arabidopsis thaliana |
| A4 | Get BED file with repeats in chr4 |
| A5 | Find the DEAR3 gene |
| A6 | Get the transcript used in Compara analyses |
| A7 | Find all orthologues of a gene |
| A8 | Get markers mapped on chr1D of bread wheat |
| A9 | Find all syntelogues among rices |
| A10 | Print all translations for other features genes |
| B1 | Check plant marts and select dataset |
| B2 | Check available filters and attributes |
| B3 | Download GO terms associated with genes |
| B4 | Get Pfam domains annotated in genes |
| B5 | Get SNP consequences from a selected variation source |
| C1 | Find RNA-seq CRAM files for a genome assembly |
| F1 | Download peptide sequences in FASTA format |
| F2 | Download CDS nucleotide sequences in FASTA format |
| F3 | Download transcripts (cDNA) |
| F4 | Download soft-masked genomic sequences |
| F5 | Upstream/downstream sequences |
| F6 | Get mappings to UniProt proteins |
| F7 | Get indexed, bgzipped VCF file with variants mapped |
| F8 | Get precomputed VEP cache files |
| F9 | Download all homologies in a single TSV file, several GBs |
| F10 | Download UniProt report of Ensembl Plants |
| F11 | Retrieve list of new species in current release |
| F12 | Get current plant species tree cladogram |
| S1 | Check currently supported Ensembl Genomes (EG) core schemas |
| S2 | Count protein-coding genes of a particular species |
| S3 | Get stable_ids of transcripts used in Compara analyses |
| S4 | Get variants significantly associated to phenotypes |
| S5 | Get Triticumaestivumhomeologous genes across A, B, and D subgenomes |
| S6 | Count the number of whole-genome alignments of all genomes |
| S7 | Extract all the mutations and consequence for a known line on triticum_aestivum |
| R1 | Create an HTTP client and helper functions |
| R2 | Get metadata for all plant species |
| R3 | Find features overlapping genomic region |
| R4 | Fetch phenotypes overlapping genomic region |
| R5 | Find homologues of selected gene |
| R6 | Get annotation of orthologous genes/proteins |
| R7 | Fetch variant consequences for multiple variant ids |
| R8 | Check consequences of single SNP within CDS sequence |
| R9 | Retrieve variation sources of a species |
| V1 | Download, install, and update VEP |
| V2 | Unpack downloaded cache file and check SIFT support |
| V3 | Predict effect of variants |
| V4 | Predict effect of variants for species not in Ensembl |
2.2. Overview of Data Content
2.2.1. Genomes and Core Data
Genome assemblies are typically imported from the European Nucleotide Archive (ENA) [11], which is part of the International Nucleotide Sequence Database Collaboration (http://www.insdc.org, INSDC). Gene model annotations are imported from the ENA [11], Phytozome [12], or provided by community members (see Note 3). For instance, the rice annotation was imported from RAP-DB [13]. After import, various computational analyses are performed for each genome. A summary of these is given in Table 2. In addition, specific datasets are imported and analyzed according to the requirements of individual communities. These datasets typically fall into two classes, markers, and variants across genotype panels.
Table 2.
Standard computational analyses that are typically run for genomes in Ensembl Plants. The full list of analyses for any species can be obtained with recipe A2
| Analysis | Description |
|---|---|
| Repeat classification and masking |
Several tools for detecting and classifying repeated elements are used: http://plants.ensembl.org/info/genome/annotation/repeat_features.html |
| RNA gene | Noncoding genes are primarily annotated by homology-based methods: http://plants.ensembl.org/info/genome/annotation/ncrna.html |
| External cross-references | Database cross-references are loaded from a predefined set of sources, using either direct mappings or sequence alignments [7, 14]: http://plants.ensembl.org/info/genome/annotation/cross_references.html |
| Ontology terms | Ontology terms are imported from external sources and also transitively annotated via InterPro [7]: http://plants.ensembl.org/info/genome/annotation/cross_references.html |
| Plant Reactome | Metabolic, transport, and hormone signaling pathways, transcriptional networks, and developmental processes [15]: https://plantreactome.gramene.org |
| Protein features | InterProScan provides protein domain and feature annotations: http://plants.ensembl.org/info/genome/annotation/protein_features.html |
| Gene trees | Comparative genomics pipeline that computes phylogenetic trees of protein-coding genes [16]: http://plants.ensembl.org/info/genome/compara/peptide_compara.html |
| Whole-genome alignment (WGA) |
Whole-genome alignments are computed for selected pairs of species. When both genomes permit, synteny calculations are also performed. See http://plants.ensembl.org/info/genome/compara/whole_genome_alignment.html and http://plants.ensembl.org/info/genome/compara/synteny.html |
| Variation coding consequences |
The consequences of polymorphisms in species with variation datasets are computed for each transcript with the Ensembl Variant Effect Predictor [14]: http://plants.ensembl.org/info/docs/tools/vep |
The genomes currently included in Ensembl Plants are listed in Table 3. A summary of UniProt coverage of proteins encoded by genes within these genomes is given in Table 4 [17]. In all cases, genomes are identified by their Ensembl production name, which is usually binomial but can also include a strain name to distinguish particular cultivars or ecotypes, such as malus_domestica_golden. Details of other datasets incorporated can be found through the homepage for each species (see Note 3).
Table 3.
