Abstract
Development of effective inhibitors of Golgi α-manosidase II (GMII, E.C.3.2.1.114) with minimal off-target effect toward lysosomal α-manosidase (LMan, E.C.3.2.1.24) is a complex task due to complicated structural and chemical nature of their active sites. The pKa values (and also protonation forms in some cases) of several ionizable amino acids, as Asp, Glu, His or Arg of enzymes, can be changed upon the binding of the inhibitor. Moreover, GMII and LMan work at different pH conditions. The pKa calculations on large enzyme-inhibitor complexes as well FMO-PIEDA energy decomposition analysis were performed on structures of selected inhibitors obtained from docking and hybrid QM/MM calculations. The used inhibitors derived from imino-D-lyxitol were synthesized in this work. Based on the calculations a role of amino group incorporated in the ring of the inhibitor and some ionizable amino acids of Golgi-type (Asp270-Asp340-Asp341 of Drosophila melanogaster α-manosidase dGMII) and lysosomal-type enzymes (His209-Asp267-Asp268 of Canavalia ensiformis α-manosidase, JBMan) were explained in connection with the inhibitory properties of the imino-D-lyxitols. The pyrrolidine ring of the imino-D-lyxitols prefer at the active site of dGMII the neutral form while in JBMan the protonated form. In opposite, imino-L-lyxitols preferred the protonation form in both enzymes. The calculations indicate that the binding mechanism of inhibitors to the active-site of α-manosidases can differ going from one to other structural type of the inhibitors. This could allow to design selective inhibitors of GMII and tune their potency.
A series of novel synthetic N-substituted imino-D-lyxitols have were evaluated with four enzymes from the glycoside hydrolase GH-38 family (two of Golgi-type, Drosophila melanogaster GMIIb and Caenorhabditis elegans AMAN-2, and two of lysosomal-type, Drosophila melanogaster LManII and Canavalia ensiformis JBMan, enzymes). The most potent structures [N-9-amidinononyl and N-2-(1-naphtyl)ethyl derivatives] inhibited GMIIb (Ki = 40 nM) and AMAN-2 (Ki = 780 nM) with a weak selectivity index (SI) toward Golgi-type enzymes of IC50(LManII)/IC50(GMIIb) = 27 or IC50(JBMan)/IC50(AMAN-2) = 86. On the other hand, weaker micromolar inhibitors, as N-2-naphtylmethyl or 4-iodobenzyl derivatives [IC50(GMIIb) = 2.4 μM and IC50 (AMAN-2) = 7.6 μM], showed a significant SI in a range from 111 to 812.
Keywords: pyrrolidine, Golgi mannosidase, glycosidase inhibitor, pKa, QM/MM calculations, FMO-PIEDA analysis
1. Introduction
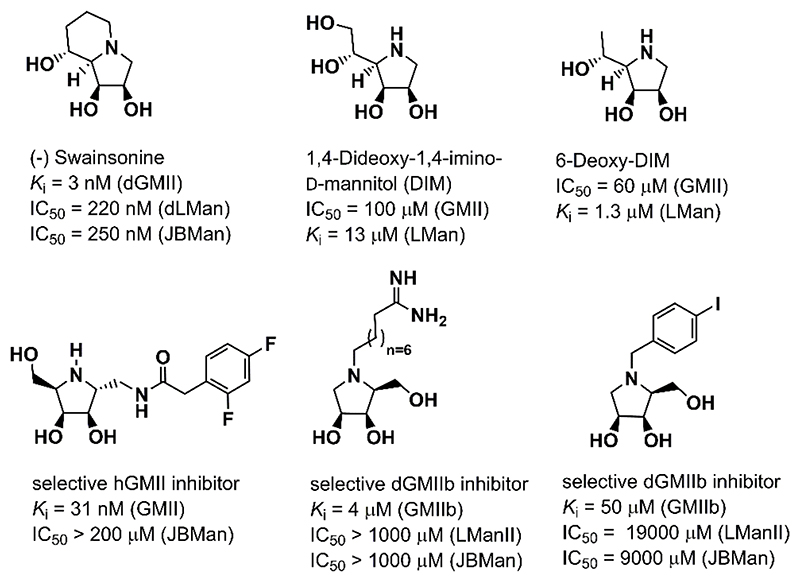
Swainsonine [(1S,2R,8R,8aR)-trihydroxy-indolizidine, Figure 1] is a natural alkaloid which interferes with the glycosylation pathway where it specifically inhibits glycoside hydrolases from the family 38 (GH38).1–6 Swainsonine has attracted attention as a potential anticancer agent as it inhibits tumor growth and metastasis, augments natural killer and macrophage-mediated tumor cell killing, and stimulates bone marrow cell proliferation.7–9 Inhibitors of the GH38 enzymes are potential cancer therapeutics, but their usefulness is limited by off-target toward lysosomal α-manosidase. Recently, selective inhibitors of GMII were prepared and tested in enzyme assays (Figure 1).10–13 Development of an effective GMII inhibitors was reviewed in detail.14–16 Other class of promising α-glycosidase inhibitors are multivalent compounds17, 18 and allosteric-site inhibitors.19
Figure 1.
Natural iminosugars (swainsonine and 1,4-dideoxy-1,4-imino-D-mannitol) and three examples of synthesized inhibitors selective toward Golgi α-mannosidase II.10, 11, 13
Golgi α-mannosidase II (GMII, E.C.3.2.1.114), a transmembrane protein encoded by MAN2A1 gene,20 catalyzes sequential trimming of two mannosyl residues from high-mannose N-glycans (from GlcNAcMan5GlcNAc2 to GlcNAcMan3GlcNAc2) in biosynthesis of complex N-glycans in Golgi apparatus.3 It is retaining glycoside hydrolase from the GH38 family and catalysis involves sequential hydrolysis of 1,3- and 1,6-α-glycosidic bond in a double displacement SN2 mechanism via a covalent glycosyl-enzyme intermediate with a catalytic nucleophile.21, 22 As it was found in X-ray structures of complexes of recombinant Drosophila melanogaster Golgi α-mannosidase II (dGMII) with intact sugar substrates a large active site of dGMII consists of three sugar-binding sites: the catalytic, holding, and anchor sites. dGMII works almost at neutral pH conditions (pH = 6) and needs Zn2+ ion co-factor for catalytic activity. The co-factor resides at the bottom of the catalytic site and is in octahedral coordination with four amino acid residues (side chains of His90, Asp92, Asp204, His471) and with two hydroxyl group of a bidentate ligand (a terminal mannose unit of the substrate or an inhibitor).
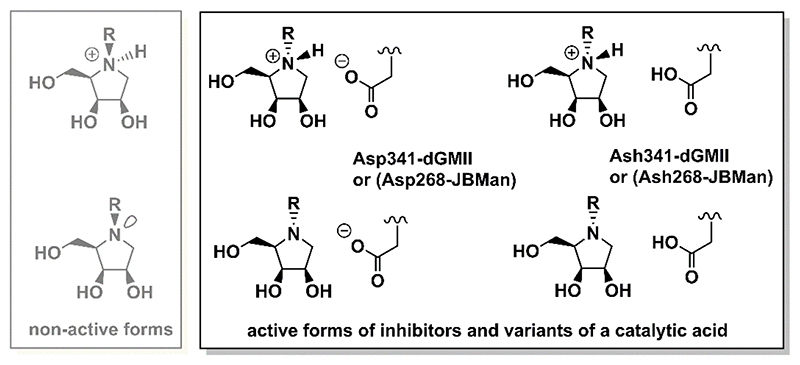
Lysosomal α-mannosidase II (LMan, E.C.3.2.1.24), encoded by MAN2B1 gene, belongs to the same GH38 family as GMII.20 It is the broad-specific and hydrolyzes 1,2- and 1,3- and 1,6- α-glycosidic bond in catabolic degradation of high-mannose, hybrid and complex type N-glycans in lysosomes. Lysosomal α-mannosidases retains a high degree of sequence similarity to Golgi α-mannosidases despite a significantly broader substrate specificity and lower pH optimum (pH = 4.5 for lysosomal enzymes versus pH = 6 for Golgi enzymes).3 Based on available X-ray structures of GH38 enzymes, dGMII,23–28 bovine lysosomal α-mannosidase II (bLMan)29 and plant Jack bean α-mannosidase (JBMan),30 the active sites of Golgi and acidic α-mannosidases are structurally identical approximately 6 Å around the Zn2+ ion.14, 19 However, protonation forms of ionizable amino acids, as Asp, Glu, His or Arg, are not known for these enzymes from X-ray data. Because the enzymes work at different pH conditions the amino acid residues placed in the same position in the active sites of the enzymes do not have to have the same protonation form. It is also not known which of amino acid residues can change protonation form upon binding of the inhibitor to the active site of the enzyme. Most potent α- mannosidase inhibitors are structurally derived from indolizidines and pyrrolidines.15 Although amino groups of these heterocyclic rings prefer protonated form (pKa > 7) in aqueous solution,31 it is not clear whether they prefer this protonation form upon binding to Golgi (at pH = 6) and lysosomal type α-mannosidases (at pH = 4.5). The previous pKa calculations11, 32 on complexes dGMII:inhibitor and bLMan:inhibitor have shown that some inhibitors prefer the protonated form at the active site of the enzymes and others prefer the neutral one. Moreover, pKa values for some inhibitors bound in dGMII were shifted to lower values compared with those measured (or calculated) in aqueous solution. For example, nanomolar GMII inhibitor swainsonine has shifted its pKa = 7.5 (in aqueous solution) to 5.0 bound in complex with dGMII. Thus, swainsonine may change protonation form to neutral one upon binding to GMII (at pH = 6). In bLMan29 for bound swainsonine the calculated pKa had a similar value11 compared with the value in aqueous solution. Thus, swainsonine may prefer the protonation form at bLMan. It should be noted that for the protonated form of the inhibitors, only a stereo configuration at which proton is attached on amino group on the same side of the ring as other attached hydroxyl groups presents a biologically active configuration (Figure 2). In this configuration a small hydrogen atom is bound at the bottom of the active site (where Asp204 and Zn2+ ion reside) and a bulky R-group attached on the opposite side of the inhibitor ring is oriented outward the active site (see also Figure 3). Similarly, for the neutral form of the inhibitor, only a configuration with a bulky R-group oriented outward the active site (and non-bonding electron pair on a nitrogen atom of the amino group oriented below the ring plane) may tightly bound to the active site of α-mannosidases presenting the biologically active configuration. For derivatives lacking N-substitution as 1,4-dideoxy-1,4-imino-D-mannitol (DIM) or 6-deoxy-DIM (Figure 1) both stereo-configurations (with hydrogen up or below the pyrrolidine ring) may present biologically active forms of the α-mannosidase inhibitors.
Figure 2.
Schematic representation of four inhibitor-enzyme complexes in which different protonation forms of inhibitor and Asp catalytic acid residue were used in theoretical calculations. Also non-active forms of the N- substituted pyrrolidine structures are shown (grey).
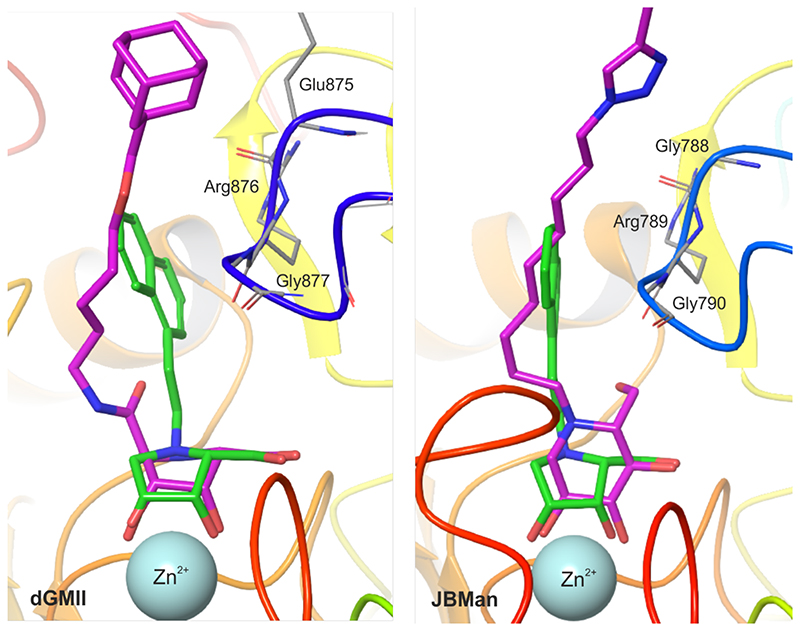
Figure 3.
