Abstract
We present a method for skill characterisation of sonographer gaze patterns while performing routine second trimester fetal anatomy ultrasound scans. The position and scale of fetal anatomical planes during each scan differ because of fetal position, movements and sonographer skill. A standardised reference is required to compare recorded eye-tracking data for skill characterisation. We propose using an affine transformer network to localise the anatomy circumference in video frames, for normalisation of eye-tracking data. We use an event-based data visualisation, time curves, to characterise sonographer scanning patterns. We chose brain and heart anatomical planes because they vary in levels of gaze complexity. Our results show that when sonographers search for the same anatomical plane, even though the landmarks visited are similar, their time curves display different visual patterns. Brain planes also, on average, have more events or landmarks occurring than the heart, which highlights anatomy-specific differences in searching approaches.
CCS Concepts: Computing methodologies → Object detection, Human-centered computing → Visual analytics
Keywords: fetal ultrasound, eye tracking, affine transformer networks, time curves
1. Introduction
During second trimester fetal anomaly ultrasound (US) scans, sonographers manipulate and adjust the position of an US probe to acquire a series of standard anatomical imaging planes. The UK Fetal Anomaly Screening Programme (FASP) guidelines require sonographers to correctly identify more than 10 specific fetal anatomical planes [Public Health England (PHE) 2018]. The time dedicated to a typical anomaly scan is 30 minutes. As the fetus is a moving target, a sonographer changes the position of the probe in response. This results in differences in position and scale of the fetal image during a full-length scan. Conversely, in other medical imaging, non-obstetric, [Nodine et al. 1996; Van der Gijp et al. 2017] or surgery [Ahmidi et al. 2010, 2012] the area of interest (AOI) is typically stationary or well-defined. The sequence of capturing these anatomical planes is not identical, and skill and experience of sonographers vary [Nicholls et al. 2014]. This makes image capture an opportunistic rather than a repeatable process. Consequently, fetal US videos present unique challenges that are not in other medical applications.
Diagnosis and interpretation of fetal ultrasound videos occur simultaneously while scanning [Drukker et al. 2021], different from radiology [Calisto et al. 2017] or mammograms [Calisto et al. 2021] where diagnosis occurs after scanning; [Calisto et al. 2017, 2021] design post-scanning specific frameworks. We have to consider instance-specific observations [Gu et al. 2021], which can be tailored to the dynamic image on the screen. Current methods such as gaze heatmaps are more useful for static images (e.g. reading a map). When used for videos, they require a non-trivial set up, for example, like that of space-time cubes [Kurzhals and Weiskopf 2013].
We are motivated to focus on skill characterisation, rather than assessment. Assessment involves grouping clinicians into different levels of expertise for classification or comparison. In skill characterisation, we aim to find features or patterns in the way clinicians search for specific fetal anatomy planes. [Droste et al. 2020; Drukker et al. 2020] showed that in fetal US, sonographers observe salient anatomical landmarks before capturing an image plane. In our work, we provide a means of identifying distinct scanning patterns using eye-tracking data. Then, we would be able to see which landmarks a sonographer has observed (or not) during their scan. From a clinical perspective, knowing which landmarks a trainee sonographer observes is important in improving their scanning skill.
In skill assessment eye-tracking studies, eye-tracking data is processed in two main ways. The first calculates AOI using raw eye-tracking data. This could be done by classical threshold algorithms that use gaze velocity to separate eye-tracking data into fixations and saccades. Alternatively, experts manually label AOIs prior to the experiment as the size of AOIs are application dependent. This method uses common evaluation metrics which include time within the AOI and time taken to reach the desired target. The second pre-processing method transforms raw eye-tracking data into different modalities such as images [Sharma et al. 2021] or discrete cluster labels [Ahmidi et al. 2010]. The transformed data is fed into classification models assessing level of skill based on specific groupings for example expert, intermediate and novice.
1.1. Related Work
Previous US studies using eye-tracking data for skill assessment either use statistical metrics or classification accuracy to assess clinician expertise. [Battah et al. 2016; Borg et al. 2018; Chen et al. 2019a,b; Harrison et al. 2016; Lee and Chenkin 2021] use number of fixations, length of fixations and total time to target. Another commonly used metric is time taken to complete the task [Nodine et al. 1996; Tatsuru et al. 2021]. In their applications, their work use simulators [Chen et al. 2019a; Harrison et al. 2016; Tatsuru et al. 2021] which do not necessarily capture real-world complexities. [Manning et al. 2006; Nodine et al. 1996] also consider static images where their AOI does not change in position or size during the experiment. Other works [Ahmidi et al. 2010; Sharma et al. 2021] use transformed raw eye-tracking data which are fed into classification models, but do not investigate landmark adherence specific behaviour for skill characterisation.