Genomes available in release 47 (April 2020) of Ensembl Plants. The chr column indicates chromosome-level assemblies. The base count of the genome golden path is given in Mbp. This table was produced with recipe R2
| Ensembl production name | Cultivar/ecotype | Assembly | chr | Base count |
|---|---|---|---|---|
| actinidia_chinensis | Red5 | GCA_003024255.1 | Y | 553.8 |
| aegilops_tauschii | AL8/78 | GCA_002575655.1 | Y | 4224.9 |
| amborella_trichopoda | NA | GCA_000471905.1 | 706.3 | |
| ananas_comosus | F153 | GCA_902162155.1 | 315.8 | |
| arabidopsis_halleri | W302 | GCA_900078215.1 | 196.2 | |
| arabidopsis_lyrata | MN47 | GCA_000004255.1 | Y | 206.7 |
| arabidopsis_thaliana | Columbia | GCA_000001735.1 | Y | 119.7 |
| beta_vulgaris | KWS2320 DH | GCA_000511025.2 | Y | 566.2 |
| brachypodium_distachyon | Bd21 | GCA_000005505.4 | Y | 271.2 |
| brassica_napus | Darmor-bzh | GCA_000751015.1 | 848.2 | |
| brassica_oleracea | TO1000 | GCA_000695525.1 | Y | 488.6 |
| brassica_rapa | Chiifu-401-42 | GCA_000309985.1 | Y | 283.8 |
| capsicum_annuum | Criollo de Morelos 334 | GCA_000512255.2 | Y | 3063.9 |
| chara_braunii | S276 | GCA_003427395.1 | 1751.2 | |
| chlamydomonas_reinhardtii | CC-503 cw92 mt+ | GCA_000002595.3 | Y | 111.1 |
| chondrus_crispus | Stackhouse | GCA_000350225.2 | Y | 105 |
| citrus_clementina | Clemenules | GCA_000493195.1 | 301.4 | |
| coffea_canephora | DH200-94 | GCA_900059795.1 | Y | 568.6 |
| corchorus_capsularis | CVL-1 | GCA_001974805.1 | 317.2 | |
| cucumis_sativus | 9930 | GCA_000004075.2 | Y | 193.8 |
| cyanidioschyzon_merolae | 10D | GCA_000091205.1 | Y | 16.7 |
| cynara_cardunculus | NA | GCA_001531365.1 | 724.7 | |
| daucus_carota | DH1 | GCA_001625215.1 | Y | 421.5 |
| dioscorea_rotundata | TDr96_F1 | GCA_002240015.2 | Y | 456.7 |
| eragrostis_curvula | Tanganyika | GCA_007726485.1 | Y | 603.1 |
| eragrostis_tef | Tsedey | GCA_000970635.1 | 607.3 | |
| galdieria_sulphuraria | 074W | GCA_000341285.1 | 13.7 | |
| glycine_max | Williams 82 | GCA_000004515.4 | Y | 978.5 |
| gossypium_raimondii | CMD 10 | GCA_000327365.1 | Y | 761.4 |
| helianthus_annuus | XRQ/B | GCA_002127325.1 | Y | 3027.8 |
| hordeum_vulgare | Morex | GCA_901482405.1 | Y | 4834.4 |
| ipomoea_triloba | NCNSP0323 | GCA_003576645.1 | Y | 461.8 |
| leersia_perrieri | IRGC:105164 | GCA_000325765.3 | Y | 266.7 |
| lupinus_angustifolius | Tanjil | GCA_001865875.1 | Y | 609.2 |
| malus_domestica | Golden Delicious | GCA_002114115.1 | Y | 703 |
| manihot_esculenta | AM560-2 | GCA_001659605.1 | Y | 582.1 |
| marchantia_polymorpha | Tak-1 | GCA_003032435.1 | 225.8 | |
| medicago_truncatula | A17 | GCA_000219495.2 | Y | 412.8 |
| musa_acuminata | DH-Pahang | GCA_000313855.1 | Y | 473 |
| nicotiana_attenuata | UT | GCA_001879085.1 | Y | 2365.7 |
| olea_europaea_sylvestris | NA | GCA_002742605.1 | Y | 1141 |
| oryza_barthii | IRGC:105608 | GCA_000182155.2 | Y | 308.3 |
| oryza_brachyantha | IRGC:101232 | GCA_000231095.2 | Y | 260.8 |
| oryza_glaberrima | CG14 | GCA_000147395.1 | Y | 316.4 |
| oryza_glumipatula | NA | GCA_000576495.1 | Y | 372.9 |
| oryza_indica | 93-11 | GCA_000004655.2 | Y | 427 |
| oryza_longistaminata | NA | GCA_000789195.1 | 326.4 | |
| oryza_meridionalis | OR44 (W2112) | GCA_000338895.2 | Y | 335.7 |
| oryza_nivara | IRGC:100897 | GCA_000576065.1 | Y | 338 |
| oryza_punctata | IRGC:105690 | GCA_000573905.1 | Y | 393.8 |
| oryza_rufipogon | W1943 | GCA_000817225.1 | Y | 338 |
| oryza_sativa | Nipponbare | GCA_001433935.1 | Y | 375 |
| ostreococcus_lucimarinus | CCE9901 | GCA_000092065.1 | Y | 13.2 |
| panicum_hallii_fil2 | FIL2 | GCA_002211085.2 | Y | 535.9 |
| panicum_hallii_hal2 | HAL2 | GCA_003061485.1 | Y | 487.5 |
| phaseolus_vulgaris | G19833 | GCA_000499845.1 | Y | 521.1 |
| physcomitrella_patens | Gransden 2004 | GCA_000002425.2 | Y | 471.9 |
| pistacia_vera | Batoury | GCA_008641045.1 | 671.2 | |
| populus_trichocarpa | Nisqually 1 | GCA_000002775.3 | Y | 434.1 |
| prunus_avium | Satonishiki | GCA_002207925.1 | 272.4 | |
| prunus_dulcis | Texas | GCA_902201215.1 | 227.5 | |
| prunus_persica | Lovell | GCA_000346465.2 | Y | 227.4 |
| saccharum_spontaneum | AP85-441 | GCA_003544955.1 | Y | 2900.2 |
| selaginella_moellendorffii | NA | GCA_000143415.1 | 212.6 | |
| setaria_italica | Yugu1 | GCA_000263155.2 | Y | 405.7 |
| solanum_lycopersicum | Heinz 1706 | GCA_000188115.3 | Y | 827.7 |
| solanum_tuberosum | DM 1-3 516 R44 | GCA_000226075.1 | Y | 810.7 |
| sorghum_bicolor | BTx623 | GCA_000003195.3 | Y | 708.7 |
| theobroma_cacao_criollo | Criollo B97-61/B2 | GCA_000208745.2 | Y | 324.7 |
| theobroma_cacao_matina | Matina 1-6 | GCA_000403535.1 | Y | 346 |
| trifolium_pratense | Milvus B | GCA_900079335.1 | Y | 304.8 |
| triticum_aestivum | Chinese spring | GCA_900519105.1 | Y | 14547.3 |
| triticum_dicoccoides | Zavitan (Atlit2015) | GCA_002162155.1 | Y | 10079 |
| triticum_turgidum | svevo | GCA_900231445.1 | Y | 10463.1 |
| triticum_urartu | G1812 (PI428198) | GCA_000347455.1 | Y | 3747.2 |
| vigna_angularis | Jingnong 6 | GCA_001190045.1 | Y | 466.7 |
| vigna_radiata | VC1973A | GCA_000741045.2 | Y | 463.1 |
| vitis_vinifera | PN40024 | GCA_000003745.2 | Y | 486.3 |
| zea_mays | B73 | GCA_000005005.6 | Y | 2135.1 |
Table 4.