Superposition of the inhibitor 18(green), docked into dGMII (left) and JBMan (right), with X-ray complexes of inhibitors (magenta) with dGMII (PDB ID: 6RRJ)28 and JBMan (PDB ID: 6B9P)30. All these inhibitors tend to bind with a loop (blue) consisted of the Glu875-Arg876-Gly877 (in dGMII) and the Gly788- Arg789-Gly790 sequences (in JBMan).
In this work, a series of synthetic N-substituted imino-D-lyxitols (N-substituted polyhydroxypyrrolidines) were evaluated with four enzymes from the glycoside hydrolase GH- 38 family (two of Golgi-type, Drosophila melanogaster GMIIb and Caenorhabditis elegans AMAN-2, and two of lysosomal-type, Drosophila melanogaster LManII and Canavalia ensiformis JBMan, enzymes). The structures of N-substituents of imino-D-lyxitols were selected based on potent and selective N-substituted imino-L-lyxitols11–13 (the structures 4, 6, 9, 16, 21, 25 and 26), while structures 17-19 [N-(2-naphtyl)methyl-, N-2-(1-naphtyl)ethyl- and N-cyclohexyl-) were designed with the aim of molecular modeling. To understand a mechanism of selective binding of inhibitors to GMII, molecular docking, hybrid quantum mechanics/molecular mechanics (QM/MM) and pKa calculations on large enzyme-inhibitor complexes and fragment molecular orbital (FMO) pair interaction energy decomposition analysis (PIEDA) of active-site clusters were performed for both enzymes, the target Golgi-type as well as the off-target lysosomal-type α-mannosidase.
2. Results and Discussion
2.1. Molecular modeling
Molecular docking and QM/MM geometry optimization. The X-ray structure of dGMII (PDB ID: 3BLB)23, 33 was used as a model of Golgi α-mannosidase II because of high sequence identity and similarity14 of the active site with human (hGMII), and Caenorhabditis elegans Golgi α-mannosidase34 (AMAN-2, used in inhibitory assays in this work). In Figure S1 of Electronic Supplementary Information (ESI) it is shown a high 3-D structural identity between the active sites of dGMII (PDB ID: 3BLB) and AMAN-2 (built homology model). For a model of lysosomal α-mannosidase, an X-ray structure of plant JBMan (PDB ID: 6B9P)30 was used.
Because protonation forms of ionizable amino acid residues of dGMII and JBMan and their bound iminolyxitol ligands (amino group of the pyrrolidine ring of the inhibitor) in X-ray structures are not known, the pKa calculations were performed for selected X-ray structures as well as for complexes dGMII:inhibitor and JBMan:inhibitor (geometries obtained from molecular docking, Tables S1 and S2 of ESI). According the calculations all inhibitors prefer the protonated form in JBMan (working at acidic pH = 4.5, the calculated pKa values of inhibitors ranges from 7.4-8.6). For dGMII a situation is more ambiguous. Although the neutral form was preferred for all bound inhibitors (pKa = 4.6 – 5.7), for some docked structural conformations the protonated form was also preferred (pKa = 7.5-7.8). (For every inhibitor two conformations were selected from molecular docking, for more details see the Experimental section). To get more accurate results, for the inhibitor 18 [N-2-(1-naphtyl)ethyl-D-lyxitol] several configurations, in which the protonation form of the catalytic acid (Asp341 in dGMII and Asp268 in JBMan) and the bound inhibitor were altered (neutral Ash0 versus ionized Asp-, neutral inhibitor I0 versus protonated I+), were calculated. In addition, two conformations of the pyrrolidine ring of the inhibitor (E1 versus 2E) found in X-ray structures of dGMII with pyrrolidines28, 35, 36 and indolizidines,23, 27, 33, and also found by molecular docking, were consider for next pKa calculations. In this case, pKa calculations were performed on optimized QM/MM geometries. The theoretical results are compile in Table 1.
Table 1. pKa values calculated for geometries of complexes of the bound inhibitor 18 [2-(1-naphtyl)ethyl derivative] in dGMII and JBMan enzymes (four different ionized combinations of the inhibitor and the amino acid Asp341-dGMII or Asp268-JBMan were included) optimized at the QM/MM level.
The preferred pyrrolidine ring conformation of the bound inhibitor and selected geometry parameters (interatomic distances between selected atoms (N1 and C5) of the inhibitor and (Zn2+ ion and Asp204) of the enzymes are also compiled.
| neutral/ionized form | Conformat ion of the ring | Interatomic distances(in Å) | pKa values and preferred ionized form | |||||
|---|---|---|---|---|---|---|---|---|
| dGMII | d(N1-Zn2+) | d(N1-O-asp204) | d(C5-O-asp204) | N1-inhibitor | Asp34l | Asp340 | Asp270 | |
| empty active site (PDB ID: 3BUB) | 5.8 (Asp-) | 4.3 (Asp-) | 1.2 (Asp-) | |||||
| swainsonine-dGMII (PDB ID: 3BLB) | E1 | 3.92 | 2.75 | 3.92 | 5.0 (N0-pyr) | 5.6 (Asp-) | 4.3 (Asp-) | 1.1 (Asp-) |
| swainsonine-dGMII (PDB ID: 1HWW) | 2 E | 4.11 | 2.88 | 3.90 | 5.3 (N0-pyr) | 5.6 (Asp-) | 3.1 (Asp-) | 6.3 (Ash0) |
| inh0-Asp341- | E 1 | 3.99 | 3.13 | 3.98 | 5.5 (N0-pyr) | 5.9 (Asp-) | 0.6 (Asp-) | 4.4 (Asp-) |
| 2E/E1 | 4.20 | 3.14 | 3.81 | 5.8 (N0-pyr) | 5.9 (Asp-) | 1.0 (Asp-) | 4.4 (Asp-) | |
| inh0-Ash3410 | E 1 | 4.13 | 3.18 | 3.83 | 5.5 (N0-pyr) | 6.0 (Ash0) | 0.1 (Asp-) | 4.6 (Asp-) |
| 2 E | 3.99 | 3.13 | 3.96 | 5.6 (N0-pyr) | 5.9 (Asp-) | 0.0 (Asp-) | 4.6 (Asp-) | |
| inh+-Asp341- | E1/2E | 3.99 | 2.74 | 3.91 | 5.1 (N0-pyr) | 6.0 (Ash0) | 1.3 (Asp-) | 4.5 (Asp-) |
| inh+-Ash3410 | E 1 | 3.95 | 2.72 | 3.94 | 5.3 (N0-pyr) | 5.7 (Asp-) | 1.0 (Asp-) | 4.3 (Asp-) |
| JBMan | N1-inhibitor | Asp268 | Asp267 | His209 | ||||
| empty active site (PDB ID: 6B9O) | 7.5 (Ash0) | -0.3 (Asp-) | 7.6 (Hip+) | |||||
| swainsonine0-JBMan | 2 E | 4.23 | 3.26 | 3.85 | 7.8 (N+-pyr) | 5.1 (Ash0) | -0.3 (Asp-) | 7.9 (Hip+) |
| swainsonine+-JBMan | 2 E | 4.09 | 2.81 | 3.86 | 8.1 (N+-pyr) | 2.7 (Asp-) | 0.1 (Asp-) | 8.9 (Hip+) |
| inh0-Asp268- | Ei | 4.23 | 3.28 | 4.01 | 7.9 (N+-pyr) | 3.5 (Ash-) | 1.5 (Asp-) | 7.8 (Hip+) |
| 2E/E1 | 4.27 | 3.30 | 3.99 | 7.9 (N+-pyr) | 5.1 (Ash0) | -0.2 (Asp-) | 7.9 (Hip+) | |
| inh0-Ash2680 | Ei | 4.21 | 3.21 | 4.04 | 8.0 (N+-pyr) | 3.7 (Asp-) | 1.5 (Asp-) | 7.6 (Hip+) |
| 2E/E3 | 4.40 | 3.42 | 3.95 | 7.5 (N+-pyr) | 5.0 (Asp0) | 1.5 (Asp-) | 7.5 (Hip+) | |
| inh+-Asp268- | E 1 | 4.06 | 2.85 | 4.07 | 8.2 (N+-pyr) | 3.3 (Asp-) | 1.5 (Asp-) | 7.9 (Hip+) |
| inh+-Ash2680 | Ei | 4.04 | 2.75 | 4.03 | 8.0 (N+-pyr) | 4.1 (Asp-) | 1.5 (Asp-) | 7.7 (Hip+) |
The above-mentioned form of the structure 18 were docked into the active site of dGMII and JBMan in different protonation forms of the pyrrolidine ring (neutral versus protonated) to see whether or not the protonization changes a binding manner of the inhibitor. As can be seen in Figure S2 of ESI, both forms of 18 bind in the active site of dGMII and JBMan in similar manner, however, some subtle structural differences were found. In the protonated form the pyrrolidine ring of the inhibitor is tightly bound at the bottom of the active site where Zn2+ ion and Asp204 are localized. Distances d(N1+I-Zn2+) = 4.27 Å and d(N1+I-OAsp204) = 3.02 Å for the protonated form of the inhibitor are slightly shorter compared to those distances of the neutral form of the bound inhibitor [d(N1I-Zn2+) = 4.29 Å and d(N1I-OAsp204) = 3.05 Å, dGMII].
To confirm this structural trend a more accurate calculations at the QM/MM level were performed for the complexes (enzyme:inhibitor). Again the protonated inhibitor is more tightly bound to the active sites of both dGMII and JBMan. The monitored interatomic distances, d(N1I-Zn2+), d(N1I-O-Asp204) and d(C5I-Zn2+) calculated at the QM/MM level are compiled in Table 1. As can be seen from this geometric parameters, the differences between the binding of the neutral and protonated forms of 18 to dGMII are very small. We have concluded that both neutral and protonation forms of the inhibitor are allowed for binding into the active site of dGMII. On the other hand, the differences in binding of 18 to JBMan is more apparent, i.e. the protonated form of the inhibitor binds to the active site of JBMan much more tightly and may be preferred. This finding is in an agreement with pKa calculations (discussed in next section) which predicted the protonated form of the bound inhibitor as preferable only in JBMan and the neutral one only in dGMII. In conclusion for the inhibitor 18, its protonation form is more important for a tight binding to JBMan (which works in acidic conditions in lysosomes), while for dGMII (which resides in almost neutral Golgi) both neutral and protonated forms can bind tightly to the active site. From the docking structures and their optimized QM/MM geometry, the preferable conformation of the protonated pyrrolidine ring was E1 (or E1/2E) while for the neutral form both E1 and 2E (or 2E/E1 and 2E/E3 for one case, see Table 1) were found in the complexes. This is in agreement with conformations found in experimental X-ray structures of pyrrolidines (for example: PDB IDs: 2F18, 2F1A, 6RRJ, 6RRN),28, 36 and indolizidines (PDB IDs: 3BLB, 1HWW, 3EJT and others)23, 27, 33 bound at dGMII. For example, the pyrrolidine moiety of swainsonine took an E1 conformation (PDB ID: 3BLB )23 or 2E (PDB ID: 1HWW).33 As it will be discussed in next sections, the conformation of the pyrrolidine ring did not change significantly neither calculated pKa value of the ring nitrogen of the inhibitor nor interaction energy between the inhibitor and the enzyme.
pKa calculations. To understand a mechanism of binding of inhibitors to dGMII and JBMan at different pH conditions (pH = 6 in Golgi for dGMII versus pH = 4.5-5 in lysosomes for LMan), pKa calculations for complexes 18:enzyme were performed for the QM/MM optimized geometries. The calculated pKa values of amino group in the pyrrolidine ring of the inhibitor 18 and selected amino acids of dGMII and JBMan are compiled in Table 1. The bound inhibitor preferred neutral form in dGMII (pKa = 5.1-5.8) while protonated one (pKa = 7.5-8.2) in JBMan. Asp341, which works as a catalytic acid in dGMII and is in a contact with the bound inhibitor, preferred for most optimized QM/MM geometries an ionized form (Asp-, with pKa = 5.7-5.9) except for the geometry of the structure inhibitor0-Ash3410-dGMII. For this geometry, pKa = 6.0 for Asp341 was predicted. It indicates that both neutral (Ash0) and ionized (Asp-) forms of Asp341 are present in the same population in pH = 6. In JBMan, Asp268 plays a role of catalytic acid. Its calculated pKa = 3.3-4.1 indicates preferred ionized form (Asp-) in pH = 5. Asp- was found for most geometries. For geometries with the inhibitor in 2E conformation of the pyrrolidine ring pKa = 5.0-5.1 was predicted. Thus, both neutral and ionized forms of Asp268 are allowed in in vivo pH = 5. In conclusion, the inhibitor 18 may prefer the different protonation state upon binding to α-mannosidases. In acidic conditions (pH = 4.5-5 in lysosomes) it may bind to JBMan in the protonation form of the pyrrolidine ring, while in almost neutral conditions (pH = 6 in Golgi) it binds in the neutral form. The same protonation forms were predicted for the nanomolar inhibitor swainsonine in previous theoretical studies (Table S1).11, 32 The preferable ring conformations of the pyrrolidine ring of 18 in both dGMII and JBMan are E1 or 2E.