1.2. Contribution
We have two main contributions. The first is an affine transformer network (ATN) for localising the anatomy circumference (example of anatomy circumference in Fig. 1, step 2) of a fetus US video which allows for scale and position invariance of the fetal image between scans. Then, we use a data visualisation methodology, time curves [Bach et al. 2016], to characterise sonographer scanning patterns for different tasks.
Figure 1. Image demonstrating the process of generating an event to represent a fixation from the original US frames.
Step 1: Original US frames. Step 2: Frames transformed using a ATN. Step 3: Selecting a representation of the event as the middle frame of the fixation. Step 4: Landmark focused on, defined as an event.
2. Methods
We present our method for localising the anatomy circumference in US frames using an ATN inspired by spatial transformer networks (STN) [Jaderberg et al. 2015]. We also show our use of time curves [Bach et al. 2016] to visualise sonographer recorded eye-tracking data. We demonstrate a variety of scanning styles while searching for anatomical planes of differing levels of complexity.
2.1. Normalisation of Eye-Tracking Data by Localising Anatomy Circumference using Affine Transformer Networks
STNs [Jaderberg et al. 2015] are differentiable modules that can be used within deep learning network architectures to remove spatial and position variance between images for a downstream task such as classification or object detection. They have been used in US for image registration [Lee et al. 2019; Roy et al. 2020]. In our work, we use the proposed architecture by [Jaderberg et al. 2015] as a simple ATN. The network localises the fetal anatomy circumference to ensure that the fetus is the object of reference for our raw eye-tracking data.
[Teng et al. 2021] has shown that normalising eye-tracking data with respect to the anatomy circumference improves the performance of using eye-tracking data for anatomy plane classification. However, they manually labelled the circumference for >300 clips. A clip is defined as a period of time spent searching for an anatomy plane. During a scan there can be multiple clips, where the anatomy plane is searched for multiple times. To reduce the manual labour involved in labelling a large-scale dataset, we used an ATN to localise the anatomy circumference. We applied the learnt transformation (Eq. 1) to our eye-tracking data for normalisation. The affine transformation contained 6 parameters (Eq. 1) and accounts for scale, translation, shear and rotation [Jaderberg et al. 2015].
| (1) |
We are inspired by the deep learning architecture [Jaderberg et al. 2015] used to learn affine transformation parameters in their localisation network. Their localisation network transforms images which become scale and position invariant for the downstream task. In our network, we used GoogleNet [Szegedy et al. 2014] and removed the last average pooling layer and last fully connected layer. On top of this, we combined (i) a 1 x 1 convolutional layer to reshape the feature channels from 1024 to 128, (ii) a fully connected layer with 128x7x7 output and (iii) a fully connected layer with 6 outputs representing our affine transformation parameters [Jaderberg et al. 2015].
We used the AdamW optimizer [Loshchilov and Hutter 2017]; our base learning rate started at 1e-4 and is reduced by a factor of 10 after 25, 50, 75 and 100 epochs. Our loss is calculated as the mean squared error between the transformed image and our ground truth at the pixel level [Roy et al. 2019]. We allowed our model to train for 150 epochs and implemented early stopping if the validation loss did not decrease by 0.001 after 10 consecutive epochs.
Our ground truth frames are US video frames which were manually cropped using the method set out in [Teng et al. 2021] to locate the anatomy circumference.
2.2. Visualisation of Eye-Tracking Data using Time Curves
For our visualisation, we focused on event-based methods which informs the user of what event has occurred and its characteristics. Each event must depict attributes of the data independently, similar to a glyph-based visualisation [Borgo et al. 2013]. We chose time curves [Bach et al. 2016] because they are informative and simple to implement and interpret. To construct our time curve we required the time spent at an event, and a similarity matrix to define how similar events are. Time curves [Bach et al. 2016] are a two-dimensional (2D) data visualisation method that preserves temporal order of defined events. First a linear timeline of events is plotted along the x axis, time. A distance matrix is used to represent the similarity between events. The distance metric chosen is application dependent. Events are mapped onto a lower dimensional space (2D) using the distance matrix. Finally, a suitable interpolation curve, for example, splines, is used to connect event points. An example is shown in Fig. 2.
Figure 2. Image demonstrating the process of developing the time curve (Illustrative purposes only, not to scale).