Protein-coding genes annotated in release 47 (April 2020) of Ensembl Plants. The last two columns indicate how many genes encode proteins computationally predicted (TrEMBL) and manually curated (SwissProt) in UniProtKB. This table was produced with recipe F10. Mappings between Ensembl and UniProt proteins can be obtained with recipe F6
| Ensembl production name | Protein-coding genes | TrEMBL | SwissProt |
|---|---|---|---|
| actinidia_chinensis | 33,044 | 33,044 | 6 |
| aegilops_tauschii | 39,614 | 24,486 | 7 |
| amborella_trichopoda | 27,313 | 27,310 | 34 |
| ananas_comosus | 25,783 | 16,219 | 8 |
| arabidopsis_halleri | 32,158 | 241 | 0 |
| arabidopsis_lyrata | 32,667 | 32,470 | 30 |
| arabidopsis_thaliana | 27,628 | 27,100 | 15,649 |
| beta_vulgaris | 26,521 | 7,405 | 37 |
| brachypodium_distachyon | 34,310 | 34,307 | 36 |
| brassica_napus | 101,040 | 62,919 | 149 |
| brassica_oleracea | 59,220 | 59,220 | 20 |
| brassica_rapa | 41,018 | 141 | 9 |
| capsicum_annuum | 35,845 | 35,845 | 52 |
| chara_braunii | 34,718 | 33,777 | 0 |
| chlamydomonas_reinhardtii | 17,743 | 17,737 | 322 |
| chondrus_crispus | 9,807 | 9,806 | 11 |
| citrus_clementina | 25,000 | 24,989 | 0 |
| coffea_canephora | 25,574 | 25,574 | 3 |
| corchorus_capsularis | 29,356 | 29,356 | 0 |
| cucumis_sativus | 23,780 | 23,780 | 65 |
| cyanidioschyzon_merolae | 4,973 | 4,640 | 97 |
| cynara_cardunculus | 26,505 | 26,504 | 6 |
| daucus_carota | 32,109 | 32,109 | 136 |
| dioscorea_rotundata | 19,023 | 13 | 0 |
| eragrostis_curvula | 55,182 | 2 | 0 |
| eragrostis_tef | 41,555 | 54 | 0 |
| galdieria_sulphuraria | 6,622 | 6,621 | 23 |
| glycine_max | 55,897 | 55,891 | 412 |
| gossypium_raimondii | 38,208 | 38,172 | 0 |
| helianthus_annuus | 52,191 | 52,191 | 315 |
| hordeum_vulgare | 37,705 | 37,636 | 292 |
| ipomoea_triloba | 31,358 | 0 | 0 |
| leersia_perrieri | 29,078 | 29,074 | 0 |
| lupinus_angustifolius | 33,074 | 14,421 | 12 |
| malus_domestica | 40,624 | 28,704 | 41 |
| manihot_esculenta | 33,044 | 33,043 | 45 |
| marchantia_polymorpha | 19,287 | 19,287 | 76 |
| medicago_truncatula | 50,444 | 50,431 | 79 |
| musa_acuminata | 36,519 | 36,519 | 11 |
| nicotiana_attenuata | 33,320 | 33,320 | 3 |
| olea_europaea_sylvestris | 50,678 | 333 | 23 |
| oryza_barthii | 34,575 | 34,564 | 0 |
| oryza_brachyantha | 32,037 | 32,032 | 0 |
| oryza_glaberrima | 33,164 | 33,161 | 1 |
| oryza_glumipatula | 35,735 | 35,721 | 0 |
| oryza_indica | 40,745 | 36,796 | 570 |
| oryza_longistaminata | 31,686 | 101 | 0 |
| oryza_meridionalis | 29,308 | 29,294 | 0 |
| oryza_nivara | 36,313 | 36,305 | 27 |
| oryza_punctata | 31,762 | 31,748 | 0 |
| oryza_rufipogon | 37,071 | 37,063 | 1 |
| oryza_sativa | 35,775 | 32,864 | 3,096 |
| ostreococcus_lucimarinus | 7,603 | 7,570 | 20 |
| panicum_hallii_fil2 | 33,805 | 33,805 | 0 |
| panicum_hallii_hal2 | 33,263 | 33,263 | 0 |
| phaseolus_vulgaris | 28,134 | 28,095 | 111 |
| physcomitrella_patens | 32,234 | 0 | 0 |
| pistacia_vera | 31,784 | 43 | 0 |
| populus_trichocarpa | 41,335 | 41,335 | 135 |
| prunus_avium | 42,794 | 219 | 8 |
| prunus_dulcis | 27,963 | 27,963 | 10 |
| prunus_persica | 26,873 | 26,873 | 17 |
| saccharum_spontaneum | 53,284 | 65 | 0 |
| selaginella_moellendorffii | 34,799 | 34,762 | 31 |
| setaria_italica | 35,831 | 35,828 | 2 |
| solanum_lycopersicum | 34,429 | 27,133 | 406 |
| solanum_tuberosum | 39,021 | 39,010 | 245 |
| sorghum_bicolor | 34,118 | 34,078 | 142 |
| theobroma_cacao_criollo | 21,146 | 4,079 | 5 |
| theobroma_cacao_matina | 29,188 | 29,188 | 5 |
| trifolium_pratense | 39,917 | 26,935 | 0 |
| triticum_aestivum | 107,545 | 107,124 | 600 |
| triticum_dicoccoides | 62,569 | 182 | 1 |
| triticum_turgidum | 66,545 | 233 | 0 |
| triticum_urartu | 33,482 | 33,479 | 1 |
| vigna_angularis | 33,860 | 33,860 | 1 |
| vigna_radiata | 22,368 | 5,978 | 0 |
| vitis_vinifera | 29,927 | 29,814 | 136 |
| zea_mays | 39,591 | 39,494 | 724 |
2.2.2. Variation Data
The variation schema can store genetic variants observed in populations or germplasm collections, alleles, and frequencies, alongside sample genotype data. Supported variant types include single nucleotide polymorphisms, indels, and structural variants. The functional consequence of variants on genes is predicted with the Ensembl Variant Effect Predictor (VEP) [14]. Linkage disequilibrium data and statistical associations with phenotypes are available for selected species. The variation datasets of release 47 of Ensembl Plants are described in Table 5. The Ensembl VEP is also a command line tool that can be used to efficiently annotate variants and we provide recipes for it as well (see Table 7).
Table 5.
Variation datasets available in release 47 (April 2020) of Ensembl Plants. The list can also be browsed interactively at https://plants.ensembl.org/species.html. This table was produced with recipe R9. The corresponding VCF files can be downloaded with recipe F7. Recipe F8 can be used to get the Ensembl VEP cache files in order to annotate variant consequences with recipes V2 and V3
| Ensembl production name |
Source |
|---|---|
| arabidopsis_thaliana | The 1001 Genomes Project [18] |
| arabidopsis_thaliana | Nordborg [19] |
| brachypodium_distachyon | Jaiswal_lab_OSU [20] |
| hordeum_vulgare | International Barley Sequencing Consortium (IBSC) [21–23] |
| hordeum_vulgare | Ensembl Plants [24] |
| hordeum_vulgare | IlluminaiSelect SNP chip [22] |
| malus_domestica | http://fruitbreedomics.com [25] |
| oryza_glaberrima | Glab (OGE) |
| oryza_glaberrima | Barthii(OGE) |
| oryza_glumipatula | Oryza Genome Evolution (OGE) |
| oryza_indica | dbSNP [26] |
| oryza_sativa | https://www.ebi.ac.uk/eva [27–30] |
| oryza_sativa | https://archive.gramene.org/qtl (Gramene_QTLdb) [6] |