FMO-PIEDA calculations
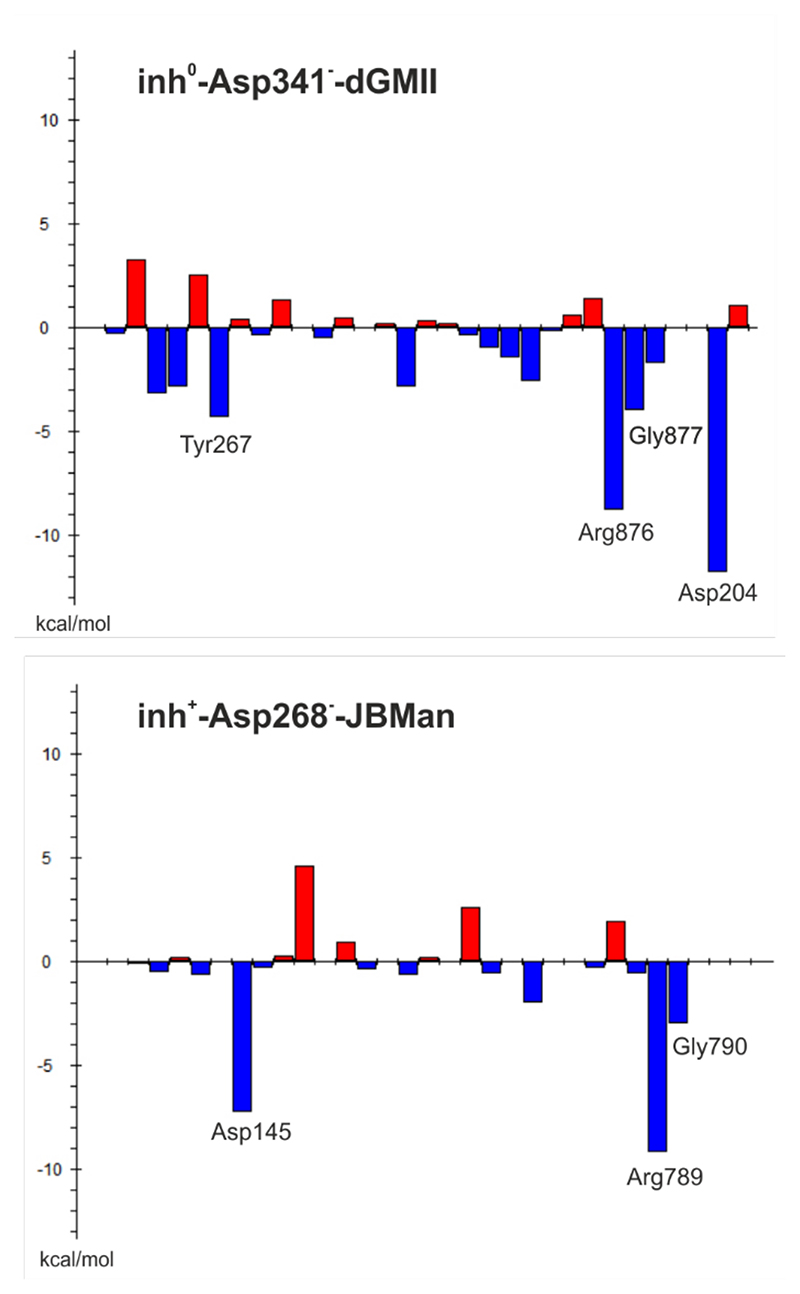
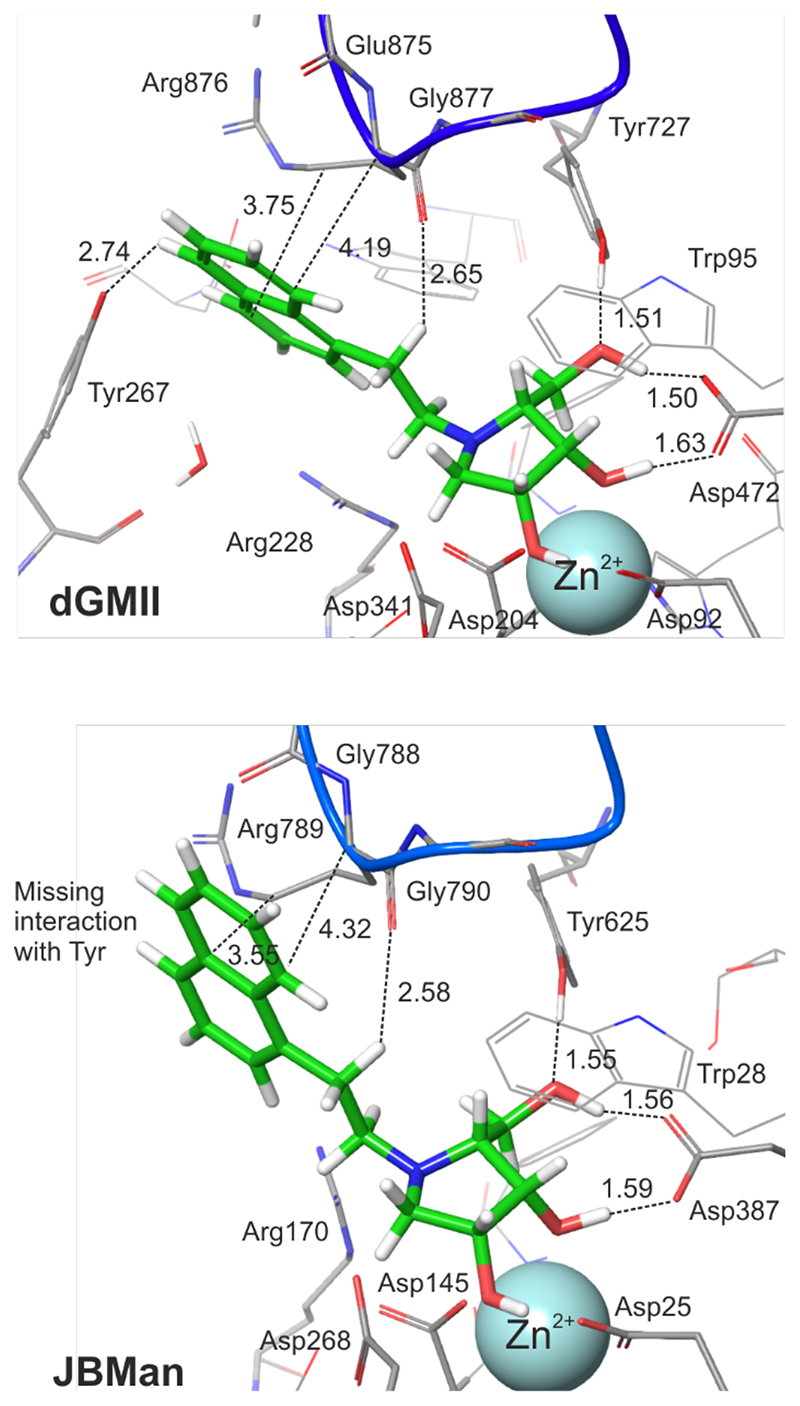
To understand an inhibitory effect of the aromatic naphtyl linker of 18 and compare differences in interactions between 18 and dGMII, and JBMan, QM calculations applying a fragmentation methodology37 were used. The inhibitor 18 was divided into two fragments, the pyrrolidine ring structure and the naphtylethyl linker. Interaction energies were calculated for most populated forms found by pKa calculations (inhibitor0-Asp341--dGMII, inhibitor+-Asp268-- JBMan) as well as for others found from docking and QM/MM calculations (Table 2). The FMO-PIEDA calculated pair interaction energy of the ring structure (ΔEring-E) of the inhibitor with the enzymes represents a major component (94-96% for dGMII and 97-98% for JBMan) of overall interaction energy (ΔEI-E). Neither a protonation form of the inhibitor (neutral versus protonated ring) nor a ring conformation (E1 versus 2E) did not influence this trend. Although the interaction energy of the aromatic linker (ΔElinker-E) represents only a small part (4-6% for dGMII and 2-3% for JBMan) of ΔEI-E, less negative values of ΔElinke-Er for 18 in the complex with JBM compared with dGMII correlated with measured Ki (or IC50) values (Table 4). While 18 inhibited JBMan only at the micromolar concentration level (6.5 μM), AMAN-2 was inhibited at the nanomolar concentration level (0.15 nM). Based on these results it could be concluded that the linker structure of an inhibitor can influence the specificity of the inhibitor toward Golgi α-mannosidase. To confirm this idea FMO-PIEDA calculations were also performed for derivative 17 (only for most populated forms inhibitor0-Asp341--dGMII,inhibitor+-Asp268--JBMan). 17 has shown a specificity toward Golgi-type enzymes (GMIIb and AMAN-2) with a high selectivity index (SI > 100). The calculated ΔEring of 17 represents a major component (94-96% for dGMII and 97-98% for JBMan) of overall ΔEI-E similarly as was found for 18 (Table 3). Smaller negative values of ΔElinker-E for 17 in the complex with JBMan compared with dGMII correlated with measured Ki (or IC50, Table 4) values for both enzymes. Thus, it can be concluded that 17 and 18 interact weaker with their linker structures with the lysosomal-type JBMan inducing the selectivity of these inhibitors toward the Golgi-type enzymes. As can be seen in Figures 5 and 3, the aromatic linker of 18 (similarly for 17 and other inhibitors in X-ray structures with GMII)28, 30 tends to interact with a loop consisted of the Glu875-Arg876-Gly877 (in dGMII) and the Gly788-Arg789-Gly790 sequences (in JBMan). Detailed analysis of energy components of ΔElinker-E of 18 (Figure 5) has shown that main contributors to ΔElinker-E for GMII are Arg876 (-8.8 kcal mol-1) and Gly877 (-4.0 kcal mol-1) of the loop, catalytic nucleophile Asp204 (-11.8 kcal mol-1) and Tyr267 (-4.3 kcal mol- 1). For JBMan, main contributors to ΔElinker-E are Arg789 (-9.2 kcal mol-1) and Gly790 (-3.0 kcal mol-1) of the loop, and catalytic nucleophile Asp145 (-7.3 kcal mol-1). In JBMan a tyrosine residue which would be mimicked Tyr267 of GMII is missing in this position (Figure 4), thus, the inhibitor linker may bind weaker in the active site of JBMan compared with GMII.
Table 2. Calculated interaction energies (ΔEI-E, in kcal mol-1) for different ionized forms of the inhibitor 18 in a complex with either dGMII or JBMan enzymes at the MP2//BP86 level.