Step 1: Creating events. Step 2: Calculating distance matrix between events. Step 3: Distance matrix reduced to 2D using multidimensional scaling (MDS). Step 4: Events connected using splines.
To construct meaningful events, we first calculated fixations and saccades using the standardised velocity threshold algorithm [Olsen 2012] based on [Cai et al. 2018]. Gaze velocity < 30 degrees per second is classified as a fixation, otherwise as saccade. We followed the procedure in [Teng et al. 2021] where we linearly interpolated any missing gaze points due to sampling discrepancies or tracking errors, and discarded any fixation which was less than 210ms.
In our work, we defined an event as a snapshot of what landmark the sonographer is focused on during a fixation. The position of a fixation is typically calculated as the average gaze point [Olsen 2012] during the fixation. Likewise, we took the middle time stamp (Fig. 1, step 2) of the fixation as the average frame representing our AOI. We cropped out a segment (20x20 pixels) of the image centered around the gaze point to create an event (Fig. 1, step 4). Since we use the resized 224x224 images, a 20x20 crop was considered to be sufficient for capturing the landmark the sonographer focused on. We adjusted these parameters based on the visibility of the structures in the frames. For frames where gaze points were outside of the anatomy circumference, we substituted the landmark with a 20x20 grey square. Snapshots which partially fell outside the anatomy circumference were zero padded with grey pixels.
We flattened all task specific event (Fig. 2, step 1) images to a 1xn dimensional vector. Then we calculated the Euclidean distance between each vector, returning a square form distance matrix (Fig. 2, step 2). We chose the Euclidean distance as [Bach et al. 2016] has shown that a naive pixel difference for normalised images still returns informative results. The distance matrix is transformed to a lower 2D space using multi-dimensional scaling (MDS). From a mxm matrix, where m is the number of total snapshots in the dataset, we return a mx2 matrix. These transformed data points are shown in Fig. 2, step 3. Finally, to connect our events in temporal order (Fig. 2, step 4), we used a 2nd order B-spline with a smoothing factor of 1 to interpolate between our transformed data points. We chose these parameters by determining whether the interpolated curve was able to pass through or near our events. We also used a colour mapping to indicate the temporal order (Fig. 2, step 3). Light orange indicates the start of the time series, and dark orange the end.
We describe gaze scanning patterns using definitions of visual patterns given by [Bach et al. 2016] in Fig. 17 of their paper. The examples are cluster, transition, cycle, u-turn, outlier, oscillation and alternation. A cluster represents events that are close to one another in space. A transition is where a cluster of events have migrated from one point in space to another. A cycle and u-turn are where the start and end events are near each other, where for cycle the time curve creates a closed circle loop. Outliers are where there are several events that are far away in position compared to majority of other events. Oscillation resembles a sine/cosine wave. Alternation is where the time curve revisits similar/identical events repeatedly.
3. Data
Our eye-tracking data recorded sonographer gaze during full-length second trimester US scans. The sonographers were recorded for the duration of the scan which takes an average of 36 minutes [Drukker et al. 2021]. We used a Tobii Eye Tracker 4C sampled at 90Hz, which was mounted underneath the US screen. Our dataset consisted of 10 operators who are either fully qualified to work as National Health Service (NHS) sonographers, trainee sonographers supervised by qualified sonographers, or fetal medicine doctors. They had between 0 and 15 years of scanning experience (at the start of the study).
3.1. Affine Transformer Network
The dataset used to train our ATN are second-trimester manually labelled abdomen, brain, and heart image planes, labelled using the protocol set out by [Drukker et al. 2021; Sharma et al. 2020]. We chose these planes based on work by [Teng et al. 2021]. The data set contained the last 100 (approximately 3 seconds at a 30Hz video sampling rate) frames just before the operator froze the video to take measurements of the captured anatomy plane. We used approximately 30,000 frames in total. We randomly shuffled all frames and use a 80/10/10 train/validation/test split. In total, there are 84, 160, 122 abdomen, brain and heart plane clips respectively and 76 unique pregnant women.
We preprocessed the image dataset by applying several transformations. For training, we (i) applied 3 random augmentations [Cubuk et al. 2019] of magnitude 3 as they were shown to be the optimal parameters for classification of standard US anatomy planes [Lee et al. 2021]. We also (ii) resized our images to 224x224 pixels, and performed (iii) image normalisation in that specified order. For our ground truth images (validation set), we applied the same 3 random augmentations that was used for our training frames by fixing the random seed in PyTorch, resized to 224x224 pixels and normalised. For our test dataset, we resized and normalised our images.