| oryza_sativa | https://archive.gramene.org/markers (gramene-marker) [6] |
| oryza_sativa | Qtaro_QTLdb [31] |
| solanum_lycopersicum | The 150 Tomato Genome ReSequencing Project [32] |
| sorghum_bicolor | Morris_2013 [33] |
| sorghum_bicolor | Database of Genomic Variants Archive (DGVa) |
| sorghum_bicolor | Mace_2013 [34] |
| sorghum_bicolor | Sorghum_EMS_mutants [35] |
| triticum_aestivum | Markers from Axiom 820K and 35K SNP Array provided (CerealsDB) [36] |
| triticum_aestivum | EMS-induced mutation [37] |
| triticum_aestivum | Inter-Homoeologous Variants (IHVs) called by alignments ofthe A, B, and D component genomes |
| triticum_turgidum | Markers from Axiom 820K, 35K, iSelect 90KSNP Infinium and TaBW280K Affymetrix array (CNR-ITB) [36, 38] |
| vitis_vinifera | CSHL/Cornell [39] |
| zea_mays | HapMap2 [40] |
| zea_mays | Panzea_2.7GBS https://www.panzea.org/genotypes |
2.2.3. Comparative Genomics Data
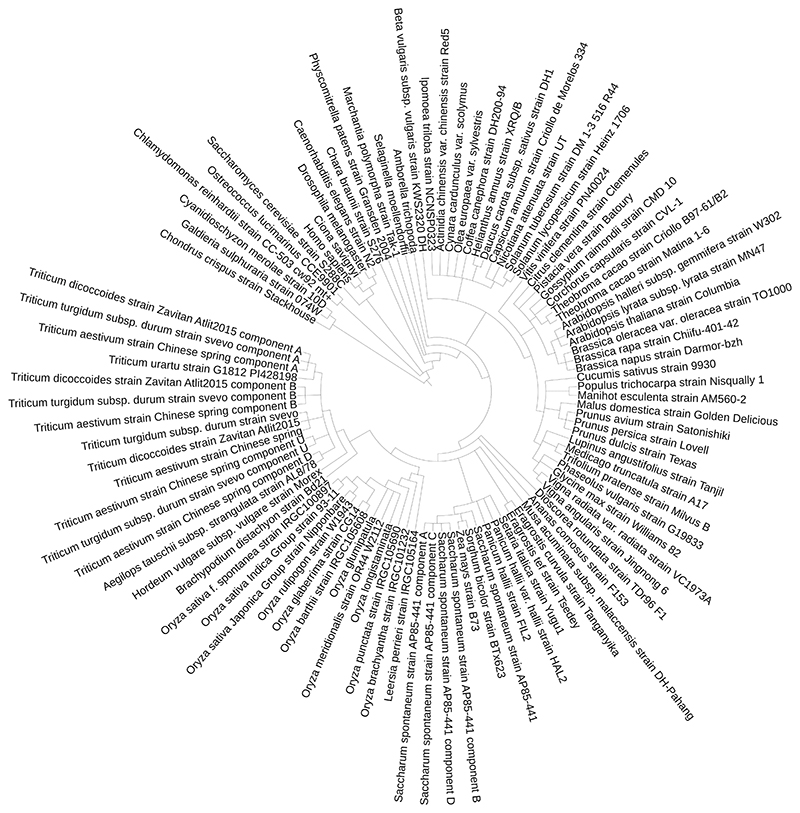
The Ensembl Gene Tree pipeline is used to calculate evolutionary relationships among members of protein families (Table 2). For each gene, the translation of the canonical transcript is selected (see Note 4). Briefly, this pipeline first finds clusters of similar proteins and then, for each cluster, attempts to reconcile the relationship between the sequences with the known species cladogram (Fig. 1), derived from the NCBI Taxonomy database [42]. The analysis also contains a few non-plant outgroups. The TreeBeST software (https://github.com/Ensembl/treebest) is used to construct a consensus tree, which allows the identification of orthologues and paralogues. As polyploid genomes are split into components, homoeologous genes are effectively defined as orthologues among subgenomes. A number of plant genomes are also included in a pan-taxonomic gene tree, containing a representative selection of sequenced genomes from all domains of life. Recipe R2 can be used to check which comparative analyses have been run for a particular species. This information is also displayed in the table at http://plants.ensembl.org/species.html.
Fig. 1.
Species cladogram of release 47 (April 2020) of Ensembl Plants. Genomes of polyploid species are decomposed into genomic components. This topology is used in the comparative genomic analyses to derive orthologous and paralogous genes. This tree was produced with the Newick file obtained with recipe F12 and visualized with iToL [41]
Other comparative analyses available in Ensembl Plants are pairwise whole-genome alignments and synteny (see Tables 2 and 6).
Table 6.
Number of pairwise whole-genome alignments and synteny analyses in release 47 (April 2020) of Ensembl Plants. Pairwise alignments are computed with LastZ [43]. Two multiple alignments are also available for Oryza species. Data obtained with recipe S6
| Ensembl production name | WGA pairwise alignments | Synteny analyses |
|---|---|---|
| actinidia_chinensis | 4 | 2 |
| aegilops_tauschii | 8 | 3 |
| amborella_trichopoda | 3 | 0 |
| ananas_comosus | 3 | 0 |
| arabidopsis_halleri | 4 | 0 |
| arabidopsis_lyrata | 5 | 2 |
| arabidopsis_thaliana | 77 | 4 |
| beta_vulgaris | 3 | 0 |
| brachypodium_distachyon | 10 | 1 |
| brassica_napus | 5 | 0 |
| brassica_oleracea | 6 | 0 |
| brassica_rapa | 6 | 1 |
| capsicum_annuum | 4 | 1 |
| chara_braunii | 0 | 0 |
| chlamydomonas_reinhardtii | 3 | 0 |
| chondrus_crispus | 3 | 0 |
| citrus_clementina | 4 | 0 |
| coffea_canephora | 4 | 2 |
| corchorus_capsularis | 5 | 0 |
| cucumis_sativus | 4 | 0 |
| cyanidioschyzon_merolae | 3 | 0 |
| cynara_cardunculus | 4 | 0 |
| daucus_carota | 4 | 0 |
| dioscorea_rotundata | 3 | 0 |
| eragrostis_curvula | 3 | 1 |
| eragrostis_tef | 3 | 0 |
| galdieria_sulphuraria | 3 | 0 |
| glycine_max | 4 | 0 |
| gossypium_raimondii | 5 | 0 |
| helianthus_annuus | 4 | 0 |
| hordeum_vulgare | 9 | 0 |
| ipomoea_triloba | 4 | 0 |
| leersia_perrieri | 11 | 2 |
| lupinus_angustifolius | 4 | 0 |
| malus_domestica | 4 | 0 |
| manihot_esculenta | 4 | 0 |
| marchantia_polymorpha | 3 | 0 |
| medicago_truncatula | 22 | 4 |
| musa_acuminata | 5 | 1 |
| nicotiana_attenuata | 4 | 0 |
| olea_europaea_sylvestris | 4 | 0 |
| oryza_barthii | 13 | 9 |
| oryza_brachyantha | 12 | 9 |
| oryza_glaberrima | 13 | 9 |
| oryza_glumipatula | 13 | 9 |
| oryza_indica | 13 | 9 |
| oryza_longistaminata | 13 | 0 |
| oryza_meridionalis | 13 | 10 |
| oryza_nivara | 13 | 9 |
| oryza_punctata | 12 | 9 |
| oryza_rufipogon | 13 | 9 |
| oryza_sativa | 77 | 20 |
| ostreococcus_lucimarinus | 3 | 0 |
| panicum_hallii_fil2 | 3 | 1 |
| panicum_hallii_hal2 | 3 | 1 |
| phaseolus_vulgaris | 4 | 1 |
| physcomitrella_patens | 4 | 0 |
| pistacia_vera | 4 | 0 |
| populus_trichocarpa | 4 | 0 |
| prunus_avium | 4 | 0 |
| prunus_dulcis | 4 | 0 |
| prunus_persica | 4 | 2 |
| saccharum_spontaneum | 3 | 0 |
| selaginella_moellendorffii | 3 | 0 |
| setaria_italica | 4 | 1 |
| solanum_lycopersicum | 13 | 4 |
| solanum_tuberosum | 4 | 2 |
| sorghum_bicolor | 6 | 1 |
| theobroma_cacao_criollo | 13 | 2 |
| theobroma_cacao_matina | 4 | 2 |
| trifolium_pratense | 4 | 0 |
| triticum_aestivum | 9 | 3 |
| triticum_dicoccoides | 9 | 3 |
| triticum_turgidum | 8 | 3 |
| triticum_urartu | 4 | 0 |
| vigna_angularis | 5 | 2 |
| vigna_radiata | 5 | 2 |
| vitis_vinifera | 77 | 9 |
| zea_mays | 8 | 1 |
2.2.4. Baseline Expression Data
Baseline gene expression reports are available as “Gene expression” on the website for selected species. An example for barley is shown at http://plants.ensembl.org/Hordeum_vulgare/Gene/ExpressionAtlas?g=HORVU5Hr1G095630;r=chr5H:599085656-599133086. The underlying curated expression data, produced by Expression Atlas [44], can be browsed and downloaded via the expression widget.