Also overall interaction energies between the enzyme and inhibitor fragment [the pyrrolidine core (ΔEring) or the 2-(1-naphtyl)ethyl linker (ΔElinker-E)] are also compiled.
| dGMII | conform | ΔEI-E | ΔEring-E | ΔElinker-E |
|---|---|---|---|---|
| inh0-Asp341- | E 1 | -591.32 | -557.14 | -34.19 |
| 2 E | -597.10 | -562.25 | -34.85 | |
| inh0-Ash3410 | E 1 | -580.41 | -548.86 | -31.55 |
| 2 E | -589.48 | -557.28 | -32.20 | |
| inh+-Asp341- | E 1 | -780.68 | -748.13 | -32.55 |
| inh+-Ash3410 | E 1 | -743.42 | -713.25 | -30.17 |
| JBMan | ||||
| inh0-Asp268- | E 1 | -585.62 | -570.46 | -15.16 |
| 2E/E1 | -586.71 | -573.96 | -12.75 | |
| inh0-Ash2680 | E 1 | -547.86 | -533.21 | -14.65 |
| 2E/E3 | -600.62 | -591.51 | -9.11 | |
| inh+ -Asp268- | E 1 | -768.16 | -752.82 | -15.33 |
| inh+ -Ash2680 | E 1 | -747.63 | -730.41 | -17.22 |
Table 4. Inhibition (IC50, Ki ,in square brackets, and selectivity index SIa,b) of class II GH38 α-mannosidases (GMIIb, AMAN-2, LManII and JBMan) by N-substituted 1,4-dideoxy-1,4-imino-D-lyxitol derivatives.
| Compound | IC50 [Ki](μM) | SIa | SIb | |||
|---|---|---|---|---|---|---|
| GMIIb | AMAN-2 | LManII | JBMan | |||
| 6 | 3.9 ± 0.1 | 2.3 ± 0.1 | 30.5 ± 3 | 10.5 ± 1.3 | 8 | 5 |
| 4 | 11.0 ± 1.3 | 8.7 ± 0.1 | 675 ± 67 | 1075 ±185 | 61 | 124 |
| 16 | 4.4 ± 0.2 | 3.5 ± 0.1 | 460 ± 53 | 940 ± 24 | 105 | 269 |
| 17 | 7.6 ± 1.1 | 2.4 ± 0.1 | 845 ±170 | 1950 ± 250 | 111 | 812 |
| [0.16 ± 0.01] | [0.15 ± 0.03] | [3.9 ± 0.1] | [6.5 ± 0.5] | |||
| 19 | 3.5 ±0.3 | 17.3 ± 0.1 | 190 ± 53 | 1290±113 | 54 | 75 |
| 20 | 15.5 ± 0.5 | 84.3 ± 0.1 | 980 ±102 | 2550±63 | 63 | 30 |
| [0.83 ± 0.08] | [10.3 ± 1.0] | [165 ± 12.5] | [52.4 ± 7.5] | |||
| 21 | 0.40 ± 0.05 | 2.3 ± 0.1 | 11.8 ± 1.2 | 20.3 ± 4.4 | 30 | 9 |
| 25 | 0.45 ± 0.10 | 2.7 ± 0.1 | 8.3 ± 2.1 | 8.3 ± 1.2 | 18 | 3 |
| [0.04 ± 0.01] | [0.78 ± 0.23] | [3.2 ± 1.2] | [1.4 ± 0.4] | |||
| [0.0027]e | 0.01c | [0.0071]e | ||||
| [0.13 ± 0.02 | [0.68 ± 0.03] | [1.95 ± 0.35] | [0.38 ± 0.03] | |||
SI = [IC50(LManII)/IC50(GMIIb)]
SI = [IC50(JBMan)/IC50(AMAN-2)]
IC50 estimated from inhibition assays measured by Paschinger et al.34 where 45% inhibition of AMAN-2 by 10nM of swainsonine was found
IC50 measuared by Poláková et al.38
IC50 and Ki measuared by Nemčovičová et al.20
1,4-dideoxy-1,4-imino- D-mannitol
Table 3. Calculated interaction energies (ΔEI-E, in kcal mol-1) for different ionized forms of the inhibitor 17 in a complex with either dGMII or JBMan enzymes at the MP2//BP86 level.
Also overall interaction energies between the enzyme and inhibitor fragment [the pyrrolidine core (ΔEring-E) or the N-2-naphtylmethyl linker (ΔElinker-E)] are also compiled.
| dGMII | conform | ΔEI-E | ΔEring-E | ΔElinker-E |
|---|---|---|---|---|
| inh0-Asp341- | E3/2E | -591.68 | -561.01 | -30.67 |
| inh0-Ash3410 | 2 E | -571.61 | -543.95 | -27.66 |
| inh+-Asp341- | 2 E | -795.53 | -765.93 | -29.61 |
| inh+-Ash3410 | 2 E | -742.24 | -718.44 | -23.80 |
| JBMan | ||||
| inh0-Asp268- | E3 | -604.14 | -586.80 | -17.34 |
| inh0-Ash2680 | 2 E | -536.59 | -526.59 | -9.50 |
| inh+ -Asp268- | 2 E | -789.23 | -773.32 | -15.91 |
| inh+ -Ash2680 | 2 E | -673.16 | -662.44 | -10.72 |
Figure 5. FMO-PIEDA total pair interaction energies (ΔElinker-AA) ( in kcal mol-1) between the inhibitor linker of 18and the active-site amino acid residues of dGMII (the form inh0-Asp341--dGMII) and JBMan (the form inh0-Asp268--JBMan). The most significant ΔElinker-AA are assigned.
Figure 4. QM/MM optimized structures of the complexes 18:dGMII and 18:JBMan.
For sake of clarity most hydrogen atoms are not visualized. Some hydrogen bonds as well as contact interactions of the naphtyl linker with a loop (in blue color, the Glu875-Arg876-Gly877 sequence in dGMII and Arg789-Gly788-Gly790 in JBMan) are shown by dash lines (values of distances in Å). Interactions between the naphtyl linker of the inhibitor and Tyr267 in dGMII is missing in JBMan where the active site is more open and solvent accessible.
2.2. Synthesis
The synthetic approach to the target derivatives has been previously based on combination of computer-assisted design with our previous findings on the potent and selective GMII inhibitors.11–13 Utilization of D-ribose as a starting material led to the derivatives having 1,4- imino-1,4-dideoxy-L-lyxitol core. These derivatives were further substituted at endocyclic nitrogen with an arylalkyl or alkyl, which were optionally terminated with a functional group, such as halogen, amidine or guanidine.
In an ongoing development of the selective GMII inhibitors in this study, L-ribose was selected as a starting material, instead of D-ribose. Upon repeating the same synthetic sequence reported for the D-ribose,11 the compounds having 1,4-imino-1,4-dideoxy-D-lyxitol core unit as in 6 were obtained. They have the same stereoconfiguration as swainsonine, which is a natural potent inhibitor of GH38 mannosidases. Computer predictions suggested that N-substitution of 6 could result in the derivatives featured with an improved potency against the target GMII enzyme.
The endocyclic nitrogen of the core 6 was substituted similarly as reported previously11, 13 e.g. with (substituted) benzyl, dodecyl, alkyl amidine and alkylguanidine function. In addition, some new structural linkers were also introduced as naphtyl and non-aromatic cyclic units such as cyclohexylalkyl and tetrahydropyranyl.
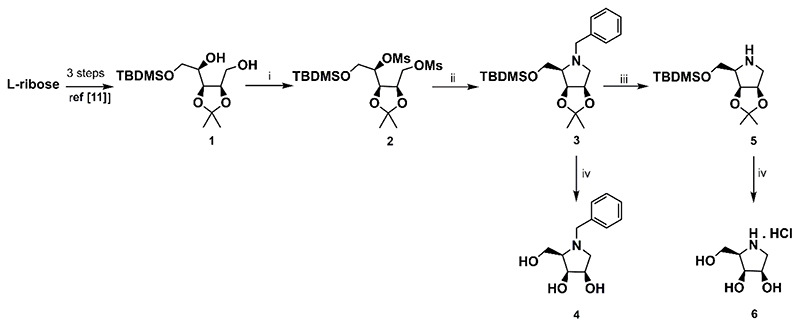
Similarly to the optimized synthesis starting from D-ribose disclosed previously,11 the strategy applied here to L-ribose also employed two key intermediates, dimesyl derivative 2 and amine 5 derived thereof (Scheme 1). The dimesylate 2 was obtained from L-ribose in three reaction steps in high overall yield. Cyclization of 2 with benzylamine smoothly afforded N-benzyl derivative 3. An exposure of 3 to catalytic hydrogenation on 10% Pd-C in MeOH yielded amine 5, another important intermediate in the synthesis of other N-substituted D-lyxitol derivatives. On the other hand, a treatment of 3 under acid condition gave the target derivative 4. Employment of the same conditions for acetonide removal from 5 resulted in 6 (Scheme 1).
Scheme 1. Synthesis of the intermediates 2, 4, and 5, and the target compounds 4 and 6.
Reagents and conditions: i) MsCl, Et3N, CH2Cl2, rt, 16 h, 89%; ii) BnNH2, 120 °C, 7 h, 88%; iii) 10% Pd-C, H2, MeOH, rt, 6 h, 81%; iv) 6M HCl/MeOH 1:2 (v/v), rt, 16 h, 68% for 5; 82% for 6.
When the dimesylate 2 was cyclized with p-xylylenediamine, it provided benzylmethylamine 7, which was subsequently transformed in two steps to desired guanidine derivative 9 (Scheme 2).
Scheme 2. Synthesis of the guanidine derivative 9.
Reagents and conditions: i) p-xylylenediamine, 95 °C, 16 h, 50%; ii) [N,N¢-bis(tert-butoxycarbonyl)]-1H- pyrazole-1-carboxamidine, 50 °C, ultrasonic irradation, 1 h, 64%; iii) 6M HCl/MeOH 1:2 (v/v), rt, 16 h, 92%.
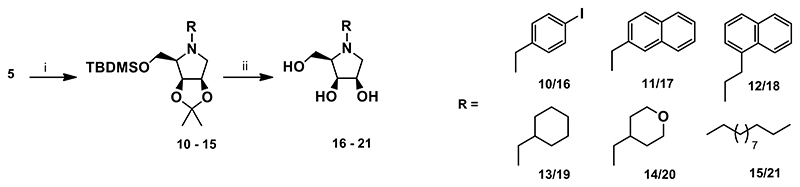
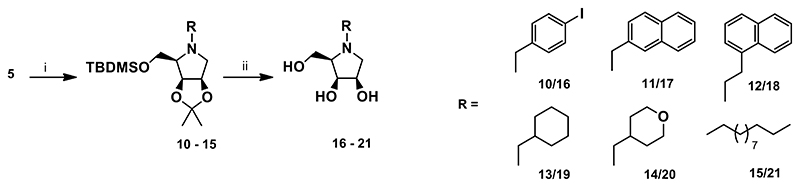
A nuclephilic substitution of the amine 5 with various bromides afforded the corresponding N-substituted derivatives 10-15 in moderate to high yields. A slight modification (a higher temperature) of alkylation conditions was required to improve the yield of some N-substituted derivatives. This was the case of 13 and 14, both having a cyclic non-aromatic unit linked to the endocyclic nitrogen. Simultaneous removal of all protective groups from 10-15 under acidic conditions afforded the target derivatives 16-21 (Scheme 3).
Scheme 3. Synthesis of the target compounds 16-21.
Reagents and conditions: i) RBr, K2CO3, DMF, 4-24 h, 45-60 °C, 90% for 10, 84% for 11, 66% for 12, 53% for 13, 49% for 14, 68% for 15; iii) 6M HCl/MeOH 1:2 (v/v), rt, 16 h, 78% for 16, 68% for 17, 60% for 18, 55% for 19, 75% for 20, 79% for 21.
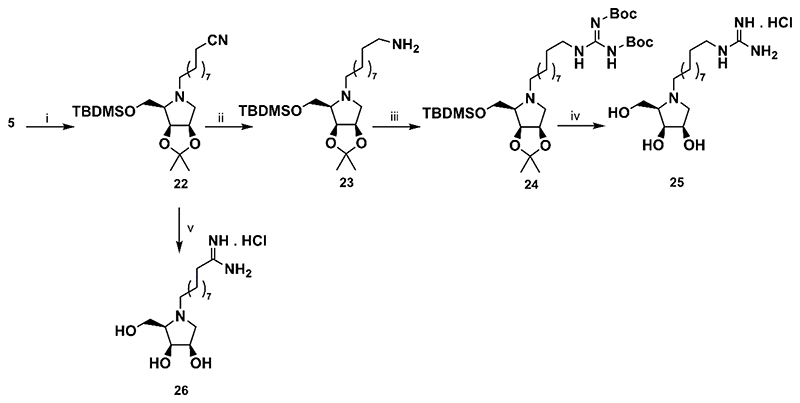
In the synthesis of N-alkylamidine and N-alkylguanidine derivatives, 10-bromodecanenitrile was used as the alkylation agent. It was reacted with the amine 5 to give nitrile 22. The latter was converted to N-alkylguanidine 25 by a three steps sequence in moderate yield. In addition, a reaction of the 22 with an excess of fresh LiHMDS followed by an acidic treatment of crude protected amidine gave smoothly required N-alkylamidine 26 (Scheme 4).