3.2. Skill Characterisation and Visualisation
For skill characterisation and visualisation, our dataset consisted of 2 brain and 5 heart planes which were manually labelled 1. Each anatomical plane is considered as a separate task. They were chosen as brain planes are considered easier to search for compared to the heart due to differences in anatomy size [Teng et al. 2021], and it was shown that operators spent the most time searching for these anatomy planes during a routine clinical second trimester scan [Sharma et al. 2019, 2020].
The 2 brain planes are: (i) head circumference (HCP) and (ii) suboccipitobregmatic (SOB) view where the transcerebellar is in view [Public Health England (PHE) 2018]. The 5 heart planes are: (i) 3 vessel and trachea view (3VT), (ii) 3 vessel view (3VV), (iii) 4 chamber view (4CH), (iv) aorta/left ventricular outflow tract (LVOT) and (v) pulmonary artery/right ventricular outflow tract (RVOT) [Public Health England (PHE) 2018]. An example of the planes are shown in Fig. 3. We did not include any scans which were used to train the ATN for generating our skill characterisation analysis. In total there were 250 unique pregnant women and 185, 188, 65, 57, 66, 50, 61 HCP, SOB, 3VT, 3VV, 4CH, LVOT and RVOT clips each. We used eye-tracking data corresponding to the last 150 frames (5 seconds at a 30Hz video sampling frequency) before the operator froze the video to take measurements for each anatomical plane.
Figure 3. Heart and brain standard planes.
Left to right: Brain planes are (i) HCP and (ii) SOB. The heart planes are (iii) 3VT, (iv) 3VV, (v) 4CH, (vi) LVOT, (vii) RVOT.
4. Results
We assessed the result of using the ATN to localise the anatomy plane visually. An example is shown in Fig. 4. Figure 4 shows that our model is able to localise the anatomy circumference for different standard brain and heart planes. The localisation effect is most noticeable in SOB, 3VT, 4CH, LVOT where the original images were off center and did not fill the screen fully. The final transformed image is scaled and centered.
Figure 4. An example showing the performance of the ATN on standard plane images.
Left to right: HCP, SOB, 3VT, 3VV, 4CH, LVOT, RVOT. Top: Original images resized to 224x224 pixels. Bottom: Images after ATN transformation, resized to 224x224 pixels.
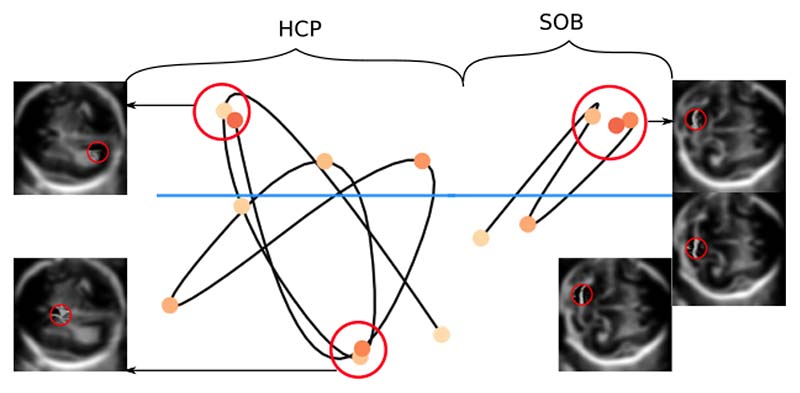
To analyse our time curves, we adopted terminology by [Bach et al. 2016], where they described some example patterns of time curves. We presented several types of scanning patterns for each standard plane below. To show as many examples as possible, we have not presented the plots with their axis. For reference, the fully labelled time curve can be seen in Fig. 2, step 4. To allow for ease of comparison between time curves, a task-specific reference line is provided in blue (Fig. 5).
Figure 5.
An example of several scanning patterns for brain planes and heart planes. From left to right, top to bottom: (i) HCP, (ii) SOB, (iii) 3VT, (iv) 3VV, (v) 4CH, (vi) LVOT, (vii) RVOT.
The average number of events for heart planes range from 1.5 - 1.9, while it ranges from 4.2 - 5.3 for brain planes. Figure 5 shows that for brain planes there are several types of scanning patterns. Many of the scans showed a combination of a u-turn, cycle, clusters and transitions where sonographers have revisited similar anatomical landmarks several times over the course of scanning. For HCP, the majority of events in Fig. 5 (i) are clustered below the reference line with differing temporal order (of events). For SOB, majority of events in Fig. 5 (ii) are clustered above the reference line. There were also patterns which showed that sonographers followed 3 - 5 distinct landmarks without revisiting them (Fig. 5, ii). There does not appear to be a pattern for when the events occurred, as shown by the scattered distribution of colours.