2.2.5. RNA-seq Tracks
RNA-seq datasets from the public INSDC archives are mapped to genome assemblies in Ensembl Plants in every release. They are handled as ENA studies and for each of them CRAM files are created with the RNA-Seq-er pipeline (https://www.ebi.ac.uk/fg/rnaseq/api) [45] and published at ftp://ftp.ensemblgenomes.org/pub/misc_data/Track_Hubs. Each study contains a separate folder for each assembly that was used for mapping. These tracks can be interactively displayed in the browser, but can be of interest for high-throughput studies as well. For instance, study SRP133995 was mapped to tomato assembly SL3.0 and the tracksDb.txt file therein indicates the full path to the relevant CRAM file next to its metadata. CRAM files for a selected assembly can be discovered with recipe C1; note that the assembly name corresponds to column “assembly_default” in recipe R2. As of May 2020 there were 89,355 CRAM files available.
3. Methods
This section describes some of the recipes listed in Table 7 in detail so that the reader can execute or modify any of them. Software dependencies required by these recipes are listed in https://github.com/Ensembl/plant-scripts/blob/master/README.md.
The different approaches are complementary. While the native Perl API is the most powerful and used extensively by Ensembl developers, it also requires some Perl knowledge and the installation of several repositories. Similarly, the Biomart and MySQL examples require knowledge of R and SQL, respectively. However, the REST endpoints can be interrogated with any programming language; however, only a defined set of queries are currently supported. The FTP recipes allow efficient bulk downloads, but with no customization. The source code for all recipes can be found at https://github.com/Ensembl/plant-scripts.
3.1. Clone the GitHub Repository and Install Dependencies
The following steps explain how to obtain a local copy of the recipes and how to test them on Linux/MacOS operating systems (OS).
Open a terminal and check whether git is installed by typing: git --version.
If required install git if using the appropriate software manager for your OS.
Clone the repository: git clone https://github.com/Ensembl/plant-scripts.git.
Navigate to the scripts directory: cd plant-scripts.
Optionally test the scripts: perl demo_test.t.
3.2. Perl API Recipes
The Ensembl Perl API enables access to all types of data from Ensembl Plants (genes, variation, comparative genomics, regulation, etc.) and it is documented extensively (see Note 5). It allows complex queries to be executed without the construction of any explicit SQL queries. The repository contains eight Perl API recipes, of which three are described here (A1, A4, and A8).
3.2.1. Get a BED File with Repeats on Chromosome 4
- Load the Registry object with details of genomes available from the public Ensembl Genomes servers (recipe A1):
Use Bio::EnsEMBL::Registry; Bio::EnsEMBL::Registry->load_registry_from_db( -USER => ‘anonymous’, -HOST => ‘mysql-eg-publicsql.ebi.ac.uk’, -PORT => ‘4157’, ); - Set species and chromosome of interest and print BED file with repeats (recipe A4). Ensembl uses 1-based inclusive coordinates internally:
my $species = ‘arabidopsis_thaliana’; my $chrname = ‘chr4’; my $slice_adaptor = Bio::EnsEMBL::Registry-> get_adaptor ($species, ‘core’, ‘Slice’); my $slice = $slice_adaptor-> fetch_by_region( ‘toplevel’, $chrname ); my @repeats = @{ $slice->get_all_RepeatFeatures() }; foreach my $repeat (@repeats) { printf(“%s\t%d\t%d\t%s\t%s\t%s\n”, $chrname, $repeat->start()-1, $repeat->end(), $repeat->analysis()->logic_name(), $repeat->repeat_consensus()->repeat_class(), $repeat->repeat_consensus()->repeat_type() ); }
3.2.2. Get Markers Mapped on Chromosome 1D of Bread Wheat
Only a few plants have markers loaded. Recipe A8 retrieves wheat KASP markers, with coordinates returned in BED format:
$species = ‘triticum_aestivum’;
$chrname = ‘1D’;
$slice_adaptor =
Bio::EnsEMBL::Registry->
get_adaptor( $species, ‘Core’, ‘Slice’ );
$slice = $slice_adaptor->
fetch_by_region( ‘chromosome’, $chrname );
foreach my $mf (@{ $slice->get_all_MarkerFeatures() }) {
my $marker = $mf->marker();
printf(“%s\t%d\t%d\t%s\t%s\t%s\t%d\n”,
$mf->seq_region_name(),
$mf->start()-1,
$mf->end(),
$mf->display_id(),
$marker->left_primer(),
$marker->right_primer(),
$marker->max_primer_dist() );
}3.3. R Biomart Recipes
The BioMart databases can be queried in many ways (see Note 6). There are five recipes in the repository written in the R language. They all use the BioConductor package BiomaRt [46], which can be installed as follows:
if (!requireNamespace(“BiocManager”, quietly = TRUE))
install.packages(“BiocManager”)
BiocManager::install(“biomaRt”)
This example corresponds to recipe R4, which queries sunflower genes to obtain annotated Pfam domains. Dataset names are abbreviations of Ensembl production names. See recipe R5 for an example querying BioMart variation databases:
EPgenes = useMart(
biomart=“plants_mart”,
host=“plants.ensembl.org”,
dataset=“hannuus_eg_gene”)
pfam = getBM(
attributes=c(“ensembl_gene_id”, “pfam”),
mart=EPgenes)3.4. FTP Recipes
There are 12 recipes in the repository that query the Ensembl Genomes FTP server. They use shell variables and the wget program to download files. The recipes refer to the Ensembl release and the Ensembl Plants release as RELEASE and EGRELEASE, respectively. Recipe F5 involves a prewritten BioMart query.