Scheme 4. Synthesis of the alkylguanidine derivative 25and alkylamidine derivative 26.
Reagents and conditions: i) Br(CH2)9CN, K2CO3, DMF, 45 °C, 16h, 87% ii) LiEt3BH, THF, ultrasonic irradiation, 50 °C, 2 h, 89% iii) N,N′-bis(tert-butoxycarbonyl)-1H-pyrazole-1-carboxamidine, THF/DMF, ultrasonic irradiation, 50 °C, 1 h, 56%; v) a) LiHMDS, Et2O, ultrasonic irradiation, 40 °C, 2 h; b) 6M HCl/MeOH 1:2 (v/v), rt, 16 h, 95%.
2.3. Enzymatic assays
A library of ten N-substituted 1,4-imino-D-lyxitols (4, 9, 16-21, 25, 26) were evaluated toward the class II α-mannosidases from the GH38 family, Golgi-type GMIIb and AMAN-2, and lysosomal-type LManII and JBMan, for their inhibition profile and ability to selectively block the Golgi-type enzyme only (Table 4). One of the requirements for a selective GMII inhibitor is to exhibit no or significantly reduced inhibition toward a lysosomal α-mannosidase. The selectivity indices (SIs) were estimated based on a ratio of IC50(LManII)/IC50(GMIIb) and IC50(JBMan)/IC50(AMAN-2). The SIs calculated from IC50(AMAN-2) are more important because an amino acid sequence and a 3-D structure of the active site of AMAN-2 is almost identical with the structure of the active site of human GMII (Figure S1 of ESI). Thus, AMAN- 2 is a better enzyme model for searching of selective inhibitors of human GMII than the GMIIb model used in our previous studies.11–13 In addition, the enzyme assays were also done for the non-substituted derivative 6, DIM and swainsonine to compare the effect of N-substitution.
All tested N-substituted derivatives except for 18 and 26 inhibited GMIIb and AMAN-2 at the micromolar concentration level. LManII and JBMan were affected weakly from the micromolar to milimolar concentration levels. The 18 [N-2-(1-naphtyl)ethyl derivative] and 26 (N-9-amidinononyl derivative) inhibited the Golgi-type mannosidases at the nanomolar concentration level [Ki(GMIIb) = 150 nM and Ki(AMAN-2) = 160 nM for 18,40 nM and 780 nM for 26], thus, they are the most potent inhibitors in this N-substituted imino-D-lyxitol series. Their potency toward Golgi-type mannosidases reached potency of natural DIM and swainsonine (Ki = 130 and 680 nM for DIM and Ki = 3 nM and 10 nM for swainsonine). On the other hand, they inhibited lysosomal-type mannosidases at the micromolar concentration level only [Ki(dLManII) = 1.4 μM and Ki(JBMan) = 6.5 μM]. 18 also exhibited a satisfactory SI = 27 and 86 (Table 4). This indicated that N-substitution may induce a selectivity toward Golgi-type mannosidases either in N-substituted imino-D-lyxitol and previously published imino-L-lyxitols.11–13 The selectivity effect was significant mainly for the compound 16(N-4- iodobenzyl derivative, SI = 269) and 17 (N-2-naphtylmethyl derivative, SI = 812). Both 16 and 17 were only micromolar inhibitors of Golgi-type mannosidases (IC50 = 2.4 – 7.6 μM).
Inhibition of Golgi-type mannosidases by derivatives with N-alkyl linker (compounds 21, 25 and 26) gave different results. These compounds inhibited GMIIb almost at the nanomolar concentration level (IC50 = 0.19 μM and 0.45 μM), while, AMAN-2 was inhibited only at the micromolar concentration level (IC50 = 1.4 μM and 2.7 μM). This may indicate some structural changes between the active sites of these enzymes.
3. Conclusion
Synthesis and inhibitory evaluation of N-substituted imino-D-lyxitol derivatives allow us to compare their activity and selectivity toward Golgi-type α-mannosidases with their N- substituted imino-L-lyxitol counterparts published previously.11–13, 39 In general, N-substituted imino-D-lyxitols are more potent toward Golgi-type α-mannosidases (inhibition at the nanomolar concentration level) with a more significant off-target effect toward lysosomal-type α-mannosidases (inhibition at the micromolar concentration level). On the other hand N-substituted imino-L-lyxitols are weaker micromolar Golgi-type α-mannosidase inhibitors with no (or very weak) inhibition toward lysosomal α-manosidases.11–13 This trend was also confirmed in this study. For example, the most potent N-2-(1-naphtyl)ethyl derivative 18 has shown excellent inhibition of GMIIb and AMAN-2 at the nanomolar concentration level [Ki(GMIIb) = 150 nM and Ki(AMAN-2) = 160 nM), however, with a weaker selectivity toward Golgi-type α-mannosidases (SI = 27 and 86). On the other hand, the derivative 17 has shown excellent selectivity toward Golgi-type α-mannosidases (SI = 111 and 812) with the inhibitory properties toward GMIIb and AMAN-2 only at the micromolar concentration level (IC50 = 2.5 μM and 7.6 μM).
Based on available X-ray structures of dGMII33 and JBMan30, complexes of N-substituted imino-D-lyxitols with both enzymes were built with aid of molecular docking and QM/MM calculations. The interaction energies between the two bound inhibitors (17 and 18) and the enzymes were predicted by quantum mechanics FMO-PIEDA calculations. The calculations have revealed that the major contribution come from an interaction energy of the pyrrolidine core structure (94-98%), while N-linker contribute only with 2-6% to the total interaction energy. We concluded that the N-linker structure may influence the overall binding affinity of an inhibitor by two ways: (i) its direct interaction with an active-site loop (predominantly with sequence Tyr267-Arg876-Gly877 in dGMII, and with Gly788-Arg789-Gly790 in JBMan); (ii) influencing of the binding position of the pyrrolidine core as well as its electronic properties. A small shift in the binding position of the pyrrolidine ring may induced a large change in binding affinity of the inhibitor due to the complex interaction network between the inhibitor and mainly Zn2+ ion, Asp92, Asp204, Asp341 and Asp472.32
Based on pKa calculations, the catalytic acid of the enzymes (Asp341 in dGMII and Asp268 in JBMan) was found either in ionized (Asp-) or neutral form (Ash0) depending on optimized structures of the enzyme-inhibitor complexes. The pyrrolidine ring of imino-D-lyxitol derivatives prefer at the active site of dGMII the neutral form while in JBMan the protonated form. This is the different outcome compared to imino-L-lyxitols which preferred the protonation form in both enzymes.11 The calculations indicate that the binding mechanism of the inhibitors to the active-site of α-manosidases can differ going from one to other structural type of the inhibitors. This could allow to design selective inhibitors of GMII and tune their potency.
4. Experimental
4.1. Molecular modeling
Docking with Glide. The X-ray structures of recombinant Drosophila melanogaster Golgi α- mannosidase II (dGMII, PDB ID: 3BLB),23, 33 and Canavalia ensiformis (Jack bean) α- mannosidase (JBMan, PDB ID: 6B9P)30 were used as 3-D enzyme models of human GMII and LMan for docking of the synthesized compounds with the GLIDE program40, 41 of the Schrödinger package. Protonation states of amino acid residues of enzymes were calculated for the pH = 6.0 (dGMII) and 5.0 (JBMan) using the Propka v.2 program.42, 43 For docking with dGMII all crystallographic molecules of water at the active site of dGMII were deleted except one (WAT1820, numbering according to PDB ID: 3BLB). This water has been shown to be conserved in crystal structures of dGMII either with intact substrates or inhibitors.24, 26, 27 In docking calculations the catalytic acid (Asp341 of dGMII or Asp268 of JBMan) was modeled either in the neutral (as Ash0) or ionized (as Asp-) form to see differences in prediction of binding poses of the docked ligands. The receptor box for the docking conformational search was centered at the Zn2+ ion co-factor at the bottom of the active site with a size of 39×39×39 Å using partial atomic charges for the receptor from the OPLS2005 force field except for the Zn2+ and side chains of His90, Asp92, Asp204, Arg228, Tyr269, Asp270, Asp340, Asp341 and His471 (analogous residues were selected for JBMan). For these structural fragments the charges were calculated at the quantum mechanics level with the DFT (Density Functional Theory) method (M06-2X)44 using a hybrid QM/MM model (M06- 2X/LACVP**:OPLS2005)45–47 with the QSite48, 49 program of the Schrödinger package. The grid maps were created with no Van der Waals radius and charge scaling for the atoms of the receptor. Flexible docking in standard (SP) precision was used. The partial atomic charges of the docked ligands were calculated at the DFT level (M06-2X/LACVP**)44, 45 using the Jaguar program50 of the Schrödinger package. All ligands were docked with the amino group at the pyrrolidine ring either in protonated and neutral forms. The ligands with N-alkyl amidine and guanidine groups were docked with these groups in protonated forms in all docking calculations). The potential for nonpolar parts of the ligands was softened by scaling the Van der Waals radii by a factor of 0.8 for atoms of the ligands with partial atomic charges less than specified cut-off of 0.15. The 5000 poses were kept per ligand for the initial docking stage with scoring window of 100 kcal mol-1 for keeping initial poses; the best 400 poses were kept per ligand for energy minimization. The ligand poses with RMS deviations less than 0.5 Å and maximum atomic displacement less than 1.3 Å were discarded as duplicates. The post-docking minimization for 10 ligand poses with the best docking score was performed and optimized structures were saved for subsequent analyses using the MAESTRO viewer51 of the Schrödinger package.
QM/MM geometry optimizations. Geometries of selected complexes (inhibitor:enzyme) from molecular docking were subsequently optimized at the QM/MM level (BP86/LACVP*:OPLS2005),45, 47, 52 using the QSite48, 49 program of the Schrödinger package. The following decomposed scheme was used: the QM part (more than 280 atoms) of the inhibitor:dGMII system consisted of Zn2+ ion, inhibitor, water molecule WAT1820 (described in the previous section) and amino acid residues (Asp92, Asp204, Asp341, Asp340, Asp270, Asp409, Asp472, Arg228, Arg876, His90, His471, Tyr267, Tyr269, Tyr727, Ser268, Trp95, Trp415, Phe206). The rest of the enzyme was included into the MM part and described by the OPLS2005 force field.47 The QM part of the inhibitor:JBMan system consisted of more than 260 atoms: Zn2+ ion, inhibitor and amino acid residues (Asp25, Asp267, Asp268, Asp327, Asp387, Arg170, Arg789, His23, His209, His386, Tyr210, Tyr625, Trp28, Phe147, Gly208, Gly788, Gly790). The QM/MM methodology (an additive scheme) with hydrogen caps on boundary QM atoms and electrostatic treatment at the interface between the QM and MM regions using Gaussian charge distributions represented on a grid (keyword HCAPESCHG=3) was employed. Only for the compounds (18 and 17) more detailed structural analysis was performed in which structural complexes with both ionized form of the pyrrolidine ring (neutral inh0 and cationic inh+) of the inhibitor and Asp341 (Asp341- and Ash3410 in complexes with dGMII) and Asp268 (Asp268- and Ash2680 in complexes with JBMan) were calculated. Thus, complexes marked as inh0-Ash3410, inh+-Ash3410, inh0-Asp341-, inh+-Asp341-, inh0- Ash2680, inh+-Ash2680, inh0-Asp268-, inh+-Asp268- were optimized at the QM/MM level and calculated in subsequent pKa calculations.