Figure 5 (iii - vii) shows that for heart planes most of the scans only had a single event occurring, corresponding to a unique landmark that the sonographer focused on during the scan. Since the event is coloured dark orange, this indicated that the landmark was followed for the full length of 5 seconds. A small minority showed patterns of loops and straight lines, where only 2-3 events occurred.
A comparison between brain and heart plane scanning patterns showed that on average there are more unique landmarks that sonographers observed and revisited for brain planes. The majority of heart planes returned a single event. Although both the brain and heart are 3-dimensional objects, the size of the heart is smaller and sonographer gaze does not need to travel as far. To get the correct heart plane view, the sonographer has to make more minor and smaller adjustments with the probe compared to the brain. These anatomical differences are reflected in the visual patterns of brain plane time curves in Fig. 5 (i, ii) and heart plane time curves in Fig. 5 (iii - vii).
5. Discussion
We performed a qualitative analysis of our results to investigate the similarity of clustered events. For brain planes, these are shown in Fig. 6. For HCP, the sonographers generally looked at choroid plexus and cavum septum pellucidem. For SOB, they looked at cerebellum where they measure the transcerebellar diameter. For heart planes, we listed some examples in Fig. 7 where the sonographer had looked at the trachea, and the intersection of the pulmonary artery (PA) and aorta (3VT), PA (3VV), crux of the ventricular septum (4CH), aorta walls (LVOT) and bifurcation of the PA (RVOT).
Figure 6. Example of landmarks which were viewed while scanning for brain planes HCP (left) and SOB (right).
Figure 7. Example of landmarks which were viewed while scanning for heart planes 3VT, 3VV, 4CH, LVOT, RVOT.
With our method, we were able to differentiate between operators at an individual level, which particular landmarks were visited during a scan. In large scale longitudinal studies such as ours, we would be able to identify outliers easily compared to the general gaze scanning patterns. For example, in heart planes operators largely followed the landmarks about a fixed axis. An outlier example would then be an operator who had looked around the image more vigorously. By studying these scans at an individual level, we can identify which particular tasks could have been more difficult for an operator based on their gaze patterns.
Our definition of an event is less able to provide defining characteristics for heart planes compared to brain planes. Many time curves returned a single event (Fig. 5) over 150 frames as their gaze points were classified as a single fixation. A more fine-grained definition of an event for the heart plane could yield an informative comparison for sonographer scanning patterns. For example, using image segmentation to fine tune the exact anatomical landmark being observed, or separating the eye movements into fixations, saccades and smooth pursuits. Suitable parameters for classification of different eye movements could be chosen using non-parametric methods [Komogortsev et al. 2010]. More experienced sonographers could also anticipate the appearance of a landmark and moved their gaze to the AOI before, instead of when it appeared instead. Our method does not account for this anticipatory type of gaze behaviour.
6. Conclusion
We present an affine transformer network to localise the anatomy circumference, which provides a reference to normalise sonographer eye-tracking data recorded while performing second trimester fetal ultrasound scans. Our normalised eye-tracking data was visualised using time curves. We distinguished task-specific sonographer scanning patterns when searching for anatomical planes of varying levels of complexity. Our work showed that using fixations occurred as events to build time curve visualisations is an effective method to demonstrate observed landmarks and scanning patterns for brain planes, but less so for heart planes.
Acknowledgments
We thank Professor Min Chen for his expertise and guidance on constructing the data visualisation. We also thank the PULSE team members Jianbo, Lok, Richard and Zeyu for providing the labelled heart and brain planes. We acknowledge the ERC (Project PULSE: ERC-ADG-2015 694581). ATP is supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the NIHR Biomedical Research Centre (BRC) funding scheme.
Footnotes
Contributor Information
Clare Teng, Email: clare.teng@eng.ox.ac.uk, Institute of Biomedical Engineering, University of Oxford Oxford, United Kingdom.
Lok Hin Lee, Email: leelokhin@gmail.com, Institute of Biomedical Engineering, University of Oxford Oxford, United Kingdom.
Jayne Lander, Email: jayne.lander@ouh.nhs.uk, Nuffield Department of Women’s and Reproductive Health, University of Oxford Oxford, United Kingdom.
Lior Drukker, Email: drukker@gmail.com, Nuffield Department of Women’s and Reproductive Health, University of Oxford Oxford, United Kingdom Women’s Ultrasound, Department of Obstetrics and Gynecology, Beilinson Medical Center, Sackler Faculty of Medicine, Tel-Aviv University Tel Aviv, Israel.