3.4.1. Download Soft-Masked Genomic Sequences
Soft-masked sequences are FASTA files with all annotated repeated elements in lower case. Using recipe F4 they can be downloaded for a chosen species and release as follows:
SERVER=ftp://ftp.ensemblgenomes.org/pub
DIV=plants
EGRELEASE=47
SPECIES=Brachypodium_distachyon
FASTASM=“${SPECIES}*.dna_sm.toplevel.fa.gz”
URL=“${SERVER}/release-${EGRELEASE}/${DIV}/fasta/${SPE-
CIES,,}/dna/${FASTASM}”
wget -c “$URL”3.4.2. Download All Homologies in a Single TSV File
Recipe F9 downloads a large file (several GB) with all homologies of a release in TSV format. Sequence identifiers correspond to canonical transcripts (see Note 4):
TSVFILE=“Compara.${RELEASE}.protein_default.homologies.tsv.gz”
URL=“${SERVER}/${DIV}/release-${EGRELEASE}/tsv/ensembl-com-para/homologies/${TSVFILE}”
wget -c “$URL”
This file can be parsed in the command line in order to extract homologies (see Note 7):
zcat “$TSVFILE” | grep triticum_aestivum | greporyza_sativa |
grep ortholog
Homologies of each species can be retrieved from a smaller, specific file:
TSVFILE=“Compara.${RELEASE}.protein_default.homologies.tsv.gz”
SPECIES=Triticum_aestivum
URL=“${SERVER}/${DIV}/release-${EGRELEASE}/tsv/ensembl-com-
para/homologies/${SPECIES,,}${TSVFILE}”
wget -c “$URL”
zcat “$TSVFILE” | grep oryza_sativa | grep ortholog
Homologies can also be downloaded in OrthoXML format [47], which renders a smaller file but requires a more complex parser.
3.5. MySQL Recipes
Direct access to the public MySQL server requires knowledge of the schemas (see Notes 1 and 8). While this approach supports complex queries with high-performance, the schemas may change in a new release and thus some queries might stop working. For this reason, API access is recommended. Three recipes are shown here, they all require the mysql-client to be installed.
3.5.1. Count Protein-Coding Genes of a Particular Species
This is recipe S2. The source code works out the current release number, but it can also be set manually as in this example:
SERVER=mysql-eg-publicsql.ebi.ac.uk
USER=anonymous
PORT=4157
EGRELEASE=47
RELEASE=$((EGRELEASE + 53))
SPECIES=arabidopsis_thaliana
SPECIESCORE=$(mysql --host $SERVER --user $USER --port $PORT \
-e “show databases” | grep \
“${SPECIES}_core_${EGRELEASE}_${RELEASE}”)
mysql --host $SERVER --user $USER --port $PORT \
$SPECIESCORE -e “SELECT COUNT(*) FROM gene \
WHERE biotype=‘protein_coding’”3.5.2. Get stable_ids of Transcripts Used in Compara Analyses
Recipe S3 gets a list of identifiers of all transcript used in the comparative genomics gene tree analysis (see Note 4):
SERVER=mysql-eg-publicsql.ebi.ac.uk
USER=anonymous
PORT=4157
EGRELEASE=47
RELEASE=$((EGRELEASE + 53))
SPECIES=arabidopsis_thaliana
mysql --host $SERVER --user $USER --port $PORT \
“ensembl_compara_plants_${EGRELEASE}_${RELEASE}” \
-e “SELECT sm.stable_id \
FROM seq_member sm, gene_member gm, genome_db gdb \
WHERE sm.seq_member_id = gm.canonical_member_id \
AND sm.genome_db_id = gdb.genome_db_id \
AND gdb.name = ‘$SPECIES’”
See recipe F3 to obtain the corresponding sequences.
3.5.3. Get Variants Significantly Associated with Phenotypes
Recipe S4 queries several tables of the variation schema (see Note 8):
SPECIESVAR=$(mysql --host $SERVER --user $USER --port $PORT \
-e “show databases” | \
grep “${SPECIES}_variation_${EGRELEASE}_${RELEASE}”)
mysql --host $SERVER --user $USER --port $PORT \
$SPECIESVAR<<SQL
SELECT f.object_id, s.name, f.seq_region_start,
f.seq_region_end, p.description
FROM phenotype p
JOIN phenotype_feature f ON p.phenotype_id = f.phenotype_id
JOIN seq_region s ON f.seq_region_id = s.name
WHERE f.type = ‘Variation’ AND f.is_significant=1
SQL3.6. REST Recipes
The following recipes, written in Python, can also be found in R and Perl languages in the repository. They communicate with the Ensembl REST service at https://rest.ensembl.org (see Note 9) using the functions get_json and get_json_post, defined in file exampleREST.py.
3.6.1. Find Features Overlapping a Genomic Region
Recipe R3 queries the endpoint overlap/region and returns all features overlapping a selected genomic region:
def get_overlapping_features(species,region):
overlap_url = (“/overlap/region/” + species + “/” + region)
# repeat or variation could have been used instead of gene
ext = (overlap_url + “?feature=gene;content-type=application/
json”)
overlap_data = get_json(ext)
for overlap_feat in overlap_data:
print(“%s\t%s\t%s” % (overlap_feat[‘id’],
overlap_feat[‘start’],
overlap_feat[‘end’]))
species = ‘triticum_aestivum’;
region = ‘3D:379400000-379540000’;
get_overlapping_features(species,region)3.6.2. Check Consequences of SNPs Within CDS Sequences
Recipe R8 queries two endpoints (map/cds/ and info/vep/:species/region). The first one translates CDS to genomic coordinates, the second one retrieves the predicted consequences of the SNP in the coding sequence. This recipe can be used to annotate genomic variants in a given gene across germplasm panels, as done in [48]:
def check_snp_consequences(species,transcript_id,SNPCDScoord,
SNPbase):
# convert CDS coords to genomic coords
ext = (“/map/cds/” + transcript_id + “/”
+ SNPCDScoord + “..” + SNPCDScoord
+ “?content-type=application/json;species=“ + species)
map_cds = get_json(ext)
if map_cds[‘mappings’][0][‘seq_region_name’]:
mapping = map_cds[‘mappings’][0]
# fetch VEP consequences for this region
SNPgenome_coord = ( mapping[‘seq_region_name’] + ‘:’ +
str(mapping[‘start’]) + ‘-’ + str(mapping[‘end’]) )
ext = (“/vep/”+ species + “/region/” + SNPgenome_coord + “/” +
SNPbase + “?content-type=application/json”)
conseq = get_json(ext)
# Print all the relevant info for the given variant
if conseq[0][‘allele_string’]:
for tcons in conseq[0][‘transcript_consequences’]:
#... some lines omitted, check exampleREST.py
values = (transcript_id, SNPCDScoord,
conseq[0][‘allele_string’],
tcons[‘biotype’],
tcons[‘codons’],
tcons[‘amino_acids’],
tcons[‘protein_start’],
tcons[‘impact’],
tcons[‘sift_prediction’],
tcons[‘sift_score’])
for val in values:
print (val, end=“\t”)
print()
species = ‘triticum_aestivum’
transcript_id = ‘TraesCS4B02G042700.1’
SNPCDScoord = ‘812’
SNPbase = ‘T’
check_snp_consequences(species,transcript_id,SNPCDScoord,
SNPbase)3.7. Annotate the Effect of Variants with the Ensembl Variant Effect Predictor
The Ensembl VEP tool can be used to predict the effect of variants on genes, transcripts, and protein sequences (see Note 10). As mentioned in Table 2, this analysis is run for all genomic variants imported into Ensembl (see Table 5). While the Ensembl VEP is available through a web interface, the advantage of a local installation is that it can be used to analyze variation sets of any species, including species that are not in Ensembl Plants. If variants are mapped to a reference genome supported in Ensembl Plants, using a cache file increases performance. However, as shown in recipe V4, it is possible to use other reference FASTA files together with the corresponding GFF/GTF annotation files. The next steps summarize how the software is installed and used following recipes F8, V1, V2, and V3.