pKa calculations with Propka. The QM/MM optimized geometries of the inhibitor-enzyme complexes were used to predict pKa values and preferred ionizable forms of amino acid residues of dGMII (for pH = 6) and JBMan (for pH = 5) as well as a bound inhibitor in the active site of the enzymes using the Propka v.2 program.42, 43
FMO-PIEDA calculations. Pair interaction energy decomposition analysis (PIEDA) was used along the two-body FMO method.37, 53, 54 From the QM/MM optimized inhibitor-enzyme complexes active-site clusters consisted of more than 30 amino acid residues, Zn2+ ion and bound inhibitor 18 (or 17) were built using the Facio program.55 The hybrid orbital projection operator (HOP) technique was used in the generation of fragments for the covalently bounded amino acids. The FMO calculations were performed using the second-order Møller-Plesset theory56, 57 (MP2) with the 6-31G(d) basis and polarizable continuum model (PCM).58 The Gamess package59, 60 [version 30 June 2019 (R1)] was used. The virtue of the FMO technique is to predict pair interactions between the two structural fragments of the molecular system embedded within the electrostatic potential of the surroundings (IFIE – inter fragment interaction energy). FMO-PIEDA enables the separation of the interaction energy into physically interpretable contributions (1).
| (1) |
The electrostatic energy Eels originates from Coulomb-like interactions between the fragments, the exchange energy Eexch arises for fermion particles, the electrons, and accounts for the Pauli repulsion of electrons between the fragments. Ect+mix is somewhat peculiar; it includes the charge transfer that results from electron transfer from occupied molecular orbitals of one fragment to the vacant virtual orbitals on the second fragment. The mixing part is basically an approximate polarization. Dispersion energy Edisp originates from interactions of instantaneous fluctuations of dipoles on the fragments due to electron correlation. This method was recently used to analyze interaction energy in different biomolecular systems.32, 61–65
To understand an inhibitory effect of the aromatic naphtyl linker of 17 and 18and compare differences in interactions between 17 (or 18)and dGMII, and JBMan, the inhibitors were divided into two fragments, the pyrrolidine ring structure (Iring) and the naphtyl linker (Ilinker). Then, the interaction energy between the inhibitor and enzyme (ΔEI-E) consists of the interaction energy between the pyrrolidine ring of the inhibitor and enzyme (ΔEring-E) and the interaction energy between the linker of the inhibitor and enzyme (ΔElinker-E):
| (2) |
4.2. Synthesis
General method for deprotection (Method A). The solution of protected d-lyxitol in MeOH (4 mL/0.3 mmol of lyxitol) was cooled to 0 °C and 6M HCl (2 mL, 6M HCl/MeOH 1:2, v/v) was added. After 15 min, the ice-water bath was removed and the stirring was continued at rt for 16 h. The reaction mixture was neutralized with NaHCO3, salts were removed by filtration, and the solvent was evaporated. The crude product was purified by column chromatography
General method for deprotection (for hydrochloride) (Method B). The solution of protected D-lyxitol in MeOH (2 mL/0.04 mmol of lyxitol) was cooled to 0 °C and 6M HCl (1 mL, 6MHCl/MeOH 1:2, v/v) was added dropwise. After 15 min, the ice-water bath was removed and the stirring was continued at rt for 16 h. The solvent was evaporated, the residue was redissolved in water (15 mL) and extracted with DCM (3 × 10 mL). Lyophilisation of the water layer afforded the target product as hydrochloride.
N-Benzyl-1,4-dideoxy-1,4-imino-d-lyxitol (4)
Deprotection of 3 (0.30 g, 0.79 mmol) was carried out following general procedure Method A. Compound 4 (121 mg, 68%), yellow oil, [α]D = - 24.7 (c 0.39, CH3OH); [5] [α]D21 = - 46.2 (c1.0, H2O). 1H NMR (600 MHz, CD3OD): δ 7.39-7.24 (m, 5H, Ar), 4.27 (dd, 1H, J 5.2 Hz, J 7.1 Hz, H-3), 4.08 (td, 1H, J 3.1 Hz, J 5.2 Hz, H-2), 3.99 (d, 1H, J 13.1 Hz, NCH2), 3.73 (dd, 1H, J 5.3 Hz, J 11.1 Hz, H-5¢), 3.65 (dd, 1H, J 3.6 Hz, J 11.1 Hz, H-5), 3.54 (d, 1H, J 13.1 Hz, NCH2), 2.91-2.83 (m, 2H, H-1¢, H-4), 2.56 (dd, 1H, J 5.2 Hz, J 10.6 Hz, H-1). 13C NMR (150 MHz, CD3OD): δ 139.7, 130.2, 129.3, 128.1 (Ar), 73.6 (C-3), 71.4 (C-2), 67.7 (C-4), 60.8 (C-5), 60.3 (NCH2), 59.0 (C-1). HRMS: m/z calcd for [C12H17NO3]H+: 224.1281, found: 224.1284.
1,4-Dideoxy-1,4-imino-d-lyxitol hydrochloride (6)
Deprotection of 5 (46 mg, 0.16 mmol) was carried out following general procedure Method B. Compound 6 (22 mg, 82%), yellowish solid, [α]D = + 19.6 (c 0.22, H2O); [6] [α]D27 = + 15.4 (c0.56, H2O);66 [α]D20 = + 19.8 (c 0.45, H2O) 1H NMR (400 MHz, CD3OD): δ 4.41 (td, 1H, J4.0 Hz, J 7.2 Hz, H-2), 4.23 (t, 1H, J 4.1 Hz, H-3), 3.97 (dd, 1H, J 4.8 Hz, J 11.8 Hz, H-5¢),3.91 (dd, 1H, J 8.7 Hz, J 11.8 Hz, H-5), 3.64 (dt, 1H, J 4.5 Hz, J 8.9 Hz, H-4), 3.42 (dd, 1H, J 7.3 Hz, J 11.7 Hz, H-1¢), 3.17 (dd, 1H, J 7.1 Hz, J 11.7 Hz, H-1). 13C NMR (100 MHz, CD3OD): δ 71.9 (C-2), 71.4 (C-3), 64.6 (C-4), 59.4 (C-5), 48.6 (C-1). HRMS: m/z calcd for [C5H11NO3]H+: 134.0812, Found: 134.0814.
1,4-Dideoxy-N-4-guanidinomethylbenzyl-1,4-imino-d-lyxitol hydrochloride (9)
Deprotection of 8 (90 mg, 0.14 mmol) was carried out following general procedure Method A. Compound 9 (42 mg, 92%), yellow oil, [α]D = + 5.6 (c 0.25, CH3OH). 1H NMR 400 MHz, CD3OD): δ 7.64 (d, 2H, J 8.2 Hz, Ar), 7.49 (d, 2H, J 8.1 Hz, Ar), 4.74 (d, 1H, J 12.9 Hz, NCH2), 4.52-4.49 (m, 2H, ArCH2guan), 4.41-4.36 (m, 3H, H-2, H-3, NCH2), 4.06 (dd, 1H, J 8.2 Hz, J12.2 Hz, H-5′), 3.92 (dd, 1H, J 4.3 Hz, J 12.2 Hz, H-5), 3.80 (ddd, 1H, J 3.6 Hz, J 5.2 Hz, J 7.2 Hz, H-4), 3.43 (dd, 1H, J 5.7 Hz, J 11.8 Hz, H-1′), 3.27 (dd, 1H, J 4.9 Hz, J 11.9 Hz, H-1). 13C NMR (100 MHz, CD3OD): δ 158.8 (C, guan), 140.0, 132.9, 131.0, 129.1 (Ar), 72.2, 71.6,70.5 (C-2, C-3, C-4), 60.6 (NCH2), 59.5 (C-5), 56.9 (C-1), 45.5 (ArCH2guan). HRMS: (ESIMS): m/z: calcd for [C14H22N4O3]H+: 295.1765, found: 295.1769.
1,4-Dideoxy-1,4-imino-N-4-iodobenzyl-d-lyxitol (16)
Deprotection of 10 (0.24 g, 0.48 mmol) was carried out following general procedure Method A. Compound 16 (0.13 g, 78%), yellow oil, [α]D = - 19.6 (c 0.29, MeOH). 1H NMR (400 MHz, CD3OD): δ 7.67 (d, 2H, J 8.0 Hz, Ar), 7.18 (d, 2H, J 8.0 Hz, Ar), 4.27 (dd, 1H, J 4.9 Hz, J 7.2 Hz, H-3), 4.09 (td, 1H, J 3.0 Hz, J 5.1 Hz, H-2), 3.96 (d, 1H, J 13.3 Hz, NCH2), 3.71 (dd, 1H, J 5.3 Hz, J 10.9 Hz, H-5¢), 3.66 (dd, 1H, J 3.7 Hz, J 11.2 Hz, H-5), 3.50 (d, 1H, J 13.3 Hz, NCH2), 2.89-2.84 (m, 2H, H-1¢, H-4), 2.54 (dd, 1H, J 5.3 Hz, J 10.6 Hz, H-1). 13C NMR (100 MHz, CD3OD): δ 140.2, 138.5, 132.2, 92.9 (Ar), 73.7 (C-3), 71.6 (C-2), 67.8 (C-4), 61.0 (C-5), 59.7 (NCH2), 59.1 (C-1). HRMS (ESI-MS): m/z: calcd for [C12H16NO3I]H+: 350.0248. Found: 350.0253.
1,4-Dideoxy-1,4-imino-N-(naphthalen-2-ylmethyl)-d-lyxitol (17)
Deprotection of 11 (0.14 g, 0.33 mmol) was carried out following general procedure Method A. Compound 17 (61 mg, 68%), yellow oil, [α]D = - 14.5 (c 0.26, CH3OH). 1H NMR (400 MHz, CD3OD): δ 7.86-7.82 (m, 4H, Ar), 7.56 (dd, 1H, J 1.7 Hz, J 8.4 Hz, Ar), 7.50-7.44 (m, 2H, Ar), 4.30 (dd, 1H, J 5.0 Hz, J 7.1 Hz, H-3), 4.16 (d, 1H, J 13.1 Hz, NCH2), 4.10 (dt, 1H, J 2.6 Hz, J 5.1 Hz, H-2), 3.77 (dd, 1H, J 5.3 Hz, J 11.2 Hz, H-5¢), 3.73-3.68 (m, 2H, NCH2, H-5), 2.95-2.91 (m, 2H, H-1¢, H-4), 2.64 (dd, 1H, J 5.2 Hz, J 10.7 Hz, H-1). 13C NMR (101 MHz, CD3OD): δ 137.6, 134.9, 134.3, 128.9, 128.7, 128.6, 128.5, 127.0, 126.7, 73.8 (C-3), 71.6 (C-2), 67.9 (C-4), 60.9 (C-5), 60.6 (NCH2), 59.2 (C-1). HRMS (ESI-MS): m/z: calcd for [C16H19NO3]H+: 274.1438, found: 274.1441.
1,4-Dideoxy-1,4-imino-N-(2-(naphthalen-1-yl)ethyl)-d-lyxitol (18)
Deprotection of 12 (98 mg, 0.22 mmol) was carried out following general procedure Method A. Compound 18 (38 mg, 60%), yellow oil, [α]D = - 65.1 (c 0.19, CH3OH). 1H NMR 400 MHz, CD3OD): δ 8.12 (dd, 1H, J 1.0 Hz, J 8.5 Hz, Ar), 7.89-7.85 (m, 1H, Ar), 7.76-7.72 (m, 1H, Ar), 7.57-7.39 (m, 4H, Ar), 4.27 (dd, 1H, J 5.0 Hz, J 6.6 Hz, H-3), 4.20 (dt, 1H, J 3.4 Hz, J 5.4 Hz, H-2), 3.81 (dd, 1H, J 5.5 Hz, J 11.2 Hz, H-5¢), 3.70 (dd, 1H, J 4.0 Hz, J 11.2 Hz, H-5), 3.33-3.26 (m, 2H, NCH2CH2), 3.23 (dd, 1H, J 3.4 Hz, J 10.5 Hz, H-1¢), 3.11 (ddd, 1H, J 7.2 Hz, J 9.5 Hz, J 12.2 Hz, NCH2), 2.82-2.72 (m, 3H, H-1, H-4, NCH2). 13C NMR (100 MHz, CD3OD): δ 137.4, 135.4, 133.2, 129.8, 127.9, 127.6, 127.0, 126.6, 126.5, 124.7 (Ar), 73.5 (C-3), 71.7 (C-2), 68.5 (C-4), 61.1 (C-5), 59.4 (C-1), 57.8 (NCH2), 32.6 (NCH2CH2). HRMS (ESI-MS):m/z: calcd for [C17H21NO3]H+: 288.1594, found: 288.1602.