Aris T. Papageorghiou, Email: aris.papageorghiou@wrh.ox.ac.uk, Nuffield Department of Women’s and Reproductive Health, University of Oxford Oxford, United Kingdom.
Alison J. Noble, Email: alison.noble@eng.ox.ac.uk, Institute of Biomedical Engineering, University of Oxford Oxford, United Kingdom.
References
- Ahmidi Narges, Hager Gregory D, Ishii Lisa, Fichtinger Gabor, Gallia Gary L, Ishii Masaru. Surgical Task and Skill Classification from Eye Tracking and Tool Motion in Minimally Invasive Surgery Narges. MICCAI 2010. 2010:295–302. doi: 10.1007/978-3-642-15711-0_37. [DOI] [PubMed] [Google Scholar]
- Ahmidi Narges, Ishii Masaru, Fichtinger Gabor, Gallia Gary L, Hager Gregory D. International forum of allergy & rhinology. Vol. 2. Wiley Online Library; 2012. An objective and automated method for assessing surgical skill in endoscopic sinus surgery using eye-tracking and tool-motion data; pp. 507–515. [DOI] [PubMed] [Google Scholar]
- Bach Benjamin, Shi Conglei, Heulot Nicolas, Madhyastha Tara, Grabowski Tom, Dragicevic Pierre. Time Curves: Folding Time to Visualize Patterns of Temporal Evolution in Data. IEEE Transactions on Visualization and Computer Graphics. 2016;22(1):559–568. doi: 10.1109/TVCG.2015.2467851. (1 2016) [DOI] [PubMed] [Google Scholar]
- Battah Mohamed, McKendrick M, Kean DE, Obregon M. Eye-tracking as a measure of trainee progress in laparoscopic training. European Journal of Obstetrics and Gynecology and Reproductive Biology. 2016;206:e13–e14. 2016. [Google Scholar]
- Borg Lindsay K, Harrison T Kyle, Kou Alex, Mariano Edward R, Udani Ankeet D, Kim T Edward, Shum Cynthia, Howard Steven K, ADAPT (Anesthesiology-Directed Advanced Procedural Training) Research Group Preliminary experience using eye-tracking technology to differentiate novice and expert image interpretation for ultrasound-guided regional anesthesia. Journal of Ultrasound in Medicine. 2018;37(2):329–336. doi: 10.1002/jum.14334. 2018. [DOI] [PubMed] [Google Scholar]
- Borgo Rita, Kehrer Johannes, Chung David HS, Maguire Eamonn, Laramee Robert S, Hauser Helwig, Ward Matthew, Chen Min. In: Eurographics 2013 - State of the Art Reports. Sbert M, Szirmay-Kalos L, editors. The Eurographics Association; 2013. Glyph-based Visualization: Foundations, Design Guidelines, Techniques and Applications. [DOI] [Google Scholar]
- Cai Y, Sharma H, Chatelain P, Noble JA. SonoEyeNet: Standardized fetal ultrasound plane detection informed by eye tracking; 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018. pp. 1475–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisto Francisco M, Ferreira Alfredo, Nascimento Jacinto C, Gonçalves Daniel. Towards Touch-Based Medical Image Diagnosis Annotation; Proceedings of the 2017 ACM International Conference on Interactive Surfaces and Spaces (Brighton, United Kingdom) (ISS ’17); New York, NY, USA: Association for Computing Machinery; 2017. pp. 390–395. [DOI] [Google Scholar]
- Calisto Francisco Maria, Santiago Carlos, Nunes Nuno, Nascimento Jacinto C. Introduction of human-centric AI assistant to aid radiologists for multimodal breast image classification. International Journal of Human-Computer Studies. 2021;150:102607. doi: 10.1016/j.ijhcs.2021.102607. 2021. [DOI] [Google Scholar]
- Chen Hong-En, Bhide Rucha R, Pepley David F, Sonntag Cheyenne C, Moore Jason Z, Han David C, Miller Scarlett R. Can eye tracking be used to predict performance improvements in simulated medical training? A case study in central venous catheterization; Proceedings of the International Symposium on Human Factors and Ergonomics in Health Care; Los Angeles, CA: SAGE Publications Sage CA; 2019a. pp. 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Hong-En, Sonntag Cheyenne C, Pepley David F, Prabhu Rohan S, Han David C, Moore Jason Z, Miller Scarlett R. Looks can be deceiving: Gaze pattern differences between novices and experts during placement of central lines. The American Journal of Surgery. 2019b;217(2):362–367. doi: 10.1016/j.amjsurg.2018.11.007. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk Ekin D, Zoph Barret, Shlens Jonathon, Le Quoc V. RandAugment: Practical data augmentation with no separate search. CoRR. 2019:abs/1909.1. (2019). http://arxiv.org/abs/1909.13719. [Google Scholar]