Clone the repository: git clone https://github.com/Ensembl/ensembl-vep.git.
Navigate to the Ensembl VEP directory: cd ensembl-vep.
Install Ensembl VEP: perl INSTALL.pl.
- Download cache file with recipe F8
SPECIES=arabidopsis_thaliana VEPCACHE=“${SPECIES,,}*.tar.gz*” URL=“${SERVER}/${DIV}/release-${EGRELEASE}/variation/vep/ ${VEPCACHE}” wget -c “$URL” - Unpack downloaded cache file and check SIFT support:
tar xfz $VEPCACHE grep sift “${SPECIES}/${EGRELEASE}_*/info.txt” - Predict effect of variants, see Note 11:
EGRELEASE=47 VCFILE=ensembl-vep/examples/arabidopsis_thaliana.TAIR10.vcf VEPOPTIONS=( --genomes # Ensembl Genomes, for Plants --species $SPECIES --cache # use local cache file, opposed to --database --dir_cache ./ # path of unpacked cache $SPECIES folder --cache_version $EGRELEASE --input_file $VCFILE --output_file ${VCFILE}.vep --check_existing # co-located known variants --distance 5000 # max dist between variant and transcript --biotype # show biotype of neighbor transcript ) ensembl-vep/vep “${VEPOPTIONS[@]”
3.8. Querying Plant Pangenomes
Upcoming Ensembl Plants releases will have an increasing number of species with multiple cultivars or ecotypes as additional assemblies are added in collaboration with the relevant communities. On the website these cultivars can be browsed from the appropriate reference genome page such as http://plants.ensembl.org/Triticum_aestivum/Info/Strains?db=core (see Note 12). Starting with several UK cultivars in release 48 (August 2020), Ensembl will host all cultivars of the first assembled wheat pangenome [49] from release 50 planned for early 2021 (see example Fig. 2). Note that related noncultivated species are often included in the pangenomes of crops. For example, Ensembl Plants hosts 11 Oryza species plus the outgroup plant Leersia perrieri. Both types of genome sets can be considered pangenomes.
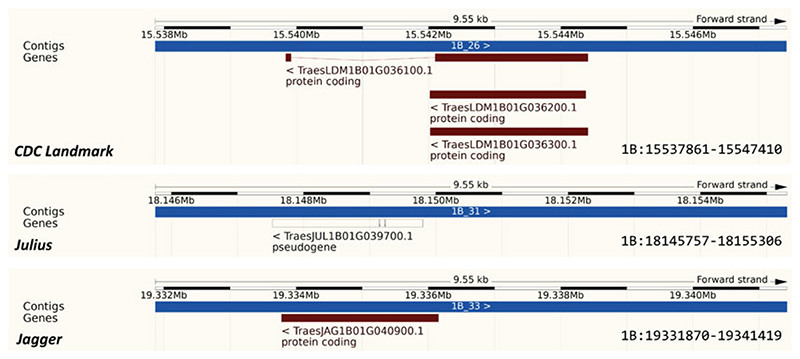
Fig. 2.
The RFL gene (TraesCS1B02G038500) lifted over from the reference landrace Chinese Spring to three wheat cultivars (CDC Landmark, Julius and Jagger). The genes are displayed in the Ensembl Plants genome browser. While in the first cultivar there are three annotated transcript isoforms including one with two exons, the others have a single transcript with one exon. Furthermore, the locus is annotated as a pseudogene in Julius
Currently, some pangenomes in Ensembl can be interrogated using gene trees and whole-genome alignments (WGAs; see Tables 2 and 6). For example, recipe A9 can be used to retrieve syntenic orthologous genes in rice or Brassicaceae species. These analyses will be available for wheat as well once de novo gene annotation and WGAs are produced.
3.9. Getting Help
Documentation for Ensembl Plants, including FAQs, tutorials, and detailed information about the project, datasets, and pipelines that we run can be found under the “Documentation” and “Website help” links at the top of every page. Detailed information for each species can be found on the species homepage. The EMBL-EBI train online website has several free courses on Ensembl, including the recently updated “Ensembl Genomes (non-chordates): Quick tour” (https://www.ebi.ac.uk/training/online/course/ensembl-genomes-non-chordates-quick-tour) and “Ensembl REST API” courses (https://www.ebi.ac.uk/training-beta/online/courses/ensembl-rest-api). Any data problems are reported on our blog http://www.ensembl.info/known-bugs. If the available documentation cannot answer your question, a helpdesk is provided (mail helpdesk@ensemblgenomes.org with your query).
Acknowledgements
We would like to thank Magali Ruffier, Ricardo Ram’rez-González, Nikolai Adamski, and Marcela Karey Tello-Ruiz for recipe suggestions and Gramene colleagues Andrew Olson, Sharon Wei, Justin Preece, Pankaj Jaiswal, and Doreen Ware for continuous support and cooperation. We also acknowledge all of the members of the Ensembl team for developing and maintaining the front-end and back-end software and infrastructure that underpins Ensembl Plants.
Funding
The UK Biosciences and Biotechnology Research Council [BB/P016855/1 and Ensembl-4-Breeders workshop support], the National Sciences Foundation [1127112], the ELIXIR implementation studies FONDUE and “Apple as a Model for Genomic Information Exchange,” and the European Molecular Biology Laboratory. Funding for open access charge: UK Biosciences and Biotechnology Research Council [BB/P016855/1].
Footnotes
The core schema is fully described at https://www.ensembl.org/info/docs/api/core/core_schema.html. There are similar documents for variation, comparative genomics, and regulation schemas: https://www.ensembl.org/info/docs/api/variation/variation_schema.html
https://www.ensembl.org/info/docs/api/compara/compara_schema.html
https://www.ensembl.org/info/docs/api/funcgen/funcgen_schema.html
The schema for the metadata database can be found at https://github.com/Ensembl/ensembl-metadata.
Instructions to set up a local Ensembl database are provided at http://plants.ensembl.org/info/docs/webcode/mirror/install/ensembl-data.html.
Check the annotation page for each species in Ensembl Plants. For Arabidopsis thaliana, this is http://plants.ensembl.org/Arabidopsis_thaliana/Info/Annotation/#genebuild.
Gene trees use canonical transcripts, defined at http://plants.ensembl.org/info/website/glossary.html. In plant species, the canonical transcript of a protein-coding gene is the transcript with the longest translation with no stop codons. This does not necessarily reflect the most biologically relevant transcript of a gene. The script https://github.com/Ensembl/plant_tools/blob/master/phylogenomics/ens_sequences.pl can be used to obtain sequences of canonical transcripts in FASTA format.
Check http://plants.ensembl.org/info/docs/Doxygen. See also debugging instructions and tutorials at http://plants.ensembl.org/info/docs/api.
See http://plants.ensembl.org/info/genome/compara/homology_method.html for the definitions of the different homology types.
The variation schema is described at http://plants.ensembl.org/info/docs/api/variation/variation_schema.html.