N-Cyclohexylmethyl-1,4-dideoxy-1,4-imino-d-lyxitol (19)
Deprotection of 13 (0.12 g, 0.31 mmol) was carried out following general procedure Method A. Compound 19 (40 mg, 55%), yellow oil, [α]D = - 55.3 (c 0.20, MeOH). 1H NMR (400 MHz, CD3OD): δ 4.24 (dd, 1H, J 5.0 Hz, J 7.2 Hz, H-3), 4.12 (td, 1H, J 3.0 Hz, J 5.2 Hz, H-2), 3.71 (dd, 1H, J 5.5 Hz, J 11.0 Hz, H-5¢), 3.62 (dd, 1H, J 3.2 Hz, J 11.0 Hz, H-5), 3.04 (dd, 1H, J 3.0 Hz, J 10.4 Hz, H-1¢), 2.66 (ddd, J 3.1 Hz, J 5.4 Hz, J 7.2 Hz, H-4), 2.53-2.47 (m, 2H, NCH2, H-1), 2.26 (dd, 1H, J 5.0 Hz, J 12.1 Hz, NCH2), 2.01-1.95 (m, 1H, Cyh), 1.78-1.69 (m, 4H, Cyh), 1.50-1.43 (m, 1H, Cyh), 1.35-1.20 (m, 3H, Cyh), 0.98-0.83 (m, 2H, Cyh). 13C NMR (100 MHz, CD3OD): δ 73.6 (C-3), 71.8 (C-2), 68.8 (C-4), 64.1 (NCH2), 60.6 (C-5), 59.7 (C-1), 38.2 (CH), 33.1, 32.6, 27.9, 27.3, 27.1 (5 × CH2). HRMS (ESI-MS): m/z: calcd for [C12H23NO3]H+: 230.1751. Found: 230.1760.
1,4-Dideoxy-1,4-imino-N-(tetrahydro-2H-pyran-4-yl)methyl)-d-lyxitol (20)
Deprotection of 14 (77 mg, 0.20 mmol) was carried out following general procedure Method A. Compound 20 (35 mg, 75%), yellow oil, [α]D = - 28.6 (c 0.21, CH3OH). 1H NMR 400 MHz, CD3OD): δ 4.25 (dd, 1H, J 5.0 Hz, J 7.1 Hz, H-3), 4.13 (dt, 1H, J 3.1 Hz, J 5.2 Hz, H-2), 3.98-3.92 (m, 2H, NCH2), 3.72 (dd, 1H, J 5.4 Hz, J 11.0 Hz, H-5¢), 3.63 (dd, 1H, J 3.3 Hz, J 11.0 Hz, H-5), 3.43 (tdd, 2H, J 2.3 Hz, J 8.4 Hz, J 12.0 Hz, THP), 3.05 (dd, 1H, J 3.1 Hz, J 10.3 Hz, H-1¢), 2.70 (ddd, 1H, J 3.3 Hz, J 5.4 Hz, J 7.1 Hz, H-4), 2.58-2.51 (m, 2H, H-1, THP), 2.35 (dd, 1H, J 5.1 Hz, J 12.2 Hz, THP), 1.90-1.85 (m, 1H, THP), 1.77-1.70 (m, 1H, THP), 1.67-1.61 (m, 1H, THP), 1.29-1.19 (m, 2H, THP). 13C NMR (100 MHz, CD3OD): δ 73.6 (C-3), 71.9 (C-2), 69.0, 68.8, 68.7 (C-4, 2 × CH2), 63.3 (CH2), 60.8 (C-5), 59.7 (C-1), 35.5, 32.8, 32.6 (3 × CH2). HRMS (ESI-MS): m/z: calcd for [C11H21NO4]H+: 232.1543. Found: 232.1544.
1,4-Dideoxy-N-dodecyl-1,4-imino-d-lyxitol (21)
Deprotection of 15 (0.14 g, 0.31 mmol) was carried out following general procedure Method A.Compound 21 (73 mg, 79%), yellowish oil, [α]D = - 40.2 (c 0.20, CH3OH). 1H NMR (400 MHz, CD3OD): δ 4.24 (dd, 1H, J 5.1 Hz, J 6.7 Hz, H-3), 4.15 (dt, 1H, J 3.6 Hz, J 5.3 Hz, H- 2), 3.80 (dd, 1H, J 5.8 Hz, J 11.0 Hz, H-5¢), 3.67 (dd, 1H, J 3.8 Hz, J 11.1 Hz, H-5), 3.04 (dd, 1H, J 3.3 Hz, J 10.7 Hz, H-1¢), 2.76 (dt, 1H, J 8.1 Hz, J 12.0 Hz, NCH2), 2.69 (dd, 1H, J 5.9 Hz, J 10.4 Hz, H-4), 2.60 (dd, 1H, J 5.7 Hz, J 10.7 Hz, H-1‧;), 2.40 (dt, 1H, J 7.5 Hz, J 11.9 Hz, NCH2), 1.54-1.50 (m, 2H, CH2), 1.31 (br s, 18H, 9 × CH2), 0.91 (t, J 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, CD3OD): δ 73.4 (C-3), 71.5 (C-2), 68.9 (C-4), 60.9 (C-5), 59.4 (C-1), 57.1 (NCH2), 33.1, 30.8(3x), 30.7, 30.6, 30.5, 29.0, 28.6, 23.7 (10 × CH2), 14.4 (CH3). HRMS (ESIMS): m/z: calcd for [C17H35NO3]H+: 302.2690, found: 302.2694.
1,4-Dideoxy-N-(10-guanidinodecyl)-1,4-imino-d-lyxitol hydrochloride (25)
Deprotection of 24 (28 mg, 0.04 mmol) was carried out following general procedure Method B. Compound 25 (13 mg, 88%), brownish solid, [α]D = + 70.6 (c 0.41, CH3OH). 1H NMR (400 MHz, CD3OD): δ 4.42-4.40 (m, 1H, H-3), 4.39 (dd, 1H, J 4.1 Hz, 5.7 Hz, H-2), 4.06-4.01 (m, 2H, H-5, H-5¢), 3.66-3.62 (m, 1H, H-4), 3.54-3.48 (m, 2H, NCH2, H-1¢), 3.38 (dd, 1H, J 5.7 Hz, J 12.0 Hz, H-1), 3.21-3.16 (m, 3H, NCH2, CH2NH), 1.80-1.72 (m, 2H, CH2), 1.65-1.58 (m, 2H, CH2), 1.39 (br s, 14H, 7 × CH2). 13C NMR (101 MHz, CD3OD): δ 158.5 (C, guan), 72.2, 72.0, 70.7 (C-2, C-3, C-4), 59.7 (C-5), 58.1 (NCH2), 57.6 (C-1), 42.5 (CH2NH), 30.5(2x), 30.3, 30.2, 29.9, 27.7(2x), 26.2 (8 × CH2). HRMS (ESI-MS): m/z calcd for [C16H34N4O3]H+: 331.2704, found: 331.2706.
N-(9-amidinononyl)-1,4-dideoxy-1,4-imino-d-lyxitol hydrochloride (26)
Nitrile 22 (71.7 mg, 0.16 mmol) was dissolved in anhydrous ether (5 mL) and LiHMDS (0.20 g, 1.21 mmol) was added. The mixture was sonicated at 40 °C for 2 h under inert atmosphere. The solvent was evaporated and the residue was stirred with water (10 mL) for 30 min. The water layer was extracted with DCM (3 × 10 mL), the combined organic phases were washed with brine (30 mL), dried (Na2SO4) and concentrated to give the crude amidine (61.6 mg) which was used in the next step without purification. 1H NMR (400 MHz, CDC13): δ 4.62-4.56 (m, 2H, H-2, H-3), 3.92 (dd, 1H, J 6.2 Hz, J 10.1 Hz, H-5¢), 3.74 (dd, 1H, J 5.3 Hz, J 10.2 Hz, H-5), 3.20 (d, 1H, J 11.0 Hz, H-1¢), 2.90 (m, 1H, NCH2), 2.21 (q, 1H, J 5.6 Hz, H-4), 2.01-1.97 (m, 2H, H-1, NCH2), 1.67-1.28 (m, 16H, 8 × CH2), 1.47 (s, 3H, C(CH3)2), 1.29 (s, 3H, C(CH3)2), 0.90 (br s, 9H, C(CH3)3), 0.07(2x) (each s, each 3H, Si(CH3)2). Deprotection of the crude amidine (61 mg, 0.14 mmol) was carried out following general procedure Method B. Compound 26 (43 mg, 95%), brownish oil, [α]D = + 31.0 (c 0.41, CH3OH). 1H NMR (400 MHz, CD3OD): δ 4.43-4.36 (m, 2H, H-2, H-3), 4.03-4.00 (m, 2H, H-5, H-5¢), 3.66-3.61 (m, 1H, H- 4), 3.51-3.47 (m, 2H, NCH2, H-1¢), 3.37-3.33 (m, 1H, H-1), 3.15-3.08 (m, 1H, NCH2), 1.75-1.30 (m, 16H, 8 × CH2). 13C NMR (100 MHz, CD3OD): δ 163.3 (C, amidine), 72.0, 70.8, 70.5(C-2, C-3, C-4), 59.5 (C-5), 57.9 (NCH2), 57.4 (C-1), 30.1(2x), 29.9(2x), 29.8(2x), 27.4, 25.9 (8 × CH2). HRMS (ESI-MS): m/z calcd for [C15H31N3O3]H+: 302.2438, found: 302.2441.
4.3. Enzyme assays
The isolation and purification of recombinant Drosophila melanogaster Golgi (GMIIb) and lysosomal (LManII) α-mannosidases was carried out as described.20 The same process was used also in case of Caenorhabditis elegans Golgi α-mannosidase AMAN-2.34 The α- mannosidase from Canavalia ensiformis (JBMan) was purchased from Sigma. The mannosidase activity of these enzyme preparations were measured using p-nitrophenyl α-D-mannopyranoside (pNP-Man; Sigma; 100 mM stock in dimethylsulphoxide) as a substrate at 2 mM final concentration in 50 mM acetate buffer of the relevant previously-defined optimal pH) (GMIIb and AMAN-2 at pH = 6.0, LManII at pH = 5.2 and JBMan at pH = 5.0) and 0.5 μL of the enzyme (0.05 μg of protein for JBMan), in a total volume of 50 μL for 1-2 h at 37 °C. GMIIb was assayed in the presence of 0.5 mM CoCl2.
The lyophilized imino-D-lyxitol derivatives were dissolved in DMSO to the final concentration 50 mM and further diluted to a desired concentration in water. The tested derivatives were preincubated with the enzyme in the buffer for 5 min at rt and the reaction was started by addition of the substrate. The reactions were terminated with two volumes (0.1 mL) of 0.5 M sodium carbonate and the production of p-nitrophenol was measured at 405 nm using a multimode reader Mithras LB943 (Berthold Technologies). The average or representative result of three independent experiments made in duplicate is presented. The IC50 value was determined with 2 mM pNP-Man. The Ki values were determined from Dixon plots of assays performed with pNP-Man (0.5-4 mM). The type of inhibition was estimated by a simple method of plotting kinetic results of the enzymatic method67 showing a competitive type of inhibition.
Supplementary Material
Acknowledgement
This work was supported by the Scientific Grant Agency of the Ministry of Education of Slovak Republic and Slovak Academy of Sciences (the project VEGA-2/0031/19) and the project implementation CEMBAM (Centre for Medical Bio-Additive Manufacturing and Research, ITMS2014+: 313011V358 supported by the Operational Programme Integrated Infrastructure funded by the European Regional Development Fund).
Footnotes
Conflicts of interest:
There are no conflicts to declare.