- Droste R, Chatelain P, Drukker L, Sharma H, Papageorghiou AT, Noble JA. Discovering Salient Anatomical Landmarks by Predicting Human Gaze. Proceedings - International Symposium on Biomedical Imaging 2020-April (2020) 2020:1711–1714. doi: 10.1109/ISBI45749.2020.9098505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker L, Droste R, Noble A, Papageorghiou AT. VP40.20: Standard bio-metric planes: what are the salient anatomical landmarks? Ultrasound in Obstetrics & Gynecology. 2020;56(S1):235. doi: 10.1002/uog.22958. 2020. [DOI] [Google Scholar]
- Drukker Lior, Sharma Harshita, Droste Richard, Alsharid Mohammad, Chate-lain Pierre, Noble J Alison, Papageorghiou Aris T. Transforming obstetric ultrasound into data science using eye tracking, voice recording, transducer motion and ultrasound video. Scientific Reports. 2021;11(1):14109. doi: 10.1038/s41598-021-92829-1. (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Hongyan, Huang Jingbin, Hung Lauren, Chen Xiang Anthony. Lessons Learned from Designing an AI-Enabled Diagnosis Tool for Pathologists. Proceedings of the ACM on Human-Computer Interaction. 2021;5(CSCW1) doi: 10.1145/3449084. 2021. [DOI] [Google Scholar]
- Harrison T Kyle, Kim T Edward, Kou Alex, Shum Cynthia, Mariano Edward R, Howard Steven K. Feasibility of eye-tracking technology to quantify expertise in ultrasound-guided regional anesthesia. Journal of anesthesia. 2016;30(3):530–533. doi: 10.1007/s00540-016-2157-6. (2016) [DOI] [PubMed] [Google Scholar]
- Jaderberg Max, Simonyan Karen, Zisserman Andrew, Kavukcuoglu Koray. Spatial Transformer Networks. CoRR. 2015:abs/1506.0. (2015) http://arxiv.org/abs/1506.02025 . [Google Scholar]
- Komogortsev Oleg V, Gobert Denise V, Jayarathna Sampath, Koh Do Hyong, Gowda Sandeep M. Standardization of Automated Analyses of Oculomotor Fixation and Saccadic Behaviors. IEEE Transactions on Biomedical Engineering. 2010;57(11):2635–2645. doi: 10.1109/TBME.2010.2057429. 2010. [DOI] [PubMed] [Google Scholar]
- Kurzhals Kuno, Weiskopf Daniel. Space-Time Visual Analytics of Eye-Tracking Data for Dynamic Stimuli. IEEE Transactions on Visualization and Computer Graphics. 2013;19(12):2129–2138. doi: 10.1109/TVCG.2013.194. (2013) - Their paper cites using mean shift for clustering. [DOI] [PubMed] [Google Scholar]
- Lee Lok Hin, Gao Yuan, Noble J Alison. In: Information Processing in Medical Imaging. Feragen Aasa, Sommer Stefan, Schnabel Julia, Nielsen Mads., editors. 2021. Principled Ultrasound Data Augmentation for Classification of Standard Planes; pp. 729–741. [Google Scholar]
- Lee Matthew CH, Oktay Ozan, Schuh Andreas, Schaap Michiel, Glocker Ben. Image-and-Spatial Transformer Networks for Structure-guided Image Registration; International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); 2019. [Google Scholar]
- Lee Wei Feng, Chenkin Jordan. Exploring Eye-tracking Technology as an Assessment Tool for Point-of-care Ultrasound Training. AEM Education and Training. 2021;5(2):e10508. doi: 10.1002/aet2.10508. (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loshchilov Ilya, Hutter Frank. Fixing Weight Decay Regularization in Adam. CoRR. 2017:abs/1711.0. (2017). http://arxiv.org/abs/1711.05101 . [Google Scholar]
- Manning David, Ethell Susan, Donovan Tim, Crawford Trevor. How do radiologists do it? The influence of experience and training on searching for chest nodules. Radiography. 2006;12(2):134–142. doi: 10.1016/j.radi.2005.02.003. (2006) [DOI] [Google Scholar]
- Nicholls Delwyn, Sweet Linda, Hyett Jon. Psychomotor skills in medical ultrasound imaging: An analysis of the core skill set. Journal of Ultrasound in Medicine. 2014;33(8):1349–1352. doi: 10.7863/ultra.33.8.1349. (2014) [DOI] [PubMed] [Google Scholar]
- Nodine Calvin F, Kundel Harold L, Lauver Sherri C, Toto Lawrence C. Nature of expertise in searching mammograms for breast masses. Academic Radiology. 1996;3(12):1000–1006. doi: 10.1016/S1076-6332(96)80032-8. (1996) [DOI] [PubMed] [Google Scholar]
- Olsen Anneli. The Tobii I-VT Fixation Filter: Algorithm description. Tobii Technology. 2012:21. (2012) https://www.tobiipro.com/siteassets/tobii-pro/learn-and-support/analyze/how-do-we-classify-eye-movements/tobii-pro-i-vt-fixation-filter.pdf . [Google Scholar]
- Public Health England (PHE) NHS Fetal Anomaly Screening Programme Handbook. 2018. Aug, (2018). https://www.gov.uk/government/publications/fetal-anomaly-screening-programme-handbook/20-week-screening-scan .