Training material to learn more about the Ensembl REST interface can be found at https://www.ebi.ac.uk/training/online/course/ensembl-rest-api and https://mybinder.org/v2/gh/Ensembl/rest-api-jupyter-course/master. The different endpoints are documented at https://rest.ensembl.org/documentation.
Ensembl VEP functionality can be extended to utilize additional data or run additional analyses using plugins, see https://www.ensembl.org/info/docs/tools/vep/script/vep_plugins.html.
The full list of options of Ensembl VEP is described at http://www.ensembl.org/info/docs/tools/vep/script/vep_options.html, there are examples at http://www.ensembl.org/info/docs/tools/vep/script/vep_example.html.
Reference genomes have binomial production names when possible, such as oryza_sativa (rice) or triticum_aestivum (bread wheat). Additional cultivars or ecotypes have longer trinomial names such as oryza_sativa_indica or triticum_aestivum_cadenza. Following this convention, theobroma_cacao_-matina and panicum_hallii_hal2 will be renamed to theobroma_cacao and panicum_hallii by release 50.
Conflict of Interest Statement
Paul Flicek is a member of the Scientific Advisory Boards of Fabric Genomics, Inc. and Eagle Genomics, Ltd.
References
- 1.Ritchie H, Roser M. Crop yields. 2013. [Accessed 1 Jul 2020]. https://ourworldindata.org/crop-yields .
- 2.Wallace JG, Rodgers-Melnick E, Buckler ES. On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu Rev Genet. 2018;52:421–444. doi: 10.1146/annurev-genet-120116-024846. [DOI] [PubMed] [Google Scholar]
- 3.Arora S, Steuernagel B, Gaurav K, et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol. 2019;37:139–143. doi: 10.1038/s41587-018-0007-9. [DOI] [PubMed] [Google Scholar]
- 4.Adamski NM, Borrill P, Brinton J, et al. A roadmap for gene functional characterisation in crops with large genomes: lessons from polyploid wheat. elife. 2020;9:55646. doi: 10.7554/eLife.55646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howe KL, Contreras-Moreira B, De Silva N, et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020;48:D689–D695. doi: 10.1093/nar/gkz890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tello-Ruiz MK, Naithani S, Stein JC, et al. Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018;46:D1181–D1189. doi: 10.1093/nar/gkx1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell AL, Attwood TK, Babbitt PC, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Gene Ontology Consortium, The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still going strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates A, Beal K, Keenan S, et al. The ensembl REST API: ensembl data for any language. Bioinformatics. 2015;31:143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database. 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amid C, Alako BTF, Balavenkataraman Kad-hirvelu V, et al. The European Nucleotide Archive in 2019. Nucleic Acids Res. 2020;48:D70–D76. doi: 10.1093/nar/gkz1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodstein DM, Shu S, Howson R, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai H, Lee SS, Tanaka T, et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naithani S, Gupta P, Preece J, et al. Plant Reactome: a knowledgebase and resource for comparative pathway analysis. Nucleic Acids Res. 2020;48:D1093–D1103. doi: 10.1093/nar/gkz996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero J, Muffato M, Beal K, et al. Ensembl comparative genomics resources. Database. 2016;2016:baw053. doi: 10.1093/database/baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium TU The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.1001 Genomes Consortium. 1,135 Genomes reveal the global pattern of polymor-phism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atwell S, Huang YS, Vilhja’lmsson BJ, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox SE, Preece J, Kimbrel JA, et al. Sequencing and de novo transcriptome assembly of Brachypodium sylvaticum (Poaceae) Appl Plant Sci. 2013;1:1200011. doi: 10.3732/apps.1200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer KFX, Waugh R, et al. International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 22.Mascher M, Muehlbauer GJ, Rokhsar DS, et al. Anchoring and ordering NGS contig assemblies by population sequencing (POP-SEQ) Plant J. 2013;76:718–727. doi: 10.1111/tpj.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariyadasa R, Mascher M, Nussbaumer T, et al. A sequence-ready physical map of bar-ley anchored genetically by two million singlenucleotide polymorphisms. Plant Physiol. 2014;164:412–423. doi: 10.1104/pp.113.228213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersey PJ, Allen JE, Allot A, et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018;46:D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianco L, Cestaro A, Linsmith G, et al. Development and validation of the Axiom(®) Apple480K SNP genotyping array. Plant J. 2016;86:62–74. doi: 10.1111/tpj.13145. [DOI] [PubMed] [Google Scholar]
- 26.Sherry ST. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.3,000 Rice Genomes Project. The 3,000 rice genomes project. GigaScience. 2014;3:7. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duitama J, Silva A, Sanabria Y, et al. Whole genome sequencing of elite rice cultivars as a comprehensive information resource for marker assisted selection. PLoS One. 2015;10:e0124617. doi: 10.1371/journal.pone.0124617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao K, Wright M, Kimball J, et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One. 2010;5:e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNally KL, Childs KL, Bohnert R, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto E, Yonemaru J-I, Yamamoto T, Yano M. OGRO: the overview of functionally characterized Genes in Rice online database. Rice. 2012;5:26. doi: 10.1186/1939-8433-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.100 Tomato Genome Sequencing Consortium. Aflitos S, Schijlen E, et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014;80:136–148. doi: 10.1111/tpj.12616. [DOI] [PubMed] [Google Scholar]
- 33.Morris GP, Ramu P, Deshpande SP, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci U S A. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mace ES, Tai S, Gilding EK, et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. 2013;4:2320. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao Y, Burke J, Chopra R, et al. A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant Cell. 2016;28:1551–1562. doi: 10.1105/tpc.16.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson PA, Winfield MO, Barker GLA, et al. CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinformatics. 2012;13:219. doi: 10.1186/1471-2105-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasileva KV, Vasquez-Gross HA, Howell T, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci U S A. 2017;114:E913–E921. doi: 10.1073/pnas.1619268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rimbert H, Darrier B, Navarro J, et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS One. 2018;13:e0186329. doi: 10.1371/journal.pone.0186329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myles S, Chia J-M, Hurwitz B, et al. Rapid genomic characterization of the genus vitis. PLoS One. 2010;5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chia J-M, Song C, Bradbury PJ, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 2012;44:803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris RS. Improved pairwise alignment of genomic DNA. The Pennsylvania State University; Pennsylvania: 2007. [Google Scholar]
- 44.Petryszak R, Keays M, Tang YA, et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petryszak R, Fonseca NA, Fullgrabe A, et al. The RNASeq-er API—a gateway to systematically updated analysis of public RNA-seq data. Bioinformatics. 2017;33:2218–2220. doi: 10.1093/bioinformatics/btx143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt T, Messina DN, Schreiber F, Sonn-hammer ELL. Letter to the editor: SeqXML and OrthoXML: standards for sequence and orthology information. Brief Bioinform. 2011;12:485–488. doi: 10.1093/bib/bbr025. [DOI] [PubMed] [Google Scholar]
- 48.Igartua E, Contreras-Moreira B, Casas AM. TB1: from domestication gene to tool for many trades. J Exp Bot. 2020;71:4621–4624. doi: 10.1093/jxb/eraa308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walkowiak S, Gao L, Monat C, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]