References
- 1.Colegate SM, Dorling PR, Huxtable CR. Aust J Chem. 1979;32:2257–2264. [Google Scholar]
- 2.Dorling PR, Huxtable CR, Colegate SM. Biochem J. 1980;191:649–651. doi: 10.1042/bj1910649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moremen KW. Biochim Biophys Acta - Gen Subj. 2002;1573:225–235. doi: 10.1016/s0304-4165(02)00388-4. [DOI] [PubMed] [Google Scholar]
- 4.Tulsiani DR, Harris TM, Touster O. J Biol Chem. 1982;257:7936–7939. [PubMed] [Google Scholar]
- 5.Tulsiani DR, Touster O. Arch Biochem Biophys. 1983;224:594–600. doi: 10.1016/0003-9861(83)90247-3. [DOI] [PubMed] [Google Scholar]
- 6.Tulsiani DRP, Broquist HP, Touster O. Arch Biochem Biophys. 1985;236:427–434. doi: 10.1016/0003-9861(85)90643-5. [DOI] [PubMed] [Google Scholar]
- 7.Goss PE, Baker MA, Carver JP, Dennis JW. Clinic Cancer Res. 1995;1:935–944. [PubMed] [Google Scholar]
- 8.Olden K, Breton P, Grzegorzewski K, Yasuda Y, Gause BL, Oredipe OA, Newton SA, White SL. Pharmacol Ther. 1991;50:285–290. doi: 10.1016/0163-7258(91)90046-o. [DOI] [PubMed] [Google Scholar]
- 9.Dennis JW, Koch K, Yousefi S, Vanderelst I. Cancer Res. 1990;50:1867–1872. [PubMed] [Google Scholar]
- 10.Cheng TJR, Chan TH, Tsou EL, Chang SY, Yun WY, Yang PJ, Wu YT, Cheng WC. ChemAsian J. 2013;8:2600–2604. doi: 10.1002/asia.201300680. [DOI] [PubMed] [Google Scholar]
- 11.Šesták S, Bella M, Klunda T, Gurská S, Džubák P, Wols F, Wilson IBH, Sladek V, Hajdúch M, Poláková M, Kóňa J. ChemMedChem. 2018;13:373–383. doi: 10.1002/cmdc.201700607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klunda T, Šesták S, Kóňa J, Poláková M. Bioorg Chem. 2019;83:424–431. doi: 10.1016/j.bioorg.2018.10.066. [DOI] [PubMed] [Google Scholar]
- 13.Klunda T, Hricovíni M, Šesták S, Kóňa J, Poláková M. New J Chem. 2021;45:10940–10951. [Google Scholar]
- 14.Lee ZY, Loo JSE, Wibowo A, Mohammat MF, Foo JB. Carbohydr Res. 2021;508:108395. doi: 10.1016/j.carres.2021.108395. [DOI] [PubMed] [Google Scholar]
- 15.Bols M, Lopez O, Ortega-Caballero F. In: Comprehensive Glycoscience. 1st. Kamerling JP, editor. Vol. 1. Elsevier; Oxford: 2007. pp. 815–884. [Google Scholar]
- 16.Yang LF, Shimadate Y, Kato A, Li YX, Jia YM, Fleet GWJ, Yu CY. Org Biomol Chem. 2020;18:999–1011. doi: 10.1039/c9ob02029b. [DOI] [PubMed] [Google Scholar]
- 17.Brissonnet Y, Mellet CO, Morandat S, Moreno MIG, Deniaud D, Matthews SE, Vidal S, Šesták S, El Kirat K, Gouin SG. J Am Chem Soc. 2013;135:18427–18435. doi: 10.1021/ja406931w. [DOI] [PubMed] [Google Scholar]
- 18.Mirabella S, D’Adamio G, Matassini C, Goti A, Delgado S, Gimeno A, Robina I, Moreno-Vargas AJ, Šesták S, Jiménez-Barbero J, Cardona F. Chem Eur J. 2017 doi: 10.1002/chem.201703011. [DOI] [PubMed] [Google Scholar]
- 19.IrsheidI L, Wehler T, Borek C, Kiefer W, BrenkI R, Ortiz-Soto ME, Seibel J, Schirmeister T. PLoS One. 2019;14:e0216132. doi: 10.1371/journal.pone.0216132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemčovičová I, Šesták S, Rendić D, Plšková M, Mucha J, Wilson IBH. Glycoconj J. 2013;30:899–909. doi: 10.1007/s10719-013-9495-5. [DOI] [PubMed] [Google Scholar]
- 21.Numao S, Kuntz DA, Withers SG, Rose DR. J Biol Chem. 2003;278:48074–48083. doi: 10.1074/jbc.M309249200. [DOI] [PubMed] [Google Scholar]
- 22.Petersen L, Ardèvol A, Rovira C, Reilly PJ. J Am Chem Soc. 2010;132:8291–8300. doi: 10.1021/ja909249u. [DOI] [PubMed] [Google Scholar]
- 23.van den Elsen JMH, Kuntz DA, Rose DR. EMBO J. 2001;20:3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah N, Kuntzt DA, Rose DR. Proc Natl Acad Sci USA. 2008;105:9570–9575. doi: 10.1073/pnas.0802206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong W, Kuntz DA, Ernber B, Singh H, Moremen KW, Rose DR, Boons GJ. J Am Chem Soc. 2008;130:8975–8983. doi: 10.1021/ja711248y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuntz DA, Zhong W, Guo J, Rose DR, Boons GJ. ChemBioChem. 2009;10:268–277. doi: 10.1002/cbic.200800538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuntz DA, Nakayama S, Shea K, Hori H, Uto Y, Nagasawa H, Rose DR. ChemBioChem. 2010;11:673–680. doi: 10.1002/cbic.200900750. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong ZCL, Kuo Lahav D, Liu B, Johnson R, Beenakker TJM, de Boer C, Wong CS, van Rijssel ER, et al. J Am Chem Soc. 2020;142:13021–13029. doi: 10.1021/jacs.0c03880. [DOI] [PubMed] [Google Scholar]
- 29.Heikinheimo P, Helland R, Leiros HKS, Leiros I, Karlsen S, Evjen G, Ravelli R, Schoehn G, Ruigrok R, Tollersrud OK, McSweeney S, et al. J Mol Biol. 2003;327:631–644. doi: 10.1016/s0022-2836(03)00172-4. [DOI] [PubMed] [Google Scholar]
- 30.Howard E, Cousido-Siah A, Lepage ML, Schneider JP, Bodlenner A, Mitschler A, Meli A, Izzo I, Alvarez HA, Podjarny A, et al. Angew Chem Int Ed. 2018;57:8002–8006. doi: 10.1002/anie.201801202. [DOI] [PubMed] [Google Scholar]
- 31.Legler G. Stütz AE. Inimosugars as Glycosidase Inhibitors: Nojirimycin and beyond. Wiley-VCH Verlag GmbH; Weinheim: 1999. pp. 31–67. ch. 3. [Google Scholar]
- 32.Sladek V, Kóňa J, Tokiwa H. Phys Chem Chem Phys. 2017;19:12527–12537. doi: 10.1039/c7cp01200d. [DOI] [PubMed] [Google Scholar]
- 33.Kuntz DA, Rose DR. Golgi Mannosidase II in complex with swainsonine at 13 Angstrom. The Research Collaboratory for Structural Bioinformatics (RCSB): RCSB-Rutgers, RCSB-San Diego Supercomputer Center, and University of Wisconsin-Madison. [Accessed June 04, 2012]; doi: 10.2210/pdb3blb/pdb. PDB ID: 3BLB., http://www.rcsb.org. [DOI] [Google Scholar]
- 34.Paschinger K, Hackl M, Gutternigg M, Kretschmer-Lubich D, Stemmer U, Jantsch V, Lochnit G, Wilson IBH. J Biol Chem. 2006;281:28265–28277. doi: 10.1074/jbc.M602878200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Kuntz DA, Hamlet T, Sim L, Rose DR, Pinto BM. Bioorg Med Chem. 2006;14:8332–8340. doi: 10.1016/j.bmc.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Englebienne P, Fiaux H, Kuntz DA, Corbeil CR, Gerber-Lemaire S, Rose DR, Moitessier N. Proteins: Struct, Funct, Bioinf. 2007;69:160–176. doi: 10.1002/prot.21479. [DOI] [PubMed] [Google Scholar]
- 37.Fedorov DG, Kitaura K. The Fragnemt Molecular Orbital Method - Practical Applications to Large Molecular Systems. CRC Press, Taylor and Francis Group; 2009. [Google Scholar]
- 38.Poláková M, Stanton R, Wilson IBH, Holková I, Šesták S, Machová E, Jandová Z, Kóňa J. Carbohydr Res. 2015;406:34–40. doi: 10.1016/j.carres.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bella M, Šesták S, Moncol J, Koóš M, Poláková M, Beilstein J. Org Chem. 2018;14:2156–2162. doi: 10.3762/bjoc.14.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 41.Glide, version 70. Schrödinger, LLC; New York, NY: 2016. [Google Scholar]
- 42.Li H, Robertson AD, Jensen JH. Proteins: Struct, Funct, Bioinf. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 43.Bas DC, Rogers DM, Jensen JH. Proteins: Struct, Funct, Bioinf. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y, Truhlar DG. Theor Chem Acc. 2008;120:215–241. [Google Scholar]
- 45.Wadt WR, Hay PJ. J Chem Phys. 1985;82:284–298. [Google Scholar]
- 46.Hay PJ, Wadt WR. J Chem Phys. 1985;82:270–283. [Google Scholar]
- 47.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 48.Murphy RB, Philipp DM, Friesner RA. J Comput Chem. 2000;21:1442–1457. [Google Scholar]
- 49.Qsite. Schrödinger, LLC; New York, NY: 2016. [Google Scholar]
- 50.Jaguar, version 91. Schrödinger, LLC; New York, NY: 2016. [Google Scholar]
- 51.Schrödinger Suite 2016 Protein Preparation Wizard; Epik v 35; Impact v 70. Schrödinger; Maestro v.10.5, LLC; New York, NY: 2016. [Google Scholar]
- 52.Becke AD. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 53.Kitaura K, Ikeo E, Asada T, Nakano T, Uebayasi M. Chem Phys Lett. 1999;313:701–706. [Google Scholar]
- 54.Fedorov DG, Nagata T, Kitaura K. Phys Chem Chem Phys. 2012;14:7562–7577. doi: 10.1039/c2cp23784a. [DOI] [PubMed] [Google Scholar]
- 55.Suenaga M. J Comput Chem-Japan. 2008;7:33–54. [Google Scholar]
- 56.Møller C, Plesset MS. Phys Rev. 1934;46:618–622. [Google Scholar]
- 57.Frisch MJ, Headgordon M, Pople JA. Chem Phys Lett. 1990;166:275–280. [Google Scholar]
- 58.Fedorov DG, Kitaura K, Li H, Jensen JH, Gordon SM. J Comput Chem. 2006;27:976–985. doi: 10.1002/jcc.20406. [DOI] [PubMed] [Google Scholar]
- 59.Barca GMJ, Bertoni C, Carrington L, Datta D, De Silva N, Deustua JE, Fedorov DG, Gour JR, Gunina AO, et al. J Chem Phys. 2020;152 doi: 10.1063/5.0005188. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, et al. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- 61.Lim H, Chun J, Jin X, Kim J, Yoon J, No KT. Sci Rep. 2019;9:16727. doi: 10.1038/s41598-019-53216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sogawa H, Sato R, Suzuki K, Tomioka S, Shinzato T, Karpov P, Shulga S, Blume Y, Kurita N. Chem Phys. 2020;530:110603 [Google Scholar]
- 63.Anan R, Nakamura T, Shimamura K, Matsushita Y, Ohyama T, Kurita N. J Mol Model. 2019;25:192. doi: 10.1007/s00894-019-4087-3. [DOI] [PubMed] [Google Scholar]
- 64.Sladek V, Tokiwa H, Shimano H, Shigeta Y. J Chem Theory Comput. 2018;14:6623–6631. doi: 10.1021/acs.jctc.8b00733. [DOI] [PubMed] [Google Scholar]
- 65.Takaya D, Niwa H, Mikuni J, Nakamura K, Handa N, Tanaka A, Yokoyama S, Honma T. J Mol Graph Model. 2020;99:107599. doi: 10.1016/j.jmgm.2020.107599. [DOI] [PubMed] [Google Scholar]
- 66.Bashyal BP, Fleet GWJ, Gough MJ, Smith PW. Tetrahedron. 1987;43:3083–3093. [Google Scholar]
- 67.Cornish-Bowden A. Biochem J. 1974;137:143–144. doi: 10.1042/bj1370143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.