- Roy Subhankar, Menapace Willi, Oei Sebastiaan, Luijten Ben, Fini Enrico, Saltori Cristiano, Huijben Iris, Chennakeshava Nishith, Mento Federico, Sentelli Alessandro, Peschiera Emanuele, et al. Deep Learning for Classification and Localization of COVID-19 Markers in Point-of-Care Lung Ultrasound. IEEE Transactions on Medical Imaging. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. (2020) [DOI] [PubMed] [Google Scholar]
- Roy Subhankar, Siarohin Aliaksandr, Sangineto Enver, Bulo Samuel Rota, Sebe Nicu, Ricci Elisa. Unsupervised Domain Adaptation Using Feature-Whitening and Consensus Loss; 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019-June; 2019. pp. 9463–9472. [DOI] [Google Scholar]
- Sharma H, Droste R, Chatelain P, Drukker L, Papageorghiou AT, Noble JA. Spatio-Temporal Partitioning And Description Of Full-Length Routine Fetal Anomaly Ultrasound Scans; 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019. pp. 987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Harshita, Drukker Lior, Chatelain Pierre, Droste Richard, Papageorghiou Aris T, Noble J Alison. Knowledge Representation and Learning of Operator Clinical Workflow from Full-length Routine Fetal Ultrasound Scan Videos. 2020 doi: 10.1016/j.media.2021.101973. (2020) [DOI] [PubMed] [Google Scholar]
- Sharma Harshita, Drukker Lior, Papageorghiou Aris T, Noble J Alison. Multi-Modal Learning from Video, Eye Tracking, and Pupillometry for Operator Skill Characterization in Clinical Fetal Ultrasound; 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI); 2021. pp. 1646–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy Christian, Liu Wei, Jia Yangqing, Sermanet Pierre, Reed Scott E, Anguelov Dragomir, Erhan Dumitru, Vanhoucke Vincent, Rabinovich Andrew. Going Deeper with Convolutions. CoRR. 2014:abs/1409.4. (2014). http://arxiv.org/abs/1409.4842 . [Google Scholar]
- Tatsuru Kaji, Keisuke Yano, Shun Onishi, Mayu Matsui, Ayaka Nagano, Masakazu Murakami, Koshiro Sugita, Toshio Harumatsu, Koji Yamada, Waka Yamada, et al. The evaluation of eye gaze using an eye tracking system in simulation training of real-time ultrasound-guided venipuncture. The Journal of Vascular Access. 2021:1129729820987362. doi: 10.1177/1129729820987362. (2021) [DOI] [PubMed] [Google Scholar]
- Teng Clare, Sharma Harshita, Drukker Lior, Papageorghiou Aris T, Noble J Alison. In: Simplifying Medical Ultrasound. Noble J Alison, Aylward Stephen, Grimwood Alexander, Min Zhe, Lee Su-Lin, Hu Yipeng., editors. 2021. Towards Scale and Position Invariant Task Classification Using Normalised Visual Scanpaths in Clinical Fetal Ultrasound; pp. 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gijp A, Ravesloot CJ, Jarodzka H, Van der Schaaf MF, Van der Schaaf IC, van Schaik Jan PJ, Ten Cate Th J. How visual search relates to visual diagnostic performance: a narrative systematic review of eye-tracking research in radiology. Advances in Health Sciences Education. 2017;22(3):765–787. doi: 10.1007/s10459-016-9698-1. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]